Abstract
Gene duplication provides raw genetic materials for evolution and potentially novel genes for crop improvement. The two seminal genomic studies of Aegilops tauschii both mentioned the large number of genes independently duplicated in recent years, but the duplication mechanism and the evolutionary significance of these gene duplicates have not yet been investigated. Here, we found that a recent burst of gene duplications (hereafter abbreviated as the RBGD) has probably occurred in all sequenced Triticeae species. Further investigations of the characteristics of the gene duplicates and their flanking sequences suggested that transposable element (TE) activity may have been involved in generating the RBGD. We also characterized the duplication timing, retention pattern, diversification, and expression of the duplicates following the evolution of Triticeae. Multiple subgenome-specific comparisons of the duplicated gene pairs clearly supported extensive differential regulation and related functional diversity among such pairs in the three subgenomes of bread wheat. Moreover, several duplicated genes from the RBGD have evolved into key factors that influence important agronomic traits of wheat. Our results provide insights into a unique source of gene duplicates in Triticeae species, which has increased the gene dosage together with the two polyploidization events in the evolutionary history of wheat.
Keywords: gene duplication, transposable elements, gene dosage, hexaploid wheat, Triticeae, agronomic traits
This study reveals a recent burst of gene duplication in Triticeae species mediated by the activity of transposable elements through examining multiple sequenced Triticeae genomes, and demonstrates the importance of these recent duplicates for differentiating the donor genomes of bread wheat and for increasing the genetic dosage, allowing for the evolution of genes that underlie important wheat agronomic traits.
Introduction
The Triticeae tribe is one of the largest taxonomic groups in the grasses and comprises many globally important food and forage crops like wheat, barley, and rye. Triticeae crop species, especially polyploid wheat, are more widely used in the agriculture of temperate regions than other cereal crops like maize and rice (He et al., 2019; Pont et al., 2019). It is known that hybridization of the diploid Triticum urartu (2n = 2x = 14, AA) and a close lineage of Aegilops speltoides (2n = 2x = 14, BB) gave rise to tetraploid wild emmer wheat (Triticum turgidum ssp. dicoccoides, BBAA), and a further hybridization of a domesticated emmer wheat with the diploid Aegilops tauschii (2n = 2x = 14, DD) formed allohexaploid common wheat (Triticum aestivum, BBAADD) (Petersen et al., 2006; Marcussen et al., 2014). Tetraploid wheat, especially durum wheat, is becoming a valuable food crop worldwide because of its versatile processing properties and high nutritional value (Maccaferri et al., 2019). Hexaploid bread wheat, which provides about a fifth of the calories consumed by humans and contributes more protein than any other food source, is the most commonly cultivated crop on earth (IWGSC et al., 2018; He et al., 2019; Pont et al., 2019).
In recent years, many large, complex, highly repetitive genomes of Triticeae species have been deciphered (Luo et al., 2017; Mascher et al., 2017; Zhao et al., 2017; IWGSC et al., 2018; Ling et al., 2018; Guo et al., 2020; Jayakodi et al., 2020; Walkowiak et al., 2020; Wang et al., 2020; Li et al., 2021; Rabanus-Wallace et al., 2021; Zhou et al., 2021). Genomes of diploid wheat species (e.g., barley and rye) range from 4.3 Gb to 7.9 Gb in size and contain more than 40,000 annotated genes (Bauer et al., 2017; Mascher et al., 2017; Zhao et al., 2017; Ling et al., 2018; Wang et al., 2020; Li et al., 2021; Rabanus-Wallace et al., 2021; Zhou et al., 2021). The tetraploid emmer genome comprises 10.5 Gb of genomic sequence and 65,012 protein-coding genes (Avni et al., 2017). The genome of the hexaploid bread wheat Chinese Spring (CS) contains 14.5 Gb of sequence and 107,891 high-confidence genes (IWGSC et al., 2018).
In the CS genome, approximately 55% of the homologous genes have been reported to exhibit 1:1:1 correspondence across the three homoeologous subgenomes, and the other 15% have more than one gene copy in at least one of the subgenomes (IWGSC et al., 2018). Furthermore, two genomics studies of Ae. tauschii, the donor of the hexaploid wheat D subgenome, revealed an apparently recent burst of gene duplications. The authors speculated that recently duplicated genes were likely to be related to the remarkable genomic enrichment of transposable elements (TEs) (Luo et al., 2017; Zhao et al., 2017). Analysis of intra-genomic synteny of Ae. tauschii clearly showed that its most recent whole genome duplication (WGD) was rho, which occurred before the divergence of Poaceae species (Tang et al., 2010; Jiao et al., 2014), and these recent duplications were independent and dispersed throughout the genome rather than derived from WGD (Zhao et al., 2017). These recent gene duplications may, at least in part, explain why so many genes in the three subgenomes of CS are not in 1:1:1 correspondence. Therefore, expanded homologous genes in wheat arise not only from polyploidization events but also from recent independent duplications. However, studies regarding the extent, timing, and mechanisms of these recent duplications in different Triticeae species are still lacking. Moreover, it remains unclear whether these duplicates are functionally important for wheat.
The proportions of TEs in these Triticeae genomes are about 80% to 90%, much higher than those of most other grasses (Mascher et al., 2017; Wicker et al., 2018). It has been proposed that TE activities can generate new genes and novel cis-regulatory elements and can also modify the epigenetic status of specific genomic regions (Deniz et al., 2019). Occasionally, such activities lead to adaptive effects. For example, Helitrons-like TEs in maize seem to produce new nonautonomous elements for the duplicative insertion of gene segments into new locations that change both the genic and nongenic fractions of the genome, profoundly affecting genetic diversity (Morgante et al., 2005).
Here, we selected a number of representative Triticeae genomes and performed a comprehensive investigation of their recently duplicated genes, classifying them into duplicates from WGD, tandem duplication (TD), proximal duplication (PD), and dispersed duplication (DD) (for definitions, see methods). We discovered a common pattern of a recent burst of gene duplications (RBGD) in these Triticeae genomes and obtained empirical evidence indicating that TEs may have been involved in generating the RBGD. Gene duplications and losses were then examined across the evolutionary history of Triticeae species diversification and allohexaploid wheat formation. Finally, we demonstrated the importance of the RBGD for differentiating the donor genomes of bread wheat and for increasing the genetic dosage, allowing for the evolution of genes that underlie important wheat agronomic traits.
