Introduction
Enzyme-mediated chemical modifications on RNA species across all domains of life have been documented over more than 50 years. A large repertoire of over 100 distinct modifications have since been described, with the first report of modified nucleotides from highly abundant RNAs dating back to 1960 [1,2]. These post-transcriptional modifications are enzymatically installed and removed in a site-specific manner by writer and eraser proteins respectively, while reader proteins can interpret modifications and transduce the signal for downstream functions. These enzymes utilize a myriad of reactions, such as methylations, deaminations, thiolations, glycosylations, isomerizations, etc. [3]. Advances in mass spectrometry and high-throughput sequencing have enabled the detection and characterization of modifications in relatively lowly expressed RNA species, revitalizing the study of RNA modifications and shaping the field of epitranscriptomics. In this review, we will introduce the chemical features and biological functions of these modifications in both coding and non-coding RNA species.
Chemical Properties and Biological Functions of tRNA Modifications
Transfer RNA (tRNA) structure
Across all organisms, tRNAs undergo the most diverse chemical modifications as part of their maturation and function. Many unique modifications have only been described in the context of tRNA and ribosomal RNA (rRNA), both of which have historically served as the frontier for discovery and characterization of RNA modifications. As such, there is no better place to begin exploring the variety of RNA modifications than in the world of tRNA. We will first extensively explore tRNA to cover a large swath of chemical diversity, and then focus on the biological implications of modifications of mRNA and other RNA species which code dynamic regulatory information.
The canonical and most common tRNA consist of 76 nucleotides and is conserved across all kingdoms of life [4]. It adopts a clover-leaf secondary structure that terminates with a CCA at the 3′ end, with the terminal adenosine 2′- or 3′- OH acting as the site of aminoacylation during tRNA charging. The arms of the clover-leaf structure are termed as the acceptor stem, the dihydrouridine stem-loop (D-loop), anticodon stem-loop, and the thymine-pseudouridine-cytosine stem-loop (TΨC-loop) (Fig. 1A). Once the tertiary L-shaped structure is adopted (Fig. 1B), the TΨC-loop and the acceptor stems stack to form the 12 base pairs (bp) acceptor-TΨC minihelix, while the stacking of the anticodon stem-loop with the D stem-loop forms the 10 bp anticodon D loop dumbbell. The tertiary right-angle L-shape is achieved largely by the conserved nucleotides between the TΨC and D stem-loop that interact to stabilize the structure. True to its name, the variable loop (V-loop) is where any additional nucleotides beyond the canonical 76 are most often found, which tends to bulge outward and avoids steric hindrances.
Figure 1.
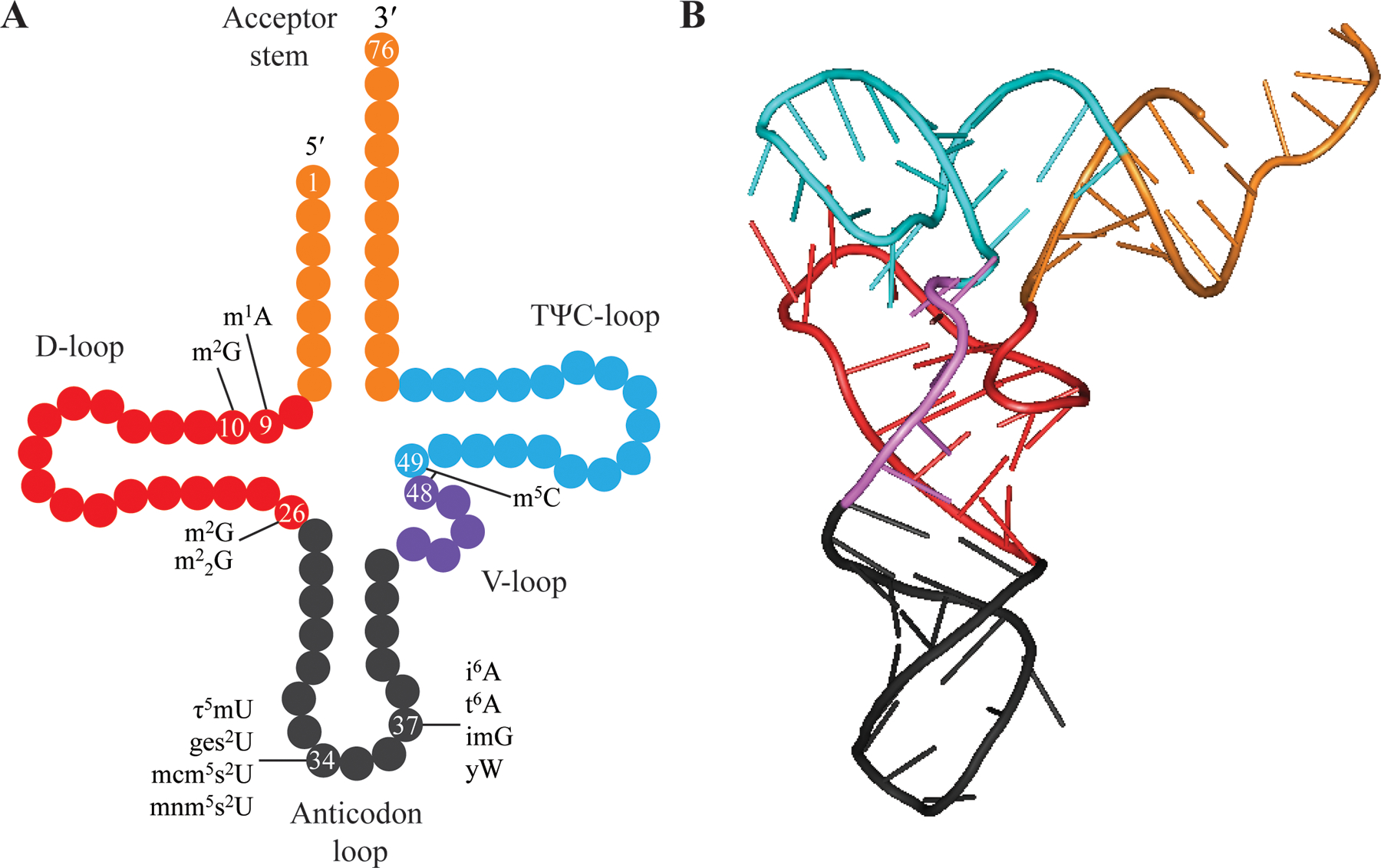
Transfer RNA secondary and tertiary structure. (A) Two-dimensional clover-leaf structure and the five domains that make up a tRNA. Specific residues whose modifications are discussed in this review are denoted. (B) Three-dimensional structure of tRNA (PDB code: 1YFG). The color code of the two-dimensional structure matches with that of the three-dimensional structure.
In addition to cytoplasmic tRNAs, animal cells also host a pool of mitochondrial tRNAs which are usually shorter than the cytosolic counterparts [5]. Interestingly, these tRNAs often have smaller loop structures and can even be missing entire domains, highlighting the versatility and high capacity for tRNAs to function after various processing events. It is also noteworthy that mitochondrial tRNAs are enriched in adenosine and uracil, resulting in reduced stability which is thought to promote base paring misalignments that are more susceptible to base substitutions, resulting in several mitochondrial tRNA-associated human diseases [5–7].
Isoacceptors and isodecoders
tRNA is the second most abundant species of RNA in cells following rRNA. There are roughly 300 tRNA genes in the human genome that have been validated for expression, and tRNA gene copy number as well as sequence diversity vary within the human population [8,9]. A large number of tRNAs in comparison to the 20 canonical amino acids used for polypeptide synthesis has given rise to tRNA isoacceptors; tRNAs with distinct sequences including different anticodon sequences that are charged with the same amino acid. In higher eukaryotes, tRNAs with matching anticodon and differing body sequences are known as isodecoders. As an example of the increased complexity of tRNAs brought about by these sequence variations, consider the large group of alanine-specific tRNAs (tRNAAla). There are 39 distinct tRNAAla in humans split into three isoacceptor families with multiple isodecoders present in each family [9,10]. This seemingly unnecessary degree of complexity in the world of tRNAs continues to underscore the importance of these molecules beyond their known functions in translation and is a trend that is taken further by tRNA modifications.
tRNA modifications and their chemical properties
The overwhelming majority of the greater than 100 unique annotated modified nucleotides have been found on tRNAs [1,11]. Some modifications, such as isopentenyl adenosine (Fig. 2A) or wybutosine (Fig. 2B) are infrequent and chemically complex, while other modifications such as dihydrouridine or pseudouridine (Fig. 2C) are present in nearly all tRNAs. These are only a small fraction of a vast array of modifications often found on tRNAs; human tRNAs on average harbor 11 to 13 modifications in a variety of combinations [12,13]. These numerous unique tRNA modifications provide a model to explore the chemical diversity of modified RNA. Detailed below is the current physio-chemical and biological understanding of several chemical modifications to specific tRNA nucleosides.
Figure 2.
