Abstract
The SARS-CoV-2 Omicron variant has evolved into four sub-lineages—BA.1, BA.1.1, BA.2, and BA.3—with BA.2 becoming dominant worldwide. We and others have reported antibody evasion of BA.1 and BA.2, but side-by-side comparisons of Omicron sub-lineages to vaccine-elicited or monoclonal antibody (mAb)-mediated neutralization are necessary. Using VSV-based pseudovirus, we report that sera from individuals vaccinated by two doses of an inactivated whole-virion vaccine shows weak to no neutralization activity, while homologous or heterologous boosters markedly improve neutralization titers against all Omicron sub-lineages. We also present neutralization profiles against a 20 mAb panel, including 10 authorized or approved, against the Omicron sub-lineages, along with mAb mapping against single or combinatorial spike mutations. Most mAbs lost neutralizing activity, while some demonstrate distinct neutralization patterns among Omicron sub-lineages, reflecting antigenic differences. Collectively, our results suggest the Omicron sub-lineages threaten the neutralization efficacy of current vaccines and antibody therapeutics, highlighting the importance of vaccine boosters.
Keywords: SARS-CoV-2, Omicron, BA.1, BA.1.1, BA.2, BA.3, antibody, vaccine, booster, BBIBP-CorV
Graphical abstract
Ai et al. report side-by-side susceptibility profiles of Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages to BBIBP-CorV vaccine-elicited neutralization, finding substantial evasion against two-dose, but not boosted, vaccinee sera. Single and combinatorial mutational analysis maps key residues along Omicron Spike that help facilitate evasion against monoclonal antibody-mediated neutralization.
Main text
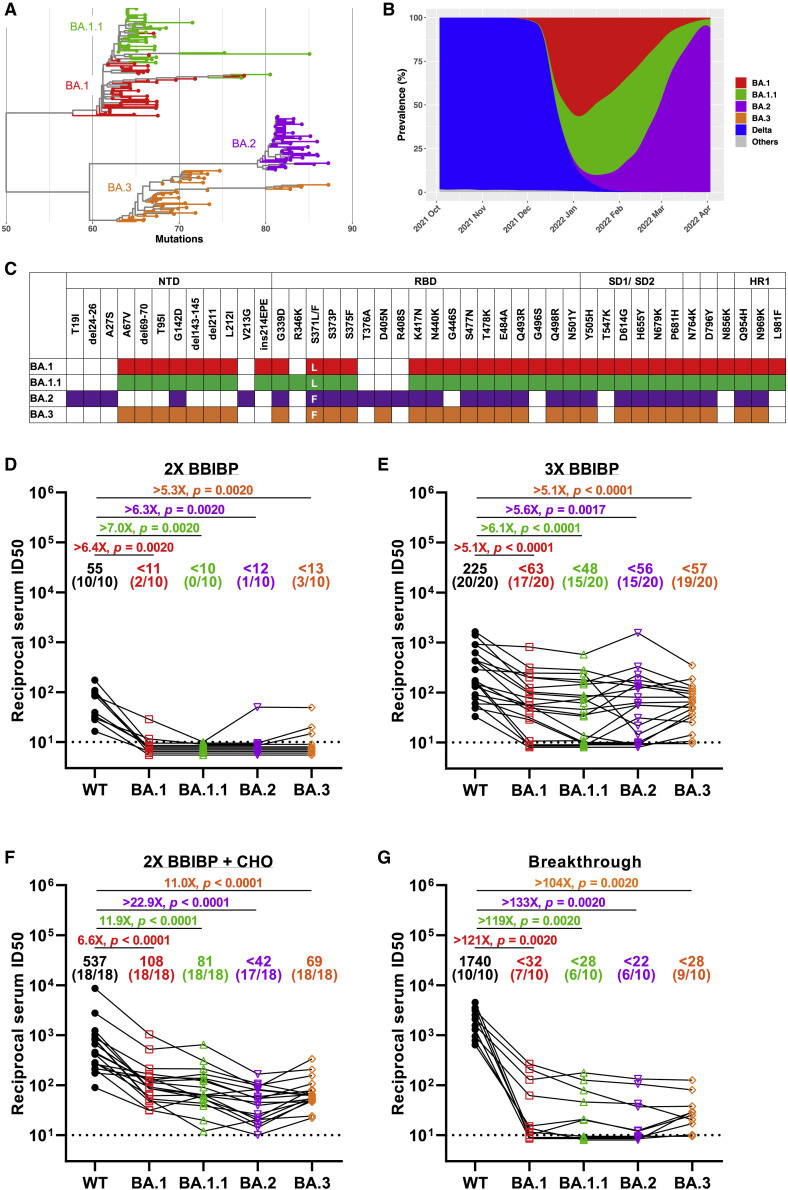
The World Health Organization has now designated five variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as variants of concern, including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529). The Omicron variant has recently been divided into four sub-lineages: BA.1, BA.1.1, BA.2, and BA.3 (Figure 1A). The original Omicron (BA.1 sub-lineage) was first identified in Botswana and South Africa in November 2021 (Viana et al., 2022) and together with its derivative BA.1.1 (containing an extra spike R346K mutation) became dominant worldwide in replacement of Delta over the span of a few weeks. But subsequently, we saw a rapid surge in the proportion of BA.2, and this sub-lineage became the dominant variant globally. Compared with the BA.1 and BA.2 sub-lineages, the prevalence of BA.3 sub-lineage is currently very low (Figure 1B).
