Abstract
What makes a mouse a mouse, and not a hamster? Differences in gene regulation between the two organisms play a critical role. Comparative analysis of gene coexpression networks provides a general framework for investigating the evolution of gene regulation across species. Here, we compare coexpression networks from 37 species and quantify the conservation of gene activity 1) as a function of evolutionary time, 2) across orthology prediction algorithms, and 3) with reference to cell- and tissue-specificity. We find that ancient genes are expressed in multiple cell types and have well conserved coexpression patterns, however they are expressed at different levels across cell types. Thus, differential regulation of ancient gene programs contributes to transcriptional cell identity. We propose that this differential regulation may play a role in cell diversification in both the animal and plant kingdoms.
INTRODUCTION
Defining the genetic mechanisms that drive species divergence is a longstanding and unachieved goal in evolutionary biology. With access to DNA sequences, comparative genomics research has uncovered associations between gene family conservation and the phenotypes that emerge in particular lineages (1–4). While this approach continues to shed light on genome evolution (5), it provides at best an incomplete picture, omitting phenotypic differences that can be driven by changes in gene regulation alone (6,7). A number of studies have used functional genomics data to find regulatory differences between species (8–12). A common approach is to compare samples that are clearly homologous, such as mammalian organs, and measure differences in gene expression as the output of changing regulatory architecture. Yet because of its dependence on shared anatomical features, this approach is necessarily limited to more closely related species: the leaves of a plant are not homologous to the limbs of an animal. How then, can we compare the conservation of gene function across the tree of life? One answer is coexpression – the patterns of gene activity that underlie organismal similarities and differences.
In a coexpression network, genes are nodes and the edges represent expression similarity between genes. Functionally related genes are often adjacent in the network as their expression is coordinated across biological conditions, allowing for the inference of gene regulatory modules through clustering (13). Where two organisms have homologous tissues, the pattern of gene variation across those tissues typically show conserved coexpression as groups of genes related to core molecular functions are expressed at varying levels across tissues, and tissue-specific genes are shared (9,10,14). But even where the tissues are not homologous, patterns of molecular variability will be conserved as gene modules jointly vary in expression across conditions (15). Evolution works, in part, by rewiring these subnetworks of genes, creating novel regulatory relationships and, by inference, changes in function (see Figure 1 for a schematic) (6). By comparing networks across a small number of model species, previous work has demonstrated that deeply conserved genes often have shared regulation (15–20). The increasing availability of data from a range of non-model species now permits investigation of shared and divergent regulation at greater resolution and breadth (21). Moreover, advances in single-cell RNA-sequencing create the opportunity to link observed patterns of conservation to mechanisms of cell type diversity.
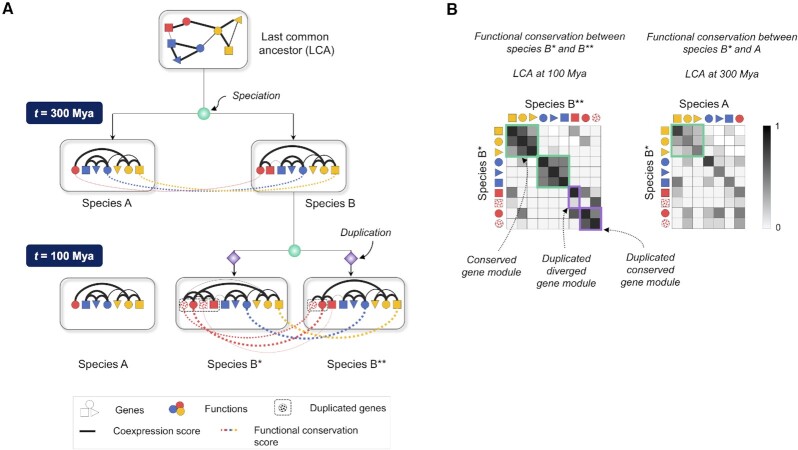
Figure 1.
Schematic to illustrate the variation of functional conservation with phylogenetic distance and lineage-specific gene gains and losses. (A) After the first speciation event, species A loses a gene (red square), while species B* and B** undergo lineage-specific gene duplications (checkered red circles and squares) after the second speciation event. Black lines indicate strength of coexpression between gene pairs within a species, and colored lines indicate the extent of functional conservation between select orthologous genes across species. (B) Heatmaps of functional conservation between every pair of genes in species B* and B** and in species B* and A indicate that functional conservation negatively correlates with phylogenetic distance. Gene modules retaining a single-copy of genes and displaying high coexpression similarity across the species pair are highlighted by green boxes. Duplicated genes are highlighted in purple boxes, and are labeled conserved or diverged based on their coexpression similarity post-duplication.
In this work, we build high-powered coexpression networks for 37 eukaryotic species and develop a measure of shared gene activity called ‘coexpression conservation’. We demonstrate that coexpression conservation tracks with evolutionary distances, enabling the reconstruction of the phylogenetic tree, and that it is significantly higher for orthologous gene pairs predicted by multiple algorithms and with higher sequence conservation. By relating coexpression conservation to single-cell expression data (22–26), we discover a fascinating association between so-called ‘ubiquitous’ genes and cell identity. In addition to their strongly conserved coexpression patterns, we find ‘ubiquitous’ genes have gradient-like expression distributions across cell types in both the mouse and model plant Arabidopsis thaliana. These results suggest a mechanism of functional diversification driven by up and down-regulation of deeply conserved modules along a continuous gradient of activity.
Coexpression conservation provides a data-driven estimate of gene functional divergence across species. To facilitate its use, we have made all of our results available for browsing and download via a webserver, CoCoBLAST (https://gillisweb.cshl.edu/CoCoBLAST/, see Supplementary Note 2 for a quick-start guide). In addition to serving conservation estimates for known orthologs, the webserver allows users to find the most closely related genes between two species based on coexpression rather than sequence.
METHODS
Analysis of public gene expression data
All analyses were performed in R version 3.6. Results are reported as means +/− standard deviations unless otherwise specified.
Aggregate coexpression networks were downloaded from CoCoCoNet (21). Networks for individual datasets are stored internally and are available on request. In brief, networks for each dataset are built by calculating the Spearman correlation between all pairs of genes based on read counts, then ranking the correlation coefficients for all gene-gene pairs, with NAs assigned the median rank. Aggregate networks are generated by averaging rank standardized networks from individual datasets. To assess the connectivity of GO groups we used the run_neighbor_voting function from the EGAD R package (74), subsetting GO to terms with 10–1000 genes. For human tissue specificity analyses, processed expression data from the GTEx project (v8, median TPM per tissue) was downloaded from the GTEx Portal (57). Tissue specificity was calculated as published (75).
The Tabula Muris Smart-seq2 expression matrix and sample metadata were downloaded from FigShare and converted to SingleCellExperiment objects for further processing. For each tissue, counts were normalized for library size and log2 transformed using the logNormCounts function from the scater package, yielding log counts per million (CPM) for each cell, with no tissue-specific scaling factors (76). Cell type specificity was calculated as published for each tissue (75), then cell type specificity scores were averaged across tissues, excluding NAs. To visualize cells, we used the t-SNE coordinates provided by the authors. Within-cell type variance was calculated using the base R function var on log CPM for each cell type in each tissue separately, then averaged for each tissue, and averaged across tissues. Across-cell type variance was calculated for each tissue separately using the mean log CPM for each cell type, then values were averaged across tissues. For Figure 6B, within and across-cell type variance were ranked and standardized between 0–1 such that the highest variance genes would have a score of 1. The limma package (77) was used to find marker genes with a log fold-change threshold > 4 between each cell type and any other within its tissue.
