Abstract
Background:
Declining proteostasis with aging contributes to increased susceptibility to neurodegenerative diseases, including Alzheimer’s disease (AD). Emerging studies implicate impairment of the endosome-lysosome pathway as a significant factor in the pathogenesis of these diseases. Previously we demonstrated that BAG3 regulates phosphorylated tau clearance. However, we did not fully define how BAG3 regulates endogenous tau proteostasis, especially in the early stages of disease progression.
Methods:
Mass spectrometric analyses were performed to identify neuronal BAG3 interactors. Multiple biochemical assays were used to investigate the BAG3-HSP70-TBC1D10B (EPI64B)-Rab35-Hrs regulatory networks. Live-cell imaging was used to study the dynamic of endosomal pathway. Immunohistochemistry and immunoblotting were performed in human AD brains and BAG3 overexpressed P301S tau transgenic mice.
Results:
The primary group of neuronal BAG3 interactors identified are involved in the endocytic pathway. Among them were key regulators of small GTPases, such as the Rab35 GTPase activating protein, TBC1D10B. We demonstrated that a BAG3-HSP70-TBC1D10B complex attenuates the ability of TBC1D10B to inactivate Rab35. Thus, BAG3 interacts with TBC1D10B to support the activation of Rab35 and recruitment of Hrs, initiating ESCRT-mediated endosomal tau clearance. Further, TBC1D10B shows significantly less co-localization with BAG3 in AD brains than in age-matched controls. Overexpression of BAG3 in P301S tau transgenic mice increased the co-localization of phospho-tau with the ESCRT III protein CHMP2B and reduced the levels of the mutant human tau.
Conclusion:
We identified a novel BAG3-TBC1D10B-Rab35 regulatory axis that modulates ESCRT-dependent protein degradation machinery and tau clearance. Dysregulation of BAG3 could contribute to the pathogenesis of AD.
Keywords: Tau, BAG3, ESCRT, Hrs, Rab35, TBC1D10B
Introduction
Maintaining proteostasis is vital for neuronal function and healthy aging. A critical aspect of proteostasis is an efficient system that recognizes and clears damaged or unnecessary proteins. Disruption of proteome homeostasis is likely a significant contributing factor to the pathogenesis of neurodegenerative diseases that are characterized by the accumulation of aggregation-prone proteins, such as Alzheimer’s disease (AD)(1, 2). One of the defining hallmarks of AD is the presence of intraneuronal aggregates of phosphorylated tau (3). The accumulation of tau, particularly in an oligomeric state, advance AD pathogenesis (4). There is increasing evidence that compromised degradative mechanisms, and in particular lysosome dysfunction, contribute to the accumulation of toxic tau species and other disease-relevant proteins (5, 6).
There is compelling evidence that impairment of the endosome-lysosome pathway is a significant contributor to the pathogenesis of neurodegenerative proteopathies such as AD and Parkinson’s disease (7, 8). The endosome-lysosome pathway is an important protein clearance mechanism that directs the engulfment of protein cargo and trafficking to the lysosome for degradation. This process depends on the endosomal sorting complex required for transport (ESCRT) machinery, which mediates multivesicular body (MVB) formation and fusion with the lysosome (9). Deficits in ESCRT and the endosome-lysosome pathway, evidenced by enlargement of the early endosome compartment and increased endosome/lysosome pH, are some of the earliest observable pathological changes in AD and other neurodegenerative diseases (10, 11). However, the mechanisms linking endosome-lysosome pathway dysregulation to AD pathogenesis have not been fully-elucidated.
Bcl-2-associated anthogene 3 (BAG3) is a stress-induced, multi-domain protein that is increased during aging (12) and plays a critical role in maintaining proteostasis, and thus neuronal health (13–17). Interestingly, neurons with higher BAG3 expression are more resistant to tau pathology in AD (18), and recent data suggest that BAG3 levels are lower in AD brain compared to age-matched controls (15). BAG3 promotes the clearance of tau (19) and other disease-relevant aggregation-prone proteins (14, 20). These findings point to the importance of BAG3 in maintaining neuronal proteostasis, which has historically been attributed to its role in autophagy (13), although the mechanisms involved have not been fully defined and it is becoming apparent that this is likely not the only degradative pathway regulated by BAG3.
In the present study, we investigated the endogenous neuronal BAG3 interactome through unbiased immunoprecipitation-coupled mass spectrometry (IP-MS). Surprisingly, the endosome-lysosome pathway was the most over-represented. Intriguingly, a key BAG3 interactor was the primary GTPase-activating protein (GAP) for Rab35, TBC1D10B ((Tre-2/USP6, BUB2, Cdc16) Domain Family 10B) (21) (also known as asEPI64B (22)). Rab35 is a prominent small GTPase which localizes to endosomes and plays a key role in protein clearance (23–25). Activated Rab35 has been shown to recruit Hrs/ESCRT0 and traffic tau as cargo to the endosome (26), suggesting that Rab35 can regulate the clearance of tau through the ESCRT-mediated endosome-lysosome pathway. However, the regulatory mechanisms underlying Rab35 in mediating tau clearance are not well understood.
In the present study, we demonstrate that the association of TBC1D10B with BAG3 disinhibits Rab35 activation, which in turn keeps Rab35 in a GTP bound, activated state resulting in enhanced ESCRT0 recruitment and Hrs mobility. BAG3 also promotes tau recognition by ESCRT machinery, resulting in greater MVB trafficking of tau, lower hyperphosphorylated tau levels, and rescued neurite density in P301S tau (PS19 line) mice. Further, TBC1D10B shows significantly less co-localization with BAG3 in AD brains than in age-matched controls. Our findings define a new role of BAG3 in regulating vacuolar-dependent protein degradation machinery through the TBC1D10B-Rab35-Hrs axis and provide a new therapeutic avenue for modulating tau clearance.
Methods and Materials
Reagents
Detailed information of plasmids and antibodies can be found in Supplemental Methods and Materials in supplement 1.
Animals
All animal procedures were approved by the University Committee on Animal Research of the University of Rochester (Protocol #:2007–023E&ER). Detailed information about the mice together with the sample preparation procedures are listed in Supplemental Methods and Materials in Supplemental Methods and Materials in supplement 1.
Other experiment procedures
Experiment procedures including cell culture, generation of lentivirus, live-cell imaging, stereotaxic surgeries, mass spectrometry analysis, Rab35 activity assay, immunoprecipitation, immunoblotting, immunohistochemical and Immunofluorescence staining can be found in Supplement 1.
Statistical analysis
All image measurements were obtained from the raw data. GraphPad Prism was used to plot graphs and perform statistical analysis. The statistical tests used are denoted in each Figure legend, and statistical significance was defined as *p<0.05 and **p<0.01. All immunoprecipitation data are repeated at least twice. For the live-cell imaging analysis, the difference of Hrs mobility curves (Figure 6D) was transformed into an accumulation curve and then analyzed using the Kolmogorov–Smirnov test.
Results
BAG3 has been shown to play a role in autophagy (19, 27). However, as a co-chaperone protein with multiple domains, BAG3 likely has regulatory functions beyond autophagy through its interaction with other proteins. To better understand the role of BAG3 in mediating these processes, we used IP-MS analysis to identify interactors of endogenous BAG3 in mature primary neuron cultures. We identified 127 high-confidence BAG3-interacting proteins in the immunoprecipitates from neurons transduced with scrambled virus compared to shBAG3-mediated knockdown neurons (Supplement Table 1). We validated several of these putative BAG3 interactors, including MAP6 and clathrin heavy chain, by immunoprecipitation (Supplemental Figure S2A&B). KEGG analysis of these BAG3 interactors revealed endocytosis as the highest over-represented pathway (Figure 1A). Protein interaction mapping using STRING identified clusters of endocytosis-related and chaperone/co-chaperone proteins (Figure 1B and Supplemental Figure S3). GAPs play crucial roles in endocytosis (28, 29). TBC1D10B (also known as EPI64B or FP2461) is one of the major GAPs for Rab35 (21, 30) and was identified as a BAG3 interactor. Rab35 regulates endocytosis, recycling of synaptic vesicles, neurite elongation (23, 31), and the targeting of tau to the endolysosome compartment (26). Given the pivotal role of Rab35 in mediating vacuolar processes, neuronal health, and tau clearance, we investigated the potential role of BAG3 in mediating endosome/lysosome pathways via interacting with TBC1D10B.
Figure 1.
