Summary
Genetic studies on telomere length are important for understanding age-related diseases. Prior GWASs for leukocyte TL have been limited to European and Asian populations. Here, we report the first sequencing-based association study for TL across ancestrally diverse individuals (European, African, Asian, and Hispanic/Latino) from the NHLBI Trans-Omics for Precision Medicine (TOPMed) program. We used whole-genome sequencing (WGS) of whole blood for variant genotype calling and the bioinformatic estimation of telomere length in n = 109,122 individuals. We identified 59 sentinel variants (p < 5 × 10−9) in 36 loci associated with telomere length, including 20 newly associated loci (13 were replicated in external datasets). There was little evidence of effect size heterogeneity across populations. Fine-mapping at OBFC1 indicated that the independent signals colocalized with cell-type-specific eQTLs for OBFC1 (STN1). Using a multi-variant gene-based approach, we identified two genes newly implicated in telomere length, DCLRE1B (SNM1B) and PARN. In PheWAS, we demonstrated that our TL polygenic trait scores (PTSs) were associated with an increased risk of cancer-related phenotypes.
Keywords: telomeres, telomere length genetics, trans-population genome-wide association study
Graphical abstract

Highlights
TOPMed sequencing-based association study identifies 36 loci for telomere length
Reliable estimation of telomere length from whole-genome sequencing data using TelSeq
Fine-mapping of sentinel variants in telomere genetic loci
Polygenic trait scores offer insight into impact of TL on disease outcomes
Taub et al. leverage TOPMed whole-genome sequencing (WGS) to estimate telomere length (TL) and report a trans-population sequencing-based association study. They identify 36 loci associated with TL and correlate polygenetic trait scores with disease outcomes.
Introduction
Telomeres shorten in replicating somatic cells, and telomere length (TL) is associated with age-related diseases.1,2 To date, 17 genome-wide association studies (GWASs) have identified 25 loci for leukocyte TL,3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 but these studies were limited to individuals of European and Asian ancestry and relied on laboratory assays of TL. The decreasing costs of high-throughput sequencing have enabled whole-genome sequencing (WGS) data generation on an unprecedented scale, including in the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) cohorts. Our analyses of TOPMed data offer the opportunity to address the limitations of prior TL GWASs with increased sample size, population diversity, and inclusion of rare variant analyses and fine-mapping of the OBFC1 locus.
In this study, we report a sequencing-based association analysis for telomere length in 109,122 ancestrally diverse individuals (European, African, Asian, and Hispanic/Latino) from the TOPMed program. We used WGS of whole blood for variant genotype calling. We used the TelSeq method for the bioinformatic estimation of telomere length from the WGS data and demonstrated that this approach has high phenotypic and genetic correlation with laboratory-based assays, providing a reliable measurement of TL. We identified 59 sentinel variants (p < 5 × 10−9) in 36 loci associated with TL; 20 of these are newly associated, and 13 replicated in external datasets. We also identified new common and rare variant associations at previously reported TL loci. Using WGS data also allowed fine-mapping approaches for OBFC1. Finally, we conducted phenome-wide association studies (PheWAS) in BioVU and identified the association of our defined polygenic trait scores (PTSs) for TL with the increased risk of cancer-related phenotypes.
Results
We selected TelSeq20 to bioinformatically estimate TL due to its computational efficiency and high correlation with Southern blot21 and flowFISH22 measurements (Figures S1A–S1C; STAR Methods). We developed a principal components-based approach to remove technical artifacts arising from the sequencing process that affected TL estimation, which further improved accuracy (Figures S1D and S1E; STAR Methods). We found high phenotypic correlation of TelSeq-derived TL with TL measured by Southern blot21 in the 2,398 TOPMed samples from the Jackson Heart Study (JHS) and with TL measured by flowFISH22 in a set of 19 TOPMed GeneSTAR samples (r = 0.68 and 0.80, respectively; Figures S1C–S1E; STAR Methods). In addition, we observed high genetic correlation between TelSeq and Southern blot assays of TL in the subset of 1,083 family-based JHS samples (ρg = 0.8069, SE = 0.05, estimated using SOLAR23). Together, the phenotypic and genetic correlations with the more traditionally used Southern blot or flowFISH assays suggest the suitability of TelSeq-based TL as a potential TL measure for large-scale genetic epidemiologic study.
Pooled trans-population association analysis was performed with n = 109,122 individuals from TOPMed (including 51,654 of European ancestry, 29,260 of African ancestry, 18,019 Hispanic/Latinos, 5,683 of Asian ancestry, and 4,506 of other, mixed, or uncertain ancestries, as determined by harmonized ancestry and race/ethnicity [HARE]24;Figure 1A; STAR Methods); 44% were male and ages ranged from <1 to 98 years old (Table S1). Genome-wide tests for association were performed across 163 million variants. Using a series of single-variant tests for association (primary to identify loci, iterative conditional by chromosome to identify additional independent variants, and joint tests, including all independent variants to summarize effect sizes; see STAR Methods), we identified 59 independently associated variants mapping to 36 loci, meeting the significance threshold of p < 5 × 10−9 (Figure 1B; Tables 1 and S2); 16 of these were previously reported and 20 were newly associated loci, as described further below.
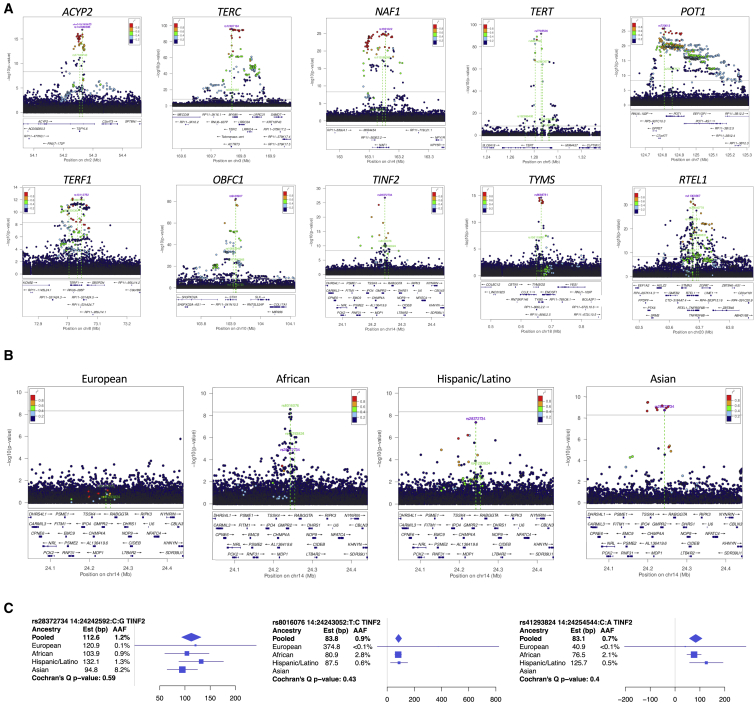
Figure 1.
Genome-wide Manhattan plot
(A) Pie chart showing population groups based on HARE for samples included in analysis: European (green, n = 51,654), African (orange, n = 29,260), Hispanic/Latino (purple, n = 18,019), Asian (red, n = 5,683), and Other/Mixed/Unknown (gray, n = 4,506).
(B) Trans-population genome-wide tests for association using 163 million sequence-identified variants on n = 109,122 samples with sequence-generated telomere length from TOPMed. All loci had a peak p < 5 × 10−9 in the pooled trans-population analysis. Previously reported loci for TL are indicated in red, and loci newly associated in the present study are indicated in blue. Note the shift in scale above the y axis break; no peak variants had a p value within the y axis break.
Table 1.
59 variants independently associated with telomere length, mapping to 36 loci, in 109,122 ancestrally diverse (African, European, Hispanic/Latino, Asian) individuals from TOPMed
| Single variant analysis in pooled trans-population sample |
p values from joint model |
Effect sizes from joint model |
Cochran’s Q (p value) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Locus | SNP | Prior GWAS | Annotation | MAC | p value | Percent variation explained | Trans- population | European | African | Hispanic/ Latino | Asian | Trans- population | European | African | Hispanic/ Latino | Asian | |
| 1 | ZMYM4 | rs11581846 | 7 | 87500 | 3.04E−10 | 0.036% | 1.74E−11 | 7.99E−5 | 8.80E−3 | 3.37E−6 | 4.13E−1 | −19.9 | −21.1 | −15.0 | −24.6 | −8.7 | 0.44 | |
| 1 | BCL2L15 | rs2296176 | 7 | 33274 | 2.84E−10 | 0.036% | 1.52E−10 | 6.57E−8 | 2.38E−3 | 3.16E−1 | 2.07E−2 | −19.5 | −22.4 | −22.0 | −7.3 | −26.9 | 0.28 | |
| 1 | PARP1 | rs1136410 | Known | missense | 36395 | 9.32E−20 | 0.076% | 9.14E−22 | 6.10E−9 | 1.02E−2 | 7.51E−7 | 2.04E−4 | −29.3 | −26.0 | −23.7 | −29.5 | −32.5 | 0.87 |
| 2 | ACYP2 | rs17189743 | Known | missense | 3741 | 7.18E−12 | 0.043% | 4.24E−11 | 2.70E−6 | 2.52E−1 | 5.20E−6 | 1.94E−1 | −55.4 | −50.7 | −30.4 | −82.4 | −46.8 | 0.34 |
| 2 | rs144980386 | deletion | 28773 | 1.32E−17 | 0.067% | 1.99E−17 | 7.78E−13 | 7.32E−4 | 1.45E−4 | 6.78E−1 | 27.5 | 33.6 | 21.2 | 30.5 | 4.8 | 0.08 | ||
| 3 | LINC00901 | rs961617801 | 6 | 12 | 1.25E−11 | 0.042% | 4.81E−11 | 2.82E−10 | – | – | – | 1009.6 | 1038.5 | – | – | – | – | |
| 3 | TERC | rs12637184 | Known | 4 | 47452 | 1.30E−96 | 0.399% | 1.58E−103 | 1.02E−50 | 1.84E−15 | 7.32E−30 | 2.33E−11 | −59.8 | −57.0 | −65.8 | −66.1 | −57.3 | 0.51 |
| 3 | rs9826466 | N/A | 4066 | 3.25E−17 | 0.065% | 3.66E−21 | 2.04E−01 | 7.28E−19 | 2.96E−03 | – | −77.0 | 253.9 | −77.6 | −77.8 | – | 0.25 | ||
| 3 | P3H2 | rs10937417 | 7 | 80209 | 6.89E−10 | 0.035% | 1.89E−10 | 9.04E−5 | 1.65E−5 | 2.33E−3 | 6.30E−1 | 14.6 | 13.5 | 17.9 | 17.1 | −4.4 | 0.16 | |
