Abstract
Clonal haematopoiesis (CH) is a phenomenon whereby somatic mutations confer a fitness advantage to haematopoietic stem and progenitor cells (HSPCs) and thus facilitate their aberrant clonal expansion. These mutations are carried into progeny leucocytes leading to a situation whereby a substantial fraction of an individual’s blood cells originate from the HSPC mutant clone. Although this condition rarely progresses to a haematological malignancy, circulating blood cells bearing the mutation have the potential to affect other organ systems as they infiltrate into tissues under both homeostatic and disease conditions. Epidemiological and clinical studies have revealed that CH is highly prevalent in the elderly and is associated with an increased risk of cardiovascular disease and mortality. Recent experimental studies in murine models have assessed the most commonly mutated ‘driver’ genes associated with CH, and have provided evidence for mechanistic connections between CH and cardiovascular disease. A deeper understanding of the mechanisms by which specific CH mutations promote disease pathogenesis is of importance, as it could pave the way for individualized therapeutic strategies targeting the pathogenic CH gene mutations in the future. Here, we review the epidemiology of CH and the mechanistic work from studies using murine disease models, with a particular focus on the strengths and limitations of these experimental systems. We intend for this review to help investigators select the most appropriate models to study CH in the setting of cardiovascular disease.
Keywords: Cardiovascular disease, Insulin resistance, CHIP, ARCH, Somatic mosaicism
1. Epidemiology of CH
With technological advances in genomic analyses, it is becoming apparent that somatic mutations can be acquired in every cell throughout the body as part of the normal ageing process. As a result of this process, individuals increasingly become a mosaic of cells with distinct genotypes; a phenomenon known as somatic mosaicism.1–3 While somatic mosaicism can occur in any tissue, it is particularly common in highly proliferative cell types, such as in the haematopoietic stem and progenitor cells (HSPCs), where the high cell production rate increases the chance occurrence of a random mutation.4 The majority of acquired somatic mutations in HSPCs have little or no impact on the cell’s phenotype. However, when a mutation occurs in a ‘driver’ gene, the mutant HSPC gains a fitness advantage, resulting in increased self-renewal, proliferation, and/or survival relative to neighbouring HSPC. This condition can lead to the clonal expansion of the mutant HSPC at the expense of HSPCs that lack a mutated driver gene. In turn, the mutation is carried through to the leucocyte progeny that are derived from this clone. Consequently, a fraction of the individual’s blood cells will carry the mutation, and this fraction can increase over time as the mutant HSPC pool expands. Notably, this process occurs in the absence of a detectable increase in blood cell counts and thus cannot simply be determined by routine blood work. The condition is generally referred to as clonal haematopoiesis (CH), and it represents an early pre-cancerous step in the progression to the pathological CH that is observed in blood cancers.5,6
CH was initially identified in studies that examined patterns of X-chromosome inactivation (XCI) in the blood cells of healthy females.7–9 In these studies, it was noted that females older than 60 years of age were more likely to exhibit skewing of their inactive X-chromosome, deviating from the 50:50 allelic ratio. Subsequently, it was found that females with skewed XCI were more likely to harbour a mutation in the blood cancer driver gene, ten-eleven translocation 2 (TET2), suggesting that this pre-leukaemic mutation was responsible for promoting the XCI skewing observed in these individuals.9 More recent epidemiological studies have expanded on these initial findings, providing consistent evidence of clonal populations of cells with pre-leukaemic driver mutations in the blood of otherwise healthy individuals.5,6 Of these known driver gene mutations, >70% of the known mutations occur in just two genes encoding epigenetic regulators, specifically DNA methyltransferase 3a (DNMT3A) and TET2. Other common driver gene mutations that have been identified include additional sex combs like 1 (ASXL1), Janus kinase 2 (JAK2), tumour protein p53 (TP53), protein phosphatase, Mg2+/Mn2+ dependent 1D (PPM1D) as well as others that are recurrently mutated in haematologic malignancies. Furthermore, these studies showed that the frequency of driver gene mutations increased sharply with age, and as such this, condition is sometimes referred to as age-related CH. The presence of a somatic mutation is associated with ∼11–13-fold increased risk of haematological malignancy, although this generally requires the acquisition of multiple oncogenic mutations. As a result, most individuals who harbour these mutations never go on to develop a haematological malignancy. Thus, this condition has also been termed CH of indeterminate potential (CHIP), which is defined by the presence of an expanded blood cell clone carrying a driver gene mutation of haematopoietic malignancies at a variant allele fraction (VAF) of at least 2%, without meeting the criteria of malignancy.
Current estimates of the prevalence of CH are largely based on the specific definition of CH and the detection limit of the DNA sequencing method utilized to assess this condition. Therefore, studies have reported different estimates on the prevalence of CH in the population. One widely cited study examined the prevalence of CH across the lifespan by sequencing the exomes of DNA from blood cells and analysing a select group of driver genes that are recurrently mutated in haematological malignancies.5 Using this approach, the VAF limit of detection was 3.5% for single nucleotide variants within exomes. It was found that driver gene mutations could be detected in ∼10% of people older than 70 years, but were rare in individuals that were <40 years of age. In contrast, a study by Zink et al.10 used whole-genome sequence analysis that allowed for the detection of aberrant clonal expansions in blood cells independent of the identification of a presumptive candidate driver gene. While this approach was relatively insensitive and could only detect clones with relatively large VAF values, the estimated prevalence of CH was substantially higher, being detected in ∼0.5% of individuals younger than 35 years old and ∼50% in those over 85 years old. Of these, only ∼20% of the observed clonal expansions in blood could be associated with a haematologic malignancy driver gene. Similarly, Genovese et al.6 found that approximately half of the clonal events could be attributed to driver genes that are recurrently mutated in haematologic malignancy. In light of these considerations, a number of studies have associated CH with mosaic chromosomal alterations, yet only a portion of these alterations coincide with loci of known haematological malignancy driver genes.11–15 Regardless of these distinctions in how CH is assessed or defined, multiple studies have shown that the prevalence of CH increases with age and is particularly common in elderly individuals.
Recent advances in sequencing technology and bioinformatic analyses have allowed investigators to assess CH at very low VAF values. For instance, studies using error-corrected sequencing, which allows the detection of mutations at a VAF as low as 0.03%, have suggested that CH is almost ubiquitous by middle age, even when the analysis is restricted to a small group of driver gene mutations. In one study, it was found that driver gene mutations, frequently found in DNMT3A and TET2, occur in 95% of individuals aged 50–70 years.16 Notably, this age group was estimated to exhibit only a ∼5% incidence of CH based upon less sensitive methods of detection.5,6,17 Collectively, these data suggest that small clones (i.e. very low VAF) may be ubiquitous in middle-aged individuals and that clone expansion occurs in a subset of individuals during the ageing process. However, the factors that control the expansion of mutant clones in some individuals and not others are generally unknown and represent an area for potential future investigation.
1.1 CH and cardiovascular disease
CH became of particular interest to investigators in the cardiovascular research field when an association was detected between this condition and increased risk of cardiovascular disease (CVD).5 In this study, The exome sequences of 160 candidate pre-leukaemic genes were analysed in the blood cells of 17 182 individuals that were unselected for haematological malignancies. They detected a strong positive association between somatic mutations in a subset of these driver genes and all-cause mortality. A similar study by Genovese et al.6 examined whole-exome sequences also found an association between CH and all-cause mortality. Interestingly, in this study, only half of the clonal events could be attributed to driver genes that are recurrently mutated in haematologic malignancies. While carriers of driver gene mutations had a markedly increased risk of haematological cancer, this risk could not account for the substantial increase in all-cause mortality.5 Unexpectedly, in a secondary analysis of the available longitudinal cohorts, it was discovered that cardiovascular causes were likely to be the main contributor to the increase of all-cause mortality observed in this study. CH carriers had a significantly increased risk of coronary-artery disease [hazard ratio (HR) =2.0, P = 0.02) and ischaemic stroke (HR = 2.6, P = 0.003) compared to non-carriers after adjusting for potential confounding factors. Since the primary aim of this study was to assess the prevalence of CH in the general population, a follow-up investigation was conducted to directly test whether CH was associated with an increase in CVD incidence.18 In this study, it was observed that individuals with CH had a 1.9 times increased risk for CVD and 4 times the risk for early-onset myocardial infarction. Notably, there was a 12-fold increased risk of an elevated coronary-artery calcification score in individuals with large mutant clones (VAF > 10%), but no statistically significant association in individuals with smaller clones, suggesting a dose–dependent relationship. More recently, Bick et al.19 analysed the exomes of 35 416 individuals from the UK Biobank for CH, resulting from mutations in DNMT3A or TET2, and incidence of CVD events including myocardial infarction, coronary revascularization, stroke, and death. In this cohort, 1079 (3%) individuals were identified with DMNT3A or TET2 mutant clones, including 432 (1.2%) individuals with a large clone size (VAF > 10%). Although an association with increased CVD event risk was observed regardless of clone size (HR = 1.27, P = 0.019), a greater CVD event risk was found in individuals with larger DNMT3A and TET2 mutant clones (with VAF >10%, HR = 1.59, P < 0.001) that was significantly different from the risk association found with smaller clones. Intriguingly, the risk escalation observed in individuals with large clones was not observed in individuals harbouring a frequent genetic polymorphism, which attenuates the interleukin-6 (IL-6) signalling pathway, suggesting that some forms of CH can promote CVD via overactivation of cytokine signalling. These findings of IL-6 activation in CH are consistent with the findings in a sub-analysis of Canakinumab Anti-Inflammatory Thrombosis Outcome Study trial, which showed that carriers of TET2-mediated CH displayed greater reductions in major adverse cardiovascular events than the unselected cohort when treated with an interleukin-1 beta (IL-1β) neutralizing antibody.20 Finally, CH was independently associated with CVD risk among post-menopausal middle-aged women (HR 1.36 with all CH, P = 0.012; HR 1.48 in CH with VAF > 10%, P = 0.005).21
While these initial clinical studies examining the relationship between CH and CVD mainly focused on the disease risk, more recent studies have examined the association between CH and prognosis in patients with chronic ischaemic heart failure (HF). In the initial report, Dorsheimer et al.22 performed deep, error-corrected targeted DNA sequencing on a cohort of 200 ischaemic HF patients that had participated in bone marrow cell therapy trials. In this study, mutations in 56 haematological cancer driver genes were analysed to determine whether they were associated with disease prognosis. They found that 18.5% of patients harboured CH mutations, with the majority of mutations occurring in either DNMT3A or TET2. Mutations in these two genes were associated with a greater risk (HR 2.1, P = 0.02) of adverse outcomes including death and HF rehospitalization. Importantly, a dose–response effect was indicated, as chronic HF patients with higher VAF (VAF > 1%) were more likely to develop worse outcomes compared to carriers with smaller clones. These findings were corroborated and extended by more recent studies. Assmus et al.23 addressed the prognostic role of clone size for DNMT3A- and TET2-driver gene mutations in a group of 419 patients with ischaemic HF. It was found that the cut-off VAF to predict 5-year survival was 1.15% and 0.73% for mutations in DNMT3A and TET2, respectively, which is substantially lower than previous estimates of pathological clone size. A recent study by Pascual-Figal et al.24 examined CH in 62 HF patients with reduced ejection fraction. They reported the adverse outcomes in HF patients carrying DNMT3A and TET2 mutations regardless of the ischaemic or non-ischaemic origin of HF. A dose–response association was also observed between clone size and HF progression in this cohort, and a 2% VAF cut-off for DNMT3A and TET2 mutations was suggested to be predictive for CVD risk. Notably, the association between DNMT3A and TET2 CH mutations and outcome became stronger when analysing for HF-related death and hospitalization, indicating that the connection between CH and adverse HF outcomes is not necessarily driven by coronary-artery disease.
To address associations between rarer driver gene mutations that are present at low VAF values and outcome, Kiefer et al.25 excluded individuals if they exhibited mutations TET2 or DNMT3A, or other CH mutations with a VAF > 2% in a cohort of ischaemic HF patients. This analysis found that somatic mutations with low VAF in a set of genes including CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, and SRSF2 are associated with mortality independently of large VAF mutations and the prevalent mutations in DNMT3A and TET2. Along these lines, Cremer et al.26 showed that ischaemic HF patients with multiple CH mutations in driver genes with relatively low VAFs also had a significantly higher risk of mortality compared to patients harbouring a single CH-mutation or non-carriers, providing evidence for a correlation between cumulative clone size and increased mortality within patient.
Beyond chronic HF and atherosclerotic CVD, the prognostic impact of CH on outcome in patients with valvular heart disease has also been investigated. Mas-Peiro et al.27 conducted targeted DNA sequencing on a cohort of 279 patients undergoing transcatheter aortic valve implantation (TAVI) for severe calcified aortic valve stenosis. This study also focused on mutations in DNMT3A and TET2. As with chronic HF, the prevalence of CH in this cohort was considerably higher than that reported in other studies for healthy individuals, suggesting that this condition is enriched in the patient population. Although cause-specific death was not provided, CH carriers were found to have a profoundly increased risk of all-cause mortality during medium-term follow-up after the TAVI procedure (HR 4.81, P = 0.009). This study excluded patients that died during the first 30 days post-TAVI to avoid potential confounding effects by the procedure itself.
The relationship between CH and thrombosis has also been examined. In a study by Wolach et al.,28 11 527 individuals (healthy control and patients with schizophrenia) were analysed, and it was found that JAK2V617F-mediated CH is strongly associated with major thrombotic events such as deep venous thrombosis or pulmonary thrombosis. Strikingly, this study reported 25% of JAK2V617F carriers experienced a thrombotic event, which was considerably higher than individuals with other CH-driver mutations (5%). Further, it was reported that JAK2V617F clones as small as 2% of VAF are associated with an increased incidence of venous thrombosis.
In addition to CH that is frequently associated with age, there is a less-studied entity of CH, referred to as therapy-associated CH (t-CH), that is prevalent in cancer survivors.29,30 t-CH is typically associated with mutations in genes that encode for DNA damage-response (DDR) proteins, such as tumour-suppressor protein 53 (TP53) and PPM1D. This form of CH occurs due to the selective pressure that the stress of radiation and/or chemotherapy exert on the HSPC, as mutations in DDR genes allow cells to survive the genotoxic stress. Although an association between PPM1D-mediated CH and ischaemic chronic HF has been reported,26 the contribution of t-CH to the long-term adverse effects of various cancer therapies on the cardiovascular system has yet to be examined in clinical studies. However, a recent study in mice suggests that t-CH could causally contribute to anthracycline-induced cardiomyopathy (AIC).31
It is worth noting that clinical studies have also found associations between CH and non-CVD conditions. Associations have been observed between CH and chronic obstructive pulmonary disease;10,32 however, this association could be the consequence of smoking on CH in this population. Natural premature menopause is also associated with CH,21 and associations have also been found between CH and ulcerative colitis and Down syndrome.33,34 Finally, Potus et al.35 reported that individuals with germ-line TET2 mutations have a 6.15-fold increased risk of pulmonary artery hypertension (PAH) compared to control subjects. Although experimental studies demonstrated a relationship between haematopoietic Tet2 depletion and a PAH phenotype, validation of this relationship in a CH cohort with TET2 somatic mutations may be warranted.
Collectively, these clinical studies reveal possible connections between somatic mutations in haematopoietic cells and the incidence and prognosis of various chronic disease processes (Table 1). However, the descriptive nature of these epidemiological studies makes it difficult to determine causality and directionality. Thus, experimental studies employing mouse models are of importance in determining causal relationships and uncovering potential mechanisms of disease progression. Here, we focus on experimental studies of CVD, with a specific focus on the practical approaches to establish murine models of CH. We also discuss the strengths and weaknesses of various models with the goal of guiding researchers to the best approaches for the experimental study of CH.
Table 1.
