Abstract
Chlorophyll (Chl) serves a number of essential functions, capturing and converting light energy as a component of photosystem supercomplexes. Chl degradation during leaf senescence is also required for adequate degeneration of chloroplasts and salvaging of nutrients from senescent leaves. In this study, we performed genetic analysis to determine the functions of BALANCE of CHLOROPHYLL METABOLISM1 (BCM1) and BCM2, which control Chl levels by regulating synthesis and degradation, and STAY-GREEN (SGR)1 (also known as NON-YELLOWING1 [NYE1]) and SGR2, which encode Mg-dechelatase and catalyze Chl a degradation in Arabidopsis (Arabidopsis thaliana). Analysis of bcm1 bcm2 revealed that both BCM1 and BCM2 are involved in the regulation of Chl levels in presenescent leaves and Chl degradation in senescing leaves. Analysis of bcm1 bcm2 nye1 nye2 suggested that BCMs repress Chl-degrading activity in both presenescent and senescing leaves by regulating SGR activity. Furthermore, transactivation analysis and chromatin immunoprecipitation (ChIP) assay revealed that GOLDEN2-LIKE1 (GLK1), a central transcription factor regulating the expression of genes encoding photosystem-related proteins, such as light-harvesting Chl a/b-binding proteins (LHCPs), directly regulates the transcription of BCM1. LHCPs are stabilized by Chl binding, suggesting that GLKs control the amount of LHCP through transcriptional and post-translational regulation via BCM-mediated Chl-level regulation. Meanwhile, we generated a mutant of the BCM ortholog in lettuce (Lactuca sativa) by genome editing and found that it showed an early yellowing phenotype, but only a slight reduction in Chl in presenescent leaves. Thus, this study revealed a conserved but slightly diversified regulation of Chl and LHCP levels via the GLK-BCM pathway in eudicots.
In Arabidopsis and lettuce, chlorophyll synthesis and degradation are regulated by a CaaX protease-like protein, expression of which is directly regulated by a light-regulated transcription factor.
Introduction
Chlorophyll (Chl) is an essential substance that captures and converts light energy and performs several functions in Chl–protein complexes (Gao et al., 2018). The major Chl, Chla, is involved in both the capture and conversion of light energy in the core complexes of photosystems I (PSI) and II (PSII), whereas Chlb, the minor Chl, contributes to light energy capture only in the peripheral light-harvesting complexes of PSI (LHCI) and PSII (LHCII).
Chl synthesis is initiated by the conversion of glutamate to 5-aminolevulinic acid (ALA), which is catalyzed by glutamyl-tRNA synthetase, glutamyl-tRNA reductase, and glutamate 1-semialdehyde aminotransferase (Tanaka et al., 2011). ALA is then metabolized to protoporphyrin IX (Proto IX). Ferrochelatase then inserts Fe2+ into Proto IX in the heme-synthesizing branch, whereas Mg-chelatase inserts Mg2+ into ProtoIX in the Chl-synthesizing branch, generating Mg-Proto IX. Mg-chelatase is composed of CHLH (GENOMES UNCOUPLED5; GUN5), CHLI, and CHLD subunits, the activity of which is regulated by the porphyrin-binding protein GUN4. Mg-Proto IX is subsequently metabolized to Chla, from which Chlb is synthesized by chlorophyllide a oxygenase (CAO).
The first step in Chla degradation involves the removal of Mg2+ from Chla by Mg-dechelatase STAY-GREEN (SGR) (Park et al., 2007; Ren et al., 2007; Shimoda et al., 2016). Phytol residues are removed from the resultant pheophytin by PHEOPHYTINASE (Morita et al., 2009; Schelbert et al., 2009), resulting in the formation of pheophorbide a, which is then metabolized into transparent substances, nonfluorescent chlorophyll catabolites (NCCs), or dioxobilin-type NCCs (DNCCs). Chlb is converted into Chla via the Chlb reductases NON-YELLOW COLORING1 (NYC1) and NYC1-LIKE (NOL) (Kusaba et al., 2007; Horie et al., 2009; Sato et al., 2009), and HYDROXYMETHYL CHLa REDUCTASE (Meguro et al., 2011) then degraded in the Chla-breakdown pathway. Mutants of Chl-degrading enzyme genes retain their green color owing to impaired Chl breakdown, although their leaves gradually die during senescence. These mutants are known as non-functional stay-green mutants and are distinguishable from delayed senescence (functional stay-green) mutants (Kusaba et al., 2013).
The amount of light-harvesting Chl a/b-binding proteins (LHCPs) (LHCI and LHCII) is controlled at both the transcriptional and posttranslational levels (Kusaba et al., 2007; Waters and Langdale, 2009; Wang and Grimm, 2015; Jia et al., 2016), with GARP (Golden2, ARR-B, Psr1) nuclear transcription factors GOLDEN2-LIKE (GLK) 1 and GLK2 acting as central transcriptional regulators of LHCP gene expression (Waters et al., 2009). LHCPs are stable only if they bind Chl into a tight fold, suggesting that LHCP content is regulated by Chl content (Paulsen et al., 1993). For example, while SGR overexpression and SGR-inducible plants show enhanced and inducible degradation of Chl and LHCP (Park et al., 2007; Wu et al., 2016; Ono et al., 2019), LHCP degradation is strongly repressed during leaf senescence in mutants of Chl-degrading enzymes (Kusaba et al., 2007; Park et al., 2007; Sato et al., 2007; Morita et al., 2009; Sato et al., 2009; Schelbert et al., 2009).
In general, Chl levels are regulated by enzymes involved in Chl synthesis and breakdown (Tanaka et al., 2011). In addition, proteins involved in the stability of Chl-binding proteins, mutants of which show a pale-green phenotype, have also been shown to play a role in regulating Chl (Kusaba et al., 2013; Wang and Grimm, 2015). Recently, a novel posttranslational regulator of Chl levels has also been reported, namely, BALANCE of CHLOROPHYLL METABOLISM (Wang et al., 2020), which is similar to CaaX proteases. Ras converting enzyme1 (RCE1)- and Sterile24 (Ste24)-class CaaX proteases are localized in the endoplasmic reticulum, where they play a role in the plasma membrane localization of proteins with a CaaX motif and translocon quality control, respectively (Majsec et al., 2017). BCMs are localized in the thylakoid membrane in chloroplasts, where they play a role in Chl synthesis and degradation (Wang et al., 2020; Zhang et al., 2020).
In Arabidopsis (Arabidopsis thaliana), there are two BCMs, BCM1 and BCM2, which have similar biochemical properties (Wang et al., 2020). BCM1 and BCM2 interact with GENOME UNCOUPLED4 (GUN4) to promote Mg-chelatase activity and Chl synthesis. They also interact with SGR to destabilize the SGR protein and repress Chl degradation. Zhang et al. (2020) suggested that CBD1 (also known as BCM1) serves as a Mg-transport protein. Although bcm1 bcm2 double mutants were described by Wang et al. (2020) and Zhang et al. (2020), detailed genetic analyses using null alleles have not been performed so far. Therefore, precise functional differences between BCM1 and BCM2 remain unrevealed. Furthermore, genetic interactions between BCMs and SGRs have not been investigated.
In this study, we analyzed the genetic interactions between BCM1 and BCM2 and between BCMs and SGRs using double and quadruple mutants. These analyses revealed the precise functions of BCM1 and BCM2 and provided evidence for the very strict regulation of SGR activity in presenescent leaves. Furthermore, we revealed that the expression of BCM1 is directly regulated by GLK1/2 transcription factors, suggesting that GLK1/2 transcription factors regulate the amount of LHCP by upregulating the transcription of LHCP genes and stabilizing LHCP by retaining Chl levels through upregulation of BCM1 transcription. In addition, we found that this scheme is conserved in Lactuca sativa L. (lettuce), although the function of the BCM ortholog may differ slightly from that of BCM1 in Arabidopsis.
