Abstract
Metal oxide nanoparticles (NPs) have shown enhanced antibacterial effects against many bacteria. Thus, understanding the potential antibacterial effects of nickel oxide nanoparticles (NiO NPs) against Gram-positive and Gram-negative pathogenic bacteria is an urgent need to enable the exploration of NiO NP use in biomedical sciences. To this end, NiO NPs were synthesized by microwave assisted hydrothermal synthesis method. The synthesized NPs were characterized by X-ray diffraction (XRD) and Fourier Transfer Infrared (FT-IR) and UV-visible spectroscopy. The morphological features of the synthesized NiO NPs were analysed using Transmission Electron Microscopy (TEM) and FE-SEM analysis. The antibacterial activity of NiO NP was explored using different antimicrobial and biophysical studies. The obtained data reveals that the NiO NP has stronger antibacterial activity against Gram-positive bacteria compared to Gram-negative bacteria. The mechanism behind the antibacterial activity of the NiO NP was explored by evaluating the amount of ROS generation at the NiO NP interface. The effect of ROS generation on the bacterial membrane was evaluated by BacLight assay and morphological analysis of the bacterial membrane using FE-SEM. The data altogether suggested that the oxidative stress generated at the NiO NP interface resulted in membrane damage leading to bacterial cell death.
Metal oxide nanoparticles (NPs) have shown enhanced antibacterial effects against many bacteria.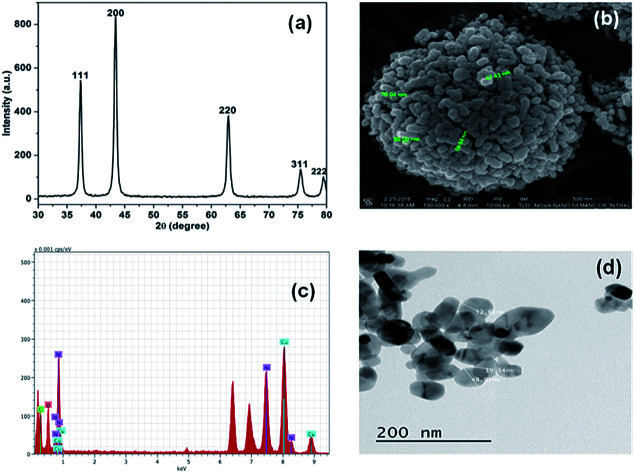
Introduction
Increasing pathogenic resistance has become a serious threat to medical science in the treatment of several human diseases.1 More than 70% of the bacterial infections are now resistant to conventional/traditional antibiotics that are used to treat the infection.2–4 Hence, the bacterial resistance towards traditional or conventional antibiotics is increasing in an alarming rate, and this has become a major global health concern in the current era.5–8 It is now important to check the development of such new resistant strains by alternative antibiotic approaches. Many research groups have recently observed that metal oxide nanoparticles (NPs) have shown better antimicrobial activity against a wide range of bacterial strains.9,10 Thus, these nanoparticles could be adopted as potential antimicrobial agents.11–17 These NPs have drawn more attention as new generation antimicrobial agents, as they are less likely to develop resistance compared to the traditional or conventional antibiotics against different bacteria.18 Additionally, metal oxide nanoparticles shape, size, morphology and physicochemical characteristics are also important, and have shown greater impacts and effects on the antimicrobial activity against many microbes.19–22 In this context, Arakha et al. have investigated the effects of different physico-chemical parameters of nanoparticles such as size, shape, band gap energy, crystallinity, interfacial potential, and functional groups at bio-nano interface on antimicrobial activity and cytotoxicity of nanoparticles.10,23–25 Additionally, Venkatesan et al. have investigated that porous chitosan–alginate–AgNPs composite can be used in antimicrobial filtration and cancer treatment.26
The size of NPs in the nano range helps the nanoparticles to have high surface area to volume ratio, hence possess high free energy content.24 The NPs maintain the stability by reducing free energy upon interaction with different interactomes inside the colloidal solution/biological milieu.24 Nanoparticle-induced oxidative stress has increased the membrane lipid peroxidation and challenged the antioxidant defence system in some bacteria. This can also be explained by decrease in intracellular glutathione (GSH) or alternatives responsible for cell viability and retardation of bacterial growth in bacteria.27 Over and above, nanoparticle induced oxidative stress could be an advantage for their use as antimicrobial agents. Recently, many such reports have mentioned and justified the exciting applications of nanoparticles. To mention a few such as Ag NP, ZnO NP, Fe2O3 NP, MgO NP and NiO NP have shown different antimicrobial activities against Gram-positive and Gram-negative bacteria.28–32
Nickel oxide nanoparticle (NiO NP) has drawn various applications both in material science and biomedical sciences.33,34 The present study investigates the potential antimicrobial activity of NiO NP against Gram-positive bacteria Bacillus subtilis (B. subtilis) and Gram-negative bacteria Escherichia coli (E. coli) to correlate and improve its biomedical applications. In this context, our investigation has explored how the reactive oxygen species generated at nickel oxide nanoparticle interface put stress on bacterial membrane determining the antibacterial activity of nickel oxide nanoparticles. Although, antimicrobial activity along with mechanism for various nanoparticles is well established, however to the best of our knowledge, oxidative and membrane stress mediated antimicrobial activity of NiO NP has not been reported till date completely. To this end, we have used different biophysical and microscopic techniques to explore the effectiveness of nickel oxide nanoparticles against Gram-positive and Gram-negative bacteria. The findings of the study explored the underlying mechanism of bacterial cell non-viability, i.e. predominantly through ROS-mediated pathway. This work is specific on its way to address the effectiveness of NiO NP and accordingly it could be adopted as an alternative antimicrobial agent for future use.
