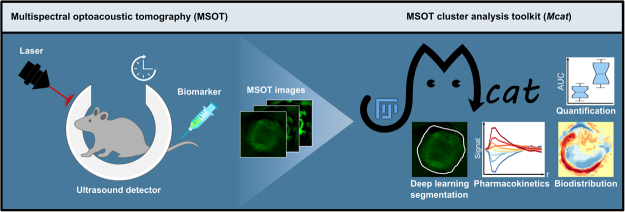
Abstract
Although multispectral optoacoustic tomography (MSOT) significantly evolved over the last several years, there is a lack of quantitative methods for analysing this type of image data. Current analytical methods characterise the MSOT signal in manually defined regions of interest outlining selected tissue areas. These methods demand expert knowledge of the sample anatomy, are time consuming, highly subjective and prone to user bias. Here we present our fully automated open-source MSOT cluster analysis toolkit Mcat that was designed to overcome these shortcomings. It employs a deep learning-based approach for initial image segmentation followed by unsupervised machine learning to identify regions of similar signal kinetics. It provides an objective and automated approach to quantify the pharmacokinetics and extract the biodistribution of biomarkers from MSOT data. We exemplify our generally applicable analysis method by quantifying liver function in a preclinical sepsis model whilst highlighting the advantages of our new approach compared to the severe limitations of existing analysis procedures.
Abbreviations: AUC, Area under the curve; DAG, Directed acyclic graph; DL, Deep learning; GUI, Graphical user interface; ICG, Indocyanine green; Mcat, MSOT cluster analysis toolkit; MSE, Mean squared error; MSOT, Multispectral optoacoustic tomography; ROI, Region of interest; PCI, Peritoneal contamination and infection; WAC, Weighted-average curve
Keywords: Multispectral optoacoustic tomography, Quantitative image analysis, Deep learning, ImageJ plugin, Biomarkers, Pharmacokinetics, Sepsis
Graphical Abstract
1. Introduction
Multispectral optoacoustic tomography (MSOT) has become increasingly recognised as a non-invasive imaging modality for preclinical and clinical research [1]. It serves as a powerful diagnostic tool for a large variety of diseases, including breast cancer [2], melanoma [3], vascular disorders [4] and Crohn’s disease [5]. In addition to the capability to conduct in vivo studies, MSOT offers the possibility to collect time-resolved, functional tissue information at high spatio-temporal resolution for multiple photoabsorbing molecules at low costs [6].
However, quantitative analysis of MSOT image data is currently based on the manual assignment of regions of interest (ROI) within specific anatomical areas from which basic signal intensity statistics are extracted and followed over time [7], [8], [9], [10], [11], [12], [13]. We will refer to this procedure by the term tissue-oriented analysis because it quantifies signal change over time within a specifically chosen tissue region. While this approach is intuitive and allows a seemingly straightforward interpretation of the measurements, it suffers from several weaknesses: (i) manual definition of ROIs can be time consuming and demands expert knowledge of the anatomy and physiology of the sample, (ii) ROI definition is prone to user bias, (iii) any bias may reduce the reproducibility of study results and (iv) hinder the comparability of results across studies. Furthermore, since the attenuation of light in deep tissue limits the penetration depth of MSOT, even homogeneous tissue structures can show highly different signal intensities at different depths [14]. Consequently, differences between experimental conditions may be entirely obscured by these uncertainties of the analysis procedure and prevent utilising MSOT where most sensitive and accurate quantification is desired, e.g. when investigating potential new drugs or aiming for predictions in personalised medicine.
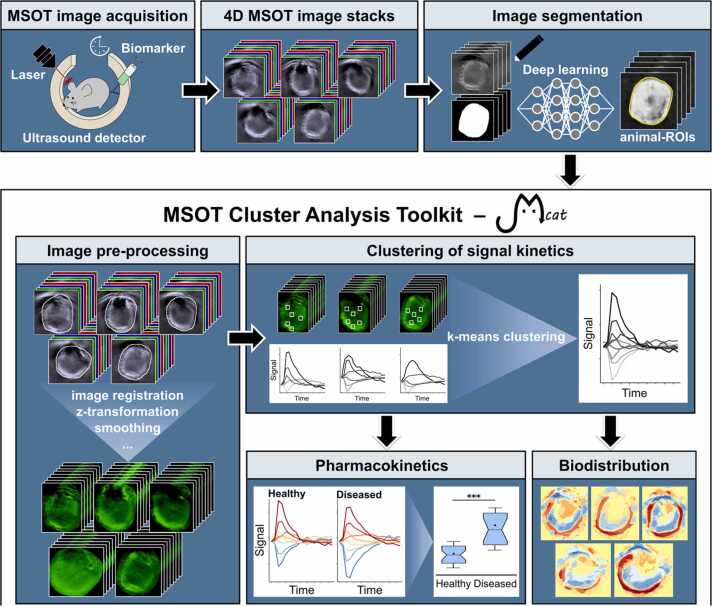
In contrast to the above-mentioned tissue-oriented analysis, we introduce a signal-oriented approach, which does not rely on the exact definition of a specific tissue region and thereby enhances the objectiveness of the analysis. Pixel-wise clustering of the observed signal kinetics is performed including the entire investigated sample, e.g. a whole mouse cross-section, to extract the pharmacokinetics and biodistribution of a particular photoabsorber. We compare the results of our method to the current standard approach of tissue-oriented analysis. Furthermore, we illustrate how the utilisation of confined ROIs can alter the outcome of quantitative MSOT image analysis, likely leading to incorrect results and erroneous conclusions. We developed the MSOT cluster analysis toolkit Mcat, which is provided as a plugin for the widely used open-source image analysis software ImageJ [15] and can be applied by users with no professional expertise in image processing. It is freely available at https://github.com/applied-systems-biology/mcat (https://doi.org/10.5281/zenodo.6046031).
We exemplify Mcat by applying it to MSOT data of mice with a life-threatening polymicrobial infection accompanied by liver dysfunction, i.e. peritoneal contamination and infection model (PCI) of sepsis [16], [17]. Liver function is quantitatively assessed by MSOT imaging of the clinically approved fluorescent chromophore indocyanine green (ICG) [18]. Hepatic ICG clearance is compared between healthy animals and those subjected to PCI. Since ICG is almost exclusively eliminated by the liver when administered intraveneously, its excretion is impaired by liver dysfunction under septic condition and it therefore serves as a marker for liver function [18]. Our fully automated signal-oriented analysis outperforms the tissue-oriented approach in distinguishing healthy from diseased animals and provides information about the spatial distribution of the biomarker. Furthermore, the presented method is not limited to any particular photoabsorber or anatomical region; thus, the main steps of this analysis can be applied to virtually any type of time-resolved image data.
2. Methods
This section will first describe the segmentation of MSOT images, their pre-processing and then introduce our newly developed signal-oriented analysis approach. Subsequently, we describe the tissue-oriented approach, which is currently the standard procedure for the quantitative analysis of MSOT image data, followed by an overview of utilised software and the statistical analysis. Finally, the experimental methods are explained that were used to validate our new signal-oriented analysis.
2.1. Image segmentation
For automated segmentation of the animal cross-sections, we utilised a deep learning (DL) approach based on the software Cellpose, an algorithm designed to identify biological objects that is based on a U-net architecture with residual blocks [19]. The complete workflow is outlined in Fig. 1.
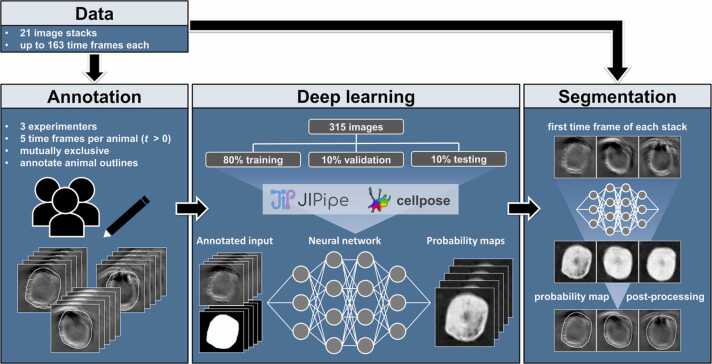
Fig. 1.