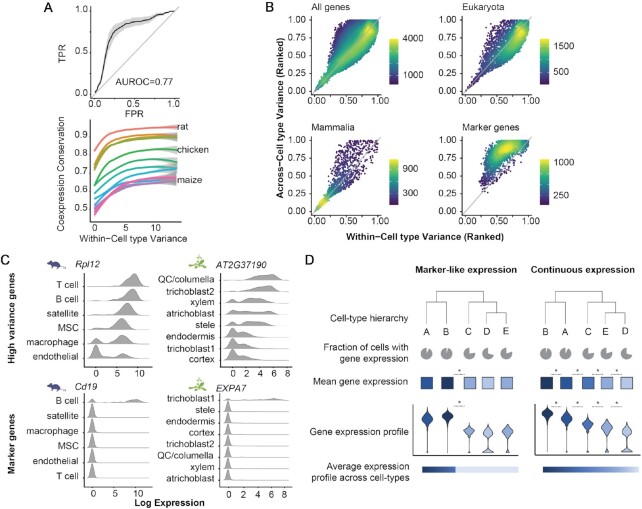
Figure 6.
Genes with conserved coexpression patterns are expressed in continuous gradients across cell types, suggesting an ancient mechanism of cell type divergence. (A) Expression variance is associated with conservation of gene neighborhoods. (Top) Expression variance across > 200 yeast datasets predicts coexpression conservation. TPR = true positive rate, FPR = false positive rate. ROC curves for all species were binned along x-axis, mean +/− SD is plotted. (Bottom) Coexpression conservation is plotted with respect to within-cell type variance in mouse, with loess fits on mean values for each species. (B) Across vs. within-cell type variance in mouse is plotted. Colors indicate local point density. The space can be broken into three regions: genes with high-within and high-across variance are typically more ancient, those that are low/low are more recent, and markers have high-across and low-within cell type variance. Due to their high conservation of coexpression patterns, high/high genes may have conserved functions vis-à-vis cell identity. (C) Examples of continuous (top) vs. marker-like (bottom) expression in mouse (left) and Arabidopsis (right). MSC = mesenchymal stem cell, QC = quiescent center. The conserved gene with high within- and across-cell type variance is more continuous across cell types, whereas the markers (high/low) are either on or off. (D) Schematic illustrating the difference between marker-like and continuous expression across cell types. See Supplementary Figure S7 for additional discussion.
Arabidopsis thaliana root single-cell RNA-seq expression matrices were downloaded from the Gene Expression Omnibus (78) (GEO IDs: GSE116614, GSE121619, GSE123818, GSE123013). Cluster assignments were downloaded from GEO for IDs GSE121619 and GSE123013, and provided by the authors for IDs GSE123981 and GSE116614. Counts were depth normalized and log2 transformed using the logNormCounts function in the scater package to yield log CPM. Cell type specificity was calculated on log CPM for each dataset separately, then averaged, excluding NAs. Within-cell type variance was calculated on log CPM for each cell type in each dataset separately, then averaged across datasets. Across-cell type variance was calculated for each dataset using the mean log CPM in each cell type, then averaged across datasets. To visualize cells from all studies, we first used MetaNeighbor (65) to find replicable clusters, and subset individual datasets to clusters replicating in at least one other study using an AUROC cut-off of 0.7. We used the multiBatchNorm function from the batchelor package for initial batch correction (79). Then, we selected variable genes using the get_variable_genes function from MetaNeighbor and used fMNN in batchelor on batch corrected data, subset to variable genes. This provided principle components which were used for the UMAP projection of all cells (80) (20 components were used).
Yeast microarray data were downloaded from the Saccharomyces Genome Database (64). We included studies with > 10 samples. After rank normalizing expression for each sample, we calculated the variance of expression for each gene that was measured in at least 80% of the datasets. Variance was calculated for each dataset separately, then averaged. For the analysis in Figure 6A, we considered all gene pairs with coexpression conservation > 0.9 to be true positives and used the ranked average variance to predict these for each species with the auroc_analytic function in EGAD (74).
Gene annotations and orthology
Gene function annotations were sourced from the Gene Ontology (27). GO terms and gene associations were obtained by merging data from the NCBI (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2go.gz) and Ensembl 99, accessed through biomaRt (81). Associations were propagated based on ‘is_a’ relationships between terms. In addition, essential gene annotations were obtained from the MacArthur lab's GitHub repository (https://github.com/macarthur-lab/gene_lists), originally sourced from Hart et al (63). Yeast-human complementation data was downloaded from the supplement of Kachroo et al (53). Mouse gene age estimates (58) were downloaded from the Marcotte lab's GitHub repository (https://github.com/marcottelab/Gene-Ages) and A. thaliana gene age estimates were collected from multiple sources (59–62).
OrthoDB (28) was used for orthology mapping. In brief, we search for the most recent phylogenetic split for each pair of species then obtain inferred orthology groups for all genes descending from a common ancestor. Orthologs were filtered to either include only one-to-one relationships, or to include all N-to-M orthologous pairs. We also downloaded orthology information from the Alliance of Genome Resources (38) for assessment with our N-to-M coexpression conservation method, detailed below. We de-duplicated the information so that each gene pair appeared only once, rather than having a direction from a source-target species. Pairs from all algorithms were considered the ‘universe’ of possible orthologs. Species divergence times were sourced from TimeTree (82).
Coexpression conservation
For each pair of species to be compared, we filter aggregate coexpression networks to include known orthologous genes (see Supplementary Figure S2 for the number of genes between each pair of species), then we compare each gene's top 10 coexpression partners across species to quantify gene similarity. We treat this as a supervised learning task, using the ranks of the coexpression values from one species to predict the top 10 coexpression partners from the second species, and then repeating this task in the opposite direction, finally averaging the scores. We refer to this as a measure of ‘coexpression conservation’ and note that it is formally equivalent to the average area under the receiver operator characteristic curve (AUROC). Note that the choice of top 10 genes is arbitrary, however we have found that our results are robust to the precise number of top genes as we can correctly reconstruct phylogenetic relationships across a wide range of top gene pairings (see Supplementary Note 1).
To generalize this to the case of N-to-M orthologs, we describe the analysis in greater detail: Consider a gene A1 in species 1 that has two orthologs in species 2 – genes B1 and B2. First, the top ten genes exhibiting the highest coexpression with A1 are chosen. All possible orthologs in species 2 for the set of top 10s of A1 are shortlisted as the ‘translated top 10s’. Note that since each gene in species 1 can now have one or more orthologs in species 2, the translated top 10s can vary in length. Additionally, some of the top ten coexpressed genes in species 1 may map to the same orthologs in species 2, but we only consider a unique list of orthologs in the translated top 10s for each gene in species 1. This task is repeated in the opposite direction, where the ranks of coexpression values of genes in species 2 are used to predict the top coexpression partners of genes from species 1. Scores from both directions are averaged, thereby providing a measure of overall coexpression conservation.
