Abstract
Objectives
Standard implementations of amyloid typing by liquid chromatography–tandem mass spectrometry use capabilities unavailable to most clinical laboratories. To improve accessibility of this testing, we explored easier approaches to tissue sampling and data processing.
Methods
We validated a typing method using manual sampling in place of laser microdissection, pairing the technique with a semiquantitative measure of sampling adequacy. In addition, we created an open-source data processing workflow (Crux Pipeline) for clinical users.
Results
Cases of amyloidosis spanning the major types were distinguishable with 100% specificity using measurements of individual amyloidogenic proteins or in combination with the ratio of λ and κ constant regions. Crux Pipeline allowed for rapid, batched data processing, integrating the steps of peptide identification, statistical confidence estimation, and label-free protein quantification.
Conclusions
Accurate mass spectrometry–based amyloid typing is possible without laser microdissection. To facilitate entry into solid tissue proteomics, newcomers can leverage manual sampling approaches in combination with Crux Pipeline and related tools.
Keywords: Amyloid, Liquid chromatography, Tandem mass spectrometry, Open source, Crux
Key Points.
Standard implementations of amyloid typing by liquid chromatography–tandem mass spectrometry use capabilities unavailable to most clinical laboratories.
Accurate mass spectrometry–based amyloid typing is possible without laser microdissection.
To facilitate entry into solid tissue proteomics, newcomers can leverage manual sampling approaches in combination with Crux Pipeline and related tools.
Introduction
Amyloidosis is a heterogenous disorder in which proteins prone to misfolding deposit in tissues and result in organ damage. Treatment often focuses on correcting the underlying disorder responsible for production of the amyloidogenic protein, and thus correctly identifying the type is critical.1 Although certain forms of amyloidosis predominate, more than 30 responsible proteins have been described, reflecting a broad range of disorders that must be considered. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) has been deployed more than two decades for the typing of amyloid deposits.2-8 Although LC-MS/MS provides advantages over other approaches,9,10 it presents significant hurdles for new users. In a typical workflow using formalin-fixed, paraffin-embedded (FFPE) tissue, samples are acquired from slides via laser microdissection (LMD), proteins are extracted and proteolytically digested, and analyses are performed using nanoflow chromatography and high-resolution mass spectrometry.11 Spectra, acquired via data-dependent acquisition (DDA), are then matched to proteins via database searching. Each component of this workflow represents a potential leap in technical demands for interested laboratories, even if already leveraging mass spectrometry for other purposes.
To increase accessibility of LC-MS/MS–based amyloid typing, we investigated whether components of the process could be substituted or streamlined. Sampling by LMD, for example, is critical in molecular genetics workflows12-14 but is costly and time-consuming.15 Ostensibly, sampling less precisely from amyloid deposits would introduce variability in the analysis,2,5 but this has not been clearly demonstrated. Replacement of DDA is similarly tempting, specifically with targeted methods run on triple quadrupole analyzers. However, as others5 have noted, amyloid proteins may be truncated and fail to generate proteotypic peptides needed for multiple-reaction monitoring. As a result, the use of untargeted acquisition methods will likely persist but may be made easier for new users by simplifying the associated data processing.
Here we describe a clinical LC-MS/MS–based amyloid typing method that uses manual microdissection in place of LMD. Sampling equipment is reduced to a needle, disposable syringe, and standard dissecting microscope. In addition, we introduce a straightforward, open-source option for the processing of complex shotgun proteomics data in clinical environments. This consists of fully scripted implementation of a mass spectrometry analysis toolkit16,17 (Crux), consolidating the steps of peptide identification, statistical confidence estimation, and label-free protein quantification for users without computational expertise.
MATERIALS AND METHODS
Specimens and Tissue Sampling
Specimens consisted of archived paraffin-embedded tissue blocks from clinical cases of amyloidosis (majority renal). All cases had a predefined type of amyloid based on previous testing by immunohistochemistry, immunofluorescence, and LC-MS/MS (using LMD), the latter performed at an outside reference laboratory. A subset of five well-characterized cases of renal amyloidosis (two λ light chain type, one κ light chain type, one serum amyloid protein A type, and one transthyretin type) was designated as control material and analyzed alongside other cases in each batch. For tissue microdissection, 10-µm tissue sections on standard glass slides (Assure plus) were deparaffinized, stored briefly (up to 1 hour) in deionized water, and scraped by hand using a 29-gauge Luer-lock needle attached to a plastic syringe (1 mL). Regions of interest were identified and scraped by a board-certified anatomic pathologist (with prior experience evaluating amyloid deposits; K.D.S.) using a dissecting microscope and guided by Congo red stains from adjacent tissue sections. For Congo red–negative samples, areas of interest were chosen based on histology and pathologic diagnosis. For studies involving residual human specimens, the Human Subjects Division of our institution has determined that the use of leftover, de-identified clinical samples for method development, method validation, and quality improvement is not considered human subjects research.
Sample Processing
A detailed standard operating procedure (SOP) is provided in the Supplemental Material (all supplemental materials can be found at American Journal of Clinical Pathology online), which contains complete descriptions of the materials, sample preparation, LC-MS/MS setup, and data analysis. In brief, deparaffinized tissue section scrapings were collected immediately into tubes containing a surfactant (RapiGest, Waters Corporation). Tissues were disrupted and denatured by heat and sonication. Proteins were reduced, alkylated, and digested with trypsin. Enzyme activity was stopped by the addition of acid and the final digested samples loaded for LC-MS/MS injection.
