Abstract
Objective
Emerging studies have demonstrated the promising clinical value of circulating tumor cells (CTCs) for diagnosis, disease assessment, treatment monitoring and prognosis in epithelial ovarian cancer. However, the clinical application of CTC remains restricted due to diverse detection techniques with variable sensitivity and specificity and a lack of common standards.
Methods
We enrolled 160 patients with epithelial ovarian cancer as the experimental group, and 90 patients including 50 patients with benign ovarian tumor and 40 healthy females as the control group. We enriched CTCs with immunomagnetic beads targeting two epithelial cell surface antigens (EpCAM and MUC1), and used multiple reverse transcription-polymerase chain reaction (RT-PCR) detecting three markers (EpCAM, MUC1 and WT1) for quantification. And then we used a binary logistic regression analysis and focused on EpCAM, MUC1 and WT1 to establish an optimized CTC detection model.
Results
The sensitivity and specificity of the optimized model is 79.4% and 92.2%, respectively. The specificity of the CTC detection model is significantly higher than CA125 (92.2% vs. 82.2%, P=0.044), and the detection rate of CTCs was higher than the positive rate of CA125 (74.5% vs. 58.2%, P=0.069) in early-stage patients (stage I and II). The detection rate of CTCs was significantly higher in patients with ascitic volume ≥500 mL, suboptimal cytoreductive surgery and elevated serum CA125 level after 2 courses of chemotherapy (P<0.05). The detection rate of CTCEpCAM+ and CTCMUC1+ was significantly higher in chemo-resistant patients (26.3% vs. 11.9%; 26.4% vs. 13.4%, P<0.05). The median progression-free survival time for CTCMUC1+ patients trended to be longer than CTCMUC1− patients, and overall survival was shorter in CTCMUC1+ patients (P=0.043).
Conclusions
Our study presents an optimized detection model for CTCs, which consists of the expression levels of three markers (EpCAM, MUC1 and WT1). In comparison with CA125, our model has high specificity and demonstrates better diagnostic values, especially for early-stage ovarian cancer. Detection of CTCEpCAM+ and CTCMUC1+ had predictive value for chemotherapy resistance, and the detection of CTCMUC1+ suggested poor prognosis.
Keywords: Circulating tumor cells, epithelial ovarian cancer, optimized detection model, diagnosis and prognosis
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecologic malignancy, with a 5-year survival rate of less than 35% (1,2). Carbohydrate antigen 125 (CA125) is the most commonly used serological marker for the clinical diagnosis of EOC, but its diagnosis rate for early-stage disease is only 50% (3). In addition, its specificity is also quite poor, as elevated serum CA125 levels can also be seen in inflammatory diseases and endometriosis (4). Therefore, there is a need to explore additional detection methods that might improve early diagnosis of ovarian cancer.
Detection of circulating tumor cells (CTCs), as a form of liquid biopsy, is a non-invasive tumor detection method that has attracted extensive attention in recent years (5) due to its advantages such as easy access, minimal invasiveness and good patient compliance. CTCs are cells that are shed into the bloodstream from a primary tumor, metastases, or recurrent tumors, and therefore possess tumor-specific characteristics such as large size, low density, less deformation, and a high nucleoplasmic ratio (N/C) (6), thus, making them good targets for liquid biopsy. The clinical value of CTCs in ovarian, breast, prostate and colorectal cancers has been confirmed in several studies and meta-analyses (5,7-13), with nearly thirty articles concerning the clinical application of CTCs to ovarian cancer being published in the last decade. Due to the variety of detection techniques used, the reported detection rates in these publications are variable, ranging from 12% to 100% (14-22). Currently, a common standard for CTC detection has not been established.
Here, through a review of relevant research databases and publications, we identified three markers (EpCAM, MUC1 and WT1) that are significantly highly expressed in ovarian cancer cells and then used these markers in our CTC detection model. In our previous study (12), we explored the diagnostic value of these markers in CTCs of ovarian cancer, however we only enrolled healthy females as controls, which lead to a sample limitation, as the actual clinical need is to differentiate ovarian cancer from benign ovarian tumors. Therefore, in this study we enrolled EOC patients as the experimental group, and benign ovarian tumor patients as well as healthy females, as the control group. We aim to analyze the diagnostic value of the above-mentioned markers in the detection of CTCs in EOC and the correlation of levels of these markers with clinical features, and to establish an optimized CTC detection model with high sensitivity and specificity to assist in clinical applications.