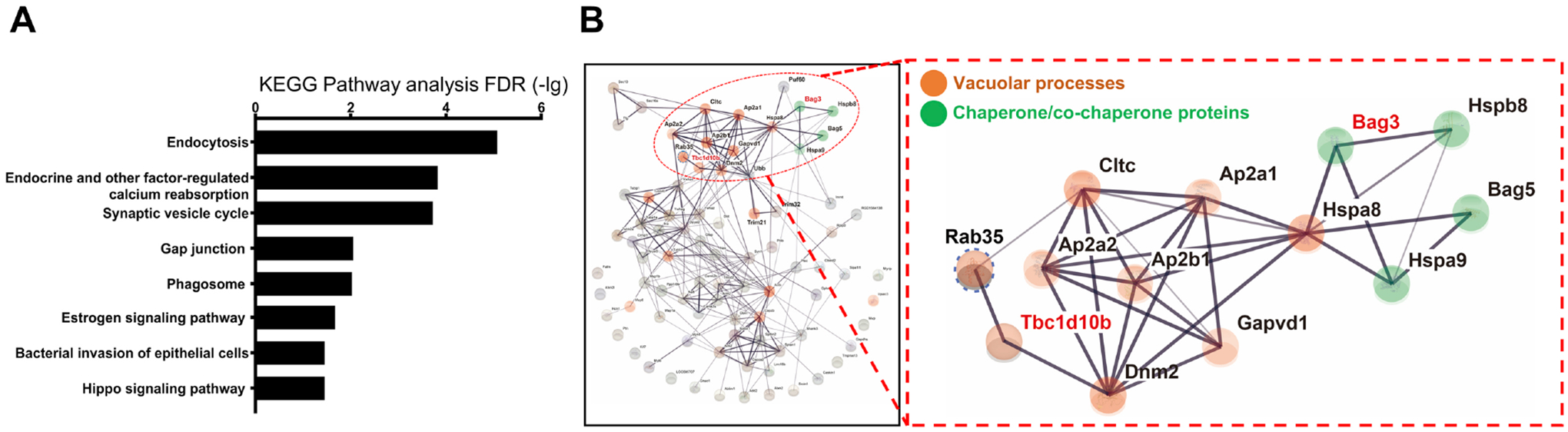
BAG3 interacts with proteins of the endocytosis pathway. Rat cortical neurons were transduced with lentivirus expressing scrambled (Scr) or BAG3 shRNA. Corresponding lysates were immunoprecipitated for BAG3 and associating proteins were run through LC-MS/MS. (A) KEGG enrichment analysis of BAG3 associated proteins with PSM ratios greater than 3 (Scr/shBAG3), FDR-adj. p-value < 0.01. (B) BAG3-associated protein interaction map using STRING. Line thickness indicates the literature interaction. Nodes directly linked to vacuolar processes are labeled in red, and nodes that are related to chaperone/co-chaperone proteins are labeled in green.
BAG3 interacts with TBC1D10B in hippocampal and cortical neurons.
To verify the interaction of TBC1D10B with BAG3, we demonstrated that endogenous BAG3 co-immunoprecipitated with TBC1D10B from rat cortical neuron lysates (Figure 2A). Immunohistochemistry (IHC) of 8-month-old wildtype WT mouse brain showed TBC1D10B puncta co-localized with BAG3 throughout neurons in the CA1 region of the hippocampus (Figure 2B). Like our in vivo observations, in rat primary neuronsTBC1D10B co-localized with BAG3 in the soma in punctate-like structures (Figure 2C–E). Co-localization analysis (32) showed overlap between TBC1D10B puncta and BAG3 in the soma (12.4 ± 2.8 %, mean ± SEM). Together, these data show BAG3 and TBC1D10B interact and co-localize in neurons. Extending this to postmortem human brains, the co-localization of BAG3 with TBC1D10B was significantly reduced in human AD brain samples compared with age-matched controls (Figure 2F–I).
Figure 2.
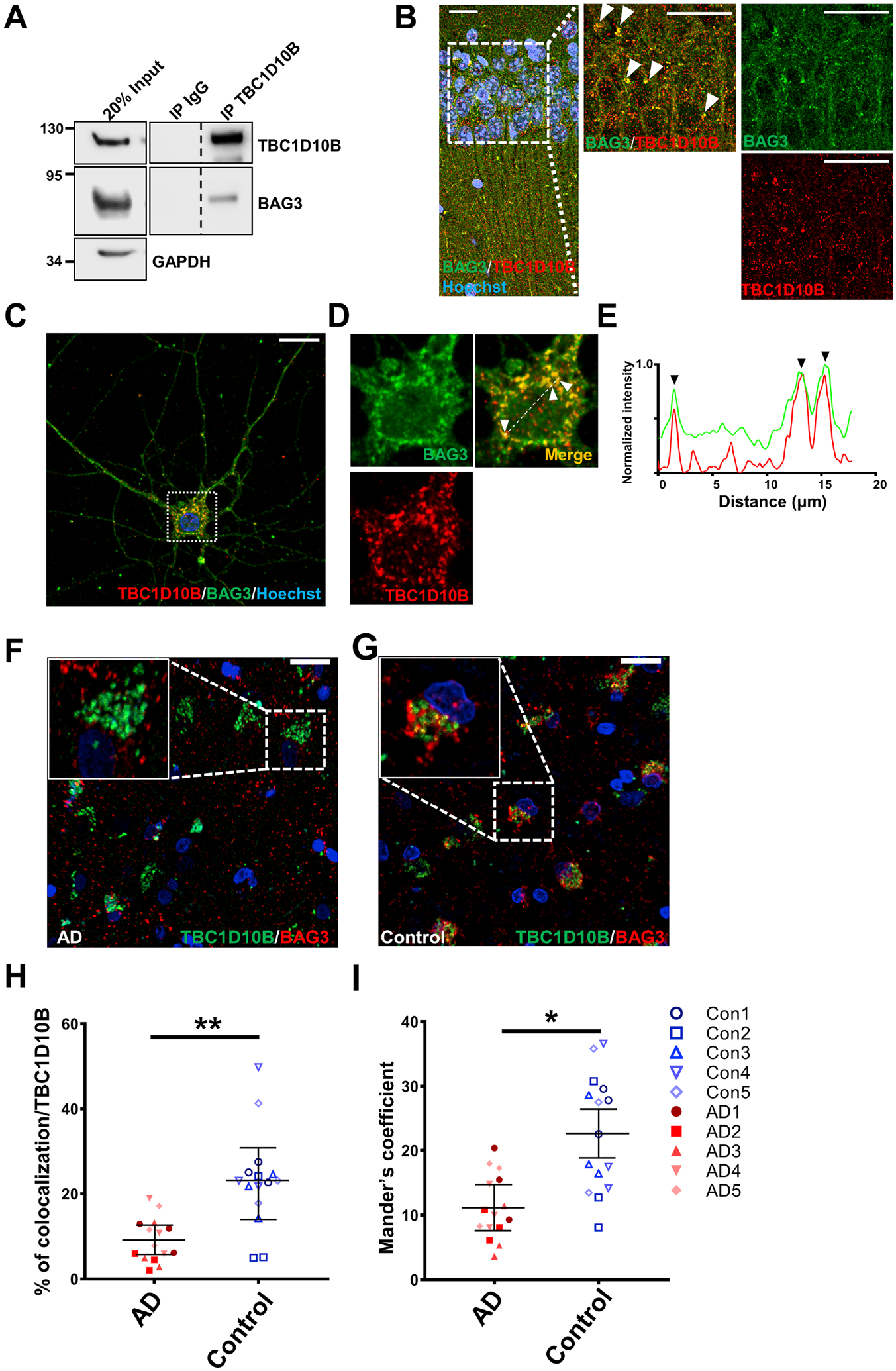
BAG3 associates with TBC1D10B in neurons. (A) Rat cortical neuron lysates were immunoprecipitated with an anti-TBC1D10B antibody and immunoblotted for BAG3. An indicated fraction of cell lysate was used as input control with GAPDH as loading control. Immunoprecipitates were probed for TBC1D10B, BAG3, and Rab35. Vertical dashed lines indicate that intervening lanes were removed. However, all images were from the same blot and exposure. (B) Representative immunostaining of BAG3 (green) and TBC1D10B (red) co-localization in the CA1, counterstained with Hoechst 33342. Scale bars, 20μm. (C-G) Neurons were immunostained for BAG3 (red) and TBC1D10B (green). Overlap of BAG3 with TBC1D10B puncta was observed in the soma (F) with corresponding line scans (E). Arrowheads indicate areas of overlap. Scale bar, 10 μm. (F, G) Representative immunostaining of BAG3 (red) and TBC1D10B (green) co-localization in human AD brain and age matched controls. Scale bars, 10 μm. (H) Quantification of the co-localization between TBC1D10B, BAG3 tau based on volume. (I) Quantification of co-occurring of BAG3 in TBC1D10B, using Mander’s coefficient. 3 slides each from 5 brains in each group were used for analysis. Data are shown as mean ± SEM with unpaired Student’s t-test;. *, P<0.05; **, P<0.01.
The interaction of BAG3 with TBC1D10B is facilitated by HSP70.
