Abstract
Background & Aims
Sporadic colorectal cancers arise from initiating mutations in APC, producing oncogenic β-catenin/TCF–dependent transcriptional reprogramming. Similarly, the tumor suppressor axis regulated by the intestinal epithelial receptor GUCY2C is among the earliest pathways silenced in tumorigenesis. Retention of the receptor, but loss of its paracrine ligands, guanylin and uroguanylin, is an evolutionarily conserved feature of colorectal tumors, arising in the earliest dysplastic lesions. Here, we examined a mechanism of GUCY2C ligand transcriptional silencing by β-catenin/TCF signaling.
Methods
We performed RNA sequencing analysis of 4 unique conditional human colon cancer cell models of β-catenin/TCF signaling to map the core Wnt-transcriptional program. We then performed a comparative analysis of orthogonal approaches, including luciferase reporters, chromatin immunoprecipitation sequencing, CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats) knockout, and CRISPR epigenome editing, which were cross-validated with human tissue chromatin immunoprecipitation sequencing datasets, to identify functional gene enhancers mediating GUCY2C ligand loss.
Results
RNA sequencing analyses reveal the GUCY2C hormones as 2 of the most sensitive targets of β-catenin/TCF signaling, reflecting transcriptional repression. The GUCY2C hormones share an insulated genomic locus containing a novel locus control region upstream of the guanylin promoter that mediates the coordinated silencing of both genes. Targeting this region with CRISPR epigenome editing reconstituted GUCY2C ligand expression, overcoming gene inactivation by mutant β-catenin/TCF signaling.
Conclusions
These studies reveal DNA elements regulating corepression of GUCY2C ligand transcription by β-catenin/TCF signaling, reflecting a novel pathophysiological step in tumorigenesis. They offer unique genomic strategies that could reestablish hormone expression in the context of canonical oncogenic mutations to reconstitute the GUCY2C axis and oppose transformation.
Keywords: Guanylin, uroguanylin, super-enhancer, Wnt signaling
Abbreviations used in this paper: cDNA, complementary DNA; cGMP, cyclic guanosine monophosphate; ChIP, chromatin immunoprecipitation; ChIP-seq, chromatin immunoprecipitation sequencing; CRISPRa, CRISPR activation; CRISPRi, CRISPR inhibition; DMEM, Dulbecco’s Modified Eagle Medium; DNase-seq, DNase hypersensitive sites sequencing; DNTCF, dominant negative TCF7L2; DOX, doxycycline; EGF, epidermal growth factor; FBS, fetal bovine serum; gRNA, guide RNA; HBSS, Hank’s Balanced Salt Solution; KO, knockout; LCR, locus control region; mRNA, messenger RNA; PBS, phosphate-buffered saline; preRNA, precursor messenger RNA; Pol II, RNA polymerase II; qRTPCR, quantitative real-time polymerase chain reaction; shRNA, short hairpin RNA; ST, heat-stable enterotoxin; TCGA, The Cancer Genome Atlas; UTR, untranslated region; WRE, Wnt recognition element
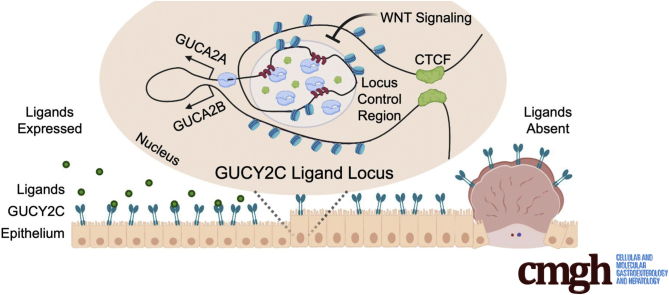
Graphical abstract
Summary.
Silencing receptor GUCY2C tumor suppressor signaling occurs widely in colorectal tumors, reflecting loss of GUCY2C ligand expression. This study identifies a novel β-catenin/TCF-sensitive locus control region that mediates ligand loss and can be targeted for gene reactivation with CRISPR activation.
In the healthy intestine, expression of the paracrine hormone Wnt at the crypt base promotes a transcriptional program driving stem cell renewal, while tapering expression up the crypt-surface axis supports epithelial differentiation. Wnt signaling destabilizes a cytosolic destruction complex, assembled on the scaffold protein APC, which targets the transcription cofactor β-catenin for proteasomal degradation. Disruption of the complex enables accumulation and nuclear translocation of β-catenin, which partners with TCF/LEF transcription cofactors to activate proliferation programs. Mutations in the APC-β-catenin-TCF axis are the most common drivers of colorectal tumorigenesis.1,2 Indeed, 80% of sporadic tumors arise from loss-of-function mutations in APC, in which acquired mutations produce allelic heterozygosity, and loss of the nonmutant allele (loss of heterozygosity) eliminates wild-type protein expression. These mutations liberate β-catenin from regulation by Wnt signaling, permitting oncogenic transcription, polyp formation, and accumulation of subsequent mutations (eg, p53, KRAS) driving tumorigenesis.1,3
While these mutational events are well defined, mechanisms contributing to tumor initiation and progression continue to evolve. Guanylyl cyclase C (GUCY2C) and its downstream effector, cyclic guanosine monophosphate (cGMP), have emerged as key intestinal tumor suppressors.4, 5, 6, 7, 8, 9, 10, 11, 12 GUCY2C, a member of the family of transmembrane receptor guanylyl cyclases, is activated by the diarrheagenic bacterial heat-stable enterotoxins (STs) and the structurally homologous endogenous hormones guanylin (GUCA2A) and uroguanylin (GUCA2B).5,13,14 While GUCA2B and GUCA2A quantitatively predominate in the small and large intestine, respectively, mechanisms regulating those levels are unknown. Peptide stimulation of GUCY2C drives cGMP accumulation, promoting electrolyte and fluid secretion, and the synthetic peptides linaclotide (ST analog; Linzess [Ironwood Pharmaceuticals, Boston, MA]) and plecanatide (GUCA2B analog; Trulance [Salix Pharmaceuticals, Bridgewater, NJ]) target this secretory function to treat chronic constipation syndromes.5,13 Beyond secretion, GUCY2C controls homeostatic processes dysregulated in tumorigenesis, including proliferation, metabolism, and differentiation programs.6, 7, 8, 9, 10,15 Silencing GUCY2C promotes crypt hyperplasia, cell cycle acceleration, DNA damage, and tumor susceptibility.6,7,9,15, 16, 17 Conversely, GUCY2C agonists and agents that elevate cGMP oppose intestinal tumorigenesis in mice and humans.11,12,18, 19, 20, 21
Silencing the GUCY2C signaling axis is a universal feature of colorectal tumorigenesis. Most tumor subtypes retain cell-surface GUCY2C expression throughout disease progression.22, 23, 24 However, transformation universally orphans the receptor through loss of hormones.7,16,21, 22, 23,25,26 Hormone loss silencing GUCY2C occurs early in dysplastic crypts and is conserved across species and tumor subtypes.16,22,23,25,26 GUCA2A is lost following APC inactivation in mouse models of conditional biallelic Apc deletion (ApcCKO/CKO) and Apc loss of heterozygosity (Apcmin/+).26 Further, silencing mutant β-catenin-TCF signaling restores GUCA2A levels.23 These observations suggest a pathophysiological model in which the earliest mutations in transformation suppress homeostatic GUCY2C signaling by eliminating its hormones. Moreover, they suggest the correlative therapeutic hypothesis that reconstituting GUCY2C signaling, by oral ligand replacement, by reactivating endogenous hormone generation, or by receptor-independent elevation of cGMP, may be a novel strategy for colorectal chemoprevention.4,5,27,28
Hormones regulating the GUCY2C axis are central to the pathophysiology of intestinal tumorigenesis. However, mechanisms regulating GUCA2A and GUCA2B levels in health and disease remain undefined. Here, comprehensive RNA sequencing (RNA-seq) analysis of unique conditional human colon cancer cell models of Wnt signaling29, 30, 31, 32 demonstrate that GUCA2A and GUCA2B are 2 of the most sensitive targets of APC-β-catenin-TCF regulation, reflecting transcriptional repression. Comparative analysis using orthogonal approaches to identify gene enhancers, including luciferase reporters, chromatin immunoprecipitation sequencing (ChIP-seq), CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/Cas9) knockout (KO), and CRISPR epigenome editing reveals a novel locus control region regulated by APC-β-catenin-TCF that silences GUCA2A and GUCA2B transcription. Moreover, these studies offer unique genomic strategies, including CRISPR epigenome editing, that could re-establish hormone expression in the context of mutant APC-β-catenin-TCF signaling to reconstitute the GUCY2C axis and oppose transformation.
Results
GUCY2C Ligands Are 2 of the Most Sensitive Targets of Wnt Signaling in Colon Cells
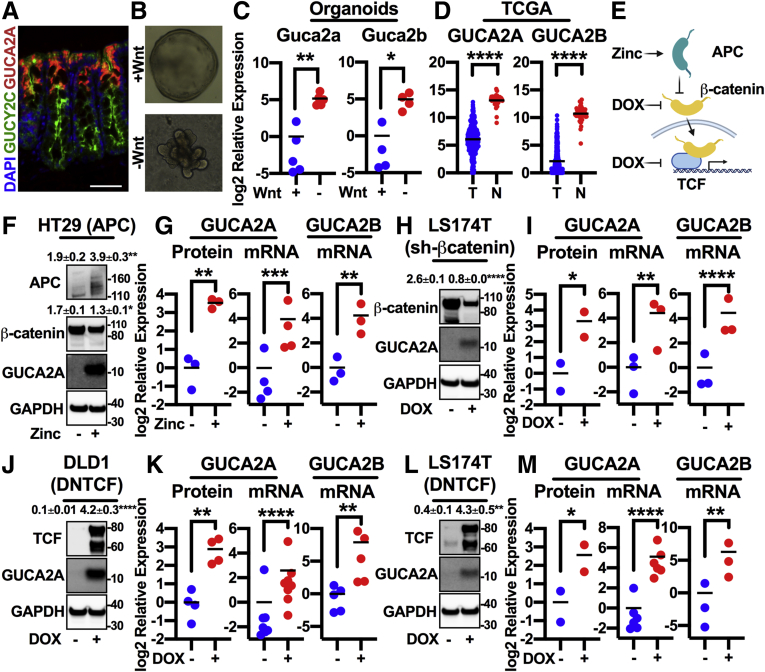
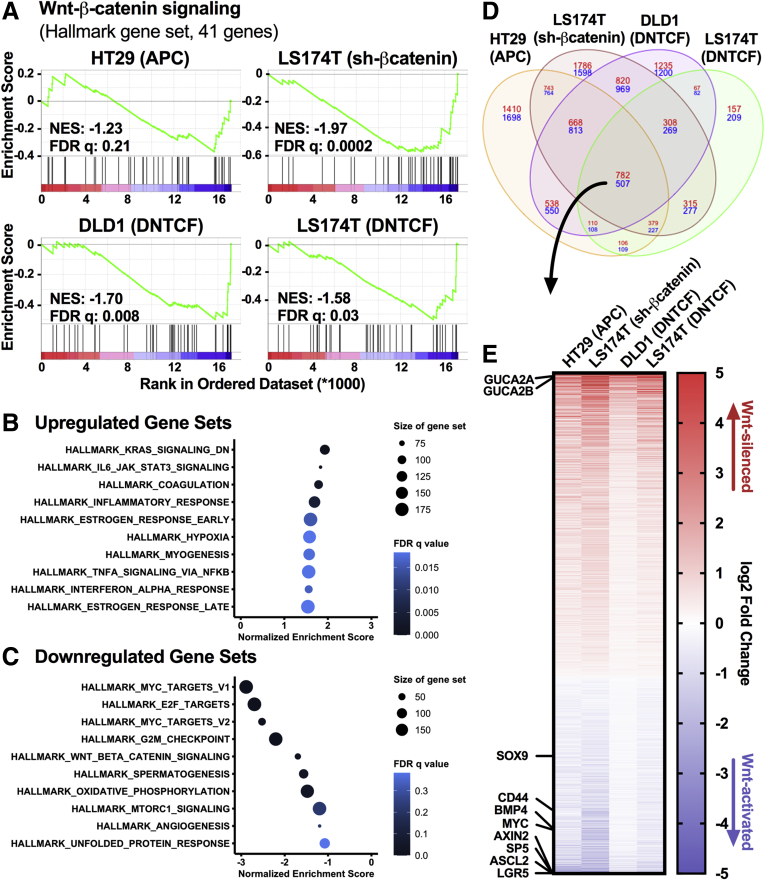
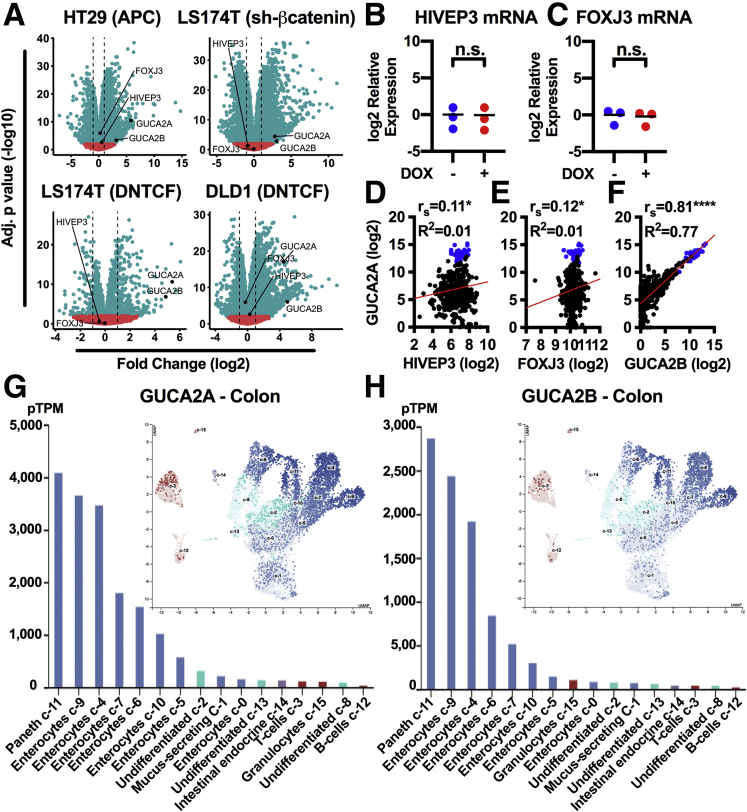