Materials and methods
Study population
This study included patients with EOC, patients with benign ovarian tumors, and healthy females who were admitted to the Department of Gynecology, Peking University Third Hospital between September 2013 and December 2020. Peripheral venous blood (5 mL) was drawn from all patients before initial treatment, with serum CA125 levels being measured at this time. All EOC patients were pathologically verified, with tumor stage determined according to the International Federation of Gynecology and Obstetrics (FIGO) staging, and patients overlapped with other malignant tumors will be excluded. Initial treatment included advanced chemotherapy or full staging of ovarian cancer surgery and tumor cytoreductive surgery, and postoperative adjuvant chemotherapy mainly based on platinum. Patients with benign ovarian tumors refer to patients with benign tumors that have been pathologically proven to be of ovarian origin, including ovarian chocolate cysts, teratomas, and serous cystadenomas, etc. The healthy female population refers to adult female patients who have no obvious abnormalities in the regular physical examination every year and whose CA125 is in the normal range. Clinical data including medical history, physical examination, laboratory examination and imaging evaluation were obtained from all enrolled patients. This study was approved by the Peking University Biomedical Ethics Committee (No. IRB00001052-16022). All patients signed the informed consent form before participation.
Study methods
We enriched CTCs with immunomagnetic beads targeting to two epithelial cell surface antigens (EpCAM and MUC1), which are not expressed on the surface of blood cells, and then used multiple reverse transcription-polymerase chain reaction (RT-PCR) to detect three markers (EpCAM, MUC1 and WT1) to quantify the enriched CTCs. The binding of two antibody-labeled immunomagnetic beads (EpCAM and MUC1) to the EOC cell line SKOV3 is shown in Supplementary Figure S1 . The primer sequences of three genes are shown in Supplementary Table S1 . A DNA 1000 LabChips (Agilent Technologies, Santa Clara, USA) was used to detect the amplified sequences using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA) according to the manufacturer’s instructions. The Agilent 2100 bioanalyzer has a list of Ladder with known DNA concentration, and polymerase chain reaction (PCR) amplification product concentration of each marker gene is obtained by comparing with Ladder.
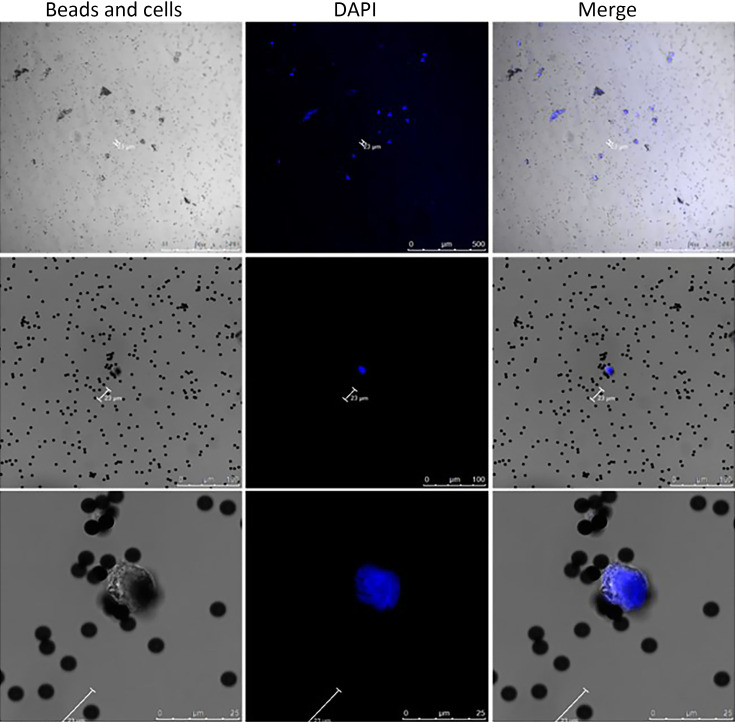
Figure S1.
Confocal laser imaging of immunomagnetic beads combined with SKOV3 cell line. Immunomagnetic beads were incubated with two monoclonal antibodies (EpCAM and MUC1) without staining; ovarian cancer cell line was SKOV3 cell line, and nucleus was stained with DAPI.
Table S1. Primer sequences of three genes.
| Genes | Forward | Reverse |
| EpCAM | TGAGCGAGTGAGAACCTA | CACAACAATTCCAGCAAC |
| MUC1 | GCACCGACTACTACCAAGAG | AAGGAAATGGCACATCACT |
| WT1 | AGTCCGCCATCACAACAT | TGGTACAATAATTCCATCCC |
| ACTIN | GAAATCGTGCGTGACATTA | AGGCAGCTCGTAGCTCTT |
Then we focused on EpCAM, MUC1 and WT1 to establish an optimized detection model. We used a binary logistic regression analysis, where we set Y as a binary dependent variable, EpCAM, MUC1 and WT1 are independent variables. The logistic regression model equation is linearly expressed as:
 |
PCEpCAM: PCR concentration of EpCAM; PCMUC1: PCR concentration of MUC1; PCWT1: PCR concentration of WT1.
Criteria for determining treatment efficacy and chemosensitivity are according to the 2021 National Comprehensive Cancer Network (NCCN) clinical practice guidelines (23). Chemo-resistance was defined as recurrence within less than 6 months when achieving complete recovery for an initial treatment, or disease progression during chemotherapy. Chemosensitive patients were defined as recurrence after more than 6 months when achieving complete recovery for an initial treatment (23).
