Abstract
The food-borne trematode Opisthorchis felineus colonizes bile ducts of the liver of fish-eating mammals including humans. Among chronically infected individuals, this opisthorchiasis involves hepatobiliary problems, including chronic inflammation, periductal fibrosis, biliary intraepithelial neoplasia, and even cholangiocarcinoma. Despite numerous studies at the pathomorphological level, the systemic response and cellular pathogenesis of these disorders are not well studied.
To conduct in-depth research and to gain insights into the mechanism by which O. felineus infection causes precancerous liver lesions, we (i) applied a next-generation-sequencing–based technology (high-throughput mRNA sequencing) to identify differentially expressed genes in the liver of golden hamsters infected with O. felineus at 1 and 3 months postinfection and (ii) verified the most pronounced changes in gene expression by western blotting and immunohistochemistry.
A total of 2151 genes were found to be differentially expressed between uninfected and infected hamsters (“infection” factor), whereas 371 genes were differentially expressed when we analyzed “time × infection” interaction. Cluster analysis revealed that sets of activated genes of cellular pathways were different between acute (1 month postinfection) and chronic (3 months postinfection) opisthorchiasis. This enriched KEGG pathways were “Cell adhesion molecules”, “Hippo signaling”, “ECM-receptor interaction”, “Cell cycle”, “TGF-beta”, and “P53 signaling”. Moreover, epithelial–mesenchymal transition was the most enriched (q-value = 2.2E-07) MSigDB hallmark in the set of differentially expressed genes of all O. felineus–infected animals. Transcriptomic data were supported by the results of western blotting and immunohistochemistry revealing the upregulation of vimentin, N-cadherin, and α-smooth muscle actin postinfection.
Our data expand knowledge about global changes in gene expression in the O. felineus–infected host liver and contribute to understanding the biliary neoplasia associated with the liver fluke infection.
Keywords: Opisthorchis felineus, Differentially expressed gene, Liver, hamster, Mesocricetus auratus
Abbreviations: DEGs, differentially expressed genes; EMT, epithelial–mesenchymal transition; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes, O. felineus; OF, Opisthorchis felineus
Highlights
-
•
A total of 2151 genes were found to be differentially expressed during the infection.
-
•
Epithelial-Mesenchymal Transition was the most enriched process in the infected animals.
-
•
Different pathways were activated between acute and chronic opisthorchiasis.
1. Introduction
Opisthorchiasis caused by the fish-borne trematodes Opisthorchis felineus (Rivolta, 1884), Opisthorchis viverrini (Poirier, 1886), and Clonorchis sinensis (Looss, 1907) is a substantial public health problem, with 40 million infected people worldwide (Beer, 2005; Pakharukova and Mordvinov, 2016; Sripa et al., 2012). These liver flukes affect bile ducts and the gall bladder of mammals including humans. People become infected after eating raw or undercooked fresh-water fish containing metacercariae of the parasites (Pakharukova and Mordvinov, 2016).
O. viverrini and Clonorchis sinensis (C. sinensis) are recognized as 1A group biological carcinogens and as a definitive cause of cholangiocarcinoma, while O. felineus (OF) is not considered a human carcinogen and is recognized as a 3A pathogen (IARC, 2012). This may be in part due to the lack of knowledge about the pathogenesis of the disease caused by OF. This fluke occurs primarily on the territory of the former USSR, with infestation outbreaks in other regions of Europe (Beer, 2005; Pakharukova and Mordvinov, 2016). According to various estimates, the infection with OF in the rural population of Western Siberia varies from 10% to 60% (Brazhnikova and Tolkaeva, 2002; Fedorova et al., 2020; Pakharukova and Mordvinov, 2016). The association of OF infection and cholangiocarcinoma development has been proven in an experimental animal model (Maksimova et al., 2017; Mordvinov et al., 2021; Pakharukova et al., 2019a). There are also reports that the incidence of cancer of the liver (e.g., of intrahepatic bile ducts) is higher in the endemic areas of the Russian Federation than in nonendemic regions (Fedorova et al., 2016; reviewed in Pakharukova and Mordvinov, 2016).
The time course of infection by these three species has been studied at the pathomorphological level, both in humans and in laboratory models. In general, there are many similarities in pathomorphological manifestations among the infections caused by these three species of flukes (Maksimova et al., 2017; Lvova et al., 2012; Sripa et al., 2012). Among chronically infected individuals, opisthorchiasis is associated with hepatobiliary problems, including chronic inflammation, periductal fibrosis, biliary intraepithelial neoplasia, cholangitis, and even cholangiocarcinoma (Brazhnikova and Tolkaeva, 2002; Kovshirina et al., 2019; Pakharukova and Mordvinov, 2016). Biliary intraepithelial neoplasia is characterized by abnormal and excessive growth of epithelial tissue, which is considered a precancerous state (Gouveia et al., 2017).
Despite numerous studies at the pathomorphological level, the systemic response and cellular mechanisms of the pathogenesis of these disorders are not well understood. Little is known about how some helminthiases are associated with cancer. What is the mechanism of this biological carcinogenesis?
To conduct in-depth research and to gain insights into the mechanism by which OF infection causes precancerous liver lesions, we (i) applied a next-generation-sequencing–based technology (high-throughput mRNA sequencing; RNA-seq) to identify differentially expressed genes (DEGs) in the liver of golden hamsters infected with OF at 1 and 3 months postinfection (p.i.) as well as (ii) verified the most pronounced changes in gene expression by western blotting and immunohistochemical analysis.
2. Materials and methods
2.1. Animals
Golden hamsters (M. auratus) from the Specific Pathogen-Free (SPF) Animal Facility of the ICG SB RAS were used for this study. For collecting OF metacercariae, a naturally infected freshwater fish (Leuciscus idus) was net-caught in the Ob River near Novosibirsk (Western Siberia, Russia) by research assistant Viktor Antonov (ICG SB RAS) and was processed without the use of chemicals. OF metacercariae were extracted as described previously (Mordvinov et al., 2017; Pakharukova et al., 2019a). After several washes with normal saline, metacercariae were identified under a light microscope.
All the procedures were in compliance with EU Directive 2010/63/EU for animal experiments. Study design protocols and standard operating procedures (concerning the hamsters and the fish) were approved by the Committee on the Ethics of Animal Experiments at the ICG SB RAS (permit number 42 of 25 May 2018).
2.2. Experimental design
Twelve 2-month-old male hamsters (M. auratus) were randomly subdivided into two groups: uninfected and infected orally with 75 metacercariae of OF by gastric intubation. One and 3 months after the infection initiation (p.i.), three hamsters from the uninfected group and three hamsters from the OF-infected group were euthanized using carbon dioxide.
All the hamsters were examined daily for signs of illness, injury, or abnormal behavior by SPF-trained personnel. Food and water availability and the macroenvironment (temperature, humidity, noise, light intensity, and cleanliness) were also evaluated daily. No unexpected deaths of hamsters were registered during this study.