While GUCY2C is expressed by colonocytes along the rostral-caudal and crypt-surface axes, GUCA2A is produced by differentiated cells bordering the lumen and is absent in crypts (Figure 1A).13,14,33 This gradient is the inverse of gene sets driven by Wnt, suggesting reciprocal regulation. Indeed, compared with undifferentiated mouse colon spheroids maintained in Wnt, mouse colonoids differentiated in the absence of Wnt exhibited a 36- and 31-fold increase in Guca2a and Guca2b messenger RNA (mRNA), respectively (Figure 1B and C). Also, 80% of colon tumors exhibit aberrant Wnt signaling arising from either APC loss-of-function or β-catenin gain-of-function mutations.1,2 Transcriptomic analysis of colon tumors (n = 380) and normal mucosa (n = 51) from The Cancer Genome Atlas (TCGA) confirmed a 131- and 385-fold higher expression of GUCA2A and GUCA2B mRNA in normal tissue, respectively (Figure 1D). Further, activating a zinc-inducible wild-type APC transgene in HT29 human colon cancer cells, which harbor mutant APC,29 degrades β-catenin, disrupts Wnt signaling, and reconstitutes GUCA2A protein (11-fold), GUCA2A mRNA (15-fold), and GUCA2B mRNA expression (18-fold) (Figure 1F and G). Activation of a doxycycline (DOX)-inducible β-catenin short hairpin RNA (shRNA) in LS174T human colon cancer cells, which harbor gain-of-function mutations in β-catenin,30,31 reduces β-catenin accumulation and reconstitutes GUCA2A protein, GUCA2A mRNA, and GUCA2B mRNA expression (Figure 1H and I). Activation of a DOX-inducible dominant negative TCF7L2 (DNTCF) transgene in DLD1 and LS174T human colon cancer cells, which harbor mutations in APC or β-catenin, respectively, also restores GUCA2A protein, GUCA2A mRNA, and GUCA2B mRNA expression (Figure 1J–M). Transcriptomic analysis of these cell lines confirmed downregulation of the Wnt-β-catenin signaling gene set upon transgene induction (Figure 2A). Indeed, MYC Targets and Wnt-β-catenin signaling were among the 5 most downregulated gene sets in all cell lines (DLD1 gene sets shown in Figure 2B and C). Across all cell lines, 1289 genes were differentially regulated (Figure 2D, center of overlap), representing the core Wnt transcriptome, including canonical β-catenin-TCF targets such as MYC, AXIN2, SP5, ASCL2, and LGR5 (Figure 2E). Conversely, GUCA2A and GUCA2B were the 7th and 12th most upregulated transcripts among the 1289 genes.
Figure 1.
Wnt signaling silences GUCY2C ligand expression. (A) Immunofluorescence of GUCA2A (red), GUCY2C (green), and nuclei (blue) in mouse colon crypts. (B) Mouse colon organoids cultured in the presence (+) or absence (–) of Wnt3a, and (C) corresponding Guca2a and Guca2b mRNA. (D) GUCA2A and GUCA2B mRNA quantified by RNA-seq from human colorectal tumor (T) (n = 380) and normal (N) (n = 51) tissue from TCGA (COAD/READ datasets). (E) Colon cancer cells express 1 of 3 inducible inhibitors of Wnt signaling: zinc-inducible wild-type APC (HT29 cells), DOX-inducible β-catenin shRNA (LS174T cells), or DOX-inducible dominant negative TCF7L2 (DNTCF; DLD1 and LS174T cells). (F, G) HT29(APC) cells treated with 300 μM zinc for 24 hours express APC, resulting in loss of β-catenin and upregulation of GUCA2A protein, GUCA2A mRNA, and GUCA2B mRNA. (H, I) LS174T(shβ-catenin) cells treated with 1 μg/mL DOX for 72 hours lose β-catenin and upregulate GUCA2A protein, GUCA2A mRNA, and GUCA2B mRNA. (J, K) DLD1(DNTCF) cells treated with 1 μg/mL DOX for 24 hours express DNTCF, upregulating GUCA2A protein, GUCA2A mRNA, and GUCA2B mRNA. (L, M) LS174T(DNTCF) cells treated with 1 μg/mL DOX for 48 hours express DNTCF, upregulating GUCA2A protein, GUCA2A mRNA, and GUCA2B mRNA. (C, F–M) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 2–8 independent experiments indicated. Significance was determined by Student’s t test with matched analysis for independent experiments on log2-transformed results. Data are presented relative to noninduced cells. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Figure 2.
The GUCY2C ligands are among the most sensitive targets of β-catenin/TCF signaling. HT29(APC) cells were treated with 300 μM zinc for 24 hours, LS174T(shβ-catenin) cells were treated with 1 μg/mL DOX for 72 hours, DLD1(DNTCF) cells were treated with 1 μg/mL DOX for 24 hours, and LS174T(DNTCF) cells were treated with 1 μg/mL DOX for 48 hours. RNA-seq and differential gene expression analysis was performed in cells treated with or without the respective inducing agent. Unbiased gene set enrichment analysis on each cell line compared the ranked log fold change of 17,229 genes to the 50 hallmark gene sets maintained by the Molecular Signatures Database (MSigDB). (A) The Wnt-β-catenin signaling gene set enrichment plot for each cell line, with the corresponding normalized enrichment score and false discovery rate (FDR) q value. (B, C) The top 10 upregulated and downregulated gene sets for DLD1(DNTCF) cells, ranked by normalized enrichment score. Color indicates FDR q value, and size of the data points indicates the number of genes in each gene set. (D) Differential gene expression in the 4 cell lines by RNA-seq reveals 782 upregulated (red) and 507 downregulated (blue) genes upon silencing Wnt signaling. (E) Heatmap of the 1289 differentially expressed genes ranked by log2 fold change. GUCA2A and GUCA2B are the 7th and 12th most upregulated transcripts. RNA-seq results represent the average of 3 replicates.
Mutant Wnt Signaling Represses GUCA2A Nuclear Transcription
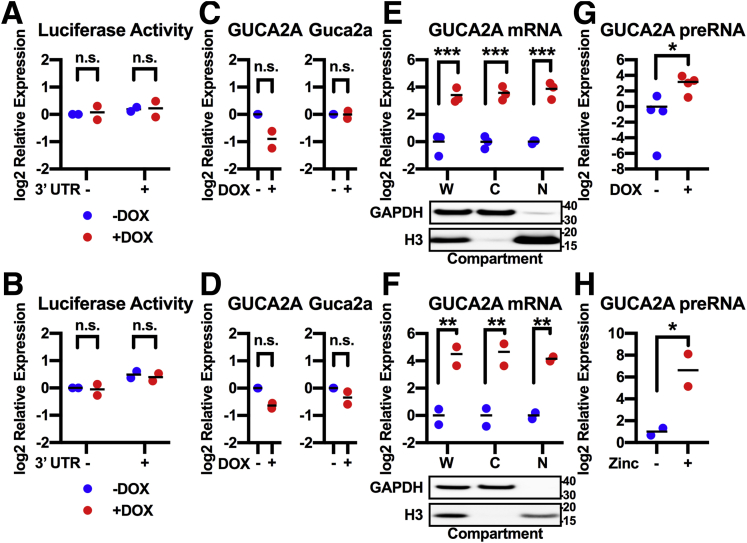
The above results show that the silencing of GUCY2C hormone expression by β-catenin-TCF reflects steady state transcript loss, agnostic to the relative contributions of transcriptional or posttranscriptional processes. Recently, we revealed that β-catenin-TCF regulated the incorporation of labeled uridine into GUCA2A mRNA, suggesting a level of transcriptional control.23 Here, silencing Wnt signaling did not regulate expression of luciferase reporter constructs containing the GUCA2A 3′ untranslated region (UTR) (Figure 3A and B) or expression constructs containing the full-length GUCA2A mRNA coding region (Figure 3C and D), suggesting little posttranscriptional regulation of steady-state transcript levels. In contrast, silencing Wnt signaling fully reconstituted nuclear GUCA2A mRNA levels, both in whole cells and their cytoplasm (Figure 3E and F). Reconstituted RNA transcripts contained GUCA2A introns, representing precursor mRNA (preRNA) (Figure 3G and H). Thus, APC-β-catenin-TCF regulation of GUCY2C hormone expression occurs at the earliest steps of transcription in the nucleus.
Figure 3.
β-catenin-TCF signaling regulates GUCA2A nuclear transcription. (A, B) Luciferase constructs driven by a constitutive (SV40) promoter and containing the guanylin 3′ UTR or no 3′ UTR, were expressed in (A) DLD1(DNTCF) or (B) LS174T(DNTCF) cells. Luciferase was quantified with (+) or without (–) 1 μg/mL DOX for 24 hours in DLD1 cells or 48 hours in LS174T cells. (C, D) Expression constructs containing the entire human GUCA2A gene from 5′ to 3′ UTR, or the entire murine Guca2a gene under the control of a constitutive (ROSA) promoter were expressed in (C) DLD1(DNTCF) or (D) LS174T(DNTCF) cells. mRNA was quantified with (+) or without (–) 1 μg/mL DOX for 24 hours in DLD1 cells, or 48 hours in LS174T cells. (E) DLD1(DNTCF) cells were treated with 1 μg/mL DOX for 24 hours and (F) HT29(APC) cells were treated with 300 μM zinc for 24 hours. GUCA2A mRNA was quantified in whole cell (W), cytoplasmic (C), or nuclear (N) fractions, with fractionation confirmed by GAPDH (cytoplasmic) or histone H3 (nuclear) protein. (G, H) GUCA2A preRNA was quantified in the (G) DLD1 and (H) HT29 whole cell fractions. (A-H) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 2–4 independent experiments indicated. Significance was determined by (A, B, E, F) 2-way analysis of variance or (C, D, G, H) Student’s t test with matched analysis for independent experiments on log2-transformed results. No significance was identified between any group in individual or combined experiments in A–D. Data are presented relative to noninduced cells. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
GUCY2C Hormones Occupy an Insulated Transcriptional Domain Regulated by Wnt Signaling
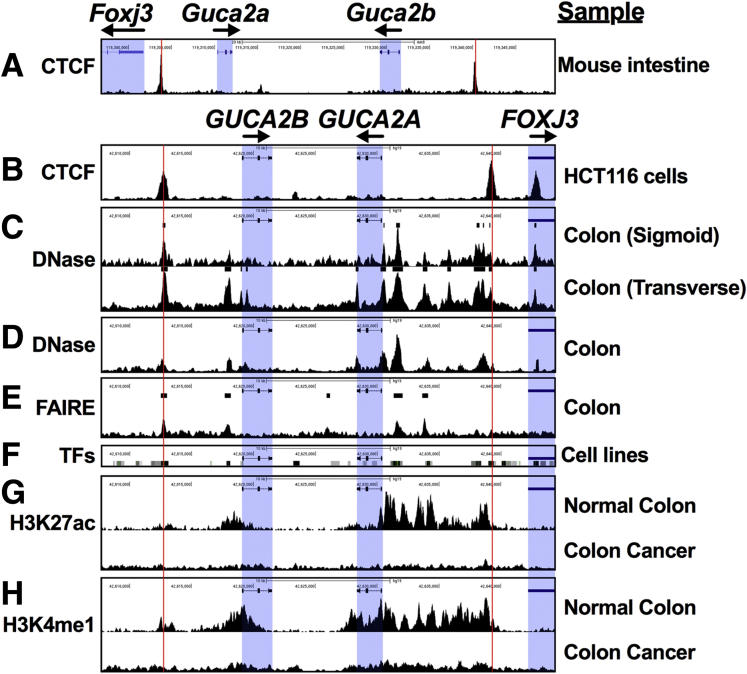
GUCA2A and GUCA2B are convergently transcribed, with their promoters separated by 21 kb in mice and 11 kb in humans. ChIP-seq suggests that these genes occupy a chromatin domain bounded by CTCF sites, an architecture that is conserved across species (Figure 4A and B).34, 35, 36 Between these CTCF sites, DNase hypersensitive sites sequencing (DNase-seq) of normal human colon identified several sites of DNase hypersensitivity, hallmarks of transcriptional activity (Figure 4C).37 Two of these sites correspond to the CTCF sites, 2 correspond to the gene promoters, and the remaining represent putative enhancers, a structure confirmed by additional DNase-seq (Figure 4C and D)38 and formaldehyde-assisted identification of regulatory elements sequencing (Figure 4E) analyses.39 These putative enhancers correspond to regions with the greatest transcription factor occupancy, as defined by ChIP-seq (Figure 4F).37 Regions upstream of both genes also harbor the enhancer-associated epigenetic marks H3K27ac (Figure 4G) and H3K4me1 (Figure 4H).38 Greater H3K27ac and H3K4me1 enrichment was observed upstream of GUCA2A than GUCA2B, consistent with greater expression of GUCA2A mRNA in the colon. Importantly, these marks were observed in normal human colon, but not colon cancer tissue (Figure 4G and H), suggesting enhancer inactivation consistent with transcriptional silencing during transformation.38
Figure 4.
