Abstract
The real-time, colorimetric detection of analytes via aptamer–gold nanoparticle technology has proven to be an important, emerging technique within the medical field. Of global health importance, the ability to detect vector mosquito species, such as the Aedes (Ae.) aegypti mosquito, and transmitted arboviruses, such as Zika virus, is paramount to mosquito control and surveillance efforts. Herein, we describe the detection of Ae. aegypti salivary protein for vector identification and the detection of Zika virus to assess mosquito infection status by aptamer–gold nanoparticle conjugates. Key to optimization of these diagnostics were gold nanoparticle capping agents and aptamer degree of labelling (i.e., the amount of aptamers per gold nanoparticle). In the present study, detection was achieved for as little as 10 ng Ae. aegypti salivary protein and 1.0 × 105 PFU live Zika virus. These aptamer–gold nanoparticle conjugate diagnostics could one day prove to be useful as deployable nano-based biosensors that provide easy-to-read optical read outs through a straightforward red-to-blue colour change either within a diagnostic solution or atop a card/membrane-based biosensor.
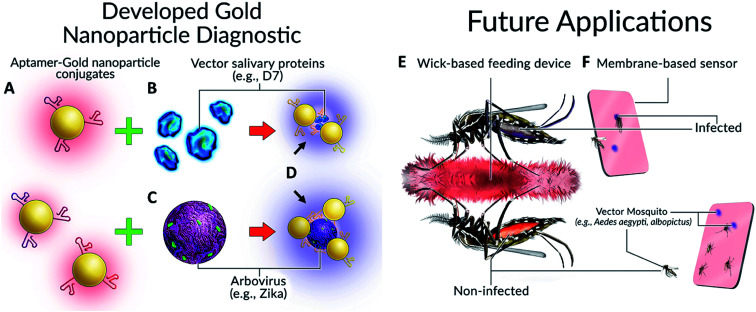
Colorimetric detection of arboviruses and vector mosquito species. (A) Aptamer-gold nanoparticle conjugates, (B) mosquito salivary protein, (C) Zika envelope protein, (D) visible color change, (E) imbibed diagnostic and (F) diagnostic cards.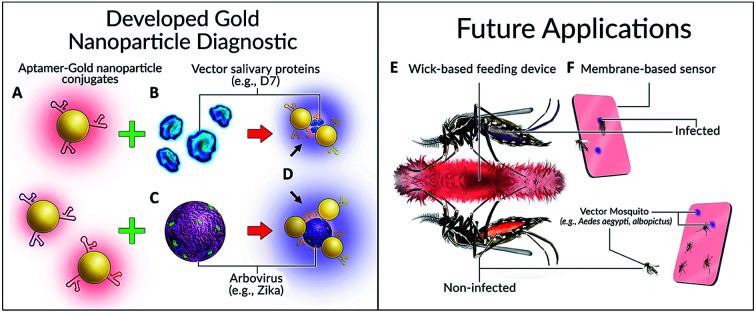
Introduction
Infectious diseases vectored by mosquitoes such as Aedes (Ae.) aegypti and Ae. albopictus are significant threats to many populations worldwide. For example, 440 000–1 300 000 new cases of Zika virus (ZIKV) infection—presumptively vectored by Ae. aegypti—were estimated in the 2015 Zika epidemic in Brazil, where a potential link to severe birth defects was observed, as well as neurological complications resulting in Guillain–Barré syndrome.1–3 To defend against such threats, new vector mosquito surveillance tools and diagnostics that enable rapid, efficient responses in the field are needed.
The timely availability of accurate surveillance information is key to the development of any mosquito-borne disease risk assessment and execution of effective and efficient mosquito control strategies. Currently however, real-time sensors and rapid diagnostics that detect vector mosquitoes and/or mosquito-borne diseases in the field are limited as the focus has largely been on the development of tools and diagnostics for clinical use in humans. We have therefore been spearheading the development of compact, lightweight, robust, self-contained tools based on DNA aptamer–gold nanoparticle conjugates (Apt–AuNPs)5 to identify and detect vector mosquitoes and arboviruses via colorimetric readouts. These new tools require little specialized training, equipment or methods to adopt, deploy and implement in field settings.
Aptamers are commonly short sequences of single-stranded deoxyribonucleic acid (DNA) developed via a synthetic process called Systematic Evolution of Ligands by Exponential Enrichment (SELEX)6,7 and can bind a wide variety of targets.6,7 Aptamers are advantageously synthesized in the laboratory, unlike antibodies,6,7 and this process allows for the creation of aptamers within weeks, whereas antibody development requires months.8 Aptamers may also be more stable and effective in a wider range of solutions and conditions than antibodies.7,8 Furthermore, aptamers can be selected that undergo large conformational changes (structure-switching) upon target binding.9 Conjugating aptamers to gold nanoparticles creates colorimetric and highly sensitive nanosensor/diagnostic tools for detection of target molecules.8 The colorimetric readout of such Apt–AuNP conjugates is a function of aggregation in solution of the gold nanoparticles.10 When no target is present, Apt–AuNPs are dispersed and the solution is red; when target is present, the aptamers coupled to the AuNPs bind, undergo a conformational change, thereby inducing aggregation of said conjugates that shifts the solution colour from red to blue/purple.11 Similar approaches have been used recently to produce colorimetric malaria diagnostic assays with aptamers targeting the lactate dehydrogenase enzymes of the Plasmodium parasites.10,12,13
Our concept, inspired in part by the work of Hall-Mendelin et al.,4 is to take advantage of mosquito sugar-feeding behaviours and interrogate expectorated saliva and/or the midguts of live vector mosquitoes with Apt–AuNPs specific for vector-identifying salivary protein(s) and/or arboviral envelope proteins. In the case of the salivary proteins, small amounts of saliva would be deposited by vector mosquitoes onto a substrate soaked with sugar solution containing Apt–AuNPs specific for vector salivary protein. If the target is present (vector salivary protein or arbovirus), the Apt–AuNPs would aggregate and the color of the membrane at that spot would change from red to blue. For the interrogation of midguts of live mosquitoes, trap-caught mosquitoes would consume a red-colored sugar solution containing Apt–AuNPs specific for an arbovirus of interest from a feeder. Non-infected (i.e., negative) mosquitoes would have red solution inside their midgut; however, infected mosquitoes (with target virus present in their midgut) would induce Apt–AuNP aggregation and thereby change the solution color from red to blue, resulting in a color-coding of mosquitoes based on disease (arboviral) status (Fig. 1).
Fig. 1. Colorimetric detection of arboviruses and vector species. (A) Aptamer–gold nanoparticle conjugates bind to either (B) mosquito salivary protein D7 or (C) Zika virus envelope protein and undergo a (D) visible color change. Possible applications of this technology for mosquito surveillance and control include field-deployable imbibed diagnostics or membrane-based sensors. (E) Upon imbibing diagnostic, mosquito abdomens are colour coded as either red (non-infected) or blue/purple colour (infected). (F) Diagnostic cards containing Apt–AuNPs specific for either arboviruses or Aedes species salivary proteins will undergo colour change following expectoration during sugar feeding.