Summary of clinical studies reporting an association between CH and CVD
| Study | ‘Driver’ genes | Population cohort | Samples | Sequence method | Association with CVD | Adjusted variables |
|---|---|---|---|---|---|---|
| Jaiswal et al. (2014)5 | DNMT3A, TET2, ASXL1 etc. | 3353 healthy individuals without prior CVD events | Whole blood | Whole-exome sequencing | CHIP carriers vs. non-carriers:
|
Age, sex, type 2 diabetes status, systolic blood pressure, BMI |
| Jaiswal et al. (2017)18 | DNMT3A, TET2, ASXL1, JAK2 etc. | 4726 CHD patients
|
Whole blood | Whole-exome sequencing | CHIP carriers vs. non-carriers:
|
|
| Wolach et al. (2019)28 | JAK2 |
4946 schizophrenia 5947 controls |
Whole blood | Whole-exome sequencing | JAK2-positive CH increased incidence to venous thrombosis | N.A. |
| Dorsheimer et al. (2019)22 | DNMT3A, TET2 | 200 ischaemic HF patients | BM & PB mononuclear cells | Error-corrected, targeted-exome sequencing | Somatic mutations in TET2/DNMT3A are independently associated with the adverse outcome of chronic HF | Baseline serum NT-proBNP levels, SHFM score tertiles |
| Mas-Peiro et al. (2020)27 | DNMT3A, TET2 | 279 severe AV stenosis patients undergoing TAVI | BM & PB mononuclear cells | Error-corrected, targeted-exome sequencing | Somatic mutations in TET2/DNMT3A increase the mortality after successful TAVI | Age, sex, baseline serum NT-proBNP levels |
| Cremer et al. (2020)26 | DNMT3A, TET2 + 7 new mutations | 419 ischaemic HF patients | BM & PB mononuclear cells | Error-corrected, targeted-exome sequencing | CHIP is an independent predictor of mortality in CHF patients | N.A. |
| Bick et al. (2020)19 | DNMT3A, TET2 | 1079 CHIP carriers | Whole blood | Whole-exome sequencing | CHIP is associated with increased incident CVD event risk | Age, genetic ancestry, sex, HDL-c, LDL-c, smoking, BMI, type 2 diabetes status, hypertension |
| Pascual-Figal et al. (2021)24 | DNMT3A, TET2 | 62 HF patients (52% non-ischaemic) | Buffy coat of blood | Error-corrected, targeted-exome sequencing | Somatic mutations in TET2/DNMT3A are associated with accelerated HF progression regardless of aetiology | Age, sex, ischaemic aetiology, LVEF, serum NT-proBNP levels |
| Kiefer et al. (2021)25 | CBL, CEBPA, EZH2, GNB1, PHF6, SMC1A, SRSF2 | 399 ischaemic HF patients | BM & PB mononuclear cells | Error-corrected, targeted-exome sequencing | Somatic mutations with low VAF in these seven genes are associated with mortality in chronic HF | N.A. |
MI, myocardial infarction; CHD, coronary heart disease; CHF, chronic heart failure; PB, peripheral blood; BM, bone marrow; TAVI, transcatheter aortic valve implantation; CVD, cardiovascular disease; CHD, coronary heart disease; MDC, Malmo Diet and Cancer cohort; ATVB/PROMIS, Atherosclerosis, Thrombosis, and Vascular Biology/the Pakistan Risk of Myocardial Infarction Study, MI, myocardial infarction; HF, heart failure; AV, aortic valve; CHIP, clonal haematopoiesis with indeterminate potential; HR, hazard ratio; CH, clonal haematopoiesis; BMI, body mass index; TC, total cholesterol; HDL-c, high-density lipoprotein cholesterol; N.A., not applied; NT-proBNP, N-terminal pro-B-type natriuretic peptide; LDL-c, low-density lipoprotein cholesterol.
a and b designate two different study types within Jaiswal et al. 2017.
2. Animal models of CH
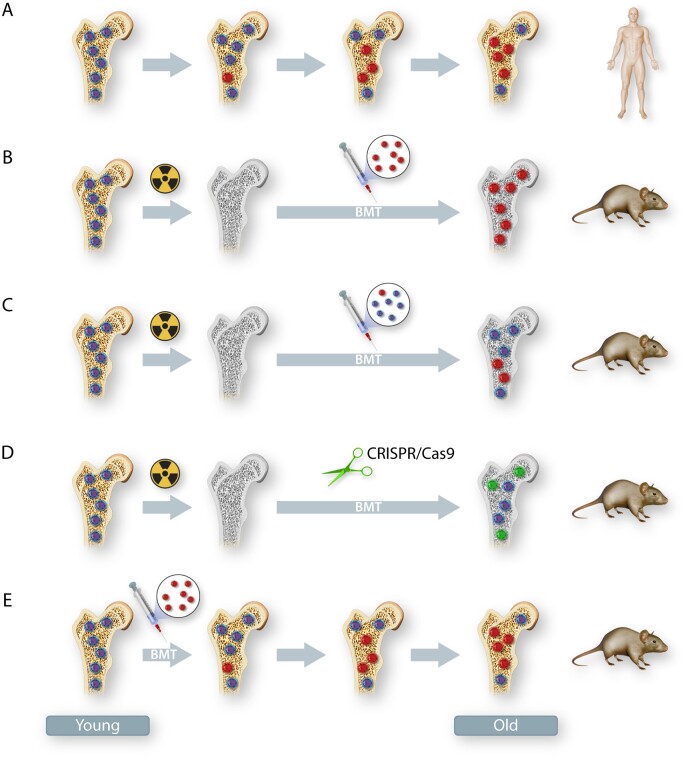
In the currently established murine models of CH, investigators have employed several methods to establish haematopoietic cell mutations in HSPC depending on the focus of their study (Figure 1).
Figure 1.
Mouse models of CH. (A) A representation of human CH. Clonal expansions of haematopoietic stem cells arise when spontaneous somatic mutations within these cells provide a selective growth, survival or self-renewal advantage. (B–E) Commonly used methods to recapitulate CH in mice include BMT with or without pre-conditioning (irradiation). (B) The experimental condition of non-competitive BMT following myeloablation. Recipient mice typically undergo lethal irradiation to remove host haematopoietic stem cells. This is followed by bone marrow reconstitution with 100% mutant cells or wild-type cells, and the recipient haematopoietic stem cells are completely replaced by donor cells. (C) The experimental condition of competitive BMT following myeloablation. Recipient mice typically undergo lethal irradiation, and haematopoietic stem cells are reconstituted with a mixture of mutant and wild-type cells. The proportion of mutant haematopoietic stem cells will increase over time if the mutation provides a selection advantage. (D) The experimental condition of driver gene editing by CRISPR/Cas9. Recipient mice typically undergo lethal irradiation, following transplantation with lineage-negative cells that have been transduced with a lentivirus expressing GFP/RFP with targeting guide RNA or non-targeting guide RNA. The targeting efficiency of candidate driver genes in haematopoietic stem cells can approach 90% in this system. (E) The experimental condition of non-myeloablative BMT. Non-conditioned recipient mice are typically transplanted with non-fractionated donor bone marrow cells on three consecutive days. If the mutated haematopoietic stem cells confer selective advantage over wild-type recipient cells, they will slowly expand over time. In panel (A–E), bone with yellow colour = non-irradiated, bone with grey colour = lethally irradiated; cells with dark blue colour = wild-type cells and cells with red colour = mutated cells. In panel (D), green colour = cells that have been transduced with genome edited cells, cells with light blue colour = cells that have not been transduced with genome edited cells.
2.1 Currently available mouse lines to study CH
Of the known CH-driver genes, DNMT3A and TET2 are most frequently mutated in elderly individuals. Multiple genetically engineered mouse lines are available to investigate these genes, and studies using these animals have offered important insights into the mechanisms by which CH impacts CVD. DNMT3A is a de novo methyltransferase that adds a methyl group to the 5′ cytosine residue at CpG dinucleotides. To date, two transgenic mouse strains have been developed to study the role of Dnmt3a. Specifically, Challen et al.36 established a mouse line whereby Dnmt3a is conditionally ablated in the haematopoietic system by crossing Mx1-Cre mice with Dnmt3afl/fl mice. To create the Dnmt3afl/fl animals, the region containing part of the catalytic domain was floxed, and deletion of this region renders the enzyme inactive. Using these mice, it was found that haematopoietic Dnmt3a deficiency results in augmented expansion of the long-term haematopoietic stem cell (HSC) (LT-HSC) pool while progressively impairing HSPC differentiation. Paradoxically, both hyper- and hypo-methylation were detected at different loci in Dnmt3a-deficient HPSC, including substantial increase in methylation at CpG islands, indicating that Dnmt3a plays a complex role in the DNA methylation process.36 Mice transplanted with Dnmt3a-deficient HSPC appeared to display a relatively benign phenotype. Another strain, namely Dnmt3afl-R878H+ mice (JAX #032289),37 was designed to express the R878H variant under the endogenous Dnmt3a promoter in Cre-dependent manner. This mouse simulates the DNMT3AR882H mutation, which is one of the most frequent somatic mutation variants observed in the human haematopoietic system.5 This variant has been shown to function in a dominant-negative manner in vitro in mouse and human cells.38,39 Further, it has been reported that the R882H mutation not only leads to CpG DNA hypo-methylation, but also impairs the CpG methylation efficiency, which differs from what is observed in the Dnmt3a-deficient mice described above.40,41 The discrepancies between the two strains may ultimately yield different phenotypes in the context of experimental CH particularly when studying its impact on CVD and other disease processes.
TET2 belongs to a family of proteins (TET1–3) that catalyse the conversion of 5-methylcytosine to 5-hydroxylcytosine, leading to DNA demethylation.42 Considering that the multitude of TET2 mutations that occur in human myeloid malignancies result in decreased enzyme catalytic activity,43 two mouse strains are available to study the impact of TET2 loss-of-function in haematopoietic cells. Ko et al.44 generated Tet2 knockout mice (JAX # 023359) that carry a mutant allele between exons 8 through 11, which leads to the deletion of the catalytic domain. Deletion of Tet2 in this manner causes a disruption to haematopoietic differentiation leading to expansion of lineage-negative Sca1+c-Kit+ (LSK) populations including HSC and multipotent progenitor cells. In addition to this strain, Moran-Crusio et al.45 created haematopoietic Tet2-deficient mice whereby Vav1-Cre mice were crossed with Tet2-floxed mice (JAX # 017573). In this mouse line, the Tet2 gene is floxed at exon 3, which is the first coding exon of Tet2, and mutations in this exon account for 41.5% of TET2 mutations observed in humans with myeloid malignancies.45 Using this particular strain of mouse, it was found that haematopoietic Tet2 deficiency results in increased HSPC self-renewal and a myeloid-skewed pattern of differentiation. Given that both Tet2-deficient strains of mice appear to recapitulate the phenotype of haematopoietic TET2 mutations in humans, studies have employed either of these strains to model CH.
In addition to DNMT3A and TET2, a mutation in JAK2 is frequently detected in individuals with CH. JAK2 is a non-receptor tyrosine kinase, which transmit signals from a variety of cytokine and growth factor receptors. JAK2V617F is the most common mutation in myeloproliferative neoplasms (MPN), which leads to constitutive activation of JAK2 signalling pathway.46 The JAK2V617F driver gene has been reported in patients with CH.5,6,18,28 To study the role of JAK2V617F in the haematopoietic system, Mullally et al.47 constructed transgenic mice that express Jak2V617F under the control of the endogenous Jak2 promoter, following Cre-mediated recombination (JAX #031658). Germ-line, heterozygous expression of Jak2V617F results in premature lethality due to MPN (median survival is 146 days). The bone marrow cells from these mice are transplantable and show similar MPN phenotypes (i.e. increased haematocrit, white blood cell, and platelet counts) in the recipient mice. This strain is advantageous for studies of MPN as it expresses a physiological level of Jak2V617F from the endogenous promoter. However, it should be noted that changes in blood cell count are generally not observed in individuals with CH, and thus this strain is generally not suitable for CH studies.
As noted previously, t-CH is associated with haematopoietic cell mutations in various genes that encode for DDR proteins including TP53 and PPM1D.29,30 Among different types of TP53 mutations, missense mutation accounts for >70% of all alterations, and >80% of these missense mutations appear to be clustered in the central DNA-binding domain of TP53.48,49 Thus, TP53 mutation variants have traditionally been considered to be loss-of-function. Considering this, two mice strains are available for studying the Trp53 loss-of-function in haematopoietic system. Jacks et al.50 generated Tp53 knockout mice (JAX # 002103) by inserting the neo locus into the murine Tp53 gene. More recently, it has been demonstrated that missense mutations in DNA-binding domain may confer the dominant-negative effect over the wild-type p53 protein in haematopoietic cells.51 Correspondingly, a Tp53 mutant strain with a R270H missense mutation is available (JAX # 008182).52
2.2 Cre-recombinase reagents used in CH research
To date, many ‘hematopoietic cell-specific’ Cre-driver strains have been developed in which the activity of Cre is either inducible or controlled by a haematopoietic cell-specific promoter. The text below describes the advantages and limitations of Cre-driver strains, and how they have been used to study CH.
2.2.1 Mx1-Cre
Mx1 is an interferon-regulated gene that is activated by type I or type III interferons. Kuhn et al.53 generated transgenic mice harbouring the Cre-recombinase transgene under the control of the Mx1-promoter as the first inducible transgenic mouse line. In these mice, Cre-expression is induced by intraperitoneal injection of polyinosinic-polycytidylic acid [poly(I: C)], a synthetic analogue of double-stranded RNA,53 which elicits a viral response and induces interferon expression. After multiple doses of poly(I: C), nearly complete recombination can be achieved in liver, spleen, and bone marrow cells.53,54 Mx1-Cre expressing mice have been commonly used in the field of haematology as a strategy to delete genes in haematopoietic cells and overcome embryonic lethality, a situation that is commonly observed in global knockout strains. However, several potential pitfalls have been identified during the use of Mx1-Cre expressing mice. First, the system is ‘leaky’, and background levels of Cre-expression can occur at levels of up to 5% the induced expression level, in almost every tissue.53,55 Furthermore, undesired recombination prior to the induction can occur in this strain, and this can cause technical issues, particularly when the recombined allele confers a growth or fitness advantage on the cell. A recent study reported that the frequency of spontaneous recombination reaches ∼20% in cultured bone marrow cells, and increases to ∼70% after bone marrow transplantation (BMT).56 The mechanism of spontaneous recombination is thought to be due to endogenous activation of the interferon signalling pathway. In addition, when performing BMT using the cells from mice, which express Mx1-Cre, careful consideration is required to select an approach to induce gene recombination. There are two commonly used approaches to induce recombination: (i) administration of the inducer [poly(I: C), etc.] to donor mice followed by BMT and (ii) administration of the inducer to recipient mice after BMT. It is generally believed that the latter approach is more preferable for studies of CH as it minimizes the effect of the gene mutation in non-haematopoietic cells and avoids accumulation of additional acquired mutations in haematopoietic cells, such as mutations in Tet2.56
2.2.2 Vav1-Cre
Vav1 is a gene that is primarily expressed in haematopoietic cells, and at least three transgenic mouse strains that express Cre-recombinase under the control of the Vav1 promoter have been generated.57–59 Of these transgenic strains, a strain carrying ‘codon-improved’ Cre (iCre) has also been developed whereby the Cre gene is modified to prevent epigenetic silencing.58 In Vav-iCre expressing mice, Cre-mediated recombination has been observed in all haematopoietic cells, which makes this strain particularly useful. However, off-target Cre-expression has also been documented in reproductive tissues and vascular endothelial cells.58,60 Due to the undesired Cre activity in endothelial cells, use of this strain could generate confounding effects, especially when studying the impact of CH on CVD. Moreover, in mice carrying both the Vav-iCre transgene and the floxed allele, the expression of Vav-iCre in reproductive tissues can possibly cause gene recombination in germ-line cells, thereby leading to systemic expression of the recombined allele in offspring mice. This situation can be particularly problematic when examining the consequences of hotspot mutations in driver genes, including DNMT3AR882C/H, SRSF2P95R/L/H, and JAK2V617F,17 as it will be technically difficult to distinguish recombined allele from WT allele in these mutant mice by standard genotyping methodology due to sequence similarities.
