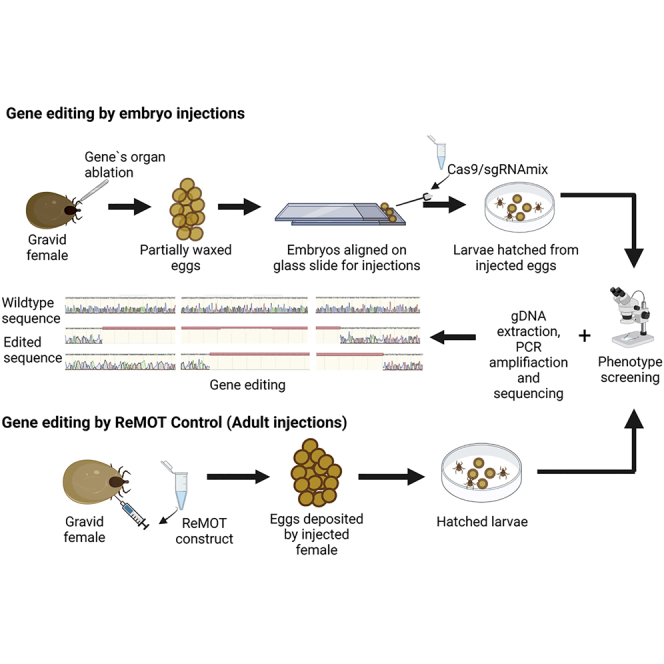
Summary
Despite their capacity to acquire and pass on an array of debilitating pathogens, research on ticks has lagged behind other arthropod vectors, such as mosquitoes, largely because of challenges in applying available genetic and molecular tools. CRISPR-Cas9 is transforming non-model organism research; however, successful gene editing has not yet been reported in ticks. Technical challenges for injecting tick embryos to attempt gene editing have further slowed research progress. Currently, no embryo injection protocol exists for any chelicerate species, including ticks. Herein, we report a successful embryo injection protocol for the black-legged tick, Ixodes scapularis, and the use of this protocol for genome editing with CRISPR-Cas9. We also demonstrate that the ReMOT Control technique could be successfully used to generate genome mutations outside Insecta. Our results provide innovative tools to the tick research community that are essential for advancing our understanding of the molecular mechanisms governing pathogen transmission by tick vectors and the underlying biology of host-vector-pathogen interactions.
Subject areas: Biological sciences, Biotechnology, Genetic engineering, Biological sciences research methodologies, Biology experimental methods
Graphical abstract
Highlights
-
•
Embryo injection protocol developed for Ixodes scapularis
-
•
CRISPR/Cas9-mediated gene editing is feasible in ticks
-
•
Tick gene editing is possible with embryo injection and adult injection (ReMOT Control)
Biological sciences; Biotechnology; Genetic engineering; Biological sciences research methodologies; Biology experimental methods
Introduction
Ticks are obligate hematophagous ectoparasites and are important vectors of a wide variety of pathogens to humans, wildlife, and domestic animals. The black-legged tick, Ixodes scapularis, is the most important vector of public health importance in the United States because it can transmit multiple pathogens, including bacteria, viruses, and protozoa (Hinckley et al., 2014). Lyme disease (LD), caused by the spirochete Borrelia burgdorferi and transmitted by Ixodes ticks, is the most prevalent vector-borne disease in the United States. Over 42,000 human LD cases were reported by the Centers for Disease Control and Prevention (Centers for Disease control and Prevention, 2015) in 2015. However, actual cases predicted by seroprevalence are estimated to be over 300,000 annually due to unrecorded cases (Rodino et al., 2020). In 2016, a 2.1 Gbp I. scapularis genome was published, the first genome for a tick species (Gulia-Nuss et al., 2016). The availability of a genome has opened the avenue for further molecular research in this species to understand critical vector-pathogen-host interactions. Despite this advance, our knowledge of the biology of ticks on a molecular level is still limited. This is in contrast to insects such as mosquitoes for which numerous tools for transgenic development and genome editing are now available. Advances in tick genomics and genetics have mainly been stymied by challenges in applying available molecular tools to the unique biology of ticks to carry out reverse genetics that can directly validate gene functions and correlate with phenotypes of interest. Progress in this area is critical for the advancement of research to solve the growing problem of tick-borne diseases.
RNAi (RNAi) is currently the only technique available for functional genomics studies in ticks. However, this technique has significant limitations and has provided only a brief insight into our understanding of these organisms (Nuss et al., 2021; Karim and Adamson, 2012; de la Fuente et al., 2007). The clustered regularly interspaced short palindromic repeats (CRISPR), CRISPR-associated protein 9 (Cas9), has revolutionized functional genetics research in many organisms (Zhang and Reed, 2017). Successful CRISPR-Cas9-mediated gene manipulation has been reported for a steadily increasing number of organisms in the arthropod subphyla Crustacea (Gui et al., 2016; Martin et al., 2016; Nakanishi et al., 2014) and Hexapoda, including Diptera (Bassett et al., 2013; Kistler et al., 2015; Macias et al., 2020; Paulo et al., 2019), Hymenoptera (Hu et al., 2019), Hemiptera (Kotwica-Rolinska et al., 2019; Zhao et al., 2019), Coleoptera (Awata et al., 2015; Gilles et al., 2015), Orthoptera (Li et al., 2017b), and diverse Lepidoptera (Sun et al., 2017).While CRISPR-Cas9-based gene-editing techniques have improved studies in mosquitoes and other arthropod disease vectors (Chaverra-Rodriguez et al., 2020; Dermauw et al., 2020; Sun et al., 2017), this technique has not yet been developed for tick vectors.
There are two components of the CRISPR-Cas9 technique: the single guide RNA (sgRNA) with a targeting sequence complementary to the sequence of the gene of interest, and a bacterial nuclease (Cas9). The Cas9 introduces double-stranded breaks in DNA at a complementary site specific to the sgRNA. In a eukaryotic cell, these double-stranded breaks (DSB) are then repaired by DSB repair mechanisms of the cell. Two major mechanisms: non-homologous end-joining (NHEJ) and homology-directed repair (HDR) have been well studied in eukaryotes. However, the alternative mechanisms, such as microhomology-mediated end joining (MMEJ) and single-strand annealing have also been shown to play a major role in DSB repair (Lieber, 2010; van Overbeek et al., 2016). To facilitate efficient genome editing in whole organisms, Cas9 and sgRNA need to be delivered to the nucleus of the oocytes (Gantz and Akbari, 2018). Therefore, most current approaches for genome editing in multicellular eukaryotes rely upon delivering the Cas9 ribonucleoprotein (RNP) complex (Cas9 protein + sgRNA) by embryonic microinjection. However, within the chelicerates, successful embryo injection has not yet been accomplished (Dermauw et al., 2020).
Tick embryos are extremely difficult to inject (Santos et al., 2013) due to multiple factors: high intra-ovular pressure, a hard chorion, and a wax layer outside the embryo that must be removed before injection. Female ticks coat their eggs with a tough wax layer using a specialized wax organ known as the Gené’s organ. The Gené’s organ is a complex dermal gland that develops in synchrony with oogenesis and oviposition. The external appendage, the horns, is an eversible balloon-like cuticular sac that manipulates the eggs and coats them in wax (Booth, 1989). This wax layer makes injection of the embryo very difficult due to the inability of a glass needle to pierce the hardened wax. Therefore, development of a successful embryo injection protocol is of critical importance. Recently, an alternative method that avoids the requirement to inject embryos has been developed, termed Receptor-Mediated Ovary Transduction of Cargo (ReMOT Control) which delivers the Cas9 RNP complex directly to the developing arthropod germline from the hemocoel, allowing targeted and heritable mutations to be generated by adult injection instead of embryo microinjection. This approach has already proven successful in several insects (Chaverra-Rodriguez et al., 2018,2020; Heu et al., 2020; Macias et al., 2020; Shirai and Daimon, 2020; Li et al., 2021), but has not yet been tested in arthropods outside Insecta.
Typically, projects involved in developing new CRISPR protocols target genes with evident phenotypes, permitting rapid screening of successful genome editing (Bassett et al., 2013; Li et al., 2017a,2020; Paulo et al., 2019). Some, such as the ortholog of the Drosophila white or scarlet genes (Bai et al., 2019; Ismail et al., 2018; Khan et al., 2017; Xue et al., 2018), or similar eye pigment genes, have been used as CRISPR-Cas9 targets for establishing proof-of-principle in other arthropods (Dermauw et al., 2020), but the absence of eyes in I. scapularis renders these genes useless for screening (Pham et al., 2021). Therefore, appendage genes, such as Proboscipedia (ProbP), that produced phenotypes in Drosophila melanogaster genome, may provide more appropriate morphological phenotypes for screening in I. scapularis. Additionally, the enzyme chitinase is without a clear screening phenotype yet is of research interest for its chitin-hydrolyzing capability and potential roles in cuticle remodeling and regulating chitin content in the peritrophic matrix lining of the midgut after a blood meal (You et al., 2003).
Here, we report a successful tick embryo injection protocol and targeted gene disruption with CRISPR-Cas9 using two methods: embryo injection and ReMOT Control. Our data show the feasibility of tick embryo injection and genetic manipulation in ticks by both methods. Our work also suggests that ticks might not efficiently use NHEJ for DSB repair, and in future work, we will focus on understanding the repair mechanisms in ticks. We expect that the availability of the I. scapularis genome along with tools that we developed here will open new research avenues that will dramatically accelerate our understanding of the molecular biology of this and related tick species. Targeted disruption of genes in tick vectors of human pathogens is a powerful method to uncover the underlying biology of tick-pathogen-host interactions that can inform the development and application of new approaches to tick-borne disease control.