The foundational studies required for the creation of these new aptamer-based colorimetric diagnostics and surveillance tools for vector mosquitoes and arbovirus is the focus of the present work. Reported here for the first time is the development and testing of novel aptamers and Apt–AuNPs that specifically detect protein in mosquito saliva (Ae. aegypti and albopictus) as well as new aptamers and Apt–AuNPs against ZIKV. The results of in vitro testing show that visible colorimetric detections of Ae. aegypti salivary protein were detectable as low as ∼10 ng of salivary gland extract (SGE) and that 105 plaque-forming units (PFUs) of active ZIKV were detectable spectrophotometrically. These results indicate that colorimetric Apt–AuNP diagnostics for mosquitoes and arboviruses have potential to be powerful new additions to the toolkits of mosquito surveillance and control communities.
Results
Capping agents influence particle stability and sensitivity of diagnostic towards mosquito salivary proteins
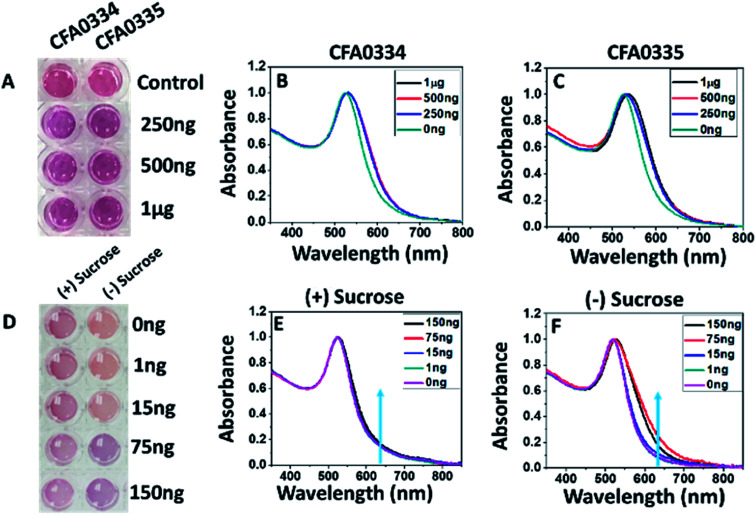
Aptamers developed against whole Ae. aegypti saliva were conjugated to citrate capped AuNPs at 2 μM. Aptamers CFA0334 and CFA0335 (aptamer IDs provided by Base Pair Biotechnologies Inc., BPB) showed the maximum colour change in the presence of analyte (Fig. 2) within a sucrose containing diagnostic buffer. 1 μg SGE (50 μL Apt–AuNP, 25 μL analyte; 75 μL total volume) caused the greatest aggregation, as indicated by a peak shift (Fig. 2B and C) followed by 500 ng and 250 ng SGE. Interestingly, recombinant Ae. albopictus D7 protein generated by Sanford Burnham Prebys (SBP) Research Institute was able to aggregate Apt–AuNPs derivatized with aptamers specific for Ae. aegypti D7 in buffer without sucrose but not with sucrose (Fig. 2D–F). For this reason, the capping agent used for our Apt–AuNP diagnostic was explored.
Fig. 2. Effects of sucrose on Apt–AuNP aggregation. (A) Visible colour change and (B and C) UV-visible spectra of Apt–AuNPs with aptamers CFA0334 and CFA0335 in the presence Ae. aegypti SGE immediately after analyte addition. The change in the peak intensity corresponds to Apt–AuNP aggregation. (D–F) To assess the effects of sucrose within the diagnostic buffer, aptamer CFA0334 was tested against recombinant Ae. albopictus D7 (E) with and (F) without sucrose. A marked increase in absorbance at 640 nm can be seen in buffer without sucrose while there is little change in the presence of sucrose. (D) A colour change in buffer solution can be seen at 75 ng of Ae. albopictus D7 without sucrose in the diagnostic buffer. Values within legends indicate amount of either (B and C) Ae. aegypti SGE or (E and F) Ae. albopictus D7 added to Apt–AuNP solutions.
Because the presence of sucrose within the diagnostic buffer had an effect on whether particles would aggregate, we explored how sucrose affected the aggregation state of bare AuNPs (i.e., no aptamers). In diagnostic buffer without sucrose, citrate-capped AuNPs remained disperse with a hydrodynamic radius of ∼30 nm (as determined by DLS) but aggregated in 10% sucrose showing a red shift in the UV spectra as well as an increase in particle size to ∼109 nm (ESI Fig. 1A and B‡). Phosphine capping with bis(p-sulfonatophenyl)phenylphosphine dihydrate dipotassium salt was explored and this prevented non-specific aggregation in the presence of sucrose (ESI Fig. 1B‡) but, when aptamers were added, the particles were too stable and never aggregated in the presence of target (data not shown).
To overcome non-specific aggregation and low specificity, we explored sucrose as a capping agent.16,20,21 Sucrose-capped AuNPs were synthesized using the two different methods (termed Type 1 and Type 2, ESI Table 1‡). The two methodologies yielded particles with sizes in the range of 49.23 nm (±0.44 nm) and 90.12 nm (±14.32) for Type 1 and Type 2 particles, respectively (ESI Fig. 1C‡). Zeta potential measurements (ESI Fig. 1D‡) showed that the particles were negatively charged and remained uniformly dispersed in the solution due to electrostatic stability. The UV-Vis spectra showed that the particles exhibited an absorbance peak at 528 nm and 522 nm for Type 1 and 2 particles, respectively (ESI Fig. 1E‡). Because the synthesis method for Type 2 particles resulted in larger and non-uniform AuNPs, the Type 1 particle synthesis method was chosen for AuNPs used for aptamer conjugation and testing against extracted Ae. aegypti salivary gland extract (SGE) protein and recombinant Ae. albopictus D7.
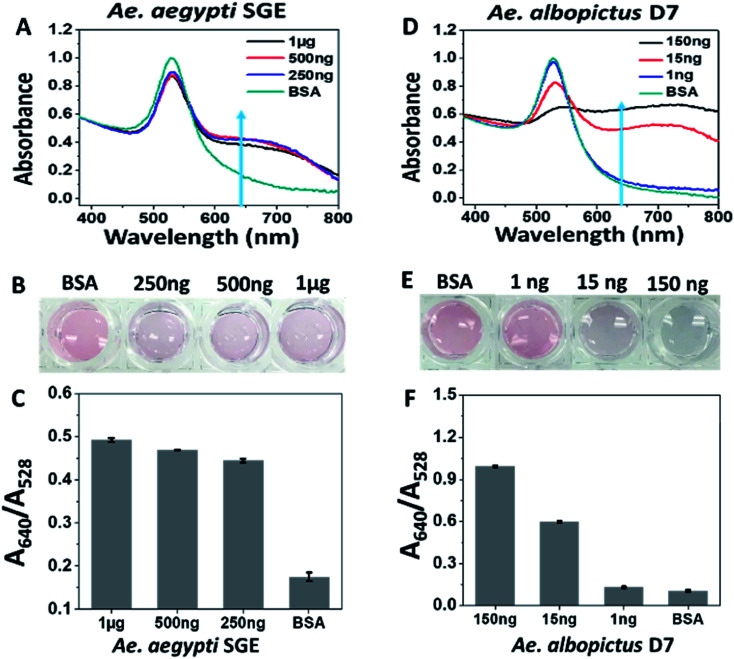
Type 1 sucrose-capped AuNPs (SAuNPs) were conjugated with 2 μM of aptamer CFA0334 and showed a dose dependent change in both the UV-Vis spectrum and visible colour in response to Ae. aegypti SGE and recombinant Ae. albopictus D7 (Fig. 3). This testing, performed in diagnostic buffer without sucrose, showed marked improvement in sensitivity with an immediate colour change observed upon addition of as low as 250 ng SGE (Fig. 3A). These particles were also tested against recombinant Ae. albopictus D7 protein with a visible colour change as low as 15 ng of protein (Fig. 3D–F).