Figure 1.
Characteristics and sera neutralization of the Omicron sub-lineages
(A) Phylogenetic tree of the BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Fifty randomly selected sequences belonging to each of the Omicron sub-lineages from GISAID were used as query sequences.
(B) Prevalence of the Omicron sub-lineages and Delta variant based on all the sequences available on GISAID over the past 6 months.
(C) Spike mutations within the Omicron sub-lineages.
(D–G) Neutralization of pseudotyped WT (D614G) and Omicron sub-lineage viruses by sera collected from individuals vaccinated with two-dose BBIBP-CorV only (D), with a BBIBP-CorV homologous booster (E), or with a ZF001 heterologous booster dose (F) following two doses of BBIBP-CorV, or from individuals infected by Delta virus after vaccination (G). For all panels, values above the symbols denote geometric mean titer, and the numbers in parentheses denote the proportion of positive sera with ID50 above the LOQ (dotted lines, >1:10). p values were determined by using a Wilcoxon matched-pairs signed-rank test (two-tailed).
See also Figures S1 and S2.
BA.1, BA.2, and BA.3 have numerous mutations in common but also have distinct sets of mutations in their spike that can differentiate these sub-lineages (Figure 1C). Although the selective advantage of BA.2 could be partially explained by its higher transmissibility than BA.1 (Lyngse et al., 2022), their relative immune evasion property could also be counted. We (Ai et al., 2021; Wang et al., 2022) and others (Cameroni et al., 2022; Cao et al., 2022; Carreño et al., 2022; Cele et al., 2022; Garcia-Beltran et al., 2022; Liu et al., 2022; Planas et al., 2022; VanBlargan et al., 2022) have reported that BA.1 demonstrated considerable escape from neutralization by monoclonal antibodies (mAbs) and sera from vaccinated individuals. BA.2 has also been reported to severely dampen antibody neutralization (Bowen et al., 2022; Iketani et al., 2022). However, evaluation and comparison of susceptibility of all the major Omicron sub-lineages to vaccine-elicited or mAb-mediated neutralization are urgently needed. In this study, therefore, we constructed the Omicron sub-lineage pseudoviruses (PsVs) and compared side by side their neutralization sensitivity to vaccinee sera as well as a panel of mAbs.
The first question we asked for the Omicron sub-lineages is their extent of immune evasion of polyclonal antibody neutralizing activity elicited in humans after vaccination or infection. To answer this, we first assessed the neutralizing activity of sera from individuals vaccinated by two doses of inactivated whole-virion vaccines (BBIBP-CorV) (Table S1). Similar to what we reported before (Wang et al., 2022), although all the sera showed neutralization activity against wild-type (WT) virus, the activity was relatively weak, with geometric mean neutralizing titers (GMTs) about 55, and when it turned to BA.1, only two of 10 vaccinees showed marginal neutralization. When we tested these sera on the three other sub-lineages, most of them showed no detectable activity, except a few had very weak neutralization against BA.2 and BA.3 (Figures 1D and S1A). These results indicate that two-dose inactivated vaccine is inadequate to provide full protection against these newly emerging Omicron variants.
Our previous study showed that a booster shot, either homologous or heterologous, can reduce Omicron BA.1 escape from neutralizing antibodies (Wang et al., 2022). To see if this is the case for the other Omicron sub-lineages, we then collected and tested 20 samples from healthy adults who had a third boosting vaccination shot with the same BBIBP-CorV vaccine (homologous booster group, Table S1). As shown in Figures 1E and S1B, the sera had a neutralizing GMT against WT of 225 with 5- to 6-fold reduction against BA.1, BA.1.1, BA.2, and BA.3 but at least 15 of 20 samples exhibiting detectable neutralizing activity against all four sub-lineages. We also collected 18 sera from individuals that received two doses of BBIBP-CorV followed by a protein subunit vaccine (ZF2001) 4–8 months later (heterologous booster group, Table S1). This cohort had higher neutralizing titers with GMTs of 537, 108, 81, 42, and 69 against WT, BA.1, BA.1.1, BA.2, and BA.3, respectively. Although these numbers amount to 7- to 23-fold reductions of potency comparing Omicron sub-lineages to WT, almost all samples maintained detectable neutralizing activity against the Omicron variants (Figures 1F and S1C). The marked improvement in serum neutralization from individuals who received a booster dose over those who did not highlights the value of vaccine boosters for eliciting neutralizing antibody responses against Omicron sub-lineages.