| 4 | SLC2A2 | rs4235345 | 6 | 44302 | 3.82E−9 | 0.032% | 1.88E−9 | 3.24E−7 | 4.39E−2 | 1.86E−2 | 5.70E−2 | 16.8 | 19.8 | 12.8 | 13.2 | 47.1 | 0.41 | |
| 4 | NAF1 | rs60735607∗ | Known | 6 | 57418 | 0.003964 | 0.008% | 4.43E−12 | 4.45E−7 | 7.20E−3 | 5.33E−4 | 9.94E−1 | −18.5 | −20.0 | −12.9 | −22.4 | −0.1 | 0.43 |
| 4 | rs113580095 | 7 | 290 | 4.72E−18 | 0.069% | 1.63E−16 | 2.57E−8 | 7.12E−2 | 3.40E−7 | – | −254.7 | −231.9 | −287.8 | −257.9 | – | 0.89 | ||
| 4 | rs1351222 | 7 | 50104 | 3.27E−26 | 0.103% | 1.60E−32 | 9.04E−16 | 1.50E−7 | 6.80E−11 | 6.58E−3 | 32.7 | 32.7 | 28.9 | 41.2 | 28.9 | 0.49 | ||
| 5 | TERT | rs192999400 | Known | 5 | 2470 | 3.10E−15 | 0.057% | 8.21E−23 | 2.15E−1 | 3.90E−17 | 3.42E−3 | 6.19E−2 | 101.3 | 88.7 | 97.4 | 96.9 | 108.4 | 1.00 |
| 5 | rs6897196 | 5 | 102081 | 1.87E−83 | 0.344% | 6.04E−13 | 7.24E−4 | 7.90E−8 | 1.87E−2 | 1.46E−1 | 20.8 | 16.2 | 25.2 | 16.8 | 23.4 | 0.55 | ||
| 5 | rs7705526∗∗ | 7 | 64162 | 1.64E−92 | 0.382% | 2.01E−18 | 1.16E−11 | 7.73E−4 | 1.46E−3 | 6.47E−2 | 30.0 | 35.5 | 22.5 | 27.1 | 34.1 | 0.47 | ||
| 5 | rs2853677∗∗ | 5 | 80905 | 8.17E−65 | 0.265% | 1.25E−19 | 9.27E−7 | 8.88E−10 | 1.33E−5 | 8.34E−2 | −23.9 | −17.7 | −33.9 | −28.9 | −22.0 | 0.08 | ||
| 5 | rs34052286 | 3a | 6173 | 1.50E−12 | 0.046% | 5.47E−22 | 3.86E−1 | 1.09E−14 | 2.39E−5 | – | −66.0 | −59.3 | −61.6 | −74.3 | – | 0.80 | ||
| 5 | rs114616103∗ | 7 | 4527 | 2.39E−7 | 0.024% | 2.03E−13 | 1.44E−9 | 2.09E−2 | 1.72E−2 | 4.93E−1 | −57.2 | −59.2 | −37.9 | −57.6 | −71.9 | 0.73 | ||
| 5 | CXXC5 | rs75903170 | 2b | 11895 | 5.69E−10 | 0.035% | 7.01E−10 | 2.83E−4 | 2.58E−6 | 8.14E−2 | 4.31E−1 | 29.6 | 25.5 | 46.7 | 22.3 | 12.6 | 0.19 | |
| 6 | HSPA1A | rs1008438 | 2b | 106908 | 3.42E−17 | 0.065% | 6.64E−19 | 3.61E−9 | 1.40E−4 | 4.10E−7 | 1.71E−2 | −20.3 | −19.7 | −18.4 | −25.7 | −20.5 | 0.73 | |
| 7 | POT1 | rs720613 | Known | 7 | 62325 | 1.27E−26 | 0.105% | 1.37E−27 | 2.59E−18 | 5.87E−6 | 3.63E−5 | 2.51E−2 | −26.3 | −31.3 | −20.4 | −24.6 | −21.0 | 0.26 |
| 7 | rs202187871 | missense | 27 | 4.89E−12 | 0.044% | 6.72E−13 | 6.42E−12 | – | – | – | 738.3 | 719.6 | – | – | – | – | ||
| 7 | TNP03 | rs7783384 | 5 | 92528 | 2.34E−12 | 0.045% | 6.10E−12 | 1.22E−7 | 1.47E−3 | 2.54E−2 | 2.04E−2 | −15.2 | −17.8 | −13.3 | −11.4 | −19.8 | 0.64 | |
| 8 | TERF1 | rs183633026 | Known | 7 | 1132 | 1.59E−10 | 0.038% | 1.45E−10 | 3.48E−2 | 7.67E−1 | 1.51E−9 | – | 99.4 | 75.4 | −22.0 | 109.6 | – | 0.18 |
| 8 | rs73687065 | 5 | 1676 | 3.10E−12 | 0.045% | 8.10E−12 | 7.74E−1 | 2.76E−10 | 5.71E−3 | – | 85.3 | 24.9 | 87.4 | 97.0 | – | 0.74 | ||
| 8 | rs10112752 | 7 | 78968 | 4.59E−13 | 0.048% | 9.83E−12 | 5.13E−9 | 5.69E−3 | 4.16E−2 | 2.83E−2 | −15.8 | −19.2 | −12.6 | −11.2 | −26.2 | 0.40 | ||
| 10 | NOC3L | rs3758526 | missense | 32511 | 6.80E−12 | 0.043% | 5.77E−13 | 1.63E−5 | 2.70E−5 | 1.38E−2 | 1.42E−2 | −22.0 | −21.1 | −22.6 | −19.8 | −23.1 | 0.99 | |
| 10 | NKX2-3 | rs10883359 | 7 | 54905 | 3.60E−12 | 0.044% | 9.34E−11 | 5.46E−5 | 5.14E−6 | 3.46E−2 | 1.70E−1 | −16.5 | −14.5 | −28.1 | −11.9 | −11.9 | 0.19 | |
| 10 | OBFC1 | rs10883948 | Known | 7 | 94489 | 2.04E−34 | 0.137% | 3.97E−12 | 1.20E−5 | 3.99E−4 | 2.18E−2 | 6.42E−4 | −18.8 | −15.7 | −24.2 | −13.9 | −46.8 | 0.11 |
| 10 | rs112163720∗ | 4 | 15559 | 0.440005 | 0.001% | 4.86E−16 | 9.57E−7 | 4.34E−7 | 3.69E−4 | 9.38E−4 | 37.1 | 48.1 | 39.1 | 35.3 | 54.9 | 0.66 | ||
| 10 | rs9420907∗∗ | 3a | 54838 | 3.90E−83 | 0.342% | 6.80E−54 | 3.65E−18 | 6.94E−19 | 6.79E−13 | 9.41E−1 | −49.2 | −44.3 | −52.4 | −53.8 | −2.4 | 0.30 | ||
| 10 | rs111447985 | 2a | 2391 | 2.29E−24 | 0.095% | 3.03E−35 | 4.94E−3 | 2.24E−2 | 5.44E−22 | 2.81E−10 | 131.9 | 120.7 | 98.1 | 143.4 | 137.2 | 0.77 | ||
| 11 | ATM | rs61380955 | Known | 7 | 105969 | 2.47E−17 | 0.066% | 1.11E−18 | 5.79E−14 | 2.97E−4 | 2.47E−3 | 9.19E−2 | −19.6 | −24.8 | −15.6 | −15.7 | −14.6 | 0.24 |
| 13 | KBTBD7 | rs1411041 | 6 | 85572 | 6.29E−14 | 0.052% | 6.65E−15 | 1.46E−8 | 6.04E−4 | 1.13E−3 | 1.77E−2 | 22.4 | 25.5 | 20.5 | 20.2 | 20.8 | 0.87 | |
| 14 | TINF2 | rs28372734 | Known | 4 | 2648 | 1.74E−27 | 0.108% | 1.27E−30 | 4.91E−2 | 3.42E−6 | 4.59E−9 | 7.26E−10 | 112.6 | 120.9 | 103.9 | 132.1 | 94.8 | 0.59 |
| 14 | rs8016076 | 2b | 1977 | 1.80E−11 | 0.041% | 4.46E−13 | 1.01E−1 | 1.70E−10 | 6.73E−3 | – | 83.8 | 374.8 | 80.9 | 87.5 | – | 0.43 | ||
| 14 | rs41293824 | 5 | 1543 | 1.31E−9 | 0.034% | 1.87E−10 | 7.43E−1 | 1.83E−7 | 2.58E−4 | – | 83.1 | 40.9 | 76.5 | 125.7 | – | 0.40 | ||
| 14 | DCAF4 | rs2572 | Known | 5 | 20731 | 5.14E−12 | 0.044% | 6.70E−14 | 2.00E−7 | 1.07E−4 | 3.42E−3 | 1.75E−3 | 28.0 | 27.7 | 36.5 | 25.5 | 33.8 | 0.80 |
| 15 | ATP8B4 | rs7172615 | 4 | 41027 | 4.31E−9 | 0.032% | 3.53E−10 | 1.14E−7 | 3.77E−3 | 1.96E−1 | 1.90E−1 | −17.8 | −20.3 | −21.4 | −8.7 | −12.0 | 0.41 | |
| 16 | TERF2 | rs9925619 | Known | 7 | 66224 | 3.01E−14 | 0.053% | 7.84E−15 | 3.03E−4 | 1.01E−7 | 1.02E−6 | 5.10E−1 | 18.6 | 13.1 | 22.5 | 27.9 | 8.9 | 0.10 |
| 16 | CLEC18C | rs62049363 | 7 | 61724 | 4.09E−10 | 0.036% | 3.25E−11 | 5.24E−7 | 1.44E−1 | 4.80E−4 | 3.52E−1 | −16.8 | −16.7 | −11.2 | −19.4 | −8.5 | 0.69 | |
| 16 | RFWD3 | rs7193541 | Known | missense | 93079 | 1.47E−16 | 0.063% | 3.18E−17 | 3.39E−12 | 5.63E−5 | 1.66E−3 | 5.21E−1 | −18.7 | −22.9 | −16.8 | −16.9 | −5.6 | 0.24 |
| 16 | MPHOSPH6 | rs2967355 | Known | 6 | 34993 | 3.96E−19 | 0.073% | 2.04E−20 | 2.69E−11 | 1.58E−4 | 4.86E−7 | 9.91E−1 | −28.2 | −26.2 | −33.8 | −36.8 | −0.1 | 0.05 |
| 16 | BANP | rs12934497 | 5 | 77109 | 9.15E−10 | 0.034% | 8.16E−0 | 1.53E−5 | 1.62E−3 | 5.66E−3 | 1.15E−1 | 14.6 | 14.1 | 15.2 | 15.2 | 31.8 | 0.86 | |
| 18 | TYMS | rs150119891∗ | 5 | 1320 | 2.27E−7 | 0.025% | 1.92E−11 | 3.69E−10 | 2.80E−1 | 2.89E−3 | – | −98.8 | −104.9 | −52.3 | −160.9 | – | 0.32 | |
| 18 | rs8088781 | 5 | 25774 | 2.49E−15 | 0.057% | 8.91E−32 | 2.78E−15 | 1.75E−8 | 8.74E−9 | 8.66E−2 | −50.6 | −56.0 | −40.8 | −59.3 | −160.9 | 0.21 | ||
| 18 | rs2612101∗ | 5 | 56194 | 0.407696 | 0.001% | 6.29E−16 | 7.76E−7 | 2.11E−4 | 5.13E−8 | 3.89E−2 | 26.0 | 29.4 | 18.4 | 35.1 | 171.4 | 0.05 | ||
| 18 | SETBP1 | rs2852770 | 7 | 46513 | 1.00E-11 | 0.042% | 1.15E-12 | 2.32E-09 | 4.37E-02 | 1.23E-04 | 7.01E-01 | −19.0 | −25.1 | −9.4 | −24.3 | −4.1 | 0.03 | |
| 19 | ZNF257/ZNF676 | rs8105767∗∗ | Known | 6 | 76591 | 3.59E−18 | 0.069% | 1.52E−18 | 6.32E−9 | 1.14E−7 | 5.00E−3 | 5.14E−3 | 20.3 | 20.8 | 22.2 | 15.0 | 25.5 | 0.68 |
| 20 | SAMHD1 | rs2342113 | 6 | 51830 | 4.62E−18 | 0.069% | 1.58E−19 | 2.50E−13 | 4.00E−4 | 1.34E−6 | 3.67E−1 | −23.7 | −33.8 | −16.1 | −27.4 | −7.7 | 0.01 | |
| 20 | RTEL1 | rs41308088 | Known | 5 | 14105 | 1.58E−10 | 0.038% | 8.42E−16 | 6.46E−15 | 6.24E−2 | 4.61E−2 | 2.05E−1 | 37.1 | 45.3 | 25.1 | 20.1 | 58.1 | 0.12 |
| 20 | rs79981941 | 6 | 21592 | 1.46E−23 | 0.092% | 1.37E−12 | 5.06E−6 | 4.73E−3 | 5.48E−6 | 9.09E−2 | −26.7 | −26.4 | −18.6 | −39.1 | −47.5 | 0.25 | ||
| 20 | rs41309367∗∗ | 5 | 71377 | 2.52E−33 | 0.133% | 1.01E−42 | 5.92E−23 | 1.49E−15 | 9.58E−9 | 6.05E−1 | −34.1 | −37.0 | −38.0 | −33.0 | −5.1 | 0.02 | ||
| 20 | rs35640778 | missense | 2354 | 2.31E−28 | 0.112% | 1.47E−38 | 3.77E−29 | 2.06E−7 | 1.11E−4 | 9.25E−1 | −140.5 | −141.8 | −176.3 | −116.7 | 13.8 | 0.42 | ||
| 20 | rs181080831 | synonymous | 675 | 6.40E−17 | 0.064% | 7.53E−19 | 8.00E−18 | 8.50E−1 | 1.39E−2 | – | 180.5 | 199.5 | 15.5 | 133.0 | – | 0.07 | ||
| 22 | TYMP | rs361725 | 3a | 101750 | 1.36E−10 | 0.038% | 8.26E−12 | 3.97E−11 | 2.32E−1 | 4.11E−2 | 2.39E−2 | −15.6 | −22.1 | −5.4 | −10.6 | −21.2 | 0.02 | |
| X | VSIG4 | rs12394264 | 4 | 50912 | 9.67E−16 | 0.059% | 5.36E−17 | 1.57E−7 | 1.94E−5 | 4.13E−5 | 7.28E−1 | 19.0 | 19.2 | 15.7 | 20.6 | 13.3 | 0.85 | |
| X | GAB3 | rs2728723 | 5 | 63517 | 1.21E−12 | 0.046% | 3.47E−12 | 4.45E−8 | 8.67E−2 | 7.96E−4 | 4.55E−2 | 13.2 | 15.7 | 6.0 | 14.4 | 17.0 | 0.17 | |
Loci are labeled as known if the sentinel variants in the locus were in LD (r2 ≥ 0.7) with previously reported GWAS association for telomere length. There are 5 variants marked with an asterisk where the primary analysis did not meet our threshold of p < 5 × 10−9; however, they reached significance after conditioning on significant variants mapping to the chromosome (detailed in Table S2). Variants marked with a double asterisk are direct matches to prior reported sentinel variants. Percentage of trait variation explained by each variant is provided from single-variant association tests. p values and effect sizes (in base pairs) are reported from a joint model including all variants. p values for effect heterogeneity across population groups were generated using Cochran’s Q statistic. MAC is the minor allele count from the full combined sample. For all exonic variants, detailed annotation is provided, while for all non-coding variants, the RegulomeDB score is given.
