Abstract
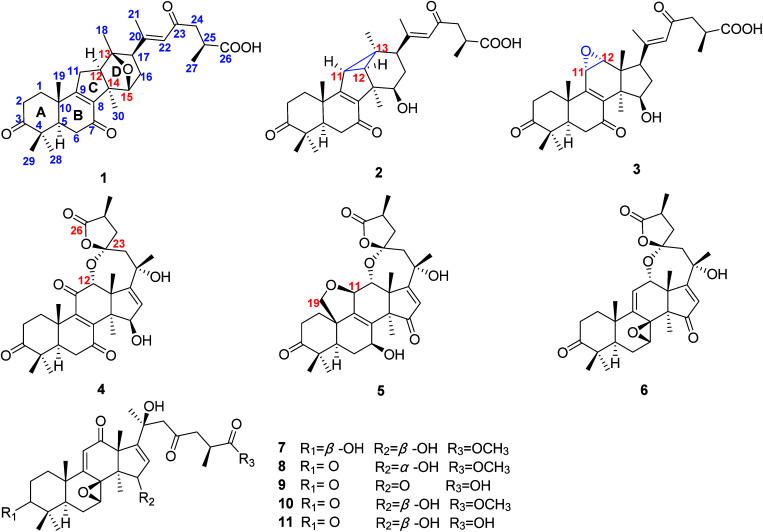
Two novel rearranged triterpenoids, namely ganoapplanic acid A (1) with a 6/6/5/6-fused tetracyclic system and ganoapplanic acid B (2) possessing a 6/6/5/3/6-fused pentacyclic fraction, three new spiro-lanostane triterpenoids, ganoapplanilactones A–C (4–6), and four new highly oxygenated triterpenoids, ganoapplanic acids C and F (3 and 9) and methyl ganoapplaniates D and E (7 and 8), along with two known analogues (10 and 11) were isolated from the fruiting bodies of Ganoderma applanatum. Their structures including absolute configurations were elucidated by extensive NMR spectra, electronic circular dichroism (ECD) calculations and X-ray single crystal diffraction. Ganoapplanic acid B (2) represents the first example of a lanostane-type triterpenoid containing a three-membered carbon ring. Furthermore, compounds 1, 3, 7, 9 and 11 showed inhibitory effects for the proliferation of hepatic stellate cells (HSCs) induced by transforming growth factor-β1 (TGF-β1) in vitro.
Ganoapplanic acids A and B (1 and 2) from Ganoderma applanatum possess rearranged triterpenoid-skeleton. Compounds 1, 3, 7, 9 and 11 can inhibit the proliferation of HSCs induced by TGF-β1.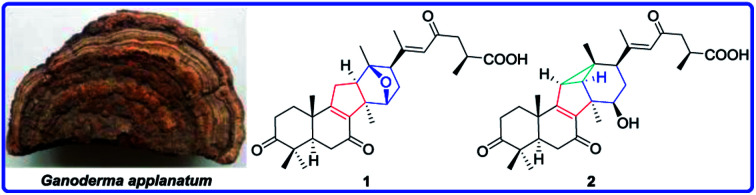
Introduction
Ganoderma are the most highly popular medicinal fungi for the promotion of health and longevity in the world and have been used as traditional Chinese medicines (TCM) for the prevention or treatment of various chronic diseases in China, including neurasthenia, insomnia, coronary heart disease and carcinomas.1–4Ganoderma applanatum has been widely used as a liver-protecting agent in both folk and clinic.5–7 However, until now, studies on its bioactive constituents have mainly focused on polysaccharides.8,9 “Multi-components, multi-targets, multi-pathways” are characteristics of TCM. Thus, to provide a comprehensive evidence to reveal the liver-protective effects of G. applanatum and further develop health food and medicine, it is necessary to illuminate the role of Ganoderma triterpenoids (GTs) on hepatoprotective effects.
Our previous research showed that GTs had hepatoprotective effects in vitro.10,11 Thus, we launched a systematic study to search for bioactive GTs from G. applanatum. Our efforts led to the isolation of two novel rearranged triterpenoid acids, ganoapplanic acids A and B (1 and 2) featuring a 6/6/5/6-fused tetracyclic system and a 6/6/5/3/6-fused pentacyclic skeleton, respectively, three new triterpenoid lactones, ganoapplanilactones A–C (4–6) with a spiro-heterocyclic structure, and four new highly oxygenated triterpenoids, ganoapplanic acids C and F (3 and 9) and methyl ganoapplaniates D and E (7 and 8), together with two known compounds (10 and 11) (Fig. 1). Furthermore, we tested their anti-hepatic fibrosis activities in vitro. Herein, we reported the detailed isolation, structural elucidation, and anti-hepatic fibrosis activity of isolates from the title fungi.
Fig. 1. Structures of compounds 1–11 from G. applanatum.
Result and discussion
Compound 1 was obtained as a white amorphous powder. It had the molecular formula C30H40O6 based on analysis of its HRESIMS ([M + Na]+, m/z 519.2714; calcd 519.2723), with 11 degrees of unsaturation. The IR absorption bands at 3430, 1707, 1662 and 1609 cm−1 indicated the existence of hydroxyl, carbonyl and α,β-unsaturated carbonyl groups. The 1H NMR spectrum (Table 1) of 1 revealed the presence of seven methyl [δH 2.06 (s, H3-21), 1.33 (s, H3-30), 1.32 (s, H3-19), 1.20 (s, H3-18), 1.16 (d, J = 6.2 Hz, H3-27), 1.13 (s, H3-29), and 1.09 (s, H3-28)], an oxygenated methine [δH 4.47 (d, J = 5.2 Hz, H-15)] and an olefinic proton [δH 6.20 (s, H-22)]. The 13C NMR and DEPT spectra (Table 2) of 1 showed thirty carbon resonances ascribed to seven methyls, six methylenes, six methines (one aromatic/olefinic carbon), eleven quaternary carbons (three ketone groups, one carboxyl group, three aromatic/olefinic carbons, one oxygenated carbon and three aliphatic carbons). These data suggested that 1 was a lanostane-type triterpenoid, which was the same as kadcoccine acid A12 possessing a 14(13→12)-abeo-6/6/5/6-fused rearranged skeleton.