2.2.3 LysM-Cre
Lysozyme M (LysM) is encoded by the Lyz2 gene in mice and is widely expressed in myeloid cells, such as macrophages, monocytes, and neutrophils. Accordingly, the Lyz2Cre strain of mice has been generated for expressing Cre-recombinase in the myeloid lineage under the control of the endogenous Lyz2 promoter/enhancer element.61 Multiple groups have crossed Lyz2Cre mice to reporter mouse lines, such as TdTomato, EYFP, EGFP, etc., to verify the efficiency and specificity of this Lyz2Cre/loxP gene recombination system. While Lyz2Cre gene efficiently recombines in myeloid cells, there is also evidence to suggest that recombination occurs in a portion of non-myeloid cells. In fact, Lyz2-Cre-mediated recombination has been observed in 5–10% of HSPC populations as well as in smaller fractions of lymphoid and erythroid cells. These data suggest that the Lyz2-promoter is active in a small subset of HSPC, and that the recombined allele is transmitted to progeny cells.62–64 Undesired recombination has also been reported in some non-haematopoietic cells. In a study by Stadtfeld et al.,65 it was demonstrated that Lyz2-Cre-mediated recombination occurred in a portion of cardiomyocytes located within the intraventricular septum and the left ventricular free wall. These cells are unlikely to be of haematopoietic origin given the low prevalence of recombination-positive cardiomyocytes in Vav1-Cre mice. In addition, another study reported that almost 25% of the lung epithelial cells show some evidence of Lyz2-Cre-mediated recombination,66 and studies have documented that some types of neurons exhibit recombination by Lyz2-Cre.67,68 As a consequence, the non-specific activity of Lyz2 promoter can lead to the misinterpretation of phenotypes in animal models of CH.
2.3 BMT considerations
Mutations that result in CH occur in HSPC, and progeny leucocytes derived from the mutant clones will carry the mutation. While CH-driver gene mutations can be seen in a variety of blood cells, different driver mutations may lead to different proportions of leucocyte subsets that carry a particular mutation. For instance, some HSPC mutations confer a myeloid skewing advantage during differentiation, and the mutations will be primarily observed in progeny cells of myeloid origin in the periphery. Thus, to investigate the molecular links between CH and clinically relevant diseases, it is desirable to develop animal models that recapitulate various human scenarios. While researchers have developed Cre/loxP systems to achieve location- and time-specific gene manipulation, as indicated above, use of these animals alone may be insufficient or confounded by a lack of specificity. To overcome these caveats, BMT methods allow investigators to engraft HSPC into recipients to investigate the function of these donor cells in models of CH and CVD.
2.4 BMT with pre-conditioning
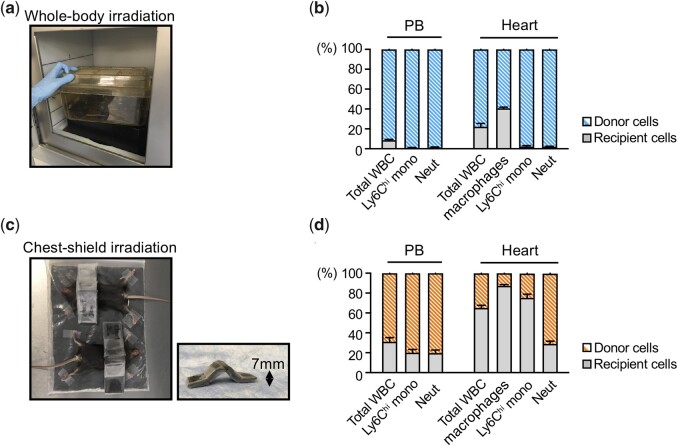
In mouse models of CH, donor cells are most commonly harvested from bone marrow of genetically manipulated mice or wild-type mice as controls. High-level engraftment can be achieved by eliminating host bone marrow cells either by cytotoxic reagents or irradiation, or a combination of both. In the absence of pre-conditioning, donor chimaerism is very low, as the bone marrow niche is limiting for donor cell engraftment. Whole-body irradiation is the most commonly used pre-conditioning method to ablate the host’s bone marrow cells (Figure 2A). The extent to which the recipient cells are destroyed will depend on the dose of the radiation. In our experience, successful reconstitution can be achieved when recipient mice are exposed to two doses of 5.5 Gy that are separated by 4 h.69 We find that a dose of 5 million bone marrow cells is the optimal number of cells to transplant during this procedure. Disease models are performed on recipient animals at ∼6–8 weeks following the BMT; a time point when haematological counts become similar to that of naive animals.70 In this procedure, the reconstituted animals carry the mutated gene only in haematopoietic cells, avoiding potential confounding factors that may occur when the mutation may be carried in cells of non-haematopoietic origin.
Figure 2.
Whole-body irradiation and chest-shielded irradiation mediated BMT. (A) Recipient mice (CD45.1+) are typically subjected to bone marrow reconstitution with donor cells CD45.2+ cells after lethal, whole-body irradiation. (B) Flow cytometry analyses of the donor chimaerism of leucocyte subsets (CD45.2+; blue) in peripheral blood (PB) and heart 1 month after BMT as described in (A). Nearly all PB leucocyte subsets and large fraction of cardiac immune cells were replaced with donor BM-derived cells. (B) Recipient CD45.1+ are subjected to chest-shielded irradiation, followed by reconstitution with CD45.2+ bone marrow cells from donor mice. (D) Flow cytometry analyses of the donor chimaerism of leucocyte subsets (CD45.1+; orange) in PB and heart 1 month after BMT as described as (C). While a large fraction of PB leucocyte subsets are replaced with donor-derived cells, the replacement of cardiac-resident macrophage by donor-derived cells is considerably less (except for CCR2+ population, data not shown). PB, peripheral blood; WBC, white blood cell; Ly6Chi Mono, Ly6Chigh monocyte; Mac, macrophage; Neut, Neutrophil. Data in this figure are from Wang et al.,81 and it is republished with permission.
To model CH, it can be helpful to employ a competitive BMT approach whereby a small proportion of mutant cells are mixed with wild-type cells prior to transplantation into lethally irradiated mice. This experimental setting can mimic the human scenario of individuals carrying CH mutations, since the mutation is only carried in a proportion of blood cells that undergo expansion. For these experiments, the use of congenic mouse strains or fluorescent reporter mice can be helpful in distinguishing donor cells from competitor wild-type and/or recipient cells. This experimental technique can be particularly useful when studying mutationsthat confer a competitive advantage over wild-type cells under homeostatic conditions. For example, studies investigating the impact of Tet2-mediated CH on CVD, have demonstrated that Tet2-mutant cells have a competitive advantage over wild-type cells.71,72 In these studies, lethally irradiated recipient mice were transplanted with a mixture of cells containing 10% Tet2-deficient (Tet2−/−) or Tet2-sufficient (Tet2+/+) bone marrow cells and 90% competitor wild-type bone marrow cells. To distinguish Tet2-deficient/-sufficient cells from competitor cells, congenic strains were used whereby Tet2-deficient/-sufficient cells were from mice with a CD45.2 allele and the competitor cells were from mice with a CD45.1 allele. Over the experimental time course, Tet2-deficient cells expanded in bone marrow, spleen, and peripheral blood (PB), and exhibited a slight myeloid bias with preferential expansion into the Ly6Chi monocyte population, which is consistent to what is observed in humans with haematopoietic TET2 mutations.73 Further, to more closely model the human scenario whereby most individuals bear a mutation in only one allele, equivalent experiments were performed using cells that were heterozygous for the Tet2 mutation.72 In these experiments, the kinetics of clonal expansion were slower for heterozygous mutant cells, indicative of a gene dose–dependent effect. Finally, in some instances, the mutant HSPC may not show a competitive advantage over wild-type cells, perhaps due to the lack of external factors that are required to drive expansion of the mutant cells in individuals with CH. In these situations, mutant cells can be mixed with wild-type cells at higher proportions to produce a more evident phenotype.
2.5 Potential pitfalls of pre-conditioning BMT experiments
While BMT techniques are commonly employed as a research tool, it is important to realize that the pre-conditioning processes that typically accompany the BMT will elicit unwanted systemic effects to the host. Beyond its primary purpose to eliminate host bone marrow cells, conditioning regimes, such as total-body irradiation or myeloablative chemotherapy, can damage multiple organs and influence the outcome of the study. For example, irradiation has been shown to induce vascular endothelial cell damage, resulting in the up-regulation of adhesion molecules and monocyte recruitment and infiltration into the intima.74 In the setting of CVD, such changes could markedly influence the pathophysiology of the disease process. Indeed, it has been reported that irradiated atherosclerosis-prone Ldlr-deficient mice develop macrophage-richer plaques at the aortic root compared to non-irradiated mice.75 Similarly, irradiation has been documented to reduce the frequency of aortic aneurysm rupture after angiotensin II (AngII) infusion to Apoe-deficient mice.76 Overall, well-controlled experiments should be designed to limit these inadvertent systemic effects in studies of CH and its impact on CVD.
The systemic conditioning of mice prior to BMT will also lead to the profound changes in the immunological cell populations of tissues. The majority of immune cells in healthy organs are tissue-resident macrophages, and in many tissues, these cells are derived from embryonic precursors.77 It is thought that these macrophages colonize the tissue prior to birth and maintain themselves mainly by local proliferation. However, in some tissue-resident macrophage populations, there may be some degree of renewal by bone marrow-derived macrophages. For example, the main resident macrophages in brain, microglia, are maintained exclusively by self-renewal while only some non-microglia resident macrophages are slowly and partially replaced by circulating cells.78 Similarly, in the normal lung, there is negligible contribution of bone marrow-derived macrophages to the resident alveolar macrophage population during adult life.79 In the heart, the extent of replacement of macrophages differs depending on the subpopulation of macrophages. Specifically, chemokine C-C motif receptor 2− (CCR2−) major histocompatibility class II− (MHCII−) and CCR2−MHCII+ cardiac macrophages are maintained locally and exhibit a slow turnover by cells from the circulation.80 In contrast, the CCR2+MHCII+ population is readily replaced by blood-borne cells. Irradiation depletes native tissue-resident macrophages and allows circulating monocyte-derived macrophages to infiltrate and reside in the tissue. In the heart following lethal irradiation and BMT, almost all of cardiac-resident macrophages are replaced by donor-derived cells, which is very different to what is observed under homeostatic conditions (Figure 2B). In the case of CH models, where lethal irradiation is used to create the chimaera, this could conceivably alter the outcome of the disease process under study. For instance, following lethal irradiation and BMT, immune cells with mutant driver genes likely replace the recipient’s cardiac-resident macrophages.81 This situation would generally be unlikely to occur in humans with CH, and the mutation would not be present in some populations of cardiac-resident macrophages (i.e. CCR2−MHCII− and CCR2−MHCII+ cardiac macrophages). This may substantially alter the disease sequelae and thus should be taken into consideration when using this approach to create models to study the impact of CH on CVD.
Shielded irradiation is a method that is being increasingly used in CH models to protect organs from radiation exposure.69 This procedure allows bone marrow to be reconstituted with donor-derived cells while protecting the tissue of interest with little or no perturbation of the tissue-resident macrophage populations. To shield the chest, and thereby protect cardiac-resident immune cells from radiation, our laboratory has found that using a curved lead plate over the chest of mice is useful (Figure 2C). Using the chest-shielded BMT procedure, the donor chimaerism of over 70% can be achieved in blood while the cardiac-resident macrophage population is largely kept intact (Figure 2D). It should be noted that the thickness of the lead shield will be dictated by the type of irradiation used to induce the conditioning. Therefore, a greater thickness of lead shield is required when using caesium source-based irradiators in comparison with using X-ray-based irradiators.
2.6 BMT without pre-conditioning
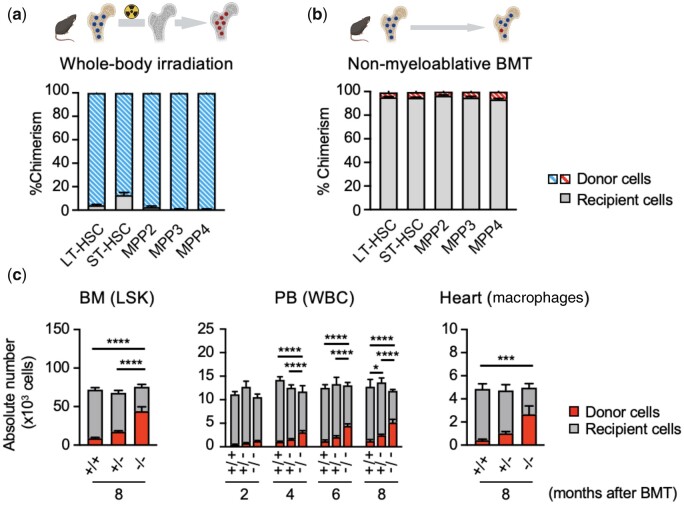
As discussed above, lethal irradiation is an effective method of depleting most of the bone marrow haematopoietic cells, but this procedure also causes permanent damage to the bone marrow niche, including the supporting stromal cells.82 The bone marrow-stromal niche has critical roles in HSPC maintenance and lineage commitment. Thus, pre-conditioning will lead to substantial changes in HSPC output with larger proportions of mutant cells being generated (Figure 3A). Non-myeloablative BMT represents a method by which HSPC is engrafted without pre-conditioning of the recipient. Studies have reported that transplantation of bone marrow cells is possible without eliminating host cells, although the total level of engraftment is considerably low.83–86 However, this limitation does not diminish its value as a tool for studying CH, particularly when studying mutations that confer a competitive advantage to HSPC under homeostatic conditions.
Figure 3.
BMT with or without pre-conditioning. Donor chimaerism of HSPC populations in bone marrow was evaluated at 1 month after BMT by flow cytometry analyses. (A) Lethally irradiated recipient mice (CD45.2+) were subjected to bone marrow reconstitution with CD45.1+ bone marrow cells (blue) after lethal whole-body irradiation. A large fraction of the different HSPC population in bone marrow were replaced with donor cells. (B) Recipient mice (CD45.2+) were subjected to BM reconstitution with CD45.1+ bone marrow cells (red) without pre-conditioning. A total of 5×106 unfractionated donor cells were injected over 3 consecutive days (total =1.5 × 107 cells). A small fraction (∼5%) of the different HSPC populations in bone marrow was replaced with donor cells. In panel (A and B), bone with yellow colour = non-irradiated, bone with grey colour = lethally irradiated; cells with dark blue colour = wild-type cells, cells with red colour = mutated cells. (C) Absolute number of donor BM-derived cells in bone marrow, PB, and heart evaluated by flow cytometry analyses. The number of Linage−Sca1+c-Kit+ (LSK) cells and cardiac macrophages are shown as representative populations of bone marrow and heart, respectively. In this experiment, Pep Boy mice (CD45.1+) were subjected to BM reconstitution with CD45.2+ bone marrow cells (red) without pre-conditioning. A total of 5×106 unfractionated CD45.2+ donor cells (Tet2+/+, Tet2+/− or Tet2−/−) were injected over 3 consecutive days (totally 1.5×107 cells). Absolute number of Tet2-deficient donor cells increased, depending on the gene dosage, in bone marrow, blood and heart, and also increased in a time-dependent manner in blood. LT-HSC, long-term haematopoietic stem cell; ST-HSC, short-term haematopoietic stem cell; MPP, multipotent progenitor cell; BM, bone marrow; LSK, lineage-negative Sca1+C-kit+ cell; PB, peripheral blood; WBC, white blood cell; BMT, bone marrow transplantation.+/+ = wild-type, +/− = heterozygous, −/− = homozygous. The data from this figure are from Wang et al.,81 and it is republished with permission.