We examined 25 previously reported loci for TL identified through GWASs, using qPCR or Southern blot assays to directly measure TL, for evidence of replication in our study. For 16 loci (PARP1, ACYP2, TERC, NAF1, TERT, POT1, TERF1, OBFC1, ATM, TINF2, DCAF4, TERF2, RFWD3, MPHOSPH6, ZNF208/ZNF257/ZNF676, and RTEL1), there was at least 1 variant with p <5 × 10−9 in our trans-population TL analysis that was in linkage disequilibrium (LD) (r2 ≥ 0.7) with a published genome-wide significant (p <5 × 10−8) variant from a previous study (Tables 1 and S3). Directionally consistent and nominal evidence for replication was noted for CTC1 (rs3027234, p = 7.97 × 10−5) and SENP7 (rs55749605, p = 0.023). A signal previously attributed to PRRC2A is located <200 kb from our signal for HSPA1A but may be distinct given the low LD (r2 = 0.26). We found no evidence of replication (all variants with p > 0.05) for the remaining previously reported TL loci (CXCR4, PXK, MOB1B, DKK2/PAPSS1, CARMIL1, and CSNK2A2; Table S3). Our comprehensive conditional analyses identified ≥1 independent sentinel variants at 9 of the 16 previously reported loci (Figure 2A; Table 1). The resolution possible with our trans-population WGS data identified a sentinel variant different from the one previously reported by tagging-based GWASs for 11 of the 16 known loci. At known loci RTEL1, RFWD3, POT1, ACYP2, and PARP1, our WGS-based sentinels included a coding missense variant in genes RTEL1, RFWD3, POT1, TSPYL6, and PARP1, respectively. For the remaining known TL loci, many of the non-coding sentinel variants are annotated as having regulatory evidence (RegulomeDB score < 7; Table 1), as illustrated further for OBFC1 below.
Figure 2.
LocusZoom plots for multi-hit loci and TINF2
(A) LocusZoom plots for all loci with >1 sentinel variant. Linkage disequilibrium (LD) was calculated from the set of samples used in the analysis with respect to the peak variant in the pooled trans-population primary analysis, thereby reflecting LD patterns specific to the TOPMed samples. For each figure, the peak sentinel variant from the pooled trans-population analysis is indexed and labeled in purple, and all of the independent variants identified through the iterative conditional approach are labeled in green and highlighted with green dotted lines.
(B) LocusZoom plots for 4 population groups for the TINF2 locus.
(C) Forest plots displaying effect sizes and standard errors, as well as minor allele frequencies, by population group for the 3 sentinel variants in TINF2.
See also Table S2.
A total of 22 independent sentinel variants were located at the 20 newly associated loci (Table 1). We examined 19 of these sentinel variants for evidence of association in 2 previously qPCR-based TL GWASs with non-overlapping subjects18,19 (Figure S2A). Variants at 10 of these loci (BCL2L15, CXXC5, HSPA1A, NOC3L, NKX2-3, ATP8B4, CLEC18C, TYMS, SAMHD1, and TYMP) had a Bonferroni-corrected p < 0.05/19 = 0.0026, and an additional 3 had variants with p < 0.05 (TNP03, KBTBD7, and BANP), as did a second variant at TYMS. The variant at SAMHD1 was previously reported at an false discovery rate (FDR) < 0.05 (p = 1.41 × 10−7),19 but here has genome-wide significance (p = 1.58 × 10−19). Proteins encoded by two of these genes have strong biological connections to TL: CXXC5, which physically interacts with ATM and transcriptionally regulates p53 levels25; two proteins implicated in telomere length regulation; and BANP (also known as SMAR1), which forms a complex with p53 and functions as a tumor suppressor.26
There is high consistency in the effect sizes at the 59 sentinel variants observed in our TelSeq-based TL GWAS compared to prior GWASs using qPCR assays of TL (Figure S2B). The Pearson correlation was 0.92 (p = 2.1 × 10−15, for 37 overlapping variants) for our study compared to Dorajoo et al.18 (n = 23,096 Singaporean Chinese; Figure S2B, upper panel) and was 0.86 (p = 1.2 × 10−13, for 43 overlapping variants) for our study compared to Li et al.19 (n = 78,592 European; Figure S2B, lower panel). However, qPCR is a relative measure of TL (relative to a single copy gene, see Note S1); it has different units from our TelSeq measurement, which is in base pairs. A direct comparison between effect sizes in base pairs from Southern blot and TelSeq in the 2,398 JHS samples (STAR Methods) confirms very high correlation (r = 0.92, p = 1.6 × 10−20 for 49 variants with minor allele frequency (MAF) ≥1% in the JHS samples; Figure S2C). We note that the effect sizes in bp for TelSeq are around half those for Southern blot (slope = 0.56, p = 1.6 × 10−20). This mirrors what we observe in a direct comparison of the TelSeq and Southern blot TL values (Figure S1D): a 1 bp increase in Southern blot TL corresponds to a 0.42 bp increase in TelSeq TL (p = 4.9 × 10−319). Furthermore, the age effects on TL in these JHS samples show a similar pattern: the estimated per year decline in TL is 22 bp (p = 1.9 × 10−114) for Southern blot compared to 11 bp (p = 1.8 × 10−69) for TelSeq (STAR Methods). As a further assessment of the general reliability of our TelSeq measurements for large-scale genetic epidemiologic analysis, we performed cross-trait LD score regression with LDSC27,28 using GWAS summary statistics from the European ancestry group (n = 51,564) in our TOPMed WGS-based analysis and the Li et al.19 analysis on n = 78,592 European ancestry individuals with qPCR-measured TL. The genetic correlation was 0.8066 (SE = 0.09, p = 1.8 × 10−17), indicating a high degree of shared genetic determinants of TL from the 2 different measurement technologies.
Each of the 59 sentinel variants individually accounted for a small percentage of phenotypic variation (Table 1), consistent with prior GWASs of TL but cumulatively accounted for 4.35% of TL variance, compared to 2%–3% from prior GWASs.3 The 37 variants mapping to 16 known loci explained 3.38% of TL variability, and an additional 0.96% was explained by the 22 variants mapping to our 20 newly associated loci, representing a sizable gain in explained variability for TL from the present study. Prior GWASs using Southern blot and qPCR report allelic effects ranging from ∼49 to 120 bp.3,4,11,13 In the TOPMed data, our estimated effect sizes for common variants (minor allele frequency, MAF ≥ 5%) ranged from 2 to 59 bp per allele. In comparison, the effect sizes were larger for rare and low-frequency variants (MAF < 5%) in the TOPMed data (40–1,063 bp per allele).
Stratified association analyses were performed in population groups with at least 5,000 samples to evaluate effect heterogeneity of the 59 sentinel variants (Table S4). Reduced sample sizes, coupled with variation in allele frequency, often limited our power to detect population-specific associations at GWAS thresholds in the individual strata (Table S4); no additional loci were identified. A major advantage of our analysis was the ability to rely on the individual-level WGS data for the iterative conditional approach to identify the final set of independent sentinel variants at each locus. The identified sentinel variants show little evidence for heterogeneity across populations (Table 1). All Cochran’s Q29 p values (Table 1) were above a Bonferroni correction threshold (p > 0.001), and the 5 with nominal significance (0.001 < p < 0.05) appear to be primarily driven by differences in the (smallest) Asian stratum. An interesting illustration of a locus with strong allele frequency differences between groups is TINF2; the evidence at the peak variant (rs28372734) in the trans-population analysis was driven by the smaller Hispanic/Latino and Asian groups (group-specific p 4.6 × 10−9 and 7.3 × 10−10, respectively), and the secondary peak (rs8016076) was driven by the African group (group-specific p 1.7 × 10−10; Figure 2B; Table 1). No association is noted in the European group, where these variants are nearly monomorphic (Figure 2C).
Gene-based tests in the combined sample of all 109,000 individuals identified 8 protein-coding genes with deleterious rare and low-frequency (MAF < 1%, including singletons) variants associated with TL (p < 1.8 × 10−6, see Figure S3; STAR Methods). Six of these genes were previously identified GWAS loci (POT1, TERT, RTEL1, CTC1, SAMHD1, and ATM), now adding support for rare variant associations in these genes. Both DCLRE1B and PARN have been implicated in short telomere syndrome (STS) patients.30, 31, 32 DCLRE1B protein localizes to the telomere via interaction with the protein of another previously implicated GWAS gene, TERF2, and contributes to telomere protection from DNA repair pathways.33,34 Notably, two PARN loss-of-function variants included in our gene-based test were previously identified in STS patients.30 Both rs878853260 and rs876661305 produce frameshift mutations; rs876661305 produces an early termination codon, truncating most of the nuclease domain.35 For each of these 8 genes, a leave-one-out approach iterating over each variant included in the aggregate test showed there were no detectable main driver variants and indicated that these gene-based association signals arise from cumulative signals across multiple rare deleterious variants (Figure S3), with the possible exception of ATM. When conditioned on the 59 sentinel variants, all of the genes, except POT1, maintained or increased statistical significance (Figure S3). For POT1, while the removal of the single variant identified in Table 1 (rs202187871) and conditioning on all 59 sentinels resulted in a decrease in significance from 1.52 × 10−24 to 5.53 × 10−18, it nonetheless remained strongly significant, meeting Bonferroni thresholds.