The emergence of the SARS-CoV-2 Delta variant led to an increasing number of breakthrough infection cases. To gain insight into their chance of re-infection by Omicron, we recruited 10 participants who were immunized with two-dose inactivated vaccines before being infected by the Delta variant (Table S1). Serum samples were obtained from them after 3–4 months of breakthrough infection and evaluated on WT and the four Omicron sub-lineage PsVs (Figures 1G and S1D). We found that breakthrough infection by Delta boosted the neutralizing antibody titers significantly to very high levels against WT virus (GMT = 1,740). However, the neutralization titers for Omicron sub-lineages were significantly reduced, more than 100-fold in comparison to WT. The reduction level was much higher than that of the homologous and heterologous vaccine booster groups, which may be associated with the antigenic difference between Delta and Omicron variants.
Taking into account of all the serum samples, we also carried out a comparison between the original Omicron BA.1 and the newly emerging sub-lineages to see if there are inherent difference regarding their immune evasion properties. BA.1.1, with an additional R346K mutation on top of BA.1, showed slightly but statistically significant lower titers than BA.1. For BA.2 and BA.3, the neutralization titers were also lower than BA.1, which was mostly contributed by the heterologous booster group, indicating the receptor binding domain (RBD) subunit vaccine booster may induce some RBD-directed antibodies which could be evaded by the BA.2/BA.3 unique mutations (Figure S2).
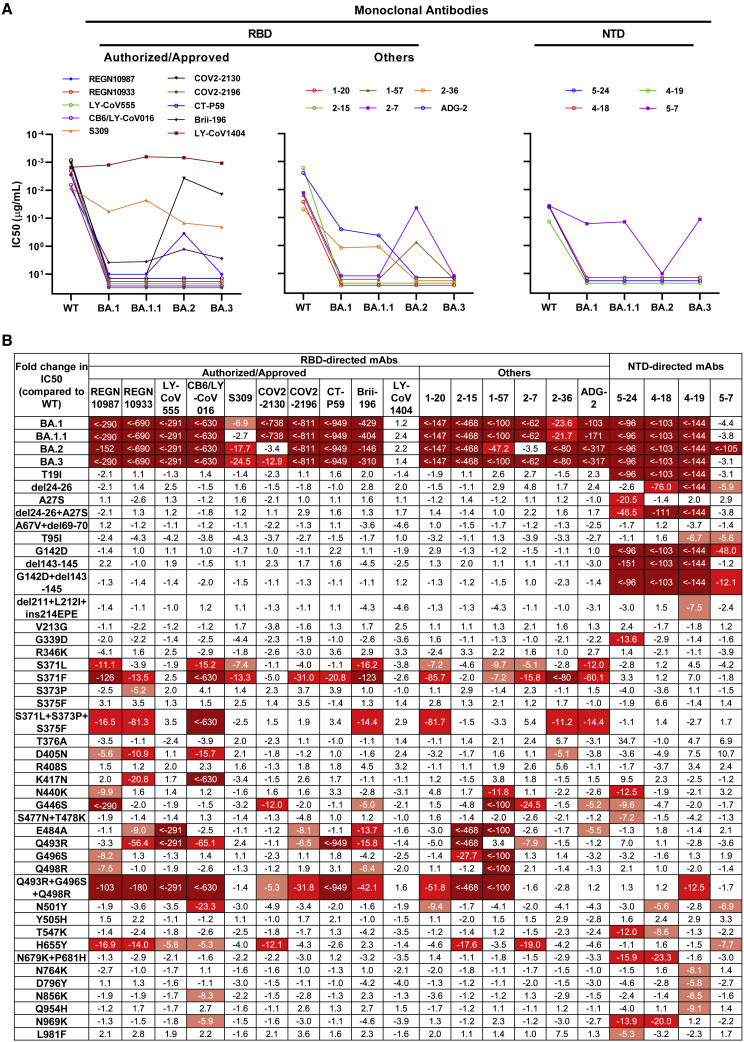
To better understand these differences and examine which types of antibodies in serum lose their activity against these Omicron sub-lineages, we further evaluated the neutralization profile of a panel of 20 mAbs targeting SARS-CoV-2 spike. These included 10 authorized or approved mAbs with sequences available: REGN10987 (imdevimab) (Hansen et al., 2020), REGN10933 (casirivimab) (Hansen et al., 2020), LY-CoV555 (bamlanivimab) (Jones et al., 2021), CB6/LY-CoV016 (etesevimab) (Shi et al., 2020), S309 (sotrovimab) (Pinto et al., 2020), COV2-2130 (cilgavimab) (Zost et al., 2020), COV2-2196 (tixagevimab) (Zost et al., 2020), CT-P59 (regdanvimab) (Kim et al., 2021), Brii-196 (amubarvimab) (Ju et al., 2020), and LY-CoV1404 (bebtelovimab) (Westendorf et al., 2022), all of which are directed to RBD. We also included some other RBD-directed mAbs of interest, including 1–20, 2–15, 1–57, 2–7 (Liu et al., 2020); 2–36 (Liu et al., 2020; Wang et al., 2021) from our own collection and ADG-2 (Rappazzo et al., 2021) from Adagio Therapeutics; and four more NTD-directed mAbs, including 5–24, 4–18, 4–19 (Cerutti et al., 2021b; Liu et al., 2020), and 5–7 (Cerutti et al., 2021a; Liu et al., 2020). Overall, all four Omicron sub-lineages had severe impact on most of the antibodies, but they also showed important differences in neutralization patterns. Among the authorized or approved mAbs, seven were either totally inactive or severely impaired in neutralizing all four sub-lineages. S309, the only approved antibody found to retain its neutralizing activity against the original form of Omicron in our previous study (Wang et al., 2022), lost more neutralizing activity against BA.2 and BA.3. COV2-2130 completely lost its neutralizing activity against BA.1 and BA.1.1 while remaining largely active against BA.2 and BA.3. Luckily, LY-CoV1404, which has been granted emergency use authorization very recently, remained potent in neutralizing all Omicron sub-lineages, continuing to broaden its coverage of SARS-CoV-2 variants (Zhou et al., 2022). For the other RBD- or NTD-directed mAbs, none of them retained full neutralizing activity against all of the Omicron