Materials and methods
Nickel chloride hexahydrate, nutrient broth and tannic acid were purchased from Himedia, India. Urea was purchased from Sigma-Aldrich, USA. Glutaraldehyde was purchased from Merck, India, and 2′,7′-dichlorodihydrofluorescein diacetate (DCHF-DA) was purchased from Cayman Chemicals, USA. All mentioned chemicals were of analytical grade and used without further purification.
Synthesis NiO NP
Nickel oxide nanoparticle (NiO NP) was synthesized from nickel chloride hexahydrate (NiCl2·6H2O) and urea (NH2CONH2) by microwave assisted hydrothermal synthesis method, following the previous protocol with some modifications.35 In brief, nickel chloride hexahydrate (107 mg) and urea (81 mg) were dissolved in 30 mL of deionized water with continuous stirring for 30 minutes at room temperature for complete homogenization. The mixture was then transferred into a Teflon-lined autoclave vessel, and subjected to microwave irradiation using a Sineo, MDS-6, China, microwave synthesizer for 25 minutes at 180 °C. The microwave-mediated product was then cool down to room temperature and the green coloured precipitate of nickel hydroxide Ni(OH)2 was separated out from the solution by centrifugation at 6000 rpm for 30 minutes. The precipitate was then rinsed thoroughly with deionized water followed by ethanol wash, and dried at 100 °C in a hot air oven prior to calcination at 450 °C for 5 hours to obtain the desired NiO NP.
Characterizations of synthesized NiO NP
Powder X-ray diffraction (XRD) study was conducted using an Ultima IV diffractometer (Rigaku, Tokyo, Japan), using Cu-Kα radiation (λ = 1.5405 Å) at 20 kV as the X-ray source. The 2θ scans were recorded at room temperature ranging from 10 to 90 degree in a continuous scan mode.
The FT-IR spectrum of synthesized NiO NP was obtained using an Attenuated Total Reflection Fourier Transfer Infrared (ATR-FTIR) spectrometer (Alpha ATR-FTIR, Bruker, Germany) in the range of 400 to 4000 cm−1 on diamond crystal. All the measurements were performed at room temperature with triplicate scanning. The data was processed using the OPUS Viewer software.
Field Emission Scanning Electron Microscope (FE-SEM) measurements of synthesized NiO NP was performed using a Nova Nano-SEM 450, FEI Netherland at an acceleration voltage of 10 kV. The High-Resolution Transmission Electron Microscopy (HR-TEM) measurement was also performed using a FEI Tecnai F20 transmission electron microscope operating at an accelerating voltage of 200 kV.
The elemental composition of prepared NiO NP was identified by Energy Dispersive X-ray (EDX) analysis. The surface potential value and the average size of the NiO NP was identified by Zeta analyzer (Malvern Zetasizer, Nano ZS90, Netherland). Spectrophotometric measurement was performed using a Carry-100 UV-Visible spectrophotometer (Agilent Technologies Pvt. Ltd., USA) at wavelength 200–800 nm.
Effects of NiO NP interface on antibacterial activity
Initially, the antimicrobial activity of NiO NP against Bacillus subtilis and Escherichia coli bacteria was studied by growth kinetics in the absence and presence of NiO NP. The NiO NP stock solutions were prepared by dispersing NiO NP in sterilized nutrient broth solution followed by sonication for 15 minutes and sterilized using UV-radiation for 30 minutes. For evaluation of antimicrobial activity, different concentrations of NiO NP were added to bacterial culture at mid log phase of growth kinetics in a 96-well cell culture plate. The reaction mixture without NiO NP was taken as control, and the growth kinetic was observed by measuring the optical density (OD) at 600 nm, using a microplate reader (Synergy H1 Hybrid Reader, BioTek, USA) at regular time intervals. Approximately in the mid-log phase of bacterial growth, NiO NP solutions of different reaction concentrations (25, 50, 100, 250 and 500 μg mL−1) were added to the respective wells and the data collection started with a dead time of 10 minutes.
Reactive oxygen species (ROS) detection
To explore the mechanism behind the antibacterial activity of NiO NP, ROS generation at NiO NP interface was estimated using 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA) dye. The DCFH-DA is a peroxynitrite dye with excitation and emission maxima at 503 nm and 523 nm respectively. This dye can detect the presence of both nitric oxide and hydrogen peroxide inside and outside the cells.36,37 Both B. subtilis and E. coli cells added with 200 μM DCFH-DA were seeded in a 96-well cell culture plate at 37 °C, and then different concentrations (25, 250, 500 μg mL−1) of NiO NP were added at the mid log phase of growth kinetics. The fluorescence emission was measured at 523 nm with an excitation at 503 nm using a micro-plate reader (Synergy H1 Hybrid Reader, Biotek, USA). The variations in ROS production was examined by comparing the fluorescence intensities with positive control.
Detection of oxidative stress mediated membrane damaged of bacteria
To explore the impact of oxidative stress generated at NiO NP interface on membrane of bacteria, we have taken the help of BacLight fluorescence assay which differentiate the dead bacterial cells resulted from membrane damage.10 The bacterial samples were prepared using the protocol adopted by Arakha et al.10,24 using bacterial viability kit (L7007, Molecular Probes, Invitrogen, USA) and examined using fluorescence microscope (Olympus IX71, Germany) with a 20× objective lens.