RESULTS
Establishing meta-analytic coexpression networks as a tool for comparative genomics
Changes in phenotype between species can be encoded in changing patterns of gene network connectivity (Figure 1). Reliable estimates of species-specific gene coexpression patterns provide a backbone for comparative analysis of regulatory divergence. In previous work, we generated high-powered coexpression networks from 14 species by aggregating RNA-sequencing data across hundreds of individual experiments (21). We have recently expanded this resource to include 37 species, greatly increasing our coverage of mammals, invertebrates and plants (Supplementary Figure S1). In the following, we establish the power and robustness of the subset of networks that have functional annotations in the Gene Ontology (GO) (27) (Figure 2).
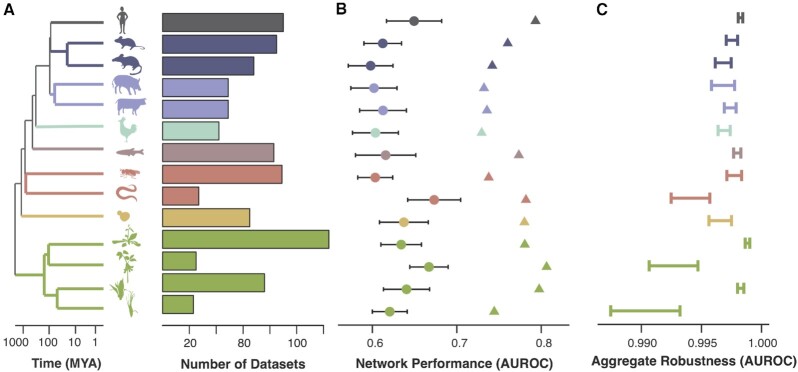
Figure 2.
Aggregate coexpression networks are a powerful tool for comparative genomics. (A) In this work we examine coexpression across species from 3 kingdoms: plants, animals and fungi. Here we focus on the subset of species with GO annotations. The dendrogram shows phylogenetic relationships between these species, and the barplots indicate the number of datasets used to build aggregate coexpression networks. (B) Circles show the mean GO prediction performance for individual networks (+/− standard deviation) while triangles indicate aggregate network performance. (C) Aggregate robustness is high across all species, with variation dependent on n.
As a first validation, we find that species-specific networks built from multiple datasets (‘aggregate’ networks) have strong connections between genes from the same GO group, and that these connections are significantly stronger than those found in networks from individual datasets (Figure 2B, neighbor voting mean Area Under the Receiver-Operating characteristic Curve (AUROC) individual networks = 0.63, mean AUROC aggregates = 0.76, Wilcoxon P < 10–8, n = 14 species). To evaluate the robustness of the aggregate networks, we bootstrapped the aggregation procedure 10 times, then used the ranked edges in the bootstrapped networks to predict the top 1% of edges in the reference aggregate networks (Figure 2C). Performance at this task was close to perfect (mean AUROC = 0.996 +/− 0.002), while variability between bootstrapped and reference aggregate networks declined as a function of the number of experiments and samples as expected (Spearman correlation coefficient = −0.83 for experiments, −0.86 for samples). We also confirmed that this held true in our larger corpus of 37 total networks (Supplementary Figure S1B). In sum, these results indicate that aggregate networks are statistically robust and biologically meaningful, with strong connections between functionally related genes.
Comparing ortholog coexpression neighborhoods quantifies the conservation of gene activity
Having established the robustness of our networks, we next explore the degree to which they can be used for cross-species comparisons. Our analysis focuses on characterizing the degree to which pairs of orthologs have retained similar coexpression patterns, or ‘neighborhoods’ in the networks. We use OrthoDB (28) to map orthologs since it is the world-leader by comprehensiveness across organisms. For each pair of species, we search for the most recent phylogenetic split, then obtain inferred orthology groups for all genes descended from the common ancestor.
To measure the similarity of ortholog neighborhoods between two species, we first subset networks to include only one-to-one orthologs between that species pair (e.g. pig and yeast as shown in the schematic, Figure 3A; note that the number of 1:1 orthologs between each pair of species can be found in Supplementary Figure S2A). Next, each gene's neighborhood is defined by ranking all edges associated with it, and then the ranks of the gene's top 10 coexpressed gene pairs are compared across species (see Methods for details). This is expressed as an AUROC and so it ranges from 0–1, with 1 meaning perfect conservation, 0.5 consistent with random re-ordering of neighborhoods, and 0 meaning that coexpression partners have inverted from being the top ranked to bottom ranked across species. We refer to this score as ‘coexpression conservation’ or ‘gene neighborhood conservation’.
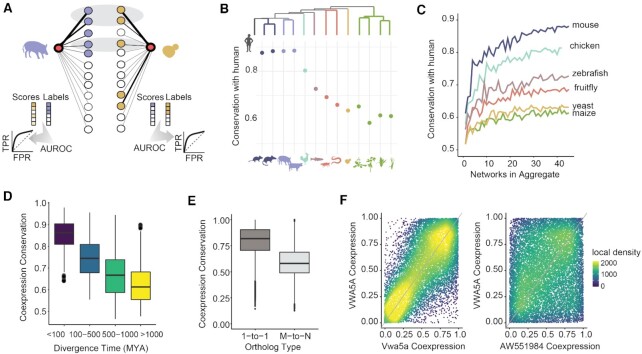
Figure 3.
Divergence in gene coexpression correlates with phylogeny. (A) Method schematic. Circles represent genes and line thickness indicates strength of coexpression between one target gene (red) and all others. For each target gene in pig, we identify the set of pig genes that are maximally coexpressed with it, shown in blue. We evaluate how conserved this coexpression pattern is in yeast, then repeat the task in the other direction. Genes with high coexpression to the target in both species are highlighted in the gray ovals. Coexpression conservation is reported as the average AUROC in both directions (i.e. pig-yeast and yeast-pig). (B) Points show mean coexpression conservation for 1-to-1 orthologs between human and each other species. Coexpression conservation is negatively correlated with phylogenetic distance (rho = −0.95, P < 10–6). (C) Mean coexpression conservation for 1-to-1 orthologs between human and each species are plotted against the number of networks included in the aggregate network. Performance increases with additional data. (D) Boxplots show coexpression conservation for 492 orthologous groups defined at the last common ancestor of all eukaryotes, plotted with respect to species divergence times. As in panel B, coexpression is more conserved among more recently diverged species. (E) Boxplots show coexpression conservation scores. 1-to-1 orthologs are more conserved than N-to-M orthologs (Wilcoxon P < 10–16). (F) Coexpression profiles for a 1-to-2 human-mouse ortholog group. The human gene VWA5A has a strongly conserved coexpression profile with mouse Vwa5a (left, conservation AUROC = 0.83) but not with mouse AW551984 (right, AUROC = 0.46).
As a first biological validation of our approach, we find that the conservation of gene neighborhoods is strongly negatively associated with phylogenetic distances between species, demonstrated in Figure 3B with respect to distance from human (Spearman correlation coefficient = −0.95). We also find that coexpression conservation is sensitive to the amount of underlying data as expected, with strong performance achieved with the inclusion of twenty datasets and scores plateauing beyond this point (Figure 3C, mean individual networks = 0.55+/−0.04, mean 20-dataset aggregate = 0.68+/−0.08, Wilcoxon P < 0.002, n = 7 species). To evaluate the sensitivity of coexpression neighborhoods to differences in data preprocessing, we used our yeast compendium as a test case. We split the data into two partitions and generated aggregate networks for each across a range of commonly used methods (see Supplementary Note 1 for details), then we assessed the degree to which the same gene's coexpression neighborhood was preserved across the two networks. While gene neighborhood preservation is generally high for all methods, we find that our previously established network building best practices (29) are among the best for this purpose. All together, these results provide strong validation of both the network building and coexpression conservation methods.