2.3. Tissue collection and RNA extraction
The liver was quickly excised and frozen in liquid nitrogen and then stored at − 80 °C until analysis. RNA was extracted from the frozen tissue with the TRI-Reagent (Sigma, USA). The total RNA was purified on Agencourt RNAClean XP beads (Beckman Coulter, Germany). The quality and quantity of the total RNA were evaluated on a NanoDrop 2000 spectrophotometer. The quality of samples for RNA-seq was assessed by means of an Agilent 2100 Bioanalyzer and the Total RNA Nano Kit (Agilent Technologies, USA). Only samples with an RNA integrity number greater than 8.0 were chosen for gene expression analysis.
2.4. Library construction
RNA-seq libraries of the hamster livers were prepared in accordance with the standard New England Biolabs protocol. Briefly, polyA-tailed mRNAs were purified from 1 μg of total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module. Then, directional cDNA libraries were created by means of the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina. Size selection of DNA fragments was performed on Agencourt AMPure XP beads (Beckman Coulter, USA). Next, PCR enrichment of the adapter-ligated library was conducted (six cycles of PCR). The size and quantity of the library were verified on the Agilent Bioanalyzer, and libraries were subjected to paired-end (2 × 150) sequencing on the Illumina HiSeq 2500 platform (Genewiz LLC, USA).
2.5. Gene expression analysis
On average, ~37 million paired-end reads (range: 32–50 million) were obtained from each sample by Illumina stranded sequencing. The sequencing data were preprocessed with the Trimmomatic 0.36 tool to remove adapters and low-quality sequences (Bolger et al., 2014). The quality of the sequencing data was assessed using FastQC tools (http://broadinstitute.github.io/picard/). The preprocessed data were mapped to the M. auratus GCF_000349665.1_MesAur1.0 reference genome assembly in HISAT2 version 2.1.0 (Kim et al., 2015; Pertea et al., 2016). Assembly, quantification, and merging of expressed genes were performed using Stringtie version 1.3.6 (Pertea et al., 2016). The data were then converted into per-gene count tables by means of StringTie version 1.3.6. Genes with at least 10 counts across all sample were then subjected to an analysis of differential gene expression via the DESeq2 R-package (Love et al., 2014). The Benjamini–Hochberg correction for multiple testing was applied to the resulting p values, and the genes with an adjusted p value (padj) < 0.1 were designated as DEGs. A heat map was constructed using the heatmap.2 (v.2.38) R package. An area-proportional Euler diagram was generated with the help of the eulerr (v.6.1.0) R package.
2.6. Gene clusters
Identification of the gene clusters manifesting specific patterns across samples was carried out using the DEGreport R-package (Pantano, 2021). Rlog-transformed normalized counts were employed to identify only DEGs (padj < 0.05), then the degPatterns function was used to find the sets of genes that show similar expression patterns across sample groups. Groups of genes from clusters 1–4 were subjected to functional analysis to identify the associated functions.
2.7. Enrichment analysis
Gene Ontology (GO) enrichment analysis was conducted using the Web-based Panther tool (Mi et al., 2013). As a reference set, we utilized a gene set from our RNA-seq dataset with counts > 10 (16,768 genes).
GO enrichment analysis was conducted by the Web-based PANTHER Overrepresentation Test (Gene Ontology database DOI: https://doi.org/10.5281/zenodo.4735677 Released 2021-05-01) and Fisher's exact test, and GO terms with a false discovery rate (FDR) < 0.05 were considered significantly enriched.
Analysis of enrichment with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was performed by means of a Web-based annotation tool, Enrichr (KEGG enriched pathways) (Kuleshov et al., 2016). KEGG pathways with padj < 0.05 were considered significantly enriched.
Analysis of enrichment with Molecular Signatures Database (MSigDB) hallmarks (gene lists) was performed by means of a Web-based annotation tool, Enrichr (MsigDB_Hallmark_2020) (Kuleshov et al., 2016). Hallmarks with padj < 0.05 were considered significantly enriched.
2.8. Histopathological analysis
Hamsters were euthanized by carbon dioxide asphyxiation. For an immunohistochemical assay, the livers were immediately fixed in 4% paraformaldehyde in phosphate-buffered saline. The fixed livers were sliced at 7 or 10 μm thickness (n = 4 to 5; 4–6 serial tissue sections per animal) on a Microm HM-505 N cryostat (Pakharukova et al., 2019b; Zaparina et al., 2021).
For histological analysis, the liver was carefully dissected and placed in 10% buffered formalin (Biovitrum, Russia). After fixation for 7 days at 4 °C, the specimens were dehydrated in a graded series of ethanol solutions and then absolute ethanol, cleared in xylene, and soaked in melted paraffin. After that, we embedded the specimens in paraffin using Microm (Microm, UK). Four-μm-thick slices were prepared by means of a microtome. The tissue slices were stained with hematoxylin and eosin by the standard methods and were examined under a light microscope (Axioskop 2 Plus; Zeiss, Germany) (Pakharukova et al., 2019b; Zaparina et al., 2021).
2.9. Immunohistochemistry
Immunohistochemical analysis was performed as described previously (Pakharukova et al., 2019b; Kovner et al., 2019). Antibodies and dilutions used in this study were as follows: anti–alpha-smooth muscle actin (α-SMA; 1:200; cat. # ab7817, Abcam), anti-MMP9 (1:200; cat. # ab228402, Abcam), anti-collagen 1 alpha (1:200; cat. # ab34710; # ab53444, Abcam), anti-E-cadherin (1:200; cat. # ab76055, Abcam), and anti–N-cadherin (1:200; # ab76011, Abcam) antibodies with the respective secondary antibodies: a cyanine 3–conjugated goat anti-mouse IgG (H + L) cross-adsorbed antibody (1:500; cat. # M30010, Invitrogen, USA) or a GFP-conjugated AffiniPure goat anti-rabbit IgG (H + L) antibody (1:1000; cat. # 111–095-003, Jackson AB, USA) and a mouse anti-rabbit IgG-CFL 555 antibody (sc-516,249, Santa Cruz, USA). The slices were coverslipped with the Fluoro-shield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; cat. # F6057, Sigma-Aldrich, USA) and examined under an Axioplan 2 microscope (Zeiss, Germany).
2.10. Western blotting
Immunoblotting was performed as described elsewhere (Pakharukova et al., 2019a). Antibodies and dilutions employed in this study were as follows: anti–beta-actin (1:2000; cat. # ab8226, Abcam), anti–α-smooth muscle actin (α-SMA; 1:2000; cat. # ab7817, Abcam), anti-vimentin (1:1000; cat. # ab8069, Abcam), anti–E-cadherin (1:2000; cat. # ab76055, Abcam), and anti–N-cadherin (1:1000; cat. # ab76011, Abcam) antibodies. Quantitative densitometric analyses were performed on digitized images of immunoblots in the Quantity One software (Bio-Rad, USA).
2.11. Statistical analysis
The data were subjected to statistical analysis in the Statistica 6.0 software (Statsoft, USA). The Shapiro–Wilk test was carried out to check whether the data distribution was normal. The one-way ANOVA F-test with Newman–Keuls post hoc analysis was applied to significant main effects and interactions to assess the differences between some sets of means.
3. Results
3.1. Gene expression changes
Principal component analysis identified two distinct clusters in the data separated along the first principal component (“infection” factor) (Fig. 1A). In the group of infected animals, we also found two clusters separated along the fourth principal component that corresponded samples at 1 and 3 months p.i., respectively (“time” factor in samples from infected animals) (Fig. 1A).
