Abstract
Photosystem I (PSI) from the thermophilic cyanobacterium Thermosynechococcus elongatus (Te) is the largest membrane protein complex to have had its structure solved by X-ray diffraction. This trimeric complex has 36 protein subunits, over 380 non-covalently bound cofactors and a molecular weight of ∼1.2 MDa. Previously, it has been isolated and characterized in a detergent micelle using the non-ionic detergent n-dodecyl-β-d-maltoside (DDM). We have now succeeded in isolating this complex without the use of detergents, using styrene–maleic acid (SMA) alternating copolymer. Intriguingly, a partially esterified copolymer formulation (SMA 1440, Cray Valley) was found to be most efficient in cyanobacterial thylakoid membranes. A host of biochemical, biophysical and functional assays have been applied to characterize this non-detergent form of PSI, referred to as a SMA Lipid Particle (SMALP). The PSI-SMALP has a lower sedimentation coefficient compared to PSI-DDM, suggesting decreased density or a more extended particle shape. We show the 77 K fluorescence maximum for PSI is red shifted in PSI-SMALP compared to PSI-DDM, suggesting a more native orientation of PsaA/B associated chlorophyll. We report that PSI-SMALPs are functional despite the selective loss of one transmembrane subunit, PsaF. This loss may reflect a more labile interaction of the PSI core and PsaF, or a selective displacement during copolymer insertion and/or assembly. PSI-SMALP exhibited decreased reduction kinetics with native recombinant cytochromes c6, while non-native horse heart cytochrome c shows faster reduction of PSI-SMALP compared to PSI-DDM. This is the largest membrane protein isolated using SMA copolymers, and this study expands the potential use of this approach for the isolation and characterization of large supramolecular complexes.
Trimeric Photosystem I (PSI) from the thermophilic cyanobacterium Thermosynechococcus elongatus (Te) is the largest membrane protein complex to be encapsulated within a SMALP to date.
Introduction
With the sequencing of thousands of prokaryotic and eukaryotic genomes it is now clear that integral membrane proteins comprise ∼30% of the proteins encoded in those genomes.1–6 Although this advance in genome science has been concurrent with the rapid increase of new high resolution crystal structures,7 only ∼3% of all the structures deposited in the PDB represent membrane proteins. This lag between genetic advances and the structural understanding of membrane proteins is due in large part to the challenges of expression, purification, and crystallization of membrane proteins.8 Traditionally membrane proteins have been isolated following solubilization with non-ionic detergents, which involves detergent insertion into the membrane exchanging with native lipids, where at high detergent levels a mixed micelle is formed containing the membrane protein(s)9,10 surrounded by detergents and remaining lipids. This approach has led to a plethora of new detergent classes, yet a slow and systematic approach is still required to determine the best means of solubilizing, stabilizing and structurally characterizing active membrane proteins.11,12 This requires an empirical approach to optimize detergent selection, and greatly obstructs efforts to understand the structure and function of membrane proteins.
In the past 5–8 years there have been many reports on non-detergent methods of membrane protein isolation. These include various peptide-based nanodiscs, utilizing amphipathic helical membrane scaffold proteins (MSPs),13 for individual proteins14,15 as well as whole proteomes.16 Most of these advances have required initial detergent solubilization followed by the addition of phospholipids and MSPs. The membrane protein(s) will self-assemble into nanodiscs when the detergent is removed. A recent investigation by Sharma and Wilkens showed that a new strategy termed “nanodisc-reconstitution before purification” resulted in a close to completely assembled vacuolar ATPase from yeast with high activity.17 These peptide-based nanodiscs were made up of native membrane lipids, however in this approach the degree to which leaflet heterogeneity of the native lipids, as well as the specific composition of lipids that interface with the isolated membrane protein is unknown. While this investigation supports our hypothesis that the native lipid environment is critical to the overall function of membrane proteins, the initial detergent solubilization step prior to insertion into MSP-based nanodiscs cannot be avoided.17 Further, though this reconstitution method may use lipids of native origin, there is no reason to believe that the lipids incorporated will conform to the same proximity or interface as the embedded complex in vivo.
A recent discovery in membrane protein research is the ability of styrene maleic acid (SMA) alternating copolymers to solubilize membranes in the form of nanodiscs, allowing extraction and purification of membrane proteins from their native environment in a single detergent-free step.18–21 The advent of SMA solubilization has three major advantages over MSP nanodiscs or other detergent-dependent isolations: (1) permits direct solubilization and purification of membrane proteins while maintaining their lipid environment; (2) avoids the empirical and laborious detergent-based procedures and their inherent risk of protein aggregation and/or denaturation; and (3) SMALPs provide a stable lipid environment for membrane proteins with small particle size, compatible with a wide range of biophysical approaches. This has important implications for membrane biology in part because it allows for the isolation and characterization of both membrane proteins and their boundary lipids in a near-native environment.22
Recently in the field of bioenergetics, two of the most well characterized classes of membrane proteins, the purple bacterial photosynthetic reaction centers23 and the plant PSI-LHCII supercomplex,24 have been characterized using SMA isolation. In the latter study, the authors report PSI-LHCII super complex was not encapsulated within a SMALP, rather the spinach thylakoid membrane fraction that remained following incubation with SMA was highly enriched in PSI-LHCII super complexes. This membrane pellet was also depleted is PSII (alone), the cytochrome b6f complex and CF1–CF0 (of the ATPase).24 This highly ordered region of plant thylakoids was described as being analogous to BBY preparation in spinach thylakoid membranes.25 This protocol utilizes the lateral heterogeneity of plant thylakoids to separate the PSII-rich stacked thylakoids and the non-stacked stromal lamellae, which contains less PSII.26
The exact reason for this apparent selectivity for SMA is unknown, however we can reason that this is either due to an unforeseen preference for particular SMA formulations to make SMALPs of a specific size range, or more likely that the isolated proteins are enriched in a particular region of the thylakoid membrane, and this region is more permissible for SMA insertion. The latter hypothesis is further supported in Cyanobacteria as well, with two recent atomic force microscopy studies that show regions of the thylakoid membrane in Te and Prochlorococcus indeed show this lateral heterogeneity, revealing that PSI trimers exist in a nearly ordered hexagonal array in parts of the thylakoid.27,28 The reason why these regions are more prone to SMA solubilization remains to be elucidated, however it is logical to assert that if the proteins are laterally heterogeneous, the lipids surrounding them may be as well, affording the possibility of lipid specificity for SMA insertion.
We have applied a selection of commercially available SMA copolymers to solubilize the PSI trimer from thylakoid membranes of the thermophilic cyanobacterium Thermosynechococcus elongatus (Te). We have characterized this PSI-SMALP biochemically to optimize the isolation method and determine subunit composition. We have also examined the biophysical activity of PSI-SMALP via chlorophyll fluorescence and reduction kinetics following photooxidation. Interestingly, this solubilization method selectively lost PsaF, a single subunit that binds to the outer edge of the complex, specifically PsaB, most distal to the center axis. It is interesting that this subunit is quantitatively lost during SMA treatment and may indicate a rather weak interaction with the PsaB core subunit. This work represents the first successful use of SMAs on a thermophilic protein, and at ∼1.47 MDa is the largest protein to be isolated using any type of non-detergent solubilization to date.
Materials
SMA copolymers
SMA copolymers used in this study were acquired from Cray Valley (formerly a Total Company, now owned by Polyscope). The physicochemical properties of the original 7 SMA formulations used in this study are included in Table 1.
Physicochemical properties of SMA copolymers used in this work.
| Copolymer | S : MA ratio | M W | M N | Đ | Counter ion | Esterification alcohol(s) |
|---|---|---|---|---|---|---|
| 1000 | 1 : 1 | 5.00 | 2.10 | 2.38 | Na+ | None |
| 2000 | 2 : 1 | 7.50 | 2.70 | 2.78 | NH4+ | None |
| 3000 | 3 : 1 | 9.50 | 3.05 | 3.11 | NH4+ | None |
| 17 352 | 1.7 : 1 | 7.00 | 2.80 | 2.50 | NH4+ | Cyclohexanol and 2-propanol |
| 2625 | 2 : 1 | 9.00 | 3.60 | 2.50 | NH4+ | 1-Propanol |
| 1400 | 1.5 : 1 | 7.00 | 2.90 | 2.41 | NH4+ | 2-Butoxyethanol |
| 10 235 | 1.5 : 1 | 7.00 | 2.90 | 2.41 | NH4+ | None |
Cytochromes
Horse heart cytochrome was purchased from Sigma-Aldrich. Cytochrome c6 from Te was expressed in E. coli as previously described.29
PsaF antibody production
To facilitate the detection of photosystem I subunits we have made many polyclonal antibodies specific to cyanobacterial subunits. In order to make a universal Cyanobacteria antibody, we selected the lumen-exposed portion of PsaF. A protein sequence alignment was done with ∼50 different Cyanobacteria that had a PsaF annotated sequence. This alignment gave two distinct consensus sequences for this lumenal exposed N-terminal domain: one of 45 residues (peptide A) and another with 50 residues (peptide B). The consensus sequences of these two peptides are shown below:
Peptide A:
VAGLT PCAES ARFQQ RASAA TTPQA KARFE MYSQA VCGED GLPHL
Peptide B:
GANLT PCSES PAFQQ KAKNA RNTTA DPQSG QKRFE RYSQA LCGPE GYPHL
Each peptide sequence was Blasted against Te and Syn. 6803 PsaF sequences to ensure recognition PsaF from these two well studied Cyanobacteria. Peptide A was 60% identical to Te, while only 37% identical to Syn. 6803. Peptide B was 73 and 71% identical to Te and Syn. 6803 PsaF sequences, respectively. Both peptides were cloned in pTYB2 and expressed in E. coli using the IMPACT System (NEB, Ipswich, MA). Following purification, the peptides were sent out for production of rabbit polyclonal antibodies at Pocono Rabbit Farm and Laboratory, Inc. (Canadensis, PA). We have shown that both antisera recognize PsaF as a single band in both Te thylakoids and PSI preparations (data not shown).