Given the plethora of domain-specific BAG3 interactors (14), we next assessed which domain(s) in BAG3 facilitate TBC1D10B interaction using a co-expression model in BAG3 knockout (KO) HEK cells. FLAG-TBC1D10B was co-expressed with WT and mutant forms of BAG3 followed by co-immunoprecipitation (Figure 3A). WT BAG3 readily associated with TBC1D10B, as did BAG3 with the GPG mutation, which is a mutation of the IPV domains that prevents association with small heat shock proteins (33, 34).. The WAWA mutant form of BAG3, which prevents BAG3 WW domain association with synaptopodin-2 (myopodin) (35) and synaptopodin (27), decreased the apparent co-immunoprecipitation of BAG3 with TBC1D10B. Strikingly, the L462P mutation in the BAG domain BAG3, which mediates binding with HSP70 (36, 37), abolished its interaction with TBC1D10B. L462P is a rare mutation in BAG3 that causes cardiomyocyte protein aggregation and dilated cardiomyopathy (37), and has been speculated to decrease BAG3-HSP70 association. To assess whether HSP70 is involved in modulating BAG3-TBC1D10B interaction, we first verified that the L462P mutation of BAG3 strongly decreased (>50%) the association with HSP70 using a co-expression model of WT or L462P BAG3 with V5-HSP70 in BAG3 KO cells (Figure 3B). Next, we examined whether modulating HSP70 levels or activity regulated BAG3-TBC1D10B interaction. HSP70 over-expression resulted in enhanced BAG3 co-immunoprecipitation with TBC1D10B compared to mock-transfected HEK cells (Figure 3C). Pharmacological inhibition of HSP70 ATPase activity and HSP70-BAG3 association using 10μM YM-01 (38) disrupted BAG3-TBC1D10B co-immunoprecipitation (Figure 3D). Pairing V5-HSP70 over-expression with YM-01 treatment prevented this loss of BAG3-TBC1D10B association, likely due to saturation of YM-01 with exogenous HSP70. Overall, these results suggest HSP70 facilitates the association of BAG3 with TBC1D10B.
Figure 3.

HSP70 facilitates the association of BAG3 with TBC1D10B at the BAG domain. (A) BAG3 null HEK293TN cells were transiently transfected with FLAG-TBC1D10B together with empty vector (Control), wild type BAG3 (WT), or BAG3 with WAWA mutation (WAWA), GPG mutation (GPG) and L462P mutation (L462P). Cell lysates were immunoprecipitated with an anti-FLAG antibody, followed by blotting for BAG3 and FLAG. Immunoprecipitates were probed for FLAG and BAG3. (B) BAG3 null HEK293TN cells were transiently transfected with V5-HSP70 together with WT BAG3 or L462P BAG3. Cell lysates were immunoprecipitated with a V5 tag antibody, followed by blotting for BAG3 and V5. (C) HEK293TN cells were transiently transfected with FLAG-TBC1D10B together with an empty vector (Control) or V5-HSP70. Cell lysates were immunoprecipitated with an anti-FLAG antibody, followed by blotting for endogenous BAG3, FLAG, and V5. (D) HEK293TN cells were transient co-expressed with FLAG-TBC1D10B together with an empty vector or V5-HSP70 and treated with 10 μM YM01 or DMSO (vehicle control). Corresponding lysates were collected and immunoprecipitated with anti-FLAG antibody, followed by blotting for BAG3, V5, and FLAG. (E) BAG3 null HEK293TN cells were transiently transfected with V5-HSP70 and FLAG-TBC1D10B. Corresponding lysates were immunoprecipitated with anti-FLAG antibody, followed by blotting for V5 and FLAG. Results of different exposure times for the same blot were separate with a vertical dotted line. Indicated fraction of cell lysate was used as input control. (F) BAG3 null HEK293TN cells were transiently transfected with FLAG-TBC1D10B, V5-HSP70 together with an empty vector (Control) or wildtype BAG3 (BAG3 WT). Cell lysates were immunoprecipitated with anti-FLAG antibody, followed by blotting for BAG3, V5, and FLAG. (G) Cell lysate from F was immunoprecipitation with the 0.5μg IgG or V5 antibody which is saturated by V5-HSP70 and blotted for FLAG. GAPDH was used as a loading control for all input lanes.
These findings prompted us to examine if HSP70 associated directly with TBC1D10B, independent of BAG3. Co-expressing FLAG-TBC1D10B and V5-HSP70 in BAG3 KO HEK cells followed by immunoprecipitation with V5 antibody revealed HSP70 associated with TBC1D10B in the absence of BAG3 (Figure 3E). BAG3 interaction prevents the degradation of the small heat shock protein HspB8 (34, 39), and we identified that the same is true for HSP70, as increased expression of BAG3 greatly increases HSP70 levels (Figure 3F). To examine if BAG3 regulated the association between HSP70 with TBC1D10B, we co-transfected V5-HSP70, FLAG-TBC1D10B, and WT BAG3 or empty vector followed by a saturated immunoprecipitation with the V5 antibody. This resulted in equal amounts of V5-HSP70 in the control and BAG3 overexpression group pulled down with V5 (Figure 3G). These data demonstrated that BAG3 greatly enhances the association of HSP70 with TBC1D10B. To determine if the interaction is direct, we used recombinant His-TBC1D10B-FLAG, His-HSP70 and GST-BAG3. Our results confirm that BAG3 directly interacts with HSP70 (Supplemental Figure S4A). TBC1D10B directly interacts with GST-BAG3, and HSP70 enhanced the association of TBC1D10B with GST-BAG3. The addition of ATP didn’t affect the association between BAG3 and TBC1D10B suggesting that HSP70 regulates the association of BAG3 and TBC1D10B through an ATP-independent mechanism (Supplemental Figure S4B). We didn’t observe an association of TBC1D10B with HSP70 without the presence of BAG3 suggesting no direct interaction between HSP70 and TBC1D10B (Supplemental Figure S4C). Overall, our data indicate that HSP70 facilitates the association of BAG3 and TBC1D10B, and that TBC1D10B and HSP70 are linked by BAG3 to form a complex.
BAG3 regulates the Rab35 activity through the association of TBC1D10B.
TBC1D10B is a GAP that specifically associates with the GTP bound, active form of Rab35 and promotes GTP hydrolysis, leading to the inactivation of Rab35 (21). To determine how BAG3 may regulate the function of TBC1D10B, neurons were transduced with lentivirus expressing scrambled or BAG3 shRNA, followed by immunoprecipitation of TBC1D10B and immunoblotting for Rab35. Neuronal BAG3 knockdown alters TBC1D10B-Rab35 association without modifying total levels of TBC1D10B (Figure 4A). This finding leads to two opposing hypotheses: 1) Because GAPs preferentially bind GTP bound Rabs (40, 41), BAG3 promotes the association of TBC1D10B with Rab35 and leads to the inactivation of Rab35 (conversion to GDP-Rab35) and thus decreases Rab35-TBC1D10B interaction; 2) Binding of BAG3 attenuates the ability of TBC1D10B to stimulate the GTPase activity of Rab35, resulting in prolonged association of TBC1D10B with Rab35 (42). To test these hypotheses, we used an established Rab35 activity assay based on GST-tagged RBD35, which specifically binds the active, GTP-bound form of Rab35 (43). Cell lysates from rat neurons transduced with lentivirus expressing scrambled or BAG3 shRNA were incubated with GST or GST-RBD35 on glutathione beads, and the precipitates blotted for Rab35. Knockdown of BAG3 did not alter total protein levels of Rab35, but instead reduced GTP-bound Rab35 (Figure 4B). This finding supports our second hypothesis that BAG3 keeps Rab35 in a GTP-bound state by associating with TBC1D10B, thereby preventing Rab35 inactivation.
Figure 4.

BAG3 associates with TBC1D10B to regulate the Rab35 activity. (A) Rat cortical neurons were transduced with lentivirus expressing shBAG3 or a scrambled (Scr) version. Cell lysates were immunoprecipitated with an anti-TBC1D10B antibody and immunoblotted for BAG3, TBC1D10B, and Rab35. An 8% fraction of cell lysate was used as input control. (B) Rat cortical neurons were transduced with lentivirus expressing scrambled (Scr) or shBAG3 shRNA. Rab35 activity was examined by incubation of cell lysates with purified GST or GST-RBD35, followed by precipitation with glutathione beads (43). The precipitated samples were blotted for Rab35 and GST. 4% of the cell lysate was used as input control. (C) BAG3 null HEK293TN cells were transiently transfected with Myc-Rab35 together with empty vector (Control), wild type BAG3 (WT BAG3) or L462P BAG3. Rab35 activity was examined by pull-down with GST-RBD35, as described in B. The precipitated samples were blot for Rab35 and GST. 1.2% of the cell lysate was used as input control. (D) Rat cortical neurons were transduced with lentivirus expressing scrambled (Scr), shBAG3, shTBC1D10B, or both shBAG3 and shTBC1D10B shRNAs. Rab35 activity was examined by pull-down with GST-RBD35. Arrow indicates the GST-RBD35 fusion protein; **, GST-RBD35 degradation products, *, GST protein. GAPDH was used as a loading control for all input lanes.