Results
Identification and characterization of recently duplicated genes
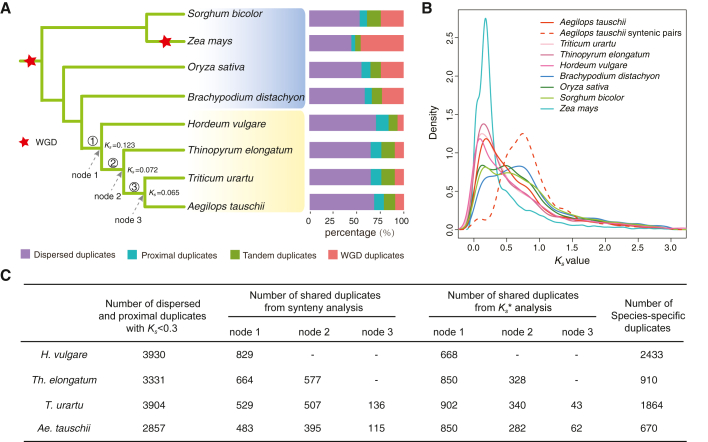
We used the best-reciprocal blast approach to retrieve paralogous gene pairs from eight sequenced diploid genomes in the Poaceae: Sorghum bicolor, Zea mays, Oryza sativa, Brachypodium distachyon, Hordeum vulgare, Thinopyrum elongatum, Ae. tauschii, and T. urartu (Figure 1A; Supplemental Table 1). To distinguish between gene duplications from historical WGD events and those from recent small-scale duplications (SSD), we performed self-genomic comparisons and classified the identified syntenic gene pairs as having arisen from WGD. The remaining duplicates were classified into three categories (TD, PD, and DD) based on the genomic distances between the gene duplicates (see methods) (Supplemental Figure 1; Supplemental Table 2). In general, the proportions of PD and DD gene pairs, which are the result of small-scale duplication events, are about two times higher in Triticeae species than in sorghum, maize, rice, and Brachypodium (Wilcoxon test, p < 0.01) (Supplemental Table 2). Specifically, we detected 9,044 to 9,787 duplicated gene pairs in the examined Triticeae species: 603 (6.3%) to 924 (10.2%) PD gene pairs and 2254 (23.4%) to 3006 (33.2%) DD gene pairs (Supplemental Table 2).
Figure 1.
Triticeae species have more recent gene duplicates than other Poaceae species.
(A) Phylogeny of representative Poaceae species (left). Red stars mark two well-acknowledged ancient WGD events. Percentage of recent duplicates classified into four duplication mechanisms in Poaceae species (right).
(B)Ks plot of recent duplicates in major Poaceae crops in (A). The Ks peaks for Triticeae species at ∼0.2 suggest a burst of recent gene duplication. The peak Ks value for Ae. tauschii syntenic pairs (dashed line) represents the rho WGD event, which closely coincides with the Ks peaks for Oryza, Sorghum, and Brachypodium.
(C) Number of recently duplicated gene pairs in H. vulgare, Th. elongatum, T. urartu, and Ae. tauschii and the phylogenetic timing of their duplications. The duplicates were dated by synteny analyses and Ks analyses. The asterisk indicates that Ks analyses were carried out after synteny analyses.
Synonymous substitution (Ks) analysis clearly showed a peak around 0.2 for all of the Triticeae species we examined (Figure 1B) and indicated that the RBGD is actually a common feature of Triticeae species. The peak Ks values for the syntenic gene pairs in the Ae. tauschii, Oryza, Sorghum, and Brachypodium genomes are around 0.75 (Figure 1B), and these duplicates resulted from the rho WGD event (Paterson et al., 2004; Tang et al., 2010; Jiao et al., 2014; Wang et al., 2015). A unique Ks peak observed for Z. mays reflected a recent WGD in the maize lineage (Schnable et al., 2009). A Gene Ontology (GO)-based analysis revealed functional enrichment of these recently duplicated genes for categories such as protein dimerization activity, xylan metabolic process, catalytic activity, and nucleobase-containing compound metabolic process (Supplemental Figure 2). These categories are distinct from those that are typically retained (and thus enriched) after WGD events in diverse sets of eukaryotes (e.g., kinases, transferases, transporters, transcription regulators, and transcription factors) (Maere et al., 2005; Freeling, 2009; Jiao et al., 2014). In addition to characterizing the distinct functional gene categories of RBGD, these results clearly suggest that RBGDs are apparently common in Triticeae genomes.
We next focused our analyses on the Triticeae by examining the duplicated gene pairs in the four diploid species in detail. We used inter-genomic synteny comparisons to determine whether both of the gene duplicates were located in inter-genomic syntenic blocks. The Ks divergences between T. urartu and H. vulgare, Th. elongatum, and Ae. tauschii were 0.123, 0.072, and 0.065, respectively (Supplemental Figure 3), and we also compared these values with the Ks values of the RBGD gene pairs to date the timing of these gene duplications. Specifically, about half of the genes (1497, 1514, 1431, and 1333 for H. vulgare, Th. elongatum, T. urartu, and Ae. tauschii, respectively) were duplicated in node 1 (before the differentiation of the Triticeae, with a Ks of approximately 0.123), and about a quarter of the genes (905, 847, and 677 for Th. elongatum, T. urartu, and Ae. tauschii, respectively) were duplicated in node 2 (before the differentiation of Th. elongatum and Triticum, with a Ks of approximately 0.072) (Figure 1C). A small number of genes (179 and 177 for T. urartu and Ae. tauschii, respectively) were duplicated in node 3 (before the differentiation of Triticum, with a Ks of approximately 0.065) (Figure 1C). These results suggest that a burst of recent gene duplications occurred before the divergence of Triticeae species and that further lineage-specific duplications have also been occurring thereafter.
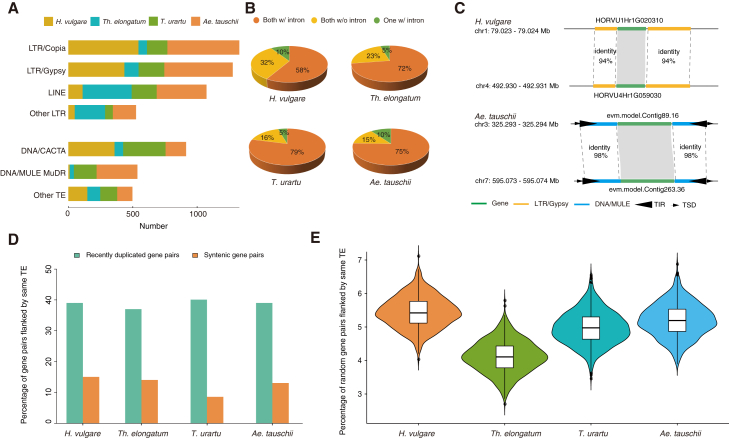
Possible mechanism of recent gene duplication
Two genomics studies of Ae. tauschii proposed that the apparent burst of recently duplicated genes in this species was probably related to the remarkable genomic enrichment of TEs (Luo et al., 2017; Zhao et al., 2017); however, empirical evidence supporting this hypothesis is still lacking. We investigated the particular types of TEs, including both long terminal repeat retrotransposons (LTR-RTs) and DNA transposons, that flanked the recently duplicated genes. Specifically, we identified any TEs that were located within 3,000 base pairs upstream and downstream of all of the recently duplicated genes. We found that the LTR-RTs were the most abundant type (68.3%), followed by the LINE and DNA/CACTA subtypes (Figure 2A). Notably, we found that about 21% to 42% of the TE-flanked, recently duplicated gene pairs possessed intronless duplicates (Figure 2B). Therefore, retrotransposition may be a major mechanism of gene duplication in these Triticeae genomes, as a conspicuous feature of retrotransposition is the formation of an intronless copy of a parental gene (Kim et al., 2017). Here, we show two examples of recently duplicated genes and their flanking sequences in H. vulgare and Ae. tauschii (Figure 2C). The H. vulgare duplicated gene pair HORVU1Hr1G020310 and HORVU4Hr1G059030 are located within TEs of the same LTR/Gypsy subtype with 94% sequence identity. Similarly, the duplicated gene pair evm.model.Contig89.16 and evm.model.Contig263.36 in Ae. tauschii are located within TEs of the same DNA/MULE subtype with 98% sequence identity (Figure 2C).
