Abstract
The structures and conformational dynamics of proteins, protein complexes, and their noncovalent interactions with other molecules are controlled specifically by the Gibbs free energy (entropy and enthalpy) of the system. For some organisms, temperature is highly regulated, but the majority of biophysical studies are carried out at room, nonphysiological temperature. In this review, we describe variable-temperature electrospray ionization (vT-ESI) mass spectrometry (MS)-based studies with unparalleled sensitivity, dynamic range, and selectivity for studies of both cold- and heat-induced chemical processes. Such studies provide direct determinations of stabilities, reactivities, and thermodynamic measurements for native and non-native structures of proteins and protein complexes and for protein–ligand interactions. Highlighted in this review are vT-ESI-MS studies that reveal 40 different conformers of chymotrypsin inhibitor 2, a classic two-state (native → unfolded) unfolder, and thermochemistry for a model membrane protein system binding lipid and its regulatory protein.
Keywords: native mass spectrometry, protein folding, protein–ligand interactions, thermodynamics, variable-temperature electrospray ionization
INTRODUCTION
Over six decades ago, Anfinsen and coworkers (29, p. 439) hypothesized that “the configuration [3-D configuration with the lowest configurational free energy] that a protein assumes, under any specific set of conditions, is the one that is thermodynamically the most stable.” This statement suggests that protein stability and structure can be modulated by environmental factors, such as temperature (27, 52), solvent (63, 65, 79), and pH (1), as well as by post-translational modifications (37, 73, 74), interactions with ligands (77, 78, 80), and formation of protein complexes; theoretical calculations support this view (7, 17, 47). Although Anfinsen’s thermodynamic hypothesis anticipates that, as a protein is denatured, many different conformations may be stabilized (19, 39-41), denaturation is normally thought of as a cooperative, two-state process whereby the well-ordered, functional (native) state becomes unstable and thus unfolds to produce distributions of non-native (denatured) states (38). In experiments that are designed to characterize a protein’s stability (e.g., pH-, solvent-, or temperature-scan studies), specific non-native species are rarely observed experimentally (5, 49). Isotopic 1H–2D exchange and chemical-cross-linking strategies, combined with nuclear magnetic resonance and mass spectrometry (MS) detection (28), reveal the existence of intermediates involved in folding (as well as other off-pathway species), providing key benchmark insights about the importance of non-native structures. Moreover, single-molecule biophysical measurements provide a fascinating look at the dynamics and the many types of non-native conformations that exist (6, 9). However, it is still the case that, for the vast number of proteoforms, far more is known about native structure than about the many non-native forms that must also exist. A straightforward analytical technology that is able to rapidly capture information about populations of native and non-native structures for large numbers of different proteins could help fill out the vast limitations in what is known about structures, stabilities, and functions of non-native structures.
It is difficult to overstate the importance of understanding the interactions that are responsible for establishing noncovalent structure. These structures are essential to every process in the life sciences. The strength of specific interactions can be described in terms of the change in Gibbs free energy upon binding (ΔG) with contributions from enthalpy (ΔH) and entropy (−TΔS) (22). The change in enthalpy describes binding modes that may involve hydrogen bonding, electrostatic interactions, and hydrophobic interactions within protein structure(s) corresponding to intramolecular (e.g., protein folding) and intermolecular (e.g., protein–ligand) interactions (42, 61). Desolvation and formation of new bonds within molecules also contribute to enthalpy because desolvation of polar groups is unfavorable (71). The change in entropy provides insight into the desolvation of polar and nonpolar groups; dynamics of proteins and ligands; and structuring of proteins, such as in refolding processes (42, 61). In some instances, protein–ligand and protein–protein interactions can be driven by large conformational entropy originating in enhanced protein motions (10, 31, 68, 69). In short, thermodynamics provides a quantitative description of the energetics and as a result is used extensively in selecting and optimizing potential drug candidates (32, 43). In some cases, the temperature dependence of the equilibrium association binding constants (KA) can follow the van’t Hoff equation (70):
In other cases, where heat capacity (ΔCp) is not constant over the selected temperature range (for a review, see 58), the nonlinear van’t Hoff equation is used to determine the ΔCp and change in enthalpy (ΔHT0) at a selected reference temperature (T0):
The magnitude and sign of ΔCp can provide insight into the change in apolar versus polar solvation, including conformational changes in proteins (58). Moreover, ΔCp imparts a temperature dependence to ΔH and ΔS that can lead to a change in their signs and in what parameter dominates. For example, consider a hypothetical protein–ligand interaction with a room-temperature (RT; 25°C or 298 K) equilibrium dissociation constant (Kd) of 1 μM (ΔG298K = −28.5 kJ/mol) (see Figure 1a). Extrapolating these RT values to those at physiological temperature (310 K) is not possible in the absence of thermodynamic parameters. More specifically, if the reaction had a positive or negative ΔH of 40 kJ/mol (ΔS back calculated using ΔG298K), then the natural log equilibrium association constant [ln(Ka)] follows a linear line with a negative or positive slope, respectively (Figure 1b). However, if ΔCp is not constant over the selected temperature range, then ln(Ka) would follow a convex or concave trend line, for example, if ΔCp was equal to +20 or −20 kJ/mol × K in our hypothetical example, respectively. Thus, measurements taken at RT provide little to no value for understanding molecular interactions at physiological temperature.
Figure 1.
