Abstract
Objective:
Several studies have recently indicated a huge shifting pattern toward early age onset cases in breast cancer (BC) patients. However, the studies exerted relatively limited to the Caucasian population. This preliminary study is aimed to investigate the genetic risk factors for young BC patients specifically in Indonesia population.
Methods:
DNA samples were extracted from 79 BC patients aged younger than 40 years old and 90 healthy samples. These DNA samples were sequenced using Illumina NextSeq 500 platform and preprocessed to extract the single-nucleotide polymorphisms (SNPs) data. Firstly, multiple univariate logistic regressions were performed to test the association between each SNP and BC incidence in young patients. Furthermore, to analyze the polygenic effects derived from multiple SNPs, we employed a multivariate logistics regression.
Results:
There were only 15 SNPs passed our 95% call rate threshold thus subsequently were used in the association test. One of these variants, rs3219493, emerged to be significantly associated with early-onset BC (p-value = 0.025, OR = 3.750, 95% CI = 1.178-11.938). This result is consistent with the multivariate logistic regression model, where the pertinent variant was found statistically significant (p-value = 0.008, OR = 8.398, 95% CI = 1.720-40.920). This variant was identified as an intronic variant within MUTYH gene which has been reported in several published studies to exhibit an association with the incidence of breast cancer in China, Italy and Sephardi Jews population. However, there is no evident this gene impacting the risk of developing early onset of BC in Indonesia population.
Conclusion:
Despite our limitation in terms of sample size analyzed in this preliminary study, our finding on significant association of intronic MUTHY with the early onset of BC in Indonesia led to a broadened insight of population-based unique aspect to being taken into an in-depth account for and advancement of chemotherapy.
Key Words: Breast cancer, MUTYH, early onset, Indonesia
Introduction
Cancer is a manifestation of bizarre and uncontrolled proliferation of the different types of cell in the body (Visvader, 2011). There are hundreds of cancer types which behave and respond distinguishably toward the given treatments (Doll, 2018). In women, breast cancer (BC) is the top leading cause of cancer death globally although its incidences and mortality rates vary in each country as a reflection of economic and lifestyle factors (Curado, 2011; Bray et al., 2018). In Eastern Africa, the incidence rate of BC was relatively low with a 19.3 rate per 100,000 women population while in Western Europe the rate was observed at the level of 89.9, both in a 100,000 women population (Curado, 2011). China as the most populated country reported in 2012 a number of 187,213 BC cases with the mortality level reached about 25.63%. In contrary, South Korea in the same year reported only 13.27% mortal cases among 17,140 patients diagnosed with BC (Youlden et al., 2014). The incidence of BC was found to increase with age in several Asian countries. In Thailand, a higher risk of developing BC was observed in women aged 45-49 years old, whereas in China the similar risk was seen in an upper age group ranged from 50-84 years old. An interesting profile is exhibited by the BC incidence in India where those women aged 20-55 showed an increased risk of having BC yet the risk declined in the age group of 60-79 (Mubarik et al., 2019).
In 2018 alone the BC incidence in Indonesia reached a number of 58,256 new cases. It constituted 30.9% of all observed cancer cases. Of the number of new cases, a total of 22,692 (38.95%) was reported to be mortal (GLOBOCAN, 2019). According to a report by the Ministry of Health of Indonesia, these far-beyond-controlled BC incidences are caused by the poor access towards early screening and treatments. Early detection is a crucial part of the cancer prevention program (Muljo et al., 2017; Muljo et al., 2018). However, only up to 5% of Indonesian women are early-screened for BC compared to about 40% screened women in some developed countries (Muljo et al., 2019; Muljo et al., 2020). Most of the BC patients are diagnosed at an advanced stage which generally affects their responsiveness upon given regiments (Departemen Kesehatan Republik Indonesia, 2010; Solikhah et al., 2017; Mardela et al., 2017). This information is usually included in the National Cancer Registry (Pardamean et al., 2016; Pardamean et al., 2018). Other than these clinical access-related factors, survivability of BC patients is highly dependent to demographic variables for instances family history (Fagerholm et al., 2018), financial and marital status (Gomez et al., 2016; Martínez et al., 2017) and educational level (Bahk et al., 2017). Poor dietary habits and an unhealthy lifestyle are also deemed to be essential reasons for the increased BC incidence (Mubarik et al., 2019).