Prognostic criteria were assessed as progression-free survival (PFS) and overall survival (OS) of patients with EOC. During follow-up, the evidence of recurrence or progression was based on the imaging or pathology confirmation on biopsy or based on serum CA125 levels according to Response Evaluation Criteriain Solid Tumors (RECIST) Version 1.1 and adhering to the Gynecological Cancer Intergroup (GCIG) definition as greater than, or equal to, two times the upper limit of the reference range on two occasions at least 1 week apart (24). OS was calculated from date of treatment (surgery or chemotherapy) to date of death or date of last follow-up (end of follow-up). PFS was defined as the time from treatment (surgery or chemotherapy) to physical, biological or radiological evidence of disease progression, or death as a result from any cause (23).
Statistical analysis
Data were statistically analyzed using IBM SPSS statistical software (Version 23.0; IBM Corp., New York, USA). Binary logistic regression was used to build an optimized CTC detection model. PCR concentration was compared using a Mann-Whitney U test. Receiver operating characteristic (ROC) curve analysis was used to evaluate the ability of different PCR marker product concentrations to distinguish EOC from non-cancerous control groups. These results were expressed as area under the ROC curve (AUC), with 95% CI and P values. Sensitivity, specificity and Youden index of the diagnostic results were calculated. The threshold corresponding to the maximum value of Youden index was taken as the best threshold for clinical diagnosis. Count data were expressed as n (%) and χ2 test was used. Multivariate Cox regressions were used on OS analysis, and P<0.05 was considered statistically significant. The survival curve was drawn by Graphpad Prism 8.4.2 (GraphPad Software, Inc., San Diego, CA, USA) for the prognostic correlation analysis.
Results
Clinical characteristics of enrolled patients
A total of 160 patients with EOC were enrolled in the experimental group, whereas 90 patients were enrolled in the non-cancerous control group, including 50 patients with benign ovarian tumor (i.e., teratoma, ovarian chocolate cysts and serous cystadenoma) and 40 healthy females. The general clinical characteristics of the patients are presented in Table 1 .
Table 1. Patients’ clinical information.
| Clinical characteristics | n (%) | |
| Experimental group (N=160) | Control group (N=90) | |
| FIGO, the International Federation of Gynecology and Obstetrics. | ||
| Age (year) | ||
| Mean (range) | 55.7 (19−82) | 44.4 (20−81) |
| CA125 level before initial treatment (U/mL) | ||
| ≤35 | 39 (24.4) | 74 (82.2) |
| >35 | 121 (75.6) | 16 (17.8) |
| Ascites volume (mL) | ||
| <500 | 93 (58.1) | 89 (98.9) |
| ≥500 | 67 (41.9) | 1 (1.1) |
| Pathological typing | ||
| Serous | 126 (78.8) | 8 (8.9) |
| Mucinous | 10 (6.2) | 4 (4.4) |
| Endometrioid | 2 (1.3) | − |
| Clear cell-like | 17 (10.6) | − |
| Mature teratoma | − | 12 (13.3) |
| Chocolate cyst | − | 10 (11.1) |
| Fibro-theca cell tumor | − | 5 (5.6) |
| Fallopian mesangial cyst | − | 4 (4.4) |
| Serous mucinous
cystadenoma |
− | 4 (4.4) |
| Others (including 40
health females) |
5 (3.1) | 43 (47.8) |
| FIGO staging | ||
| I | 34 (21.3) | − |
| II | 21 (13.1) | − |
| III | 84 (52.5) | − |
| IV | 21 (13.1) | − |
| Grading | ||
| 1 | 9 (5.6) | − |
| 2 | 8 (5.0) | − |
| 3 | 127 (79.4) | − |
| Others | 16 (10.0) | − |
Expression level differences of three marker (EpCAM, MUC1 and WT1) in EOC patients vs. non-cancerous controls
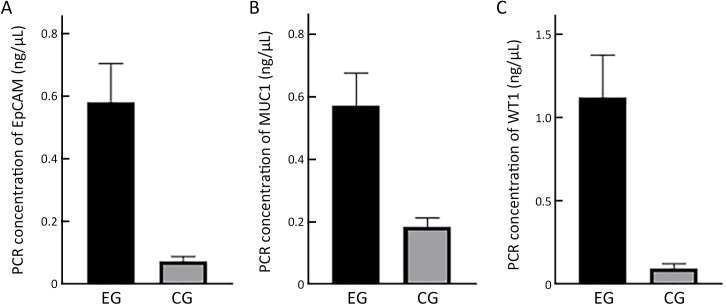
mRNA abundance of EpCAM, MUC1 and WT1 was quantified using multiplex RT-PCR. Comparison of the mRNA abundance levels of EpCAM, MUC1 and WT1 in EOC patients and non-cancerous controls showed that all of the three markers were significantly more abundant in EOC (P<0.001,Figure 1 ).