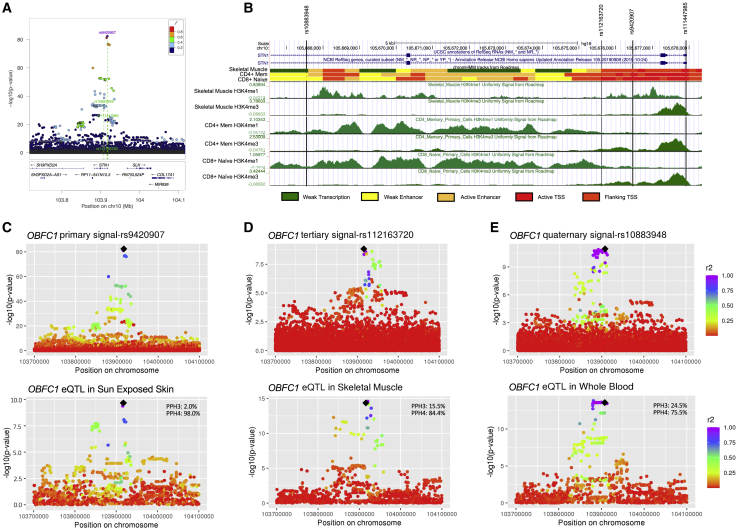
The identification of multiple independent sentinel variants for several loci offers the unique opportunity to evaluate the potential for distinct regulatory mechanisms (Figures 2A and S4). OBFC1 is part of a complex that binds single-stranded telomeric DNA36 and is expressed across multiple tissues in GTEx37 and in whole-blood studies meta-analyzed in eQTLGen.38 All four signals at the OBFC1 locus are in the promoter and early introns of OBFC1 (Figure 3A and 3B). Evidence for expression quantitative trait loci (eQTL) colocalization was detected at the primary, tertiary, and quaternary signals in various tissues (STAR Methods). While all 3 signals colocalized with OBFC1 eQTLs, the strongest colocalization evidence in each case was in a distinct tissue: sun-exposed skin from the lower leg (posterior probability of shared signal, PPH4 = 98.0%) for the primary, skeletal muscle (PPH4 = 84.4%) for the tertiary, and whole blood (GTEx PPH4 = 75.5%, eQTLGen PPH4 = 75.5%) for the quaternary signal (Figures 3C–3E and S5E; Table S5). Data from the Roadmap Epigenomics Consortium39 indicate that all 4 signals are consistent with promoter or enhancer regions across blood cells and skeletal muscle tissue (Figure 3B). We were unable to perform colocalization analysis on the secondary signal with data from either GTEx or eQTLGen as it is driven by rare variants only in the Hispanic/Latino and Asian individuals (rs111447985; Table S4).
Figure 3.
Fine-mapping of multiple OBFC1 signals
(A) LocusZoom plot of the OBFC1 locus, where green dotted lines indicate each independent signal, as in Figure 2.
(B) Roadmap Epigenomics Consortium data in hg19 coordinates for skeletal muscle tissue, Primary T CD4+ memory cells from peripheral blood, and primary T CD8+ naive cells from peripheral blood (Roadmap samples E108, E037, and E047, respectively; data were not available for sun-exposed skin). The ChromHMM state model is shown for the 18-state auxiliary model. The state model suggests the primary (rs9420907), secondary (rs111447985), and tertiary (rs112163720) signals are in the promoter region, while the quaternary signal (rs10883948) is in an enhancer region in all Roadmap blood cell types but is transcriptional for peripheral blood monocytes and CD19+ B cells.
(C–E) GWAS and eQTL results for the primary (C), tertiary (D), and quaternary (E) signals. The top panels are the GWAS summary statistics from the primary, and iterative conditional analyses that were used to perform colocalization analysis (secondary signal was rare and not available for colocalization). Bottom panels are eQTLs for OBFC1 in the indicated tissue from GTEx. The GTEx eQTLs for these tissues do not colocalize with one another (PPH4 < 4.4 × 10−7), and each signal did not significantly colocalize in the other tissues. LD was calculated from the pooled trans-population samples with respect to the sentinel (black diamond).
See also Figures S4 and S5 and Table S5.
Using individual-level data within the Vanderbilt University biobank BioVU, we performed a PheWAS (Table S6) using 49 available sentinel variants individually in addition to a TL polygenic trait score (PTS). The PTS was generated separately for European and African individuals in BioVU as a simple linear combination of the effect sizes from the stratified joint analysis in European or African individuals, respectively (Table 1, Effect sizes from joint model). PTS values were significantly higher in BioVU African Americans (AAs, mean = −217 bp, SD = 96 bp) compared to European Americans (EAs, mean = −279 bp, SD = 96 bp, p < 0.05, Welch’s 2-sample t test; Figure S6A), offering evidence that previously observed differences in TL by ancestry (longer TL in individuals of African ancestry1) may be explained in part by the genetic contribution to TL. The largest cumulative effect of the sentinel variants, as evidenced from the PTS, is for the category of neoplasms in the EAs, with higher TL PTS associated with increased risk to the individual cancer phenotypes (11 of the 14 significant results after Bonferroni correction for 1,704 tested phecodes were cancer related; Figure S6B; Table S6); associations were only nominal in the BioVU AAs, likely due to lower power from the smaller sample size. Single variant PheWAS (Table S6) in the BioVU EAs are largely replicated within the UK Biobank (UKBB; Table S7), again showing strong associations with neoplasms, and in general, demonstrating the alleles that increased TL also increased risk for these cancer-related phenotypes. In addition, analyses of both the UKBB and BioVU data identified an association between the HSPA1A locus (rs1008438) and type 1 diabetes-related endocrine/metabolism phenotypes (BioVU p 1.2 × 10−8 to 4.1 × 10−28, UKBB p 6.7 × 10−6 to 2.6 × 10−27 for a range of phecodes grouped under 250.1); here, the allele decreasing TL increased risk for these phenotypes (BioVU odds ratios 1.4–2.1, UKBB odds ratios 1.4–2.2). This agrees with prior associations between shorter TL and increased risk of type 1 diabetes,40 and between the protein product of HSPA1A (Hsp72) and diabetic ketoacidosis.41
Discussion
Leveraging WGS available through the NHLBI TOPMed program, we have illustrated the value of a large, trans-population WGS study for a harmonized phenotype of broad interest, bioinformatically estimated TL, to identify new loci associated with TL. The well-powered study enabled identification of rare deleterious variants with estimated effect sizes larger than those of common variants. Using WGS allowed us the unique opportunity to hone in on causal variants using fine-mapping approaches for one locus, OBFC1, and begin to characterize tissue-specific genetic effects for this locus. We were also able to establish that for most population groups, effects are highly consistent at sentinel variants, despite differences in association strength at loci such as TINF2 and OBFC1, in which allele frequencies varied among populations.
One of the main limitations to the interpretation of human genetic studies of TL pertains to the heterogeneity and lack of standardization of various TL assays, and therefore comparability of results (including the genetic effect sizes) between studies42,43 (Note S1). To date, there is a paucity of data directly comparing WGS TL estimates with laboratory-based TL measurements in large-scale genetic epidemiologic studies.44 In the present study, we have performed an in-depth analysis of the robustness of our TelSeq-derived TL and the resulting GWAS statistics and report the following: (1) we confirm previous observations that TelSeq estimates are consistently shorter than Southern blot (mTRF), but that the 2 values are highly correlated44; (2) we demonstrate a high degree of shared heritability (i.e., genetic correlation) between TelSeq-derived and Southern blot-derived TL using phenotype-on-phenotype measures of heritability in the same subjects; (3) we see similarly high genetic correlation using GWAS summary statistic measures between qPCR- and TelSeq-derived TL GWASs in Europeans; (4) we show high correlation of effect sizes at sentinel variants between TelSeq- and qPCR-derived GWASs; and (5) we show that effect sizes from TelSeq were consistently ∼50% lower compared to Southern blot at the same variants measured on the same subjects mirroring the correlation patterns noted in the original phenotypes themselves.
Limitations of the study
The limitations of this study include the lack of datasets that include TL measurements in diverse populations for replication of the newly associated loci and genes; external studies are limited to largely European and Asian ancestry. For example, the identified effects at the ZMYM4 and P3H2 loci may be larger in Hispanic/Latino and African ancestry populations, respectively, than European. In both, the strength of the association, the effect sizes, and percent variation explained in the context of relative sample size in our data are larger in these non-European groups. Our lack of replication described here may be overcome with an ability to evaluate these loci in additional studies with greater population representation.
In addition, in this study, we evaluate statistical significance for association using p values that could result in anunequal ability to define significance thresholds across allele frequencies (lower allele frequencies need higher effect sizes, for example). Alternative approaches that consider effect sizes as a prioritization scheme could be applied in the future.
Finally, our fine-mapping approach based on tissue expression is limited for many of the associated loci, due to their lack of expression in GTEx tissues. Here, we followed up on the OBFC1 locus because the gene of interest is expressed in multiple adult tissues readily available in eQTL resources such as GTEx. In contrast, while TERT and TERC are important components of telomerase, they have low to undetectable expression in most GTEx tissue samples. As discussed by others, to adequately fine-map these loci, data on stem cell and/or developmental tissues will be important.45
While our TelSeq-based TL measurements and the resulting genetic effect sizes appear to be robust based on our comparison to laboratory-based assays (summarized above), caveats described (Note S1) necessitate attention when interpreting and comparing the results between large-scale TL genetic studies, especially from the perspective of clinical risk quantification. Nonetheless, the ability to implement sequence-based TL phenotype estimation in a large, trans-population WGS dataset creates opportunities to meaningfully expand our ability to evaluate the role of genes influencing TL in human health and disease, to dissect the genetic basis to TL differences across populations, and to set in place a model to leverage preexisting resources of WGS to bioinformatically quantify TL.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| TOPMed genetic variant calls from whole-genome sequencing data, by study | Taliun et al., 202146 | See Table S8 for dbGaP study phs IDs |
| TOPMed phenotype data, by study | Taliun et al., 202146 | See Table S8 for dbGaP study phs IDs |
| TOPMed batch-adjusted telomere length calls, by study | This paper | See Table S8 for dbGaP study phs IDs |
| TOPMed GWAS summary statistics for trans-population and population subgroup analyses | This paper | dbGaP: phs001974.v3.p1 |
| Software and algorithms | ||
| All original computer code | This paper | Zenodo: https://doi.org/10.5281/zenodo.5360775 |
| TelSeq | Ding et al., 201420 | https://github.com/zd1/telseq |
| HARE | Fang et al., 201924 | https://github.com/tanglab/HARE |
| GENESIS (R package) | Gogarten et al., 201947 | https://bioconductor.org/packages/release/bioc/html/GENESIS.html 10.18129/B9.bioc.GENESIS |
| CAVIAR | Hormozdiari et al., 201448 | http://genetics.cs.ucla.edu/caviar/download.html |
| coloc | Giambartolomei et al., 201449 | https://chr1swallace.github.io/coloc/articles/a01_intro.html |
| PheWAS (R package) | Carroll et al., 201450 | https://github.com/PheWAS/PheWAS |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rasika Mathias (rmathias@jhmi.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
TOPMed study populations
Our study involves human subjects only. To perform this genome-wide association study of telomere length, we leveraged the whole genome sequence samples available through the NHLBI Trans Omics for Precision Medicine (TOPMed) program. The program currently consists of more than 80 participating studies,51 with a range of study designs as described in Taliun et al.46 Participants are mainly U.S. residents with diverse ancestries (self-reported European, African, Hispanic/Latino, Asian, and Other). Smaller representation comes from non-US populations including Samoan, Brazilian, and Asian studies. Details on the specific samples included for telomere length analysis are outlined below, details on the population groupings using HARE (harmonized ancestry and race/ethnicity) are described in detail below, and final categories summarized in Table S1; additional information is also described by TOPMed.51 Counts of subjects by sex are also included in Table S1. While sex is included as a covariate in all relevant models in our analysis, we do not specifically investigate the effect of sex on telomere length in this work.
TOPMed whole-genome sequencing (WGS)
WGS was performed to an average depth of 38X using DNA isolated from blood, PCR-free library construction, and Illumina HiSeq X technology. Details for variant calling and quality control are described in Taliun et al.46 Briefly, variant discovery and genotype calling was performed jointly, across all the available TOPMed Freeze 8 studies, using the GotCloud52 pipeline resulting in a single multi-study genotype call set.
