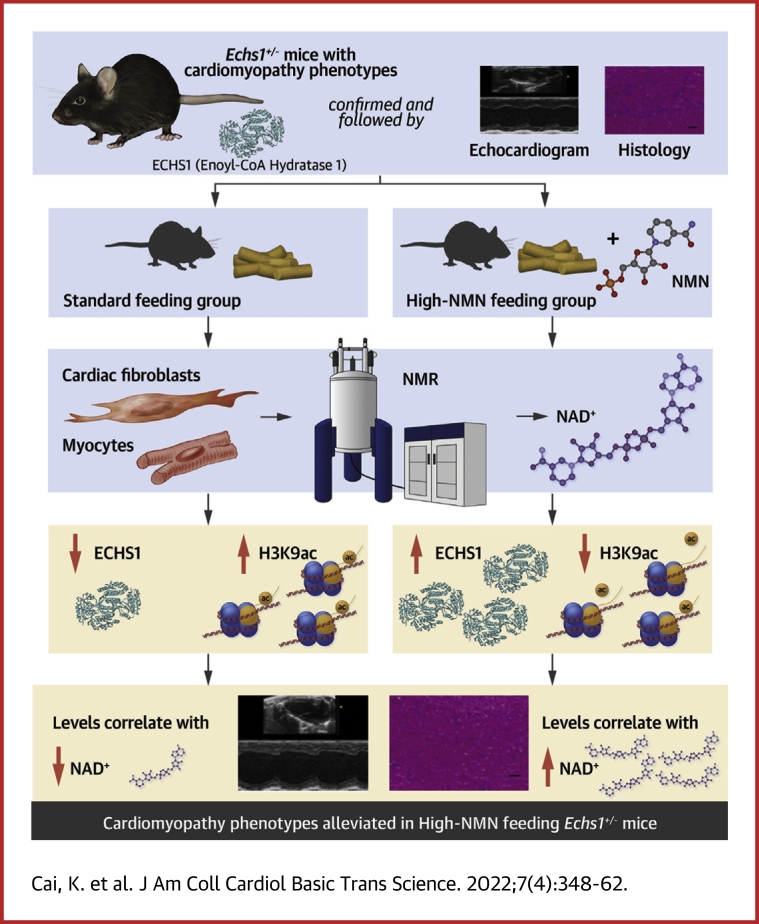
Visual Abstract
Key Words: acetylation of H3K9, cardiomyopathy, enoyl-CoA hydratase 1, nicotinamide mononucleotide, p300
Abbreviations and Acronyms: α-SMA, smooth muscle actin-α; ANP, atrial natriuretic peptide; BCAA, branched-chain amino acid; BNP, brain natriuretic peptide; DCM, dilated cardiomyopathy; ECHS1, short-chain enoyl-CoA hydratase 1; FA, fatty acid; HCM, hypertrophic cardiomyopathy; HFF, human foreskin fibroblast; IVSd, interventricular septum in end-diastole; IVSs, interventricular septum in end-systole; LVEF, left ventricular ejection fraction; LVFS, left ventricular fractional shortening; LVIDd, left ventricular internal dimension in end-diastole; LVIDs, left ventricular internal dimension in end-systole; LVPWd, left ventricular posterior wall in end-diastole; LVPWs, left ventricular posterior wall in end-systole; NMN, nicotinamide mononucleotide
Highlights
-
•
ECHS1 hydrates medium- and short-chain enoyl CoAs and catalyzes the oxidation of fatty acids and branched-chain amino acids.
-
•
The mechanism driving ECHS1 deficiency–associated cardiomyopathy was investigated using conventional biochemistry and molecular biology methods, including immunoprecipitation and polymerase chain reaction.
-
•
Echs1 heterogeneous knockout mice displayed cardiac dysfunction as evaluated by echocardiography.
-
•
ECHS1 deficiency causes cardiomyopathy by enhancing p300-mediated H3K9ac.
-
•
ECHS1 deficiency–induced cardiomyopathy can be prevented using an intervention approach targeting H3K9ac.
Summary
Short-chain enoyl-CoA hydratase 1 (ECHS1) deficiency plays a role in cardiomyopathy. Whether ECHS1 deficiency causes or is only associated with cardiomyopathy remains unclear. By using Echs1 heterogeneous knockout (Echs1+/-) mice, we found that ECHS1 deficiency caused cardiac dysfunction, as evidenced by diffuse myocardial fibrosis and upregulated fibrosis-related genes. Mechanistically, ECHS1 interacts with the p300 nuclear localization sequence, preventing its nuclear translocation in fibroblasts. ECHS1 deficiency promotes p300 nuclear translocation, leading to increased H3K9 acetylation, a known risk factor for cardiomyopathy. Nicotinamide mononucleotide–mediated acetylation targeting suppressed ECHS1 deficiency–induced cardiomyopathy phenotypes in Echs1+/- mice. Thus, enhancing p300-mediated H3K9ac is a potential interventional approach for preventing ECHS1 deficiency–induced cardiomyopathy.
Cardiomyopathy is a genetically heterogeneous disease that involves multiple alterations in several causative genes, threatening human health with a high incidence of 1 in 10,000 children or 5,000 adults.1,2 Moreover, it can also result from coronary artery abnormalities, tachyarrhythmias, exposure to infection or toxins, secondary or other confounding comorbidities such as atherosclerosis, hypertension, renal dysfunction, diabetes, as well as exposure to alcohol, anabolic steroids, and chemotherapeutic agents (eg, anthracyclines, cyclophosphamide, and doxorubicin).3,4
Recently, cardiomyopathy has received increasing attention because of the elucidation of its genetic causes in terms of genes encoding components of the sarcomere or costamere, related binding proteins, Z-band, nuclear membrane, desmosome, mitochondrial and calcium-handling proteins, as well as the availability of sensitive imaging and genetic testing.5,6 However, less than one-third of cases of early-onset cardiomyopathy can be diagnosed based on alterations associated with myocardial contraction, and increasing evidence suggests that nutrient changes and metabolic-related gene mutants (such as alanyl-tRNA synthetase 2, inorganic pyrophosphatase 2, tafazzin, and BAG cochaperone 3) that account for >35% of cases of hypertrophic cardiomyopathy (HCM) or dilated cardiomyopathy (DCM) in children, play a significant role in the occurrence of cardiomyopathy.7,8
Furthermore, nutritional deficiencies can also lead to cardiomyopathies, the phenotypes of which can be observed in patients with Kwashiorkor. Individuals with L-carnitine, niacin, selenium, thiamine, vitamin C, and selenium deficiencies are more likely to possess these phenotypes.9,10 The number of patients with cardiomyopathy harboring metabolic gene mutants, such as solute carrier family 25 member 20, carnitine palmitoyltransferase 2, solute carrier family 22 member 5, acyl-CoA dehydrogenase very long chain, hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha/beta, and enoyl-CoA hydratase 1 (ECHS1), is on the increase; the products of the expression of these genes are involved in carnitine shuttle and fatty acid (FA) β-oxidation pathways, indicating the close connection between FA metabolism and the occurrence of cardiomyopathy.11 Furthermore, 40% of congenital ECHS1 defects (with a rare incidence of 1 in 1,000,000 in human newborns or children) contribute to cardiomyopathy, and decreased expression and loss-of-function of ECHS1 are frequently observed in adults with HCM or DCM, albeit the underlying mechanism remains unknown.12, 13, 14
In addition to gene alteration–induced protein inactivation, environmental factors were recently established as regulators of the intracellular protein activity. A few metabolic enzymes are capable of responding to environmental signals, thereby regulating intracellular metabolism and facilitating cellular accommodation of environment changes. Our previous studies on FA metabolism revealed that ECHS1, the key enzyme in the oxidation of FAs and branched-chain amino acids (BCAAs), sensed nutrient and energy abundance and regulated cell growth and apoptosis.15,16 ECHS1 levels are closely associated with the risk of human HCM, and ECHS1 links metabolism with the environment.14 Therefore, we investigated the role of ECHS1 deficiency in the development of cardiomyopathy and explored the underlying mechanisms using a transgenic mouse model.