Liquid Chromatography–Tandem Mass Spectrometry
Nanoflow liquid chromatography was performed on a Easy nLC 1000 chromatography system coupled to a Q–Exactive Plus mass spectrometer (Thermo Scientific). Tandem mass spectrometric analysis consisted of data-dependent acquisition with dynamic exclusion, performed over a 60-minute gradient.
Data Processing
A human protein database was constructed in FASTA format using the Swiss-Prot human proteome with appended sequences for common amyloid variants (database available upon request). Tandem mass spectrometric data files (Thermo RAW format) were converted to MS2 format using MSConvert (64-bit)18 (available from http://proteowizard.sourceforge.net/tools.shtml). Crux 3.2-a761451 was installed on a Linux server running Ubuntu 18.04.5 LTS and Python 2.7.17. Crux analysis consisted of (1) searching the set of MS2 files using Comet,19 (2) processing search results using Percolator,20,21 (3) aggregating counts for different isoforms, and (4) computing protein abundance as normalized spectral abundance factors (NSAFs).22,23 NSAFs were used to reduce bias in quantification toward larger proteins. Steps 1 to 4 were scripted in Python (termed Crux Pipeline, available as Supplemental Material). Further instructions for setup are provided in the SOP. NSAF data were plotted in Excel (Microsoft) and Prism (GraphPad Software) to compare protein abundances between samples with different pathologic diagnoses and to perform statistical analyses.
RESULTS
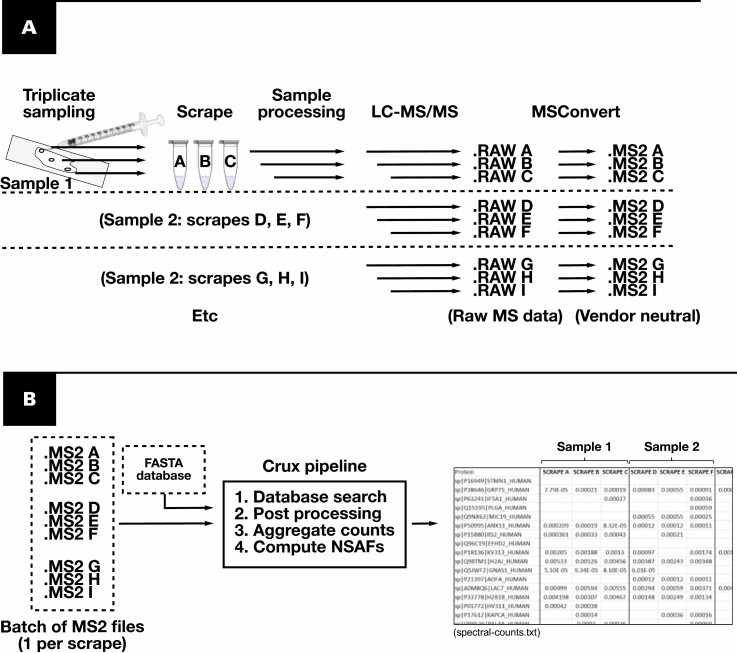
The assay was deployed as depicted in Figure 1, using a needle for sampling under a microscope in place of LMD. Analyses were performed in batches of 12 to 24 scrapings, each processed separately. Up to four replicate scrapes were performed per slide, targeting different Congo red–positive areas. Crux Pipeline enabled batched processing of the generated shotgun proteomics data. Starting with the database searching, only two inputs were required from the user: (1) a directory containing mass spectrometry result files and (2) a FASTA protein database. Data processing from 24 scrapes (ie, database searching, postprocessing of peptide spectrum matches, and quantification of proteins using spectral counting methods) could be completed in approximately 1 hour.
Figure 1.
Workflow of amyloid typing using manual microdissection and Crux Pipeline. A, Samples are acquired in triplicate from slides using a needle, targeting areas containing amyloid deposits. Each scrape is processed and analyzed separately by liquid chromatography–tandem mass spectrometry (LC-MS/MS). B, Crux Pipeline requires only two user inputs (dotted lines), a FASTA database, and a directory of mass spectrometry result files. The output (“spectral-counts”) is a tab-delimited text file containing a list of all proteins detected and the normalized spectral abundance factor (NSAF) results for each scrape.