Results
BCM1- and BCM2-mediated regulation of Chl content in presenescent leaves
Plastid CaaX protease-like proteins constitute a plant-specific family (Majsec et al., 2017), of which only a few have been characterized in detail. In soybean, the CaaX protease-like protein gene G regulates seed coat coloring and plays an important role in soybean (Glycine max) domestication (Wang et al., 2018). Meanwhile, Arabidopsis contains two co-orthologs, G, BCM1, and BCM2, which show high similarity (82% amino acid identity) with each other and retain chloroplast transit peptides and six transmembrane domains, although BCM2 has a longer stretch at its N-terminal, suggesting that they are chloroplast membrane proteins (Wang et al., 2020; Supplemental Figure S1A). To determine the subcellular localization of BCM1 in detail, we produced transgenic plants expressing the BCM1 protein with a 4× MYC-tag. Isolated chloroplasts from the transgenic plants were then fractionated into thylakoid membrane and stroma + envelope fractions using density gradient centrifugation (Supplemental Figure S1B). SDS-PAGE and western blot analysis revealed signals for BCM1 in the thylakoid membrane fraction, similar to those of other thylakoid membrane proteins, such as YELLOW VARIEGATED2 (VAR2; Chen et al., 2000; Takechi et al., 2000) and LHCII. This result confirms previous studies suggesting that BCM1 is localized in the thylakoid membrane (Wang et al., 2020; Zhang et al., 2020).
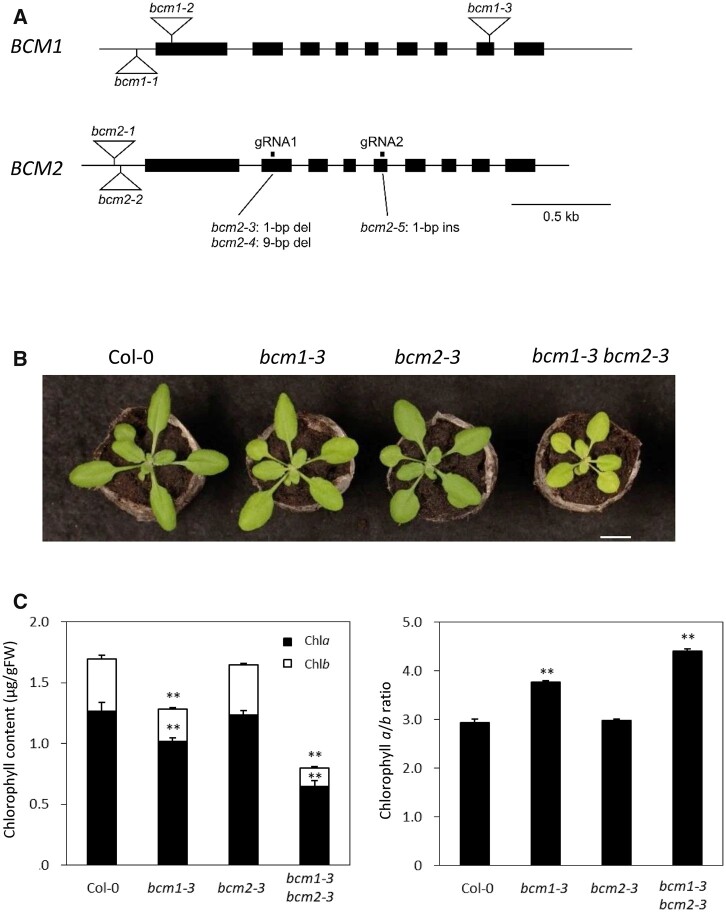
Wang et al. (2020) reported that both BCM1 and BCM2 are involved in Chl synthesis through the regulation of Mg-chelatase activity. To genetically determine the function of BCM1 and BCM2 in Chl synthesis, we generated a bcm2 mutant via genome editing using the CRISPR-Cas9 system, as only bcm2 mutants with T-DNA insertions in the 5′ upstream or untranslated regions were available (bcm2-1 and bcm2-2; Figure 1A). For genome editing of BCM2, we designed gRNAs at the second and fourth exons and isolated bcm2-3, bcm2-4, and bcm2-5, which contained a 1-bp deletion in the second exon, a 9-bp deletion in the second exon, and a 1-bp insertion in the fifth exon, respectively (Figure 1A and Supplemental Figures S1A and S2A). Mutations in bcm2-3 and bcm2-5 cause frameshifts, suggesting that they are null mutants. Moreover, while the null mutant bcm1-3 presented a weak pale green/low Chl content phenotype, no changes in leaf color/Chl content were observed in bcm2-3 (Figure 1, B and C). Meanwhile, bcm1-3 bcm2-3 showed a strong pale green/low Chl content and a retarded growth phenotype (Figure 1, B and C). Similarly, bcm1-3 bcm2-4 and bcm1-3 bcm2-5 showed a strong pale green/low Chl content and retarded growth, confirming the redundant functions of BCM1 and BCM2 in regulating Chl content, although BCM2 contributed less than BCM1 (Supplemental Figure S2B). This result was consistent with that reported by Zhang et al. (2020). However, Wang et al. (2020) did not observe an enhanced reduction in the Chl content in bcm1-3 bcm2-2. This is likely because bcm2-2, which has a T-DNA insertion at the 5′ upstream region of BCM2, is a weak allele of bcm2. The low Chl phenotypes of bcm1-3 bcm2-3 were also complemented by BCM1 promoter-driven BCM1-4× MYC and BCM2-4× MYC constructs, suggesting that BCM1 and BCM2 have the same protein function (Supplemental Figure S3, A and B), which is further supported by lines of biochemical evidence (Wang et al., 2020). Interestingly, bcm1-3 and bcm1-3 bcm2-3 had a higher Chla/b ratio than Col-0, as observed by Zhang et al. (2020), suggesting that the reduction in Chlb was more severe than that of Chla in both mutants (Figure 1C).
Figure 1.
Characterization of the alleles of bcm1 and bcm2. A, Structures of the bcm1 and bcm2 alleles. bcm2-3, bcm2-4, and bcm2-5 were generated using the CRISPR-Cas9 system. bcm2-3 and bcm2-4 were obtained using gRNA1 in the second exon, while bcm2-5 was obtained using gRNA2 in the fifth exon. Triangles indicate the T-DNA insertions. B, bcm1-2, bcm2-3, and bcm1-2 bcm2-3 plants were grown for 20 days under long-day conditions at 22°C. Scale bar: 1 cm. C, Chl contents and the Chl a/b ratio of presenescent leaves of bcm1-2, bcm2-3, and bcm1-2 bcm2-3. Eighth leaves from the top of 27-day-old plants grown under short-day conditions were used. Error bars indicate the standard error (n = 4). **P < 0.01 (Student’s t test).
Wang et al. (2020) reported a slight reduction of Lhca1 and Lhcb1 in presenescent leaves of the bcm1-3 single mutant. We extensively examined chloroplast proteins in bcm1 bcm2 double mutants (Supplemental Figure S4). Accordingly, bcm1-3 and bcm2-3 showed a reduction in the protein content of PSI and PSII subunits in presenescent leaves. Moreover, this reduction was particularly prominent in LHCI and LHCII. The LHC1 level was 61%, 65%, 54%, and 72% of wild-type levels in Lhca1, Lhca2, Lhca3, and Lhca4, respectively, and the LHCII level was 46%, 33%, and 54% of wild-type levels in Lhcb1, Lhcb3, and Lhcb4, respectively. Lhca and Lhcb are the only Chlb-containing proteins in plant cells, which may explain the higher Chla/b ratio in bcm1 and bcm1 bcm2 mutants (Figure 1C). PSI core complex subunits were also reduced in bcm1 bcm2 mutants, although the PSII core complex subunits were largely unchanged. Previous studies reported that the degradation of PSII core complex subunits is regulated by THYLAKOID FORMATION1/NYC4 rather than by Chl degradation (Yamatani et al., 2013; Chen et al., 2021). It is, therefore, possible that the reduced Chl content in the bcm1 bcm2 mutant did not affect the protein levels of the core PSII subunits. In contrast, the levels of chloroplast proteins localized in the envelope, such as TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 40 (TIC40) and TIC110, showed a slight increase, whereas those in the stroma, such as HIGH-CHLOROPHYLL-FLUORESCENCE 101 (HCF101) and Rubisco large subunit, were unchanged in the bcm1 bcm2 double mutants. Meanwhile, mRNA levels of LHCI and LHCII genes Lhca1 and Lhcb1.1, and the PSI core subunit gene PsaF, showed no prominent reduction in presenescent leaves of bcm1-3 bcm2-3 (Supplemental Figure S5). Overall, these findings suggest that the reduction in LHCPs and PsaF in the presenescent leaves of bcm1-3 bcm2-3 was mainly caused by a posttranslational mechanism.