One-to-one orthologs are frequently used for comparative genomics analyses (e.g.30,31), however, as species grow more distant to one another, there are fewer one-to-one orthologs for comparison, particularly in the plant kingdom, where genome duplication events are common (32). To explore more distant and complex relationships, we generalized our method to be able to compare groups of orthologs, i.e. all genes descended from a single gene in a common ancestor, including lineage-specific duplicates (see Supplementary Figure S2B for the number of many-to-many orthologs between all species pairs and Supplementary Figure S3 for a schematic). As validation, we assessed the conservation of groups of orthologs descended from the last common ancestor of all eukaryotes. We find that coexpression conservation scores for many-to-many (aka N-to-M) orthologs are strongly associated with phylogenetic distances between species (Figure 3D). Expanding out to the entire set of species in our compendium, we find that the average N-M score lets us reconstruct the phylogenetic tree (Supplementary Figure S4), though distances among plant species are not preserved. We note that these results are robust to the precise size of the gene neighborhoods used to calculate coexpression conservation, with neighborhood sizes ranging from 5–500 yielding nearly identical phylogenetic trees (Supplementary Note 1).
We next evaluated all N-to-M orthologs defined in the last common ancestor between each pair of species, finding that one-to-one orthologs have higher coexpression conservation than N-to-M orthologs on average (Figure 3E, mean 1-to-1 = 0.79 +/− 0.14, mean N-to-M = 0.59 +/− 0.14, Wilcoxon P < 10–16). Notably, we also find cases where N-to-M orthologs differ strongly by coexpression conservation, with ∼7% of all N-to-M groups containing orthologs with scores > 0.7 and <0.5. An example of this is shown in Figure 3F. Here, we see that human VWA5A shares a large fraction of its coexpression neighborhood with mouse Vwa5a, but that it is quite distinct from mouse AW551984 which exists only in the muroid lineage.
These results indicate that coexpression neighborhoods can be usefully compared across species to provide a measure of ortholog conservation, and even of species divergence. Distinguishing orthologs by differential coexpression conservation may provide a route forward for discovering genes with ‘the same’ function across species (a.k.a. ‘functional analogs’), an idea we discuss in the following section.
Conservation of gene activity is associated with ortholog confidence and can predict functional analogs
Our conservation analysis is made possible by the use of OrthoDB to define orthologous genes. However, orthology prediction is an active area of research in genomics (33,34), and relying on a single algorithm has known limitations (35–37). How would our results change if we used a different reference? In the following, we take advantage of orthology information from the Alliance of Genome Resources (38) which has predictions from 12 independent sources (39–50) for humans and 6 common model organisms. We assess algorithms with respect to gene neighborhood conservation, and we explore how coexpression conservation can be applied to predict functional analogs between human and yeast.
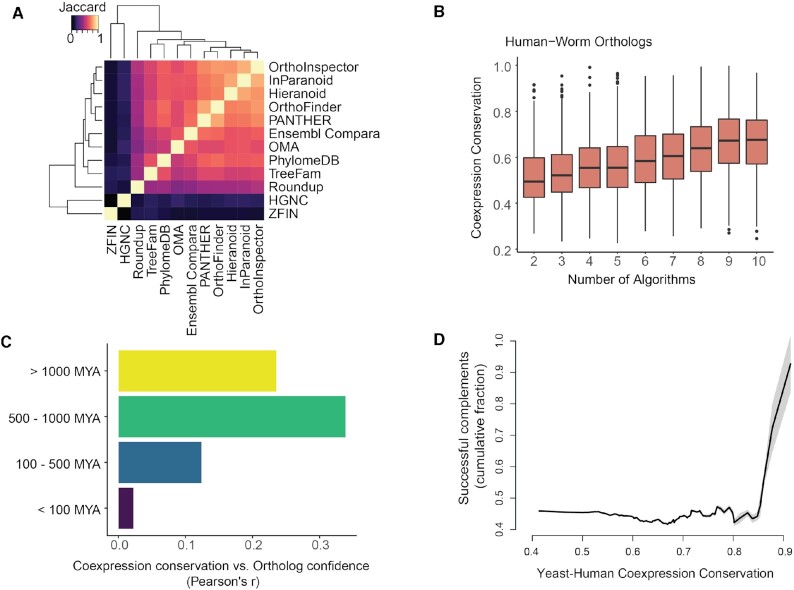
The majority of algorithms within the Alliance of Genome Resources database (9/12) have high concordance across their orthology predictions (Figure 4A, mean Jaccard = 0.7), with exceptions attributable to differences in species coverage (e.g. HGNC only includes human orthology information). In keeping with their overall similarity, we find that average coexpression conservation scores for these 9 algorithms are close to tied (mean = 0.74 +/− 0.009, Supplementary Table S1) and although OrthoDB is fairly distinct in its predictions (mean Jaccard index = 0.18), it performs very close to this average (0.73 +/− 0.13). Remarkably, even though the algorithms are tied on average, we find that where they agree, conservation of gene coexpression is preferentially high: for almost all pairs of species, ortholog neighborhood conservation is correlated with the number of algorithms that predict the relationship (human-worm shown as an example in Figure 4B, all correlations in Figure 4C). The only exceptions to this rule are among the three mammals, where conservation of gene coexpression is almost uniformly high. Because orthology prediction is easier for long genes and those under stronger selection (51), we tested the association between ortholog commonality across species and these features. Using human and worm as an example, we compared percent sequence similarity and gene length to ortholog commonality across algorithms. As expected, we find that sequence similarity is positively associated with commonality across algorithms (Supplementary Figure S5), but we do not see a relationship between length and ortholog commonality. Together, this indicates that constrained genes of varying lengths are among those that are predicted across multiple algorithms. It is interesting to note that this also correlates with conservation of coexpression relationships.
Figure 4.
Gene coexpression conservation is associated with ortholog concordance across algorithms and can predict human-yeast functional analogs. (A) Heatmap of algorithm concordance. The majority of algorithms (9/12) make similar predictions, with outliers arising from selection biases (i.e. inclusion of only a subset of species). (B) Mean conservation for human-worm orthologs is plotted against the number of algorithms predicting the relationship. Conservation of gene neighborhoods correlates with ortholog confidence. (C) Bars show the correlation between the number of algorithms and conservation of neighborhoods for each gene pair, binned into three divergence times. Conservation correlates with ortholog confidence for pairs of species that diverged > 100MYA but not for more recently diverged species. (D) Cumulative success of human-yeast complementation is plotted as a function of gene activity conservation. Human genes with conserved gene neighborhoods are likely to compensate for loss of their yeast orthologs.