Results
Ixodes scapularis embryo injection protocol
To eliminate the deposition of wax on eggs, the Gené’s organ of gravid I. scapularis was ablated prior to oviposition. Ablation of the Gené’s organ prevented wax deposition on the eggs but did not negatively impact tick survival or oviposition. Ticks resumed oviposition within 24 h after Gené’s organ removal. Approximately 14- to 18-h-old eggs were collected for injections. This time point was selected based on our calculations of early embryology events. In insects, the first 10% of embryo development time is well suited for injections (1–2 h in insects that hatch around 24 h post oviposition). Embryo development and larval hatching take ∼30 days in the I. scapularis reared under our laboratory conditions; therefore, 48–72 h post egg-laying was estimated to be an ideal window. The un/partially waxed eggs were treated with benzalkonium chloride and NaCl for removal of chorion and egg desiccation to decrease the intraoval pressure (Figures S1 and S2A). We designed a slide platform by adhering two microscope slides in a way that created a backstop to keep the spherical eggs from rolling off during injection (Figure S2B). We first used food color or dye for embryo injection. The dye injections demonstrated successful needle penetration and also ensured that no reflux occurred (dye can be easily seen in the embryo) (Figures S2C and S2D). Expression of dsRed from the injected CMV/CAAGS-dsRed promoter (Kurtti et al., 2008) plasmid construct was determined by fluorescence under a TRITC filter (Figure S2E) which further supported successful embryo injection. On average, ∼7% of the injected embryos hatched (Table 1). Treated but un-injected embryos had better survival (∼60%–70% hatching), suggesting that further refinement in the injection protocol might increase hatch rates.
Table 1.
Summary of gene editing by embryo injection and ReMOT Control in Ixodes scapularis
| Construct | Cas9/sgRNA concentration (ng/ul) | Total injections | Hatched larvae (% of total) | No. of larvae sequenced (% larvae sequenced) | No. edited larvae (% editing efficiency) |
|---|---|---|---|---|---|
|
Proboscipedia ISCW021086 |
300/120a | 865 | 35 (4.0%) | 35 (100%) | 0 |
| 150/60 | 600 | 61 (10.2%) | 57 (93.4%) | 0 | |
| 300/120 | 950 | 113 (11.9%) | 112 (99.1%) | 2 (1.8%) | |
| 600/240 | 240 | 24 (10%) | 24 (100%) | 0 | |
| 800/500 | 480 | 36 (7.5%) | 35 (97.2%) | 0 | |
|
Chitinase ISCW003950 |
300/120 |
2460 |
139 (5.6%) |
60 (43.1%) |
40 (66.6%) |
|
ReMOT Control |
Saponin concentration (μM) |
Total Eggs |
Hatched larvae (% of total) |
No. of larvae sequenced (% larvae sequenced) |
No. edited larvae (% editing efficiency) |
|
bProboscipedia ISCW021086 |
24 | 176 (N = 1) | 168 (95.4%) | 168 (100%) | 7 (4.2%) |
|
cProboscipedia ISCW021086 |
36 | 773 (N = 4) | 230 (30%) | 230 (100%) | 4 (1.7%) |
Edited larvae were detected by ICE analysis and confirmed by manual analysis of sequences.
Synthetic modified sgRNA; N = number of females used for ReMOT Control.
Abdomen injections.
Spiracle injections.
sgRNA-Cas9 embryo injections and transformation
The gene Proboscipedia (ProbP) was selected because of its expression in the legs in the spider Cupiennius salei (Schwager et al., 2007) and high sequence similarity in ticks. In D. melanogaster, ProbP mutants had either completely transformed proboscises to legs or severely shortened maxillary palps (Tayyab et al., 2004). Considering that ProbP is one of the Hox genes that is not required for embryonic development in other organisms, we predicted that mutation in IsProbP would not be embryonic lethal and might lead to a phenotype similar to D. melanogaster. Another gene, Chitinase, was selected due to its importance during molting and porosity of the peritrophic membrane in Haemaphysalis longicornis making it a potential candidate for tick control (You et al., 2003). Isolation of mutant tick lines was not attempted in this study because the generation time for ticks is up to 7 months or longer in the laboratory (Kocan et al., 2015). G0 larvae sequenced using the Sanger sequencing platform was first analyzed by Synthego Inference of CRISPR Edits (ICE) analysis (Hsiau et al., 2018). Although useful for detecting small edits, no large deletions were determined by ICE, and poor sequence quality was frequently flagged as mutations. Manual annotation was required to detect large deletions and confirm mutated individuals (Table 1).
ProbP editing
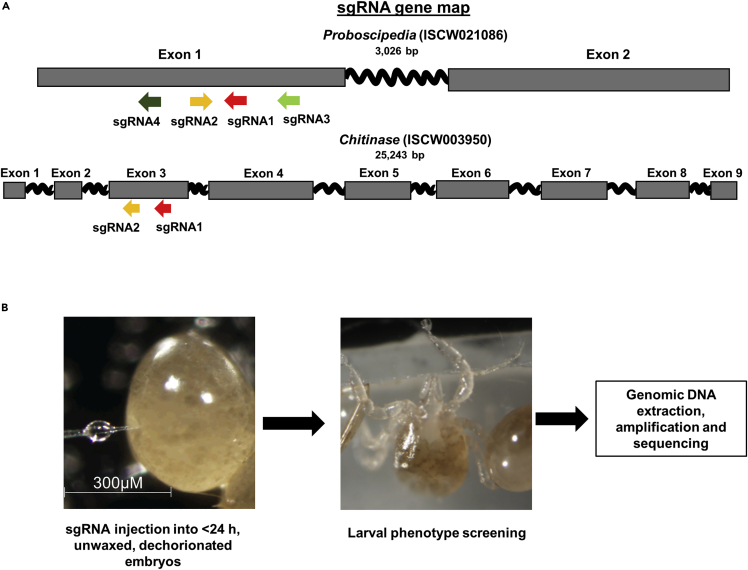
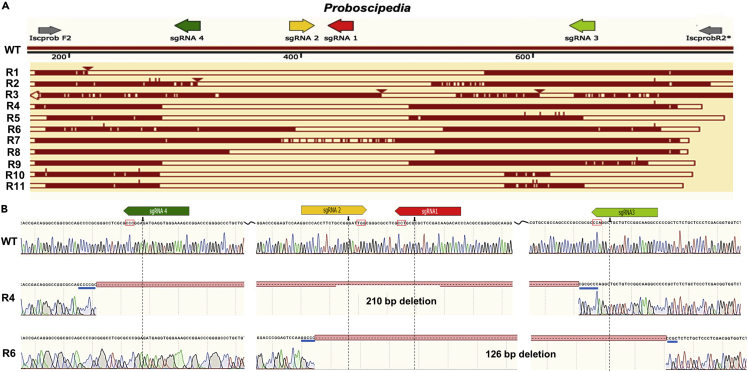
Four sgRNAs (sgRNA 1–4) were synthesized by in vitro transcription (IVT) in our laboratory at UNR, and three modified sgRNAs were synthesized commercially by Integrated DNA Technologies (IDT; sgRNAs 5–7) (Figure 1A and Table S1).
Figure 1.
Gene editing by embryo injections
(A) Gene maps showing exons (gray boxes) and introns (wavy lines) and designed sgRNAs targeting the first exon for Proboscipedia (ISCW021086), and the third exon for Chitinase (ISCW003950) genes used for embryo injections in I. scapularis.
(B) Workflow schematic for embryo injections against target genes.
Approximately 2,300 embryos from multiple tick cohorts were injected with Cas9 and multiplexed IVT sgRNAs with varying concentrations of Cas9/sgRNA (Figure1B). Out of these, 7%–12% of eggs hatched. Two larvae (1.8%) were edited at concentrations 300/120 ng/μL of Cas9 and sgRNA; however, no other concentration resulted in clear editing (Table 1). Modified sgRNAs (IDT) were multiplexed and used at a single concentration of 300/120 ng/μL of Cas9 and sgRNA. Out of 865 total injections with IDT modified sgRNAs, 4% of injected embryos hatched, and none were edited (Table 1).