sub-lineages. Two class 4 antibodies, ADG-2 and 2–36, retained decent activity against BA.1 and BA.1.1 but lost their neutralizing activity completely against BA.2 and BA.3. Interestingly, 2-7, one of our class 3 antibodies, completely lost its neutralizing activity against BA.1, BA.1.1, and BA.3 while remaining largely active against BA.2. A similar pattern was seen for another approved class 3 antibody, REGN10987. On the contrary, we found that the activity of 5-7, the non-supersite-directed NTD antibody, was partially retained against BA.1, BA.1.1, and BA.3 but totally abolished against BA.2 (Figures 2A and S3 ).
Figure 2.
Neutralization of pseudotyped WT (D614G) and Omicron sub-lineage viruses by mAbs targeting different epitopes
(A) Changes in neutralization IC50 of select RBD and NTD mAbs against Omicron sub-lineage pseudoviruses.
(B) Fold increase or decrease in neutralization IC50 of mAbs against Omicron sub-lineage as well as single- and combinatorial-spike mutation pseudoviruses, relative to WT, presented as a heatmap with darker colors implying greater change.
See also Figure S3.
To dissect the key mutations conferring antibody resistance and the specific mutations leading to the different neutralization patterns of Omicron sub-lineages, we constructed PsVs with each of the single-spike mutations alone or in combination, if they are spatially close, and tested them using the same panel of 20 mAbs. In total, 40 specific mutation viruses were tested, and their comprehensive neutralization profiles by these 20 mAbs are summarized in Figure 2B as fold change in 50% inhibitory concentration (IC50) relative to WT virus. For mutations affecting antibody activity, the first ones that caught our attention were S371L and S371F. Both broadly affected most of the RBD-directed mAbs, with S371F having a greater negative impact. Intriguingly, when we tested S371L, S373P, and S375F in combination, as they form a loop adjacent to a lipid-binding pocket (Dejnirattisai et al., 2022), we indeed observed synergistic effect of the triple serine mutations in the reduction of neutralization potency of some mAbs. Q493R appears to be another key mutation responsible for the loss in potency of many RBD antibodies, and again, when it was tested in combination with G496S and Q498R, apparent synergistic effect was seen for some mAbs (Table S2). G446S, which is lacking in BA.2 but presented in the other sub-lineages, may explain why COV2-2130 and 2-7 are not much affected by BA.2. Other mutations, such as D405N, K417N, N440K, E484A, and N501Y, distinctly affected the activity of different RBD-directed mAbs, most of which could be explained by the mutations falling into the antibody epitopes. For LY-CoV1404, as we saw for the Omicron sub-lineages, none of the single mutations significantly affected its neutralization potency, indicating that despite the constellation of spike mutations present in these viruses, there is still a patch within LY-CoV1404’s binding region that is not affected. For the NTD-directed mAbs, it was mostly the mutations falling into the NTD of the spike, including T19I, del24-26+A27S, and G142D+del142-145, that are responsible for the neutralization activity loss, as expected.
The SARS-CoV-2 Omicron variant immediately raised alarms after its identification, and the scenario seems to getting worse, with the emerging Omicron sub-lineages, like BA.2, which has been reported to be inherently substantially more transmissible than BA.1 (Lyngse et al., 2022). Many research articles have been published studying the original Omicron BA.1 virus, but less is known about the BA.2 and other sub-lineages. Here in this study, we constructed all the major Omicron sub-lineage viruses to date—BA.1, BA.1.1, BA.2, and BA.3—and investigated their antibody evasion property in parallel.