Figure 1.
Expression of three markers in EOC patients vs. non-cancerous controls. mRNA abundance levels are quantified by PCR. Expression of (A) EpCAM (P<0.001); (B) MUC1 (P<0.001) and (C) WT1 (P<0.001) in EOC patientsvs. non-cancerous controls, respectively. EOC, epithelial ovarian cancer; PCR, polymerase chain reaction; EG, experimental group; CG, control group.
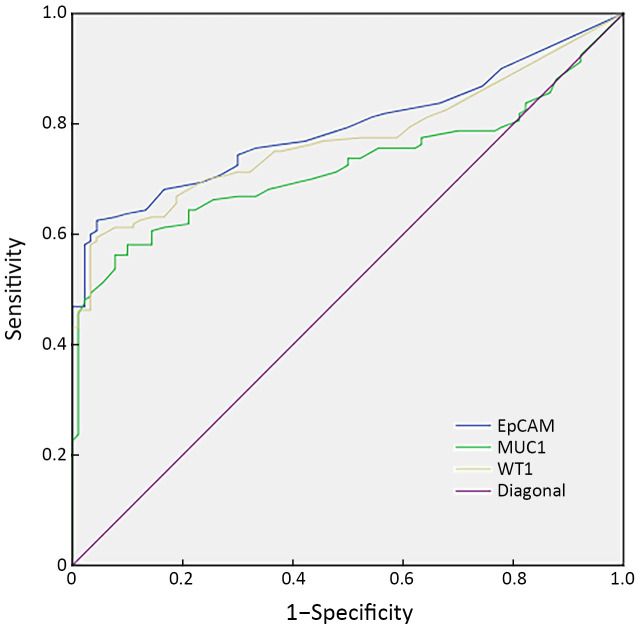
We plotted ROC curves (Supplementary Figure S2 ) to find the sensitivity and specificity of single marker (EpCAM, MUC1 and WT1) in distinguishing EOC from non-cancerous controls. The thresholds, sensitivity and specificity of each single marker are shown in Table 2 . The results of this analysis showed that each marker gene had a high specificity (92.1%−95.5%), but relatively low sensitivity (56.1%−62.6%) (Table 2 ).
Figure S2.
ROC curves for three markers to distinguish EOC patients from the non-cancerous control group. AUC of EpCAM, MUC1 and WT1 is 0.794, 0.721 and 0.791, respectively. ROC, receiver operating characteristic; EOC, epithelial ovarian cancer; AUC, area under the curve.
Table 2. Detection thresholds, sensitivity and specificity of single marker to distinguish EOC patients from non-cancerous control group.
| Marker | Cut-off value (ng/μL) | Sensitivity (%) | Specificity (%) | AUC | 95% CI |
| EOC, epithelial ovarian cancer; AUC, area under the curve; 95% CI, 95% confidence interval. | |||||
| EpCAM | 0.195 | 62.6 | 95.5 | 0.794 | 0.739−0.849 |
| MUC1 | 0.355 | 56.1 | 92.1 | 0.721 | 0.658−0.784 |
| WT1 | 0.305 | 61.3 | 95.5 | 0.791 | 0.736−0.846 |
Establishment of an optimized detection model for CTCs that distinguishes EOC from non-cancerous controls
To assess the contributions of three marker genes more comprehensively, we used a binary logistic regression analysis. The regression coefficients of EpCAM, MUC1 and WT1 are shown in Supplementary Table S2 . The regression equation of the optimized CTC detection model is:
Table S2. Regression coefficients.
| Variables | Estimate | SE | Z (Estimate/se) | P |
| SE, standard error. | ||||
| (Intercept) | −1.917 | 0.3369 | −5.691 | <0.001 |
| EpCAM | 5.413 | 1.5547 | 3.482 | <0.001 |
| MUC1 | 2.463 | 0.8748 | 2.815 | <0.01 |
| WT1 | 3.586 | 0.8758 | 4.094 | <0.001 |
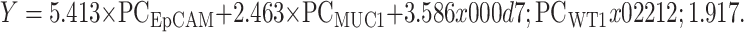 |
PCEpCAM: PCR concentration of EpCAM; PCMUC1: PCR concentration of MUC1; PCWT1: PCR concentration of WT1.
