Abstract
Ribonucleic acid (RNA) is involved in translation and transcription, which are the mechanisms in which cells express genes.1 The three classes of RNA discussed are transfer RNA (tRNA), messenger RNA (mRNA), and ribosomal RNA (rRNA). mRNA is the transcript encoded from DNA, rRNA is associated with ribosomes, and tRNA is associated with amino acids and is used to read mRNA transcripts to make proteins.2 Interestingly, the function of tRNA, rRNA, and mRNA can be significantly altered by chemical modifications at the co-transcriptional and post-transcriptional levels, and there are over 171 of these modifications identified thus far.3,4 Several of these modifications are linked to diseases such as cancer, diabetes, and neurological disorders. In this review, we will introduce a few RNA modifications of biological functions and how dysregulation of these RNA modifications linking to human disease.
Keywords: RNA, Modifications, Disease
N1-methyladenosine in tRNA
N1-methyladenosine (m1A) was first discovered occurring at positions 9, 14, and 58 in tRNA and is important for the tRNA 3-dimensional structure.5,6 m1A is also important for structure stabilization of tRNA due to its positive electrostatic charge and also assists in correct folding of tRNA.6,7 Recently evidence suggests that m1A in tRNA is under reversible regulation that is installed by a tRNA m1A methyltransferase (MTase) and is erased by AlkB Homologue 1 (ALKBH1) and AlkB Homologue 3 (ALKBH3).5 Studies have shown ALKBH1 catalyzed demethylation of tRNA can result in a reduction in translation initiation as well as a decrease in use of the demethylated tRNAs in protein synthesis.8
Methylated Guanosines in tRNA
One of the most conserved modifications in tRNA is 7-methylguanosine (m7G), found at position 46 in the variable loop of tRNA, and is installed by tRNA m7G46 methyltransferase.9,10 This modification forms a tertiary base pair with C13 and G22 and stabilizes the structure of the tRNA.11
An additional guanosine modification in tRNA is N 2-methylguanosine (m2G).12 m2G forms stable interactions with cytosine as well as uridine.13,14 The modification is isoenergetic with guanosine in the Watson-Crick G-C base pair at RNA duplexes with cytosine, and does not play a significant role in secondary structure stability.12 It is located in the dihydrouridine loop (position 10) of tRNA, the CCA stem of tRNAMet (position 6), as well as position 26 in tRNALys.13
N2, N2-dimethylguanosine (m22G) and N2, N2, 2’O-trimethylguanosine (m22Gm) are highly conserved modifications at positions 10 and 26.15 At position 26, this modification is installed by tRNA(m22G) dimethyltransferase, and at position 10, this modification is installed by the Trm-m22G10 enzyme in archaea.16,17 m22G is located in the bend between the dihydro-uridine stem and the anticodon stem in tRNA, and prevents the folding of tRNA into unusual structures, often adopted by mitochondrial tRNA (mtRNAs).18 The double methylation of m22G allows the guanosine to base pair with adenosine, uridine, or guanosine at position 44, and prevents the base from pairing with cytosine.18
Methylation at position N-1 of guanosine occurs at position 37 and position 9 in tRNA.19 TRMT10A methylates tRNA guanosines at position 9 with S-adenosylmethionine as a methyl donor.19,20 At position 37, the methyl group is added to guanosine by TrmD from AdoMet.21 Without this modification at position 37, there can be effects on translation efficiency due to frameshift.21
Pseudouridine om tRNA
Pseudouridine (Ψ) is that most abundant modified type of nucleoside across all species of RNA.22 Ψ is an isomer of uridine, where rather than the usual uracil attached via a nitrogen-carbon glyosidic bond, uracil is attached via a carbon-carbon bond. This modification alters the secondary structure of the RNA by increasing base stacking, making more ridged sugar-phosphate backbones, and improving the base pairing.23
5-methylcytosine (m5C) in tRNA
5-methylcytosine is found at position 34, 38, 40, 48, 49, and 50 in tRNA.24 m5C at positions 48 and position 49 are located in the TΨC loop of tRNA, and contributes to the stability of tRNA as well as protein synthesis.24 This modification is established via Dnmt2 (for cytosine 38) and NSun2 (for cytosine 34, 40, 48, 49, 50).24 m5C has been found to cooperate with other modifications on tRNA to protect the tRNA from degradation and is also linked to stress signaling.24
Dihydrouridine in tRNA