Method details
Estimating telomere length for WGS samples
A variety of computational tools exist that leverage WGS data to generate an estimate of telomere length.44 Here, we performed a thorough comparison of two leading methods for estimating telomere length from WGS data to choose the preferred scalable method for performing the estimation on all available samples from TOPMed. The first method, TelSeq,20 calculates an estimate of individual telomere length using counts of sequencing reads containing a fixed number of repeats of the telomeric nucleotide motif TTAGGG. Given that 98% of our data was sequenced using read lengths of 151 or 152 (as confirmed from the SEQ field in the analyzed CRAM files), we chose to use a repeat number of 12. These read counts are then normalized according to the number of reads in the individual WGS dataset with between 48% and 52% GC content to adjust for potential technical artifacts related to GC content. The second method, Computel53 uses an alignment-based method to realign all sequenced reads from an individual to a “telomeric reference sequence.” Reads aligning to this reference sequence are considered to be telomeric and are included in the estimate of telomere length. Because Computel performs a complete realignment, additional computational steps are involved compared to those needed for TelSeq.
To compare the results and scalability from these two methods, we first directly compared estimates obtained from TelSeq and Computel on 2,398 samples from the Jackson Heart Study (JHS) and found them to be highly correlated with one another (Pearson correlation r = 0.98, Figure S1A). We also compared computational time to generate the telomere length estimates on these samples and show that Computel is around ten times more time-consuming (Figure S1B). This is in part due to the fact that Computel requires CRAM-formatted files (as the WGS data are currently stored) to first be converted back to Fastq format (while TelSeq requires a CRAM to BAM conversion), but also due to the computationally expensive step of realignment to the telomeric reference genome that the Computel algorithm employs.
TelSeq generates an estimate of TL in bp similar to laboratory assays such as Southern blot21 and flowFISH;22 in contrast qPCR approaches are represented as T/S ratios.54,55 As a further comparison to orthogonally measured telomere length values, we used data on the same 2,398 samples from JHS with Southern blot21 telomere length estimates.56 For these samples, the Southern blot assay was performed on the same source DNA sample that was used to generate the WGS in TOPMed. The Pearson correlation values between the TelSeq and Computel estimates and the Southern blot estimates did not differ (r = 0.58 and 0.56 for TelSeq and Computel, respectively, Figure S1C). Based on our observation that both Computel and TelSeq showed similar correlation to the Southern blot estimates and high correlation with each other, and that TelSeq was an order of magnitude more computationally efficient, we chose to use TelSeq to perform telomere length estimation on our data. Final telomere length estimation was performed on a set of 128,901 samples whose CRAM-files were available for analysis at the TOPMed IRC at the time of analysis.
Batch adjustment to correct for confounders
To account for technical sources of variability in our telomere length estimates, both within a study (see, for example, colors in Figures S1A and S1C which indicate grouping by shared 96-well plate for shipment to the sequencing center) and across studies, we developed a method to estimate components of technical variability in our samples. We estimated these covariates using the sequencing data itself, similar to methods developed for other multivariate genomics data types (SVA or PEER factors57,58), using aligned sequencing reads and relying on the fact that genomic coverage patterns of aligned reads can reflect technical variation.
We computed average sequencing depth for every 1,000 bp genomic region (“bin”) genome-wide using mosdepth.59 We removed bins known to be problematic: those containing repetitive DNA sequence with difficulty mapping (mappability < 1.0 using 50bp k-mers in GEMTools v1.75960) or that overlap the list of known problematic SVs61 or overlap known CNVs in the Database of Genomic Variants. To avoid overcorrecting for sex, bins were limited to autosomes. After normalizing the approximately 150,000 remaining bin counts within sample, we performed Randomized Singular Value Decomposition62 (rSVD), a scalable alternative to principal components analysis, to generate batch principal components (bPCs). We included increasing numbers of bPCs in a linear regression model predicting TelSeq TL, and computed the correlation of the resulting residuals with external data measurements, including Southern blot measurements for JHS (n = 2,398) and the Women’s Health Initiative (WHI; n = 596) and age at blood draw (JHS n = 3,294; WHI n = 10,708). Based on the observed correlation, the final decision was to include the first 200 bPCs across all samples. Using the n = 2,398 JHS samples described above, we compared TL estimates before and after batch correction. The percent of variance in TL explained by sequencing plate reduced from 21.9% (baseline) to 10.5% (200 bPCs), and the variance explained by age increased from 8.0% (baseline) to 10.3% (200 bPCs), evidence that the signal-to-noise ratio was improved. Overall, the correlation between the bPC corrected TL and Southern blot data improved from r = 0.58 to 0.68 (Figure S1D) in the JHS data and from r = 0.54 to 0.72 for the WHI data. Further, we compared TelSeq estimates of 19 samples within a single sequencing batch from the GeneSTAR study to the clinical gold standard of flowFISH22 (Figure S1E) and observed a correlation of 0.80 in both granulocytes and lymphocytes. Therefore, our data show that we are able to reduce the sequencing artifacts stemming from batch variability to attain correlation of TelSeq to Southern blot similar to the correlation of TelSeq to flowFISH.
Samples included in genetic analysis
All samples with telomere length estimated from the WGS data from TOPMed Freeze 8 were considered for inclusion, provided they had consent that allowed for genetic analysis of telomere length. Only samples with sequencing read lengths of 151 or 152 base pairs and having age at blood draw data available were included. For the set of samples that were part of a duplicate pair/group (either part of the intended duplicates designed by TOPMed, or a duplicate identified across the studies through sample QC) only one sample from each duplicated pair/group was retained. The final counts and demographic summary statistics for subjects grouped by TOPMed study for all 54 studies included in our analysis are shown in Table S1.
While self-reported race (Asian, Black and White) and Hispanic ethnicity group (Central American, Costa Rican, Cuban, Dominican, Mexican, Puerto Rican, South American) data are available in TOPMed, these data have limitations for analysis that include individuals with missing information or non-specific responses (e.g., ‘other’ or ‘multiple’) and high variability in genetically inferred measures of ancestry among individuals with the same reported race/ethnicity. To overcome these limitations, we used a computational method called HARE (harmonized ancestry and race/ethnicity), a newly developed machine learning approach for jointly leveraging reported and genetic data in the definition of population strata for GWAS.24 HARE uses provided race/ethnicity labels and genetic ancestry principal component (PC) values to compute probability estimates for each individual’s membership in each race/ethnicity stratum. For our HARE analysis, we used provided race (Asian, Black, White) or Hispanic ethnicity group (Central American, Costa Rican, Cuban, Dominican, Mexican, Puerto Rican, South American) as input labels to define population strata, and we used 11 PCs computed with PC-AiR63 using 638,486 LD-pruned (r2 < 0.1) autosomal variants with minor allele frequency > 1% to represent genetic ancestry. Genetic outliers for population strata were identified as individuals for whom their maximum stratum probability was more than 5 times greater than their reported stratum probability. Stratum values for genetic outliers and individuals with missing or non-specific race/ethnicity were imputed as the stratum for which they had the highest membership probability.
Our primary analysis allowed for heterogeneous residual variance (see Primary single variant tests for association for details) among groups defined jointly by study and HARE-based population stratum assignment, with minor study-specific modifications to account for small strata. We required at least 30 individuals within a study-HARE grouping and collapsed individuals into merged HARE groups within a study as necessary to retain everyone for analysis. For our population-specific analyses, we used HARE assignment to stratify individuals into the following population groups: African (corresponding to the Black HARE stratum), Asian (Asian), European (White), and Hispanic/Latino (Central American, Costa Rican, Cuban, Dominican, Mexican, Puerto Rican, and South American). To better preserve genetic ancestry similarity among individuals in population-specific stratified analyses, we restricted to individuals for whom their HARE population stratum membership probability was at least 0.7; the population stratum counts in Table S1 reflect the counts in the stratified analyses, where individuals not meeting this criterion are labeled as “Other/Uncertain.”
Samoan individuals from the Samoan Adiposity Study and Brazilian individuals from the Reds-III Brazil study were excluded from the HARE analyses due to their unique ancestry in the TOPMed dataset; these studies were treated as their own population groups for analyses.
Quantification and statistical analysis
Primary single variant tests for association
Genome-wide tests for association were performed using the R Bioconductor package GENESIS.47 The primary analysis included all available trans-population TOPMed samples (n = 109,122). A secondary analysis was performed for all population groups with n > 5,000, which included European (n = 51,654), African (n = 29,260), Hispanic/Latino (n = 18,019) and Asian (n = 5,683) groups as defined above using HARE. Prior to genetic modeling, we generated residuals from a linear regression model on all 109,122 samples with 200 batch principal components (bPCs), as described above; for clarity we call these residuals below. For the pooled trans-population analysis, we used a fully adjusted two-stage model, as described in the next two bullets.64 For each population-specific analysis, the same approach was used, limited to samples within that population group.
Stage 1: We fit a linear mixed model (LMM) on n = 109,122 samples, using as the outcome; adjusting for age, sex, study, sequencing center, and 11 PC-AiR63 PCs of ancestry as fixed effect covariates; including a random effect with covariance matrix proportional to a sparse empirical kinship matrix computed with PC-Relate65 to account for genetic relatedness among samples; and allowing for heteroskedasticity of residual variance across study-HARE groups as defined above. The marginal residuals from this Stage 1 model were then inverse-normalized and rescaled by their original standard deviation. This rescaling restores values to the original trait scale, providing more meaningful effect size estimates from subsequent association tests.66
Stage 2: We fit a second LMM on all n = 109,122 samples, using the inverse-normalized and rescaled residuals from Stage 1 as the outcome; all other aspects of the model including fixed effects adjustment, random effects, and residual variance structure were identical to the model in Stage 1. This two-stage covariate adjustment has been shown to be most effective at controlling for false-positives and increasing statistical power in this setting.64 The output of this Stage 2 model was then used to perform both single variant and gene-based tests for association.
Single variant tests for association
We used the output of the two-stage LMM to perform score tests of association for each variant with minor allele count (MAC) 5 that passed TOPMed Informatics Research Center (IRC) at the University of Michigan quality filters46 and which had < 10% of samples with read depth < 10. Genotype effect size estimates and percent of variability explained (PVE) were approximated from the score test results.67
Significance, conditional analysis, locus definitions
A p value cutoff of 5x10−9 was used to determine genome-wide significance in the primary trans-ethnic analysis. We identified our set of independent significant variants (as reported in Table 1) through an iterative conditioning process within each chromosome. For a given chromosome, if at least one variant from the primary analysis crossed the genome-wide significance cutoff, this peak variant was included as an additional fixed-effect covariate in a new two-stage LMM (see Stages 1 and 2 described above), and score test results were examined to see if any remaining variants crossed the 5x10−9 threshold. If so, we performed a second round of conditioning, including both the original peak variant and the new conditional peak variant as fixed-effect covariates in another two-stage LMM; and so on, adding conditional peak variants for additional rounds (Table S2). For each chromosome, the conditioning procedure was completed when no additional variants crossed the genome-wide threshold (p < 5x10−9) on that chromosome. At each step, all variants passing the p < 5x10−9 threshold were examined in BRAVO68 to assess quality, and 334 variants were filtered out due to variant call quality issues. In the case where a current peak variant was flagged for quality, the next most significant variant, provided its p value was below the 5x10−9 cutoff, was considered the peak variant instead. Variants were grouped into loci based on physical distance and an examination of linkage disequilibrium (LD) patterns, and locus names were determined using a combination of previous literature, known telomere biology, and physical location.
Cumulative percent of variability explained (PVE)
Through the iterative conditional approach, we identified a total of 59 variants (Table 1) that met our genome-wide significance threshold of p < 5x10−9. The cumulative PVE values for this full set of 59 variants (4.35%), the set of 37 variants mapping to known loci (3.38%), and the set of 22 variants mapping to previously un-identified loci (0.96%, see Overlap with prior published GWAS below for definition of previously un-identified variants) were each estimated jointly using approximations from multi-parameter score tests. This joint PVE approximation is similar to the single variant PVE approximation described above, except that the set of variants is tested jointly, accounting for covariance among the estimated variant effect sizes. This approach avoids inadvertently double counting any partially shared signal among the set of identified variants.