Recently, Wang et al.81 and Fuster et al.87 used non-conditioning BMT approaches to study the effect of Tet2-mediated CH on the cardiovascular and metabolic systems, respectively, under homeostatic conditions. In these studies, 5 ×106 unfractionated CD45.2+ bone marrow cells from either Tet2-deficient or wild-type mice were transplanted into recipient mice (CD45.1+) without pre-conditioning over 3 consecutive days (a total of 1.5 ×107 cells per mouse). Congenic markers were used for tracking the donor-derived cells in the PB as well as in tissues. The successful transplantation of wild-type bone marrow cells into recipient mice can be observed at 6 weeks post-BMT (Figure 3B), revealing a relatively low but stable engraftment of donor-derived cells into LSK and other HSPC fractions at 8 months post-BMT. In line with these findings, there is minimal contribution of the wild-type donor cells to circulating leucocyte subsets and monocyte-derived cardiac macrophages. In contrast, the adoptive transfer of Tet2-deficient bone marrow cells leads to a much higher chimaerism of LSK cells and robust expansion into other haematopoietic cell populations. A higher degree of chimaerism was observed following transplantation of homozygous Tet2-deficient cells compared to heterozygous Tet2-deficient cells, indicating a dose–dependent relationship (Figure 3C). Notably, there is negligible replacement of the embryonic-derived populations of cardiac-resident macrophages (MHCII+CCR2− and MHCII−CCR2−) by donor-derived cells using the non-conditioning method. This finding differs from what is observed in irradiated animals whereby donor cells replace a substantial population of resident macrophages in the heart. Collectively, these data show that BMT without pre-conditioning is a suitable method to engraft HSPC, without damage to the bone marrow niche or organs, and this represents a useful model to study CH and its contribution to tissue pathologies under homeostatic conditions.
2.7 Potential pitfalls of congenic markers in CH-CVD research
As indicated in the sections above, mouse strains, such as the CD45.1 and CD45.2 variants, are commonly used to distinguish mutant cells from wild-type cells in competition assays. The congenic strain with the Ptprca allele (CD45.1; B6.SJL-Ptprca Pepcb/BoyJ), commonly referred to as ‘Pep Boy’ mice, was created by extensively back-crossing the SJL/J strain (Ptprca) with the C57BL/6 strain (Ptprcb; CD45.2). These animals are therefore on a C57BL/6 background and have been extensively used in studies involving competitive BMT experiments. The CD45.1 and CD45.2 variants differ by five amino acids within the extracellular domain, which enables these variants to be distinguished by flow cytometric analysis using monoclonal antibodies directed against these epitopes. A competitive BMT is typically performed by transplanting a fraction of CD45.2+ test cells mixed with CD45.1+ competitor cells into recipient mice. While useful for studies of CH, there are several potential issues associated with the use of these strains. Investigators have found that HSPC from Pep Boy mice (CD45.1+) have an inherent disadvantage over HSPC from C57BL/6 mice (CD45.2+), due to a decrease in homing efficiency, reduced numbers of LT-HSC, and an intrinsic cell-engraftment defect.88,89 Further, a study by Jang et al.90 reported that C57BL/6 and Pep Boy respond differently to viral challenge, whereby Pep Boy mice are more resistant to cytomegalovirus and more susceptive to influenza virus than C57BL/6 mice. Emerging evidence also suggests that C57BL/6 and Pep Boy mice exhibit genetic differences in the region of the Ptprc allele.89 These studies suggest that genetic differences between C57BL/6 and Pep Boy mice could confound studies of experimental CH and therefore should be taken into account when designing well-controlled experiments.
2.8 Ex vivo genome editing of HSPC
Experimental studies employing transgenic mice have provided detailed information about the pathogenesis of a small number of driver gene mutations in HSPC.91,92 However, dozens of other candidate driver genes have yet to be studied. Analyses of the mechanistic relationships between the large number of driver gene mutations and disease are hampered by the cost and time required to maintain the relevant transgenic mouse strains. In contrast to standard transgenesis approaches, the clustered regularly interspaced short palindromic repeats (CRISPR)-based gene editing system enables greater flexibility and more rapid gene manipulation in studies of CH. Briefly, CRISPR-based genome editing system requires two major components: single-guide RNA (sgRNA) that targets a putative driver gene and a Cas nuclease (e.g. Cas9).93 The sgRNA directs Cas9 to generate a double-stranded break in the target sequence of the genomic DNA. This DNA damage triggers DNA repair systems to fix the break, creating a series of random mutations at the targeted sequence.
Heckl et al.94 utilized CRISPR to introduce mutations in mouse primary HSPC to study myeloid malignancies. In this study, lentivirus vectors were used to deliver both a sgRNA and Cas9 to HSPC, and a variety of driver genes were evaluated for their ability to promote haematologic malignancies following HSPC transplantation to lethally irradiated mice. Sano et al.95 repurposed this method to study the impact of CH on CVD, and demonstrated that either Tet2 or Dnmt3a loss-of-function mutations in haematopoietic cells could accelerate AngII-induced HF. In these studies, wild-type, lineage-negative bone marrow cells from mice were transduced ex vivo with a lentivirus that used different promoters to encode Cas9, the cell marker protein eGFP and sgRNAs that target the driver gene (e.g. Tet2 or Dnmt3a). These cells were then transplanted into lethally irradiated mice to establish the mouse model of CH. In subsequent studies, it was shown that the efficiency of target gene editing could be dramatically improved by separating the sgRNA and Cas9 components.96 In this refinement, donor HSPCs are derived from Cas9-transgenic mice and the lentivirus vector is solely used to deliver the sgRNA.
The introduction of CRISPR technology enables the rapid and relatively affordable manipulation of many candidate driver genes. However, there are concerns that should be considered. For example, many CH-driver genes exhibit ‘hotspot’ sites of mutations. An example is the missense mutations at R882 residue in DNMT3A gene, most commonly resulting in arginine to histidine change (i.e. DNMT3AR882H), that is suggested to differentially impact protein function relative to other mutations in DNMT3A.17,97 This highlights the need to recreate somatic mutations at a single-base resolution. Whereas the widely used CRISPR systems typically result in the stochastic introduction of mutations that result in an array of different mutations in neighbouring cells, technological advancements in CRISPR-Cas toolkits, such as homology-directed repair, should allow for highly specific gene editing in the near future.98
Other limitations associated with the CRISPR/Cas system should be noted. In addition to off-target effects, which introduces double strand break in unintended target sites, it has been reported that CRISPR/Cas system activates the DDR pathway resulting in growth disadvantage or cell cycle arrest.99,100 Moreover, Cas9 expression itself is reported to activate p53 and increase the transcription of p53 target genes, although this effect has yet to be examined in HSPC.100 Therefore, employing CRISPR/Cas-mediated genome editing in the haematopoietic system requires appropriate experimental controls. For example, controls should include the use of gRNAs that produce double strand breaks in non-essential regions of the genome to evaluate possible confounding effects. These precautions are particularly important when applying CRISPR/Cas technology to studies of mutations in DDR genes, such as TP53 and PPM1D, which are enriched in individuals with t-CH.29,30
3. Experimental finding in disease models
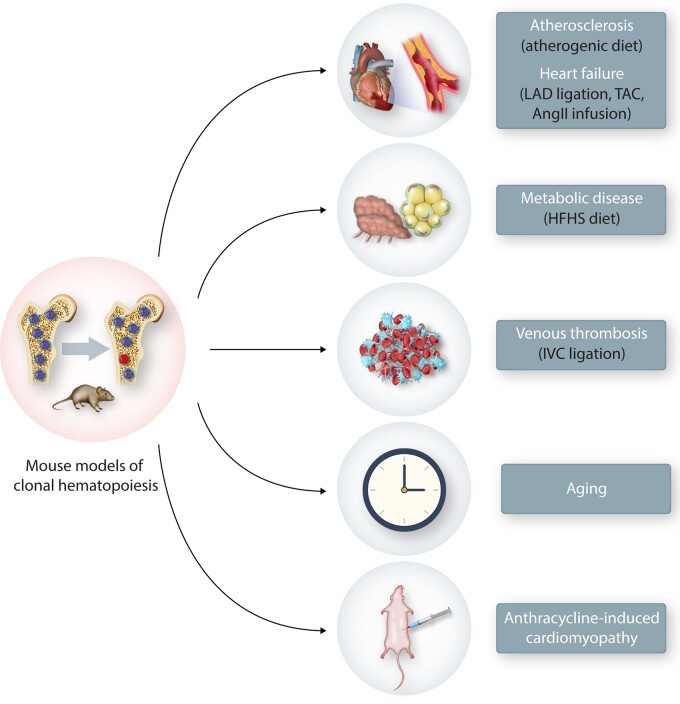
Numerous studies have reported an association between CH and various cardiovascular disorders. However, epidemiological studies are inherently descriptive and generally lack information about causality, directionality, and mechanism. Thus, a series of studies in model systems have been employed to elaborate the details of the relationships between CH and CVD (Figure 4). The following sections and Table 2 describe the mouse models used to assess these relationships and summarize the current findings.
Figure 4.
Currently reported animal models of CH and CVD. Using various BMT models of CH, mice were subjected to various models of CVD. To generate the atherosclerosis model, Ldlr-deficient hypercholesterolemic mice were used as recipients and fed a western diet. HF models were performed with operations, such as TAC, LAD artery ligation or AngII infusion pump implantation. To establish metabolic disease models, such as insulin resistance and obesity, recipient mice were fed on a high-fat/high-sucrose diet (HFHS). Venous thrombosis model was performed by ligation of the IVC. For ageing studies, non-conditioned mice underwent adoptive BMT and were maintained on a normal chow diet as they aged (>1 year). AIC model was performed by the intraperitoneal injection of an anthracycline (such as Dox) delivered in cycles. Donor cell chimaerism of PB is typically assessed by flow cytometry analysis, and CVD phenotypes were evaluated at various time points depending on the disease model.
Table 2.
Summary of experimental studies reporting a causal connection between CH and CVD
| Driver gene | Clonal haematopoiesis model | CVD model | Disease phenotype | Suggested mechanisms | Reference |
|---|---|---|---|---|---|
| Tet2 | Myeloablative BMT | Atherosclerosis | ↑Atherosclerosis | ↑Pro-inflammatory transcripts | 18 , 72 |
| KO donor | ↑Nlrp3, Il1b, Il6 in macrophages | ||||
| Tet2 | Myeloablative BMT | MI (LAD ligation) | ↑Cardiac remodelling | ↑Nlrp3, Il1b, Il6 in macrophages | 71 |
| KO donor | PO (TAC) | ||||
| Tet2 | Myeloablative BMT | PO (AngII) | ↑Cardiac remodelling | ↑Il1b, Il6 in J774.1 cells | 95 |
| ex vivo CRISPR | ↑Renal fibrosis | ||||
| Tet2 | Non-myeloablative BMT | Ageing | ↑Cardiac remodelling | ↑Pro-inflammatory transcripts in cardiac macrophages | 81 |
| KO donor | |||||
| Dnmt3a | Myeloablative BMT | PO (AngII) | ↑Cardiac remodelling | ↑Il6, Cxcl1/2, Ccl5 in J774.1 cells | 95 |
| ex vivo CRISPR | ↑Renal fibrosis | ||||
| Jak2 | Myeloablative BMT | Venous thrombosis | ↑Thrombosis | ↑NET formation | 28 |
| Jak2V617F donor | ↑MPN | ||||
| Jak2 | Myeloablative BMT | Atherosclerosis | ↑Atherosclerosis | ↓Erythrophagocytosis | 106 |
| Jak2V617F donor | ↑MPN | ↑Nlrp3, Il1b, Il6 in macrophages | |||
| JAK2 | Myeloablative BMT | MI (LAD ligation) | ↑Cardiac remodelling | ↑IFNGR1 mediated AIM2, IL1B, IL6, TNF in THP-1 cells | 104 |
| ex vivo myeloid-promoter | PO (TAC) | ||||
| Jak2 | Myeloablative BMT | Atherosclerosis | ↑Atherosclerosis | ↑Proliferation and glycolytic metabolism | 108 |
| Jak2V617F donor | ↑MPN | ↑Aim2, Il1b in macrophages | |||
| Tp53 | Non-myeloablative BMT | AIC | ↑Cardiac remodelling | ↑Prolonged neutrophil infiltration | 31 |
| Myeloablative BMT | ↑ROS in neutrophils | ||||
| Het donor |
CVD, cardiovascular disease; KO, knock out; Het, heterozygous; MI, myocardial infarction; LAD, left anterior descending; PO, pressure-overload; AngII, angiotensin II; MPN, myeloproliferative neoplasms; AIC, anthracycline-induced cardiomyopathy.
3.1 Atherosclerosis
Genetically modified hypercholesterolemic mice (Ldlr-deficient mice or Apoe-deficient mice) are typically used to study atherosclerosis, and plaque buildup is accelerated in these models by feeding these animals a high-fat high-cholesterol (HFHC) diet.101 For studies involving BMT procedures, the HFHC diet is typically started ∼4 weeks after BMT in conditioned mice, providing sufficient time for the donor haematopoietic cells to reconstitute in the bone marrow. Depending on the experiment, the atherosclerotic lesion can be evaluated at ∼2–3 months after HFHC diet feeding. Studies have detailed sex differences in the pathogenesis of atherosclerosis in this model, highlighting the importance of including animals of both sexes in experimental studies.102 Nevertheless, studies predominantly use female mice as they develop atherosclerotic lesions faster than male mice.
To determine whether a causal connection exists between CH and CVD, our laboratory assessed the impact of Tet2-mediated CH on the development of atherosclerosis.72 To create a mouse model of CH, the congenic competitive BMT approach was used, as described in the sections above. Briefly, lethally irradiated female Ldlr−/− mice were transplanted with a mixture of bone marrow cells containing 10% CD45.2 (Tet-deficient or wild-type) and 90% CD45.1 competitor cells. Recipient mice were then fed a normal diet or a HFHC diet for 9 weeks to induce atherosclerosis. Following BMT, Tet2-deficient cells expanded progressively in the bone marrow, spleen and blood exhibited a slight myeloid bias with preferential expansion into the Ly6Chi monocyte population. Importantly, there was no effect of haematopoietic Tet2 deficiency on the total numbers of white blood cells, which is similar to what is observed in cancer-free humans that exhibit CH. The expansion of Tet2-deficient cells in this fashion accelerated atherosclerosis, leading to a 60% increase in plaque size at 12 weeks post-BMT. Notably, there were no differences observed in plasma cholesterol, blood glucose, and body or spleen weight between mice that received Tet2-deficient cells and those that received wild-type cells. To more closely model the human condition, whereby most individuals have a mutation at a single TET2 allele, the impact of Tet2 haploinsufficiency was also examined using the similar congenic competitive BMT strategy. In these experiments, heterozygosity of Tet2 also accelerated the development of atherosclerosis, although the phenotype was milder than what was observed following transplantation of homozygous cells. It was also noticed that the kinetics of clonal expansion was slower for heterozygous cells indicating a gene dose–dependent effect. As it was observed that haematopoietic Tet2 deficiency led to myeloid skewing and a greater proportion of CD45.2 macrophages within the vessel wall, the contribution of myeloid cells to the atherosclerotic phenotype was also investigated. Thus, BMT and Lyz2-Cre/LoxP strategies were used to generate atherosclerosis-prone mice exhibiting Tet2 deficiency restricted to myeloid cells. More precisely, Ldlr-deficient mice were transplanted with Tet2flox/floxLyz2Cre/+ (myeloid-specific Tet2-deficient) cells or Tet2flox/floxLyz2Cre/− (wild-type) and fed an HFHC diet for 9 weeks. Mice that received Tet2-deficient myeloid cells exhibited larger atherosclerotic plaques, demonstrating myeloid cells are likely responsible for the pro-atherosclerotic phenotype.