Figure 2.
Recently duplicated genes tend to be surrounded by TEs.
(A) Histogram showing the number of TE-flanked, dispersed duplicate gene pairs for each TE type.
(B) Pie chart showing three types of intron distribution within duplicated gene pairs.
(C) Two examples of gene duplicates embedded in TEs of the same subtype with high sequence identity.
(D) Percentage of dispersed and syntenic duplicate gene pairs that are flanked by the same TE type (e.g., Copia, Gypsy, LINE).
(E) Percentage of randomly selected gene pairs that are flanked by the same type of TE. We rarely found two genes flanked by the same type of TE (approximately 5% by chance).
Given the prevalence of TEs throughout the genomes of Triticeae, we next investigated the chances of two genes duplicated in a WGD being flanked by similar types of TEs. The results revealed a clear trend: a large proportion (38%–42%) of the recently duplicated genes in the four diploid genomes were flanked by TEs of the same subtype (e.g., GYPSY, COPIA, etc.), whereas only ∼10% of the syntenic gene pairs (generated by WGD) were flanked by TEs of the same subtype in the four diploid genomes (Figure 2D). When we randomly selected two genes from individual Triticeae genomes, only ∼5% of them were flanked by TEs of the same subtype (Figure 2E). Thus, genome-wide empirical evidence supports a major functional contribution of TEs to the generation of RBGDs in Triticeae.
The retention and conservation of the recently duplicated genes
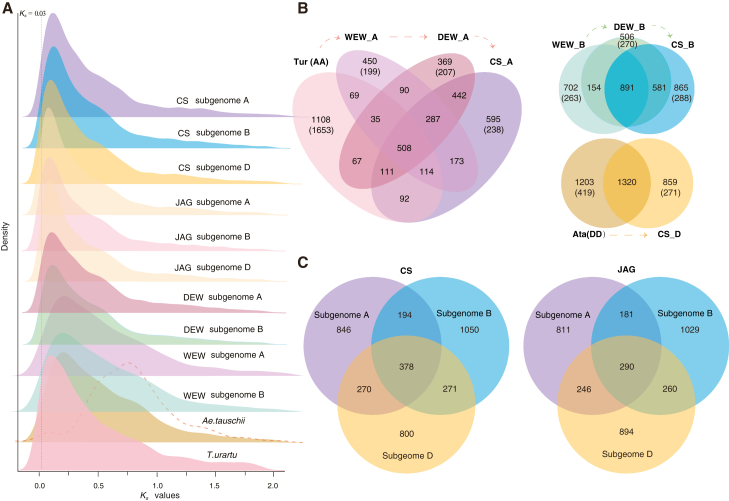
To further understand the genetic contribution of these recently duplicated genes to polyploid wheats, we investigated the retention and diversification of the RBGDs after the formation and diversification of allohexaploid wheat. First, we identified and compared recent duplicates in the genomes of T. urartu and Ae. tauschii to the corresponding subgenomes of wild and cultivated tetraploid wheat and the subgenomes of hexaploid bread wheat cultivars (Figure 3). We found 1,925 and 2,010 duplicates in subgenome A and B of wild emmer wheat (WEW), and 2,116 and 2,402 duplicates in subgenomes A and B of durum wheat (DEW) (Figure 3B). For hexaploid wheat, there are 2,560, 2,625, and 2,450 duplicates in subgenomes A, B, and D of CS and 2,374, 2,642, and 2,497 duplicates in subgenomes A, B, and D of Jagger (JAG) (Figure 3B and Supplemental Figure 4). JAG and CS are two representative hexaploid wheats that originated in the West and the East, respectively. We found that JAG and CS have about 2,000 co-retained gene pairs in each subgenome (i.e., more than 80% are shared) (Supplemental Figure 4).
Figure 3.
The retention and conservation of the recently duplicated genes in CS.
(A)Ks plot of recent duplicates in CS and JAG that originated in the West and their progenitor species. Dashed line on the Ae. tauschii plot represents the Ks distribution of the syntenic gene pairs that arose from the rho WGD event.
(B) Venn diagrams show the retention pattern of the recent duplicates following the evolution of the diploid progenitor, wild emmer, domesticated emmer, and CS wheat. The numbers in parentheses show the number of newly duplicated gene pairs in the progenitors or wheat subgenomes for which no corresponding orthologs were identified in other genomes.
(C) Venn diagrams show the commonly retained recent gene duplicates in the three subgenomes of CS and JAG. These retained gene pairs were duplicated prior to the diversification that led to the diploid AA, BB, and DD species, based on Ks analysis.
We next investigated the retention patterns of these recent gene duplicates after the two successive polyploidization events using CS as a representative hexaploid wheat (Figure 3B and Supplemental Figure 5; Supplemental Tables 3–5). We found that 508, 891, and 1,320 gene pairs were co-retained in the A, B, and D subgenomes after polyploidization events (Figure 3B). We also investigated the duplication times by comparing Ks divergence of these RGBDs with the corresponding species divergence times to separate the species-specific gene pairs into specifically retained or newly duplicated gene pairs in each species. For the A subgenome, 1,108, 450, 369, and 595 gene pairs were specifically retained, and 1,635, 199, 207, and 238 gene pairs were newly duplicated in T. urartu, emmer wheat, durum wheat, and CS, respectively. For the B subgenome, 702, 506, and 865 gene pairs were specifically retained, and 263, 270, and 288 gene pairs were newly duplicated in emmer wheat, durum wheat, and CS, respectively. For the D subgenome, 1,203 and 859 gene pairs were specifically retained, and 419 and 217 gene pairs were newly duplicated in Ae. tauschii and CS, respectively (Figure 3B). We further compared the particularly well-retained subset with the specifically retained gene pairs and found that the well-retained gene pairs were characterized by their typically higher Ks values (Wilcoxon test, p < 0.01) (Supplemental Figure 6). Moreover, genes in the well-retained subset had clearly undergone stronger purifying selection than genes of other duplicated pairs in common wheat that showed no obvious synteny to progenitor genomes (Wilcoxon test, p < 0.01) (Supplemental Figure 6).