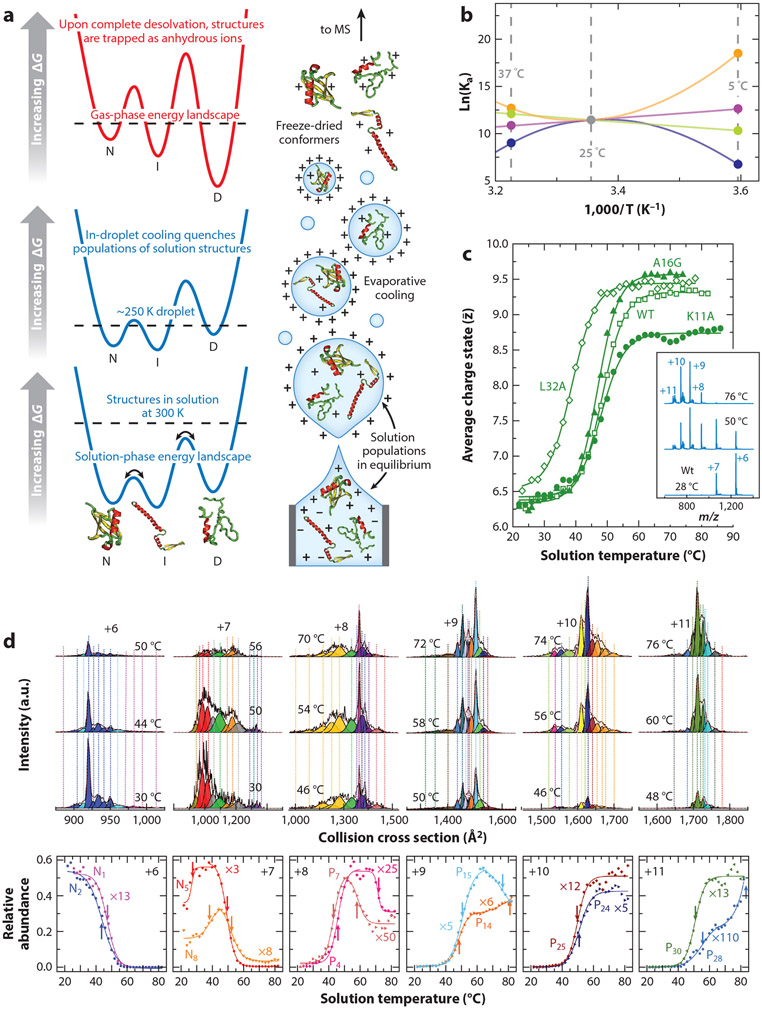
Thermodynamics, electrospray ionization (ESI), and capturing non-native structures. (a) Illustration of the ESI process beginning with formation of nanodroplets. In solution, rapid interconversion of protein structures [native (N), intermediate (I), and denatured (D)] is mediated by solvent and other chemical species, e.g., buffer, pH, temperature, and ligands. During early states of ESI, the droplet contains the solution populations of conformers (bottom). As the droplet dries, the solution equilibrium is quenched by evaporative cooling (middle), and during the late stages of solvent removal, protein structures are kinetically trapped on a new, gas-phase free energy landscape associated with the solvent-free ion (top). T-dependent charge state is shown as a function; water pH is 2.6 and Tm = 48.5 ± 0.3°C, in good agreement with traditional calorimetry studies (34) and mass spectra. (b) Temperature dependence of equilibrium association constants (Ka). Ka can follow linear (negative: green line; positive: purple line) and non-linear (concave: orange line; convex: blue line) trends depending on the sign and magnitude of thermodynamic parameters (ΔH and TΔS). (c) Melting for chymotrypsin inhibitory 2 (CI-2). (d) A plot of relative abundance as a function of solution temperature for each of the six individual charge states of CI-2 (56). For a free-energy landscape depicting ΔG as a function of collision cross section (CCS) (for each charge state) showing 41 unique solution structures, the reader is referred to Reference 56. (Top) T-dependent CCS distributions of wild-type (WT) 6+ through 11+ charge states. Vertical lines represent peak centers for each Gaussian function, and the color represents the corresponding conformational group determined from grouping analysis of the WT and mutant (L32A, A16G, K11A) data. (Bottom) Scaled plots of the relative abundance of representative groups as a function of solution temperature. The arrows indicate the respective Tm or Tf values for each conformer family. Panels a, b, and d adapted with permission from Reference 59, copyright 2021 American Chemical Society. Panel c adapted with permission from Reference 56, copyright 2020 American Chemical Society.
EXPERIMENTAL ELECTROSPRAY IONIZATION MASS SPECTROMETRY-BASED APPROACH FOR STUDYING NATIVE AND NON-NATIVE STRUCTURES
Electrospray ionization (ESI) (see Figure 1a) makes it possible to introduce intact biomolecules into the gas phase for analysis by MS. It is now routine to determine the molecular weights of macromolecules to within fractions of a Dalton (34, 62); MS-based fragmentation techniques provide rapid, sensitive means of identifying unknowns, even when species exist as low-abundance components of complex mixtures. Since its inception, ESI has been described as a soft ionization source. However, the extent of its softness is sometimes hard to appreciate. A complete, intact sequence of amino acids, with no evidence of fragments, is remarkable; however, it turns out that, with care and under conditions referred to as native ESI, native conformation and noncovalent complexes, including complexes involving specific hydrophilic domains of the conformations, can be studied. This is possible for several reasons, as illustrated in Figure 1. The initial ionized droplets produced by ESI are evaporatively cooled as they shrink during the dehydration process. This cooling traps the distribution of structures that were present in solution. Because the response of protons to changes in the environment is much faster than the timescales necessary for conformational changes (and because no additional protons can be added when the anhydrous ion has been formed), the charge state of the protein directly reflects the protein’s solution conformation—specifically, what types of sites were accessible for protonation on a conformation in solution. Thus, relatively simple m/z distributions can be used to assess differences in protein conformation. Additionally, in the absence of lubricating solvent, conformation types appear to be stable on the ms timescales required for mobility measurements, which provide information about the overall shape of the anhydrous form of the solution conformation that has been trapped, and in cases where structures are isolated for long times [for example, Fourier transform ion cyclotron resonance (FTICR) MS isotopic 1H-2D exchange measurements of gaseous ions trapped for many minutes]. We refer to these anhydrous structures, which have been trapped into a conformational type in the absence of lubricating solvent, as being freeze dried (59). While we cannot rule out the possibility that, in the absence of water, proteins may also undergo folding and unfolding structural transitions (which has been reported), removal of the protein dynamics associated with solvation (4) allows for a unique preservation of structure(s) that can be studied by means of ion mobility spectrometry (IMS) and MS. Of particular note, a carefully tuned instrument is required to preserve the trapped structures, and introduction of activation can lead to unwarranted gas-phase dissociation and unfolding of protein complexes.
The sensitivity, dynamic range, and fidelity of component assignments by MS raise new opportunities for characterizing stabilities. Some of these have transformative potential. Arguably, we cannot fully understand the factors that stabilize native structures unless we understand the nature of non-native or intermediate states. A noteworthy example is chymotrypsin inhibitor 2 (CI-2), a classic example of a cooperative two-state native → denatured transition (38). As shown in Figure 1c, an MS-based ensemble stability analysis provides a melting temperature (Tm) of 48.5°C, in excellent agreement with that obtained by calorimetric studies (38). Evidence that the unfolding transition of CI-2 yields a population of multiple conformers is seen in the high-resolution IMS distributions within each charge state (Figure 1d). For example, several conformers are observed for the +9 charge state, and their abundances change with temperature. More generally, increasing solution temperature provides further evidence for many (approximately 40 in total) stable states (in solution) near the melting transition temperature (3, 7, 17) (Figure 1d). The extraordinary sensitivity and dynamic range of IMS analysis have allowed researchers to resolve and characterize detailed Gibbs free energy landscapes of these structures across approximately 30 temperatures, providing an unprecedented measurement of changes in ΔG (Figure 1c), ΔH, ΔS, and ΔCp with temperature for each state. The level of experimental detail in the free energy diagram (Figure 1c) is unprecedented and begins to resemble the schematic representations of free energy landscapes that have been published for decades (19). However, in this case, it is possible to capture how the stabilities of each of the non-native and native structures contribute to the structural transition. This type of detailed information using IMS-MS-based analyses is now accessible for many types of systems, including peptides, proteins, and complexes in a range of environments. Such data can be obtained rapidly (within a few minutes) on minute (MS detection limits) quantities of sample and for individual components analyzed as a mixture (26).