Methods
Isolation of thylakoid membranes
Te was cultured in BG-11 medium at 45° with aeration. Cells were harvested at late log phase. Cell pellets were stored at −80 °C or directly washed in buffer A (50 mM MES–NaOH, pH = 6.5, 5 mM CaCl2, and 10 mM MgCl2) for thylakoid membrane isolation.30 After pelleting the cell suspension, cell pellets were re-suspended in buffer A containing 500 mM sorbitol and lysed using French Press. Thylakoid membranes were precipitated by spinning down at 180 000g for 1 h. The pellets were again re-suspended in buffer A containing 12.5% glycerol and stored at −80 °C. Chlorophyll concentration was determined as described previously.31 Thylakoid membranes were washed by 3 times by dounce homogenization and pelleting at 180 000 × g in buffer A (for DDM isolation). Buffer S (50 mM Tris–Cl, pH = 9.5 at room temperature) was used for SMA isolation with varied amounts of KCl and NaCl. Following the last wash, the thylakoid membranes were brought up to a chlorophyll concentration of 1 mg mL−1 prior to all solubilization trials.
Photosystem analysis using BN-PAGE and SDS-PAGE
Thylakoid membranes were solubilized in DDM (Glycon) or SMA (Cray Valley) at 40 °C for 3 h. Insoluble material was removed by centrifugation at 180 000g for 5 min at 4 °C. Supernatants were taken off for analysis by blue native polyacrylamide gel electrophoresis (BN-PAGE) of isolated PSI. 4 to 16% BN-PAGE gels (Invitrogen) were used to analyze solubilized thylakoids or isolated photosystems according to the user manual and ref. 32 and 33. For photosystem identification, SDS-PAGE using 10% acrylamide gel was used as described previously,34 and silver staining was performed using the fast stain method.35 PSI-subunit profile for DDM extracted photosystem was confirmed using a different SDS-PAGE method, using a 18–24% gradient gel.36
Sedimentation analysis using analytical ultracentrifugation
PSI-SMALP and PSI-DDM were first dialyzed against buffer (0.05% SMA 1440 or DDM, Tris–HCl, pH 9.0) using a 12–14 kDa molecular mass cutoff membrane (Spectrum Labs) with three buffer changes. Analytical ultracentrifugation (AUC) sedimentation velocity was performed in a Beckman Coulter ProteomeLab™ XL-I analytical ultracentrifuge using a double sector Epon, charcoal-filled centerpiece, quartz windows, and Ti50 rotor (Beckman/Coulter). Absorption measurements at 680 nm were made every minute at 30 000 rpm and 20 °C, using the appropriate dialysis buffer as the reference. The buffer density and viscosity were determined by SEDNTERP to be 0.71006 g mL−1, 0.99823 g mL−1, and 0.001002 pascal s respectively. Measurements were analyzed by Sedfit v.13.0b using the continuous c(s) analysis model.37
77 K chlorophyll fluorescence
After PSI was solubilized by SMA or DDM from Te cell membranes, measurements of chlorophyll concentration were made at 650 nm using an ultraviolet-visible light spectrophotometer, and chlorophyll concentration was standardized between both extraction methods. The extracts were then transferred into glass electron paramagnetic resonance (EPR) tubes and were slowly frozen in liquid nitrogen. Chlorophyll fluorescence spectra were obtained using a PTI Quantamaster dual-channel fluorometer. Excitation light of 420 nm was used. The emission spectrum was measured by scanning from 550–800 nm with 0.5 nm steps, with a slit width of 1 nm. The resulting spectra was the average of 4 traces and the emission maxima was recorded.
PsaF immunoblotting
The isolated PSI and sucrose density gradient fractions were separated by SDS-PAGE and transferred to PVDF (Immobilon, EMD Millipore, Burlington, MA). This blot was then blocked and probed with the rabbit anti-PsaB {Peptide b} antisera. The immunoblot was visualized using a GAR HRP conjugate and detected using the SuperSignal West Dura extended duration chemiluminescent substrate (Thermo Scientific, Waltham, MA).
P700 photooxidation and reduction by native and non-native cytochromes
Laser flash photolysis was conducted using a Joliot Type Spectrophotometer (JTS-100), equipped with an actinic LED source emitting a short excitation pulse at 630 nm and a probe beam of infrared light at 810 nm. Upon photoexcitation, P700 becomes oxidized and the absorbance of 810 nm light disappears. P700+ reduction rate is then monitored as the return of 810 nm absorbance. Fitting was done with a single exponential decay function in Prism 7. Constraints of plateau equal to 0 and Kobs > 0 were used. 1000 iterations were used for fitting. Observed rates are plotted against molar ratio of cytochrome used.
Molecular modeling and analysis
The simulation system was built starting from the PDB of the PSI complex from Te (PDB ID: 1JB0). Missing residues 263 to 265 in PsaA were added. PsaK, for which resolution was too poor to assign side chains, was removed. Chlorophylls, carotenoids, 1,2-dipalmitoyl-phosphatidylcholine (DPPC) and 1,2-distearoyl-monogalactosyl-diglyceride (MGDG) lipids, iron–sulfur clusters, water, and calcium ions present in the crystal structure were retained, and missing atoms (e.g., chlorophyll tails), were added using VMD.38 The trimer was created by successive 120° rotations of the monomer. The protein was then placed in a 30% POPG/70% POPE hexagonal membrane using the CHARMM-GUI Membrane Builder39 and solvated above and below with a 150 mM NaCl solution. The resulting system is 588 444 atoms.
Molecular Dynamics (MD) simulations were carried out using NAMD 2.11 (ref. 40) and the CHARMM36 force field.41,42 Additional force field parameters for PSI-specific components came from Harris et al.43 All simulations used a 2 fs time step with bonded and short-range non-bonded interactions evaluated every time step; long-range non-bonded interactions were evaluated every other time step with the particle-mesh Ewald method. van der Waals interactions were cutoff at 12 Å, with a force-based switching function starting at 10 Å. After a 5 ns staged relaxation of the membrane and protein side chains, the system was equilibrated for 30 ns.44
Results
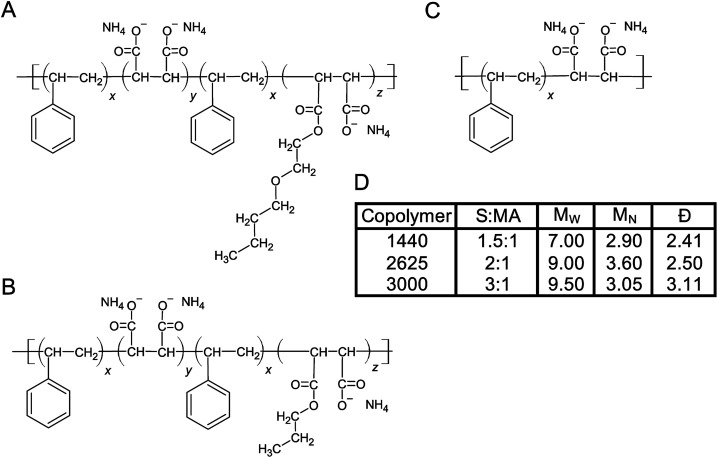
We began this investigation with an initial set of 7 copolymers including 1000, 2000, 3000, 1440, 17 352, 2625 and 10 235. Not all of these were effective for liberating chlorophyll containing protein complexes and we thus reduced the study to a smaller number. SMA 1440 has been found to be most efficient in forming trimeric PSI-SMALPs from Te, yielding ∼40% the amount of PSI trimer compared to DDM extraction (based on BCA protein assay and chlorophyll absorbance). This copolymer formulation is distinct with regard to the functionalization of a portion of the maleic acid groups with butoxyethanol. Therefore, the SMA alternating copolymers chosen to be the focus of this work differed with respect to functionalization of the maleic acid moiety (Fig. 1A–C). Fig. 1A shows that SMA 1440 has a butoxyethanol functional group added to one maleic acid moiety via a dehydration reaction. Similarly, SMA 2625 is functionalized with propanol and SMA 3000 has no alkoxy chain added (Fig. 1B and C). Fig. 1D lists differences in styrene to maleic acid ratio (S : MA), number weighted molecular weight (MN), weight weighted molecular weight (MW) and dispersity (Đ), which is the quotient of Mw/MN.
Fig. 1. Linear structures of SMA copolymers used in this work. The three SMA copolymers used in this study were obtained from Cray Valley. (A) SMA 1440, (B) SMA 2625 and (C) SMA 3000. (D) Table listing physicochemical properties of each copolymer formulation, the ‘H’ identifier is used by the manufacturer to indicate the copolymer has been solubilized (maleic anhydride ring opened following basic reflux). S : MA, MN, MW and Đ are shown.
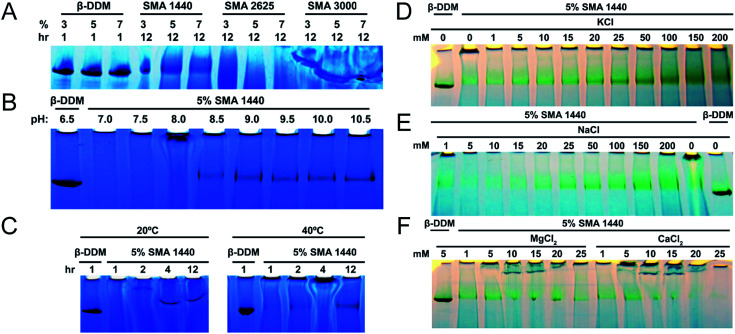
To test SMA copolymers with various physicochemical properties and optimize extraction methods, solubilization of thylakoid membranes were conducted at various concentrations (v/v) of these three SMA formulations (Fig. 2A). The starting concentrations of neat SMA solutions obtained from Cray Valley were established gravimetrically following lyophilization. Solid content of the obtained SMA copolymers were determined to be 36.47%, 20.57% and 14.48% (w/v) for SMAs 1440, 2625 and 3000, respectively. In all solubilization experiments, trimeric PSI extracted with DDM was used as a control and size marker. Following incubation with thylakoid membranes for 12 hours at 25 °C, BN-PAGE shows that SMA 1440 is the most efficient formulation of the three tested for the solubilization of trimeric PSI out of the Te thylakoid membrane (Fig. 2A). It can also be seen that trimeric PSI extracted with SMA 1440 shows a slight decrease in mobility by BN-PAGE, indicating a slightly larger size compared to PSI trimer isolated using DDM. Additionally, 5% of the neat SMA 1440 solution (1.82% (w/v) total solids) was found to be the optimal concentration for PSI trimer extraction by SMA 1440. SMA 2625 is unable to isolate trimeric PSI across all conditions tested. Further, SMA 3000 did demonstrate efficacy in solubilizing a protein complex at similar size to the PSI trimer, however this result is inconclusive due to salting effect encountered in the gel when using this formulation (Fig. 2A). Overall, we did not observe effective solubilization with SMA formulations 3000 or 2626, as is shown in Fig. 2A. The chemical basis for this inactivity is not known, yet we have also confirmed decreased solubilization efficacy of these SMAs on protein extraction from spinach chloroplast thylakoid membranes.45 Therefore, PSI-SMALP encapsulated using SMA 1440 was used exclusively for subsequent experiments in this study. It is worth noting, Te thylakoid membranes solubilized with DDM liberate more chlorophyll containing protein complexes than SMA 1440 in all trials. We find these solubilizations to yield approximately 62–84% chlorophyll for DDM and 30–40% chlorophyll for SMA 1440 respective to the starting chlorophyll content of the isolated thylakoid membranes. We suspect based on the BN-PAGE that this additional chlorophyll from the DDM solubilization comes not only from PSI trimer but also from solubilized PSII and PSI monomers. Interestingly, as is shown in this work the SMA 1440 is highly selective for only the PSI trimer.