Regulatory elements in the GUCA locus are silenced in colorectal cancer. Public datasets reveal regulatory features in the GUCA2A locus. (A) CTCF ChIP-seq from mouse intestinal epithelial cells (GSE98724; mm9, chr4:119,296,885–119,349,526). (B) CTCF ChIP-seq from human HCT116 colon cancer cells (GSE92879; hg19, chr1:42,607,745–42,644,356). (C) DNase-seq from sigmoid (top; GSE90366) and transverse colon (bottom; GSE90398). (D) DNase-seq from normal colon crypts (GSE77737). (E) Formaldehyde-assisted identification of regulatory elements (FAIRE) sequencing from normal colon (GSE94935). (F) Clusters of transcription factor density, representing ChIP-seq of 338 factors in 130 cell types (UCSC Genome Browser, ENCODE Transcription Factor Binding track). (G) H3K27ac ChIP-seq and (H) H3K4me1 ChIP-seq identified poised enhancers in normal colon (top) but not in colon cancer (bottom) (GSE77737). Red lines denote CTCF binding sites. Blue boxes denote gene bodies. Sequencing datasets were obtained from NCBI Gene Expression Omnibus and visualized in the UCSC genome browser.
This locus architecture, in which DNase, H3K27ac, and H3K4me1 enrichment terminate at the CTCF sites between GUCA2A and GUCA2B and their adjacent genes, FOXJ3 and HIVEP3, respectively, is consistent with a role for CTCF in transcriptional insulation.34 RNA-seq in the 4 cell lines revealed that GUCA2A and GUCA2B were among the most upregulated genes upon silencing Wnt signaling, while FOXJ3 and HIVEP3 were not significantly changed (Figure 5A). Quantitative real-time polymerase chain reaction (qRTPCR) analysis in all 4 cell lines confirmed that silencing APC/β-catenin signaling did not alter FOXJ3 or HIVEP3 expression (DLD1 cells shown in Figure 5B and C). These results recapitulate those from human normal colon and tumor samples in TCGA, in which HIVEP3 and FOXJ3 expression does not correlate with GUCA2A (Figure 5D and E). In contrast, expression of GUCA2A and GUCA2B directly correlate, and both are lost in colon tumors (Figure 5F). Indeed, GUCA2B is the gene most correlated with GUCA2A in colon tumors in TCGA, and single cell transcriptomics reveal that these transcripts are coexpressed in normal colon epithelial cells as well (Figure 5G and H).40,41 Thus, GUCA2A and GUCA2B are transcriptionally coregulated within an insulated domain sensitive to APC-β-catenin-TCF signaling.
Figure 5.
Transcriptional coregulation of GUCA2A and GUCA2B within an insulated domain sensitive to Wnt signaling. (A) RNA-seq volcano plots illustrate the fold change of 17,229 genes upon silencing Wnt signaling in the 4 inducible cell lines, with GUCA2A, GUCA2B, HIVEP3, and FOXJ3 indicated. Each gene is represented as a data point, dotted lines denote 2-fold change, and significance is indicated in red (P > .01) or blue (P ≤ .01). (B-C) qRTPCR of (B) HIVEP3 and (C) FOXJ3 in DLD1(DNTCF) cells treated with (+) or without (–) 1 μg/mL DOX for 24 hours. (D–F) GUCA2A mRNA compared with that of (D) HIVEP3, (E) FOXJ3, or (F) GUCA2B, quantified by RNA-seq from human colorectal tumor (n = 380; black) and normal (n = 51; blue) tissue from TCGA (COAD/READ datasets). Significance was determined by 2-tailed Spearman rank correlation (rs), and linear regression (R2) is indicated by the red line. (G, H) Single-cell gene expression data retrieved from The Human Protein Atlas reveals coexpression of (G) GUCA2A and (H) GUCA2B in normal colon epithelial cell clusters (insets show corresponding colon UMAP plots of cell clusters). (B, C) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 3 independent experiments indicated. Significance was determined by Student’s t test with matched analysis for independent experiments on log2-transformed results. Data are presented relative to noninduced cells. ∗P < .05; ∗∗∗∗P < .0001.
Regulation of the GUCA2A-GUCA2B Locus by Wnt Signaling Is Mediated by an Upstream RNA Polymerase II–Rich Super-Enhancer
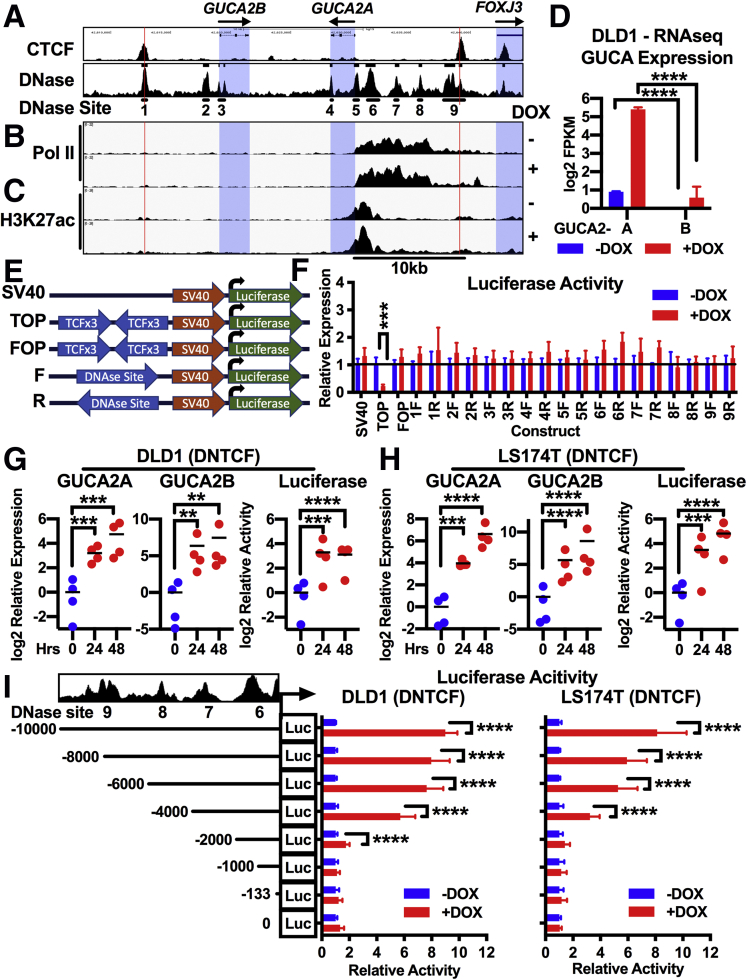
There are 9 (#1-#9) DNase hypersensitive sites within the GUCA2A and GUCA2B insulated domain representing putative regulatory DNA (Figure 6A). ChIP-seq in DLD1(DNTCF) cells revealed enrichment of RNA polymerase II (Pol II) in sites #5-#8, extending from the GUCA2A promoter to 6kb upstream (Figure 6B). In contrast, the GUCA2B 5′ upstream region was devoid of Pol II enrichment (Figure 6B). This differential Pol II enrichment is consistent with greater GUCA2A, compared with GUCA2B, transcript expression in this cell line (Figure 6D). While silencing β-catenin-TCF signaling induced GUCY2C hormone expression and Pol II loss at Wnt target genes such as LEF1 and ASCL2 (Supplementary Table 1), Pol II recruitment to the GUCA2A locus was unaffected, consistent with poised transcriptional machinery independent of Wnt status. Additionally, H3K27ac was enriched at sites #5–#6, and increased 1.3-fold (P = .008) after inducing DNTCF expression, consistent with transcriptional activation of this enhancer region (Figure 6C). The existence of a discrete β-catenin-TCF-sensitive element within this enhancer region was tested by cloning DNA fragments encompassing individual sites #1-#9 into luciferase reporters (Figure 6E). Unexpectedly, although DNTCF silenced luciferase expression in cells expressing the TOPflash TCF site-containing reporter,2 it did not activate expression in cells expressing any of the individual GUCA2A DNase site reporters (Figure 6F). In contrast, DNTCF induced the coordinated expression of GUCA2A and GUCA2B mRNA, as well as luciferase driven by a region extending from the GUCA2A 5′ UTR (+15) to beyond the CTCF site (–10,000; Figure 6G and H). Truncation analysis revealed a region between –1000 and –6000 that conferred maximum DNTCF sensitivity (Figure 6I) encompassing DNase hypersensitive sites #6 and #7.
Figure 6.
(See previous page). A Pol II-rich super-enhancer upstream of GUCA2A confers Wnt sensitivity. (A) Nine regions of DNase hypersensitivity were identified from sequencing tracks in Figure 4: CTCF ChIP-seq from human HCT116 colon cancer cells (GSE92879) and DNase-seq from transverse colon (GSE90398; hg19, chr1:42,607,745–42,644,356). ChIP-seq of (B) Pol II and (C) H3K27ac in DLD1(DNTCF) cells with (+) or without (–) 1 μg/mL DOX for 24 hours. (D) Corresponding RNA-seq FPKM quantification illustrating greater transcript expression of GUCA2A than GUCA2B in DLD1(DNTCF) cells. (E, F) Nine regions of DNase hypersensitivity were selected and cloned into luciferase reporters. (E) Diagram of enhancer-luciferase reporters driven by a constitutive (SV40) promoter with no upstream enhancer, upstream TCF sites (TOP), upstream mutant TCF sites (FOP), or upstream GUCA2A DNase-sensitive sites in forward (5′-3′) or reverse (3′-5′) orientation. (F) Luciferase constructs were expressed in DLD1(DNTCF) cells, and luciferase was quantified with (+) or without (–) 1 μg/mL DOX for 24 hours. (G, H) A luciferase construct driven by the region from +15 to –10,000 relative to the GUCA2A transcription start site was expressed in (G) DLD1(DNTCF) and (H) LS174T (DNTCF) cells. GUCA2A mRNA, GUCA2B mRNA, and luciferase activity were quantified following 1 μg/mL DOX for 0–48 hours. (I) Luciferase constructs containing 5′ truncations of the region from +15 to –10,000 relative to the GUCA2A transcription start site were expressed in DLD1(DNTCF) and LS174T (DNTCF) cells and luciferase activity was quantified following 1 μg/mL DOX for 24 hours (DLD1) or 48 hours (LS174T). (B, C) ChIP-seq tracks represent the average of 2 replicate immunoprecipitations. (D) RNA-seq results represent the average of 3 replicates. (G, H) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 4 independent experiments indicated. (F, I) Bars represent the average of 4 independent experiments ± SEM. Significance was determined by (G, H) 1-way, or (F, I) 2-way analysis of variance with matched analysis for independent experiments on log2-transformed results. Data are presented relative to noninduced cells. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
A 2683 bp Element Mediates β-Catenin-TCF-Sensitive Control of the GUCY2C Hormone Locus
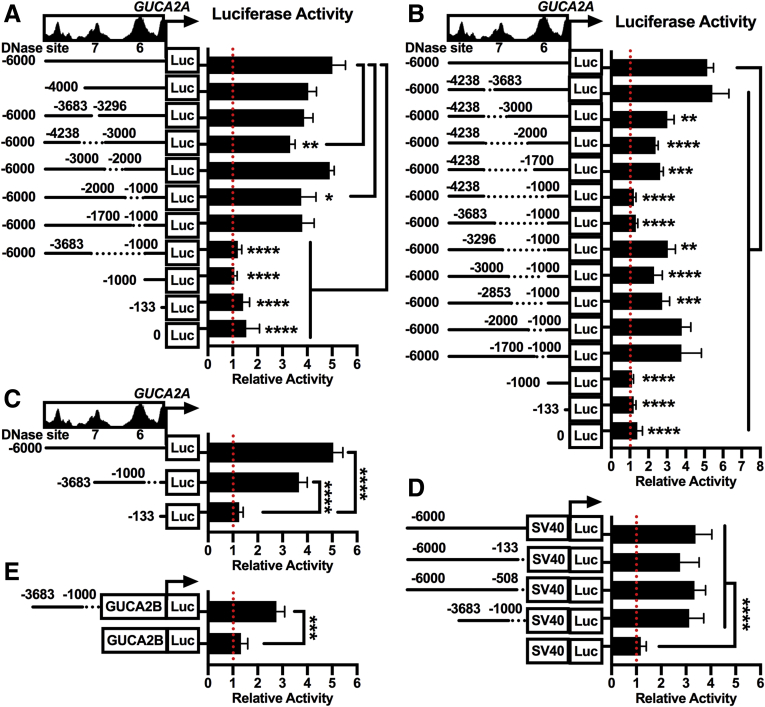
Mutational analysis to define the β-catenin-TCF-sensitive element revealed that deletion of sites #6 (1000-2000) or #7 (3000-4238) only modestly decreased, while a 2683-bp deletion spanning both sites (1000-3683) abolished, DNTCF-regulated luciferase expression (Figure 7A). Sequential truncation of either edge of this region partially reduced, but did not eliminate, DNTCF sensitivity (Figure 7B), demonstrating the contribution of the entire region to maximum β-catenin-TCF regulation. Expression of a luciferase construct containing the 2683 bp region with a minimal GUCA2A promoter conferred full β-catenin-TCF-sensitivity (Figure 7C). Moreover, while individual DNase sites failed to activate a constitutive (SV40) promoter (Figure 6F), the 2683 bp region was sufficient (Figure 7D), identifying this region as a bona fide β-catenin-TCF-sensitive enhancer. Finally, the 2683-bp enhancer conferred β-catenin-TCF-sensitivity to a minimal GUCA2B promoter (Figure 7E), supporting its role in coregulating GUCA2A and GUCA2B expression.