The dihydrouridine modification is catalyzed by dihydrouridine synthases and is found in the D-loop of tRNA.25 Dihydrouridine synthases catalyze the reduction of the carbon-carbon double bond at positions 5 and 6 on the uridine base. It is thought that this base modification supports structural flexibility by destabilizing C3’-endoribose conformation and stabilizing the C2’-endoribose form.26 This conformation allows flexibility and dynamic motion in areas of tertiary interactions and loop formation in tRNA.26 Dihydrouridine causes the formation of a stable hairpin structure in the tRNA and is important in the folding and stability of tRNA.27
tRNA Modifications Link to Human Disease
It has been found that nonsense mutations in TRMT10A, resulting in loss of function of the methyltransferase, are linked with familial syndromes including intellectual disabilities, microcephaly, and faults in glucose metabolism.20,28 Mutations in NSun2, a m5C tRNA methyltransferase, have been found to be associated with intellectual disabilities with a mutation that causes NSun2 to not localize to the nucleus.29,30 NSun2 has also been linked to Dubowitz syndrome, an autosomal recessive disorder that is characterized by small structure, mild microcephaly, mental retardation, eczema, and distinct facial expression.31 Patients that have this disorder lack NSun2 specific methylation at nucleotides 47 and 48 on tRNA.31 The loss of m5C in tRNA, results in endonucleolytic cleavage by angiogenin and causes the accumulation of 5’tRNA fragments.28 Additionally, mutations to genes that are associated with the m7G tRNA methyltransferase complex can lead to microcephalic primordial dwarfism.10
Myc, a known proto-oncogene, heterodimerizes with another protein, Max.32 Together, these Myc/Max heterodimers recruit histone acetyltransferases to activate gene transcription.33 The overexpression of Myc is linked to cancer, and has been found to activate the RNA methyltransferase NSun2, leading to NSun2 overexpression.34 An epigenetic cancer therapy, azacytidine, is found to inhibit the m5C methyltransferase, DNMT2, that catalyzes the m5C modification on tRNAAsp.35 This has led to the idea that DNMT2 is associated with tumorigenesis.35 It is suggested that since DNMT2 methylation leads to stabilization of tRNA, the de-methylated tRNAs may be misfolded and impact the rate of protein synthesis in cancer.35,36
2’-O-methylation in rRNA
2’-O-methylation (2’-O-me) in rRNA replaces the normal 2’-OH in rRNA with 2’-O-me, and is added postranscriptionally.37 This modification protects the rRNA from hydrolysis, additionally, 2’-O-me also stabilizes the rRNA into a C3’-endo conformation.38,37 Additionally, this modification restricts the rRNA strand conformation and flexibility as a result of phosphate restriction.37 Ribosome structure and function are impacted by 2’-O-me, and this modification is located in more flexible regions of rRNA.39,40 2’-O-me plays a role in protein synthesis by regulating translational activity of the ribosome.37
Pseudouridine (Ψ) in rRNA
Pseudouridine (Ψ) is found in both the small and large subunit rRNAs, and is the most common single modified nucleoside in rRNA.22,41 The N-1 positions in Ψ are thought to catalyze the transfer of aminoacyl residues from the peptidyl tRNA to aminoacyl tRNA (P site to A site in ribosome).42 In the large subunit of the ribosome, Ψ is clustered in areas that are connected to the peptidyl transferase center.41 There is no known clustering of Ψ in the small subunit of the ribosome.41
N1-methyladenosine (m1A) in rRNA
In addition to tRNA and mRNA, m1A has been found in rRNA.5 m1A is catalyzed by the human nucleolar protein, nucleomethylin (NML) in 28S rRNAs.43 Rrp8, a NML yeast homolog, methylates m1A at position 645 of 25S rRNA. This modification impacts the structure of the large subunit as well as translation and ribosome synthesis.44 In yeast studies, the loss of m1A modification resulted in anisomycin and peroxide sensitivity as well as defects in ribosomal subunit joining in cells.45 Additionally, in bacteria, m1A at position 1408 in 16S rRNA confers aminoglycoside resistance.46
N6-methyladenosine (m6A) in mRNA
N6-methyladenosine (m6A) is the most prevalent modification on messenger RNA (mRNA). This methylation at position N6 on mRNA is installed by METTL3 and METTL14, cooperatively.47 Conversely, this modification is erased by Fat mass and obesity-associated protein (FTO) and/or AlkB Homologue 5 (ALKBH5).48,49 The m6A modification is present 3 to 5 times per mRNA and is one of the most common modifications on RNA.50,51 m6A can be in syn and anti-conformations depending on the structure of the RNA.52 The syn conformation is energetically favorable in single stranded RNA and the anti-conformation is adopted in the double stranded RNA in order to accommodate Watson-Crick base pairing.53 The methyl group installed onto the 6th position of RNA is positioned in the major groove of the RNA helix, which leads to the destabilization of the transcript and is a means for the mRNA to switch structurally between double stranded RNA and single stranded RNA.53 The presence of m6A changes the way that reader proteins recognize the mRNA transcripts, as well as the transcript’s fate.54