To examine if disrupting the association of BAG3 with TBC1D10B could also affect Rab35 activity, we co-transfected Myc-Rab35 with WT BAG3, L462P BAG3, or empty vector in BAG3 KO HEK cells. Repeating the Rab35 activity assay with these conditions showed that loss of BAG3 substantially decreased the level of GTP-bound Rab35 without affecting its expression level, consistent with our findings in rat cortical neurons (Figure 4C). Interestingly, the L462P mutation of BAG3, which greatly disrupted BAG3 association with TBC1D10B, also decreased the level of GTP-bound Rab35 (Figure 4C).
Our findings suggest that BAG3 associates with TBC1D10B in order to maintain Rab35 in an active state. In contrast, depletion of BAG3 or freeing TBC1D10B from BAG3 will increase the inactivation of Rab35. This finding further leads to another hypothesis that a primary mechanism by which BAG3 regulates the Rab35 activity is through mediating the function of TBC1D10B. To test whether BAG3 regulates Rab35 activity through TBC1D10B, we examined Rab35 activity in conditions with BAG3, TBC1D10B, or both were knocked down using shRNA. Resulting data demonstrated that, compared to scrambled shRNA conditions, GTP-bound Rab35 levels are increased when TBC1D10B is knocked down, but decreased when BAG3 is knocked down. The decreased GTP-bound Rab35 levels observed with BAG3 knockdown was alleviated when TBC1D10B was knocked down as well (Figure 4D). Our findings suggest a model where BAG3 inhibits the function of TBC1D10B, maintaining active Rab35 levels. Conditions with depleted BAG3, such as in AD(15), could therefore lead to inactivated Rab35 and impaired endocytosis.
TBC1D10B has other Rab targets, including Rab8a (44) and Rab27B (45). Therefore, we examined if Rab35 is able to rescue the effect of shBAG3 on tau clearance (Supplemental Figure S5A). Overexpression of either WT Rab35 or Rab35Q67L significantly reduced the increased p-Ser396/404 and p-Thr231 tau levels in response to BAG3 depletion (Supplemental Figure S5B–D).
BAG3 interacts with TBC1D10B to regulate tau sorting into the endocytic pathway through the ESCRT system.
Rab35 is a GTPase that plays an essential role in the endocytic pathway and facilitates tau clearance in neurons (26). BAG3 promotes tau degradation (19) and interacts with TBC1D10B (Figure 2A). BAG3-TBC1D10B association regulates the activity of Rab35 (Figure 4B), and thus TBC1D10B likely plays a role in phosphorylated tau clearance. Indeed, our data show that depletion of TBC1D10B in neurons significantly decreased the levels of p-Thr231, p-Ser262, and p-Ser396/Ser404 tau in mature neurons (Figure 5A&B). We next examined whether combining knockdown of TBC1D10B and BAG3 rescued phosphorylated tau levels (Figure 5C). First, we supported our previous results showing BAG3 knockdown increased the levels of p-Thr231, p-Ser262, and p-Ser396/Ser404 tau in mature neurons (19). Consistent with our proposed model, simultaneous depletion of TBC1D10B and BAG3 negated this effect on p-tau levels compared with BAG3 knockdown alone (Figure 5D&E). These data suggested that TBC1D10B is downstream of BAG3 in regulating tau clearance. Since Rab35 clears tau through the ESCRT system (26), we hypothesized that BAG3 acts in concert with TBC1D10B to regulate phosphorylated tau clearance by similar means. We assessed whether BAG3 knockdown altered the co-localization between CHMP2B, an ESCRT III component (46), and p-Ser396/404 tau in neuronal processes. In BAG3 knockdown neurons, CHMP2B co-localized with p-Ser396/404 tau significantly less compared with the scrambled group. Depletion of both BAG3 and TBC1D10B significantly increased the co-localization of p-Ser396/404 tau with CHMP2B (Figure 6 and Supplemental Figure S6A–C). Further, 29.6±4.5% of the p-Ser396/404 tau/ CHMP2B co-localized volume is Rab7 positive (Supplemental Figure S6D&E), indicating that BAG3 acts with TBC1D10B to regulate phosphorylated tau clearance through the ESCRT system.
Figure 5.

BAG3 cooperates with TBC1D10B to regulate tau levels in neurons. (A) Rat cortical neurons were transduced with lentivirus expressing scrambled (Scr) or shTBC1D10B shRNA. Cell lysates were immunoblotted for TBC1D10B, p-Ser262, p-Ser396/404, and p-Thr231 tau. GAPDH is used as a loading control. (B) Relative level of p-tau to total tau. Data are shown as mean ± SEM with unpaired Student’s t-test; *, P<0.05, **, P<0.01. (C-D) Rat cortical neurons were transduced with lentivirus expressing scrambled (Scr) or shBAG3 or both shBAG3 and shTBC1D10B shRNAs. Cell lysates were either immunoblotted for BAG3 and TBC1D10B for validating the knockdown (C) or p-Ser262, p-Ser396/404, and p-Thr231 tau (D). (E) Relative level of p-tau to total tau. Data are shown as mean ± SEM with one-way ANVOA and Tukey’s multiple comparisons test. *, P<0.05, **, P<0.01. For all experiment n=3 for each group. Vertical dotted lines indicate that intervening lanes were removed, however, all images were from the same blot and exposure.
Figure 6. BAG3 interacts with TBC1D10B to regulate the tau sorting into the endocytic pathway through the ESCRT system.

(A) Rat cortical neurons were transduced with lentivirus expressing scrambled (Scr), shBAG3, shTBC1D10B or both shBAG3 and shTBC1D10B shRNAs. Neurons were co-immunostained for CHMP2B (red) and p-Ser396/404 tau (green). The corresponding line scans are shown below for neurites. Arrowheads indicate areas of overlap. Scale bar, 5 μm. (B) Quantification of the co-localization between CHMP2B and p-Ser396/404 tau based on volume. (C) Quantification of co-occurring of CHMP2B in p-Ser396/404 tau using Mander’s coefficient. Data were presented as mean ± SD, N=10 neurites from 6+ cells each with one-way ANVOA and Tukey’s multiple comparisons test. *, P<0.05, **, P<0.01.
BAG3 regulates the recruitment of Hrs to Rab35.
When activated, Rab35 recruits Hrs to the surface of the endosome to facilitate the recruitment of other ESCRT machinery to form intraluminal vesicles and in directing cargo, such as phospho-tau species (26), to the endosome for engulfment (31). Since our data suggest that BAG3 regulates the activity of Rab35 through interacting with TBC1D10B, we hypothesize that BAG3, by promoting active Rab35, should facilitate the recruitment of Hrs to Rab35. In primary neurons, Rab35 co-localizes with Hrs both in the soma and neuronal processes, and that this co-localization was significantly decreased in BAG3 knockdown neurons (Figure 7A–F). Additionally, 60.8±2.6% of the Rab35/Hrs co-localized volume is Rab7 positive and thus at the late endosome (Supplemental Figure S7). The recruitment of Hrs to Rab35 to initiate the ESCRT pathway is a dynamic process; Hrs must be both recruited and released for intraluminal vesicle formation of MVBs (47). Therefore we examined eGFP-Rab35 and Hrs-RFP dynamics using live-cell imaging with and without BAG3. In BAG3 KO HEK cells, the majority of Hrs puncta were stationary; 4.5% of the Hrs puncta moved more than 1μm over 10 minutes (Figure 7G&H). Expressing WT-BAG3 rescued Hrs puncta mobility, with 23% of the Hrs puncta moving more than 1μm over 10 minutes (Figure 7G&H). Additionally, the co-localization of Rab35 with Hrs was greatly increased in the presence of BAG3 compared with the BAG3 KO condition (Figure 7I). These findings suggest BAG3 enhances the mobility of Hrs and boosts the recruitment dynamics of Hrs to Rab35. Next, we examined the regulatory function of BAG3 and TBC1D10B on the association of Hrs and Rab35 by expressing Myc-Rab35 with and without overexpression of BAG3 and TBC1D10B. Co-immunoprecipitation analysis revealed that Rab35 and Hrs minimally associate in BAG3 KO cells, but returned to a strong association when BAG3 was re-introduced (Figure 7J). Further, overexpression of TBC1D10B disrupts the association of Rab35 with Hrs in the presence of BAG3 (Figure 7J). These findings suggest BAG3 promotes the recruitment of Hrs to Rab35, and TBC1D10B acts downstream of BAG3 to inhibit this recruitment.
Figure 7. BAG3 and TBC1D10B regulate the mobility of Hrs and its interaction with Rab35.