Joint tests for association, cross-population heterogeneity
We then performed joint association analyses for the full multi-ethnic sample (n = 109,122), as well as each of the four population groups with n > 5000, to determine effect sizes and p values when all 59 variants were considered together. Using the inverse-normalized and rescaled residuals from the primary analysis Stage 1 LMM as the outcome, we fit a new Stage 2 LMM that was the same as described above, except with the additional inclusion of the genotypes for these 59 variants as additive genetic fixed effects. Given this joint modeling framework, the variant effect size estimates are all adjusted for one another. These estimates were used as input for calculation of a polygenic trait score used for the PheWAS described below. Finally, we tested for heterogeneity of effect sizes from these analyses among the population groups by adapting Cochran’s Q statistic and its p value,69 commonly used to test for effect heterogeneity in meta-analysis (Table 1). For each variant, the effect size estimates and standard errors from each population group analysis were used to calculate Q, and a Bonferroni threshold of 0.001 (0.05/59) was used to assess significance.
Overlap with prior published GWAS
For each of the 59 variants identified, we examined the linkage disequilibrium (LD) with previously reported sentinel variants from 17 published GWAS. Only sentinel variants with p < 5x10−8 in their published study were considered, which included a total of 56 variants (Table S3). If one of our variants had LD ≥ 0.7 with a published variant, it was labeled as a known variant/part of a known locus in Table 1. Within a locus, we then compared each independent variant to the prior GWAS reported sentinel variant. If they were identical, the variant was labeled as a known sentinel variant in Table 1. Additionally, locus names for the final set of independent variants were selected based on (i) prior GWAS study definition for known loci, and (ii) the specific gene annotation for each variant mapping directly to a gene for previously un-identified loci.
Replication of newly associated loci
To determine whether the loci newly associated in the current study are supported by findings from prior studies, we considered the two largest most recent studies of telomere genetics in European19 (Li et al., n = 78,592) and Asian18 (Dorajoo et al., n = 26,875) ancestry individuals. These studies both used telomere length as measured by qPCR. For all newly associated variants in Table 1, we pulled the effect size estimates, standard errors, and p values, where available (Figure S2A). These results were available in at least one of the two studies for 19 of our 22 previously un-identified variants, so we considered a p value cutoff of 0.05/19 = 0.0026 to be replicated, after multiple testing correction. We also labeled variants where at least one study reported p < 0.05 as suggestive. We compared effect sizes between the qPCR results and our TelSeq results, assessing correlation of all overlapping variants (n = 37 for Dorajoo et al.,18 n = 43 for Li et al.,19 Figure S2B).
Comparison of Southern blot and TelSeq effect sizes
Using the 2,398 samples from JHS with both TelSeq and Southern blot TL measurements, we used the same fully-adjusted two-stage LMM framework to perform tests for genetic association at the 49 of 59 sentinel variants with MAF 1% in this group. We calculated Pearson correlation between the estimated effect sizes and fit a linear regression to relate them to one another as an estimate of difference in effect size magnitude (Figure S2C). The Stage 1 models from our LMM framework also provided estimated effect sizes for the average change in Southern blot and TelSeq TL estimates in basepairs for a one-year difference in age at blood draw.
Genetic correlation of TelSeq with other TL estimates
We assessed genetic correlation of our TelSeq estimates with other TL estimates in two ways: (1) Using a subset of 1,083 of our 2,398 JHS samples with both Southern blot and TelSeq TL estimates who were either participants in the nested family cohort portion of JHS or unselected 1st or 2nd degree relatives from the remaining samples,70 we measured genetic correlation () of the Southern blot and TelSeq TL estimates using SOLAR23; and (2) We performed cross-trait LD Score regression using LDSC27,28 [ to estimate genetic correlation using genetic association summary statistics from our European ancestry group (n = 51,564) and summary statistics from the Li et al. study,19 which used qPCR to measure TL on 78,592 individuals of European ancestry. We used pre-computed LD scores from 1000 Genomes European data (downloaded from https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2), and used the SNP list from https://data.broadinstitute.org/alkesgroup/LDSCORE/w_hm3.snplist.bz2 to match up alleles across studies and LD scores. After preprocessing and SNP checks (using defaults from LDSC), we were left with a set of 1,171,171 SNPs for the LD Score regression analysis.
Gene-based coding variant tests - variant annotation
For its use in gene-based tests for association, annotation based variant filtering and GENCODE v29 gene model-based71 aggregation was performed using the TOPMed freeze 8 WGSA Google BigQuery-based variant annotation database on the BioData Catalyst powered by Seven Bridges platform (http://doi.org/10.5281/zenodo.3822858). The annotation database was built using variant annotations for TOPMed freeze 8 variants gathered by Whole Genome Sequence Annotator (WGSA) version v0.872 and formatted by WGSAParsr version 6.3.8 (https://github.com/UW-GAC/wgsaparsr). Variants were annotated as exonic, splicing, transcript ablation/amplification, ncRNA, UTR5, UTR3, intronic, upstream, downstream, or intergenic using Ensembl Variant effect predictor (VEP).73 Exonic variants were further annotated as frameshift insertion, frameshift deletion, frameshift block substitution, stop-gain, stop-loss, start-loss, non-frameshift insertion, non-frameshift deletion, non-frameshift block substitution, nonsynonymous variant, synonymous variant, or unknown. Additional scores used included REVEL,74 MCAP75 or CADD76 effect prediction algorithms.
Gene-based coding variant tests - tests for association
Gene-based association testing was performed on the pooled trans-population dataset (n = 109,122). To improve the power to identify rare variant associations in coding regions, we aggregated deleterious rare coding variants in all protein-coding genes and then tested for association with telomere length. To enrich for likely functional variants, only variants with a “deleterious” consequence for its corresponding gene or genes,77 were included. For each protein-coding gene, a set of rare coding variants (MAF < 0.01, including singletons where MAC = 1, restricted to variants which passed IRC quality filters46 and which had < 10% of samples with read depth < 10) was constructed, which was composed of all stop-gain, stop-loss, start-loss, transcript ablation, transcript amplification, splice acceptor variants, splice donor variants and frameshift variants, as well as the exonic missense variants that fulfilled one of these criteria: 1) REVEL score > 0.5, 2) predicted M_CAP value was “Damaging,” or 3) CADD PHRED-scaled score > 30. We applied the variant Set Mixed Model Association Test (SMMAT)78 as implemented in GENESIS, using the genesis_tests app on the Analysis Commons,79 with MAF based variant weights given by a beta-distribution with parameters of 1 and 25, as proposed by Wu et al.,80 and using the same two-stage LMM output as used in the primary single variant analysis. Only genes with a cumulative MAC ≥ 5 over all variants were evaluated, leaving a total of 27,558 genes, and significance was evaluated after a Bonferroni correction for multiple testing (p < 0.05 / 27,558 = 1.815x10−6) (Figure S3).
Next, we sought to determine the influence of each rare deleterious variant in each significant gene on the association signal. We iterated through the variants, removing one variant at a time (leave-one-out approach),81 and repeated the SMMAT analysis. If a variant made a large contribution to the original association signal, one would expect the signal to be significantly weakened with the removal of the variant from the set (Figure S3).
Finally, we further tested for independence of the gene-based and single variant signals by performing a conditional SMMAT analysis that included the 59 genome-wide significant variants from our primary analysis as fixed-effect covariates in the two-stage LMM. These 59 variants were also removed from the aggregated set of rare variants for a gene if they had been previously included (e.g., rs202187871 in POT1). All other analysis parameters were the same as described above (Figure S3).
Colocalization of OBFC1 signals using GTEx and eQTLGen
Iterative conditional analysis was repeated for chromosome 10 focusing on a 2Mb window centered on the primary signal near OBFC1 (rs10883948). The original pooled GWAS results (n = 109,122) were used for colocalization analysis with the primary signal while the appropriate round of conditional analysis was used for each subsequent signal (e.g., the output of the second round of conditional analysis was used for colocalization analysis with the tertiary signal). Credible set analysis was performed using CAVIAR on primary signal data and the output of each conditional analysis each with a single assumed causal variant.48 For each independent OBFC1 signal, the credible set contained the top sentinel variant (Figures S5A–S5D). Colocalization analysis was performed using coloc, a Bayesian posterior probability method that estimates the probability of shared signal across testing modalities at each variant.49 We report the posterior probability that the two signals are independent (PPH3) and the posterior probability that the two signals overlap (PPH4). The sentinel variants from each signal were assayed as expression quantitative trait loci (eQTLs) in both GTEx82 and eQTLGen38 datasets. For each sentinel, significant gene-tissue pairs for that sentinel were identified from GTEx v8 (FDR < 0.05) and assayed for colocalization comparing the beta and standard error of the beta from our GWAS results and the eQTL results. For colocalization analysis in the eQTLGen dataset, all eGenes within a 2Mb window of the sentinel were identified and assayed for colocalization comparing the MAF, p value, and number of observations. MAF was estimated for eQTLGen data using the TOPMed MAF. Colocalization analysis was not possible for the OBFC1 secondary signal as that variant is absent in both datasets and a representative proxy variant was not available. Roadmap39 data was accessed July, 2020 using the hg19 (February, 2009 release) UCSC genome browser83 track data hubs.84,85
Phenome-wide association tests (PheWAS)
Using individual level data within the Vanderbilt University biobank BioVU, PheWAS86 (tests for association between genotype and phenotype) were performed using the 49 (of 59) sentinel variants available in the multi-ethnic genotyping array (MEGA) chip results imputed to the Haplotype Reference Consortium.87 Single variant tests using SNP dosage values were performed for all available phecodes (number of cases at least 20), including the covariates age, sex, genotype batch and the first ten ancestry principal components. Analysis was performed separately in BioVU self-identified African Americans (AA, n = 15,174) and BioVU self-identified European Americans (EA, n = 70,439). In addition, European and African specific effect sizes from the joint analysis from Table 1 were combined to create separate polygenic trait scores (PTS) for each population group which were then tested for association with available phecodes, again including the covariates age, sex, genotype batch and the first ten ancestry principal components. Results were evaluated at a Bonferroni threshold corrected for the number of informative phecodes for each variant (range n = 1,114-1,361) or the PTS (n = 1,704) (Figure S6; Table S6). Analysis was performed using the PheWAS R package.50
We queried United Kingdom Biobank (UKBB) GWAS results using the University of Michigan PheWeb web interface (http://pheweb.sph.umich.edu/SAIGE-UKB/). The UKBB PheWeb interface contains results from a SAIGE88 genetic analysis of 1,403 ICD-based traits of 408,961 UKBB participants of European ancestry. PheWeb is a publicly accessible database that allows querying genome-wide association results for 28 million imputed genetic variants. 47 out of our 59 sentinel variants were present in PheWeb. We report all hits passing a Bonferroni correction for the number of tests performed for each variant (0.05/1403 = 3.6x10−5, Table S7).
Acknowledgments
We thank Chen Li, Claudia Langernberg, and Veryan Codd for providing summary statistics from their TL GWAS for replication analysis. The whole-genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung, and Blood Institute (NHLBI). Specific funding sources for each study and genomic center are given in the supplemental information. Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Phenotype harmonization, data management, sample-identity QC, and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The full study specific acknowledgments, as well as individual acknowledgments, are detailed in the supplemental information. The BioVU projects at Vanderbilt University Medical Center are supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH-funded Shared Instrumentation Grant S10OD017985 and S10RR025141 and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711. and additional funding sources listed at https://victr.vumc.org/biovu-funding/. Support for this work was provided by the National Institutes of Health, National Heart, Lung, and Blood Institute, through the BioData Catalyst program (awards 1OT3HL142479-01, 1OT3HL142478-01, 1OT3HL142481-01, 1OT3HL142480-01, and 1OT3HL147154). F.D.M. is supported by grants from NIH/NHLBI (HL139054, HL091889, HL132523, HL130045, HL098112, and HL056177), the NIH/NIEHS (ES006614), the NIH/NIAID (AI126614), and the NIH/Office of Director (OD023282). Vifor Pharmaceuticals provided medicine and additional funding to support recruitment for HL130045. P.T.E. is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular diseases. Any opinions expressed in this document are those of the authors and do not necessarily reflect the views of NHLBI, the National Institutes of Health, the US Department of Health and Human Services, individual BioData Catalyst Consortium members, or affiliated organizations and institutions.
