Abstract
Poly(lactic-co-glycolic acid) (PLGA) particles are very widely used, particularly for drug delivery, including commercial clinical formulations. Adding perfluorocarbon (PFC) enables in vivo imaging and quantification of the PLGA particles through 19F NMR, MRS or MRI. PFCs are both hydrophobic and lipophobic at the same time. This property makes their encapsulation in particles challenging, as it requires the addition of a third immiscible phase during the emulsification process. Here we explore how different parameters affect the miniemulsion formation of particles loaded with perfluoro-15-crown-5-ether (PFCE). By changing the concentration of surfactant and type of solvent, we were able to control the radius of synthesized particles, between 85–200 nm. We assessed stability and release from the particles at different pH values, showing that hydrophobic agents are released from the particles by diffusion rather than degradation. With cell experiments, we show that primary human dendritic cells take up the particles without any apparent effect, including on cell migration. In summary, the control of synthesis conditions leads to particles with sufficient PFCE encapsulation, which are suitable for drug loading and cell labeling, and do not affect cell viability or functionality. Finally, these nanoparticles can be produced at GMP-grade for clinical use.
The influence of different synthesis parameters on the characteristics of polymeric particles with a third perfluorocarbon phase.
Introduction
Recent developments in the field on nanomedicine have focused on multifunctional theranostic devices. Such theranostic devices, encapsulating both a contrast agent for imaging, e.g. magnetic resonance imaging (MRI), and a drug for therapy, allow not only for the delivery of a therapeutic compound to the site of interest, but also give the ability to visualize and monitor therapy and progression of the disease.1 For these purposes, the use of biodegradable polymeric particles is of great interest due to their ease of processing, biological stability, low toxicity, and effectiveness as delivery agents.2 Poly(lactic-co-glycolic acid) (PLGA) is particularly attractive for the synthesis of nanoparticles, as PLGA is biocompatible, biodegradable, and shows long-term storage stability.3 PLGA nanoparticles can be modified, for example with contrast agents and drugs, by either covalent coupling or non-covalent loading using the hydrophobic effect.4–6 PLGA protects the drug from fast degradation and the release time of the encapsulated cargo from the particles can be modulated by altering the synthesis parameters and polymer composition.7
Loading the particles with liquid perfluorocarbons (PFCs) for fluorine (19F) MRI is advantageous for biomedical use, as it can be used for in vivo non-invasive imaging. Fluorine is essentially absent from biological tissues, therefore, 19F MRI results in high specificity of images and moreover, enables the quantification of the signal, including determination of 19F content and particularly quantification of 19F-labeled cells directly from image data.8,9 PFCs are simultaneously hydrophobic and lipophobic synthetic organic compounds with a high payload of fluorine atoms,10 with a long history of clinical application, initially as blood substitutes and more recently for imaging applications.11,12 To overcome the immiscibility of PFCs, liquid PFCs are usually delivered as surfactant-stabilized PFC emulsions for 19F MRI. However, the colloidal stability of PFC emulsions is relatively low, especially at high concentrations required for cells loading, which further may lead to accumulation in organs and poor biodistribution.9,13–16 The stability can be improved with the use of phospholipids or poloxamers, however both types of surfactants exhibit some disadvantages, e.g. they are prone to hydrolysis and oxidation (phospholipids) or they tend to form highly viscous dispersions (poloxamers).17
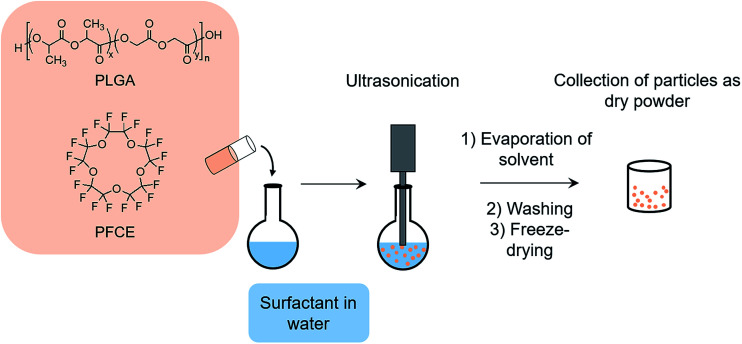
Encapsulation of PFCs into polymeric nanoparticles is relatively new to the field and has some specific advantages, particularly for cell loading and imaging. Here, perluoro-15-crown-5-ether (PFCE) is of special interest, as it is chemically and biologically inert and consists of 20 chemically equivalent 19F nuclei, leading to a singlet in NMR spectra, which is an important factor for quantification and MR imaging.18,19 PFCE-loaded PLGA particles can be synthesized using a miniemulsion formulation technique (Fig. 1).3 It has been previously shown that parameters such as type of solvent and surfactant, and concentration of surfactant and polymer can influence the characteristics of PLGA nanoparticles prepared by the solvent evaporation method.20
Fig. 1. Schematic representation of miniemulsion formulation method. PLGA and PFCE are added to surfactant solution under sonication, followed by solvent evaporation. Next, particles are washed, freeze-dried, and collected as a dry powder.
Aiming at different applications, such as cell tracking and imaging of drug delivery, it is important to control the properties of PLGA–PFC particles, including diameter, diameter distribution, fluorine content, and encapsulation efficiency of hydrophobic drug or other agents. Particle size and size distribution influence the in vivo distribution, cell uptake, biological fate, toxicity and the targeting ability. For example, Waiczies et al. reported on fluorine-rich emulsion particles, where larger particles (280 nm in radius) were more readily taken up by dendritic cells (DCs) and promoted the DCs maturation process.21 These particle characteristics can also affect drug loading, drug release, and stability of particles.22 However, systematic study of perfluorocarbon-loaded PLGA nanoparticles have not been reported thus far. Hence, there is little prior work describing a systematic study of triphasic PLGA–PFC systems, although these have been shown to have a very high loading and excellent stability.3,23 In this work, we explore the influence of different synthesis parameters on the radius, homogeneity, stability and encapsulation efficiency of PLGA–PFCE nanoparticles and the influence of the PFC phase on particle characteristics. We focus only on PLGA–PFCE nanoparticles, which are very different from PLGA particles that are extensively described in the literature. We also investigate the impact of PFC-encapsulation on the release characteristics of PLGA particles. For this, we introduce nile blue dye into the nanoparticles, as a model hydrophobic drug. Finally, we compare the cellular uptake of different particles in primary human monocyte-derived dendritic cells (moDCs), and the effects of uptake on cell viability and functionality. Overall, the objective of the present study is to show how different synthesis parameters influence the synthesis of PLGA particles with a third PFC phase, with a view to biomedical applications.