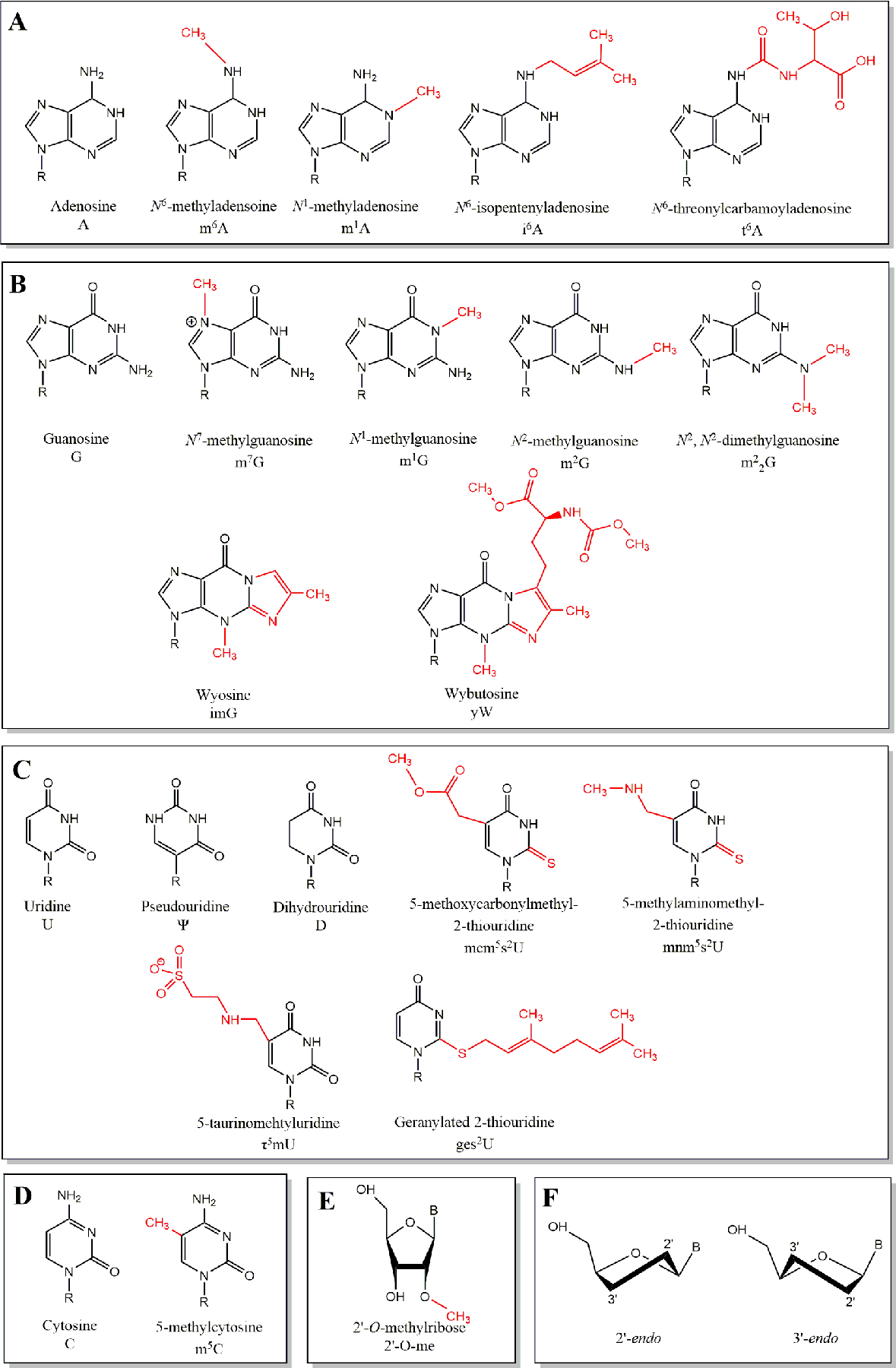
The chemical structures of nucleoside and ribose modifications addressed in this review. (A) Adenosine and its modified derivatives. (B) Guanosine and its modified derivatives. (C) Uridine and its modified derivatives. (D) Cytosine and its modified derivatives. (E) Modified 2′-O-methylribose. (F) Chair representations of ribose sugar pucker 2′ and 3′ endo conformations. B = base. R = ribose
N1-methyladenosine in tRNA
In the 2-dimensional clover-leaf structure of tRNAs, nucleoside 9 has been shown to be critical for proper folding, most notably in the case of smaller mitochondrial tRNAs. N1-methyladenosine (Fig. 2A) at A9 (m1A9) can disrupt the Watson-Crick base pairing of an intra-stem interaction during maturation of mitochondrial tRNALys, favoring the functional, canonical clover-leaf secondary structure [14–16]. Lack of the modification favors the intra-stem interaction, shifting the equilibrium towards a stable alternative extended hairpin structure that is not suitable for aminoacylation nor translation. During tRNALys maturation, m1A9 modification confers 0.7 – 1 kcal/mol of stability, dependent on Mg2+ concentration [17]. This structural dependence on m1A9 modification and indeed many other base modifications is a common theme among animal mitochondrial tRNA biogenesis [18].
Methylated Guanosines
As was just noted, methylation of nucleosides can play an enormous role in the maturation of tRNAs. One of the critical steps of tRNA maturation is the convergence of the inward ends of each stem on the clover-leaf structure, forming the characteristic clover-leaf junction in the secondary structure and leading to the formation of an internal loop characteristic of tRNA’s L-shaped tertiary structure. Methylations within this junction are crucial in preventing each stem from forming a spatially tight interaction, keeping the core open enough to allow proper folding. The family of 2-methylated guanosines, N2-methylguanosine (m2G), N2, N2-dimethylguanosine (m22G), and N2, N2, 2′-O-trimethylguanosine (m22Gm), are highly conserved at tRNA positions 10 and 26 across all domains of life, and several other tRNA positions also bare methylguanosine modifications (Fig. 2B) [19]. The methyl groups are located on the Watson-Crick face, disrupting proper base pairing and in the case of positions 10 and 26, terminating the duplexes in tRNA. It is noteworthy that m2G has been shown to be energetically identical to guanosine in the context of a G to C pairing in a synthesized RNA duplex [20], but likely has a more disruptive role in the context of tRNA. Additionally, m22G has an undisputed effect on duplex stability and base pairing properties, most notably that m22G stably forms a pseudo-Watson-Crick pair with adenosine (see below). Interestingly, substitution experiments replacing guanosine with inosine indicate that the 2-methylamino group of guanosines at tRNA positions 2, 3, and 10 play an important role in tRNA-protein recognition [21]. These studies highlight how conserved methylated guanosine nucleosides can act as critical points of identification and recognition during aminoacylation.
Methylation of guanosine 26 occurs in roughly 80% of eukaryotic tRNAs that contain guanosine at position 26. However there are documented unmodified G26 nucleosides such as yeast tRNAAsp [22]. A crystal structure of an RNA duplex containing two m22G:A pairs has shown that the modified guanosine adopts an imino-hydrogen bond with adenosine which explains the mismatch pairing of G26 and A44 in these tRNAs [4,18,23]. A study on the biological effects of tRNA m22G26 showed that a reduction in RNA polymerase III production of tRNAs correlates with an increasing trend of m22G tRNA modification in yeast and human cells [24]. It is known that m22G is important for decoding during translation, acting as an “activator” of tRNAs and increasing tRNA usage in the ribosome. It is thought that there is an inverse link between the rate of RNA polymerase III activity and m22G26-linked activation of tRNA, perhaps as a compensatory mechanism for the slowdown in tRNA production [24,25].
Pseudouridine in tRNA
Pseudouridine (Ψ) is one of the most abundant modifications of RNA and is found in a variety of different RNA species such as tRNA, rRNA, mRNA, and some small RNA species [26–28]. Ψ is an isomer of uridine that replaces the N1-C1′ glycosidic bond between the nucleobase and the sugar with a C5′-C1′ bond (Fig. 2C). The slight alteration of uridine to Ψ can induce drastic differences in the structure and function of RNAs. The unique C-C bond between the sugar and base afford greater rotational freedom to Ψ, and the free N1-H imino group can potentially act as an additional hydrogen donor outside of the Watson-Crick face. In the context of tRNAs, Ψ can be found on all loops of tRNAs. However it most frequently plays a structural role as part of the TΨC or anticodon loop due to the striking effect on RNA stability conferred by the sugar of Ψ through a property known as sugar puckering.
The helix of an RNA containing a Ψ can be thermodynamically stabilized by the C3′-endo sugar pucker (Fig. 2F) [26,29,30]. The puckered sugar intrinsically has increased stacking ability and can induce a localized increase in helix stability. This sugar conformation restricts the base to an axial anti conformation which also confers additional rigidity and stability to local helices, resulting in an additive, doubly stabilized Ψ residue [31–33]. Computational and NMR studies indicate that Ψ may fluidly adopt several different puckered conformations, but it is apparent that the context of Ψ in an RNA sequence contributes to the particular bond conformations that appear at a given residue [34–36].
Ψ in tRNA generally do not affect the overall 3-dimensional structure and have been shown to be non-essential for cell viability and aminoacylation. However, Ψ’s stabilizing effects on the local structure is critical for the maintenance of the anticodon loop and in turn proper binding and recognition of tRNAs by the ribosome. It is believed that the stability conferred to the anticodon loop by Ψ may induce a tighter binding of the appropriate tRNA to the ribosome and facilitate codon-anticodon interactions [31,33,37,38]. This less remarkable role while not essential may nonetheless have a meaningful impact on maintaining proper, homeostatic levels of translational accuracy by slowing the rate of peptide synthesis and facilitating the rejection of improper tRNAs [39].
Dihydrouridine in tRNA
As the name may suggest, the most common and conserved dihydrouridine (D) modification (Fig. 2C) in tRNAs can be found within the D-loop and is conserved in nearly all life. Reduction of the double bond between C5 and C6 of uridine generates D, and the presence of this modification in the D-loop is known to promote tRNA secondary and tertiary structure [40–42]. The nucleoside structure of D indicates that the loss of aromaticity prevents D from making base stacking interactions [43]. Additionally, the ribose sugar of D prefers the C2′-endo conformation over C3′-endo (Fig. 2F), unlike uridine which does not favor either of these two conformations. This conformation preference is due to the loss of a pi orbital interaction between uridine and its cognate sugar that is prohibited after conversion to D [44,45]. This induction of ribose C2′-endo conformation by D can ultimately have a dynamic effect on tRNA structure and stability. Diyhdrouridine has been shown to both destabilize RNA helices and decrease melting temperatures in vitro [46], but is also capable of inducing stable hairpin loop structures within the D-loop as well [42]. The destabilizing nature of D proves critical in mediating dynamic, flexible sugar arrangements necessary for proper D-loop folding.
5-methylcytosine in tRNA
The presence of 5-methylcytosine (m5C) (Fig. 2D) is more varied than some other modifications among the tree of life. It can be found in tRNA and rRNA of eukaryotes and archaea but is absent in tRNAs from E. coli despite also being found in E. coli rRNA [47]. In tRNAs, m5C is most consistently found at positions 48 and 49 as part of the junction between the variable loop and the TΨC loop and is known to promote tRNA stability and protein synthesis in eukaryotes and prokaryotes [48,49]. The methyl groups of m5C48 and m5C49 are not located on the Watson-Crick interface of the nucleobase and thus do not interfere with canonical base pairing interactions. A ubiquitous component of all tRNAs is a non-canonical base pairing between nucleotides 15 and 48 known as the Levitt pair (Fig. 3) [50]. This pair assumes a reverse Watson-Crick base pairing geometry or a trans arrangement that helps to join the D-loop and the variable loop. Nucleoside 48 and 49 are modified to m5C in 26% and 80% of tRNAs respectively, but what this modification contributes to this system is not known [51,52]. A study of the thermodynamic parameters of m5C40 in yeast tRNAPhe noted an increase in the melting temperature to a similar extent as Ψ in the same tRNA, although the contribution of m5C to the free energy of the tRNA was negligible [52]. When investigating the biological functions of m5C in tRNAs, one group has shown that growth and development of mice and flies are disturbed following the loss of tRNA m5C modifications conferred by two methyltransferases, DNA methyltransferase 2 (Dnmt2) and NOP2/Sun RNA methyltransferase family member 2 (Nsun2) [53,54]. The biological roles of m5C are more apparent in the realm of mRNA as will be discussed.
Figure 3.
