Abstract
Ulvans, complex polysaccharides found in the ulvales (green seaweed) cell wall, contain predominantly 3-sulfated rhamnose (Rha3S) linked to either d-glucuronic acid, l-iduronic acid or d-xylose. The ulvan lyase endolytically cleaves the glycoside bond between Rha3S and uronic acid via a β-elimination mechanism. Ulvan lyase has been identified as belonging to the polysaccharide lyase family PL24 or PL25 in the carbohydrate active enzymes database, in which fewer members have been characterized. We present the cloning and characterization of a novel ulvan lyase from Pseudoalteromonas sp. strain PLSV (PsPL). The enzymes were heterologously expressed in Escherichia coli BL21 (DE3) and purified as the His-tag fusion protein using affinity chromatography, ion-exchange chromatography and size-exclusion chromatography. The degradation products were determined by thin-layer chromatography (TLC), liquid chromatography-mass spectrometry (LC-MS) to be mainly disaccharides and tetrasaccharides. Ulvan lyase provides an example of degrading ulvales into oligosaccharides. Arg265, His152 and Tyr249 were considered to serve as catalytic residues based on PsPL structural model analysis.
Ulvans, complex polysaccharides found in the ulvales (green seaweed) cell wall, contain predominantly 3-sulfated rhamnose (Rha3S) linked to either d-glucuronic acid, l-iduronic acid or d-xylose.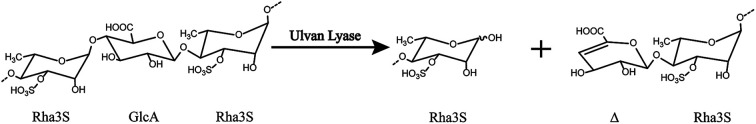
Introduction
There is a large amount of polysaccharides in nature and many enzymes have been studied for their synthesis, processing and degradation [Carbohydrate-Active enzymes database (CAZy)].1,2 These enzymes cleave the bond between the bridging oxygen of the glycosidic linkage and the anomeric carbon. The predominant catalytic mechanism for the degradation of oligosaccharides is hydrolysis and more than 140 different glycosyl hydrolase (GH) families have been identified to date (CAZy database). The polysaccharide lyase (PL) family has been divided into 27 subfamilies.3 Only two general catalytic mechanisms have been identified among PL families and several different protein folds have been observed.4,5
Ulvales are common seaweeds that are related to proliferations in eutrophicated coastal waters.6,7 This kind of green alga represents an important biomass that is grown and collected for food or feed. They are generally used as sea-lettuce for rich dietary fibers. Therefore, they play an important role in the purification and recycling of water.8–11 It is necessary to perform further research on them for the potential industrial application and environmental improvement. Ulvan is the main polysaccharide components of cell wall (29% wt),12 and it has a potential application in the fields of food, agriculture, pharmaceuticals and chemistry.4,13 Ulvan constitutes of d-glucuronic acid (GlcA), d-xylose (Xyl), l-iduronic acid (IdoA) and 3-sulfated rhamnose (Rha3S).14–16 The repeating disaccharides of ulvan are composed of Rha3S linked with either GlcA or IdoA, which is named as ulvanobiuronic acid A or B respectively.17 However, the disaccharide portion of Rha3S linked to Xyl occurs in a lower amount.
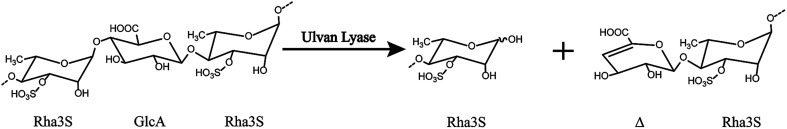
Ulvan lyase cleaves the β(1-4) glycosidic bond between Rha3S and uronic acid via the beta-elimination mechanism (Fig. 1).18 Lahaye and his colleagues found and identified the first ulvan lyase, which was isolated from Gram-negative bacteria.8 Recently, Nyvall Collen identified the Gram-negative Nonlabens ulvanivorans PLR19 and Kopel sequenced its genome.20 Ulvan lyases isolated from the Flavobacteria Persicivirga ulvanivorans are identified by Collén.21 Here we presented the characterization of ulvan lysas from Pseudoalteromonas sp. strain PLSV (PsPL). The recombinant protein was overexpressed in E. coli BL21 (DE3), and purified by Ni2+ affinity chromatography and ion exchange chromatography. The products were determined by using TLC, LC-MS analysis.