Fig. 2. Optimized conditions for thylakoid membrane solubilization by SMA. PSI trimer isolated using DDM is shown as control (0.6% DDM, B–F). (A–C) PSI solubilization conditions with regard to SMA formulation, pH and temperature, respectively. (D–F) PSI extraction efficiency as a function of ion concentration (mM) and valency, photos were taken prior to CBB staining to increase contrast of chlorophyll containing PSI. (A) PSI extraction with DDM and various SMAs as a function of varied amphiphilic molecule concentration. Solubilization time of thylakoid membranes listed as 1 hour for DDM and 12 hours for SMAs. (B) Extraction of PSI using 5% SMA 1440 over varied pH. (C) PSI extraction at 20 °C and 40 °C over time. Thylakoid membranes were solubilized with increasing concentrations of KCl (D) and NaCl (E). Thylakoid membranes were also washed in this buffer (devoid of amphiphilic molecule), prior to solubilization. (F) Effect of divalent ions on solubilization with 5% SMA 1440.
Previous investigations of SMA copolymers have reported insolubility at pH values approaching neutral.46,47 Similarly, we find that SMA 1440 becomes inefficient in PSI solubilization below pH = 8.5. Further, at pH values ranging from 9.5–10.5 there appears to be no increase in PSI trimer liberated from thylakoid membranes (Fig. 2B). Without agitation, increasing temperature from 20 °C to 40 °C enhanced solubilization efficiency of PSI by SMA 1440, with more PSI trimer being isolated with increasing time from 1 to 12 hours (Fig. 2C). At 20 °C, very large complexes are shown to enter the gel at 2 hours, with smaller SMALPs (still larger than trimeric PSI) being produced at 4 and 12 hours of incubation (Fig. 2C).
The requirement for 100–500 mM monovalent ions to facilitate protein extraction using SMA copolymers has previously been reported in the literature.48–51 Our results corroborate these findings showing an increase in PSI extracted as concentrations of KCl (Fig. 2D) and NaCl (Fig. 2E) increase from 1 to 200 mM. Our results also agree with previous investigations that report divalent ion sensitivity of SMA copolymers decreasing extraction efficiency.51–53 In Fig. 2F, significant reductions in solubilized PSI can be seen at CaCl2 and MgCl2 concentrations above 10 mM. Considering all data from Fig. 2, we determined the optimum solubilization conditions for trimeric PSI from Te to be: SMA 1440 in 50 mM Tris–Cl (pH 9.5 at room temperature) with 125 mM KCl, incubated at 40 °C for 3 hours with agitation (shaker at 350 rpm). These conditions were used in all subsequent analyses.
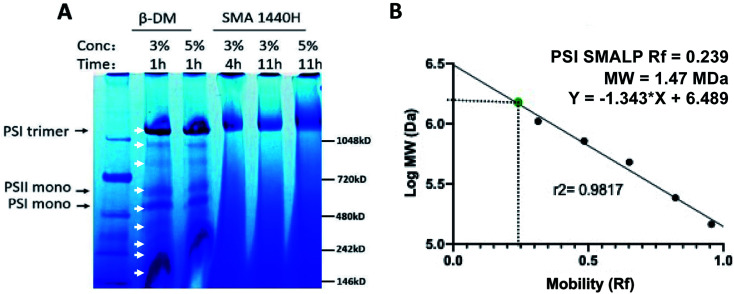
Following solubilization of Te thylakoid membranes with DDM, many proteins can be seen by BN-PAGE (Fig. 3A, white arrows), whereas SMA solubilization preferentially extracts trimeric PSI alone at all concentrations and times tested (Fig. 3A). The streaking toward the bottom of the gel in the SMA lanes can be attributed to unbound SMA copolymer. The relative mobility of this PSI-SMALP reveals the size of this complex to be slightly larger than trimeric PSI-DDM, at 1.47 MDa (Fig. 3B).
Fig. 3. Full protein profile following solubilization of Thermosynechococcus elongatus thylakoid membranes with SMA 1440 and DDM by blue native PAGE. (A) White arrows indicate proteins extracted by DDM, PSI trimer, PSI monomer and PSI dimer are labeled. (B) Relative mobility of PSI-SMALP as determined by linear regression of molecular weight standards.
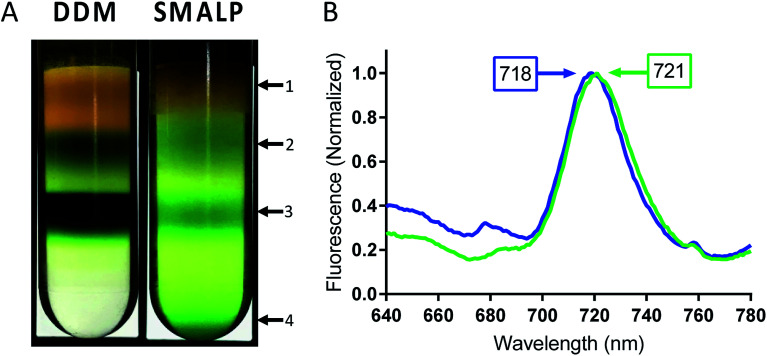
Following incubation with copolymer or detergent, insolubilized material was spun down and the supernatant was then separated using sucrose density gradient centrifugation. The sucrose gradients show distinct bands for free carotenoids (band 1), free chlorophyll (band 2) and trimeric PSI (band 3), in both DDM and SMA extracted thylakoids (Fig. 4A). In DDM preparations of cyanobacterial thylakoid membranes, band 2 also contains PSII and monomeric PSI.54 Interestingly, trimeric PSI seems to be exclusively isolated in SMA preparations. Further, a pellet can be seen in the SMALP sucrose gradient, suggesting that even larger particles are being encapsulated within SMALPs (Fig. 4A, band 4). These particles may be larger thylakoid membrane fragments, or potentially multi-protein supermolecular complexes, which have long been suspected to exist within the thylakoid membrane.55,56 Future investigations will focus on elucidating the composition of these particles.
Fig. 4. Low temperature fluorescence of PSI-trimer in SMALPs and DDM. (A) Bands of PSI extracted by SMA 1440 and DDM in sucrose gradient centrifugation. Arrows point to discernable bands 1–4 and are designated as follows: 1 – liberated carotenoids, 2 – free chlorophyll, 3 – trimeric PSI and 4 – larger thylakoid membrane fragments or supercomplexes (absent in DDM preparation). (B) Low temperature fluorescence spectrum of band 3 (trimeric PSI) isolated from sucrose gradient for PSI-DDM (blue) and PSI-SMALP (green).
Fluorescence of Te trimeric PSI can occur at wavelengths as long as ∼730 nm in intact cells, dependent on the solvation state of the chlorophyll. It has previously been shown that as PSI goes from trimeric to monomeric form in Te, a blue shift in chlorophyll fluorescence of 3–6 nm can be seen.57 In addition, DDM isolation causes a blue-shift in fluorescence maximum of ∼3 nm compared to that of intact cells. Together, this data confirms that the 77 K fluorescence emission spectra becomes more red-shifted relative to how native the setting of the protein (PSI in intact cells > DDM PSI trimer > DDM PSI monomer).28,57 Low temperature fluorescence of band 3 from both preparations, performed in liquid nitrogen at 77 K, shows a significant 3 nm bathochromic shift in chlorophyll fluorescence for PSI-SMALP (Fmax = 721 nm) compared to that of PSI-DDM (Fmax = 718 nm, Fig. 4B). This red shift signifies a fluorescence at lower energy in the PSI-SMALP, which has been suggested to represent a more native conformation of the chlorophyll antennae structure within the reaction center.58 Further, it is accepted that the local environment of far-red chlorophylls are key to their emission wavelength and that a blue shift reflects more solvation and exposure to a more polar environment.59
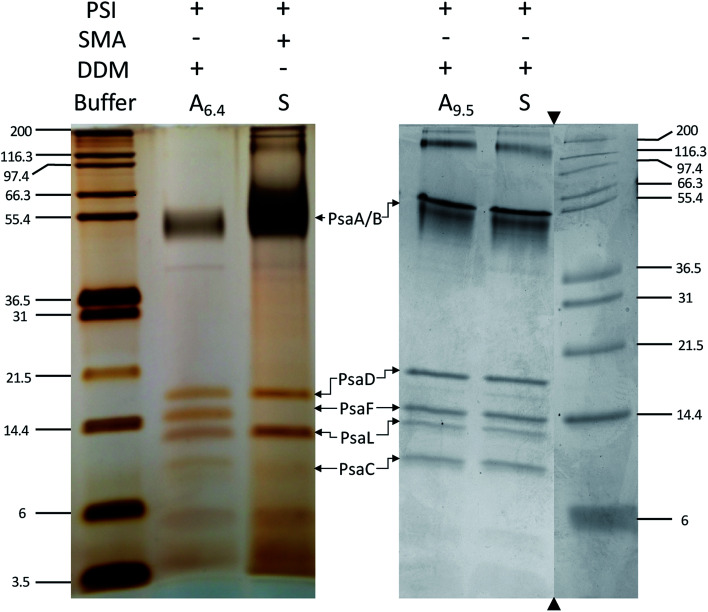
Next, the subunit profile of the PSI-SMALPs was investigated using sodium dodecyl sulfate PAGE (SDS-PAGE). The overall subunit profile for the SMALP band 3 matched with PSI-DDM, further confirming that this band contains PSI-SMALP (Fig. 5). Interestingly, the PsaF subunit was missing in the PSI-SMALP, while all other subunits above 3 kDa in size are present in the SMA and DDM extracted PSI trimer (Fig. 5). To confirm this loss of PsaF was not an effect of the harsh buffer conditions required for SMA copolymers, we performed an SDS-PAGE analysis on DDM solubilized thylakoid membranes using buffer A at pH = 9.5 (A9.5) and DDM solubilization in SMA buffer (S). PsaF is shown to be retained by DDM extraction across all buffer conditions tested (Fig. 5).