Table 1 provides a partial summary of MS-based thermodynamic measurements from our laboratories for several model systems, including small peptides, for which detailed mechanisms of complex structural transitions have been elucidated (12, 13, 25, 33, 64, 65); model proteins and protein complexes, for which complex equilibria are observed (23, 27, 60); and protein–cofactor complexes, for which the relationships among multiple coupled events, such as cofactor loss and multiple formation of non-native structures, have been discerned (76-78). These details are not accessible with other technologies, and MS-based thermodynamic measurements have led to substantial new insights. For example, at high temperatures, our laboratories observed the formation of non-native disulfide bonds, oxidation of methionine residues, and decomposition of proteins. High-definition studies at low temperatures reveal cold denaturation events (to form specific non-native states), as well as the formation of multimeric forms that have not been reported previously. In addition to the analysis of individual molecules in solution, our laboratories’ early results indicate that it will be possible to examine the stabilities of multiple species during a single experiment. Early studies of mixtures of ribosomal proteins and fractions of proteins from cell lysates indicate that simple mixtures of 10 to 30 proteins are already accessible for an unbiased simultaneous thermal analysis with MS–MS detection. This type of analysis is ideal for characterizing mixtures of species differing in post-translational modification; ligand (drug) binding; and solution, excipient, or lipid membrane composition on native and non-native structure and stabilities. Implementation of MS–MS techniques for identification is straightforward and opens the door for stability characterization in an untargeted manner (very simple omics-like experiments).
Table 1.
Summary of IMS-MS derived thermochemistry of model systems
| System | Type of measurement |
Insight about mechanism | Derived thermochemistry |
Reference(s) |
|---|---|---|---|---|
| Pro peptides (Pro-13, Pro-7, HisPro-13) | Equil, kinetics | PPI↔PPII; trans→cis isomerization; configurationally coupled proton transfer | ΔG, ΔH, ΔS: multiple intermediates; ΔG‡, ΔH‡, ΔS‡ for TS | 25, 64, 65 |
| Peptides (bradykinin, Sub P) | Kinetics | Diketopiperazine formation; trans→cis isomerization; hidden intermediates | ΔG‡, ΔH‡, ΔS‡ for TS | 12, 13, 33, 55 |
| Single-domain proteins (CI-2, Ub) | Equil, LASER heating (μs) | Two-state transition involving multiple native and denatured conformations | Tm for many states; ΔG, ΔH, ΔS, ΔCp, energy landscapes (CI-2) | 24, 27, 60 |
| Multidomain protein complexes (Ub-dimer, Con A) | Equil | Complex dissociation; monomer subunits unfold independently (Ub); gradual structural change with increasing T (Con A) | Tm, ΔG, ΔH, ΔS, and ΔCp for many states | 23 |
| Protein–ligand, metal cofactors (myoglobin, myo-hemerythrin, hemoglobin) | Equil, LASER heating (μs) | Loss of α-helical structure; loss of heme and oxygen; presence of partially unfolded state at μs timescales; formation of non-native disulfide bond; dioxidation modification | Tm for multiple states and processes; tetramer states | 76-78 |
| Antibodies (IgG) | Kinetics | Light chain loss; non-native disulfide bond | ΔG‡, ΔH‡, ΔS‡ for loss of light chain | 8 |
| Escherichia coli lysate | Equil | Complex mixture; complexes dissociate | Tm for many unknown complexes | 26 |
| K-Ras and oncogenic mutants | Kinetics | Transition-state thermodynamics of the intrinsic GTPase activity | ΔG‡, ΔH‡, ΔS‡ for intrinsic GTPase activity | 53 |
| Membrane protein–lipid and protein–protein interactions (AmtB, AmtB–GlnK) | Equil | Binding thermodynamics for membrane protein–lipid and membrane protein–soluble protein interactions | ΔG, ΔH, ΔS, and ΔCp | 14, 15 |
Abbreviations: CI-2, chemotrypsin inhibitor 2; IMS, ion mobility spectrometry; MS, mass spectrometry; TS, transition state.
BEYOND PROTEIN FOLDING THERMODYNAMICS
Native MS, coupled with variable temperature (vT)-ESI-MS, has also been used to deduce transition-state thermodynamics for the intrinsic GTPase activity of K-RAS, a protein in the RAS GTPase family, whose members are the most commonly mutated of all discovered oncogenes (11, 20, 35).
The resolution afforded by modern mass spectrometers yielded an isotopically resolved native mass spectrum of purified K-RAS, which revealed several molecular species bound to the enzyme. The most abundant signal corresponds to guanosine 5′-diphosphate (GDP) bound to K-RAS. Interestingly, additional signals correspond to K-RAS bound to 2′-deoxyguanosine 5′-diphosphate (dGDP), 2′-deoxyguanosine 5′-triphosphate (dGTP), and guanosine 5′-triphosphate (GTP). Oncogenic mutants [G12C, G13D, and Q61H, selected based on their occurrence in cancer (49)] also show different abundances of the guanosine nucleotide forms. Notably, information about the different nucleotide-bound forms, including their abundances, would be difficult to ascertain or remain unknown using traditional biophysical approaches.