Fig. 1. The overall reaction catalyzed by ulvan lyase. Ulvan lyase mode of action. The β-eliminative cleavage lead to the formation of an unsaturated ring (Δ, 4-deoxy-l-threo-hex-4-enopyranosiduronic acid) on the non-reducing end of one fragment and a reducing end on the other fragment.
Experimental procedures
Strain and plasmid constructions
The PsPL gene (GenBank No. AMA19992.1) was amplified by polymerase chain reaction (PCR) with the forward primer 5′-GGAATTCCATATGATGAGCAAAGCCAACGGCGTG-3′ and the reverse primer 5′-CGGAATTCTTAGCGGCTCACGCCCAGATC-3′. The PCR fragment was digested with Nde I and EcoR I and inserted into the Nde I/EcoR I sites of plasmid pET-28a including His6-tag in the same open reading frame. The constructed recombinant plasmids were verified by restriction digestion enzymes, and sequencing. The plasmid harboring PsPL gene were subsequently transformed into the protein expression host E. coli BL21(DE3).
Overexpression and enzyme purification
The strains were grown in 200 mL LB medium with shaking at 220 rpm at 37 °C. When OD600 reached 0.6–0.8, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM for induction for 16–18 h at 16 °C. Cells were harvested by centrifugation at 5000 rpm for 15 min, storing at −80 °C.
The cells were resuspended in 20 mL of 20 mM Tris–HCl buffer (pH 8.0) containing 0.5 M NaCl, 1 mM dithiothreitol (DTT), 20 mM imidazole. They were then disrupted by sonication using an ultrasonicator, set at 1.5 s pulse, 1 s output and 40% duty cycle for 20 min on ice. After that, the cell debris was removed by centrifugation at 40 000 × g at 4 °C for 30 min. Cleared lysate was loaded onto an open column and the PsPL was trapped on 1 mL of Ni-NTA Superflow resin (Qiagen, Hilden, Germany). PsPL was washed with 10 mL wash buffer (20 mM Tris–HCl pH 8.0, 0.5 M NaCl, 1 mM DTT, 30 mM imidazole). After washing, the protein was eluted with 15 mL using elution buffer (20 mM Tris–HCl pH 8.0, 0.3 M NaCl, 1 mM DTT, 0.5 M imidazole). The soluble protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit. The molecular mass and purity of target protein were analyzed by SDS-PAGE.
Activity assay
The enzymatic activity was determined by the DNS method that reducing end-residue of saccharides was measured.22–25 The reaction systems were composed of 3 g L−1 ulvan in 0.1 M Tris–HCl, 0.2 M NaCl at pH 8.0 and 0.1 g L−1 purified enzyme. The reaction was carried out at 37 °C for 30 min in a final volume of 50 μL and stopped with 150 μL of DNS reagent. The reaction mixture was boiled at 100 °C for 5 min and immediately cooled on ice for 2 min. The absorbance values were determined at 540 nm. One unit (1 U) was defined as the amount of enzyme required to liberate 1 μmol of reducing sugar per minute.26 The kinetic parameter of PsPL was determined by measuring the enzyme activity with substrate at different concentrations (0.1–5.0 mg mL−1).
To determine the effect of metal ions on the activity of PsPL, 0.1 mM final concentrations of Mg2+, Ca2+, Cu2+, Mn2+, Co2+, Fe3+, Ni2+, Zn2+ and EDTA were added to the reaction mixture. The optimal pH of the purified enzyme was determined in 20 mM MES buffer (pH 5.5–6.5), 20 mM Tris–HCl buffer (pH 7.5–8.5) and 20 mM Gly-NaOH (pH 9.5). The optimal temperature was determined by incubating PsPL at different temperatures ranging from 25 to 70 °C. The activity was then measured under standard reaction conditions. All assays were repeated three times.