BCM1- and BCM2-mediated regulation of Chl degradation during leaf senescence
Previous studies have shown that BCMs are also involved in leaf yellowing (Wang et al., 2020). Therefore, we verified the functions of BCM1 and BCM2 during leaf senescence using null single and double mutants (Supplemental Figure S6A). While the Chl content of bcm1-3 and bcm2-3 decreased in a similar manner to Col-0 during dark incubation, an apparent early yellowing phenotype was observed during leaf senescence in bcm1-3 bcm2-3, in addition to the lower Chl content phenotype in presenescent leaves (Supplemental Figure S6, B and C). These observations suggest that single mutants of bcm1 and bcm2 do not exhibit an early yellowing phenotype, with BCM1 and BCM2 redundantly repressing leaf yellowing. Although presenescent leaves of the BCM1 overexpressing plants (BCM1-OE) did not have a higher Chl content than Col-0, a high Chl content was retained during dark incubation (Supplemental Figure S6, A and B). There was no statistical difference in the decrease in Fv/Fm values, reflecting PSII activity between BCM1-OE and Col-0 during dark incubation, suggesting that BCM1 represses Chl degradation, but not leaf senescence (Supplemental Figure S6D). Thus, BCM1-OE plants represent a nonfunctional stay-green line, in which Chl degradation is primarily repressed during leaf senescence, which is consistent with the observations of Wang et al. (2020). Meanwhile, in bcm1-3 bcm2-3, Fv/Fm values decreased much earlier than in Col-0 during dark incubation, which is congruent with its early yellowing phenotype (Supplemental Figure S6D). Consistent with this, induction of senescence-associated genes (SAGs), such as SGR1, ORESARA1 (ORE1), AtNAP, and NYC1, occurred sooner in bcm1-3 bcm2-3 than in the wild-type during dark incubation (Supplemental Figure S7A). These findings suggest that leaf senescence was synchronous with Chl degradation in bcm1-3 bcm2-3, in contrast to BCM1-OE plants. Ono et al. (2019) previously reported that dexamethasone-induced SGR expression induces leaf senescence (i.e. increased expression of ORE1 and SAGs). Although BCMs primarily act in the degradation of Chl, accelerated Chl degradation may also have induced leaf senescence in bcm1-3 bcm2-3.
Genetic interactions between BCM and SGR
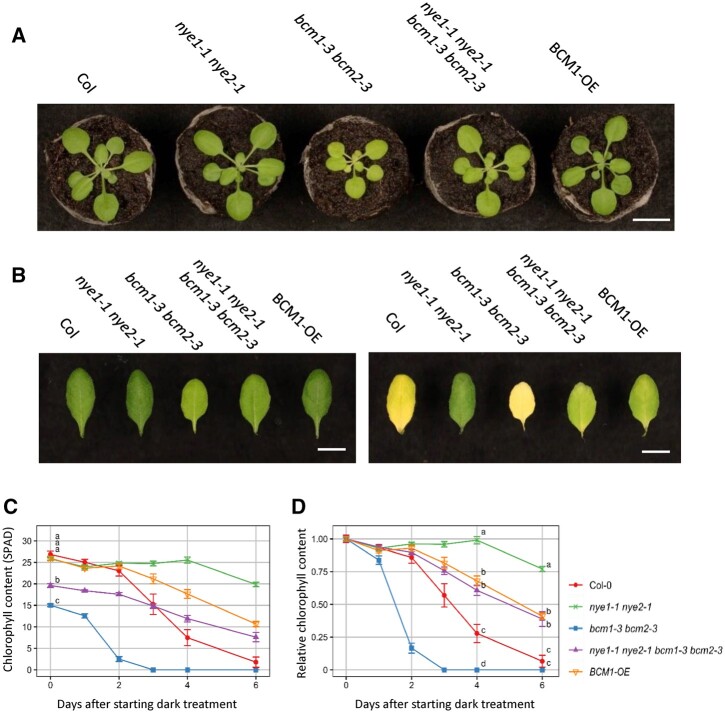
Wang et al. (2020) previously revealed that BCM represses Chl degradation through SGR destabilization during leaf senescence. To investigate the genetic interactions between BCM and SGR, we generated a bcm1-3 bcm2-3 nye1-1 nye2-1 mutant (nye1-1 and nye2-1 represent SGR1 and SGR2 mutants, respectively [Wu et al., 2016; Figure 2A]). Unexpectedly, the Chl content of the quadruple mutant plants was significantly higher than that of bcm1-3 bcm2-3, although nye1-1 nye2-1 had a similar Chl content to that of Col-0 (Figure 2, A and C). This observation suggests that the reduced Chl content in bcm1-3 bcm2-3 was partly caused by the degradation of Chl via SGR, although the expression of SGR was very low in presenescent leaves. In addition, nye1-1 nye2-1 partially reversed the retarded growth of bcm1-3 bcm2-3, suggesting that the increased Chl content resulting from SGR impairment contributed to the restoration of photosynthesis/biomass to some extent (Figure 2A).
Figure 2.
Leaf color and senescence phenotypes of the bcm nye quadruple mutant. A, Twenty-two-day-old plants were grown at 22°C under short-day conditions. B, Leaves before and 6 days after dark incubation. Seventh leaves from the top of 28-day-old plants grown at 22°C under short-day conditions were detached and incubated at 22°C in the dark. C and D, Changes in Chl content with time during dark incubation. Data indicate SPAD values and relative values to presenescent leaves. Scale bar: 1 cm.
Next, we examined the degradation of Chl during dark incubation in bcm1-3 bcm2-3 nye1-1 nye2-1 (Figure 2, B–D). Accordingly, the relative SPAD value of nye1-1 nye2-1 decreased to ∼75% that of the presenescent leaves 6 days after the beginning of dark treatment, while that of bcm1-3 bcm2-3 nye1-1 nye2-1 decreased to ∼40%, which was substantially lower than that of nye1-1 nye2-1. While BCMs are reportedly involved in the destabilization of SGR protein (Wang et al., 2020), this result suggests that BCMs repress Chl degradation through SGR-dependent and SGR-independent pathways.
Transcriptional regulation of BCM1 by GLK
As mentioned above, it has been previously suggested that BCM1 and BCM2 act as regulators of Chl content. Therefore, we analyzed light- and senescence-induced expression of BCM1 and BCM2. As a result, the expression of BCM1 was drastically reduced within 6 h of dark treatment, similar to that of Lhcb1.1 and CAO (Supplemental Figure S7B), both of which are direct targets of GLK transcription factors (Waters et al., 2009). Expression of GLK1 and GLK2 was also drastically reduced within 6 h of dark treatment (Supplemental Figure S7B). Meanwhile, expression of BCM2 also decreased to 57.2% within 6 h of dark treatment, although an increase was observed from 4 days (Supplemental Figure S7B). Overall, the expression of BCM2 increased during extended dark incubation, suggesting that expression of BCM2 is regulated by not only light but also senescence.
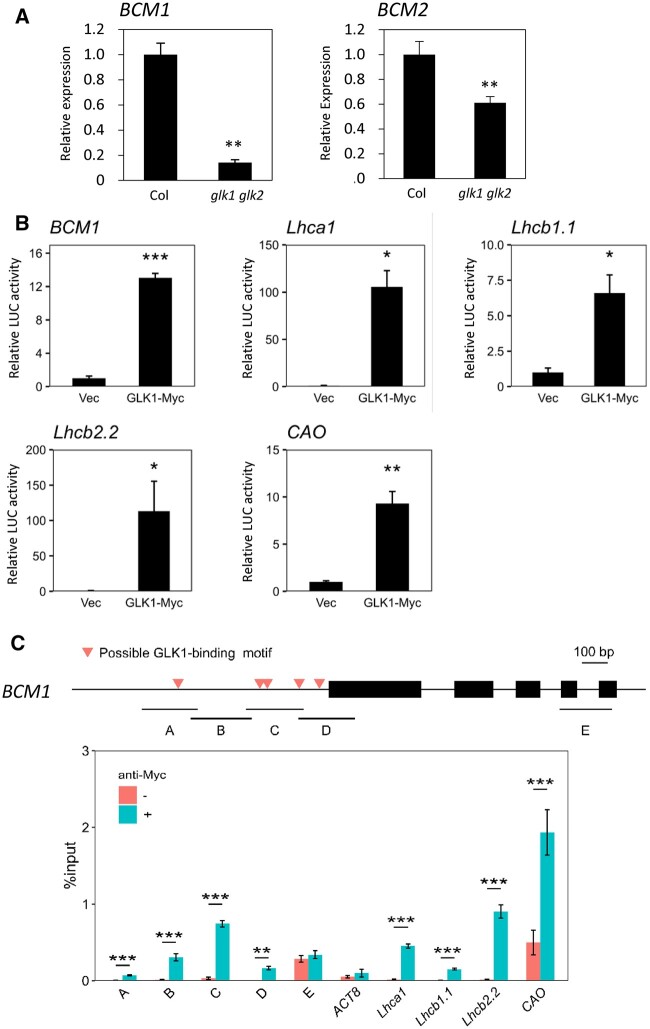
To determine the transcriptional regulation of BCM1 and BCM2 by GLKs, we analyzed the expression of BCM1 and BCM2 in the glk1 glk2. Accordingly, the expression was reduced to 14.1% that of the wild-type, suggesting an important contribution of GLK1 and GLK2 to the expression of BCM1 in presenescent leaves (Figure 3A). Meanwhile, the expression of BCM2 was reduced to only 61.1% that of the wild-type, suggesting less of contribution to BCM2 expression (Figure 3A). These findings were also consistent with the difference in expression between BCM1 and BCM2 in response to dark treatment (Supplemental Figure S7B).