A primary application of orthology prediction is to infer shared function across species. Benchmarks recurrently find a precision-recall trade-off across algorithms (37,52), with little evidence that any one approach outperforms another (34). By incorporating functional information directly, coexpression conservation scores may improve sequence-based inference. For example, recent work has found that human genes with similar coexpression patterns can compensate for the loss of their yeast orthologs in complementation screens (53,54). Here we find that coexpression conservation scores from our independent data (RNA-seq rather than microarray) and our analysis method are predictive of this effect (Figure 4D). However, we also find that certain pairs of complementing orthologs have very low coexpression conservation, with the two lowest scoring pairs excluded from the Alliance of Genome Resources database (Supplementary Table S2).
Altogether, these results highlight that confidence in sequence-based orthology is reflected by similarity in gene coexpression relationships. Although ortholog confidence may also be associated with underlying gene properties, such as mutational constraint, these results suggest that a wisdom-of-the-crowds approach may be beneficial for ortholog prediction. Since our results rely only on OrthoDB, they likely represent a lower limit for gene activity conservation which would only improve with the use of meta-orthology methods (55).
Genes expressed in all cell types have conserved coexpression relationships
Above, we established that the conservation of gene coexpression tracks with phylogeny as expected, and that one-to-one orthologs are more likely to have similar coexpression neighborhoods than those that have duplicated. We also find evidence of strong divergence within orthogroups, which has long been postulated to be an evolutionary mechanism for cell diversification (56).
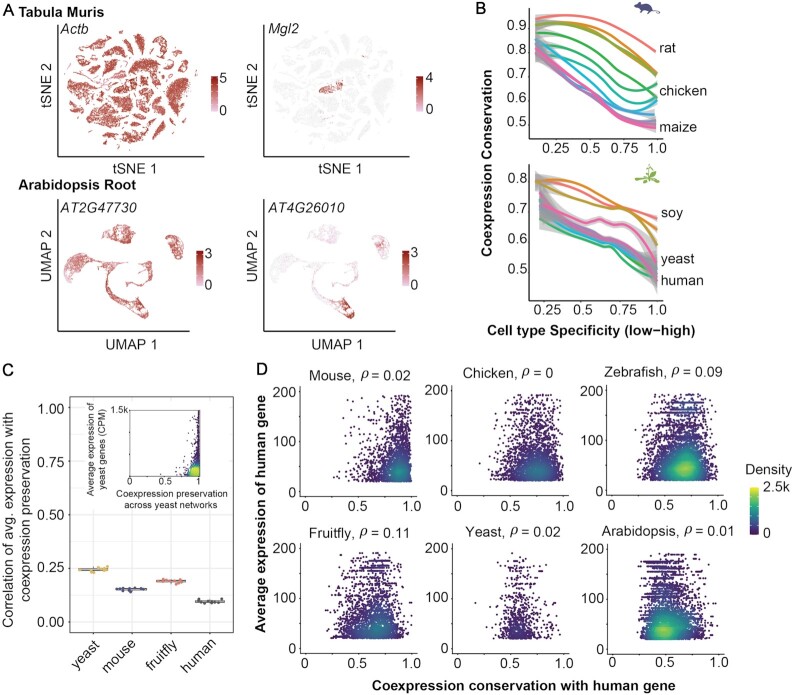
By leveraging coexpression relationships we can 1) extend these observations to any species pair of interest without requiring knowledge of homologous tissues or cell types, and 2) identify conserved relationships between genes, reflecting conserved regulation and function. Importantly, we can investigate the role of conserved gene activity in cell phenotypes by taking advantage of single-cell RNA-seq data. We use comprehensive single-cell RNA-seq data from the Tabula Muris project for mouse (22), and four single-cell RNA-seq datasets from A. thaliana root (23–26) as references for cell-type specific expression (Figure 5A, see Methods for details), and we use data from the Genotype-Tissue Expression Project (GTEx) (57) as a reference for human tissue-specific expression.
Figure 5.
Ubiquitously expressed genes have strongly conserved coexpression patterns that are not explained by expression level alone. (A) Plots of mouse and Arabidopsis scRNA-seq data, with examples of constitutive (left) vs. cell-type specific expression (right), color indicates expression level. (B) Coexpression conservation is plotted with respect to cell type specificity for mouse (top) and Arabidopsis (bottom). Lines are loess fits on mean values for each species, +/− SD. Cell type specificity is negatively associated with conservation of gene neighborhoods. (C) Pearson correlation of within-species average expression with neighborhood preservation. (Inset) Representative scatterplot showing the relationship between expression level and coexpression preservation within yeast. Because most genes show strong coexpression preservation (x-axis) there is a weak relationship between expression level and coexpression preservation. (D) Human gene expression level is plotted with respect to coexpression conservation for six representative species. No relationship is observed.
Consistent with previous research, we find that genes expressed ubiquitously across tissues have higher coexpression conservation than those with tissue-specific expression (Supplementary Figure S6, Spearman correlation coefficient = −0.37 +/− 0.08). Expanding this analysis to cell types, we again see the same pattern, with cell-type specificity associated with decreased coexpression conservation in both A. thaliana and mouse (Figure 5B, mouse Spearman correlation coefficient = −0.50 +/− 0.04, Arabidopsis = −0.26 +/− 0.07). We note that this trend holds across all species pairs used to calculate conservation of coexpression neighborhoods. We also confirm that cell type-specific expression and coexpression conservation are both associated with estimates of gene age in both kingdoms (Supplementary Figure S6, mouse cell-type specificity vs. gene age (58) Spearman correlation coefficient = 0.47, coexpression conservation vs. gene age = −0.15 +/− 0.1 across 13 species pairs, Arabidopsis cell-type specificity vs. gene age (59–62) Spearman correlation = 0.27 +/− 0.1, coexpression conservation vs. gene age = −0.23 +/− 0.05, averaged across 6 gene age estimates and 13 species pairs). Conservation of gene neighborhoods is also significantly higher for orthologs of genes known to be essential in human (63) (Supplementary Figure S6, essential = 0.83 +/− 0.1, non-essential = 0.69 +/− 0.08, Wilcoxon P < 0.01, n = 13 species compared to human).
Ubiquitous genes are likely to have higher expression levels than those that are tissue- or cell type-specific. To determine whether expression level is sufficient to explain the differences in coexpression conservation between ubiquitous and cell type specific genes, we performed two control experiments. Because orthologous genes can have different expression levels across species, in the first experiment we compared gene neighborhoods within species after splitting our compendium into 10 partitions and generating aggregate networks for each. We find that gene neighborhood preservation is weakly but positively associated with gene expression level (mean Pearson correlation = 0.2, Figure 5C and Supplementary Note 1). Genes which are highly expressed have preserved coexpression neighborhoods, but because most genes show relatively high coexpression preservation, the relationship is not that strong. As a second test, we used human as an index case, and compared the relationship between human gene expression levels and coexpression conservation with mouse, chicken, zebrafish, fruitfly, yeast and arabidopsis. We find no relationship between human expression levels and coexpression conservation (mean Spearman correlation = 0.04, Figure 5D and Supplementary Note 1).
In summary, gene-gene relationships are less well conserved among younger and more cell type-specific genes, and this cannot be explained by their expression level alone.
Conserved coexpression links ubiquitous genes to cellular diversity
Our compendium of networks includes one single-celled organism, the budding yeast Saccharomyces cerevisiae. To explore how these results might generalize to a species without cell types, we performed a meta-analysis of yeast microarray expression data (64), using bulk expression variance as an analog for expression in all cell states since they are strongly correlated (Supplementary Figure S4). Consistent with our findings in plants and animals, we find that expression variability in yeast is predictive of coexpression conservation (Figure 6A, mean AUROC = 0.77 +/− 0.04 SD).