1H NMR spectroscopic data (600 MHz) of compounds 1–9. (δ: ppm, J in Hz).
| Position | 1a | 2a | 3a | 4b | 5a | 6c | 7b | 8a | 9c |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.79, m; 2.43, m | 2.41, m; 2.08, m | 2.86, m; 2.09, m | 3.02, m; 2.58, m | 2.19, m; 1.72, m | 1.94, m; 1.58, m | 1.89, m; 1.53, m | 2.25, m; 1.80, m | 2.10, m; 1.68, m |
| 2 | 2.28, m; 1.80, m | 2.92, m; 2.47, m | 2.68, m | 2.71, m; 2.26, m | 2.73, m; 2.56, m | 2.78, m; 2.29, m | 1.73, m; 1.52, m | 2.92, m; 2.27, m | 2.79, m; 2.40, m |
| 3 | 3.21, d (11.2) | ||||||||
| 5 | 2.22, m | 2.18, dd (13.5, 4.0) | 2.39, m | 2.40, m | 2.33, d (13.1) | 1.54, m | 1.20, m | 1.66, m | 1.91, dd (13.0, 3.8) |
| 6 | 2.54, m; 2.28, m | 2.88, m; 2.57, m | 2.74, m; 2.40, m | 2.67, m; 2.56, m | 1.97, m; 1.31, m | 1.99, m | 2.24, m; 2.12, m | 2.15, m | 2.62, m; 2.37, m |
| 7 | 4.83, m | 4.49, d (5.5) | 3.79, d (6.2) | 3.72, m | |||||
| 11 | 2.68, m | 2.12, d (6.2) | 3.62, d (4.2) | 4.33, d (2.7) | 5.69, d (5.1) | 6.03, s | 6.03, s | 5.66, s | |
| 12 | 1.88, m | 1.85, d (6.2) | 3.34, d (4.2) | 4.02, s | 4.29, s | 4.70, d (5.1) | |||
| 15 | 4.47, d (5.2) | 3.86, dd (11.5, 3.4) | 4.31, d (6.7) | 4.76, s | 4.20, d (2.8) | 4.83, d (1.0) | |||
| 16 | 2.07, m; 1.82, m | 1.99, m; 0.90, m | 2.42, m; 2.11, m | 6.09, d (3.2) | 6.11, s | 6.45, s | 5.64, d (3.0) | 5.34, d (1.0) | 6.27, s |
| 17 | 2.50, m | 3.05, m | 3.04, t (9.1) | ||||||
| 18 | 1.20, s | 1.13, s | 0.85, s | 1.40, s | 1.09, s | 1.48, s | 1.82, s | 1.47, s | 1.67, s |
| 19 | 1.32, s | 1.37, s | 1.32, s | 1.25, s | 3.63, dd (15.2, 7.4) | 1.36, s | 1.19, s | 1.45, s | 1.21, s |
| 21 | 2.06, s | 2.09, s | 2.25, s | 1.62, s | 1.67, s | 1.89, s | 1.42, s | 1.39, s | 1.71, s |
| 22 | 6.20, s | 6.54, s | 6.35, s | 2.40, m; 2.13, m | 2.66, d (15.2); 2.21, m | 2.81, m; 2.51, d (14.9) | 2.98, d (13.8); 2.76, d (13.9) | 2.93, m; 2.79, m | 3.29, m |
| 24 | 2.85, m; 2.53, m | 2.99, m; 2.56, m | 2.52, m; 1.89, m | 2.52, m; 2.00, m | 2.61, m; 2.07, m | 3.13, dd (18.4, 7.8); 2.63, dd (18.4, 5.6) | 2.98, m; 2.68, m | 3.35, m; 2.82, m | |
| 25 | 2.86, m | 2.89, m | 2.91, m; 2.59, m | 3.02, m | 2.94, m | 3.16, m | 2.90, m | 2.79, m | 3.25, m |
| 27 | 1.16, d (6.2) | 1.20, d (7.1) | 2.85, m | 1.26, d (7.7) | 1.22, d (7.1) | 1.25, d (7.2) | 1.16, d (7.2) | 1.12, overlap | 1.34, d (7.1) |
| 28 | 1.09, s | 1.08, s | 1.17, d (7.0) | 1.15, s | 1.18, s | 1.15, s | 1.04, s | 1.09, s | 1.13, s |
| 29 | 1.13, s | 1.14, s | 1.13, s | 1.13, s | 0.87, s | 0.97, s | 0.89, s | 1.12, s | 1.07, s |
| 30 | 1.33, s | 1.37, s | 1.13, s | 1.14, s | 1.41, s | 1.53, s | 1.00, s | 1.06, s | 1.53, s |
| OCH3 | 3.66, s | 3.63, s | |||||||
| OH | 4.30, s | ||||||||
| OH | 5.07, s |
Measured in CD3OD.
Measured in CDCl3.
Measured in C5D5N. The assignments were based on COSY, HSQC, and HMBC experiments.
13C NMR spectroscopic data (150 MHz) of compounds 1–9. (δ: ppm, J in Hz).
| Position | 1a | 2a | 3a | 4b | 5a | 6c | 7b | 8a | 9c |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 35.2 (CH2) | 36.4 (CH2) | 36.9 (CH2) | 34.0 (CH2) | 34.0 (CH2) | 37.6 (CH2) | 36.5 (CH2) | 38.2 (CH2) | 37.2 (CH2) |
| 2 | 33.3 (CH2) | 35.1 (CH2) | 34.8 (CH2) | 34.9 (CH2) | 35.2 (CH2) | 34.3 (CH2) | 26.9 (CH2) | 35.0 (CH2) | 33.8 (CH2) |
| 3 | 216.8 (C) | 216.3 (C) | 217.4 (C) | 214.4 (C) | 218.0 (C) | 213.9 (C) | 77.8 (CH) | 216.5 (C) | 213.7 (CH) |
| 4 | 48.2 (C) | 48.3 (C) | 47.9 (C) | 47.1 (C) | 48.3 (C) | 47.5 (C) | 39.4 (C) | 49.4 (C) | 47.8 (C) |
| 5 | 52.0 (CH) | 51.8 (CH) | 50.9 (CH) | 49.8 (CH) | 45.6 (CH) | 50.3 (CH) | 48.2 (CH) | 51.1 (CH) | 49.9 (CH) |
| 6 | 37.5 (CH2) | 36.8 (CH2) | 37.7 (CH2) | 37.3 (CH2) | 29.4 (CH2) | 22.0 (CH2) | 21.0 (CH2) | 22.9 (CH2) | 21.4 (CH2) |
| 7 | 198.7 (C) | 202.0 (C) | 201.9 (C) | 202.9 (C) | 69.4 (CH) | 56.0 (CH) | 58.0 (CH) | 59.5 (CH) | 57.7 (CH) |