Although human disease-associated genetic variants identification demonstrated a huge advancement thus far, heredity-driven risks have yet to be elucidated. Hence, genome-wide studies are necessitated to allow an in-depth understanding of, particularly, less common alleles in a broad range of ancestry (The International HapMap 3 Consortium, 2010). The international HapMap project (International HapMap Consortium, 2003), the human genome project (Lander et al., 2001) and SNP consortium (Sachidanandam et al., 2001) were built to mainly map and characterize gigantic information provided within the DNA sequences. Approximately 10 million common DNA variants have been identified hitherto (The International HapMap 3 Consortium, 2010). However, they were limited to some well-represented population such as Asian, Europe and African (International HapMap Consortium, 2003).
Cancers are considered a manifestation of disadvantageous genome alteration and when the alterations are derived from ancestors this would worsen the condition. Among cancer subtypes that mostly are fatally dead, breast cancer (BC) emerges as the most frightening cancer for females although the most killer disease is heart diseases (P&T Snapshot, 2014). BC, in Indonesia, to most recent data accounts for 30.5% of all cancers diagnosed and 21.5% of cancer-related deaths among females. The increasing rates of mortality of the cancer are primarily attributable to the tardiness of getting an initial diagnosis (Bray et al., 2019).
Globally, BC incidence is associated with either deleterious BRCA1/2 mutations (Henouda et al., 2016). A study conducted at UT MD Anderson Cancer Center (MDACC) revealed that, in BRCA1/2 mutation carriers, breast cancer, other than ovarian cancer, the incidence was reported to be increasing in rates (Mersch et al., 2015). Separately elsewhere, Henouda et al., (2016) demonstrated 3 deleterious mutations as well as a large genomic rearrangement being that exon 2 deletion in BRCA 1 gene and 4 mutations in BRCA 2 gene with the novel mutation was discovered within Algerian population (Henouda et al., 2016).
Besides BRCA1/2 gene, MUTYH gene mutations were detected in breast cancer patients and high-risk groups in China and contributed to an increased risk of developing on male breast cancer in Italy (Jian et al., 2017; Rizzolo et al., 2018). Hitherto we figured out that it was very little information regarding the association of MUTYH mutation and BC incidences. One of which is a study conducted in the Sephardi Jews population where MUTYH variants observed were G396D and Y179C variants. Of these 2, only G396D variant that yielded significance to the augmented BC risk healthy controls (Rennert et al., 2012).
A pilot genome-wide association study (GWAS) in 89 Indonesia women with BC and 46 healthy women as referral population has been recently performed using 443,813-covering single nucleotide polymorphism (SNP) array 5.0 from Affymetrix. It was recognized 11 highly-suggestive chromosomes with BC risk, of those 4 loci were with identified genes: 2p.12 with the CTNNA2 gene; chromosome 18p11.2 with the SOGA2 gene; chromosome 5q14.1 with the SSBP2 gene and chromosome 9q31.1 with the TEX10 gene (Haryono et al., 2015). The study, however, did not exclusively plot the possible influence of hereditary aspect or family history the BC participants.
We, henceforward, were interested in evaluating if there is a particular mutation in the women with early-onset BC case. Therefore, we compared the genotypic-profile of two groups, women with early-onset breast cancer (age < 40 years old) and healthy women as a control group. The 40-years-old is our baseline age as suggested by a previously published meta-analysis study by Nelson et al., (2012) who concluded that within the range of 40 - 49 years old the first degree relative with BC was seen to be strongly associated with higher than 2-fold increase of BC risk. In this particular study, we employed the Next Generation Sequencing (NGS) approach through which we could anticipate novel BC-associated SNP findings. Significant factor(s) found in this study can be harnessed as a predictor to calculate early-onset breast cancer risk in women in Indonesia or, in a wider scope, South-East Asian population.