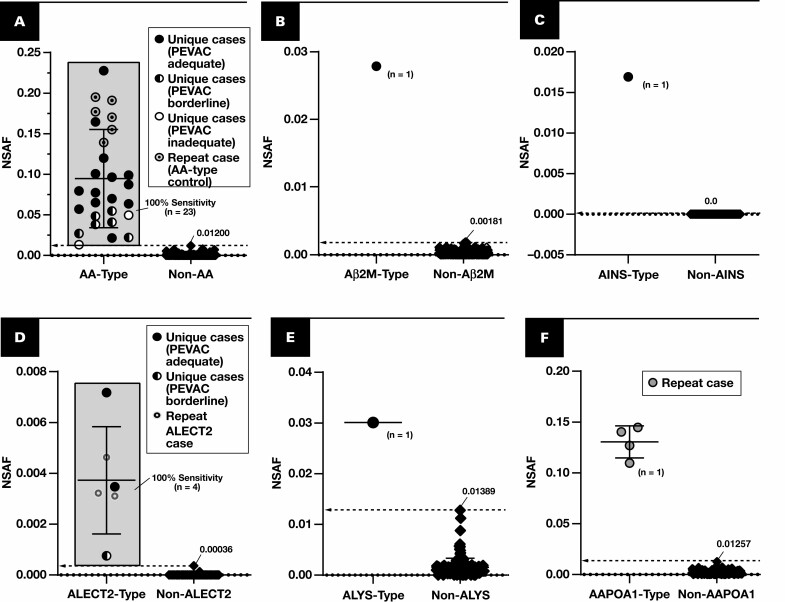
We first analyzed a “training” set of 121 unique, paraffin-embedded tissue blocks (a majority from renal tissue), including 87 from cases of amyloidosis Table 1. From these specimens, a total of 468 scrapes were analyzed (see Supplemental Table 1). Eighty-four of the amyloidosis cases had at least triplicate scrapings performed (see Supplemental Table 2). The first 120 scrapes of designated control material provided cutoffs of amyloid sampling sufficiency based on the abundance of five proteins commonly associated with amyloid deposition: serum amyloid P, apolipoprotein E, victronectin, apoliprotein A4, and clusterin24-28 (“PEVAC”). The cutoff for each was set as its minimum NSAF in the 120 analyses (Supplemental Figure 1). For all other scrapes, then, the number of PEVAC proteins with NSAF above these minimum cutoffs constituted (in sum) an integer score of 0 to 5 (a PEVAC score). The rules in Table 2 were used to designate amyloid sampling in the other individual scrapes as “sufficient” or “insufficient” and in cases (with triplicate scrapes per slide) as “adequate,” “borderline,” or “inadequate.” The results of these categorizations are shown in Supplemental Table 1 and Supplemental Table 2. The established cutoffs designated 14 (5%) of 280 of noncontrol, putative amyloid scrapings as “insufficient.” Comparatively, for unique cases of amyloid with at least three analyses per slide, 6 (7%) of 84 putative amyloid cases were designated inadequate (or not amyloid) by the PEVAC rules. These results exclude reanalyses of scrapings from the same five control blocks, none of which demonstrated PEVAC scores less than 5 in any replicate in subsequent retesting.
Table 1.
Training Set Specimens
| Diagnosis | Unique Blocksa | Unique Blocksa Scraped in Triplicateb |
|---|---|---|
| AA-type amyloidosis | 23 (16 kidney, 7 lymph node) | 23 |
| Aβ2MG-type amyloidosis | 1 (bone marrow) | 1 |
| AINS-type amyloidosis | 1 (subcutaneous fat) | 1 |
| ALECT2-type amyloidosis | 5 (3 kidney, 1 spleen, 1 adrenal) | 4 (1 spleen, 3 kidney) |
| ALYS-type amyloidosis | 1 | 1 |
| AAPOA1-type amyloidosis | 1 | 1 |
| AAPOA-IV-type amyloidosis | 1 | 1 |
| ATTR-type amyloidosis | 8 (7 heart, 1 subcutaneous fat) | 8 |
| AL(κ)-type amyloidosis | 13 (7 kidney, 4 lung, 1 heart, 1 stomach) | 13 |
| AL(λ)-type amyloidosis | 31 (21 kidney, 4 heart, 2 colon, 1 heart and kidney, 1 subcutaneous fat, 1 skin, 1 bone) | 29 (19 kidney, 4 heart, 2 colon, 1 heart and kidney, 1 subcutaneous fat, 1 skin, 1 bone) |
| AHL(IgG- λ)-type amyloidosis | 1 | 1 |
| Amyloid NOS | 1 (thyroid) | 1 |
| Subtotal | 87 | 84 |
| Diabetic | 2 | 2 |
| Fibrillary glomerulonephritis | 19 | 7 |
| Light chain deposition disease | 4 | 3 |
| Immunoglobulin deposition disease | 1 | 1 |
| Myeloma cast nephropathy | 2 | 2 |
| Arterionephrosclerosis | 1 | 1 |
| Membranous nephropathy | 3 | 0 |
| No diagnosis/unknown | 2 (kidney, aortic valve) | 2 |
| Subtotal | 34 | 18 |
| Total | 121 | 102 |
aUnique “blocks” reflected unique surgical pathology or autopsy cases with two exceptions (one AL(λ) case and one AL(κ) case each contributed two blocks). Samples are from kidney unless otherwise indicated. Each case was from a different patient.
bRefers to blocks for which scraping in triplicate occurred at least once over the course of method validation.
Table 2.
Amyloid Sampling Sufficiency Rules
| Designation | PEVAC Rules |
|---|---|
| Individual scrape | |
| “Sufficient” | Both of the following: |
| (A) At least two PEVAC proteins above minimum threshold (PEVAC ≥2) | |
| (B) APOE detected (NSAF >0) | |
| “Insufficient” | All others |
| Case (or slide)a | |
| “Adequate” | At least two sufficient scrapes, including either: |
| (A) At least two scrapes with PEVAC ≥4 | |
| (B) One scrape with PEVAC = 5 and a second with PEVAC = 3 | |
| “Borderline” | At least two sufficient scrapes, including either: |
| (A) At least two scrapes with PEVAC = 3 | |
| (B) One scrape with PEVAC = 4 and a second with PEVAC = 2 | |
| “Inadequate” | All other cases |
NSAF, normalized spectral abundance factor; PEVAC, serum amyloid P, apolipoprotein E, victronectin, apoliprotein A4, and clusterin.
aApplicable with sampling in at least three areas of the slide (leading to three separate liquid chromatography–tandem mass spectrometry analyses and averaging of NSAF results across replicates).