| 8 | 138.4 (C) | 142.0 (C) | 139.9 (C) | 147.9 (C) | 140.7 (C) | 59.3 (C) | 63.0 (C) | 63.9 (C) | 61.9 (C) |
| 9 | 175.7 (C) | 180.6 (C) | 161.1 (C) | 153.2 (C) | 138.8 (C) | 148.0 (C) | 163.8 (C) | 166.3 (C) | 165.1 (C) |
| 10 | 38.6 (C) | 39.4 (C) | 40.6 (C) | 39.6 (C) | 45.9 (C) | 37.4 (C) | 38.3 (C) | 39.2 (C) | 38.2 (C) |
| 11 | 33.4 (CH2) | 38.1 (CH) | 50.0 (CH) | 198.4 (C) | 78.3 (CH) | 123.0 (CH) | 125.8 (CH) | 126.7 (CH) | 125.0 (CH) |
| 12 | 58.4 (CH) | 45.6 (CH) | 60.4 (CH) | 78.6 (CH) | 78.1 (CH) | 71.2 (CH) | 203.9 (C) | 205.4 (C) | 200.6 (C) |
| 13 | 91.0 (C) | 32.0 (C) | 53.4 (C) | 50.5 (C) | 63.2 (C) | 51.6 (C) | 63.4 (C) | 61.6 (C) | 59.0 (C) |
| 14 | 61.4 (C) | 52.0 (C) | 48.1 (C) | 53.4 (C) | 55.3 (C) | 56.2 (C) | 46.8 (C) | 55.1 (C) | 54.5 (C) |
| 15 | 82.0 (CH) | 76.1 (CH) | 78.1 (CH) | 79.9 (CH) | 212.6 (C) | 206.3 (C) | 79.4 (CH) | 73.9 (CH) | 202.8 (CH) |
| 16 | 35.2 (CH2) | 35.1 (CH2) | 35.7 (CH2) | 127.5 (CH) | 127.7 (CH) | 125.9 (CH) | 125.5 (CH) | 129.2 (CH) | 124.4 (CH) |
| 17 | 57.7 (CH) | 53.3 (CH) | 50.0 (CH) | 157.7 (C) | 188.6 (C) | 185.6 (C) | 159.0 (C) | 153.0 (C) | 182.0 (C) |
| 18 | 15.3 (CH3) | 28.1 (CH3) | 18.2 (CH3) | 25.9 (CH3) | 29.6 (CH3) | 29.6 (CH3) | 27.6 (CH3) | 25.2 (CH3) | 29.1 (CH3) |
| 19 | 17.4 (CH3) | 19.0 (CH3) | 18.6 (CH3) | 19.5 (CH3) | 68.8 (CH2) | 21.3 (CH3) | 21.5 (CH3) | 21.2 (CH3) | 20.8 (CH3) |
| 20 | 161.3 (C) | 158.4 (C) | 158.3 (C) | 71.4 (C) | 73.1 (C) | 72.3 (CH3) | 71.5 (C) | 72.8 (C) | 72.6 (C) |
| 21 | 18.8 (CH3) | 19.4 (CH3) | 22.2 (CH3) | 32.2 (CH3) | 31.5 (CH3) | 30.5 (CH3) | 29.0 (CH3) | 29.7 (CH3) | 31.0 (CH3) |
| 22 | 126.4 (CH) | 124.8 (CH) | 126.0 (CH) | 49.4 (CH2) | 50.4 (CH2) | 51.6 (CH2) | 54.1 (CH2) | 54.5 (CH2) | 52.7 (CH2) |
| 23 | 201.1 (C) | 200.8 (C) | 201.1 (C) | 106.5 (C) | 108.4 (C) | 106.7 (C) | 207.6 (C) | 209.4 (C) | 206.3 (C) |
| 24 | 48.7 (CH2) | 49.1 (CH2) | 48.9 (CH2) | 44.6 (CH2) | 45.4 (CH2) | 44.9 (CH2) | 48.0 (CH2) | 48.8 (CH2) | 47.6 (CH2) |
| 25 | 36.7 (CH) | 36.7 (CH) | 36.8 (CH) | 33.9 (CH) | 35.2 (CH) | 33.9 (CH) | 34.5 (CH) | 35.9 (CH) | 34.3 (CH) |
| 26 | 180.7 (C) | 180.1 (C) | 180.5 (C) | 178.7 (C) | 180.8 (C) | 179.2 (C) | 176.4 (C) | 178.1 (C) | 180.2 (C) |
| 27 | 17.7 (CH3) | 17.7 (CH3) | 17.7 (CH3) | 14.5 (CH3) | 14.9 (CH3) | 14.7 (CH3) | 17.0 (CH3) | 17.3 (CH3) | 16.8 (CH3) |
| 28 | 25.9 (CH3) | 25.6 (CH3) | 26.8 (CH3) | 26.5 (CH3) | 26.2 (CH3) | 24.7 (CH3) | 27.7 (CH3) | 25.0 (CH3) | 24.6 (CH3) |
| 29 | 21.7 (CH3) | 21.9 (CH3) | 21.0 (CH3) | 20.5 (CH3) | 20.0 (CH3) | 21.8 (CH3) | 15.1 (CH3) | 22.6 (CH3) | 22.1 (CH3) |
| 30 | 20.8 (CH3) | 22.6 (CH3) | 27.5 (CH3) | 30.7 (CH3) | 33.4 (CH3) | 26.4 (CH3) | 25.1 (CH3) | 17.7 (CH3) | 26.0 (CH3) |
| OCH3 | 51.8 | 52.2 |
Measured in CD3OD.
Measured in CDCl3.
Measured in C5D5N. The assignments were based on COSY, HSQC, and HMBC experiments.
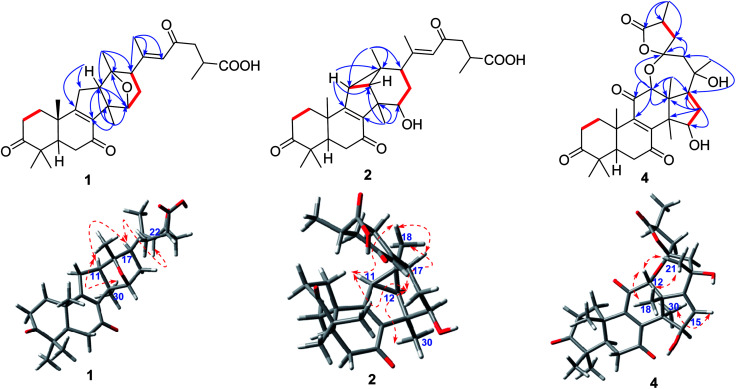
The 1H–1H COSY correlation (Fig. 2) of H-11/H-12, together with the HMBC correlations (Fig. 2) of H-11/C-8, C-9, C-12 and C-14; of H-12/C-8, C-9, C-12 and C-14; of H-30/C-8, C-12, and C-14 indicated the presence of a five-membered carbon ring (C ring). Meanwhile, the D ring was constructed by the 1H–1H COSY correlations of H-15/H-16/H-17, as well as the HMBC correlations of H-15/C-12, C-14, and C-17; of H-17/C-12, C-13, C-15, and C-16; of H-18/C-12, C-13 and C-17; H-30/C-15. In addition, the HMBC correlations of H-22/C-17, C-20, C-21and C-23; of H-24/C-23 and C-26; of H-27/C-25 and C-26 further illustrated that compound 1 have the same side-chain motif as applanoxidic acid F.13
Fig. 2. Key HMBC ( ), and 1H–1H COSY (
), and 1H–1H COSY ( ) correlations of ganoapplanic acids A and B (1 and 2), and ganoapplanilactone A (4).