Because variability occurs in the absence of cell type variation in yeast (it may reflect temporal or state variation), we wondered whether we could find a similar effect in multicellular organisms when holding cell type constant. Our expectation is that cells of the same type may vary by cell cycle phase, nascency, or activation state, for example. Strikingly, after calculating expression variation within each cell type in Tabula Muris, we find that the most variable genes have the most strongly conserved coexpression patterns (Figure 6A). In other words, while their coexpression partners remain consistent, their expression levels vary dramatically within and across cells. These properties suggest that such genes contribute to aspects of cell identity that require tightly coordinated signaling, like differences in size or metabolic activity. As deeply conserved features, they would differ between cell types by degree, rather than being present or absent, meaning that cells of different types would have overlapping expression distributions but with different mean values or ‘set points’ (e.g. see Schematic, Figure 6D and Supplementary Figure S7). These features would be expected to generalize across kingdoms more than poorly conserved marker genes (Figure 6B).
One example of a gene that might contribute to cell identity by dialing its expression up and down is Rpl12, a component of the large ribosomal subunit 60S in mouse. This gene is among the most variably expressed genes within and across cell types (mean standardized rank within = 0.83, across = 0.97) but its expression distribution across cell types is continuous, with different cell types having distinct mean levels of the gene and overlapping expression distributions (Figure 6C). Given that Rpl12 is a component of the ribosome, we speculate that differences in protein synthesis rate could contribute to differences in cell phenotypes. Notably, we find a similar pattern of continuous expression when we look at an Arabidopsis ortholog of Rpl12 (Figure 6C). This pattern is in stark contrast to more typical marker genes, which tend to have high variance across cell types, and lower variance within cell types because they are switch-like in their expression patterns. Two examples are shown in Figure 6C: Cd19, which is exclusively expressed by mouse B-cells and is conserved only among mammals (mean standardized rank within = 0.55, across = 0.97); and EXPA7, which is exclusive to Arabidopsis root trichoblasts and is conserved only among eudicots (mean standardized rank within = 0.77, across = 0.99).
In summary, our combined analysis of single-cell RNA-seq and yeast expression variation shows that while cell type- or tissue-specific marker genes are generally not well conserved, genes with high expression variability are deeply conserved at both a sequence level and in terms of their coexpression patterns. These genes may contribute to continuous aspects of cell transcriptional identity in both single-celled and multicellular organisms.
DISCUSSION
By combining sequence-based orthology predictions with robust estimates of gene coexpression neighborhoods, we have developed a measure of gene conservation that can be calculated between any pair of species. Our study is the largest of its kind to date, making use of data from hundreds of individual studies, and measuring the conservation of gene-gene relationships across very long diverged species. We find that coexpression conservation is associated with phylogenetic distances, expectations of conservation based on gene family size, and sequence similarity. Moreover, taking advantage of the recent explosion in single-cell data, we identify commonalities between the forces that drive conservation in both single-celled and multi-cellular organisms. The genes that vary most – both within cells of the same type and across cells of different types or states – show deeply conserved patterns of coexpression, suggesting their fundamental role in eukaryotic cell function and identity.
Previous research to investigate the changing gene expression landscape across species highlighted the relative lack of conservation among tissue and cell type-specific genes (9,12). Here, we confirm these previous findings, but also extend them. Genes with cell type specific expression have more poorly conserved coexpression patterns than those that are expressed in cells of all types, as previously described. Yet genes expressed in all cell types not only have strongly conserved coexpression patterns, but they also show differences in expression level across cell types. Variation in activity across cell types is likely what drives their strong coexpression within networks.
What does this mean for cellular evolution and multicellularity? We hypothesize that these genes work in tightly coordinated modules to tune non-negotiable aspects of cellular identity, like cell size, or metabolic rate, generating diversity that allows cells to respond to varying environments. With these diverse populations established, evolution could then use novel genomic variation to mark cell types and refine their organismal roles. Further work to explore this hypothesis is necessary, including additional analyses of single-cell data from long diverged species. However, we note that this will require targeted investigation, as typical single-cell analyses are designed to identify genes that are strongly variable across cell types (i.e. markers) rather than subtler continuous signals. Cell identity is known to be broadly distributed across the transcriptome, and this allows low-depth single-cell RNA-seq to find expected cell clusters even when individual marker genes are not sampled (65–67). Determining the relative contributions of switch-like versus continuous expression to cell identity will be informative for updating empirical and mechanistic models of cell type.
This work is at the intersection of transcriptomics and orthology prediction, and improvements in both will allow for increasing precision of coexpression conservation. To generate robust co-expression networks, we require a minimum of 20 independent datasets with at least 10 samples each. Based on our current estimates of available data, we expect that a modest number of species can be added on an ongoing basis as these thresholds are met. The ability to move substantially beyond that will require efforts to profile the transcriptomes of a greater diversity of organisms, similar to the goals of the Genome10K project for genome sequences (68). However, a necessary consequence of using bulk RNA-seq data from heterogenous samples is that networks are better powered for genes that are expressed in all cells, and our results make it clear that tissue-specific networks (69) cannot overcome this as genes expressed in all cell types will continue to dominate. Instead, future work to develop cell-type specific coexpression networks (70), including methods to map cell identities across distantly related species (71), and/or to selectively sample the transcriptome for genes with lower expression (72), could be routes forward. Regarding orthology, we find that our conservation measure is correlated with the number of algorithms that predict the orthology relationship. Orthology algorithms are notoriously difficult to benchmark given the lack of gold-standard data (i.e. we lack genomes for the last common ancestors of extant species (37)). Our results suggest that a wisdom-of-the-crowds approach combined with robust functional genomics data could improve on baseline predictions (73). Functional genomics data is likely to be particularly valuable for orthologs with low sequence similarity.
Defining the regulatory mechanisms that allow for evolutionary divergence is a central goal in biology. Our method and data now provide a clear route forward for investigating these mechanisms with both breadth (across species) and depth (genome-wide). We invite users to explore and re-use our results through our webserver, CoCoBLAST (https://gillisweb.cshl.edu/CoCoBLAST/). Inspired by BLAST, it uses Conserved Coexpression to find genes with the most similar coexpression patterns across the tree of life.
MATERIALS & CORRESPONDENCE
Correspondence and material requests should be addressed to Jesse Gillis.
DATA AVAILABILITY
Data and code to reproduce the coexpression conservation analysis are available through CoCoBLAST (https://gillisweb.cshl.edu/CoCoBLAST/, see Supplementary Note 2 for a guided tutorial).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Trygve Bakken, Dave Jackson, Ken Birnbaum and Stephan Fischer for thoughtful feedback on earlier drafts of this manuscript.
Author contributions: J.G. conceived the project. M.C. and J.G. wrote the paper, designed analyses and interpreted results. M.C., H.S. and J.L. performed computational experiments, with H.S. contributing to algorithmic design and figure generation, and J.L. contributing webserver integration. All authors read and approved the final manuscript.
Notes
Present address: Megan Crow, Genentech, 1 DNA Way, South San Francisco CA, USA.
Contributor Information
Megan Crow, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor NY, USA.
Hamsini Suresh, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor NY, USA.
John Lee, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor NY, USA.