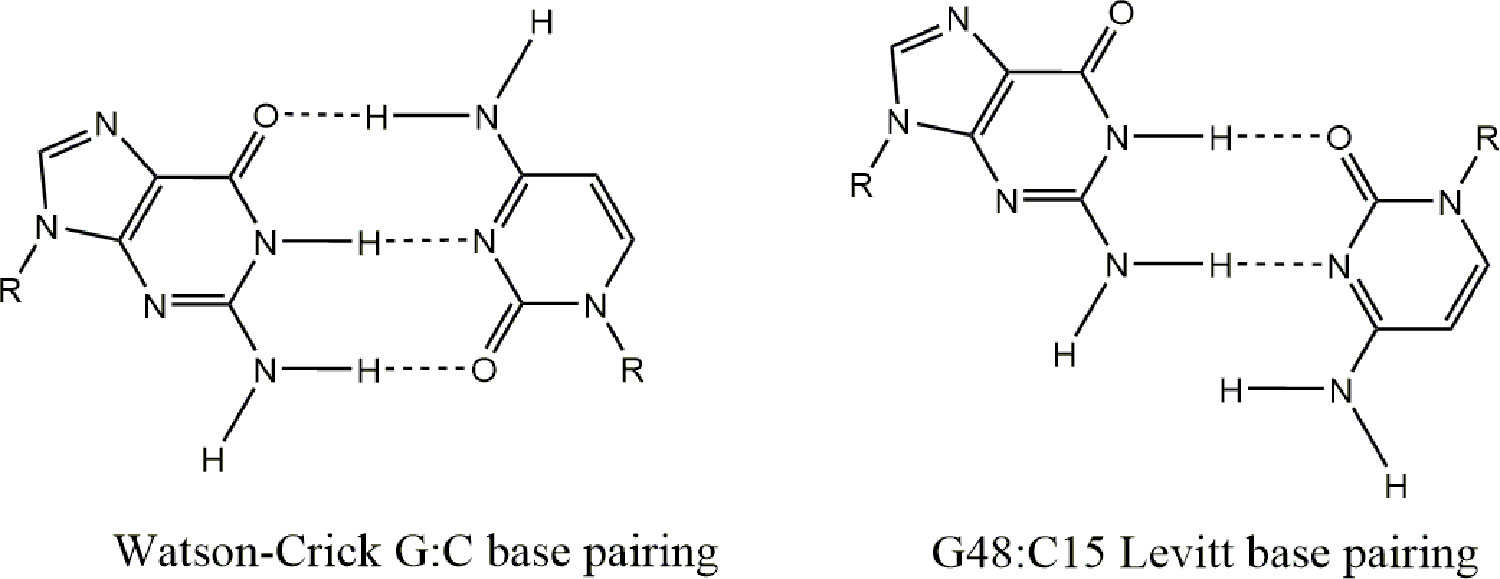
The chemical structure of a canonical Watson-Crick base pairing between G and C (left) and the reverse Watson-Crick base pairing between G48 and C15 which form the Levitt pair (right).
The chemical variety of tRNA anticodon loop
The anticodon loop, responsible for decoding during translation, is the most chemically varied portion of tRNA, especially positions 34 and 37 [4,11,47]. Chemical modifications at these two positions have implications in a diverse set of roles related to RNA biology such as the maturation, stability, and conformational variety of tRNAs [55–59], enhancement of ribosomal A-site entry and binding [57,59–65], enhancement of mRNA translocation during translation [64], and modulation of mRNA decoding, frame maintenance, and frameshifting [66–68].
Position 34 of tRNA
Position 34 is often called the wobble position due to Francis Crick’s Wobble Hypothesis which states that a guanosine, uridine, or inosine at position 34 can pair with two or three different nucleosides [69]. This idea has since been updated to account for the dynamic changes in codon recognition conferred by chemical modifications at the wobble position [70]. Modifications at position 34, especially in the case of uridines, can both expand and contract the decoding range of a given tRNA, and over 80% of cytosolic tRNAs across species carry a modified nucleoside at position 34 [57,71,72]. Surprisingly, the majority of modifications to nucleosides at position 34 occur at position 5 of pyrimidines, opposite of the Watson-Crick face, indicating that modifications are having an indirect effect on the chemistry of the anticodon-codon interface [11,69].
A common effect of modifications to position 5 of a pyrimidine is the induction of either a C3′- or C2′- endo ribose sugar pucker conformation which has been shown to modulate decoding capabilities of tRNAs (Fig. 2F) [70,73–75]. An example of a modification that usually restricts decoding is the 2-thio-5-methyluridine family of modifications at position 34. The van der Waals radius of the sulfur atom sterically clashes with the ribose 2′-oxygen atom, favoring the C3′-endo conformation, ultimately restricting decoding of the third position of the codon to adenosine [75]. Conversely, modification families that lack the additional sulfur group such as 5-hydroxyuridine or 5-methyluridine can adopt both sugar pucker forms and therefore have an expanded wobble recognition repertoire that includes adenosine, guanosine, and uridine [75]. The 5-taurinomethylurdine modification at position 34 (τ5mU34) is a considerably larger modification that is common to several mitochondrial tRNAs (Fig. 2C). This modification has been shown to contribute to decoding by stabilizing the wobble base pairing of U34 and G3. Through crystallographic and computational studies, τ5mU34 in mitochondrial tRNALeuUAA have been shown to stabilize hydrogen bonding with adjacent nucleosides as well as contribute to the wobble pairing of U34 and G3 via increased stacking interactions [63,76]. Similarly, bacterial geranylated 2-thiouridine (ges2U34) (Fig. 2C) facilitates this same wobble pairing due to the large hydrophobic geranyl group modified at the two thio position [77–79].
An additional layer of mismatch pairing at the wobble position is conferred by modifications that can allow for tautomerization of the nucleobase. Modification of uridine 34 to 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U34) or 5-methylaminomethyl-2-thiouridine modification (mnm5s2U34) permits both human and E. coli tRNALysUUU to recognize the wobble codon AAG (Fig. 2C). Crystal structures have shown that these modifications can shift the keto-enol equilibrium to favor the enol form which permits a guanosine-uridine base pairing that maintains Watson-Crick geometry within the ribosome [80,81]. This permission of Watson-Crick geometry despite a mismatch base pairing is thought to help such mismatches avoid the penalty by translational fidelity machinery and therefore allow tRNAs to utilize an expanded wobble recognition [80,82].
Position 37 of tRNA
Modifications at purine-invariant position 37, directly 3′-adjacent to the anticodon, can have varying effects on translational accuracy and efficiency [60,65,83,84]. Generally speaking, modifications at position 37 most often form a hydrophobic platform that supports the presentation of the anticodon to the codon and also proactively structures the anticodon loop for chemically favorable recycling [81,85–87]. When the first base of a mRNA codon is a uridine or an adenosine, position 37 of the tRNA is nearly always a modified adenosine which can often sport complex chemistry, such as N6-isopentenyladenosine (i6A) or N6-threonylcarbamoyladenosine (t6A) (Fig. 2A). Phenylalanine tRNAs from archaea and eukarya have a G37 in place of A37 which can respond to codons starting with U. In the case of archaeal, eukaryotic, and mitochondrial tRNAs containing a G37 in place of A37, tricyclic wyosine (imG) or it’s derivative wybutosine (yW) are often found (Fig. 2B) [88,89]. Each of these tRNAs also carries an invariant U33 that would form a stable cross- loop H-bond with A37 if not for the elaborate N6 modifications of A37 and G37 [57,85]. The rotation of t6A and i6A modifications induces steric hindrance that prevents the nucleoside from slipping into the helix, effectively disrupting the hydrogen bond between A37 and U33 [90]. Modification of G37 to imG37 or yW37 can provide a large platform above the first codon-anticodon interaction. Altogether, the complex structures of these modifications can provide considerable energetic stability and accuracy to the codon-anticodon interaction through van der Waals forces, hydrophobic effects, and solvent displacement [81,91,92].
There are numerous other modifications to tRNA, more than can be feasibly discussed here in any significant detail. Even so, the variety and plasticity of modifications in tRNA make apparent the critical and dynamic role they play in RNA structure and biology. From here, we will discuss the variety and current understanding of modifications to mRNA.
Chemical Properties and Biological Effects of mRNA Modifications
The discovery of reversible mRNA modifications as well as improvements in transcriptome-wide high-throughput sequencing has generated a wave of interest and research towards understanding the role these modifications play in a wide range of cellular processes. Chemical modifications to mRNA may interact directly with specific reader proteins to recruit machinery for cellular processes such as translation, decay, or localization. In addition to direct reader protein recognition, these modifications can also have effects on the secondary structure of mRNA transcripts and can be interpreted via indirect reader protein-RNA substrate recognition [93]. Of the various modifications, mRNAs especially utilize methylation which provides unique regulatory signaling capacities while also minimally perturbing mRNA nucleosides, thus maintaining translational efficiency. The use of the S-adenosylmethionine (SAM) cofactor, whose synthesis requires the net investment of 12–13 ATP, supports the importance of this modification in cell function. The discovery of the mRNA modification-specific reader, writer, and eraser proteins has continued to expand (Fig. 4), yet there are still several important modifications that have either partial or no known enzymes associated with their deposition and regulation. However, we now know that the intricate and expanding network of readers, writers, and erasers that regulate the modification of mRNA confer a dynamic, combinatorial layer of complexity to what was once thought of as solely a gene expression intermediate.
Figure 4.
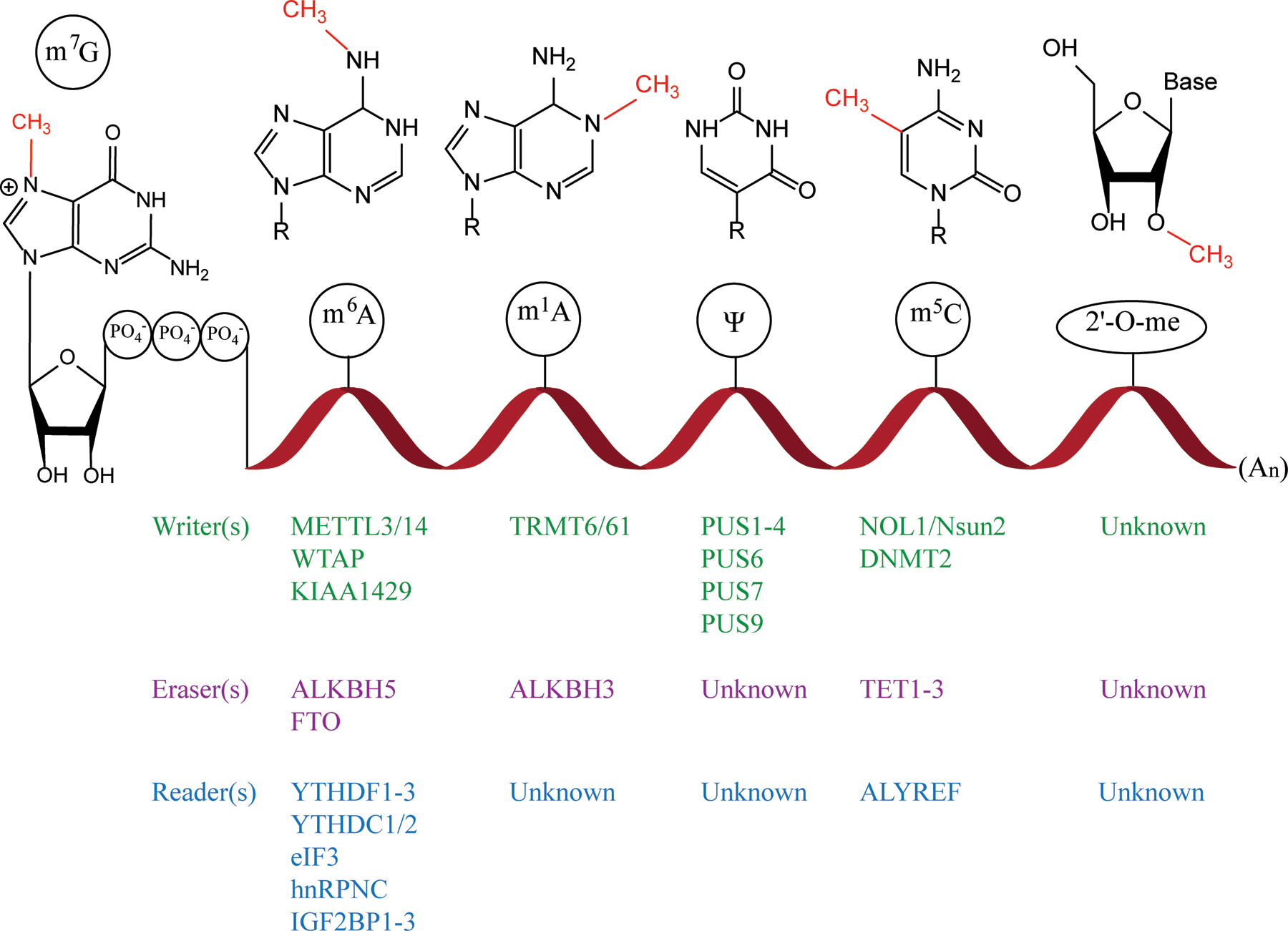
Modifications and proteins related to mRNA modifications. Diagramed from the left is the 5′ cap structure consisting of m7G, ribose sugar, and the 5′ to 5′ triphosphate linkage. Following this are the five internal mRNA modification structures as well as their related writers (green), erasers (purple) and readers (blue) described in this review listed in columns below. An = Polyadenylated tail.