Fig. 5. PsaF is lost in SMALPs. SDS-PAGE of PSI extracted from T. elongatus BP-1, stained with silver (left) and Coomassie (right). DDM solubilization of PSI was performed with buffer A at pH 6.4 (A6.4), at elevated pH (A9.5) and with SMA bummer (S). The subunit profile was confirmed using two different gel systems and gradients. Triangles indicate where the gel image was cropped, molecular weight markers and associated lanes for both images are from the same gel, respectively.
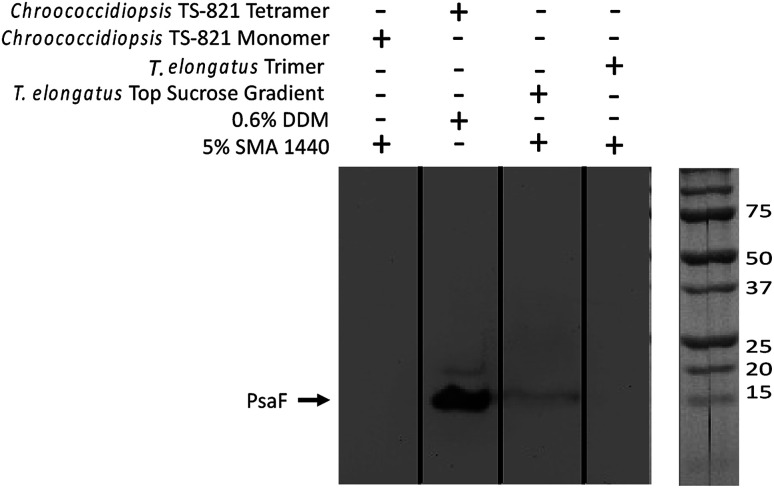
Immunoblot analysis using a PsaF specific antibody, α-PsaF, confirms this loss of PsaF from the PSI trimer and showed that PsaF is left behind at the top of the SMALP sucrose gradient (Fig. 6). The presence of this faint band suggests that PsaF has dissociated from the complex at some point during SMALP formation, and due to its small sedimentation coefficient, does not enter the sucrose gradient. This PsaF signal from the top of the sucrose gradient is diffuse due to dilution into the sucrose buffer. To confirm this loss of PsaF from thermophilic cyanobacterial PSI following SMA extraction, a second organism Chroococcidiopsis sp. TS-821 was used and similarly probed with α-PsaF. TS-821 has been shown to produce tetrameric PSI oligomeric complexes, which have been proposed to be an intermediate conformation in the monomerization of PSI from Cyanobacteria to land plants.57 Following SMA extraction, PSI SMALPs isolated from TS-821 contained PSI monomers rather than tetramers, which are obtained when TS-821 thylakoids are extracted with DDM. This monomerization may be due to thylakoid washing steps in high pH buffer devoid of divalent ions for SMALP extraction. These monomeric PSI-SMALPs also lack PsaF, whereas intact tetrameric PSI isolated using DDM retains this subunit (Fig. 6).
Fig. 6. Western blot confirms PsaF is lost in PSI-SMALPs. In the far-left lane, PsaF is shown to dissociate from Chroococcidiopsis TS-821 PSI monomer when extracted with SMA 1440. In the second lane from left, when extracted with DDM PSI from this organism retains both a tetrameric oligomeric state and PsaF. The third lane from left shows PsaF was found at the top of the sucrose density gradient, following solubilization. The far-right lane shows PsaF is missing in the trimeric PSI-SMALP.
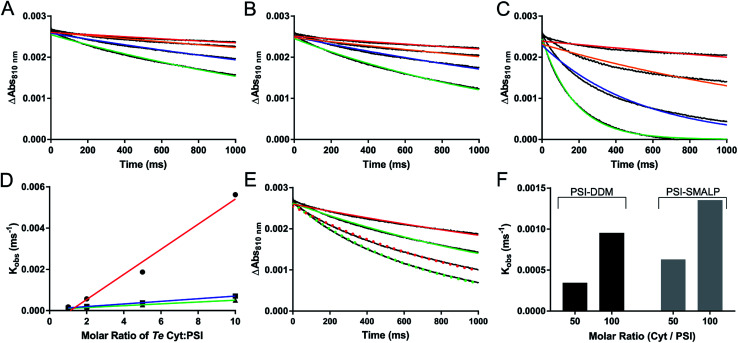
Reduction kinetics of PSI-SMALP and PSI-DDM from Te using native cytochrome c6 (cyt6) and non-native cytochrome from horse heart (cytHH) was observed using a Joliot type spectrometer (JTS-100, Fig. 7). Significantly slower reduction kinetics for PSI-SMALP compared to PSI-DDM were observed when using cyt6 as the electron donor when analyzed in their respective buffer systems (Fig. 7A and C). However, when both PSI-SMALP and PSI-DDM are analyzed in buffer S, consisting of 50 mM Tris–Cl and 125 mM KCl at pH = 9.5 (room temperature), PSI reduction occurs at comparable rates for both particles (Fig. 7A–D). Interestingly, when cytHH is used, the reduction rate of PSI-SMALP exceeds that of PSI-DDM (Fig. 7E and F). The ability for PSI-SMALP to become both photooxidized and reduced by cyt6 and cytHH indicates that the core of the PSI complex remains intact and the electron transfer chain within the reaction center remains functional, despite the loss of PsaF.
Fig. 7. Photochemical activity of PSI-SMALP. (A–C) Reduction kinetics of P700 reaction center of PSI by various ratios of native cytochrome c6 soluble electron carrier; Cyt : PSI at 1 : 1 (red), 2 : 1 (orange), 5 : 1 (blue) and 10 : 1 (green) are shown. The color curves represent single exponential decay traces overlaid on raw absorbance difference data shown in black (A–D). (A) PSI-SMALP in 50 mM Tris–Cl with 150 mM KCl at pH = 9.5 (SMA buffer). P700+ reduction kinetics of PSI-DDM in buffer S (B) and in buffer A (C). (D) Observed reduction rate constants from single exponential decay curves plotted against molar ratio of cytochrome c6 for PSI-DDM in DDM buffer (red) and in SMA buffer (blue), and for PSI-SMALP in SMA buffer (green). (E) P700 reduction curves for PSI-DDM (red) and PSI-SMALP (green) using horse heart cytochrome at 50 : 1 (solid lines) and 100 : 1 (dotted lines) ratios of Cyt/PSI. (F) Observed P700 reduction rate of PSI-DDM and PSI-SMALP with horse heart cytochrome. Results shown in panels A–C and E represent the average of 7 technical replicates of the same sample.
The rate of P700+ reduction seen in Fig. 7 also suggests that the iron sulfur cluster containing subunit PsaC (specifically FA and FB) is intact and functional. Without these FeS clusters, charge recombination to reduce P700+ would dominate. This process has been previously shown to occur on the timescale of 750 μs, which would be shown via ultrafast reduction of P700+.60,61Fig. 7 clearly shows reduction of P700+ occurring at the millisecond timescale, indicative of externally facilitated reduction of the reaction center. The stromal subunits (PsaC, D and E) have also been shown to be very tightly associated to the PSI core complex, requiring treatment with 3.5–6.8 M chaotropic agents (depending on strength of chaotrope) to dissociate these subunits.62 The back reaction observed by flash photolysis of the PSI-SMALP in this paper has the characteristic slow back reaction kinetics (Fig. 7), suggesting the presence of FA and FB, and is not similar to the stromal-subunit depleted core complex. This is also supported by the presence of the PsaD and E subunits in the silver stain. It would be very surprising of PsaC was lost without the accompanied loss of PsaD and E which are even more peripherally associated on the stromal surface. For these reasons, though it is difficult to resolve these small molecular weight subunits by SDS-PAGE (Fig. 5), we are confident these subunits are retained in the PSI-SMALP.
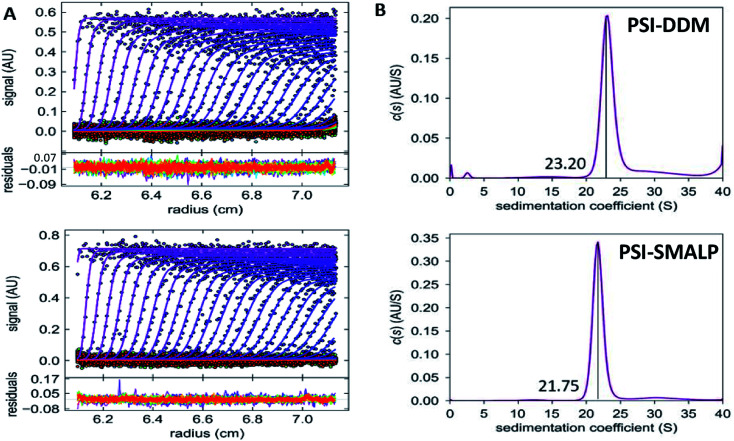
AUC was used to determine the difference in sedimentation coefficient between PSI-SMALP and PSI-DDM (Fig. 8). Particle movement was tracked by monitoring chlorophyll absorbance at 680 nm. From sedimentation velocities for PSI-DDM (Fig. 8A, top) and PSI-SMALP (Fig. 8A, bottom), sedimentation coefficients of 23.20 and 21.75 svedbergs for PSI-DDM (Fig. 8B, top) and PSI-SMALP (Fig. 8B, bottom), respectively. These data imply that the PSI-SMALP is either less dense or experiences more resistance while migrating through the chamber. The former possibility is in line with previous studies that report increased lipid to protein ratios in SMALPs, which would cause a decrease in particle density. The latter may be caused by extended geometry, or larger overall particle size. Further investigation is needed to discern which of these (or both) is the case for PSI-SMALP, but these sedimentation coefficients correlate well previously reported values for trimeric PSI and differ from monomeric, dimeric and tetrameric forms.63,64 Further, the AUC results in Fig. 8 also suggest the protein preparations are clean, containing only trimeric PSI particles.