Jesse Gillis, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor NY, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for the work was provided by the National Institutes of Health (R01 LM012736 and R01MH113005 for H.S., J.L. and J.G.; K99 MH120050 for M.C.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Rubin G.M., Yandell M.D., Wortman J.R., Gabor Miklos G.L., Nelson C.R., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W.et al.. Comparative genomics of the eukaryotes. Science. 2000; 287:2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant arabidopsis thaliana. Nature. 2000; 408:796–815. [DOI] [PubMed] [Google Scholar]
- 3. Copley R.R., Schultz J., Ponting C.P., Bork P.. Protein families in multicellular organisms. Curr. Opin. Struct. Biol. 1999; 9:408–415. [DOI] [PubMed] [Google Scholar]
- 4. Hutter H., Vogel B.E., Plenefisch J.D., Norris C.R., Proenca R.B., Spieth J., Guo C., Mastwal S., Zhu X., Scheel J., Hedgecock E.M.. Conservation and novelty in the evolution of cell adhesion and extracellular matrix genes. Science. 2000; 287:989–994. [DOI] [PubMed] [Google Scholar]
- 5. Fernández R., Gabaldón T.. Gene gain and loss across the metazoan tree of life. Nat. Ecol. Evol. 2020; 4:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. King M.-C., Wilson A.C.. Evolution at two levels in humans and chimpanzees. Science. 1975; 188:107–116. [DOI] [PubMed] [Google Scholar]
- 7. Gilad Y., Oshlack A., Smyth G.K., Speed T.P., White K.P.. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006; 440:242–245. [DOI] [PubMed] [Google Scholar]
- 8. Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A.et al.. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002; 99:4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brawand D., Soumillon M., Necsulea A., Julien P., Csárdi G., Harrigan P., Weier M., Liechti A., Aximu-Petri A., Kircher M.et al.. The evolution of gene expression levels in mammalian organs. Nature. 2011; 478:343–348. [DOI] [PubMed] [Google Scholar]
- 10. Patel R.V., Nahal H.K., Breit R., Provart N.J.. BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species. Plant J. 2012; 71:1038–1050. [DOI] [PubMed] [Google Scholar]
- 11. Cardoso-Moreira M., Halbert J., Valloton D., Velten B., Chen C., Shao Y., Liechti A., Ascenção K., Rummel C., Ovchinnikova S.et al.. Gene expression across mammalian organ development. Nature. 2019; 571:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alam T., Agrawal S., Severin J., Young R.S., Andersson R., Arner E., Hasegawa A., Lizio M., Ramilowski J.A., Abugessaisa I.et al.. Comparative transcriptomics of primary cells in vertebrates. Genome Res. 2020; 30:951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eisen M.B., Spellman P.T., Brown P.O., Botstein D.. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998; 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yanai I., Peshkin L., Jorgensen P., Kirschner M.W.. Mapping gene expression in two xenopus species: evolutionary constraints and developmental flexibility. Dev. Cell. 2011; 20:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stuart J.M., Segal E., Koller D., Kim S.K.. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003; 302:249–255. [DOI] [PubMed] [Google Scholar]
- 16. Bergmann S., Ihmels J., Barkai N.. Similarities and differences in genome-wide expression data of six organisms. PLOS Biol. 2003; 2:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruprecht C., Proost S., Hernandez-Coronado M., Ortiz-Ramirez C., Lang D., Rensing S.A., Becker J.D., Vandepoele K., Mutwil M.. Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 2017; 90:447–465. [DOI] [PubMed] [Google Scholar]
- 18. Gerstein M.B., Rozowsky J., Yan K.-K., Wang D., Cheng C., Brown J.B., Davis C.A., Hillier L., Sisu C., Li J.J.et al.. Comparative analysis of the transcriptome across distant species. Nature. 2014; 512:445–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dutilh B.E., Huynen M.A., Snel B.. A global definition of expression context is conserved between orthologs, but does not correlate with sequence conservation. BMC Genomics. 2006; 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chikina M.D., Troyanskaya O.G.. Accurate quantification of functional analogy among close homologs. PLoS Comput. Biol. 2011; 7:e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J., Shah M., Ballouz S., Crow M., Gillis J.. CoCoCoNet: conserved and comparative co-expression across a diverse set of species. Nucleic Acids Res. 2020; 48:W566–W571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., Darmanis S., Batson J., Botvinnik O., Chen M.B.et al.. Single-cell transcriptomics of 20 mouse organs creates a tabula muris. Nature. 2018; 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jean-Baptiste K., McFaline-Figueroa J.L., Alexandre C.M., Dorrity M.W., Saunders L., Bubb K.L., Trapnell C., Fields S., Queitsch C., Cuperus J.T.. Dynamics of gene expression in single root cells of arabidopsis thaliana. Plant Cell. 2019; 31:993–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denyer T., Ma X., Klesen S., Scacchi E., Nieselt K., Timmermans M.C.P.. Spatiotemporal developmental trajectories in the arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell. 2019; 48:840–852. [DOI] [PubMed] [Google Scholar]
- 25. Ryu K.H., Huang L., Kang H.M., Schiefelbein J.. Single-Cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019; 179:1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shulse C.N., Cole B.J., Ciobanu D., Lin J., Yoshinaga Y., Gouran M., Turco G.M., Zhu Y., O’Malley R.C., Brady S.M.et al.. High-Throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019; 27:2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kriventseva E.V., Kuznetsov D., Tegenfeldt F., Manni M., Dias R., Simão F.A., Zdobnov E.M.. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019; 47:D807–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ballouz S., Verleyen W., Gillis J.. Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics. 2015; 31:2123–2130. [DOI] [PubMed] [Google Scholar]
- 30. Nehrt N.L., Clark W.T., Radivojac P., Hahn M.W.. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLOS Comput. Biol. 2011; 7:e1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Z.-Y., Leushkin E., Liechti A., Ovchinnikova S., Mößinger K., Brüning T., Rummel C., Grützner F., Cardoso-Moreira M., Janich P.et al.. Transcriptome and translatome co-evolution in mammals. Nature. 2020; 588:642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams K.L., Wendel J.F.. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005; 8:135–141. [DOI] [PubMed] [Google Scholar]
- 33. Altenhoff A.M., Garrayo-Ventas J., Cosentino S., Emms D., Glover N.M., Hernández-Plaza A., Nevers Y., Sundesha V., Szklarczyk D., Fernández J.M.et al.. The quest for orthologs benchmark service and consensus calls in 2020. Nucleic Acids Res. 2020; 48:W538–W545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deutekom E.S., Snel B., van Dam T.J.P.. Benchmarking orthology methods using phylogenetic patterns defined at the base of eukaryotes. Brief. Bioinform. 2021; 22:bbaa206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E.. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011; 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Glover N., Dessimoz C., Ebersberger I., Forslund S.K., Gabaldón T., Huerta-Cepas J., Martin M.-J., Muffato M., Patricio M., Pereira C.et al.. Advances and applications in the quest for orthologs. Mol. Biol. Evol. 2019; 36:2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altenhoff A.M., Boeckmann B., Capella-Gutierrez S., Dalquen D.A., DeLuca T., Forslund K., Huerta-Cepas J., Linard B., Pereira C., Pryszcz L.P.et al.. Standardized benchmarking in the quest for orthologs. Nat. Methods. 2016; 13:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. The Alliance of Genome Resources Consortium Alliance of genome resources portal: unified model organism research platform. Nucleic Acids Res. 2020; 48:D650–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bruford E.A., Lush M.J., Wright M.W., Sneddon T.P., Povey S., Birney E.. The HGNC database in 2008: a resource for the human genome. Nucleic Acids Res. 2008; 36:D445–D448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DeLuca T.F., Cui J., Jung J.-Y., St. Gabriel K.C., Wall D.P. Roundup 2.0: enabling comparative genomics for over 1800 genomes. Bioinformatics. 2012; 28:715–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schreiber F., Sonnhammer E.L.L.. Hieranoid: hierarchical orthology inference. J. Mol. Biol. 2013; 425:2072–2081. [DOI] [PubMed] [Google Scholar]