Results and discussion
Change in surfactant concentration and type of solvent influences the final properties of particles
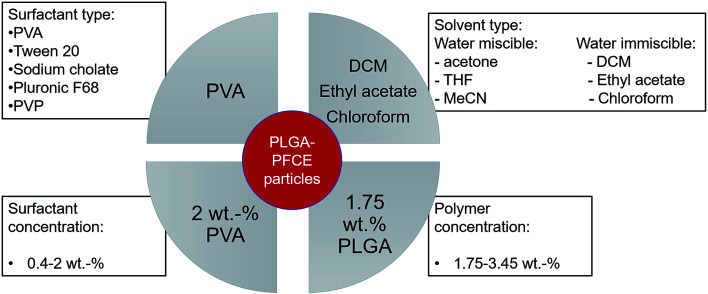
The main parameters, which affect the synthesis of PLGA particles are type and concentration of surfactant, concentration of polymer, and type of solvent for the organic phase (Scheme 1).3,21,24–26
Scheme 1. Schematic representation of parameters, which affect properties of nanoparticles, tested in this study, including factors that resulted in nanoparticles with low PDI, narrow size distribution and high PFCE content.
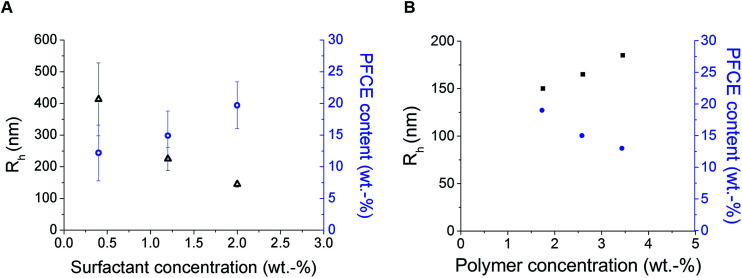
Loading the particles with PFC alters the properties of miniemulsions, as PFCs form their own phase in solutions. Among different surfactants, polyvinyl alcohol (PVA) with low molecular weight (10 kDa) resulted in nanoparticles with a small hydrodynamic radius (Rh), and high PFCE loading. Thus, by reducing the concentration of surfactant, in this case PVA, which is perhaps the most often used surfactant for synthesis of PLGA particles, we could obtain particles with a larger size. This result is similar to PLGA particles without PFC (Fig. 2).20,26,27
Fig. 2. Results of DLS (black) and NMR (blue) measurements of nanoparticles prepared with different concentration of surfactant (A) or PLGA (B). (A) Influence of PVA concentration on particle radius and PFCE encapsulation. Particles were prepared using following concentrations of surfactant: 0.4 wt%, 1.2 wt% and 2 wt%. Here, the concentration of polymer in organic phase was the same for all samples, 1.75 wt%. Better reproducibility of particle radius was achieved with higher concentration of surfactant. Furthermore, higher concentration of PVA resulted in nanoparticles with higher encapsulation efficiency of PFCE. Error bars represent the standard deviation (n = 3) (B) influence of concentration of PLGA on particle radius and PFCE encapsulation. Higher concentration of polymer resulted in slightly larger particles, but lower encapsulation efficiency. For all samples concentration of surfactant at 2 wt% was used. DLS results are presented in black and NMR results in blue. DLS: c(NP) = 0.1 mg ml−1, NMR with TFA as internal reference.
However, the polydispersity of particles also increased, while the encapsulation yield of PFCE dropped, perhaps because at lower concentrations of surfactant, stabilization of the PFCE-droplets was not achieved. In a miniemulsion, diameter and diameter distribution of droplets directly depend on the concentration of surfactant. A higher concentration of surfactant in the aqueous phase (in this case 2.0 wt%) leads to a sufficient coverage of the interface between droplet and aqueous phase by surfactant, which stabilizes the emulsion droplets and prevents the coalescence. Hence, a higher concentration of surfactant results in more homogenous particles with a narrower size distribution. This effect is reflected in lower PDI values (Fig. S1†), as well as in lower standard deviation between measurements. Thus, particles prepared with 2 wt% of surfactant have lower PDI values and the diameter measurements also display better reproducibility than at lower concentration of surfactant. Interestingly, several other surfactants that are commonly used in synthesis of PLGA particles without PFC,27,28 were not able to stabilize particles when the PFCE phase was added (Table S1†).
Furthermore, increasing concentration of polymer, while keeping the volume of organic phase and other parameters constant, resulted in larger particles with lower PFCE loading (Fig. 2B). Typically, increasing concentration of polymer leads to the formation of bigger droplets due to increased viscosity of the organic phase.20,29,30 The total amount of obtained nanoparticle yield was slightly higher when we increased the concentration of PLGA, while the overall amount of PFCE in the whole batch was at around 20 mg for all three concentrations of polymer (Table 1). Finally, the type of solvent used for miniemulsion process did not affect the properties of the final particles. Thus, using chloroform or ethyl acetate, which are both higher polarity solvents resulted in similar particles as with dichloromethane (DCM) (Table S2†).
Characterization of nanoparticles prepared with combination of different solvents.
| Nanoparticle (solvent type) | R (nm) ± SD (cryoSEM) | R h ± SD (DLS at 173°) | PFCE encapsulation (wt%) | Dye concentration μg ml−1 |
|---|---|---|---|---|
| DCM | 120 ± 20 | 93 ± 5 | 22 | 8 |
| DCM + MeCN | 90 ± 20 | 85 ± 2 | 19 | 6 |
| DCM + toluene | 253 ± 62, 110 ± 25 | 117 ± 32 | 19 | 12 |
Along with the miniemulsion technique, nanoprecipitation is another common technique for the synthesis of PLGA nanoparticles. We investigated nanoprecipitation to better understand the formation of nanoparticles. As nanoprecipitation does not involve the use of shear forces; it is not suitable for the encapsulation of PFCE. However, we modified a typical nanoprecipitation procedure by applying sonication during the addition of the organic to aqueous phase; this resulted in increased size and polydispersity of nanoparticles, and low PFCE encapsulation (Table S2†).
Combination of DCM with other solvents with different polarities influences the size of particles
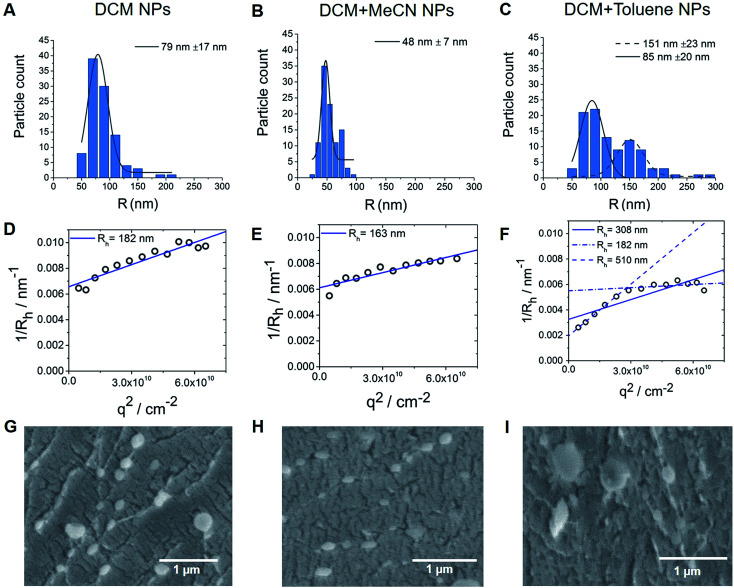
Interestingly, in contrast to replacing DCM with another water immiscible solvent (Table S2†), mixing DCM with solvents of higher or lower polarity, while keeping the concentration of surfactant and polymer constant (2 and 1.75 wt% respectively), resulted in altered size of particles, with efficient encapsulation of PFCE (Table S3†). Thus, the combination of DCM with the higher polarity solvent acetonitrile (MeCN, relative polarity 0.460) or lower relative polarity solvent toluene (relative polarity 0.099) resulted in smaller or bigger size respectively, according to Transmission Electron Microscopy (TEM) (Fig. 3B and C). The differences between solvents result from the interfacial tension between aqueous and organic phase. The interfacial tension between aqueous and organic phase is higher for solvents with lower polarity, such as DCM + toluene mixture.31 Bigger droplets have lower surface-to-volume ratio. Therefore, for oil-in-water miniemulsion the formation of bigger droplets is facilitated, when solvents with lower polarity are used for dispersed phase, compared to solvents with higher polarity.31 An additional reason for the smaller size of particles prepared with DCM + MeCN mixture could be a diffusion of water miscible MeCN into aqueous phase directly after the formation of the droplets. This diffusion results in faster shrinking of the droplets and thus in smaller size of the final particles. However, further experiments are necessary to confirm this mechanism.