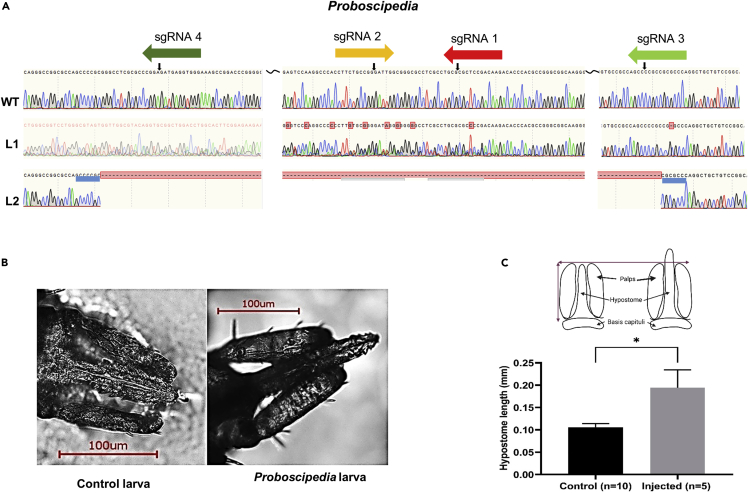
The edited larvae had long deletions [Larva 1(L1) and L2] corresponding to cuts with sgRNAs 1, 2, and 4 (Figure 2A). L1 is heterozygous with a deletion of indefinite length. The L1 sequence alignment in NCBI BLAST suggests an inversion near sgRNA 2 (Figure S3). When we BLAST searched the Sanger sequence of L1, it matched 1–353 bp with the IsProbP sequence on plus/plus orientation while another match was suggested from 353 to 562 bp on the plus/minus strand of ProbP (Figures S3A–S3C). Another larva with long deletion (L2) had a homozygous in-frame deletion (Figure 2A) and the deleted sequence was flanked by a short GC-rich sequence (Figure 2A). A significantly longer hypostome was observed in five larvae (all from the same cohort and injected with 300/120 ng/μL of Cas9 and sgRNA) (Figures 2B and2C). Out of these, only L1 had confirmed edits (Figures 2A and 2B). A separate biological cohort was injected with sgRNA 1–4 at 300/120 ng/μL, and one-week post injections eggs were used for Illumina Miseq, resulting in <1% editing (Figures S4 and Table S3). We did not see editing corresponding to sgRNA3 in our Sanger sequencing data; however, pooled deep sequencing data suggested some editing by sgRNA3. This discrepancy could be due to sequencing embryos one week after injection (deep sequencing) versus larvae hatched from the injected eggs. It is also possible that all sgRNA activity was not captured due to technical issues such as DNA quality, sequencing quality, or mortality of eggs before hatching.
Figure 2.
Sequences of Proboscipedia G0 larvae edited by embryo injection aligned to wildtype (WT)
(A) Sequences and chromatograms of wildtype (WT), heterozygous deletion (L1), and homozygous deletion (L2). GC-rich regions flanking the homozygous deletions are underlined in blue. Black squiggly lines represent an additional deleted sequence in that region.
(B) Control and mutant larva (L1) showing extended hypostome phenotype.
(C) Illustration of tick mouthparts and measurements (top), a graph showing hypostome length in wildtype (control) and injected larvae (bottom).Data are presented as mean ±SEM. An unpaired Student’s t test was used to calculate significance ∗ = p> 0.05
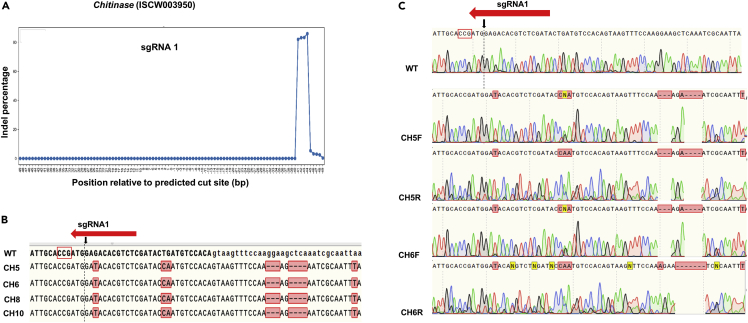
Chitinase editing
Two sgRNAs were designed targeting the third exon of Chitinase and were synthesized by IVT (sgRNA 1–2) (Figure 3A). Out of 2,460 injected eggs, 5.6% of eggs hatched. 48 larvae were Sanger sequenced and 66.6% had deletions 40 bp from the predicted cut site for sgRNA 1 in the intron region 3′ from the third exon (Table 1, Figures 3A–3C, and Table S4). The remaining hatched larvae were analyzed using Illumina Miseq, which confirmed the Sanger sequencing data. The percentage of indels in deep sequencing samples was approximately 80% for sgRNA 1 (Figure 3A) and less than 1% for sgRNA 2 (see Table S4). Both Sanger and deep sequencing showed that the sgRNA 1 was active (see Table S4 and original sequencing data on Mendeley). There were base substitutions near the cut sites for sgRNA 1 and sgRNA 2; however, these substitutions are possibly single nucleotide polymorphism (SNP) because two of these matched in our updated genome (Nuss et al., 2018). These include a G to T SNP 2 bp from the predicted cut site for sgRNA 1 and immediately 3 bp from the start of sgRNA 1 (G to A) that results in a change from aspartic acid to glycine. The intron deletions 40 bp from the predicted cut site are not present in the VectorBase sequence (ISCW003950), our wild-type Sanger sequencing, and our updated genome, providing a solid reason to believe that these are due to CRISPR edits.
Figure 3.
Sequences of Chitinase G0 larvae edited by embryo injection aligned to wildtype (WT)
(A) Summary of deep sequencing data of animals injected with sgRNA 1. Indel percentage is on the Y axis. The X axis depicts predicted Cas9 cut site (position 0) and sequences up- and downstream of the cut site.
(B) Representative sequences containing deletions corresponding to sgRNA 1. The predicted cut site is shown by a black arrow and dotted line. PAM site is indicated by a red rectangle outline. Alignments were generated using Snapgene.
(C) Forward (F) and Reverse (R) chromatograms of WT, CH5, and CH6 larvae. Additional sequences, chromatograms, and indel/substitution percentages calculated by deep sequencing are shown in Figure S5 and Table S4.
Delivery of protein cargo into tick oocytes by ReMOT Control
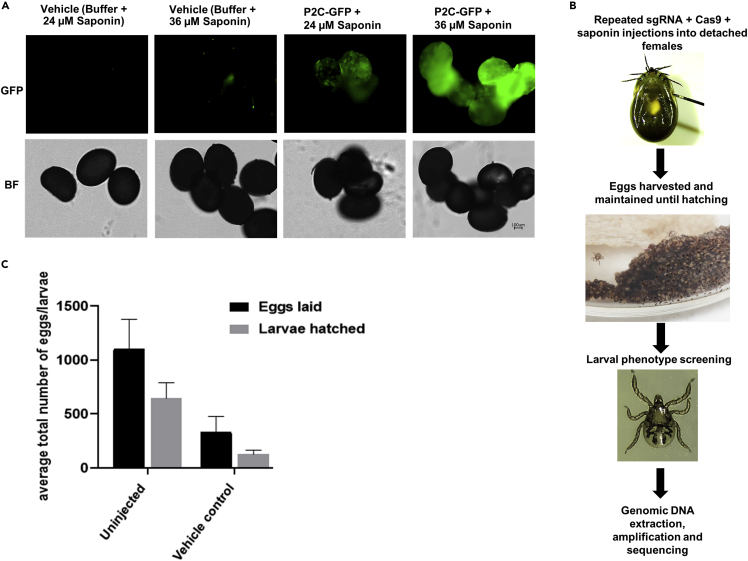
Ixodes scapularis IsVg8 (predicted I. scapularis vitellogenin receptor binding peptide, NFTKTKNY)-tagged mCherry, D. melanogaster P2C (yolk protein-peptide 2C)-tagged EGFP, or P2C-Cas9 protein were used for injecting engorged ticks in the preliminary ReMOT Control experiments. Both IsVg8 and P2C-tagged fluorescent protein injections had similar fluorescence intensity suggesting that ticks were able to take up both ligands (Data not shown). Therefore, we used P2C-tagged EGFP or Cas9 in subsequent experiments as it has been successfully used in multiple distantly related insect species (Chaverra-Rodriguez et al., 2018, 2020; Macias et al., 2020; Shirai and Daimon, 2020; Li et al., 2021). We used saponin as an endosomal reagent because of the higher gene-editing efficiency in mosquitoes with saponin (Macias et al., 2020). Saponin was used at 24, 36, or 48 μM concentrations. P2C-EGFP uptake was confirmed by visualizing eggs from injected females within 24 h of egg-laying (Figure 4A). An increase in saponin concentration had a direct correlation with increased fluorescence intensity (Figure 4A). Control females were injected with vehicle buffer and 36-μM saponin (vehicle control), and the same injection schedule was followed. Injections of the ReMOT Control components in the lower quadrant of the engorged females caused mortality in both controls and P2C-Cas9-injected females. 48-μM saponin was toxic and resulted in 100% mortality. However, no mortality was recorded when the females were injected near the spiracle with sgRNA + Cas9+saponin (24 or 36 μM). The first injection was carried out 2 days after detaching from the host, a second injection followed once the female started laying eggs (6–7 days post detachment), and subsequent injections were performed every two days until the female stopped laying eggs (∼21 days). Injection timing was based on hemolymph vitellogenin levels reported in I. scapularis (James and Oliver, 1996), and our observations that EGFP fluorescence decreased by 50% in eggs 2 days post injection. The injections with P2C-Cas9, sgRNA, and 24-μM saponin were carried out at the lower right quadrant (Kocan et al., 2011)); however, due to high mortality by injections at this site, we injected the remaining females at the spiracle (Nijhof et al., 2007). Consequently, all subsequent injections with 36-μM saponin were carried out at the right spiracle (ventral side; Figure 4B). Multiple injections during oviposition significantly decreased fecundity. The vehicle control-injected females deposited an average of 330 eggs whereas un-injected ticks deposited ∼1,100 eggs (Figure4C).
Figure 4.
Delivery of injection components to ovaries via ReMOT Control
(A) Visualization of delivery of injection components to ovaries via fluorescent imaging following injection of P2C-GFP.
(B) Workflow schematic of blood-fed adult female injections with P2C-Cas9 protein, saponin, and pooled sgRNAs.