Methods
All the data and analytic and study materials supporting the findings of our study are available from the corresponding author upon reasonable request. All supporting data are available with the article.
Reagents and antibodies
The details about all the chemicals and antibodies used are listed in Supplemental Table 1.
Cell culture
Human foreskin fibroblasts (HFFs) (American Type Culture Collection [ATCC] number: SCRC-1041) and Rattus norvegicus myoblasts (H9c2) (ATCC number: CRL-1446) were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (HyClone) supplemented with 10% fetal bovine serum (Gibco), 100 units/mL penicillin (Invitrogen), and 100 μg/mL streptomycin (Invitrogen). The cells were incubated in 5% CO2 at 37°C.
Plasmid constructs and transfection
Full-length human p300 and ECHS1, truncated-p300-N, p300-M, and p300-C were amplified using HEK293T cDNA and cloned into the Xho I and EcoR I restriction sites of the pcDNA3.1-Flag/HA vector using the CloneExpress MultiS One Step Cloning Kit (#C113-02; Vazyme). Mutant p300 were generated by site-directed mutagenesis using the Mut Express MultiS Fast Mutagenesis kit (#C215-01, Vazyme) according to the manufacturer’s instructions and transformed to DH5α cells (relief 5α chemically competent cell, TSC-C01; Tsingke) to amplify. Each plasmid was transfected using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The primers used are listed in Supplemental Table 2.
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 generation of knockout cells
ECHS1 and p300 knockout (KO) HFF cells were generated according to standard clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) gene editing protocols using the guide sequence targeting the human ECHS1 and p300 listed in Supplemental Table 2.17
Small RNA interference
Synthetic oligos were used for small interfering RNA (siRNA)–mediated silencing of ECHS1, and scramble siRNA was used as control.18 Cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Knockdown efficiency was verified by western blotting. The siRNA sequences used in this study are listed in Supplemental Table 2.
Animal model
All animal procedures and experiments were approved by the Animal Care Committee of Fudan University. The C57BL/6J mice were housed (with free access to standard diet and water) in a specific pathogen-free animal room at 20 to 26 °C with a 12:12-hour light:dark cycle. All animal experiments were performed using both male and female littermates at 3 to 96 weeks of age. Mice were randomized into groups of 6. The heterozygous Echs1 KO (Echs1+/-) mice were generated using the CRISPR/Cas 9 system as previously described, and mated to produce control littermates and study mice with C57BL/6 background carrying heterozygous Echs1 KO alterations19; their genotypes were confirmed by polymerase chain reaction (PCR). For the high-FAs/high-BCAAs mouse model, wild-type and Echs1+/- mice were fed with high-FA/high-BCAA chow (570 g of fat, 200 g of carbohydrate, 200 g of protein, 10 g of valine, 10 g of leucine, and 10 g of isoleucine per kilogram). For the short-term feeding, we increased BCAAs and FAs in the infant mouse by feeding the maternal mice with high-FA/high-BCAA chow from the first day of mating to postdelivery day 21. For the long-term feeding, the mice born during short-term feeding were fed high-BCAA/high-FA chow from ablactation to 16 weeks of age. All the mice were anesthetized with 2% isoflurane gas and sacrificed; the heart, brain, liver, and kidneys were removed and washed in 0.9% sodium chloride. Then, they were processed as required for the different analyses. For BCAA analyses, different tissues were homogenated with 80% methanol. For hematoxylin and eosin (HE), Masson’s trichrome, Sirius red, and Nissl staining, wheat germ agglutinin (WGA), immunohistochemistry, and immunofluorescence, formalin-fixed, paraffin-embedded tissue blocks were created from the heart, brain, liver, and kidney tissues of wild-type and Echs1+/- mice. Liver and heart tissues were placed in an optimal cutting temperature compound (Thermo Fisher Scientific Inc) in a peel-away mold and frozen at -80 °C for further experiments (Oil Red O staining). Samples were processed blindly during the experiments and assessments.
Echocardiography
Transthoracic echocardiography was performed using a Vevo3100 system (FUJIFILM VisualSonics Inc) with a MS400 transducer under 2% isoflurane gas and 0.8 L/min oxygen anesthesia. Surface electrocardiograms were recorded during isoflurane inhalation and the heart rate was monitored by continuous electrocardiogram. Data were analyzed in a blinded fashion.
Mouse cardiomyocyte and cardiac fibroblast isolation
Mouse primary cardiomyocytes and cardiac fibroblasts were isolated from heart tissues according to published methods.19,20 Briefly, the heart was removed and perfused with a buffer (130 mM NaCl, 5 mM KCl, 0.5 mM NaH2PO4, 10 mM HEPES, 10 mM glucose, 10 mM 2, 3-butanedione monoxime, 10 mM taurine, and 1.5 mM EDTA), perfusion buffer (130 mM NaCl, 5 mM KCl, 0.5 mM NaH2PO4, 10 mM HEPES, 10 mM glucose, 10 mM 2, 3-butanedione monoxime, 10 mM taurine, and 1 mM MgCl2), and an enzyme solution containing collagenase II (245 U/mg, Worthington Biochemical), collagenase IV (235 U/mg, Worthington Biochemical), and protease XIV (0.05 mg/mL, Sigma), at 80 mL/h.