m6A is involved in maturation of pre-mRNA into mRNA as well as nuclear export of mRNA.55 When in the cytosol, this modification can enhance translation efficiency of the mRNA through identification by the reader protein, YTH-domain family member 1 (YTHDF1).56 Additionally, the IGF2BP class of m6A reader proteins can promote translation and increase the stability and storage of mRNA targets.56
m6A can also contribute to destabilization and degradation of mRNA through targeting by YTH-domain family member 2 (YTHDF2), another reader of mRNA m6A.57 YTHDF2 selectively binds mRNA with m6A methylation and decays the mRNA, based on this modification. YTHDF2 may also play a role in stability and/or translation since it targets the stop codon region, the 3’ untranslated region, and the coding region.57 Furthermore, the YTHDF2 gene has been thought to be associated with human longevity.58
m6A has been linked to many diseases including cancer, type 2 diabetes, obesity, leukemia, infertility, neuropsychiatric behaviors and depressive disorders.59,60,61 These diseases have been found to be associated with mutations in FTO, ALKBH2, METTL14, and METTL3.62 m6A associated proteins and their link to disease will be discussed in detail.
FTO is known to oxidatively demethylate N6-methyladenosine residues in mRNA.48 This demethylase has been found to be associated with obesity and energy homeostasis.48 Single nucleotide polymorphisms in the FTO gene affects body mass index and predisposes people to type 2 diabetes.63 In addition to an association with obesity, FTO is also associated with DNA damage repair and cancer.64 FTO accumulates at DNA damage sites after ultraviolet irradiation and induces m6A demethylation, regulating mRNA m6A along METTL3.64 In relation to cancer, FTO is also highly expressed in acute myeloid leukemia and enhances the leukemia oncogenic-mediated cell transformation.65 This is done through regulating expression of targets by reducing the levels of m6A in mRNA.65
METTL3 and METTL14 are associated with promoting tumorigenesis in many cancers, however these methylases have also shown tumor suppressive functions in glioblastoma and hepatocellular carcinoma.66 The knockdown of METTL3 or METTL14 results in a decrease in m6A modification, and in glioblastoma stem cells, results in the upregulation and expression of oncogenes that downregulate tumor suppressor expression.67 This results in enhanced glioblastoma stem cell growth, tumorigenesis, and self-renewal.67 METTL3 upregulation in glioblastoma tumor tissue has also been associated with poor patient survival according to recent studies.68
In patients with acute myeloid leukemia (AML), there is an increase in METTL3 as well as an increase in protein expression.69 METTL3 catalyzes m6A formation, and in AML, this promotes the translation of c-MYC, BCL2, and PTEN, which are proteins that are important for proliferation, survival, and differentiation.69 When AML cells are depleted of METTL3, the translation of these mRNA is repressed, which results in increase AKT activation and subsequent increase in cell differentiation and apoptosis.69
It has been shown that METTL3 is up-regulated in hepatocellular carcinoma tissues and this is associated with poor patient prognosis.70 Knockdown of METTL3 results in reduced cell proliferation, migration, and colony formation in vitro, and restrained hepatocellular carcinoma growth and metastasis in vitro and in vivo.70,69 Conversely, overexpression of METTL3 results in growth, metastasis, and tumor growth of hepatocellular carcinoma, in vivo.69 In hepatocellular carcinoma, METTL3 methylates SOCS2, a known tumor suppressor, and decreases SOCS2 stability via a YTHDF2-dependent pathway.70 It is also shown that there is a decrease in m6A in hepatocellular carcinoma tissue, and METTL14 is associated with this aberrant m6A modification.71 Consequently, METTL14 knockdown leads to metastasis of hepatocellular carcinoma and overexpression of METTL14 results in less invasiveness and metastasis of hepatocellular carcinoma.69 The contribution of METTL3 and METTL14 to this cancer is a bizarre methylation tug-of-war.