Fig. 1.
Differentially expressed genes (DEGs) in the liver of Opisthorchis felineus–infected hamsters at 1 and 3 months postinfection. A. Principal component analysis identified two distinct clusters in the data separated along the first principal component. B. The most enriched KEGG pathways in the set of DEGs (interaction of factors: “infection × time”). C. A Euler diagram illustrating overlaps of the sets of genes matching the enriched KEGG pathways among the DEGs upregulated (>2-fold) in the OF-infected hamsters Mesocricetus auratus (“infection” factor). ECM: extracellular matrix.
A total of 2151 genes were found to be differentially expressed (padj < 0.1) between uninfected and infected hamsters (1330 upregulated and 822 downregulated as compared to the uninfected group) (“infection” factor). The list of top 10 DEGs found to be significantly downregulated and top 10 DEGs upregulated in the liver of the OF-infected hamsters compared with uninfected ones presented in a Table 1.
Table 1.
List of top 10 DEGs found to be significantly downregulated and top 10 DEGs upregulated in the liver of the OF-infected hamsters compared with uninfected animals.
| Top 10 downregulated genes |
Top 10 upregulated genes |
||||
|---|---|---|---|---|---|
| Gene name | Description | Log2FoldChange | Gene name | Description | Log2FoldChange |
| Ogn | Osteoglycin | −8.44 | Ccl17 | C-C motif chemokine ligand 17 | 21.98 |
| Adam33 | ADAM metallopeptidase domain 33 | −7.94 | Cdhr5 | cadherin related family member 5 | 19.22 |
| Tmem108 | transmembrane protein 108 | −7.89 | Dhrs9 | dehydrogenase/reductase 9 | 10.64 |
| Tnnt3 | troponin T3, fast skeletal type | −6.66 | Fgf19 | fibroblast growth factor 19 | 10.30 |
| Sbk2 | SH3 domain binding kinase family member 2 | −6.61 | Cldn18 | claudin 18 | 9.44 |
| Chgb | chromogranin B | −6.18 | Adgrf4 | adhesion G protein-coupled receptor F4 | 9.34 |
| Slc34a1 | solute carrier family 34 member 1 | −6.08 | Cxcl11 | C-X-C motif chemokine ligand 11 | 8.82 |
| Rgs8 | regulator of G protein signaling 8 | −5.65 | Lexm | lymphocyte expansion molecule | 8.81 |
| Ly6g6c | lymphocyte antigen 6 family member G6C | −4.96 | Cdhr3 | cadherin related family member 3 | 8.78 |
| LOC101822511 | alcohol dehydrogenase 6 | −4.83 | Rbp2 | retinol binding protein 2 | 8.75 |
Seventy-one genes proved to be differentially expressed between groups 1 month and 3 months p.i. (“time” factor). Forty-five genes were upregulated, and 26 genes were downregulated. No significantly enriched terms were found in KEGG and GO (FDR < 0.05) enrichment analysis.
When we analyzed the interaction of the two factors (time × infection), 371 genes (padj < 0.1) turned out to be differentially expressed (252 genes: more than twofold). Among them, 186 genes were upregulated and 66 were downregulated.
Functional enrichment analysis of the genes upregulated in the liver of OF-infected animals was conducted on the Enrichr website and via the Web-based Panther tool (Fig. 1B and C). We found 15 significantly enriched (padj < 0.05) KEGG pathways in the set of genes upregulated in OF-infected animals (Fig. 1C; Supplementary Table ST6), 31 significantly over-represented (FDR < 0.05) GO biological processes (Table 2, Supplementary Table ST5), and 13 significantly enriched (q-value < 0.05) gene sets (hallmarks) according to the Molecular Signatures Database (MSigDB) (Table 2, Supplementary Table ST7).
Table 2.
Gene sets (Molecular Signatures Database hallmarks) enriched and GO terms over-represented in the DEG set from the liver of the OF-infected hamsters.
| MsigDB Hallmark enrichment analysis |
GO Overrepresentation Test |
|||||
|---|---|---|---|---|---|---|
| Expression | Hallmark | p-value | q-value | GO | Fold enrichment | FDR |
| Up-regulated | Epithelial Mesenchymal Transition | 1.22E-08 | 5.99E-07 | neutrophil degranulation (GO:0043312) | > 100 | 1.48E-37 |
| Apoptosis | 8.03E-07 | 1.31E-05 | neutrophil activation involved in immune response (GO:0002283) | 59.54 | 1.38E-34 | |
| IL-6/JAK/STAT3 Signaling | 8.05E-07 | 1.31E-05 | Fc receptor mediated stimulatory signaling pathway (GO:0002431) | 43 | 1.19E-10 | |
| TNF-alpha Signaling via NF-kB | 3.27E-06 | 4.00E-05 | Fc-epsilon receptor signaling pathway (GO:0038095) | 39.42 | 1.48E-09 | |
| Inflammatory Response | 2.38E-05 | 2.34E-04 | stimulatory C-type lectin receptor signaling pathway (GO:0002223) | 38.7 | 8.98E-08 | |
| G2-M Checkpoint | 6.09E-05 | 4.97E-04 | ERBB2 signaling pathway (GO:0038128) | 35.83 | 3.76E-04 | |
| IL-2/STAT5 Signaling | 1.37E-04 | 9.61E-04 | keratan sulfate metabolic process (GO:0042339) | 35.83 | 3.75E-04 | |
| Down-regulated | Xenobiotic metabolism | 2E-04 | 9E-03 | protein activation cascade (GO:0072376) | 12.14 | 1.83E-02 |
| triglyceride metabolic process (GO:0006641) | 6.2 | 1.05E-02 | ||||
| ATP synthesis coupled electron transport (GO:0042773) | 5.44 | 3.53E-02 | ||||
| neutral lipid metabolic process (GO:0006638) | 5.37 | 6.20E-03 | ||||
| acylglycerol metabolic process (GO:0006639) | 5.02 | 1.79E-02 | ||||
| cholesterol metabolic process (GO:0008203) | 4.94 | 1.07E-02 | ||||
GO: Gene Ontology. We performed Fisher's exact test, false discovery rate calculation (Panther), and PANTHER Overrepresentation Test (Released 20,210,224). Gene Ontology database DOI: https://doi.org/10.5281/zenodo.4735677 released 2021-05-01.
The most enriched MSigDB hallmarks were Epithelial Mesenchymal Transition (EMT), Apoptosis, IL6/JAK/STAT3 Signaling, TNF-alpha Signaling, and Inflammatory Response (Table 2). The most enriched KEGG-pathways included Cell adhesion molecules, Hippo signaling, ECM-receptor interaction, Cell cycle, TGF-beta signaling, and P53 signaling. A more detailed analysis of the genes of these pathways indicated that the official gene lists of major pathways overlapped substantially. In particular, Fig. 1C presents an Euler diagram of upregulated DEGs (log2 fold change >1; padj < 0.05) in overlaps among major KEGG pathways.