We next investigated the retention pattern of gene duplicates that were generated before the diversification of the Triticum and Aegilops species in multiple hexaploid wheat genomes, including JAG, CS, and nine other newly available wheat genomes. In CS, we found that 5,300 of 7,821 gene pairs (1,688, 1,893, and 1,719 in the three subgenomes, respectively) from RBGD were duplicated before the divergence of the Triticum and Aegilops species. Among these 5,300 duplicated genes, 378 pairs of genes were well retained after two allopolyploidization events (each set of homologous genes contains six copies in CS), and 846, 1,050, and 800 gene pairs were specifically retained in the A, B, and D subgenomes of CS, respectively (Figure 3C). Similarly, in JAG, 4,978 of 8,573 gene pairs were duplicated before the divergence of the Triticum and Aegilops species; 290 duplicates were retained in the three subgenomes of JAG, and 811, 1,029, and 894 gene pairs were specifically retained in the A, B, and D subgenomes of JAG, respectively (Figure 3C). Similarly, we identified about 300 co-retained gene pairs and approximately 800, 1,000, and 800 specifically retained gene pairs in the three subgenomes of the other nine sequenced wheat genomes (Supplemental Figure 7). Among these co-retained gene pairs, about 70% to 80% were shared among the hexaploid wheat genomes, whereas CS and JAG shared only 60% of these co-retained gene pairs (Supplemental Tables 6 and 7). A GO-based analysis revealed functional enrichment of these co-retained pairs (∼300 pairs) in CS and JAG in categories such as aminoacyl-tRNA ligase activity, tRNA aminoacylation, and tRNA metabolic process. Categories of chromatin modification and histone modification were only enriched in the CS retained duplicates, and the category of transporter activity was specifically enriched in JAG retained duplicates (Supplemental Figure 8).
The diversification patterns of the recently duplicated genes
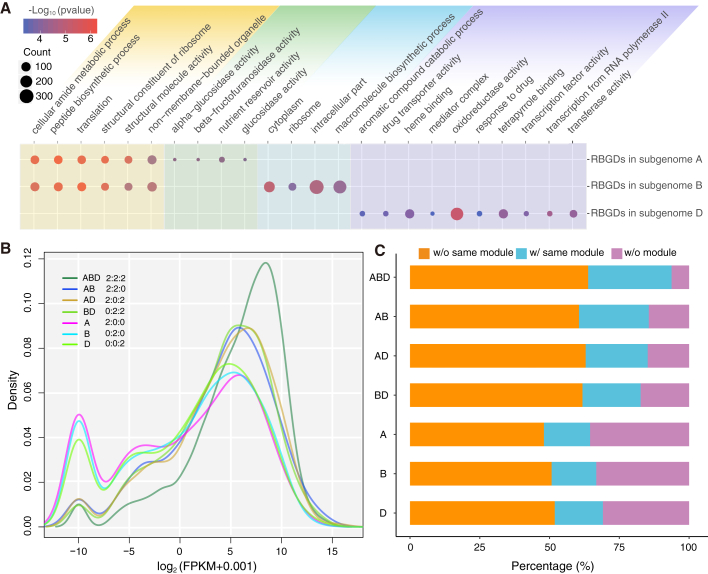
Given the allohexaploid nature of wheat, we also performed multiple subgenome-specific comparisons of the duplicated gene pairs to investigate any differential regulation and related functional diversity among such pairs in the three subgenomes. First, patterns of functional category enrichment (GO categories) among the retained duplicates differed among the three CS subgenomes; for example, nutrient reservoir activity was enriched in the gene pairs of the A subgenome, macromolecule biosynthetic process was enriched in the gene pairs of the B subgenome, and oxidoreductase activity was enriched in the gene pairs of the D subgenome (Figure 4A). Second, the basic trend from an RNA-sequencing (RNA-seq)-based analysis showed weaker expression for genes of pairs present in a single subgenome compared with genes of pairs whose orthologous gene pairs were retained in two or three subgenomes (Figure 4B). We found that 38% of the subgenome-specific retained duplicates exhibited no expression, a larger percentage than that of the non-subgenome-specific gene pairs (Figure 4B). It was notable that the 378 gene pairs common to all three subgenomes exhibited the highest expression levels (Figure 4B). Third, after reconstructing co-expression modules using the RNA-seq data, we found that about 25% of the subgenome-specific duplicates were not clustered into any modules, compared with only about 10% of the multi-subgenome retained pairs (Figure 4C). Further co-expression network analysis revealed that a larger percentage of the duplicates common to all subgenomes diverged into different modules compared with the subgenome-specific duplicates (73% versus 55%), indicating possible sub- or neo-functionalization of the duplicates over evolutionary time (Figure 4C). Collectively, these analyses emphasize that distinguishing among ancient versus recent duplicates and among subgenome-specific duplicated gene pairs is a viable analytical strategy for isolating specific trends in the regulation and attendant expression divergence of these genes and thus their potential sub- and neo-functionalization.
Figure 4.
The diversification patterns of the recently duplicated genes in CS.
(A) Different patterns of GO enrichment for recently duplicated genes in the three subgenomes of CS.
(B) Distribution of expression levels for differentially retained genes among the three subgenomes of CS. “ABD” indicates that the three subgenomes all retained duplicates; “AB” indicates that only the A and B subgenomes retained duplicates; and “A” indicates that only the A subgenome retained duplicates.
(C) Differentially retained duplicates assigned to particular modules (same, divergent, or none) in a co-expression network analysis. w/o same module indicates divergent co-expression networks; w/same module indicates the same network; w/o module indicates that neither gene was assigned to a network.
Evolutionary and expression analyses of NAC genes
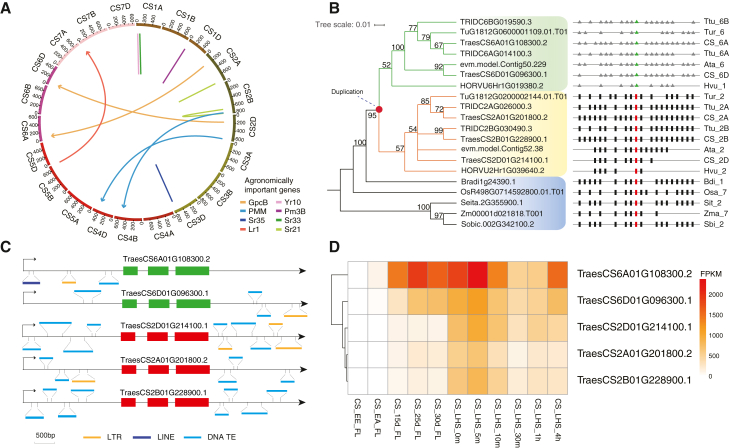
Several genes derived from the RBGD have been previously identified as agronomically important genes in wheat, e.g., Sr21, Sr33, and Sr35, which specify stem rust resistance (Periyannan et al., 2013; Saintenac et al., 2013; Chen et al., 2018), Yr10, which specifies stripe rust resistance (Liu et al., 2014), Lr1, which specifies leaf rust resistance (Feuillet et al., 1995), Pm3B, which specifies powdery mildew resistance (Brunner et al., 2011), GPC, which controls the contents of proteins and health-promoting minerals (iron and zinc) in the grain (Uauy et al., 2006), and phosphomannomutase (PMM), which functions in temperature adaptability (Yu et al., 2010) (Figure 5A). In addition, we found that most of these duplicates were derived from the ancestor of the Triticeae (Supplemental Figure 9). We conducted a more systematic study of the evolutionary history of GPC genes (encoding NAC transcription factors), among which NAM-B1 is well studied for its function in accelerating leaf senescence and increasing grain protein content in wheat (Uauy et al., 2006). Through phylogenetic and syntenic analyses, we found that a duplication belonging to RBGDs occurred before the divergence of Triticeae species, creating the NAM-B1 on chromosome 6 from its parental gene on chromosome 2 (Figure 5B). We identified five NAM homologs in CS and found that the copy on chromosome 6B was lost. Moreover, we found that similar types of TEs flanked the five NAM homologs (two homoeologous pairs in the A and D subgenomes plus one singleton in the B subgenome), indicating potential involvement of TE activity in generating the duplicated functional NAM-B1 allele before the divergence of Triticeae (Figure 5B and 5C).