N1-methyladenosine (m1A) in mRNA
N1-methyladenosine (m1A) is influential in tRNA and rRNA both structurally and functionally and it has also been identified as an important modification in mRNA.72 This modification is installed by S-adenosylmethionine (SAM) and is removed by ALKBH3.72 m1A gives the adenosine a positive charge, which can affect the structure as well as protein-RNA interactions.72 m1A is also believed to promote translation initiation.72
ALKBH3 is found to be highly expressed in human prostate cancer and non-small cell lung cancer.73,74 Additionally, this demethylase functions in angiogenesis and apoptotic resistance, and proliferation in pancreatic cancers.75 Silencing of ALKBH3 expression induces apoptosis, suppresses cell proliferation, and inhibits angiogenesis in pancreatic cancer.75 Conversely, overexpression of ALKBH3 increases anchorage-independent growth and invasiveness in pancreatic cancer.75
5-methlcytosine (m5C) in mRNA
The 5-methylcytosine (m5C) modification is installed by Nsun2 and is thought to be erased by TET enzymes.76 The m5C methylation and demethylation pattern suggests that the mRNA could be involved in many biological pathways regulating several functions of an organism.76 For instance, the m5C modification is found to play a role in nuclear export of mRNA transcripts.76 Furthermore, this modification is thought to affect translation efficiency as well as have an effect on codon specificity.77
Pseudouridine in mRNA
The Pseudouridine (Ψ) modification is found more frequently in tRNA and rRNA, however it is also found in mRNA. This modification is linked to cell stress, as well as growth under different environmental cues.78,23 Ψ plays a role in stabilizing RNA structure and could be involved in processes such as enhancing translation initiation efficiency, ribosome pausing, RNA localization, as well as RNA interference.79
Diseases associated with mutations in Pseudouridine synthases include mitochondrial myopathy and sideroblastic anemia (MLASA), dyskeratosis congenita, and lung cancer.79 MLASA is a autosomal recessive oxidative phosphorylation disorder expressed bone marrow and skeletal muscle that is caused when there is a missense mutation in Pseudouridine Synthase 1 (PUS1).80
Conclusion
Dynamic regulation on RNA modifications significantly impact the features of the RNA molecules. In mRNA, modifications play a role in stability, translation, decay, location, structure, and export of the transcript. In rRNA, modifications are important for the stability, function, and translation activity of the ribosome. In tRNA, modifications are important for the translation efficiency, structure, flexibility of the tRNA. Additionally, modifications to tRNA can affect protein synthesis in the cell. RNA modifications may seem like a small part of the bigger picture; however, the impact of these modifications is vast. Misregulation of RNA modifications can lead to a plethora of illnesses, from cancer to neurodegenerative disease. However, there remains much to be done, as more quantitative sequencing technologies will be required to precisely map and understand the roles of modifications to different RNA species. Furthermore, the biological functions of site-specific modifications require a deeper understanding. Recent efforts have been made to integrate CRISPR technology to study specific sites of RNA modifications which hold the promise to advance the field.