Cardiomyocytes were pelleted and suspended in a calcium buffer containing CaCl2 (0.125 μmol/L), with gradual increments in the concentration (5, 10, 30, and 50 μL) of the CaCl2 solution (100 mM) and a 4-minute delay between each increment. The cardiomyocytes were seeded into a dish, centrifuged for 5 minutes at 1,000 × g and 4 °C to increase adherence; then they were cultured in DMEM media at 37 °C for 1 hour. The culture medium was changed to a CM medium (MEM; HyClone with 0.2% bovine serum albumin, 10 mM HEPES, 4 mM NaHCO3, 10 mM creatine monohydrate, 1% penicillin/streptomycin, and 0.5% insulin-selenium-transferrin and blebbistatin) for further treatment.
Cardiac fibroblasts from the supernatant were pelleted by centrifugation at 1,000 × g and 4°C for 5 minutes and plated in a 4 to 5 mL cardiac fibroblast media (DMEM with 10% fetal bovine serum and 1% penicillin/streptomycin). After 2 to 3 hours, they were washed vigorously 3 to 5 times with 2 mL phosphate-buffered saline (PBS), and fresh media was added.
Nuclei extraction
For nuclear metabolite and protein localization analyses, nuclei were extracted as described previously.21,22 A brief description is as follows: 1 × 108 cells were harvested in hypotonic buffer (10 mM HEPES-NaOH [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, and 0.5 mM beta-mercaptoethanol) and incubated on ice for 20 minutes. Then, 0.5% NP-40 was added and vortexed for 2 minutes, followed by centrifugation at 600 × g for 10 minutes. The pellet contained the nuclear fraction and the supernatant contained the postnuclear fraction.
Nuclear and postnuclear metabolite extraction
Metabolites were extracted from nuclear and postnuclear fractions by adapting previously described methods with minor modifications.21,22 A brief description is as follows: immediately following nuclear isolation, isolated nuclear and postnuclear fractions were subjected to metabolite extraction using prechilled (-80°C) 80% (v/v) methanol. Metabolites extracted in the solvent phase were freeze-dried and subjected to further analysis.
Metabolite measurement by liquid chromatography – tandem mass spectrometry
Metabolites were extracted using a previously described protocol.21, 22, 23 Briefly described, the samples were resuspended in high-performance liquid chromatography–grade H2O before flow injection and analysis in a 6500 QTRAP mass spectrometer (AB SCIEX, Thermo Fisher Scientific) coupled to a Shimadzu high-performance liquid chromatography system (LC 20AB, Waters Corporation) via multiple reaction monitoring mode.
Metabolite measurement by nuclear magnetic resonance
Nicotinamide adenine dinucleotide (NAD+) was quantified in the extracted metabolites by nuclear magnetic resonance (NMR) as previously described.24 A brief description is as follows: the samples were dissolved in a phosphate buffer containing 10% deuterium oxide and trimethylsilyl propanoic acid (TSP) in NMR tubes. The mixture was then centrifuged at 20,817 × g for 10 minutes at 4°C. Then, the 600-μL supernatant was transferred to a standard 5-mm NMR tube for analysis. One-dimensional 1H NMR spectra were obtained at 298 K on a Bruker Advance spectrometer using dried extracts that were dissolved in a phosphate buffer containing 10% deuterium oxide and TSP in NMR tubes. Sixty-four transients, each with a spectral width of 20 ppm, were collected into 32 k data points. The signal intensities were normalized against that of the TSP signal at 0 ppm. The absolute concentration of NAD+ was calculated using the known concentration of TSP.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from tissue samples of wild-type and Echs1+/- mice preserved in RNA and then converted to cDNA using random hexamers, oligo (dT) primers, and Moloney murine leukemia virus reverse transcriptase (TaKaRa). The ECHS1, atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), β-catenin, connexin43, and myosin heavy chain β mRNA levels were measured by quantitative real-time PCR (qRT-PCR) using the ABI Prism 7900 sequence detection system (Applied Biosystems), with actin as an internal reference gene. Each reaction was performed in triplicate. The primers used are listed in Supplemental Table 2.
Western blot analyses
Cultured cells or mouse tissues were homogenized with 0.5% NP-40 buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40, and a mixture of protease inhibitors (Sigma-Aldrich). After centrifugation at 12,000 g and 4°C for 15 minutes, the supernatant was collected for western blotting according to the standard procedures described previously.25 Bands were detected by measuring chemiluminescence on a Typhoon FLA 9500 (GE Healthcare). All western blotting experiments were repeated at least 3 times.
Immunoprecipitation assays
Cells or tissues were homogenized in 0.5% NP-40 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin, and 1 mM phenylmethylsulfonyl). Each sample was incubated with an anti-p300 antibody. The antigen-antibody complexes were washed with a 0.5% NP-40 buffer and mixed with a loading buffer for sodium dodecyl sulphate–polyacrylamide gel electrophoresis.
Immunofluorescence assays
Cultured cells were harvested and washed twice with PBS to remove the remaining medium. Paraformaldehyde (4%) was used to fix the cells at room temperature (20-25°C), followed by incubation with 0.5% Triton X-100 in PBS for 20 minutes at room temperature. Then, the cells were incubated with 3% bovine serum antigen for 30 minutes at room temperature. The primary antibodies used were as follows: anti-rabbit α–smooth muscle actin (SMA), anti-rabbit α-actinin, anti-mouse ECHS1, and anti-rabbit p300. Alexa Fluor 488 donkey anti-rabbit and Alexa Fluor 546 donkey anti-mouse secondary antibodies were used. 4’,6-diamidino-2-phenylindole (DAPI) was subsequently added for nuclear staining. Cells were observed under a fluorescence microscope, and images were obtained in the multitracking mode on a confocal laser scanning microscope (LSM880-Airyscan; Carl Zeiss). The details about the antibodies used are listed in Supplemental Table 1.
Paraffin-embedded heart tissue sections were rehydrated and their antigens retrieved with citrate-assisted heat. Sections were then blocked with 30% goat serum and incubated overnight at 4°C with primary antibodies, anti-rabbit α-SMA, anti-rabbit α-actinin, and anti-mouse ECHS1. The following day, sections were washed in 1× Tris-buffered saline, 0.1% Tween 20 detergent, incubated with corresponding Alexa Fluor 594 donkey anti-rabbit and Alexa Fluor 488 goat anti-mouse secondary antibodies (Invitrogen). The details about the antibodies used are listed in Supplemental Table 1.