Figure 7.
Identification of a 2683-bp β-catenin/TCF-sensitive super-enhancer. (A–E) Luciferase constructs driven by the indicated GUCA2A DNA regions were expressed in DLD1(DNTCF) cells. Luciferase activity was quantified with (+) or without (–) 1 μg/mL DOX for 24 hours, and data are presented relative to noninduced cells. (A-B) Luciferase constructs were driven by the region from +15 to –6000 relative to the GUCA2A transcription start site, and harbored deletions of the indicated DNA positions, revealing a 2683-bp region responsible for β-catenin-TCF sensitivity. (C) Luciferase constructs were driven by the GUCA2A promoter (+15 to –133) and core enhancer (–1000 to –3683). (D) Luciferase constructs were driven by a constitutive (SV40) promoter and truncations of the GUCA2A upstream region from +15 to –6000. (E) Luciferase constructs were driven by the GUCA2B promoter (+33 to –100) and GUCA2A core enhancer (–1000 to –3683). Bars represent the average of 4 independent experiments ± SEM. Significance was determined by 2-way analysis of variance with matched analysis for independent experiments on log2-transformed results. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
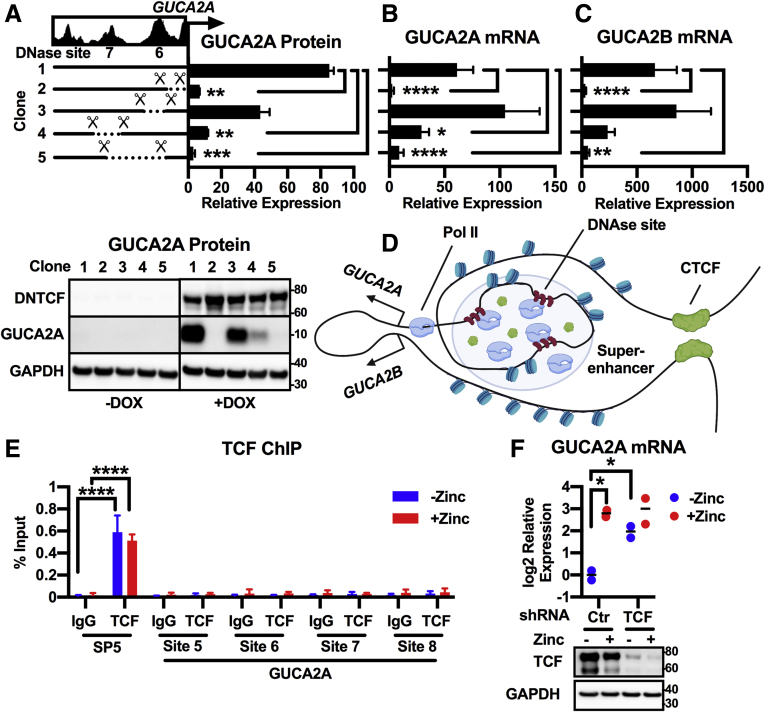
To validate the role of this enhancer in mediating β-catenin-TCF-sensitivity to GUCA2A and GUCA2B in the context of the chromosomal locus, DNase sites were eliminated with CRISPR-Cas9. Deletion of the GUCA2A promoter abolished GUCA2A mRNA and protein expression, and deletion of single DNase sites only incompletely reduced DNTCF sensitivity (Figure 8A and B). In contrast, deletion of both sites almost completely abolished DNTCF sensitivity. Moreover, GUCA2B mRNA expression paralleled that of GUCA2A, revealing that GUCA2A promoter-proximal and upstream super-enhancer elements synchronize the expression of both genes (Figure 8C). Collectively, these data support a paradigm involving a 2683 bp Pol II-rich locus control region (LCR) that confers β-catenin-TCF-sensitivity to the GUCA2A and GUCA2B promoters (Figure 8D). This sensitivity is conveyed indirectly by β-catenin/TCF. ChIP analysis did not identify TCF enrichment in the GUCA2A LCR (Figure 8E), indicating that TCF does not directly interact with the GUCA2A LCR to regulate transcription. Furthermore, elimination of TCF expression with shRNA restored GUCA2A mRNA expression (Figure 8F), mimicking the effect of DNTCF expression and indicating that TCF is not required for GUCA2A expression.
Figure 8.
Cas9 deletion confirms a super-enhancer necessary for β-catenin-TCF-sensitive locus control. DLD1(DNTCF) cells expressing Cas9 and untargeted (clone #1) or GUCA2A-locus-targeted gRNAs harbor biallelic deletions encompassing the GUCA2A promoter (clone #2), DNase site #6 (clone #3), DNase site #7 (clone #4), or both DNase sites (clone #5). Expression of (A) GUCA2A protein, (B) GUCA2A mRNA, and (C) GUCA2B mRNA was quantified with (+) or without (–) 1 μg/mL DOX for 24 hours. (D) Hypothetical model of GUCA2A and GUCA2B promoter positioning relative to a Pol II-rich super-enhancer region upstream of GUCA2A, consisting of multiple DNase sites. (E) ChIP-pcr in HT29(APC) cells treated with (+) or without (–) 300 μM zinc for 24 hours reveals enrichment of TCF at the promoter of a Wnt target gene, SP5, with a TCF-specific antibody, but not with control IgG. In contrast, TCF was not detected at sites within 6kb of the GUCA2A TSS: DNase site #5 (GUCA2A promoter), site #6, site #7, or site #8. (F) GUCA2A mRNA expression in HT29(APC) cells stably expressing an untargeted (Ctr) or TCF-targeted shRNA, and treated with (+) or without (–) 300 μM zinc for 24 hours. GUCA2A mRNA expression is retained despite TCF knockdown, illustrated by Western blot. (A–C) Bars represent the average of (A) 2 or (B, C) 3 independent experiments ± SEM, and data are presented relative to noninduced cells. Significance was determined by 2-way analysis of variance with matched analysis for independent experiments on log2-transformed results. (E) Bars represent the mean ± SD of 3 IPs, and data are presented relative to input DNA. Significance was determined by 2-way analysis of variance. (F) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 2 independent experiments indicated. Significance was determined by 2-way analysis of variance with matched analysis for independent experiments on log2-transformed results. Data are presented relative to noninduced cells receiving control shRNA. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Epigenetic Modifiers Targeted to the GUCA2A LCR Mediate Wnt-Independent Control of Gene Expression
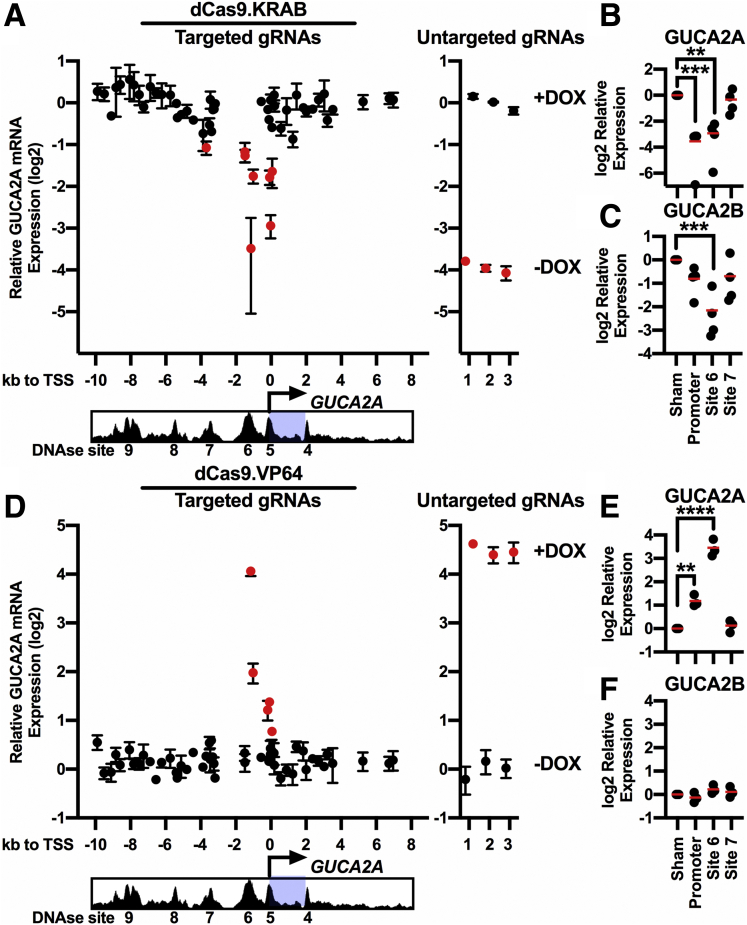
For unbiased validation of the results with luciferase and Cas9 KO, monoclonal DLD1(DNTCF) cells were generated stably expressing dCas9 fused to the SID4X and KRAB transcriptional repressor domains (dCas9.KRAB),42 or the VP64 transcriptional activation domain (dCas9.VP64).43 The ability to target dCas9.KRAB to the GUCA2A promoter was confirmed by ChIP, and knockdown of mRNA expression with guide RNA (gRNAs) targeting multiple genes was confirmed by qRTPCR (data not shown). Fifty-four gRNAs spanning the GUCA2A gene body, promoter, and enhancer regions were expressed individually in DLD1(DNTCF).dCas9.KRAB cells with DNTCF to identify sites controlling GUCA2A expression (Figure 9A). Indeed, individual gRNAs targeting the gene promoter, DNase site #6, or DNase site #7 repressed DNTCF-sensitive GUCA2A expression, supporting previous observations. Further, gRNA pools targeting the GUCA2A promoter or upstream enhancers also inhibited GUCA2A expression (Figure 9B). Importantly, pools targeting the enhancer (DNase site #6) also repressed DNTCF-sensitive GUCA2B expression, consistent with a role for the LCR in gene coregulation (Figure 9C).
Figure 9.