The functional enrichment analysis of the set of genes downregulated in the liver OF-infected animals showed a smaller number of significantly enriched KEGG terms. Among the downregulated genes (Supplementary Table ST8), we found only one significantly enriched (FDR < 0.05) KEGG pathway (Histidine metabolism), 27 significantly over-represented (padj < 0.1) GO biological processes (Table 2, Supplementary Table ST4), and one enriched MSigDB hallmark (Xenobiotic metabolism) (q-value <0.05). Among the most enriched GO biological processes, there were many pathways associated with the metabolism of fatty acids, cholesterol, neutral lipids, mitochondrial electron transport, fatty-acyl-CoA metabolic processes, and cholesterol biosynthesis processes (Table 2, Supplementary Table ST4).
The most interesting finding after functional annotation of 371 DEGs (interaction of factors: “time × infection”) (Fig. 1B; Supplementary Table ST3) was that only three KEGG pathways were significantly enriched (padj < 0.05): Protein digestion and absorption (hsa04974), TGF-beta signaling pathway (hsa04350), and AMPK signaling pathway (hsa04152). The TGF-β (TGFB) signaling pathway is involved in many cellular processes and is the primary factor that drives fibrosis. This pathway also plays critical roles in EMT, which was the most enriched term in all OF-infected animals (Table 2). We found 51 significantly over-represented (FDR < 0.05) GO biological processes (Supplementary Table ST10).
3.2. Gene clusters
We were interested in in-depth analysis of the DEGs (interaction of factors: “time × infection”) to identify gene clusters that have specific patterns across the groups of liver samples. To identify these genes, we used a clustering tool, degPatterns, which groups genes based on their changes in expression across sample groups. Thus, the genes were subdivided into four clusters (gene sets), which are shown in Fig. 2 (Supplementary Table ST11). In particular, cluster 1 (DEGs downregulated at 1 month p.i., 77 genes), cluster 2 (DEGs upregulated at 1 month p.i., 87 genes); cluster 3 (DEGs upregulated only at 3 months p.i., 148 genes); and cluster 4 (DEGs downregulated only at 3 months p.i., 51 genes).
Fig. 2.
Clustering analysis of 376 DEGs (padj < 0.1; interaction of factors: “infection × time”) found in the liver of hamsters (Mesocricetus auratus) infected with Opisthorchis felineus (at 1 and 3 months p.i.).
GO terms that were found to be over-represented (FDR < 0.05) in cluster 2 (upregulated at 1 month p.i.) are related to apoptotic signaling, negative regulation of transcription factors, response to endogenous stimulus, and other terms. The most enriched KEGG pathways in this cluster included TGF-beta signaling and Hippo signaling. More detailed examination revealed that this cluster included such genes as Id1, Id2, Id3, Inhbb, Neo1, Smad7, Fzd1, Ccnd1, and Tnfrsf1a (Fig. 2).
In contrast to cluster 2, GO terms that were found to be over-represented (FDR < 0.05) in cluster 3 (upregulated at 3 months p.i.) were biological adhesion, growth, immune system process, and metabolic process, among others. It was this module that contained many genes (148 genes) whose products are associated with protein digestion and absorption, AMPK signaling pathway, extracellular matrix (ECM)-receptor interaction, and fructose and mannose metabolism. Closer inspection revealed that this cluster included six collagen-coding genes: Eln, Lamc1, Lama3, Pfkfb2, and Frem1. All collagen genes proved to be over-represented only at 3 months p.i.
In the GO over-representation test, we did not find any terms over-represented in cluster 1 (downregulated at 1 month p.i.) and cluster 4 (downregulated at 3 months p.i.).
In total, RNA-seq analysis revealed that the majority of DEGs were associated with the “infection” factor. At the molecular signaling level, the pathways most significantly enriched among upregulated DEGs were EMT, Cell Adhesion, Hippo signaling, ECM receptor interaction, Cell cycle, TGF-beta signaling, and P53 signaling. Furthermore, the sets of activated genes of cellular pathways were different between acute (1 month p.i.) and chronic (3 months p.i.) opisthorchiasis.
3.3. Epithelial Mesenchymal transition
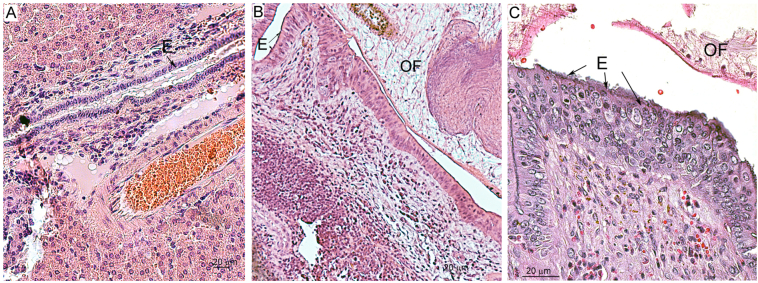
Fig. 3 presents a typical picture of epithelial neoplasia, which can be observed after 3 months of opisthorchiasis in hamsters. A transition from the normal duct epithelium to an atypical epithelium was noticeable. The epithelium became multirow; cell contacts, cellular phenotype, and the presence of nucleoli were affected; and there was a loss of polarity. Cholangiocyte hyperplasia (multirow bile duct epithelium consisting of cholangiocytes that appear normal) was noted too.
Fig. 3.
Histopathological analysis of a hamster liver at 3 months postinfection (p.i.) with Opisthorchis felineus metacercariae. A. The liver of an uninfected animal; B and C: biliary intraepithelial neoplasia at 3 months p.i. Biliary neoplasia was detectable, including the presence of nucleoli and loss of polarity with a transition from the normal duct epithelium to an atypical epithelium. E: epithelial cells; OF: O. felineus.
EMT was the most enriched (q-value = 2.2E-07) MSigDB hallmark in the DEG set of all OF-infected animals. The activation of processes from this pathway was evidenced by upregulation of the TGF-β pathway, which was the most enriched term in all OF-infected animals, owing to its critical role in EMT. In addition, it was important to confirm the RNA-seq data at the protein level. Therefore, we decided to take a deeper look at these EMT processes by immunohistochemical (Fig. 4A) and western blot analyses (Fig. 4C and D) by examining not only proteins whose genes were differentially expressed during the infection but also the entire gene list that is considered a signature of EMT (Chen et al., 2020). This list includes 15 markers of both mesenchymal (Fn1, Cdh2, Twist1, Zeb1, Zeb2, Mmp2, Vim, Foxc2, Snai2, Snai1, and Mmp9) and epithelial tissues (Tjp1, Ocln, Dsp, and Cdh1) (Chen et al., 2020). The mRNA abundance of these genes is presented as a heat map (Fig. 4B). Samples from uninfected and infected animals clustered well into two groups. Fig. 4B shows that both mesenchymal and epithelial genes were upregulated after the infection.
Fig. 4.