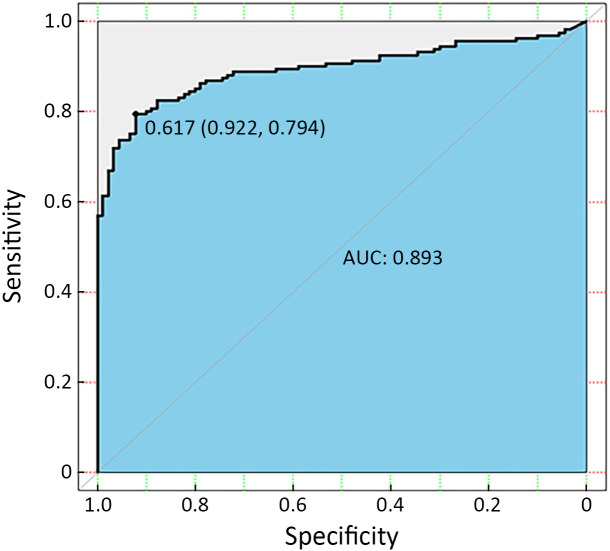
As shown in Figure 2 , the optimal threshold for Y is 0.617, the corresponding sensitivity is 79.4%, and the specificity is 92.2%. So we define Y≥0.617 as positive for CTC detection. And we also performed ten-fold cross validation on the dataset (Supplementary Table S3 ).
Figure 2.
ROC curve for three combined markers to distinguish EOC patients from the non-cancerous control group (AUC=0.893; Y=0.617; specificity=0.922; sensitivity=0.794). ROC, receiver operating characteristic; EOC, epithelial ovarian cancer; AUC, area under the curve.
Table S3. Ten-fold cross validation results.
| Cross validation | Accuracy | Sensitivity | Precision |
| SE, standard error. | |||
| 1.00 | 0.84 | 0.81 | 0.93 |
| 2.00 | 0.84 | 0.83 | 0.91 |
| 3.00 | 0.84 | 0.84 | 0.91 |
| 4.00 | 0.84 | 0.83 | 0.91 |
| 5.00 | 0.82 | 0.83 | 0.89 |
| 6.00 | 0.83 | 0.83 | 0.90 |
| 7.00 | 0.84 | 0.84 | 0.90 |
| 8.00 | 0.85 | 0.85 | 0.92 |
| 9.00 | 0.82 | 0.83 | 0.89 |
| 10.00 | 0.85 | 0.84 | 0.92 |
| Average | 0.84 | 0.83 | 0.91 |
| SE | 0.01 | 0.01 | 0.01 |
Correlation analysis between CTC detection model and clinical features in EOC patients
In comparison with CA125, the specificity of the optimized model was significantly higher than that of CA125 (92.2% vs. 82.2%, P=0.044), while the sensitivity was also slightly higher than that of CA125 (79.4% vs. 75.6%, P>0.05). Especially in early-stage EOC (stage I and II), the detection rate of CTCs was significantly higher than the positive rate for CA125 (74.5%vs. 58.2%, P=0.069). But in advanced-stage EOC (stage III and IV), the detection rate of CTCs was close to CA125 (81.9% vs. 84.8%, P=0.579).
Univariate analysis showed that the detection rate of CTCs was significantly higher in patients with ascitic volume ≥500 mL, suboptimal cytoreductive surgery and elevated serum CA125 levels after 2 courses of chemotherapy (P<0.05,Table 3 ).
Table 3. Univariate analysis of factors affecting CTC detection in EOC.
| Factors | N | n (%) | χ 2 | P | |
| CTCs positive rate | CTCs negative rate | ||||
| CTC, circulating tumor cell; EOC, epithelial ovarian cancer; *, R0 refers to no residual disease; R1 refers to residual disease with tumors <1 cm; R2 refers to residual disease with tumors ≥1 cm. | |||||
| Age (year) | 0.666 | 0.414 | |||
| ≤55 | 78 | 64 (82.1) | 14 (17.9) | ||
| >55 | 82 | 63 (76.8) | 19 (23.2) | ||
| CA125 (U/mL) | 1.919 | 0.166 | |||
| ≤35 | 39 | 34 (87.2) | 5 (12.8) | ||
| >35 | 121 | 93 (76.9) | 28 (23.1) | ||
| Ascites (mL) | 5.311 | 0.021 | |||
| <500 | 93 | 68 (73.1) | 25 (26.9) | ||
| ≥500 | 67 | 59 (88.1) | 8 (11.9) | ||
| Cytoreductive surgery* | 5.732 | 0.017 | |||
| R0−R1 | 126 | 95 (75.4) | 31 (24.6) | ||
| R2 | 34 | 32 (94.1) | 2 (5.9) | ||
| Staging | 1.194 | 0.275 | |||
| I−II | 55 | 41 (74.5) | 14 (25.5) | ||
| III−IV | 105 | 86 (81.9 ) | 19 (18.1) | ||
| Grading | 0.354 | 0.552 | |||
| 1−2 | 17 | 15 (88.2) | 2 (11.8) | ||
| 3 | 127 | 100 (78.7) | 27 (21.3) | ||
| Lymph node metastasis | 0.023 | 0.880 | |||
| Yes | 60 | 48 (80.0) | 12 (20.0) | ||
| None | 100 | 79 (79.0) | 21 (21.0) | ||
| Normalized CA125 with the course of chemotherapy | 4.323 | 0.038 | |||
| ≤2 | 72 | 50 (69.4 ) | 22 (30.6) | ||
| >2 | 39 | 34 (87.2) | 5 (12.8) | ||
| Chemosensitivity | 1.913 | 0.167 | |||
| Sensitivity | 121 | 93 (76.9) | 28 (23.1) | ||
| Resistance | 33 | 29 (87.9) | 4 (12.1) | ||
Application value of CTC optimized detection model and CTCs characterized by single marker genes in predicting chemo-resistance and prognosis in EOC patients.