Fig. 3. (A–C) Nanoparticle size distribution based on TEM images. Bimodal size distribution can be observed for particles made with the combination of DCM with toluene solvent (3C). (D–F) Multi-angle LS results, where extrapolation q → 0 results in inverse z-average radii <Rh>1/z of 182, 163 and 308 nm for DCM, DCM + MeCN and DCM + toluene nanoparticles respectively. (G–I) CryoSEM images, here both fractions of particles are visible for particles made with DCM + toluene (cNP = 10 mg ml−1). Also, some larger particles could be found with samples made either with DCM only or with DCM + MeCN solvents.
TEM data analysis of these nanoparticles differed from the results of Dynamic Light Scattering (DLS) (Table S3†). TEM results showed a bimodal size distribution (151 nm and 85 nm fractions) of nanoparticles prepared with DCM + toluene as solvent (Fig. 3C), while DLS measurements of these particles showed a radius of 195 nm. These DLS measurements were done with backscatter detection at 173°, which is less sensitive to the presence of larger particles, as larger particles primarily scatter light at forward angles. For particles larger than λ/20 (λ is wavelength of the laser) the normalized single particle scattering intensity the particle form factor P(q) becomes dependent on scattering angle, represented by the scattering vector q. Thus, the values of the diffusion coefficient at single measurement angles are apparent values. Therefore, to obtain the absolute z-average of the diffusion coefficient for calculation of absolute hydrodynamic radius, the apparent values of the diffusion coefficient need to be extrapolated q → 0, which corresponds to P(q) = 1.32–34 To investigate this discrepancy between TEM and single-angle DLS, we characterized the particles with multi-angle DLS.
Fig. 3D–F shows the angular dependency of the inverse hydrodynamic radius 1/Rh, which was obtained from apparent diffusion coefficients with the Stokes–Einstein equation. Extrapolation q → 0 results in inverse z-average of radii <Rh>1/z of 182, 163 and 308 nm for DCM, DCM + MeCN and DCM + toluene nanoparticles respectively, with the μ2 values of second cumulant from cumulant analysis of autocorrelation function at θ = 90°of 0.13, 0.08 and 0.27 (Fig. 3D–F).32 Typically, particles with μ2 values smaller than 0.05 are considered as monodisperse, while values higher than 0.2 are described as polydisperse.32 As the size distribution of particle seems to be non-Gaussian according to TEM, especially for toluene particles, we did not recalculate μ2 values into PDI value. Further analysis of angular dependency of DCM + toluene particles indicates the presence of two size fractions, which were also visualized by TEM. Thus, when only apparent values from θ = 30°–70° are used, the resulting <Rh>1/z = 510 nm, while extrapolation of bigger angles results in <Rh>1/z = 182 nm. Generally, the hydrodynamic radii obtained from DLS are bigger than the radii from TEM, indicating that particles shrink when dried for TEM. Especially, in the case of DCM + MeCN particles this effect seems to be quite pronounced, as the hydrodynamic radius is three times higher than the TEM-determined radius. To study this behaviour further we measured cryogenic Scanning Electron Microscopy (cryoSEM), which enables imaging of particles in a solution-like state. CryoSEM demonstrates that the majority of DCM + MeCN particles have an approximate radius around 65–90 nm, however, some particles have a radius around 125–175 nm. With LS, only this fraction of bigger particles could be detected. The results from cryoSEM on DCM + toluene particles confirm the presence of both sizes 90–130 nm and 180–230 nm. We also observed two fractions of particles in multi-angle LS as presented in Fig. 3C. Thus, cryoSEM confirms the results, which were obtained with LS.
The size of particles affects the release rate of hydrophobic agent
The release rate is a very important parameter when developing systems suitable for different biomedical applications, as it determines whether the nanoparticles will be able to reach the site of interest, before significant degradation and loss of active component has occurred. Factors influencing the release rate include e.g. particle radius and pH of the environment. The pH of the medium has a significant influence on PLGA degradation, as a lower pH increases the degradation rate of polymer chains.35
To study the effect of size and PFCE-loading on release, we synthesized particles with the addition of fluorescent dye, nile blue, as a model compound for hydrophobic drugs, such as paclitaxel, which have previously been encapsulated in PLGA nanoparticles.36 Concentration of dye varied between different nanoparticles and increased with an increase in nanoparticle radius (Table 1), as determined by ultraviolet/visible (UV/vis) spectroscopy. The particles were incubated at 37 °C in solutions with pH values of 4.0, 5.5 and 8.5 to compare the release in acidic and alkaline conditions. These pH values were chosen based on the pH of different subcellular compartments, namely lysosomes, endosomes and mitochondria respectively. A study by Panyam et al. showed that PLGA nanoparticles of approximate mean radius of 0.05 μm, incubated at 37 °C in pH 7.4 maintained their structural integrity for up to 20–30 days, followed by the formation of pores, deformation and fusion of particles.37 Thus, we expect PLGA–PFCE particles to remain stable after in vivo injection, until they are taken up the immune cells and reach the targeted site.
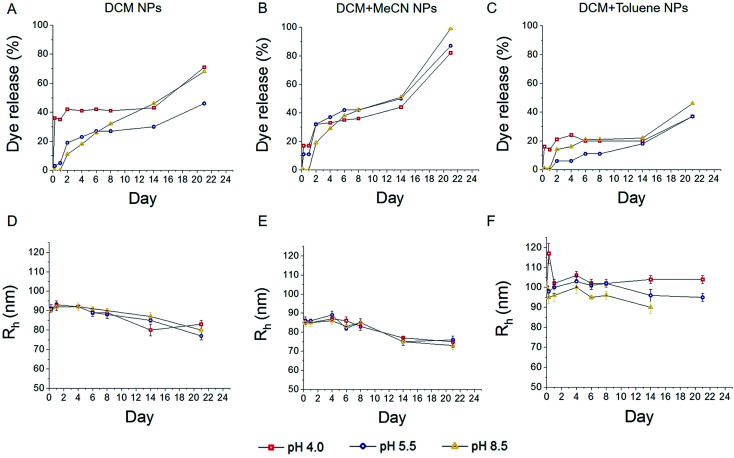
The percentage of released dye was calculated based on the UV/vis results (Fig. 4A–C). Mid-size and bigger particles, which were synthesized with DCM (Fig. 4A) and DCM + toluene (Fig. 4C), showed higher burst release of encapsulated dye at pH 4 after 6 hours. Even at later time points the release remained higher than at higher pH. In case of DCM particles, the release at pH 8.5 seems gradual, while at pH 5.5 faster release was observed within first two days and saturated afterwards. Larger particles display faster release within first two days, followed by saturation of both release curves. The release of dye from smaller particles was generally higher compared to both other samples. This trend can be explained by the fact that smaller particles have a larger surface area to volume ratio. With a larger surface area, water has better accessibility to enter the particles, resulting in an increase of degradation and release of the encapsulated compound. In the case of smaller particles, the release at pH 4 was also faster than at higher pH, although the difference was less pronounced than with larger particles.