Figure 1:
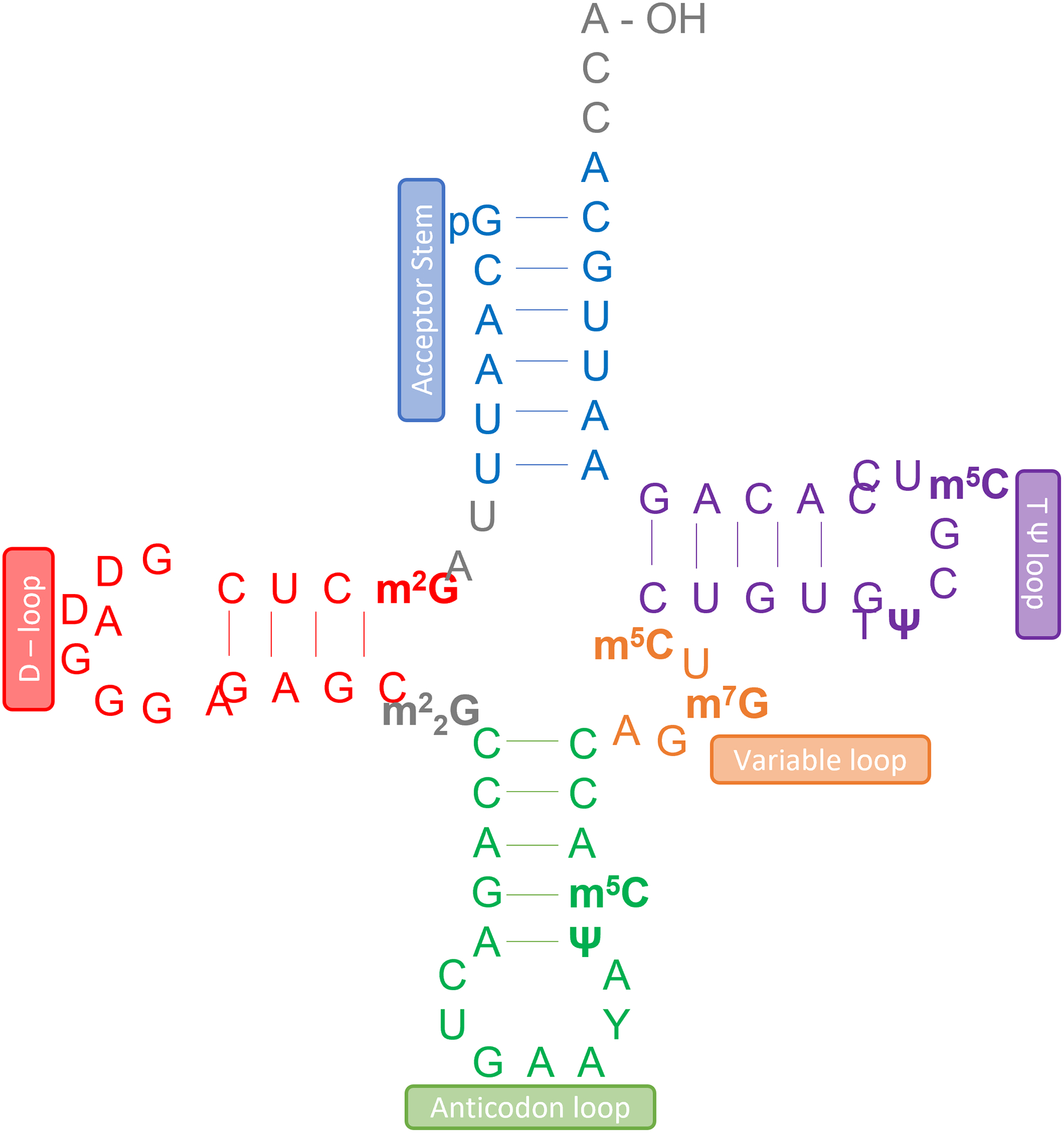
Modifications made to tRNA.