DL-based segmentation of MSOT images. For the manual annotation, three experimenters drew animal outlines in five random and unique time frames (t > 0) for each of the 21 animals. Based on these 315 annotated images, a Cellpose neural network was trained from scratch using the visual programming language JIPipe[20], where 80% of the images were used for training and 10% each for validation and testing. The final segmentation of the first time frame (t = 0) of each MSOT image stack was then performed with the trained neural network. The resulting probability maps were post-processed with basic image processing functions to obtain the final ROIs outlining the animals.
The training dataset was provided by three experimentalists who drew ROIs around animal outlines for five random and unique time frames per animal (see Supplementary Fig. A1 for more details on data selection). The manual annotation and subsequent training of the Cellpose neural network model were performed with the visual programming language JIPipe [20]. The final dataset consisted of 315 images from which 80% were used for training, 10% for validation, and 10% for testing of the model. We configured Cellpose to utilise no pre-trained model, run for 1000 epochs and assume the object diameter to be 210 pixels. The default settings were kept for all other parameters: loss function = L2 and cross-entropy, learning rate = 0.2, batch size = 0.8, augmentation = [random rotation (0–360°), random scaling (0.5x to 1.5x), random translations].
The trained model was then applied to the first time frame (t=0) of each of the image stacks. The resulting probability maps were segmented in Mcat with ImageJ functions. The probability maps were first convolved with a median filter of radius 3, the contrast was enhanced with saturation set to 0.3 and then binarised with Otsu thresholding. The remaining holes were filled, noise was removed, and only the largest foreground object was kept. Next, morphological closing and opening with radii of 50 pixels from the plugin MorphoLibJ [21] were applied to smoothen the ROI outline. The canvas size was doubled before this step to avoid artefacts at the image borders and afterwards reduced to the original image size. Finally, the convex hull of the foreground object was created and used as the final ROI, which will be from now on referred to as animal-ROI.
2.2. Image pre-processing
The multidimensional MSOT image stacks (see Methods Section 2.6.2 for details on image acquisition) were pre-processed prior to image analysis. The water and ICG image channels were extracted, and rigid registration was performed on the water channel. The resulting transformation matrix was then applied to the ICG channel to reduce motion artefacts. To compensate for signal intensity differences between MSOT scans, the ICG channel was z-transformed for each animal according to
| (1) |
where is the z-transformed pixel value at position , is the original pixel intensity value at position , is the mean signal intensity value of the first time frame of the ICG channel, and is the corresponding standard deviation. The image stacks were then smoothed by downsampling in the z-direction to compensate for animal breathing. This factor is denoted as smoothing factor and was set to s = 4, i.e. the signal intensity values of four subsequent time frames were averaged into one new time frame. Finally, the pixel intensity values were transformed into time derivatives of pixel intensities between consecutive time frames. Thus, the analysis was performed on signal intensity change over time, to avoid dealing with negative signal intensity values that may be introduced by image reconstruction and spectral unmixing.
2.3. Signal-oriented analysis
The details on our newly developed signal-oriented analysis approach are presented along with the results in Section 3.2. In brief, it automatically identifies connected regions of similar signal kinetics from the animal-ROIs that were obtained by automated image segmentation. We therefore utilised k-means clustering, where the parameter k defines the number of clusters with different signal kinetics. All animals from one group were mixed together to compare treatment groups, and clustering was performed on these pooled datasets. The overall extent of biomarker signal increase over time was calculated for each animal based on the kinetic clusters reflecting a signal net increase and their corresponding pixel frequencies. Additionally, colour-coded images of the spatial distribution of all kinetic clusters were reconstructed for each animal to visualise the biodistribution of ICG.
2.4. Tissue-oriented analysis
For the tissue-oriented analysis, an expert labelled the liver tissue in the water channel, referred to as liver-ROIs (see Results Section 3.3). We then extracted the mean, maximum and 95th-percentile intensity values within the liver-ROIs from the ICG channel of the pre-processed images. These signal statistics were calculated for each time frame independently, yielding time curves for each animal. Since the number of time frames per MSOT image stack was not always identical, excess time frames were excluded from the analysis. We then calculated the area under the curve (AUC) for the time curves of the three signal statistics to compare treatment groups.
To investigate the sensitivity of ROI placement in MSOT images, we placed a circular ROI with a diameter of ten pixels in the outer region of liver tissue. The ROI was then shifted four times in the y-direction in 5-pixel steps and three times in the x- and y-direction in 5-pixel steps, respectively, to obtain multiple samples of liver tissue within close proximity (5 pixels ≈ 0.38 mm; see Results Section 3.3). As before, the mean, maximum and 95th-percentile signal intensity values were extracted from each of these ROIs and the liver-ROI, and they were followed over time to assess the homogeneity of the ICG signal within the liver.
2.5. Utilised software and statistical analysis
The signal-oriented analysis, including the pre-processing of MSOT images, was implemented in Java (see Supplementary Table A2 for details on utilised software and libraries). The tissue-oriented analysis and the investigation of its sensitivity towards ROI placement were performed with the open-source image analysis software Fiji (ImageJ v1.52p) [15]. Statistical analysis was performed using the statistics software R v3.6.1. Differences between groups were evaluated using a two-tailed Wilcoxon rank sum test. Effect sizes were calculated as Hedges' g using the cohen.d function of the R package effsize (parameters: pooled = TRUE, hedges.correction = TRUE, paired = FALSE), where g is interpreted as indicative for a low effect for values around g = 0.2, as a medium effect for values around g = 0.5 and as a large effect for values around g = 0.8 and larger. All p-values and effect sizes were calculated for = 9 and = 12 animals. Boxplot centre lines denote median values. Mean values are represented as black triangles.
We screened a range of values for the two main parameters smoothing factor s and k-means k to perform parameter estimation of the signal-oriented analysis. The whole analysis was repeated five times for each parameter combination to also consider the random initialisation step of k-means clustering. The robustness of a specific parameter combination was evaluated by calculating the weighted mean squared error (MSE) of the mean effect size between treatment groups of the five runs with regard to of the neighbouring parameter combinations (see Results Section 3.4). The MSE of parameter combination was calculated as
| (2) |
where are all 4-connected nearest-neighbour parameter combinations of , are all 8-connected nearest-neighbour parameter combinations of that are not included in and is the mean effect size of parameter combination . MSE values were not calculated for parameter combinations at the borders of the parameter space because the numbers of connected neighbours are reduced for these combinations. To find the best parameter combination in terms of and MSE, we scaled both values to the same range [0,1] and subtracted the resulting MSE values from 1. The scaled values of were calculated as
| (3) |
where is the scaled value of , is the mean effect size for the parameter combination , and and are the minimum and maximum values of all . Similarly, the scaled MSE values were calculated as
| (4) |
where is the scaled value of , is the MSE for the parameter combination , and and are the minimum and maximum values of all .
Descriptive statistics for all group comparisons can be found in the Supplementary Information A3.
2.6. Experimental confirmation
To confirm the advantage of our signal-oriented approach over the tissue-oriented analysis, we applied both concepts to an established preclinical sepsis model and quantitatively evaluated the liver function of healthy and diseased animals.