Materials and Methods
Study participants
A total of 79 breast cancer patients aged below 40 years old were recruited from Cipto Mangunkusumo Hospital, Jakarta, Indonesia (Panigoro et al., 2020). These participants (case) were subsequently matched with 90 healthy donors (control) recruited by Genetics Indonesia (GI) as a part of their Hereditary Cancer Screening (HCS) service. All participants have previously well written-consented and agreed to participate in this study. This study protocol was approved by the ethic committees of Faculty of Medicine, Universitas Indonesia with the reference number of 450/UN2.F1/ETIK/2018.
DNA Extraction
DNA samples were extracted from the whole blood of our participated BC patients and a buccal swab of our healthy donors. Prior to DNA purification with QIAmp DNA Mini Kit (Qiagen), the blood samples were centrifugated at 2500 x g at room temperature for 10 minutes to obtain the buffy coat. Separately, the buccal swab samples were ethanol-precipitated (prepIT-L2P Kit, DNA Genotek) prior to DNA isolation using OCR-100 Kit (DNA Genotek). DNA concentrations were gauged using the Qubit dsDNA HS Assay Kit (Invitrogen). We took a 10 ng/µl of DNA per sample for our sequencing.
Next-Generation Sequencing
Sequencing was performed on the Illumina NextSeq 500 platform with a High Output Kit v2.5 (300 Cycles). Library construction was conducted following the Hereditary Cancer Screening protocol by Kailos which consisted of steps elucidated as follows. The first step was targeting, where two parts oligos contained a region complementary to the DNA sequence outside the region to capture plus a recognition sequence for restriction enzyme to cut DNA in a specific location. The second step was patch ligation where the oligos, barcodes, and adapters were ligated onto the ends of the targeted region. The last step was amplification where the results of ligated regions were amplified to yield sufficient amount of amplicons necessitated by the sequencer.
Statistical Analysis
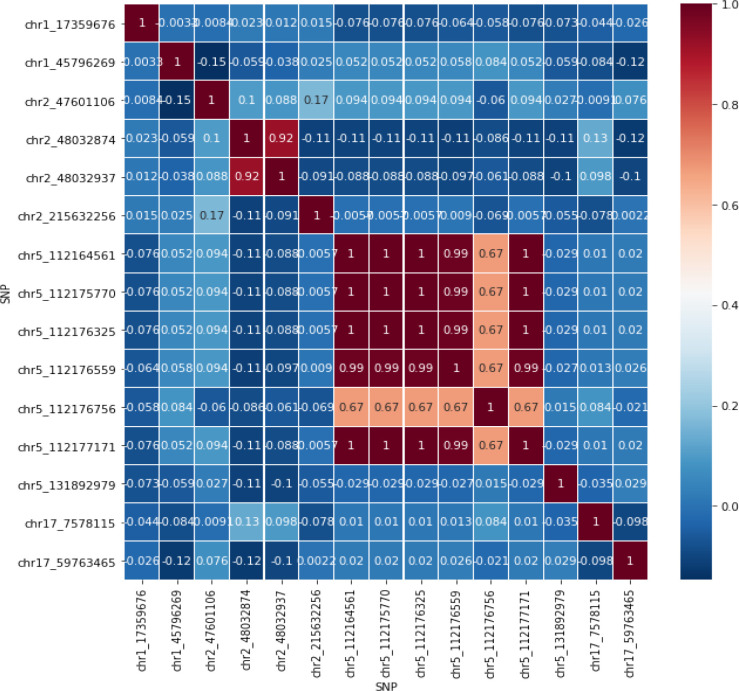
We filtered the SNPs in which we only used the SNPs that passed our 95% call rate. Next, based on these SNPs, we carried out two case-control association analyses using logistic regression. The first analysis was regressing each individual SNP with call rate > 95% between case and control group. In the second analysis, 95%-call rate-passing SNPs were tested in the same polynomial logistic regression model. To avoid singular matrix problem in polynomial regression, the redundant SNPs that share a similar proportion between samples were removed and only represented by only one SNP. The similarity of this proportion was measured with a correlation score. Those SNPs which correlate with other SNP that higher or lower than 0.7 and -0.7 and have a significant level below 0.05 were filtered.