Figure 5.
Evolutionary and expression analyses of NAC genes.
(A) Circos plot showing eight previously identified important genes that have experienced the RBGD. The known agronomically important genes are associated with stem rust resistance (Sr21, Sr33, Sr35), stripe rust resistance (Yr10), leaf rust resistance (Lr1), powdery mildew resistance (Pm3B), phosphomannomutase (PMM), and earlier senescence and higher grain protein, iron, and zinc content (GPC). The arrow/line represents the direction of gene duplication from the ancestral gene to the newly duplicated copy.
(B) Maximum likelihood phylogeny of the NAC genes and the syntenic regions that contain NAC genes in other Poaceae genomes. A red solid circle in the phylogenetic tree represents one of the RBGD duplication events that created a duplicated copy in chromosome 6 of the common ancestor of Triticeae species. The right side of the phylogenetic tree presents the syntenic regions with NAC genes. The identified syntenic relationships among genes shown as black and red rectangles suggest that the genes in group I are positionally conserved, and therefore the ancestral copies, whereas the genes in group II that are illustrated as green triangles surrounded by gray triangles are the new, duplicated copies.
(C) Schematic diagrams showing the gene structure and flanking TEs around NAC genes in CS. Different types of TEs are indicated by bars with different colors.
(D) Expression levels of NAC genes in CS. The duplicated copy of TraesCS6A01G108300 has the highest expression in the flag leaf among the five NAC genes. EE, ear emergence; EA, anthesis; LHS, leaf under heat stress.
We examined the expression pattern of the remaining five NAM genes in CS using 100 RNA-seq samples (Ramírez-González et al., 2018). The expression of the NAM-A1 gene (TraesCS6A01G108300), which resulted from duplication, was significantly higher in the flag leaf than that of other NAM genes (Figure 5D). This result may reflect the modification of regulatory elements because of the removal of TEs downstream of TraesCS6A01G108300 or variations in TEs in the upstream region (Figure 5C). Further functional experiments to identify and test the regulatory elements around TraesCS6A01G108300 may help to unravel the underlying mechanisms that cause increased expression of the novel duplicated gene. However, the case study of NAM indicates that the RBGDs may have quickly increased the dosage of agronomically important wheat genes, in addition to the two consecutive allopolyploidization events.
Discussion
Gene duplicates and their duplication mechanisms
Gene duplication provides raw genetic material for evolution and adaptation and is considered to be a driving force in evolution (Ohno, 1970; Adams and Wendel, 2005). Multiple mechanisms have been proposed to generate gene duplicates (Panchy et al., 2016; Qiao et al., 2019; Zhang et al., 2020). Polyploidization is a major source of large-scale gene duplication because it involves the doubling of the entire genome (Soltis et al., 2015; Van de Peer et al., 2017). In this study, we observed a large number of recent gene duplications in all sequenced Triticeae species, a finding that is commonly, if sometimes mistakenly, interpreted as evidence for a WGD event. Genomic synteny comparisons clearly showed that these gene duplicates are the result of independent SSDs rather than a WGD event. However, it is challenging to determine the mechanism if a reference genome is not available, and that is why there are such active controversies (Wang et al., 2019; Zwaenepoel et al., 2019).
In addition to the genomic positions of the duplicated genes, their functional categories can provide another perspective on their possible origins. In an extremely diverse set of eukaryotes, retention of gene duplicates after WGD events was shown to be biased toward certain categories, such as kinases, transferases, transporters, transcription regulators, and transcription factors (Davis and Petrov, 2005; Maere et al., 2005; Freeling, 2009; Jiao et al., 2011). If no chromosomal genome assembly is available, we can compare the enriched GO categories of the identified gene duplicates with those typically enriched in the duplicates retained after WGD events. In this study, we found apparently distinct functional categories for the RGBD genes in Triticeae species, thus clearly excluding the possibility of their WGD origin. Therefore, the enriched GO pattern of duplicates can serve as complementary evidence to determine whether duplications are the result of an SSD or WGD event.
TE-mediated gene duplication
TEs are widespread components of plant genomes, and expansion in TE numbers can cause dramatic differences in the overall architecture of plant genomes (Arabidopsis Genome Initiative, 2000; Tenaillon et al., 2010; Lisch, 2013; IWGSC et al., 2018). TE activity can cause a broad range of changes in gene expression and function, as well as the evolution of entirely new genes (Kaessmann et al., 2009; Lisch, 2013; Tan et al., 2016; Cerbin and Jiang, 2018). In this study, we found that RBGDs in Triticeae genomes were clearly associated with TEs: a large proportion (38%–42%) of the recently duplicated genes in the four diploid genomes were flanked by TEs of the same subtype and obviously did not result from tandem duplications. We also found that 59% of TEs from the same subtype associated with gene duplications had high identities, greater than 90%. Notably, we found that about 21% to 42% of these same TE-flanked recently duplicated gene pairs had intronless duplicates, which is also powerful evidence, especially for LTR-RT-mediated duplicates. For example, TraesCS1B01G041800 and TraesCS6B01G016300 are located beside TEs of the same subtype with 91% sequence identity; the duplicated copy (TraesCS6B01G016300) lacks introns (Supplemental Figure 10). These findings suggest that the abundant TEs in Triticeae may have created a large number of new genes via previously reported mechanisms, although other mechanisms such as haplotype recombination may also have contributed to some of these duplications (Jiang et al., 2004; Wang et al., 2006; Kaessmann et al., 2009; Kim et al., 2017).