(C) Measurements of survival: egg-laying and larval hatching following injections with varying concentrations of saponin and sgRNAs. Vehicle control = dialysis buffer + saponin (36 μM). Data are presented as mean ±SEM.
Verification of gene editing by ReMOT control
ProbP (also used in embryo injections above) was used to test the potential of ReMOT Control for tick gene-editing (Figure 1A).
ProbP editing
The sgRNA 1–4 for ProbP were the same as mentioned above in the embryo injections section. A total of 11 mutants [ReMOT (R); R1-R11] were confirmed. 24-μM saponin resulted in 4.2% overall editing efficiency compared to 1.7% with 36-μM saponin (Table 1). 24-μM saponin was better tolerated by ticks as evident from higher larval hatch rate [168/176 (N = 1) to 230/773 (N = 4 females)] in comparison to that of 36 μM (Table1). The mutations were characterized by long deletions (R4-R6, R8-R11). We observed large (>100bp) homozygous deletions in seven larvae: three with 24-μM saponin (R4-R6) and four with 36-μM saponin (R8-R11) (Figure 5A). These homozygous deletion mutants had GC-rich sequences flanking the deletions (Figure 5A). R4-R6 and R11 had in-frame deletions of the predicted amino acid sequence (Figure S5A). R7, R9, and R10 deletions resulted in frameshift mutations with premature stop codons (R10: Figure S5B).
Figure 5.
Sequences of Proboscipedia G0 larvae CRISPR edited by ReMOT Control aligned to wildtype (WT)
(A) Map depicting aligned homozygous and heterozygous deletions.
(B) Representative chromatogram alignment of WT and two homozygous mutants (R4 and R6). GC-rich flanking regions are underlined in blue. PAM sites are indicated by a red rectangle. The approximate cut sites are shown by black arrows and dotted lines. PAM sites are indicated by a red rectangle. The approximate cut sites are shown by black arrows. Alignments were generated using Snapgene.
Discussion
In this study, we have developed a tick embryo injection protocol and describe CRISPR-Cas9-directed mutagenesis protocols in I. scapularis using embryo injection and an alternative approach, ReMOT Control. This is the first demonstration of embryo injections in any chelicerate species and the first guided CRISPR/Cas9 mutagenesis in ticks. We also demonstrate that ReMOT Control can induce mutations in a non-insect arthropod, and both embryo injections and ReMOT Control had comparable editing efficiency for the gene tested. Our data hints toward a DSB repair mechanism different from NHEJ in ticks because of the long deletions and less common edits distal to the cut site; however, more work is needed to understand the DSB repair mechanisms in ticks.
Our embryo injection protocol was able to successfully deliver plasmid and Cas9 RNPs for gene expression and gene editing, respectively. Manipulation of the Gene's organ was a simple and effective method to obtain un- or partially waxed eggs for injections. Benzalkonium chloride was used to degrade the chorion and also provided controlled desiccation, necessary for needle penetration (Zhang and Reed, 2016, 2017). The fungicidal properties of benzalkonium chloride may additionally have helped prevent fungal growth on embryos (Bundgaard-Nielsen and Nielsen, 1996). A survival rate of ∼10% of injected embryos is comparable to the well-established insect models, Aedes aegypti (18.6% at 500 ng/μL Cas9) (Kistler et al., 2015) and Anopheles gambiae (10%) (Dong et al., 2018). Other researchers have reported a much lower survival rate for Ae. aegypti (Dong et al., 2015) ranging from 4.5% to 11.7%.
Editing efficiency varied among different sgRNA/Cas9 concentrations and target genes. Of the two genes targeted for embryo injections, Chitinase sgRNA injections led to the higher editing efficiency (up to 80%), compared to ProbP (1.8%). However, deep sequencing of ProbP-injected eggs showed less than 1% editing efficiency. Several possibilities could lead to differences in editing efficiency, such as 1) ProbP might be essential for tick survival, and therefore most edited eggs die before hatching, 2) the sgRNA sequence of Chitinase might have been more efficient than the sgRNA sequences of ProbP, 3) presence of SNPs in our wild-type control larvae such as one 6 bp from the PAM sequence in sgRNA4 might have affected editing efficiency, or 4) edits in an intron region for Chitinase may not affect protein expression. More work with additional gene knockout (KO) will help understand these differences.
For ReMOT Control, injections near the spiracle into engorged adult females were well tolerated (100% survival of injected ticks), likely due to decreased tissue reflux and hemolymph loss than the injections in the lower right quadrant. Co-injection with 36-μM saponin significantly increased EGFP fluorescence and the overall percentage of homozygous mutants compared to 24-μM saponin and had similar editing efficiency to 24-μM saponin. The G0 gene-editing efficiency in ticks with ReMOT Control (1.7%–4.1%) is comparable to mosquitoes such as Ae. aegypti (∼1.5% with chloroquine) (Chaverra-Rodriguez et al., 2018) and An. stephensi (3.7% with saponin concentrations 11–20 μM) (Macias et al., 2020). The ability to edit genes by injecting gravid females instead of embryos that require sophisticated infrastructure and expertise makes ReMOT Control convenient for many labs interested in gene KO studies in ticks. However, gene knockin is currently not possible with ReMOT; therefore, any knockin experiments would still require embryo injections.
Gene-editing frequency was higher for ProbP in ReMOT Control (1.7%–4.1%) compared to embryo injections for the same gene. The higher efficiency is possibly due to the age of embryos because the injected embryos were older when edited than the developing embryos within ovaries targeted by ReMOT Control. The timing of cellularization in tick embryos remains unclear, but our preliminary work and work in other Ixodid ticks (Wagner, 1894; Pressesky, 1952) suggest that the nuclear division occurs between 18 and 24 h post egg-laying (Pham et al., 2021). Therefore, our choice of an early time point (14–18 h) might have resulted in embryos with a single nucleus or very few nuclei resulting in homozygous deletion in a few injected embryos.
The transformation efficiency of ProbP by embryo injection in ticks is lower than the mosquito, Ae.aegypti, where G0mutation rate has been shown to vary and range from 5.5% (Dong et al., 2015) to 24.8% (Kistler et al., 2015). Although the editing efficiency of 80% in Chitinase is higher than most reports, similar editing efficiencies have been shown in D. melanogaster (88%) (Bassett et al., 2013) and primary human cells (>90%) (Martufi et al., 2019). In future experiments, it will be worthwhile to compare the efficiency of Chitinase sgRNA in embryo injection with ReMOT Control injections. The higher transformation efficiency for Chitinase was 40 bp away from the predicted cut site and raises questions about active DSB repair mechanisms, which will be explored in future work.
The presence of long deletions (>100 bp) and short indels distal to the cut site in the tick genome was unexpected, although similar observations have been made in other systems. DNA breaks introduced by sgRNA/Cas9 were resolved into deletions extending over many kilobases and complex genomic rearrangements at the targeted sites in mouse embryonic stem cells, mouse hematopoietic progenitors, and human differentiated cell lines (Kosicki et al., 2018). Furthermore, lesions distal to the cut site were also identified (Kosicki et al., 2018). A single sgRNA has also been shown to cause deletions that partially alter splicing or unexpected larger deletions that remove exons in lung adenocarcinoma cell lines (Mou et al., 2017). Based on the pattern of deletions in our data, the mechanism of DSB repair appears to be different in ticks compared to well-studied models such as mosquitoes. Our data show the presence of long deletions and deletions distal from the cut site. In most cases, the deletions in our data were flanked by GC-rich short segments that have some degree of homology between the 5′ and 3′ ends. We hypothesize that these long deletions are caused by MMEJ, a less understood mechanism of DSB repair. In this repair mechanism, ends are resected until there are small microhomologies, ranging from 1 to 16 bp, dependent on the organism (Sfeir and Symington, 2015). Reduction in GC content or additional mismatches at the ends impedes MMEJ success in yeast (Sfeir and Symington, 2015).While NHEJ rapidly ligates compatible ends, ends that need further processing eventually undergo resection and become available for MMEJ or HDR (Truong et al., 2013). MMEJ has been suggested to contribute to the stability of repetitive DNA (Sfeir and Symington, 2015) and might be a preferred mechanism for DSB repair in tick embryos because approximately 50% of the 2.1 Gbp I. scapularis genome is comprised of repetitive elements (terminal repeats, tandem repeats, and transposable elements) (Gulia-Nuss et al., 2016; Nuss et al., 2018). Interestingly, recent data in human cells suggest that many repair outcomes, if not the majority of those attributed to NHEJ, are due to MMEJ (Yeh et al., 2019). However, the precise mechanism will be hard to distinguish unless inhibition assays are employed. Therefore, more work is needed to understand the mechanism of DSB repair in ticks. Additionally, genome editing occurs in the context of chromatin which is heterogeneous across the genome. Mutagenesis in mouse embryonic stem cells was impeded by up to 7-fold when Cas9 exposure was brief and when intracellular Cas9 expression was low in imprinted genes with identical DNA sequences situated in the same nucleus but differ in epigenetic mechanisms. In contrast, the outcome of mutagenic DNA repair was unaffected for HDR (Kallimasioti-Pazi et al., 2018). Therefore, we will compare HDR efficiency in tick genome manipulation in future experiments. If our hypothesis that the tick genome frequently uses MMEJ for DSB repair is correct, this will allow us to use short (micro) homologies to insert sequences in the genome and compare the MMEJ and HDR-based insertions.