Among the 160 EOC patients, 115 were found to be chemo-sensitive and 28 were chemo-resistant. The results showed that the detection of CTCs through the optimized detection model was not correlated with chemotherapy resistance (P>0.05,Supplementary Table S4 ). But the detection rate of CTCEpCAM+ (defined as EpCAM PCR concentration ≥0.195 ng/μL) and CTCMUC1+ (defined as MUC1 PCR concentration ≥0.355 ng/μL) were significantly higher in chemo-resistant EOC patients (P<0.05,Supplementary Table S4 ).
Table S4. Detection of CTCs through optimized detection model and CTCs characterized by single marker gene (EpCAM, MUC1 and WT1) in EOC patients with different levels of chemo-sensitivity .
| Marker | N | n (%) | χ 2 | P | |
| Sensitivity | Resistance | ||||
| CTC, circulating tumor cell; EOC, epithelial ovarian cancer. CTCEpCAM positive defined as EpCAM PCR concentration ≥0.195 ng/μL; CTCMUC1 positive defined as MUC1 PCR concentration ≥0.355 ng/μL; CTCWT1 positive defined as WT1 PCR concentration ≥0.305 ng/μL. | |||||
| CTCs | |||||
| Positive | 121 | 93 (76.9) | 28 (23.1) | 1.913 | 0.167 |
| Negative | 33 | 29 (87.9) | 4 (12.1) | ||
| CTCEpCAM | |||||
| Positive | 95 | 70 (73.7) | 25 (26.3) | 4.617 | 0.032 |
| Negative | 59 | 52 (88.1) | 7 (11.9) | ||
| CTCMUC1 | |||||
| Positive | 87 | 64 (73.6) | 23 (26.4) | 3.888 | 0.049 |
| Negative | 67 | 58 (86.6) | 9 (13.4) | ||
| CTCWT1 | |||||
| Positive | 91 | 72 (79.1) | 19 (20.9) | 0.001 | 0.971 |
| Negative | 63 | 50 (79.4) | 13 (20.6) | ||
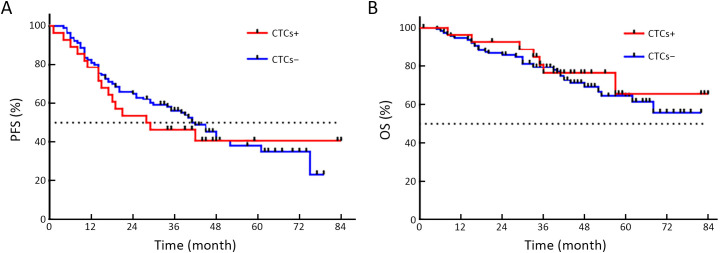
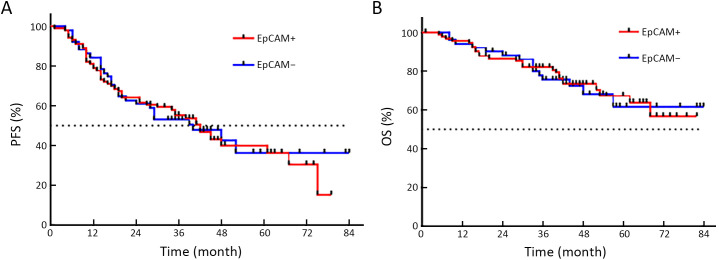
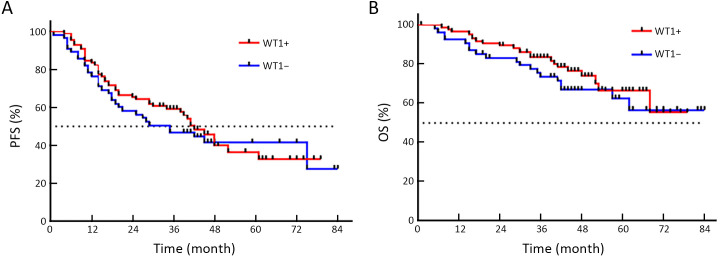
The time distribution of PFS was 4−84 months, with a median PFS of 34 months. The time distribution of OS was 5−84 months, with a median OS of 42 months. While no statistical difference was found, PFS of CTCMUC1+ patients trended to be shorter than that of CTCMUC1− patients (P=0.357, Figure 3A ). But OS of CTCMUC1+ patients was significantly shorter (P=0.043, Figure 3B ). In addition, PFS and OS of CTC+ (detected by the optimized model), CTCEpCAM+ and CTCWT1+ patients did not show any significant difference with their corresponding negative patients (P>0.05,Supplementary Figure S3−5 ).