(A-F) Rat cortical neurons that were transduced with lentivirus expressing scrambled (Scr) or shBAG3 shRNAs were co-immunostained for Rab35(Green) and Hrs (Red). Overlap of Rab35 with Hrs puncta was observed in the soma (A) and neurites (D). Scale bars, 10 μm. The corresponding line scans are shown at the bottom for A or right for D. Arrowheads indicate areas of overlap. (B, E) Quantification of the co-localization between Rab35 and Hrs based on volume. (C, F) Quantification of co-occurring Rab35 in Hrs using Mander’s coefficient. N=12 neurites from 6 different neurons in each group. Data was presented as mean±SD with unpaired Student’s t-test., *P<0.05, **P<0.01. (G) BAG3 null HEK293TN cells were transfected with BAG3 WT (+BAG3) or empty vector (BAG3 KO), RFP-Hrs, and GFP-Rab35. Live-cell imaging was imaged at 0.1 Hz for 10 minutes. G1 and G2, are representative snapshots of HEKS with or without BAG3. G1’, G1” and G2’, G2” are 3D kymographs of G1 and G2. The white color indicates the co-localized region of Rab35 and Hrs over time. Scale bars, 20μm. (H) The graph shows the track distance of Hrs puncta in 10 minutes. The distance of the track was binned every 0.1 μm. The percentage of events is defined as the percentage of tracks of Hrs in a certain distance range in one cell. At least 10 cells from each group were analyzed. (p < 0.01, Kolmogorov-Smirnov test). (I) Comparison of co-occurring Rab35 in Hrs using Mander’s co-localization coefficient over time in BAG3 expressing and BAG3 KO cells. Mean±SD, N=12, with unpaired Student’s t-test. **, P<0.01. (J) BAG3 null HEK293TN cells were transfected with Myc-Rab35 and with/without BAG3 and TBC1D10B. Corresponding cell lysates were immunoprecipitated with anti-Myc antibody and immunoblotted for Hrs and Myc. Four percent of the lysates were used for input control. (K-M) BAG3 null HEK293TN cells stably expressing tau were transfected with Myc-Rab35 together with empty vector (Control), wildtype BAG3 (BAG3 WT), or BAG3 mutants. (K) Corresponding lysates were immunoblotted for total tau, Hrs and BAG3 and quantified in (L). N=3 for each group. Data are shown as mean ± SEM with one-way ANOVA and Tukey’s multiple comparisons test. **, P<0.01. n.s. not significant. (M) Cell lysates were immunoprecipitated with Myc antibody and immunoblotted for Hrs. For immunoblots shown in F, G, and I, GAPDH was used as a loading control.
We next examined the role of the different BAG3 domains in mediating the recruitment of Hrs to Rab35. Since Hrs recruitment is important to initiate the ESCRT pathway to sort tau into endosome-lysosome for degradation (26, 47, 48), we also examined tau levels with the different BAG3 mutants. To test these hypotheses, we generated stable tau-expressing BAG3 KO HEKs cells, followed by co-transfection with Hrs, Myc-Rab35, and WT BAG3 or BAG3 with different mutations. As expected, WT BAG3, WAWA BAG3, and GPG BAG3 expression significantly reduced tau levels in BAG3 KO HEKs (Figure 7K&L). Expression of L462P BAG3 failed to decrease tau levels, further supporting the importance of BAG3-HSP70 interactions in tau degradation (Figure 7G&H). Next, we immunoprecipitated Myc-Rab35 and blotted for Hrs. We found that expressing all BAG3 constructs, except L462P BAG3, enhanced the association of Hrs with Rab35 compared with the BAG3 KO condition (Figure 7M). Since the L462P mutation of BAG3 disrupted the association of BAG3 and TBC1D10B, our findings indicate that BAG3 interacts with TBC1D10B to regulate the Hrs recruitment to Rab35.
Hippocampal overexpression of BAG3 alleviates the tau pathology development in P301S mice.
Recent reports have suggested BAG3 maintains neuronal proteostasis and acts as a protector against tau aggregation (12, 15, 19). We tested whether BAG3 over-expression reduces tau pathology in male P301S tau transgenic mice (PS19 line) as it has been reported that they develop tau pathology more consistently than females (49–51). Two-month-old P301S mice were subjected to bilateral intrahippocampal AAV injections of T2A-hBAG3 (with an N-terminal FLAG-Myc tag) or eGFP control and collected at 6 months of age. Resulting IHC data showed endogenous BAG3 is expressed both in the soma and neurites of the CA1 region (Figure 8A). BAG3 was overexpressed at approximately four-fold of that of the control group and found largely limited to the hippocampal region (Figure 8B&C). BAG3 was found unaltered in the cerebellum, indicating the regional specificity of BAG3 overexpression (Supplemental Figure S8A&B). Total human tau (recognized by the human tau-specific antibodies HT7 (52) and 5A6 (53)) was significantly decreased in the BAG3 overexpression group (Figure 8D&E). Phosphorylated tau species including p-Ser262 tau, p-Thr231 tau and p-Ser396/404 tau were similarly significantly decreased in the BAG3 overexpression groups compared with the AAV control groups (Figure 8F&G), validating our in vitro studies. As expected, total human tau levels were not significantly changed in the cerebellum, showing that BAG3-mediated tau clearance is dependent on the regional overexpression of BAG3 (Supplemental Figure S8C&D). Quantitative IHC also showed p-Ser262 tau and p-Ser396/404 tau were significantly decreased in the CA1 region in the BAG3 overexpression brains (Figure 8H and I). Moreover, the percentage of oligomeric tau (recognized by T22 antibody (54)) positive cells was significantly reduced in the BAG3 overexpression group compared with the control group (Figure 8J). Staining with the conformation-dependent MC1 tau antibody (55) was also decreased in the CA1 in the BAG3 overexpression group (Figure 8K). These findings suggest that overexpression of BAG3 reduced the level of pathogenic tau species, indicating an essential role for BAG3 in tau clearance in vivo. To extend these findings, AAV9-control or AAV9 shBAG3 particles were injected into CA1 of the hippocampus of 15-month-old C57Bl/6 WT mice and collected 5 months post-injection. Immunofluorescence staining, as well as immunoblotting, demonstrated that expression of BAG3 was greatly reduced in the hippocampus in the shBAG3 group (Supplemental Figure S9C–E).). In addition, we observed a significant increase p-Ser262 tau in the shBAG3 hippocampi supporting a role for BAG3 in tau clearance in vivo (Supplemental Figure S9D&E).
Figure 8.

Overexpression of BAG3 in P301S mice in the hippocampus reduced the tau levels. Two-month-old P301S mice were intrahippocampally injected with AAV2/9-eGFP as control or AAV2/9-eGFP-hBAG3 (with a FLAG-Myc N terminal tag) for BAG3 overexpression (BAG3 O/E), and animals were collected at 6 months of age. (A) Images of the AAV injected brains were co-immunostained for BAG3 (red) and Myc tag (purple). The immunofluorescence images showed increased levels of BAG3 in the hippocampal region in the BAG3 O/E brain. Scale bars, 200μm. (B) Hippocampal lysates from control and BAG3 O/E brains were immunoblotted for BAG3. GAPDH is used as a loading control. (C) The graph shows the level of BAG3 normalized to GAPDH and relative to control. N=4 for control, n=5 for BAG3 O/E, Data are shown as mean ± SEM with Student’s t-test, *P<0.05. (D, E) Hippocampal lysates from control and BAG3 O/E brains were blot for total human tau (HT7 and 5A6). The graphs on the right show the quantitative analysis of tau normalized to GAPDH and relative to control. N=4 for control, n=5 for BAG3 O/E. (F) Representative blots of phosphorylated tau (p-Thr231, p-Ser262, and p-Ser396/Ser404) in the hippocampus lysate of control and BAG3 O/E brains. (G) Quantification of the levels of phosphorylated tau in control and BAG3 O/E hippocampal lysates. Data were normalized to the loading control GAPDH and then compared to controls, N=4 for control, n=5 for BAG3 O/E. (H, I) Phosphorylated tau immunofluorescence (p-Ser262 and p-Ser396/Ser404, Green) staining in the CA1 with corresponding intensity quantification relative to controls, N=4. (J) Oligomeric tau (T22 tau, purple) immunofluorescence staining in the CA1 with corresponding quantification, N=3. (K) Pre-tangle and tangle conformation tau (MC1 tau, green) immunofluorescence staining in the CA1 with corresponding quantification relative to controls, N=3. For all images, scale bars, 20μm and data are shown as mean ± SEM with unpaired Student’s t-test;. *, P<0.05; **, P<0.01.
To demonstrate BAG3 promotes the recruitment of Hrs to Rab35 in vivo, we co-immunostained P301S mouse brains for Hrs and Rab35 and found significantly more Rab35 co-localized with Hrs in the CA1 region in the BAG3 overexpression group compared with the control group (Figure 9A–C). Co-localization of p-Ser396/404 tau with the ESCRT III component CHMP2B was also significantly increased in the BAG3 overexpression group (Figure 9D–F). Finally, we examined the role of BAG3 in the synapse loss of P301S mice (49). IHC staining for MAP2 and PSD95 was used to visualize dendrites and postsynaptic compartments, respectively (56, 57). To quantify the density of MAP2 and PSD95, the images were reconstructed using the Imaris Surface Rendering Model. Our data showed the MAP2 and PSD95 density was significantly increased in the BAG3 overexpression group (Figure 10A–C).