- 42. Huerta-Cepas J., Capella-Gutiérrez S., Pryszcz L.P., Marcet-Houben M., Gabaldón T.. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014; 42:D897–D902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonnhammer E.L.L., Östlund G.. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015; 43:D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Altenhoff A.M., Glover N.M., Train C.-M., Kaleb K., Warwick Vesztrocy A., Dylus D., de Farias T.M., Zile K., Stevenson C., Long J.et al.. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 2018; 46:D477–D485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruzicka L., Howe D.G., Ramachandran S., Toro S., Van Slyke C.E., Bradford Y.M., Eagle A., Fashena D., Frazer K., Kalita P.et al.. The zebrafish information network: new support for non-coding genes, richer gene ontology annotations and the alliance of genome resources. Nucleic Acids Res. 2019; 47:D867–D873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nevers Y., Kress A., Defosset A., Ripp R., Linard B., Thompson J.D., Poch O., Lecompte O.. OrthoInspector 3.0: open portal for comparative genomics. Nucleic Acids Res. 2019; 47:D411–D418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emms D.M., Kelly S.. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019; 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R.et al.. Ensembl 2020. Nucleic Acids Res. 2020; 48:D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D.. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019; 47:D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ruan J., Li H., Chen Z., Coghlan A., Coin L.J.M., Guo Y., Hériché J.-K., Hu Y., Kristiansen K., Li R.et al.. TreeFam: 2008 update. Nucleic Acids Res. 2008; 36:D735–D740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jain A., Perisa D., Fliedner F., von Haeseler A., Ebersberger I.. The evolutionary traceability of a protein. Genome Biol. Evol. 2019; 11:531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hulsen T., Huynen M.A., de Vlieg J., Groenen P.M.. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006; 7:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kachroo A.H., Laurent J.M., Yellman C.M., Meyer A.G., Wilke C.O., Marcotte E.M.. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015; 348:921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laurent J.M., Garge R.K., Teufel A.I., Wilke C.O., Kachroo A.H., Marcotte E.M.. Humanization of yeast genes with multiple human orthologs reveals functional divergence between paralogs. PLOS Biol. 2020; 18:e3000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chorostecki U., Molina M., Pryszcz L.P., Gabaldón T.. MetaPhOrs 2.0: integrative, phylogeny-based inference of orthology and paralogy across the tree of life. Nucleic Acids Res. 2020; 48:W553–W557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohno S. Evolution by Gene Duplication. 1970; Springer-Verlag. [Google Scholar]
- 57. Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N.et al.. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liebeskind B.J., McWhite C.D., Marcotte E.M.. Towards consensus gene ages. Genome Biol. Evol. 2016; 8:1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mustafin Z.S., Zamyatin V.I., Konstantinov D.K., Doroshkov A.V., Lashin S.A., Afonnikov D.A.. Phylostratigraphic analysis shows the earliest origination of the abiotic stress associated genes in a. thaliana. Genes. 2019; 10:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Drost H.-G., Gabel A., Grosse I., Quint M.. Evidence for active maintenance of phylotranscriptomic hourglass patterns in animal and plant embryogenesis. Mol. Biol. Evol. 2015; 32:1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Quint M., Drost H.-G., Gabel A., Ullrich K.K., Bönn M., Grosse I.. A transcriptomic hourglass in plant embryogenesis. Nature. 2012; 490:98–101. [DOI] [PubMed] [Google Scholar]
- 62. Arendsee Z.W., Li L., Wurtele E.S.. Coming of age: orphan genes in plants. Trends Plant Sci. 2014; 19:698–708. [DOI] [PubMed] [Google Scholar]
- 63. Hart T., Brown K., Sircoulomb F., Rottapel R., Moffat J.. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol. Syst. Biol. 2014; 10:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R.et al.. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012; 40:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Crow M., Paul A., Ballouz S., Huang Z.J., Gillis J.. Characterizing the replicability of cell types defined by single cell RNA-sequencing data using metaneighbor. Nat. Commun. 2018; 9:884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Crow M., Gillis J.. Co-expression in single-cell analysis: saving grace or original sin?. Trends Genet. 2018; 34:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Heimberg G., Bhatnagar R., El-Samad H., Thomson M.. Low dimensionality in gene expression data enables the accurate extraction of transcriptional programs from shallow sequencing. Cell Syst. 2016; 2:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Koepfli K.-P., Paten B., Brien S.J.. The genome 10K project: a way forward. Annu. Rev. Anim. Biosci. 2015; 3:57–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guan Y., Gorenshteyn D., Burmeister M., Wong A.K., Schimenti J.C., Handel M.A., Bult C.J., Hibbs M.A., Troyanskaya O.G.. Tissue-Specific functional networks for prioritizing phenotype and disease genes. PLOS Comput. Biol. 2012; 8:e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Crow M., Paul A., Ballouz S., Huang Z.J., Gillis J.. Exploiting single-cell expression to characterize co-expression replicability. Genome Biol. 2016; 17:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tarashansky A.J., Musser J.M., Khariton M., Li P., Arendt D., Quake S.R., Wang B.. Mapping single-cell atlases throughout metazoa unravels cell type evolution. Elife. 2021; 10:e66747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vallejo A.F., Davies J., Grover A., Tsai C.-H., Jepras R., Polak M.E., West J.. Resolving cellular systems by ultra-sensitive and economical single-cell transcriptome filtering. iScience. 2021; 24:102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pereira C., Denise A., Lespinet O.. A meta-approach for improving the prediction and the functional annotation of ortholog groups. BMC Genomics. 2014; 15:S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ballouz S., Weber M., Pavlidis P., Gillis J.. EGAD: ultra-fast functional analysis of gene networks. Bioinformatics. 2017; 33:612–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E.et al.. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005; 21:650–659. [DOI] [PubMed] [Google Scholar]
- 76. McCarthy D.J., Campbell K.R., Lun A.T.L., Wills Q.F.. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017; 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smyth G.K. Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S.. limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Statistics for Biology and Health. 2005; NY: Springer; 397–420. [Google Scholar]
- 78. Edgar R., Domrachev M., Lash A.E.. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C.. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018; 36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Konopka T. umap: Uniform Manifold Approximation and Projection. 2020; [Google Scholar]
- 81. Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W.. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005; 21:3439–3440. [DOI] [PubMed] [Google Scholar]
- 82. Kumar S., Stecher G., Suleski M., Hedges S.B.. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017; 34:1812–1819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code to reproduce the coexpression conservation analysis are available through CoCoBLAST (https://gillisweb.cshl.edu/CoCoBLAST/, see Supplementary Note 2 for a guided tutorial).