Product analysis using TLC, LC-MS
The products were determined by using thin-layer chromatography (TLC) on a 2 mm silica gel preparative TLC plate (Kieselgel 60F254, Merck AG, Darmstadt, Germany), with butyl alcohol : acetic acid : water = 2 : 1 : 1 as the solvent phase and p-anisaldehyde : methyl alcohol : H2SO4 = 1 : 15 : 2 as the chromogenic agent. A liquid chromatograph-mass spectrometer assay (LC-MS) was applied to analyze products catalyzed by PsPL. Samples were injected on a Prevail Carbohydrate ES column-W (5 μm, 4.6 × 250 mm, Agela Technologies, China), with an acetonitrile : water (75 : 25) mobile phase. Detection of products was performed with a UV detector operating at 235 nm.27 The MS analysis was performed in positive ion mode to determine the molecular weight of the expected product.
Structure modeling of the PsPL
The three-dimensional (3D) homology model of PsPL was generated on Swiss-Model (https://swissmodel.expasy.org/). The crystal structure of ulvan lyase PLSV_3936 from Pseudoalteromonas sp. strain PLSV (PDB ID: 5UAM) was chosen as the template. The substrate was docked based on the alignment of PsPL model and PLSV_3936 crystal structure.28
Results and discussion
Overexpression and purification of recombinant pET28a-PsPL
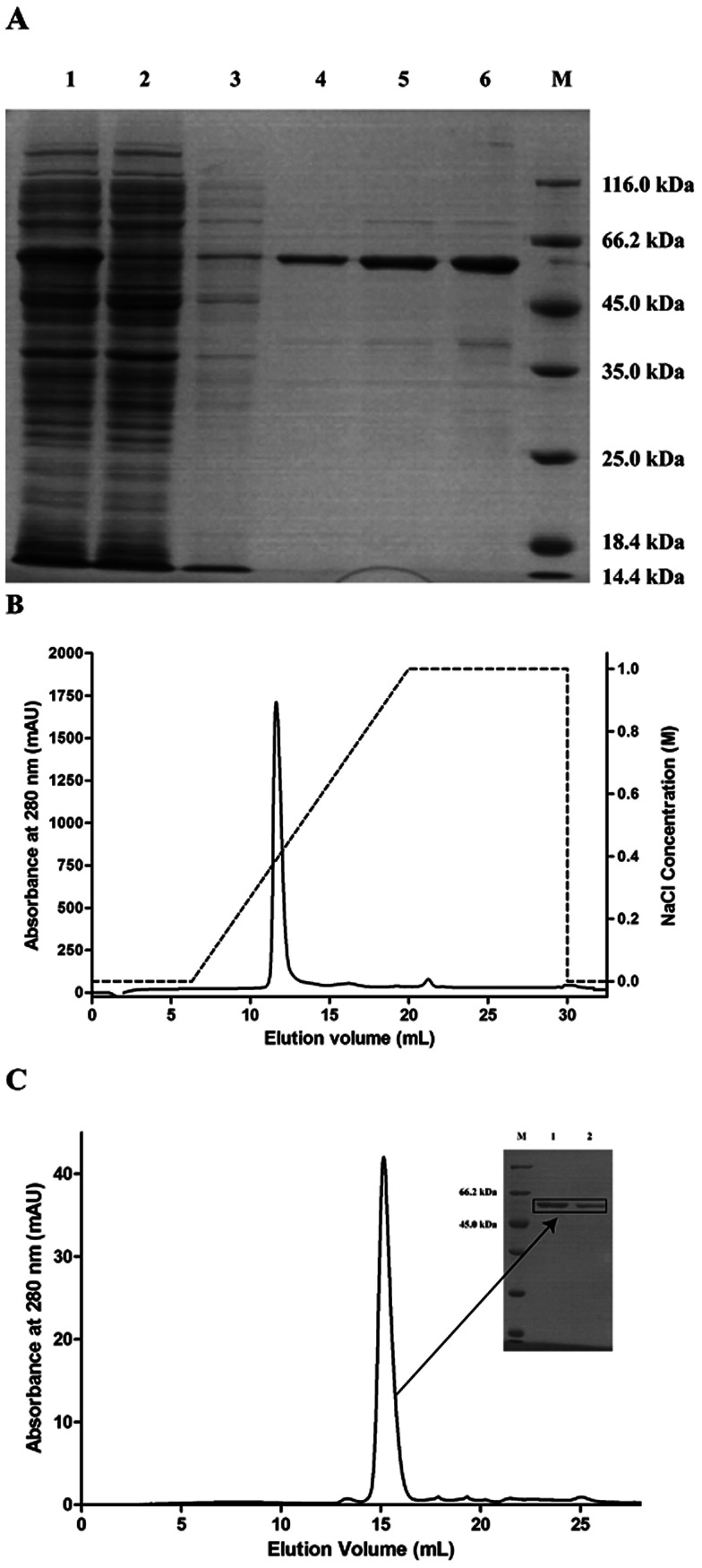