Expression of epithelial–mesenchymal transition (EMT) genes in the liver of hamsters (Mesocricetus auratus) at 1 and 3 months postinfection (p.i.) with Opisthorchis felineus (OF). A. Immunohistochemical analysis of expression levels of MMP9, E-cadherin, N-cadherin, type 1 collagen, and α-smooth muscle actin in an OF-infected M. auratus liver at 1 and 3 months p.i. BD: bile duct, PF: periductal fibrosis, BV: blood vessel, Inf: inflammatory infiltrate, and E: epithelial cells. B. A heat map illustrating z-scored mRNA abundance of 16 EMT signature genes was constructed using the heatmap.2 (v.2.38) R-package. Z-scoring normalization was conducted using DESeq2 R-package). C. E-cadherin, N-cadherin, vimentin, and α-smooth muscle actin protein levels assessed by western blotting. α-SMA: α-smooth muscle actin. D. Protein levels were determined by densitometry of immunoblots (E), and the results were normalized to β-actin. The data are presented as mean ± SD; *p < 0.05. SD was calculated from the results of three independent experiments.
Using immunohistochemistry, we evaluated how the expression of some of these markers was distributed in the liver. For instance, we performed MMP9 staining of randomly chosen cells in the area around the bile ducts and periductal fibrosis. Although the number of stained cells was significantly higher in the infected than in uninfected animals, there was no significant difference between 1 and 3 months p.i.
Vimentin turned out to be significantly overexpressed in the infected animals and remained upregulated (P < 0.05) at 1 and 3 months p.i. It was shown by immunohistochemistry that α-smooth muscle actin (α-SMA) expression was elevated at 1 month p.i. and was even higher at 3 months p.i. Moreover, although in the uninfected animals and at 1 month p.i., the signal was mainly present in the walls of blood vessels in the liver, at 3 months p.i., diffuse presence of this protein in the liver was revealed too. According to western blotting, however, the amount of this protein significantly increased only at 3 months p.i. In the liver, the location of expression of another mesenchymal marker, N-cadherin, was more difficult to determine after the infection, although its expression according to western blotting was significantly higher at 1 and 3 months p.i. For example, although N-cadherin was present in the uninfected animals mainly in the walls of blood vessels (red signal, upper panel in Fig. 3A), after the infection, the signal was also detectable in some epithelial cells of the bile ducts (3 months p.i., upper panel).
After the infection, the expression of epithelial marker E-cadherin, which stained green in the epithelial cells of the bile ducts, reflected enhanced proliferation of bile ducts and their multiplication in the liver (Fig. 3A). Nevertheless, at the same time, according to western blot analysis, a significant increase in this protein's amount in the liver was not detectable.
4. Discussion
We for the first time determined the DEGs and cellular pathways in acute and chronic opisthorchiasis caused by OF. We demonstrated that EMT is the most enriched term among MSigDB hallmarks in the infected animals. The TGF-β pathway was also one of the most enriched terms in the set of DEGs associated with this infection. The activation of EMT pathways, as a general systemic response, is in good agreement with the data that we obtained earlier in the analysis of chronic opisthorchiasis in humans (Kovner et al., 2019).
The large number of DEGs can probably be explained by the fact that we isolated RNA from a big piece of the liver. The liver contains many types of cells, including cholangiocytes, hepatocytes, fibroblasts, cells of inflammatory infiltration, immune cells, and endothelial cells of blood vessels. This seems to explain the variation between liver RNA samples in the analysis of mRNA abundance of EMT signature genes (Figs. 4B and 5). According to the mRNA abundance and protein expression of EMT signature genes, there were no specific features of EMT in the liver. Probably, if we had included in the analysis only the areas containing the epithelium of the bile ducts, then EMT would have been detectable more clearly.
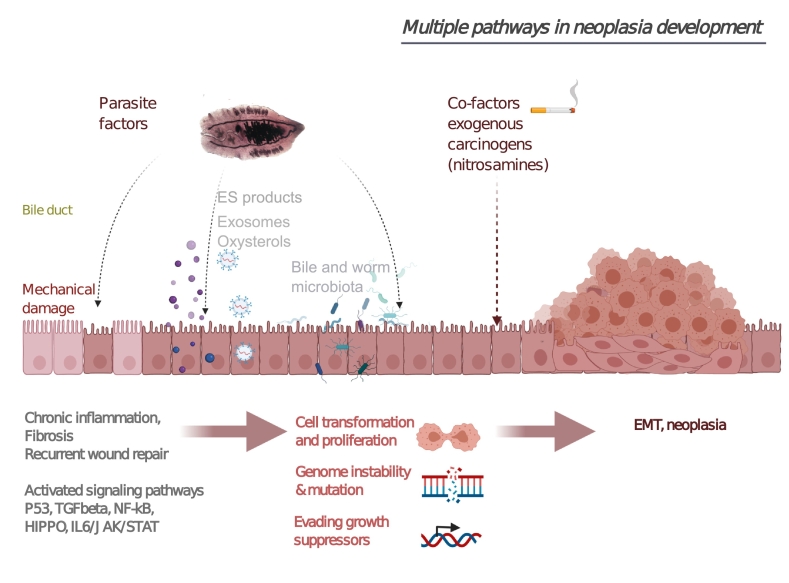
Fig. 5.
Multiple pathways implicated in neoplasia formation. Chronic inflammation during Opisthorchis felineus infection leads to the activation of signaling cascades including p53, NF-κB, JAK/STAT, and TGF-β pathways. Fluke-derived substances and metabolites secreted into the host microenvironment (oxysterols and excretory-secretory [ES] products) may induce metabolic processes (including oxidative stress) that facilitate damage to the chromosomal DNA of cholangiocytes. In addition, physical damage to host tissues by parasites together with the wound healing process cause recurrent wound healing and cell proliferation. Such cofactors as dietary nitrosamines or changes in the microbiome may promote the pathology. Combined events in the parasite–host interaction (chronic inflammation, parasite-derived substances, and physical damage) and their effects on host chromosomes alter cell growth, proliferation, and survival. Created with BioRender.com.
Pathways associated with inflammatory processes such as the terms “AMPK signaling pathway,” “HIPPO-signaling,” “Cell-cycle,” “NF-κB,” “JAK/STAT,” and “Cell adhesion molecules” were also over-represented in the experimental opisthorchiasis. The inflammatory response at the level of differential gene expression has been confirmed by pathomorphological analyses both in laboratory animals and in samples from patients (Kovner et al., 2019; Lvova et al., 2012; Pakharukova et al., 2019a, Pakharukova et al., 2019b). In addition, comparing chronic and acute inflammation in the liver during various infectious and noninfectious diseases (e.g., hepatitis B virus infection, nonalcoholic fatty liver diseases, and disorders caused by administration of CCl4 or tunicamycin), one can see similar upregulated cellular pathways associated with inflammation (Campos et al., 2020). For example, MAPK and TNF pathways are reported to be upregulated, followed by the genes corresponding to such GO terms as “Cell cycle,” “ECM organization,” and “Immune response” and the KEGG term “DNA replication” (Campos et al., 2020).
Moreover, here, metabolic genes were significantly downregulated at 3 months p.i., indicating a general impairment of liver metabolism. These data overall are well consistent with our previously published results on hepatocyte changes in the form of fatty degeneration and excessive accumulation of lipids in the liver (Zaparina et al., 2021). The excessive accumulation of neutral lipids in the liver may be related to pathological processes, including those taking place in response to toxic compounds (Campos et al., 2020; Chaidee et al., 2018). Overall, our data are in line with reports that Opisthorchis viverrini infection causes fatty liver and liver cirrhosis in gerbils (Wonkchalee et al., 2011). Our findings are also consistent with the data on common features of all acute and chronic liver conditions (Campos et al., 2020) that are characterized by pronounced downregulation of metabolic genes.