The logistic regression was conducted in Python with Statsmodel library (Seabold, 2010). The significant SNPs or SNPs with a p-value less than 0.05 from the logistic regression were subsequently re-evaluated using fisher test whenever the SNPs comprised less than 5 samples in one of its segments.
Results
We successfully gathered 79 respondents of under 40 years old breast cancer patients and 90 samples of healthy respondent aged between 20 and 61 years old. The mean age at diagnosis in cases of breast cancer was 41.033 ± SD 8.703 years. DNA 260/280 ratio and breast cancer DNA concentration represented the quality control index of this study. Both of case and control DNA concentration are range between 80 ng/µl and 100 ng/µl.
The next-generation platform was employed to assess polymorphisms in the case and control samples. Eventually, our analysis yielded 15 autosomal SNPs which association of each to BC incidence was next evaluated via logistics regression analysis. The result of the univariate logistic regression analysis on the 15 SNPs can be found in Table 1.
Table 1.
Association of the 15 Autosomal Polymorphisms and BC Risk in Indonesian Women Population
| SNPs | Location | Gene | p-values | OR | CI 0.05 | CI 0.95 | n | Variants |
| rs1022580 | chr1:17359676 | SDHB | 0.555 | 0.787 | 0.356 | 1.742 | 163 | Intron |
| rs3219493 | chr1:45796269 | MUTYH | 0.025* | 3.75 | 1.178 | 11.938 | 163 | Intron |
| rs1126497 | chr2:47601106 | EPCAM | 0.193 | 1.558 | 0.799 | 3.037 | 163 | Coding sequence |
| rs751033188 | chr2:48032874 | MSH6 | 0.269 | 0.689 | 0.355 | 1.335 | 162 | Intron |
| rs3136359 | chr2:48032937 | MSH6 | 0.868 | 0.944 | 0.481 | 1.856 | 162 | Intron |
| rs386654966 | chr2:215632256 | BARD1 | 0.138 | 2.057 | 0.793 | 5.339 | 169 | Coding sequence |
| rs351771 | chr5:112164561 | APC | 0.806 | 0.918 | 0.464 | 1.816 | 163 | Synonymous |
| rs41115 | chr5:112175770 | APC | 0.806 | 0.918 | 0.464 | 1.816 | 163 | Synonymous |
| rs42427 | chr5:112176325 | APC | 0.806 | 0.918 | 0.464 | 1.816 | 163 | Synonymous |
| rs780061589 | chr5:112176559 | APC | 0.747 | 0.893 | 0.448 | 1.778 | 161 | Synonymous |
| rs459552 | chr5:112176756 | APC | 0.797 | 1.117 | 0.482 | 2.588 | 163 | Missense |
| rs587779801 | chr5:112177171 | APC | 0.806 | 0.918 | 0.464 | 1.816 | 163 | Missense & Synonymous |
| rs4526098 | chr5:131892979 | RAD50 | 0.77 | 1.31 | 0.213 | 8.051 | 168 | 5-Prime UTR |
| rs1625895 | chr17:7578115 | TP53 | 0.396 | 2.69 | 0.274 | 26.394 | 169 | Intron |
| rs4986765 | chr17:59763465 | BRIP1 | 0.916 | 1.043 | 0.475 | 2.289 | 163 | Coding sequence |
Only rs3219493 appeared as a significant factor from the logistic regression analysis. Since this SNP contain less than 5 samples in one of its segments, the SNP was analyzed using the Fisher Exact test. The cross table of rs3219493 between sample group can be found in Table 2. The p-value of the Fisher Exact test on rs3219493 cross table is 0.024.
Table 2.
Contingency Table of rs3219493
| Case | Control | |
|---|---|---|
| Monoallelic | 4 (5.06%) | 14 (16.67%) |
| Biallelic | 75 (94.94%) | 70 (83.33%) |
In the second analysis, we run a polynomial analysis to the 15 SNPs. However, similar predictors may cause a poor result. Therefore, SNPs that correlate higher or lower than ±0.6 with p-value smaller than 0.05 were represented with only one SNP. The correlation heatmap and polynomial logistic regression result can be seen in Figure 1 and Table 3.