Figure 9.

Overexpression of BAG3 in tau transgenic mice promotes Rab35-mediated recruitment of Hrs and tau sorting into the endocytic pathway. Two-month-old P301S tau mice were intrahippocampally injected with AAV2/9-GFP as control or AAV2/9-GFP-hBAG3 (with a FLAG-Myc N terminal tag) as BAG3 overexpression (BAG3 O/E) and collected at 6-month-old. (A) Representative immunofluorescence staining and co-localization of Rab35 (Green) and Hrs (Red) in the CA1. (B) Quantification of co-occurring Rab35 in Hrs using Mander’s co-localization coefficient. (C) Quantification of the co-localization between Rab35 and Hrs based on volume. N=3 with unpaired Student’s t-test.. (D) Representative immunofluorescence staining of p-Ser396/404 tau (green) co-localization with CHMP2B (red) in the CA1. (E) Quantification of co-occurring of CHMP2B in p-Ser396/404 tau using Mander’s coefficient (F) Quantification of the co-localization between p-Ser396/404 tau and CHMP2B. All samples were counterstained with Hoechst 33342 (blue) to visualize the nuclei. Scale bars denote 20 μm. N=4 with unpaired Student’s t-test. *P<0.05, **P<0.01.
Figure 10.

Overexpression of BAG3 in tau transgenic mice in the hippocampus increased the density of synapses and dendrites. (A) Representative immunofluorescence images of MAP2+ dendrites (green) and PSD95+ postsynaptic compartments (red). CA1 region of the hippocampus was imaged with Hoechst 33342 (blue) counterstaining for nuclei visualization. The border of each neuron was outlined using the ‘surface’ IMARIS function. Scale bars denote 20μm. (B&C) Relative density of MAP2 (B) and PSD95 (C) in the depicted area. The density of MAP2 or PSD95 were defined as the volume of fluorescence positive area/total area. Data are shown as mean ± SEM, N=4 using unpaired Student’s t-test, **, P<0.01.
Discussion
Proteostatic dysregulation occurs during normal aging and could contribute to the pathogenesis of neurodegenerative diseases involving the pathological accumulation of protein, such as in AD, which is defined by the presence of aggregates of abnormally phosphorylated tau (58–61). The cellular machinery involved in clearing toxic tau species is not fully understood, though evidence suggests that these pathways have strong therapeutic potential for the treatment of AD (62). BAG3 is ubiquitously expressed and has numerous interacting partners ranging from synaptic proteins to many chaperone/co-chaperone proteins (27, 63, 64). The function of BAG3 is likely governed by the expression of cell-specific interactors and could be sensitive to over-expression, which may explain disparities in the BAG3 interactome literature (15, 65).
In this study, we assessed the endogenous neuronal BAG3 interactome and uncovered a novel mechanism of BAG3-mediated tau clearance through the endosome-lysosome pathway. BAG3 interacts with TBC1D10B, one of the main regulators of Rab35, to control the trafficking and clearance of toxic tau species. Mutations in the different domains of BAG3 had little effect on tau clearance and TBC1D10B interaction apart from the L462P mutation, which impacts BAG3-HSP70 complexing (66), further supporting the importance of BAG3-HSP70 association in neuronal proteostasis. This BAG3-HSP70-TBC1D10B complex has important implications for the endosome-lysosome pathway. Since TBC1D10B inactivates Rab35 (23, 31), this complex implies BAG3-HSP70 monitoring of neuronal activities controlled by Rab35, such as presynaptic protein homeostasis, neuritic outgrowth, among others (31, 67, 68).
Besides TBC1D10B, BAG3 interacted with other vacuolar-regulating proteins, including clathrin heavy chain and AP2a. Interestingly, these interactors may cooperate with TBC1D10B and Hrs. TBC1D10B has been found at the level of clathrin-coated pits where AP2 is recruited (69). Hrs recruits clathrin to define the timing and morphology of intraluminal vesicle formation (47). Furthermore, IQSEC3, a guanine nucleotide exchange factor (GEF) (70), was also identified as a BAG3 interactor. Interestingly one of the targets of IQSEC3 is Arf6 which is also a regulator for Rab35 through TBC1D10B(30). Therefore, a larger BAG3 network may exist to modulate the endocytic pathway.
Our findings support previous studies showing Rab35-mediated tau clearance (26) by demonstrating that BAG3 disinhibits Rab35 activity through its interaction with TBC1D10B and initiates downstream ESCRT activity through Hrs recruitment, MVB formation, and the trafficking of tau species to the late endosome for degradation in an HSP70-dependent manner (26, 71, 72). Moreover, we provide the first evidence that upregulating BAG3 in disease-relevant regions of the brain in vivo ameliorates tau pathology by increasing engulfment of tau by ESCRT machinery and rescuing neuritic and synaptic morphology. Overall, the present study reports a novel TBC1D10B-BAG3-HSP70 signaling axis for further study as a modulator of ESCRT-mediated endosomal protein clearance in aging and neurodegenerative diseases (Figure 11).
Figure 11.

BAG3-TBC1D10B-Rab35 signaling axis regulates ESCRT and endosomal tau clearance. (A) During normal aging, BAG3 level increases (represented by large green shape), promoting the association of TBC1D10B-BAG3-HSP70. This prevents TBC1D10B from inactivating Rab35, leading to Hrs recruitment and initiation of ESCRT-mediated endosomal tau clearance. (B) In AD brains, however, this increase of BAG3 may be attenuated (represented by small green shape) (15), which would release TBC1D10B from the TBC1D10B-BAG3-HSP70 complex. Free TBC1D10B would thus be able to inactivate Rab35 and prevents endosomal clearance of tau, leading to accumulation over time.
Supplementary Material
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | ||||
| BAG3 Rabbit | Proteintech | 10599–1-AP | ||
| TBC1D10B Rabbit | Invitrogen | PA5–61832 | ||
| V5 Tag Rabbit | Cell Signaling Technology | 13202 | ||
| Rab35 | Proteintech | 11329–2-AP | ||
| Rab7 Alexa Fluor® 647 Conjugated Rabbit | Cell Signaling Technology | 94298 | ||
| HSP70 Rabbit | Proteintech | 10995–1-AP | ||
| β-Actin Rabbit | Cell Signaling Technology | 4970 | ||
| PSD95 Rabbit | Cell Signaling Technology | 3450S | ||
| Hrs Rabbit | Cell Signaling Technology | 15087 | ||
| tau Rabbit | Dako | A0024 | ||
| phospho-Tau (Thr231) Rabbit | Thermo Fisher | OPA-03156 | ||
| CHMP2B Rabbit | Proteintech | 12527-I-AP | ||
| Normal Rabbit IgG Rabbit | EMD Millipore | 12–370 | ||
| Hrs Mouse | Santa Cruz | sc-271455 | ||
| Myc Tag Mouse | Cell Signaling Technology | 2276 | ||
| FLAG Tag Mouse | Cell Signaling Technology | 8146 | ||
| clathrin Mouse | Santa Cruz | sc-271178 | ||
| MAP6 Mouse | Biolegend | 824701 | ||
| HT7 tau Mouse | ThermoFisher | MN1000B | ||
| T22 tau Mouse | Millipore Sigma | ABN454 | ||
| MAP2 AP14 Mouse | Dr. L.I. Binder (PMID: 6591209,3510221) | |||
| MC1 Mouse | Dr. P. Davies | |||
| 5A6 Mouse | DSHB, Univ. Iowa | |||
| tau5 Mouse | Dr L.I. Binder (PMID: 6591209,3510221) | |||
| phospho-Tau (Ser262) 12E8 Mouse | Dr. P. Seubert | |||
| phospho-Tau (Ser396/404) PHF1 Mouse | Dr. P. Davies | |||
| Normal mouse IgG Mouse | EMD Millipore | 12–371 | ||
| Bacterial or Viral Strain | ||||
| Lemo21(DE3) Competent E. coli | New England Biolabs | C2528J | ||
| BL21 E. coli | New England Biolabs | C2530H | ||
| AAV9-SYN1-eGFP | Vector Biolabs | |||
| AAV9-SYN1-GFP-T2A-BAG3 | Vector Biolabs | |||
| AAV9-CMV-mCherry | SignaGen Laboratories | |||
| AAV9-U6-shBAG3-CMV-mCherry | SignaGen Laboratories | |||
| Biological Sample | ||||
| Human brain samples | University of Florida Brain Bank | See supplimental material and method for detailed information | ||
| Cell Line | ||||
| HEK293T cells | System Biosciences | LV900A-1 | ||
| Chemical Compound or Drug | ||||
| YM01 | Sigma | SML0943 | ||
| poly-D-lysine | Sigma | P6407 | ||
| Commercial Assay Or Kit | ||||
| EZ-Link™ Sulfo-NHS-LC-Biotinylation Kit | Thermo Fisher Scientific | 21435 | ||
| HisPurTM Ni-NTA Spin Columns | Thermo Fisher Scientific | 88224 | ||
| Glutathione S-transferase beads | Ge healthcare | 17–5132 | ||
| Deposited Data; Public Database | ||||
| Mass Spectrometry Analysis data | See supplimental table for detailed information | |||
| Organism/Strain | ||||
| B6;C3-Tg(Prnp-MAPT*P301S)PS19Vle/J | Jackson laboratories | Stock No: 008169 | ||
| Peptide, Recombinant Protein | ||||
| GST-BAG3 | Abnova | H00009531-P01 | ||
| Software; Algorithm | ||||
| GraphPad Prism 7 | GraphPad Software | |||
| Imaris | Oxford Instruments | |||
| Image J | NIH | |||
| Transfected Construct | ||||