The pET28a-PsPL recombinant vector was transformed in the expression host E. coli BL21(DE3) for protein expression. The recombinant protein with a N-terminal His6-tagged fusion was solubly expressed at 16 °C and 0.2 mM IPTG. The cell-free extracts were trapped on a NI-NTA-agarose column. By increasing the imidazol concentration to 500 mM, the elution of the recombinant proteins from the affinity columns was completed. The eluted fractions were further purified by ion exchange chromatography using Mono Q (GE Healthcare) (Fig. 2B) and size-exclusion chromatography using a Superdex 200 HR 10/30 column. A single major peak was detected at 280 nm in the purified protein fraction (Fig. 2C). SDS-PAGE analysis showed that the molecular mass of the recombinant protein was about 59 kDa (Fig. 2A), which was consistent with the theoretic value. The purified proteins were collected, concentrated, and used for enzymatic analysis.
Fig. 2. Purification of pET28a-PsPL. (A) pET28a-PsPL was purified and analyzed by SDS-PAGE. Lane 1: supernatant; Lane 2: precipitant; Lane 3: flowthrough; Lane 4, 5, 6: elution. The recombinant protein was further purified by ion exchange chromatography (B) and size-exclusion chromatography (C).
Enzyme characteristics of ulvan lyase PsPL
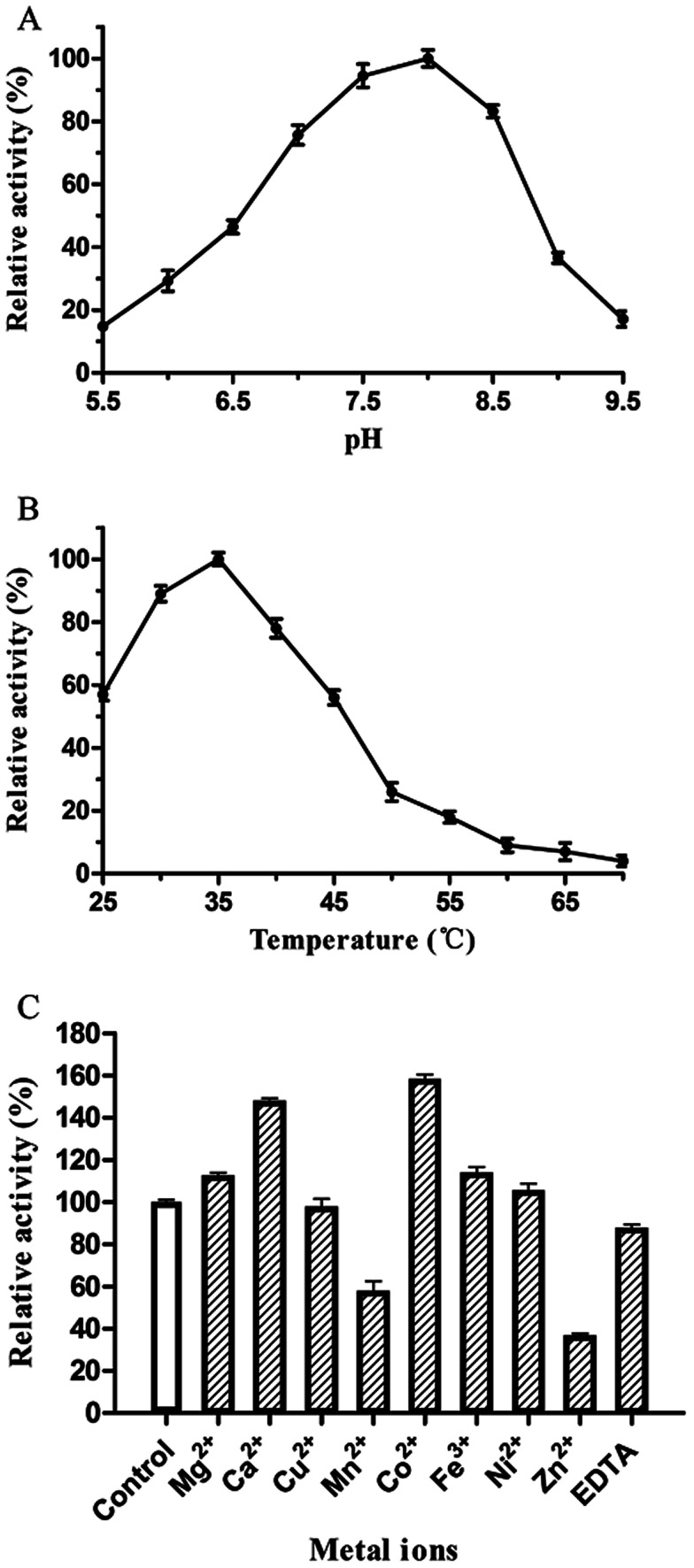
The catalytic activity of PsPL was related to the buffer composition, pH and temperature. PsPL was active at pH 5.5–9.5 and the maximal activity was observed in Tris–HCl buffer pH 8.0 (Fig. 3A). The optimal temperature for PsPL activity was 35 °C (Fig. 3B). The activity of PsPL was affected by several metal ions (Fig. 