Fig. 3. Response of Type 1 sucrose-capped AuNPs with aptamer CFA0334 to salivary proteins. (A–C) Ae. aegypti SGE caused changes in both UV-Vis and visual colour change in response to as low as 250 ng SGE. (D–F) 15 ng of recombinant Ae. albopictus D7 induces colour change. BSA used for control. Testing done in diagnostic buffer without sucrose.
To explore further Type 1 SAuNPs synthesis, reagent concentrations were varied in order to obtain differently sized particles with the aim of obtaining the smallest particles and greatest possible red shift.22 ESI Table 2‡ shows the characterization results of the different synthesized particles. The smallest size obtained was for SAuNPs Type 1–4, which was 36.4 nm (±0.95 nm) with a maximum surface charge of −41.2 mV (±1.1 mV). The higher surface charge results in repulsion and ensures a homogenous dispersion of AuNPs. Apt–AuNPs with optimized SAuNPs detect mosquito salivary proteins with greater sensitivity. Aptamer CFA0334 was conjugated at 2 μM to SAuNP Type 1.4 particles (properties listed in ESI Table 2‡) and tested for stability in buffer with sucrose (referred to as Apt–SAuNPs henceforth). To increase the colour saturation of these particles, the Apt–SAuNPs were used at 3× concentration of the previous experiments.
ESI Fig. 2‡ details when Apt–SAuNPs were dispersed in diagnostic buffer with and without sucrose. ESI Fig. 2A and B‡ show that there was no change in the spectrum or colour of the solution after adding sucrose. Aptamer conjugated SAuNPs in diagnostic solution without sucrose exhibited a size of 45.49 nm. Addition of sucrose immediately caused the hydrodynamic radius of Apt–SAuNPs to further increase from 45.49 nm to 49.93 nm (ESI Fig. 2C‡); this size remained essentially constant over a 48 h period. These observations are consistent with the changes in zeta potential (ESI Fig. 2D‡). The particles possessed a high negative charge, −42.9 mV, when suspended in buffer without sucrose and became less negatively charged in buffer with sucrose (−27.9 mV) (ESI Fig. 2D‡). Zeta potentials remained consistent after 48 h, indicating that the particles continued to repel each other and remained uniformly dispersed. This is likely due to sucrose molecules interacting with and suppressing the negatively charged Apt–SAuNPs, causing a decrease in zeta potential magnitude and increasing the size by ∼5 nm.
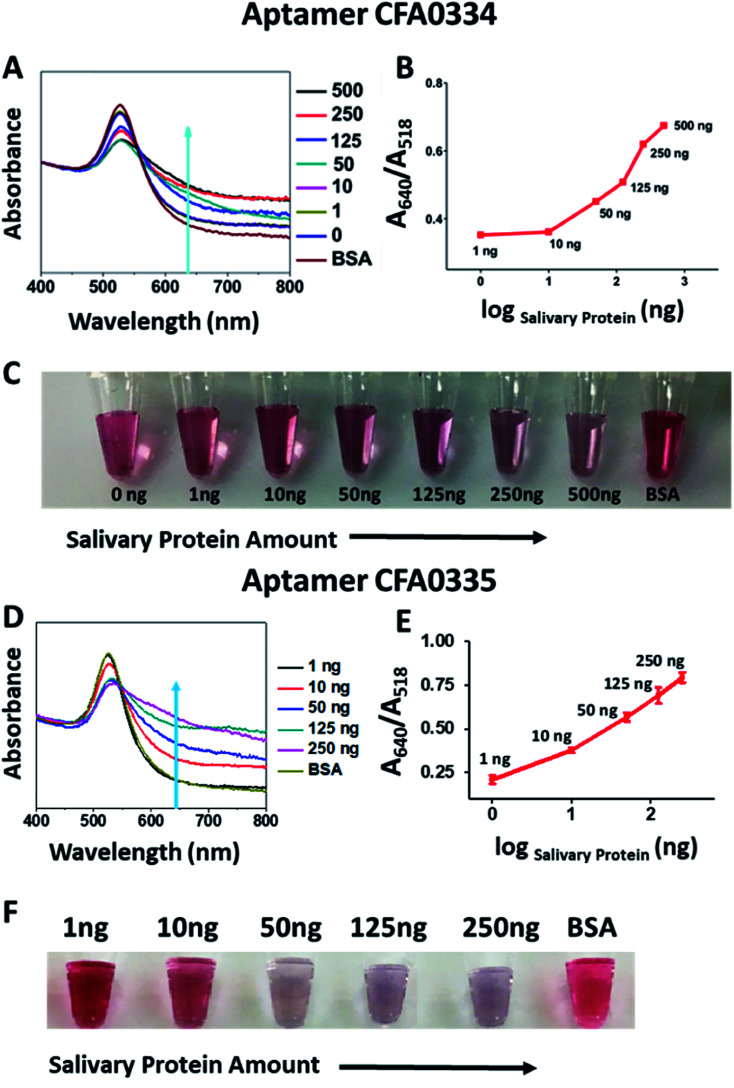
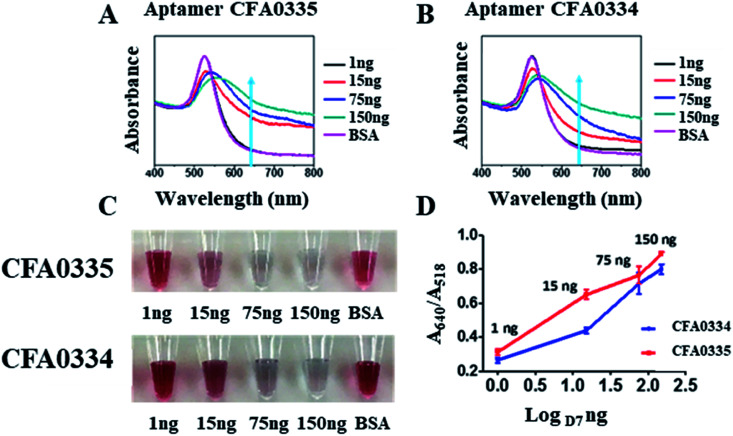
Following sucrose capping experimentation, Apt–SAuNPs derivatized with aptamer CFA0334 were tested against Ae. aegypti SGE in buffer with sucrose (Fig. 4A–C). A marked improvement in detection was achieved with optimized Apt–SAuNPs with detection as low as 50 ng SGE. The change in colour corresponded to an increase in the A640/A518 ratio (Fig. 4B). Maximum aggregation was observed for 500 ng SGE, followed by 250 ng and 125 ng (Fig. 4A). Similarly, this testing with Ae. aegypti SGE was done with aptamer CFA0335. The minimum amount of SGE detected improved to 10 ng (Fig. 4D–F), several orders of magnitude more sensitive than citrate capped AuNPs (Fig. 2). Finally, Apt–SAuNPs conjugated with either aptamer CFA0334 or CFA0335 were tested against recombinant Ae. albopictus D7 in buffer with sucrose to see if these aptamers could also detect this recombinant target (Fig. 5). Recombinant D7 was tested at 1 ng, 15 ng, 75 ng and 150 ng with detection as low as 15 ng for aptamer CFA0335.