Fig. 4. Cumulative dye release and changes in particle diameter after incubation at different pH. Faster dye release was observed for smallest particles (B), when compared to particles emulsified with DCM solvent (A), while larger particles resulted in slower release (C). DLS analysis of incubated samples, showed a slow gradual decrease in particles radius (D–F). Lines indicate trend of the data. NPs = nanoparticles.
It has been previously demonstrated that smaller nanoparticles display higher polymer degradation and release rates during the initial phase compared to larger particles.38 In our experiment, we also observed faster release of dye from DCM + MeCN particles (Fig. 4B) during the first 4 days of incubation when compared to DCM + toluene particles with larger radius (Fig. 4C). This trend can be explained by the fact that smaller particles have a larger surface area to volume ratio; with a larger surface area, water has better accessibility can more readily enter the particle, which further results in increased release of the encapsulated compound. While replicating the pH of various cell compartments, our model/assay lacks the presence of various enzymes that can influence the release.
To detect any changes in the average radius of nanoparticles and PDI upon incubation, we measured samples at various time points using DLS. The results are presented in Fig. 4D–F, and Fig. S3† (PDI). We observed a slow gradual decrease in particle radius (Fig. 4D–E) with an average reduction of about 10 nm within 24 days, for particles prepared with DCM and DCM + MeCN solvents (93 to 80 nm and 85 to 75 nm respectively). No significant changes in the radius of particles formulated with DCM + toluene mixture were detected (Fig. 4F).
In general, the size of particles decreased only slightly during the whole period, indicating that diffusion of the dye rather than particle degradation caused release. Note, however, that the dye itself has an adsorption peak close to the wavelength of the laser and could affect size determination. The increase in radius of DCM-toluene particles at 0 h time point could be due to agglomeration of the sample. However, the backscattering set up, which we used for size control during the release experiment, was not able to detect the fraction of bigger size fraction, which we detected with TEM and with multi-angle DLS. With smaller particles, a low fraction of larger particles affected determination of size, as shown in the previous section. Therefore, the limitations of a backscattering set up should be taken into account when looking at the results of these size measurements.
Overall, we found that PFCE-loaded particles show similar trends in release as PLGA nanoparticles without PFCE. In literature, PLGA particles do not show significant changes in size until up to 14 days of incubation in ref. 7, 37 and 39. For MRI applications, PFCs are usually used as an emulsion, which are stabilized by surfactants. The colloidal stability of these emulsions is usually low, in a range of hours to days especially at higher concentrations, as the stabilization of the PFC phase is a challenge due to solubility properties of PFCs. By encapsulation of this third PFC phase in the PLGA matrix, we obtained particles that were stable for at least 21 days at 37 °C, which is a significant improvement over many formulations of PFC emulsions. Furthermore, the stability of the particles, which were lyophilized and stored in a freezer, is at least 6 months.
To assess the influence of the third phase on the encapsulation of the hydrophobic agent, we synthesized particles encapsulating nile blue, and with or without the addition of PFCE. The highest encapsulation of nile blue was achieved with nanoparticles prepared with the use of DCM as solvent, both with and without the addition of PFCE, as presented in the Fig. S4.† Furthermore, the difference in nile blue encapsulation between these particles was minimal. Thus, we conclude that the incorporation of even relatively large amounts of PFC does not significantly affect the encapsulation and release of hydrophobic agents from the PLGA particles.
Nanoparticles are suitable for the loading of cells
To study the uptake of PFCE–PLGA nanoparticles, we loaded human moDCs, as used in clinical trials,40 with the same particles that were used for the dye release experiment, followed by either 1 or 24 hours of incubation at 37 °C, depending on the assay. We chose DCs as a model for a therapeutic cell type, due to their use in cancer immunotherapy a fast-growing field. We have previously shown that nanoparticle-loaded DCs can be imaged in vivo, using 19F MRI.3,41 However, here we focused on the chemical optimization of the particles and ensuring that they are located intracellularly.
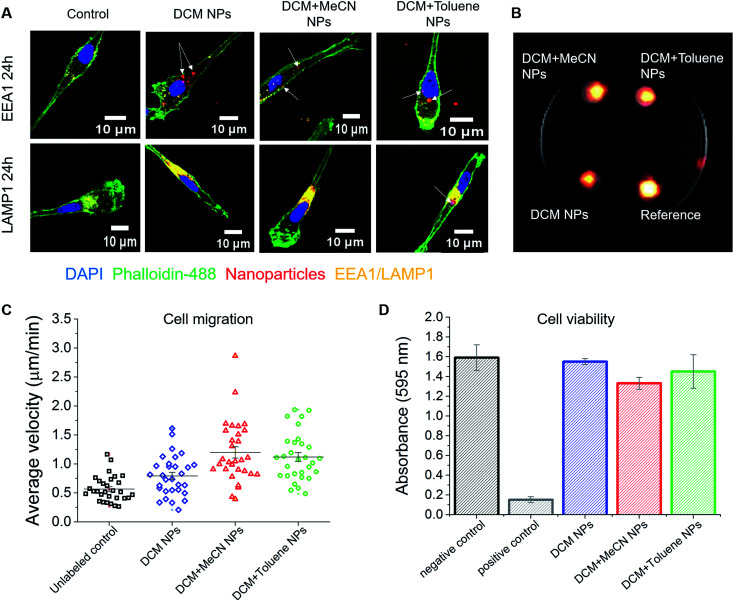
We used confocal microscopy to determine the intracellular presence of PFCE–PLGA particles in moDCs. After incubation with particles for 24 hours, cells were stained for the early endosomal marker EEA1 and the late endosomal/lysosomal marker LAMP1 to study the potential intracellular localization of nanoparticles. As presented in Fig. 5A and S3A and B†, fluorescent signal from the particles (arrows) partially overlaps with the EEA1 signal. More co-localization of nanoparticles was observed with the LAMP1 fluorescent signal. Part of the fluorescent signal coming from the nanoparticles did not localize with neither EEA1 nor LAMP1 signal. As explained in a previous study,42 lack of co-localization could mean that some nanoparticles escape from the endosomal pathway. Another explanation could be that nile blue leaked out of the particles resulting in the signal detection outside of the endosomes or lysosomes.