3C). Notably, Ca2+ and Co2+ increased the activity by 45 and 60%, respectively. The Michaelis–Menten constant (Km) and the catalytic rate constant (kcat/Km) of PsPL toward ulvan were 2.10 mg mL−1 and 3.37 mL mg−1 s−1, respectively (Table 1 and Fig. S1†).
Fig. 3. Effect of pH (A), temperature (B) and metal ions (C) on activity of PsPL. All assays were repeated three times.
Kinetic parameter of PsPL.
| Substrate | K m (mg mL−1) | V max (U mg−1) | k cat (s−1) | k cat/Km (mL mg−1 s−1) |
|---|---|---|---|---|
| Ulvan | 2.10 ± 0.31 | 12.74 ± 0.89 | 7.08 ± 0.12 | 3.37 |
Characterization of reaction product confirmation
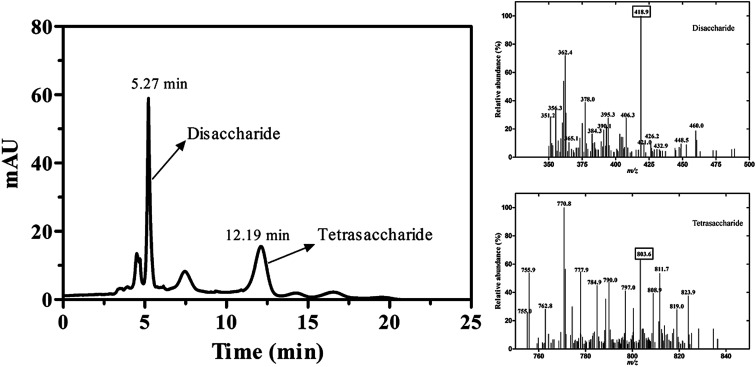
The reaction mixture was analyzed by TLC on Silica F254 plates. The new spot was shown above ulvan in the reaction mixture (Fig. S2†). For the qualitative analysis of the new compound, samples were analyzed by LC-MS, whereas new peaks appeared at retention time of 5.27 min and 12.19 min. The MS analysis of the compound in positive mode showed that the ion fragments of m/z were 418.9 and 803.6, which was consistent with the molecular weight of disaccharide and tetrasaccharide products (Fig. 4), respectively. It also implied that the substrate (Ulvan) was mainly cleaved by ulvan lyase into disaccharide.
Fig. 4. The LC-MS analysis of reaction products catalyzed by PsPL. Two peaks appeared at retention time of 5.27 min and 12.19 min (left). The ion fragments of m/z were 418.9 and 803.6 (right).