Jaiswal et al.18 reported similar findings in a mouse model of atherosclerosis. In this study, female Ldlr-deficient mice on Pep Boy (CD45.1) background were lethally irradiated and transplanted with CD45.2-expressing Tet2flox/floxVav1Cre/+ (Tet2-deficient) or Tet2flox/floxVav1Cre/− (wild-type) cells. Mice were then placed on an HFHC diet at 4 weeks post-BMT for either 5, 9, or 17 weeks. Recipients that received the Tet2flox/floxVav1Cre/+ donor cells exhibited enhanced atherosclerotic lesions in the aortic root after 5 or 9 weeks on the HFHC diet. It was also noted that the atherosclerotic lesion was larger in the descending portion of the aorta at 17 weeks after HFHC diet feeding. In this study, a conventional BMT approach was employed, whereby all transplanted haematopoietic cells were deficient in Tet2. In contrast to what was observed in the study by Fuster et al.,72 the authors of this study noted a large infiltration of macrophages-2 positive cells in the spleen, kidney, lung and liver, and widespread xanthomas of recipients who received Tet2flox/floxVav1Cre/+ donor cells; observations that are not consistent with what is observed in individuals with CH without an overt haematological condition. This study also found that myeloid-specific deletion of Tet2 led to an augmented atherosclerotic lesion, providing additional evidence that the effects of haematopoietic Tet2 loss-of-function are mediated through myeloid cells. Collectively, these studies consistently show that experimental Tet2-loss-of-function in HSPC accelerates atherosclerosis, suggesting that the relationship between CH and CVD is causal.
A recent study by Heyde et al.103 proposed the possibility that CH might be a symptom rather than a cause of atherosclerosis. Specifically, these investigators found that HSPC proliferation was markedly increased in both atherosclerotic mice and patients with atherosclerosis compared to controls. This atherosclerosis-induced HSPC proliferation was predicted to promote the somatic evolution and expansion of mutant clones in a mathematical model. Evidence for this hypothesis was provided by mouse models of atherosclerosis and sleep fragmentation in which the expansion Tet2-deficient bone marrow cells were accelerated, suggesting a reverse causality between CH and disease progression. However, these findings are generally inconsistent with previous experimental studies showing that neither hypercholesterolemia-induced atherosclerosis nor ischaemic heart diseases accelerated the clonal expansion of Tet2 mutant cells.71,72 On the other hand, ischaemic heart disease has been shown to promote the expansion of haematopoietic cells harbouring the JAK2V617F mutation in a murine model.104 Taken together, these discrepancies highlight the need for additional well-designed experimental studies in mice and longitudinal studies in human cohorts.105
As noted above, mechanistic studies suggest that pro-inflammatory macrophages function as key players in mediating the accelerated atherosclerosis phenotype caused by Tet2 loss-of-function in haematopoietic cells.18,72 In support to this hypothesis, enhanced expression of pro-inflammatory cytokines and chemokines, particularly the over-production of IL-1β, were detected both in plaques of atherosclerotic mice transplanted with Tet2-deficient cells and in the isolated Tet2-deficient macrophages with lipopolysaccharide stimulation, suggesting the pivotal role of IL-1β. Further, elevated IL-1β expression was found to promote endothelial cell activation and SELP expression, which further promotes monocyte recruitment to the lesion site and perpetuates vascular inflammation and atherosclerosis.72 This study also found that wild-type Tet2 inhibits Il1b transcription via modulating the recruitment of histone deacetylase to the Il1b promoter and that TET2 regulates components of NLR family pyrin domain-containing 3 (NLRP3) inflammasome, which is responsible for cleavage of pro-IL-1β and secretion of active IL-1β. Treating mice with MCC950, a selective NLRP3 inflammasome inhibitor, reduced atherosclerotic plaque burden in Tet2-deficient mice and eliminated the differences in plaque size between genotypes. Collectively, these data highlight the crucial role of IL-1β/NLRP3 signalling in mediating the effect of CH on the pathogenesis of atherosclerosis.
In addition to TET2, investigations on the impact of JAK2V617F-mediated CH on CVD has been of interest since an association between individuals that harbour this type of mutation and CVD has been reported.18 Initially, Wang et al.106 investigated the role of the haematopoietic Jak2V617F mutation on the development of atherosclerosis, albeit in a mouse model that also displays haematologic malignancy phenotypes. The authors utilized mice expressing the physiological level of mouse form of the mutation, Jak2V617F, following Mx1-driven, Cre-mediated recombination.47 To create the atherosclerosis model, Ldlr-deficient mice were lethally irradiated and transplanted with Jak2V617F/+Mx1Cre/+ or control bone marrow cells, and subsequently placed on an HFHC diet. At 7 weeks of HFHC diet feeding, recipient mice transplanted with Jak2V617F-positive haematopoietic cells displayed an increased plaque burden compared with control mice despite lower serum cholesterol and triglyceride levels. The lower indices of plasma cholesterol seen in mice transplanted with Jak2V617F mutant cells are consistent with what is observed in humans with the analogous mutation.107 At 12 weeks of HFHC diet feeding, the recipient mice transplanted with Jak2V617F mutant cells exhibited a tendency for an increased lesion size and necrotic core area, together with the NLRP3 and Caspase11 dependent macrophage inflammasomes activation observed in Jak2V617F macrophage and thereby exacerbated pro-inflammatory signalling, likely contribute to the plaque instability.106 Furthermore, analysis of plaque composition revealed that there was increased erythrophagocytosis and decreased efferocytosis in the plaque. Although these studies provided evidence that JAK2V617F-mediated CH could increase the risk of atherosclerotic CVD, it should be noted that the pan-haematopoietic Jak2V617F mouse model also displayed abnormalities associated with MPN including increased haematocrit that are not observed in individuals with CH.
In a more recent study, Fidler et al.108 utilized two models to investigate the potential mechanisms of how Jak2V617F CH promotes atherosclerosis. Specifically, they transplanted bone marrow from tamoxifen inducible, macrophage-specific Jak2V617F expression (Cx3cr1-cre Jak2V617F) mice to irradiated Ldlr-deficient mice. After 15 weeks of HFHC diet, mice with macrophage expression of Jak2V617F exhibited significantly more atherosclerotic lesion size, macrophage proliferation, and necrotic core formation compared to WT controls. This finding was further corroborated in a second Ldlr-deficient model using competitive BMT to mimic CH, where donor bone marrow was comprised of 20% wild-type or Mx1-cre Jak2V617F mice. Consistent with the previous report,106 the Jak2V617F mutation increased IL-1β expression and activated both NLRP3 and Caspase-1/11 dependent inflammasomes in macrophages. The Jak2V617F-expressing macrophage was also shown to have metabolic alterations, which lead to mitochondrial reactive oxygen species over-production, double-stranded DNA breaks, and activation of the AIM2 inflammasome. Up-regulation of AIM2 inflammasome expression was also observed in JAK2V617F macrophages in a previous report,104 and activation was shown to be dependent upon the interferon receptor gamma 1 upstream of STAT signalling. Notably, Fidler et al.108 showed AIM2 inflammasome depletion largely reversed the prominent atherosclerosis phenotype driven by Jak2V617F in haematopoietic cells. Taken together, the involvement of the NLRP3 and AIM2 inflammasomes by mutant forms of JAK2/Jak2 further supports the concept that inflammation is a critical component of CH-mediated cardiovascular pathogenesis.
3.2 Heart failure
Myocardial damage can eventually lead to defects in ventricular pumping and/or filling. This common condition, referred to as HF, can be modelled in mice by various procedures. To model HF of ischaemic origin, temporary or permanent surgical ligation of the left anterior descending (LAD) artery is commonly performed to recapitulate a myocardial infarction. To investigate non-ischaemic HF, a pressure-overload model involving transverse aortic constriction (TAC) has been employed. Alternatively, since the renin-angiotensin system is a key determinant of arterial blood pressure, the implantation of an osmotic pump to continuously deliver AngII can be employed to produce cardiac hypertrophy and non-ischaemic HF. All of these models induce injury to the heart and lead to the progressive decline of cardiac function over time.
Sano et al.71 examined whether the comorbidity of Tet2-mediated CH promotes HF phenotypes in experimental models. In this study, the LAD ligation and TAC models of HF were employed at ∼8 weeks following a competitive BMT procedure with mutant and wild-type cells. In both models, mice with haematopoietic Tet2 deficiency displayed significantly worse features of HF outcome as demonstrated by poorer cardiac contractility and greater tissue fibrosis, inflammation, and cardiac hypertrophy compared to mice transplanted with wild-type bone marrow cells. Further, it was shown that myeloid-specific Tet2 ablation was sufficient to promote the HF phenotypes in both models, highlighting the key roles that these immune cells exert in the pathogenesis of various forms of HF. Evidence was provided that the over-production of IL-1β by Tet2-deficient macrophages and subsequent hyper-inflammation contributes to the exacerbated HF phenotypes. Specifically, the detrimental effects of haematopoietic Tet2 deficiency could be alleviated by treatment with a NLRP3 inflammasome inhibitor, supporting the central role of macrophage IL-1β signalling in the pathologies attributed to haematopoietic Tet2 deficiency. A subsequent study by Sano et al.95 employed lentivirus-mediated delivery of CRISPR/Cas9 components to edit driver genes in a model of HF. In this study, Tet2 editing in HSPC by CRISPR/Cas9 methodology replicated the effects of a competitive BMT with Tet2-deficient cells in an HF model that employed the continuous infusion of AngII. Sano et al.95 further employed the lentiviral/CRISPR-Cas9 approach to investigate the effects of haematopoietic loss-of-function of Dnmt3a gene on HF in mice. Under these experimental conditions, Dnmt3a-deficient HSPC did not display clonal expansion properties, whereas similar editing of Tet2 in HSPC led to expansion. Notably, a recent study reported that Interferon-γ signalling induced by chronic mycobacterial infection could drive the clonal expansion of Dnmt3a-deficient HSPC, highlighting a potential environmental driver of Dnmt3a-mediated CH.109 Regardless, haematopoietic cell Dnmt3a deficiency generated by lentiviral/CRISPR-Cas9 approach only achieved a mutation VAF of 5–10%, yet this was sufficient to promote greater cardiac dysfunction, hypertrophy, and fibrosis.95 These experiments also observed increased myocardial inflammation by assessing markers of monocytes (Cd68) and T cell (Cd3e, Cd4, and Cd8) infiltration in the AngII infusion model. A similar pro-inflammatory signature was subsequently observed in chronic ischaemic HF patients with DNMT3A-mediated CH.110 In this study, single-cell RNA sequencing of PB revealed a pro-inflammatory monocyte profile and evidence of monocyte-T cell interactions in HF patients. Collectively, these studies provided initial evidence that TET2 and DNMT3A-mediated CH could contribute to the pathogenesis of HF through the overactivation of inflammatory pathways. Subsequent studies of CH in patients with chronic ischaemic HF revealed that TET2 and DNMT3A-mediated CH was indeed associated with worse prognosis.22
As noted previously, the creation of a mouse model of JAK2V617F-mediated CH is challenging because mice engrafted with these mutant HSPC display MPN phenotypes that can confound the cardiovascular phenotype under investigation. Notably, this confounding phenotype can be apparent even when using cell type-specific promoters. For example, Lyz2Cre/Jak2V617F mice are reported to exhibit an MPN phenotype, perhaps due to Cre-expression in small populations of HSPC,62 further highlighting the issue of specificity with this traditional ‘myeloid-specific’ Cre driver. To overcome these challenges, Sano et al.104 employed the ex vivo lentivirus-mediated, HSPC transduction method to create myeloid-specific JAK2V617F mice using a synthetic enhancer SP146 and myeloid-specific gp91-promoter combination to express this mutant driver gene. Notably, this experimental study focused on the effect of JAK2V617F in myeloid-lineage cells because JAK2V617F expressed from the Vav1 promoter was observed to result in a strong myeloid cell expansion bias in a competitive BMT assay.104 Under control of this synthetic promoter/enhancer the transgene is silent in the transduced HSPC, but expressed following differentiation to neutrophils, monocytes, and macrophages. This strategy avoids the MPN phenotype, and mice display normal counts of red blood cells, platelets, and white blood cells. Accordingly, these myeloid-specific, JAK2V617F-expressing mice showed accelerated cardiac dysfunction following experimental myocardial infarction or pressure overload-mediated hypertrophy, which is associated with cytokine activation in the myocardium.
3.3 Anthracycline-induced cardiomyopathy
Anthracyclines, such as doxorubicin (Dox), are conventional components of chemotherapeutic regimens for both solid and haematological cancers. However, both short and long-term AIC is a prominent adverse event that is associated with the use of these drugs.111 As noted previously, t-CH is often observed in cancer survivors where genotoxic stress of cancer therapy can accelerate the clonal expansion of pre-existing Tp53 or Ppm1d mutations in HSPC.29,112–114 In light of findings, it is tempting to speculate if the clonal expansion of HSPC with mutations in genes encoding DDR pathway regulators may contribute to the long-term risk of AIC.31
To model AIC in a clinically relevant manner, intraperitoneal injections of Dox are typically administered in cycles to mice.113 Using this model, Sano et al.31 recently investigated the potential role of t-CH in Dox-induced cardiomyopathy using Tp53 as a test case. Notably, in distinct adoptive BMT models using either heterozygous Tp53 deficiency or Tp53R270H as donor cells, Dox administration promoted the expansion of pre-existing Tp53 mutant cells, and these models of clonal HSPC expansion were associated with significantly worse cardiac outcome as demonstrated by greater functional impairments and myocardial wall thinning, greater fibrosis and a reduced capillary density in the myocardium compared to mice transplanted with wild-type bone marrow cells. Mechanistically, Dox administration led to the prolonged infiltration of neutrophils as a source of oxidative/nitrosative stress damage in the myocardium. These adverse effects were augmented by Tp53-deficiency, and Tp53-mutant neutrophils displayed substantial transcriptional changes associated with neutrophil recruitment and activation. Notably, the depletion or pharmacological inhibition of neutrophils led to an amelioration of the Dox-induced cardiomyopathy in both control mice and in mouse models of TP53-mediated t-CH. Collectively, these data suggest that t-CH could contribute to the risk of HF that develops in cancer survivors. Indeed, the clinical application of these experimental findings awaits future validation in studies of cancer survivors.
3.4 Venous thrombosis
Venous thrombosis is an age-dependent disorder that is associated with mortality. While the incidence is <0.005% per year among individuals under the age of 15 years, it increases sharply after age 60 and reaches 0.5% per year among those over the age of 80 years.115 To model venous thrombosis, ligation of the inferior vena cava (IVC) is widely employed.116 Full or partial ligation of IVC just caudal to the left renal vein creates a stasis or stenosis venous thrombosis model, respectively. These models resemble features of human thrombus,117 and they rely on different components of the coagulation system. The stenosis model involves neutrophil extracellular traps (NET) formation, whereas the stasis model is NET-independent.118
JAK2V617F-mediated CH is associated with a pro-thrombotic phenotype and this could put individuals with this condition at a greater risk of thrombotic cardiovascular events.28 To elucidate potential causal connections between JAK2V617F-mediated CH and venous thrombosis, Wolach et al.28 performed both full ligation and partial ligation of the IVC in recipient mice following a non-competitive, myeloablative BMT with donor bone marrow c-Kit+ cells from Jak2V617F/+ × Vav1Cre mice and Jak2WT mice. In the stenosis model, but not stasis model, mice transplanted with cells from Jak2V617F/+ × Vav1Cre mice displayed increased venous thrombosis compared to mice transplanted with wild-type cells. This venous thrombosis phenotype was associated with greater NET formation, and inhibition of JAK-STAT signalling with ruxolitinib reversed NET formation and diminished the incidence of venous thrombosis attributed to the haematopoietic Jak2V617F mutation. The mouse model employed in these experiments displays MPN phenotypes, and this is a limitation of these studies as these phenotypes are not observed in human CH carriers. However, these data provide potential clues to the mechanisms by which JAK2V617F-mediated CH may increase the incidence of CVD. Additionally, Edelmann et al.119 found that granulocytes expressing the activating JAK2 (or Jak2) mutation are more adhesive to vascular endothelial cells due to the enhanced activation of β1 and β2 integrins, thereby promoting pathological thrombus formation in the stenosis model. In aggregate, experimental studies in different venous thrombosis models indicate that JAK2V617F-mediated CH could contribute to an increased risk of venous thrombosis through multiple mechanisms. It should also be noted that CH involving other driver genes is also associated with an increased risk of venous thrombosis,28 and mechanistic details await further studies.