Figure 3.
Direct regulation of BCM1 expression by GLK1. A, RT-qPCR analysis of BCM expression in presenescent leaves of glk1 glk2. **P < 0.01 (n = 4). Seventh to eighth leaves from the top of 25-day-old plants grown at 22°C under short-day conditions were used. B, Transactivation of photosynthesis-related genes by GLK1. Constructs containing luciferase genes driven by promoters of BCM1 and known GLK1-target genes were transiently introduced into Arabidopsis mesophyll protoplasts together with GLK1-4 × MYC constructs (GLK1-Myc). “Vec” represents a control into which the luciferase reporter construct and an empty effector vector were introduced. C, ChIP-qPCR analysis of GLK1 binding to the BCM1 promoter was performed using GLK1-4 × MYC overexpressing plants. The upper panel shows the gene structure of BCM1. Red triangles indicate the positions of possible GLK1-binding sequences. Black boxes represent exons. A–E represent DNA regions used in ChIP-qPCR analysis. The lower panel shows the results of ChIP-qPCR analysis. Fold enrichment is indicated as the % input. ** and ***P < 0.01 and P < 0.001, respectively (Student’s t test).
To investigate the transcriptional regulation of BCM1 by GLK1, transactivation analysis using luciferase as a reporter gene was performed, along with a chromatin immunoprecipitation (ChIP) assay (Figure 3, B and C). When the BCM1 promoter-LUC construct was transiently introduced into mesophyll protoplasts together with the GLK1-4× MYC construct, luciferase activity was elevated 13.0-fold compared to the noneffector controls. The luciferase activities of Lhca1, Lhcb1.1, Lhcb2.2, and CAO, which are reported to be directly regulated by GLK1, were also notably elevated by 105.6-, 6.6-, 113.3-, and 9.3-fold, respectively, following the introduction of the GLK1-4× MYC construct (Figure 3B; Waters et al., 2009). A ChIP assay was subsequently performed using GLK1-4× MYC overexpressing plants. Accordingly, GLK1-binding was observed with promoters of known GLK1-target genes, Lhca1, Lhcb1.1, Lhcb2.2, and CAO, but not with the promoter of the non-GLK1 target gene ACTIN8, confirming the observation by Waters et al. (2009). The BCM1 promoter did not contain a perfect GLK1-binding consensus sequence (CCAATC), but instead contained five 1-bp-substituted sequences (Figure 3C). Therefore, we analyzed four fragments of the promoter region (designated A–D) and one fragment of the structural gene region (E) for GLK1 binding. All four fragments in the promoter region, but not in the structural gene region, were enriched for GLK1 binding (Figure 3C), suggesting that GLK1 selectively binds to the promoter region of BCM1. A similar result was obtained in an independent experiment (Supplemental Figure S8). These observations suggested that BCM1 is a direct target of GLK1, consistent with a previous study that identified BCM1 as an early GLK1-inducible gene (Waters et al., 2009). We also performed a ChIP analysis of the BCM2 promoter for GLK1 binding. The BCM2 promoter harbored possible GLK1-binding motifs, but no statistically significant enrichment was observed in any of the promoter fragments examined (Supplemental Figure S9).
Analysis of the BCM ortholog in lettuce
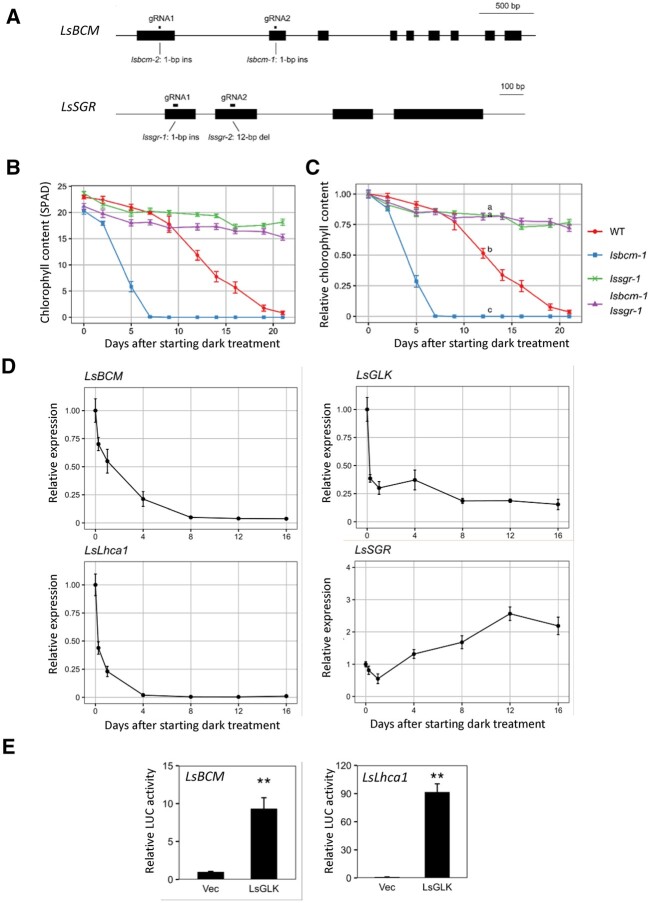
To determine whether the function of BCM is conserved in other species, we analyzed lettuce mutants of BCM and SGR. Lettuce contains only single genes for BCM (LsBCM), SGR (LsSGR), and GLK (LsGLK) in its genome (Supplemental Figures S10, A–C and S11). Therefore, we generated LsBCM and LsSGR mutants via CRISPR-Cas9, using two guide RNAs (Figure 4A). The resulting lsbcm-1 and lsbcm-2 mutants contained 1-bp insertions at the 411th and 216th nucleotides from the initiation codon, respectively. These insertions caused frameshifts, suggesting that they were null mutants. lssgr-1 and lssgr-2 contained a 1-bp insertion and 12-bp deletion at the 39th and 199–210th nucleotides from the initiation codon, respectively. lssgr-1 was also considered a null mutant due to frameshifts (Figure 4A).
Figure 4.
Functional analysis of the BCM ortholog in lettuce. A, Structures of lsbcm-1, lsbcm-2, lssgr-1, and lssgr-2 mutants generated using CRISPR-Cas9-based genome editing. lsbcm-1 and lsbcm-2 were obtained using gRNA1 in the first exon and gRNA2 in the second exon. lssgr-1 and lssgr-2 were obtained using gRNA1 in the first exon and gRNA2 in the second exon. B and C, Changes in Chl content with time during dark incubation. Third leaves of three-week-old plants incubated at 22°C in the dark were used. B and C indicate SPAD values and relative values to presenescent leaves, respectively. D, Changes in gene expression of LsBCM, LsGLK, LsLhca1, and LsSGR with time. Lettuce leaves were incubated in the dark as described in panel B. E, Transactivation of LsBCM and LsLhca1 expression by LsGLK1. Constructs carrying luciferase genes driven by promoters of LsBCM1 and LsLhca1 were transiently introduced into lettuce mesophyll protoplasts together with LsGLK1 constructs. “Vec” represents a control into which the luciferase reporter construct and an empty effector vector were introduced. n = 3. **P < 0.01 (Student’s t test).