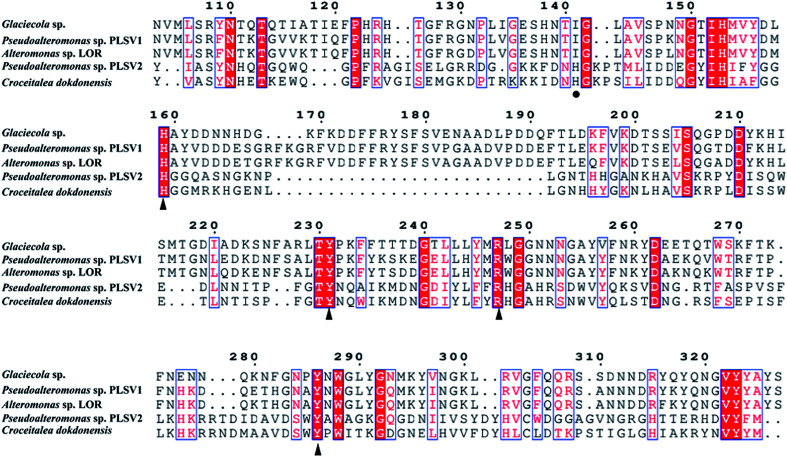
Phylogenetic tree analysis and amino acid sequence alignments of PsPL
The enzymes with different amino acid sequences in the CAZy database PL24 and PL25 families were blasted and their sequences were compared with PsPL (Fig. S3†). PsPL shares 43.44%, 38.47%, 31.58% sequence identities with Alteromonas sp. LOR (AMA 19991.1), Catenovulum agarivorans DS2 15349 (EWH08847.1) and Glaciecola sp. (BAY00693.1), respectively. Although the crystal structure of ulvan lyases (PLSV3936) from Pseudoalteromonas sp. PLSV2 has been determined, PsPL only shares sequence identity of 11.5% with it. The sequence alignment result showed that His173, Tyr249, Arg265 and Tyr304 of PsPL [His159, Tyr231, Arg247 and Tyr286 of ulvan lyases (PLSV 3936)] are conserved in ulvan lyase family (Fig. 5, marked with ▲). Among these residues, Tyr231 and Arg247 were reported to make contacts with the substrate GlcA residue.
Fig. 5. Amino acid sequence alignments of PsPL from different strains. The speculated conserved residues marked with ▲, the amino acids associated with the binding of the substrate marked with ●.
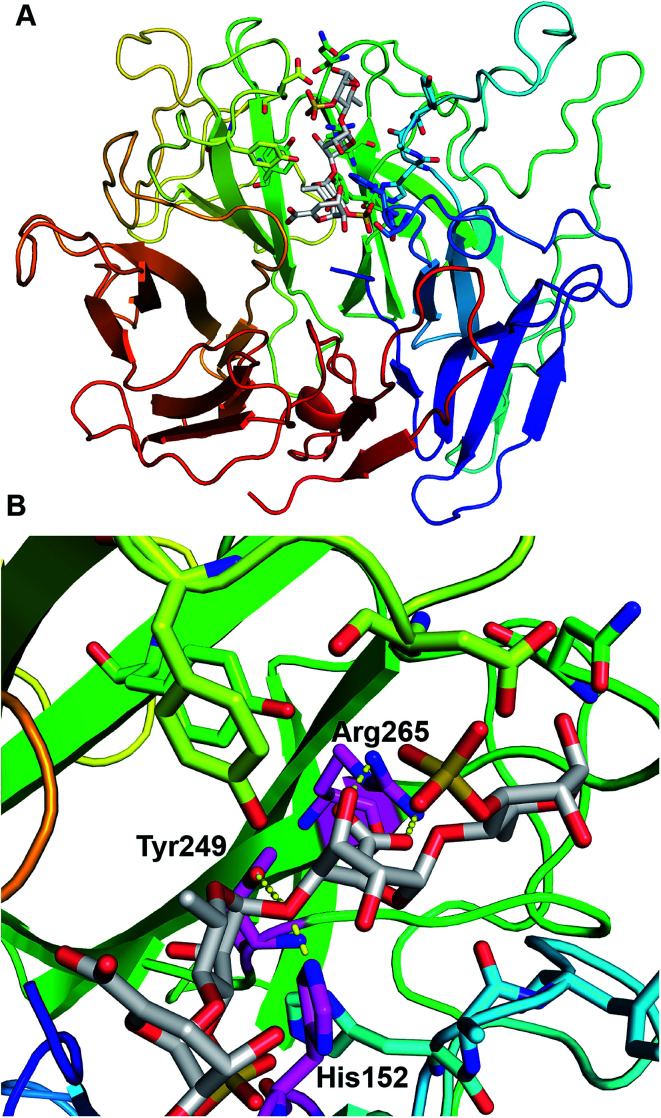
Structure modeling of PsPL and the active site
PsPL adopts the fold of a seven-bladed β-propeller, whereas each propeller consists of four antiparallel β-strands (Fig. 6A). The active site is located at top of propellers. Arg265 neutralizes the charge of the carboxylic group of GlcA with the aid of His152 and Tyr249, which implying these three residues are considered to serve as catalytic residues (Fig. 6B). Additional, Ile155, Tyr249 and Arg265 of PsPL [His141, Tyr231 and Arg247 of ulvan lyases (PLSV 3936)] were associated with the substrate binding (Fig. 6, marked with ●).