N6-methyladenosine in mRNA
N6-methyladenosine (m6A) (Fig. 2) is the most prevalent internal modification in eukaryotic mRNA, occurring on an average of 3 sites per given mRNA molecule [94,95]. The methyl group of m6A can adopt both the syn and anti conformations based on the structural context of the given RNA. In single-stranded RNA (ssRNA), the syn conformation is the most energetically favorable geometry adopted by the nucleotide [96,97]. In double-stranded RNA (dsRNA), the methyl group must adopt the anti conformation to accommodate A to U Watson-Crick base pairing, positioning the methyl group in the major groove of an RNA helix and resulting in a 0.5 – 1.7 kcal/mol destabilization compared to an unmodified dsRNA [97,98]. This property has proven important as a means for m6A to induce localized structural switches from dsRNA to ssRNA [93,99]. These m6A switches allow for the indirect reading of m6A by readers that recognize not the modification itself but the primary sequence exposed or otherwise modulated by the structural switch. This includes the indirect reader ribonucleoproteins HNRNPA2B1 and HNRNPC [93,100]. The orientation of m6A towards the Watson-Crick geometry also explains why most reverse transcriptase enzymes are unimpeded by m6A modifications, as many polymerases can likely accommodate the anti methyl group in the major groove [98].
The heterodimeric enzyme complex of METTL3 and METTL14 is considered the canonical N6-adenosine methyltransferase complex responsible for the majority of mRNA m6A modifications [101–103]. Notably, crystallographic and biochemical studies have shown that METTL3/METTL14 function cooperatively, in which the enzymatic activity is greatly enhanced by the structural support of METTL14 [101]. Additionally, WTAP serves as an associated subunit of METTL3/METTL14 that increases localization of the complex into nuclear speckles, while KIAA1429 has been shown to be critical in maintaining complex activity and cellular m6A profiles [104,105]. Other interactors of the complex include RBM15/RBM15B which bind U rich regions near DRACH, the consensus sequence of m6A, to recruit the m6A writer complex for m6A methylation [106].
m6A is known to play an essential role in the processing and maturation of pre-mRNAs in the nucleus. An essential step of the mRNA maturation process is splicing, in which the excision of introns and the retention and ligation of exons from pre-mRNA transcripts is enzymatically driven and combined with additional processing mechanisms to form mature mRNAs. Additionally, alternative splicing can differentially include or exclude exons from a given mRNA. The mixture of splicing events allows for the generation of different mature mRNA which encodes different protein isomers. This process allows for additional levels of regulation and diversity within the proteome. Several m6A motifs have been found within introns and studies on several writers and erasers of the m6A modification have shown effects on alternative splicing [107,108]. Knockout of the METTL3 in mice embryonic stem cells has shown trends of intron retention and exon skipping in transcripts while depletion of the putative m6A eraser enzyme Fat mass and obesity-associated protein (FTO) results in increased levels of m6A near 5′ and 3′ splicing sites as well as enhanced recruitment of the serine/arginine-rich splicing factor 2 (SRSF2) and increased exon inclusion [109,110]. The m6A reader protein YTHDC1 has been shown to associate which SRSF3 to inhibit binding of SRSF10 and promote exon inclusion rather than skipping [111].
A similar process to alternative splicing is alternative polyadenylation (APA) in which alternative polyadenylation sites within coding or non-coding regions of transcripts can be cleaved and subsequently polyadenylated, effectively modulating transcript length. Transcript isoforms enriched with m6A are correlated with generally shorter 3′ UTRs, although what this functionally means and whether m6A is a causative agent in APA-dependent 3′UTR shortening is unclear [112].
Along with the maturation of pre-mRNA into mRNA, m6A is also involved in nuclear export. Studies have suggested that depletion of m6A results in loss of nuclear export of some mRNAs. Knockdown of METTL3 was shown to inhibit mRNA export and knockdown of the eraser enzyme ALKBH5 conversely enhanced mRNA export [113,114]. This evidence suggests reader proteins play a crucial role in identifying specific m6A motifs on mRNAs and facilitate their export into the cytosol.
Once finally exported to the cytosol, m6A plays further roles in enhancing the translation of mRNAs. The m6A reader protein YTHDF1 has shown the ability to enhance the translation efficiency of m6A-containing transcripts by binding m6A motifs and recruiting translation initiation machinery such as eIF3 [115]. The IGF2BP class of m6A reader protein also promote translation and increases stability and storage of target mRNAs [116]. In another completely independent pathway, METTL3 is also shown to recruit eIF3 to enhance eIF4E-dependent translation [117].
In addition to the effective translation of transcripts, m6A promotes the destabilization and degradation of mRNA. It has been shown that, in addition to the recruitment of YTHDF1 for translation efficiency, m6A is also targeted by YTHDF2 which targets transcripts for degradation [115]. Though not entirely understood, evidence suggests that YTHDF3 also promotes translation efficiency and degradation by cooperating with YTHDF1 and YTHDF2 [118]. This quality of m6A allows for swift and short bursts of translation and dynamic changes in gene expression in which modified transcripts are quickly translated and degraded. A recent study demonstrated that YTHDC2 might be capable of chaperoning both aspects of m6A bursts by interacting with both XRN exoribonuclease and the small subunit of the ribosome [119]. This quality of m6A allows for additional levels of control and regulation so that cells only invest energy in translating crucial transcripts and is often employed during dynamic cellular physiological events such as differentiation.
N1-methyladenosine (m1A) in mRNA
m1A was initially only found in tRNA and rRNA [120]. The ignorance of this modification’s existence was due to its extremely low abundance in mRNA. In fact, a single transcript may only possess a single m1A modification which usually lies near the translation start site and the first splicing site [121,122].
The chemistry of m1A endows a positive charge which could play an essential role in reader protein interaction and potentially affect base pairing and secondary structure. As was discussed for tRNAs, m1A can disrupt canonical Watson-crick base pairing and induces local RNA duplex melting [123], but little is known about the biological role of m1A although it is well known for its inherent ability to stall reverse transcription [124]. mRNA and tRNA share the same writer protein of m1A, TRMT6/61-complex, which acts upon the same consensus motif in both RNA species [125]. ALKBH3 is responsible for m1A demethylation on both RNA and DNA species. Transcriptome-wide mapping of m1A allowed for the identification of several dynamic sites of ALKBH3 demethylation [122]. Also, gene ontology analysis using the transcriptome mapping data indicated that the m1A modification is involved in a wide variety of cell functions and mainly enriched in the 5′ UTR or near the start codon. Currently, m1A is believed to most likely play a role in promoting translation, but more evidence will need to be shown such as the identification of additional m1A-specific writers, readers, and erasers.
5-methylcytosine (m5C) in mRNA
m5C is most well known as a DNA modification and its role in epigenetics. However, this modification exists in mRNA as well although its biological role has never been well studied until recent years. The classic bisulfite treatment which has been used to characterize the distribution and function of this modification in DNA has also been used to characterize m5C’s role in RNAs. The tRNA methyltransferase Nsun2 (Fig. 4) was shown to methylate cytosine in mRNAs and non-coding RNAs in addition to tRNA [126–128]. A few studies identified oxidative derivatives of m5C mediated by TET eraser enzymes (Fig. 4)[129,130]. The identification of these derivatives suggests that the nucleotide could be involved in several signaling pathways related to its sequential modification and break down. There is also evidence that m5C plays a role in promoting the nuclear export of target transcripts. The m5C reader-cytoplasmic shuttle, ALYREF (Fig. 4), has been shown to work with Nsun2 to promote the export of m5C transcripts [131]. Knockdown of ALYREF was shown to result in an accumulation of nuclear m5C transcripts which could be rescued by reconstitution of wild-type ALYREF.
Pseudouridine (Ψ) in mRNA
Ψ is the most abundant of all RNA modifications. Though mainly abundant in rRNA and tRNA, this modification has also been found in mRNA transcripts. Mapping of Ψ in mRNA has been accomplished at a single base resolution, and hundreds of mammalian and yeast transcripts have been identified too with this modification [27,132,133]. In particular, it was shown that pseudouridylation is dynamic and can change in response to several factors including stress and cell growth state. Analysis of yeast modifications at log phase and post-diauxic growth demonstrated significant changes in identified pseudouridylation sites in RBS28B, MRPS12, CDC33, U5 snRNA, RNase MRP, and snR37. HeLa cells grown in the presence or absence of serum showed dynamic changes in known modification sites of RPL19, ATP5E, MALAT1, and RN7SK. It is postulated that, due to Ψ’s ability to stabilize RNA structure, this modification could serve a role in several biological processes including enhance translation initiation efficiency, ribosome pausing, RNA localization, and RNA interference [27]. Several tRNA Ψ writer proteins have been shown to modify mRNAs [134]. This is indicative of the role that secondary structure plays in the identification of pseudouridylation sites in mRNA rather than a specific consensus motif. Ψ tRNA writers such as Pus1 and Pus2 has no sequence similarity in their mRNA targets while other writer enzymes like Pus4 and Pus7 showed very defined consensus motifs [27]. Also, several target RNAs were identified for Pus3, Pus6, and Pus9.