We next evaluated the performance of amyloid type–specific proteins in the discrimination of scrapes of different amyloid forms. Supplemental Figure 2 shows the NSAF results for type-specific proteins from all scrapes with PEVAC of 2 or more in the training set. Scrapes of non–light chain forms of amyloidosis were distinguishable from other forms (including light chain amyloidoses) with areas under the receiver operating characteristic curve (AUC) of 0.997 or greater in all cases. Unexpected elevation of the “wrong” amyloid type–specific protein in rare scrapes generally reflected carryover, which could be corrected for via triplicate evaluations from each slide and using averaging of NSAF results across replicates (described below). For the light chain λ and light chain κ types, the λ constant region (LAC) demonstrated an AUC of 0.89 and κ constant region (IGKC) an AUC of 0.97, respectively. In discriminating λ -type cases from κ, the ratio of λ constant region to κ constant region demonstrated an AUC of 0.98.
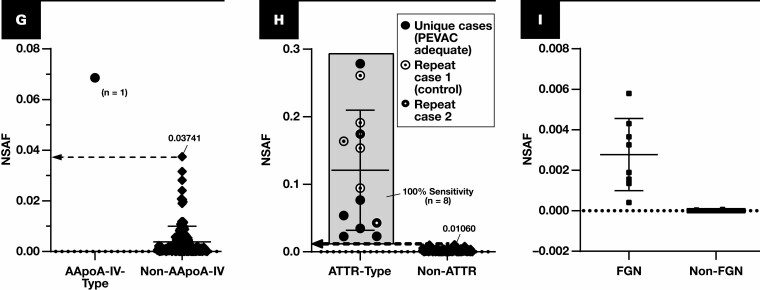
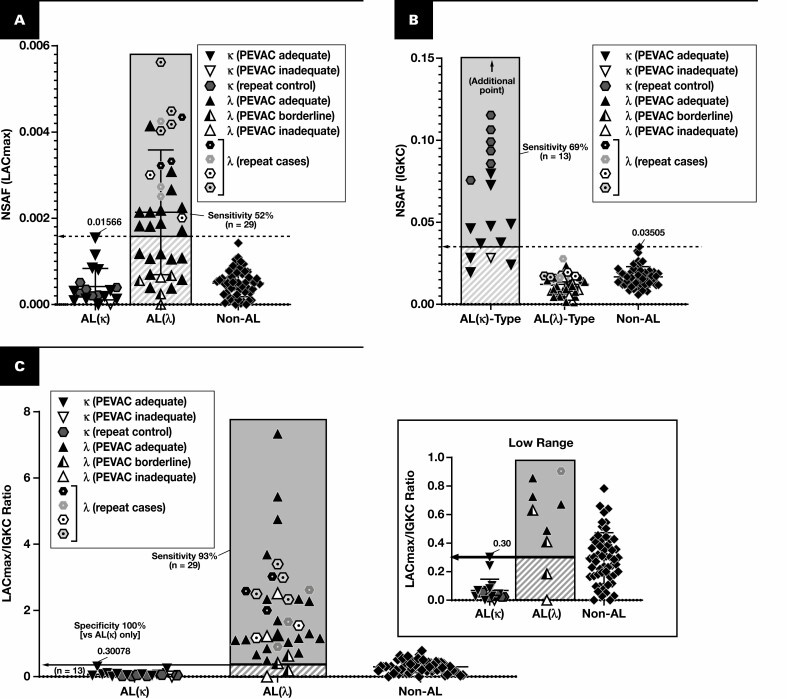
The results for the training set when using average NSAF results from triplicate analyses are shown in Figure 2 and Figure 3. The following performance metrics focus on unique cases that were tested rather than all analyses. Using the marked cutoffs, each non–light chain case was distinguishable with both 100% sensitivity and 100% specificity based on the assessment of a single protein—serum amyloid A-1, β2-microglobulin, insulin, leukocyte cell–derived chemotaxin 2, lysozyme C, apolipoprotein A-I, apolipoprotein A-IV, or transthyretin Figure 2. None of the immunoglobulin light chain amyloid cases exhibited NSAF results above these thresholds. Instead, a majority (24 of 42 unique cases) demonstrated λ or κ light chain in excess of that observed for other amyloid forms Figure 3A and FIGURE 3B, and most (40 of 42) could otherwise be clarified (or confirmed) based on the ratio between λ and κ (using the cutoff marked in Figure 3C). Two λ light chain cases did not demonstrate an increase in λ constant regions, the λ to κ ratio, or other amyloidogenic protein, but neither were PEVAC adequate and thus would not be reported clinically without further investigation and possible resampling. Preliminary results were also available for detection of some nonamyloid conditions. For example, cases of fibrillary glomerulonephritis were discernible through the detection of DNAJB9.29,30
Figure 2.