To support the findings obtained from BacLight fluorescence assay, we have analyzed the bacterial membrane using Field Emission Scanning Electron Microscope (FE-SEM), which visualises the morphological variation of bacterial membrane upon NiO NP treatment. The sample for FE-SEM analysis was prepared following the protocol adopted by Arakha et al.24
Results
Characterization of synthesized NiO NP nanoparticles
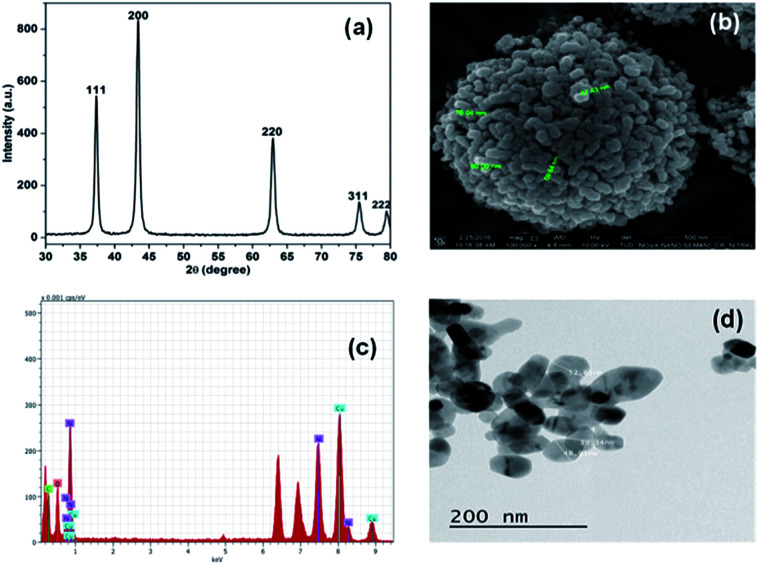
Initially, the synthesis of NiO NP was confirmed using X-ray diffraction spectroscope. The X-ray diffraction (XRD) spectra with no additional peaks for any impurities in Fig. 1(a) indicates the crystalline nature of the synthesize NiO NP. The intense diffraction peaks at different 2θ, i.e. angle values of 37, 43, 63, 75 and 79 correspond to different crystal planes such as (111), (200), (220), (311) and (222) respectively.
Fig. 1. Characterization of synthesized NiO NP. (a) XRD pattern, (b) FE-SEM images of synthesized NiO NP, (c) EDX analysis and (d) TEM images of NiO NP.
These diffraction peaks are in good agreements with the reported diffraction patterns of NiO NP.2 Further, the average crystallite of NiO NP was calculated to be 22.8 nm using the Scherer's equation:Particle size = Kλ/β cos θwhere, K (=0.9) represents proportionality coefficient (shape factor), λ (=1.540 × 10−10 m) wavelength of X-ray, β is the full-width at half-maxima of diffraction peak (in radians) and θ is the Bragg's angle. The band gap energy of the NiO NP was determined to be 3.71 eV by using the equation, Ebg = 1240/λ (eV),10 where Ebg and λ stands for band gap energy (eV) and the wavelength (nm) respectively.
The FE-SEM image in Fig. 1(b) shows the surface morphology features of the synthesized NiO NP. As shown in the figure, most of the synthesized NiO NPs are spherical in shape along with some rod. The size of the particles varies from 40–100 nm. The EDX spectrum in Fig. 1(c) reveals the elemental composition of the synthesized NiO NP. The presence of nickel and oxygen signals in the prepared NiO NP of 77.65% and 22.35%, respectively confirmed the synthesis of NiO NP. The TEM image in Fig. 1(d) shows that NiO NPs are spherical and rod shape with diameter in the range of 10–80 nm.
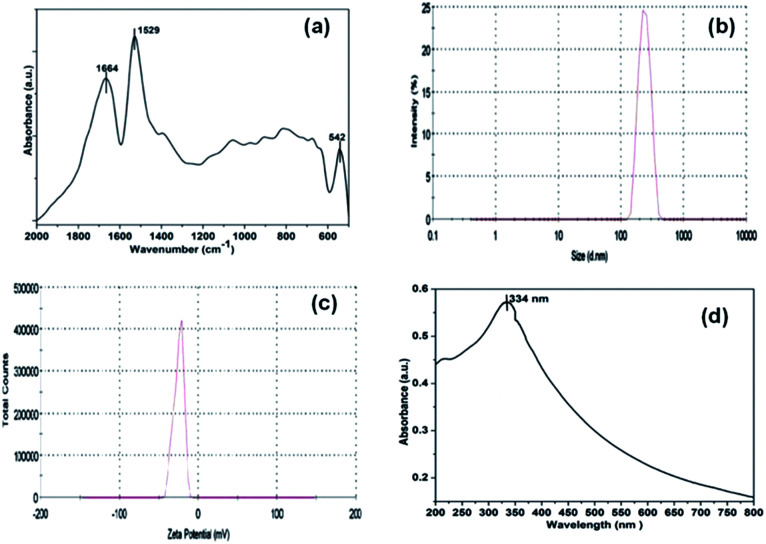
The FT-IR spectrum of NiO NP in Fig. 2(a) shows two distinct strong characteristics bands at 1529 cm−1 and 1664 cm−1 indicating N–H bending and C O stretching vibrations, respectively. The absorption peaks below 800 cm−1 provides important information about internal metal–oxygen bond vibrations.38 The band near 542 cm−1 identifies the presence of internal nickel–oxygen bond. As the sample was thoroughly washed with deionized water, there were no additional peaks indicating the absence of impurities.