Statistical analysis
Animal groups were predefined according to genotype and active treatment vs vehicle, and were age- and sex-matched. Age groups represent distinct animals and not the same animals followed over time. Values are expressed as mean ± SEM. One-way and two-way analysis of variance tests were used to compare parameters among ≥3 independent groups, whereas the Student t-test was used to compare 2 groups. Pairwise comparisons between groups were made using Tukey's post hoc multiple comparisons test when analysis of variance yielded significant differences. Values were considered significant at P < 0.05. All statistical tests were performed using GraphPad PRISM software version 8.3.1 (GraphPad Software, Inc).
Results
Cardiac dysfunction in Echs1 heterogeneous KO (Echs1+/-) mice
To determine whether Echs1+/- mice developed cardiac dysfunction, we continuously monitored the heart function of wild-type and Echs1+/- mice born in the same litter using echocardiography, starting from week 3 when they were capable of independent feeding, until week 96. We found impaired left ventricular systolic function in both male (Figure 1A) and female (Supplemental Figure 1A) Echs1+/- mice, especially the older ones. Echocardiography revealed decreased left ventricular ejection fraction (LVEF) (Figure 1B, Supplemental Figure 1B), left ventricular fractional shortening (LVFS) (Figure 1C, Supplemental Figure 1C), increased left ventricular internal dimension in end-systole (LVIDs) (Figure 1D, Supplemental Figure 1D) and end-diastole (LVIDd) (Figure 1E, Supplemental Figure 1E), and thicker interventricular septum in end-systole (IVSs) (Figure 1F, Supplemental Figure 1F) and end-diastole (IVSd) (Figure 1G, Supplemental Figure 1G) in Echs1+/- mice from week 3 to week 96; these changes were more prominent in older mice. In contrast, the left ventricular posterior wall in end-systole (LVPWs) and end-diastole (LVPWd) did not exhibit significant changes between the Echs1+/- and control groups (Supplemental Figures 2A to 2D). Moreover, increased heart weight/body weight and heart weight/tibia length ratios (Figures 1H and 1I, and Supplemental Figures 1H and I) were observed in Echs1+/- mice despite the lack of significant difference in body weight at all time points between the 2 groups (Supplemental Figures 2E and 2F), indicating significantly higher heart weight gain in the Echs1+/- mice than wild-type mice.
Figure 1.
Cardiac Dysfunction in Echs1 Heterogeneous Knockout (Echs1+/-) Male Mice
(A) An echocardiographic parasternal long-axis view and M-mode echocardiographic images of the wide-type and Echs1+/- male mice at different ages. (B–G) Various indicators of cardiac dysfunction in Echs1+/- mice. (H,I) Heart weight/body weight and heart weight/tibia length ratios in Echs1+/- and wild-type mice. Complete echocardiographic and morphometric measurements are presented in Supplemental Table 3. Data are presented as mean ± SEM. N = 6 mice/group. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 using Student t-test and one-way analysis of variance with Tukey’s post hoc test for 2 and multiple group comparisons, respectively. IVSd = interventricular septum in end-diastole; IVSs = interventricular septum in end-systole; LVEF = left ventricular ejection fraction; LVFS = left ventricular fractional shortening; LVIDd = left ventricular internal dimension in end-diastole; LVIDs = left ventricular internal dimension in end-systole; ns = no significance.
Diffuse myocardial fibrosis and upregulated fibrosis-related genes in Echs1+/- mice
To investigate whether enlarged cardiomyocytes or increased number of myofibroblasts contributed to cardiac hypertrophy in Echs1+/- mice, heart tissues of Echs1+/- and wild-type mice aged 3 and 96 weeks were stained with HE, WGA, Masson’s trichrome, and sirius red. Diffuse myocardial fibrosis but normal-sized cardiomyocytes were observed in male (Figure 2A) and female (Supplemental Figure 3A) Echs1+/- mice. Further immunohistochemical analysis showed increased α-SMA, collagen I, and collagen Ⅲ levels in the hearts of male (Figure 2B) and female (Supplemental Figure 3B) Echs1+/- mice, compared with that in wild-type male and female mice aged 3 and 96 weeks, which were validated by western blot (Figure 2C, Supplemental Figure 3C). We detected an increase in the expression of ANP, BNP, β-catenin, connexin43 (Cx43), and myosin heavy chain β (β-MHC) by qRT-PCR in male (Figure 2D) and female (Supplemental Figure 3D) Echs1+/- mice, compared with wild-type male and female mice. These findings indicate that ECHS1 inhibition might affect cardiac contractile function by increasing the expression of fibrosis-related genes in the heart.
Figure 2.
Diffuse Myocardial Fibrosis and Upregulated Fibrosis-Related Genes in Echs1+/- Male Mice
(A) Hematoxylin and eosin (HE) staining showing diffused cardiac fiber arrangement; wheat germ agglutinin (WGA) staining showing similar cardiomyocyte areas; Masson’s trichrome and sirius red staining of cardiac cross-sections showing increased collagen deposition in heart tissues of Echs1+/- mice compared with that of wild-type mice at weeks 3 and 96. Scale bar: 50 μm. N = 3 mice/group. (B) Cardiac cross-sections showing increased α–smooth muscle actin (α-SMA), collagen i, collagen III, and H3K9ac expression in Echs1+/- hearts at weeks 3 and 96. Scale bar: 50 μm. N = 3 mice/group. (C) Western blot showing increased α-SMA, collagen i, collagen III expression in Echs1+/- hearts at weeks 3 and 96. N = 3 mice/group. (D) Increased mRNA levels of atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), β-catenin, connexin43 (Cx43), and myosin heavy chain β (β-MHC), detected by quantitative real-time polymerase chain reaction (qRT-PCR), indicating failing hearts in Echs1+/- male mice. N = 6 mice/group. Scale bar: 50 μm. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 using Student t-test and one-way analysis of variance with Tukey’s post hoc test for 2 and multiple group comparisons, respectively. Abbreviation as in Figure 1.
We observed significant accumulation of BCAAs and FAs in blood and heart tissues of Echs1+/- mice compared with that of wild-type mice (Supplemental Figures 4A and 4B). Therefore, we evaluated the relationship between the cardiac phenotypes in Echs1+/- mice and the accumulation of these metabolic substrates. To investigate whether increased BCAA and FA levels contribute to the cardiac phenotypes, we generated a high-BCAA/high-FA mouse model through short- and long-term feeding of the wild-type mouse with high-FA/high-BCAA diet. We evaluated the cardiac function of mice that received high-BCAA/high-FA feeding for 3 and 16 weeks using echocardiography and examined cardiac fibrosis through tissue staining. We found that although a high-BCAA/high-FA diet caused high levels of FA and BCAA in tissues (Supplemental Figures 5A and 5B), neither FAs nor BCAAs caused the same cardiac phenotypes as in Echs1+/- mice (Supplemental Figures 5C to 5H). Moreover, we fed Echs1+/- mice with high-BCAA/high-FA chow for 3 and 16 weeks, respectively. Although increased BCAAs and FAs were observed in heart tissues (Supplemental Figures 6A and 6B), they did not alter the cardiac phenotypes as in Echs1+/- mice (Supplemental Figures 6C to 6H). These results excluded the contribution of the traditional metabolic role of ECHS1 in the development of cardiac fibrosis.