Spectral abundance of non-AL type-specific proteins. Results are plotted for cases of amyloidosis (circles and diamonds) with at least triplicate analyses (scrapes) per slide. Each point represents the average of normalized spectral abundance factor (NSAF) results from scrapes with PEVAC (serum amyloid P, apolipoprotein E, victronectin, apoliprotein A4, and clusterin) of 2 or more and detection of APOE. Circles represent unique cases unless otherwise indicated in each key (number of unique samples indicated in each plot). Repeat cases typically reflected designated control material, potential control material under evaluation, or cases for amyloid types of which few unique cases were available (eg, ALECT2, AAPOA1). Cutoff points are marked (dotted line), indicating a 100% specificity (and 100% sensitivity) level when comparing with other forms of amyloidosis, including the AL forms. A, Serum amyloid A-1 (SAA1) (P0DJI8) as a marker for AA type. B, β2-Microglobulin (B2MG) (P61769) for Aβ2M type. C, Insulin (INS) (P01308) for AINS type. D, Leukocyte cell–derived chemotaxin 2 (LECT2) (O14960) for ALECT2 type. E, Lysozyme C (LYSC) (P61626) for ALYS type. F, Apolipoprotein A-I (APOA1) (P02647). G, APOA4 (P06727), apolipoprotein A-IV. H, TTHY (P02766), transthyretin. I, DnaJ homolog subfamily B member 9 (DNJAB9) (Q9UBS3) for cases of fibrillary glomerulonephritis (FGN, squares). Because FGN is not a form of amyloidosis, PEVAC scores were not considered when computing average NSAF for DNJAB9. UniProt entry IDs are indicated in parentheses.
Figure 3.
Discrimination of AL(κ) and AL(λ). Results are plotted for cases of amyloidosis with at least triplicate analyses (scrapes) per slide. Each point represents the average of normalized spectral abundance factor (NSAF) results from scrapes with PEVAC (serum amyloid P, apolipoprotein E, victronectin, apoliprotein A4, and clusterin) of 2 or more. At the level of 100% specificity, the sensitivity for λ (A) and κ types (B) is marked. Cases that were repeated (hexagons, reflecting the control blocks) are counted only once in the sensitivity calculations. When replotting λ and κ light chain amyloidoses (AL(λ) and AL(κ), respectively) by the ratio of λ constant region to κ constant region (C), only two cases of putative AL(λ) type did not have a measured ratio greater than all cases of AL(κ), but neither were PEVAC adequate. LAC, immunoglobulin λ constant; LACmax, maximum of UniProt entries P0CG04, P0CG0, P0CG06, P0CF74, A0M8Q6.
We applied the cutoffs derived from the training set to typing additional samples in a “test” set (n = 27) Table 3. Twenty-three of these 27 specimens were from cases of amyloidosis of known type. Five were repeats from the training set but included to assess whether typing could be replicated. Typing was based on finding the highest NSAF for an amyloidogenic protein relative to the 100% specificity threshold defined with the training set. For example, in case 1, the NSAF of transthyretin (TTHY) was 0.06466, a level corresponding to 610% of the specificity threshold (0.01060) in Figure 2. In the absence of a protein above its specificity threshold, the λ to κ ratio can clarify typing as suspected AL(λ) or AL(κ). Twenty-two (96%) of 23 cases of known diagnosis typed as expected using this approach (one was PEVAC inadequate), or 18 (95%) of 19 when excluding the repeats. Of note, within this set, we uniquely detected a calcitonin-related peptide (CALA, UniProt entry P06881), consistent with the underlying ACal-type amyloid. CALA was not detected in any other samples in either the training or test set. Typing in this case signified the method’s potential to capture additional forms of amyloidosis that may be suspected as they arise clinically. Complete NSAF results for the test set appear in Supplemental Table 3.
Table 3.
Test Set Specimens
| Blocka | Diagnosis | Highest Amyloid Protein Above Cutoffb | Other Amyloidogenic Proteinsc | λ/κd | LC-MS/MS Typing |
|---|---|---|---|---|---|
| 1 | ATTR | TTHY (0.06466, 610%) | ATTR | ||
| 2 | ACal | CALCA (0.05698, NA e ) | APOA4 (0.04200) | ACal | |
| IGKC (0.01950) | |||||
| 3 | AL(κ) | IGKC (0.06541, 187%) | 0.1 | AL(κ) | |
| 4 | AL(κ) | IGKC (0.05909, 169%) | 0.1 | AL(κ) | |
| 5 | Amyloid type indeterminate | LAC (0.06485, 414%) | APOA1 (0.01413) | 2.2 | AL(λ) |
| IGKC (0.02995) | |||||
| 6 | AA | SAA1 (0.04145, 345%) | AA type | ||
| 7 | AL(λ) | None | LAC (0.01483, 95%) | 0.9 | AL(λ) |
| 8* | FGN | NA | NA | FGN (DNAJB9 detectedf) | |
| 9* | ALECT2 | LECT2 (0.00274, 762%) | ALECT2 | ||
| 10 | AL(λ) | LAC (0.03176, 203%) | 2.5 | AL(λ) | |
| 11 | AL(λ) | LAC (0.02729, 174%) | 10.7 | AL(λ) | |
| 12* | ATTR | TTHY (0.19490, 1,839%) | IGKC (0.01869) | ATTR | |