Despite the fact that it is currently impossible to compare liver transcriptomes in the same model during infection with different liver flukes due to the lack of available mRNAseq data, nevertheless, we compared our findings with some published research. Thus, TLR2 and TLR4 were reported to be upregulated in a mouse model of clonorchiasis [Yan et al., 2015]. We also found the significant upregulation of Tlr2 and Tlr4 genes in the liver of the OF-infected animals. A cell cycle regulatory cyclin-dependent kinase CDK4, as well as CDK4 inhibitor p16INK4 showed upregulation the relative expression of mRNA by real-time PCR in the C. sinensis-infected hamsters [Uddin et al., 2015]. In contrast to this, we found other genes of cyclin-dependent kinases and cyclin-dependent kinase inhibitors to be significantly elevated during OF-infection (Cdk1, Cdkn2b, Cdkn2a, Cdkn1a genes).
Expression of some of EMT signature genes, such as collagens I and III, matrix metalloproteinase (MMP)-2, MMP-13, MMP-14, and tissue inhibition of matrix metalloproteinase (TIMP)-1 genes were increased in O. viverrini-infected hamster liver at 1 and 2 months p.i. [Prakobwong et al., 2009]. Moreover, collagen I, MMP9, MMP2, α-SMA, TIMP1, TIMP2, and urokinase plasminogen activator gene as well as cytokines TNF-α and TGF-β were found to be significantly elevated in hamster CCA model [Prakobwong et al., 2010], suggesting the involvement of EMT processes in CCA development. Furthermore, O. viverrini infection activates PI3K–AKT–PTEN and WNT–β-catenin signaling pathways in hamster CCA model (Yothaisong et al., 2014).
At the genomic level, analysis of the mutation profiles of O. viverrini-related cholangiocarcinomas and non—O. viverrini-related human samples of cholangiocarcinomas revealed significant differences in mutation patterns (Chan-On et al., 2013). In particular, somatic mutations in O. viverrini-related CCA samples occured more frequently in TP53, KRAS, SMAD4, BRAF, MLL3, ARID1A, PBRM1 and BAP1 genes, which are known to be involved in cell cycle control, cell signaling pathways and chromatin dynamics. Mutations in the tumor suppressor genes p53 and SMAD4 directly affect the related cellular signaling pathways p53 and TGF- β (Jusakul et al., 2015).
Opisthorchiidae species have strong proinflammatory properties (Maeng et al., 2016; Pak et al., 2016; Pakharukova et al., 2019a, Pakharukova et al., 2019b). During chronic inflammation, inflammatory cells are recruited to injury sites, thereby increasing a release and accumulation of free radicals, resulting in the formation of lipid peroxidation byproducts (Pakharukova et al., 2019a; Zaparina et al., 2021) as well as oxidative DNA lesions like 8-OHdG (Maeng et al., 2016; Pakharukova et al., 2019a). It was previously shown that TNF-α and CD163 protein levels increase in a strongly time-dependent manner in OF-infected golden hamsters (Pakharukova et al., 2019a). During chronic liver fluke infection, persistent activation of CD163+ macrophages (Thanee et al., 2015) presumably causes continuous production of TGF-β and growth factors that promote proliferation of myofibroblasts, activation of EMT, extracellular-matrix deposition, and fibrosis development. During the experimental opisthorchiasis, elevated α-SMA expression and excessive amounts of the extracellular matrix were revealed in our study, which manifested themselves as fibrogenesis and collagen deposition as determined by histopathological examination.
Several mechanisms may account for the involvement of OF infection in neoplasia and cancer (Fig. 5). Proposed mechanisms behind the carcinogenicity of liver fluke infection include biliary-epithelium damage by parasites, long-lasting immune-system–mediated pathogenesis, and effects of parasite-derived substances that are internalized by the bile duct epithelium (Pakharukova et al., 2019b; Petrenko et al., 2017; Smout et al., 2015).
Together with the liver fluke infection, such cofactors as environmental or exotic microbes in the biliary system that are resistant to host inflammatory responses may contribute to carcinogenesis (Chng et al., 2016; Pakharukova et al., 2021; Sripa et al., 2012). In particular, it was demonstrated that Helicobacter pylori infection enhances the severity of hepatobiliary abnormalities in hamsters during O. viverrini infection (Dangtakot et al., 2017).
Other co-factors as dietary nitrosamines could strongly facilitate the carcinogenicity (Maksimova et al., 2017; Mordvinov et al., 2021; Sripa et al., 2012). In such studies, hamsters infected with OF and receiving subcarcinogenic doses of dimethylnitrosamine develop cholangiocarcinoma, whereas those subjected to OF infection alone do not.
Additionally, evidence indicates that helminths produce metabolites that might interact with host DNA thereby triggering the carcinogenic process associated with opisthorchiasis. In particular, investigation of adult-worm lysates (O. viverrini and OF) has identified novel oxysterol derivatives that are potential promutagenic compounds (Gouveia et al., 2017, Gouveia et al., 2020; Vale et al., 2013).
The mechanisms of biological carcinogenesis need to be investigated further. Is chronic inflammation alone a necessary and sufficient condition for the formation of cholangiocarcinoma? If so, then three epidemiologically significant species of liver flukes O. viverrini, C. sinensis, and OF would cause carcinogenesis in the same way. Although at present, there are no comparative bias-free studies on their carcinogenicity under identical conditions, it can already be said (judging from indirect evidence) that these fluke species have pronounced differences in human carcinogenicity. For instance, when ~60% of a rural population in OF endemic regions are infected, liver cancer ranks only 18th among all cancers (Petrova et al., 2019). In contrast, liver cancer in Korea ranked 5th among cancers and 2nd in terms of mortality in 2016 (Jung et al., 2019).
The development of studies on comparative analysis, in particular, high-throughput mRNA sequencing, in order to identify differences in host response and differentially expressed genes during the infection caused by closely related species having moderate and high carcinogenicity, will significantly elucidate the mechanisms underlying helminth-associated carcinogenesis and shed light on the pathogenesis of biological carcinogenesis.
Authors' contributions
O.Z. and N.V.B. conducted the histological and microscopic experiments; M.Y.P. performed the RNA extraction, library preparation, and bioinformatic analyses. M.Y.P. and V.A.M. designed the study protocol and supervised the study. M.Y.P. and V.A.M. analyzed the data and wrote the manuscript. All the authors participated in the drafting of the manuscript, contributed to its revisions, and approved the final version.
Availability of data and materials
The sequence data were deposited in NCBI BioProject under accession number PRJNA789062.
Funding
This work was supported financially by the Russian Science Foundation (grant number # 18-15-00098 to V.A.M.). Imaging was performed on the equipment maintained by the Center for Microscopy, the ICG SB RAS (FWNR-2022-0021). The funding agencies had no role in this study, e.g., in study design, data collection, or decision to publish the results.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are thankful to Nikolai Shevchuk (http://shevchuk-editing.com) for the English-language editing of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fawpar.2022.e00159.
Contributor Information
Maria Y. Pakharukova, Email: pmaria@yandex.ru.
Nina V. Baginskaya, Email: bagin@bionet.nsc.ru.