Targeting the GUCA2A LCR for Wnt-independent gene control. (A–C) CRISPRi in DLD1(DNTCF) cells stably expressing SID4X.dCas9.KRAB (dCas9.KRAB). (A) GUCA2A mRNA was quantified after stable transduction with 54 individual gRNAs targeting the GUCA2A locus and induction of DNTCF with 1 μg/mL DOX for 24 hours. gRNAs producing significant (P < .05) inhibition of GUCA2A mRNA relative to that of untargeted gRNAs are indicated in red. (B) GUCA2A mRNA and (C) GUCA2B mRNA were quantified after stable transduction with 3–5 gRNAs targeting nothing (sham), the GUCA2A promoter, DNase site #6, or DNase site #7, followed by treatment with 1 μg/mL DOX for 24 hours. (D–F) CRISPRa in DLD1(DNTCF) cells stably expressing dCas9.VP64. (D) GUCA2A mRNA was quantified after stable transduction with 54 gRNAs, as in panel A, in the absence of DOX. (E) GUCA2A mRNA and (F) GUCA2B mRNA were quantified after stable transduction with 3–5 gRNAs, as in panels B and C, in the absence of DOX. Data points represent the average of (A) 2 independent experiments ± SEM or (B) 1 experiment ± SEM. (B, C, E, F) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 3 independent experiments indicated. Significance was determined by 1-way analysis of variance with matched analysis for independent experiments on log2-transformed results. Data are presented relative to cells expressing untargeted gRNAs. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Finally, overcoming GUCA2A transcript loss in the context of mutant β-catenin-TCF-signaling represents a therapeutic goal to reconstitute hormone expression. As proof of principle, the 54 gRNAs were expressed in DLD1 (DNTCF).dCas9.VP64 cells in the absence of DNTCF. Compared with untargeted gRNAs, those targeting the GUCA2A promoter and DNase site #6 reconstituted GUCA2A expression, with greater reconstitution at site #6 than at the promoter (Figure 9D and E). In contrast to the effectiveness of the dCas9.KRAB system, dCas9.VP64 did not reconstitute GUCA2B expression (Figure 9F), possibly reflecting differences in the physical locus topology in the context of repressive Wnt signaling (VP64 experiments) compared with the absence of Wnt signaling (KRAB experiments). Nevertheless, these experiments confirm the potential to target the LCR to reconstitute GUCY2C hormone expression in the context of mutant β-catenin-TCF-signaling.
Discussion
The tumor suppressive axis controlled by the intestinal receptor GUCY2C and its paracrine ligands GUCA2A and GUCA2B is among the first pathways dysregulated in transformation. Loss of GUCY2C hormones is an early feature of tumorigenesis in mice and humans, occurring in dysplastic crypts in temporal proximity to APC loss.23 Here, we reveal that the GUCY2C hormones share an insulated genomic locus containing an LCR that mediates gene silencing by APC-β-catenin-TCF signaling. These mechanistic insights support the pathophysiologic hypothesis that silencing of GUCY2C hormone gene transcription is a key step in tumorigenesis, overcoming tumor suppressive signaling by the GUCY2C-cGMP axis.4,5,13 While hormone loss represents a well-recognized point of potential therapeutic intervention, studies here add reconstitution of gene expression to GUCY2C agonist replacement and cGMP-elevating agents as strategies to transform a disease of irreversible genetic mutations in APC or β-catenin into a reversible condition that restores cGMP signaling.4,5,11, 12, 13,18,19,21,28
To explore mechanisms linking mutant APC-β-catenin-TCF-signaling to GUCY2C ligand loss, we assembled human colon cancer cells engineered to define intestinal Wnt target genes.29, 30, 31, 32,44,45 These cells express inducible transgenes for wild-type APC, β-catenin shRNA, or dominant negative TCF7L2, which interrupt Wnt signaling at 3 discrete steps. Silencing Wnt signaling at any step reconstituted GUCA2A and GUCA2B expression, consistent with ligand loss in tumors in mice and humans.16,21,22,25 Interestingly, these genes have not been identified previously as Wnt targets in these models,32,44, 45, 46 likely reflecting the insensitivity of array-based approaches to genes with low transcript abundance in the context of Wnt signaling. Here, RNA-seq, applied to the 4 cell models for the first time, identified a common set of 1289 differentially expressed genes in which GUCA2A and GUCA2B mRNA were among the most sensitive readouts of Wnt signaling.
RNA-seq identifies changes in steady-state mRNA levels, reflecting either (or both) transcriptional (synthesis) and posttranscriptional (stability) regulation. With respect to GUCY2C hormones, silencing Wnt signaling increased nascent transcripts,23 transcript levels in the nucleus, and preRNA transcripts. Further, APC-β-catenin-TCF sensitivity was recapitulated with luciferase reporters containing the LCR. These observations are consistent with primarily transcriptional, rather than posttranscriptional, regulation of GUCY2C hormones by Wnt signaling. In that context, stable enrichment of H3K27ac and Pol II in the LCR, seen in both the presence and the absence of Wnt signaling, suggest dynamic regulation of hormone mRNA synthesis beyond the histone modification and Pol II recruitment steps preceding transcription initiation. To our knowledge, this represents the first example of a β-catenin/TCF–dependent transcriptional silencing switch controlling the events between Pol II recruitment to an LCR and activation of transcription at a gene promoter. Interestingly, in contrast to results here and in normal colon, H3K27ac is depleted at the GUCA2A LCR in colorectal cancer (Figure 4).38 These observations suggest that at least 2 mechanisms contribute to GUCY2C hormone loss in tumorigenesis. Acutely, APC-β-catenin-TCF signaling dynamically regulates hormone mRNA transcription at the LCR, a mechanism likely operating in normal colon to establish the ascending gradient of hormone expression along the crypt-surface axis.14 Chronically, during the evolution of a tumor, removal of activating chromatin marks and closure of chromatin domains produces LCR atrophy, reflected by H3K27ac ChIP-seq.38 Elucidating these phases of regulation will inform therapeutic strategies focused on reconstitution of gene expression in existing tumors.
While mechanisms activating gene expression by Wnt signaling are defined, those silencing gene expression remain incompletely understood. In Wnt-activated genes, TCF at consensus DNA Wnt recognition elements (WREs) in target gene promoters or enhancers binds β-catenin, which recruits chromatin remodeling enzymes and coactivators required for transcription.2 Paradoxically, in Drosophila, Wnt-repressed genes also include TCF binding sites, which differ from canonical WREs and are required for repression.47 In that context, there are no canonical WREs in the GUCA2A LCR (Supplementary File 1). Further, TCF was not enriched in the GUCA2A LCR by ChIP analysis (Figure 8E), ruling out binding to noncanonical WREs. Moreover, shRNA inhibition of TCF expression restored GUCA2A mRNA production (mimicking DNTCF), eliminating the possibility that TCF directly regulates GUCY2C hormone transcription (Figure 8F). Thus, Wnt regulation of GUCY2C hormone mRNA synthesis appears indirect, wherein β-catenin-TCF controls expression of the transcriptional machinery required for hormone regulation. The 1289 Wnt target gene set revealed by RNA-seq includes 95 transcription factors (Supplementary Table 2), many of which are known intermediates of Wnt (ie, MYC, SP5, and ASCL2). Ongoing analyses are exploring interactions between this Wnt transcriptional network and the GUCA2A LCR.
This is the first report to reveal transcriptional coregulation of GUCY2C hormones and its sensitivity to Wnt repression mediated by cis elements within an insulated genomic locus. This regulation is mediated by an LCR containing 4 DNase hypersensitive sites. Deletion of individual sites from luciferase constructs or from the genomic locus did not eliminate gene regulation, suggesting functional redundancy that might reflect evolutionary pressure to retain GUCY2C hormone expression in healthy intestine. Indeed, loss of GUCY2C hormone expression has been implicated in the pathophysiology of colorectal cancer, inflammatory bowel disease, irritable bowel disease, and obesity.4,5,13 DNase sites clustered in LCRs form phase-separated transcriptional condensates rich in transcriptional machinery.48 The region of Pol II and H3K27ac enrichment upstream of GUCA2A is consistent with this model. Further, LCRs integrate multiple gene control elements.49 Here, site #9 associates with CTCF and borders H3K27ac, supporting its role as an insulator, while sites #6 and #7 together form a 2683-bp Wnt-sensitive region. This 2683-bp region is a bona fide enhancer, with the ability to confer Wnt sensitivity to luciferase reporters driven by the GUCA2A, GUCA2B, or SV40 promoters. Further, deleting the enhancer with Cas9, or targeting it with the repressor dCas9.KRAB, eliminates Wnt sensitivity from both genes. Together, these observations are consistent with a model in which the LCR forms a transcriptional condensate which concentrates the machinery mediating Wnt-sensitive coregulation of GUCA2A and GUCA2B expression (Figure 8D). While speculative, ongoing chromosome conformation capture studies will clarify this working model.
Enhancer DNA sequences share common features, including sensitivity to DNase, enrichment with transcriptional machinery (eg, Pol II), and histone modifications (eg, H3K27ac). Here, putative enhancers with these characteristics were identified from public databases, and enhancer activity was validated with traditional reporter plasmids. Complementary analytic strategies included a candidate approach, incorporating individual DNase sites into luciferase reporters, and an unbiased approach, incorporating serial truncations of the 10-kb sequence spanning the GUCA2A TSS and nearest CTCF site (Figure 6). While the candidate approach was not yielding, and no single DNase site regulated luciferase expression, the unbiased approach revealed regulatory activity corresponding to sites #6 and #7, underscoring a pitfall of overly reductionist approaches to examining DNA elements. These considerations highlight the importance of applying comprehensive strategies to capture regulatory elements that function as small units independent of native chromatin, as well as those that function as large interdependent units, which can include super-enhancers and LCRs capable of forming transcriptional condensates.
Complementary CRISPR approaches were used to examine Wnt sensitivity of the GUCA2A 5′ super-enhancer, including CRISPR/Cas9 KO, CRISPR inhibition (CRISPRi), and CRISPR activation (CRISPRa). In that context, there was concordance between luciferase reporter and CRISPR KO analyses. However, luciferase analyses only revealed regulation of reporter gene expression, while CRISPR KO analyses revealed regulation of both GUCA2A and GUCA2B mRNA expression, identifying the super-enhancer as an LCR. Pitfalls of CRISPR KO include unanticipated alterations to chromatin architecture and the creation of new DNA motifs at recombination sites. CRISPRi and CRISPRa tiling screens have emerged as a less disruptive tool to interrogate enhancers through epigenetic modulation. Thus, to validate CRISPR KO analyses, CRISPRi and CRISPRa were targeted to the GUCA2A locus to identify sites contributing to hormone expression. CRISPRi targeting site #6 and site #7 repressed GUCA2A and GUCA2B expression, confirming LCR activity. Conversely, CRISPRa targeting site #6 reconstituted GUCA2A mRNA expression. Interestingly, CRISPRa targeting the LCR failed to reconstitute GUCA2B expression, likely reflecting the relatively weak transactivation capacity of the VP64 domain,50 and barriers imposed by active repressive Wnt signaling on interactions between the LCR and GUCA2B promoter. Notably, this is the first example of GUCY2C hormone mRNA reconstitution in the context of repressive mutant APC-β-catenin-TCF signaling. Indeed, LCR-targeted CRISPRa produced greater reconstitution than CRISPRa targeted to the GUCA2A promoter.
These observations provide a compelling rationale for pathologic enhancer identification and targeted epigenome modulation to overcome active oncogenic signaling, and proof-of-principle for reconstitution of GUCY2C ligand expression. In the context of colorectal cancer, β-catenin-TCF–mediated decommissioning of the GUCY2C hormone LCR represents a previously unrecognized pathophysiologic step in tumorigenesis. This mechanistic insight suggests several future pathways for clinical translation. Ongoing efforts to identify the intermediate factors conveying the β-catenin-TCF signal to the LCR should reveal novel targets for small molecule inhibitors, which could be employed for hormone reconstitution to oppose tumorigenesis. We also envision future applications for direct LCR targeting with CRISPRa, enabling gene activation without a priori knowledge of the transcription factor network acting on the locus. Indeed, CRISPRa has recently been employed to increase in situ gene expression in tumor stroma to augment antitumor immune responses.51 Given the sustained expression of the orphaned GUCY2C receptor on transformed cells, it is tempting to speculate that intratumoral reactivation of GUCY2C ligand expression with CRISPRa could have a therapeutic role. Ultimately, the present study reveals a mechanistic basis for GUCY2C hormone loss in colorectal cancer, creating a unique opportunity to reverse GUCY2C silencing and oppose tumorigenesis in the context of mutant Wnt signaling.
Materials And Methods
Cell and Organoid Culture Reagents
McCoy’s 5A (#10050CV), Dulbecco’s Modified Eagle Medium (DMEM) (#10013CV), DMEM/F12 (#10092CV), Matrigel (#354230), and cell recovery solution (#354253) were from Corning (Corning, NY). Ca2+/Mg2+-free Hank’s Balanced Salt Solution (HBSS) (#14175095), GlutaMax supplement (#35050061), penicillin-streptomycin (#15140122), HEPES (#15630080), HyClone fetal bovine serum (#SH3007103), hygromycin B (#10687010), zeocin (#R25005), blasticidin hydrochloride (#A1113903), geneticin/G418 sulfate (#10131027), doxycycline hydrochloride (#BP2653-1), puromycin (#A1113803), N-2 supplement (#17502048), and B-27 supplement (#12587010) were from Thermo Fisher Scientific (Waltham, MA). N-acetyl-l-cysteine (#A9165), A83-01 (#SML0788), SB202190 (#S7067), nicotinamide (#NO636), and zinc chloride (#Z0152) were from Sigma-Aldrich (St Louis, MO). Wnt3a-, R-Spondin-, and Noggin- conditioned medium (from L-WRN cells; #CRL-3276) was from the American Type Culture Collection (Manassas, VA). Mouse recombinant epidermal growth factor (EGF) (#315-09) and Noggin (#250-38) were from PeproTech (Rocky Hill, NJ). Human recombinant R-spondin-1 (#4645-RS-025/CF) was from R&D Systems (Minneapolis, MN). CHIR99021 (#129830-38-2) and Y-27632 (#252917-06-9) were from StemCell Technologies (Vancouver, Canada).