Fig. 8. Sedimentation velocity of SMALPs and DDM-PSI. (A) Sedimentation velocity data for PSI-DDM (top) and PSI-SMALP (bottom). (B) c(s) distributions for PSI-DDM (top) and PSI-DDM (bottom), sedimentation coefficients are labeled.
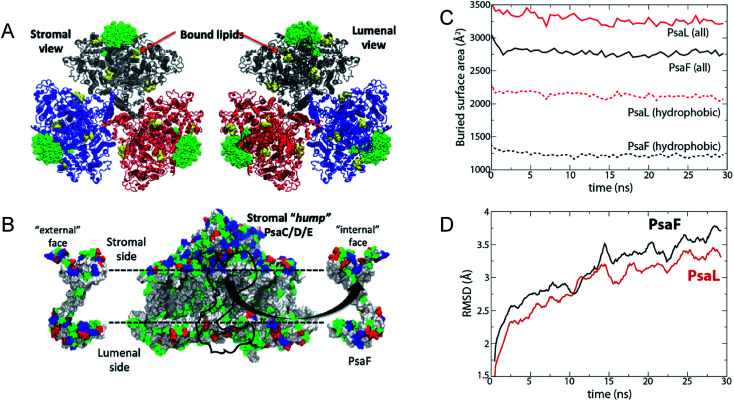
Molecular dynamics simulations demonstrate that the buried surface area of the PsaF subunit when bound to PsaB is much lower than that of an interior subunit such as PsaL (∼2750 Å2 for each PsaF copy on average and ∼3250 Å2 for each PsaL (Fig. 9C)). The hydrophobic buried surface area shows an even greater difference with that for PsaL being nearly twice as large as that for PsaF (∼2000 Å2 for PsaL and ∼1250 Å2 for PsaF). At 15 cal mol−1 Å−2 buried hydrophobic area, this difference would lead to as much as an 11 kcal mol−1 lower binding free energy for a single copy of PsaF compared to PsaL. The root mean-square deviation (RMSD) of PsaF is also slightly higher than PsaL over the 30 ns simulation (Fig. 9D). The reduced buried surface area and increased RMSD suggest that PsaF is less tightly associated to the PSI complex than other subunits, and therefore dissociates more easily.
Fig. 9. Simulated interaction energy analysis of PsaF bound to PSI. Simulated system. (A) The three units of the PSI trimer are shown in blue, grey, and red, respectively. PsaF from each unit is highlighted in a green, space-filling representation. Co-crystallized lipids are shown in yellow; although they were present in the simulation, chlorophylls, carotenoids, and iron–sulfur clusters are not shown. (B) PsaF interactions. On the left and right are the outward- and inward-facing sides of PsaF, respectively. Its position on the rest of the PSI monomer is indicated by a black outline on the central image. Residues are colored according to polarity: blue (basic), red (acidic), green (polar, uncharged), and white (hydrophobic). The horizontal dotted lines indicate the position of the membrane. (C) Total (solid lines) and hydrophobic (dotted lines) buried surface area averaged over three copies of PsaF (black) and PsaL (red). (D) Root mean-square deviation (RMSD) of PsaF (red) and PsaL (black).
Discussion
The crystal structure for PSI trimer from Te (PDB ID: 1JB0) shows galactolipids bound in the space between the monomers of the protein complex, which raises some very interesting questions.65 It has long been suspected that these membrane lipids may serve a functional purpose to the overall activity of the protein complex.66 In the case of PSI specifically, this reaction center resides within a specialized membrane, the thylakoid membrane, that is enriched in galactolipids. Evidence that these unique lipids may be vital to the function of these embedded protein complexes has become more prevalent in recent decades. For example, in higher plants, MGDG has been shown to be critical for the dissociation of violaxanthin from LHC-II (and subsequent deepoxidation) to create the NPQ active form, zeaxanthin.67 Further, the removal or replacement of thylakoid lipids by detergent extraction has been shown to cause the loss of oligomeric structure and activity of membrane protein complexes.68–70 Therefore, thinking of these membrane bound reaction centers as “lipoproteins” or “proteo-lipid assemblies” may be more accurate when attempting to describe their function. In the SMALP field, it's recently been shown that GPCRs exhibit higher activity in vitro when extracted with SMA system as compared to detergent isolation methods, supporting the hypothesis that preservation of the native membrane environment influences activity of membrane bound proteins.48,66,71
The SMA buffer used in this study (pH = 9.5), lack of divalent ions and high monovalent ionic strength (125 mM KCl), make this buffer system non-physiological for most proteins. Beyond that, it makes the direct comparison of DDM and SMA extracted proteins in certain downstream analyses difficult, as seen in our P700+ reduction kinetics (Fig. 7). The nature of this experiment allowed us to put the fully formed PSI-DDM particle in the SMA buffer system and directly measure this buffer effect. However, when looking at the complex picture of protein isolation from membrane systems, surfactant lipid exchange may be hindered at the conditions required for SMA solubilization. For example, it has been shown that at constant ionic strength, surface charge density of DDM foams increases, which may change the way in which DDM–lipid mixed micelles formed, thereby altering their action during protein isolation.72In lieu of re-optimizing DDM extraction methods at elevated pH, the current study has focused on the properties of the fully formed particles, in their respective buffer systems. For these reasons, it is worth noting the recent creation of amphiphilic copolymers that are able to operate at physiological pH, such as SMA-ED and SMAd-A from the Ramamoorthy group.51 It's of the utmost importance to expand upon these pioneering formulations, and this feat will be more easily achieved once the mechanism of SMALP formation is fully elucidated.
We have shown that SMA 1440 performs most efficiently for the extraction of trimeric PSI-SMALP from Te. There are 3 ways in which the 1440 formulation differs from 2625 and 3000; it has the shortest copolymer length, the least amount of styrene compared to maleic acid, and it's partially functionalized (esterified) with butoxyethanol. More specifically, SMA 1440 as shown in Fig. 1A is ∼75% monoester, meaning the ratio of z : y moieties is approximately 3 : 1. Overall, this translates to roughly 37.5% conversion of the carboxylates to esters along the length of the polymer. Of these physicochemical properties, SMA 1440 stands apart from most previously reported SMA copolymer formulations that have been used for protein isolation.
To our knowledge, trimeric PSI is the largest membrane protein complex to be encapsulated within a SMALP to date. These findings suggest that smaller copolymer fragments, with low S : MA ratio and increased hydrophobicity through ester formation with alkoxy groups are optimum for reaction center isolation from highly saturated, galactolipid rich membrane systems. This finding is in line with the 2016 study from Morrison et al. with regard to copolymer length and a styrene rich S : MA ratio.73 A recent investigation published by Brady et al. in 2018 posits that the supramolecular structure of SMA copolymer aggregates free in solution may be important in the interaction of these copolymers with native membranes. In this study, the SMA 1440 particles were found to be significantly larger in size compared to SMA 1000 (Cray Valley), 25 010 and 30 010 (Polyscope). These 1440 particles were less stable at elevated temperature, presumably due to the butyl tail of the ester needing to be buried in the core.74 It's possible that these larger, less stable SMA 1440 aggregates are more prone to unraveling after partitioning to the interface, but why this would be more pronounced in galactolipid membranes rather than phospholipid membranes requires further investigation. However, this hypothesis of the SMA supramolecular aggregate structure playing a role at the membrane interface lends a possible explanation for the lack of direct correlation between physicochemical properties of SMA copolymers and protein solubilization efficacy.
The loss of PsaF in the PSI-SMALP, and its presence in the top of the sucrose gradient (Fig. 7) suggests that this subunit is lost following SMALP formation. This loss of PsaF, coupled with the significant decrease in reduction rate for the PSI-SMALP compared to PSI-DDM in their respective buffer systems seen in Fig. 7, initially seemed to be correlated. Cyanobacterial PsaF contains three helices protruding from the lumenal face of the PSI complex.65 In the green algae Chlamydomonas reinhardtii it has been shown that the lumenal extension in algal PsaF is necessary for recognition, binding and fast electron transfer from cytochrome and plastocyanin to PSI.75–79 Hippler et al. went a step further in a study published in 1999, showing that insertion of the N-terminal extension from algal PsaF into the cyanobacterium Synechococcus elongatus facilitated binding of algal cyt and plastocyanin in this Cyanobacteria.80 However, the lack of PsaF in Cyanobacteria has been shown in numerous studies to have no effect on reduction of the complex via native or non-native cytochromes, indicating that PsaF is not involved in cytochrome docking or electron transfer in Cyanobacteria.81–85
As shown in Fig. 2, SMA copolymers possess a well-documented drawback, most formulations exhibit a sensitivity to divalent cations and become insoluble as pH drops, with more hydrophobic (less charged) polymers aggregating closer to neutral pH.51–53,74,86,87 When using a buffer consisting of 150 mM KCl, devoid of divalent ions and pH = 9.5 (room temperature) for laser flash photolysis measurements, the reduction rate decreased for both PSI-SMALP and PSI-DDM (Fig. 7A and B). Recently in Te, binding energy between PSI and cyt6 has been shown to be repulsive at low ionic strength and pH 8.84,88 Kölsch et al. demonstrate that this repulsion is caused in part by negatively charged side chains on the lumenal loop of PsaA and cyt6. Further, this shielding effect is more pronounced for divalent ions compared to monovalent ions of equivalent ionic strength, suggesting the ability of Mg2+ can bridge negative charges on PSI sidechains.88 We conclude that the decrease in reduction kinetics for PSI-SMALP and PSI-DDM in this buffer system are attributed to elevated pH and a lack of divalent ions, not the absence of the PsaF subunit (Fig. 7A and B). Elevated pH causes acidic residues (specifically D and E) to be fully deprotonated, causing the luminal surface of PSI to be negatively charged. In the absence of divalent ions, which can bridge the negative charges of adjacent carboxylates, the electron laden cyt6 is repelled from the luminal face of PSI, slowing reduction kinetics. Our finding that non-native cytHH reduces the PSI-SMALP at a faster rate as compared to PSI-DDM is very interesting. The non-specific binding and lack of native electrostatics driving the interaction of these two proteins explains why the buffer effect seen with native cty6 is absent in this experiment.88 However, the fact that mitigating this variable allows for the reduction of PSI in the SMALP at nearly twice the rate as PSI-DDM may suggest that the translocation of the high energy electron through the protein complex following photoexcitation may be faster in the SMALP compared to DDM isolation, further investigation is needed to evaluate this hypothesis.