Fig. 2. FT-IR spectra (a), DLS analysis (b), zeta potential measurements (c) and UV-visible absorption spectrum (d) of synthesized NiO NP.
Fig. 2(b), displays the averaged hydrodynamic size of synthesized nickel oxide nanoparticles determined in the deionized water by dynamic light scattering (DLS) was found to be 296 nm. Further, the zeta potential of NiO NP in deionized water in Fig. 2(c) has negative surface potential of −25 mV. The prepared NiO NP gives a characteristic absorption peak at 334 nm (Fig. 2(d)) using a Carry-100 UV-visible spectrophotometer scanned in the wavelength range of 200–800 nm at room temperature.
Effect of the NiO NP interfaces on bacterial cell viability
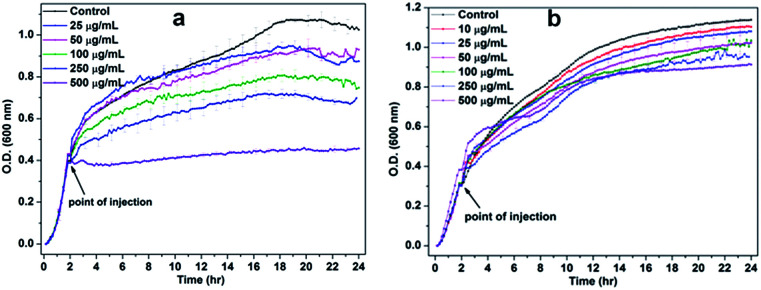
To study the effect of NiO NP interface on bacterial cell viability, the growth kinetic experiments were carried out. In Fig. 3(a), the lower concentrations of NiO NP do not show any significant inhibition against B. subtilis, whereas higher concentrations above 100 μg mL−1 to 250 μg mL−1 shows significant inhibition effects and at 500 μg mL−1 the growth kinetic of B. subtilis was completely suppressed. Whereas in the case of E. coli, we did not observe any significant growth inhibition in the presence of NiO NP, except at 500 μg mL−1 as shown in Fig. 3(b). Thus, the data revealed that NiO NP has strong antimicrobial propensity against studies B. subtilis bacterial strains compared to E. coli.
Fig. 3. Growth kinetics of (a) B. subtilis and (b) E. coli in the absence and presence of different concentrations of NiO NP.
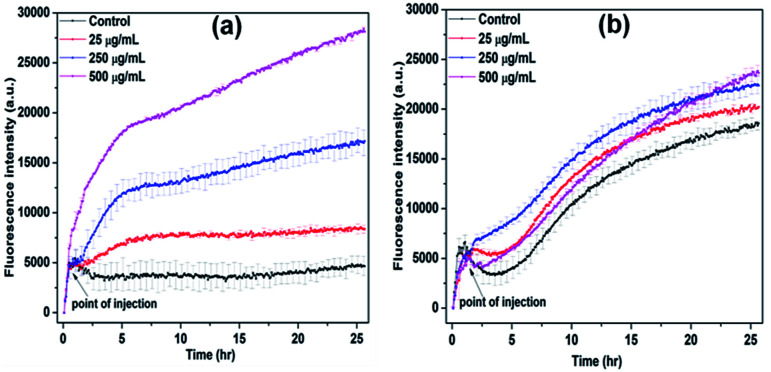
It is known that the production of reactive oxygen species (ROS) is responsible for the lipid, protein and DNA damages and resulted increasing non-viable bacterial population.39 The change in ROS production in the presence of NiO NP was observed using DCFH-DA fluorescence dye. In Fig. 4(a) and (b), it is evident that at higher concentration of NiO NP, i.e. from 250 μg mL−1 to 500 μg mL−1 the production of ROS in the case of B. subtilis was enhanced by five-fold compared to control, whereas in the case of E. coli, it increased by 5% only. The results obtained from growth kinetic study and ROS detection indicates strong antimicrobial activity of NiO NP against Gram-positive bacteria compare to Gram-negative bacteria.40,41
Fig. 4. ROS detection of (a) B. subtilis and (b) E. coli in the presence of different concentrations of NiO NP at room temperature.