Mouse and Organoid Studies
Animal protocols were approved by the Thomas Jefferson University Institutional Animal Care and Use Committee. Colon organoids from wild-type C57B/6(J) mice (6–12 weeks old) were cultured according a protocol modified from Fan et al.52 Mouse colon was dissected, flushed with cold HBSS, everted onto a gavage needle, and vortexed with six 5-second pulses in cold HBSS supplemented with 0.5% penicillin-streptomycin. The colon was incubated in HBSS supplemented with 20 mM EDTA at 37°C for 30 minutes, then vortexed in cold HBSS, with eight 5-second pulses to release crypts. Crypts were centrifuged at 125 g for 3 minutes and resuspended in cold complete medium (DMEM/F12 supplemented with 1% Glutamax, 1% penicillin-streptomycin, and 1% HEPES). A total of 500 crypts were plated in 50 μL of Matrigel in 24-well plates, incubated for 15 minutes at 37°C, and then overlaid with 500 μL of complete medium supplemented with Wnt3a-, R-spondin–, and Noggin-conditioned medium (L-WRN, 1:1 dilution), 1× N-2, 1× B-27, 100 μM N-acetyl-l-cysteine, 50 ng/mL EGF, 500 nM A83-01, 10 μM SB202190, 10 mM nicotinamide, 10 μM CHIR99021, and 10 μM Y-27632. Medium was replaced every 3 days, and organoids were passaged weekly. For organoid differentiation studies, medium was replaced with differentiation medium (complete medium supplemented with 1 μg/mL R-spondin-1, 100 ng/mL Noggin, 1× N-2, 1× B-27, 100 μM N-acetyl-l-cysteine, 50 ng/mL EGF, 500 nM A83-01, and 10 μM Y-27632). Differentiation medium was replaced daily for 3 days. To harvest, Matrigel containing organoids was dissolved in cell recovery solution for 1 hour at 4°C. Organoid pellets were flash frozen and stored at –80°C for biochemical analyses.
Cell Lines
DLD1 and LS174T cells stably expressing Tet-inducible dominant negative TCF7L2 (DNTCF), and LS174T cells stably expressing Tet-inducible shRNA to β-catenin were provided by Dr. H. Clevers in November 2013.30,31 Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 500 μg/mL zeocin, and 10 μg/mL blasticidin. Cells were induced with 1 μg/mL DOX for 24 hours (DLD1-DNTCF), 48 hours (LS174T-DNTCF), or 72 hours (LS174T- shβ-catenin) for mRNA analyses, or an additional 24 hours for protein analyses. HT29 cells stably expressing zinc-inducible wild-type APC were provided by Dr. B. Vogelstein in September 2013.29 HT29 cells were cultured in McCoy’s-5A supplemented with 10% FBS and 600 μg/mL hygromycin. Cells were induced with 300 μM zinc chloride for 24 hours for mRNA, or 48 hours for protein analyses. Conditional cell lines were authenticated at each use by testing their genetic inducibility, and all cell lines were confirmed to be free of mycoplasma semiannually.
Immunofluorescence
Following deparaffinization and rehydration in a xylene-ethanol-water gradient, 4μm sections of wild-type mouse [C57BL/6(J)] intestines underwent antigen retrieval by heating at 100°C for 15 minutes in a pressure cooker in pH 9.0 DAKO antigen retrieval buffer (Agilent Technologies, Santa Clara, CA; S236784-2). The monoclonal antibody to GUCY2C (MS20) was described previously,20 and the antisera to GUCA2A (#2538) was kindly provided by Dr. M. Goy.33 GUCY2C and GUCA2A were detected by tyramine signal amplification, as described previously.53 Images were captured using an EVOS FL auto cell imaging system (Thermo Fisher Scientific).
Immunoblots
Nuclear and cytoplasmic cell fractions were separated with the Nuclei EZ Prep kit (Sigma-Aldrich; #NUC-101), per kit instructions. Protein was extracted from nuclear, cytoplasmic, or total cell lysates using M-Per (Thermo Fisher Scientific; #78501) supplemented with protease inhibitors (Thermo Fisher Scientific; #a32955). Lysates were prepared in 4X LDS Sample buffer (Invitrogen, Waltham, MA; #NP0007) supplemented with 2.75 mM 2-Mercaptoethanol (Thermo Fisher Scientific; #21985023), boiled at 100°C for 8 minutes, loaded onto 4%–12% Bis-Tris gels (20 μg/lane; Thermo Fisher Scientific; #NP0336), and transferred to nitrocellulose membranes with an iBlot station (Thermo Fisher Scientific; #IB301031). Blots were blocked for 1 hour in phosphate-buffered saline (PBS) containing 0.1% Tween 20 and 10% milk, then incubated overnight at 4°C with primary antibodies at the indicated dilutions targeting GAPDH (#2118, 1:5000), APC (#2504, 1:1000), β-catenin (#8480, 1:1000), TCF (#2569, 1:1000), Histone H3 (#4499, 1:1000), or Cas9 (#19526, 1:1000) from Cell Signaling Technology (Danvers, MA), or GUCA2A (#HPA018215, 1:250) from Sigma-Aldrich. Secondary antibodies conjugated to horseradish peroxidase were used at a 16 pg/μL (Jackson Immuno Research Laboratories, West Grove, PA; #111-035-144 or #115-035-062). Blots were developed with Dura or Femto Chemiluminescent Substrates (Thermo Fisher Scientific; #34075, #34095) and imaged on a ChemiDoc MP Imaging Station (Bio-Rad, Hercules, California). Bands were quantified by densitometry normalized to that of GAPDH using FIJI software (ImageJ version 2.0.0-rc-49).
RNA Analysis
Samples were lysed in RLT buffer supplemented with 550 μM 2-mercaptoethanol, genomic DNA was removed, and RNA was purified on spin columns using the RNeasy Plus kit (Qiagen, Hilden, Germany; #74134) according to kit directions. RNA concentration and purity were measured with a Nanodrop 1000 (Thermo Fisher Scientific), and 200 ng of RNA was reverse transcribed to complementary DNA (cDNA) using the TaqMan Reverse Transcription kit according to kit directions (Thermo Fisher Scientific; #N8080234). Transcripts were quantified by qRTPCR using Taqman primer probes on an ABI PRISM 7000 System (Thermo Fisher Scientific), with TaqMan Universal PCR Master Mix (Thermo Fisher Scientific; #4318157), per kit instructions. For analysis of preRNA transcripts, primer pairs recognizing introns were used at 125–500 nM per reaction, and qRTPCR was performed with SYBR Green Master Mix (Thermo Fisher Scientific; #A2572), per kit instructions. Transcript abundance was calculated by the delta-delta Ct method, normalized to that of GAPDH mRNA or GAPDH preRNA. Primers and probes are listed in Supplementary Table 3.
RNA Sequencing
Total RNA was extracted as above (triplicate samples from each of 4 inducible cell lines, treated with and without respective inducing agents; 24 samples total), and library preparation, sequencing, alignment, and read counting was performed by Novogene Corporation (Tianjin, China). Briefly, libraries were prepared from 1 μg RNA with the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA; #E7530L) according to kit directions, and index codes were added to attribute sequences to each sample. mRNA was purified from total RNA using poly-T oligo magnetic beads and fragmentated with divalent cations in 5X NEBNext First Strand Synthesis Reaction Buffer. First-strand cDNA was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA was synthesized with DNA polymerase I and RNase H. Fragments were end-repaired, blunt-ended, 3′ adenylated, and ligated to hairpin loop NEBNext Adaptors. Then, 150- to 200-bp cDNA fragments were selected with the AMPure XP system (Beckman Coulter, Brea, CA), then incubated with USER Enzyme (New England Biolabs) at 37°C for 15 minutes, followed by 5 minutes at 95°C. Library amplification by PCR used Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. Libraries were sequenced on an Illumina Novaseq 6000 platform (Illumina, San Diego, CA), generating 150-bp paired-end reads. Raw reads were processed with in-house perl scripts to remove low-quality reads containing adapter and poly-N sequences. Clean reads were aligned to the human (hg19) reference genome using TopHat (v2.0.12)54 and reads mapping to each gene were counted with HTSeq (v0.6.1).55 Genes with counts per million >0.5 in at least 3 samples were included in subsequent analyses (17,728 genes). Differential gene expression (adjusted P value <.05) was determined between cells treated with or without respective inducing agents, using the limma (v3.40.6) and edgeR (v3.26.8) packages in R (R Foundation for Statistical Computing, Vienna, Austria, version 1.2.1335).56,57 Gene set enrichment analysis was performed for each cell line, comparing the 17,728 genes (ranked by log-fold change) to the H:Hallmark Gene Sets in MSigDB (v.7.2),58 and results were plotted using the ggplot2 (v.3.3.0) package in R.59
Plasmids and Cloning
For GUCA2A luciferase reporter assays, DNA fragments were amplified by PCR from the RP11-799L22 BAC (BACPAC Genomics), which contains the human GUCA2A/GUCA2B locus. Control inserts containing TCF or mutant TCF binding sites were amplified from the TOPflash and FOPflash luciferase reporter plasmids (Sigma-Aldrich; #21-170, #21-169). The PGL3-Basic and PGL3-Promoter (Promega, Madison, WI; #E1751, #E1761) luciferase reporter plasmids were linearized with NcoI (#R3193)/XhoI (#R0146) or SacI (#R0156)/XhoI, respectively, from New England Biolabs. Gibson assembly of inserts and linearized plasmids was performed with the Geneart Seamless Cloning and Assembly kit (Thermo Fisher Scientific; #A13288) per package instructions. CRISPR-Cas9 plasmids were obtained from Addgene (Watertown, MA): SID4X.dCas9.KRAB was a gift from Jason Gertz (#106399),42 pcDNA-dCas9-VP64 was a gift from Charles Gersbach (#47107),43 and Lentiguide-Puro (#52963) was a gift from Feng Zhang.60 The active Cas9 plasmid, pLv5-Cas9-Neo, was from Sigma (#CAS9NEO). Protospacer-adjacent motifs in the GUCA2A locus were identified with the online tool, CHOPCHOP,61 and gRNAs without off-target binding sites were selected. Forward and reverse gRNA oligos were designed, annealed, and ligated into the Esp3I (New England Biolabs; #R0734)-digested Lentiguide-Puro vector according to directions provided by the Zhang lab on Addgene. Plasmid DNA was prepared from overnight E-coli cultures using HiPure Plasmid Midiprep kits (Thermo Fisher Scientific; #K210005). Plasmids were validated by Sanger sequencing by Eurofins Genomics. All cloning primers and gRNA sequences are listed in Supplement Table 1.
Luciferase Reporter Assay
Cells were seeded in white 96-well dishes at 20,000 cells/well. The following day, cells were cotransfected with 250 ng of Firefly luciferase plasmid and 50 ng of Renilla luciferase plasmid (pRLTK; Promega; #E2241), using Fugene HD (Promega; #E2311), according to kit instructions. DOX was added to induce cells for 24 hours (DLD1) or 48 hours (LS174T). A total of 48 hours after transfection, cells were lysed with the Dual-Glo Luciferase Assay kit (Promega; #E2940), following kit instructions. Luminescence was read on a PolarStar Omega Plate Reader (BMG Labtech, Ortenberg, Germany). Firefly luciferase activity was normalized to Renilla luciferase activity in each well.
CRISPR Cell Line Generation
DLD1(DNTCF) cells were transfected with SID4X.dCas9.KRAB or pcDNA-dCas9-VP64 using Fugene HD (Promega; #E2311), or were transduced with pLv5-Cas9-Neo lentiviral supernatants. After 48 hours, cells were selected with 1mg/mL Geneticin (Thermo Fisher Scientific; #10131027) until death of nontransduced cells. Stable cells were passaged into 96-well plates by limiting dilution. Single colonies were identified, expanded for 1 month, and probed for Cas9 protein expression. A single subclone was selected for subsequent experiments. For CRISPRi and CRISPRa experiments, DLD1(DNTCF).dCas9.KRAB (clone E9) or DLD1(DNTCF).dCas9.VP64 (clone A4) cells were transduced with lentiviral gRNA supernatants, incubated for 48 hours, and selected with 1 μg/mL Puromycin for 72 hours. Cells were induced with or without 1 μg/mL DOX for 24 hours and collected for mRNA analysis. For CRISPR KO experiments, DLD1(DNTCF).Cas9 (clone C1) cells were transduced with lentiviral gRNA pools targeting the GUCA2A DNase site #6, DNase site #7, both DNase sites, or nothing (scrambled gRNAs). After 48 hours, cells were selected with 1 μg/mL puromycin until death of nontransduced cells. Stable cells were passaged into 96-well plates by limiting dilution. Single colonies were identified, expanded for 1 month, genomic DNA was harvested, and target loci were amplified by PCR to identify deletions. A single subclone from each group was selected for subsequent experiments, including a control clone identified with a deletion encompassing the GUCA2A promoter and a portion of the 5′ coding region. No clone was initially identified with a deletion spanning both DNase sites. The clone harboring a deletion of DNase site #6 was retransduced with gRNAs spanning both DNase sites. Subclones were again isolated and expanded, and 1 clone was identified with a deletion spanning both sites.