Figure 1.
Correlation Heatmap
Table 3.
Polynomial Logistic Regression Result
| Location | p-values | coefficient | OR | OR CI 0.05 | OR CI 0.95 |
|---|---|---|---|---|---|
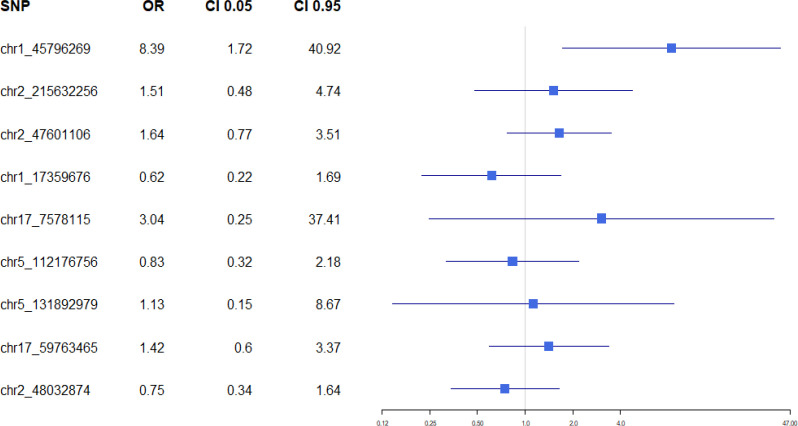
| chr1:45796269 | 0.008523 | 2.126925 | 8.389027 | 1.719838 | 40.92002 |
| chr2:215632256 | 0.478549 | 0.4131 | 1.511497 | 0.48217 | 4.738212 |
| chr2:47601106 | 0.199444 | 0.496889 | 1.6436 | 0.769436 | 3.51091 |
| chr1:17359676 | 0.344962 | -0.48597 | 0.615097 | 0.224352 | 1.686386 |
| chr17:7578115 | 0.384991 | 1.112375 | 3.041573 | 0.247273 | 37.41281 |
| chr5:112176756, | 0.710557 | -0.18196 | 0.833634 | 0.318872 | 2.179389 |
| chr5:112164561, | |||||
| chr5:112175770, | |||||
| chr5:112176325, | |||||
| chr5:112176559, | |||||
| chr5:112177171 | |||||
| chr5:131892979 | 0.90842 | 0.119757 | 1.127222 | 0.146502 | 8.673152 |
| chr17:59763465 | 0.430882 | 0.348143 | 1.416434 | 0.595634 | 3.368319 |
| chr2:48032874, | 0.46613 | -0.29196 | 0.746801 | 0.340572 | 1.637571 |
| chr2:48032937 |
To help visualize the result in Table 3, a forest plot of this polygenic analysis was generated and can be seen in Figure 2.
Figure 2.
Polygenic Analysis Forest Plot
Discussion
Both the univariate and multivariate analysis output the same significant variant associated with BC cases. This intronic variant MUTYH DNA glycosylase without strand mutation was found in all our BC patient cohort where, based on a coding DNA reference sequence (LRG_220), nucleotide guanine is substituted by cytosine. The mutation was detected only in chromosome 1 at position g.45796269 (rs3219493). Although an alternative position at g.45330597 has also been listed in https://www.ncbi.nlm.nih.gov/snp, we were unable to confirm it with our datasets. It suggests that the alternative form of the substitution is ethnical-associated. However, further assessment is necessary to confirm this speculation. Taking into account no previous data related with the risk of BC in Indonesian women with family history, this information emerges intriguing. Especially when, on the other hand, BRCA1/2, which, more globally, are known as highly-associated and are suggestive genes to BC incidence are insignificant to the cancer case.