2′-O-methylation in mRNA
Little is known about the biological effects of 2′-O-methylation (2′-O-me) in the internal region of mRNA. In vitro studies of 2′-O-me of RNA have shown an ability to inhibit A to I editing [135]. It is also suggested that small nucleolar RNAs (snoRNAs) play a role in site-directed 2′-O-me modification of specific mRNA transcripts [136,137].
2′-O-me is involved in discrimination of self and non-self mRNA [138]. Dafis et al. (2010) demonstrated that West Nile virus mutants lacking 2′-O-methyltransferase activity were attenuated but retained pathogenicity in cells lacking type I interferon [129]. Similarly, Case JB et al. (2016) observed that when mutations are inserted into the SAM-binding region of the N7-methyltransferase of coronavirus, there is a loss of viral replication and enhanced sensitivity to immune response [139]. Taken together, this indicates that viruses may have adapted 2′-O-methyltransferase activity as a mechanism of evading host recognition systems. Given these findings, it is understandable why incorporation of 2′-O-me into small interference RNA (siRNA) optimizes stability and immunogenic properties [140]. RNA capping could potentially be a new target for antiviral therapies as we continue to discover more about the nature of this relationship of host vs. pathogen recognition.
Modifications in the 5′ cap
All eukaryotic mRNA contains N7-methylguanosine (m7G) (Fig. 2B) linked by a reverse 5′ to 5′ triphosphate linkage. This capping is carried out co-transcriptionally by RNA triphosphatase, RNA guanylyltransferase, and guanine-N7 methyltransferase [141]. The 5′ cap of mRNA plays essential roles in nearly every step of mRNA life cycle including splicing, export, protection from degradation, and translation initiation [142,143]. The 5′ cap protects transcripts from degradation by 5′ to 3′ exonucleases due to its conformation being similar to that of a 3′ end. Additionally, enzymes recruited to the 5′ cap such as CBC and eIF4E/eIF4G block decapping enzymes from acting on the cap. These factors increase the half-life of mRNA and are essential to allow for proper processing and translation to occur. The 5′ cap works synergistically with the poly(A) tail to stimulate translation by recruitment of translation initiation machinery [144]. eIF4G, bound to the 5′ cap, interacts with PABP1, bound to the poly(A) tail, to create a pseudocircular structure which is thought to ensure that mature mRNA are translated and assist in ribosome advancement along the transcript [144,145]. The poly(A) tail and 5′ cap alone has been shown to be insufficient to drive efficient translation [144]. This synchrony between the poly(A) tail and 5′ cap is especially important in development where translation is known to be regulated by the lengthening and shortening of the poly(A) tail [146].
In addition to m7G , several other variations of 5′ caps have been found. 5′-NAD+ capping is one unique pathway in which RNAs may be capped [147,148].It was recently shown that eukaryotes may possess this type of cap and that these RNAs are unstable and inefficiently translated. The NAD+ cap plays a role in RNA degradation by recognition by DXO protein [149]. It was demonstrated that DXO will preferentially favor the removal of NAD+ caps rather than m7G, which was thought to be DXO’s primary function in the past. Another unique capping mechanism is the +1 and +2 ribose 2′-O-me. These reactions are carried out in humans by hMTr1 and hMTr2 respectively [150,151]. Details of the biological role of these modifications have yet to be elucidated, but they likely serve to modify efficiency of transcript processing, translation, and stability. It is also possible that 2′-O-me of the +1 and +2 cap serves a role in viral vs. host mRNA recognition; a possibility discussed earlier in this review.
rRNA Modifications
Ribosomes consist of both protein and RNA components whose subunits come together to form a functional complex responsible mRNA translation and polypeptide production. Mapping has shown that rRNA modifications are mainly concentrated in functional regions of the ribosome such as the peptidyl transferase center (PTC). In total, eukaryotic rRNA has been mapped to have 91 Ψ, 105 2′-O-me of backbone sugars, and ten methylated bases [152]. These modifications are still not well understood in their role or function in rRNA, but there have been some studies performed which provide some insight of their activity along with what we know about the chemistry of these modifications. The modifications most likely play a role in fine-tuning the structure of the ribosome and play a role in the processing of rRNA. The chemical activity of the ribosome is classically understood to be the responsibility of the protein components which make up the functional regions of the ribosome. There has been some controversy surrounding the possibility that rRNA modifications may be involved in the peptidyl transferase activity [153]. This is based on the evidence of modification mapping however there is little evidence supporting any actual role which modifications may play in the catalysis of the reaction.
2′-O-methylation in rRNA
Modifications of the rRNA are proposed to play a role in modifying ribosome structure and function by altering the structure or the molecular interactions taking place within functionally relevant regions such as the PTC. Based on our understanding of the chemistry, 2′-O-me prevented hydrolysis of the phosphate backbone and caused the ribose sugar to favor the 3′ endo conformation [26,29,30]. Williams, D.J. et al. demonstrated the effects of 2′-O-me modifications in synthetic UUGC tetraloops and found changes in the stability and flexibility of the stem-loops [154]. The observed effects of the substitutions were due to differences in hydrogen bonding, solvation effects, and intrinsic puckering of the hairpins. This evidence further supports how the modification of 2′-O-me in rRNA may play a role in shaping the secondary and tertiary structures necessary for proper ribosome function.
Pseudouridine in rRNA
In rRNA, Ψ modifications have been mapped and found clustered in functionally important regions of the large subunit. [153,155]. The Ψ residues clustered within domain V, which constitutes the PTC, have been debated over as to whether or not these modifications play a role catalytically in peptidyl transferase activity of the ribosome. It is hypothesized that the N1 position of Ψ can allow for high group transfer potential for acyl moieties [153,156]. Currently, there is no support of Ψ direct role in peptidyl transferase activity. Mutational studies in yeast have shown that deletions of the 8 residues which reside in the PTC do not affect growth or viability alone [157] However, collectively, these modifications are essential as introducing several mutations in yeast ribosome PTC results in changes in tRNA binding and peptidyl transferase rates [157,158]. It was also shown that the collective loss of these modifications results in several defects including cell growth and translation [157]. Ψ residues have been found in the small subunit, but none as of yet have been seen in functionally important regions [157,159]. It is classically accepted that this modification mainly aids in forming the secondary and tertiary structure required for ribosome biogenesis and proper formation of protein-RNA interactions.
rRNA biogenesis and processing
Modification of rRNA is involved not only in its function and structure of ribosomal subunits but also in rRNA processing and cleavage [160]. Similar to mRNA, the modifications of rRNA are performed by snoRNA-proteins complexes known as small nucleolar ribonucleoproteins (snoRNPs) in which the snoRNA act as a guide to target specific sequences on the rRNA and ensure that the exact target base gets modified [161]. The Ψ residues of eukaryotic rRNA are synthesized by coordination between guiding H/ACA snoRNP complexes. Researchers can study the loss of Ψ as well as modifications of other RNA species by disrupting this specific snoRNPs and observing the outcome in the transcriptome. Several snoRNPs are involved in RNA processing and cleavage. The modifications which these essential snoRNPs make on pre-rRNA serve as markers for cleavage sites. These cleavages occur within the nucleus before being exported to the cytosol except the 18S unit in Saccharomyces which usually requires an additional cytosolic cleavage step of the 20S precursor [162]. These structural rearrangements and cleavages mediated by rRNA modification give rise to mature ribosomal subunits. One particularly interesting snoRNA is snR35 which is involved in the processing of the site 1191 of 18S rRNA. This particular modification is a hypermodified Ψ m1acp3C1191. The process of forming this modification involves several steps — loss of this modification in the P-site by disruption of snR35 results in defects in pre-rRNA processing and translation due to loss of D-site cleavage during processing [163]. Several nucleolar enzymes which make up the snRNPs have been identified and associated with their target modification. In some cases, the exact catalytic mechanism of the modification is not well understood due to the lack of sequence similarity to other well-characterized modifying enzymes [164–166]. Compared to our understanding and characterization of snoRNA, there is still much left to be discovered about the protein factors which make up pre-ribosomal complexes including the protein components of the snoRNPs, trafficking machinery [167–170], helicases [171,172], and GTPases [173–175].
snoRNAs can be separated into two classes based on their sequence elements; C/D box snoRNA or H/ACA box snoRNA [176,177]. These two classes are responsible for most rRNA modifications, and some may have several target sites [178]. The H/ACA family is responsible for methylation modifications while C/D box snoRNA are responsible for pseudouridylation modifications. The nucleolytic cleavages and nucleolar structures of pre-rRNA processing are entirely dependent on proper modifications by the snoRNP complexes, and it is widely believed that these steps in rRNA processing serve as complex levels of quality control to ensure proper ribosome formation and healthy cell function. Future studies should attempt to characterize the complex dynamics and machinery involved in this quality control and the process of rRNA processing.
Modification of other Non-coding RNAs
It has become greatly appreciated that eukaryotic cells transcribe a wide variety of RNA species with varies regulatory functions, such as micro RNA (miRNA), small interfering RNA (siRNA), P-element-induced wimpy testis (PIWI) interacting RNA (piRNA), small nuclear RNA (snRNA), long non-coding RNA (lncRNA), and tRNA-derived small RNA (tsRNA). What has more recently emerged is the importance of modifications to these RNA species and the information they carry. Here, we will briefly describe some known modifications that modulate the functions of several unique RNA species.
miRNAs are critical components in RNA silencing pathways that induce mRNA degradation and inhibit translation [179]. It has been found that 2′-O-me at the 3′ end of plant miRNAs prevents poly-3′-uridylation, a known marker for small RNA degradation pathways [180–182]. The same protective mechanism using 2′-O-me has also been described in several mammalian small RNAs such as siRNAs and piRNAs [182].
The snRNAs that make up the family of spliceosomal RNAs are extensively modified, mainly bearing Ψ and 2′-O-me [183,184]. Modifications in snRNAs can influence several structural and mechanistic aspects pre-mRNA splicing, and thus can influence the expression and function of many genes. snRNA modifications can modulate RNA-RNA interactions, the interaction of spliceosomal snRNAs with spliceosomal proteins, and the direct catalysis of the splicing reactions [185]. Although the exact role of all snRNA modifications has not been fully dissected, it is thought that the ten 2′-O-me and 13 Ψ present in the U2 snRNA can influence spliceosome assembly and splicing efficiency via the mechanisms just described [186].
As was stated in the tRNA section, Dnmt2 and Nsun2-mediated methylation of cytosine support tRNA stability. Additionally, loss of m5C can increase angiogenin-mediated cleavage of tRNAs, resulting in tsRNAs [48,187,188]. Accumulation of tsRNAs combined with m5C-deficient tRNAs alter protein expression due to codon mistranslation, resulting in disrupted body growth and neuronal cell dysfunction [48,187–189]. These studies hint at the complex interplay between the levels of methylated tRNAs, production of tsRNAs, and protein synthesis. Lastly, lncRNA are known to have a diverse and expanding array of functions in mRNA processing, transcription regulation, and chromatin remodeling [190–192]. Several lncRNAs have been shown to contain multiple m6A sites, such as MALAT1, TUG1, XIST, and NEAT1 [103,193–195]. It is known that XIST-mediated X-inactivation is in part accomplished by YTHDC1 recognition of m6A residues [195]; however, what RNA modifications contribute to the functional role of lncRNAs remains unclear.