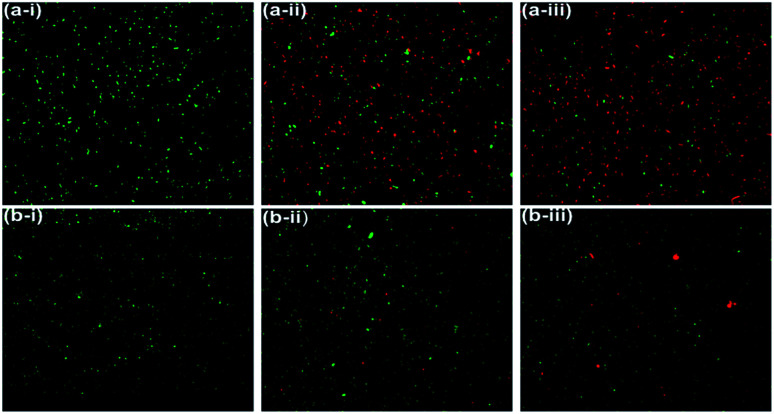
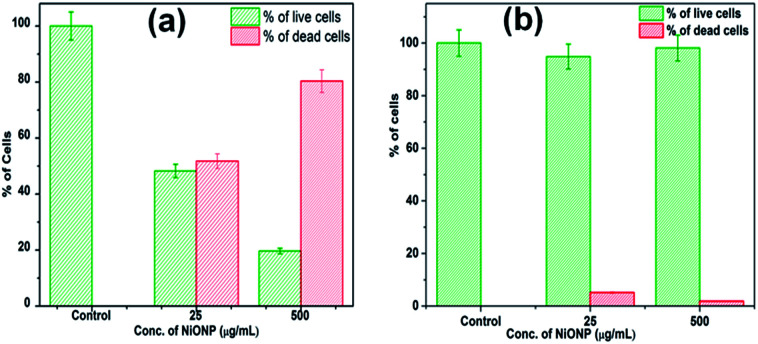
The LIVE/DEAD BacLight fluorescence assay was used further to distinguish and to quantify the viable and non-viable cells resulted from the membrane damage due to oxidative stress at NiO NP interface. When NiO NP was interacted with the bacterial strains, some of cells with integral membrane remain viable, upon treatment give green fluorescence as they are stained by membrane permeable Syto9 dye, whereas non-viable bacteria cells with ruptured/damaged membrane give red fluorescence as they are strained by propidium iodide (PI). In Fig. 5(a-i) and (b-i), the untreated B. subtilis and E. coli cells exhibited green fluorescence indicating the presence of 100% viable cells. Whereas, in presence of 25 μg mL−1 of NiO NP in Fig. 5(a-ii) and (b-ii), the fraction of red cells for B. subtilis was 52% and 6% for E. coli. At highest concentrations of NiO NP (500 μg mL−1) the fraction of red cells increases to 81% for B. subtilis and there was no significant difference in the percentages of red cells at this concentration for E. coli [Fig. 5(a-iii) and (b-iii)]. The percentage of live and dead cells of B. subtilis and E. coli in presence of the NiO NP was estimated by statistical data analysis and presented in Fig. 6(a) and (b), which confirmed the strong antimicrobial activity of NiO NP against B. subtilis and compare to E. coli.
Fig. 5. Fluorescence microscopic images of B. subtilis (a-i) control, (a-ii) 25 μg mL−1 (a-iii) 500 μg mL−1 of NiO NP and E. coli (b-i) control, (b-ii) 25 μg mL−1, and (b-iii) 500 μg mL−1 of NiO NP.
Fig. 6. Percentage of live and dead cells in the presence of NiO NP, (a) B. subtilis and (b) E. coli.
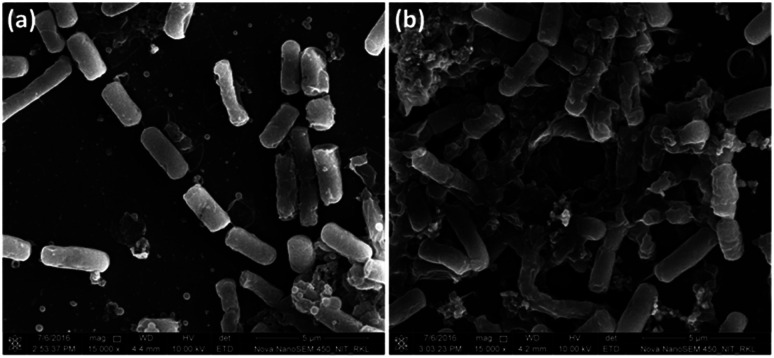
To collect strong evidence about oxidative stress mediated bacterial cell membrane damage, we have visualised the morphology of B. subtilis cells using FE-SEM upon NiO NP treatment (Fig. 7). The NiO NP upon interaction with bacterial cells resulted in ROS formation at the interface. Thus, the generated ROS put stress on the bacterial membrane resulting in bacterial membrane damage. Hence, the FE-SEM image of NiO NP treated bacterial cells show abnormal textures like membrane rupture, membrane damage (Fig. 7(b)) compared to control (Fig. 7(a)). We did the statistics of intact membrane bacteria by taking total 50 cells from 5–7 different FE-SEM micrographs. In control, we found approximately 80% cells with intact membrane, whereas in NiO NP treated cells, we found approximately 10% cells with intact membrane, demonstrating the membrane damaging/antibacterial activity of NiO NP. The ruptured cells no longer remain intact and often found in aggregates or clumps. The results obtained from FE-SEM analysis are in good agreement with the results obtained from BacLight fluorescence assay confirming that the oxidative stress generated at the NiO NP interface resulted in bacterial membrane damage leading to bacterial cell death.
Fig. 7. FE-SEM analysis of NiO NP treated B. subtilis. (a) Control (without NiO NP treatment), and (b) showing membrane damage in NiO NP treated cells.