Some previous studies have reported, MUTYH gene monoallelic and biallelic mutation variations escalated risk of developing cancer in the absence of the BRCA1/2 mutation (Nielsen et al., 2005; Vogt et al., 2009; Wasielewski et al., 2010; Rennert et al., 2012). However, on some research related to breast cancer in SNP mutation profiling, mutations of MUTYH and TP53 genes were found with a significant BRCA1/2 mutation frequently (Lin et al., 2016; Rummel et al., 2017; Nones et al., 2019; Ow et al., 2019). A study by Jian et al. (2017) regarding the genetic characteristics of breast cancer in the Chinese population showed mutations in the MUTYH gene in breast cancer cases and high-risk groups. MUTYH mutation variant rate of c.892-2A> G in high-risk women who have a family history of breast cancer higher than breast cancer cases based on East Asians in 1000G. Several cases of breast and colorectal cancer in Dutch families occurred an increase in the frequency of MUTYH mutations (Wasielewski et al., 2010). From 153 Dutch families, mutations have been observed withinside the MUTYH p.Tyr179Cys, p.Gly396Asp and p. Pro405Leu genotypes in breast and colorectal cancer cases. However, the correlation between MUTYH gene mutations and breast cancer still needs to be explored considering this gene also has a complex correlation with colorectal cancer, especially in polyposis.
Based on the KEGG pathway, the MUTYH gene plays a role in the DNA repair process, especially through Base Excision Repair. MUTYH as an early function gene that recognizes and removes the presence of mismatched, alkylated, and deaminated Long BER patches. Gene MUTYH is located in the short arm of chromosome 1 (1p34.1) and 11.2 kb long. It is a DNA glycosylase that is pivotally involved in fixing post-replicative mispairs (Kashfi et al., 2013). The effect of the MUTYH gene activates the DNA glycosylase enzyme to detect peculiar A: 8-oxoG and 2-OH-A: G mispairs on DNA helix (AP site) (Slupska et al., 1996; Ohtsubo et al., 2000; Gu, 2001). The mutation on the MUTYH gene possible to interference with the DNA repair process in breast cancer cases. DNA glycosylase mutations especially those influenced by the MUTYH gene were also found to affect the increase in colorectal cancer cases (Farrington et al., 2005). The ability of DNA glycosylase to remove bases, especially methylated ones, correlates with increasing age. According to age growth, the base excision repair process decreases due to a lack of the DNA glycosylase enzyme (Fernandes et al., 2017). This theory allows for the association of high-risk groups for breast cancer, which is generally middle-aged women (over 50 years). However, there are very few studies on the effect of variations in MUTYH mutations on age-related breast cancer. Even though several studies have shown mutations in MUTYH in some cancer cases, the study of this gene which is particularly related to the initiation of the carcinogenesis process or early onset of cancer still requires more massive data to validate the effect of this gene, especially in breast cancer (Kairupan and Scott, 2007).
In this preliminary study, we found out a statistically significant variant associated with early-onset breast cancer case, tested in both univariate and multivariate analysis. It is the intronic variant of the MUTYH gene in chromosome 1. MUTYH is associated with several cancers including breast cancer from other populations but, to our knowledge, there are no reports of the gene’s significance in the incidence of early onset of BC in Indonesia or South Asian countries thus far. Surprisingly, variants from BRCA1 and BRCA2 genes did not emerge as significant genetical aspect in our observed population. Despite our limitation in the sense of assayed sample size in our study, our results demonstrated a potential clue of the harnessing MUTYH gene as a novel target for chemotherapy, yet it seemed to be population restricted. However, there is still a long way to go to confirm our finding and thus in our future study, we are going to set as much clinical data as we can extract as our confounding variable in the model.
Author Contribution Statement
All authors contributed and approved the final manuscript. SSP and EL coordinated and supervised the clinical sampling. IN drafted and structured the manuscript. BM, AAH, AB and DS equally conducted data analysis. DA carried out the genetic sequencing. S supervised the sequencing work. JB and BP supervised the data analysis.
Conflict of interest
All authors declare that there is no conflict of interests in the entire processes in the study.
Acknowledgements
This study was jointly funded Bina by Nusantara University and Genetics Indonesia. This study protocol was approved by the ethic committees of Faculty of Medicine, University of Indonesia with the reference number of 450/UN2.F1/ETIK/2018.
We do not register our dataset in order to assure safety of some sensitive information regarding clinical trials and meta-analysis.