Concluding Remarks
The chemical diversity of RNA modifications found throughout the tree of life is staggeringly complex, and how these modifications are regulated and interpreted is even more daunting. In the early years of modification research, most believed that RNA modifications served a relatively static role in much the same way the scientific community once viewed tRNA or ribosomes. Recent discoveries of the dynamic and reversible nature of RNA modifications across many RNA species have caused a resurgence of interest and appreciation for these chemical variants. There remains much to be done, as more quantitative sequencing technologies will be required to precisely map and understand the roles of modifications to different RNA species. That being said, we now know that the chemical structure, occupancy, reversibility, and enzyme selectivity associated with different RNA modifications all play key roles in determining the combinatorial output that ultimately results in exquisite fine-tuning of gene expression and organismal physiology.
References
- 1.Boccaletto P, MacHnicka MA, Purta E, Pitkowski P, Baginski B, Wirecki TK, De Crécy-Lagard V, Ross R, Limbach PA, Kotter A, et al. (2018) MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res, Oxford University Press 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohn WE (1960) Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J. Biol. Chem 235, 1488–1498. [PubMed] [Google Scholar]
- 3.Jackman JE and Alfonzo JD (2013) Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rich A and Rajbhandary UL (1976) Transfer RNA: Molecular structure, sequence, and properties. In Annu. Rev. Biochem 45th ed., pp 805–860. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki T, Nagao A and Suzuki T (2011) Human Mitochondrial tRNAs: Biogenesis, Function, Structural Aspects, and Diseases. Annu. Rev. Genet 45, 299–329. [DOI] [PubMed] [Google Scholar]
- 6.Kelley SO, Steinberg SV and Schimmel P (2001) Fragile T-stem in Disease-associated Human Mitochondrial tRNA Sensitizes Structure to Local and Distant Mutations. J. Biol. Chem, JBC Papers in Press 276, 10607–10611. [DOI] [PubMed] [Google Scholar]
- 7.Kelley SO, Steinberg SV and Schimmel P (2000) Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nat. Struct. Biol, Nature Publishing Group 7, 862–865. [DOI] [PubMed] [Google Scholar]
- 8.Parisien M, Wang X and Pan T (2013) Diversity of human tRNA genes from the 1000-genomes project. RNA Biol, Taylor & Francis 10, 1853–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iben JR and Maraia RJ (2014) tRNA gene copy number variation in humans. Gene 536, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geslain R and Pan T (2010) Functional Analysis of Human tRNA Isodecoders. J. Mol. Biol 396, 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D and Agris PF (2011) The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saikia M, Fu Y, Pavon-Eternod M, Chuan H and Pan T (2010) Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan T (2018) Modifications and functional genomics of human transfer RNA. Cell Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helm M, Giegé R and Florentz C (1999) A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNA(Lys). Biochemistry 38, 13338–13346. [DOI] [PubMed] [Google Scholar]
- 15.Sakurai M, Ohtsuki T and Watanabe K (2005) Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucleic Acids Res, Oxford University Press 33, 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oerum S, Dégut C, Barraud P and Tisné C (2017, February 21) m1A post-transcriptional modification in tRNAs. Biomolecules, Multidisciplinary Digital Publishing Institute. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobitski AY, Hengesbach M, Helm M and Nienhaus GU (2008) Sculpting an RNA conformational energy landscape by a methyl group modification - A single-molecule FRET study. Angew. Chemie - Int. Ed, Wiley-Blackwell 47, 4326–4330. [DOI] [PubMed] [Google Scholar]
- 18.Motorin Y and Helm M (2010) TRNA stabilization by modified nucleotides. Biochemistry [DOI] [PubMed] [Google Scholar]
- 19.Saenger W (1984) Intercalation. In Principles of Nucleic Acid Structure, pp 350–367, Springer, New York. [Google Scholar]
- 20.Rife JP, Cheng CS, Moore PB and Strobel SA (1998) N2-methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res, Oxford University Press 26, 3640–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayase Y, Jahn M, Rogers MJ, Sylvers LA, Koizumi M, Inoue H, Ohtsuka E and Söll D (1992) Recognition of bases in Escherichia coli tRNA(Gln) by glutaminyl-tRNA synthetase: a complete identity set. EMBO J 11, 4159–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edqvist J, Grosjean H and Stråby KB (1992) Identity elements for N2-dimethylation of guanosine-26 in yeast tRNAs. Nucleic Acids Res 20, 6575–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallan PS, Kreutz C, Bosio S, Micura R and Egli M (2008) Effects of N2,N2-dimethylguanosine on RNA structure and stability: Crystal structure of an RNA duplex with tandem m2 2G:A pairs. RNA, Cold Spring Harbor Laboratory Press 14, 2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arimbasseri AG, Blewett NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M and Maraia RJ (2015) RNA Polymerase III Output Is Functionally Linked to tRNA Dimethyl-G26 Modification. PLoS Genet (Hopper A, ed.), Public Library of Science; 11, e1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori H (2014) Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray, Michael Charette MW (2000) Pseudouridine in RNA: What, Where, How, and Why. IUBMB Life (International Union Biochem. Mol. Biol. Life), Wiley-Blackwell 49, 341–351. [DOI] [PubMed] [Google Scholar]
- 27.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM and Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature, Nature Publishing Group 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert WV, Bell TA and Schaening C Messenger RNA modifications: Form, distribution, and function [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis DR (1995) Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23, 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spenkuch F, Motorin Y and Helm M (2014) Pseudouridine: Still mysterious, but never a fake (uridine)! RNA Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis DR, Veltri CA and Nielsen L (1998) An rna model system for investigation of pseudouridine stabilization of the codon-anticodon interaction in trnaLys, tRNAHisand tRNATyr. J. Biomol. Struct. Dyn 15, 1121–1132. [DOI] [PubMed] [Google Scholar]
- 32.Davis DR (1998) Biophysical and conformational properties of modified nucleosides in RNA (Nuclear Magnetic Resonance Studies). In Modification and editing of RNA, pp 85–102, American Society of Microbiology. [Google Scholar]
- 33.Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A and Agris PF (1999) Structural and functional roles of the N1- and N3-protons of Ψ at tRNA’s position 39. Nucleic Acids Res 27, 3543–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Vanommeslaeghe K, Aleksandrov A, MacKerell AD and Nilsson L (2016) Additive CHARMM force field for naturally occurring modified ribonucleotides. J. Comput. Chem, Wiley-Blackwell 37, 896–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanda RK, Tewari R, Govil G and Smith ICP (1974) The Conformation of β-Pseudouridine about the Glycosidic Bond as Studied by 1 H Homonuclear Overhauser Measurements and Molecular Orbital Calculations. Can. J. Chem 52, 371–375. [Google Scholar]
- 36.NEUMANN JM, TRAN‐DINH S, BERNASSAU JM and GUÉRON M (1980) Comparative Conformations of Uridine and Pseudouridine and Their Derivatives. Eur. J. Biochem, Wiley/Blackwell (10.1111) 108, 457–463. [DOI] [PubMed] [Google Scholar]
- 37.Durant PC and Davis DR (1999) Stabilization of the anticodon stem-loop of tRNA(Lys,3) by an A+-C base-pair and by pseudouridine. J. Mol. Biol, Academic Press 285, 115–131. [DOI] [PubMed] [Google Scholar]
- 38.Dao V, Guenther R, Malkiewicz A, Nawrot B, Sochacka E, Kraszewski A, Jankowska J, Everett K and Agris PF (1994) Ribosome binding of DNA analogs of tRNA requires base modifications and supports the “extended anticodon.” Proc. Natl. Acad. Sci 91, 2125–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrington KM, Nazarenko IA, Uhlenbeck OC, Dix DB and Thompson RC (1993) In Vitro Analysis of Translational Rate and Accuracy with an Unmodified tRNA. Biochemistry, American Chemical Society 32, 7617–7622. [DOI] [PubMed] [Google Scholar]
- 40.Malkiewicz A, Sierzputowska-Gracz H and Agris PF (1995) Rna Modified Uridines VII: Chemical Synthesis and Initial Analysis of tRNA D-Loop Oligomers with Tandem Modified Uridines. Nucleosides and Nucleotides, Taylor & Francis Group 14, 143–165. [Google Scholar]
- 41.Dalluge J (1996) Conformational flexibility of RNA: the role of dihydrouridine. Nucleic Acids Res 24, 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyubankova N, Sochacka E, Kraszewska K, Nawrot B, Herdewijn P and Lescrinier E (2015) Contribution of dihydrouridine in folding of the D-arm in tRNA. Org. Biomol. Chem, The Royal Society of Chemistry 13, 4960–4966. [DOI] [PubMed] [Google Scholar]
- 43.Suck D, Saenger W and Zechmeister K (1972) Molecular and crystal structure of the tRNA minor constituent dihydrouridine. Acta Crystallogr. Sect. B Struct. Crystallogr. Cryst. Chem 28, 596–605. [Google Scholar]
- 44.Uhl W, Reiner J and Gassen HG (1983) On the conformation of 5-substituted uridines as studied by proton magnetic resonance. Nucleic Acids Res, Oxford University Press 11, 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egert E, Lindner HJ, Hillen W and Buhm MC (1980) Influence of Substituents at the 5 Position on the Structure of Uridine. J. Am. Chem. Soc, Wiley 102, 3707–3713. [Google Scholar]
- 46.Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G and Nawrot B (2007) Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA, Cold Spring Harbor Laboratory Press 13, 1301–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF and Pütz J (2009) tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37, 159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G and Lyko F (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol, Nature Publishing Group 19, 900–905. [DOI] [PubMed] [Google Scholar]
- 49.Burgess AL, David R and Searle IR (2015) Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levitt M (1969) Detailed molecular model for transfer ribonucleic acid. Nature 224, 759–763. [DOI] [PubMed] [Google Scholar]
- 51.Oliva R, Tramontano A and Cavallo L (2007) Mg2+ binding and archaeosine modification stabilize the G15-C48 Levitt base pair in tRNAs. RNA 13, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agris PF, Guenther R, Sochacka E, Newman W, Czerwińska G, Liu G, Ye W and Malkiewicz A (1999) Thermodynamic contribution of nucleoside modifications to yeast tRNAPhe anticodon stem loop analogs. Acta Biochim. Pol 46, 163–172. [PubMed] [Google Scholar]
- 53.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G and Lyko F (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol 19, 900–905. [DOI] [PubMed] [Google Scholar]
- 54.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M and Lyko F (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24, 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cabello-Villegas J, Winkler ME and Nikonowicz EP (2002) Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAphe. J. Mol. Biol 319, 1015–1034. [DOI] [PubMed] [Google Scholar]
- 56.Denmon AP, Wang J and Nikonowicz EP (2011) Conformation effects of base modification on the anticodon stem-loop of Bacillus subtilis tRNATyr. J. Mol. Biol 412, 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A and Agris PF (2000) Modified nucleoside dependent Watson - Crick and wobble codon binding by tRNA(Lys)(UUU) species. Biochemistry 39, 13390–13395. [DOI] [PubMed] [Google Scholar]
- 58.Stuart JW, Koshlap KM, Guenther R and Agris PF (2003) Naturally-occurring Modification Restricts the Anticodon Domain Conformational Space of tRNAPhe. J. Mol. Biol 334, 901–918. [DOI] [PubMed] [Google Scholar]