Both lssgr-1 and lssgr-2 showed a strong stay-green phenotype similar to the nye1-1 nye2-1 double mutant in Arabidopsis during both dark-induced and natural senescence, confirming that LsSGR is a single-copy gene with the same function in lettuce (Figure 4, B and C and Supplemental Figure S12, A and B). Meanwhile, lsbcm-1 and lsbcm-2 also showed an early yellowing phenotype during dark-induced and natural senescence, similar to bcm1-3 bcm2-3 in Arabidopsis. Furthermore, the lsbcm-1 lssgr-1 and lsbcm-2 lssgr-2 double mutants showed a stay-green phenotype similar to that of the bcm1-3 bcm2-3 nye1-1 nye2-1 quadruple mutant in Arabidopsis. These observations suggest that the function of BCM in Chl degradation during leaf senescence is conserved between lettuce and Arabidopsis plants. However, the early yellowing phenotype of lsbcm-1 was strongly suppressed by lssgr-1, suggesting that LsBCM represses Chl degradation almost exclusively through downregulation of SGR function in lettuce, unlike in Arabidopsis (Figure 4, B and C and Supplemental Figure S12B).
The bcm1-3 bcm2-3 mutant in Arabidopsis showed a prominent pale green phenotype (Figure 1B), whereas presenescent leaves of lsbcm-1 did not (Supplemental Figure S12B). Detailed analysis using nonsenescent leaves from nine individuals further revealed a slight but significantly lower Chl content in presenescent leaves of lsbcm-1compared with the wild-type (P < 0.05, Tukey’s multiple comparison test) (Supplemental Figure S13A). These observations suggest that BCM in lettuce is involved in Chl synthesis in presenescent leaves, but its contribution is limited. In addition, there was no significant difference in Chl content between presenescent leaves of lsbcm-1 and lsbcm-1 lssgr-1, suggesting that SGR does not play a role in Chl degradation in presenescent lettuce leaves (Supplemental Figure S13, A and B).
Next, we analyzed the changes in expression of LsBCM, LsGLK, LsLhca1, and LsSGR over time during dark incubation (Figure 4D). Similar to GLK1 and GLK2 in Arabidopsis, the expression of LsGLK decreased sharply in response to light depletion treatment, while downregulation of LsBCM was slightly slower than that of BCM1 in Arabidopsis. One LHCP gene, LsLhca1, also showed a very fast response to light depletion, whereas the induction rate of LsSGR was low (2.6-fold) during dark incubation. In contrast, the expression of SGR1 in Arabidopsis increased more than 10-fold even after 4 days of dark treatment, when leaves were yet to become fully senescent (Supplemental Figure S7A). To investigate the role of GLK in LsBCM and LsLhca1 expression, transactivation assays were performed using lettuce mesophyll protoplasts. Transient introduction of the LsGLK-4× MYC construct significantly increased luciferase activity from the LsBCM and LsLhca1 promoters, suggesting that expression of BCM and Lhca1 is also directly regulated by GLK in lettuce (Figure 4E).
Discussion
In this study, we investigated the functions of BCM1 and BCM2 in Chl regulation using null double mutants. We found that bcm2 null mutations prominently enhanced the pale green phenotype of bcm1, suggesting that both BCM1 and BCM2 play a role in regulating Chl levels in presenescent leaves, although the single bcm2 mutant showed no such phenotype. Wang et al. (2020) did not observe such a function of BCM2, likely because the bcm2-2 allele that they used was a weak allele. During leaf senescence, bcm1 and bcm2 single mutants showed no obvious early yellowing phenotype, in contrast to their double mutants, which did so prominently, suggesting that BCM1 and BCM2 redundantly repress Chl degradation during leaf senescence, which is consistent with the conclusion of Wang et al. (2020).
Analysis of a quadruple mutant carrying mutations in SGR1 and SGR2 (nye1-1 nye2-1) as well as bcm1-3 bcm2-3 further revealed that loss-of-function mutations in SGRs suppressed the early yellowing phenotype of bcm1 bcm2, suggesting that SGRs act downstream of BCMs. However, retention of Chl content in the quadruple mutant was lower than that in nye1-1 nye2-1, suggesting that BCMs repress SGR-dependent and SGR-independent Chl degradation pathways.
It is well accepted that the upregulation of SGR during leaf senescence induces Chl degradation and subsequent LHCP degradation during dark-induced leaf senescence (Park et al., 2007; Ren et al., 2007; Sato et al., 2007; Ono et al., 2019). Our study suggests that the reduced expression of BCM1 is also one of the initial steps in leaf yellowing. A reduction in BCM expression in response to dark treatment may stabilize SGR proteins and facilitate Chl degradation by SGR. Particularly in lettuce, this regulation seemed more important because the induction rate of SGR expression during dark incubation was only 2.6-fold (Supplemental Figure S4D). Since the expression of BCM2 tended to increase during the late stage of leaf senescence in Arabidopsis, BCM2 is also thought to contribute to the fine-tuning of Chl degradation during late senescence.
Unexpectedly, presenescent leaves of the quadruple mutant bcm1-3 bcm2-3 nye1-1 nye2-1 had a higher Chl content than those of bcm1-3 bcm2-3, implying that SGR contributed to the low Chl content of presenescent leaves in bcm1-3 bcm2-3. Although the expression of SGR in the presenescent leaves was very low, it is thought that a small amount of SGR protein resulted in the degradation of Chl in the presenescent leaves of bcm1-3 bcm2-3. Meanwhile, the Chl content of the presenescent leaves of nye1 nye2 was no higher than that of Col-0, suggesting that BCMs eliminated SGR activity completely in wild-type presenescent leaves. Although it has been hypothesized that there is almost no SGR expression in presenescent leaves and that induction of SGR expression causes Chl degradation during leaf senescence, our results suggest that the Chl-degrading activity of SGR is strictly and securely regulated in presenescent leaves.
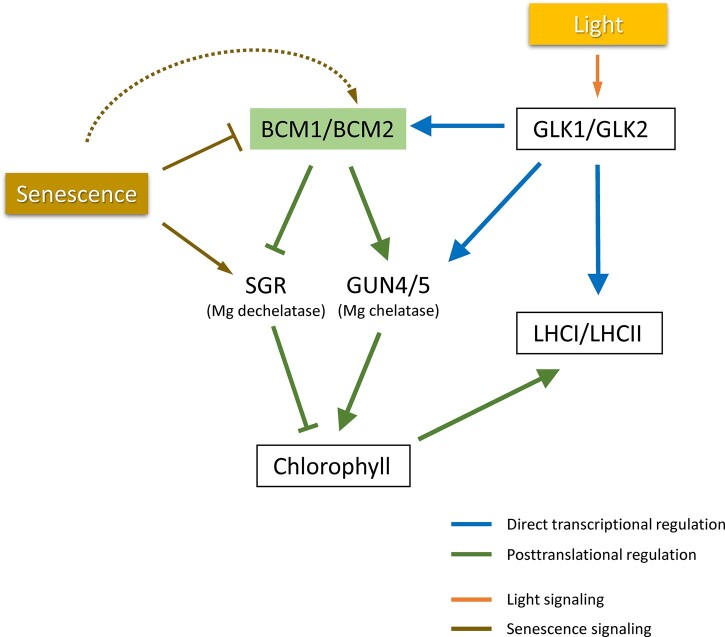
In this study, we revealed that GLK1, the central transcription factor in regulation of expression of photosynthesis-related genes, directly regulates expression of BCM1 in Arabidopsis, confirming that BCM1 is one of the important machineries regulating photosynthesis activity. Figure 5 summarizes the proposed Chl/photosynthesis regulation via the GLK-BCM pathway. Expression of BCMs is directly regulated by GLK transcription factors, which are known to regulate the expression of nuclear photosynthetic genes in Arabidopsis (Waters et al., 2009). Light activates the transcription of BCMs via GLK1 and GLK2, resulting in Chl synthesis via the Mg-chelatase GUN5 and its activator GUN4 and repression of Chl degradation by destabilization of the Mg dechelatase SGR, securing photosynthetic performance. Furthermore, considering that the stability of LHCPs (LHCI/LHCII) is regulated by Chl content (Kusaba et al., 2013), GLKs are thought to regulate the amount of LHCP through transcriptional and posttranslational pathways: direct transcriptional activation of LHCP genes and stabilization of LHCP proteins via regulation of Chl synthesis/degradation by BCMs.
Figure 5.
Schematic depiction of the GLK-BCM pathway that regulates Chl and LHCP levels in response to environmental factors in Arabidopsis. BCM is required for full activation of GUN4/5 (Chl synthesis) and destabilization of the SGR protein (repression of Chl degradation; Wang et al., 2020). GLKs control the amount of LHCP (LHCI/LHCII) via direct transcriptional regulation of (i) LHCP apoprotein genes and (ii) BCM genes, regulating levels of Chl and stabilizing LHCP. During dark-induced leaf senescence, the reduction in BCM expression caused by the rapid decrease in GLK expression in response to dark treatment is also thought to play a role in the onset of leaf yellowing. Meanwhile, expression of BCM2 is upregulated during the late stage of leaf senescence, thereby fine-tuning the Chl content.