2.6.1. Sepsis animal model
All experimental protocols involving animals were approved by the State Administration Office of Thuringia, Germany. Eight to twelve weeks old male and female FVB/NRj inbred mice were housed under specific pathogen-free conditions in the animal facility of the Jena University Hospital under an artificial 12 h/12 h dark/light cycle with 20 min dim phases at 21 ± 2 °C and humidity of 55% ± 10%. Animals had access to standard rodent chow and drinking water ad libitum. Food soaked in water was additionally offered on the ground during infection. A life-threatening infection accompanied by an early liver failure, i.e. sepsis, was induced in 12 animals by the peritoneal contamination and infection model (PCI group). Therefore, approximately 60 µl of a characterised human stool suspension were injected per animal [16]. A control group of 9 animals (Sham group) received an intraperitoneal injection with an equal volume of a sterile salt solution (Ringer acetate, Fresenius Kabi). Approximately 2.5 mg metamizole dissolved in drinking water was given orally as drops on the animals’ tongue at infection and then every 6 h for pain relief. Further, both groups received either the broad-spectrum antibiotic meropenem subcutaneously 6 h and 18 h after PCI or a salt solution treatment (2.5 mg−1 kg body weight dissolved in Ringer acetate to a volume of 2.5 mg mL−1). The liver function was evaluated by MSOT imaging 24 h post infection.
2.6.2. Multispectral optoacoustic tomography
MSOT imaging was performed 24 h after either PCI induction or injection of sterile salt solution using the inVision 256-TF MSOT system (iTheraMedical, Munich, Germany). Animals were anaesthetised throughout MSOT preparation and image acquisition with 1.5–2% of isoflurane vaporised in oxygen. The abdominal imaging area was shaved using an electric shaver and commercial hair removal cream. Animals were positioned in the imaging chamber to scan a cross-section of the liver. Images were acquired at six wavelengths (715 nm, 730 nm, 760 nm, 800 nm, 850 nm and 900 nm) per time frame. Ten images were averaged for each wavelength and time frame to reduce animal motion and breathing artefacts. Approximately 20 μg ICG (Verdye, Diagnostic Green, USA) were administered intravenously through a tail-vein catheter 2 min after image acquisition was started. Imaging lasted for 20 min in total. Raw MSOT images were reconstructed by model-based backprojection (filter range: 50 kHz to 6.5 MHz) and spectrally unmixed into four channels (water, ICG, deoxygenated haemoglobin, oxygenated haemoglobin) by linear regression using the proprietary software ViewMSOT v3.8.1.04 (iTheraMedical, Munich, Germany).
3. Results
This section will first address the accuracy of automated image segmentation, followed by a detailed description of our new signal-oriented analysis of MSOT images and its performance in quantification of liver function in comparison with the standard approach. Subsequently, we will highlight the robustness of our new approach regarding parameter choice and segmentation accuracy and finally introduce the implementation as a readily available software toolkit.
3.1. Deep learning-based approach enables accurate automated segmentation of MSOT image data
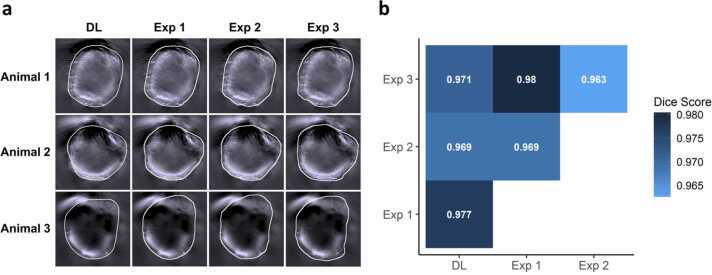
The DL-based segmentation extracted the animal outline for each of the 21 MSOT image stacks. The identified animal-ROIs were visually very close to the manual annotations of each of the three experimenters, as exemplified for three representative animals in Fig. 2a. The pairwise Dice coefficient, which measures the similarity of two annotations in terms of overlap, shows a high concordance between the manual annotations and the automated segmentation (Fig. 2b). All pairwise comparisons reached high Dice coefficients ranging from 0.963 to 0.98, indicating an overlap of over 95% for each of the annotations. The lowest agreement was found between experimenters two and three and the highest agreement between experimenters one and three. The agreement between all the manual annotations and the DL-based segmentation reached intermediate values ranging from 0.969 to 0.977.
Fig. 2.
Accuracy of DL-based segmentation in comparison to manual annotation. (a) The manual annotations of three experimenters and the DL-based segmentation are shown for three representative animals as white overlays on the first time frame of each of the image stacks. Besides minor variations, all outlines are visually very close and there is no apparent difference between the automated and manual segmentations. (b) The pairwise Dice coefficients show a high concordance between all the segmentations, where the lowest agreement was observed between two of the three experimenters (n = 21 for each pairwise comparison).
3.2. Signal-oriented analysis determines biomarker pharmacokinetics and biodistribution
Our new signal-oriented approach automatically identifies connected regions within the segmented area that exhibit similar signal kinetics of the analysed biomarker (Fig. 3).
Fig. 3.
Scheme of new signal-oriented analysis. The biomarker ICG was injected shortly after the start of the MSOT image acquisition, and the collected time-resolved MSOT image stacks were spectrally unmixed into four channels (water, ICG, deoxygenated haemoglobin and oxygenated haemoglobin). ROIs were derived with a DL-based approach and are denoted as animal-ROIs (yellow outline). The image stacks were then pre-processed, and the primary signal kinetics present in the ICG channel were extracted by k-means clustering on the pixel level. The obtained pharmacokinetics were then used to compare treatment groups quantitatively. Additionally, the biodistribution of the extracted signal kinetics were reconstructed and visualised per animal.
All pixels outside the animal-ROIs determined by DL-based segmentation are set to zero, and the image stacks are cropped to the bounding boxes of the animal-ROIs. These steps reduce the impact of potentially noisy background regions, which can otherwise also comprise a varying proportion of the image area depending on the animal size, during the following analysis. Each pixel position in the pre-processed ICG image stacks holds a vector of time derivative values describing the ICG signal change over time for this spatial position. These vectors of length n serve as n-dimensional feature vectors for k-means clustering to obtain clusters reflecting the most prominent signal kinetics. The parameter k is used to control how many of these kinetic clusters are derived. Alongside, colour-coded images of their spatial distribution are reconstructed to visualise the biomarkers’ distribution. To this end, a new two-dimensional image is created for each animal where each pixel value is set to a predefined colour, depending on which kinetic cluster has the smallest Euclidean distance to the time derivative vector of this pixel position. To compare treatment groups, all vectors from all animals in one treatment group, i.e. Sham or PCI, are mixed and clustering is performed on these combined vector sets. The clustering is executed 100 times, and the result with the lowest overall Euclidean distance of the kinetic clusters to the corresponding data points is picked for subsequent analysis, accounting for the random initialisation step of the k-means algorithm.
Since we aim to analyse ICG uptake and clearance through the liver, we focus on kinetic clusters that reflect a signal net increase over time. This is assumed to be the case if the cumulative value of the curve of a kinetic cluster is larger than ε. The definition of this offset is necessary to ensure that only curves with a signal increase are considered and that curves with only slight deviations from the zero line are excluded as these are associated with regions where the ICG signal is not substantially changing over time. We set ε = 0.1 based on the observed typical variations around the zero line. For each animal, we calculate a normalised weighted-average-curve (WAC) according to
| (5) |
where is the weighted-average-curve of animal , is the curve of the -th kinetic cluster with a signal net increase, is the frequency of for animal , is the width of the image stack of animal , and is the corresponding height. AUC values are then calculated based on the WACs to compare treatment groups.
3.3. New signal-oriented analysis outperforms current standard approach
Mice were treated either with PCI or a sterile salt solution 24 h before MSOT imaging to compare the liver function between diseased and healthy animals. No animal died or reached an experimental endpoint, e.g. lethargy, within this 24 h period. In addition, all PCI animals exhibited clinical signs of infection, including reduced activity and reactivity to external stimuli, unconditioned fur and diarrhoea, while the appearance and behaviour of animals in the Sham group were within normal limits.