An extended hypostome phenotype was observed in ProbP knockout and four additional larvae from ProbP injected embryos that did not show a clear editing. We have screened several thousand larvae and this phenotype was never present in wildtype or any other gene knockout, strongly suggesting that this is an actual phenotype due to CRISPR editing.
Here, we show the embryo injection protocol for a chelicerate species and tools for tick genome editing to expand the I. scapularis molecular toolbox. The ease of injections into adults with ReMOT Control makes it an excellent tool for KO studies, but currently, only embryo injection is suitable for future knockin experiments (Pham et al., 2021). Our work also suggests that ticks may not preferentially use NHEJ as a DSB repair mechanism, and future studies will help understand the ideal repair mechanism. This work provides the foundation for future studies on the generation of tick mutant lines and the HDR or MMEJ-mediated integration of donor constructs. These tools will open new avenues for tick biology and tick-pathogen-host studies.
Limitations of the study
In this study, we have developed an embryo injection protocol for ticks that we used to demonstrate successful gene editing by CRISPR/Cas 9. In addition, we showed successful gene KO by ReMOT Control, a less labor-intensive method of gene editing in arthropods. While these tools will accelerate tick genetic research, improvements are needed in embryo injection protocol for better survival and larval hatching and KO efficiencies. One of the significant limitations of this study is potential somatic editing because we did not attempt to generate heritable mutant lines due to the long lifecycle of ticks. Another limitation is the fewer genes used for KO in this study. More work is needed to understand the active mechanisms of DSB repair during the early embryogenesis in ticks for better experimental design for KO and possibly knockins.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| BL21 E. coli | New England Biolabs, City, MA | Cat # C2530H |
| Biological samples | ||
| Tick eggs | UNR colony, Oklahoma State University | N/A |
| Ticks | UNR colony, Oklahoma State University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Benzalkonium Chloride | SIGMA Aldrich | Cat # 63449-41-2 |
| Sodium Chloride | SIGMA Aldrich | Cat # S3014 |
| Tris | Fluka | Cat # 93362 |
| KCl | SIGMA Aldrich | Cat # P9541 |
| EDTA | SIGMA Aldrich | Cat # E5134 |
| PMSF | SIGMA Aldrich | Cat # P20270 |
| DTT | SIGMA Aldrich | Cat # D9779 |
| Saponin | EMD Millipore | Cat # 558255, 9.65% Ash |
| SlowFade™ Gold Antifade Mountant | Invitrogen | Cat # S36973 |
| Chelex® 100 sodium form | Sigma-Aldrich | Cat # C7901 |
| Recombinant Streptococcus pyogenes Cas9 | PNA Bio, CA, USA | Cat # CP01 |
| pET28a-IsVg8-EGFP | Rasgon lab, Penn State | |
| pET28a-P2C-Cas9 | Rasgon lab, Penn State | Chaverra-Rodriguez et al. (2018) |
| pRSET-P2C-EGFP | Rasgon lab, Penn State | Chaverra-Rodriguez et al. (2018) |
| Critical commercial assays | ||
| GeneArt™ Precision gRNA Synthesis Kit | Invitrogen | Cat # A29377 |
| QIAseq 1-Step amplicon library kit | Qiagen | Cat # 180412 |
| GeneRead Adapter I Set B 12-plex | Qiagen | Cat # 180986 |
| Deposited data | ||
| Raw and Analyzed data | This paper; Mendeley Data | https://doi.org/10.17632/wzj67n7gz6.2 |
| Experimental models: Organisms/strains | ||
| Ticks: Ixodes scapularis | UNR, Oklahoma State University | N/A |
| Oligonucleotides | ||
| Primers for sequencing | This paper, Integrated DNA Technology | N/A |
| sgRNA for Proboscipedia and Chitinase | This paper, Integrated DNA Technology | N/A |
| Modified sgRNA | This Paper, Integrated DNA Technology | N/A |
| Recombinant DNA | ||
| pCis CAGGS LifeAct cherry promoter | Kurtti lab, University of Minnesota | Oliver et al. (2015) |
| Software and algorithms | ||
| CRISPOR | Haeussler et al. (2016) | http://crispor.tefor.net |
| ChopChop | Labun et al. (2019) | https://chopchop.rc.fas.harvard.edu |
| ICE analysis | Hsiau et al. (2018) | https://ice.synthego.com/ |
| Deep Sequencing Data Analysis | Kistler et al. (2015) | https://github.com/bnmtthws/crispr_indel |
| Other | ||
| 3M Tegaderm® film | EZ Med, Amazon Inc., USA | Ref#1628 |
| Aluminosilicate needles | Sutter instruments; CA, USA | AF100-64-10 |
| MSP, 15°, 5.0 mm depth | Beaver-Visitec International, Inc. | Cat# 377515 |
Resource availability
Lead contact
Further information about the protocols and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Monika Gulia-Nuss (mgulianuss@unr.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Ticks
Ixodes scapularis adults were either purchased from Oklahoma State University (OSU) or reared at the University of Nevada, Reno (UNR) (IACUC protocol #21-001-1118). Ticks were reared at 27°C, 95% relative humidity (RH), and 12h light: 12h dark in an environmental chamber. Larvae and nymphs were blood-fed on mice (Nuss et al., 2017), and adults were fed on a rabbit until engorgement (Reyes et al., 2020). Both male and female vertebrate animals were used for tick feeding. Once replete, a batch of females was kept individually at 4°C until ready for egg-laying.
Method details
Preparation of embryos for microinjection
Engorged females were recovered from 4°C and kept at 27°C for egg-laying. At this temperature, females start laying eggs in approximately 4–5 days and keep depositing for 21–28 days. Once females start laying eggs (first day of egg-laying), the Gene’s organ (the wax gland) was either dissected using fine Tungsten needles or emptied of wax so that little or no wax was deposited on the eggs (Schöl et al., 2001) (Figure S2A). In the latter condition, the Gene’s organ was emptied of wax every 3–4 days to ensure un/partially waxed eggs. Freshly deposited eggs were collected within <24 h (∼14–18 h) for injections. To remove the chorion, embryos were submerged in 5% benzalkonium chloride for 5 min followed by two washes with deionized (DI) H2O. The washed eggs were then treated with 5% NaCl for 3–5 min, washed twice with DI water, and kept in 1% NaCl until injected (Figure S1).
Food color and plasmid injection mix preparation
Liquid green food dye was purchased from a local grocery store and diluted 1:100 with DNAse/RNAse free water. The diluted dye was filtered through a 0.45 μm filter prior to injection. A plasmid encoding red fluorescent protein (RFP) controlled by the CAGGS promoter (Oliver et al., 2015) (a gift from Dr. Timothy Kurtti, University of Minnesota, USA) was injected into the embryos at a concentration of 1 μg/μL. 24–36 h post injections, RFP expression was detected in embryos with an Olympus MVX 10 macro zoom fluorescence microscope system (Olympus, USA) at 586 nm wavelength.
Single guide RNA (sgRNA) design and construction
Two genes were selected for sgRNA design: Proboscipedia (ProbP, ISCW021086, XM_002406373.1) and Chitinase (CHI, ISCW003950, XM_002399485.1). The gene sequences were obtained from VectorBase and further confirmed with our assembly (Nuss et al., 2018). The sgRNAs were designed by manually searching genomic regions for the presence of protospacer-adjacent motifs (PAMs) with the sequence NGG, where N is any nucleotide. Sequences of sgRNA were required to be 18–20 bp in length, excluding the PAM, and contain one or two 5′ terminal guanines to facilitate transcription by T7 RNA polymerase. sgRNA sequences were checked for potential off-target binding using the following two web tools: CRISPOR (http://crispor.tefor.net/) (Haeussler et al., 2016), and ChopChop (https://chopchop.rc.fas.harvard.edu/) (Labun et al., 2019). 3-4 prospective sgRNA sequences were identified with PAM regions from the first exon of ProbP and third exon of Chitinase (Table S1). Both sgRNAs were designed from the Exon 3. sgRNAs were prepared using a PCR template amplified by CRISPR_F primers designed for each target gene and CRISPR_R primers. Two methods were used for sgRNAs synthesis: 1) short primer design and in vitro transcription (IVT) in the laboratory using the PCR template and GeneArt™ Precision gRNA Synthesis Kit (Thermo Fisher) per the manufacturer’s instructions and 2) modified synthetic sgRNA (Integrated DNA Technology, Coralville, IA, USA).
CRISPR-Cas9 mix
The synthesized sgRNAs were mixed with injection-ready Cas9 protein in the injection buffer. Recombinant Streptococcus pyogenes Cas9 protein was obtained commercially (CP01, PNA Bio, CA, USA). The injection mix comprised of different concentrations of sgRNAs and Cas9 was used to identify the best concentration for editing efficiency and survival.
All sgRNAs were individually mixed with Cas9 RNP and then multiplexed in equal ratios before injections. Most injections were carried out using an injection mix consisting of 100 ng/μL sgRNA (multiplexed three sgRNAs to make the final concentration) and 300 ng/μL Cas9.
Embryo microinjections
Treated eggs were aligned on a glass slide platform. To make this platform, two glass slides were adhered with double-sided tape, leaving a gap of about 0.5cmat the edge. In the gap, a piece of 3M Tegaderm® film (EZ Med, Amazon Inc.,USA) was inserted to mount the eggs (Figure S2B).