Author contributions
M.A.T. and R.A.M. conceived of and led the study. M.A.T., M.P.C., R. Keener, K.R.I., L.R.Y., C.P.M., D.J., S.M.G., C.A.L., A.K., M. Arvanitis, J.C.B., E.G.B., J.C.C., Y.C.C., L.M.R., M.H.C., J.E.C., M.D., B.M.P., C.C.L., D.L., I.R., T.W.B., J.A.P., M. Armanios, A.B., A.P.R., and R.A.M. drafted the manuscript. M.A.T., M.P.C., R. Keener, J.S.W., J.A.B., D.J., A.K., C.C.L., G.A., D.A.N., J.G.W., S.S.R., D.L., I.R., A.A., T.W.B., T.T., J.O., N.J.C., J.A.P., M. Armanios, A.B., N.P., A.P.R., and R.A.M. contributed substantive analytical guidance. M.A.T., M.P.C., R. Keener, K.R.I., J.S.W., L.R.Y., J. Lane, T.W.M., J.A.B., C.P.M., D.J., S.M.G., C.A.L., A.K., M. Arvanitis, A.V.S., B.H., T.T., N.J.C., M. Armanios, A.B., N.P., A.P.R., and R.A.M. performed and led the analysis. L.R.Y., L.B., L.C.B., J.C.B., J.B., E.R.B., E.G.B., J.C.C., Y.C.C., B.C., D.D., L.d.l.F., D.L.D., B.I.F., M.E.G., M.T.G., S.R.H., B.A.H., M.R.I., T.I., W.C.J., S. Kaab, L.L., J. Lee, S.L., A.M., K.E.N., P.A.P., N.R., L.M.R., C.S., D.E.W., M.M.W., L.W., I.V.Y., W.Z., S.A., P.L.A., D.W.B., B.E.C., Z.C., M.H.C., L.A.C., J.E.C., M.D., R.D., C.E., T.E.F., X.G., L.H., S.H., J.M.J., E.E.K., A.M.L., C.L., R.L.M., T.N., M.N., M.S.R., E.C.S., J.A.S., N.L.S., J.L.S., M.J.T., H.K.T., R.P.T., M.J.W., Y.Z., K.L.W., S.T.W., R.S.V., K.D.T., M.F.S., E.K.S., M.B.S., W.H.-H.S., F.S., D.A.S., J.I.R., D.R., S.R., B.A.R., B.M.P., J.M.P., N.D.P., S.N., C.G.M., B.D.M., D.A.M., S.T.M., F.D.M., A.C.Y.M., R.J.F.L., R. Kumar, C.K., B.A.K., S. Kelly, S.L.R.K., R. Kaplan, J.H., H.G., F.D.G., B.D.G., M.F., P.T.E., M.d.A., A.C., Y.-D.I.C., E.B., K.C.B., A.E.A.-K., D.K.A., C.A., N.J.C., and M. Armanios were involved in the guidance, collection, and analysis of one or more of the studies that contributed data to this article. All of the authors read and approved the final draft.
Declaration of interests
J.C.C. has received research materials from GlaxoSmithKline and Merck (inhaled steroids) and Pharmavite (vitamin D and placebo capsules) to provide medications free of cost to participants in NIH-funded studies, unrelated to the current work. B.I.F. is a consultant for AstraZeneca Pharmaceuticals and RenalytixAI. L.K.W. is on the advisory board of GlaxoSmithKline and receives grant funding from NIAID, NHLBI, and NIDDK (NIH). I.V.Y. is a consultant for ElevenP15. S.A. receives equity and salary from 23andMe. M.H.C. receives grant support from GlaxoSmithKline. S.T.W. receives royalties from UpToDate. E.K.S. received grant support from GlaxoSmithKline and Bayer in the past 3 years. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. F.D.M. is a council member for the Council for the Developing Child. P.T.E. has served on the advisory boards of or consulted for Bayer AG, Quest Diagnostics, and Novartis. K.C.B. receives royalties from UpToDate. G.A. is an employee of Regeneron Pharmaceuticals and owns stock and stock options in Regeneron Pharmaceuticals. A.M. is an employee of Regeneron Pharmaceuticals and owns stock and stock options in Regeneron Pharmaceuticals. A.B. is a consultant for Third Rock Ventures, and holds stock in Google. D.A.S. is the founder and chief scientific officer of Eleven P15, a company focused on the early diagnosis and treatment of pulmonary fibrosis.
Published: January 12, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2021.100084.
Supplemental information
Data and code availability
TOPMed genomic data and pre-existing parent study phenotypic data are made available to the scientific community in study-specific accessions in the database of Genotypes and Phenotypes (dbGaP) (https://www.ncbi.nlm.nih.gov/gap/?term=TOPMed). Telomere length calls were derived from the raw sequence data as described in the Method details, and the phenotype covariates of age, sex, and ancestry are available through the study-specific dbGaP accession IDs as listed in the supplementary information, Table S8. This TOPMed work includes multiple studies, some of which are based on sensitive populations, precluding the unrestricted sharing of GWAS summary statistics. The TOPMed dbGaP accession (dbGaP: phs001974.v3.p1) provides a mechanism for sharing sensitive results, using the controlled-access mechanism of dbGaP to provide protections for sensitive populations. The full results from this GWAS for the full group, European, African, Asian and Hispanic/Latino subgroup analyses have all been deposited at phs001974.v3.p1 and are available publicly through dbGaP access.
All original code has been deposited at Zenodo (Zenodo: https://doi.org/10.5281/zenodo.5360775) and is publicly available.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Aviv A., Shay J.W. Reflections on telomere dynamics and ageing-related diseases in humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20160436. doi: 10.1098/rstb.2016.0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNally E.J., Luncsford P.J., Armanios M. Long telomeres and cancer risk: the price of cellular immortality. J. Clin. Invest. 2019;129:3474–3481. doi: 10.1172/JCI120851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Codd V., Nelson C.P., Albrecht E., Mangino M., Deelen J., Buxton J.L., Hottenga J.J., Fischer K., Esko T., Surakka I., et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 2013;45:422–427. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Codd V., Mangino M., van der Harst P., Braund P.S., Kaiser M., Beveridge A.J., Rafelt S., Moore J., Nelson C., Soranzo N., et al. Wellcome Trust Case Control Consortium Common variants near TERC are associated with mean telomere length. Nat. Genet. 2010;42:197–199. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delgado D.A., Zhang C., Chen L.S., Gao J., Roy S., Shinkle J., Sabarinathan M., Argos M., Tong L., Ahmed A., et al. Genome-wide association study of telomere length among South Asians identifies a second RTEL1 association signal. J. Med. Genet. 2018;55:64–71. doi: 10.1136/jmedgenet-2017-104922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu J., Chen M., Shete S., Amos C.I., Kamat A., Ye Y., Lin J., Dinney C.P., Wu X. A genome-wide association study identifies a locus on chromosome 14q21 as a predictor of leukocyte telomere length and as a marker of susceptibility for bladder cancer. Cancer Prev. Res. (Phila.) 2011;4:514–521. doi: 10.1158/1940-6207.CAPR-11-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.H., Cheng R., Honig L.S., Feitosa M., Kammerer C.M., Kang M.S., Schupf N., Lin S.J., Sanders J.L., Bae H., et al. Genome wide association and linkage analyses identified three loci-4q25, 17q23.2, and 10q11.21-associated with variation in leukocyte telomere length: the Long Life Family Study. Front. Genet. 2014;4:310. doi: 10.3389/fgene.2013.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D., Neuhausen S.L., Hunt S.C., Kimura M., Hwang S.J., Chen W., Bis J.C., Fitzpatrick A.L., Smith E., Johnson A.D., et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc. Natl. Acad. Sci. USA. 2010;107:9293–9298. doi: 10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Cao L., Li Z., Zhou D., Liu W., Shen Q., Wu Y., Zhang D., Hu X., Wang T., et al. A genome-wide association study identifies a locus on TERT for mean telomere length in Han Chinese. PLoS ONE. 2014;9:e85043. doi: 10.1371/journal.pone.0085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangino M., Christiansen L., Stone R., Hunt S.C., Horvath K., Eisenberg D.T., Kimura M., Petersen I., Kark J.D., Herbig U., et al. DCAF4, a novel gene associated with leucocyte telomere length. J. Med. Genet. 2015;52:157–162. doi: 10.1136/jmedgenet-2014-102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangino M., Hwang S.J., Spector T.D., Hunt S.C., Kimura M., Fitzpatrick A.L., Christiansen L., Petersen I., Elbers C.C., Harris T., et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 2012;21:5385–5394. doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangino M., Richards J.B., Soranzo N., Zhai G., Aviv A., Valdes A.M., Samani N.J., Deloukas P., Spector T.D. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J. Med. Genet. 2009;46:451–454. doi: 10.1136/jmg.2008.064956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pooley K.A., Bojesen S.E., Weischer M., Nielsen S.F., Thompson D., Amin Al Olama A., Michailidou K., Tyrer J.P., Benlloch S., Brown J., et al. A genome-wide association scan (GWAS) for mean telomere length within the COGS project: identified loci show little association with hormone-related cancer risk. Hum. Mol. Genet. 2013;22:5056–5064. doi: 10.1093/hmg/ddt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prescott J., Kraft P., Chasman D.I., Savage S.A., Mirabello L., Berndt S.I., Weissfeld J.L., Han J., Hayes R.B., Chanock S.J., et al. Genome-wide association study of relative telomere length. PLoS ONE. 2011;6:e19635. doi: 10.1371/journal.pone.0019635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxena R., Bjonnes A., Prescott J., Dib P., Natt P., Lane J., Lerner M., Cooper J.A., Ye Y., Li K.W., et al. Genome-wide association study identifies variants in casein kinase II (CSNK2A2) to be associated with leukocyte telomere length in a Punjabi Sikh diabetic cohort. Circ. Cardiovasc. Genet. 2014;7:287–295. doi: 10.1161/CIRCGENETICS.113.000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh K.M., Codd V., Smirnov I.V., Rice T., Decker P.A., Hansen H.M., Kollmeyer T., Kosel M.L., Molinaro A.M., McCoy L.S., et al. ENGAGE Consortium Telomere Group Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat. Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeiger A.M., White M.J., Eng C., Oh S.S., Witonsky J., Goddard P.C., Contreras M.G., Elhawary J.R., Hu D., Mak A.C.Y., et al. Genetic Determinants of Telomere Length in African American Youth. Sci. Rep. 2018;8:13265. doi: 10.1038/s41598-018-31238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorajoo R., Chang X., Gurung R.L., Li Z., Wang L., Wang R., Beckman K.B., Adams-Haduch J., M Y., Liu S., et al. Loci for human leukocyte telomere length in the Singaporean Chinese population and trans-ethnic genetic studies. Nat. Commun. 2019;10:2491. doi: 10.1038/s41467-019-10443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C., Stoma S., Lotta L.A., Warner S., Albrecht E., Allione A., Arp P.P., Broer L., Buxton J.L., Da Silva Couto Alves A., et al. Genome-wide Association Analysis in Humans Links Nucleotide Metabolism to Leukocyte Telomere Length. Am. J. Hum. Genet. 2020;106:389–404. doi: 10.1016/j.ajhg.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Z., Mangino M., Aviv A., Spector T., Durbin R., UK10K Consortium Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 2014;42:e75. doi: 10.1093/nar/gku181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M., Stone R.C., Hunt S.C., Skurnick J., Lu X., Cao X., Harley C.B., Aviv A. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protoc. 2010;5:1596–1607. doi: 10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
- 22.Alder J.K., Hanumanthu V.S., Strong M.A., DeZern A.E., Stanley S.E., Takemoto C.M., Danilova L., Applegate C.D., Bolton S.G., Mohr D.W., et al. Diagnostic utility of telomere length testing in a hospital-based setting. Proc. Natl. Acad. Sci. USA. 2018;115:E2358–E2365. doi: 10.1073/pnas.1720427115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almasy L., Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang H., Hui Q., Lynch J., Honerlaw J., Assimes T.L., Huang J., Vujkovic M., Damrauer S.M., Pyarajan S., Gaziano J.M., et al. VA Million Veteran Program Harmonizing Genetic Ancestry and Self-identified Race/Ethnicity in Genome-wide Association Studies. Am. J. Hum. Genet. 2019;105:763–772. doi: 10.1016/j.ajhg.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M., Wang R., Wang Y., Diao F., Lu F., Gao D., Chen D., Zhai Z., Shu H. The CXXC finger 5 protein is required for DNA damage-induced p53 activation. Sci. China C Life Sci. 2009;52:528–538. doi: 10.1007/s11427-009-0083-7. [DOI] [PubMed] [Google Scholar]