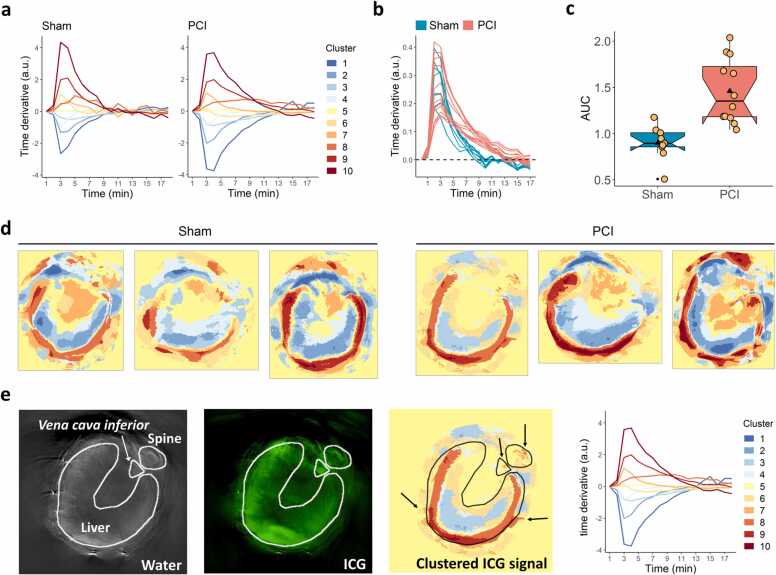
With our new signal-oriented analysis, we extracted ten kinetic clusters for both Sham and PCI treatment groups, capturing the most prominent signal kinetics present in the ICG channels of the respective treatment group (Fig. 4a; see Results Section 3.4 for information on parameter choice). For Sham animals, one more of the ten kinetic clusters reflected a signal increase compared to PCI animals. Additionally, one kinetic cluster of the PCI group exhibited a constant signal increase over time (cluster 8), which could not be observed in the Sham group.
Fig. 4.
Results from the signal-oriented analysis. (a) Main ICG signal kinetics were obtained by k-means clustering with k = 10 for Sham and PCI animal groups. (b) For each animal, a WAC of all curves of kinetic clusters with a net increase, i.e. curves reflecting ICG uptake, was calculated with the weights being the pixel frequencies of the respective kinetic clusters for this animal. The dashed line marks the change from a signal increase (positive values) to signal decrease (negative values). (c) AUC values were calculated for the WACs and are visualised as boxplots for Sham and PCI groups, with the black triangles denoting mean values and the yellow dots being individual data points (two-tailed Wilcoxon rank sum test p = 1.3 ∙ 10−4, Hedges' g = 1.83). (d) Colour-coded images reveal the heterogeneous distribution of ICG signal across liver tissue (three representative images are depicted for each treatment group). (e) ROIs were drawn around liver tissue, spine and Vena cava inferior in the water channel by an expert and were overlaid on the ICG channel and the colour-coded visualisation of ICG biodistribution. The right panel depicts the corresponding kinetic curves. The liver tissue was strongly heterogeneous with respect to ICG kinetics, and smaller clusters of ICG signal increase fitted to the positions of the spine, the Vena cava inferior and vessels (arrows).
The curves of the kinetic clusters and corresponding pixel frequencies were used to calculate a WAC for each animal (Fig. 4b), and the difference between Sham and PCI treatments was quantified by calculating the AUC values of the WACs (Fig. 4c). PCI animals had significantly higher AUC values than Sham animals, which was also reflected by a very large effect size between the two groups (p = 1.3 ∙ 10−4, g = 1.83).
The WACs showed a substantial initial increase of ICG signal for both Sham and PCI animals. This increase peaked at around 4 min after the acquisition was started and slowed down subsequently. We note that ICG signals are always reported as time derivatives and not as absolute signal intensity values. For some animals, signal change turned into a signal decrease, which is expected as a result of the ICG clearance by the liver.
In this phase, PCI animals exhibited constantly higher values of ICG signal than Sham animals, which led to a higher overall ICG signal intensity detected in the liver tissue. The plateau phase of the ICG signal was reached later for PCI animals: starting at around 9 min for Sham and 13 min for PCI (see dashed line in Fig. 4b). The ICG signal decreased at this time point for healthy animals, whereas it continued to rise for seven out of the twelve PCI mice. Generally, these animals were characterised by lower peak signal intensity, albeit higher signal increase in the second half of image acquisition than other animals from this group. This difference became even more apparent when looking at the time curves of integrated signal intensity (see Supplementary Fig. A4).
Representative images of the spatial distribution of kinetic clusters revealed that only the outer part of liver tissue showed an increase of ICG signal. In contrast, deeper parts were characterised by a signal decrease or stable ICG signal over time (Fig. 4d). The signal increase was not only present in liver tissue but also in other regions, which are consistent with blood vessels and regions of increased perfusion (Fig. 4e). The overlaid outline of a manual annotation of the liver in Fig. 4e clearly highlights the heterogeneity of the MSOT signal of ICG within the liver. Sham animals were visually not different from PCI animals with regard to the spatial distribution of kinetic clusters (Supplementary Fig. A5).
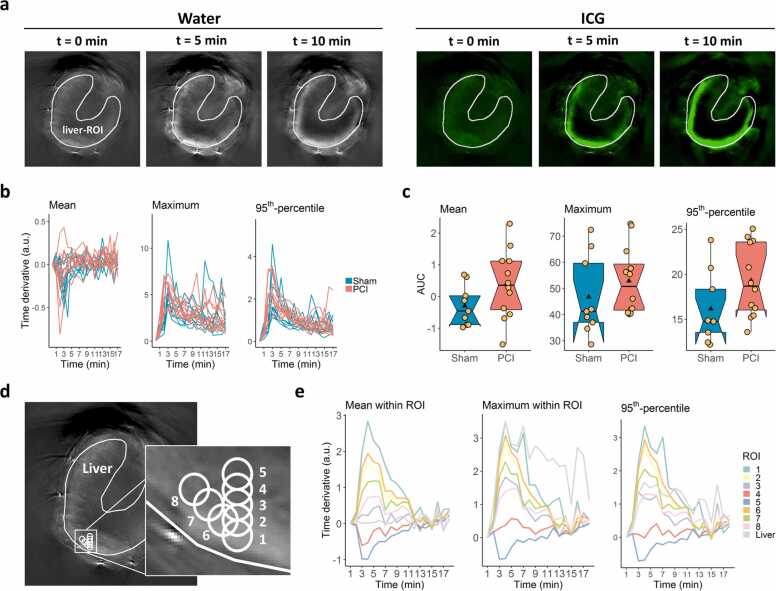
Liver-ROIs containing the entire liver tissue were drawn by an expert (Fig. 5a) to compare the results of our new approach with the tissue-oriented analysis. Time-resolved mean, maximum and 95th-percentile signal intensity values within the liver-ROIs were extracted from the pre-processed MSOT images (Fig. 5b). For each of the three features, the time curves from Sham and PCI animals were strongly overlapping, making visual separation of the two treatment groups impossible. After integrating the time derivative values, which did amplify group differences for the signal-oriented analysis, the time curves of Sham and PCI animals were still not visually distinguishable (Supplementary Fig. A6). The results from individual animals of the same treatment group showed a substantial variation, especially for maximum and 95th-percentile values, which was also reflected by the corresponding AUC values (Fig. 5c). No significant differences were found between treatments for the mean and maximum signal intensity values, whereas the effect sizes ranged from low to medium (mean: p = 0.09, g = 0.71; maximum: p = 0.22, g = 0.42). Only for the 95th-percentile signal intensity values, the difference between Sham and PCI was just below the level of significance (p = 0.049, g = 0.75).
Fig. 5.
Tissue-oriented image analysis. (a) Liver-ROIs were drawn manually by an expert in the first time frame of the water channel of the MSOT image stacks (grey coloured images) and used to analyse signal intensity in the ICG channel (green coloured images). (b) The mean, maximum and 95th-percentile time derivative values within the liver-ROIs were extracted from the ICG channel. (c) AUC values of the time curves were calculated for each animal in the two treatment groups and are shown as boxplots for mean, maximum and 95th-percentile signal intensity values, with the black triangle denoting the average value and the yellow dots representing individual data points (mean: p = 0.09, g = 0.71; maximum: p = 0.22, g = 0.42; 95th-percentile: p = 0.049, g = 0.75; p-values were derived by two-tailed Wilcoxon rank sum test; g-values denote Hedges' g). (d) A ROI with a diameter of 10 pixels was drawn in the liver tissue region (ROI 1) and then shifted by 5-pixel steps in the y-direction (ROIs 2–5) or in the x-y-direction (ROIs 6–8) to obtain a set of ROIs within close proximity. (e) The mean, maximum and 95th-percentile signal intensity values from ROIs 1–8 and the liver-ROI were extracted from the pre-processed MSOT image and plotted for the whole time range.