Fig. 6. Structural analysis of the PsPL-substrate complex model (A) and catalytic residues at active site (B).
Proposed catalytic mechanism
The ulvan lyase cleavages the glycoside bond between Rha3S and uronic acid via β-elimination mechanism. The ulvan lyase cleaves the glycosidic bond adjacent to the carboxylate group, this reaction results in the formation of new reducing and an unsaturated sugar. The specific process can be divided into three steps. Firstly, neutralization of the C-5 carboxyl group that is a prerequisite for C5 protons to be vulnerable to base attacks. Secondly, an enolate intermediate is formed. Finally, the elimination of glycosidic bond is accompanied by the transfer of electron from the carboxylate groups to form double bonds between C4 and C5. This elimination reaction requires a Brønsted base to accept H-5 protons and a Brønsted acid to donate proton to re-form a new reducing end. In addition, these catalytic mechanisms fall into two general categories, one is based on Lys or Arg as a Brønsted base and a water molecule as a Brønsted acid, a metal-assisted neutralizing acidic group, and the other is Tyr or His as Bronsted base, Tyr as Bronsted acid, Asn/Gln or a protonated Asp/Glu neutralizing acidic groups.29,30
Conclusion
We characterised a novel ulvan lyase from Pseudoalteromonas sp. strain PLSV, which is responsible for the endolytic cleavage of the glycoside bond between Rha3S and uronic acid. The enzyme could be solubly expressed and purified. The disaccharide was determined as the main reaction product by LC-MS. Further investigation on structural analysis and the degrading mechanism of PsPL is essential to explore the potential industrial catalyst.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by National Key Research and Development Project (2016YFD0400803), National Natural Science Foundation of China (31771911), Natural Science Foundation of Tianjin (16JCQNJC09200) and the Overseas High-level Talents Program of Tianjin University of Science and Technology to H.-M. Qin, China.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12294b
References
- Lombard V. Golaconda Ramulu H. Drula E. Coutinho P. M. Henrissat B. Nucleic Acids Res. 2014;42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrapon N., Lombard V., Drula E., Coutinho P. M. and Henrissat B., The CAZy database/the carbohydrate-active enzyme (CAZy) database: principles and usage guidelines, Springer, Japan, 2017, pp. 117–131 [Google Scholar]
- Lombard V. Bernard T. Rancurel C. Brumer H. Coutinho P. M. Henrissat B. Biochem. J. 2010;432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- Charnock S. J. Brown I. E. Turkenburg J. P. Black G. W. Davies G. J. Proc. Natl. Acad. Sci. 2002;99:12067–12072. doi: 10.1073/pnas.182431199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garron M. L. Cygler M. Glycobiology. 2010;20:1547–1573. doi: 10.1093/glycob/cwq122. [DOI] [PubMed] [Google Scholar]
- Lahaye M. Robic A. Biomacromolecules. 2007;8:1765–1774. doi: 10.1021/bm061185q. [DOI] [PubMed] [Google Scholar]
- Morand P. Briand X. Bot. Mar. 1996;39:491–516. [Google Scholar]