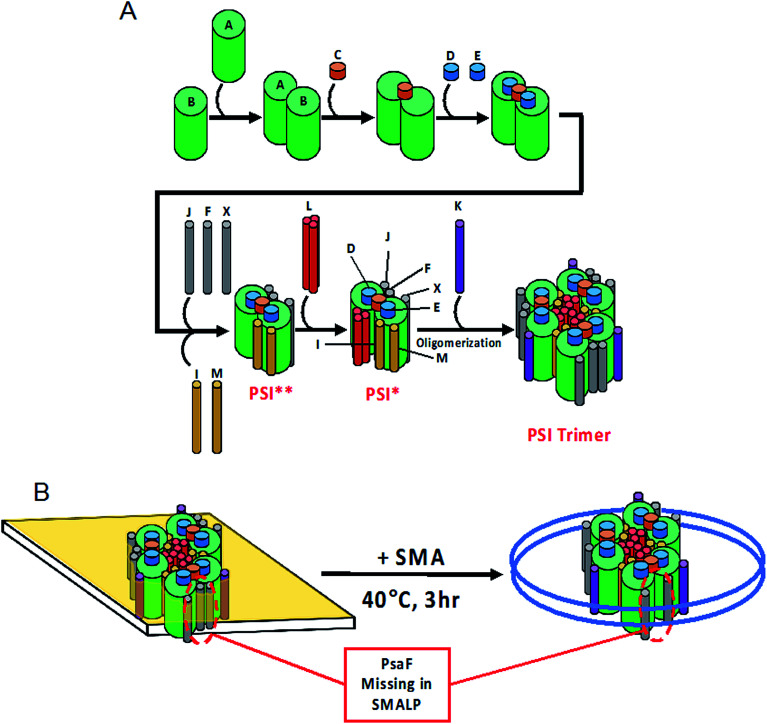
As far back as 2003, it has been known that PsaF could be partially lost during detergent isolation of PSI.89 Our findings suggest that lost more readily from SMALPs as compared to DDM isolation, leading us to speculate further into the dynamic interaction of PsaF to the PSI core complex. The crystal structure for PSI from Te shows that PsaF and the PsaB core subunit both associate to the same carotenoid molecule located at the binding interface.65 This carotenoid may act as a molecular glue, holding PsaF to PsaB by relatively weak hydrophobic interactions. The MD simulation shown here (Fig. 9) indicates that the buried hydrophobic surface area of PsaF bound to the outer edge of the PSI monomers is lower (less favorable) compared to that of PsaL, which is buried in the interior of the complex. The exact order of assembly for trimeric PSI in Cyanobacteria is still unclear. A combination of hypotheses from Hippler et al.,90 and Dühring et al.,91 and based on the crystal structure for PSI from Te (1JB0 (ref. 65)), is depicted in Fig. 10A. Following PSI-SMALP formation, PsaF dissociates from the particle (Fig. 10B). We hypothesize that the lipid annulus, predicted to be present in the PSI-SMALP, allows space for PsaF to dissociate from the PSI complex. In the case of PSI-DDM, PsaF dissociation occurs much more slowly due to the very small hydrophobic cage in the PSI_DDM toroidal micelle. It has been proposed that large amounts of PsaF may exist unbound in the thylakoid membrane and that PsaF may freely partition between this pool and the bound state.91 This would set up a dynamic equilibrium, in which the PsaF content of the thylakoid would drive the amount of PsaF that is bound to the PSI complex, thus potentially regulating PSI activity, possibly via facilitating attachment of the phycobilisome antennae complex, though the exact function of this subunit has yet to be discovered. Taken together, these results suggest a mechanism by which PsaF associates very dynamically, and the amount of bound PsaF is dependent on the concentration of free PsaF in the thylakoid, as described by Le Chatelier's principle. Therefore, concentration of PsaF in the thylakoid may act as a transcriptionally driven rheostat to modulate PSI activity, by means other than docking of electron cyt6 to the lumenal face of PSI, as our data agrees with previous studies that suggest PsaF is not involved in this process. The addition of SMA technology as a means to study these complex membrane environments has great import in photosynthetic research, serving as an indispensable tool in the elucidation of these multicomponent systems.
Fig. 10. Model of PSI-SMALPs. (A) Proposed order of assembly for PSI subunits. Asterisks denote short-lived intermediate complexes. (B) Upon incubation of native thylakoid membranes with SMA 1440 at 40 °C for at least 3 hours, PSI-SMALPs are formed. These particles lack PsaF subunit normally associated to the outer edge of the protein complex.
Following with the long-standing paradigm in protein chemistry, structure gives rise to function, the preservation of the native environment of PSI in the SMALP system is very exciting. The key difference between SMA and DDM extractions with regard to this structure/function relationship deals with the preservation of native lipids surrounding and throughout membrane protein complexes. Decreased migration of PSI-SMALP into BN-PAGE suggest this particle is larger in size than PSI-DDM (Fig. 3). Further, decreased sedimentation of PSI-SMALP compared to PSI-DDM by AUC suggests PSI-SMALPs either contain more lipids, resulting in a less dense complex, and/or exhibit a larger or more extended shape than DDM extracted PSI (Fig. 7). This finding agrees with the overall consensus in the field that proteins embedded within SMALPs are disc shaped, retaining an annulus of native lipids. However, the lipid profile PSI-SMALPs compared to PSI-DDM is yet to be determined.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations
- 1,2-Dipalmitoyl-phosphatidylcholine
DPPC
- 1,2-Distearoyl-monogalactosyl-diglyceride
MGDG
- Analytical ultracentrifugation
AUC
- Blue native polyacrylamide gel electrophoresis
BN-PAGE
- Coomassie Brilliant Blue stain
CBB
- Cytochrome c6
cyt6
- Dispersity
Đ
- Electron paramagnetic resonance
EPR
- Horse heart cytochrome c
cytHH
- Joliot type spectrophotometer
JTS-100
- Membrane scaffold proteins
MSPs
- n-Dodecyl-β-d-maltoside
DDM
- Number weighted molecular weight
M N
- Photosystem I
PSI
- Root mean-square deviation
RMSD
- SMA lipid particle
SMALP
- Sodium dodecyl sulfate PAGE
SDS-PAGE
- Styrene to maleic acid ratio
S : MA
- Styrene–maleic acid
SMA
- Thermosynechococcus elongatus
Te
- Weight weighted molecular weight
M W
Supplementary Material
Acknowledgments
We would like to thank Dr Khoa Nguyen for production of the polyclonal antibody against PsaF and contributions of the SDS-PAGE and immunoblot (Fig. 5 and 6) to this work. We would also like to thank Alexandra Teodor and Dr Michael Vaughn (Bio-logics, USA) for their assistance and training on the JTS-100 for reduction kinetics analysis (Fig. 7). Additionally, we thank Jyotirmoy Mondal and Jon Nguyen for culturing and harvesting Te used in this work. Support has been provided from the Gibson Family Foundation, the Bredesen Center for Interdisciplinary Research and Education, the Dr Donald L. Akers Faculty Enrichment Fellowship, and National Science Foundation (DGE-0801470 and EPS-1004083) support to B. D. B. In addition, N. G. B. and B. D. B. have been supported via a JDRD Award from UTK/ORNL Science Alliance to B. D. B., M. L. has been supported as a CIRE Fellow at University of Tennessee, Knoxville. N. G. B. has also been supported via a Penley Fellowship. J. C. G. acknowledges support from the National Science Foundation (MCB-1452464). Computational resources were provided via the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF Grant (OCI-1053575).