3.5 Metabolic disease
Insulin resistance is a risk factor for CVD.120 Some studies have reported associations between CH and the prevalence of diabetes,5,19 and a higher incidence of vascular complications was observed in diabetes patients with clonal expansions of chromosomal structural variants in blood cells.121 Therefore, the question of whether CH is causally associated with metabolic disease is of particular interest. To understand the metabolic consequences of CH at a molecular level, Fuster et al.87 evaluated the impact of Tet2-mutation-driven CH on insulin resistance employing two murine model systems. Because the myeloablative pre-conditioning BMT approach has limitations due to the confounding long-term side effects of γ-radiation, particularly in metabolic studies where mice do not develop robust diet-induced obesity,122–124 the adoptive transfer strategy in non-irradiated mice was employed.81 These mice were then monitored for the effects of the mutation on haematopoietic cell expansion and metabolic parameters in the context of ageing. Following BMT, mice were monitored until approximately 1.6 years with periodic evaluation of basic haematologic and metabolic parameters. Adoptive transfer led to stable engraftment of donor haematopoietic cells, and Tet2−/− cells expanded progressively, whereas wild-type cells displayed little or no selective expansion.87 Consistent with the clinical paradigm of CH, the expansion of Tet2-deficient cells did not affect absolute numbers of WBCs or specific blood cell populations. There was a trend towards increased mortality in mice that received the Tet2-deficient bone marrow cells, and the expansion of Tet2-deficient haematopoietic cells had no effect on body weight or composition in mice that survived to the end of the experiment. However, these mice developed a progressive aggravation of systemic insulin resistance with ageing that was accompanied by increased fasting blood glucose levels and a significant reduction of insulin-stimulated Akt Ser473-phosphorylation in epididymal white adipose tissue, which is indicative of reduced insulin signalling.
In a second model, Fuster et al.87 evaluated the metabolic effects of TET2-mediated CH by employing competitive BMT to mice that received relatively low doses of radiation (two doses of 400 rad, 3 h apart). Following recovery from irradiation and BMT, mice were fed a high-fat/high-sucrose obesogenic diet for 14 weeks. As expected from prior studies,71,72 Tet2-deficient bone marrow cells expanded progressively into all blood cell lineages but did not affect body weight or composition. However, the obesity-induced systemic insulin resistance was enhanced, and this was paralleled by increased fasting blood glucose levels and reduced insulin-induced Akt phosphorylation in adipose tissue. Collectively these data indicate that TET2 loss-of-function-driven CH aggravates insulin resistance induced by ageing or diet-induced obesity. At a mechanistic level, changes in Il1b transcript levels were observed in the adipose tissues of obese mice, whereas no significant change was detected in the transcript expression of a selected group of other pro-inflammatory cytokines and chemokines known to play a role in adipose tissue inflammation. Based on these findings, the effects of Tet2 inactivation-mediated CH on diet-induced obesity were evaluated in mice treated with the NLRP3 inflammasome inhibitor MCC950. Treatment with this drug suppressed IL-1β protein expression, but had no effects on body weight. However, MCC950-treatment suppressed the increased hyperglycaemia and insulin resistance associated with the expansion of Tet2-deficient cells. These data demonstrate that the NLRP3 inflammasome has a key role in the metabolic derangement associated with Tet2-mediated CH. Combined with findings in the models of CVD,71,72 these studies provide further evidence for an association between TET2 deficiency in the haematopoietic cells and the overactivation of the NLRP3 inflammasome in disease processes.
3.6 Age-associated cardiomyopathy
Ageing is associated with multiple pathological changes in the cardiovascular system.125 Mouse hearts undergo progressive age-dependent cardiac dysfunction that is associated with elevated levels of myocardial inflammation. In C57BL/6J mice, cardiac myopathy is typically observed by 18 months, and it is characterized by systolic dysfunction, left ventricle hypertrophy, and fibrosis.126 Using the adoptive transfer model, Wang et al.81 provided evidence that Tet2-mediated CH can accelerate age-related cardiomyopathy. In mice receiving Tet2-deficient HSPC, contractile dysfunction could be observed as early as 8 months after BMT through non-invasive echocardiography. At the 18-month termination of the experiment, systolic dysfunction, left ventricular hypertrophy, and fibrosis were observed to a significantly greater extent in the mice receiving Tet2-deficient vs. wild-type bone marrow.
Using the non-conditioned BMT model, that mimics the immune cell composition and dynamics in the heart, the study of Wang et al.81 also evaluated how Tet2 deficiency in HSPC affects the immune cell composition of the ageing heart. In this study, the expanding population of bone marrow-derived, Tet2-deficient cells display differential expansion kinetics into various cardiac immune cell populations.81 For example, the degree of Tet2-deficient cell chimaerism in the short-lived neutrophils was essentially the same in blood and cardiac tissue. In contrast, the expansion of Tet2-deficient cells into the cardiac macrophage population was relatively attenuated due in large part to the very slow turnover of the embryo-derived, cardiac macrophage population. In contrast, this adoptive transfer study found higher levels of chimaerism in the macrophage compartments of the kidney and liver, presumably because these tissues display higher levels of turnover by monocyte-derived macrophages.81 Collectively, the adoptive transfer approach represents a relatively benign bone marrow approach that can enable studies of how mutant HSPC expand in the bone marrow and how their progeny leucocytes interact with tissue-resident immune cells in disease processes and during the ageing process. Overall, it appears that the adoptive BMT to non-conditioned mice represents a superior means to model the consequences of CH on the organism.
Finally, while age is a major contributor to the development of CH, the studies of Wang et al.81 and Fuster et al.87 indicate that CH can also impact processes that contribute to biological ageing per se. In both of these adoptive transfer studies, Tet2-mediated HSPC expansion is sufficient to promote age-related cardiac dysfunction and insulin resistance in the absence of external injurious stimuli. In relation to these findings, CH has been associated with age acceleration in humans as assessed by the examination of CpG methylation patterns that represent the ‘epigenetic clock’.127 In this longitudinal study, individuals with any form of driver gene-associated CH displayed ∼4 years of advanced epigenetic age, and individuals with TET2-mediated CH displayed ∼6 years of epigenetic age acceleration. In toto, these data raise the possibility that CH may promote biological ageing and diminishing health span, perhaps by promoting the condition of ‘inflammaging’.128
4. Conclusions and future directions
The past few years have led to an explosion in interest of how CH impacts CVD. Accumulating studies show that individuals with CH exhibit a higher risk of mortality due largely to an increased prevalence of CVD. With the development of novel mouse model systems, investigators have been able to elaborate the mechanistic links between this relatively common haematological condition and disease processes. Collectively, these epidemiological and experimental studies suggest that CH is a new causal risk factor for CVD that may be as consequential as the traditional risk factors that have been known for decades.
Despite substantial advances in our understanding of how CH promotes CVD, many questions remain unanswered. Numerous driver genes are recurrently mutated in individuals with CH, yet relatively few have been evaluated in experimental studies. In this regard, the known CH-driver genes encode proteins with diverse functions that are likely to exert divergent effects in disease processes. Most experimental studies have focused on the TET2, DNMT3A, and JAK2V617F driver genes. However, we know very little about how the haematopoietic and cardiovascular systems are impacted by mutations in the dozens of other, less-studied CH-driver genes. Furthermore, it appears that the known driver genes (that are recurrently mutated in haematologic malignancies) can only account for a relatively small portion of the CH observed in the population,10 and we have little understanding of the mechanistic details associated with these other clonal expansion events. Thus, broader investigative approaches, enabled by advances in experimental models, will be likely required to develop a comprehensive view of the impact of CH on disease processes.
Conflict of interest: none declared.
Funding
This work was supported by National Institutes of Health grants HL138014, HL139819, HL141256 and AG072095 to K.W., HL152174 to S.S. and K.W., American Heart Association grant 20POST35210098 to M.A.E., Japan Heart Foundation grant to H.O., and the Chongqing Innovation Support Program for Returned Overseas Scholars cx2020010 to Y.W.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Contributor Information
Ying Wang, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA; Department of Cardiology, Xinqiao Hospital, Army Medical University, 183 Xinqiao Main Street, Chongqing 400037, P.R. China.
Soichi Sano, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA; Department of Cardiology, Osaka City University Graduate School of Medicine, -4-3 Asahi-machi, Abeno-ku, Osaka 545-8585, Japan.
Hayato Ogawa, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA.
Keita Horitani, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA.
Megan A Evans, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA.
Yoshimitsu Yura, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA.
Emiri Miura-Yura, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA.
Heather Doviak, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA.
Kenneth Walsh, Hematovascular Biology Center, Robert M. Berne Cardiovascular Research Center, University of Virginia School of Medicine, 415 Lane Road, PO Box 801394, Suite 1010, Charlottesville, VA 22908, USA.
References
- 1. Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, Shiozawa Y, Sato Y, Aoki K, Kim SK, Fujii Y, Yoshida K, Kataoka K, Nakagawa MM, Inoue Y, Hirano T, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Nishikawa Y, Amanuma Y, Ohashi S, Aoyama I, Horimatsu T, Miyamoto S, Tsunoda S, Sakai Y, Narahara M, Brown JB, Sato Y, Sawada G, Mimori K, Minamiguchi S, Haga H, Seno H, Miyano S, Makishima H, Muto M, Ogawa S. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019;565:312–317. [DOI] [PubMed] [Google Scholar]
- 2. Sano S, Wang Y, Walsh K. Somatic mosaicism: implications for the cardiovascular system. Eur Heart J 2020;41:2904–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, Kandoth C, Fulton RS, McLellan MD, Dooling DJ, Wallis JW, Chen K, Harris CC, Schmidt HK, Kalicki-Veizer JM, Lu C, Zhang Q, Lin L, O'Laughlin MD, McMichael JF, Delehaunty KD, Fulton LA, Magrini VJ, McGrath SD, Demeter RT, Vickery TL, Hundal J, Cook LL, Swift GW, Reed JP, Alldredge PA, Wylie TN, Walker JR, Watson MA, Heath SE, Shannon WD, Varghese N, Nagarajan R, Payton JE, Baty JD, Kulkarni S, Klco JM, Tomasson MH, Westervelt P, Walter MJ, Graubert TA, DiPersio JF, Ding L, Mardis ER, Wilson RK. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012;150:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landen M, Hoglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Gronberg H, Hultman CM, McCarroll SA. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gale RE, Fielding AK, Harrison CN, Linch DC. Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. Br J Haematol 1997;98:512–519. [DOI] [PubMed] [Google Scholar]
- 8. Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, Gilliland DG. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 1996;88:59–65. [PubMed] [Google Scholar]
- 9. Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, Hassimi M, Socci N, Bhatt PK, Gonen M, Mason CE, Melnick A, Godley LA, Brennan CW, Abdel-Wahab O, Levine RL. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 2012;44:1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, Thorgeirsson TE, Sigurdsson A, Gudjonsson SA, Gudmundsson J, Jonasson JG, Tryggvadottir L, Jonsson T, Helgason A, Gylfason A, Sulem P, Rafnar T, Thorsteinsdottir U, Gudbjartsson DF, Masson G, Kong A, Stefansson K. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017;130:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner M-J, Cullen M, Epstein CG, Burdett L, Dean MC, Chatterjee N, Sampson J, Chung CC, Kovaks J, Gapstur SM, Stevens VL, Teras LT, Gaudet MM, Albanes D, Weinstein SJ, Virtamo J, Taylor PR, Freedman ND, Abnet CC, Goldstein AM, Hu N, Yu K, Yuan J-M, Liao L, Ding T, Qiao Y-L, Gao Y-T, Koh W-P, Xiang Y-B, Tang Z-Z, Fan J-H, Aldrich MC, Amos C, Blot WJ, Bock CH, Gillanders EM, Harris CC, Haiman CA, Henderson BE, Kolonel LN, Le Marchand L, McNeill LH, Rybicki BA, Schwartz AG, Signorello LB, Spitz MR, Wiencke JK, Wrensch M, Wu X, Zanetti KA, Ziegler RG, Figueroa JD, Garcia-Closas M, Malats N, Marenne G, Prokunina-Olsson L, Baris D, Schwenn M, Johnson A, Landi MT, Goldin L, Consonni D, Bertazzi PA, Rotunno M, Rajaraman P, Andersson U, Beane Freeman LE, Berg CD, Buring JE, Butler MA, Carreon T, Feychting M, Ahlbom A, Gaziano JM, Giles GG, Hallmans G, Hankinson SE, Hartge P, Henriksson R, Inskip PD, Johansen C, Landgren A, McKean-Cowdin R, Michaud DS, Melin BS, Peters U, Ruder AM, Sesso HD, Severi G, Shu X-O, Visvanathan K, White E, Wolk A, Zeleniuch-Jacquotte A, Zheng W, Silverman DT, Kogevinas M, Gonzalez JR, Villa O, Li D, Duell EJ, Risch HA, Olson SH, Kooperberg C, Wolpin BM, Jiao L, Hassan M, Wheeler W, Arslan AA, Bueno-de-Mesquita HB, Fuchs CS, Gallinger S, Gross MD, Holly EA, Klein AP, LaCroix A, Mandelson MT, Petersen G, Boutron-Ruault M-C, Bracci PM, Canzian F, Chang K, Cotterchio M, Giovannucci EL, Goggins M, Hoffman Bolton JA, Jenab M, Khaw K-T, Krogh V, Kurtz RC, McWilliams RR, Mendelsohn JB, Rabe KG, Riboli E, Tjønneland A, Tobias GS, Trichopoulos D, Elena JW, Yu H, Amundadottir L, Stolzenberg-Solomon RZ, Kraft P, Schumacher F, Stram D, Savage SA, Mirabello L, Andrulis IL, Wunder JS, Patiño García A, Sierrasesúmaga L, Barkauskas DA, Gorlick RG, Purdue M, Chow W-H, Moore LE, Schwartz KL, Davis FG, Hsing AW, Berndt SI, Black A, Wentzensen N, Brinton LA, Lissowska J, Peplonska B, McGlynn KA, Cook MB, Graubard BI, Kratz CP, Greene MH, Erickson RL, Hunter DJ, Thomas G, Hoover RN, Real FX, Fraumeni JF, Caporaso NE, Tucker M, Rothman N, Pérez-Jurado LA, Chanock SJ. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012;44:651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, Ling H, Hetrick KN, Pugh EW, Amos C, Wei Q, Wang LE, Lee JE, Barnes KC, Hansel NN, Mathias R, Daley D, Beaty TH, Scott AF, Ruczinski I, Scharpf RB, Bierut LJ, Hartz SM, Landi MT, Freedman ND, Goldin LR, Ginsburg D, Li J, Desch KC, Strom SS, Blot WJ, Signorello LB, Ingles SA, Chanock SJ, Berndt SI, Le Marchand L, Henderson BE, Monroe KR, Heit JA, de Andrade M, Armasu SM, Regnier C, Lowe WL, Hayes MG, Marazita ML, Feingold E, Murray JC, Melbye M, Feenstra B, Kang JH, Wiggs JL, Jarvik GP, McDavid AN, Seshan VE, Mirel DB, Crenshaw A, Sharopova N, Wise A, Shen J, Crosslin DR, Levine DM, Zheng X, Udren JI, Bennett S, Nelson SC, Gogarten SM, Conomos MP, Heagerty P, Manolio T, Pasquale LR, Haiman CA, Caporaso N, Weir BS. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012;44:642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Terao C, Suzuki A, Momozawa Y, Akiyama M, Ishigaki K, Yamamoto K, Matsuda K, Murakami Y, McCarroll SA, Kubo M, Loh PR, Kamatani Y. Chromosomal alterations among age-related haematopoietic clones in Japan. Nature 2020;584:130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loh PR, Genovese G, McCarroll SA. Monogenic and polygenic inheritance become instruments for clonal selection. Nature 2020;584:136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loh PR, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, Birmann BM, Talkowski ME, Bakhoum SF, McCarroll SA, Price AL. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 2018;559:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016;7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, Fisher DS, Blundell JR. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020;367:1449–1454. [DOI] [PubMed] [Google Scholar]