Figure 2:
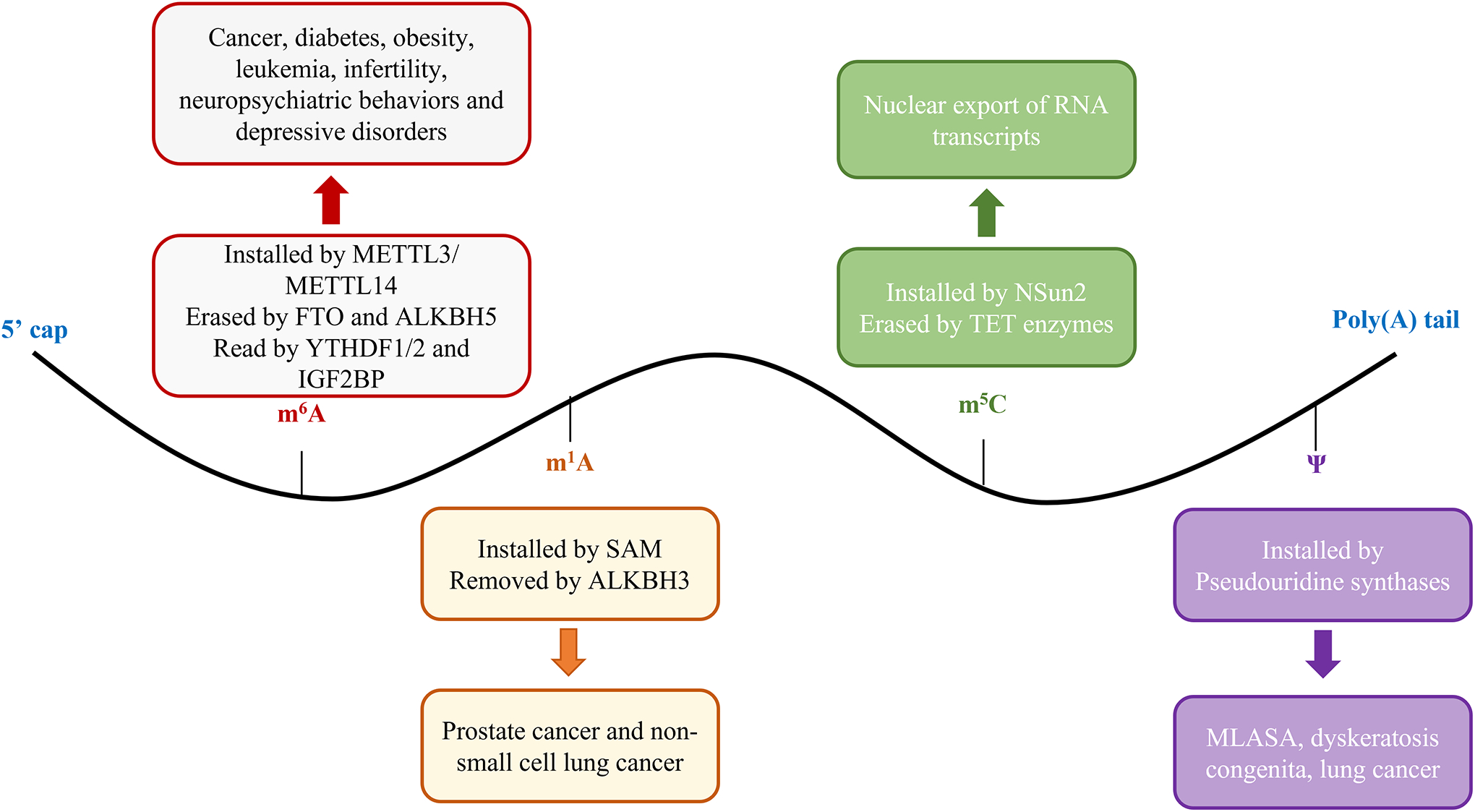
Modifications made to mRNA. The readers, writers, and erasers associated with these modifications, and their link to different disease.