- 59.Lusic H, Gustilo EM, Vendeix FAP, Kaiser R, Delaney MO, Graham WD, Moye VA, Cantara WA, Agris PF and Deiters A (2008) Synthesis and investigation of the 5-formylcytidine modified, anticodon stem and loop of the human mitochondrial tRNAMet. Nucleic Acids Res 36, 6548–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agris PF (2008) Bringing order to translation: The contributions of transfer RNA anticodon-domain modifications. EMBO Rep 9, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gustilo EM, Vendeix FA and Agris PF (2008) tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A and Agris PF (2002) Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem 277, 16391–16395. [DOI] [PubMed] [Google Scholar]
- 63.Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V and Suzuki T (2008) Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U·G wobble pairing during decoding. J. Biol. Chem 283, 18801–18811. [DOI] [PubMed] [Google Scholar]
- 64.Phelps SS, Malkiewicz A, Agris PF and Joseph S (2004) Modified nucleotides in tRNALys and tRNAVal are important for translocation. J. Mol. Biol 338, 439–444. [DOI] [PubMed] [Google Scholar]
- 65.Väre VYP, Eruysal ER, Narendran A, Sarachan KL and Agris PF (2017, March 16) Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules, Multidisciplinary Digital Publishing Institute. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Björk GR, Durand JMB, Hagervall TG, Leipuviene R, Lundgren HK, Nilsson K, Chen P, Qian Q and Urbonavičius J (1999, June 4) Transfer RNA modification: Influence on translational frameshifting and metabolism. FEBS Lett, Wiley-Blackwell. [DOI] [PubMed] [Google Scholar]
- 67.Brierley I, Meredith MR, Bloys AJ and Hagervall TG (1997) Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: Influence of tRNA anticodon modification on frameshifting. J. Mol. Biol 270, 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Urbonavičius J, Qian Q, Durand JMB, Hagervall TG and Björk GR (2001) Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 20, 4863–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crick FHC (1966) Codon—anticodon pairing: The wobble hypothesis. J. Mol. Biol 19, 548–555. [DOI] [PubMed] [Google Scholar]
- 70.Agris PF (1991) Wobble position modified nucleosides evolved to select transfer RNA codon recognition: A modified-wobble hypothesis. Biochimie 73, 1345–1349. [DOI] [PubMed] [Google Scholar]
- 71.Rogalski M, Karcher D and Bock R (2008) Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol, Nature Publishing Group 15, 192–198. [DOI] [PubMed] [Google Scholar]
- 72.Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FAP, Agris PF and Ramakrishnan V (2007) Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol, Nature Publishing Group 14, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agris PF, Guenther R, Ingram PC, Basti MM, Stuart JW, Sochacka E and Malkiewicz A (1997) Unconventional structure of tRNA(Lys)SUU anticodon explains tRNA’s role in bacterial and mammalian ribosomal frameshifting and primer selection by HIV-1. RNA 3, 420–8. [PMC free article] [PubMed] [Google Scholar]
- 74.Agris PF, Söll D and Seno T (1973) Biological function of 2-thiouridine in Escherichia coli glutamic acid transfer ribonucleic acid. Biochemistry 12, 4331–4337. [DOI] [PubMed] [Google Scholar]
- 75.Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S and Miyazawa T (1985) Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci 82, 4905–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamble AS, Kumbhar BV, Sambhare SB, Bavi RS and Sonawane KD (2015) Conformational Preferences of Modified Nucleoside 5-Taurinomethyluridine, τm5U Occur at ‘wobble’ 34th Position in the Anticodon Loop of tRNA. Cell Biochem. Biophys, Springer US 71, 1589–1603. [DOI] [PubMed] [Google Scholar]
- 77.Wang R, Vangaveti S, Ranganathan SV, Basanta-Sanchez M, Haruehanroengra P, Chen A and Sheng J (2016) Synthesis, base pairing and structure studies of geranylated RNA. Nucleic Acids Res 44, 6036–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sierant M, Leszczynska G, Sadowska K, Dziergowska A, Rozanski M, Sochacka E and Nawrot B (2016) S-Geranyl-2-thiouridine wobble nucleosides of bacterial tRNAs; Chemical and enzymatic synthesis of S-geranylated-RNAs and their physicochemical characterization. Nucleic Acids Res 44, 10986–10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang R, Ranganathan SV, Basanta-Sanchez M, Shen F, Chen A and Sheng J (2015) Synthesis and base pairing studies of geranylated 2-thiothymidine, a natural variant of thymidine. Chem. Commun 51, 16369–16372. [DOI] [PubMed] [Google Scholar]
- 80.Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M and Yusupova G (2016) Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun, Nature Publishing Group 7, 10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vendeix FAP, Murphy IV FV, Cantara WA, Leszczyńska G, Gustilo EM, Sproat B, Malkiewicz A and Agris PF (2012) Human tRNALys3UUUis pre-structured by natural modifications for cognate and wobble codon binding through Keto-Enol tautomerism. J. Mol. Biol 416, 467–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rozov A, Westhof E, Yusupov M and Yusupova G (2016) The ribosome prohibits the G•U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res 44, 6434–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW and Al-Hashimi HM (2015) Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature, Nature Publishing Group 519, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manickam N, Joshi K, Bhatt MJ and Farabaugh PJ (2015) Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res 44, 1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A and Agris PF (2000) Functional anticodon architecture of human tRNA(Lys3) includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 39, 13396–13404. [DOI] [PubMed] [Google Scholar]
- 86.Durant PC, Bajji AC, Sundaram M, Kumar RK and Davis DR (2005) Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALysanticodon loop: The effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44, 8078–8089. [DOI] [PubMed] [Google Scholar]
- 87.Witts RN, Hopson EC, Koballa DE, Van Boening TA, Hopkins NH, Patterson EV and Nagan MC (2013) Backbone-base interactions critical to quantum stabilization of transfer RNA anticodon structure. J. Phys. Chem. B, UTC 117, 7489–7497. [DOI] [PubMed] [Google Scholar]
- 88.Sample PJ, Kořený L, Paris Z, Gaston KW, Rubio MAT, Fleming IMC, Hinger S, Horáková E, Limbach PA, Lukeš J, et al. (2015) A common tRNA modification at an unusual location: The discovery of wyosine biosynthesis in mitochondria. Nucleic Acids Res 43, 4362–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Crécy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, et al. (2010) Biosynthesis of wyosine derivatives in tRNA: An ancient and highly diverse pathway in archaea. Mol. Biol. Evol 27, 2062–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murphy FV, Ramakrishnan V, Malkiewicz A and Agris PF (2004) The role of modifications in codon discrimination by tRNALysUUU. Nat. Struct. Mol. Biol, Nature Publishing Group 11, 1186–1191. [DOI] [PubMed] [Google Scholar]
- 91.Ponnuswamy PK and Michael Gromiha M (1994) On the conformational stability of oligonucleotide duplexes and tRNA molecules. J. Theor. Biol 169, 419–432. [DOI] [PubMed] [Google Scholar]
- 92.Kim SH, Sussman JL, Suddath FL, Quigley GJ, McPherson A, Wang AH, Seeman NC and RICH A (1974) The general structure of transfer RNA molecules. Proc. Natl. Acad. Sci. U. S. A 71, 4970–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu N, Dai Q, Zheng G, He C, Parisien M and Pan T (2015) N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perry RP and Kelley DE (1974) Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42. [Google Scholar]
- 95.Desrosiers R, Friderici K and Rottman F (1974) Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci 71, 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Engel JD and von Hippel PH (1974) Effects of methylation on the stability of nucleic acid conformations: Studies at the monomer level. Biochemistry 13, 4143–4158. [DOI] [PubMed] [Google Scholar]
- 97.Engel JD and von Hippel PH (1978) Effects of methylation conformations on the stability of nucleic acid. J. Biol. Chem 253, 927–934. [PubMed] [Google Scholar]
- 98.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY and Kool ET (2015) Structure and Thermodynamics of N 6 -Methyladenosine in RNA: A Spring-Loaded Base Modification. J. Am. Chem. Soc, American Chemical Society 137, 2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. (2015) Structural imprints in vivo decode RNA regulatory mechanisms. Nature, Nature Publishing Group 519, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S and Tavazoie SF (2015) HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang P, Doxtader KA and Nam Y (2016) Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell, Cell Press 63, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. (2016) Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature, Nature Publishing Group 534, 575–578. [DOI] [PubMed] [Google Scholar]
- 103.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol, Nature Publishing Group 10, 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res, Nature Publishing Group 24, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. (2014) Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep 8, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M and Jaffrey SR (2016) M6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carroll SM, Narayan P and Rottman FM (1990) N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol. Cell. Biol 10, 4456–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stoltzfus CM and Dane RW (1982) Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J. Virol, American Society for Microbiology (ASM) 42, 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-haim MS, et al. (2015) m6 mRNA methylation facilitates resolution of naive pluripotence toward differentiation. Science (80-. ) 3, 1002–1006. [DOI] [PubMed] [Google Scholar]
- 110.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. (2014) FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res, Nature Publishing Group 24, 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, Sun B-F, Sun H-Y, Li A, Ping X-L, Lai W-Y, et al. (2016) Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell, Cell Press 61, 507–519. [DOI] [PubMed] [Google Scholar]
- 112.Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, et al. (2016) M6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods, Nature Publishing Group 13, 692–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. (2013) ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. (2013) RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. [DOI] [PubMed] [Google Scholar]
- 115.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H and He C (2015) N6-methyladenosine modulates messenger RNA translation efficiency. Cell, Elsevier 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. (2018) Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol, Nature Publishing Group 20, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin S, Choe J, Du P, Triboulet R and Gregory RI (2016) The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C and He C (2017) YTHDF3 facilitates translation and decay of N 6-methyladenosine-modified RNA. Cell Res 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C and Bohnsack MT (2018) The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′−3′ exoribonuclease XRN1. RNA 24, 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dunn DB (1961) The occurence of 1-methyladenine in ribonucleic acid. BBA - Biochim. Biophys. Acta, Elsevier 46, 198–200. [DOI] [PubMed] [Google Scholar]
- 121.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. (2016) The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature, Nature Publishing Group 530, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S and Yi C (2016) Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol, Nature Publishing Group 12, 311–316. [DOI] [PubMed] [Google Scholar]