Fig. 4. Sucrose-capped Apt–AuNPs detection of Ae. aegypti SGE. (A) UV-visible spectroscopy, (B) A640/A518 ratios and (C) corresponding digital image of the CFA0334-derivated Apt–SAuNPs in the presence of SGE show a dose dependent increase in particle aggregation with detection as low as 50 ng Ae. aegypti SGE. BSA used as control. Corresponding (D) UV-visible spectroscopy, (E) A640/A518 ratios and (F) corresponding digital image of CFA0335-derivatized Apt–SAuNPs showing a dose dependent increase in aggregation in response to SGE as low as 10 ng. BSA used as control.
Fig. 5. Detection of Ae. albopictus D7 with Apt–SAuNPs. (A and B) UV-visible spectroscopy showing response to as little as 15 ng D7 for aptamers Ae. aegypti salivary aptamers CFA0335 and CFA0334. (C) Digital image of Apt–SAuNPs in the presence of D7. (D) A640/A518 ratio shows a dose dependent increase in response to D7.
Degree of gold nanoparticle labelling with aptamers influences sensitivity of diagnostic towards ZIKV
Following sucrose-capping optimization, testing continued with another target integral to mosquito surveillance and control: Zika virus (ZIKV). Aptamers developed by BPB against recombinant ZIKV envelope protein (ZIKV-E) were conjugated to SAuNPs and absorbance was recorded at intervals up to 42 h in response to 200 nM ZIKV-E (ESI Fig. 3‡). Aggregation among the Apt–SAuNPs in the presence of the ZIKV-E increased over time and maximum aggregation occurred with aptamers CFA0201, CFA0205, CFA0206, CFA0208, CFA0212 and CFA0213 after 21 h and 42 h (ESI Fig. 3B, D and F‡). These aptamers showed minimal response towards control, 600 nM BSA. A digital image of SAuNPs conjugated with aptamers (ESI Fig. 3E‡) shows the colour change in response to 200 nM ZIKV-E.
Degree of labelling (DOL) is the number of aptamers immobilized onto each AuNP; the extent of aptamer derivatization on the gold surface has significant effects on diagnostic sensitivity.23 Using qPCR, concentrations of aptamers on Apt–SAuNPs were calculated by comparing cycle threshold (CT) values of SAuNPs against those values generated from standard curves (ESI Fig. 4‡). DOL of ZIKV-specific aptamers CFA0201 or CFA0204 following conjugation is depicted in ESI Table 3.‡ In brief, the standard curves for each sample were generated from CT values and measured concentrations of standards (0.5–5000 pM). Aptamer concentrations for each SAuNPs were calculated by applying the mean CT of each sample to the standard curve. The estimated nanoparticles per mL (nps per mL) were calculated by multiplying the measured optical density (OD), obtained by Nanodrop quantification at 525 nM, with the estimated number of 1.64 × 1012 nps per mL of SAuNP at OD 1. Finally, the DOL was calculated by multiplying the measured concentrations (pM) with Avogadro's number and divided by the estimated nanoparticles per mL. The DOL value for CFA0201 indicated a 10-fold difference between the three concentrations used during conjugation of aptamer to bare SAuNP.
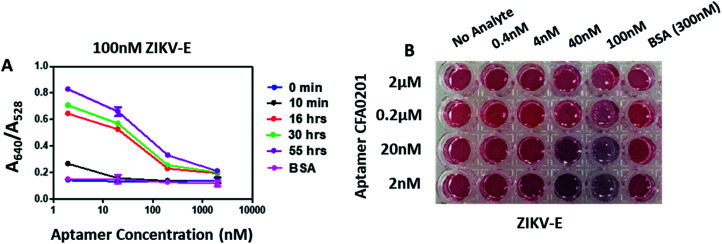
By sequentially reducing the conjugated aptamer concentration to 2 nM, the A640/A528 ratio increased substantially and the appreciable visible colour change was apparent with as little as 4 nM ZIKV-E (Fig. 6); control 300 nM BSA showed no colour change. Aggregation increased over time for all particles with the maximum ratio recorded with 2 nM DOL for CFA0201 (Fig. 6A). Particles with a lower DOL may show increased sensitivity due to lesser repulsive forces from the aptamer coating thus making aggregation more favorable in the presence of target analyte, such as ZIKV.
Fig. 6. Effect of degree of labelling on detection of ZIKV-E. (A) Absorbance ratio (A640/A528) in the presence of 100 nM ZIKV-E at time intervals 0 min, 10 min, 16 h, 30 h and 55 h. Apt–SAuNPs with a DOL of 2 nM showed significant changes in the absorbance ratio within 10 min of analyte addition. (B) Digital image showing visible color change of Apt–SAuNPs with DOLs of 2 nM to 2 μM in the presence of ZIKV-E at 0.4 nM to 100 nM. Visible color change was observed as low as 4 nM ZIKV-E for CFA0201 at 2 nM DOL.
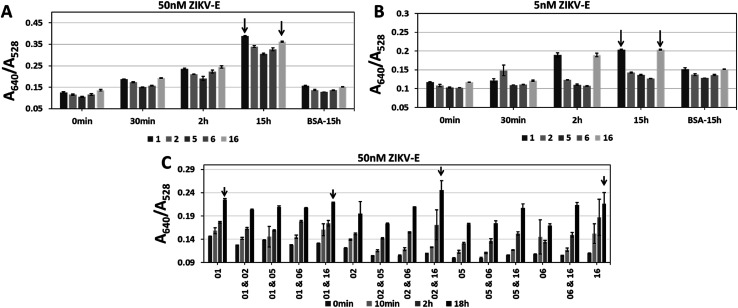
Aptamers CFA0201, CFA0202, CFA0205, CFA0206, CFA0216 were conjugated to SAuNPs (Apt–SAuNP) at a concentration of 2 nM and tested against ZIKV-E. To test aggregation of particles over time, the absorbance was recorded at intervals up to 15 h in response to 50 nM or 5 nM ZIKV-E (Fig. 7). In addition to individually screening the best ZIKV-E Apt–SAuNPs from the five shortlisted top performing Apt–SAuNP reagents, a 5 × 5 matrix utilizing different combinations was performed (e.g., AuNPs conjugated with aptamer CFA0201 were mixed with AuNPs conjugated with aptamer CFA0202, etc.). A640/A528 ratio increased over time with a maximum value being observed for aptamers CFA0201 alone, CFA0201 & CFA0216, CFA0202 & CFA0216 and CFA0216 alone (Fig. 7C).
Fig. 7. Single and sandwich Apt–SAuNP morphologies for aptamers CFA0201, CFA0202, CFA0205, CFA0206 and CFA0216 (termed 1, 2, 5, 6 and 16 for short, respectively). Single or combinations of aptamers were tested to find the best individuals and working pairs (black arrows) against (A and C) 50 nM or (B) 5 nM ZIKV-E. A620/A528 ratios increased with prolonged inoculation for all aptamers.