Once eggs were ready for injection, 10–15 embryos were aligned on the Tegaderm of the slide platform using a single-hair paint brush under a dissecting scope. Since tick eggs are spherical and have no visible anterior/posterior poles, embryos were aligned so that the widest part of the embryo (middle) touched the slide base. This orientation was well tolerated by the embryos and helped minimize embryo bursting during the injection. Aluminosilicate needles (Sutter instruments; CA, USA; AF100-64-10) were pulled using a Sutter P1000 needle puller (Heat = 575, Pull = 20, Velocity = 50, Time = 200 and Pressure = 700) and used with an injector (Shutter injector MPC-200) and Sutter micromanipulator (ROE-200). Needles were filled with the injection mix using a needle filler (Eppendorf #930001007) and opened by rubbing it slightly against the embryo using high-pressure settings on the microinjector (>5000 hPa). Depending on the opening of the needle, the pressure of the injector was reduced (2000–3500 hPa), and embryos were injected.
Following injection, the embryo slides were kept temporarily in a Petri dish (180 mm) lined with wet filter paper inside an airtight box lined with damp paper towels to maintain high humidity. After 3–4 h (enough time to seal off the injection wound), a small drop of water was placed on the Tegaderm® to transfer the injected embryos to another Petri dish (100 mm). Embryos were submerged in water and kept in an incubator at 27°C and >90% RH. Ixodes larvae in our lab can survive submerged for over three weeks; however, we collected larvae using a paintbrush to a dry filter paper soon after hatching (within 24 h) and kept them in scintillation vials with a fine screen adhered to the cap. The larvae (generation 0, G0) were screened for phenotype and used for DNA extraction for Sanger sequencing. Control embryos were treated similarly but were not injected.
Adult injection mix preparation
The I. scapularis vitellogenin targeting peptide sequence (IsVg8) is NFTKTKNY. IsVg8-EGFP, P2C-Cas9, and P2C-EGFP proteins were expressed from pET28a-IsVg8-EGFP, pET28a-P2C-Cas9, and pRSET-P2C-EGFP, respectively, by recombinant BL21 E. coli (New England Biolabs, City, MA) and purified as described in detail elsewhere (Chaverra-Rodriguez et al., 2018). The selected sgRNAs were generated using the GeneArt™ Precision gRNA Synthesis Kit, as described above. Two to four volumes of the in vitro transcription reaction were set up with a 4 h reaction time to achieve the high quantities of sgRNA required for adult injections. In addition to P2C-Cas9 and sgRNAs, saponin (EMD Millipore 558255, 9.65% Ash) was included as an endosomal escape reagent in injection mixes. Saponin dilutions were prepared fresh prior to each injection. We used different concentrations (24, 36, and 48 μM, adjusted for ash content) of saponin in each injection. Each sgRNA was individually complexed with P2C-Cas9 then mixed to produce solutions of 2.2 μg/μL P2C-Cas9 and 1.2 μg/μL of total sgRNAs to achieve a 1:3 M ratio. Protein dialysis buffer (vehicle buffer: 50 mM Tris-HCL pH 8, 300 mM KCl, 0.1 mM EDTA, 0.5 mM PMSF +0.1 mM DTT +24 or 36 μM saponin) was used as a vehicle control for injections.
Adult injections
Fully engorged adult females were either raised at UNR or purchased from the tick rearing facility at OSU. When purchased from OSU, females were shipped to UNR as soon as they dropped off the host. Females were either stored at 4°C or held at 20°C for immediate use. Egg-laying initiated within 6–7 days post host detachment (at 20°C) or 4–5 days after ticks were moved from 4°C to the 27°C incubator. The injection site was cleaned with a small paintbrush dipped in a 3% hydrogen peroxide solution prior to injection.
For injection, a sterilized insulin needle (30G 1cc 1/2") was first used to pre-puncture the injection site. A Hamilton syringe (Hamilton Inc, Reno, NV) containing 2 μL of injection material was then inserted into the injection site, and the mix was injected after the needle opening was completely inside the tick. Needles and syringes were cleaned between injections by rinsing first in sterile water (3x), then 3% hydrogen peroxide (Aaron Industries, Inc., CA, USA), then with nuclease-free or DEPC water. The first injection was carried out 2 days after detaching from the host, a second injection followed once the female started laying eggs (6–7 days post detachment), and subsequent injections were performed every two days until the female stopped laying eggs (∼21 days). Injection timing was based on hemolymph vitellogenin levels reported in a previous study in I. scapularis (James and Oliver, 1996), and our observations that EGFP fluorescence decreased by 50% in eggs 2 days post-injection. 24 μM saponin injections were carried out at the lower right quadrant previously used to inject unfed ticks. Due to high mortality in injections at the lower quadrant, 36 μM injections were done at the spiracle. All subsequent injections were carried out at the right spiracle (ventral side). Control females were injected with the buffer alone, and un-injected females were also kept from the same cohort to test the effect of injections on tick survival. Injected ticks were then placed in small plastic deli cups with clear perforated lids and allowed to recover for 5–10 min. These deli cups were rinsed with hydrogen peroxide and nuclease-free water prior to tick placement. Individual cups were kept inside a humid box within an incubator for egg-laying. Larvae hatched from eggs within 3–4 weeks and were observed for phenotype or used to extract genomic DNA (gDNA) for PCR and Sanger sequencing.
To test the receptor-mediated uptake of the IsVg8-Cas9-mCherry mix by the developing oocytes in the ovaries, we dissected ovaries 72 h post-blood meal (drop off from host), mounted on a slide in SlowFade™ Gold Antifade Mountant (Invitrogen) and covered by the plastic coverslip. The coverslip was sealed in place using nail polish on the edges of the slip, and ovaries were visualized on an Olympus (BX41 epifluorescent) microscope. To detect whether P2C tagged ligands could be directed to the developing eggs, P2C-EGFP (1 μg/μL) was injected along with either 24 or 36 μM saponin into blood-fed adult females. The eggs were collected within 24 h after deposition and visualized on a glass slide or petri dish using a Keyence BZX700 fluorescent microscope (Keyence America). The green fluorescent was observed in the eggs to test uptake efficiencies. Eggs from (Vehicle/Buffer) injected females were used as a negative control.
Larval screening and imaging
G0 larvae were immobilized by pressing them slightly on double-sided tape. Larvae were kept close to a moist filter paper to avoid desiccation during screening under a Leica dissecting scope or a Keyence (BZX700 fluorescent) microscope. Larval images were taken on a Leica M125 stereo microscope at 0.6x.
Measurement of palps and hypostome length
The ProbP mutants with a phenotype were further analyzed using a Leica M125 stereo microscope equipped with a digital camera. The gnathosoma of all mutants and 10 control individuals were examined under the microscope and the distance between the tips and the base at the basis capituli of palps and hypostome was measured using Leica Application Suite software version 3.4.0 (Build:272). The basis capituli connects the mouth parts (hypostome and palps) to the body. Unpaired t-test (95% confidence interval) for mean length of control and ProbP injected larvae were used to calculate the statistical significance.
Molecular analysis of mutant individuals
Putative mutant G0 larvae were collected individually from both embryo injections and adult injections, frozen, and stored at −20°C. Genomic DNA was extracted from individual larvae hatched either from injected embryos or from eggs from the injected adult females (ReMOT Control). For gDNA isolation, a 10% aqueous solution of Chelex® 100 sodium form (Sigma-Aldrich, St. Louis, MO, USA) (Richlen and Barber, 2005) was used. For Chelex, the larvae were cut into halves using a sterile surgical blade (MSP, 15°, 5.0 mm depth) and were transferred into 0.2 mL PCR tubes containing 200 μL of Chelex solution. Tubes were vortexed and spun for 20 s before incubating in a Thermocycler for 20minat 95°C and the resulting supernatant was used directly as a template for PCR (1–5 μL in 20 μL PCR reaction).
A nested PCR protocol was used to amplify regions surrounding the putative CRISPR-Cas9 cut site from gDNA of G0 larvae using primers in Table S2. PCR products were sent for PCR clean up and high-volume Sanger sequencing at the University of Arizona Genomics Core facility. The sequences from injected individuals were aligned to the wild-type reference sequence and examined for the presence of insertions, deletions, or other polymorphisms. ICE analyses (https://www.synthego.com/products/bioinformatics/crispr-analysis) were also used to determine gene-editing.
Amplicon library preparation and analysis
The larvae hatched from embryo injections of Chitinase and ProbP CRISPR construct were pooled together and genomic DNA was extracted. The PCR amplicons surrounding predicted cut sites were generated using primers (Table S2). The deep sequencing library was prepared using QIAseq 1-Step amplicon library kit (Qiagen, 180412) following the manufacturer's recommended protocol and purified amplicons were sequenced on Illumina MiSeq (Novogene Inc, Davis, CA).Data were analyzed using a published script (Kistler et al., 2015) available at https://github.com/bnmtthws/crispr_indel.
Quantification and statistical analysis
Graph Pad (version 8.0) was used for all graphs. Unpaired t-test (95% confidence interval) for mean length of control and ProbP injected larvae were used to calculate the statistical significance (Figure 2C).
Acknowledgments
We thank Prof. Timothy Kurtti for sharing the CMV/CAGGS plasmid. We acknowledge Channa-Aluvihare and Yonas Gebremicale, Insect Transformation Center, for their insight during the initial phase of embryo injection protocol development. We also thank Drs. Juli Petereit and Hans Vasquez-Gross, the Nevada Bioinformatics Center, for deep sequencing data analysis. This project was funded by NIH-NIAID R21AI128393 and Plymouth Hill Foundation, NY to MG-N, Startup funds from the University of Nevada to AN, Peer-to-Peer Grant from IGTRCN to AS, NIH-NIAID R21AI111175, USDA-NIFA 2014-10320, USDA Hatch funds (Project #PEN04769), and NSF-BIO 1645331 to JLR. This work was in part supported by the NV INBRE (GM103440) and MW CTR-IN (5 U54 GM104944) grant from the National Institute of General Medical Sciences from the National Institutes of Health (NIH).