Fig. 5. Effect of nanoparticles on monocyte-derived DCs. (A) Confocal microscope images of nanoparticle uptake by DCs. Fluorescent signal coming from the nanoparticles partially overlaps with the signal of the early endosomal marker EEA1. Higher colocalization of nanoparticles was observed with the signal of late endosomal/lysosomal marker LAMP1. (B) 1H/19F MRI image showing the signal of 5 × 106 cells loaded with different nanoparticles. Here, PFCE–PLGA particles, at 10 mg ml−1 concentration, were used as a reference. (C) Nanoparticles do not affect the migration of dendritic cells. The graph represents average velocity of unloaded cells and cells loaded with nanoparticles. (D) The graph shows the viability of cells after incubation with various nanoparticles for 72 hours. No significant influence on cell viability was observed. Error bars represent the standard deviation (n = 3).
Size of particles is an important factor in biodistribution, and in obtaining therapeutic effectiveness. For example, PLGA nanoparticles smaller than 50 nm in radius have lower plasma protein adsorption on their surface and lower hepatic filtration.43 It has been shown that larger particles can be taken up more efficiently by phagocytic cells than particles of smaller radius.44 Particles larger than 250 nm in radius are typically taken up via phagocytosis, while the uptake of smaller nanoparticles occurs via endocytosis. Other particle characteristics which can affect the cellular uptake include charge, shape and surface topology.21 Here, we show that all three sizes of particle were readily taken up by the cells. However, uptake may differ with cell type, including other DC subsets, macrophages or non-phagocytic cells. Thus, synthesizing particles of different sizes is crucial, as it gives the ability to target different types of cells.
One of the applications of PFCE–PLGA nanoparticles is their use as cell tracking agents for 19F MRI. To investigate whether nanoparticle-loaded cells can be detected with 19F MRI, we first incubated moDCs in the presence of DCM, DCM + MeCN or DCM + toluene particles and then imaged them after 3 days of incubation. As shown is Fig. 5B, we were able to detect the fluorine signal from all three samples, using a biocompatible imaging sequence and within a reasonable imaging time (15 minutes). The differences in the signal intensity are due to different encapsulation of PFCE among the nanoparticles used for this assay. To investigate whether uptake affects cell functionality, we studied cell migration; both the velocity and the distance covered by the labelled cells. The results of the measurements of average cell velocity are presented in Fig. 5C. We observed an increase in velocity of cells incubated with particles when compared to control cells (in red), which corresponds to the fact that nanoparticle uptake can induce the migration of DCs.45 The mean velocity was 0.56 ± 0.22, 0.79 ± 0.34, 1.19 ± 0.53 and 1.11 ± 0.41 μm min−1 for unloaded controls, DCM nanoparticle, DCM + MeCN nanoparticle or DCM + toluene nanoparticle-loaded cells respectively. The trajectory of migrating cells for all four conditions is presented in Fig. S5.† We further confirmed the cell viability using standard viability assay (Fig. 5D). Our results, show that the particles are suitable for cell labelling and imaging with 19F MRI, and have no significant effect on viability or functionality.
Conclusions
The size of particles, size distribution and degradation are important factors for various biomedical applications, including controlled delivery of drugs, drug release and cell uptake. Control of these properties of nanoparticles is crucial for optimal particle performance. In this study, we have shown that a narrow size distribution and high 19F loading in PFCE–PLGA nanoparticles can be achieved by using an optimal concentration of surfactant and the right solvent. We also showed that nanoparticle size has a strong influence on the encapsulation and release of the encapsulated cargo. Here, we used nile blue as a model for a hydrophobic agent, such as paclitaxel. PFCE–PLGA nanoparticles can be further adapted for targeting by the addition of targeting ligands.42 The use of such multifunctional nanoparticles for biomedical applications in cancer or other diseases has great potential for diagnosis and targeted treatment, such as targeted delivery of hydrophobic anticancer drug combined with quantitative 19F MRI, or cell tracking.46 In particular, 19F MRI is a quantitative technique that is not limited by penetration depth in vivo; therefore, it would be possible to use non-invasive in vivo imaging to quantify drug delivery using these nanoparticles. Finally, we have shown that this method of preparation of PLGA particles is robust, and not significantly affected even by the addition of large amounts of a third immiscible phase, the PFC. Overall, our results could provide a guideline in the rational design of polymer-based particles loaded with a third PFC phase for imaging and drug delivery, where size, encapsulation efficiency and release can be controlled.
Materials and methods
Materials
Poly(d,l-lactide-co-glycolide) (PLGA) resomer RG 502H, lactide : glycolide molar ratio 50 : 50 was obtained from Evonik Industries AG, Essen, Germany. Solvents: dichloromethane (DCM), chloroform (Merck, Darmstadt, Germany), ethyl acetate (AcOEt), acetone, acetonitrile (MeCN) (all from VWR, Netherlands), tertrahydrofuran (THF) (Sigma-Aldrich, St. Louis, MO, USA). Perluoro-15-crown-5-ether (PFCE) was purchased from Exfluor, Round Rock USA, and Prohance from Bracco, Konstanz Germany. Surfactants: polyvinyl alcohol (PVA, 9000–10 000 Mw, 80% hydrolized), pluronics F68, sodium cholate, polyvinylpyrrolidone (PVP) were purchased from Sigma-Aldrich St. Louis, MO, USA, and Tween20 from Merck, Darmstadt, Germany. Some nanoparticles were prepared with Nile blue dye (Acros Organics, Belgium) For cell culture X-VIVO 15 medium (Lonza, Belgium) or RPMI 1640 (Gibco, Thermo Fisher, The Netherlands) was used. Phosphate-buffer saline (PBS) (Braun, Germany) was used to resuspend nanoparticles for cell experiments and for cell washing.
Particle synthesis
Particles were prepared using a miniemulsion synthesis technique. Briefly, PLGA (100–200 mg), was dissolved in 3 ml of various solvents, followed by the addition of 900 μL PFCE and 1.8 ml Prohance. For dye release and cellular uptake experiments, Nile blue dye (0.2 mg) was also included in the organic phase. The organic phase was added to 25.5 g of aqueous solution of surfactant (0.4–2 wt%) and emulsified for 3 minutes with the use of probe sonicator (Sonifier 250, microtip 6.3 mm, Branson Sonic Power, St. Louis, USA) at 40% amplitude. Sonication was followed by overnight evaporation of solvent at room temperature. Next, the particles were collected by centrifugation (15 000 rpm, 20 minutes), washed several times with deionized water, and then freeze-dried. Several parameters tested in this protocol, including: the type of surfactant, the type of solvent, polymer concentration (3.3, 5, 6.6 wt%) and surfactant concentration (0.4, 1.2, 2 wt%). All particle characterization was done on the freeze-dried samples which.
Particle characterization
Dynamic light scattering was measured at Malvern zeta sizer NS nano (Malvern, Worcestershire, United Kingdom), using disposable ZEN0040 cuvettes, at concentrations of 0.1 mg ml−1. Data evaluation was done with Malvern software (Zetasizer software, Ink) using cumulant fitting. The values are the intensity mean of 4 independent measurements.