| pHUUG-shBAG3 | PMID: 25212465 | 5’-AAGGTTCAGACCATCTTGGAA-3’ | ||
| pHUUG-scrRNA | PMID: 25212465 | 5’-CAGTCGCGTTTGCGACTGG-3’ | ||
| pHUUG-shTBC1D10B | This paper | 5’-GCTGTCTTAATTTGCCTTTGG-3’ | ||
| pHUUG-shRab35 ORF | This paper | 5’-CGATTGTGGTGATGTAGCTG-3’ | ||
| pHUUG-shRab35 3’UTR | This paper | 5’-ATTTGTTAAGAGAATGCTCC-3’ | ||
| pcDNA3 TBC1D10B | Genscript | |||
| Myc-Rab35 | Addgene | 47433 | ||
| Myc-Rab35 Q67L | Addgene | 47434 | ||
| eGFP-Rab35 | Addgene | 49552 | ||
| pCS2 Hrs-RFP | Addgene | 29685 | ||
| V5-HSP70 | Addgene | 19510 | ||
| PCDH | System Biosciences | CD516-B2 | ||
| pCDH-WT BAG3 | This paper | |||
| pCDH-GPG BAG3 | This paper | |||
| pCDH-WAWA BAG3 | This paper | |||
| pCDH-L462P BAG3 | This paper | |||
| pCDH-T4 tau-T2A-RFP | This paper | |||
| pGEX-RBD35 | This paper | |||
| FigB-Myc-Rab35 | This paper | |||
| FigB-Myc-Rab35Q67L | This paper | |||
| psPAX2 | Addgene | 12260 | ||
| VSV-G | Addgene | 12259 | ||
| PET28a-TBC1D10B | This paper | |||
| PET28a-HSP70 | This paper |
Acknowledgments:
We thank Dr. C.Pröschel, University of Rochester for providing us with the pHUUG vector; Dr. J. Höfeld, University of Bonn for providing us with BAG3, L462P BAG3 and WAWA BAG3 in pcDNA; Dr. L.I. Binder for the gift of the MAP2 (AP14) and Tau5 antibodies [31, 32]; Dr. P. Davies for the gift of MC1 and phospho-Tau (Ser396/404) (PHF1) antibodies; Dr. P. Dolan for the gift of phospho-Tau (Ser262) antibody. Brain samples were obtained the UF Neuromedicine Human brain and Tissue bank (UF HBTB) at the University of Florida with informed consent of the patients or their relatives and the approval of the local institutional review boards. The UF HBTB is supported by the Florida ADRC (P30AG066506). This work was supported by National Institute of Health (NIH) grants R56 AG067739, R01 NS098769 and R01 AG073121. This research has been facilitated by a fee for service provided by the University of Rochester Mass Spectrometry Resource Laboratory and NIH instrument grant (S10OD021486). This manuscript has been posted on bioRxiv (2021.01.25.428055; doi: https://doi.org/10.1101/2021.01.25.428055).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts
Conflict of interest: The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Hipp MS, Kasturi P, Hartl FU (2019): The proteostasis network and its decline in ageing. Nature reviews Molecular cell biology. 20:421–435. [DOI] [PubMed] [Google Scholar]
- 2.Kurtishi A, Rosen B, Patil KS, Alves GW, Moller SG (2019): Cellular Proteostasis in Neurodegeneration. Mol Neurobiol. 56:3676–3689. [DOI] [PubMed] [Google Scholar]
- 3.Long JM, Holtzman DM (2019): Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell. 179:312–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen XQ, Mobley WC (2019): Alzheimer Disease Pathogenesis: Insights From Molecular and Cellular Biology Studies of Oligomeric Abeta and Tau Species. Front Neurosci. 13:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nixon RA (2020): The aging lysosome: An essential catalyst for late-onset neurodegenerative diseases. Biochim Biophys Acta Proteins Proteom. 1868:140443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y, Sato Y, Im E, Berg M, Bordi M, Darji S, et al. (2019): Lysosomal Dysfunction in Down Syndrome Is APP-Dependent and Mediated by APP-betaCTF (C99). The Journal of neuroscience : the official journal of the Society for Neuroscience. 39:5255–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winckler B, Faundez V, Maday S, Cai Q, Guimas Almeida C, Zhang H (2018): The Endolysosomal System and Proteostasis: From Development to Degeneration. The Journal of neuroscience : the official journal of the Society for Neuroscience. 38:9364–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pensalfini A, Kim S, Subbanna S, Bleiwas C, Goulbourne CN, Stavrides PH, et al. (2020): Endosomal Dysfunction Induced by Directly Overactivating Rab5 Recapitulates Prodromal and Neurodegenerative Features of Alzheimer’s Disease. Cell reports. 33:108420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majumder P, Chakrabarti O (2016): ESCRTs and associated proteins in lysosomal fusion with endosomes and autophagosomes. Biochem Cell Biol. 94:443–450. [DOI] [PubMed] [Google Scholar]
- 10.Hu YB, Dammer EB, Ren RJ, Wang G (2015): The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA (2000): Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. The American journal of pathology. 157:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang M, Ji C, Pallo S, Rahman I, Johnson GVW (2018): Nrf2 mediates the expression of BAG3 and autophagy cargo adaptor proteins and tau clearance in an age-dependent manner. Neurobiology of aging. 63:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behl C (2016): Breaking BAG: The Co-Chaperone BAG3 in Health and Disease. Trends Pharmacol Sci. 37:672–688. [DOI] [PubMed] [Google Scholar]
- 14.Sturner E, Behl C (2017): The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front Mol Neurosci. 10:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Chow HM, Liu Y, Wu D, Shi M, Li J, et al. (2020): Cyclin-Dependent Kinase 5-Dependent BAG3 Degradation Modulates Synaptic Protein Turnover. Biol Psychiatry. 87:756–769. [DOI] [PubMed] [Google Scholar]
- 16.Chakraborty D, Felzen V, Hiebel C, Sturner E, Perumal N, Manicam C, et al. (2019): Enhanced autophagic-lysosomal activity and increased BAG3-mediated selective macroautophagy as adaptive response of neuronal cells to chronic oxidative stress. Redox Biol. 24:101181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin H, Koren SA, Cvetojevic G, Girardi P, Johnson GVW (2021): The role of BAG3 in health and disease: A “Magic BAG of Tricks”. J Cell Biochem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu H, Possenti A, Freer R, Nakano Y, Hernandez Villegas NC, Tang M, et al. (2019): A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nature neuroscience. 22:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei Z, Brizzee C, Johnson GV (2015): BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiol Aging. 36:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao YL, Yang YP, Mao CJ, Zhang XQ, Wang CT, Yang J, et al. (2017): A role of BAG3 in regulating SNCA/alpha-synuclein clearance via selective macroautophagy. Neurobiology of aging. 60:104–115. [DOI] [PubMed] [Google Scholar]
- 21.Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, et al. (2010): Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 189:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A (2006): EPI64 regulates microvillar subdomains and structure. J Cell Biol. 175:803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinkert K, Echard A (2016): Rab35 GTPase: A Central Regulator of Phosphoinositides and F-actin in Endocytic Recycling and Beyond. Traffic. 17:1063–1077. [DOI] [PubMed] [Google Scholar]
- 24.Mignogna ML, D’Adamo P (2018): Critical importance of RAB proteins for synaptic function. Small GTPases. 9:145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehan P, Waites CL (2019): Coordination of synaptic vesicle trafficking and turnover by the Rab35 signaling network. Small GTPases. 10:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaz-Silva J, Gomes P, Jin Q, Zhu M, Zhuravleva V, Quintremil S, et al. (2018): Endolysosomal degradation of Tau and its role in glucocorticoid-driven hippocampal malfunction. EMBO J. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji C, Tang M, Zeidler C, Hohfeld J, Johnson GV (2019): BAG3 and SYNPO (synaptopodin) facilitate phospho-MAPT/Tau degradation via autophagy in neuronal processes. Autophagy. 15:1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granger E, McNee G, Allan V, Woodman P (2014): The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 31:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wandinger-Ness A, Zerial M (2014): Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol. 6:a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chesneau L, Dambournet D, Machicoane M, Kouranti I, Fukuda M, Goud B, et al. (2012): An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr Biol. 22:147–153. [DOI] [PubMed] [Google Scholar]
- 31.Sheehan P, Zhu M, Beskow A, Vollmer C, Waites CL (2016): Activity-Dependent Degradation of Synaptic Vesicle Proteins Requires Rab35 and the ESCRT Pathway. J Neurosci. 36:8668–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aaron JS, Taylor AB, Chew TL (2018): Image co-localization - co-occurrence versus correlation. J Cell Sci. 131. [DOI] [PubMed] [Google Scholar]