References
- Bahk J, Jang SM, Jung-Choi K. Increased breast cancer mortality only in the lower education group: age-period-cohort effect in breast cancer mortality by educational level in South Korea, 1983-2012. Int J Equity Health. 2017;16:56. doi: 10.1186/s12939-017-0554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Chen HL, Zhou MQ, Tian W, et al. Effect of age on breast cancer patient prognoses: A Population-Based Study Using the SEER 18 Database. PLoS One. 2016;11:e0165409. doi: 10.1371/journal.pone.0165409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado MP. Breast cancer in the world: Incidence and mortality. Salud Publica Mex. 2011;53:372–84. [PubMed] [Google Scholar]
- GLOBOCAN Indonesia Fact Sheet. The Global Cancer Observatory. 2019;256: 1–2. [Google Scholar]
- Fagerholm R, Faltinova M, Aaltonen K, et al. Family history influences the tumor characteristics and prognosis of breast cancers developing during postmenopausal hormone therapy. Fam Cancer. 2018;17:321–31. doi: 10.1007/s10689-017-0046-2. [DOI] [PubMed] [Google Scholar]
- Farrington S M, Tenesa A, Barnetson R, et al. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am J Hum Genet. 2005;77:112–9. doi: 10.1086/431213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Arida RM, Gomez-Pinilla F. Physical exercise as an epigenetic modulator of brain plasticity and cognition. Neurosci Biobehav Rev. 2017;80:443–56. doi: 10.1016/j.neubiorev.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez SL, Hurley S, Canchola AJ, et al. Effects of marital status and economic resources on survival after cancer: A population-based study. Cancer. 2016;122:1618–25. doi: 10.1002/cncr.29885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y. Differential DNA recognition and glycosylase activity of the native human MutY homolog (hMYH) and recombinant hMYH expressed in bacteria. Nucleic Acids Res. 2001;29:2666–74. doi: 10.1093/nar/29.12.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haryono SJ, Gusti Bagus Datasena I, Santosa , et al. A pilot genome-wide association study of breast cancer susceptibility loci in Indonesia. Asian Pac J Cancer Prev. 2015;16:2231–5. doi: 10.7314/apjcp.2015.16.6.2231. [DOI] [PubMed] [Google Scholar]
- Henouda S, Bensalem A, Reggad R, et al. Contribution of BRCA1 and BRCA2 germline mutations to early Algerian breast cancer. Dis Markers. 2016;2016:7869095. doi: 10.1155/2016/7869095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Jian W, Shao K, Qin Q, et al. Clinical and genetic characterization of hereditary breast cancer in a Chinese population. Hered Cancer Clin Pract. 2017;15:19. doi: 10.1186/s13053-017-0079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kairupan C, Scott RJ. Base excision repair and the role of MUTYH. Hered Cancer Clin Pract. 2007;5:199–209. doi: 10.1186/1897-4287-5-4-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashfi SMH, Golmohammadi M, Behboudi F, et al. MUTYH the base excision repair gene family member associated with polyposis colorectal cancer. Gastroenterol Hepatol Bed Bench. 2013;6:1–10. [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lin PH, Kuo WH, Huang AC, et al. Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget. 2016;7:8310–20. doi: 10.18632/oncotarget.7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardela AP, Maneewat K, Sangchan H. Breast cancer awareness among Indonesian women at moderate-to-high risk. Nurs Health Sci. 2017;19:301–6. doi: 10.1111/nhs.12345. [DOI] [PubMed] [Google Scholar]
- Martínez ME, Unkart JT, Tao L, et al. Prognostic significance of marital status in breast cancer survival: A population-based study. PloS One. 2017;12:e0175515. doi: 10.1371/journal.pone.0175515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersch J, Jackson M, Park M, et al. Cancers Associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015;121:269–75. doi: 10.1002/cncr.29041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubarik S, Malik SS, Wang Z, et al. Recent insights into breast cancer incidence trends among four Asian countries using age-period-cohort model. Cancer Manag Res. 2019;11:8145–55. doi: 10.2147/CMAR.S208323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo HH, Perbangsa AS, Yulius , Pardamean B. The implementation of online learning for early detection of cervical cancer. J Comput Sci. 2017;13:600–7. [Google Scholar]