Apt–AuNPs developed against Zika envelope protein aggregate in the presence of infectious ZIKV
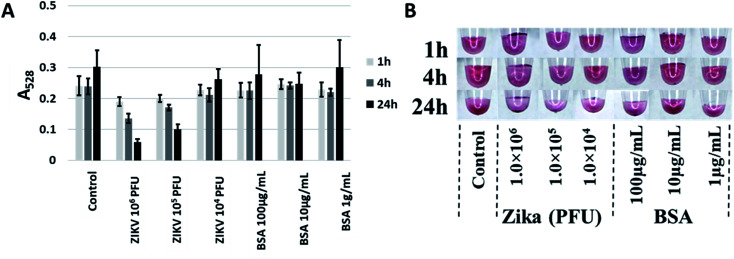
After testing against the recombinant envelope protein (i.e., ZIKV-E), testing against infectious virus was performed. ZIKV virions24 over three orders of magnitude (104 to 106 PFU) were tested at three time points (1 h, 4 h and 24 h) with BSA at the same amount of total protein (as determined by A280) used as a control. Though aptamer CFA0204 and the corresponding Apt–SAuNP conjugate was initially ruled out in preliminary testing, following DOL optimization and testing against active virus, this conjugate showed the greatest aggregation in response to active ZIKV with negligible aggregation in the presence of BSA (Fig. 8A). Interestingly, CFA0204 and CFA0201 showed the highest binding to ZIKV-E during microscale thermophoresis (MST) analysis, Kd = 8.75 nM and Kd = 2.23 nM, respectively (ESI Fig. 6‡), though the slightly lower binding CFA0204 outperformed CFA0201 against live virus in terms of decreasing the absorbance at 528 nm (data not shown).
Fig. 8. Best performing aptamer, CFA0204, detecting Zika virus. (A) ZIKV at 105 PFU was able to induce aggregation as evidenced by a decrease in A528. (B) While precipitation was apparent, no shift to a blue color was induced by ZIKV.
Discussion
Mosquitoes continue to be the largest natural threat to global human health, accounting for the most deaths from any animal via the disease agents they vector. The ability to rapidly identify the presence of medically important vector species or determine a mosquito's infection status would prove an indispensable tool for mosquito surveillance and control specialists around the world. This project sought to assess the potential of using Apt–AuNP conjugate technology for rapid, colorimetric detection of targets pertinent to mosquito surveillance and control (i.e., Ae. aegypti salivary proteins and ZIKV). The ultimate goals are to detect these targets via end-use card-based diagnostics or observing a colour change of the mosquito abdomen after imbibing Apt–AuNP solution (Fig. 1).
Apt–AuNPs first were optimized by changing the capping agent from citrate to a sucrose, which produced particles that were not only stable but also responsive to analyte within a specially formulated diagnostics solution. The components of this diagnostic buffer (outlined in Experimental) entice mosquitoes to feed and are therefore critical to the deployment of our diagnostic in the field either on a card or as a mosquito imbibed diagnostic (Fig. 1). As citrate-capped AuNPs either non-specifically aggregated and Apt–AuNPs were poorly responsive to Ae. aegypti SGE (Fig. 2), sucrose was explored as a capping agent. This lead to an improvement in sensitivity down to 10 ng protein by UV-Vis and 50 ng by visual inspection (Fig. 4). Aptamers raised against Ae. aegypti salivary gland extracts (rich in D7, the most abundant protein in mosquito saliva14,25,26) were able to detect recombinant Ae. albopictus D7, possibly making the diagnostic a “pan-Aedes” detection tool as Ae. aegypti and Ae. albopictus D7 proteins share significant sequence homology.25,26 Only two Aedes mosquito species were tested so further validation is needed to determine whether salivary gland proteins from other Aedes species react similarly. Regardless, Ae. aegypti and Ae. albopictus in the subgenus Stegomyia are regarded as the main vectors of Zika, chikungunya, dengue and yellow fever viruses and so their detection is most relevant as a diagnostic tool for surveillance.27,28 Following Apt–AuNP optimization, we were able to detect as low as 15 ng of recombinant Ae. albopictus D7 protein (Fig. 5).
In addition to modifying AuNP capping, aptamer derivatization (degree of labelling, DOL) upon the AuNP surface was explored with aptamers sensitive to Zika virus envelope (ZIKV-E) protein. Initially, ZIKV-E specific Apt–SAuNPs showed no visible colour change against 200 nM ZIKV-E (ESI Fig. 3‡). By lowering the DOL, detection was improved to 4 nM ZIKV-E by UV-Vis with a vibrant colour change against 40 nM ZIKV-E (Fig. 7). This improvement in sensitivity is likely due to the presence of fewer negatively charged DNA-based aptamers on the surface of the AuNP that reduces the repulsion and facilitates aggregation after analyte binding.22,29–31 Also, the packing density of the aptamers play an important role in the aptamer–analyte binding.32,33 The higher packing density might interfere with adequate aptamer folding, thus affecting the aptamer–analyte interaction.34 A lower packing density results in less binding sites, thus decreasing the sensitivity of the approach.35
Following these optimizations, active ZIKV was tested and the top performing ZIKV-specific Apt–SAuNP was able to undergo aggregation in the presence of 105 PFU of ZIKV in 10 μL of buffer (Fig. 8). This sensitivity would be sufficient to detect Zika virus found in infected mosquitos as the salivary gland and midgut tissues have been shown to contain arboviral titers as high as 104 and 105 PFU, respectively.38–40 No visible color shift was noted in the presence of active ZIKV, likely due to the altered aggregation architecture of AuNPs with the intact ZIKV. A previous study slowly increased the AuNP–AuNP interparticle distance and reported the disappearance of the absorbance band at 540 nm when the distance approached a magnitude similar to or greater than the AuNP diameter.36 Because the Zika virion is similar in size to AuNPs,24 the AuNPs may not be approaching the minimum distance needed for interparticle plasmon–plasmon interactions to ensue. Speaking to the specificity of the Apt–AuNP system, previous work in our lab has shown that ZIKV-E aptamers and aptamers generated against chikungunya envelope protein 1 (CHIKV-E1) show no cross reactivity in a microfluidic-based assay37 and, as stated in the Experimental section, our ZIKV-E binding aptamer was negatively selected against recombinant dengue envelope protein during SELEX. This shows that we can distinguish between flavivirus and alphavirus envelope proteins though future work will incorporate live flaviviruses like dengue or West Nile to see if this assay can distinguish whole virions of related viruses.
Conclusions
This study serves as the critical first step in adapting colorimetric aptamer–gold nanoparticle technology for use within deployable tools for mosquito surveillance and control. Herein we show the development and testing of novel aptamers and Apt–AuNPs that specifically detect protein in vector mosquito saliva (Ae. aegypti and albopictus) as well as new aptamers and Apt–AuNPs against ZIKV. This initial exploration into the colorimetric detection of vector mosquito species and arboviruses will hopefully lead to future in-field, colorimetric diagnostics with the potential to be powerful, integral components of mosquito surveillance and control. Exploration of maximum analyte size that may be detected with a colorimetric aptamer–gold nanoparticle system would also improve the understanding technological limits when detecting targets such as whole virions. Such nanoscale level tools and diagnostics will further expand the impact of nanoscience techniques for the betterment of global health.