Next, we examined whether ECHS1 deficiency also had an effect on other organs, including liver, kidneys, and brain. We observed accumulation of total FAs in these organs in Echs1+/- mice but not in the wild-type mice (Supplemental Figure 4B). Moreover, we did not detect abnormalities in the cerebral tissues of Echs1+/- and wild-type mice on HE and Nissl staining (Supplemental Figure 7A). Similarly, HE and Masson staining of liver and kidney tissues of Echs1+/- mice did not reveal fibrosis (Supplemental Figures 7B and 7C). Although Oil Red O staining revealed lipid droplet accumulation in the liver of the Echs1+/- mouse (Supplemental Figures 7B), no cardiac steatosis was observed in the heart tissue (Supplemental Figure 7D). These findings suggest the specificity of ECHS1 in regulating fibrosis in the heart.
Increased H3K9 acetylation in primary cardiac fibroblasts of Echs1+/- mice
Because ECHS1 is involved in acetyl-CoA synthesis, we analyzed protein acetylation in the wild-type and Echs1+/- mice. Increased pan-acetylation of proteins weighing approximately 20 kDa in the heart tissue lysates of Echs1+/- mice (Figure 3A) was observed. Because the sizes of histones H3 and H4 were approximately 20 kDa and histone H3/H4 played a role in the development of cardiomyopathy, we examined whether acetylation of histones H3 and H4 was affected by ECHS1 deficiency.26,27 Using H3K4ac-, H3K9ac-, H3K18ac-, H3K27ac-, H4K5ac-, H4K8ac-, H4K12ac-, and H4K16ac-targeting antibodies, we found increased H3K9ac levels, a pathogenic factor in cardiac fibroblasts for fibrosis, in the Echs1+/- mice heart tissues (Figure 3B).28 Similarly, immunohistochemistry showed increased H3K9ac levels in heart tissues of Echs1+/- mice compared with that in wild-type mice at week 3 and week 96 (Figure 2B, Supplemental Figure 3B). To investigate whether cardiac fibroblasts or cardiomyocytes contributed to the level of alteration of histone 3 acetylation, we assessed the pan-acetylation in both cells from wild-type and Echs1+/- mice; only primary cardiac fibroblasts from Echs1+/- mice showed increased pan-acetylation around 20 kDa similar to that in whole-heart tissues (Figures 3C and 3D). Thus, a specific increase in H3K9ac and decrease in ECHS1 in mouse cardiac fibroblasts contributed to cardiac fibrosis in Echs1+/- mice (Figures 3E and 3F).
Figure 3.
H3K9 Acetylation Specifically Increased in Primary Cardiac Fibroblasts of Echs1+/- Male Mice
(A) Western blot showing increased pan-acetylation of proteins weighing ∼20 kDa in heart tissue lysates of Echs1+/- mice compared with that of wild-type aged 8 weeks, suggesting increased histones 3/4 acetylation. (B) Increased H3K9ac levels in heart tissues of Echs1+/- mice. N = 3 mice/group. (C to F) Western blot showing pan-acetylation and histone acetylation levels in mouse primary cardiac fibroblasts and primary cardiomyocytes. N = 3 mice/group. (G)ECHS1 knockdown increased H3K9ac levels in human foreskin fibroblasts (HFFs) but not in rat cardiomyocytes (H9c2). (H,I) Carnitine palmitoyltransferase 2 (CPT2) or hydroxyacyl-CoA dehydrogenase (HADH) knockdown did not elevate H3K9ac in HFF and H9c2 cells. (J to M) ECHS1 deficiency did not alter acetyl-CoA levels in the nucleus, but decreased them in the mitochondria of mouse primary cardiac fibroblasts and cardiomyocytes, HFFs, and H9c2 cells. N = 3 to 6/group. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 using Student t-test and one-way analysis of variance with Tukey’s post hoc test for 2 and multiple group comparisons, respectively. CBB, coomassie brilliant blue; siRNA, small interfering RNA; other abbreviation as in Figure 1.
We confirmed that knockdown of ECHS1 led to increased levels of H3K9ac in HFF but not H9c2 cells (Figure 3G). However, knockdown of either CPT2 or HADH, upstream and downstream enzymes, respectively, of ECHS1 in FA oxidation pathway, did not result in elevated H3K9ac levels in both HFF and H9c2 cells (Figures 3H and 3I). These results suggest that altered levels of H3K9ac might not be induced by altered levels of acetyl-CoA caused by beta-oxidation blockade. To validate this, we analyzed the acetyl-CoA level in the nucleus and mitochondria of mouse primary cardiac fibroblasts and HFFs and found that ECHS1 deficiency did not alter acetyl-CoA levels in the nucleus but decreased them in the mitochondria (Figures 3J and 3K); a similar trend was observed in mouse primary cardiomyocytes and H9c2 cells (Figures 3L and 3M). Because ECHS1 is located primarily in the mitochondria and cytosol, these findings suggested that ECHS1 upregulated H3K9ac levels by an unknown mechanism.
Decreased ECHS1 prompted p300 translocation into the nucleus, enhancing p300-mediated increase in H3K9ac levels
To investigate the molecular and cellular mechanisms underlying the ECHS1- and H3K9ac-mediated detrimental effects on cardiac fibrosis, we analyzed our previous database of ECHS1-interacting proteins that was generated by tandem affinity purification.16 The results showed that p300, a histone acetyltransferase, strongly interacted with ECHS1.16 This interaction was validated by coimmunoprecipitation assays performed using either exogenous ECHS1 and p300 proteins in HFFs (Figure 4A) or endogenous proteins in mouse primary cardiac fibroblasts (Figure 4B). Moreover, immunofluorescence assays confirmed that a decrease in ECHS1 enhanced the translocation of p300 into the nucleus of cultured HFFs (Figure 4C). Furthermore, reduced p300 levels in the cytoplasm and elevated p300 and H3K9ac levels in the nucleus were observed in primary cardiac fibroblasts isolated from heart tissues of Echs1+/- mice at 8 weeks of age; the levels of other enzymes, such as KAT2A, SIRT1, SIRT6, HDAC1, and HDAC3, that reportedly regulate H3K9 acetylation, remained unchanged (Figure 4D).29, 30, 31 Similarly, siRNA-mediated ECHS1 knockdown, but not p300 knockdown, in HFFs led to elevated levels of H3K9ac (Figure 4E). Although ECHS1 interacts with p300 in mouse primary cardiomyocytes (Supplemental Figure 8A), ECHS1 deficiency did not alter the translocation of p300 and the acetylation levels of H3K9 in cardiomyocytes significantly (Supplemental Figure 8B), probably because of very low expression levels of p300 in cardiomyocytes (Supplemental Figures 8C and 8D). These results suggest that, instead of p300, other kinds of histone acetyltransferases determined the acetylation of H3K9 in cardiomyocytes.