To evaluate the impact of tissue heterogeneity on the tissue-oriented analysis, we placed a small circular ROI within the liver tissue. We shifted this ROI multiple times in 5-pixel steps (≈ 0.38 mm) in x- and y-direction (Fig. 5d). The time curves of these ROIs' mean, maximum and 95th-percentile signal intensity values are shown in Fig. 5e. Although all ROIs were in very close vicinity and within the liver, the time curves of the three features followed a gradient from the outer to the inner parts of liver tissue. Furthermore, they turned from a signal increase to a signal decrease at the innermost ROI, demonstrating that signal statistics of specific tissue regions may lead to contradictory results in the tissue-oriented approach and obscure group differences due to spatial inhomogeneities of the signal.
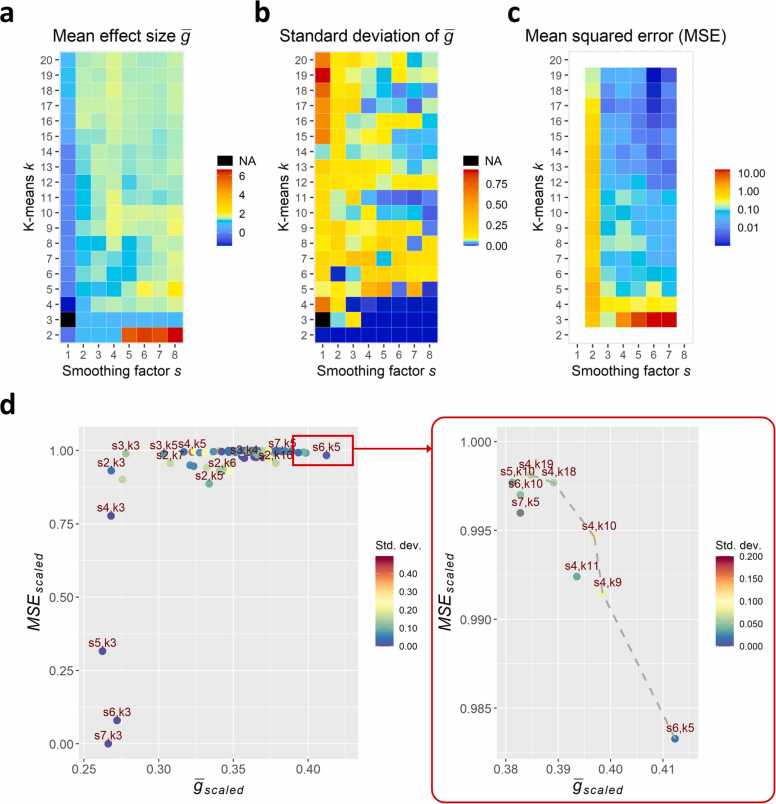
3.4. Signal-oriented analysis is robust across parameter combinations
The proposed signal-oriented analysis approach depends on two main parameters: the smoothing factor s and the expected number of clusters k in the k-means clustering algorithm. We screened a range of values for s (from s = 1 to s = 8) and k (from k = 2 to k = 20). The impact of s on the resulting main kinetic curves is shown in Supplementary Fig. A7. With s = 1, i.e. no smoothing, the resulting kinetic curves were dominated by breathing artefacts. The two lowest levels of smoothing (s = 2 and s = 3) could not fully compensate for the breathing artefact, especially for k > 7. Starting from s = 4, breathing artefacts were sufficiently reduced, while higher s resulted in a disproportionate loss of time resolution. The screened value range of k spread from k = 2 to k = 20, taking into account that large values of k will generally promote overfitting, i.e. the curves of the kinetic clusters become fluctuating also for relatively large values of s, as is apparent from Supplementary Fig. A7.
To evaluate the robustness of our signal-oriented analysis, we compared all parameter combinations with regard to two main properties: (i) high effect size between treatments and (ii) high tolerance towards minor changes of s and k (see Methods Section 2.5 for details on calculations). To take into account the randomised initialisation step of k-means clustering, the analysis was repeated five times for each parameter combination. Regarding property (i), we calculated the mean effect size () and the corresponding standard deviation between AUC values from Sham and PCI treatment groups across the five separate runs for each parameter combination (Fig. 6a and b). The difference between treatment groups in terms of effect size was comparable across a large range of combinations for s and k. Generally, combinations with s > 1 and k > 3 yielded a mean effect size larger than 0.8, which is considered to represent a large effect, highlighting the robustness of our new approach.
Fig. 6.
Parameter estimation for smoothing factor s and k-means k. (a) The mean effect size between Sham and PCI AUC values of five independent runs is highest for k = 2 and shows, in addition, several local maxima. (b) Similarly, the corresponding standard deviation of is lowest for small values of k and shows several local minima. (c) The mean squared error (based on the weighted 8-connected neighbourhood of the matrix of values) denotes the robustness towards changes of s and k in the ranges ± 1 (not calculated for border values because of the uneven number of neighbouring cells). (d) The best parameter combinations of s and k are found at the upper right part in a scatterplot of and . Pareto-optimal parameter combinations are highlighted by the dashed line in the zoomed-in panel on the right.
Concerning property (ii), we calculated the MSE of for each parameter combination with regard to of neighbouring parameter combinations (i.e. all possible changes of s and k in the ranges ± 1; Fig. 6c).
We scaled both and MSE values to the same value range [0,1], and the scaled MSE values were subtracted from 1. The resulting measures are denoted by and . The best parameter combinations are located in the upper right part of a scatterplot of versus (Fig. 6d). We identified multiple Pareto-optimal solutions, i.e. solutions where one feature cannot improve without worsening another, as indicated by the dashed line in the zoomed-in panel of Fig. 6d. These combinations are characterised by comparably high effect size and a relatively low MSE. We decided to use the parameter combination (s = 4, k = 10) from all Pareto-optimal solutions, representing intermediate values for s and k. Thus, the loss of details in the kinetic curves with s = 4 is less compared to other candidates with s = 6 or s = 7, whereas using k = 10 at the same time offers a suitably high number of kinetic clusters to identify the different tissue types of the cross-section without promoting overfitting.
3.5. Signal-oriented analysis is robust towards slight changes of ROIs
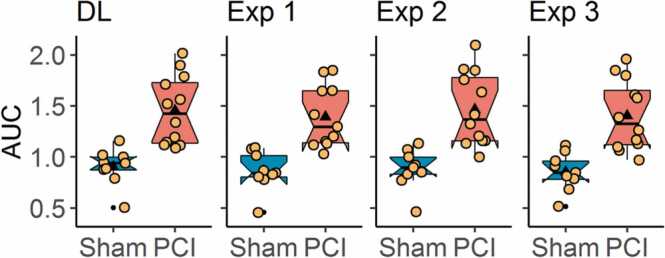
The DL approach for image segmentation was trained on a comparably low number of annotated images. Therefore, we assessed the robustness of our new signal-oriented analysis towards slight changes of the underlying animal-ROIs by performing the analysis for the animal-ROIs derived from the DL-based segmentation as well as for the annotations by the three experimentalists. All animal-ROIs from the three experimenters and the automated segmentation were in visual agreement, as shown in Results Section 3.1. Similarly, the quantitative analysis yielded very similar results for each of the four sets of animal-ROIs (Fig. 7; DL-based segmentation: p = 8.2 ∙ 10−5, g = 1.89; Experimenter 1: p = 2.7 ∙ 10−5, g = 1.88; Experimenter 2: p = 3.8 ∙ 10−4, g = 1.86; Experimenter 3: p = 8.2 ∙ 10−5, g = 1.85).
Fig. 7.