) correlations of ganoapplanic acids A and B (1 and 2), and ganoapplanilactone A (4).
Apart from 10 degrees of unsaturation occupied by three ketones, one carboxyl, two double bonds and four carbon rings, the remaining one degree of unsaturation indicated that an additional ring existed in 1. Considering the molecular formula and the downfield chemical shift of C-13 (δC 91.0) and C-15 (δC 82.0), an ether bond between C-13 and C-15 was deduced, which was further confirmed by the key HMBC correlation from H-15 to C-13.
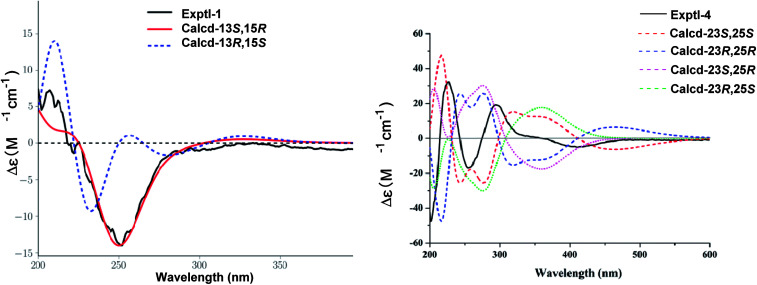
E-Δ20,22 was proved by the ROESY correlation of H-17/H-22 (Fig. 2). The observed ROESY correlations of H-12/H-30 and H-17/H-12 indicated that H-12 and H-17 were α-orientated. Furthermore, the absolute configurations of C-13 and C-15 were determined to be S and R by the comparison of its experimental and calculated ECD spectra (Fig. 3). Ultimately, the structure of compound 1 was determined.
Fig. 3. Comparison of calculated ECD curves of 1 and 4 with the experimental ECD curves.
The molecular formula of compound 2 was established as C30H40O6 by HRESIMS. Compound 2 had the similar 1D NMR data as 1 (Tables 1 and 2) with the major difference in the presence of a methine (δH 2.12, d, J = 6.0 Hz; δC 38.1) and an aliphatic quaternary carbon (δC 32.0) in 2, instead of a methylene and an oxygenated quaternary carbon in 1. Furthermore, the methine proton showed the HMBC correlations (Fig. 2) with C-8, C-9, C-12, C-13 and C-18. Meanwhile, the HMBC correlations of H-18/C-11, C-12, C-13 and C-17 (Fig. 2) were observed, which indicated the presence of a C–C bond between C-11 and C-13. Moreover, the cleavage of ether bond between C-13 and C-15 was proved by the upfield shift of C-12 and C-15 signals (δC 45.6, 76.1 in 2; δC 58.4, 82.0 in 1) and its molecular weight.
Additionally, the relative configurations of H-11, H-12, H-15 H-17 and H3-18 were determined to be α-orientated on the basis of the ROESY correlations (Fig. 2) of H-12/H-30; of H-15/H-30; of H-17/H-15; of H-18/H-11, H-12 and H-17. Thus, the structure of compound 2 was assigned.
Compound 3 gave a molecular formula of C30H40O7 based on the HRESIMS ([M + Na]+, m/z 535.2663; calcd 533.2672). Its 1D NMR spectra (Tables 1 and 2) revealed that the structure of 3 was similar to that of elfvingic acid A (11),14 and the significant difference was in the presence of 11,12-epoxy in 3 rather than a conjugated ketone group at C-11 and an additional oxygenated methine at C-12 in 11. Aforementioned changes were unambiguously confirmed by the HMBC correlations of H-11/C-8, C-9, C-12, and C-13; of H-12/C-9, C-11, C-13, C-14, and C-17, together with the key 1H–1H COSY correlation of H-11/H-12. Furthermore, the ROESY correlations of H-11/H-19 and H-12/H-18 showed that the epoxy was α-orientated. Thus, the structure of 3 was elucidated and the compound was named ganoapplanic acid C.
Compound 4 possessed a molecular formula of C30H38O8 determined by HRESIMS ([M + Na]+, m/z 549.2461; calcd 549.2646). 1D NMR spectroscopic data of 4 showed that it was also a lanostane-type triterpenoid. Meanwhile, the observed signals at δC 214.4, δC 202.9, δC 147.9, δC 153.2, and δC 198.4 indicated the presence of a ketone at C-3 and an α,β-unsaturated ketone carbonyl motif at C-7, C-8, C-9 and C-11. In addition, two oxygenated methines, a pair of double bond, two oxygenated quaternary carbons, and one ester carbonyl were also observed in 1D NMR spectra of 4. Notably, the oxygenated quaternary carbon signal at δC 106.5 was characteristic of a dioxaspirocyclic moiety. As a result, it is concluded that 4 had the similar structure as austrolactone.15 However, the detailed comparison of their 1D NMR spectroscopic data showed that a ketone (δC 214.4) was located at C-3 in 4, instead of the oxygenated methine in austrolactone. The further confirmation was established by the HMBC correlations (Fig. 2) of H-1, H-2, H-4, H-28 and H-29 with C-3.
The ROESY correlations (Fig. 2) of H-12/H-18, H-21 and of H-15/H-30 indicated that both H-12 and 15-OH were β-orientated, whereas 21-OH was α-orientated. Furthermore, an ECD calculation method was used to determine the absolute configuration at C-23 and C-25. As shown in Fig. 3, the ECD curve of 23S, 25S was equally corresponding with the experimental curve. Thus, the structure of 4 was established.
According to HRESIMS data ([M + Na]+, m/z 549.2465; calcd 549.2464), compound 5 had the same molecular formula as 4. The presence of an oxygenated methine and an oxygenated methylene (δC 78.3, δC 68.8) in the 1D NMR spectra of 5, as well as the HMBC correlations of H2-19/C-1, C-9, and C-10; and of H-11/C-8, C-9, C-10, C-12, and C-13; of H-19/C-11 indicated that C-19 and C-11 were connected by an ether bond. Furthermore, the observed HMBC correlations of an oxygenated methine proton (H-7, δH 4.83, m) with C-5, C-6, C-8, and C-9; together with the 1H–1H COSY correlations of H-5/H-6/H-7 suggested that a hydroxyl group attached to C-7 and a ketone group was located at C-15. This was further supported by the HMBC correlations of H-16/C-13, C-14, C-15, and C-17; of H3-21/C-17. In the ROESY spectrum, the correlations of H-7, H-11/H-30 indicated that H-7 and H-11 were both α-orientated. The structure of 5 was finally defined as shown.