- 18. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, Baber U, Mehran R, Fuster V, Danesh J, Frossard P, Saleheen D, Melander O, Sukhova GK, Neuberg D, Libby P, Kathiresan S, Ebert BL. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bick AG, Pirruccello JP, Griffin GK, Gupta N, Gabriel S, Saleheen D, Libby P, Kathiresan S, Natarajan P. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation 2020;141:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Svensson EC, Madar A, Campbell CD, He Y, Sultan M, Healey ML, D’Aco K, Fernandez A, Wache-Mainier C, Ridker PM, Beste MT, Basson CT. Abstract 15111: TET2-driven clonal hematopoiesis predicts enhanced response to canakinumab in the CANTOS trial: an exploratory analysis. Circulation 2018;138:A15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Honigberg MC, Zekavat SM, Niroula A, Griffin GK, Bick AG, Pirruccello JP, Nakao T, Whitsel EA, Farland LV, Laurie C, Kooperberg C, Manson JE, Gabriel S, Libby P, Reiner AP, Ebert BL, Program N-O, Natarajan P; NHLBI Trans-Omics for Precision Medicine Program. Premature menopause, clonal hematopoiesis, and coronary artery disease in postmenopausal women. Circulation 2021;143:410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, Schmid T, Brune B, Wagner S, Serve H, Hoffmann J, Seeger F, Dimmeler S, Zeiher AM, Rieger MA. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol 2019;4:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Assmus B, Cremer S, Kirschbaum K, Culmann D, Kiefer K, Dorsheimer L, Rasper T, Abou-El-Ardat K, Herrmann E, Berkowitsch A, Hoffmann J, Seeger F, Mas-Peiro S, Rieger MA, Dimmeler S, Zeiher AM. Clonal haematopoiesis in chronic ischaemic heart failure: prognostic role of clone size for DNMT3A- and TET2-driver gene mutations. Eur Heart J 2021;42:257–265. [DOI] [PubMed] [Google Scholar]
- 24. Pascual-Figal DA, Bayes-Genis A, Díez-Díez M, Hernández-Vicente Á, Vázquez-Andrés D, de la Barrera J, Vazquez E, Quintas A, Zuriaga MA, Asensio-López MC, Dopazo A, Sánchez-Cabo F, Fuster JJ. Clonal hematopoiesis and risk of progression of heart failure with reduced left ventricular ejection fraction. J Am Coll Cardiol 2021;77:1747–1759. [DOI] [PubMed] [Google Scholar]
- 25. Kiefer KC, Cremer S, Pardali E, Assmus B, Abou-El-Ardat K, Kirschbaum K, Dorsheimer L, Rasper T, Berkowitsch A, Serve H, Dimmeler S, Zeiher AM, Rieger MA. Full spectrum of clonal haematopoiesis-driver mutations in chronic heart failure and their associations with mortality. ESC Heart Fail 2021;8:1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cremer S, Kirschbaum K, Berkowitsch A, John D, Kiefer K, Dorsheimer L, Wagner J, Rasper T, Abou-El-Ardat K, Assmus B, Serve H, Rieger M, Dimmeler S, Zeiher AM. Multiple somatic mutations for clonal hematopoiesis are associated with increased mortality in patients with chronic heart failure. Circ Genom Precis Med 2020;13:e003003. [DOI] [PubMed] [Google Scholar]
- 27. Mas-Peiro S, Hoffmann J, Fichtlscherer S, Dorsheimer L, Rieger MA, Dimmeler S, Vasa-Nicotera M, Zeiher AM. Clonal haematopoiesis in patients with degenerative aortic valve stenosis undergoing transcatheter aortic valve implantation. Eur Heart J 2020;41:933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolach O, Sellar RS, Martinod K, Cherpokova D, McConkey M, Chappell RJ, Silver AJ, Adams D, Castellano CA, Schneider RK, Padera RF, DeAngelo DJ, Wadleigh M, Steensma DP, Galinsky I, Stone RM, Genovese G, McCarroll SA, Iliadou B, Hultman C, Neuberg D, Mullally A, Wagner DD, Ebert BL. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Sci Transl Med 2018;10:eaan8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, Patel M, Berthon A, Syed A, Yabe M, Coombs CC, Caltabellotta NM, Walsh M, Offit K, Stadler Z, Mandelker D, Schulman J, Patel A, Philip J, Bernard E, Gundem G, Ossa JEA, Levine M, Martinez JSM, Farnoud N, Glodzik D, Li S, Robson ME, Lee C, Pharoah PDP, Stopsack KH, Spitzer B, Mantha S, Fagin J, Boucai L, Gibson CJ, Ebert BL, Young AL, Druley T, Takahashi K, Gillis N, Ball M, Padron E, Hyman DM, Baselga J, Norton L, Gardos S, Klimek VM, Scher H, Bajorin D, Paraiso E, Benayed R, Arcila ME, Ladanyi M, Solit DB, Berger MF, Tallman M, Garcia-Closas M, Chatterjee N, Diaz LA Jr, Levine RL, Morton LM, Zehir A, Papaemmanuil E. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 2020;52:1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, Arcila ME, Ladanyi M, Tallman MS, Levine RL, Berger MF. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 2017;21:374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sano S, Wang Y, Ogawa H, Horitani K, Sano M, Polizio Ah Kour A, Yura Y, Doviak H, Walsh K. TP53-mediated therapy-related clonal hematopoiesis contributes to doxorubicin-induced cardiomyopathy by augmenting a neutrophil-mediated cytotoxic response. JCI Insight 2021;6:146076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lepine G, Mollica L, Szuber N, Dube MP, Busque L. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 2017;130:753–762. [DOI] [PubMed] [Google Scholar]
- 33. Zhang CRC, Nix D, Gregory M, Ciorba MA, Ostrander EL, Newberry RD, Spencer DH, Challen GA. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp Hematol 2019;80:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liggett LA, Galbraith MD, Smith KP, Sullivan KD, Granrath RE, Enriquez-Estrada B, Kinning KT, Shaw JR, Rachubinski AL, Espinosa JM, DeGregori J. Precocious clonal hematopoiesis in Down syndrome is accompanied by immune dysregulation. Blood Adv 2021;5:1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Potus F, Pauciulo MW, Cook EK, Zhu N, Hsieh A, Welch CL, Shen Y, Tian L, Lima P, Mewburn J, D’Arsigny CL, Lutz KA, Coleman AW, Damico R, Snetsinger B, Martin AY, Hassoun PM, Nichols WC, Chung WK, Rauh MJ, Archer SL. Novel mutations and decreased expression of the epigenetic regulator TET2 in pulmonary arterial hypertension. Circulation 2020;141:1986–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JP, Godley LA, Li W, Goodell MA. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2011;44:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loberg MA, Bell RK, Goodwin LO, Eudy E, Miles LA, SanMiguel JM, Young K, Bergstrom DE, Levine RL, Schneider RK, Trowbridge JJ. Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis. Leukemia 2019;33:1635–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 2013;122:4086–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, Ley TJ. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 2014;25:442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anteneh H, Fang J, Song J. Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation. Nat Commun 2020;11:2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang ZM, Lu R, Wang P, Yu Y, Chen D, Gao L, Liu S, Ji D, Rothbart SB, Wang Y, Wang GG, Song J. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 2018;554:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013;502:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010;468:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA 2011;108:14566–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen E, Mullally A. How does JAK2V617F contribute to the pathogenesis of myeloproliferative neoplasms? Hematology Am Soc Hematol Educ Program 2014;2014:268–276. [DOI] [PubMed] [Google Scholar]
- 47. Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, Paktinat M, Haydu JE, Housman E, Lord AM, Wernig G, Kharas MG, Mercher T, Kutok JL, Gilliland DG, Ebert BL. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 2010;17:584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 2007;26:2157–2165. [DOI] [PubMed] [Google Scholar]
- 49. Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol 2018;15:13–30. [DOI] [PubMed] [Google Scholar]
- 50. Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol 1994;4:1–7. [DOI] [PubMed] [Google Scholar]
- 51. Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, Giacomelli AO, Wong W, Kim J, Chao S, Kurppa KJ, Yang X, Milenkowic K, Piccioni F, Root DE, Rucker FG, Flamand Y, Neuberg D, Lindsley RC, Janne PA, Hahn WC, Jacks T, Dohner H, Armstrong SA, Ebert BL. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 2019;365:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004;119:847–860. [DOI] [PubMed] [Google Scholar]
- 53. Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science 1995;269:1427–1429. [DOI] [PubMed] [Google Scholar]
- 54. Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 1999;10:547–558. [DOI] [PubMed] [Google Scholar]
- 55. Kemp R, Ireland H, Clayton E, Houghton C, Howard L, Winton DJ. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res 2004;32:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Velasco-Hernandez T, Sawen P, Bryder D, Cammenga J. Potential pitfalls of the Mx1-Cre system: implications for experimental modeling of normal and malignant hematopoiesis. Stem Cell Reports 2016;7:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity 2004;20:153–165. [DOI] [PubMed] [Google Scholar]
- 58. de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol 2003;33:314–325. [DOI] [PubMed] [Google Scholar]
- 59. Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, Print CG. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 2002;34:251–256. [DOI] [PubMed] [Google Scholar]
- 60. Ueda T, Yokota T, Okuzaki D, Uno Y, Mashimo T, Kubota Y, Sudo T, Ishibashi T, Shingai Y, Doi Y, Ozawa T, Nakai R, Tanimura A, Ichii M, Ezoe S, Shibayama H, Oritani K, Kanakura Y. Endothelial cell-selective adhesion molecule contributes to the development of definitive hematopoiesis in the fetal liver. Stem Cell Reports 2019;13:992–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 1999;8:265–277. [DOI] [PubMed] [Google Scholar]
- 62. Wang J, Hayashi Y, Yokota A, Xu Z, Zhang Y, Huang R, Yan X, Liu H, Ma L, Azam M, Bridges JP, Cancelas JA, Kalfa TA, An X, Xiao Z, Huang G. Expansion of EPOR-negative macrophages besides erythroblasts by elevated EPOR signaling in erythrocytosis mouse models. Haematologica 2018;103:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood 2000;96:719–726. [PubMed] [Google Scholar]
- 64. Ye M, Iwasaki H, Laiosa CV, Stadtfeld M, Xie H, Heck S, Clausen B, Akashi K, Graf T. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 2003;19:689–699. [DOI] [PubMed] [Google Scholar]
- 65. Stadtfeld M, Ye M, Graf T. Identification of interventricular septum precursor cells in the mouse embryo. Dev Biol 2007;302:195–207. [DOI] [PubMed] [Google Scholar]
- 66. McCubbrey AL, Allison KC, Lee-Sherick AB, Jakubzick CV, Janssen WJ. Promoter specificity and efficacy in conditional and inducible transgenic targeting of lung macrophages. Front Immunol 2017;8:1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Orthgiess J, Gericke M, Immig K, Schulz A, Hirrlinger J, Bechmann I, Eilers J. Neurons exhibit Lyz2 promoter activity in vivo: implications for using LysM-Cre mice in myeloid cell research. Eur J Immunol 2016;46:1529–1532. [DOI] [PubMed] [Google Scholar]
- 68. Wang J, Wegener JE, Huang T-W, Sripathy S, De Jesus-Cortes H, Xu P, Tran S, Knobbe W, Leko V, Britt J, Starwalt R, McDaniel L, Ward CS, Parra D, Newcomb B, Lao U, Nourigat C, Flowers DA, Cullen S, Jorstad NL, Yang Y, Glaskova L, Vingeau S, Vigneau S, Kozlitina J, Yetman MJ, Jankowsky JL, Reichardt SD, Reichardt HM, Gärtner J, Bartolomei MS, Fang M, Loeb K, Keene CD, Bernstein I, Goodell M, Brat DJ, Huppke P, Neul JL, Bedalov A, Pieper AA. Wild-type microglia do not reverse pathology in mouse models of Rett syndrome. Nature 2015;521:E1–E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Park E, Evans MA, Doviak H, Horitani K, Ogawa H, Yura Y, Wang Y, Sano S, Walsh K. Bone marrow transplantation procedures in mice to study clonal hematopoiesis. J Vis Exp 2021;In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. de Winther MP, Heeringa P. Bone marrow transplantations to study gene function in hematopoietic cells. Methods Mol Biol 2011;693:309–320. [DOI] [PubMed] [Google Scholar]
- 71. Sano S, Oshima K, Wang Y, MacLauchlan S, Katanasaka Y, Sano M, Zuriaga MA, Yoshiyama M, Goukassian D, Cooper MA, Fuster JJ, Walsh K. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1beta/NLRP3 inflammasome. J Am Coll Cardiol 2018;71:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu CL, Sano S, Muralidharan S, Rius C, Vuong J, Jacob S, Muralidhar V, Robertson AA, Cooper MA, Andres V, Hirschi KK, Martin KA, Walsh K. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017;355:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Buscarlet M, Provost S, Zada YF, Bourgoin V, Mollica L, Dube MP, Busque L. Lineage restriction analyses in CHIP indicate myeloid bias for TET2 and multipotent stem cell origin for DNMT3A. Blood 2018;132:277–280. [DOI] [PubMed] [Google Scholar]
- 74. Venkatesulu BP, Mahadevan LS, Aliru ML, Yang X, Bodd MH, Singh PK, Yusuf SW, Abe JI, Krishnan S. Radiation-induced endothelial vascular injury: a review of possible mechanisms. JACC Basic Transl Sci 2018;3:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schiller NK, Kubo N, Boisvert WA, Curtiss LK. Effect of gamma-irradiation and bone marrow transplantation on atherosclerosis in LDL receptor-deficient mice. ATVB 2001;21:1674–1680. [DOI] [PubMed] [Google Scholar]
- 76. Patel J, Douglas G, Kerr AG, Hale AB, Channon KM. Effect of irradiation and bone marrow transplantation on angiotensin II-induced aortic inflammation in ApoE knockout mice. Atherosclerosis 2018;276:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 2016;44:439–449. [DOI] [PubMed] [Google Scholar]