Mutants of BCM orthologs in soybean and tomato (Solanum lycopersicum) showed pale green and early yellowing phenotypes similar to bcm1 bcm2 in Arabidopsis, suggesting that the function of BCM in Chl degradation is conserved during evolution (Liu et al., 2020, 2021). However, the BCM-mediated regulation of Chl levels in lettuce was somewhat different from that in Arabidopsis. lsbcm showed an early yellowing phenotype during leaf senescence, similar to the bcm1 bcm2 mutant in Arabidopsis. However, it showed only a very slight reduction in Chl content in presenescent leaves, suggesting that the contribution of BCM to Chl synthesis and degradation in presenescent leaves is limited in lettuce. Moreover, mutants of BCM orthologs in soybean and tomato showed an obvious pale green phenotype, suggesting that the functions of BCM are similar to those of Arabidopsis (Liu et al., 2020, 2021).
Arabidopsis contains several chloroplast-localized CaaX protease-like proteins other than BCMs. Of these, only SNOWY COTYLEDON4 (SCO4) has been analyzed so far, the mutant of which has albino cotyledons (Albrecht-Borth et al., 2013). Although Wang et al. (2020) reported that BCMs bind GUN4 and GUN5 to promote Mg-chelatase activity and SGR to destabilize the SGR protein, the precise biochemical function of BCMs has yet to be elucidated, including whether BCMs possess protease activity. Yeast, mammals, and plants share the CaaX proteases RCE1 and STE24, whereas BCM does not exist in the green alga Chlamydomonas. Therefore, it has been suggested that BCM evolved in association with the acquisition of the Chl degradation function of SGR. However, SGR is involved in pheophytin synthesis rather than Chl degradation in Chlamydomonas (Chen et al., 2019; Wang et al., 2020). Chloroplast-localized CaaX protease-like proteins are highly divergent; therefore, their biochemical functions may also be diverse. Therefore, further studies on the function of this protein family are warranted.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) accession Col-0 was used as the wild-type. The bcm1-3 (SALK_058830C), glk1 glk2 carrying LhGR-N (4c) inducible expression transgene (CS9907) and GLK1 overexpression line (CS9905) were obtained from the Arabidopsis Biological Resource Center. glk1 glk2 was obtained by backcrossing CS9907 with Col-0 to remove the LhGR-N (4c) transgene. Arabidopsis plants were grown on Jiffy-7 peat pellets (33 mm in diameter; Jiffy Products International AS, Norway) at 22°C under long- (16 h light/8 h dark; 16L/8D) or short-day conditions (10L/14D) with 100 µmol photons s−1 · m−2. Lettuce (L. sativa) cultivar Greenwave (Takii seed, Japan) was used as the wild-type lettuce. Plants were grown on Jiffy-7 peat pellets (33 or 44 mm in diameter) at 22°C under long-day conditions (16L/8D) with 170–200 µmol photons s−1 · m−2.
For dark treatment of Arabidopsis plants, leaves were detached and incubated in 24-well plates under high humidity in the dark at 22°C. For dark treatment of lettuce plants, leaves were detached and incubated in square petri dishes under high humidity in the dark at 26°C.
Generation of mutant and transgenic plants
bcm2-3, bcm2-4, and bcm2-5 mutants were generated using the CRISPR-Cas9 system. Two guide RNA, gRNA1 and gRNA2, were designed using the “Focas” website (Osakabe et al., 2016) and CRISPR direct (https://crispr.dbcls.jp/). The oligonucleotides for gRNA1 or gRNA2 were cloned into the BsaI site of pEgP126_Paef1-2A-GFBSD2 (Osakabe et al., 2016) and the AarI site of pKI1.1R (Tsutsui and Higashiyama, 2016), respectively. Null segregants of the CRISPR-Cas9 transgene were used as bcm2 mutants. The primers used for gRNA are shown in Supplemental Table S1.
For complementation analysis, constructs expressing BCM1–4× Myc or BCM2–4× Myc were generated under control of the BCM1 promoter as follows. The multiple cloning site and NOS terminator of pMOE (Fukazawa et al., 2021) was cloned into the NotI and AscI sites of pENTR (Thermo Fisher Scientific, Waltham, MA, USA), giving pENTR-MCS-NOST. The coding region of BCMs was then amplified by PCR using cDNA as a template and subcloned between the NotI and XbaI sites of pMOE 4× Myc, giving pMOE-BCM1–4× Myc and pMOE-BCM2-4× Myc, respectively. DNA fragments containing BCM and 4× Myc were then amplified and cloned between the HindIII and XbaI sites of pENTR-MCS-NOST, giving pENTR-BCM1–4× Myc and pENTR-BCM2–4× Myc, respectively. The promoter regions of BCM1 (−979 to −1 bp; +6, translation start site) were amplified by PCR using the genome DNA as a template then cloned between the NotI and HindIII sites of pENTR-BCM1–4× Myc and pENTR-BCM2–4× Myc, giving pENTR-BCM1pro:BCM1–4× Myc and pENTR-BCM1pro:BCM2–4× Myc, respectively. These entry constructs were then recombined into the destination binary vector pGWB601 (Nakamura et al., 2010) using Gateway LR Clonase II Enzyme mix (Thermo Fisher Scientific). To generate BCM1 overexpression plants (BCM1-OE), the promoter region of AtUBQ10 (−636 to −1 bp; +1, translation start site) was amplified by PCR and assembled into pENTR BCM1–4× Myc using NEBuilder (New England Biolabs, Ipswich, MA, USA). These entry constructs were then recombined into the destination binary vector pGWB601 using Gateway LR Clonase II Enzyme mix. Arabidopsis plants were transformed based on the floral dip method using Agrobacterium tumefaciens strain EHA105.lsbcm, lssgr, and lsbcm lssgr mutants were generated using the CRISPR-Cas9 system as follows. Oligonucleotides of gRNAs designed using CRISPR direct were cloned into the BbsI site of pENTR_AtU6gRNA2 (Nobusawa et al., 2021). The gRNA expression cassette of pENTR_AtU6gRNA2 with gRNA LsBCM was then digested with PacI and PvuI, and inserted into the PacI site of pENTR_AtU6gRNA2 with LsSGR gRNA in the same direction to generate an entry vector carrying gRNA expression cassettes of both LsBCM and LsSGR. The constructs were then recombined using Gateway technology into the destination vector pGWB401_AtRPS5A-Cas9, which was constructed by subcloning a RPS5A promoter:Cas9:HSP terminator cassette from pGWB601_AtRPS5A-Cas9 into pGWB401 (Nakamura et al., 2010). Lettuce transformation was performed as described by Sun et al. (2006) with slight modifications. In this study, A. tumefaciens strain EHA105 and carbenicillin were used instead of GV2260 and augmentin. The primers used for gRNA are shown in Supplemental Table S1.
Measurement of senescence parameters
The foliar Chl content was measured nondestructively using a SPAD-502 Chl meter (Konica Minolta, Tokyo, Japan). For measurement of Chla and Chlb contents, pigments were extracted from leaves using 80% (v/v) acetone after leaves were frozen and pulverized to a powder using Tissuelyser II (Qiagen, Hilden, Germany). Chla and Chlb levels were determined according to Wellburn (1994). Fv/Fm values were measured using a Junior PAM Chl fluorometer (Walz, Hilden, Germany) according to Kohzuma et al. (2017).
RNA extraction and quantitative reverse-transcriptase PCR analysis
Total RNA was extracted using TRI reagent (Molecular Research Center) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 100 to 500 ng total RNA using ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan). Quantitative reverse-transcriptase PCR (RT-qPCR) was performed using KAPA SYBR FAST qPCR kit (Roche, Basel, Switzerland) and Rotor-Gene Q 2PLEX (Qiagen). RT-qPCR conditions were as follows: initial denaturation at 95°C for 30 s, followed by 35–50 cycles at 95°C for 5 s, ending with 60°C for 30 s. Transcript levels were normalized to those of ACTIN8 (ACT8) and LsACT7 in Arabidopsis and lettuce, respectively. Data analysis was performed using the Comparative CT Method (also known as the ΔΔCT Method). The primers used for RT-qPCR are shown in Supplemental Table S1.