Multi-angle LS measurements were performed using an ALV7004 correlator, ALV/LSE-5004 Goniometer, ALV/Dual High QE APD detector unit with fibre splitting device with a set-up of 2 off detection system and a Uniphase Model 1145P He–Ne Laser. The laser wavelength and power were 632.8 nm and 22 mW, respectively and the temperature was controlled by a Julabo CF41 thermostatic bath. Water for dilutions was filtered with 0.45 μm hydrophilic filters. The data analysis was done with HDRC software, which was kindly provided by Prof. Manfred Schmidt, University of Mainz, Germany. The apparent radii of hydrodynamic radii at different angles were calculated using from a biexponential fitting of autocorrelation function, as it provides a good approximation to calculate the size of monomodal samples with a non-monodisperse size distribution. The absolute inverse z-averages of hydrodynamic radii were obtained by extrapolating q → 0. The second cumulant values μ2 were obtained from cumulant fitting at θ = 90°. The μ2 value is the second cumulant, obtained from the cumulant analysis of autocorrelation function and provides a quantitative measure for the polydispersity of the diffusion coefficient distribution function:μ2 = (〈Ds2〉 − 〈Ds〉2)q4where Ds is the diffusion coefficient and q the scattering vector.
Transmission electron microscopy (TEM) was done on JEOL TEM1010 transmission electron microscope with 35 mm film camera, Kodak megaplus 4CCD camera. For TEM analysis, samples were placed on copper grids and left o/n to dry. TEM images were then processed in ImageJ program, where 100 nanoparticles were measured to study the size distribution.
Nuclear Magnetic Resonance spectroscopy was done on Bruker Avance III 400 MHz NMR spectrometer equipped with a BBFO + probe. To measure the fluorine content of nanoparticles, defined amount of nanoparticles was dissolved in 500 μL deuterium oxide (D2O) and mixed 100 μL 1 vol% trifluoroacetic acid (TFA), as an internal reference, and transferred to NMR tubes. The interscan relaxation delay was set to 5T1,TFA* = 20 s for quantification. Data analysis was carried out with Mestrenova 10.0.2.
Cryogenic Scanning Electron Microscopy was done at JEOL 6330 Cryo Field Emission Scanning Electron Microscope (FESEM). For cryo-SEM analysis, the samples (8 μl at concentration 10 mg ml−1) were pipetted in 2 rivets, which were then placed together. Next, the samples were frozen in liquid nitrogen slush and placed in an Oxford Alto 2500 cryo station with a cryo-transfer device. There, the top rivet was broken and the sample was heated to −95 °C for 5 minutes, followed by a coating of 60/40 Au/Pd and transfer to the Cryo-SEM.
Dye release
To test dye release, PLGA–PFCE nanoparticles with Nile blue, were incubated at 37 °C for several time points in pH solutions (0.3% PVA solution with either 1 M HCl or NaOH) of 4.0, 5.5, 8.5, followed by the DLS measurements (concentration 0.1 mg ml−1) to detect any changes in nanoparticle size and polydispersity. The calibration of UV/vis absorption was done with nile blue reference at 8 different concentrations using same solvent as for particles. PVA was added to nile blue samples and particles, as it was not possible to dissolve nile blue for calibration without adding PVA. The DLS values should be treated carefully, as the absorption of Nile blue overlaps with the DLS laser wavelength. However, they still provide a good indication of the size trend, as fluctuations due to Brownian motion still can be detected. Additionally, dye release from nanoparticles was tested at each incubation time point by centrifuging the samples, collecting the supernatant and measuring its absorption (JASCO V-630 UV-vis spectrophotometer, JASCO Benelux, B.V.). Obtained data were converted to concentrations by means of calibration curves.
Cell culture and labeling
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy individuals after informed consent, using ficoll density centrifugation (Lymphoprep, STEMCELL Technologies, Vancouver, Canada). Adherent monocytes were cultured in X-VIVO 15 medium supplemented with 2% human serum or in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum, and in the presence of interleukin-4 (300 U ml−1) and granulocyte-monocyte colony stimulating factor (450 U ml−1) to obtain immature dendritic cells (DCs). Day 3 cells were harvested, counted and labeled with PLGA nanoparticles (resuspended in PBS shortly prior the labelling) at concentration 2 mg of nanoparticles/1 × 106 cells and incubated at 37 °C for 1 and 24 h for confocal microscopy, 24 h for cell migration, 72 h for viability assay and 72 h for MRI imaging.
Cell staining and confocal microscopy
The uptake of various nanoparticles containing nile blue was confirmed with confocal microscopy. Day 3-moDCs were incubated at 37 °C for 1 and 24 h on glass coverslips (20 000 cells per coverslip). At each time point the excess of the label was removed, the coverslips with cells were washed gently with PBS, and then the cells were fixed with 2% paraformaldehyde (PFA) for further testing with confocal microscopy. Labelled cells on coverslips were permeabilized in CLSM buffer + 0.1% Saponin, stained first with LAMP-1 or EEA1-specific primary antibody (Ab), followed by staining with isotype-specific AlexaFluor-conjugated secondary Ab for intracellular compartments, phalloidin-488 for cell membrane, and DAPI for nucleus. Stained cells were then imaged with Olympus FV1000 Confocal Laser Scanning Microscope. Acquired images were then processed in the ImageJ program.
Cell migration
To study cell migration and velocity, nanoparticles were resuspended in PBS shortly before the labelling and added to day-3 moDCs at concentration 2 mg of nanoparticles per 1 × 106 cells. Cells with nanoparticles were then added to 24 well plates and imaged for 24 hours using Okolab 2.0 microscope (Nikon Diaphot 300 with Hamamatsu C8484-05 G CCD Camera) with Okolab 4 well CO2 stage incubator and Okolab 2D time lapse software. Acquired images were then processed using ImageJ program to measure the distance and velocity.
Cell viability
To investigate the influence of labelling on cells viability with performed an MTT assay. For this, 0.5 × 106 cells were incubated in the presence of DCM, DCM + MeCN or DCM + toluene nanoparticles for 72 hours. Each condition was done in triplicate. After incubation, the excess of the label was removed by gentle washing with PBS. Next, the cells were collected and placed in flat-bottom 96 well plates, washed twice in PBS (100 μl per well) with centrifugation for 2 min between each wash. Next, 60 μl RPMI medium with 10 μl MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (concentration 4 mg ml−1) was added to each well, followed by 30 minutes incubation at 37 °C. After incubation, the plate was centrifuged for 2 min and 100 μl of lysis buffer (isopropanol, 10% SDS, 2 N HCL, deionized water) was added to each well, and the plate was incubated for 15 minutes in dark at room temperature. Before the measurements, samples were resuspended to remove any precipitate of crystals. The plate was measured with iMark™ microplate reader (Bio-Rad, The Netherlands) at 595 nm. Cells not loaded with nanoparticles were used as negative control and cells treated with 0.2% Triton X-100 were used a positive control.