We previously reported the markedly reduced neutralizing activity against BA.1 of convalescent or BBIBP-CorV vaccination sera (Ai et al., 2021; Wang et al., 2022). Here, we showed that all polyclonal sera also had a substantial loss in neutralizing activity against the other Omicron sub-lineages, with drops comparable to or even greater than that of BA.1, indicating that all these sub-lineages have a very far antigenic distance from the WT virus. Our results are quite comparable to studies on the mRNA vaccines (Iketani et al., 2022; Yu et al., 2022), showing that neutralizing antibody titer against BA.2 was similar to or lower than that against BA.1. Based on these, we suggest that the selective advantage of BA.2 over BA.1 should be mainly contributed by its higher transmissibility rather than by immune evasion. On the other hand, we showed that a third homologous inactivated vaccine booster or a heterologous protein subunit vaccine booster could significantly elevate neutralization titer against BA.1 (Wang et al., 2022). This is also true for the other Omicron sub-lineages. Most recently, three recombinant lineages (XD, XE, and XF) have been reported (UK_Health_Security_Agency, 2022), but their antibody evasion should not be significantly different from the Omicron sub-lineages studied here, since their spikes are identical to either BA.1 or BA.2. Therefore, promotion and popularization of vaccine booster injection is still an effective means to prevent SARS-CoV-2 transmission.
We also investigated the immune evasion capacity of Omicron sub-lineages with mAbs. Similar to what we reported for BA.1 (Wang et al., 2022), most mAbs lost their neutralizing activity against BA.1.1, BA.2, and BA.3 completely or substantially. But we did observe some distinct neutralization patterns for certain mAbs among these sub-lineages, reflecting their different mutations. For example, S309 and 5-7, targeting some unique sites in RBD (Pinto et al., 2020) or NTD (Cerutti et al., 2021a), were the two mAbs reported to retain largely activity against BA.1 (Liu et al., 2020; Wang et al., 2022), but their activity was further abolished by BA.2. On the contrary, some mAbs such as COV2-2130 and 2-7 totally lost activity against BA.1 but regained activity against BA.2. The good news is that LY-CoV1404 or bebtelovimab kept its potent neutralization activity against all Omicron sub-lineages and other major SARS-CoV-2 variants (Westendorf et al., 2022; Zhou et al., 2022). Our data are in good consistency with others (Iketani et al., 2022; Liu et al., 2022) regarding the mAb neutralization profile of the Omicron sub-lineages and single mutations, but we had more sub-lineage—BA.3—and combined some mutations in proximity to investigate their synergistic actions.
Although LY-CoV1404 currently remains as our hope for SARS-CoV-2 therapeutic antibodies, resistance may arise sometime if it is administered as mono-therapy for a prolonged period, given the error-prone property of RNA virus. Therefore, it is advisable to develop more potent and broad neutralizing antibodies to be administered as a cocktail to contain this ever-evolving pathogen. Meanwhile, vaccine boosters, either homologous or heterologous, could elicit neutralizing antibodies that help reduce the viral escape and should be pushed forward.
Limitations of the study
Limitations of the study included inaccessibility to mRNA or adenovirus-based vaccines, which have been widely used in the world, and we have not tested the effect of interval between the first two doses on vaccine-elicited humoral responses (Chatterjee et al., 2022; Tauzin et al., 2022). We only focused on neutralization activities without touching any other aspects of vaccine efficacy, like Fc-effector functions and cellular responses, which have been reported to play a role in protection from severe outcome (Carazo et al., 2021; Tauzin et al., 2021).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 2-36 |
Liu et al. (2020) Wang et al. (2021) |
N/A |
| 2-15 | Liu et al. (2020) | N/A |
| 2-7 | Liu et al. (2020) | N/A |
| 1-57 | Liu et al. (2020) | N/A |
| 1-20 | Liu et al. (2020) | N/A |
| 4-18 | Liu et al. (2020) | N/A |
| 5-24 | Liu et al. (2020) | N/A |
| 4-19 | Liu et al. (2020) | N/A |
| 5-7 | Liu et al. (2020) | N/A |
| REGN10987 | Hansen et al. (2020) | N/A |
| REGN10933 | Hansen et al. (2020) | N/A |
| LY-CoV555 | Jones et al. (2021) | N/A |
| CB6 | Shi et al. (2020) | N/A |
| CT-P59 | Kim et al. (2021) | N/A |
| S309 | Pinto et al. (2020) | N/A |