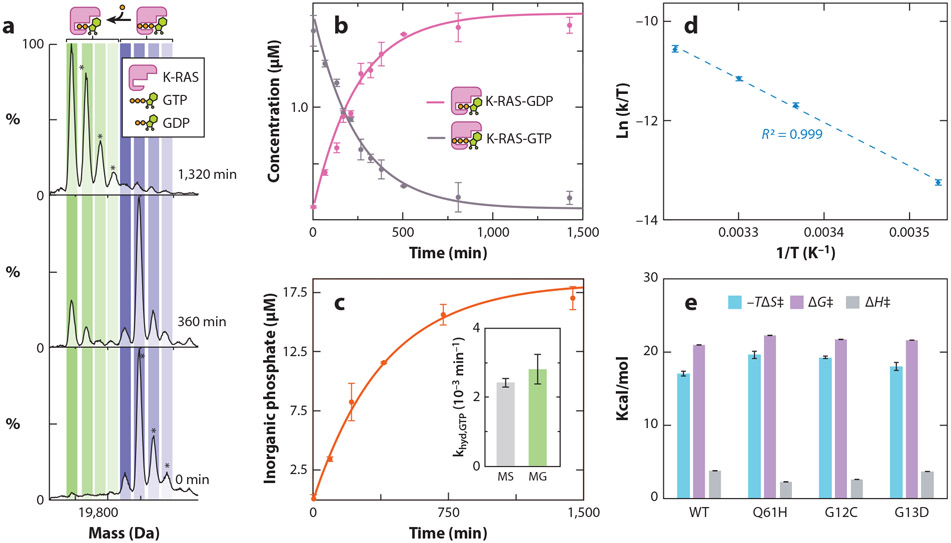
To determine the transition-state thermodynamics for the intrinsic GTPase activity of K-RAS, the enzyme initially loaded with GTP incubated at a given temperature is monitored over time using native MS (Figure 2). The protein starts out with signals corresponding to the K-RAS bound to GTP (Figure 2c). As the enzyme hydrolyzes GTP to GDP and inorganic phosphate is released, peaks emerge corresponding to K-RAS-GDP. The native MS data at each time point are then used to determine the rate constant for the K-RAS GTPase activity (Figure 2d). Importantly, the intrinsic hydrolysis rates determined using traditional solution-based inorganic phosphate assays are in direct agreement with those determined by native MS (Figure 2e). Eyring analysis (30) allows for the determination of transition-state thermodynamics (Figure 2f,g). For K-RAS and selected mutants, the transition-state barrier is dominated by ΔH‡. The transition-state thermodynamics determined for GTPase activity of K-RAS are in excellent agreement with those determined using other approaches, such as molecular dynamics simulations (2, 44, 72, 75).
Figure 2.
Transition-state thermodynamics for the intrinsic GTPase activity of K-RAS and its oncogenic mutants. (a) Deconvoluted native mass spectra of K-RAS-GTP incubated at 25°C and recorded at multiple time points. Concurrent with the K-RAS-mediated hydrolysis of GTP is the occurrence of K-RAS binding to GDP. (b) Plot of the concentration of K-RAS bound to either GDP or GTP from deconvolution of native MS data (dots) and fit to a first-order rate constant model (solid lines). (c) Plot of inorganic phosphate concentration determined for K-RAS using a solution MG assay. The inset shows the rate constants determined by native MS and an MG assay, which are statistically indistinguishable. (d) Eyring plot for intrinsic GTPase activity of K-RAS. (e) Transition-state enthalpy (ΔH‡), entropy (ΔS‡), and change in Gibbs free energy (ΔG‡) determined by Eyring analysis (T = 298 K). Reported are the mean and standard deviation (n = 3). Figure adapted with permission from Reference 53, copyright 2019 American Chemical Society. Abbreviations: GDP, guanosine 5′-diphosphate; GTP, guanosine 5′-triphosphate; MG, malachite green; MS, mass spectrometry; WT, wild type.
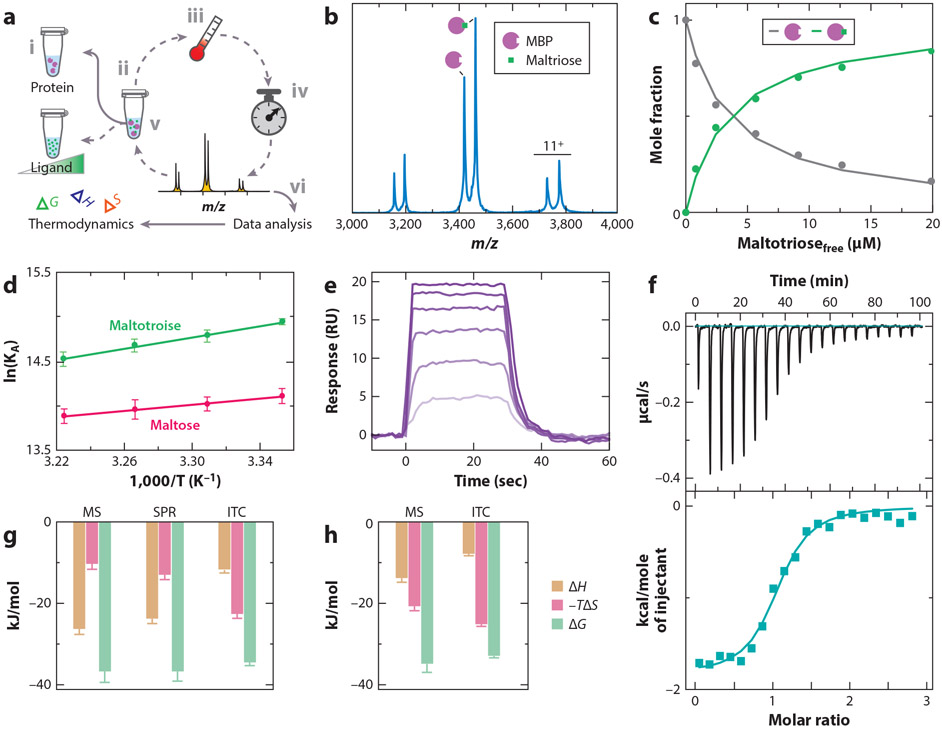
Native MS has also been employed to determine thermodynamics for protein–ligand interactions (18). For example, Cong et al. (15) selected three soluble protein–ligand systems: maltose binding protein (MBP) from Escherichia coli binding either maltose or maltotriose; hen egg white lysozyme binding N, N′, N″-triacetyl-chitotriose; and the nitrogen regulatory protein (GlnK) from E. coli, a trimeric protein that binds up to three adenosine diphosphate (ADP) molecules. The protein held at a constant concentration is titrated with the ligand and loaded into a nano-ESI emitter (Figure 3a). The emitter incubated at a temperature on the instrument for some time to allow the sample to reach equilibrium, and a mass spectrum was then recorded (Figure 3a). For example, mass spectra of MBP titrated with maltotriose at a given temperature is used to determine the equilibrium binding association constant (KA) (Figure 3b,c). The procedure is repeated for samples incubated at different temperature, and thermodynamic parameters are deduced from van’t Hoff analysis (70) (Figure 3d). Binding thermodynamics can be determined for these protein systems using surface plasmon resonance (SPR), where the protein is immobilized on the sensor surface, and isothermal calorimetry (ITC) in the same buffer used in native MS studies (Figure 3e,f). Thermodynamics found by ITC and SPR (see also 18, 51, 67) were similar to those obtained using the native MS approach. However, it is important to note that Cong et al. used 20,0000-fold less protein for native MS studies compared to ITC.
Figure 3.