- 123.Zhou H, Kimsey IJ, Nikolova EN, Sathyamoorthy B, Grazioli G, McSally J, Bai T, Wunderlich CH, Kreutz C, Andricioaei I, et al. (2016) M1A and m1G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol, Nature Publishing Group 23, 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hauenschild R, Tserovski L, Schmid K, Thüring K, Winz ML, Sharma S, Entian KD, Wacheul L, Lafontaine DLJ, Anderson J, et al. (2015) The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res 43, 9950–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ozanick S, Krecic A, Andersland J and Anderson JT (2005) The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Khoddami V and Cairns BR (2013) Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol 31, 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. (2013) NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM and Preiss T (2012) Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, et al. (2014) Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc, UTC 136, 11582–11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huber SM, Van Delft P, Mendil L, Bachman M, Smollett K, Werner F, Miska EA and Balasubramanian S (2015) Formation and abundance of 5-hydroxymethylcytosine in RNA. ChemBioChem 16, 752–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. (2017) 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m 5 C reader. Cell Res 27, 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li X, Zhu P, Ma S, Song J, Bai J, Sun F and Yi C (2015) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol 11, 592–597. [DOI] [PubMed] [Google Scholar]
- 133.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. (2014) Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karijolich J and Yu YT (2011) Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Beal PA, Maydanovych O and Pokharel S (2007) The Chemistry and Biology of RNA editing by Adenosine Deaminases. Nucleic Acids Symp. Ser 51, 83–84. [DOI] [PubMed] [Google Scholar]
- 136.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie J-P, Brosius J and Huttenhofer A (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bachellerie JP, Cavaillé J and Hüttenhofer A (2002, August 1) The expanding snoRNA world. Biochimie, Elsevier. [DOI] [PubMed] [Google Scholar]
- 138.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. (2010) 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Case JB, Ashbrook AW, Dermody TS and Denison MR (2016) Mutagenesis of S -Adenosyl-l-Methionine-Binding Residues in Coronavirus nsp14 N7-Methyltransferase Demonstrates Differing Requirements for Genome Translation and Resistance to Innate Immunity. J. Virol 90, 7248–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Judge AD, Bola G, Lee ACH and MacLachlan I (2006) Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther, Cell Press 13, 494–505. [DOI] [PubMed] [Google Scholar]
- 141.Moteki S and Price D (2002) Functional coupling of capping and transcription of mRNA. Mol. Cell, Cell Press 10, 599–609. [DOI] [PubMed] [Google Scholar]
- 142.Topisirovic I, Svitkin YV, Sonenberg N and Shatkin AJ (2011, March) Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA [DOI] [PubMed] [Google Scholar]
- 143.Cougot N, Van Dijk E, Babajko S and Séraphin B (2004, August 1) “Cap-tabolism.” Trends Biochem. Sci, Elsevier Current Trends. [DOI] [PubMed] [Google Scholar]
- 144.Preiss T and Hentze MW (1998) Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392, 516–520. [DOI] [PubMed] [Google Scholar]
- 145.Preiss T and Hentze MW (1999) From factors to mechanisms: Translation and translational control in eukaryotes. Curr. Opin. Genet. Dev, Elsevier Current Trends 9, 515–521. [DOI] [PubMed] [Google Scholar]
- 146.Wickens M (1990) In the beginning is the end: regulation of poly(A) addition and removal during early development. Trends Biochem. Sci, Elsevier Current Trends 15, 320–324. [DOI] [PubMed] [Google Scholar]
- 147.Bird JG, Zhang Y, Tian Y, Panova N, Barvík I, Greene L, Liu M, Buckley B, Krásný L, Lee JK, et al. (2016) The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature, Nature Publishing Group 535, 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kiledjian M (2018, June) Eukaryotic RNA 5′-End NAD+Capping and DeNADding. Trends Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jiao X, Doamekpor SK, Bird JG, Nickels BE, Tong L, Hart RP and Kiledjian M (2017) 5′ End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding. Cell 168, 1015–1027.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bélanger F, Stepinski J, Darzynkiewicz E and Pelletier J (2010) Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem 285, 33037–33044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, Zamudio JR, Sturm NR, Jaworski J and Bujnicki JM (2011) 2′-O-ribose methylation of cap2 in human: Function and evolution in a horizontally mobile family. Nucleic Acids Res 39, 4756–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP and Fournier MJ (2008) Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem, American Society for Biochemistry and Molecular Biology 283, 26026–26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lane BG, Ofengand J and Gray MW (1992) Pseudouridine in the large-subunit (23 S-like) ribosomal RNA The site of peptidyl transfer in the ribosome? FEBS Lett 302, 1–4. [DOI] [PubMed] [Google Scholar]
- 154.Williams DJ, Boots JL and Hall KB (2001) Thermodynamics of 2-O-ribose substitutions in UUCG tetraloops. RNA 7, 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bakin A, Lane BG and Ofengand J (1994) Clustering of Pseudouridine Residues around the Peptidyltransferase Center of Yeast Cytoplasmic and Mitochondrial Ribosomes. Biochemistry 33, 13475–13483. [DOI] [PubMed] [Google Scholar]
- 156.Lane BG, Ofengand J and Gray MW (1995) Pseudouridine and O2-methylated nucleosides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bonds in proteins. Biochimie 77, 7–15. [DOI] [PubMed] [Google Scholar]
- 157.Fournier MJ and Ofengand J (1998) The Pseudouridine Residues of rRNA: Number, Location, Biosynthesis, and Function. In Modification and Editing of RNA, pp 229–253, American Society of Microbiology. [Google Scholar]
- 158.Liang XH, Liu Q and Fournier MJ (2009) Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA, Cold Spring Harbor Laboratory Press 15, 1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ofengand J, Bakin A, Wrzesinski J, Nurse K and Lane BG (1995) The pseudouridine residues of ribosomal RNA. Biochem. Cell Biol 73, 915–924. [DOI] [PubMed] [Google Scholar]
- 160.Kiss T (2001, July 16) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J, European Molecular Biology Organization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Galardi S, Fatica A, Bachi A, Scaloni A, Presutti C and Bozzoni I (2002) Purified box C/D snoRNPs are able to reproduce site-specific 2’-O-methylation of target RNA in vitro. Mol. Cell. Biol 22, 6663–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vanrobays E, Gleizes PE, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M and Gélugne JP (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rp10p, an essential non-ribosomal cytoplasmic protein. EMBO J, European Molecular Biology Organization 20, 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liang XH, Liu Q and Fournier MJ (2009) Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15, 1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Koonin EV (1996) Pseudouridine syntheses: Four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res 24, 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bousquet-Antonelli C, Henry Y, Gélugne JP, Caizergues-Ferrer M and Kiss T (1997) A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J 16, 4770–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lafontaine DLJ and Tollervey D (2000) Synthesis and Assembly of the Box C+D Small Nucleolar RNPs. Mol. Cell. Biol 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kufel J, Allmang C, Petfalski E, Beggs J and Tollervey D (2003) Lsm proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem 278, 2147–2156. [DOI] [PubMed] [Google Scholar]
- 168.Lee MS, Henry M and Silver PA (1996) A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev 10, 1233–1246. [DOI] [PubMed] [Google Scholar]
- 169.Gadal O, Strau D, Kessl J, Trumpower B, Tollervey D and Hurt E (2001) Nuclear Export of 60S Ribosomal Subunits Depends on Xpo1p and Requires a Nuclear Export Sequence-Containing Factor, Nmd3p, That Associates with the Large Subunit Protein Rpl10p. Mol. Cell. Biol 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stage-Zimmermann T, Schmidt U and Silver PA (2000) Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell, American Society for Cell Biology 11, 3777–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kressler D, Linder P and de La Cruz J (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.De la Cruz J, Kressler D and Linder P (1999, May) Unwinding RNA in saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci [DOI] [PubMed] [Google Scholar]
- 173.Gelperin D, Horton L, Beckman J, Hensold J and Lemmon SK (2001) Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nissan TA, Baßler J, Petfalski E, Tollervey D and Hurt E (2002) 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J, European Molecular Biology Organization 21, 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fromont-Racine M, Senger B, Saveanu C and Fasiolo F (2003, August 14) Ribosome assembly in eukaryotes. Gene [DOI] [PubMed] [Google Scholar]
- 176.Bachellerie JP and Cavaillé J (1997, July) Guiding ribose methylation of rRNA. Trends Biochem. Sci [DOI] [PubMed] [Google Scholar]
- 177.Decatur WA and Fournier MJ (2002, July) rRNA modifications and ribosome function. Trends Biochem. Sci [DOI] [PubMed] [Google Scholar]
- 178.Bonnerot C, Pintard L and Lutfalla G (2003) Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2′-O-ribose methylation of a conserved rRNA position in yeast. Mol. Cell 12, 1309–1315. [DOI] [PubMed] [Google Scholar]
- 179.Jonas S and Izaurralde E (2015, July 16) Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet, Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 180.Ren G, Xie M, Zhang S, Vinovskis C, Chen X and Yu B (2014) Methylation protects microRNAs from an AGO1-associated activity that uridylates 5’ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci 111, 6365–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kishi M, Pan YA, Crump JG and Sanes JR (2005) Mammalian SAD kinases are required for neuronal polarization. Science (80-. ), MIT Press 307, 929–932. [DOI] [PubMed] [Google Scholar]
- 182.Ji L and Chen X (2012, April 13) Regulation of small RNA stability: Methylation and beyond. Cell Res, Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Massenet S and Branlant C (1999) A limited number of pseudouridine residues in the human atac spliceosomal UsnRNAs as compared to human major spliceosomal A limited number of pseudouridine residues in the human atac spliceosomal UsnRNAs as compared to human major spliceosomal UsnRNAs. Methods [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Reddy R and Busch H (1988) Small Nuclear RNAs: RNA Sequences, Structure, and Modifications. Struct. Funct. Major Minor Small Nucl. Ribonucleoprotein Part, Springer Berlin Heidelberg, Berlin, Heidelberg: 1–37. [Google Scholar]
- 185.Karijolich J and Yu YT (2010) Spliceosomal snRNA modifications and their function. RNA Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dönmez G, Hartmuth K and Lührmann R (2004) Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA, Cold Spring Harbor Laboratory Press 10, 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. (2014) Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33, 2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza N, et al. (2016) Stem cell function and stress response are controlled by protein synthesis. Nature, Nature Publishing Group 534, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Gro ne H-J, et al. (2015) The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 34, 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lee JT and Bartolomei MS (2013) X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell [DOI] [PubMed] [Google Scholar]
- 191.Rinn JL and Chang HY (2012) Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem 81, 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mercer TR, Dinger ME and Mattick JS (2009, March 1) Long non-coding RNAs: Insights into functions. Nat. Rev. Genet, Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 193.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE and Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, Nature Publishing Group 485, 201–206. [DOI] [PubMed] [Google Scholar]
- 195.Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M and Jaffrey SR (2016) m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature, Nature Publishing Group 537, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]