| 13* | AL(λ) | None | TTHY (0.00990) | 0.6 | AL (λ) |
| LAC (0.01285, 82%) | |||||
| IGKC (0.02156) | |||||
| 14* | AL(λ) | LAC (0.02488, 159%) | 1.1 | AL(λ) | |
| 15 | AA type | SAA1 (0.02289, 191%) | AA type | ||
| 16 | AL(λ) | None | LAC (0.01038, 66%) | 1.3 | AL (λ) |
| 17 | AA-type | SAA1 (0.073963, 616%) | IGKC (0.01939) | AA type | |
| 18 | AL(λ) | LAC (0.01811, 116%) | 15.5 | AL(λ) | |
| 19 | AL(λ) | LAC (0.03816, 244%) | 4.7 | AL(λ) | |
| 20 | AL(κ) | None | IGKC (0.01968, 56%) | 0.0 | AL(κ) |
| 21 | AL(λ) | None (PEVAC inadequate) | NA | ||
| 22 | AA type | SAA1 (0.03080, 257%) | IGKC (0.03776) | AA type | |
| LAC (0.01012) | |||||
| 23 | AA type | SAA1 (0.14968, 1,247%) | AA type | ||
| 24 | AL(λ) | None | LAC (0.01476, 94%) | 3.1 | AL(λ) |
| SAA1 (0.00734) | |||||
| 25 | FGN | NA | NA | FGN (DNAJB9 detectedf) | |
| 26 | AL(λ) | None | LAC (0.00809, 52%) | 1.6 | AL(λ) |
| 27 | Amyloid type indeterminate | TTHY (0.02464, 232%) | APOA1 (0.017062) | ATTR type | |
| LAC (0.01988) | |||||
| IGKC (0.04088) |
AA, serum protein A type; ACal, calcitonin type; ALECT2, LECT2 type; AL(κ), κ light chain type; AL(λ), λ light chain type; APOA4, apolipoprotein A-IV (UniProt P06727); ATTR, transthyretin type; CALCA, calcitonin gene–related peptide 1 (UniProt P06881); DNAJB9, DnaJ heat shock protein family member B9 (UniProt Q9UBS3); FGN, fibrillary glomerulonephritis; IGKC, immunoglobulin κ constant (UniProt P01837); LAC, λ constant region (UniProt entry P0CG04, P0CG05, P0CG06, P0CF74, or A0M8Q6); LC-MS/MS, liquid chromatography–tandem mass spectrometry; LECT2, leukocyte cell–derived chemotaxin 2; NA, not applicable; SAA1, serum amyloid A-1 (UniProt P0DJI8); PEVAC, serum amyloid P, apolipoprotein E, victronectin, apoliprotein A4, and clusterin; TTHY, transthyretin (Uniprot P02766).
aFive of 27 formalin-fixed, paraffin-embedded blocks were reevaluations (*) of cases included in the training set. All blocks above reflected unique cases/patients.
bAmyloidogenic protein with highest normalized spectral abundance factor (NSAF) relative to its 100% specificity threshold (bolded, if a protein met this criteria). Values in parentheses are the actual NSAF result and its proportion compared to the cutoff, expressed as a percentage: (NSAFResult / NSAFCutoff) * 100%.
cOther amyloidogenic proteins measured with NSAF of at least 50% of its 100% specificity cutoff.
dLess than 0.30 favors κ type; more than 0.30 favors λ type.
eAn ACal case was only encountered in the test set. CALCA protein was not detected in any other samples from the training or test set, and thus the proportion of the NSAF result to a specificity cutoff cannot be calculated.
fNon–Congo red positive but subjected to similar processing for the detection of DNAJB9 and confirmation of FGN. For case 8, average NSAF of DNABJ9 was 0.00367 (5,241% of cutoff). For case 25, DNAJB9 NSAF was 0.0091379839 (13,054% of cutoff).
Discussion
Here we described LC-MS/MS–based amyloid protein typing using manual microdissection in place of LMD. The approach enabled discrimination between the major amyloid types, and all cases in a test set with known underlying amyloid form were correctly typed based on the decision cutoffs established in the training set. LMD has become standard in mass spectrometry–based characterization of amyloid deposits because less precise sampling allows for the dilution from nonamyloid tissue.5,31 Initial method iterations involving FFPE tissue2,3 in fact did not employ LMD and were potentially subject to these problems. However, it must be noted that the methods preceding use of LMD did not appear to use microdissection either, instead employing a razor blade to loosen deparaffinized tissue nonspecifically from slides (up to 30 sections per case). In addition, extensive prefractionation was used, which could have contributed to variability in the results. Finally, both chromatography and mass spectrometry have changed remarkably over the past two decades, particularly with the introduction hybrid orbitrap mass spectrometers.32,33 Thus, the possibility of using a more intermediate sampling approach (ie, manual microdissection) remained. Others have attempted to avoid LMD by other means. For example, Kamiie et al15 described using organic solvents for selective extraction of amyloid proteins from FFPE tissue. This strategy does not require a pathologist for the selection of amyloid areas (an advantage over our method and methods employing LMD) and avoids heating of the samples in surfactant. However, it is not yet clear whether such an approach could be leveraged in a clinical environment or if it adapts well in the face of an ever-increasing number of known amyloid proteins.