| COV2-2130 | Zost et al. (2020) | N/A |
| COV2-2196 | Zost et al. (2020) | N/A |
| Brii-196 | Ju et al. (2020) | N/A |
| LY-CoV1404 | Westendorf et al. (2022) | N/A |
| ADG-2 | Rappazzo et al. (2021) | N/A |
| Bacterial and virus strains | ||
| VSV-G pseudo-typed ΔG-luciferase | Kerafast | Cat# EH1020-PM |
| Biological samples | ||
| Convalescent human plasma samples | Huashan Hospital, Fudan University | N/A |
| Serum samples from BBIBP-CorV Vaccine | Huashan Hospital, Fudan University | N/A |
| Serum samples from ZF2001 Vaccine | Huashan Hospital, Fudan University | N/A |
| Critical commercial assays | ||
| Luciferase Assay System | Beyotime | Cat# RG066M |
| Experimental models: Cell lines | ||
| Vero E6 | ATCC | Cat# CRL-1586 |
| Expi293F | ThermoFisher | Cat# A14527 |
| I1 mouse hybridoma | ATCC | Cat# CRL-2700 |
| Recombinant DNA | ||
| pCMV3-SARS-CoV-2-spike D614G | Wang et al. (2021) | N/A |
| FC4122-SARS-CoV-2-spike BA.1 | Wang et al. (2022) | N/A |
| FC4122-SARS-CoV-2-spike BA.1.1 | This study | N/A |
| pCMV3-SARS-CoV-2-spike BA.2 | This study | N/A |
| pCMV3-SARS-CoV-2-spike BA.3 | This study | N/A |
| pCMV3-SARS-CoV-2-spike T19I | This study | N/A |
| pCMV3-SARS-CoV-2-spike del24-26 | This study | N/A |
| pCMV3-SARS-CoV-2-spike A27S | This study | N/A |
| pCMV3-SARS-CoV-2-spike del24-26+A27S | This study | N/A |
| pCMV3-SARS-CoV-2-spike A67V+del69-70 | This study | N/A |
| pCMV3-SARS-CoV-2-spike T95I | This study | N/A |
| pCMV3-SARS-CoV-2-spike G142D | This study | N/A |
| pCMV3-SARS-CoV-2-spike del143-145 | This study | N/A |
| pCMV3-SARS-CoV-2-spike G142D+del143-145 | This study | N/A |
| pCMV3-SARS-CoV-2-spike del211+L212I+ins214EPE | This study | N/A |
| pCMV3-SARS-CoV-2-spike V213G | This study | N/A |
| pCMV3-SARS-CoV-2-spike G339D | This study | N/A |
| pCMV3-SARS-CoV-2-spike R346K | This study | N/A |
| pCMV3-SARS-CoV-2-spike S371L | This study | N/A |
| pCMV3-SARS-CoV-2-spike S371F | This study | N/A |
| pCMV3-SARS-CoV-2-spike S373P | This study | N/A |
| pCMV3-SARS-CoV-2-spike S375F | This study | N/A |
| pCMV3-SARS-CoV-2-spike S371L+S373P+S375F | This study | N/A |
| pCMV3-SARS-CoV-2-spike T376A | This study | N/A |
| pCMV3-SARS-CoV-2-spike D405N | This study | N/A |
| pCMV3-SARS-CoV-2-spike R408S | This study | N/A |
| pCMV3-SARS-CoV-2-spike K417N | This study | N/A |
| pCMV3-SARS-CoV-2-spike N440K | This study | N/A |
| pCMV3-SARS-CoV-2-spike G446S | This study | N/A |
| pCMV3-SARS-CoV-2-spike S477N+T478K | This study | N/A |
| pCMV3-SARS-CoV-2-spike E484A | This study | N/A |
| pCMV3-SARS-CoV-2-spike Q493R | This study | N/A |
| pCMV3-SARS-CoV-2-spike G496S | This study | N/A |
| pCMV3-SARS-CoV-2-spike Q498R | This study | N/A |
| pCMV3-SARS-CoV-2-spike Q493R+G496S+Q498R | This study | N/A |
| pCMV3-SARS-CoV-2-spike N501Y | This study | N/A |
| pCMV3-SARS-CoV-2-spike Y505H | This study | N/A |
| pCMV3-SARS-CoV-2-spike T547K | This study | N/A |
| pCMV3-SARS-CoV-2-spike H655Y | This study | N/A |
| pCMV3-SARS-CoV-2-spike N679K+P681H | This study | N/A |
| pCMV3-SARS-CoV-2-spike N764K | This study | N/A |
| pCMV3-SARS-CoV-2-spike D796Y | This study | N/A |
| pCMV3-SARS-CoV-2-spike N856K | This study | N/A |
| pCMV3-SARS-CoV-2-spike Q954H | This study | N/A |
| pCMV3-SARS-CoV-2-spike N969K | This study | N/A |
| pCMV3-SARS-CoV-2-spike L981F | This study | N/A |
| Software and algorithms | ||
| GraphPad Prism Software | GraphPad Prism Software, Inc. | N/A |
| MEGA 11 | Tamura et al. (2021) | https://www.megasoftware.net/ |
| CLUSTAL W | Thompson et al. (1994) | N/A |
| Ggtree | Yu et al. (2017) | https://bioconductor.org/packages/release/bioc/html/ggtree.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pengfei Wang (pengfei_wang@fudan.edu.cn).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Experimental model and subject details
Serum samples
Peripheral blood samples were collected from individuals who received two or three doses of BBIBP-CorV or ZF2001 vaccine were collected at Huashan Hospital, Fudan University 14 days after the final dose. Blood samples were also obtained from patients after 3–4 months of SARS-CoV-2 breakthrough infection caused by Delta variant after immunizing with two-dose inactivated vaccines (CoronaVac). Sera were isolated from centrifuged blood samples and then stored at −80°C. All collections were conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Ethics Committee of Huashan Hospital (2020-688, 2021-041 and 2021-749). All the participants provided written informed consents.