Robustness of signal-oriented analysis towards slight changes of ROIs. Animal-ROIs were derived by automated segmentation via DL and by manual annotation of three experimenters. The signal-oriented analysis was applied separately for each of the four sets of animal-ROIs. AUC values were calculated for Sham and PCI animals as well as the corresponding p-values and effect sizes, confirming high compliance between the different ROI sets (DL: p = 8.2 ∙ 10−5, g = 1.89; Experimenter 1: p = 2.7 ∙ 10−5, g = 1.88; Experimenter 2: p = 3.8 ∙ 10−4, g = 1.86; Experimenter 3: p = 8.2 ∙ 10−5, g = 1.85; p-values were derived by two-tailed Wilcoxon rank sum test; g-values denote Hedges' g). The black triangles indicate mean values, and the yellow dots represent individual data points.
3.6. Mcat enables automated and user-friendly application of signal-oriented analysis
To make our new approach easily available and applicable for other researchers, we implemented it in our MSOT cluster analysis toolkit Mcat, a Java-based plugin for ImageJ that builds upon MISA+ + [22] and JGraphT [23]. It provides an easy-to-use graphical user interface (GUI) for loading data (Supplementary Fig. A8a) and defining parameter sets (Supplementary Fig. A8b).
Data can be imported either manually by specifying input images and, if desired, existing annotations of ROIs, or with the help of a batch importer functionality using a customisable pattern-matcher. If no annotations are provided, the inbuilt DL model will derive the ROIs as described in the Methods Section 2.1. Additionally, custom Cellpose models can be imported to adjust the segmentation to other sample types. After the data have been imported, users can adjust the pre-processing, clustering and post-processing parameters. This allows the specification of the channel of interest, the indication of which output data should be generated, the determination of how clustering is applied and which quantitative measurements will be performed. Multiple parameter sets can be defined within one project, which are then automatically executed sequentially.
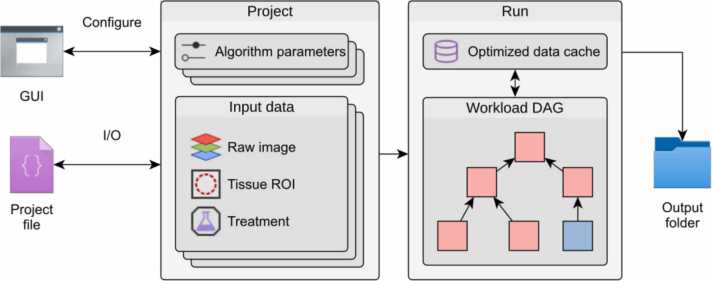
The software contains components that organise input data and parameters and generates results by applying processing steps. Its two main components are termed project and run (Fig. 8). A project contains all parameters and settings on how to load input data. A run, generated from a project, includes all necessary functions to perform the analysis with the desired parameter settings. In addition, it contains the data processing steps, which are generated based on the user-defined parameters and the available data. The processing steps are organised in a directed acyclic graph (DAG), where the nodes are the processing steps and edges represent dependencies between these steps. Upon running the analysis, processing steps are executed in topological order. Input and output data are internally organised in a cache structure that is accessed by the processing steps during programme execution. The cache uniquely assigns data, e.g. images, ROIs or results, to the set of parameters and input data. This prevents the repetition of equal workloads and improves the efficiency of the programme. After generating the results, they are automatically saved in the output folder, alongside the parameter file of the project that generated the run.
Fig. 8.
Internal structure of the Mcat software toolkit. Mcat organises data in projects that store all parameter sets and groups raw images and ROIs into annotated input data sets. The algorithm parameters can be configured in a graphical user interface (GUI). Upon running a project, a directed acyclic graph (DAG) is generated that models the dependencies between the individual algorithms. In conjunction with the optimised data cache, it prevents duplication of workloads when running multiple parameter sets. The generated result data is written to an output folder together with a project file that includes the parameters and data used in this run.
The documentation for all parameters used in Mcat is provided in Supplementary Information A9. A detailed user manual including installation instructions, the software toolkit and example data is available online at https://github.com/applied-systems-biology/mcat (https://doi.org/10.5281/zenodo.6046031).
4. Discussion and conclusions
This study presented an automated approach to objectively and quantitatively analyse biomarker uptake and distribution from MSOT image data. We utilised and validated our approach by assessing liver function in a well-characterised murine model of sepsis-associated liver failure [16], [24], based on the uptake and clearance of the clinically established biomarker ICG [18], [25]. We have shown multiple advantages of our signal-oriented approach compared to the widely used tissue-oriented analysis [7], [8], [9], [10], [11], [12], [13].
In this study, the tissue-oriented approach as used in its most common form (time-resolved extraction of mean and maximum signal intensities from a ROI) could not detect significant differences between healthy and diseased animals. Only the 95th-percentile signal intensity analysis yielded a group difference that was just below the level of significance (p = 0.049). Thus, the results also depended on the analysed intensity feature (Fig. 5c). By placing multiple small ROIs within the region of liver tissue, we could show that confined ROIs are highly sensitive to minor positional changes, and that even those structures that are expected to constitute homogeneous signal kinetics can be characterised by pronounced signal heterogeneity (Fig. 5e). A possible reason for this observation is the limited penetration depth of MSOT. Consequently, quantitative results based on confined ROIs may be misleading, and differences between treatment groups may be overlooked due to tissue heterogeneity, as was the case in the present study for the tissue-oriented analysis. Additionally, the tissue-oriented analysis approach can yield biased results if the locally observed biomarker signal gives rise to the selective placment of ROIs.
In contrast, our signal-oriented approach revealed significant differences in liver function between healthy and diseased animals without estimating the liver areas responsible for drug uptake and removal. This analysis provided a quantitative measure of liver function and enabled the extraction of information about spatial ICG distribution. In contrast to the tissue-oriented approach, our signal-oriented analysis is more objective, since the entire sample is considered, and the regions of photoabsorber uptake are determined by the algorithm itself. DL-based segmentation aids to analyse future data easily without the need for manual annotation, eliminating possible user bias (see Supplementary information A10 for example on application of inbuilt DL-based segmentation model to different MSOT data). However, we want to note that retraining plays an essential role when transferring a neural network to new data and ensures that satisfying results are achieved. Thus, we included both the possibility to use a retrained model for segmentation, as well as the option to provide manual annotations. Additionally, we could show that our approach is not sensitive to small changes of the ROIs or parameters but provides robust results. At the same time, the complete automation of all processing steps ensures high data throughput. Furthermore, it is not restricted to studying ICG as photoabsorber. It is also possible to analyse other photoabsorbers, as demonstrated for the channel of oxygenated haemoglobin from another MSOT study in Supplementary information A10. For such an analysis, the photoabsorber has to exhibit sufficient resolution in space and time when imaged by MSOT. Our approach is in general limited to the analysis of time kinetics instead of analysing endpoint experiments. The current version of our analysis is also restricted to one anatomical cross-section, with plans to provide multi-slice analysis in future versions of Mcat. As the principles of our approach are not specific for MSOT images, we expect it to be transferrable to other types of image data to analyse signal kinetics, e.g. spatio-temporal characterisation of fluorescently labelled molecules from time-resolved microscopy data. However, such an application is beyond the scope of this manuscript and will have to be the subject of future studies.
The fact that we extracted connected areas of comparable signal kinetics shows that the results of our signal-oriented approach are not governed by image noise or signal fluctuations. The analysis of the spatial distribution of these areas was solely based on a visual assessment in this study. However, a quantitative analysis, e.g. by measuring scatteredness, area or homogeneity of specific kinetic clusters, could help to better characterise the liver function of individual animals and to learn the link to particular patterns of the spatial distribution of kinetic clusters. Together with the sample-specific kinetic curves, our approach offers a much more comprehensive analysis of the investigated samples than the tissue-oriented approach and could help to better discriminate severe liver failure from milder impairment of liver function. Such differentiation is a key towards personalised medicine and targeted treatment not only with regard to sepsis [26], [27], but also critically ill patients [28], acute liver failure [29] and liver transplantation [30]. Although there already exist methods to assess the ICG plasma disappearance rate as a predictor of organ failure [31], these either involve invasive blood sampling or depend on unimpeded peripheral perfusion, which can be problematic in various cases [26], [32]. Non-invasive whole-organ imaging with MSOT, combined with robust and quantitative image analysis, could help identify patients with a critical health status, initiate proper treatment and monitor its effectiveness longitudinally.