Chloroplast isolation and fractionation
Intact chloroplasts were isolated according to Olinares et al. (2010) with slight modifications. Mature rosette leaves from 4-week-old Arabidopsis plants were briefly homogenized in grinding medium (50 mM HEPES-KOH, pH 8.0, 330 mM sorbitol, 2 mM EDTA) then filtered through two layers of miracloth (Merck Millipore, Burlington, MA, USA). The crude plastids were purified by centrifugation at 2,380 × g on 30%–80% Percoll cushions (Percoll in 0.6% Ficoll, 1.8% polyethylene glycol) for 15 min then washed with five volumes of the grinding medium. After lysing in chloroplast isolation buffer (10 mM Tricine-KOH, pH 7.5, 0.6 M sucrose), the chloroplasts were disrupted by freeze–thaw treatment (Chu and Li, 2011). The sucrose concentration of the chloroplast isolation buffer was then reduced to 0.2 M, and the lysates were fractionated into thylakoid and stroma + envelope fractions.
Protein analysis
Frozen leaf samples were pulverized into a powder using Tissuelyser II (Qiagen) and suspended in four volumes of 2 × SDS sample buffer (125 mM Tris–HCl, pH 6.8, 4% [w/v] SDS, 4% [v/v] β-mercaptoethanol, 1% [w/v] bromophenol blue, 20% [v/v] glycerol). Samples were then diluted 10-fold with 1× SDS sample buffer and subjected to SDS-PAGE analysis (Tris/Gly buffer) with or without thermal denaturation, and transferred onto an Immobilon-P transfer membrane (Millipore) (Yamatani et al., 2018). Membranes were incubated with anti-Lhca1, anti-Lhcb1, anti-Lhcb3, anti-Lhcb4, anti-PsaL, anti-D2, anti-TIC40, anti-HCF101 (Agrisera), anti-Myc (MBL), anti-CP1 (Tanaka et al., 1991), anti-PsaF (provided by Y. Takahashi), anti-D1 (Kato et al., 2012), anti-VAR2 (Sakamoto et al., 2003), or anti-TIC110 antibody (Kikuchi et al., 2013), followed by horseradish peroxidase-conjugated secondary antibody. Chemiluminescence was detected using ECL Prime western blotting detection reagents (Cytiva) and quantified using Odyssey Fc imaging system and Image Studio software (LI-COR biosciences). The Rubisco large subunit was visualized using Coomassie brilliant blue G-250 staining on SDS-PAGE gel.
Transient transfection and reporter gene assay
Arabidopsis mesophyll protoplasts were isolated according to Yoo et al. (2007) with slight modifications. Lettuce mesophyll protoplasts were isolated from the third true leaf of 3-week-old plants. Protoplasts were transfected using reporter (4 μg), effector (4 μg), and reference plasmids (1 μg) then incubated at 22°C for 16 h under continuous light in 12-well tissue culture plates. Firefly and Renilla luciferase activities were then measured using the dual-luciferase reporter assay system and GloMax 20/20 luminometer (Promega, Madison, WI, USA). Coding regions of GLK1 and LsGLK were amplified by PCR and cloned into pMOE then used as effector plasmids. A multiple cloning site, luciferase gene, and NOS terminator of the promoter-less LUC vector were amplified by PCR then subcloned into pENTR, giving pENTR-LUC-NOST. To construct reporter plasmids, the promoter regions of BCM1 (−979 to +6 bp relative to the translation start site), Lhca1 (−1000 to +6 bp), Lhcb1.1 (−1014 to +6 bp), Lhcb2.2 (−1001 to +6 bp), CAO (−1045 to +6), LsBCM (−1811 to +6), and LsLhca1 (−1055 to +27) were amplified by PCR then cloned into pENTR-LUC-NOST. pPTRL, which expresses Renilla LUC under control of the CaMV 35S promoter, was used as a reference plasmid.
ChIP
ChIP was performed according to Saleh et al. (2008) with slight modifications. Samples were sonicated using Microson XL 2000 (Misonix Incorporated, USA). Dynabeads Protein G (Thermo Fisher Scientific; 100 µL/sample) was used for immunoprecipitation instead of Protein A agarose beads and pre-equilibrated with nuclei lysis buffer with 5 mg/mL BSA instead of salmon sperm. Anti-Myc antibody (MBL, 5 µL) was added to the 10-fold diluted samples and incubated overnight at 4°C. qPCR was performed using a KAPA SYBR FAST qPCR kit (Roche) and Rotor-Gene Q 2PLEX (Qiagen). qPCR conditions were as follows: initial denaturation at 95°C for 30 s, followed by 45–50 cycles at 95°C for 5 s, ending with 60°C for 30 s. qPCR data were normalized using the percentage of input method. The primers used for RT-qPCR are shown in Supplemental Table S1.
Prediction of transit peptide and transmembrane regions
Transit peptide sequences were predicted using ChloroP version 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/). Transmembrane regions of BCM1 were predicted using TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The amino acid sequences of the BCMs were aligned (Supplemental Figure S11) then regions corresponding to the transmembrane regions of BCM1 were regarded as the transmembrane regions of BCM2.
Phylogenetic tree construction
Neighbor-Joining phylogenetic trees of the BCM, SGR, and GLK proteins were constructed using MEGA X and CLUSTAL W for amino acid sequence alignment. The evolutionary distances were computed using the Poisson correction method in units of the number of amino acid substitutions per site.
Statistical analysis
Student’s t test and Tukey’s multiple comparison analysis were performed using R software (version 3.6.3 or 4.1.0).
Accession numbers
Sequence data were deposited in the Arabidopsis Genome Initiative or National Center for Biotechnology Information under the following accession numbers: ACT8 (AT1G49240), APRR2 (AT4G18020), ARR1 (AT3G16857), BCM1 (AT2G35260), BCM2 (AT4g17840), CAO (AT1g44446), GLK1 (AT2G20570), GLK2 (AT5G44190), Lhca1 (AT3g54890), Lhcb1.1 (AT1G29920), Lhcb2.2 (AT2g05070), NAP (AT1G69490), NYC1 (AT4G13250), ORE1 (AT5G39610), PsaF (AT1G31330), RCE1 (AT2G36305), SGR1/NYE1 (AT4G22920), SGR2/NYE2 (AT4G11910), SGRL (AT1G44000), STE24 (AT4G01320), LsACT7 (LOC111897580, LOC111909684, LOC111882438), LsBCM (LOC111918094), LsCAO (LOC111911963), LsGLK (LOC111886262), LsLhca1 (LOC111877237), LsLhcb1 (LOC111888054), LsSGR (LOC111920775), LsSGRL (LOC111901931), LsAPRR2-like (LOC111897177), LsARR1-like (LOC111914139), GmG (Glyma.01G198500), GmGL (Glyma.11G043400), GmGLK1 (Glyma.13G294300, Glyma.12G206600, Glyma.06G289300, Glyma.12G117700), GmSGR1 (Glyma.11G027400), GmSGR2 (Glyma.01G214600), SlBCM/G (LOC101260158), SlGLK1 (LOC101055587), SlGLK2 (LOC101055613), SlSGR1 (LOC778212), and SlSGR2 (LOC101268214).
Supplemental data
The following materials are available in the online version of this article.
SupplementalFigure S1. Protein structures and subcellular localization of BCM1 and 2.
SupplementalFigure S2. Mutations and phenotypes of the bcm mutants.
SupplementalFigure S3. Complementation of the bcm1 bcm2 phenotype by BCM1 and BCM2.
SupplementalFigure S4. Western blot analysis of chloroplast proteins in presenescent leaves of bcm1 bcm2.
SupplementalFigure S5. Expression of photosynthesis-related genes in presenescent leaves of bcm1 bcm2.
SupplementalFigure S6. Chlorophyll degradation in bcm mutants and BCM1 overexpressing plants during leaf senescence.
SupplementalFigure S7. Expression of senescence- and photosynthesis-related genes during dark incubation.
SupplementalFigure S8. ChIP-qPCR analysis of GLK1 binding to the BCM1 promoter.
SupplementalFigure S9. ChIP-qPCR analysis of GLK1 binding to the BCM2 promoter.
SupplementalFigure S10. Neighbor-joining trees of BCM, SGR, and GLK families.
SupplementalFigure S11. Amino acid sequence alignment of BCM proteins in lettuce, Arabidopsis, soybean and tomato.
SupplementalFigure S12. Natural senescence of lsbcm, lssgr, and lsbcm lssgr mutants in lettuce.
SupplementalFigure S13. Characterization of lsbcm and lssgr mutants.
SupplementalTable S1. Primers used in genome editing and RT-qPCR.