Figure 4.
Decreased ECHS1 Promoted P300 Translocation Into the Nucleus Enhancing P300-Mediated H3k9ac Acetylation in Primary Cardiac Fibroblasts
(A,B) Co-immunoprecipitation assays using exogenous ECHS1 and p300 proteins in HFF cells (A) and endogenous proteins in mouse primary cardiac fibroblasts (B) validated the interaction between p300 and ECHS1. (C) Immunofluorescence experiment showing that decreased ECHS1 promotes p300 translocation into the nucleus in HFFs. Scale bar: 20 μm. (D) Western blot showing reduced cytoplasmic p300 levels and elevated nuclear p300 and H3K9ac levels in primary cardiac fibroblasts isolated from heart tissues of wild-type and Echs1+/- mice aged 8 weeks. N = 3 mice/group. (E) siRNA-mediated ECHS1 knockdown, but not p300 knockout, caused elevated H3K9ac levels in HFFs. (F) N, M, and C parts of p300 plasmid, mutant p300, and FLAG-p300 nuclear localization sequence-Del. (G) Co-immunoprecipitation assays showing the interaction between the N part and ECHS1. (H) ECHS1 did not interact with p300-NLS-Del. DAPI = 4’,6-diamidino-2-phenylindole; IgG = immunoglobulin G; IP-HA = immunoprecipitation - hemagglutinin; other abbreviations as in Figure 3.
To elucidate how the interaction between ECHS1 and p300 induces p300 translocation into the nucleus, we divided the p300 plasmid into 3 parts (N, M, and C) and constructed them into the same vector (Figure 4F). On assessing their individual interactions with ECHS1, we found that only the N part interacted with ECHS1 (Figure 4G). Because the nuclear localization sequence (NLS) is located in the N part, we speculated that ECHS1 might be interacting with the NLS in p300 to inhibit its translocation into the nucleus.32 Indeed, NLS deletion (Figure 4F) prevented the interaction between ECHS1 and a p300 mutant (Figure 4H). Together, these results indicate that ECHS1 interacts with the N terminal of p300 and occupies the NLS region, thereby blocking its nuclear translocation.
Nicotinamide mononucleotide inhibited H3K9ac and prevented the development of ECHS1 deficiency–induced cardiac phenotypes
Previously, we found that nutrient abundance promoted ECHS1 degradation by inhibiting SIRT3-mediated ECHS1 deacetylation, whereas nutrient restriction activated ECHS1 transcription.15,16 Interestingly, SIRT1 and SIRT6 can regulate H3K9ac, suggesting a potential treatment strategy for ECHS1 deficiency–induced cardiac phenotypes, which involves controlling the deacetylation function of the SIRT family. Here, we further tested whether targeting histone acetylation with NMN, which activates intracellular deacetylase activities by increasing NAD+ levels, prevented the development of ECHS1 deficiency–induced cardiac phenotypes.33,34
The addition of NMN to control and siRNA-mediated ECHS1 knockdown HFFs resulted in elevated ECHS1 levels in both the cytoplasm and lysate, but reduced p300 and H3K9ac levels in the nucleus, especially in ECHS1 knockdown HFFs (Figure 5A). Notably, increased NAD+ levels after NMN addition were observed in control and ECHS1 knockdown HFFs (Supplemental Figure 9A). Echs1+/- and wild-type mice were fed standard or high-NMN chow from week 3 to week 8. We verified that high-NMN chow feeding caused increased ECHS1 protein levels in mouse heart, as evidenced by western blotting of mouse primary cardiac cells (Supplemental Figure 9B), coimmunostaining assay in mouse primary cardiac cells (Supplemental Figure 9C), and immunohistology assay in mouse heart tissues (Supplemental Figure 9D). Moreover, we found, in keeping with the observation in HFFs, that high-NMN chow feeding resulted in decreased nuclear localization of p300 and decreased acetylation levels of H3K9 (Figure 5B). Interestingly, the impaired left ventricular systolic function observed in F1 Echs1+/- mice at 8 weeks was prevented by consumption of high-NMN chow (Figure 5C). Functional analyses of the echocardiography showed normal LVEF, LVIDd, LVIDs, and IVSd in NMN-fed F1 Echs1+/- mice, compared with the wild-type mice (Figures 5D to 5G). In addition, cardiac fibrosis was decreased in NMN-fed F1 Echs1+/- mouse hearts (Figures 5H and 5I), indicating that NMN supplementation prevented the cardiomyopathy phenotypes resulting from ECHS1 deficiency. Further, we confirmed the increase in NAD+ levels in Echs1+/- and wild-type mice after NMN supplementation at 8 weeks (Supplemental Figure 9E). Thus, the evidence indicated that targeting acetylation via NMN-mediated SIRT activation reversed the effects of ECHS1 deficiency in cultured fibroblasts and suppressed ECHS1 deficiency–induced cardiomyopathy in the mouse model.
Figure 5.
NMN Inhibited H3k9ac And Prevented The Development Of ECHS1 Deficiency–Induced Cardiac Phenotypes
(A) Nicotinamide mononucleotide (NMN) treatment caused elevation in cytoplasmic and lysate ECHS1 levels and reduction in nuclear p300 and H3K9ac levels, especially in siECHS1-transfected HFFs. (B) High-NMN diet resulted in decreased nuclear localization of p300 and acetylation levels of H3K9 in heart tissues. (C-G) Echocardiogram showing that high-NMN diet rescued diastolic function in Echs1+/- male mice aged 8 weeks, as evidenced by normal LVEF, LVIDd, LVIDs, and IVSd in NMN-fed F1 Echs1+/- mice compared with control. N = 6 to 10 mice/group. (H) HE, WGA, Masson’s trichrome, and sirius red staining of cardiac cross-sections indicated normal collagen deposition in Echs1+/- hearts after high-NMN chow consumption. Scale bar: 50 μm. N = 3 mice/group. (I) Fibrosis-related protein (α-SMA, collagen I, and collagen III) and H3K9ac levels decreased in Echs1+/- hearts after high-NMN chow consumption. Scale bar: 50 μm. N = 3 mice/group. (J) Summary diagram showing the relationship between ECHS1 and histone acetylation for increasing the risk of cardiomyopathy. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 using Student t-test and one-way analysis of variance with Tukey’s post hoc test for 2 and multiple group comparisons, respectively. Abbreviations as in Figures 1 and 3.