Our MSOT cluster analysis toolkit Mcat is implemented as an ImageJ plugin, which is freely available and easy to use for scientists without programming skills, coming from various research areas (https://github.com/applied-systems-biology/mcat; https://doi.org/10.5281/zenodo.6046031). Furthermore, the software is easy to adapt to various study setups via its user-friendly parameter editor. In combination with the non-invasive imaging modality MSOT, our signal-oriented analysis approach will play an essential role in the objective evaluation of organ function in critically ill patients. It will support the prediction of disease progression and the success of therapy in personalised medicine.
Author Contributions
Z.C., O.S., A.T.P., M.B. and M.T.F. conceptualised the study. W.F., O.S. and A.T.P. designed and performed animal experiments. B.H., R.G., Z.C. and M.T.F. developed image analysis methodology. B.H. and R.G. developed the presented software. B.H. performed the quantitative image analysis. B.H., R.G. and Z.C. validated the image analysis methodology and analysed the data. B.H. and R.G. wrote the original draft and prepared figures. A.T.P., M.B. and M.T.F. supervised the study. M.B. and M.T.F. did project administration and funding acquisition. All authors revised the manuscript and approved it for submission.
Code availability
The presented software toolkit Mcat, a user manual and example data are freely available (licensed under BSD 2-clause) at https://github.com/applied-systems-biology/mcat (https://doi.org/10.5281/zenodo.6046031).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was funded by the German Research Foundation (DFG), Germany, through the Collaborative Research Centre PolyTarget 1278 “Polymer-based nanoparticle libraries for targeted anti-inflammatory strategies” (Project Z01 to MTF and Project C03 to MB) [grant number 316213987]; the Leibniz ScienceCampus InfectoOptics Jena, Germany, which is financed by the funding line Strategic Networking of the Leibniz Association; the Federal Ministry of Education and Research (BMBF), Germany, through the Centre for Sepsis Control and Care (CSCC) [grant number 01EO1502] and the International Leibniz Research School for Microbial and Biomolecular Interactions Jena (ILRS Jena), Germany. ATP acknowledges the Interdisciplinary Centre for Clinical Research, Germany, [grant number AMSP05].
Biographies

Bianca Hoffmann is working in the field of image-based systems biology. She studied Bioinformatics at the Friedrich-Schiller-University Jena, Germany, and completed her Ph.D. in natural sciences in the research group Applied Systems Biology at the Leibniz Institute of Natural Product Research and Infection Biology e.V. Hans Knöll Institute Jena in 2018. She is now working as a postdoctoral researcher in the group Applied Systems Biology with the research focus being on automated analysis and processing of image data. Her special interest is the extraction of quantitative features from microscopy and tomography image data.

Ruman Gerst is a doctoral student in the group Applied Systems Biology at the Leibniz Institute of Natural Product Research and Infection Biology e.V. – Hans Knöll Institute, Jena, Germany. From 2012–2018 he studied Bioinformatics (B.Sc. and M.Sc.) at the Friedrich-Schiller University Jena, Germany. Since 2018 he is pursuing his PhD with the research focus on quantitative image analysis of infections.

Zoltán Cseresnyés received his university diploma in physics in 1985 in Hungary, at the University of Debrecen. His Ph.D. work in cell biology focused on the confocal microscopy studies of calcium signalling in peripheral neurons. He worked for many years in the USA, Great Britain and Germany. His work interest has been in quantitative microscopy, both from the experimental and analytical point of view. He is currently a postdoctoral fellow at the Hans Knöll Institute in Jena, where he aids the collaborative efforts between microscopists and image analysts. His own research projects focus on label-free image acquisition and analysis methods.

WanLing Foo is currently pursuing her doctoral grade degree in University Hospital Jena, Germany. She is working on targeted drug and gene delivery systems and translational medicine for anti-inflammation. She received her M.Eng. degree in Bioengineering from The University of Tokyo, Japan and B.Eng. degree in Materials Science from Nanyang Technological University, Singapore. Her master focused on targeted gene delivery and systemic gene therapy for anticancer treatments. In between the studies, she has also involved in the research of intrauterine stem cell transplantation for Thalassaemia therapy.

Oliver Sommerfeld is Consultant for Anaesthesiology and Intensive Care. He studied medicine at Friedrich-Schiller University Jena (2004–2011). He completed his specialist training at the Clinic for Anaesthesiology and Intensive Care at the Jena University Hospital (2011–2018). After he received his M.D. in 2013, he completed a research rotation at the Center for Sepsis Control Care (2014–2015). His research is in the field of innate immune response and complement activation in sepsis, transnational medicine and animal models.

Adrian T. Press, studied molecular medicine for his bachelor’s and master’s degrees at Furtwangen and the Friedrich Schiller University in Jena, Germany. He then obtained his doctoral degree at the medical faculty in Jena researching novel theranostic nanoparticles. During his postdoc, he specialised in researching biophotonic technologies to characterise and model septic organ failure and translate this knowledge to generate novel personalised therapeutics and diagnostics. Since 2021 he is intensifying his research at Friedrich Schiller University as Junior professor for Molecular Medicine of Life-Threatening Infection.

Michael Bauer is Professor and Chair of Anesthesiology and Intensive Care and Chief Executive Director of the Center for Sepsis Control Care (CSCC) at University Hospital of Jena, Germany. He received his M.D. from Saarland University and postgraduate training in molecular biology at Johns Hopkins (1993–1995). After Board Certification in Anesthesiology and Intensive Care, he was visiting professor at the University of North Carolina in Charlotte (1998–1999). Dr. Bauer has published extensively in the fields of molecular mechanisms of sepsis and shock-related organ dysfunction with a focus on biophotonic strategies to visualise liver perfusion and function.

Marc Thilo Figge is Professor of Applied Systems Biology at the Friedrich Schiller University Jena and in the Center for Systems Biology of Infection at the Leibniz Institute for Natural Product Research and Infection Biology in Jena. He received his Ph.D. in Theoretical Physics at the University of Groningen and became Junior Fellow in Systems Immunology at the Frankfurt Institute for Advanced Studies (FIAS) before setting up his own research group in Jena in 2011. His research focuses on image-based systems biology, combining automated quantification of microscopy and spectroscopy data with mathematical modelling and computer simulation.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.pacs.2022.100361.
Appendix A. Supplementary material
Supplementary material
.
Data availability
All MSOT image data, ROI files used in this study as well as code and data used for the DL model training are available at https://doi.org/10.6084/m9.figshare.13567265.v3.