Given the adjustment in sampling technique, we adopted a semiquantitative measure of sufficiency, the PEVAC score. Its introduction required two features common to LC-MS/MS–based amyloid typing. The first is preliminary, repetitive testing of a set of well-characterized cases of amyloidosis (process-level control material5) spanning the major forms of amyloidosis. This testing is performed as part of validation and offers a source of quality control material moving forward. The second is coanalysis of amyloid-associated proteins measured alongside the type-specific proteins of interest.31,34 The specificity of these proteins for amyloid appears tissue dependent. For example, Vrana et al6 previously described a “universal” signature for amyloid in subcutaneous fat aspirates. There, detection of APOE, SAP, and/or APOA4 was essentially diagnostic for amyloidosis. In our analyses involving a broader range of tissues (predominantly renal but also cardiac, lymphatic, endocrine, pulmonary, and dermal, among others), the PEVAC proteins showed some specificity for amyloid but were also present in nonamyloid tissue (evident in Supplemental Table 1) and thus were not diagnostic for amyloid. The utility of PEVAC scoring is more evident in Figure 2 and FIGURE 3. Samples with lower PEVAC scoring (half-filled and empty circles in Figure 2) were in the bottom half of NSAF for the diagnostic protein in all cases. Given that the amyloidogenic protein was above the specificity target for most of these cases, corresponding typing would have been correct for these cases. Thus, the primary benefit of PEVAC scoring is to provide caution where appropriate, and suggest resampling if the ordering pathologist determines this to be necessary within the context of other data.
Constructing an assay of this type is an ongoing process. The expected NSAF thresholds for amyloidogenic proteins were determined over time and may be reevaluated as additional cases are processed. Although the major types of amyloidosis were addressed in this study, additional types1 will need to be considered as they arise clinically. Furthermore, limited cases were available for the rarer amyloid forms. Additional cases with these purported subtypes will need to be evaluated to generate robust cutoffs for clinical use. A potential format for reporting data to the requesting pathologist is shown in Supplemental Figures 3 and 4. Such a report compares NSAFs with the determined specificity cutoffs, expressed as a percentage (NSAFResult/NSAFCutoff × 100%). As noted above, carryover was recognized as a potential confounder in interpreting the results for individual scrapes. A visualization of the carryover affect is provided in Supplemental Figure 5, showing results may be affected in the absence of triplicate analyses and use of blank injection between cases.
It is important to point out that this method relies on nanoflow chromatography and a hybrid quadrupole-orbitrap tandem mass spectrometer, which are not readily available in most clinical laboratories. For other solid tissue applications, such as targeted assays for Her2/Neu, modifications to sample preparation (including immunoaffinity enrichment) may facilitate the use of less sophisticated analyzers, such as triple quadrupoles, and higher flow rates.35 However, such adjustments may not be possible for all solid tissue applications, particularly those benefiting most from untargeted acquisition approaches. Where untargeted methods cannot be eliminated, low-cost, accessible options for data processing will be critical to expanding use. A plethora of open-source options36 exists for the purposes of matching spectra to peptides (and therefore proteins), but many are too cumbersome “out of the box” to easily validate in a clinical setting. Crux Pipeline provides a “plug-and-play” option for users new to proteomics, integrating existing free tools to bypass potential challenges. Of note, Crux “pipeline” exists as a command in the Crux toolbox (described here: http://crux.ms/commands/pipeline.html), but the scripted functionality here has been further extended to be useful in clinical settings and has not been previously described. The expanded functionality specifically includes aggregation of isoforms and the computation of NSAF values, as well as the ability to process batches of results rather than individual files.
Conclusions
In conclusion, amyloid typing by LC-MS/MS is possible without LMD. Critical in this endeavor is the development of approaches to assessing adequacy of amyloid sampling. Laboratories seeking to deploy tissue mass spectrometry should consider alternatives to all potential hurdles in launching new assays, even if substitutions are without clear precedent. Scripted utilization of Crux may facilitate implementation of data processing for untargeted proteomics for laboratories starting without this capability.
Supplementary Material
Acknowledgments
This work is supported in part by the University of Washington’s (UW’s) Proteomics Resource (UWPR95794) and by National Institutes of Health awards R01 GM121818 and P41 GM103533. Priska von Haller, PhD (UWPR), and Richard Johnson, PhD (UW Genome Sciences), provided significant technical guidance. Elisha Goonatilleke, PhD, assisted in the setup and operation of mass spectrometry. Database searching was performed on UW Department of Laboratory Medicine and Pathology server infrastructure provided through collaboration with the Division of Laboratory Informatics. Kaipo Tamura developed the initial version of the Crux Pipeline.