- 78. Cronk JC, Filiano AJ, Louveau A, Marin I, Marsh R, Ji E, Goldman DH, Smirnov I, Geraci N, Acton S, Overall CC, Kipnis J. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J Exp Med 2018;215:1627–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 2013;210:1977–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, Schilling JD, Schwendener R, Sergin I, Razani B, Forsberg EC, Yokoyama WM, Unanue ER, Colonna M, Randolph GJ, Mann DL. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 2014;40:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang Y, Sano S, Yura Y, Ke Z, Sano M, Oshima K, Ogawa H, Horitani K, Min KD, Miura-Yura E, Kour A, Evans MA, Zuriaga MA, Hirschi KK, Fuster JJ, Pietras EM, Walsh K. Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight 2020;5:e135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Abbuehl JP, Tatarova Z, Held W, Huelsken J. Long-term engraftment of primary bone marrow stromal cells repairs niche damage and improves hematopoietic stem cell transplantation. Cell Stem Cell 2017;21:241–255. [DOI] [PubMed] [Google Scholar]
- 83. Nilsson SK, Dooner MS, Tiarks CY, Weier HU, Quesenberry PJ. Potential and distribution of transplanted hematopoietic stem cells in a nonablated mouse model. Blood 1997;89:4013–4020. [PubMed] [Google Scholar]
- 84. Bhattacharya D, Czechowicz A, Ooi AG, Rossi DJ, Bryder D, Weissman IL. Niche recycling through division-independent egress of hematopoietic stem cells. J Exp Med 2009;206:2837–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bhattacharya D, Rossi DJ, Bryder D, Weissman IL. Purified hematopoietic stem cell engraftment of rare niches corrects severe lymphoid deficiencies without host conditioning. J Exp Med 2006;203:73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Czechowicz A, Kraft D, Weissman IL, Bhattacharya D. Efficient transplantation via antibody-based clearance of hematopoietic stem cell niches. Science 2007;318:1296–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fuster JJ, Zuriaga MA, Zorita V, MacLauchlan S, Polackal MN, Viana-Huete V, Ferrer-Pérez A, Matesanz N, Herrero-Cervera A, Sano S, Cooper MA, González-Navarro H, Walsh K. TET2-loss-of-function-driven clonal hematopoiesis exacerbates experimental insulin resistance in aging and obesity. Cell Rep 2020;33:108326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mercier FE, Sykes DB, Scadden DT. Single targeted exon mutation creates a true congenic mouse for competitive hematopoietic stem cell transplantation: the C57BL/6-CD45.1(STEM) mouse. Stem Cell Reports 2016;6:985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Waterstrat A, Liang Y, Swiderski CF, Shelton BJ, Van Zant G. Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment. Blood 2010;115:408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jang Y, Gerbec ZJ, Won T, Choi B, Podsiad A, B Moore B, Malarkannan S, Laouar Y. Cutting edge: check your mice-a point mutation in the Ncr1 locus identified in CD45.1 congenic mice with consequences in mouse susceptibility to infection. J Immunol 2018;200:1982–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Evans MA, Sano S, Walsh K. Cardiovascular disease, aging, and clonal hematopoiesis. Annu Rev Pathol 2020;15:419–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Min KD, Kour A, Sano S, Walsh K. The role of clonal haematopoiesis in cardiovascular diseases: epidemiology and experimental studies. J Intern Med 2020;288:507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 2017;46:505–529. [DOI] [PubMed] [Google Scholar]
- 94. Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 2014;32:941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res 2018;123:335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sano S, Wang Y, Evans MA, Yura Y, Sano M, Ogawa H, Horitani K, Doviak H, Walsh K. Lentiviral CRISPR/Cas9-mediated genome editing for the study of hematopoietic cells in disease models. J Vis Exp 2019;10.3791/59977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, Bowman M, Famulare C, Patel MA, Mendez P, Ainali C, Demaree B, Delley CL, Abate AR, Manivannan M, Sahu S, Goldberg AD, Bolton KL, Zehir A, Rampal R, Carroll MP, Meyer SE, Viny AD, Levine RL. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ogawa H, Sano S, Walsh K. Employing the CRISPR-Cas system for clonal hematopoiesis research. Int J Phys Med Rehabil 2020;9:1–5. [PMC free article] [PubMed] [Google Scholar]
- 99. Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 2018;24:927–930. [DOI] [PubMed] [Google Scholar]
- 100. Enache OM, Rendo V, Abdusamad M, Lam D, Davison D, Pal S, Currimjee N, Hess J, Pantel S, Nag A, Thorner AR, Doench JG, Vazquez F, Beroukhim R, Golub TR, Ben-David U. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat Genet 2020;52:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Getz GS, Reardon CA. Diet and murine atherosclerosis. Arterioscler Thromb Vasc Biol 2006;26:242–249. [DOI] [PubMed] [Google Scholar]
- 102. Daugherty A, Tall AR, Daemen M, Falk E, Fisher EA, Garcia-Cardena G, Lusis AJ, Owens AP III, Rosenfeld ME, Virmani R, American Heart Association Council on Arteriosclerosis Thrombosis, Vascular Biology, Council on Basic Cardiovascular Sciences. Recommendation on design, execution, and reporting of animal atherosclerosis studies: a scientific statement from the American Heart Association. Arterioscler Thromb Vasc Biol 2017;37:e131–e157. [DOI] [PubMed] [Google Scholar]
- 103. Heyde A, Rohde D, McAlpine CS, Zhang S, Hoyer FF, Gerold JM, Cheek D, Iwamoto Y, Schloss MJ, Vandoorne K, Iborra-Egea O, Munoz-Guijosa C, Bayes-Genis A, Reiter JG, Craig M, Swirski FK, Nahrendorf M, Nowak MA, Naxerova K. Increased stem cell proliferation in atherosclerosis accelerates clonal hematopoiesis. Cell 2021;184:1348–1361.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sano S, Wang Y, Yura Y, Sano M, Oshima K, Yang Y, Katanasaka Y, Min KD, Matsuura S, Ravid K, Mohi G, Walsh K. JAK2 (V617F) -mediated clonal hematopoiesis accelerates pathological remodeling in murine heart failure. JACC Basic Transl Sci 2019;4:684–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sanchez-Cabo F, Fuster JJ. Clonal haematopoiesis and atherosclerosis: a chicken or egg question? Nat Rev Cardiol 2021;18:463–464. [Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- 106. Wang W, Liu W, Fidler T, Wang Y, Tang Y, Woods B, Welch C, Cai B, Silvestre-Roig C, Ai D, Yang YG, Hidalgo A, Soehnlein O, Tabas I, Levine RL, Tall AR, Wang N. Macrophage inflammation, erythrophagocytosis, and accelerated atherosclerosis in Jak2 (V617F) mice. Circ Res 2018;123:e35–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Liu DJ, Peloso GM, Yu H, Butterworth AS, Wang X, Mahajan A, Saleheen D, Emdin C, Alam D, Alves AC, Amouyel P, Di Angelantonio E, Arveiler D, Assimes TL, Auer PL, Baber U, Ballantyne CM, Bang LE, Benn M, Bis JC, Boehnke M, Boerwinkle E, Bork-Jensen J, Bottinger EP, Brandslund I, Brown M, Busonero F, Caulfield MJ, Chambers JC, Chasman DI, Chen YE, Chen YI, Chowdhury R, Christensen C, Chu AY, Connell JM, Cucca F, Cupples LA, Damrauer SM, Davies G, Deary IJ, Dedoussis G, Denny JC, Dominiczak A, Dube MP, Ebeling T, Eiriksdottir G, Esko T, Farmaki AE, Feitosa MF, Ferrario M, Ferrieres J, Ford I, Fornage M, Franks PW, Frayling TM, Frikke-Schmidt R, Fritsche LG, Frossard P, Fuster V, Ganesh SK, Gao W, Garcia ME, Gieger C, Giulianini F, Goodarzi MO, Grallert H, Grarup N, Groop L, Grove ML, Gudnason V, Hansen T, Harris TB, Hayward C, Hirschhorn JN, Holmen OL, Huffman J, Huo Y, Hveem K, Jabeen S, Jackson AU, Jakobsdottir J, Jarvelin MR, Jensen GB, Jorgensen ME, Jukema JW, Justesen JM, Kamstrup PR, Kanoni S, Karpe F, Kee F, Khera AV, Klarin D, Koistinen HA, Kooner JS, Kooperberg C, Kuulasmaa K, Kuusisto J, Laakso M, Lakka T, Langenberg C, Langsted A, Launer LJ, Lauritzen T, Liewald DCM, Lin LA, Linneberg A, Loos RJF, Lu Y, Lu X, Magi R, Malarstig A, Manichaikul A, Manning AK, Mantyselka P, Marouli E, Masca NGD, Maschio A, Meigs JB, Melander O, Metspalu A, Morris AP, Morrison AC, Mulas A, Muller-Nurasyid M, Munroe PB, Neville MJ, Nielsen JB, Nielsen SF, Nordestgaard BG, Ordovas JM, Mehran R, O'Donnell CJ, Orho-Melander M, Molony CM, Muntendam P, Padmanabhan S, Palmer CNA, Pasko D, Patel AP, Pedersen O, Perola M, Peters A, Pisinger C, Pistis G, Polasek O, Poulter N, Psaty BM, Rader DJ, Rasheed A, Rauramaa R, Reilly DF, Reiner AP, Renstrom F, Rich SS, Ridker PM, Rioux JD, Robertson NR, Roden DM, Rotter JI, Rudan I, Salomaa V, Samani NJ, Sanna S, Sattar N, Schmidt EM, Scott RA, Sever P, Sevilla RS, Shaffer CM, Sim X, Sivapalaratnam S, Small KS, Smith AV, Smith BH, Somayajula S, Southam L, Spector TD, Speliotes EK, Starr JM, Stirrups KE, Stitziel N, Strauch K, Stringham HM, Surendran P, Tada H, Tall AR, Tang H, Tardif JC, Taylor KD, Trompet S, Tsao PS, Tuomilehto J, Tybjaerg-Hansen A, van Zuydam NR, Varbo A, Varga TV, Virtamo J, Waldenberger M, Wang N, Wareham NJ, Warren HR, Weeke PE, Weinstock J, Wessel J, Wilson JG, Wilson PWF, Xu M, Yaghootkar H, Young R, Zeggini E, Zhang H, Zheng NS, Zhang W, Zhang Y, Zhou W, Zhou Y, Zoledziewska M, Charge Diabetes Working G, Consortium EP-I, Consortium E-C, Consortium G, Program VAMV, Howson JMM, Danesh J, McCarthy MI, Cowan CA, Abecasis G, Deloukas P, Musunuru K, Willer CJ, Kathiresan S, VA Million Veteran Program. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet 2017;49:1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fidler TP, Xue C, Yalcinkaya M, Hardaway B, Abramowicz S, Xiao T, Liu W, Thomas DG, Hajebrahimi MA, Pircher J, Silvestre-Roig C, Kotini AG, Luchsinger LL, Wei Y, Westerterp M, Snoeck HW, Papapetrou EP, Schulz C, Massberg S, Soehnlein O, Ebert B, Levine RL, Reilly MP, Libby P, Wang N, Tall AR. The AIM2 inflammasome exacerbates atherosclerosis in clonal haematopoiesis. Nature 2021;592:296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hormaechea-Agulla D, Matatall KA, Le DT, Kain B, Long X, Kus P, Jaksik R, Challen GA, Kimmel M, King KY. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNgamma signaling. Cell Stem Cell 2021;S1934-5909(21)00108-9. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Abplanalp WT, Cremer S, John D, Hoffmann J, Schuhmacher B, Merten M, Rieger MA, Vasa-Nicotera M, Zeiher AM, Dimmeler S. Clonal hematopoiesis-driver DNMT3A mutations alter immune cells in heart failure. Circ Res 2021;128:216–228. [DOI] [PubMed] [Google Scholar]
- 111. Stoltzfus KC, Zhang Y, Sturgeon K, Sinoway LI, Trifiletti DM, Chinchilli VM, Zaorsky NG. Fatal heart disease among cancer patients. Nat Commun 2020;11:2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, Heath S, Baty JD, Klco JM, Ding L, Mardis ER, Westervelt P, DiPersio JF, Walter MJ, Graubert TA, Ley TJ, Druley T, Link DC, Wilson RK. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015;518:552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hsu JI, Dayaram T, Tovy A, De Braekeleer E, Jeong M, Wang F, Zhang J, Heffernan TP, Gera S, Kovacs JJ, Marszalek JR, Bristow C, Yan Y, Garcia-Manero G, Kantarjian H, Vassiliou G, Futreal PA, Donehower LA, Takahashi K, Goodell MA. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell 2018;23:700–713.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kahn JD, Miller PG, Silver AJ, Sellar RS, Bhatt S, Gibson C, McConkey M, Adams D, Mar B, Mertins P, Fereshetian S, Krug K, Zhu H, Letai A, Carr SA, Doench J, Jaiswal S, Ebert BL. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood 2018;132:1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Heit JA. Epidemiology of venous thromboembolism. Nat Rev Cardiol 2015;12:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Diaz JA, Saha P, Cooley B, Palmer OR, Grover SP, Mackman N, Wakefield TW, Henke PK, Smith A, Lal BK. Choosing a mouse model of venous thrombosis. Arterioscler Thromb Vasc Biol 2019;39:311–318. [DOI] [PubMed] [Google Scholar]
- 117. von Bruhl ML, Stark K, Steinhart A, Chandraratne S, Konrad I, Lorenz M, Khandoga A, Tirniceriu A, Coletti R, Kollnberger M, Byrne RA, Laitinen I, Walch A, Brill A, Pfeiler S, Manukyan D, Braun S, Lange P, Riegger J, Ware J, Eckart A, Haidari S, Rudelius M, Schulz C, Echtler K, Brinkmann V, Schwaiger M, Preissner KT, Wagner DD, Mackman N, Engelmann B, Massberg S. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J Exp Med 2012;209:819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Meng H, Yalavarthi S, Kanthi Y, Mazza LF, Elfline MA, Luke CE, Pinsky DJ, Henke PK, Knight JS. In vivo role of neutrophil extracellular traps in antiphospholipid antibody-mediated venous thrombosis. Arthritis Rheumatol 2017;69:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Edelmann B, Gupta N, Schnoeder TM, Oelschlegel AM, Shahzad K, Goldschmidt J, Philipsen L, Weinert S, Ghosh A, Saalfeld FC, Nimmagadda SC, Müller P, Braun-Dullaeus R, Mohr J, Wolleschak D, Kliche S, Amthauer H, Heidel FH, Schraven B, Isermann B, Müller AJ, Fischer T. JAK2-V617F promotes venous thrombosis through beta1/beta2 integrin activation. J Clin Invest 2018;128:4359–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol 2011;11:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bonnefond A, Skrobek B, Lobbens S, Eury E, Thuillier D, Cauchi S, Lantieri O, Balkau B, Riboli E, Marre M, Charpentier G, Yengo L, Froguel P. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat Genet 2013;45:1040–1043. [DOI] [PubMed] [Google Scholar]
- 122. Ablamunits V, Weisberg SP, Lemieux JE, Combs TP, Klebanov S. Reduced adiposity in ob/ob mice following total body irradiation and bone marrow transplantation. Obesity (Silver Spring) 2007;15:1419–1429. [DOI] [PubMed] [Google Scholar]
- 123. Katiraei S, Hoving LR, van Beek L, Mohamedhoesein S, Carlotti F, van Diepen JA, Rensen PCN, Netea MG, Willems van Dijk K, Berbée JFP, van Harmelen V. BMT decreases HFD-induced weight gain associated with decreased preadipocyte number and insulin secretion. PLoS One 2017;12:e0175524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Poglio S, Galvani S, Bour S, Andre M, Prunet-Marcassus B, Penicaud L, Casteilla L, Cousin B. Adipose tissue sensitivity to radiation exposure. Am J Pathol 2009;174:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dai DF, Chen T, Johnson SC, Szeto H, Rabinovitch PS. Cardiac aging: from molecular mechanisms to significance in human health and disease. Antioxid Redox Signal 2012;16:1492–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Boyle AJ, Shih H, Hwang J, Ye J, Lee B, Zhang Y, Kwon D, Jun K, Zheng D, Sievers R, Angeli F, Yeghiazarians Y, Lee R. Cardiomyopathy of aging in the mammalian heart is characterized by myocardial hypertrophy, fibrosis and a predisposition towards cardiomyocyte apoptosis and autophagy. Exp Gerontol 2011;46:549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Robertson NA, Hillary RF, McCartney DL, Terradas-Terradas M, Higham J, Sproul D, Deary IJ, Kirschner K, Marioni RE, Chandra T. Age-related clonal haemopoiesis is associated with increased epigenetic age. Curr Biol 2019;29:R786–R787. [DOI] [PubMed] [Google Scholar]
- 128. Campisi J, Kapahi P, Lithgow GJ, Melov S, Newman JC, Verdin E. From discoveries in ageing research to therapeutics for healthy ageing. Nature 2019;571:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.