- Muljo HH, Perbangsa AS, Yulius , Pardamean B. Mobile learning for early detection cancer. Int J Interact Mob Technol. 2018;12:39–53. [Google Scholar]
- Muljo HH, Perbangsa AS, Yulius , Pardamean B. Improving early cancer detection knowledge through mobile learning application. Int J Online Eng. 2019;15:60–70. [Google Scholar]
- Muljo HH, Pardamean B., Perbangsa AS, et al. TAM as a model to understand the intention of using a mobile-based cancer early detection learning application. Int J Online Eng. 2020;16:80–93. [Google Scholar]
- Nelson HD, Zakher B, Cantor A, et al. Risk factors for breast cancer for women aged 40 to 49 years: A systematic review and meta-analysis a systematic review and meta-analysis. Ann Intern Med. 2012;156:635–48. doi: 10.1059/0003-4819-156-9-201205010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Franken PF, Reinards TH, et al. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYHassociated polyposis coli (MAP) J Med Genet. 2005;42:e54. doi: 10.1136/jmg.2005.033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nones K, Johnson J, Newell F, et al. Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann Oncol. 2019;30:1071–9. doi: 10.1093/annonc/mdz132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo T, Nishioka K, Imaiso Y, et al. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–64. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow SGW, Ong PY, Lee SC. Discoveries beyond BRCA1/2: Multigene testing in an Asian multi-ethnic cohort suspected of hereditary breast cancer syndrome in the real world. PLoS One. 2019;14:e0213746. doi: 10.1371/journal.pone.0213746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigoro SS, Siswiandari KM, Paramita RI, et al. Targeted genome sequencing data of young women breast cancer patients in Cipto Mangunkusumo national hospital. Data In Brief. 2020;32:2352–3409. doi: 10.1016/j.dib.2020.106138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardamean B, Suparyanto T. Hospital-based cancer registry application Proceedings of 2017 International Conference on Information Management and Technology . ICIMTech. 2018: 44–8. [Google Scholar]
- Rennert G, Lejbkowicz F, Cohen I, et al. MutYH mutation carriers have increased breast cancer risk. Cancer. 2012;118(8):1989–93. doi: 10.1002/cncr.26506. [DOI] [PubMed] [Google Scholar]
- Rizzolo P, Silvestri V, Bucalo A, et al. Contribution of MUTYH variants to male breast cancer risk: Results from a multicenter study in Italy. Front Oncol. 2018;8:583. doi: 10.3389/fonc.2018.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel SK, Lovejoy L, Shriver CD, Ellsworth RE. Contribution of germline mutations in cancer predisposition genes to tumor etiology in young women diagnosed with invasive breast cancer. Breast Cancer Res Treat. 2017;164:593–601. doi: 10.1007/s10549-017-4291-8. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1 42 million single nucleotide polymorphisms. Nature. 2001;409:928–33. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Snapshot P. Breast cancer by the numbers. P and T. 2014;39:213–4. [PMC free article] [PubMed] [Google Scholar]
- Slupska MM, Baikalov C, Luther WM, et al. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli MutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178:3885–92. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solikhah S, Promthet S, Rakkapao N, Hurst CP. Validation of an Indonesian Version of the Breast Cancer Awareness Scale (BCAS-I) Asian Pac J Cancer Prev. 2017;18:515–22. doi: 10.22034/APJCP.2017.18.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations: The International HapMap 3 Consortium. Nature. 2010;467:52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–22. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Vogt S, Jones N, Christian D, et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology. 2009;137:1976–85. doi: 10.1053/j.gastro.2009.08.052. [DOI] [PubMed] [Google Scholar]
- Wasielewski M, Out AA, Vermeulen J, et al. Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat. 2010;124:635–41. doi: 10.1007/s10549-010-0801-7. [DOI] [PubMed] [Google Scholar]
- Youlden DR, Cramb SM, Yip CH, Baade PD. Incidence and mortality of female breast cancer in the Asia-Pacific region. Cancer Biol Med. 2014;11:101–15. doi: 10.7497/j.issn.2095-3941.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]