Thermodynamics of soluble protein–ligand interactions. (a) An overview of the native mass spectrometry (MS) to determine binding thermodynamics. The protein at a fixed concentration (i) is titrated with ligand (ii) and loaded into a nano-electrospray ionization (ESI) emitter. The temperature is set (iii), and the sample is incubated for a given time to allow equilibrium to be reached (iv), followed by recording of a native mass spectrum (v). This procedure is repeated for different ligand concentrations and temperatures. The MS data are analyzed (vi) and used to determine thermodynamics, as described in panels b–d. (b) Representative mass spectrum of maltose binding protein (MBP) in the presence of maltotriose at 29°C. (c) Plot of mole fraction of apo- and maltotriose-bound MBP (dots) as a function of free ligand concentration. A protein–ligand binding model is fit to the data (solid lines) to determine equilibrium binding constants. (d) van’t Hoff plot for MBP binding to maltose and maltotriose. (e) Representative binding and dissociation surface plasmon resonance (SPR) profiles at 29°C for different concentrations of maltotriose injected onto a sensor surface containing immobilized MBP. (f) Isothermal calorimetry (ITC) power-versus-time plot for the titration (top) and integrated heat plotted as function of molar ratio (bottom). (g,h) Binding thermodynamics for MBP binding to (g) maltotriose or (h) maltose reported at a temperature of 298 K. Figure adapted with permission from Reference 15, copyright 2016 American Chemical Society.
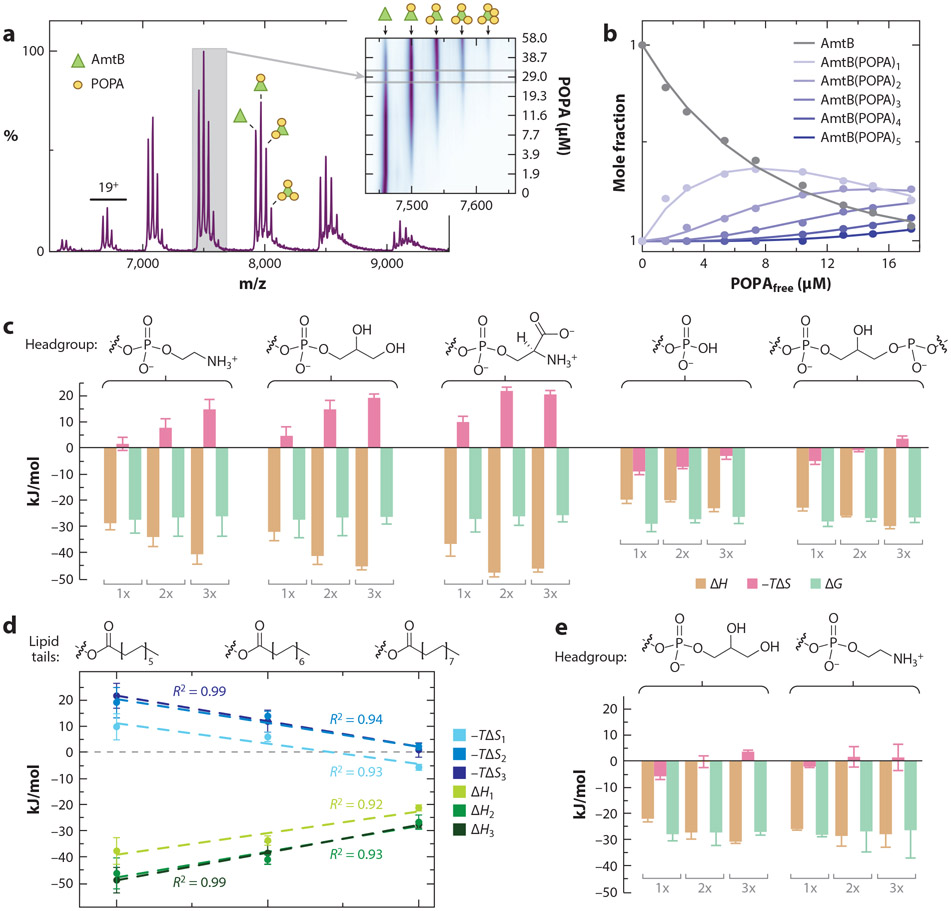
The unique ability of native MS to resolve individual ligand binding events has created new opportunities to characterize membrane protein–lipid interactions. To determine binding thermodynamics for protein–lipid interactions, mass spectra are recorded at different temperatures for the ammonia channel (AmtB) from E. coli titrated with different lipids. For example, individual 1-palmitoyl-2-oleoyl phosphatidic acid (POPA) binding events can be resolved (Figure 4a), and the mole fraction of each species can directly be determined across the titration series (Figure 4b). Distinct thermodynamic signatures are observed for the binding of lipids with different headgroups or acyl chain lengths (Figure 4c,d). Thermodynamics for the binding of lipids with longer acyl chains displays enthalpy–entropy compensation, and the binding thermodynamics becomes more hydrophobic, indicating that the acyl chains contribute to and form interactions with the membrane protein (Figure 4d). A phosphatidylglycerol (PG)-binding site has been identified in AmtB (46) and, when a mutant form of AmtB (AmtBN72A/N79A) is engineered to abolish the PG-binding site, shows altered thermodynamic signatures for PG binding to AmtB. These results demonstrate the unique opportunity to obtain thermodynamics signatures for individual binding events, which is not possible using traditional approaches.
Figure 4.
Thermodynamic signatures for the interaction of lipids with the ammonia channel (AmtB), an integral membrane protein, from Escherichia coli. (a) Native mass spectrum of AmtB in the C8E4 detergent with 1-palmitoyl-2-oleoyl phosphatidic acid (POPA). The inset shows a contour plot of the 17+ charge state as functions of POPA concentration. (b) Plot of mole fraction of AmtB and lipid-bound states in the titration series (dots) and fit of a sequential lipid binding model (solid lines). (c) Thermodynamic signatures of AmtB binding phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidic acid (PA) containing 1-palmitoyl-2-oleoyl (PO) tails, and 1,1′,2,2′-tetraoleoyl-cardiolipin (TOCDL) determined through van’t Hoff analysis. Binding of the first, second, and third lipid is shown as 1x, 2x, and 3x, respectively. Headgroup structures are shown above. (d) Binding thermodynamics for PG with different acyl chain length: 12 (1,2-dilauroyl), 14 (1,2-dimyristoyl), and 16 (1,2-dipalmitoyl). The first, second, and third trendlines are plotted. (e) AmtB double mutant (N72A/N79A) binding thermodynamics for POPG and POPE. Figure adapted with permission from Reference 15, copyright 2016 American Chemical Society.