- 33.Fuchs M, Poirier DJ, Seguin SJ, Lambert H, Carra S, Charette SJ, et al. (2009): Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 425:245–255. [DOI] [PubMed] [Google Scholar]
- 34.Carra S, Seguin SJ, Lambert H, Landry J (2008): HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. The Journal of biological chemistry. 283:1437–1444. [DOI] [PubMed] [Google Scholar]
- 35.Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, et al. (2013): Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol. 23:430–435. [DOI] [PubMed] [Google Scholar]
- 36.Doong H, Rizzo K, Fang S, Kulpa V, Weissman AM, Kohn EC (2003): CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: accumulation of poly-ubiquitinated Hsp90 client proteins. The Journal of biological chemistry. 278:28490–28500. [DOI] [PubMed] [Google Scholar]
- 37.Arimura T, Ishikawa T, Nunoda S, Kawai S, Kimura A (2011): Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum Mutat. 32:1481–1491. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Colvin T, Rauch JN, Acosta-Alvear D, Kampmann M, Dunyak B, et al. (2015): Validation of the Hsp70-Bag3 protein-protein interaction as a potential therapeutic target in cancer. Molecular cancer therapeutics. 14:642–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchs M, Luthold C, Guilbert SM, Varlet AA, Lambert H, Jette A, et al. (2015): A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis. PLoS genetics. 11:e1005582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivera-Molina FE, Novick PJ (2009): A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proceedings of the National Academy of Sciences of the United States of America. 106:14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhen Y, Stenmark H (2015): Cellular functions of Rab GTPases at a glance. Journal of cell science. 128:3171–3176. [DOI] [PubMed] [Google Scholar]
- 42.Nottingham RM, Pfeffer SR (2009): Defining the boundaries: Rab GEFs and GAPs. Proceedings of the National Academy of Sciences of the United States of America. 106:14185–14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi H, Etoh K, Marubashi S, Ohbayashi N, Fukuda M (2015): Measurement of Rab35 activity with the GTP-Rab35 trapper RBD35. Methods Mol Biol. 1298:207–216. [DOI] [PubMed] [Google Scholar]
- 44.Hokanson DE, Bretscher AP (2012): EPI64 interacts with Slp1/JFC1 to coordinate Rab8a and Arf6 membrane trafficking. Mol Biol Cell. 23:701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou Y, Chen X, Tolmachova T, Ernst SA, Williams JA (2013): EPI64B acts as a GTPase-activating protein for Rab27B in pancreatic acinar cells. J Biol Chem. 288:19548–19557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, et al. (2005): Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nature genetics. 37:806–808. [DOI] [PubMed] [Google Scholar]
- 47.Wenzel EM, Schultz SW, Schink KO, Pedersen NM, Nahse V, Carlson A, et al. (2018): Concerted ESCRT and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat Commun. 9:2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bache KG, Brech A, Mehlum A, Stenmark H (2003): Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. The Journal of cell biology. 162:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, et al. (2007): Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 53:337–351. [DOI] [PubMed] [Google Scholar]
- 50.Makani V, Zhang B, Han H, Yao Y, Lassalas P, Lou K, et al. (2016): Evaluation of the brain-penetrant microtubule-stabilizing agent, dictyostatin, in the PS19 tau transgenic mouse model of tauopathy. Acta Neuropathol Commun. 4:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang B, Carroll J, Trojanowski JQ, Yao Y, Iba M, Potuzak JS, et al. (2012): The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 32:3601–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, et al. (2009): Transmission and spreading of tauopathy in transgenic mouse brain. Nature cell biology. 11:909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson GV, Seubert P, Cox TM, Motter R, Brown JP, Galasko D (1997): The tau protein in human cerebrospinal fluid in Alzheimer’s disease consists of proteolytically derived fragments. Journal of neurochemistry. 68:430–433. [DOI] [PubMed] [Google Scholar]
- 54.Lasagna-Reeves CA, Castillo-Carranza DL, Sengupta U, Sarmiento J, Troncoso J, Jackson GR, et al. (2012): Identification of oligomers at early stages of tau aggregation in Alzheimer’s disease. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 26:1946–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jicha GA, Bowser R, Kazam IG, Davies P (1997): Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. Journal of neuroscience research. 48:128–132. [DOI] [PubMed] [Google Scholar]
- 56.Hunt CA, Schenker LJ, Kennedy MB (1996): PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. The Journal of neuroscience : the official journal of the Society for Neuroscience. 16:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernhardt R, Matus A (1984): Light and electron microscopic studies of the distribution of microtubule-associated protein 2 in rat brain: a difference between dendritic and axonal cytoskeletons. The Journal of comparative neurology. 226:203–221. [DOI] [PubMed] [Google Scholar]
- 58.Klaips CL, Jayaraj GG, Hartl FU (2018): Pathways of cellular proteostasis in aging and disease. The Journal of cell biology. 217:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kruger L, Mandelkow EM (2016): Tau neurotoxicity and rescue in animal models of human Tauopathies. Current opinion in neurobiology. 36:52–58. [DOI] [PubMed] [Google Scholar]
- 60.Yin Y, Gao D, Wang Y, Wang ZH, Wang X, Ye J, et al. (2016): Tau accumulation induces synaptic impairment and memory deficit by calcineurin-mediated inactivation of nuclear CaMKIV/CREB signaling. Proceedings of the National Academy of Sciences of the United States of America. 113:E3773–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kimura T, Yamashita S, Fukuda T, Park JM, Murayama M, Mizoroki T, et al. (2007): Hyperphosphorylated tau in parahippocampal cortex impairs place learning in aged mice expressing wild-type human tau. The EMBO journal. 26:5143–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jadhav S, Avila J, Scholl M, Kovacs GG, Kovari E, Skrabana R, et al. (2019): A walk through tau therapeutic strategies. Acta Neuropathol Commun. 7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki M, Iwasaki M, Sugio A, Hishiya A, Tanaka R, Endo T, et al. (2011): BAG3 (BCL2-associated athanogene 3) interacts with MMP-2 to positively regulate invasion by ovarian carcinoma cells. Cancer letters. 303:65–71. [DOI] [PubMed] [Google Scholar]
- 64.Feldman AM, Gordon J, Wang J, Song J, Zhang XQ, Myers VD, et al. (2016): BAG3 regulates contractility and Ca(2+) homeostasis in adult mouse ventricular myocytes. Journal of molecular and cellular cardiology. 92:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y, Yang LN, Cheng L, Tu S, Guo SJ, Le HY, et al. (2013): Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol Cell Proteomics. 12:2804–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I (2001): Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 291:1553–1557. [DOI] [PubMed] [Google Scholar]
- 67.Kouranti I, Sachse M, Arouche N, Goud B, Echard A (2006): Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Current biology : CB. 16:1719–1725. [DOI] [PubMed] [Google Scholar]
- 68.Chevallier J, Koop C, Srivastava A, Petrie RJ, Lamarche-Vane N, Presley JF (2009): Rab35 regulates neurite outgrowth and cell shape. FEBS Lett. 583:1096–1101. [DOI] [PubMed] [Google Scholar]
- 69.Cauvin C, Rosendale M, Gupta-Rossi N, Rocancourt M, Larraufie P, Salomon R, et al. (2016): Rab35 GTPase Triggers Switch-like Recruitment of the Lowe Syndrome Lipid Phosphatase OCRL on Newborn Endosomes. Curr Biol. 26:120–128. [DOI] [PubMed] [Google Scholar]
- 70.Um JW, Choii G, Park D, Kim D, Jeon S, Kang H, et al. (2016): IQ Motif and SEC7 Domain-containing Protein 3 (IQSEC3) Interacts with Gephyrin to Promote Inhibitory Synapse Formation. J Biol Chem. 291:10119–10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frankel EB, Audhya A (2018): ESCRT-dependent cargo sorting at multivesicular endosomes. Semin Cell Dev Biol. 74:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamashita Y, Kojima K, Tsukahara T, Agawa H, Yamada K, Amano Y, et al. (2008): Ubiquitin-independent binding of Hrs mediates endosomal sorting of the interleukin-2 receptor beta-chain. Journal of cell science. 121:1727–1738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