Discussion
The use of metal oxide NP in medical applications remains a challenge. However, advances in nanoparticle research have addresses many such applications and improved conditions for their use in medicine. Recent studies have revealed that nanoparticles being smaller in size display greater antimicrobial activity against many infectious microorganisms.42 NiO NPs having advanced physico-chemical properties such as electron transfer capability, high chemical stability, super capacitance properties, electro catalysis have drawn the attention of various research groups for different biomedical applications.2 In this context, various research groups have suggested NiO NP as prospective antimicrobial agent and antitumor agent.2 However, the exact mechanism behind antimicrobial activity of NiO NP has not been explored till yet. Although various mechanisms of antimicrobial activity such as generation of ROS, release of metal ions, cell wall damage and dissemination of cell envelop have been reported for various metal oxide NPs against various Gram-positive and Gram-negative bacteria,43–45 but still the mechanism remains unexplored and need extensive evaluation for safe use of nanoparticles as modern antimicrobial agents. Here, we have tried to explore the mechanism to understand the antimicrobial activity of NiO NP against Gram-positive (B. subtilis) and Gram-negative (E. coli) bacteria using different antimicrobial and biophysical techniques. From various antimicrobial and biophysical studies, our investigation intended to explore the role of oxidative stress generated at NiO NP interface resulting in membrane damage leading to bacterial cell death. For the same, we have synthesized small size and stable NiO NP with negative surface potential to explore its antimicrobial activity against selective Gram-positive and Gram-negative bacteria. In the case of Gram-negative bacteria, the presence of additional negatively charged lipopolysaccharides (LPS) makes the surface potential more negative compare to the Gram-positive bacteria.43 It is reported that the interaction between NPs and bacteria membrane is primarily due to the electrostatic interactions at the nanoparticle bacteria interface.24 In this context, many research groups have reported that the electrostatic interaction between the bacterial membrane and different chemical entity is mediated by bacterial membrane surface charge neutralization.10,46,47 The surface charge neutralization is a common biological phenomenon which is attributed in balancing the interaction between positively charged fractions of nanoparticles with negatively charged fractions like phosphates and carboxylates present in LPS of bacteria, altering the membrane permeability, leading to antimicrobial activity.46,47 Here, we observed that synthesized NiO NP with negative surface potential of −25 mV showed stronger antimicrobial propensity towards the partially less negative surface potential of Gram-positive bacteria (B. subtilis) which might be due to stronger electrostatic interactions between two entities. The above findings are supported by the results obtained by Khashan et al.48 where they have investigated that NiO NP has significant antimicrobial activity against wide range of Gram positive bacteria in comparison to Gram negative bacteria. In addition, in our study, we have explored the mechanism behind the antimicrobial activity of NiO NP using different antimicrobial and biophysical experiment. At very low NP concentrations, the interaction was very week as no such specific changes in the growth kinetic were observed. We further observed that the growth kinetic of B. subtilis was strongly inhibited in the presence of higher concentrations of NiO NP than in case E. coli. The Gram-positive bacteria, B. subtilis possess a partially less negative surface potential, thus have strong surface interactions with negative potential NiO NP compared to the negative surface potential Gram-negative E. coli bacteria. The finding from the growth kinetic studies was also validated from ROS detection and backlight assay.
Although many research groups are trying to find out the mechanism behind the antimicrobial activity of different nanoparticles, still it is a matter of intensive research, since exploration of this concept is important to increase the application domain of nanoparticles as modern antibiotics. In this context, many research groups claim that liberation of metal ions from metal oxide nanoparticles to be one of the mechanisms for antimicrobial activity.10,29 We have also investigated the effect of nickel ions on growth of bacteria to explore the antimicrobial activity of nickel ions (Fig. S1, ESI†). The experiment indicated that with increasing nickel ions concentration in growth medium, the growth of both bacteria (B. subtilis and E. coli) increases, and the observed results are in accordance with the findings obtained by Mulrooney et al.49 The nickel is an essential nutrient for some microorganisms which is taken up by ATP-binding cassette-type transport systems or nickel-specific permeases.49
The generated ROS put oxidative stress on the bacterial membrane causing bacterial membrane damage leading to bacterial cell death which was confirmed from BacLight fluorescence assay and FE-SEM analysis. The Fig. 4(a) and (b) clearly indicates that reactive oxygen species (ROS) also produced in the absence of NiO NP, i.e. in control culture, since the dye is showing increasing quantum yield with bacterial growth (black line, control). The produced ROS in non-stress condition is countered by different ROS scavenging enzymes present in bacteria. However, in presence of 250 and 500 μg mL−1 of NiO NP, the ROS production enhanced five folds, which is likely beyond the capacity of ROS scavenging system of the bacteria. The production of ROS in the presence of NiO NP on the interface of B. subtilis mainly results in non-viability of bacterial populations. The ROS production in the case of B. subtilis is significantly high which caused the cell membrane damage.
The effect of oxidative stress generated at NiO NP interface on bacterial cell membrane was explored using BacLight fluorescence assay. The bacterial membrane potential facilitates the interaction of NiO NP on bacterial membrane resulting in generation of ROS leading to bacterial cell membrane damage. This hypothesis is highly supported by the results obtained from BacLight assay and FE-SEM analysis. This observation rationalized that NiO NP shows greater antimicrobial activity towards B. subtilis than E. coli in a concentration dependence manner.
Exploring the physico-chemical phenomena occurring at bio-nano interface, resulted due to interaction of nanomaterials with biological entity inside the biological milieu, is a key challenge to understand the antimicrobial, cytotoxic, cytocompatibility propensities of nanomaterial for their safe use. Hence, this manuscript intends to explore the antimicrobial activity of NiO NP and its underlying mechanism by understanding the physico-chemical phenomena occurring at bacteria-NiO NP interface. Initial findings from growth kinetic analysis suggested that NiO NPs have significant antimicrobial activity against Gram-positive bacteria in comparison to Gram-negative bacteria. However, the evaluation of ROS at interface suggested that the generated ROS at NiO NP interface put stress on bacterial membrane leading to membrane damage as suggested by LIVE/DEAD BacLight bacterial viability assay, SEM analysis, resulting in non-viable bacterial cells. Hence, the oxidative stress generated at NiO NP interface is the principal mechanism behind the antimicrobial activity of NiO NP.