References
- Wallin E. von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Engelman D. M. Gerstein M. Genomic analysis of membrane protein families: abundance and conserved motifs. Genome Biol. 2002;3(10):research0054. doi: 10.1186/gb-2002-3-10-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert U. Xia Y. Royce T. E. Goh C. S. Liu Y. Senes A. Yu H. Zhang Z. L. Engelman D. M. Gerstein M. Computational analysis of membrane proteins: genomic occurrence, structure prediction and helix interactions. Q. Rev. Biophys. 2004;37(2):121–146. doi: 10.1017/S003358350400397X. [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D. Binnewies T. T. Hallin P. F. Ussery D. W. Genome update: prediction of membrane proteins in prokaryotic genomes. Microbiology. 2005;151(Pt 7):2119–2121. doi: 10.1099/mic.0.28181-0. [DOI] [PubMed] [Google Scholar]
- Arinaminpathy Y. Khurana E. Engelman D. M. Gerstein M. B. Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov. Today. 2009;14(23–24):1130–1135. doi: 10.1016/j.drudis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. V. Gianoulis T. A. Bjornson R. D. Yip K. Y. Engelman D. M. Gerstein M. B. Analysis of membrane proteins in metagenomics: networks of correlated environmental features and protein families. Genome Res. 2010;20(7):960–971. doi: 10.1101/gr.102814.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski M. Niedzialkowska E. Zimmerman M. D. Minor W. The impact of structural genomics: the first quindecennial. J. Struct. Funct. Genomics. 2016;17(1):1–16. doi: 10.1007/s10969-016-9201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbus L. Post-expression strategies for structural investigations of membrane proteins. Curr. Opin. Struct. Biol. 2015;32:131–138. doi: 10.1016/j.sbi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavito R. M. Ferguson-Miller S. Detergents as tools in membrane biochemistry. J. Biol. Chem. 2001;276(35):32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- Arnold T. Linke D. The use of detergents to purify membrane proteins. Curr. Protoc. Protein Sci. 2008:4.8.1–4.8.30. doi: 10.1002/0471140864.ps0408s53. [DOI] [PubMed] [Google Scholar]
- Prive G. G. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41(4):388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Arachea B. T. Sun Z. Potente N. Malik R. Isailovic D. Viola R. E. Detergent selection for enhanced extraction of membrane proteins. Protein Expression Purif. 2012;86(1):12–20. doi: 10.1016/j.pep.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Bayburt T. H. Sligar S. G. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584(9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civjan N. R. Bayburt T. H. Schuler M. A. Sligar S. G. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35(3):556–563. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- Ritchie T. K. Grinkova Y. V. Bayburt T. H. Denisov I. G. Zolnerciks J. K. Atkins W. M. Sligar S. G. Chapter 11 – reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty M. T. Wilcox K. C. Klein W. L. Sligar S. G. Nanodisc-solubilized membrane protein library reflects the membrane proteome. Anal. Bioanal. Chem. 2013;405(12):4009–4016. doi: 10.1007/s00216-013-6790-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. Wilkens S. Biolayer interferometry of lipid nanodisc-reconstituted yeast vacuolar H+-ATPase. Protein Sci. 2017;26(5):1070–1079. doi: 10.1002/pro.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. J. Finka R. Smith C. Lin Y. P. Dafforn T. Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009;131(22):7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- Jamshad M. Lin Y. P. Knowles T. J. Parslow R. A. Harris C. Wheatley M. Poyner D. R. Bill R. M. Thomas O. R. Overduin M. Dafforn T. R. Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem. Soc. Trans. 2011;39(3):813–818. doi: 10.1042/BST0390813. [DOI] [PubMed] [Google Scholar]
- Rajesh S. Knowles T. Overduin M. Production of membrane proteins without cells or detergents. New Biotechnol. 2011;28(3):250–254. doi: 10.1016/j.nbt.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Dorr J. M. Koorengevel M. C. Schafer M. Prokofyev A. V. Scheidelaar S. van der Cruijsen E. A. Dafforn T. R. Baldus M. Killian J. A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl. Acad. Sci. U. S. A. 2014;111(52):18607–18612. doi: 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr J. M. Scheidelaar S. Koorengevel M. C. Dominguez J. J. Schafer M. van Walree C. A. Killian J. A. The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J. 2016;45(1):3–21. doi: 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury D. J. Scheidelaar S. van Grondelle R. Killian J. A. Jones M. R. Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability. Angew. Chem., Int. Ed. Engl. 2014;53(44):11803–11807. doi: 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. J. Frankel L. K. Bricker T. M. High Yield Non-detergent Isolation of Photosystem I-Light-harvesting Chlorophyll II Membranes from Spinach Thylakoids: Implications for the Organization of the Ps I Antennae in Higher Plants. J. Biol. Chem. 2015;290(30):18429–18437. doi: 10.1074/jbc.M115.663872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold D. A. Babcock G. T. Yocum C. F. A highly resolved, oxygen-evolving photosystem II preparation from spinach thylakoid membranes. FEBS Lett. 1981;134(2):231–234. doi: 10.1016/0014-5793(81)80608-4. [DOI] [Google Scholar]
- Schiller H. Dau H. Preparation protocols for high-activity photosystem II membrane particles of green algae and higher plants, pH dependence of oxygen evolution and comparison of the S2-state multiline signal by X-band EPR spectroscopy. J. Photochem. Photobiol., B. 2000;55(2):138–144. doi: 10.1016/S1011-1344(00)00036-1. [DOI] [PubMed] [Google Scholar]
- MacGregor-Chatwin C. Sener M. Barnett S. F. H. Hitchcock A. Barnhart-Dailey M. C. Maghlaoui K. Barber J. Timlin J. A. Schulten K. Hunter C. N. Lateral Segregation of Photosystem I in Cyanobacterial Thylakoids. Plant Cell. 2017;29(5):1119–1136. doi: 10.1105/tpc.17.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor-Chatwin C. Jackson P. J. Sener M. Chidgey J. W. Hitchcock A. Qian P. Mayneord G. E. Johnson M. P. Luthey-Schulten Z. Dickman M. J. Scanlan D. J. Hunter C. N. Membrane organization of photosystem I complexes in the most abundant phototroph on Earth. Nat. Plants. 2019;5(8):879–889. doi: 10.1038/s41477-019-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K. Vaughn M. Frymier P. Bruce B. D. J. P. R. In vitro kinetics of P700+ reduction of Thermosynechococcus elongatus trimeric photosystem I complexes by recombinant cytochrome c6 using a Joliot-type LED spectrophotometer. Photosynth. Res. 2017;131(1):79–91. doi: 10.1007/s11120-016-0300-8. [DOI] [PubMed] [Google Scholar]
- Sakthivel K. Watanabe T. Nakamoto H. A small heat-shock protein confers stress tolerance and stabilizes thylakoid membrane proteins in Cyanobacteria under oxidative stress. Arch. Microbiol. 2009;191(4):319–328. doi: 10.1007/s00203-009-0457-z. [DOI] [PubMed] [Google Scholar]
- Iwamura T. Nagai H. Ichimura S.-E. Improved Methods for Determining Contents of Chlorophyll, Protein, Ribonucleic Acid, and Deoxyribonucleic Acid in Planktonic Populations. Int. Rev. Gesamten Hydrobiol. Hydrogr. 1970;55(1):131–147. doi: 10.1002/iroh.19700550106. [DOI] [Google Scholar]
- Schägger H. von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199(2):223–231. doi: 10.1016/0003-2697(91)90094-A. [DOI] [PubMed] [Google Scholar]
- Wittig I. Braun H.-P. Schägger H. Blue native PAGE. Nat. Protoc. 2006;1:418. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Schägger H. Tricine–SDS-PAGE. Nat. Protoc. 2006;1:16. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Nesterenko M. V. Tilley M. Upton S. J. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods. 1994;28(3):239–242. doi: 10.1016/0165-022X(94)90020-5. [DOI] [PubMed] [Google Scholar]
- Kubota H. Sakurai I. Katayama K. Mizusawa N. Ohashi S. Kobayashi M. Zhang P. Aro E.-M. Wada H. Purification and characterization of photosystem I complex from Synechocystis sp. PCC 6803 by expressing histidine-tagged subunits. Biochim. Biophys. Acta, Bioenerg. 2010;1797(1):98–105. doi: 10.1016/j.bbabio.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W. Dalke A. Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Wu E. L. Cheng X. Jo S. Rui H. Song K. C. Dávila-Contreras E. M. Qi Y. Lee J. Monje-Galvan V. Venable R. M. Klauda J. B. Im W. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35(27):1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C. Braun R. Wang W. Gumbart J. Tajkhorshid E. Villa E. Chipot C. Skeel R. D. Kale L. Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda J. B. Venable R. M. Freites J. A. O'Connor J. W. Tobias D. J. Mondragon-Ramirez C. Vorobyov I. MacKerell Jr A. D. Pastor R. W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R. B. Zhu X. Shim J. Lopes P. E. Mittal J. Feig M. MacKerell Jr A. D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le R. K. Harris B. J. Iwuchukwu I. J. Bruce B. D. Cheng X. Qian S. Heller W. T. O'Neill H. Frymier P. D. Analysis of the solution structure of Thermosynechococcus elongatus photosystem I in n-dodecyl-beta-D-maltoside using small-angle neutron scattering and molecular dynamics simulation. Arch. Biochem. Biophys. 2014;550–551:50–57. doi: 10.1016/j.abb.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Gumbart J. Schulten K. Molecular dynamics studies of the archaeal translocon. Biophys. J. 2006;90(7):2356–2367. doi: 10.1529/biophysj.105.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotych O. Mondal J. Gattás-Asfura K. M. Hendricks J. Bruce B. D. Evaluation of commercially available styrene-co-maleic acid polymers for the extraction of membrane proteins from spinach chloroplast thylakoids. Eur. Polym. J. 2018;114:485–500. doi: 10.1016/j.eurpolymj.2018.10.035. [DOI] [Google Scholar]
- Esmaili M. Overduin M. Membrane biology visualized in nanometer-sized discs formed by styrene maleic acid polymers. Biochim. Biophys. Acta, Biomembr. 2017;1860(2):257–263. doi: 10.1016/j.bbamem.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshad M., Lin Y.-P., Knowles T. J., Parslow R. A., Harris C., Wheatley M., Poyner D. R., Bill R. M., Thomas O. R. and Overduin M., Surfactant-free purification of membrane proteins with intact native membrane environment, Portland Press Limited, 2011 [DOI] [PubMed] [Google Scholar]
- Lee S. C. Knowles T. J. Postis V. L. Jamshad M. Parslow R. A. Lin Y.-p. Goldman A. Sridhar P. Overduin M. Muench S. P. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11(7):1149. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- Hall S. C. L. Tognoloni C. Price G. J. Klumperman B. Edler K. J. Dafforn T. R. Arnold T. Influence of Poly(styrene-co-maleic acid) Copolymer Structure on the Properties and Self-Assembly of SMALP Nanodiscs. Biomacromolecules. 2018;19(3):761–772. doi: 10.1021/acs.biomac.7b01539. [DOI] [PubMed] [Google Scholar]