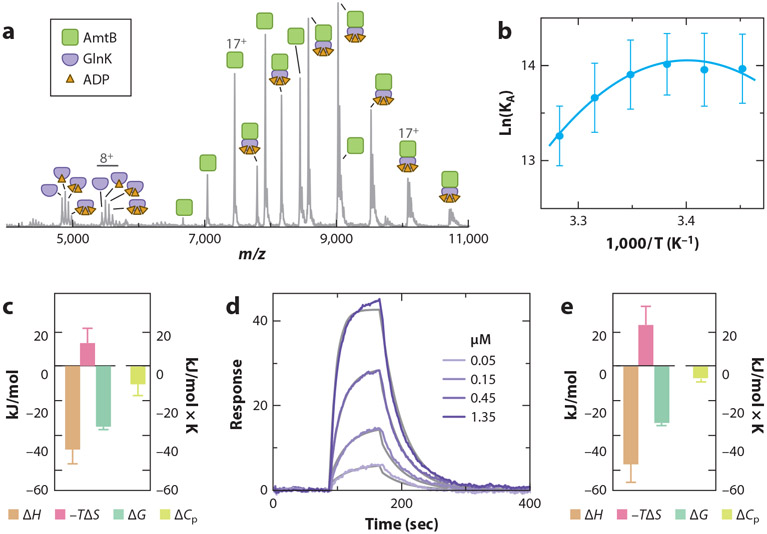
Another recent application of vT-ESI native MS has deduced the thermodynamics for the interaction of AmtB (membrane protein) with the soluble regulator protein GlnK (14). Formation of the AmtB–GlnK complex is dependent on ADP concentration, in which ADP binds to each subunit of the trimeric GlnK complex, displacing a loop that interacts with a subunit of the AmtB complex (16, 36). When the concentration of ADP is adjusted, an equal population of AmtB and AmtB–GlnK can be observed in the native mass spectrum (Figure 5a). The KA can be computed using the abundances determined after deconvoluting the mass spectrum. The plot of the natural log of KA measured at different temperatures gives rise to a curve that can be fit with the nonlinear form of the van’t Hoff equation (54) (Figure 5b,c). This result indicates that heat capacity is not constant over the temperature range, as well as demonstrating temperature-dependent conformational changes in AmtB and/or GlnK (for a review, see 58). The large heat capacity suggests interconverting conformations (21), and the negative sign indicates a change in polar solvation (50, 58), which is consistent with desolvation of the loop in GlnK upon binding a pocket in AmtB. The molecular interaction of AmtB binding to the GlnK sensor surface, determined using SPR (Figure 5d), displayed similar temperature-dependent binding (Figure 5e). Importantly, binding thermodynamics for this protein–protein interaction determined by SPR and native MS are in direct agreement.
Figure 5.
Thermodynamics of the interaction between AmtB and GlnK. (a) Mass spectrum of purified AmtB–GlnK complex, with the concentration of ADP adjusted to obtain a near equal population of AmtB and AmtB in complex with GlnK. (b) Plot of the natural log of KA (dots) and fit of the non-linear van’t Hoff equation (lines). (c) Native mass spectrometry (MS)–derived thermodynamics of the AmtB–GlnK interaction. The reference temperature is 298 K. (d) Sensorgrams for different concentrations of AmtB injected over a GlnK sensor surface. The fit of a Langmuir 1:1 binding model is shown (grey lines). (e) Surface plasmon resonance (SPR)-derived thermodynamics of the AmtB–GlnK interaction. Figure adapted with permission from Reference 14.
FUTURE DIRECTIONS
As discussed above, implicit in Anfinsen’s thermodynamic hypothesis is the idea that, in addition to the folded native conformation, many different non-native structures should be favored when conditions that influence stability are varied. However, non-native states are rarely observed using traditional experimental approaches. Because of this, little is known about the structure and function of such species. The ability to trap non-native conformers or protein–ligand bound states from solution upon introduction of biopolymers into the gas phase for MS-based analyses offers a new means of directly studying such species. The resolution afforded by modern MS and IMS instrumentation has led to the discovery of specific copurified molecules. For example, recent discoveries have shown zinc copurifying with transthyretin (56, 57, 66) and IscU (48); in both cases, the bound zinc directly impacts structure and function. The presence of these adducts would remain hidden if one were using traditional gel-based technology (45) to assess protein quality. In our opinion, native IMS and MS should take center stage in understanding protein structure and function, including how these processes are impacted by a change in the local environment, such as changes in temperature, pH, and the presence of small-molecule osmolytes.
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences (NIGMS) and National Cancer Institute (NCI) of the National Institutes of Health (NIH) (grants DP2GM123486, R01GM121751, P41GM128577, R01GM138863, and R01GM139876).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Anderson DE, Becktel WJ, Dahlquist FW. 1990. pH-Induced denaturation of proteins: A single salt bridge contributes 3–5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry 29:2403–8 [DOI] [PubMed] [Google Scholar]
- 2.Åqvist J, Kamerlin SCL. 2016. Conserved motifs in different classes of GTPases dictate their specific modes of catalysis. ACS Catal. 6:1737–43 [Google Scholar]
- 3.Baldwin RL. 2008. The search for folding intermediates and the mechanism of protein folding. Annu. Rev. Biophys 37:1–21 [DOI] [PubMed] [Google Scholar]
- 4.Barron LD, Hecht L, Wilson G. 1997. The lubricant of life: a proposal that solvent water promotes extremely fast conformational fluctuations in mobile heteropolypeptide structure. Biochemistry 36:13143–47 [DOI] [PubMed] [Google Scholar]
- 5.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. 2008. Biomolecule analysis by ion mobility spectrometry. Annu. Rev. Anal. Chem 1:293–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgia A, Williams PM, Clarke J. 2008. Single-molecule studies of protein folding. Annu. Rev. Biochem 77:101–25 [DOI] [PubMed] [Google Scholar]
- 7.Brockwell DJ, Radford SE. 2007. Intermediates: ubiquitous species on folding energy landscapes? Curr. Opin. Struct. Biol 17:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown CJ, Woodall DW, El-Baba TJ, Clemmer DE. 2019. Characterizing thermal transitions of IgG with mass spectrometry. J. Am. Soc. Mass Spectrom 30:2438–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustamante C, Alexander L, Maciuba K, Kaiser CM. 2020. Single-molecule studies of protein folding with optical tweezers. Annu. Rev. Biochem 89:443–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caro JA, Harpole KW, Kasinath V, Lim J, Granja J, et al. 2017. Entropy in molecular recognition by proteins. PNAS 114:6563–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colicelli J 2004. Human RAS superfamily proteins and related GTPases. Sci. STKE 2004:RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conant CR, Fuller DR, El-Baba TJ, Zhang Z, Russell DH, Clemmer DE. 2019. Substance P in solution: trans-to-cis configurational changes of penultimate prolines initiate non-enzymatic peptide bond cleavages. J. Am. Soc. Mass Spectrom 30:919–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conant CR, Fuller DR, Zhang Z, Woodall DW, Russell DH, Clemmer DE. 2019. Substance P in the gas phase: conformational changes and dissociations induced by collisional activation in a drift tube. J. Am. Soc. Mass Spectrom 30:932–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong X, Liu Y, Liu W, Liang X, Laganowsky A. 2017. Allosteric modulation of protein-protein interactions by individual lipid binding events. Nat. Commun 8:2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong X, Liu Y, Liu W, Liang X, Russell DH, Laganowsky A. 2016. Determining membrane protein-lipid binding thermodynamics using native mass spectrometry. J. Am. Chem. Soc 138:4346–49 [DOI] [PubMed] [Google Scholar]