- Lahaye M. Brunel M. Bonnin E. Carbohydr. Res. 1997;304:325–333. doi: 10.1016/S0008-6215(97)00270-X. [DOI] [PubMed] [Google Scholar]
- Cohen I. Neori A. Bot. Mar. 1991;34:475–482. [Google Scholar]
- van der Meulen H. Gordin H. J. Phycol. 1990;2:363–374. doi: 10.1007/BF02180927. [DOI] [Google Scholar]
- Guiry M. D. and Blunden G., Seaweed Resources in Europe : Uses and Potential, John Wiley, Chichester, U.K., 1991, pp. 149–168 [Google Scholar]
- Robic A. Gaillard C. Sassi J. F. Lerat Y. Lahaye M. Biopolymers. 2009;91:652–664. doi: 10.1002/bip.21195. [DOI] [PubMed] [Google Scholar]
- Lahaye M. Jegou D. J. J. Appl. Phycol. 1993;5:195–200. doi: 10.1007/BF00004017. [DOI] [Google Scholar]
- Percival E. and McDowell R. H., Chemistry and Enzymology of Marine Algal Polysaccharides, Academic Press, London, 1967, p. 219 [Google Scholar]
- Ray B. Lahaye M. Carbohydr. Res. 1995;274:251–261. doi: 10.1016/0008-6215(95)00138-J. [DOI] [Google Scholar]
- Quemener B. Lahaye M. Bobin Dubigeon C. J. Appl. Phycol. 1997;9:179–188. doi: 10.1023/A:1007971023478. [DOI] [Google Scholar]
- Lahaye M. Inizan F. Vigouroux J. Carbohydr. Polym. 1998;314:1–12. doi: 10.1016/S0008-6215(98)00293-6. [DOI] [Google Scholar]
- Wong T. Y. Preston L. A. Schiller N. L. Annu. Rev. Microbiol. 2000;54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- Nyvall Collen P. Sassi J. F. Rogniaux H. Marfaing H. Helbert W. J. Biol. Chem. 2011;286:42063–42071. doi: 10.1074/jbc.M111.271825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopel M. Helbert W. Henrissat B. Doniger T. Banin E. Genome Announc. 2014;2:e00793–14. doi: 10.1128/genomeA.00793-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collén P. N. Sassi J. F. Rogniaux H. Marfaing H. Helbert W. J. Biol. Chem. 2011;286:42063–42071. doi: 10.1074/jbc.M111.271825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. L. Anal. Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- El-Katatny M. H. Hetta A. M. Shaban G. M. El-Komy H. M. Food Technol. Biotechnol. 2003;41:219–225. [Google Scholar]
- Zhu B. Chen M. Yin H. Du Y. Ning L. Mar. Drugs. 2016;14:108. doi: 10.3390/md14060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista D. E. de Araújo E. A. Neto M. O. Kadowaki M. A. S. Polikarpov I. New Biotechnol. 2018;40:268–274. doi: 10.1016/j.nbt.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Qin H.-M. Miyakawa T. Inoue A. Nakamura A. Nishiyama R. Ojima T. Tanokura M. Sci. Rep. 2017;7:11425. doi: 10.1038/s41598-017-11542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopel M. Helbert W. Belnik Y. Buravenkov V. Herman A. Banin E. J. Biol. Chem. 2016;291:5871–5878. doi: 10.1074/jbc.M115.673947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulaganathan T. Boniecki M. T. Foran E. Buravenkov V. Mizrachi N. Banin E. Helbert W. Cygler M. ACS Biol. Chem. 2017;12:1269–1280. doi: 10.1021/acschembio.7b00126. [DOI] [PubMed] [Google Scholar]
- Huang W. Boju L. Tkalec L. Su H. Yang H. O. Gunay N. S. Lihardt R. J. Kim Y. S. Matte A. Cygler M. Biochemistry. 2001;40:2359–2372. doi: 10.1021/bi0024254. [DOI] [PubMed] [Google Scholar]
- Gacesa P. FEBS Lett. 1987;212:199–202. doi: 10.1016/0014-5793(87)81344-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.