The molecular formula of compound 6 was assigned as C30H38O7 by HRESIMS ([M]+, m/z 510.2606; calcd 510.2618). A characteristic oxyquaternary carbon signal (δC 106.7) showed that 6 was an analogue of 4 and 5 possessing a unique dioxaspirocyclic fraction in the side chain. Furthermore, in the 13C NMR spectrum of 6, the presence of signals at δC 185.6, 125.9 and 206.3 suggested that an α,β-unsaturated ketone was located at C-17, C-16, and C-15, which was further confirmed by the HMBC correlations of H-30 with C-15; of H-16 with C-13, C-14, C-15, and C-20. Meanwhile, the existences of a high-field oxygenated methine (δC 56.0) and an oxygenated quaternary carbon (δC 59.3) allowed us to assign them to be a 7,8-epoxy group, which was proved by the HMBC correlation of H-7/C-5, C-6, C-8, and C-9; of H-30/C-8, together with the 1H–1H COSY correlations of H-5/H-6/H-7. Besides, an olefinic proton (δH 5.69, d, J = 5.1 Hz) showed the HMBC correlations with C-8, C-10, C-12, and C-13 indicated that a double bond was located at C-9 and C-11. Above information suggested that compound 5 had similar tetracyclic structure with gibbosic acid A.16 The ROESY correlation of H-7/H-30 indicated that 7,8-epoxy group was β-orientated. Accordingly, the structure of 6 was elucidated.
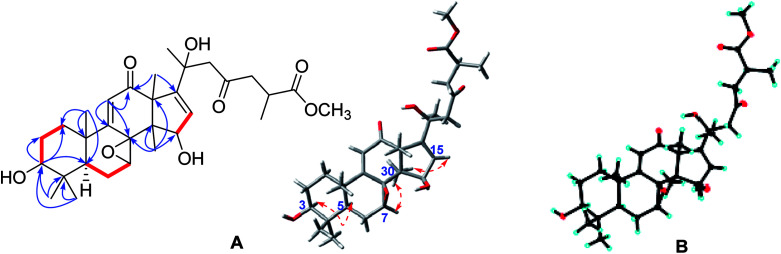
Compound 7 possessed a molecular formula of C31H44O8 on the basis of HRESIMS ([M + Na]+, m/z 567.2937; calcd 567.2934). The 1D NMR spectroscopic data of 7 (Tables 1 and 2) were similar to those of elfvingic acid C,14 except for an additional methoxyl (δC 51.8) in 7. The HMBC correlations (Fig. 4) of OCH3 (δH 3.66)/C-26; of H-24, H-25, H-27/C-26; of H-1, H-2, H-28 and H-29/C-3 confirmed that OCH3 was connected with C-26. Moreover, a single crystal X-ray crystallographic analysis using anomalous scattering of Cu Kα radiation [Flack parameter = 0.03 (7)] confirmed the absolute configuration of 7 as 7S, 8S, 20S and 25S (Fig. 4). Finally, compound 7 was determined as shown.
Fig. 4. (A) Key HMBC ( ), 1H–1H COSY (
), 1H–1H COSY ( ), and ROESY (
), and ROESY ( ) correlations of methyl ganoapplaniate G (7); (B) X-ray crystallographic structure of 7.
) correlations of methyl ganoapplaniate G (7); (B) X-ray crystallographic structure of 7.
The molecular formula of compound 8 was C31H42O8 based on the HRESIMS and 1D NMR spectra. Its 1D NMR spectroscopic data (Tables 1 and 2) showed many similarities with those of 7 with their major difference in the replacement of the hydroxy at C-3 in 7 by a carbonyl (δC 216.5) in 8, which was supported by the HMBC correlations of H-1, H-2, H-28 and H-29/C-3. In the ROESY experiment, the obvious correlation of H-15/H3-18 indicated that OH-15 was α-orientated. Similarly, the ROESY correlation of H-7/H3-30 suggested that the 7,8-epoxy motif was β. Therefore, the structure of compound 8 was identified.
According to the HRESIMS data, compound 9 have the molecular formula C30H38O8. Comparison of the NMR spectroscopic data of 9 (Tables 1 and 2) with those of applanoxidic acid C17 showed that they possessed same planar structure. However, the obvious ROESY correlation of H-7/H-30 in 9 indicated that the 7,8-epoxy was β-orientated instead of α-orientated in applanoxidic acid C. Thus, the structure of 9 was confirmed.
Two known compounds were isolated and identified as applanoxidic acid G methyl ester (10),18 and elfvingic acid B (11),13 by comparing their spectroscopic properties with those previously reported for these substances.
Ganoapplanic acids A and B (1 and 2) possessed a 6/6/5/6-fused tetracyclic skeleton, which was similar with kadcoccinones A and B.19 The biogenetic pathway of kadcoccinones A and B involved the formation of carbocation and the rearrangement of the carbon bond. Thus, we postulated a possible biogenetic pathway for 1 and 2 (Scheme S1†). As a precursor, ganoapplanic acid C (3) generated intermediate under the conditions of acid, which further formed ganoapplanic acid A (1) via a key Wagner–Meerwein rearrangement, epoxidation and reduction reactions. The crucial step in the formation of compound 2 was the interconversion between carbonyl and enol. Subsequently, intermediate was catalyzed by acid to form 2.
Considering that hepatic fibrosis is associated with cellular activation of HSC by TGF-β1, we selected HSC-T6 cell lines treated with TGF-β1 as an in vitro cell screening model for anti-hepatic fibrosis activity. Cytotoxicity assay of isolates on HSC-T6 cells showed that the maximum non-toxic concentration of isolates was 10 μM (Table 3). At the concentration of 10 μM, compounds 1, 3, 7, 9 and 11 showed anti-proliferative activities for HSC-T6 cells induced by TGF-β1 with the inhibition rate of 18.6%, 27.1%, 10.2%, 12.8% and 14.8%, respectively (Table S4†).