In this study, we found similar TEs near ∼40% of the RBGD genes, and we suspect that the rest of the duplicates may have been generated from other mechanisms or their flanking TEs may have undergone sequence divergence during evolution. In fact, we found that the Ks values of the duplicated genes that were not flanked by TEs of the same subtype were larger than those of duplicates with similar TEs (Wilcoxon test, p < 0.01) (Supplemental Figure 11A). Moreover, we also found that the larger the Ks values of the duplicated genes, the lower the identity of their flanking TEs (Supplemental Figure 11B). This trend is consistent with previous reports that only relatively young duplications via TEs can be detected (Jiang et al., 2004; Morgante et al., 2005; Wang et al., 2006; Xiao et al., 2008; Kim et al., 2017; Cerbin and Jiang, 2018). Notably, our reported RBGD includes some duplications that occurred nearly 10 million years ago, and we expect that many other sequence divergences may have occurred and thus erased the signature of the similar TEs (if they existed) over such a long evolutionary period.
Polyploidy advantage of bread wheat and RBGD
Bread wheat has a large, redundant, and allohexaploid genome, making it by far the largest and most complex genome of all sequenced plant species. The genome of the wheat cultivar CS contains 14.5 Gb of sequence and 107,891 high-confidence genes, a larger number of genes than any other sequenced diploid genome. The complexity of the wheat genome is due not only to its allohexaploid nature but also to its enrichment in repetitive sequences and TEs. These features may make a large contribution to its genetic diversity and innovation during evolutionary history, making wheat one of the most complicated genomes.
The advantage of wheat polyploidy may be associated, at least in part, with the increased gene dosage produced by genome merging (Ramírez-González et al., 2018), and the resulting redundant genes may go through mutation robustness, differential gene loss, subgenomic expression dominance, or divergence, which often lead to novel functional molecular networks and ultimately to phenotypic innovations (Wu et al., 2020). As reported previously, 55% of genes exhibit perfect 1:1:1 correspondence across the three subgenomes of CS (Ramírez-González et al., 2018). As we reported here, a recent burst of small-scale gene duplications also occurred during the evolutionary history of speciation and diversification of Triticeae, probably because of TE enrichment in the Triticeae genomes. Thus, in bread wheat, certain functional genes dramatically increased in dosage through both allopolyploidization events and RBGD, and the resulting increased gene dosage may have contributed to the polyploidy advantage of bread wheat. Many previously identified agronomic genes in polyploid wheat species have experienced recent duplications, a finding that highlights the genetic contribution and general importance of RBGD for common wheat.
In conclusion, we revealed a common, recent burst of numerous gene duplications in the Triticeae species, a novel feature of Triticeae that has not been reported for any other clades of green plants. We also provided evidence suggesting that the RBGD resulted from the abundant TEs in Triticeae genomes. By investigating the birth and death patterns of the recently duplicated genes in the Triticeae species, we found that the RBGD began after the origin of Triticeae species, and a large number of young genes may have contributed to their species diversification. Probably because of increased dosage or sub-/neo-functionalization of gene duplicates, several genes have evolved into key factors that function in agronomically important traits of wheat.
Methods
Genomic data resources
We selected 10 taxa in the Poaceae clade that have whole-genome assemblies: H. vulgare (Mascher et al., 2017), Th. elongatum (Wang et al., 2020), Ae. tauschii (Zhao et al., 2017), T. urartu (Ling et al., 2018), T. turgidum (Avni et al., 2017; Maccaferri et al., 2019), T. aestivum (IWGSC et al., 2018; Walkowiak et al., 2020), O. sativa (Goff et al., 2002), B. distachyon (Vogel et al., 2010), Z. mays (Jiao et al., 2017), and S. bicolor (Paterson et al., 2009). Genomic data were downloaded from public repositories or specific project websites (Supplemental Table 1).
Genomic synteny analyses
We performed self-alignment of the protein sequences using BLASTP (Altschul et al., 1997) with parameters “-outfmt 6 -evalue 1e-5”, and the top 15 hits were extracted as an input file for MCScanX (Wang et al., 2012). The intra-genome syntenic blocks were detected using MCScanX with parameters “-e 1e-5 –m 25 –w 5” (Wang et al., 2012). Gene pairs in collinear blocks were identified as whole-genome duplicates.
Paralogous gene detection and classification
We performed genome-wide, all-by-all BLASTP (Altschul et al., 1997) with parameters “-outfmt 6 -evalue 1e-5”, and the best reciprocal matches were then extracted as the paralogous genes. For all of the examined Poaceae genomes, we classified the paralogous genes into four categories: tandem duplicated pairs (located within five genetic loci of each other), proximal duplicated pairs (within 5–10 genetic loci), dispersed duplicated pairs (more than 10 genetic loci apart), and duplicated pairs from WGD (gene pairs with evidence of genomic synteny).
Statistical test
The Wilcoxon test was used to evaluate differences between groups (Supplemental Figures 6 and 11A). Taking Supplemental Figure 6 as an example, we divided the duplicates into two groups based on whether they were conserved. We then tested the significance of differences in Ka, Ks, and Ka/Ks between these two groups of data. A p value of <0.05 was considered to be statistically significant: NS (not significant) p > 0.05, ∗p < 0.05, ∗∗p < 0.01.
Synonymous substitution (Ks) analysis
For each pair of homologous genes, protein sequences were aligned using MUSCLE (Edgar, 2004) with default parameters, and nucleotide sequences were then forced to fit the amino acid alignments using PAL2NAL (Suyama et al., 2006). Finally, Ks values were calculated using the Nei-Gojobori algorithm (Nei and Gojobori, 1986) implemented in the codeml package of PAML (Yang, 1997).
TE annotation
The repetitive sequences were identified using a combination of repeat homology searching and ab initio prediction approaches. For homology searching, Repbase (2018) (Bao et al., 2015) was used to search against the genome using RepeatMasker (Tarailo-Graovac and Chen, 2009) with default parameters. For ab initio predictions, a consensus sequence library was built using RepeatModeler (http://repeatmasker.org/RepeatModeler/) with the parameters “-engine ncbi.” Then LTR_harvest (Ellinghaus et al., 2008), LTR_finder (Xu and Wang, 2007), and LTR_retriever (Ou and Jiang, 2018) were used to build an LTR library with default parameters. Both libraries were then used to annotate the genome using RepeatMasker, and the detected TEs were combined to obtain the final TE annotation. A wheat TE reference library named ClariTeRep, described previously (Daron et al., 2014), was also used to annotate the TEs of Triticum genomes.
Phylogenetic analysis
A phylogenetic tree was constructed for the Poaceae homologs of the T. turgidum NAC gene (GenBank accession No. ABI94352.1). To identify the homologs in other species, the amino acid sequences of the T. turgidum NAC genes were used as a query to search against the other eight species with a previously reported method (Jiao et al., 2014). Protein sequences were aligned using MUSCLE (Edgar, 2004) with default parameters. The maximum likelihood trees were then constructed using the JTT+G4 model implemented in IQ-TREE, and bootstrap supports were evaluated by ultrafast bootstrapping testing (1,000 replicates) (Nguyen et al., 2015).