Lentiviral Production and Transduction
Lentiviral packaging and transfer plasmids were from Addgene: pCMV-dR8.2-dvpr (#8455) and pCMV-VSV-g (#8454) were gifts from Bob Weinberg,62 and pPGS-DNTCF4(deltaN41) was a gift from E. Fearon (#19284).63 TCF and β-catenin shRNA transfer plasmids were obtained as bacterial glycerol stocks from Sigma (sequences listed in Supplementary Table 1). Two million HEK293T cells were plated in 10-cm dishes and cultured in DMEM supplemented with 10% FBS. The following day, cells were cotransfected with 3 μg VSV-g, 6 μg dR8.2-dvpr, and 6 μg of lentiviral transfer plasmids using the Profection Mammalian Transfection reagents (Promega; #E1200) according to the kit protocol. Media was replaced the following day. Viral supernatants were collected 48 hours after transfection and filtered through 0.45 μm syringe filters (Sigma-Aldrich; #SLHV033RS). Cancer cell lines were transduced with viral supernatants supplemented with 0.8 μg/mL Polybrene (Sigma-Aldrich; #TR-1003-G), incubated for 48 hours, then selected with 1 μg/mL Puromycin (Thermo Fisher Scientific; #A1113803). Cells were collected for analysis 72 hours after selection (upon death of nontransduced cells), or propagated to produce stable cell lines.
Chromatin Immunoprecipitation
ChIP was performed according to a protocol modified from Schmidt et al.64 Cells grown to confluence in 15-cm dishes were fixed with 1% formaldehyde (Thermo Fisher Scientific; #28906) for 10 minutes at room temperature, then quenched with 180 mM glycine (Thermo Fisher Scientific; #15527013). Cells were washed with ice-cold PBS, collected, resuspended in lysis buffer 1 (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and protease inhibitors), and rotated for 10 minutes at 4°C. Cells were pelleted, resuspended in lysis buffer 2 (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitors), and rotated for 5 minutes at 4°C. Cells were pelleted, resuspended in lysis buffer 3 (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, and protease inhibitors), and sonicated to an average length of 1kb in a Q800R2 sonicator (QSonica, Newtown, CT) at 4°C. Sheared chromatin was frozen at –80°C. Protein G Dynabeads (Thermo Fisher Scientific; #100003D) were washed 3 times in blocking buffer (0.5% bovine serum albumin in PBS; Sigma-Aldrich; #BP1605). A total of 25 μL of bead slurry was incubated with 5 μg of ChIP antibody per IP, rotating overnight at 4°C. ChIP antibodies targeted TCF7L2 (Sigma-Aldrich; #17-10109), Cas9 (Active Motif, Carlsbad, CA; #61757), or were IgG control (Abcam, Cambridge, United Kingdom; #ab171870). The following morning, unbound antibody was washed from the beads, and antibody-coated beads were incubated with 25 μg of chromatin diluted in lysis buffer 3 supplemented with 1% Triton X-100 and protease inhibitors. A 2% sample of input chromatin was set aside at –20°C, and beads were rotated with chromatin at 4°C overnight. The following morning, beads were washed 3 times with wash buffer 1 (0.1% SDS, 1% Triton X-100, 10mM EDTA, 150mM NaCl, 20mM Tris HCl, pH 8.0) and once with wash buffer 2 (0.1% sodium dodecyl sulfate, 1% Triton X-100, 10 mM EDTA, 500 mM NaCl, 20 mM Tris HCl, pH 8.0). Beads were eluted for 30 minutes at 65°C with frequent vortexing in elution buffer (1% sodium dodecyl sulfate, 100 mM NaHCO3), and the supernatants were collected in fresh, low-adherence tubes (Eppendorf, Hamburg, Germany; #022431021). Input and IP samples were incubated at 37°C for 30 minutes with 5 μg RNase A (Thermo Fisher Scientific; #AM2271), followed by reverse crosslinking for 5 hours at 65°C with 100 μg proteinase K. ChIP DNA was purified with MinElute PCR Purification columns (Qiagen; #28004) according to kit instructions.
ChIP Sequencing
ChIP-seq samples were prepared as previously described, but chromatin was sonicated to an average size of 250–300 bp. IPs were scaled up to 50 μL of bead slurry, 10 μg of ChIP antibody, and 100 μg of chromatin per IP. ChIP antibodies targeting H3K27ac (#ab4729) and RNA Pol II (#ab26721) were from Abcam. Library preparation, sequencing, and analysis was performed by the MetaOmics Shared Resource Facility of the Sidney Kimmel Cancer Center of Thomas Jefferson University. Briefly, ChIP-seq libraries were prepared using Swift Biosciences ACCEL-NGS 2S Plus DNA library kit following the manufacturer’s protocol. Single-end 75-bp sequencing was performed using a high-output flowcell on the Illumina NextSeq 500 at a depth of 40–55M reads per sample. Sequence read fragments were aligned to the GRCh38 human genome using the BWA-MEM aligner. Samtools was used to sort the resulting alignments and merge replicate .bam files for consensus peak calling.65 Peak calling was performed for each replicate in addition to the merged alignments using macs2, first with default settings, and then using the --call-summits options to call subpeaks.66 Differential binding analysis was performed on the consensus peaks and again on the subpeak summits using the DiffBind package (v3.0.14) in R/Bioconductor.67 Consensus peaks and differentially bound summits were annotated for genomic features using the ChIPpeakAnno package (v3.24.1) in R/Bioconductor.68
Dataset Acquisition and Analyses
RNA-seq gene expression data from normal mucosa (n = 51) and primary colon tumors (n = 380) was downloaded from the TCGA COAD/READ dataset on Xenabrowser on January 30, 2021.3,69 Analyses of single cell RNA-seq gene expression data from human colon tissue were retrieved from The Human Protein Atlas on February 23, 2021.41,70 ChIP-seq datasets with the indicated accession numbers were accessed from the National Center for Biotechnology Information Gene Expression Omnibus71 and ENCODE,37 and were visualized with the UCSC Genome Browser on December 21, 2018.72 Transcription factor consensus sequences in the DNA region –1000- to –3683-bp upstream of the GUCA2A transcription start site were identified with the TRANSFAC Match tool on July 21, 2020, using the minimize false positives cutoff parameter and vertebrate_non_redundant_minFP matrix profile (Supplementary File 1).73
Statistical Analysis
Except where noted, experiments were performed with triplicate measures for each condition (ie, 3 wells of cells with independent treatment, collection of samples, and measurement of analytes). Experiments were repeated at least twice (on separate days), and results are presented as the mean of experiments, where each data point represents the mean of the replicates from a single experiment. Where graph space is limited, bars represent the mean of independent experiments ± SEM. Statistical significance was determined by 2-tailed Student’s t test or 1- or 2-way analysis of variance (as appropriate), on log-transformed data, with matched analysis accounting for independent experiments. Statistical tests were calculated using GraphPad Prism (GraphPad Software, San Diego, CA, version 9.0) (∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001).
All authors had access to the study data and have reviewed and approved the final manuscript.
Acknowledgments
The results published here are based upon data from The Cancer Genome Atlas Research Network (https://www.cancer.gov/tcga), the UCSC genome browser (http://genome.ucsc.edu), the UCSC Xenabrowser (http://xenabrowser.net), the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo), and the Human Protein Atlas (http://www.proteinatlas.org). Figures were created with BioRender.com. LS174T(shβcatenin), LS174T(DNTCF), and DLD1(DNTCF) cell lines were kindly provided by H. Clevers. HT29(APC) cells were kindly provided by B. Vogelstein. Antiserum to mouse guanylin was a gift from M. Goy. Sequencing analysis was provided by the MetaOmics Shared Resource Facilities of the Sidney Kimmel Cancer Center at Jefferson Health and was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA056036. Additional bioinformatic analyses were provided by S. Peri and E. Londin. ChIP advice was kindly provided by M. Farkas and T. Fenstermaker. CRISPR advice and resources were provided by H. Deng. The authors would like to thank J. Brody for his insights; E. Leong and M. Weindorfer for administrative and technical support; and H. Wensil-Strow for editing assistance.
CRediT Authorship Contributions
Jeffrey A Rappaport, BS (Conceptualization: Equal; Data curation: Lead; Formal analysis: Equal; Funding acquisition: Supporting; Investigation: Equal; Methodology: Equal; Validation: Equal; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Ariana A Entezari, BS (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Validation: Equal; Visualization: Supporting; Writing – review & editing: Supporting)
Adi Caspi, BS (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Signe Caksa, BS (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Aakash V Jhaveri, BS (Data curation: Supporting; Investigation: Supporting)
Timothy J Stanek, PhD (Formal analysis: Supporting; Writing – review & editing: Supporting)
Adam Ertel, PhD (Formal analysis: Supporting; Software: Supporting; Writing – review & editing: Supporting)
Joan Kupper, BS (Investigation: Supporting; Writing – review & editing: Supporting)
Paolo M Fortina, MD, PhD (Resources: Supporting; Writing – review & editing: Supporting)
Steven B McMahon, PhD (Conceptualization: Supporting; Resources: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
James B Jaynes, PhD (Conceptualization: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Adam E Snook, PhD (Conceptualization: Supporting; Funding acquisition: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Scott A. Waldman, MD, PhD (Conceptualization: Equal; Funding acquisition: Lead; Project administration:
Footnotes
Conflicts of interest These authors disclose the following: Scott A. Waldman is a member of the Board and Chair of the Scientific Advisory Board of, and Adam E. Snook is a consultant for, Targeted Diagnostics and Therapeutics, Inc., which provided research funding that, in part, supported this work and has a license to commercialize inventions related to this work. The other authors disclose no conflicts.
Funding This work was supported by grants to Scott A. Waldman from the National Institutes of Health (1R01 CA204881, 1R01 CA206026), the Department of Defense Congressionally Directed Medical Research Program (W81XWH-17-PRCRP-TTSA), the Courtney Ann Diacont Memorial Foundation, and Targeted Diagnostic and Therapeutics, Inc.; to Adam E. Snook from the Department of Defense Congressionally Directed Medical Research Program (W81XWH-19-1-0263, W81XWH-19-1-0067, and W81XWH-17-1-0299), the DeGregorio Family Foundation, and David and Lorraine Swoyer; to Jeffrey A. Rappaport from a predoctoral fellowship from the PhRMA Foundation and a National Institutes of Health Ruth Kirschstein Individual Research Fellowship Award (F30 CA232469; and to the Sidney Kimmel Cancer Center of Thomas Jefferson University (P30 CA56036).