- Scheidelaar S. Koorengevel M. C. Pardo J. D. Meeldijk J. D. Breukink E. Killian J. A. Molecular model for the solubilization of membranes into nanodisks by styrene maleic acid copolymers. Biophys. J. 2015;108(2):279–290. doi: 10.1016/j.bpj.2014.11.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravula T. Hardin N. Z. Ramadugu S. K. Ramamoorthy A. pH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs. Langmuir. 2017;33(40):10655–10662. doi: 10.1021/acs.langmuir.7b02887. [DOI] [PubMed] [Google Scholar]
- Gulati S. Jamshad M. Knowles T. J. Morrison K. A. Downing R. Cant N. Collins R. Koenderink J. B. Ford R. C. Overduin M. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J. 2014;461(2):269–278. doi: 10.1042/BJ20131477. [DOI] [PubMed] [Google Scholar]
- Morrison K. A. Akram A. Mathews A. Khan Z. A. Patel J. H. Zhou C. Hardy D. J. Moore-Kelly C. Patel R. Odiba V. Membrane protein extraction and purification using styrene–maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem. J. 2016;473(23):4349–4360. doi: 10.1042/BCJ20160723. [DOI] [PubMed] [Google Scholar]
- Rögner M. Nixon P. J. Diner B. A. Purification and characterization of photosystem I and photosystem II core complexes from wild-type and phycocyanin-deficient strains of the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 1990;265(11):6189–6196. [PubMed] [Google Scholar]
- Iwai M. Takizawa K. Tokutsu R. Okamuro A. Takahashi Y. Minagawa J. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature. 2010;464:1210. doi: 10.1038/nature08885. [DOI] [PubMed] [Google Scholar]
- Minagawa J. Dynamic reorganization of photosynthetic supercomplexes during environmental acclimation of photosynthesis. Frontiers in Plant Science. 2013;4:513. doi: 10.3389/fpls.2013.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Semchonok D. A. Boekema E. J. Bruce B. D. Characterization and evolution of tetrameric photosystem I from the thermophilic cyanobacterium Chroococcidiopsis sp. TS-821. Plant Cell. 2014;26(3):1230–1245. doi: 10.1105/tpc.113.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosinotto T. Breton J. Bassi R. Croce R. The nature of a chlorophyll ligand in Lhca proteins determines the far red fluorescence emission typical of photosystem I. J. Biol. Chem. 2003;278(49):49223–49229. doi: 10.1074/jbc.M309203200. [DOI] [PubMed] [Google Scholar]
- Franck J. Remarks on the long-wave-length limits of photosynthesis and chlorophyll fluorescence. Proc. Natl. Acad. Sci. U. S. A. 1958;44(9):941–948. doi: 10.1073/pnas.44.9.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüneberg J. Fromme P. Jekow P. Schlodder E. J. F. l. Spectroscopic characterization of PS I core complexes from thermophilic Synechococcus sp.: identical reoxidation kinetics of A−1 before and after removal of the iron-sulfur-clusters FA and FB. FEBS Lett. 1994;338(2):197–202. doi: 10.1016/0014-5793(94)80364-1. [DOI] [PubMed] [Google Scholar]
- Kruip J. Boekema E. J. Bald D. et al., Isolation and Structural Characterization of Monomeric and Trimeric Photosystem I Complexes (P700*FA/FB and P700*fFrxo)m the Cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 1993;268:23353–23360. [PubMed] [Google Scholar]
- Parrett K. G. Mehari T. Warren P. G. Golbeck J. H. Purification and properties of the intact P-700 and F x-containing photosystem I core protein. Biochim. Biophys. Acta, Bioenerg. 1989;973(2):324–332. doi: 10.1016/S0005-2728(89)80439-6. [DOI] [PubMed] [Google Scholar]
- Iwuchukwu I. J. Vaughn M. Myers N. O'neill H. Frymier P. Bruce B. D. Self-organized photosynthetic nanoparticle for cell-free hydrogen production. Nat. Nanotechnol. 2010;5(1):73. doi: 10.1038/nnano.2009.315. [DOI] [PubMed] [Google Scholar]
- Li M. Semchonok D. A. Boekema E. J. Bruce B. D. Characterization and evolution of tetrameric photosystem I from the thermophilic cyanobacterium Chroococcidiopsis sp. TS-821. Plant Cell. 2014;26(3):1230–1245. doi: 10.1105/tpc.113.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. Fromme P. Witt H. T. Klukas O. Saenger W. Krauß N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature. 2001;411:909. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- Hasan S. S. Cramer W. A. Internal lipid architecture of the hetero-oligomeric cytochrome b6f complex. Structure. 2014;22(7):1008–1015. doi: 10.1016/j.str.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller S. Latowski D. Jemiola-Rzeminska M. Wilhelm C. Strzalka K. Goss R. The main thylakoid membrane lipid monogalactosyldiacylglycerol (MGDG) promotes the de-epoxidation of violaxanthin associated with the light-harvesting complex of photosystem II (LHCII) Biochim. Biophys. Acta. 2010;1797(3):414–424. doi: 10.1016/j.bbabio.2009.12.011. [DOI] [PubMed] [Google Scholar]
- le Maire M. Champeil P. MÖller J. V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta, Biomembr. 2000;1508(1):86–111. doi: 10.1016/S0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Esmann M. Skou J. C. Christiansen C. Solubilization and molecular weight determination of the (Na+ + K+)-ATPase from rectal glands of Squalus acanthias. Biochim. Biophys. Acta, Enzymol. 1979;567(2):410–420. doi: 10.1016/0005-2744(79)90127-X. [DOI] [PubMed] [Google Scholar]
- Breyton C. Tribet C. Olive J. Dubacq J.-P. Popot J.-L. Dimer to Monomer Conversion of the Cytochrome b6f Complex Causes and Consequences. J. Biol. Chem. 1997;272(35):21892–21900. doi: 10.1074/jbc.272.35.21892. [DOI] [PubMed] [Google Scholar]
- Wheatley M. Charlton J. Jamshad M. Routledge S. J. Bailey S. La-Borde P. J. Azam M. T. Logan R. T. Bill R. M. Dafforn T. R. Poyner D. R. GPCR-styrene maleic acid lipid particles (GPCR-SMALPs): their nature and potential. Biochem. Soc. Trans. 2016;44(2):619–623. doi: 10.1042/BST20150284. [DOI] [PubMed] [Google Scholar]
- Stubenrauch C. Cohen R. Exerowa D. J. L. A pH-Study of n-Dodecyl-β-d-maltoside Foam Films. Langmuir. 2007;23(4):1684–1693. doi: 10.1021/la062310m. [DOI] [PubMed] [Google Scholar]
- Morrison K. A. Akram A. Mathews A. Khan Z. A. Patel J. H. Zhou C. Hardy D. J. Moore-Kelly C. Patel R. Odiba V. Membrane protein extraction and purification using styrene-maleic acid (SMA) co-polymer: effect of variations in polymer structure. Biochem. J. 2016;473:4349–4360. doi: 10.1042/BCJ20160723. [DOI] [PubMed] [Google Scholar]
- Brady N. G. Qian S. Bruce B. D. Analysis of Styrene Maleic Acid Alternating Copolymer Supramolecular Assemblies in Solution by Small Angle X-Ray Scattering. Eur. Polym. J. 2018;111:178–184. doi: 10.1016/j.eurpolymj.2018.11.034. [DOI] [Google Scholar]
- Hippler M. Drepper F. Farah J. Rochaix J.-D. Fast Electron Transfer from Cytochrome c6 and Plastocyanin to Photosystem I of Chlamydomonas reinhardtii Requires PsaF. Biochemistry. 1997;36(21):6343–6349. doi: 10.1021/bi970082c. [DOI] [PubMed] [Google Scholar]
- Hippler M. Drepper F. Haehnel W. Rochaix J.-D. The N-terminal domain of PsaF: precise recognition site for binding and fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 1998;95(13):7339–7344. doi: 10.1073/pnas.95.13.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengis C. Nelson N. Subunit Structure of Chloroplast Photosystem I Reaction Center. J. Biol. Chem. 1977;252(13):4564–4569. [PubMed] [Google Scholar]
- Farah J. Rappaport F. Choquet Y. Joliot P. Rochaix J.-D. Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunit. EMBO J. 1995;14(20):4976–4984. doi: 10.1002/j.1460-2075.1995.tb00180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler M. Reichert J. Sutter M. Zak E. Altschmied L. Schröer U. Herrmann R. Haehnel W. The plastocyanin binding domain of photosystem I. EMBO J. 1996;15(23):6374–6384. doi: 10.1002/j.1460-2075.1996.tb01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler M. Drepper F. Rochaix J.-D. Mühlenhoff U. Insertion of the N-terminal part of PsaF from Chlamydomonas reinhardtii into photosystem I from Synechococcus elongatus enables efficient binding of algal plastocyanin and cytochrome c6. J. Biol. Chem. 1999;274(7):4180–4188. doi: 10.1074/jbc.274.7.4180. [DOI] [PubMed] [Google Scholar]
- Xu Q. Yu L. Chitnis V. P. Chitnis P. R. Function and organization of photosystem I in a cyanobacterial mutant strain that lacks PsaF and PsaJ subunits. J. Biol. Chem. 1994;269(5):3205–3211. [PubMed] [Google Scholar]
- Sun J. Xu W. Hervás M. Navarro J. A. Miguel A. Chitnis P. R. Oxidizing Side of the Cyanobacterial Photosystem I Evidence For Interaction Between The Electron Donor Proteins And A Luminal Surface Helix Of The Psab Subunit. J. Biol. Chem. 1999;274(27):19048–19054. doi: 10.1074/jbc.274.27.19048. [DOI] [PubMed] [Google Scholar]
- Molina-Heredia F. P. Díaz-Quintana A. Hervás M. Navarro J. A. Miguel A. Site-directed Mutagenesis of Cytochrome c6 from Anabaena Species PCC 7119 Identification Of Surface Residues Of The Hemeprotein Involved In Photosystem I Reduction. J. Biol. Chem. 1999;274(47):33565–33570. doi: 10.1074/jbc.274.47.33565. [DOI] [PubMed] [Google Scholar]
- Hatanaka H. Sonoike K. Hirano M. Katoh S. Small subunits of Photosystem I reaction center complexes from Synechococcus elongatus. I. Is the psaF gene product required for oxidation of cytochrome c-553? Biochim. Biophys. Acta, Bioenerg. 1993;1141(1):45–51. doi: 10.1016/0005-2728(93)90187-K. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U. Chauvat F. Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol. Gen. Genet. MGG. 1996;252(1–2):93–100. doi: 10.1007/BF02173209. [DOI] [PubMed] [Google Scholar]
- Scheidelaar S. Koorengevel M. C. van Walree C. A. Dominguez J. J. Dörr J. M. Killian J. A. Effect of Polymer Composition and pH on Membrane Solubilization by Styrene-Maleic Acid Copolymers. Biophys. J. 2016;111(9):1974–1986. doi: 10.1016/j.bpj.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury D. J. K. Scheidelaar S. Foster N. van Grondelle R. Killian J. A. Jones M. R. The effectiveness of styrene-maleic acid (SMA) copolymers for solubilisation of integral membrane proteins from SMA-accessible and SMA-resistant membranes. Biochim. Biophys. Acta, Biomembr. 2017;1859(10):2133–2143. doi: 10.1016/j.bbamem.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch A. Hejazi M. Stieger K. R. Feifel S. C. Kern J. F. Müh F. Lisdat F. Lokstein H. Zouni A. Insights into the binding behavior of native and non-native cytochromes to photosystem I from Thermosynechococcus elongatus. J. Biol. Chem. 2018;293(23):9090–9100. doi: 10.1074/jbc.RA117.000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S., Methods and results in crystallization of membrane proteins, Internat'l University Line, 2003, vol. 4 [Google Scholar]
- Hippler M. Rimbault B. Takahashi Y. Photosynthetic complex assembly in Chlamydomonas reinhardtii. Protist. 2002;153(3):197. doi: 10.1078/1434-4610-00098. [DOI] [PubMed] [Google Scholar]
- Dühring U. Ossenbühl F. Wilde A. Late assembly steps and dynamics of the cyanobacterial photosystem I. J. Biol. Chem. 2007;282(15):10915–10921. doi: 10.1074/jbc.M609206200. [DOI] [PubMed] [Google Scholar]