Cell lines
Expi293F cells (Thermo Fisher Cat# A14527) were cultured in the serum free SMM 293-TI medium (Sino Biological Inc.) at 37°C with 8% CO2 on an orbital shaker platform. Vero E6 cells (cat# CRL-1586) were from ATCC and cultured in 10% Fetal Bovine Serum (FBS, GIBCO cat# 16140071) supplemented Dulbecco’s Modified Eagle Medium (DMEM, ATCC cat# 30-2002) at 37°C, 5% CO2. I1 mouse hybridoma cells (ATCC, cat# CRL-2700) were cultured in Eagle’s Minimum Essential Medium (EMEM, ATCC cat# 30-2003)) with 20% FBS.
Method details
Monoclonal antibodies
Monoclonal antibodies tested in this study were constructed and produced at Fudan University. For each antibody, variable genes were codon optimized for human cell expression and synthesized by HuaGene™ (Shanghai, China) into plasmids (gWiz or pcDNA3.4) that encode the constant region of human IgG1 heavy or light chain. Antibodies were expressed in Expi293F (ThermoFisher, A14527) by co-transfection of heavy and light chain expressing plasmids using polyethylenimine and cells were cultured at 37 °C with shaking at 125 rpm and 8% CO2. On day 5, antibodies were purified using MabSelect™ PrismA (Cytiva, 17549801) affinity chromatography.
Construction and production of variant pseudoviruses
Plasmids encoding the WT (D614G) SARS-CoV-2 spike and Omicron sub-lineage spikes, as well as the spikes with single or combined mutations were synthesized. Expi293F cells were grown to 3×106/mL before transfection with the indicated spike gene using Polyethylenimine (Polyscience). Cells were cultured overnight at 37°C with 8% CO2 and VSV-G pseudo-typed ΔG-luciferase (G∗ΔG-luciferase, Kerafast) was used to infect the cells in DMEM at a multiplicity of infection of 5 for 4 h before washing the cells with 1×DPBS three times. The next day, the transfection supernatant was collected and clarified by centrifugation at 300g for 10 min. Each viral stock was then incubated with 20% I1 hybridoma (anti-VSV-G; ATCC, CRL-2700) supernatant for 1 h at 37°C to neutralize the contaminating VSV-G pseudotyped ΔG-luciferase virus before measuring titers and making aliquots to be stored at −80°C.
Pseudovirus neutralization assays
Neutralization assays were performed by incubating pseudoviruses with serial dilutions of monoclonal antibodies or sera, and scored by the reduction in luciferase gene expression. In brief, Vero E6 cells were seeded in a 96-well plate at a concentration of 2×104 cells per well. Pseudoviruses were incubated the next day with serial dilutions of the test samples in triplicate for 30 min at 37°C. The mixture was added to cultured cells and incubated for an additional 24 h. The luminescence was measured by Luciferase Assay System (Beyotime). IC50 was defined as the dilution at which the relative light units were reduced by 50% compared with the virus control wells (virus + cells) after subtraction of the background in the control groups with cells only. The IC50 values were calculated using nonlinear regression in GraphPad Prism.
Sequence alignment and phylogenetic tree
This analysis involved 200 nucleotide sequences, including 50 samples for each lineage randomly selected from the GISAID database. Sequence alignment was carried out using ClustalW progress (Thompson et al., 1994) and corrected manually. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The optimal tree is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method (Nei and Kumar, 2001) and are in the units of the number of base differences per site. All positions with less than 50% site coverage were eliminated. There was a total of 29743 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021) and visualized with the package 'ggtree' in R. The current snapshot of COVID-19 data was taken from GISAID between Oct 2021 and Mar 2022 in weekly basis. Lineage level prevalence rate was summarized using cubic spline interpolation.
Quantification and statistical analysis
The statistical analyses for the pseudovirus virus neutralization assessments were performed using GraphPad Prism for calculation of mean value and SEM for each data point. Each specimen was tested in triplicate. Antibody neutralization IC50 values were calculated using a five-parameter dose-response curve in GraphPad Prism. For comparing the serum neutralization titers, statistical analysis was performed using a Wilcoxon matched-pairs signed rank test. Two-tailed p values are reported. No statistical methods were used to determine whether the data met assumptions of the statistical approach.
Acknowledgments
We thank Dr. Li Jin (Fudan University) for his support and thoughtful discussions. This study was supported by funding from National Natural Science Foundation of China (82002141 to J.A.), Shanghai Science and Technology Committee (21Y11902100 to J.A.), Shanghai Youth Science and Technology Talents Sailing Project (20YF1404300 to J.A.), Shanghai Science and Technology Committee (20dz2260100, 20Z11901100, 20dz2210400 to W.Z.), and Key Discipline Construction Plan from Shanghai Municipal Health Commission (GWV-10.1-XK01 to W.Z.). This research was also partially supported by Shanghai Municipal Science and Technology Major Project (2017SHZDZX01) and the major project of Study on Pathogenesis and Epidemic Prevention Technology System (2021YFC2302500) by the Ministry of Science and Technology of the People's Republic of China.