References
- 1.B A, A J, J L & Al. E Molecular Biology of the Cell 4th Edition. (Garland Science, 2002). [Google Scholar]
- 2.Lodish H, Berk A, Zipursky SL, et al. Molecular Cell Biology 4th edition. W. H. Freeman; (2000). [Google Scholar]
- 3.Boccaletto P et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res 46, D303–D307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modomics - Modified bases Available at: http://modomics.genesilico.pl/modifications/. (Accessed: 24th May 2019)
- 5.Zhang C & Jia G Reversible RNA Modification N1-methyladenosine (m1A) in mRNA and tRNA. Genomics, Proteomics and Bioinformatics 16, 155–161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson JT & Droogmans L Biosynthesis and function of 1-methyladenosine in transfer RNA 121–139 (2016). doi: 10.1007/b106364 [DOI] [Google Scholar]
- 7.Oerum S, Dégut C, Barraud P & Tisné C m1A post-transcriptional modification in tRNAs. Biomolecules 7, 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Clark W & Klungland A ALKBH1-Mediated tRNA Demethylation Regulates Translation In Brief Reversible tRNA methylation facilitates translation response to nutrient availability. Data Resources GSE65299 Liu et al. Cell 167, 816–828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomikawa C 7-methylguanosine modifications in transfer RNA (tRNA). Int. J. Mol. Sci 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin S et al. Mettl1/Wdr4-Mediated m7G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol. Cell 71, 244–255.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hori H Methylated nucleosides in tRNA and tRNA methyltransferases. Frontiers in Genetics 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rife JP, Cheng CS, Moore PB & Strobel SA N2-methylguanosine is isoenergetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res 26, 3640–3644 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginell SL & Parthasarathy R Conformation of N2-methylguanosine, A modified nucleoside of tRNA. Biochem. Biophys. Res. Commun 84, 886–894 (1978). [DOI] [PubMed] [Google Scholar]
- 14.Armengaud J et al. N 2-Methylation of Guanosine at Position 10 in tRNA Is Catalyzed by a THUMP Domain-containing, S-Adenosylmethionine-dependent Methyltransferase, Conserved in Archaea and Eukaryota. The Journal of Biological Chemistry. 279, 37142–37152 (2004). doi: 10.1074/jbc.M403845200 [DOI] [PubMed] [Google Scholar]
- 15.Saenger W Intercalation. in Principles of Nucleic Acid Structure 350–367 (Springer, 1984). [Google Scholar]
- 16.Liu J & Stråby KB The human tRNA(m22G26)dimethyltransferase: functional expression and characterization of a cloned hTRM1 gene. Nucleic Acids Res 28, 3445–3451 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urbonavičius J, Armengaud J & Grosjean H Identity Elements Required for Enzymatic Formation of N2,N2-dimethylguanosine from N2-monomethylated Derivative and its Possible Role in Avoiding Alternative Conformations in Archaeal tRNA. J. Mol. Biol 357, 387–399 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Pallan PS, Kreutz C, Bosio S, Micura R & Egli M Effects of N2,N2-dimethylguanosine on RNA structure and stability: Crystal structure of an RNA duplex with tandem m2 2G:A pairs. RNA 14, 2125–2135 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackman JE, Montange RK, Malik HS & Phizicky EM Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9, 574–85 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igoillo-Esteve M et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet 9, e1003888 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian M, Srinivasan T & Sudarsanam D Examining the Gm18 and m(1)G Modification Positions in tRNA Sequences. Genomics Inform 12, 71–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray MW, Charette Michael, Pseudouridine in RNA: What, Where, How, and Why. IUBMB Life (International Union Biochem. Mol. Biol. Life) 49, 341–351 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Zhao BS & He C Pseudouridine in a new era of RNA modifications. Cell Research 25, 153–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuorto F et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nature Structural and Molecular Biology 19, 900–905 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Yao M et al. Molecular basis of dihydrouridine formation on tRNA. Proc. Natl. Acad. Sci 108, 19593–19598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalluge J Conformational flexibility of RNA: the role of dihydrouridine. Nucleic Acids Res 24, 1073–1079 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyubankova N et al. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org. Biomol. Chem 13, 4960–4966 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Swinehart WE & Jackman JE Diversity in mechanism and function of tRNA methyltransferases. RNA Biol 12, 398–411 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan MA et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet 90, 856–863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bednářová A et al. Lost in Translation: Defects in Transfer RNA Modifications and Neurological Disorders (2017). doi: 10.3389/fnmol.2017.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gleeson JG et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet 49, 380–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackwood E & Eisenman R Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science (80-.) 251, 1211–1217 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Frank SR, Schroeder M, Fernandez P, Taubert S & Amati B Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15, 2069–82 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frye M & Watt FM The RNA Methyltransferase Misu (NSun2) Mediates Myc- Induced Proliferation and Is Upregulated in Tumors. Curr. Biol 16, 971–981 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Schaefer M, Hagemann S, Hanna K & Lyko F Azacytidine Inhibits RNA Methylation at DNMT2 Target Sites in Human Cancer Cell Lines. Cancer Res 69, 8127–8159 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Helm M Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34, 721–33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo Monaco P, Marcel V, Diaz JJ & Catez F 2′-O-methylation of ribosomal RNA: Towards an epitranscriptomic control of translation? Biomolecules 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prusiner P, Yathindra N & Sundaralingam M Effect of ribose O(2′)-methylation on the conformation of nucleosides and nucleotides. Biochim. Biophys. Acta - Nucleic Acids Protein Synth 366, 115–123 (1974). [DOI] [PubMed] [Google Scholar]