Experimental
Mosquito rearing (University of Central Florida College of Medicine Mosquito Colony)
Ae. aegypti eggs obtained from the United States Department of Agriculture-Agricultural Research Service, Center for Medical, Agricultural and Veterinary Entomology (USDA-ARS-CMAVE, Gainesville, FL) were kept on seed germination paper in humidified containers in the dark. For each batch, 800 eggs (by weight; ∼8 mg) were brushed off the cards and dispersed in 3 L of spring water (Zephyrhills, FL) and 7.5 mL of 50 g L−1 larval food consisting of a 60 : 40 mixture of liver powder (MP Biologics, Santa Ana, CA) and brewer's yeast (https://www.Insectsales.com). Rearing trays were incubated at 29 °C and larval food added at days four (7.5 mL), five and six (10 mL). At d6 or at a time when there was greater than 50% pupae, the larvae/pupae were collected and weighed. Up to 4 g of pupae/larvae were added to an 8 oz specimen cup with 200 mL water. Adult mosquitoes were provided 10% sucrose ad libitum via a saturated cotton ball placed atop the holding cage (8 × 8′ Bioquip, Rancho Dominguez, CA).
Extraction of salivary glands
Mosquitoes were cold anesthetized and had the wings and legs removed; a small drop of buffer, ∼10 μL of phosphate-buffered saline (PBS), was then applied at the head. The salivary glands were then dissected away and collected. Glands were stored at −80 °C until further processing for protein isolation.
Protein isolation, purification and characterization
Salivary glands from approximately 200 mosquitos were pooled within gentleMACS™ C Tubes (Miltenyi Biotec Inc., San Diego, CA) and dissociated with pre-set template B twice. The subsequent salivary gland extracts (SGE) were clarified via centrifugation at 120 × g for 5 min at 10 °C and filtered through a 70 μm cell strainer. SGEs were centrifuged again within a 1.5 mL microcentrifuge tube at 10k × g for 5 min at 4 °C to pellet remaining debris. Supernatants were transferred to new 1.5 mL tubes and trichloroacetate (TCA) was added to yield a final concentration of 15–20% TCA. Samples were centrifuged at 13.5k × g for 10 min at 4 °C to pellet protein. Ice-cold acetone was added (500 μL) and pellets were dislodged. Samples were stored in acetone for at least 1 h at −20 °C before centrifugation at 13.5k × g for 10 min at 4 °C and repeated washings. Pellets were air dried for 3–5 min and resuspended in 1 mM PBS at 37 °C for 20–30 min with agitation every 5 min and overnight incubation at 4 °C. Samples were centrifuged at 10k × g for 1 min at RT to remove insoluble fractions. 10 μL of 1 mM PBS was added and pipetted to wash pellets before centrifugation at 10k × g for 1 min at RT. Spin-filter concentrators with molecular weight cut-offs of 30 kDa and 50 kDa were employed to create focused extracts presumably rich in salivary protein D7, Mw ∼ 37–39 kDa (ref. 14) (ESI Fig. 5‡). Protein concentrations were determined by Bradford assay (Thermo Fisher) and purified protein samples were ran on 4–12% bis-Tris acrylamide gels (Thermo Fisher) (50–500 ng). Gels were stained with Pierce™ Silver Stain kits (Thermo Fisher) and dried with a DryEase® System (Thermo Fisher).
Diagnostic buffer solution (i.e., aptamer working solution)
The developed aptamer-based diagnostics within this project will be eventually used in the field and therefore must be within a solution that mosquitoes feed upon. Consequently, all experiments were conducted within a diagnostic buffer formulated within the lab that both aids in aptamer stability and entices mosquitoes to feed.5 The formulation consisted of: 10% sucrose, 15 mM NaCl, 1 mM NaHCO3, 1 μM MgCl2 and 5 μM β, γ-methylene-ATP, all at an unadjusted solution pH of ∼8.3.
Aptamer discovery
SGE from adult female Ae. aegypti and recombinant Zika envelope protein (ZKV-005; ProSpec Bio, Ness-Ziona, Israel) were sent to Base Pair Biotechnologies, Inc. (BPB, Pearland, TX) as targets to discover novel aptamers. The diagnostic buffer described above was used during the selection of the aptamers to ensure optimal function in the intended end-use chemical environment. Targets were input into multiple rounds of SELEX processing followed by high-throughput microarray analysis of thousands of aptamer candidates against respective targets. A commercially available dengue envelope protein (DEN-036; ProSpec Bio, Ness-Ziona, Israel) was utilized during selection of ZIKV-E binding aptamers for negative selection to enhance specificity towards a single flavivirus envelope protein.
Preparation and characterization of gold nanoparticles (AuNPs)
Citrate capped gold nanoparticles (∼13 nm) were prepared using and adapting methods described elsewhere.15 Briefly, aqueous solutions of HAuCl4 (100 mL, 1 mM) were reduced using trisodium citrate solution (2 mL, 194 mM) at 90 °C. The colour of the solution changes from faint yellow to a reddish wine-like colour indicating nanoparticle formation. Solutions were then passed through a 0.22 μm syringe filter to obtain nanoparticles with homogenous size.
Sucrose capped AuNPs (Type 1) were created using an adapted method described previously.16 Briefly, gold(iii) chloride trihydrate aqueous solution (10 mM) was mixed with adequate ascorbic acid and sucrose used as capping and reducing agent (ESI Table 2‡). The solution was allowed to stir for 20 min before filtering with a 0.22 μm syringe filter. Synthesis parameters and sizes listed in ESI Table 1.‡
A separate synthesis method (Type 2) was also utilized.17 Briefly, sucrose (0.05 g) was dissolved in 3 mL of 10 mM HAuCl4 solution. The solution was allowed to dry at room temperature until a solid residue was obtained. The residue was heated at 80 °C for 15 min which resulted in a change of colour from yellow to deep green. After cooling to room temperature, 100 mL deionized water was added and stirred vigorously for 1 h. While stirring, the colour of the solution changed from a yellow into a dark red indicating the formation of AuNPs. After mixing, solutions were refluxed for 20 min and allowed to cool to room temperature. Solutions were then passed through a 0.22 μm syringe filter to obtain nanoparticles with homogenous size. Dynamic light scattering (DLS) and high-resolution transmission electron microscopy (HR-TEM) were used to measure size; surface charge was analysed using DLS.18,19
Preparation and characterization of salivary aptamer–gold nanoparticle conjugates (Apt–AuNPs)
Cryopreserved aptamers obtained from BPB were brought to room temperature and diluted using the provided folding buffer as described in the BPB best-practices documentation. Aptamers were folded by incubating at 95 °C for 5 min and kept at room temperature for another 15 min. Apt–AuNPs conjugates were prepared by mixing AuNPs with different concentrations of aptamers (2 μM, 200 nM, 20 nM and 2 nM) to assess the effects degree of labelling has of diagnostic sensitivity. Different concentrations of soluble Ae. aegypti SGE, recombinant Ae. albopictus D7 or ZIKV-E were used to screen each Apt–AuNP. Bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO) was used a negative control. Aggregation of Apt–AuNPs was evaluated using UV-Vis spectroscopy (PerkinElmer Lambda-750S) and visual inspection. For all experiments, data was processed in Origin Pro (OriginLab Corporation; Northampton, MA) and baseline corrected prior to plotting and analysis. For these absorbance studies, a sample of each Apt–AuNP was measured three (3) times and the standard deviations were calculated from these values. Representative spectra are presented.