Viatcheslav A. Mordvinov, Email: mordvin@bionet.nsc.ru.
Appendix A. Supplementary data
Supplementary material 1: Results of Agilent analysis of total RNA extracted from the liver samples
Supplementary material 2: Results of High Sensitivity DNA Assay (Agilent 2100) of DNA libraries constructed
Supplementary material 3: Principal component analysis scree plot (Percentage of Variance for each by PC)
Supplementary Tables
References
- Beer S.A. KMK Scientific Press Ltd.; Moscow: 2005. Biology of the Agent of Opisthorchiasis. (in Russian) (in Russian) [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazhnikova N.A., Tolkaeva M.V. Cancer of liver, biliary tracts and pancreas at chronic opisthorchiasis (in Russian) Bull. Sib. Med. 2002;2:71–77. [Google Scholar]
- Campos G., Schmidt-Heck W., De Smedt J., Widera A., Ghallab A., Pütter L., et al. Inflammation-associated suppression of metabolic gene networks in acute and chronic liver disease. Arch. Toxicol. 2020;94:205–217. doi: 10.1007/s00204-019-02630-3. [DOI] [PubMed] [Google Scholar]
- Chaidee A., Onsurathum S., Intuyod K., Pannangpetch P., Pongchaiyakul C., Pinlaor P., et al. Co-occurrence of opisthorchiasis and diabetes exacerbates morbidity of the hepatobiliary tract disease. PLoS Negl. Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-On W., Nairismägi M.L., Ong C.K., Lim W.K., Dima S., Pairojkul C., Lim K.H., McPherson J.R., Cutcutache I., Heng H.L., Ooi L., Chung A., Chow P., Cheow P.C., Lee S.Y., Choo S.P., Tan I.B., Duda D., Nastase A., Myint S.S., Teh B.T. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat. Genet. 2013;45:1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- Chen T., Zhao Z., Chen B., Wang Y., Yang F., Wang C., et al. An individualized transcriptional signature to predict the epithelial-mesenchymal transition based on relative expression ordering. Aging. 2020;12:13172–13186. doi: 10.18632/aging.103407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng K.R., Chan S.H., Ng A.H., Li C., Jusakul A., Bertrand D., et al. Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in Opisthorchis viverrini associated cholangiocarcinoma. EBioMedicine. 2016;8:195–202. doi: 10.1016/j.ebiom.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangtakot R., Pinlaor S., Itthitaetrakool U., Chaidee A., Chomvarin C., Sangka A., et al. Coinfection with helicobacter pylori and Opisthorchis viverrini enhances the severity of hepatobiliary abnormalities in hamsters. Infect. Immun. 2017;85:e00009–e00017. doi: 10.1128/IAI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O.S., Kovshirina Y.V., Kovshirina A.E., Fedotova M.M., Deev I.A., Petrovsky F.I., et al. Analysis of the incidence of Opisthorchis felineus infection and malignant neoplasms of the hepatobiliary system in the Russian Federation. Bul. Siberian Med. 2016;15:147–158. doi: 10.20538/1682-0363-2016-5-147-158. [DOI] [Google Scholar]
- Fedorova O.S., Fedotova M.M., Zvonareva O.I., Mazeina S.V., Kovshirina Y.V., Sokolova T.S., et al. Opisthorchis felineus infection, risks, and morbidity in rural Western Siberia. Russian Feder. PLoS Negl. Trop. Dis. 2020;14(6) doi: 10.1371/journal.pntd.0008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia M.J., Pakharukova M.Y., Laha T., Sripa B., Maksimova G.A., Rinaldi G., et al. Infection with Opisthorchis felineus induces intraepithelial neoplasia of the biliary tract in a rodent model. Carcinogenesis. 2017;38:929–937. doi: 10.1093/carcin/bgx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia M.J., Pakharukova M.Y., Rinaldi G., Mordvinov V.A., Brindley P.J., Gärtner F., et al. Helminth infection-induced carcinogenesis: spectrometric insights from the liver flukes, Opisthorchis and Fasciola. Experimental Results. 2020;1:E40. doi: 10.1017/exp.2020.38. [DOI] [Google Scholar]
- IARC IARC working group on the evaluation of carcinogenic risks to humans. Biological agents. A review of human carcinogens. IARC Monogr. Eval. Carcinog. Risks Hum. 2012;100:1–441. [PMC free article] [PubMed] [Google Scholar]
- Jung K.W., Won Y.J., Kong H.J., Lee E.S. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res. Treat. 2019;51:417–430. doi: 10.4143/crt.2019.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusakul A., Kongpetch S., Teh B.T. Genetics of Opisthorchis viverrini-related cholangiocarcinoma. Curr. Opin. Gastroenterol. 2015;31:258–263. doi: 10.1097/MOG.0000000000000162. [DOI] [PubMed] [Google Scholar]
- Kim D., Landmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–U121. doi: 10.1038/Nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovner A.V., Pakharukova M.Y., Maksimova G.A., Mordvinov V.A. Characteristics of liver fibrosis associated with chronic Opisthorchis felineus infection in Syrian hamsters and humans. Exp. Mol. Pathol. 2019;104274 doi: 10.1016/j.yexmp.2019.104274. [DOI] [PubMed] [Google Scholar]
- Kovshirina Y.V., Fedorova O.S., Vtorushin S.V., Kovshirina A.E., Ivanov S.D., Chizhikov A.V., et al. Case report: two cases of cholangiocarcinoma in patients with Opisthorchis felineus infection in Western Siberia, Russian Federation. Am. J. Trop. Med. Hyg. 2019;100:599–603. doi: 10.4269/ajtmh.18-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1. doi: 10.1186/S13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lvova M.N., Tangkawattana S., Balthaisong S., Katokhin A.V., Mordvinov V.A., Sripa B. Comparative histopathology of Opisthorchis felineus and Opisthorchis viverrini in a hamster model: an implication of high pathogenicity of the European liver fluke. Parasitol. Int. 2012;61:167–172. doi: 10.1016/j.parint.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Maeng S., Lee H.W., Bashir Q., Kim T.I., Hong S.J., Lee T.J., et al. Oxidative stress-mediated mouse liver lesions caused by Clonorchis sinensis infection. Int. J. Parasitol. 2016;46(195–204):March. doi: 10.1016/j.ijpara.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Maksimova G.A., Pakharukova M.Y., Kashina E.V., Zhukova N.A., Kovner A.V., Lvova M.N., et al. Effect of Opisthorchis felineus infection and dimethylnitrosamine administration on the induction of cholangiocarcinoma in Syrian hamsters. Parasitol. Int. 2017;66:458–463. doi: 10.1016/j.parint.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. (Epub 2013 Jul 18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordvinov V.A., Ershov N.I., Pirozhkova D.S., Pakharukov Y.V., Pakharukova M.Y. ABC transporters in the liver fluke Opisthorchis felineus. Mol. Biochem. Parasitol. 2017;216:60–68. doi: 10.1016/j.molbiopara.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Mordvinov V.A., Minkova G.A., Kovner A.V., Ponomarev D.V., Lvova M.N., Zaparina O., et al. A tumorigenic cell line derived from a hamster cholangiocarcinoma associated with Opisthorchis felineus liver fluke infection. Life Sci. 2021;119494 doi: 10.1016/j.lfs.2021.119494. [DOI] [PubMed] [Google Scholar]