Author contributions
P.W., W.Z., and Z.H. conceived and supervised the project. J.A., X.W., X.Z., Y.Z., Y.J., M.L., Y.Cui, Y.Chen, R.Q., L.L., and L.Y. conducted the biological experiments. X.H., Y.L., and Z.H. conducted the bioinformatics analysis. J.A., X.W., X.Z., Y.Z., Y.J., Z.H., W.Z., and P.W. analyzed the results and wrote the manuscript. All the authors reviewed, commented, and approved the manuscript.
Declaration of interests
P.W. is an inventor on patent applications on some of the antibodies described in this manuscript, including 1–20, 2–15, 1–57, 2–7, 2–36, 5–24, 4–18, 4–19, and 5–7. All other authors have no conflict of interest.
Published: May 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.05.001.
Supplemental information
Data and code availability
All the data are provided in the paper or the supplemental information. Lineage submission statistics is available at GISAID. The sequence of virus is available in the GISAID data base with the sample id listed at https://github.com/wenrurumon/GISAID/blob/main/2022.04.07.487489/map.csv, visualization code are available at plot.R in the same location.
References
- Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., Song J., Fu Z., et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2021;11:337–343. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.E., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., Franko N.M., Ahmed K., Shariq A., Cameroni E., Gori A., et al. Omicron BA.1 and BA.2 neutralizing activity elicited by a comprehensive panel of human vaccines. bioRxiv. 2022 doi: 10.1101/2022.03.15.484542. Preprint at. [DOI] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo S., Talbot D., Boulianne N., Brisson M., Gilca R., Deceuninck G., Brousseau N., Drolet M., Ouakki M., Sauvageau C., et al. Single-dose mRNA vaccine effectiveness against SARS-CoV-2 in healthcare workers extending 16 weeks post-vaccination: a test-negative design from Quebec, Canada. Clin. Infect. Dis. 2021:ciab739. doi: 10.1093/cid/ciab739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Wang P., Nair M.S., Wang M., Huang Y., Yu J., Liu L., Katsamba P.S., Bahna F., et al. Neutralizing antibody 5-7 defines a distinct site of vulnerability in SARS-CoV-2 spike N-terminal domain. Cell Rep. 2021;37:109928. doi: 10.1016/j.celrep.2021.109928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D., Tauzin A., Marchitto L., Gong S.Y., Boutin M., Bourassa C., Beaudoin-Bussieres G., Bo Y., Ding S., Laumaea A., et al. SARS-CoV-2 Omicron Spike recognition by plasma from individuals receiving BNT162b2 mRNA vaccination with a 16-week interval between doses. Cel Rep. 2022;38:110429. doi: 10.1016/j.celrep.2022.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradnik J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kim C., Ryu D.-K., Lee J., Kim Y.-I., Seo J.-M., Kim Y.-G., Jeong J.-H., Kim M., Kim J.-I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., Skov R.L., Krause T.G., Rasmussen M., Sieber R.N., et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. medRxiv. 2022 doi: 10.1101/2022.01.28.22270044. Preprint at. [DOI] [Google Scholar]
- Nei M., Kumar S. Molecular evolution and phylogenetics. Genet. Res. 2001;77:117–120. [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Gong S.Y., Beaudoin-Bussieres G., Vezina D., Gasser R., Nault L., Marchitto L., Benlarbi M., Chatterjee D., Nayrac M., et al. Strong humoral immune responses against SARS-CoV-2 Spike after BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Host Microbe. 2022;30:97–109.e5. doi: 10.1016/j.chom.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussieres G., Brassard N., Laumaea A., Vezina D., Prevost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150.e6. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK_Health_Security_Agency . UK Health Security Agency; 2022. SARS-CoV-2 Variants of Concern and Variants under Investigation in England: Technical Briefing 39. [Google Scholar]
- VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Yu J., Guo Y., Wang M., Chan J.F.W., Cerutti G., Iketani S., Liu L., et al. A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses. Emerg. Microbes Infect. 2021;11:147–157. doi: 10.1080/22221751.2021.2011623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao X., Song J., Wu J., Zhu Y., Li M., Cui Y., Chen Y., Yang L., Liu J., et al. Homologous or heterologous booster of inactivated vaccine reduces SARS-CoV-2 Omicron variant escape from neutralizing antibodies. Emerg. Microbes Infect. 2022;11:477–481. doi: 10.1080/22221751.2022.2030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf K., Wang L., Zentelis S., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. bioRxiv. 2022 doi: 10.1101/2021.04.30.442182. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Collier A.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 variants. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D., Zhu H., Guan Y., Lam T.T. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- Zhou T., Wang L., Misasi J., Pegu A., Zhang Y., Harris D.R., Olia A.S., Talana C.A., Yang E.S., Chen M., et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science. 2022;376:eabn8897. doi: 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schafer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are provided in the paper or the supplemental information. Lineage submission statistics is available at GISAID. The sequence of virus is available in the GISAID data base with the sample id listed at https://github.com/wenrurumon/GISAID/blob/main/2022.04.07.487489/map.csv, visualization code are available at plot.R in the same location.