Testing Apt–AuNPs against Zika virus
Zika strain MR-766 was produced by infecting C6/36 insect cells and collecting cell culture media. This media was clarified by centrifugation at 13k × g before being placed atop a 30% glycerol cushion and centrifuged at 100k × g in an SW-28 rotor. Viral pellets were resuspended in diagnostic buffer, had the plaque forming units (PFU) determined and were stored at −80 °C until use. ZIKV-specific Apt–AuNPs were tested against 104 to 107 PFU ZIKV and the absorbance at 528 nm and 640 nm were recorded at 1 h, 4 h and 24 h on a Nanodrop 8000 (Thermo Scientific; Waltham, MA). A digital camera was used to capture colour images of tubes containing samples. For controls, viral titres were matched with a BSA concentration equal to the total protein content of virus (determined by A280). This experiment was performed in triplicate and the means and corresponding standard deviations were calculated using the obtained absorbance data. Subsequent time points were sampled from the same tube (i.e., each sample had 2 μL removed for imaging on the spectrophotometer at each time point).
Degree of labelling determination
Aptamers CFA0201, CFA0204, and their PCR primers were created by BPB. Apt–AuNPs were created by derivatizing bare sucrose-capped gold nanoparticle (SAuNP) surfaces with either aptamer CFA0201 or CFA0204 at three concentrations of aptamer (20, 2 and 0.2 nM). Following aptamer conjugation as described above, direct quantitation of the amount of aptamer remaining in the sample (i.e., conjugated to the AuNP surface) was determined by quantitative PCR (qPCR). In brief, a 10-fold serial dilution of free aptamers CFA0201 and CFA0204 was created from 10 nM to 1 pM using dH2O. The solutions were then mixed with bare SAuNP (OD 0.2) at 1 : 1 volume ratio to generate a set of standards at concentrations from 5 nM to 0.5 pM in bare SAuNP at OD 0.1. Sample Apt–SAuNPs were diluted to OD 0.1 using dH2O. qPCR reactions were prepared by mixing 10 μL of Applied Biosystems Power SYBR Green PCR Master Mix (ThermoFisher Scientific, cat# 4367659), 0.6 μL of 10 μM forward primer, 0.6 μL of 10 μM reverse primer, 4 μL of template (standard or sample), and 4.8 μL of dH2O. Reactions were run on QuantStudio 3 Real-Time PCR System (ThermoFisher Scientific) with protocol of 95 °C for 10 min, then 35 cycles of 95 °C, 15 s; 60 °C, 1 min. Data were analysed using Excel software (Microsoft).
Conflicts of interest
Authors GWJ, RHB, TDNH and VS are full-time employees of Base Pair Biotechnologies, Inc.
Supplementary Material
Acknowledgments
Financial support for this project was provided via a grant from the Florida Department of Agriculture and Consumer Services (FDACS #022392, #023593, #24236). We would also like to acknowledge and thank the following people, organizations and agencies for aiding, supporting and/or otherwise enabling this project: the University of Central Florida College of Medicine (UCF-COM), the UCF-COM Department of Internal Medicine, the Florida Coordinating Council on Mosquito Control (FCCMC), the USDA-ARS CMAVE, (Gainesville, FL), Daniel Kline, Joyce Urban and team (USDA-ARS-CMAVE) for help collecting saliva and salivary glands as well as providing work space, adult mosquitoes, mosquito eggs and mosquito rearing guidance, Matthew Longo from the Parks Lab for aid in the production of purified ZIKV, Dr Darrin Kuystermans and Sanford Burnham Prebys Medical Discovery Institute at Lake Nona for recombinant protein production, Ryan Dickerson from UCF-COM for helping with artwork and Christopher S. Bibbs (Education Specialist, Anastasia Mosquito Control District of St. Johns County), Dennis Moore (Director, Pasco County Mosquito Control District) and Kelley Deutsch (Orange County Mosquito Control) for supporting the project and educating the Willenberg Lab on mosquito control.
Parts presented at the 2018 Florida Mosquito Control Annual Meeting in St. Petersburg, FL and the 2019 American Mosquito Control Annual Meeting in Orlando, FL.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02089f
References
- Pan American Health Organization and World Health Organization, Zika - Epidemiological Report. United States of America, PAHO/WHO, Washington, D.C., June 2017 [Google Scholar]
- Mlakar J. Korva M. Tul N. Popovic M. Poljsak-Prijatelj M. Mraz J. Kolenc M. Rus K. R. Vipotnik T. V. Vodusek V. F. Vizjak A. Pizem J. Petrovec M. Zupanc T. A. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016;374(10):951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control, Rapid risk assessment: Zika virus epidemic in the Americas: potential association with microcephaly and Guillain-Barré syndrome, ECDC, Stockholm, 10 December 2015 [Google Scholar]
- Hall-Mendelin S. Ritchie S. A. Johansen C. A. Zborowski P. Cortis G. Dandridge S. Hall R. A. van den Hurk A. F. Exploiting mosquito sugar feeding to detect mosquito-borne pathogens. Proc. Natl. Acad. Sci. U. S. A. 2010;107(25):11255–11259. doi: 10.1073/pnas.1002040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal S. and Willenberg B. J., Colorimetric Sensor Devices Using Aptamer-Gold Nanoparticle Conjugates for Field Surveillance of Mosquito-Borne Diseases. FDACS 2015 Final Report. Contract #022392, retrived from FDACS website: https://www.freshfromflorida.com/content/download/73603/2112338/Willenberg_Colorimetric_Sensor_Device_Final_Report, accessed September 14th, 2018 [Google Scholar]
- Tuerk C. Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ellington A. D. Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Song K. M. Lee S. Ban C. Aptamers and their Biological Applications. Sensors. 2012;12(1):612–631. doi: 10.3390/s120100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. Z. Huang P. J. J. Ding J. S. Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139(11):2627–2640. doi: 10.1039/C4AN00132J. [DOI] [PubMed] [Google Scholar]
- Jeon W. Lee S. Manjunatha D. H. Ban C. A colorimetric aptasensor for the diagnosis of malaria based on cationic polymers and gold nanoparticles. Anal. Biochem. 2013;439(1):11–16. doi: 10.1016/j.ab.2013.03.032. [DOI] [PubMed] [Google Scholar]
- Liu J. Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat. Protoc. 2006;1(1):246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- Lee S. Manjunatha D. H. Jeon W. Ban C. Cationic Surfactant-Based Colorimetric Detection of Plasmodium Lactate Dehydrogenase, a Biomarker for Malaria, Using the Specific DNA Aptamer. PLoS One. 2014;9(7):e100847. doi: 10.1371/journal.pone.0100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y., Development of aptamer-nanoparticle conjugates as a new approach to malaria diagnosis, University of Hong Kong, 2012 [Google Scholar]