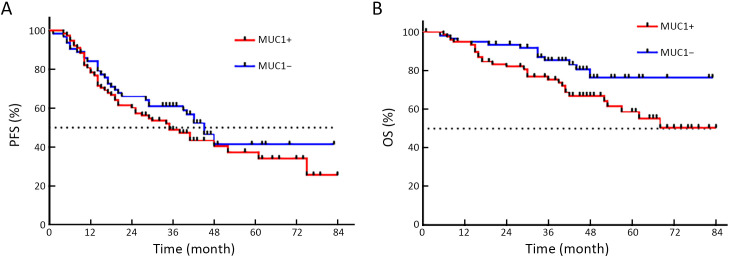
Figure 3.
Comparison of PFS and OS curves in CTCMUC1+ and CTCMUC1− patients. (A) Median PFS of CTCMUC1+ and CTCMUC1− patients was 35.0 and 45.0 months, respectively, but there was no statistical difference (HR=1.242; 95% CI: 0.787−1.959; P=0.357); (B) OS of CTCMUC1+ patients tended to be significantly shorter than CTCMUC1− patients (HR=1.900; 95% CI: 1.020−3.540; P=0.043). PFS, progression-free survival; OS, overall survival; CTC, circulating tumor cell; HR, hazard ratio; 95% CI, 95% confidence interval.
Figure S3.
Comparison of PFS and OS between CTC+ and CTC− EOC patients. (A) Median PFS of CTC+ and CTC− patients was 28.5 and 48.0 months, respectively, but there was no statistical difference (HR=1.137; 95% CI: 0.638−2.025; P=0.386); (B) OS of CTC+ and CTC− patients was not statistically different (HR=0.838; 95% CI: 0.388−1.808; P=0.652). PFS, progression-free survival; OS, overall survival; CTC, circulating tumor cell; HR, hazard ratio; 95% CI, 95% confidence interval.
Multivariate Cox regression analysis was performed on the effects of CTCMUC1+, staging, the level of CA125, ascites volume, cytoreductive surgery, lymph node metastasis, and chemotherapy sensitivity on the prognosis of patients. The results showed that advanced staging (P=0.006) and chemotherapy resistance (P<0.001) were independent risk factors affecting PFS of patients (Supplementary Table S5 ). The volume of ascites ≥500 mL (P=0.006), suboptimal cytoreductive surgery (P=0.046) and chemotherapy resistance (P<0.001) were independent risk factors affecting OS of patients (Supplementary Table S6 ).
Table S5. Multivariate Cox regression analysis on PFS in patients with EOC.
| Factors | B | SE | Exp (B) | 95% CI | P |
| PFS, progression-free survival; EOC, epithelial ovarian cancer; SE, standard error; 95% CI, 95% confidence interval. | |||||
| Staging | 1.060 | 0.387 | 2.887 | 1.353−6.159 | 0.006 |
| Level of CA125 | 0.395 | 0.275 | 1.485 | 0.866−2.545 | 0.151 |
| Ascites | 0.241 | 0.275 | 1.273 | 0.743−2.181 | 0.379 |
| Cytoreductive surgery | 0.097 | 0.276 | 1.101 | 0.641−1.893 | 0.727 |
| Lymph node metastasis | 0.116 | 0.258 | 1.123 | 0.677−1.862 | 0.653 |
| Chemosensitivity | 2.613 | 0.305 | 13.634 | 7.499−24.790 | <0.001 |
Table S6. Multivariate Cox regression analysis on OS in patients with EOC.
| Factors | B | SE | Exp (B) | 95% CI | P |
| OS, overall survival; EOC, epithelial ovarian cancer; SE, standard error; 95% CI, 95% confidence interval. CTCMUC1+ defined as MUC1 PCR concentration ≥0.355 ng/μL. | |||||
| CTCMUC1+ | 0.284 | 0.372 | 1.328 | 0.640−2.775 | 0.446 |
| Staging | 0.681 | 0.593 | 1.976 | 0.618−6.320 | 0.251 |
| Level of CA125 | −0.354 | 0.363 | 0.702 | 0.344−1.430 | 0.330 |
| Ascites | 1.107 | 0.405 | 3.025 | 1.367−6.691 | 0.006 |
| Cytoreductive surgery | 0.742 | 0.371 | 2.100 | 1.015−4.346 | 0.046 |
| Lymph node metastasis | 0.179 | 0.350 | 1.196 | 0.603−2.373 | 0.609 |
| Chemosensitivity | 2.211 | 0.354 | 9.121 | 4.555−18.266 | <0.001 |
Discussion
CTCs have become popular as it is expected that they will become screening markers for early diagnosis, treatment monitoring, chemotherapy reaction and prognostic evaluation (12,25). A large amount of research concerning ovarian cancer CTCs has been reported in the recent 5 years (Table 4 ). Though promising evidence for their clinical application in assessing patient prognosis, real-time monitoring of treatment effects and disease recurrence has been reported (12,25), many researchers still focus on continuous optimization of CTC detection technologies.