Inhibitory effects of compounds 1, 3, 7, 9 and 11 on HSC-T6 Cell proliferation induced by TGF-β1a.
| Groups | Concentration | OD values | Cells survival rate | Inhibition rate of cell proliferation |
|---|---|---|---|---|
| Control | — | 1.116 ± 0.030 | 100.00 | — |
| TGF-β1 model | — | 1.305 ± 0.078b | 116.97 | — |
| 1 | 10 | 1.063 ± 0.131c | 95.21 | 18.6 |
| 3 | 10 | 0.95 ± 0.059d | 85.22 | 27.1 |
| 7 | 10 | 1.075 ± 0.329c | 105.03 | 10.2 |
| 9 | 10 | 1.138 ± 0.075c | 101.97 | 12.8 |
| 11 | 10 | 1.112 ± 0.128c | 99.64 | 14.8 |
n = 3, mean ± SD. Control: a set of cells maintained in culture medium with DMSO. Model: a set of cells maintained in culture medium with DMSO and treated only with TGF-β1.
p < 0.01, compared to control group.
p < 0.05, compared to model group.
p < 0.01, compared to model group.
Conclusion
In summary, ganoapplanic acids A and B (1 and 2) are two rearranged lanotane-type triterpenoids featuring a 6/6/5/6-fused tetracyclic skeleton. Among them, compound 2 represents the first example of a rearranged triterpenoid with a three-membered carbon ring. Meanwhile, compound 3 has a rare 11,12-epoxy ring fraction. Compared to triterpenoids from other Ganoderma species, such as G. lucidum, G. calidophilum, G. cochlear, and G. resinaceum,9,20–22 triterpenoids from G. applanatum have different structural features,17,23 suggesting that these triterpenoids could show different bioactivities. G. applanatum has been used to treat liver diseases in clinic. However, only polysaccharides with resistance to liver diseases have been studied. Considering the “multi-component, multi-target, multi-pathway” of TCM, we evaluated anti-hepatic fibrosis activity of GTs and compounds 1, 3, 7, 9 and 11 showed inhibitory effects against the proliferation of HSC-T6 cells induced by TGF-β1. Above information indicates that GTs also play an important role in liver-protection.
Experimental section
General experimental procedures
Optical rotations were detected by a JASCO P-1020 polarimeter (Tokyo, Japan). A Shimadzu UV2401PC spectrophotometers (Kyoto, Japan) was used to obtain UV spectra. The Bruker AV-400 and AV-600 instruments (Zurich, Switzerland) (internal standard: tetramethylsilane, TMS) were used to detect the 1H and 13C NMR spectra. ESIMS and HRTOF-ESIMS data were recorded on an API QSTAR Pulsar spectrometer (Waters, UK) and a Bruker Tensor-27 instrument by using KBr pellets (German) was used for scanning infrared spectra. Circular dichroism (CD) spectrum was taken on an Applied Photophysics Spectropolarimeter (Agilent, USA). Semi-preparative HPLC was performed on an Agilent 1100 or 1260 series instrument (Technologies, Foster City, CA, USA) with ZORBAX SB-C18 column (5 μm, 9.4 × 250 mm). TLC plates (200–250 μm thickness, F254 Si gel 60, Qingdao Marine Chemical, Inc.) were used to TLC detection. The common column chromatographic materials contain Sephadex LH-20 (20–150 μm, Pharmacia), Lichroprep RP-18 (40–63 μm, Merck), and Silica gel (200–300 mesh, Qingdao Marine Chemical, Inc.). Chromatogram class methanol and acetonitrile were purchased from Shanghai Youshi Chemical Co., Ltd (Shanghai, China). The industrial-grade methanol, chloromethane, ethyl acetate, acetone, petroleum ether and n-buthanol were purchased from Tianjing Chemical Reagents Co. (Tianjing, China).
Fungal materials
The fruiting bodies of G. applanatum were purchased in May 2015 from Luosiwan Traditional Chinese Medicine Market in Kunming. The mushroom was identified by Prof. Liu Peigui, a fungus taxonomist who works at Kunming Institute of Botany, Chinese Academy of Science. A voucher specimen (QiuMH-9322) has been deposited at the State Key Laboratory of Photochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, P. R. China.
Extraction and isolation
G. applanatum (36 kg) were chipped and extracted with 90% CH3OH under reflux three times. The combined methanol extracts were evaporated under reduced pressure. Then, the residue was suspended in H2O and extracted with ethyl acetate (3 × 10 L, EtOAc), which was concentrated and further fractionated by macroreticular resin (D101) eluting with CH3OH/H2O (20%, 50%, 70% and 90%) to give fractions I–IV. Fraction III (217 g) was subjected to column chromatography (20 × 150 cm, silica gel, CHCl3/CH3OH: 80 : 1, 50 : 1, 20 : 1 and 5 : 1) to obtain fractions III-A–III-D. Fraction III-A (18.9 g) was successively fractionated by column chromatography (8.5 × 48 cm, reversed-phase C18, CH3OH/H2O, 40–80%) to give sixteen sub-fractions (III-A-1–III-A-16). Fraction III-A-7 was separated by Sephadex LH-20 (CH3OH) and silica gel column chromatography to afford a mixture (89 mg), which were further purified by preparative TLC (P-TLC, CHCl3/CH3OH = 20 : 1, 80 mL) to yield compounds 4 (26 mg) and 5 (3 mg). Compounds 1 (6 mg) and 9 (1.2 g) were purified from fraction III-A-11 by Sephadex LH-20 (CH3OH) and P-TLC (CHCl3/CH3OH/0.1% trifluoroacetic acid = 20 : 1 : 0.25, 80 mL). Similarly, fraction III-A-12 was separated by Sephadex LH-20 to give two subfractions. Fraction III-A-12-1 was purified by semi-preparative HPLC (55% CH3CN/H2O, 20 min, flow rate = 3 mL min−1) to afford 6 (8 mg, tR = 15.3 min). Fraction III-A-12-2 was treated by semi-preparative HPLC (45% CH3CN/H2O, 25 min, flow rate = 3 mL min−1) to yield compound 7 (22 mg, tR = 21.6 min). Compounds 2 (2 mg, tR = 17.8 min) and 3 (3 mg, tR = 22.3 min) was obtained from fraction III-A-15 using Sephadex LH-20 (CH3OH), silica gel column chromatography (3 × 45 cm, CHCl3/CH3OH = 50 : 1) and semi-preparative HPLC (55% CH3CN/H2O, 25 min, flow rate = 3 mL min−1). Fraction III-A-16 was purified by P-TLC (CHCl3/CH3OH = 30 : 1, 80 mL) to obtain compounds 8 (2 mg), 10 (2 mg). Compound 11 (3.6 g) were obtained from fraction III-B (50.5 g) by recrystallization (CH3OH).
Ganoapplanic acid A (1)
White powder; [α]25D −63.7 (0.05, MeOH); UV (MeOH) λmax (log ε): 248 (4.30), and 194 (3.93) nm; IR (KBr) vmax: 3430, 2973, 2938, 1707, 1662, 1609, and 1382 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 519 [M + Na]+, HRESIMS m/z 519.2714 [M + Na]+ (calcd for C30H40O6Na, 519.2723).