- Valenzuela J. G. Pham V. M. Garfield M. K. Francischetti I. M. B. Ribeiro J. M. C. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2002;32(9):1101–1122. doi: 10.1016/S0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Storhoff J. J. Elghanian R. Mucic R. C. Mirkin C. A. Letsinger R. L. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998;120(9):1959–1964. doi: 10.1021/ja972332i. [DOI] [Google Scholar]
- Hurtado R. B. Cortez-Valadez M. Ramírez-Rodríguez L. P. Larios-Rodriguez E. Alvarez R. A. Rocha-Rocha O. Delgado-Beleño Y. Martinez-Nuñez C. E. Arizpe-Chávez H. Hernández-Martínez A. R. Flores-Acosta M. Instant synthesis of gold nanoparticles at room temperature and SERS applications. Phys. Lett. A. 2016;380(34):2658–2663. doi: 10.1016/j.physleta.2016.05.052. [DOI] [Google Scholar]
- Qi Z. M. Zhou H. S. Matsuda N. Honma I. Shimada K. Takatsu A. Kato K. Characterization of gold nanoparticles synthesized using sucrose by seeding formation in the solid phase and seeding growth in aqueous solution. J. Phys. Chem. B. 2004;108(22):7006–7011. doi: 10.1021/jp035972i. [DOI] [Google Scholar]
- Das S. Singh S. Dowding J. M. Oommen S. Kumar A. Sayle T. X. T. Saraf S. Patra C. R. Vlahakis N. E. Sayle D. C. Self W. T. Seal S. The induction of angiogenesis by cerium oxide nanoparticles through the modulation of oxygen in intracellular environments. Biomaterials. 2012;33(31):7746–7755. doi: 10.1016/j.biomaterials.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnihotri N. P. Bhide M. Design of Aptamer-Gold Nanoparticles Based Colorimetric Assay for the Early Diagnosis of Breast Tumor. Int. J. Sci. Res. 2012:1703–1707. [Google Scholar]
- Katti K. K. Kattumuri V. Bhaskaran S. Katti K. V. Kannan R. Facile and general method for synthesis of sugar-coated gold nanoparticles. Int. J. Green Nanotechnol. 2009;1(1):B53–B59. doi: 10.1080/19430850902983848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvarna S. Das U. Sunil K. C. Mishra S. Sudarshan M. Das Saha K. Dey S. Chakraborty A. Narayana Y. Synthesis of a novel glucose capped gold nanoparticle as a better theranostic candidate. PLoS One. 2017;12(6):1–15. doi: 10.1371/journal.pone.0178202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraf N. Bosak A. Willenberg A. Das S. Willenberg B. J. Seal S. Colorimetric detection of epinephrine using an optimized paper-based aptasensor. RSC Adv. 2017;7(77):49133–49143. doi: 10.1039/C7RA10272K. [DOI] [Google Scholar]
- Aliakbarinodehi N. Jolly P. Bhalla N. Miodek A. De Micheli G. Estrela P. Carrara S. Aptamer-based Field-Effect Biosensor for Tenofovir Detection. Sci. Rep. 2017;7:1–10. doi: 10.1038/srep44409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D. Chen Z. G. Sun L. Klose T. Pierson T. C. Rossmann M. G. Kuhn R. J. The 3.8 angstrom resolution cryo-EM structure of Zika virus. Science. 2016;352(6284):467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E. Mans B. J. Andersen J. F. Ribeiro J. M. C. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 2006;281(4):1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Ribeiro J. M. C. Arca B. Lombardo F. Calvo E. Phan V. M. Chandra P. K. Wikel S. K. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:1–27. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S. V. Tesh R. B. Vasilakis N. The emergence of arthropod-borne viral diseases: a global prospective on dengue, chikungunya and zika fevers. Acta Trop. 2017;166:155–163. doi: 10.1016/j.actatropica.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C. Charlier C. Vasilakis N. Lecuit M. Zika, Chikungunya, and Other Emerging Vector-Borne Viral Diseases. Annu. Rev. Med. 2018;69:395–408. doi: 10.1146/annurev-med-050715-105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C. Wen W. Lin F. G. Zhang X. H. Gu H. S. Wang S. F. Simplified aptamer-based colorimetric method using unmodified gold nanoparticles for the detection of carcinoma embryonic antigen. RSC Adv. 2015;5(15):10994–10999. doi: 10.1039/C4RA14833A. [DOI] [Google Scholar]
- Javier D. J. Nitin N. Levy M. Ellington A. Richards-Kortum R. Aptamer-targeted gold nanoparticles as molecular-specific contrast agents for reflectance imaging. Bioconjugate Chem. 2008;19(6):1309–1312. doi: 10.1021/bc8001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. A. Chiuman W. Lam J. C. F. McManus S. A. Chen W. Cui Y. G. Pelton R. Brook M. A. Li Y. F. DNA aptamer folding on gold nanoparticles: from colloid chemistry to biosensors. J. Am. Chem. Soc. 2008;130(11):3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- White R. J. Phares N. Lubin A. A. Xiao Y. Plaxco K. W. Optimization of electrochemical aptamer-based sensors via optimization of probe packing density and surface chemistry. Langmuir. 2008;24(18):10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. Brook M. A. Li Y. F. Design of Gold Nanoparticle-Based Colorimetric Biosensing Assays. ChemBioChem. 2008;9(15):2363–2371. doi: 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- Saraf N. Woods E. R. Peppler M. Seal S. Highly selective aptamer based organic electrochemical biosensor with pico-level detection. Biosens. Bioelectron. 2018;117:40–46. doi: 10.1016/j.bios.2018.05.031. [DOI] [PubMed] [Google Scholar]
- Cheng A. K. H. Ge B. Yu H. Z. Aptamer-based biosensors for label-free voltammetric detection of lysozyme. Anal. Chem. 2007;79(14):5158–5164. doi: 10.1021/ac062214q. [DOI] [PubMed] [Google Scholar]
- Schmitt J. Decher G. Dressick W. J. Brandow S. L. Geer R. E. Shashidhar R. Calvert J. M. Metal nanoparticle/polymer superlattice films: fabrication and control of layer structure. Adv. Mater. 1997;9(1):61–65. doi: 10.1002/adma.19970090114. [DOI] [Google Scholar]
- Saraf N. Villegas M. Willenberg B. J. Seal S. Multiplex Viral Detection Platform Based on a Aptamers-Integrated Microfluidic Channel. ACS Omega. 2019;4:2234–2240. doi: 10.1021/acsomega.8b03277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvez E. O'Connor O. Pol M. Faye O. Richard V. Tarantola A. Dupont-Rouzeyrol M. Differential transmission of Asian and African Zika virus lineages by Aedes aegypti from New Caledonia. Emerging Microbes Infect. 2018;7(1):159. doi: 10.1038/s41426-018-0166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotiwan N. Andre B. G. Sanchez-Vargas I. Islam M. N. Grabowski J. M. Hopf-Jannasch A. Gough E. Nakayasu E. Blair C. D. Belisle J. T. Hill C. A. Kuhn R. J. Perera R. Dynamic remodeling of lipids coincides with dengue virus replication in the midgut of Aedes aegypti mosquitoes. PLoS Pathog. 2018;14(2):e1006853. doi: 10.1371/journal.ppat.1006853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglero-Rodriguez Y. I. MacLeod H. J. Kang S. Carlson J. S. Jupatanakul N. Dimopoulos G. Aedes aegypti molecular responses to zika virus. Modulation of infection by the Toll and Jak/Stat immune pathways and virus host factors. Front. Microbiol. 2017;8:2050. doi: 10.3389/fmicb.2017.02050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.