Author contributions
MG-N, ABN, JLR, and RH conceived and designed the experiments. ABN provided technical and intellectual support. AS developed an embryo injection protocol and carried out CRISPR embryo injections. JR assisted in screening the larvae hatched from the injected eggs, DNA extraction, and sequencing. MP developed the ReMOT Control adult injection protocol in consultation with DCR, DK, and JLR, and screened larvae hatched from the adult injections. RC (with MP) optimized the Chelex DNA extraction protocol and assisted in DNA extractions. WCY assembled the new tick genome and identified full-length sequences of genes. DCR, DK, CCH, and JLR generated ReMOT Control reagents. AS and MP wrote the initial drafts of the manuscript. MG-N, ABN, AS, MP, and JLR wrote the final draft. MG-N, ABN, AS, and MP revised the manuscript.
Declaration of interests
JLR, DCR, and CCH have filed for patent protection on the ReMOT Control technology. No other authors have any competing interests.
Published: February 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103781.
Contributor Information
Andrew B. Nuss, Email: nussab@unr.edu.
Monika Gulia-Nuss, Email: mgulianuss@unr.edu.
Supplemental information
Data and code availability
Original sequencing data for figures in the paper are available at Mendeley data (https://data.mendeley.com/datasets/wzj67n7gz6/2). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the LEAD Contact upon request.
References
- Awata H., Watanabe T., Hamanaka Y., Mito T., Noji S., Mizunami M. Knockout crickets for the study of learning and memory: dopamine receptor Dop1 mediates aversive but not appetitive reinforcement in crickets. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Zeng T., Ni X.Y., Su H.A., Huang J., Ye G.Y., Lu Y.Y., Qi Y.X. CRISPR/Cas9-mediated knockout of the eye pigmentation gene white leads to alterations in colour of head spots in the oriental fruit fly, Bactrocera dorsalis. Insect Mol. Biol. 2019;28:837–849. doi: 10.1111/imb.12592. [DOI] [PubMed] [Google Scholar]
- Bassett A.R., Tibbit C., Ponting C.P., Liu J.L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundgaard-Nielsen K., Nielsen P.V. Fungicidal effect of 15 disinfectants against 25 fungal contaminants commonly found in bread and cheese manufacturing. J. Food Prot. 1996;59:268–275. doi: 10.4315/0362-028x-59.3.268. [DOI] [PubMed] [Google Scholar]
- Booth T.F. Wax lipid secretion and ultrastructural development in the egg-waxing (Gene's) organ in ixodid ticks. Tissue Cell. 1989;21:113–122. doi: 10.1016/0040-8166(89)90026-8. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention How many people get Lyme disease? | Lyme Disease | CDC. 2015. https://www.cdc.gov/lyme/stats/humancases.html
- Chaverra-Rodriguez D., Macias V.M., Hughes G.L., Pujhari S., Suzuki Y., Peterson D.R., Kim D., McKeand S., Rasgon J.L. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-05425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaverra-Rodriguez D., Benetta E.D., Heu C.C., Rasgon J.L., Ferree P.M., Akbari O.S. Germline mutagenesis of Nasonia vitripennis through ovarian delivery of CRISPR-Cas9 ribonucleoprotein. Insect Mol.Biol. 2020;29:569–577. doi: 10.1111/imb.12663. [DOI] [PubMed] [Google Scholar]
- de la Fuente J., Kocan K.M., Almazán C., Blouin E.F. RNA interference for the study and genetic manipulation of ticks. Trends Parasitol. 2007;23:427–433. doi: 10.1016/j.pt.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Dermauw W., Jonckheere W., Riga M., Livadaras I., Vontas J., Van Leeuwen T. Targeted mutagenesis using CRISPR-Cas9 in the chelicerate herbivore Tetranychus urticae. Insect Biochem.Mol. Biol. 2020;120:103347. doi: 10.1016/j.ibmb.2020.103347. [DOI] [PubMed] [Google Scholar]
- Dong S., Lin J., Held N.L., Clem R.J., Passarelli A.L., Franz A.W.E. Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PLoS One. 2015;10:e0122353. doi: 10.1371/journal.pone.0122353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Simões M.L., Marois E., Dimopoulos G. CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLoS Pathog. 2018;14:e1006898. doi: 10.1371/journal.ppat.1006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V.M., Akbari O.S. Gene editing technologies and applications for insects. Curr.Opin. Insect Sci. 2018;28:66–72. doi: 10.1016/j.cois.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles A.F., Schinko J.B., Averof M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development. 2015;142:2832–2839. doi: 10.1242/dev.125054. [DOI] [PubMed] [Google Scholar]
- Gui T., Zhang J., Song F., Sun Y., Xie S., Yu K., Xiang J. CRISPR/Cas9-mediated genome editing and mutagenesis of EcChi4 in Exopalaemon carinicauda. G3 Genes Genomes Genet. 2016;6:3757–3764. doi: 10.1534/g3.116.034082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulia-Nuss M., Nuss A.B., Meyer J.M., Sonenshine D.E., Roe R.M., Waterhouse R.M., Sattelle D.B., De La Fuente J., Ribeiro J.M., Megy K., et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016;7:1–13. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heu C.C., McCullough F.M., Luan J., Rasgon J.L. CRISPR-Cas9-Based genome editing in the silverleaf whitefly (Bemisia tabaci) Cris. J. 2020;3:89–96. doi: 10.1089/crispr.2019.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinckley A.F., Connally N.P., Meek J.I., Johnson B.J., Kemperman M.M., Feldman K.A., White J.L., Mead P.S. Lyme disease testing by large commercial laboratories in the United States. Clin. Infect. Dis. 2014;59:676–681. doi: 10.1093/cid/ciu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T., Maures T., Waite K., Yang J., Kelso R., Holden K., Stoner R. Inference of CRISPR edits from sanger trace data. BioRxiv. 2018:251082. doi: 10.1101/251082. [DOI] [PubMed] [Google Scholar]
- Hu X.F., Zhang B., Liao C.H., Zeng Z.J. High-efficiency CRISPR/Cas9-mediated gene editing in honeybee (Apis mellifera) embryos. G3 Genes, Genomes, Genet. 2019;9:1759–1766. doi: 10.1534/g3.119.400130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N.I.B., Kato Y., Matsuura T., Watanabe H. Generation of white-eyed Daphnia magna mutants lacking scarlet function. PLoS One. 2018;13:e0205609. doi: 10.1371/journal.pone.0205609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A.M., Oliver J.H. Vitellogenin concentrations in the haemolymph and ovaries of Ixodes scapularis ticks during vitellogenesis. Exp. Appl. Acarol. 1996;20:639–647. doi: 10.1007/BF00053327. [DOI] [PubMed] [Google Scholar]
- Kallimasioti-Pazi E.M., Thelakkad Chathoth K., Taylor G.C., Meynert A., Ballinger T., Kelder M., Lalevée S., Sanli I., Feil R., Wood A.J. Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 2018;16:e2005595. doi: 10.1371/journal.pbio.2005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S., Adamson S.W. In: Advances in Insect Physiology. Jockusch E.L., editor. Academic Press; 2012. RNA interference in ticks.A functional genomics tool for the study of physiology; pp. 119–154. [Google Scholar]
- Khan S.A., Reichelt M., Heckel D.G. Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations. Sci. Rep. 2017;7:1–14. doi: 10.1038/srep40025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler K.E., Vosshall L.B., Matthews B.J. Genome engineering with CRISPR-Cas9 in the mosquito aedes aegypti. Cell Rep. 2015;11:51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocan K.M., Blouin E., de la Fuente J. RNA interference in ticks. J. Vis. Exp. 2011:2474. doi: 10.3791/2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocan K.M., De La Fuente J., Coburn L.A. Insights into the development of Ixodes scapularis: a resource for research on a medically important tick species. Parasites and Vectors. 2015;8:592. doi: 10.1186/s13071-015-1185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwica-Rolinska J., Chodakova L., Chvalova D., Kristofova L., Fenclova I., Provaznik J., Bertolutti M., Wu B.C.-H., Dolezel D. CRISPR/Cas9 genome editing introduction and optimization in the non-model insect Pyrrhocoris apterus. Front. Physiol. 2019;10:891. doi: 10.3389/fphys.2019.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti T.J., Mattila J.T., Herron M.J., Felsheim R.F., Baldridge G.D., Burkhardt N.Y., Blazar B.R., Hackett P.B., Meyer J.M., Munderloh U.G. Transgene expression and silencing in a tick cell line: a model system for functional tick genomics. Insect Biochem. Mol. Biol. 2008;38:963–968. doi: 10.1016/j.ibmb.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Au L.Y.C., Douglah D., Chong A., White B.J., Ferree P.M., Akbari O.S. Generation of heritable germline mutations in the jewel wasp Nasonia vitripennis using CRISPR/Cas9. Sci. Rep. 2017;7:1–7. doi: 10.1038/s41598-017-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Y., Liu G.-C., Sheng W.-J., Dong Z.-W., Chen L., Zhao R.-P., Wang W. Genome editing in the butterfly type-species Papilio machaon. Insect Sci. 2017;24:708–711. doi: 10.1111/1744-7917.12421. [DOI] [PubMed] [Google Scholar]