References
- 1.Attia A.B.E., Balasundaram G., Moothanchery M., Dinish U.S., Bi R., Ntziachristos V., Olivo M. A review of clinical photoacoustic imaging: current and future trends. Photoacoustics. 2019;16 doi: 10.1016/j.pacs.2019.100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diot G., Metz S., Noske A., Liapis E., Schroeder B., Ovsepian S.V., Meier R., Rummeny E., Ntziachristos V. Multispectral Optoacoustic Tomography (MSOT) of human breast cancer. Clin. Cancer Res. 2017;23:6912–6922. doi: 10.1158/1078-0432.CCR-16-3200. [DOI] [PubMed] [Google Scholar]
- 3.Stoffels I., Morscher S., Helfrich I., Hillen U., Leyh J., Burton N.C., Sardella T.C., Claussen J., Poeppel T.D., Bachmann H.S., Roesch A., Griewank K., Schadendorf D., Gunzer M., Klode J. Metastatic status of sentinel lymph nodes in melanoma determined noninvasively with multispectral optoacoustic imaging. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aad1278. [DOI] [PubMed] [Google Scholar]
- 4.Wildgruber M., Masthoff M., Helfen A., Claussen J., Karlas A., Markwardt N.A., Ntziachristos V., Eisenblätter M., Wildgruber M. Use of multispectral optoacoustic tomography to diagnose vascular malformations. JAMA Dermatol. 2018;154:1457–1462. doi: 10.1001/jamadermatol.2018.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waldner M.J., Knieling F., Egger C., Morscher S., Claussen J., Vetter M., Kielisch C., Fischer S., Pfeifer L., Hagel A., Goertz R.S., Wildner D., Atreya R., Strobel D., Neurath M.F. Multispectral optoacoustic tomography in Crohn’s disease: noninvasive imaging of disease activity. Gastroenterology. 2016;151:238–240. doi: 10.1053/j.gastro.2016.05.047. [DOI] [PubMed] [Google Scholar]
- 6.Ntziachristos V., Razansky D. Molecular imaging by means of multispectral optoacoustic tomography (MSOT) Chem. Rev. 2010;110:2783–2794. doi: 10.1021/cr9002566. [DOI] [PubMed] [Google Scholar]
- 7.Taruttis A., Morscher S., Burton N.C., Razansky D., Ntziachristos V. Fast multispectral optoacoustic tomography (MSOT) for dynamic imaging of pharmacokinetics and biodistribution in multiple organs. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharkey J., Ressel L., Brillant N., Scarfe L., Wilm B., Park B.K., Murray P. A noninvasive imaging toolbox indicates limited therapeutic potential of conditionally activated macrophages in a mouse model of multiple organ dysfunction. Stem Cells Int. 2019;2019 doi: 10.1155/2019/7386954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song W., Tang Z., Zhang D., Burton N., Driessen W., Chen X. Comprehensive studies of pharmacokinetics and biodistribution of indocyanine green and liposomal indocyanine green by multispectral optoacoustic tomography. RSC Adv. 2015;5:3807–3813. doi: 10.1039/c4ra09735a. [DOI] [Google Scholar]
- 10.Morscher S., Driessen W.H.P., Claussen J., Burton N.C. Semi-quantitative multispectral optoacoustic tomography (MSOT) for volumetric PK imaging of gastric emptying. Photoacoustics. 2014;2:103–110. doi: 10.1016/j.pacs.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y., Huang S., Wang J., Sun L., Zeng F., Wu S. Activatable probes for diagnosing and positioning liver injury and metastatic tumors by multispectral optoacoustic tomography. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06499-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y., Qi Y., Zhan C., Zeng F., Wu S. Diagnosing drug-induced liver injury by multispectral optoacoustic tomography and fluorescence imaging using a leucine-aminopeptidase-activated probe. Anal. Chem. 2019;91:8085–8092. doi: 10.1021/acs.analchem.9b00107. [DOI] [PubMed] [Google Scholar]
- 13.Scarfe L., Rak-Raszewska A., Geraci S., Darssan D., Sharkey J., Huang J., Burton N.C., Mason D., Ranjzad P., Kenny S., Gretz N., Lévy R., Park B.K., García-Fiñana M., Woolf A.S., Murray P., Wilm B. Measures of kidney function by minimally invasive techniques correlate with histological glomerular damage in SCID mice with adriamycin-induced nephropathy. Sci. Rep. 2015;5 doi: 10.1038/srep13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taruttis A., Ntziachristos V. Advances in real-time multispectral optoacoustic imaging and its applications. Nat. Photonics. 2015;9:219–227. doi: 10.1038/nphoton.2015.29. [DOI] [Google Scholar]
- 15.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonnert F.A., Recknagel P., Seidel M., Jbeily N., Dahlke K., Bockmeyer C.L., Winning J., Lösche W., Claus R.A., Bauer M. Characteristics of clinical sepsis reflected in a reliable and reproducible rodent sepsis model. J. Surg. Res. 2011;170:123–134. doi: 10.1016/j.jss.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Seemann S., Zohles F., Lupp A. Comprehensive comparison of three different animal models for systemic inflammation. J. Biomed. Sci. 2017;24:1–17. doi: 10.1186/s12929-017-0370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levesque E., Saliba F. ICG clearance monitoring in ICU patients. Yearb. Intensive Care Emerg. Med. 2007;2009:646–657. doi: 10.1007/978-0-387-92278-2_60. [DOI] [Google Scholar]
- 19.Stringer C., Wang T., Michaelos M., Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- 20.R. Gerst, Z. Cseresnyés, J.P.M.T. Figge. 〈www.jipipe.org〉. (Accessed 18 January 2022).
- 21.Legland D., Arganda-Carreras I., Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- 22.Gerst R., Medyukhina A., Figge M.T. MISA++: a standardized interface for automated bioimage analysis. SoftwareX. 2020;11 doi: 10.1016/j.softx.2020.100405. [DOI] [Google Scholar]
- 23.Michail D., Kinable J., Naveh B., Sichi J.V. JGraphT - a java library for graph data structures and algorithms. ACM Trans. Math. Softw. 2020;46:1–33. doi: 10.1145/3381449. [DOI] [Google Scholar]
- 24.Sommerfeld O., Medyukhina A., Neugebauer S., Ghait M., Ulferts S., Lupp A., König R., Wetzker R., Schulz S., Figge M.T., Bauer M., Press A.T. Targeting complement C5a receptor 1 for the treatment of immunosuppression in sepsis. Mol. Ther. 2021;29:338–346. doi: 10.1016/j.ymthe.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kortgen A., Paxian M., Werth M., Recknagel P., Rauchfu F., Lupp A., Krenn C.G., Müller D., Claus R.A., Reinhart K., Settmacher U., Bauer M. Prospective assessment of hepatic function and mechanisms of dysfunction in the critically ill. Shock. 2009;32:358–365. doi: 10.1097/SHK.0b013e31819d8204. [DOI] [PubMed] [Google Scholar]
- 26.Kaffarnik M.F., Lock J.F., Vetter H., Ahmadi N., Lojewski C., Malinowski M., Neuhaus P., Stockmann M. Early diagnosis of sepsis-related hepatic dysfunction and its prognostic impact on survival: a prospective study with the LiMAx test. Crit. Care. 2013;17:R259. doi: 10.1186/cc13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inal M.T., Memiş D., Kargi M., Sut N. Prognostic value of indocyanine green elimination assessed with LiMON in septic patients. J. Crit. Care. 2009;24:329–334. doi: 10.1016/j.jcrc.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Sakka S.G. Assessment of liver perfusion and function by indocyanine green in the perioperative setting and in critically ill patients. J. Clin. Monit. Comput. 2018;32:787–796. doi: 10.1007/s10877-017-0073-4. [DOI] [PubMed] [Google Scholar]
- 29.Merle U., Sieg O., Stremmel W., Encke J., Eisenbach C. Sensitivity and specificity of plasma disappearance rate of indocyanine green as a prognostic indicator in acute liver failure. BMC Gastroenterol. 2009;9:91. doi: 10.1186/1471-230X-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y., Yu L., Liu Y. Predictive value of indocyanine green plasma disappearance rate on liver function and complications after liver transplantation. Med. Sci. Monit. 2018;24:3661–3669. doi: 10.12659/MSM.907783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vos J.J., Wietasch J.K.G., Absalom A.R., Hendriks H.G.D., Scheeren T.W.L. Green light for liver function monitoring using indocyanine green? An overview of current clinical applications. Anaesthesia. 2014;69:1364–1376. doi: 10.1111/anae.12755. [DOI] [PubMed] [Google Scholar]
- 32.Pantanali C.A., Esteban G.D., LA D., W A., P B., AM A. Lessons learned with the LiMON method of indocyanine green elimination. EC Gastroenterol. Dig. Syst. 2018;4:297–304. doi: 10.31080/ecgds.2018.05.00182. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All MSOT image data, ROI files used in this study as well as code and data used for the DL model training are available at https://doi.org/10.6084/m9.figshare.13567265.v3.