Discussion
As a metabolic enzyme involved in FA and BCAA metabolism, ECHS1 is associated with the risk of cardiomyopathy. Despite the gene alterations and decreased ECHS1 expression observed in cardiomyopathy, the causality between ECHS1 deficiency and cardiomyopathy and the underlying mechanism remain unknown.11,12 This study showed the pathological role of ECHS1 deficiency in the development of the cardiomyopathy phenotypes and, more importantly, our findings suggest a new strategy for preventing and treating ECHS1 deficiency–related human cardiomyopathy. We previously found that nutrients promoted ECHS1 degradation, whereas nutrient restriction activated ECHS1 transcription, suggesting nutrition status control as a potential intervention in ECHS1 deficiency–induced cardiac dysfunction.16 In this study, we further validated that NMN-mediated targeting of histone acetylation prevented the development of ECHS1 deficiency–induced cardiac phenotypes.
Although alterations in several genes encoding enzymes involved in the beta-oxidation pathway are associated with the onset of cardiomyopathy, we found that high-FA diets did not induce cardiomyopathy in the mouse model (see Supplemental Figures 5 and 6). Notably, we found a new function of ECHS1, which was the regulation of histone acetylation; it may contribute to the risk of cardiomyopathy via this mechanism (Figure 5J). Our findings suggest that other enzymes involved in beta-oxidation may also induce cardiomyopathy through atypical function during beta-oxidation. However, further studies are required to investigate whether alterations in other beta-oxidation enzyme-encoding genes cause cardiomyopathy and to elucidate the underlying mechanisms.
Epigenetic alternations, including chromatin remodeling, DNA methylation, histone modification, and non–coding RNA–mediated transcriptional suppression or activation, increase the risk of cardiomyopathy.35 Among these epigenetic factors, increased acetylation of H3K9 is an established epigenetic factor for HCM and DCM.28,36 Here, we showed that ECHS1 deficiency caused cardiomyopathy by enhancing p300-mediated H3K9ac. Our results suggest that dysregulated metabolic enzymes could regulate the H3K9 acetylation levels through their noncanonical role. ECHS1 targets the NLS region of p300, regulating its nuclear translocation efficiency and, thus, the H3K9 acetylation levels. Taken together, these findings confirm that cellular metabolism regulates epigenetic signals.
Study limitations
Because of the homozygous lethality of Echs1 KO in mice, we were unable to validate the cardiac phenotypes in Echs1-/- mice. ECHS1 deficiency specifically regulates H3K9 acetylation in cardiac fibroblasts but not cardiomyocytes; the underlying mechanism requires further study in the future. Considering the genetically heterogeneous nature of patients with cardiomyopathy, more studies in different patient cohorts with ECHS1 mutants are warranted to verify the causal relationship between ECHS1 mutants and cardiomyopathy.
Conclusions
We found that ECHS1 deficiency caused cardiomyopathy by enhancing p300-mediated H3K9 acetylation in Echs1+/- KO mouse model. Our findings emphasize NMN as a potential intervention approach for preventing ECHS1 deficiency–induced cardiomyopathy.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE 1: Alterations in genes encoding metabolic enzymes play a crucial role in the pathogenesis of inherited forms of HCM and DCM. HCM and DCM can be caused by alterations in genes which regulate epigenetic modification.
COMPETENCY IN MEDICAL KNOWLEDGE 2: Identification of the metabolism-related gene mutants in HCM and DCM would facilitate prenatal and early postnatal identification of alteration carriers and thus provide options for earlier intervention therapies.
TRANSLATIONAL OUTLOOK: By potentially rectifying the abnormal epigenetic alternations through interventions at an early stage, the development and severity of cardiomyopathy may be prevented and mitigated, respectively, in later life.
Funding Support and Author Disclosures
This work was supported by the grants from Key Development Programs of Basic Research of China (Nos. 2020YFA0803601, 2019YFA0801900 (Dr Zhao), and SQ2018YFC100242 (Dr Zhao)), and the National Natural Science Foundation of China (Nos. 81771627 (Dr Zhao), 31871432, 81971061 (Dr Zhao), 81770312, 82170305 (Dr Wang), and 81873481 (Dr Gui). The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental tables and figures, please see the online version of this paper.
Contributor Information
Yong-Hao Gui, Email: yhgui@fudan.edu.cn.
Jian-Yuan Zhao, Email: zhaojy@fudan.edu.cn.