19F Magnetic resonance imaging
19F MRI was performed on a pre-clinical 11.7 T MRI scanner (Biospec 117/16, 500 MHz, Bruker, Ettlingen, Germany). For in vitro imaging, 5 × 106 dendritic cells were co-incubated with particles at a concentration of 10 mg ml−1. Nanoparticle loaded cells were were extensively washed to remove excess label, before being placed in tubes, left to sediment and imaged with the zero echo time (ZTE) sequence with the following parameters: TR = 2 ms, flip angle 4°, matrix size = 32 isotropic voxels, FOV = 60 × 60 × 60 mm, (voxel size = 1.87 mm), number of averages = 128, scan time = 15 minutes. Sedimented particles (5 mg in 5 μl) in PBS were used as the reference.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
Authors would like to acknowledge Ad Swolfs for his support during NMR measurements. Prof. Manfred Schmidt and Georg Conrad for providing a HDRC software for multi-angle light scattering. Jorieke Weiden for the help with cell migration assay. Andor Veltien for the help with 19F MRI imaging. Dr Oya Tagit for the consult and writing support. The MR measurements were supported by NWO Investment Grants 91106021 and BIG (VISTA). This work was supported by the European Research Council (ERC) Starting Grant (CoNQUeST grant no. 336454) and grant 14716 from the Netherlands organization for scientific research NWO-TTW grant. to MS. IJMdV received NWO-Vici 918.14.655. CGF received the NWO Spinoza Grant, ERC Adv Grant PATHFINDER (269019) and Dutch Cancer Society Grant KUN2009-4402.
Electronic supplementary information (ESI) available: Supporting information consists of: a table listing all the surfactants tested in this study, a table of solvents tested during the synthesis of particles, a figure on the influence of PVA concentration on particle radius, PDI and PFCE encapsulation with, TEM images of particles formulated with different PLGA concentration and different solvents, a figure showing PDI of samples from the dye release experiment, a figure showing nile blue encapsulation in nanoparticles synthesized with and without the addition of PFCE, images from confocal microscopy showing the uptake of particles by human monocyte-derived DCs, and MRI 3D image of nanoparticle-loaded cells. See DOI: 10.1039/c7ra13062g
References
- Fuchs A. V. Bapat A. P. Cowin G. J. Thurecht K. J. Switchable 19F MRI polymer theranostics: towards in situ quantifiable drug release. Polym Chem. 2017;8(34):5157–5166. doi: 10.1039/C7PY00345E. [DOI] [Google Scholar]
- Kumari A. Yadav S. K. Yadav S. C. Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf., B. 2010;75(1):1–18. doi: 10.1016/j.colsurfb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Srinivas M. Cruz L. J. Bonetto F. Heerschap A. Figdor C. G. et al., Customizable, multi-functional fluorocarbon nanoparticles for quantitative in vivo imaging using 19F MRI and optical imaging. Biomaterials. 2010;31(27):7070–7077. doi: 10.1016/j.biomaterials.2010.05.069. [DOI] [PubMed] [Google Scholar]
- Srinivas M. Boehm-Sturm P. Figdor C. G. de Vries I. J. M. Hoehn M. Labeling cells for in vivo tracking using (19)F MRI. Biomaterials. 2012;33(34):8830–8840. doi: 10.1016/j.biomaterials.2012.08.048. [DOI] [PubMed] [Google Scholar]
- Tel J. Sittig S. P. Blom R. A. M. Cruz L. J. Schreibelt G. et al., Targeting uptake receptors on human plasmacytoid dendritic cells triggers antigen cross-presentation and robust type I IFN secretion. J. Immunol. 2013;191(10):5005–5012. doi: 10.4049/jimmunol.1300787. [DOI] [PubMed] [Google Scholar]
- Sun D. Li N. Zhang W. Zhao Z. Mou Z. et al., Design of PLGA-functionalized quercetin nanoparticles for potential use in Alzheimer's disease. Colloids Surf., B. 2016;148:116–129. doi: 10.1016/j.colsurfb.2016.08.052. [DOI] [PubMed] [Google Scholar]
- Makadia H. K. Siegel S. J. Poly Lactic-co-Glycolic Acid (PLGA) as Biodegradable Controlled Drug Delivery Carrier. Polymers. 2011;3(3):1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas M. Morel P. A. Ernst L. A. Laidlaw D. H. Ahrens E. T. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn. Reson. Med. 2007;58(4):725–734. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- Srinivas M. Heerschap A. Ahrens E. T. Figdor C. G. de Vries I. J. M. (19)F MRI for quantitative in vivo cell tracking. Trends Biotechnol. 2010;28(7):363–370. doi: 10.1016/j.tibtech.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri H. Srinivas M. Veltien A. van Uden M. J. de Vries I. J. M. et al., Cell tracking using 19F magnetic resonance imaging: Technical aspects and challenges towards clinical applications. Eur. Radiol. 2015;25(3):726–735. doi: 10.1007/s00330-014-3474-5. [DOI] [PubMed] [Google Scholar]
- Swider E. and Srinivas M., Spinning to a Different Beat: 19F Agents for “Hot-Spot” Cellular MR Imaging, in Design and Applications of Nanoparticles in Biomedical Imaging, Springer International Publishing, Cham, 2017, pp. 153–170 [Google Scholar]
- Díaz-López R. Tsapis N. Fattal E. Liquid perfluorocarbons as contrast agents for ultrasonography and (19)F-MRI. Pharm. Res. 2010;27(1):1–16. doi: 10.1007/s11095-009-0001-5. [DOI] [PubMed] [Google Scholar]
- Yoon J. K. Burgess D. J. Interfacial properties as stability predictors of lecithin-stabilized perfluorocarbon emulsions. Pharm. Dev. Technol. 1996;1(4):333–341. doi: 10.3109/10837459609031428. [DOI] [PubMed] [Google Scholar]
- Kuznetsova I. N. Perfluorocarbon Emulsions: Stability in vitro and in vivo (A Review) Pharm. Chem. J. 2003;37(8):415–420. doi: 10.1023/A:1027355913348. [DOI] [Google Scholar]
- Landfester K. Miniemulsion polymerization and the structure of polymer and hybrid nanoparticles. Angew. Chem., Int. Ed. 2009;48(25):4488–4508. doi: 10.1002/anie.200900723. [DOI] [PubMed] [Google Scholar]
- Solans C. Izquierdo P. Nolla J. Azemar N. Garciacelma M. Nano-emulsions. Curr. Opin. Colloid Interface Sci. 2005;10(3–4):102–110. doi: 10.1016/j.cocis.2005.06.004. [DOI] [Google Scholar]
- Grapentin C., Mayenfels F., Barnert S., Süss R. and Schubert R., et al., Optimization of perfluorocarbon nanoemulsions for molecular imaging by 19F MRI, Nanomedicine, One Central Press (OCP), 2014, pp. 268–286 [Google Scholar]
- Tirotta I. Dichiarante V. Pigliacelli C. Cavallo G. Terraneo G. et al., 19F magnetic resonance imaging (MRI): From design of materials to clinical applications. Chem. Rev. 2015;115:1106–1129. doi: 10.1021/cr500286d. [DOI] [PubMed] [Google Scholar]
- Grapentin C., Mayenfels F., Barnert S., Süss R. and Schubert R., et al., Optimization of perfluorocarbon nanoemulsions for molecular imaging by 19F MRI, Nanomedicine, One Central Press (OCP), 2014, pp. 268–286 [Google Scholar]