Conservation of the recently duplicated gene pairs
We used both inter-genomic synteny comparisons and Ks analysis to date all of the recently duplicated gene pairs detected in the three subgenomes of CS. The inter-genome syntenic blocks were detected using MCScanX with the default parameters. Then, if a pair of duplicated genes in CS had collinear genes in the genomes of progenitors of CS or other early diverging species (e.g., H. vulgare), we considered that this pair of genes was duplicated before the speciation and were therefore retained and conserved duplicates. If no syntenic relationship was detected, we further dated the duplication by calculating the Ks value and comparing it with the Ks values of speciation among the Triticeae species.
GO enrichment analysis
To find the enriched GO terms in dispersed duplicates and syntenic genes, we used the R package topGO and calculated the p values of GO terms with the default method “weight01.” Fisher's exact test in combination with the “classic” algorithm of this R package was used to test for overrepresented GO terms. Statistical enrichment of GO terms was evaluated by comparing the sample (duplicated genes) with the background (all annotated genes) based on Fisher’s exact test, and adjusted p values (p < 0.01) were calculated by the Benjamini and Hochberg (false-discovery rate) method (Ashburner et al., 2000).
Gene expression analysis and co-expression module construction
RNA-seq data for 100 diverse CS samples from different tissues, growth conditions, and developmental stages were mapped to the CS genome using STAR with default parameters (Dobin et al., 2013), and RSEM was used to estimate gene expression levels (Li and Dewey, 2011). Read counts for each gene were normalized to the sequencing depth of the samples using DESeq2 with default parameters (Love et al., 2014).
All expressed genes were used to build a co-expression network with the WGCNA R package (Langfelder and Horvath, 2008). A soft power threshold of five was used because it was the lowest power for which the scale-free topology fit index reached 0.9. The blockwise module function in WGCNA was used to construct blockwise in two blocks, with a maximum block size of 46,000 genes. Other parameters for the blockwise module function were set as follows: maxPOutliers = 0.05, TOMType = “unsigned,” mergeCutHeight = 0.15, and minimum module size ≥30. The most highly correlated genes identified by the signedKME() function were considered central to the module.
Funding
We thank the National Natural Science Foundation of China (Grant number 31870209) and the Key Science and Technology Program of Henan Province (201300110800) for research funding.
Author contributions
Y.J. and J.J. initiated and conceived the study; Y.J. and X.W. performed the principal gene duplication data analyses; Y.H., X.Y., and L.Q. performed some preliminary analyses and helped with the discussion of the research and final figures; Y.J., X.W., X.Y., and Y.H. drafted the manuscript; D.W. contributed to the discussion and editing of the manuscript. All authors contributed to and approved the final manuscript.
Acknowledgments
The authors declare no competing interests.
Published: December 11, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information can be found online at Plant Communications Online.
Contributor Information
Daowen Wang, Email: dwwang@henau.edu.cn.
Jizeng Jia, Email: jiajizeng@caas.cn.
Yuannian Jiao, Email: jiaoyn@ibcas.ac.cn.
Supplemental information
References
- Adams K.L., Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (WGSC) IWGSC RefSeq principal investigators. Appels R., Eversole K., Feuillet C., Keller B., Rogers J., Stein N., IWGSC Whole-Genome Assembly Principal Investigators. Pozniak C.J., et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:eaar7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R., Nave M., Barad O., Baruch K., Twardziok S.O., Gundlach H., Hale I., Mascher M., Spannagl M., Wiebe K., et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 2017;357:93–97. doi: 10.1126/science.aan0032. [DOI] [PubMed] [Google Scholar]
- Bao W., Kojima K.K., Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E., Schmutzer T., Barilar I., Mascher M., Gundlach H., Martis M.M., Twardziok S.O., Hackauf B., Gordillo A., Wilde P., et al. Towards a whole-genome sequence for rye (Secale cereale L.) Plant J. 2017;89:853–869. doi: 10.1111/tpj.13436. [DOI] [PubMed] [Google Scholar]
- Brunner S., Hurni S., Herren G., Kalinina O., von Burg S., Zeller S.L., Schmid B., Winzeler M., Keller B. Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol. J. 2011;9:897–910. doi: 10.1111/j.1467-7652.2011.00603.x. [DOI] [PubMed] [Google Scholar]
- Cerbin S., Jiang N. Duplication of host genes by transposable elements. Curr. Opin. Genet. Dev. 2018;49:63–69. doi: 10.1016/j.gde.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang W., Bolus S., Rouse M.N., Dubcovsky J. Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PLoS Genet. 2018;14:4. doi: 10.1371/journal.pgen.1007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daron J., Glover N., Pingault L., Theil S., Jamilloux V., Paux E., Barbe V., Mangenot S., Alberti A., Wincker P., et al. Organization and evolution of transposable elements along the bread wheat chromosome 3B. Genome Biol. 2014;15:546. doi: 10.1186/s13059-014-0546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.C., Petrov D.A. Do disparate mechanisms of duplication add similar genes to the genome? Trends Genet. 2005;21:548–551. doi: 10.1016/j.tig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Deniz Ö., Frost J.M., Branco M.R. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019;20:417–431. doi: 10.1038/s41576-019-0106-6. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C., Messmer M., Schachermayr G., Keller B. Genetic and physical characterization of the LR1 leaf rust resistance locus in wheat (Triticum aestivum L.) Mol. Gen. Genet. 1995;248:553–562. doi: 10.1007/BF02423451. [DOI] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Guo W., Xin M., Wang Z., Yao Y., Hu Z., Song W., Yu K., Chen Y., Wang X., Guan P., et al. Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat. Commun. 2020;11:5085. doi: 10.1038/s41467-020-18738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Pasam R., Shi F., Kant S., Keeble-Gagnere G., Kay P., Forrest K., Fritz A., Hucl P., Wiebe K., et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 2019;51:896–904. doi: 10.1038/s41588-019-0382-2. [DOI] [PubMed] [Google Scholar]
- Jayakodi M., Padmarasu S., Haberer G., Bonthala V.S., Gundlach H., Monat C., Lux T., Kamal N., Lang D., Himmelbach A., et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature. 2020;588:284–289. doi: 10.1038/s41586-020-2947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Li J., Tang H., Paterson A.H. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell. 2014;26:2792–2802. doi: 10.1105/tpc.114.127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Peluso P., Shi J., Liang T., Stitzer M.C., Wang B., Campbell M.S., Stein J.C., Wei X., Chin C.S., et al. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524–527. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H., Vinckenbosch N., Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Park J., Yeom S.I., Kim Y.M., Seo E., Kim K.T., Kim M.S., Lee J.M., Cheong K., Shin H.S., et al. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 2017;18:210. doi: 10.1186/s13059-017-1341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang L., Yang J., He H., Jin H., Li X., Ren T., Ren Z., Li F., Han X., et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 2021;53:574–584. doi: 10.1038/s41588-021-00808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H.Q., Ma B., Shi X., Liu H., Dong L., Sun H., Cao Y., Gao Q., Zheng S., Li Y., et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature. 