Conclusions
Findings from the above study concluded that ROS generation at the NiO NP interface play important role in determining the antibacterial activity of NiO NP. NiO NPs were successfully synthesized using microwave assisted hydrothermal synthesis method and were characterized using different biophysical techniques. The growth kinetics study initially confirmed the strong antibacterial activity of NiO NP against Gram-positive bacteria compared to Gram-negative bacteria. The mechanism behind this antibacterial activity was explored using ROS detection study and BacLight assay, which clearly demonstrated that, the ROS generated at NiO NP interface put oxidative stress on bacterial cell membrane followed by membrane damage leading to bacterial cell death.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We are thankful to the Department of Ceramic Engineering and Department of Life Science, NIT Rourkela, Odisha, India for supporting the FE-SEM facility and imaging facilities. We are all so thankful to DBT, Govt. of India for financial support to Biraja S. Pattanayak to carry this research work.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02082a
References
- Arakha M. Borah S. M. Saleem M. Jha A. N. Jha S. Free Radical Biol. Med. 2016;101:434. doi: 10.1016/j.freeradbiomed.2016.11.016. [DOI] [PubMed] [Google Scholar]
- Saleem S. Ahmed B. Khan M. S. Al-Shaeri M. Musarrat J. Microb. Pathog. 2017;111:375. doi: 10.1016/j.micpath.2017.09.019. [DOI] [PubMed] [Google Scholar]
- Kaittanis C. Nath S. Perez J. M. PLoS One. 2008;3:e3253. doi: 10.1371/journal.pone.0003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T. Kirketerp Moller K. Jensen P. O. Madsen K. G. Phipps R. Krogfelt K. Hoiby N. Givskov M. Wound Repair Regen. 2008;16:2. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- Hajipour M. J. Fromm K. M. Ashkarran A. A. de Aberasturi D. J. de Larramendi I. R. Rojo T. Serpooshan V. Parak W. J. Mahmoudi M. Trends Biotechnol. 2012;30:499. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Pestova E. Millichap J. J. Noskin G. A. Peterson L. R. J. Antimicrob. Chemother. 2000;45:583. doi: 10.1093/jac/45.5.583. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A. Clin. Infect. Dis. 2005;41:S120. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- Briones E. Colino C. I. Lanao J. M. J. Controlled Release. 2008;125:210. doi: 10.1016/j.jconrel.2007.10.027. [DOI] [PubMed] [Google Scholar]
- Tiwari A. Prince A. Arakha M. Jha S. Saleem M. Nanoscale. 2018;10:3369. doi: 10.1039/C7NR08351C. [DOI] [PubMed] [Google Scholar]
- Arakha M. Saleem M. Mallick B. C. Jha S. Sci. Rep. 2015;5:9578. doi: 10.1038/srep09578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkodi C. Rajeshkumar S. Paulkumar K. Vanaja M. Gnanajobitha G. Annadurai G. Bioinorg. Chem. Appl. 2014;2014:1. doi: 10.1155/2014/347167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller J. C. Liao C.-J. Lewis K. Klibanov A. M. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5981. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Klibanov A. M. Trends Biotechnol. 2005;23:343. doi: 10.1016/j.tibtech.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Stoimenov P. K. Klinger R. L. Marchin G. L. Klabunde K. J. Langmuir. 2002;18:6679. doi: 10.1021/la0202374. [DOI] [Google Scholar]
- Panda S. Yadav K. K. Nayak P. S. Arakha M. Jha S. Bull. Mater. Sci. 2016;39:397. doi: 10.1007/s12034-016-1181-3. [DOI] [Google Scholar]
- Sharma M. Nayak P. S. Asthana S. Mahapatra D. Arakha M. Jha S. IET Nanobiotechnol. 2018;12:626. doi: 10.1049/iet-nbt.2017.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak P. S. Pradhan S. Arakha M. Kumar D. Saleem M. Mallick B. Jha S. IET Nanobiotechnol. 2018;13:193. doi: 10.1049/iet-nbt.2018.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. T. Ahamed M. Alhadlaq H. A. Musarrat J. Al-Khedhairy A. Arch. Oral Biol. 2013;58:1804. doi: 10.1016/j.archoralbio.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Seil J. T. Webster T. J. Int. J. Nanomed. 2012;7:2767. doi: 10.2147/IJN.S24805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi G. Valizadeh H. Barzegar-Jalali M. Lotfipour F. Adibkia K. Milani M. Azhdarzadeh M. Kiafar F. Nokhodchi A. Colloids Surf., B. 2010;80:34. doi: 10.1016/j.colsurfb.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Buzea C. Pacheco I. I. Robbie K. Biointerphases. 2007;2:MR17. doi: 10.1116/1.2815690. [DOI] [PubMed] [Google Scholar]