References
- 1. Picken MM. Diagnosis of amyloid beyond Congo red. Curr Opin Nephrol Hypertens. 2021;30:303-309. [DOI] [PubMed] [Google Scholar]
- 2. Murphy CL, Eulitz M, Hrncic R, et al. . Chemical typing of amyloid protein contained in formalin-fixed paraffin-embedded biopsy specimens. Am J Clin Pathol. 2001;116:135-142. [DOI] [PubMed] [Google Scholar]
- 3. Murphy CL, Wang S, Williams T, et al. . Characterization of systemic amyloid deposits by mass spectrometry. Methods Enzymol. 2006;412:48-62. [DOI] [PubMed] [Google Scholar]
- 4. Rodriguez FJ, Gamez JD, Vrana JA, et al. . Immunoglobulin derived depositions in the nervous system: novel mass spectrometry application for protein characterization in formalin-fixed tissues. Lab Invest. 2008;88:1024-1037. [DOI] [PubMed] [Google Scholar]
- 5. Theis JD, Dasari S, Vrana JA, et al. . Shotgun-proteomics-based clinical testing for diagnosis and classification of amyloidosis. J Mass Spectrom. 2013;48:1067-1077. [DOI] [PubMed] [Google Scholar]
- 6. Vrana JA, Theis JD, Dasari S, et al. . Clinical diagnosis and typing of systemic amyloidosis in subcutaneous fat aspirates by mass spectrometry–based proteomics. Haematologica. 2014;99:1239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mollee P, Boros S, Loo D, et al. . Implementation and evaluation of amyloidosis subtyping by laser-capture microdissection and tandem mass spectrometry. Clin Proteomics. 2016;13:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holub D, Flodrova P, Pika T, et al. . Mass spectrometry amyloid typing is reproducible across multiple organ sites. Biomed Res Int. 2019;2019:3689091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gonzalez Suarez ML, Zhang P, Nasr SH, et al. . The sensitivity and specificity of the routine kidney biopsy immunofluorescence panel are inferior to diagnosing renal immunoglobulin-derived amyloidosis by mass spectrometry. Kidney Int. 2019;96:1005-1009. [DOI] [PubMed] [Google Scholar]
- 10. Gilbertson JA, Theis JD, Vrana JA, et al. . A comparison of immunohistochemistry and mass spectrometry for determining the amyloid fibril protein from formalin-fixed biopsy tissue. J Clin Pathol. 2015;68:314-317. [DOI] [PubMed] [Google Scholar]
- 11. Payto D, Heideloff C, Wang S. Sensitive, simple, and robust nano-liquid chromatography–mass spectrometry method for amyloid protein subtyping. Methods Mol Biol. 2016;1378:55-60. [DOI] [PubMed] [Google Scholar]
- 12. Yazdi AS, Puchta U, Flaig MJ, et al. . Laser-capture microdissection: applications in routine molecular dermatopathology. J Cutan Pathol. 2004;31:465-470. [DOI] [PubMed] [Google Scholar]
- 13. Espina V, Wulfkuhle JD, Calvert VS, et al. . Laser-capture microdissection. Nat Protoc. 2006;1:586-603. [DOI] [PubMed] [Google Scholar]
- 14. Golubeva Y, Salcedo R, Mueller C, et al. . Laser capture microdissection for protein and NanoString RNA analysis. Methods Mol Biol. 2013;931:213-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamiie J, Aihara N, Uchida Y, et al. . Amyloid-specific extraction using organic solvents. Methodsx. 2020;7:100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park CY, Klammer AA, Käll L, et al. . Rapid and accurate peptide identification from tandem mass spectra. J Proteome Res. 2008;7:3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McIlwain S, Tamura K, Kertesz-Farkas A, et al. . Crux: rapid open source protein tandem mass spectrometry analysis. J Proteome Res. 2014;13:4488-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chambers MC, Maclean B, Burke R, et al. . A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22-24. [DOI] [PubMed] [Google Scholar]
- 20. Käll L, Canterbury JD, Weston J, et al. . Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923-925. [DOI] [PubMed] [Google Scholar]
- 21. Käll L, Storey JD, MacCoss MJ, et al. . Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7:29-34. [DOI] [PubMed] [Google Scholar]
- 22. Neilson KA, Keighley T, Pascovici D, et al. . Label-free quantitative shotgun proteomics using normalized spectral abundance factors. Methods Mol Biol. 2013;1002:205-222. [DOI] [PubMed] [Google Scholar]
- 23. McIlwain S, Mathews M, Bereman MS, et al. . Estimating relative abundances of proteins from shotgun proteomics data. BMC Bioinformatics. 2012;13:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cathcart ES, Comerford FR, Cohen AS. Immunologic studies on a protein extracted from human secondary amyloid. N Engl J Med. 1965;273:143-146. [DOI] [PubMed] [Google Scholar]
- 25. Cathcart ES, Wollheim FA, Cohen AS. Plasma protein constituents of amyloid fibrils. J Immunol. 1967;99:376-385. [PubMed] [Google Scholar]
- 26. Lee KW, Lee DH, Son H, et al. . Clusterin regulates transthyretin amyloidosis. Biochem Biophys Res Commun. 2009;388:256-260. [DOI] [PubMed] [Google Scholar]
- 27. Winter M, Tholey A, Krüger S, et al. . MALDI-mass spectrometry imaging identifies vitronectin as a common constituent of amyloid deposits. J Histochem Cytochem. 2015;63:772-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pilling D, Gomer RH. The development of serum amyloid P as a possible therapeutic. Front Immunol. 2018;9:2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andeen NK, Yang HY, Dai DF, et al. . DnaJ homolog subfamily B member 9 is a putative autoantigen in fibrillary GN. J Am Soc Nephrol. 2018;29:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nasr SH, Vrana JA, Dasari S, et al. . DNAJB9 is a specific immunohistochemical marker for fibrillary glomerulonephritis. Kidney Int Rep. 2018;3:56-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dogan A. Amyloidosis: insights from proteomics. Annu Rev Pathol. 2017;12:277-304. [DOI] [PubMed] [Google Scholar]
- 32. Zubarev RA, Makarov A. Orbitrap mass spectrometry. Anal Chem. 2013;85:5288-5296. [DOI] [PubMed] [Google Scholar]
- 33. Michalski A, Damoc E, Hauschild JP, et al. . Mass spectrometry–based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10:M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gertz MA. Immunoglobulin light chain amyloidosis diagnosis and treatment algorithm 2018. Blood Cancer J. 2018;8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kennedy JJ, Whiteaker JR, Kennedy LC, et al. . Quantification of human epidermal growth factor receptor 2 by immunopeptide enrichment and targeted mass spectrometry in formalin-fixed paraffin-embedded and frozen breast cancer tissues. Clin Chem. 2021;67:1008-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fenyö D. Identifying the proteome: software tools. Curr Opin Biotechnol. 2000;11:391-395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.