Appendix
References
- 1.Lee T.M., Hsu D.T., Kantor P., et al. Pediatric cardiomyopathies. Circ Res. 2017;121:855–873. doi: 10.1161/CIRCRESAHA.116.309386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKenna W.J., Maron B.J., Thiene G. Classification, epidemiology, and global burden of cardiomyopathies. Circ Res. 2017;121:722–730. doi: 10.1161/CIRCRESAHA.117.309711. [DOI] [PubMed] [Google Scholar]
- 3.Braunwald E. Cardiomyopathies: an overview. Circ Res. 2017;121:711–721. doi: 10.1161/CIRCRESAHA.117.311812. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura M., Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol. 2018;15:387–407. doi: 10.1038/s41569-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 5.Fatkin D., Calkins H., Elliott P., James C.A., Peters S., Kovacic J.C. Contemporary and future approaches to precision medicine in inherited cardiomyopathies: JACC focus seminar 3/5. J Am Coll Cardiol. 2021;77:2551–2572. doi: 10.1016/j.jacc.2020.12.072. [DOI] [PubMed] [Google Scholar]
- 6.McKenna W.J., Judge D.P. Epidemiology of the inherited cardiomyopathies. Nat Rev Cardiol. 2021;18:22–36. doi: 10.1038/s41569-020-0428-2. [DOI] [PubMed] [Google Scholar]
- 7.Kindel S.J., Miller E.M., Gupta R., et al. Pediatric cardiomyopathy: importance of genetic and metabolic evaluation. J Card Fail. 2012;18:396–403. doi: 10.1016/j.cardfail.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasilescu C., Ojala T.H., Brilhante V., et al. Genetic basis of severe childhood-onset cardiomyopathies. J Am Coll Cardiol. 2018;72:2324–2338. doi: 10.1016/j.jacc.2018.08.2171. [DOI] [PubMed] [Google Scholar]
- 9.Brieler J., Breeden M.A., Tucker J. Cardiomyopathy: an overview. Am Fam Physician. 2017;96:640–646. [PubMed] [Google Scholar]
- 10.Oropeza-Moe M., Wisløff H., Bernhoft A. Selenium deficiency associated porcine and human cardiomyopathies. J Trace Elem Med Biol. 2015;31:148–156. doi: 10.1016/j.jtemb.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Houten S.M., Violante S., Ventura F.V., Wanders R.J. The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu Rev Physiol. 2016;78:23–44. doi: 10.1146/annurev-physiol-021115-105045. [DOI] [PubMed] [Google Scholar]
- 12.Bedoyan J.K., Yang S.P., Ferdinandusse S., et al. Lethal neonatal case and review of primary short-chain enoyl-CoA hydratase (SCEH) deficiency associated with secondary lymphocyte pyruvate dehydrogenase complex (PDC) deficiency. Mol Genet Metab. 2017;120:342–349. doi: 10.1016/j.ymgme.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masnada S., Parazzini C., Bini P., et al. Phenotypic spectrum of short-chain enoyl-Coa hydratase-1 (ECHS1) deficiency. Eur J Paediatr Neurol. 2020;28:151–158. doi: 10.1016/j.ejpn.2020.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Tang X., Chen X.F., Sun X., et al. Short-chain enoyl-CoA hydratase mediates histone crotonylation and contributes to cardiac homeostasis. Circulation. 2021;143:1066–1069. doi: 10.1161/CIRCULATIONAHA.120.049438. [DOI] [PubMed] [Google Scholar]
- 15.Qu Y.Y., Zhao R., Zhang H.L., et al. Inactivation of the AMPK-GATA3-ECHS1 pathway induces fatty acid synthesis that promotes clear cell renal cell carcinoma growth. Cancer Res. 2020;80:319–333. doi: 10.1158/0008-5472.CAN-19-1023. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y.K., Qu Y.Y., Lin Y., et al. Enoyl-CoA hydratase-1 regulates mTOR signaling and apoptosis by sensing nutrients. Nat Commun. 2017;8:464. doi: 10.1038/s41467-017-00489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran F.A., Hsu P.D., Wright J., et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzmann J.L., Packard C.J., Chapman M.J., et al. Targeting RNA with antisense oligonucleotides and small interfering RNA: JACC State-of-the-Art Review. J Am Coll Cardiol. 2020;76:563–579. doi: 10.1016/j.jacc.2020.05.070. [DOI] [PubMed] [Google Scholar]
- 19.Ackers-Johnson M., Li P.Y., Holmes A.P., et al. A Simplified, Langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ Res. 2016;119:909–920. doi: 10.1161/CIRCRESAHA.116.309202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Subbaiah K.C.V., Xie L.H., et al. Glutamyl-prolyl-tRNA synthetase regulates proline-rich pro-fibrotic protein synthesis during cardiac fibrosis. Circ Res. 2020;127:827–846. doi: 10.1161/CIRCRESAHA.119.315999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shechter D., Dormann H.L., Allis C.D., et al. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 22.Sutendra G., Kinnaird A., Dromparis P., et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 23.Ankney J.A., Muneer A., Chen X. Relative and absolute quantitation in mass spectrometry–based proteomics. Annu Rev Anal Chem. 2018;11:49–77. doi: 10.1146/annurev-anchem-061516-045357. [DOI] [PubMed] [Google Scholar]
- 24.Wang X.H., Xu S., Zhou X.Y., et al. Low chorionic villous succinate accumulation associates with recurrent spontaneous abortion risk. Nat Commun. 2021;12:3428. doi: 10.1038/s41467-021-23827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Yao C.F., Xu F.J., et al. APC/C(CDH1) synchronizes ribose-5-phosphate levels and DNA synthesis to cell cycle progression. Nat Commun. 2019;10:2502. doi: 10.1038/s41467-019-10375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palomer X., Román-Azcona M.S., Pizarro-Delgado J., et al. SIRT3-mediated inhibition of FOS through histone H3 deacetylation prevents cardiac fibrosis and inflammation. Signal Transduct Target Ther. 2020;5(1):14. doi: 10.1038/s41392-020-0114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilić P., Guillemin N., Kovačević A., et al. Serum proteome profiling in canine idiopathic dilated cardiomyopathy using TMT-based quantitative proteomics approach. J Proteomics. 2018;15(179):110–121. doi: 10.1016/j.jprot.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh A.K. P300 in cardiac development and accelerated cardiac aging. Aging Dis. 2020;11:916–926. doi: 10.14336/AD.2020.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenantin A.C., Jebeniani I., Leschik J., et al. Targeting the histone demethylase LSD1 prevents cardiomyopathy in a mouse model of laminopathy. J Clin Invest. 2021;131 doi: 10.1172/JCI136488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiano C., Benincasa G., Franzese M., et al. Epigenetic-sensitive pathways in personalized therapy of major cardiovascular diseases. Pharmacol Ther. 2020;210:107514. doi: 10.1016/j.pharmthera.2020.107514. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y., Chen W., Lian J., et al. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ. 2020;27:695–710. doi: 10.1038/s41418-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Tu K., Liu D., et al. p300 acetyltransferase is a cytoplasm-to-nucleus shuttle for SMAD2/3 and TAZ nuclear transport in transforming growth factor β-stimulated hepatic stellate cells. Hepatology. 2019;70:1409–1423. doi: 10.1002/hep.30668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kane A.E., Sinclair D.A. Sirtuins and NAD+ in the development and treatment of metabolic and cardiovascular diseases. Circ Res. 2018;123:868–885. doi: 10.1161/CIRCRESAHA.118.312498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C.F., Chavez J.D., Garcia-Menendez L., et al. Normalization of NAD+ redox balance as a therapy for heart failure. Circulation. 2016;134:883–994. doi: 10.1161/CIRCULATIONAHA.116.022495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Ren J. Epigenetics and obesity cardiomyopathy: from pathophysiology to prevention and management. Pharmacol Ther. 2016;161:52–66. doi: 10.1016/j.pharmthera.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Bagul P.K., Deepthi N., Sultana R., Banerjee S.K. Resveratrol ameliorates cardiac oxidative stress in diabetes through deacetylation of NFkB-p65 and histone 3. J Nutr Biochem. 2015;26:1298–1307. doi: 10.1016/j.jnutbio.2015.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.