- Besinis A. De Peralta T. Handy R. D. Nanotoxicology. 2014;8:1. doi: 10.3109/17435390.2012.742935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakha M. Roy J. Nayak P. S. Mallick B. Jha S. Free Radical Biol. Med. 2017;110:42. doi: 10.1016/j.freeradbiomed.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Arakha M. Pal S. Samantarrai D. Panigrahi T. K. Mallick B. C. Pramanik K. Mallick B. Jha S. Sci. Rep. 2015;5:14813. doi: 10.1038/srep14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakha M., Mallick B. C. and Jha S., in Magnetic Nanostructures, Springer, 2019, pp. 287 [Google Scholar]
- Venkatesan J. Lee J.-Y. Kang D. S. Anil S. Kim S.-K. Shim M. S. Kim D. G. Int. J. Biol. Macromol. 2017;98:515. doi: 10.1016/j.ijbiomac.2017.01.120. [DOI] [PubMed] [Google Scholar]
- Nel A. Xia T. Mädler L. Li N. Science. 2006;311:622. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- Tsuji J. S. Maynard A. D. Howard P. C. James J. T. Lam C.-w. Warheit D. B. Santamaria A. B. Toxicol. Sci. 2005;89:42. doi: 10.1093/toxsci/kfi339. [DOI] [PubMed] [Google Scholar]
- Nayak P. S. Arakha M. Kumar A. Asthana S. Mallick B. C. Jha S. RSC Adv. 2016;6:8232. doi: 10.1039/C5RA21281B. [DOI] [Google Scholar]
- Mahmoudi K. Hadjipanayis C. G. Front. Chem. 2014;2:109. doi: 10.3389/fchem.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek Y.-W. An Y.-J. Sci. Total Environ. 2011;409:1603. doi: 10.1016/j.scitotenv.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Gong N. Shao K. Feng W. Lin Z. Liang C. Sun Y. Chemosphere. 2011;83:510. doi: 10.1016/j.chemosphere.2010.12.059. [DOI] [PubMed] [Google Scholar]
- Ezhilarasi A. A. Vijaya J. J. Kaviyarasu K. Maaza M. Ayeshamariam A. Kennedy L. J. J. Photochem. Photobiol., B. 2016;164:352. doi: 10.1016/j.jphotobiol.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Kaviyarasu K. Manikandan E. Kennedy J. Jayachandran M. Ladchumananandasiivam R. De Gomes U. U. Maaza M. Ceram. Int. 2016;42:8385. doi: 10.1016/j.ceramint.2016.02.054. [DOI] [Google Scholar]
- Faisal M. Saquib Q. Alatar A. A. Al-Khedhairy A. A. Hegazy A. K. Musarrat J. J. Hazard. Mater. 2013;250:318. doi: 10.1016/j.jhazmat.2013.01.063. [DOI] [PubMed] [Google Scholar]
- Zhang Q. Kusaka Y. Sato K. Nakakuki K. Kohyama N. Donaldson K. J. Toxicol. Environ. Health, Part A. 1998;53:423. doi: 10.1080/009841098159169. [DOI] [PubMed] [Google Scholar]
- Ogami A. Morimoto Y. Myojo T. Oyabu T. Murakami M. Todoroki M. Nishi K. Kadoya C. Yamamoto M. Tanaka I. Inhalation Toxicol. 2009;21:812. doi: 10.1080/08958370802499022. [DOI] [PubMed] [Google Scholar]
- Xiong H. M. Shchukin D. G. Möhwald H. Xu Y. Xia Y. Y. Angew. Chem., Int. Ed. 2009;48:2727. doi: 10.1002/anie.200805590. [DOI] [PubMed] [Google Scholar]
- Possel H. Noack H. Augustin W. Keilhoff G. Wolf G. FEBS Lett. 1997;416:175. doi: 10.1016/S0014-5793(97)01197-6. [DOI] [PubMed] [Google Scholar]
- Rajan P. I. Vijaya J. J. Jesudoss S. Kaviyarasu K. Kennedy L. J. Jothiramalingam R. Al-Lohedan H. A. Vaali-Mohammed M.-A. Mater. Res. Express. 2017;4:85030. doi: 10.1088/2053-1591/aa7e3c. [DOI] [Google Scholar]
- Ezhilarasi A. A. Vijaya J. J. Kaviyarasu K. Kennedy L. J. Ramalingam R. J. Al-Lohedan H. A. J. Photochem. Photobiol., B. 2018;180:39. doi: 10.1016/j.jphotobiol.2018.01.023. [DOI] [PubMed] [Google Scholar]
- Wang Q. Xu Y.-F. Xu G.-L. Su H. Shen S.-Y. Tu T.-T. Huang L. Li J.-T. Sun S.-G. J. Alloys Compd. 2015;648:59. doi: 10.1016/j.jallcom.2015.06.221. [DOI] [Google Scholar]
- Dutta R. Nenavathu B. P. Gangishetty M. K. Reddy A. Colloids Surf., B. 2012;94:143. doi: 10.1016/j.colsurfb.2012.01.046. [DOI] [PubMed] [Google Scholar]
- Leung Y. Chan C. Ng A. Chan H. Chiang M. Djurišić A. Ng Y. Jim W. Guo M. Leung F. Nanotechnology. 2012;23:475703. doi: 10.1088/0957-4484/23/47/475703. [DOI] [PubMed] [Google Scholar]
- Xie Y. He Y. Irwin P. L. Jin T. Shi X. Appl. Environ. Microbiol. 2011;77:2325. doi: 10.1128/AEM.02149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves C. S. Melo M. N. Franquelim H. G. Ferre R. Planas M. Feliu L. Bardají E. Kowalczyk W. Andreu D. Santos N. C. J. Biol. Chem. 2010;285:27536. doi: 10.1074/jbc.M110.130955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder S. Yadav K. K. Sarkar R. Mukherjee S. Saha P. Haldar S. Karmakar S. Sen T. SpringerPlus. 2015;4:672. doi: 10.1186/s40064-015-1476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khashan K. S. Sulaiman G. M. Ameer A. Kareem F. A. Napolitano G. Pak. J. Pharm. Sci. 2016;29:541. [PubMed] [Google Scholar]
- Mulrooney S. B. Hausinger R. P. FEMS Microbiol. Rev. 2003;27:239. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.