Supplementary Material
Acknowledgments
We thank Yumi Nagashima for technical assistance, Kazuya Kiyokawa for helpful advice on the Agrobacterium strains, Benke Kuai (Fudan University) and Ayumi Tanaka (Hokkaido University) for providing the nye1-1 nye2-1 double mutant, Yuichiro Takahashi (Okayama University) for providing the anti-PsaF antibody, and Masato Nakai (Osaka University) for providing the anti-TIC110 antibodies.
Funding
This research was supported by Research Fellowships from the Project of the Bio-oriented Technology Research Advancement Institution (Research Program on Development of Innovative Technology) awarded to M.K. and the Joint Research Program implemented by the Institute of Plant Science and Resources, Okayama University, awarded to M.K. and T.I.
Conflict of interest statement. None declared.
Contributor Information
Hiroshi Yamatani, Graduate School of Integrated Sciences for Life, Hiroshima University, Hiroshima 739-8526, Japan.
Takeshi Ito, Graduate School of Integrated Sciences for Life, Hiroshima University, Hiroshima 739-8526, Japan.
Kenji Nishimura, Institute of Plant Science and Resources, Okayama University, Kurashiki 710-0046, Japan.
Tetsuya Yamada, Graduate School of Agriculture, Hokkaido University, Sapporo 060-8589, Japan.
Wataru Sakamoto, Institute of Plant Science and Resources, Okayama University, Kurashiki 710-0046, Japan.
Makoto Kusaba, Graduate School of Integrated Sciences for Life, Hiroshima University, Hiroshima 739-8526, Japan.
These authors contributed equally (H.Y. and T.I.)
M.K. and H.Y. conceived the original research plans. T.Y., W.S., and K.N. supervised the experiments. H.Y. and T.I. designed and performed the experiments. H.Y., T.I., and M.K. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Makoto Kusaba (akusaba@hiroshima-u.ac.jp).
References
- Albrecht-Borth V, Kauss D, Fan D, Hu Y, Collinge D, Marri S, Liebers M, Apel K, Pfannschmidt T, Chow WS, et al. (2013) A novel proteinase, SNOWY COTYLEDON4, is required for photosynthetic acclimation to higher light intensities in Arabidopsis. Plant Physiol 163: 732–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Choi YD, Voytas DF, Rodermel S (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J 22: 303–313 [DOI] [PubMed] [Google Scholar]
- Chen Y, Shimoda Y, Yokono M, Ito H, Tanaka A (2019) Mg-dechelatase is involved in the formation of photosystem II but not in chlorophyll degradation in Chlamydomonas reinhardtii. Plant J 97: 1022–1031 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yamori W, Tanaka A, Tanaka R, Ito H (2021) Degradation of the photosystem II core complex is independent of chlorophyll degradation mediated by Stay-Green Mg2+ dechelatase in Arabidopsis. Plant Sci 307: 110902. [DOI] [PubMed] [Google Scholar]
- Chu CC, Li HM (2011) Determining the location of an Arabidopsis chloroplast protein using in vitro import followed by fractionation and alkaline extraction. Methods Mol Biol 774: 339–350 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Ohashi Y, Takahashi R, Nakai K, Takahashi Y (2021) DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 33: 2258–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang H, Yuan Q, Feng Y (2018) Structure and function of the photosystem supercomplexes. Front Plant Sci 9: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A (2009) Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem 284: 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T, Ito H, Tanaka A (2016) Simultaneous regulation of antenna size and photosystem I/II stoichiometry in Arabidopsis thaliana. Planta 244: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sun X, Zhang L, Sakamoto W (2012) Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol 159: 1428–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplastinner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kohzuma K, Sato Y, Ito H, Okuzaki A, Watanabe M, Kobayashi H, Nakano M, Yamatani H, Masuda Y, Nagashima Y, et al. (2017) The non-mendelian green cotyledon gene in soybean encodes a small subunit of photosystem II. Plant Physiol 173: 2138–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell Online 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Tanaka A, Tanaka R (2013) Stay-green plants: What do they tell us about the molecular mechanism of leaf senescence. Photosynth Res 117: 221–234 [DOI] [PubMed] [Google Scholar]
- Liu G, Yu H, Yuan L, Li C, Ye J, Chen W, Wang Y, Ge P, Zhang J, Ye Z, et al. (2021) SlRCM1, which encodes tomato Lutescent1, is required for chlorophyll synthesis and chloroplast development in fruits. Hortic Res 8: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang Y, Nie Z, Gai J, Bhat JA, Kong J, Zhao T (2020) Double mutation of two homologous genes YL1 and YL2 results in a leaf yellowing phenotype in soybean [Glycine max (L.) Merr]. Plant Mol Biol 103: 527–543 [DOI] [PubMed] [Google Scholar]
- Majsec K, Bhuiyan NH, Sun Q, Kumari S, Kumar V, Ware D, van Wijk KJ (2017) The plastid and mitochondrial peptidase network in Arabidopsis thaliana: a foundation for testing genetic interactions and functions in organellar proteostasis. Plant Cell 29: 2687–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A (2011) Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23: 3442–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59: 940–952 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Mano S, Tanaka Y, Ohnishi M, Nakamori C, Araki M, Niwa T, Nishimura M, Kaminaka H, Nakagawa T, et al. (2010) Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci Biotechnol Biochem 74: 1315–1319 [DOI] [PubMed] [Google Scholar]
- Nobusawa T, Kamei M, Ueda H, Matsushima N, Yamatani H, Kusaba M (2021) Highly pleiotropic functions of CYP78As and AMP1 are regulated in non-cell-autonomous/organ-specific manners. Plant Physiol 186: 767–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PDB, Ponnala L, Van Wijk KJ (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Kimura M, Matsuura H, Tanaka A, Ito H (2019) Jasmonate production through chlorophyll a degradation by Stay-Green in Arabidopsis thaliana. J Plant Physiol 238: 53–62 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep 6: 26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen H, Finkenzeller B, Kühlein N (1993) Pigments induce folding of light-harvesting chlorophyll a/b-binding protein. Eur J Biochem 215: 809–816 [DOI] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Zaltsman A, Adam Z, Takahashi Y (2003) Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M (2009) Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57: 120–131 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Nishimura M, Yamaguchi H, Kusaba M (2007) Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci USA 104: 14169–14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during Leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Ito H, Tanaka A (2016) Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Cui ML, Ma B, Ezura H (2006) Functional expression of the taste-modifying protein, miraculin, in transgenic lettuce. FEBS Lett 580: 620–626 [DOI] [PubMed] [Google Scholar]
- Takechi K, Sodmergen, Murata M, Motoyoshi F, Sakamoto W (2000) The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol 41: 1334–1346 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Yamamoto Y, Tsuji H (1991) Formation of chlorophyll–protein complexes during greening. 2. Redistribution of chlorophyll among apoproteins. Plant Cell Physiol 32: 195–204 [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole Metabolism in Arabidopsis thaliana. Arab B 9: e0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Higashiyama T (2016) pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol pcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Grimm B (2015) Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res 126: 189–202 [DOI] [PubMed] [Google Scholar]
- Wang M, Li W, Fang C, Xu F, Liu Y, Wang Z, Yang R, Zhang M, Liu S, Lu S, et al. (2018) Parallel selection on a dormancy gene during domestication of crops from multiple families. Nature Genet 50: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Wang P, Richter AS, Kleeberg JRW, Geimer S, Grimm B (2020) Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat Commun 11: 1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Langdale JA (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144: 307–313 [Google Scholar]
- Wu S, Li Z, Yang L, Xie Z, Chen J, Zhang W, Liu T, Gao S, Gao J, Zhu Y, et al. (2016) NON-YELLOWING2 (NYE2), a close paralog of NYE1, plays a positive role in chlorophyll degradation in Arabidopsis. Mol Plant 9: 624–627 [DOI] [PubMed] [Google Scholar]
- Yamatani H, Kohzuma K, Nakano M, Takami T, Kato Y, Hayashi Y, Monden Y, Okumoto Y, Abe T, Kumamaru T, et al. (2018) Impairment of Lhca4, a subunit of LHCI, causes high accumulation of chlorophyll and the stay-green phenotype in rice. J Exp Bot 69: 1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A, et al. (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-Protein complexes during leaf senescence. Plant J 74: 652–662 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang B, Mu B, Zheng X, Zhao F, Lan W, Fu A, Luan S (2020) A thylakoid membrane protein functions synergistically with GUN5 in chlorophyll biosynthesis. Plant Commun 1: 100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.