- Li M., Li T., Liu N., Raban R.R., Wang X., Akbari O.S. Methods for the generation of heritable germline mutations in the disease vector Culex quinquefasciatus using clustered regularly interspaced short palindrome repeats-associated protein 9. Insect Mol. Biol. 2020;29:214–220. doi: 10.1111/imb.12626. [DOI] [PubMed] [Google Scholar]
- Li X., Xu Y., Zhang H., Yin H., Zhou D., Sun Y., Ma L., Shen B., Zhu C. ReMOT control delivery of CRISPR-Cas9 ribonucleoprotein complex to induce germline mutagenesis in the disease vector mosquitoes Culex pipiens pallens (Diptera: Culicidae) J. Med. Entomol. 2021;58:1202–1209. doi: 10.1093/jme/tjab016. [DOI] [PubMed] [Google Scholar]
- Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias V.M., McKeand S., Chaverra-Rodriguez D., Hughes G.L., Fazekas A., Pujhari S., Jasinskiene N., James A.A., Rasgon J.L. Cas9-Mediated gene-editing in the malaria mosquito Anopheles stephensi by ReMOT control. G3 Genes Genomes Genet. 2020;10:1353–1360. doi: 10.1534/g3.120.401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Serano J.M., Jarvis E., Bruce H.S., Wang J., Ray S., Barker C.A., O’Connell L.C., Patel N.H. CRISPR/Cas9 mutagenesis reveals versatile roles of Hox genes in Crustacean limb specification and evolution. Curr. Biol. 2016;26:14–26. doi: 10.1016/j.cub.2015.11.021. [DOI] [PubMed] [Google Scholar]
- Martufi M., Good R.B., Rapiteanu R., Schmidt T., Patili E., Tvermosegaard K., New M., Nanthakumar C.B., Betts J., Blanchard A.D., Maratou K. Single-Step, high-efficiency CRISPR-Cas9 genome editing in primary human disease-derived fibroblasts. Cris. J. 2019;2:31–40. doi: 10.1089/crispr.2018.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Smith J.L., Peng L., Yin H., Moore J., Zhang X.O., Song C.Q., Sheel A., Wu Q., Ozata D.M., et al. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017;18:108. doi: 10.1186/s13059-017-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T., Kato Y., Matsuura T., Watanabe H. CRISPR/Cas-mediated targeted mutagenesis in Daphnia magna. PLoS One. 2014;9:e98363. doi: 10.1371/journal.pone.0098363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhof A.M., Taoufik A., de la Fuente J., Kocan K.M., de Vries E., Jongejan F. Gene silencing of the tick protective antigens, Bm86, Bm91 and subolesin, in the one-host tick Boophilus microplus by RNA interference. Int. J. Parasitol. 2007;37:653–662. doi: 10.1016/j.ijpara.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss A., Sharma A., Gulia-Nuss M. Genetic manipulation of ticks: a paradigm shift in tick and tick-borne diseases research. Front. Cell. Infect. Microbiol. 2021;11:678037. doi: 10.3389/fcimb.2021.678037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss A.B., Mathew M.G., Gulia-Nuss M. Rearing Ixodes scapularis, the black-legged tick: feeding immature stages on mice. J. Vis. Exp. 2017:e55286. doi: 10.3791/55286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss A.B., Sharma A., Gulia-Nuss M. Chicago and Dovetail Hi-C proximity ligation yield chromosome length scaffolds of Ixodes scapularis genome. bioRxiv. 2018:392126. doi: 10.1101/392126. [DOI] [Google Scholar]
- Oliver J.D., Chávez A.S., Felsheim R.F., Kurtti T.J., Munderloh U.G. An Ixodes scapularis cell line with a predominantly neuron-like phenotype. Exp. Appl. Acarol. 2015;66:427–442. doi: 10.1007/s10493-015-9908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo D.F., Williamson M.E., Arp A.P., Li F., Sagel A., Skoda S.R., Sanchez-Gallego J., Vasquez M., Quintero G., Pérez de León A.A., et al. Specific gene disruption in the major livestock pests Cochliomyia hominivorax and Lucilia cuprina using CRISPR/Cas9. G3 Genes Genomes Genet. 2019;9:3045–3055. doi: 10.1534/g3.119.400544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M., Sharma A., Nuss A., Gulia-Nuss M. In: Transgenic Insects: Techniques and Applications. Second Edition. Benedict M., Scott M., editors. CABI Biotechnology; 2021. Chapter 9: germline transformation of ticks. [Google Scholar]
- Pressesky M. University of Saskatchewan; 1952. A Study of the Embryonic Development of the Rocky Mountain Spotted Fever Tick Dermacentor Andersoni (Stiles) MS Thesis. [Google Scholar]
- Reyes J., Ayala-Chavez C., Sharma A., Pham M., Nuss A.B., Gulia-Nuss M. Blood digestion by trypsin-like serine proteases in the replete lyme disease vector tick, Ixodes scapularis. Insects. 2020;11:201. doi: 10.3390/insects11030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richlen M.L., Barber P.H. A technique for the rapid extraction of microalgal DNA from single live and preserved cells. Mol. Ecol. Notes. 2005;5:688–691. [Google Scholar]
- Rodino K.G., Theel E.S., Pritt B.S. Tick-borne diseases in the United States. Clin. Chem. 2020;66:537–548. doi: 10.1093/clinchem/hvaa040. [DOI] [PubMed] [Google Scholar]
- Santos V.T., Ribeiro L., Fraga A., de Barros C.M., Campos E., Moraes J., Fontenele M.R., Araújo H.M., Feitosa N.M., Logullo C., et al. The embryogenesis of the Tick Rhipicephalus (Boophilus) microplus: the establishment of a new chelicerate model system. Genesis. 2013;51:803–818. doi: 10.1002/dvg.22717. [DOI] [PubMed] [Google Scholar]
- Schöl H., Sieberz J., Göbel E., Gothe R. Morphology and structural organization of Gené’s organ in Dermacentor reticulatus (Acari: ixodidae) Exp. Appl. Acarol. 2001;25:327–352. doi: 10.1023/a:1017963531560. [DOI] [PubMed] [Google Scholar]
- Schwager E.E., Schoppmeier M., Pechmann M., Damen W.G.M. Duplicated Hox genes in the spider Cupiennius salei. Front. Zool. 2007;4:10. doi: 10.1186/1742-9994-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Symington L.S. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem.Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai Y., Daimon T. Mutations in cardinal are responsible for the red-1 and peach eye color mutants of the red flour beetle Tribolium castaneum. Biochem.Biophys. Res. Commun. 2020;529:372–378. doi: 10.1016/j.bbrc.2020.05.214. [DOI] [PubMed] [Google Scholar]
- Sun D., Guo Z., Liu Y., Zhang Y. Progress and prospects of CRISPR/Cas systems in insects and other arthropods. Front. Physiol. 2017;8:608. doi: 10.3389/fphys.2017.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayyab I., Hallahan H.M., Percival-Smith A. Analysis of Drosophila proboscipedia mutant alleles. Genome. 2004;47:600–609. doi: 10.1139/g03-133. [DOI] [PubMed] [Google Scholar]
- Truong L.N., Li Y., Shi L.Z., Hwang P.Y.H., He J., Wang H., Razavian N., Berns M.W., Wu X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. U S A. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M., Capurso D., Carter M.M., Thompson M.S., Frias E., Russ C., Reece-Hoyes J.S., Nye C., Gradia S., Vidal B., et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol.Cell. 2016;263:633–646. doi: 10.1016/j.molcel.2016.06.037. [DOI] [PubMed] [Google Scholar]
- Wagner J. Die Embryonalentwicklung von Ixodes calcaratus. Bir Trav. Soc. Nat. St Petersb. 1894;24:214–246. [Google Scholar]
- Xue W.H., Xu N., Yuan X.B., Chen H.H., Zhang J.L., Fu S.J., Zhang C.X., Xu H.J. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) Insect Biochem. Mol. Biol. 2018;93:19–26. doi: 10.1016/j.ibmb.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Yeh C.D., Richardson C.D., Corn J.E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019;21:1468–1478. doi: 10.1038/s41556-019-0425-z. [DOI] [PubMed] [Google Scholar]
- You M., Xuan X., Tsuji N., Kamio T., Taylor D.M., et al. Identification and molecular characterization of a Chitinase from the hard tick Haemaphysalis longicornis. J. Biol. Chem. 2003;278:8556–8563. doi: 10.1074/jbc.M206831200. [DOI] [PubMed] [Google Scholar]
- Zhang L., Reed R.D. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat. Commun. 2016;7:1–7. doi: 10.1038/ncomms11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Reed R.D. In: Diversity and Evolution of Butterfly Wing Patterns: An Integrative Approach. Sekimura T., Nijhout H., editors. Springer Singapore; 2017. A practical guide to CRISPR/Cas9 genome editing in Lepidoptera; pp. 155–172. [Google Scholar]
- Zhao Y., Huang G., Zhang W. Mutations in NlInR1 affect normal growth and lifespan in the brown planthopper Nilaparvata lugens. Insect Biochem.Mol. Biol. 2019;115:103246. doi: 10.1016/j.ibmb.2019.103246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original sequencing data for figures in the paper are available at Mendeley data (https://data.mendeley.com/datasets/wzj67n7gz6/2). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the LEAD Contact upon request.