2018;557:424–428. doi: 10.1038/s41586-018-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013;14:49–61. doi: 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- Liu W., Frick M., Huel R., Nykiforuk C.L., Wang X., Gaudet D.A., Eudes F., Conner R.L., Kuzyk A., Chen Q., et al. The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS-LRR sequence in wheat. Mol. Plant. 2014;7:1740–1755. doi: 10.1093/mp/ssu112. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Gu Y., Puiu D., Wang H., Twardziok S., Deal K., Huo N., Zhu T., Wang L., Wang Y., et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2017;551:498–502. doi: 10.1038/nature24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M., Harris N.S., Twardziok S.O., Pasam R.K., Gundlach H., Spannagl M., Ormanbekova D., Lux T., Prade V.M., Milner S.G., et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019;51:885–895. doi: 10.1038/s41588-019-0381-3. [DOI] [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U S A. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T., Sandve S.R., Heier L., Spannagl M., Pfeifer M., International Wheat Genome Sequencing Consortium. Jakobsen K.S., Wulff B.B., Steuernagel B., Mayer K.F., et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;102:5454–5459. doi: 10.1126/science.1250092. [DOI] [PubMed] [Google Scholar]
- Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S.O., Wicker T., Radchuk V., Dockter C., Hedley P.E., Russell J., et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544:427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Springer; Berlin: 1970. Evolution by Gene Duplication. [Google Scholar]
- Ou S., Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N., Lehti-Shiu M., Shiu S.H. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Chapman B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U S A. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., Hellsten U., Mitros T., Poliakov A., et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- Periyannan S., Moore J., Ayliffe M., Bansal U., Wang X., Huang L., Deal K., Luo M., Kong X., Bariana H., et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341:786–788. doi: 10.1126/science.1239028. [DOI] [PubMed] [Google Scholar]
- Petersen G., Seberg O., Yde M., Berthelsen K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum) Mol. Phylogenet. Evol. 2006;39:70–82. doi: 10.1016/j.ympev.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Pont C., Leroy T., Seidel M., Tondelli A., Duchemin W., Armisen D., Lang D., Bustos-Korts D., Goué N., Balfourier F., et al. Tracing the ancestry of modern bread wheats. Nat. Genet. 2019;51:905–911. doi: 10.1038/s41588-019-0393-z. [DOI] [PubMed] [Google Scholar]
- Qiao X., Li Q., Yin H., Qi K., Li L., Wang R., Zhang S., Paterson A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019;20:38. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanus-Wallace M.T., Hackauf B., Mascher M., Lux T., Wicker T., Gundlach H., Baez M., Houben A., Mayer K., Guo L., et al. Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 2021;53:564–573. doi: 10.1038/s41588-021-00807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-González R.H., Borrill P., Lang D., Harrington S.A., Brinton J., Venturini L., Davey M., Jacobs J., van Ex F., Pasha A., et al. The transcriptional landscape of polyploid wheat. Science. 2018;361:eaar6089. doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- Saintenac C., Zhang W., Salcedo A., Rouse M.N., Trick H.N., Akhunov E., Dubcovsky J. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science. 2013;341:783–786. doi: 10.1126/science.1239022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., Ware D., Fulton R.S., Stein J.C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T.A., et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Marchant D.B., Van de Peer Y., Soltis D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Cardoso-Moreira M., Shi W., Zhang D., Huang J., Mao Y., Jia H., Zhang Y., Chen C., Shao Y., et al. LTR-mediated retroposition as a mechanism of RNA-based duplication in metazoans. Genome Res. 2016;26:1663–1675. doi: 10.1101/gr.204925.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Paterson A.H. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. U S A. 2010;107:472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi0410s25. Chapter 4. [DOI] [PubMed] [Google Scholar]
- Tenaillon M.I., Hollister J.D., Gaut B.S. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010;15:471–478. doi: 10.1016/j.tplants.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.P., Garvin D.F., Mockler T.C., Schmutz J., Rokhsar D., Bevan M.W., Barry K., Lucas S., Harmon-Smith M., Lail K., et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Walkowiak S., Gao L., Monat C., Haberer G., Kassa M.T., Brinton J., Ramirez-Gonzalez R.H., Kolodziej M.C., Delorean E., Thambugala D., et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588:277–283. doi: 10.1038/s41586-020-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zheng H., Fan C., Li J., Shi J., Cai Z., Zhang G., Liu D., Zhang J., Vang S., et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang J., Jin D., Guo H., Lee T.H., Liu T., Paterson A.H. Genome Alignment spanning major poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol. Plant. 2015;8:885–898. doi: 10.1016/j.molp.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Wang H., Guo C., Ma H., Qi J. Reply to Zwaenepoel et al.: meeting the challenges of detecting polyploidy events from transcriptomic data. Mol. Plant. 2019;12:137–140. doi: 10.1016/j.molp.2018.12.020. [DOI] [PubMed] [Google Scholar]
- Wang H., Sun S., Ge W., Zhao L., Hou B., Wang K., Lyu Z., Chen L., Xu S., Guo J., et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:eaba5435. doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- Wicker T., Gundlach H., Spannagl M., Uauy C., Borrill P., Ramírez-González R.H., De Oliveira R., International Wheat Genome Sequencing Consortium. Mayer K., Paux E., et al. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 2018;19:103. doi: 10.1186/s13059-018-1479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Han B., Jiao Y. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant. 2020;13:59–71. doi: 10.1016/j.molp.2019.10.012. [DOI] [PubMed] [Google Scholar]
- Xiao H., Jiang N., Schaffner E., Stockinger E.J., van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yu C., Li Y., Li B., Liu X., Hao L., Chen J., Qian W., Li S., Wang G., Bai S., et al. Molecular analysis of phosphomannomutase (PMM) genes reveals a unique PMM duplication event in diverse Triticeae species and the main PMM isozymes in bread wheat tissues. BMC Plant Biol. 2010;10:214. doi: 10.1186/1471-2229-10-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li X., Zhao R., Zhou Y., Jiao Y. Evolutionary strategies drive a balance of the interacting gene products for the CBL and CIPK gene families. New Phytol. 2020;226:1506–1516. doi: 10.1111/nph.16445. [DOI] [PubMed] [Google Scholar]
- Zhao G., Zou C., Li K., Wang K., Li T., Gao L., Zhang X., Wang H., Yang Z., Liu X., et al. The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants. 2017;3:946–955. doi: 10.1038/s41477-017-0067-8. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Bai S., Li H., Sun G., Zhang D., Ma F., Zhao X., Nie F., Li J., Chen L., et al. Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat. Plants. 2021;7:774–786. doi: 10.1038/s41477-021-00934-w. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A., Li Z., Lohaus R., Van de Peer Y. Finding evidence for whole genome duplications: a reappraisal. Mol. Plant. 2019;12:133–136. doi: 10.1016/j.molp.2018.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.