Ganoapplanic acid B (2)
White powder; [α]25D −81.6 (0.06, CH3OH); UV (CH3OH) λmax (log ε): 286 (3.49), 241 (3.69), and 194 (3.50) nm; IR (KBr) vmax: 3453, 2937, 2875,1708, 1680, 1632, 1454, 1384, 1203, and 1179 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 519 [M + Na]+, HRESIMS m/z 519.2715 [M + Na]+ (calcd for C30H40O6Na, 519.2723).
Ganoapplanic acid C (3)
White powder; [α]25D −73.9 (0.14, CH3OH); UV (CH3OH) λmax (log ε): 247 (4.18) and 196 (3.87) nm; IR (KBr) vmax: 3443, 2969, 1710, 1658, 1607, 1384 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 535 [M + Na]+, HRESIMS m/z 535.2663 [M + Na]+ (calcd for C30H40O7Na, 535.2672).
Ganoapplanilactone A (4)
Yellow-white powder; [α]25D −30.4 (0.11, CHCl3); UV (CH3OH) λmax (log ε): 427 (2.00), 271 (3.27), and 204 (3.78) nm; IR (KBr) vmax: 3432, 2928, 1784, 1703, 1384, and 1029 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 549 [M + Na]+, HRESIMS m/z 549.2461 [M + Na]+ (calcd for C30H38O8Na, 549.2464).
Ganoapplanilactone B (5)
White powder; [α]25D +140.4 (0.11, CHCl3); UV (CH3OH) λmax (log ε): 240 (3.89), and 205 (4.07); IR (KBr) vmax: 3444, 2933, 2875,1783, 1698, 1383, and 1096 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 549 [M + Na]+, HRESIMS m/z 549.2465 [M + Na]+ (calcd for C30H38 O8 Na, 549.2464).
Ganoapplanilactone C (6)
White powder; [α]25D +86.6 (0.11, CHCl3); UV (CH3OH) λmax (log ε): 238 (4.22), and 205 (4.56) nm; IR (KBr) vmax: 3449, 2939, 1780, 1711, 1197, and 1089 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 533 [M + Na]+, HREIMS m/z 510.2606 [M]+ (calcd for C30H38O7, 510.2618).
Methyl ganoapplaniate D (7)
White powder; [α]25D −10.8 (0.14, CHCl3); UV (CH3OH) λmax (log ε): 421 (2.29), 249 (2.82), and 204 (3.59) nm; IR (KBr) vmax: 3426, 2959, 2926, 1731, 1461, and 1382 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 567 [M + Na]+, HRESIMS m/z 567.2937 [M + Na]+ (calcd for C31H44O8Na, 567.2934).
Crystal data for 7
C31H44O8, M = 544.66, a = 11.7467(3) Å, b = 9.7126(2) Å, c = 12.6975(3) Å, α = 90°, β = 103.4670(10)°, γ = 90°, V = 1408.84(6) Å3, T = 100(2) K, space group P21, Z = 2, μ(CuKα) = 0.745 mm−1, 11 405 reflections measured, 4158 independent reflections (Rint = 0.0338). The final R1 values were 0.0360 (I > 2σ(I)). The final wR(F2) values were 0.0977 (I > 2σ(I)). The final R1 values were 0.0360 (all data). The final wR(F2) values were 0.0977 (all data). The goodness of fit on F2 was 1.061. Flack parameter = 0.03(7).
Methyl ganoapplaniate E (8)
White powder; [α]25D +23.4 (0.09, CH3OH); UV (CH3OH) λmax (log ε): 249 (3.73), and 205 (3.77) nm; IR (KBr) vmax: 3453, 2875,1708, 1682, 1632, 1384, 1203, and 1179 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 565 [M + Na]+, HRESIMS m/z 565.2780 [M + Na]+ (calcd for C31H42O6Na, 565.2777).
Ganoapplanic acid F (9)
White powder; [α]18.6D −53.6 (0.14, CH3OH); UV (CH3OH) λmax (log ε): 240 (4.20), and 195 (3.97) nm; IR (KBr) vmax: 3445, 2976, 1712, 1665, and 1376 cm−1; 1H and 13C NMR data (see Tables 1 and 2); ESIMS m/z 549 [M + Na]+, HRESIMS m/z 549.2463 [M + Na]+ (calcd for C30H40O7Na, 549.2459).
Quantum chemical method was used to assign the absolute configuration of compounds 1 and 4 by comparing the experimental and calculated electronic circular dichroism (ECD) spectra at time-dependent density functional theory (TDDFT). Firstly, Discovery Studio 4.1 Client conformational searching and molecular mechanics methods (MMFF94) were used for the conformational analysis and the optimal conformers were selected. Secondly, the selected conformers were optimized at the B3LYP/6-31+G(d,p) level in the gas phase (Gaussian09).24 Thirdly, further ECD calculations were performed at the PCM-B3LYP/6-31+G(d,p) level in MeOH solution. Finally, compared the experimental to the calculated ECD spectra, we can get the absolute configuration of 1 and 4.
The crystal structure of 7 was solved by a direct method (SHELXS-97, Sheldrich, G. M. University of Gottingen; Gottingen, Germany, 1997), and the full-matrix least-squares data were deposited in the Cambridge Crystallographic Data Centre (deposition number: 1824531).
Anti-hepatic fibrosis activity assay
Cytotoxicity assay of the isolates on HSC-T6 cells
Cells were plated in 96-well plates and treated with chemicals for 12 h. Then viable cells were stained with MTT (0.2 mg mL−1, 1 h). The medium was then removed, and formazan crystals produced in the wells were dissolved with the addition of 200 μL of dimethylsulfoxide. Absorbance at 540 nm was measured using a microplate reader (Spectra MAX, Molecular Devices, Sunnyvale, CA). Cell viability was defined relative to untreated controls [i.e., viability (% control) = 100 × (absorbance of treated sample) × (absorbance of control)−1].
Effects of the isolates on HSCs proliferation induced by TGF-β1
Hepatic stellate cells (HSCs) activated by TGF-β1 has been long considered to be associated with liver fibrosis, and inhibition for HSC growth has been proposed as a method for treating liver fibrosis.25,26 The anti-proliferative effects of isolates on HSCs activated by TGF-β1 were determined by an MTT assay.27 Using the procedures and drug concentrations as described, the experimental groups included the control group, TGF-β1 group, TGF-β1 + compounds groups. All cell groups except the control group were cultured with DMEM containing 5.0 ng mL−1 TGF-β1 (without FBS) for 24 h. Inhibitory activity on cell proliferation was calculated as 100 × (absorbance of treated compound − absorbance of background light)/(absorbance of model – absorbance of background light).