- 16.Conroy MJ, Durand A, Lupo D, Li XD, Bullough PA, et al. 2007. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. PNAS 104:1213–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daggett V, Fersht A. 2003. The present view of the mechanism of protein folding. Nat. Rev. Mol. Cell Biol 4:497–502 [DOI] [PubMed] [Google Scholar]
- 18.Daneshfar R, Kitova EN, Klassen JS. 2004. Determination of protein-ligand association thermochemistry using variable-temperature nanoelectrospray mass spectrometry. J. Am. Chem. Soc 126:4786–87 [DOI] [PubMed] [Google Scholar]
- 19.Dill KA, Chan HS. 1997. From Levinthal to pathways to funnels. Nat. Struct. Biol 4:10–19 [DOI] [PubMed] [Google Scholar]
- 20.Downward J 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11–22 [DOI] [PubMed] [Google Scholar]
- 21.Eftink MR, Anusiem AC, Biltonen RL. 1983. Enthalpy-entropy compensation and heat capacity changes for protein-ligand interactions: general thermodynamic models and data for the binding of nucleotides to ribonuclease A. Biochemistry 22:3884–96 [DOI] [PubMed] [Google Scholar]
- 22.Eisenberg DS, Crothers DM. 1979. Physical Chemistry: With Applications to the Life Sciences. Menlo Park, CA: Benjamin/Cummings Publ. [Google Scholar]
- 23.El-Baba TJ, Clemmer DE. 2019. Solution thermochemistry of concanavalin A tetramer conformers measured by variable-temperature ESI-IMS-MS. Int. J. Mass Spectrom 443:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Baba TJ, Fuller DR, Woodall DW, Raab SA, Conant CR, et al. 2018. Melting proteins confined in nanodroplets with 10.6 μm light provides clues about early steps of denaturation. Chem. Commun 54:3270–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Baba TJ, Kim D, Rogers DB, Khan FA, Hales DA, et al. 2016. Long-lived intermediates in a cooperative two-state folding transition. J. Phys. Chem. B 120:12040–46 [DOI] [PubMed] [Google Scholar]
- 26.El-Baba TJ, Raab SA, Buckley RP, Brown CJ, Lutomski CA, et al. 2021. Thermal analysis of a mixture of ribosomal proteins by vT-ESI-MS: toward a parallel approach for characterizing the stabilitome. Anal. Chem 93:8484–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Baba TJ, Woodall DW, Raab SA, Fuller DR, Laganowsky A, et al. 2017. Melting proteins: evidence for multiple stable structures upon thermal denaturation of native ubiquitin from ion mobility spectrometry-mass spectrometry measurements. J. Am. Chem. Soc 139:6306–9 [DOI] [PubMed] [Google Scholar]
- 28.Englander SW, Mayne L, Kan Z-Y, Hu W. 2016. Protein folding—how and why: by hydrogen exchange, fragment separation, and mass spectrometry. Annu. Rev. Biophys 45:135–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epstein CJ, Goldberger RF, Anfinsen CB. 1963. The genetic control of tertiary protein structure: studies with model systems. Cold Spring Harb. Symp. Quant. Biol 28:439–49 [Google Scholar]
- 30.Eyring H. 1935. The activated complex in chemical reactions. J. Chem. Phys 3:107–15 [Google Scholar]
- 31.Frederick KK, Marlow MS, Valentine KG, Wand AJ. 2007. Conformational entropy in molecular recognition by proteins. Nature 448:325–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freire E. 2006. Overcoming HIV-1 resistance to protease inhibitors. Drug Discov. Today Dis. Mech 3:281–86 [Google Scholar]
- 33.Fuller DR, Conant CR, El-Baba TJ, Brown CJ, Woodall DW, et al. 2018. Conformationally regulated peptide bond cleavage in bradykinin. J. Am. Chem. Soc 140:9357–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gault J, Donlan JA, Liko I, Hopper JT, Gupta K, et al. 2016. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat. Methods 13:333–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goitre L, Trapani E, Trabalzini L, Retta SF. 2014. The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol. Biol 1120:1–18 [DOI] [PubMed] [Google Scholar]
- 36.Gruswitz F, O’Connell J 3rd, Stroud RM. 2007. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. PNAS 104:42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houde D, Peng Y, Berkowitz SA, Engen JR. 2010. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol. Cell. Proteom 9:1716–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson SE, Fersht AR. 1991. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry 30:10428–35 [DOI] [PubMed] [Google Scholar]
- 39.Karplus M, McCammon JA. 2002. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol 9:646–52 [DOI] [PubMed] [Google Scholar]
- 40.Karplus M, Sali A. 1995. Theoretical studies of protein folding and unfolding. Curr. Opin. Struct. Biol 5:58–73 [DOI] [PubMed] [Google Scholar]
- 41.Kazmirski SL, Wong KB, Freund SM, Tan YJ, Fersht AR, Daggett V. 2001. Protein folding from a highly disordered denatured state: the folding pathway of chymotrypsin inhibitor 2 at atomic resolution. PNAS 98:4349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keserü G, Swinney DC, Mannhold R, Kubinyi H, Folkers G. 2015. Thermodynamics and Kinetics of Drug Binding. Hoboken, NJ: Wiley [Google Scholar]
- 43.Klebe G 2015. Applying thermodynamic profiling in lead finding and optimization. Nat. Rev. Drug Discov 14:95–110 [DOI] [PubMed] [Google Scholar]
- 44.Kötting C, Gerwert K. 2004. Time-resolved FTIR studies provide activation free energy, activation enthalpy and activation entropy for GTPase reactions. Chem. Phys 307:227–32 [Google Scholar]
- 45.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–85 [DOI] [PubMed] [Google Scholar]
- 46.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, et al. 2014. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510:172–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lazaridis T, Karplus M. 1997. “New view” of protein folding reconciled with the old through multiple unfolding simulations. Science 278:1928–31 [DOI] [PubMed] [Google Scholar]
- 48.Lin CW, McCabe JW, Russell DH, Barondeau DP. 2020. Molecular mechanism of ISC iron-sulfur cluster biogenesis revealed by high-resolution native mass spectrometry. J. Am. Chem. Soc 142:6018–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lumry R, Eyring H. 1954. Conformation changes of proteins. J. Phys. Chem 58:110–20 [Google Scholar]