- 26.Kaul R., Mukherjee S., Ahmed F., Bhat M.K., Chhipa R., Galande S., Chattopadhyay S. Direct interaction with and activation of p53 by SMAR1 retards cell-cycle progression at G2/M phase and delays tumor growth in mice. Int. J. Cancer. 2003;103:606–615. doi: 10.1002/ijc.10881. [DOI] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R., Patterson N., Robinson E.B., et al. ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cochran W.G. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 30.Stuart B.D., Choi J., Zaidi S., Xing C., Holohan B., Chen R., Choi M., Dharwadkar P., Torres F., Girod C.E., et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet. 2015;47:512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tummala H., Walne A., Collopy L., Cardoso S., de la Fuente J., Lawson S., Powell J., Cooper N., Foster A., Mohammed S., et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 2015;125:2151–2160. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Touzot F., Callebaut I., Soulier J., Gaillard L., Azerrad C., Durandy A., Fischer A., de Villartay J.P., Revy P. Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA. 2010;107:10097–10102. doi: 10.1073/pnas.0914918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Overbeek M., de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Lenain C., Bauwens S., Amiard S., Brunori M., Giraud-Panis M.J., Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 35.Wu M., Reuter M., Lilie H., Liu Y., Wahle E., Song H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart J.A., Wang Y., Ackerson S.M., Schuck P.L. Emerging roles of CST in maintaining genome stability and human disease. Front. Biosci. 2018;23:1564–1586. doi: 10.2741/4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods Groups—Analysis Working Group. Enhancing GTEx (eGTEx) Groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration & Visualization—EBI. Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California, Santa Cruz. Lead Analysts. Laboratory, Data Analysis & Coordinating Center (LDACC) NIH Program Management. Biospecimen Collection. Pathology. eQTL Manuscript Working Group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 38.Vosa U., Claringbould A., Westra H.-J., Bonder M.J., Deelen P., Zeng B., Kirsten H., Saha A., Kreuzhuber R., Yazar S., et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. Nat. Genet. 2021;53:1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Januszewski A.S., Sutanto S.S., McLennan S., O’Neal D.N., Keech A.C., Twigg S.M., Jenkins A.J. Shorter telomeres in adults with type 1 diabetes correlate with diabetes duration, but only weakly with vascular function and risk factors. Diabetes Res. Clin. Pract. 2016;117:4–11. doi: 10.1016/j.diabres.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 41.Oglesbee M.J., Herdman A.V., Passmore G.G., Hoffman W.H. Diabetic ketoacidosis increases extracellular levels of the major inducible 70-kDa heat shock protein. Clin. Biochem. 2005;38:900–904. doi: 10.1016/j.clinbiochem.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Nussey D.H., Baird D., Barrett E., Boner W., Fairlie J., Gemmell N., Hartmann N., Horn T., Haussmann M., Olsson M., et al. Measuring telomere length and telomere dynamics in evolutionary biology and ecology. Methods Ecol. Evol. 2014;5:299–310. doi: 10.1111/2041-210X.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aubert G., Hills M., Lansdorp P.M. Telomere length measurement-caveats and a critical assessment of the available technologies and tools. Mutat. Res. 2012;730:59–67. doi: 10.1016/j.mrfmmm.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee M., Napier C.E., Yang S.F., Arthur J.W., Reddel R.R., Pickett H.A. Comparative analysis of whole genome sequencing-based telomere length measurement techniques. Methods. 2017;114:4–15. doi: 10.1016/j.ymeth.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Demanelis K., Jasmine F., Chen L.S., Chernoff M., Tong L., Delgado D., Zhang C., Shinkle J., Sabarinathan M., Lin H., et al. GTEx Consortium Determinants of telomere length across human tissues. Science. 2020;369:eaaz6876. doi: 10.1126/science.aaz6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gogarten S.M., Sofer T., Chen H., Yu C., Brody J.A., Thornton T.A., Rice K.M., Conomos M.P. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35:5346–5348. doi: 10.1093/bioinformatics/btz567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hormozdiari F., Kostem E., Kang E.Y., Pasaniuc B., Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carroll R.J., Bastarache L., Denny J.C. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.NHLBI Trans-Omics for Precision Medicine . 2021. TOPMed Projects and Their Parent Studies.https://www.nhlbiwgs.org/group/project-studies [Google Scholar]
- 52.Jun G., Wing M.K., Abecasis G.R., Kang H.M. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nersisyan L., Arakelyan A. Computel: computation of mean telomere length from whole-genome next-generation sequencing data. PLoS ONE. 2015;10:e0125201. doi: 10.1371/journal.pone.0125201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aviv A., Hunt S.C., Lin J., Cao X., Kimura M., Blackburn E. Impartial comparative analysis of measurement of leukocyte telomere length/DNA content by Southern blots and qPCR. Nucleic Acids Res. 2011;39:e134. doi: 10.1093/nar/gkr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Callaghan N.J., Fenech M. A quantitative PCR method for measuring absolute telomere length. Biol. Proced. Online. 2011;13:3. doi: 10.1186/1480-9222-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mwasongwe S., Gao Y., Griswold M., Wilson J.G., Aviv A., Reiner A.P., Raffield L.M. Leukocyte telomere length and cardiovascular disease in African Americans: the Jackson Heart Study. Atherosclerosis. 2017;266:41–47. doi: 10.1016/j.atherosclerosis.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pedersen B.S., Quinlan A.R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Derrien T., Estellé J., Marco Sola S., Knowles D.G., Raineri E., Guigó R., Ribeca P. Fast computation and applications of genome mappability. PLoS ONE. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.sv_blacklist.bed. http://cf.10xgenomics.com/supp/genome/GRCh38/sv_blacklist.bed.
- 62.Halko N., Martinsson P.G., Tropp J.A. Finding Structure with Randomness: Probabilistic Algorithms for Constructing Approximate Matrix Decompositions. SIAM Rev. 2011;2:217–288. [Google Scholar]
- 63.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sofer T., Zheng X., Gogarten S.M., Laurie C.A., Grinde K., Shaffer J.R., Shungin D., O’Connell J.R., Durazo-Arvizo R.A., Raffield L., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium A fully adjusted two-stage procedure for rank-normalization in genetic association studies. Genet. Epidemiol. 2019;43:263–275. doi: 10.1002/gepi.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free Estimation of Recent Genetic Relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Z.Z., Lin D.Y. Meta-analysis for Discovering Rare-Variant Associations: Statistical Methods and Software Programs. Am. J. Hum. Genet. 2015;97:35–53. doi: 10.1016/j.ajhg.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou B., Shi J., Whittemore A.S. Optimal methods for meta-analysis of genome-wide association studies. Genet. Epidemiol. 2011;35:581–591. doi: 10.1002/gepi.20603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.The NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program . 2018. BRAVO variant browser: University of Michigan and NHLBI.https://bravo.sph.umich.edu/freeze5/hg38/ [Google Scholar]
- 69.Cochran W.G. The Combination of Estimates from Different Experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 70.Wilson J.G., Rotimi C.N., Ekunwe L., Royal C.D., Crump M.E., Wyatt S.B., Steffes M.W., Adeyemo A., Zhou J., Taylor H.A., Jr., et al. Study design for genetic analysis in the Jackson Heart Study. Ethn. Dis. 2005;15 S6-30-37. [PubMed] [Google Scholar]
- 71.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X., White S., Peng B., Johnson A.D., Brody J.A., Li A.H., Huang Z., Carroll A., Wei P., Gibbs R., et al. WGSA: an annotation pipeline for human genome sequencing studies. J. Med. Genet. 2016;53:111–112. doi: 10.1136/jmedgenet-2015-103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahn D.H., Ozer H.G., Hancioglu B., Lesinski G.B., Timmers C., Bekaii-Saab T. Whole-exome tumor sequencing study in biliary cancer patients with a response to MEK inhibitors. Oncotarget. 2016;7:5306–5312. doi: 10.18632/oncotarget.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 76.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Graham G. Disparities in cardiovascular disease risk in the United States. Curr. Cardiol. Rev. 2015;11:238–245. doi: 10.2174/1573403X11666141122220003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen H., Huffman J.E., Brody J.A., Wang C., Lee S., Li Z., Gogarten S.M., Sofer T., Bielak L.F., Bis J.C., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. TOPMed Hematology and Hemostasis Working Group Efficient Variant Set Mixed Model Association Tests for Continuous and Binary Traits in Large-Scale Whole-Genome Sequencing Studies. Am. J. Hum. Genet. 2019;104:260–274. doi: 10.1016/j.ajhg.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brody J.A., Morrison A.C., Bis J.C., O’Connell J.R., Brown M.R., Huffman J.E., Ames D.C., Carroll A., Conomos M.P., Gabriel S., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. TOPMed Hematology and Hemostasis Working Group. CHARGE Analysis and Bioinformatics Working Group Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat. Genet. 2017;49:1560–1563. doi: 10.1038/ng.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keramati A.R., Yanek L.R., Iyer K., Taub M.A., Ruczinski I., Becker D.M., Becker L.C., Faraday N., Mathias R.A. Targeted deep sequencing of the PEAR1 locus for platelet aggregation in European and African American families. Platelets. 2019;30:380–386. doi: 10.1080/09537104.2018.1447659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li D., Hsu S., Purushotham D., Sears R.L., Wang T. WashU Epigenome Browser update 2019. Nucleic Acids Res. 2019;47(W1):W158–W165. doi: 10.1093/nar/gkz348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D., Kent W.J. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dey R., Schmidt E.M., Abecasis G.R., Lee S. A Fast and Accurate Algorithm to Test for Binary Phenotypes and Its Application to PheWAS. Am. J. Hum. Genet. 2017;101:37–49. doi: 10.1016/j.ajhg.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TOPMed genomic data and pre-existing parent study phenotypic data are made available to the scientific community in study-specific accessions in the database of Genotypes and Phenotypes (dbGaP) (https://www.ncbi.nlm.nih.gov/gap/?term=TOPMed). Telomere length calls were derived from the raw sequence data as described in the Method details, and the phenotype covariates of age, sex, and ancestry are available through the study-specific dbGaP accession IDs as listed in the supplementary information, Table S8. This TOPMed work includes multiple studies, some of which are based on sensitive populations, precluding the unrestricted sharing of GWAS summary statistics. The TOPMed dbGaP accession (dbGaP: phs001974.v3.p1) provides a mechanism for sharing sensitive results, using the controlled-access mechanism of dbGaP to provide protections for sensitive populations. The full results from this GWAS for the full group, European, African, Asian and Hispanic/Latino subgroup analyses have all been deposited at phs001974.v3.p1 and are available publicly through dbGaP access.
All original code has been deposited at Zenodo (Zenodo: https://doi.org/10.5281/zenodo.5360775) and is publicly available.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.