- Pak J.H., Son W.C., Seo S.B., Hong S.J., Sohn W.M., Na B.K., Kim T.S. Peroxiredoxin 6 expression is inversely correlated with nuclear factor-κB activation during Clonorchis sinensis infestation. Free Radic. Biol. Med. 2016;99:273–285. doi: 10.1016/j.freeradbiomed.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Pakharukova M.Y., Mordvinov V.A. The liver fluke Opisthorchis felineus: biology, epidemiology, and carcinogenic potential. Trans. R. Soc. Trop. Med. Hyg. 2016;110:28–36. doi: 10.1093/trstmh/trv085. [DOI] [PubMed] [Google Scholar]
- Pakharukova M.Y., Zaparina O.G., Kapushchak Y.K., Baginskaya N.V., Mordvinov V.A. Opisthorchis felineus infection provokes time-dependent accumulation of oxidative hepatobiliary lesions in the injured hamster liver. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0216757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakharukova M.Y., Zaparina O.G., Kovner A.V., Mordvinov V.A. Inhibition of Opisthorchis felineus glutathione-dependent prostaglandin synthase by resveratrol correlates with attenuation of cholangiocyte neoplasia in a hamster model of opisthorchiasis. Int. J. Parasitol. 2019;49(12):963–973. doi: 10.1016/j.ijpara.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Pakharukova M.Y., Zaparina O., Hong S.J., Sripa B., Mordvinov V.A. A comparative study of helicobacter pylori infection in hamsters experimentally infected with liver flukes Opisthorchis felineus, Opisthorchis viverrini, or Clonorchis sinensis. Sci. Rep. 2021;11(1):7789. doi: 10.1038/s41598-021-87446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano L. DEGreport: report of DEG analysis. R package version 1.30.0. 2021. http://lpantano.github.io/DEGreport/
- Pertea M., Kim D., Pertea G., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko V.A., Pakharukova M.Y., Kovner A.V., L’vova M.N., Lyakhovich V.V., Mordvinov V.A. Secretion of thioredoxin peroxidase protein of cat liver fluke Opisthorchis felineus during modeling of experimental opisthorchiasis. Bull. Exp. Biol. Med. 2017;162(6):773–776. doi: 10.1007/s10517-017-3710-5. [DOI] [PubMed] [Google Scholar]
- Petrova G.V., Starinskiy V.V., Gretsova O.P., Shakhzadova A.O., Samsonov Y.V. The state of cancer care to the population of Russia in 2017 according to the federal statistical observation. Oncology. Journal P.A. Herzena. 2019;1:32–40. [Google Scholar]
- Prakobwong S., Pinlaor S., Yongvanit P., Sithithaworn P., Pairojkul C., Hiraku Y. Time profiles of the expression of metalloproteinases, tissue inhibitors of metalloproteases, cytokines and collagens in hamsters infected with Opisthorchis viverrini with special reference to peribiliary fibrosis and liver injury. Int. J. Parasitol. 2009;39:825–835. doi: 10.1016/j.ijpara.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Prakobwong S., Yongvanit P., Hiraku Y., Pairojkul C., Sithithaworn P., Pinlaor P., Pinlaor S. Involvement of MMP-9 in peribiliary fibrosis and cholangiocarcinogenesis via Rac1-dependent DNA damage in a hamster model. Int. J. Cancer. 2010;127:2576–2587. doi: 10.1002/ijc.25266. [DOI] [PubMed] [Google Scholar]
- Smout M.J., Sotillo J., Laha T., Papatpremsiri A., Rinaldi G., Pimenta R.N., et al. Carcinogenic parasite secretes growth factor that accelerates wound healing and potentially promotes neoplasia. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripa B., Brindley P.J., Mulvenna J., Laha T., Smout M.J., Mairiang E., et al. The tumorigenic liver fluke Opisthorchis viverrini–multiple pathways to cancer. Trends Parasitol. 2012;28:395–407. doi: 10.1016/j.pt.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanee M., Loilome W., Techasen A., Namwat N., Boonmars T., Pairojkul C., et al. Quantitative changes in tumor-associated M2 macrophages characterize cholangiocarcinoma and their association with metastasis. Asian Pac. J. Cancer Prev. 2015;16:3043–3050. doi: 10.7314/apjcp.2015.16.7.3043. [DOI] [PubMed] [Google Scholar]
- Uddin M.H., Choi M.H., Kim W.H., Jang J.J., Hong S.T. Involvement of PSMD10, CDK4, and tumor suppressors in development of intrahepatic cholangiocarcinoma of Syrian golden hamsters induced by Clonorchis sinensis and N-nitrosodimethylamine. PLoS NTDs. 2015;9 doi: 10.1371/journal.pntd.0004008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale N., Gouveia M.J., Botelho M., Sripa B., Suttiprapa S., Rinaldi G., et al. Carcinogenic liver fluke Opisthorchis viverrini oxysterols detected by LC-MS/MS survey of soluble fraction parasite extract. Parasitol. Int. 2013;62:535–542. doi: 10.1016/j.parint.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonkchalee O., Boonmars T., Kaewkes S., Chamgramol Y., Pairojkul C., Wu Z., et al. Opisthorchis viverrini infection causes liver and biliary cirrhosis in gerbils. Parasitol. Res. 2011;109:545–551. doi: 10.1007/s00436-011-2282-y. [DOI] [PubMed] [Google Scholar]
- Yan C., Li X.Y., Li B., Zhang B.B., Xu J.T., Hua H., Yu Q., Liu Z.Z., Fu L.L., Tang R.X., Zheng K.Y. Expression of toll-like receptor (TLR) 2 and TLR4 in the livers of mice infected by Clonorchis sinensis. J. Infect. Dev. Ctries. 2015;9:1147–1155. doi: 10.3855/jidc.6698. [DOI] [PubMed] [Google Scholar]
- Yothaisong S., Thanee M., Namwat N., Yongvanit P., Boonmars T., Puapairoj A., et al. Opisthorchis viverrini infection activates the PI3K/AKT/PTEN and Wnt/beta-catenin signaling pathways in a Cholangiocarcinogenesis model. Asian Pac. J. Cancer Prev. 2014;15:10463–10468. doi: 10.7314/apjcp.2014.15.23.10463. [DOI] [PubMed] [Google Scholar]
- Zaparina O.G., Rakhmetova A.S., Kolosova N.G., Cheng G., Mordvinov V.A., Pakharukova M.Y. Antioxidants resveratrol and SkQ1 attenuate praziquantel adverse effects on the liver in Opisthorchis felineus infected hamsters. Acta Trop. 2021;105954 doi: 10.1016/j.actatropica.2021.105954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1: Results of Agilent analysis of total RNA extracted from the liver samples
Supplementary material 2: Results of High Sensitivity DNA Assay (Agilent 2100) of DNA libraries constructed
Supplementary material 3: Principal component analysis scree plot (Percentage of Variance for each by PC)
Supplementary Tables
Data Availability Statement
The sequence data were deposited in NCBI BioProject under accession number PRJNA789062.