- Sharma N. Madan P. Lin S. Effect of process and formulation variables on the preparation of parenteral paclitaxel-loaded biodegradable polymeric nanoparticles: A co-surfactant study. Asian J. Pharm. Sci. 2016;11(3):404–416. doi: 10.1016/j.ajps.2015.09.004. [DOI] [Google Scholar]
- Waiczies H. Lepore S. Janitzek N. Hagen U. Seifert F. et al., Perfluorocarbon particle size influences magnetic resonance signal and immunological properties of dendritic cells. PLoS One. 2011;6(7):e21981. doi: 10.1371/journal.pone.0021981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. Lillard Jr J. W. Nanoparticle-based targeted drug delivery. Exp. Mol. Pathol. 2009;86(3):215–223. doi: 10.1016/j.yexmp.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte H. Geers B. Liang S. Himmelreich U. Demeester J. et al., Design and evaluation of theranostic perfluorocarbon particles for simultaneous antigen-loading and 19F-MRI tracking of dendritic cells. J. Controlled Release. 2013;169(1–2):141–149. doi: 10.1016/j.jconrel.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Cheng J. Teply B. Sherifi I. Sung J. Luther G. et al., Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28(5):869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. J., Strohm E. M., Sun Y., Niu C. and Zheng Y., et al., PLGA/PFC particles loaded with gold nanoparticles as dual contrast agents for photoacoustic and ultrasound imaging, Proc. SPIE 8943, Photons Plus Ultrasound: Imaging and Sensing, 89433M, 2014 [Google Scholar]
- Mao S. Shi Y. Li L. Xu J. Schaper A. et al., Effects of process and formulation parameters on characteristics and internal morphology of poly(d,l-lactide-co-glycolide) microspheres formed by the solvent evaporation method. Eur. J. Pharm. Biopharm. 2008;68(2):214–223. doi: 10.1016/j.ejpb.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Menon J. U. Kona S. Wadajkar A. S. Desai F. Vadla A. et al., Effects of surfactants on the properties of PLGA nanoparticles. J. Biomed. Mater. Res., Part A. 2012;100(8):1998–2005. doi: 10.1002/jbm.a.34040. [DOI] [PubMed] [Google Scholar]
- Barwal I. Sood A. Sharma M. Singh B. Yadav S. C. Development of stevioside Pluronic-F-68 copolymer based PLA-nanoparticles as an antidiabetic nanomedicine. Colloids Surf., B. 2013;101(11):510–516. doi: 10.1016/j.colsurfb.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Halayqa M. Domańska U. PLGA biodegradable nanoparticles containing perphenazine or chlorpromazine hydrochloride: effect of formulation and release. Int. J. Mol. Sci. 2014;15(12):23909–23923. doi: 10.3390/ijms151223909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T. Tran T. H. Kim J. O. Yong C. S. Nguyen C. N. Enhancing the in vitro anti-cancer efficacy of artesunate by loading into poly-d,l-lactide-co-glycolide (PLGA) nanoparticles. Arch. Pharmacal Res. 2015;38(5):716–724. doi: 10.1007/s12272-014-0424-3. [DOI] [PubMed] [Google Scholar]
- Song K. C. Lee H. S. Choung I. Y. Cho K. I. Ahn Y. et al., The effect of type of organic phase solvents on the particle size of poly(D,L-lactide-co-glycolide) nanoparticles. Colloids Surf., A. 2006;276(1–3):162–167. [Google Scholar]
- Bantz C. Koshkina O. Lang T. Galla H.-J. Kirkpatrick C. J. et al., The surface properties of nanoparticles determine the agglomeration state and the size of the particles under physiological conditions. Beilstein J. Nanotechnol. 2014;5(i):1774–1786. doi: 10.3762/bjnano.5.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärtl W., Light Scattering from Polymer Solutions and Nanoparticle Dispersions, Springer, 2007 [Google Scholar]
- Fischer K. Schmidt M. Pitfalls and novel applications of particle sizing by dynamic light scattering. Biomaterials. 2016;98:79–91. doi: 10.1016/j.biomaterials.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Silva A. T. C. R. Cardoso B. C. O. Silva M. E. S. R. Freitas R. F. S. Sousa R. G. Synthesis, Characterization, and Study of PLGA Copolymer in Vitro Degradation. J. Biomater. Nanobiotechnol. 2015;6(1):8–19. doi: 10.4236/jbnb.2015.61002. [DOI] [Google Scholar]
- Fonseca C. Simões S. Gaspar R. Paclitaxel-loaded PLGA nanoparticles: preparation, physicochemical characterization and in vitro anti-tumoral activity. J. Controlled Release. 2002;83(2):273–286. doi: 10.1016/S0168-3659(02)00212-2. [DOI] [PubMed] [Google Scholar]
- Panyam J. Dali M. M. Sahoo S. K. Ma W. Chakravarthi S. S. et al., Polymer degradation and in vitro release of a model protein from poly(d,l-lactide-co-glycolide) nano- and microparticles. J. Controlled Release. 2003;92(1):173–187. doi: 10.1016/S0168-3659(03)00328-6. [DOI] [PubMed] [Google Scholar]
- Joshi V. B. Geary S. M. Salem A. K. Biodegradable particles as vaccine delivery systems: size matters. AAPS J. 2013;15(1):85–94. doi: 10.1208/s12248-012-9418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotts G. Park T. G. Protein delivery from poly(lactic-co-glycolic acid) biodegradable microspheres: Release kinetics and stability issues. J. Microencapsulation. 1998;15(6):699–713. doi: 10.3109/02652049809008253. [DOI] [PubMed] [Google Scholar]
- https://clinicaltrials.gov/ct2/results?term=perfluorocarbons&Search=Search. https://clinicaltrials.gov/ct2/results?term=perfluorocarbons&Search=Search
- Srinivas M. Tel J. Schreibelt G. Bonetto F. Cruz L. J. et al., PLGA-encapsulated perfluorocarbon nanoparticles for simultaneous visualization of distinct cell populations by 19F MRI. Nanomedicine. 2015;10(15):2339–2348. doi: 10.2217/nnm.15.76. [DOI] [PubMed] [Google Scholar]
- Cruz L. J. Tacken P. J. Bonetto F. Buschow S. I. Croes H. J. et al., Multimodal Imaging of Nanovaccine Carriers Targeted to Human Dendritic Cells. Mol. Pharmaceutics. 2011;8(2):520–531. doi: 10.1021/mp100356k. [DOI] [PubMed] [Google Scholar]
- Acharya S. Sahoo S. K. PLGA nanoparticles containing various anticancer agents and tumour delivery by EPR effect. Adv. Drug Delivery Rev. 2011;63(3):170–183. doi: 10.1016/j.addr.2010.10.008. [DOI] [PubMed] [Google Scholar]
- He C. Hu Y. Yin L. Tang C. Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31(13):3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- Li N. Peng L.-H. Chen X. Zhang T.-Y. Shao G.-F. et al., Antigen-loaded nanocarriers enhance the migration of stimulated Langerhans cells to draining lymph nodes and induce effective transcutaneous immunization. Nanomedicine Nanotechnology, Biol Med. 2014;10(1):215–223. doi: 10.1016/j.nano.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Aarntzen E. H. J. G. Srinivas M. Walczak P. Janowski M. Heerschap A. et al., In vivo tracking techniques for cellular regeneration, replacement, and redirection. J. Nucl. Med. 2012;53(12):1825–1828. doi: 10.2967/jnumed.112.106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.