Supplementary Material
References
- 1.Fearon E.R. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 2.Nusse R., Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rappaport J.A., Waldman S.A. The guanylate cyclase C-cGMP signaling axis opposes intestinal epithelial injury and neoplasia. Front Oncol. 2018;8:299. doi: 10.3389/fonc.2018.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waldman S.A., Camilleri M. Guanylate cyclase-C as a therapeutic target in gastrointestinal disorders. Gut. 2018;67:1543–1552. doi: 10.1136/gutjnl-2018-316029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitari G.M., Di Guglielmo M.D., Park J., Schulz S., Waldman S.A. Guanylyl cyclase C agonists regulate progression through the cell cycle of human colon carcinoma cells. Proc Natl Acad Sci U S A. 2001;98:7846–7851. doi: 10.1073/pnas.141124698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinbrecher K.A., Wowk S.A., Rudolph J.A., Witte D.P., Cohen M.B. Targeted inactivation of the mouse guanylin gene results in altered dynamics of colonic epithelial proliferation. Am J Pathol. 2002;161:2169–2178. doi: 10.1016/S0002-9440(10)64494-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P., Lin J.E., Chervoneva I., Schulz S., Waldman S.A., Pitari G.M. Homeostatic control of the crypt-villus axis by the bacterial enterotoxin receptor guanylyl cyclase C restricts the proliferating compartment in intestine. Am J Pathol. 2007;171:1847–1858. doi: 10.2353/ajpath.2007.070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P., Schulz S., Bombonati A., Palazzo J.P., Hyslop T.M., Xu Y., Baran A.A., Siracusa L.D., Pitari G.M., Waldman S.A. Guanylyl cyclase C suppresses intestinal tumorigenesis by restricting proliferation and maintaining genomic integrity. Gastroenterology. 2007;133:599–607. doi: 10.1053/j.gastro.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 10.Lin J.E., Li P., Snook A.E., Schulz S., Dasgupta A., Hyslop T.M., Gibbons A.V., Marszlowicz G., Pitari G.M., Waldman S.A. The hormone receptor GUCY2C suppresses intestinal tumor formation by inhibiting AKT signaling. Gastroenterology. 2010;138:241–254. doi: 10.1053/j.gastro.2009.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam B.N., Sharman S.K., Hou Y., Bridges A.E., Singh N., Kim S., Kolhe R., Trillo-Tinoco J., Rodriguez P.C., Berger F.G., Sridhar S., Browning D.D. Sildenafil suppresses inflammation-driven colorectal cancer in mice. Cancer Prev Res (Phila) 2017;10:377–388. doi: 10.1158/1940-6207.CAPR-17-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharman S.K., Islam B.N., Hou Y., Singh N., Berger F.G., Sridhar S., Yoo W., Browning D.D. Cyclic-GMP-elevating agents suppress polyposis in Apc(Min) mice by targeting the preneoplastic epithelium. Cancer Prev Res (Phila) 2018;11:81–92. doi: 10.1158/1940-6207.CAPR-17-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn M. Molecular physiology of membrane guanylyl cyclase receptors. Physiol Rev. 2016;96:751–804. doi: 10.1152/physrev.00022.2015. [DOI] [PubMed] [Google Scholar]
- 14.Ikpa P.T., Sleddens H.F., Steinbrecher K.A., Peppelenbosch M.P., de Jonge H.R., Smits R., Bijvelds M.J. Guanylin and uroguanylin are produced by mouse intestinal epithelial cells of columnar and secretory lineage. Histochem Cell Biol. 2016;146:445–455. doi: 10.1007/s00418-016-1453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitari G.M., Zingman L.V., Hodgson D.M., Alekseev A.E., Kazerounian S., Bienengraeber M., Hajnoczky G., Terzic A., Waldman S.A. Bacterial enterotoxins are associated with resistance to colon cancer. Proc Natl Acad Sci U S A. 2003;100:2695–2699. doi: 10.1073/pnas.0434905100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinbrecher K.A., Tuohy T.M., Heppner Goss K., Scott M.C., Witte D.P., Groden J., Cohen M.B. Expression of guanylin is downregulated in mouse and human intestinal adenomas. Biochem Biophys Res Commun. 2000;273:225–230. doi: 10.1006/bbrc.2000.2917. [DOI] [PubMed] [Google Scholar]
- 17.Wang R., Kwon I.K., Thangaraju M., Singh N., Liu K., Jay P., Hofmann F., Ganapathy V., Browning D.D. Type 2 cGMP-dependent protein kinase regulates proliferation and differentiation in the colonic mucosa. Am J Physiol Gastrointest Liver Physiol. 2012;303:G209–G219. doi: 10.1152/ajpgi.00500.2011. [DOI] [PubMed] [Google Scholar]
- 18.Huang W., Sundquist J., Sundquist K., Ji J. Use of phosphodiesterase 5 inhibitors is associated with lower risk of colorectal cancer in men with benign colorectal neoplasms. Gastroenterology. 2019;157:672–681.e4. doi: 10.1053/j.gastro.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Huang W., Sundquist J., Sundquist K., Ji J. Phosphodiesterase-5 inhibitors use and risk for mortality and metastases among male patients with colorectal cancer. Nat Commun. 2020;11:3191. doi: 10.1038/s41467-020-17028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J.E., Colon-Gonzalez F., Blomain E., Kim G.W., Aing A., Stoecker B., Rock J., Snook A.E., Zhan T., Hyslop T.M., Tomczak M., Blumberg R.S., Waldman S.A. Obesity-induced colorectal cancer is driven by caloric silencing of the guanylin-GUCY2C paracrine signaling axis. Cancer Res. 2016;76:339–346. doi: 10.1158/0008-5472.CAN-15-1467-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shailubhai K., Yu H.H., Karunanandaa K., Wang J.Y., Eber S.L., Wang Y., Joo N.S., Kim H.D., Miedema B.W., Abbas S.Z., Boddupalli S.S., Currie M.G., Forte L.R. Uroguanylin treatment suppresses polyp formation in the Apc(Min/+) mouse and induces apoptosis in human colon adenocarcinoma cells via cyclic GMP. Cancer Res. 2000;60:5151–5157. [PubMed] [Google Scholar]
- 22.Bashir B., Merlino D.J., Rappaport J.A., Gnass E., Palazzo J.P., Feng Y., Fearon E.R., Snook A.E., Waldman S.A. Silencing the GUCA2A-GUCY2C tumor suppressor axis in CIN, serrated, and MSI colorectal neoplasia. Hum Pathol. 2019;87:103–114. doi: 10.1016/j.humpath.2018.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blomain E.S., Rappaport J.A., Pattison A.M., Bashir B., Caparosa E., Stem J., Snook A.E., Waldman S.A. APC-beta-catenin-TCF signaling silences the intestinal guanylin-GUCY2C tumor suppressor axis. Cancer Biol Ther. 2020;21:441–451. doi: 10.1080/15384047.2020.1721262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrithers S.L., Barber M.T., Biswas S., Parkinson S.J., Park P.K., Goldstein S.D., Waldman S.A. Guanylyl cyclase C is a selective marker for metastatic colorectal tumors in human extraintestinal tissues. Proc Natl Acad Sci U S A. 1996;93:14827–14832. doi: 10.1073/pnas.93.25.14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson C., Lin J.E., Li P., Snook A.E., Gong J., Sato T., Liu C., Girondo M.A., Rui H., Hyslop T., Waldman S.A. The paracrine hormone for the GUCY2C tumor suppressor, guanylin, is universally lost in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2014;23:2328–2337. doi: 10.1158/1055-9965.EPI-14-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pattison A.M., Barton J.R., Entezari A.A., Zalewski A., Rappaport J.A., Snook A.E., Waldman S.A. Silencing the intestinal GUCY2C tumor suppressor axis requires APC loss of heterozygosity. Cancer Biol Ther. 2020;21:799–805. doi: 10.1080/15384047.2020.1779005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovani M., Chan A.T. Are phosphodiesterase-5 inhibitors a new frontier for prevention of colorectal cancer? Gastroenterology. 2019;157:602–604. doi: 10.1053/j.gastro.2019.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg D.S., Lin J.E., Foster N.R., Della'Zanna G., Umar A., Seisler D., Kraft W.K., Kastenberg D.M., Katz L.C., Limburg P.J., Waldman S.A. Bioactivity of oral linaclotide in human colorectum for cancer chemoprevention. Cancer Prev Res (Phila) 2017;10:345–354. doi: 10.1158/1940-6207.CAPR-16-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin P.J., Vogelstein B., Kinzler K.W. Apoptosis and APC in colorectal tumorigenesis. Proc Natl Acad Sci U S A. 1996;93:7950–7954. doi: 10.1073/pnas.93.15.7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A.P., Tjon-Pon-Fong M., Moerer P., van den Born M., Soete G., Pals S., Eilers M., Medema R., Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 31.van de Wetering M., Oving I., Muncan V., Pon Fong M.T., Brantjes H., van Leenen D., Holstege F.C., Brummelkamp T.R., Agami R., Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van der Flier L.G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M.E., Suijkerbuijk S., Van de Wetering M., Marra G., Clevers H. The intestinal Wnt/TCF signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 33.Qian X., Prabhakar S., Nandi A., Visweswariah S.S., Goy M.F. Expression of GC-C, a receptor-guanylate cyclase, and its endogenous ligands uroguanylin and guanylin along the rostrocaudal axis of the intestine. Endocrinology. 2000;141:3210–3224. doi: 10.1210/endo.141.9.7644. [DOI] [PubMed] [Google Scholar]
- 34.Hnisz D., Day D.S., Young R.A. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016;167:1188–1200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuijers J., Manteiga J.C., Weintraub A.S., Day D.S., Zamudio A.V., Hnisz D., Lee T.I., Young R.A. Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep. 2018;23:349–360. doi: 10.1016/j.celrep.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxena M., Roman A.K.S., O'Neill N.K., Sulahian R., Jadhav U., Shivdasani R.A. Transcription factor-dependent 'anti-repressive' mammalian enhancers exclude H3K27me3 from extended genomic domains. Genes Dev. 2017;31:2391–2404. doi: 10.1101/gad.308536.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen A.J., Saiakhova A., Corradin O., Luppino J.M., Lovrenert K., Bartels C.F., Morrow J.J., Mack S.C., Dhillon G., Beard L., Myeroff L., Kalady M.F., Willis J., Bradner J.E., Keri R.A., Berger N.A., Pruett-Miller S.M., Markowitz S.D., Scacheri P.C. Hotspots of aberrant enhancer activity punctuate the colorectal cancer epigenome. Nat Commun. 2017;8 doi: 10.1038/ncomms14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lickwar C.R., Camp J.G., Weiser M., Cocchiaro J.L., Kingsley D.M., Furey T.S., Sheikh S.Z., Rawls J.F. Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., Sanli K., von Feilitzen K., Oksvold P., Lundberg E., Hober S., Nilsson P., Mattsson J., Schwenk J.M., Brunnstrom H., Glimelius B., Sjoblom T., Edqvist P.H., Djureinovic D., Micke P., Lindskog C., Mardinoglu A., Ponten F. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 42.Carleton J.B., Berrett K.C., Gertz J. Multiplex enhancer interference reveals collaborative control of gene regulation by estrogen receptor alpha-bound enhancers. Cell Syst. 2017;5:333–344-e5. doi: 10.1016/j.cels.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W., Guilak F., Crawford G.E., Reddy T.E., Gersbach C.A. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mokry M., Hatzis P., de Bruijn E., Koster J., Versteeg R., Schuijers J., van de Wetering M., Guryev V., Clevers H., Cuppen E. Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mokry M., Hatzis P., Schuijers J., Lansu N., Ruzius F.P., Clevers H., Cuppen E. Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes. Nucleic Acids Res. 2012;40:148–158. doi: 10.1093/nar/gkr720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herbst A., Jurinovic V., Krebs S., Thieme S.E., Blum H., Goke B., Kolligs F.T. Comprehensive analysis of beta-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/beta-catenin signaling. BMC Genomics. 2014;15:74. doi: 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blauwkamp T.A., Chang M.V., Cadigan K.M. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. Embo j. 2008;27:1436–1446. doi: 10.1038/emboj.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boija A., Klein I.A., Sabari B.R., Dall'Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M., Abraham B.J., Afeyan L.K., Guo Y.E., Rimel J.K., Fant C.B., Schuijers J., Lee T.I., Taatjes D.J., Young R.A. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855-e16. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q., Peterson K.R., Fang X., Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li K., Liu Y., Cao H., Zhang Y., Gu Z., Liu X., Yu A., Kaphle P., Dickerson K.E., Ni M., Xu J. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat Commun. 2020;11:485. doi: 10.1038/s41467-020-14362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang G., Chow R.D., Bai Z., Zhu L., Errami Y., Dai X., Dong M.B., Ye L., Zhang X., Renauer P.A., Park J.J., Shen L., Ye H., Fuchs C.S., Chen S. Multiplexed activation of endogenous genes by CRISPRa elicits potent antitumor immunity. Nat Immunol. 2019;20:1494–1505. doi: 10.1038/s41590-019-0500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Y.Y., Davidson L.A., Chapkin R.S. Murine Colonic Organoid Culture System and Downstream Assay Applications. Methods Mol Biol. 2019;1576:171–181. doi: 10.1007/7651_2016_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li P., Wuthrick E., Rappaport J.A., Kraft C., Lin J.E., Marszalowicz G., Snook A.E., Zhan T., Hyslop T.M., Waldman S.A. GUCY2C signaling opposes the acute radiation-induced GI syndrome. Cancer Res. 2017;77:5095–5106. doi: 10.1158/0008-5472.CAN-17-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickham H. Springer-Verlag; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 60.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S., Hahn W.C., Sharp P.A., Weinberg R.A., Novina C.D. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolligs F.T., Hu G., Dang C.V., Fearon E.R. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt D., Wilson M.D., Spyrou C., Brown G.D., Hadfield J., Odom D.T. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., Ali S., Chin S.F., Palmieri C., Caldas C., Carroll J.S. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goldman M.J., Craft B., Hastie M., Repecka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., Zhu J., Haussler D. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karlsson M., Zhang C., Mear L., Zhong W., Digre A., Katona B., Sjostedt E., Butler L., Odeberg J., Dusart P., Edfors F., Oksvold P., von Feilitzen K., Zwahlen M., Arif M., Altay O., Li X., Ozcan M., Mardinoglu A., Fagerberg L., Mulder J., Luo Y., Ponten F., Uhlen M., Lindskog C. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7 doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.