- 39.Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I & Klaholz BP Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Stage-Zimmermann T, Schmidt U & Silver PA Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11, 3777–89 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ofengand J, Bakin A, Wrzesinski J, Nurse K & Lane BG The pseudouridine residues of ribosomal RNA. Biochem. Cell Biol 73, 915–924 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Lane BG, Ofengand J & Gray MW Pseudouridine in the large-subunit (23 S-like) ribosomal RNA The site of peptidyl transfer in the ribosome? FEBS Lett 302, 1–4 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Waku T et al. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner (2016). doi: 10.1242/jcs.183723 [DOI] [PubMed] [Google Scholar]
- 44.Sharma S et al. A single N1-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci. Rep 8, 11904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma S, Watzinger P, Kötter P & Entian K-D Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res 41, 5428–5443 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prammananan T et al. A Single 16S Ribosomal RNA Substitution Is Responsible for Resistance to Amikacin and Other 2‐Deoxystreptamine Aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis 177, 1573–1581 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Wang P, Doxtader KA & Nam Y Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 63, 306–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez-Perez V, Zeng XH, Henzler-Wildman K, L. C N6-Methyladenosine in Nuclear RNA is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol 7, 885–887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng G et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dignam JD, Lebovitz RM & Roeder RG Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11, 1475–1489 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei CM, Gershowitz A & Moss B Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 4, 379–86 (1975). [DOI] [PubMed] [Google Scholar]
- 52.Engel JD & von Hippel PH Effects of methylation on the stability of nucleic acid conformations: Studies at the monomer level. Biochemistry 13, 4143–4158 (1974). [DOI] [PubMed] [Google Scholar]
- 53.Engel JD & von Hippel PH Effects of methylation conformations on the stability of nucleic acid. J. Biol. Chem 253, 927–934 (1978). [PubMed] [Google Scholar]
- 54.Zhang C, Fu J & Zhou Y A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front. Immunol 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao BS, Roundtree IA & He C Post-transcriptional gene regulation by mRNA modifications. Nature Reviews Molecular Cell Biology 18, 31–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardelli M et al. A polymorphism of the YTHDF2 gene (1p35) located in an Alu-rich genomic domain is associated with human longevity. Journals Gerontol. - Ser. A Biol. Sci. Med. Sci 61, 547–556 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Zhao X et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24, 1403–1419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hess ME et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci 16, 1042–1048 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Rao S et al. An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect. Disord 183, 279–286 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Kadumuri RV & Janga SC Epitranscriptomic Code and Its Alterations in Human Disease. Trends Mol. Med 24, 886–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindgren CM et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science (80-.) 316, 889–894 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiang Y et al. RNA m 6 A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lan Q et al. The Critical Role of RNA m 6 A Methylation in Cancer. Cancer Res 79, 1285–1292 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Cui Q et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 18, 2622–2634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visvanathan A et al. Essential role of METTL3-mediated m 6 A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Luo J, Liu H, Luan S, He C & Li Z Aberrant Regulation of mRNA m6A Modification in Cancer Development. Int. J. Mol. Sci 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen M et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Ma JZ et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N 6 -methyladenosine-dependent primary MicroRNA processing. Hepatology 65, 529–543 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Dominissini D et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konishi N, Nakamura M, Ishida E, Shimada K, Mitsui E, Yoshikawa R, et al. High expression of a new marker PCA-1 in human prostate carcinoma. Clin Cancer Res 11, 5090–7 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Tasaki M, Shimada K, Kimura H, Tsujikawa K & Konishi N ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. British Journal of Cancer 104, 700–706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamato I et al. PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis. Cancer Res 72, 4829–4839 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Yang X et al. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m 5 C reader. Cell Res 27, 606–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trixl L & Lusser A The dynamic RNA modification 5‐methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 10, e1510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ge J & Yu Y-T RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci 38, 210–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carlile TM et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bykhovskaya Y, Casas K, Mengesha E, Inbal A & Fischel-Ghodsian N Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet 74, 1303–8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]