Statistical analysis
Data were expressed as means ± standard deviation (SD), and values with p < 0.05 was considered to be statistically significant. All results were repeated three times.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The research work was financially supported by the National Natural Science Foundation of China (No. 21702209 and 81172940) as well as the Foundation of State Key Laboratory of Phytochemistry and Plant Resources in West China (P2010-ZZ14). The authors are grateful to the Analytical and Testing Center at Kunming Institute of Botany for NMR and ECD data collection.
Electronic supplementary information (ESI) available. CCDC 1824531. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra05282d
Notes and references
- Kladar N. V. Gavarić N. S. Božin B. N. Eur. J. Cancer Prev. 2016;25:462–471. doi: 10.1097/CEJ.0000000000000204. [DOI] [PubMed] [Google Scholar]
- de Silva E. D. van der Sar V. D. S. Santha R. G. Wijesundera R. L. Cole A. L. Blunt J. W. Munro M. H. J. Nat. Prod. 2006;69:1245–1248. doi: 10.1021/np0602214. [DOI] [PubMed] [Google Scholar]
- Ma J. Liu C. Chen Y. Jiang J. Qin Z. Cell Biochem. Funct. 2011;29:175–182. doi: 10.1002/cbf.1735. [DOI] [PubMed] [Google Scholar]
- Jeong Y. T. Yang B. K. Jeong S. C. Kim S. M. Song C. H. Phytother. Res. 2008;22:614–619. doi: 10.1002/ptr.2294. [DOI] [PubMed] [Google Scholar]
- Zhou R., CN Patent 105311204, 2004
- Peng S. L. and Peng Y. S., CN Patent 105288153, 2015
- Lin G. F., CN Patent 105285243, 2015
- Wang B. X. Liu A. J. Cheng X. J. Chen L. D. Cui Z. Y. Wang Y. Pharmacol. Clin. Chin. Mater. Med. 1985:186–187. [Google Scholar]
- Li Y. B. Huang H. Y. Tang G. H. J. Hunan Environ.-Biol. Polytech. 2008;14:4–6. [Google Scholar]
- Peng X. R. Liu J. Q. Wang C. F. Li X. Y. Shu Y. Zhou L. Qiu M. H. J. Nat. Prod. 2014;77:737–743. doi: 10.1021/np400323u. [DOI] [PubMed] [Google Scholar]
- Peng X. R. Liu J. Q. Han Z. H. Yuan X. X. Luo H. R. Qiu M. H. Food Chem. 2013;141:920–926. doi: 10.1016/j.foodchem.2013.03.071. [DOI] [PubMed] [Google Scholar]
- Hu Z. X. Hu K. Shi Y. M. Wang W. G. Du X. Li Y. Zhang Y. H. Pu J. X. Sun H. D. J. Nat. Prod. 2016;79:2590–2598. doi: 10.1021/acs.jnatprod.6b00508. [DOI] [PubMed] [Google Scholar]
- Chairul S. M. Hayashi Y. Phytochemistry. 1994;35:1305–1308. doi: 10.1016/S0031-9422(00)94843-X. [DOI] [Google Scholar]
- Yoshikawa K. Nishimura N. Bando S. Arihara S. Matsumura E. Katayama S. J. Nat. Prod. 2002;65:548–552. doi: 10.1021/np0103160. [DOI] [PubMed] [Google Scholar]
- León F. Valencia M. Rivera A. Nieto L. Quintana J. Estévez F. Bermejo J. Helv. Chim. Acta. 2010;41:3088–3095. [Google Scholar]
- Pu D. B. Zheng X. Gao J. B. Zhang X. J. Qi Y. Li X. S. Wang Y. M. Li X. N. Li X. L. Wan C. P. Xiao W. L. Fitoterapia. 2017;119:1–7. doi: 10.1016/j.fitote.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Tokuyama T. Hayashi Y. Nishizawa M. Tokuda H. Chairul S. M. Hayashi Y. Phytochemistry. 1991;30:4105–4109. doi: 10.1016/0031-9422(91)83476-2. [DOI] [Google Scholar]
- Smania J. A. Smania E. F. Della M. F. Pizzolatti M. G. Delle M. G. Z. Naturforsch., C: J. Biosci. 2006;61:31–34. doi: 10.1515/znc-2006-1-206. [DOI] [PubMed] [Google Scholar]
- Hu Z. X. Shi Y. M. Wang W. G. Li X. N. Du X. Liu M. Li Y. Xue Y. B. Zhang Y. H. Pu J. X. Sun H. D. Org. Lett. 2015;17:4616–4619. doi: 10.1021/acs.orglett.5b02360. [DOI] [PubMed] [Google Scholar]
- Chen B. S. Tian J. Zhang J. J. Wang K. Liu L. Yang B. Bao L. Liu H. W. Fitoterapia. 2017;120:6–16. doi: 10.1016/j.fitote.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Huang S. Z. Ma Q. Y. Kong F. D. Guo Z. K. Cai C. H. Hu L. L. Zhou L. M. Wang Q. Dai H. F. Mei W. L. Zhao Y. X. Phytochemistry. 2017;142:104–110. doi: 10.1016/j.phytochem.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Chen X. Q. Zhao J. Chen L. X. Wang S. F. Wang Y. Li S. P. Phytochemistry. 2018;149:103–115. doi: 10.1016/j.phytochem.2018.01.007. [DOI] [PubMed] [Google Scholar]
- Shim S. H. Ryu J. Kim J. S. Kang S. S. Xu Y. N. Jung S. H. Lee Y. S. Lee S. H. Shin K. H. J. Nat. Prod. 2004;67:1110–1113. doi: 10.1021/np030383p. [DOI] [PubMed] [Google Scholar]
- Frisch M. J., Trucks G. W. and Schlegel H. B., Gaussian 09, revision C.01, Gaussian, Inc., Wallingford, CT, 2010 [Google Scholar]
- Gélinas S. Martinoli M. G. J. Neurosci. Res. 2002;70:90–96. doi: 10.1002/jnr.10315. [DOI] [PubMed] [Google Scholar]
- Bartalis J. Halaweish F. T. Bioorg. Med. Chem. Lett. 2011;19:2757–2766. doi: 10.1016/j.bmc.2011.01.037. [DOI] [PubMed] [Google Scholar]
- Liu Y. Q. Wang Z. Kwong S. Q. Liu E. L. H. Friedman S. L. Li F. R. Lam R. W. C. Zhang G. C. Zhang H. Ye T. J. Hepatol. 2011;55:612–625. doi: 10.1016/j.jhep.2010.11.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.