- 50.Makhatadze GI, Privalov PL. 1990. Heat capacity of proteins. I. Partial molar heat capacity of individual amino acid residues in aqueous solution: hydration effect. J. Mol. Biol 213:375–84 [DOI] [PubMed] [Google Scholar]
- 51.Maple HJ, Scheibner O, Baumert M, Allen M, Taylor RJ, et al. 2014. Application of the Exactive Plus EMR for automated protein-ligand screening by non-covalent mass spectrometry. Rapid Commun. Mass Spectrom 28:1561–68 [DOI] [PubMed] [Google Scholar]
- 52.Mirza UA, Cohen SL, Chait BT. 1993. Heat-induced conformational changes in proteins studied by electrospray ionization mass spectrometry. Anal. Chem 65:1–6 [DOI] [PubMed] [Google Scholar]
- 53.Moghadamchargari Z, Huddleston J, Shirzadeh M, Zheng X, Clemmer DE, et al. 2019. Intrinsic GTPase activity of K-RAS monitored by native mass spectrometry. Biochemistry 58:3396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naghibi H, Tamura A, Sturtevant JM. 1995. Significant discrepancies between van’t Hoff and calorimetric enthalpies. PNAS 92:5597–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pierson NA, Clemmer DE. 2015. An IMS–IMS threshold method for semi-quantitative determination of activation barriers: interconversion of proline cis↔trans forms in triply protonated bradykinin. Int. J. Mass Spectrom 377:646–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poltash ML, McCabe JW, Shirzadeh M, Laganowsky A, Russell DH. 2019. Native IM-Orbitrap MS: resolving what was hidden. Trends Analyt. Chem 124:115533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poltash ML, Shirzadeh M, McCabe JW, Moghadamchargari Z, Laganowsky A, Russell DH. 2019. New insights into the metal-induced oxidative degradation pathways of transthyretin. Chem. Commun 55:4091–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prabhu NV, Sharp KA. 2005. Heat capacity in proteins. Annu. Rev. Phys. Chem 56:521–48 [DOI] [PubMed] [Google Scholar]
- 59.Raab SA, El-Baba TJ, Laganowsky A, Russell DH, Valentine SJ, Clemmer DE. 2021. Protons are fast and smart; proteins are slow and dumb: on the relationship of electrospray ionization charge states and conformations. J. Am. Soc. Mass Spectrom 32:1553–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raab SA, El-Baba TJ, Woodall DW, Liu W, Liu Y, et al. 2020. Evidence for many unique solution structures for chymotrypsin inhibitor 2: a thermodynamic perspective derived from vT-ESI-IMS-MS measurements. J. Am. Chem. Soc 142:17372–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raffa RB. 2001. Drug-Receptor Thermodynamics: Introduction and Applications. Hoboken, NJ: Wiley [Google Scholar]
- 62.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. 2012. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat. Methods 9:1084. [DOI] [PubMed] [Google Scholar]
- 63.Shi H, Clemmer DE. 2014. Evidence for two new solution states of ubiquitin by IMS-MS analysis. J. Phys. Chem. B 118:3498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi L, Holliday AE, Glover MS, Ewing MA, Russell DH, Clemmer DE. 2016. Ion mobility-mass spectrometry reveals the energetics of intermediates that guide polyproline folding. J. Am. Soc. Mass Spectrom 27:22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi L, Holliday AE, Khanal N, Russell DH, Clemmer DE. 2015. Configurationally-coupled protonation of polyproline-7. J. Am. Chem. Soc 137:8680–83 [DOI] [PubMed] [Google Scholar]
- 66.Shirzadeh M, Boone CD, Laganowsky A, Russell DH. 2019. Topological analysis of transthyretin disassembly mechanism: Surface-induced dissociation reveals hidden reaction pathways. Anal. Chem 91:2345–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Telmer PG, Shilton BH. 2003. Insights into the conformational equilibria of maltose-binding protein by analysis of high affinity mutants. J. Biol. Chem 278:34555–67 [DOI] [PubMed] [Google Scholar]
- 68.Tzeng S-R, Kalodimos CG. 2009. Dynamic activation of an allosteric regulatory protein. Nature 462:368–72 [DOI] [PubMed] [Google Scholar]
- 69.Tzeng S-R, Kalodimos CG. 2012. Protein activity regulation by conformational entropy. Nature 488:236–40 [DOI] [PubMed] [Google Scholar]
- 70.van’t Hoff MJH. 1884. Etudes de dynamique chimique. Recl. Trav. Chim. Pays.-Bas 3:333–36 [Google Scholar]
- 71.Velazquez-Campoy A, Freire E. 2006. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat. Protoc 1:186–91 [DOI] [PubMed] [Google Scholar]
- 72.Villà J, Štrajbl M, Glennon TM, Sham YY, Chu ZT, Warshel A. 2000. How important are entropic contributions to enzyme catalysis? PNAS 97:11899–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vouret-Craviari V, Grall D, Chambard J-C, Rasmussen UB, Pouysségur J, Van Obberghen-Schilling E. 1995. Post-translational and activation-dependent modifications of the G protein-coupled thrombin receptor. J. Biol. Chem 270:8367–72 [DOI] [PubMed] [Google Scholar]
- 74.Walsh G, Jefferis R. 2006. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol 24:1241–52 [DOI] [PubMed] [Google Scholar]
- 75.Warshel A 1998. Electrostatic origin of the catalytic power of enzymes and the role of preorganized active sites. J. Biol. Chem 273:27035–38 [DOI] [PubMed] [Google Scholar]
- 76.Woodall DW, Brown CJ, Raab SA, El-Baba TJ, Laganowsky A, et al. 2020. Melting of hemoglobin in native solutions as measured by IMS-MS. Anal. Chem 92:3440–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woodall DW, El-Baba TJ, Fuller DR, Liu W, Brown CJ, et al. 2019. Variable-temperature ESI-IMS-MS analysis of myohemerythrin reveals ligand losses, unfolding, and a non-native disulfide bond. Anal. Chem 91:6808–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woodall DW, Henderson LW, Raab SA, Honma K, Clemmer DE. 2021. Understanding the thermal denaturation of myoglobin with IMS-MS: evidence for multiple stable structures and trapped pre-equilibrium states. J. Am. Soc. Mass Spectrom 32:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wyttenbach T, Bowers MT. 2011. Structural stability from solution to the gas phase: Native solution structure of ubiquitin survives analysis in a solvent-free ion mobility–mass spectrometry environment. J. Phys. Chem. B 115:12266–75 [DOI] [PubMed] [Google Scholar]
- 80.Xie Y, Zhang J, Yin S, Loo JA. 2006. Top-down ESI-ECD-FT-ICR mass spectrometry localizes noncovalent protein-ligand binding sites. J. Am. Chem. Soc 128:14432–33 [DOI] [PubMed] [Google Scholar]