Table 4. Overview of studies on detection methods for CTCs in ovarian cancer.
| References | Year | No. of
patients |
Tumor
stage |
Isolation method | Detection method | Targeted antigen/
targeted gene |
Positivity
rate (%) |
HR (95% CI) |
| ND, new disease; RD, recurrent disease; NR, not reported; RT-PCR, real-time polymerase chain reaction; RT-qPCR, real-time quantitative polymerase chain reaction; ICC, immunocytochemistry; BT, before treatment; AT, after treatment; HR, hazard ratio; 95% CI, 95% confidence interval; PFS, progression-free survival; OS, overall survival; NS, not significant; | ||||||||
| Fan,
et al. (26) |
2009 | 66 | I−IV | CAMt (functional
enrichment) |
ICC | EpCAM; CK4, 5, 6, 8, 10, 13, 18 | 60.6 | PFS: 1.44 (0.78−2.64), P=0.040;
OS: 0.89 (0.40−1.95), NS |
| Aktas,
et al. (27) |
2011 | 122 | NR | Immunomagnetic
(Adnatest®) |
RT-PCR
(Adnatest®) |
EpCAM, MUC-1,
CA-125, HER-2 |
19.0 (BT)
27.0 (AT) |
PFS: 1.58 (0.86−2.88), NS
OS: 4.56 (1.94−10.73), P=0.050 |
| Sang,et al.(28) | 2014 | 80 | I−IV | Red blood cells lysis | RT-PCR | MAGE-As | 47.5 | OS: P=0.002 |
| Pearl,
et al. (29) |
2014 | 88 | I−IV | CAMt (functional
enrichment) |
ICC | EpCAM, CA-125,
DPP4 and CKs |
88.6 | PFS: 1.21 (0.49−2.97), P=0.0024;
OS: 1.06 (0.41−2.73), P=0.022 |
| Kuhlmann,
et al. (30) |
2014 | 143 | I−IV | Immunomagnetic
(Adnatest®) |
RT-PCR
(Adnatest®) |
EpCAM, MUC1,
MUC6, ERCC1 |
14.0 | PFS: 1.50 (0.81−2.79), NS
OS: 1.85 (1.03−3.32), P=0.041 |
| Pearl,
et al.(18) |
2015 | 31 | I−IV | CAMt (functional
enrichment) |
ICC/RT-PCR | EpCAM, CA-125, CD44, seprase. Gene expression: EpCAM, CD44, MUC16, FAP | 100 | Risk for PD, P<0.001
OR=121.3 |
| Blassl,
et al.(31) |
2015 | 10 | NR | Immunomagnetic
(Adnatest) |
Multiplex RT-PCR (Adnates) | 19 gene transcripts: epithelial, EMT and stem cell markers | 30.0 | NR |
| Kolostova,
et al. (32) |
2016 | 56 | NR | MetaCell® | ICC/RT-PCR | EpCAM, MUC1, MUC16,
KRT18, KRT19, ERCC1, WT1 |
58.0 | NR |
| Liu,
et al. (33) |
2016 | 10 | NR | Immunomagnetic
(Folic acid) |
ICC | HE4 and FITC AffiniPure | 50.0 | NR |
| Chebouti,
et al. (34) |
2017 | 91 | I−IV | Immunomagnetic
(Adnatest) |
Multiplex RT-PCR (Adnatest) | EpCAM, ERCC1, MUC1,
MUC16, PI3Kα, Akt-2, Twist |
82.0 | I−III:
PFS: 2.35 (1.06−8.74), P=0.042 OS: 7.22 (3.21−111.5), P=0.001 |
| Suh,
et al.(35) |
2017 | 31 | I−IV | Tapered-slit membrane filters | ICC | EpCAM, CK9 | 77.4 | NR |
| Lee,
et al.(36) |
2017 | 24 (ND)
30 (RD) |
I−IV | Microfluidic device | ICC | EpCAM, TROP-2, EGFR, CD45 vimentin, N-cadherin, DAPI | 98.1 | PFS: 1.3 (0.230−7.145), P=0.035
OS: 1.3 (0.350−2.298), NS |
| Rao,
et al.(37) |
2017 | 23 | I−IV | Microfluidics plus immunomagnetic beads (EpCAM) | ICC | EpCAM, CK3–6H5, panCK | 87.0 | NR |
| Obermayr,
et al. (38) |
2017 | 102 (BT)
78 (AT) |
II−IV | Density gradient centrifugation | ICC and FISH | ICC: EpCAM, Cytokeratins,
EGFR, MUC1, HER2 FISH: MECOM, HHLA1 |
26.5 (BT)
7.7 (AT) |
PFS: 5.671 (1.560−20.618),
P=0.008 OS: 3.305 (1.386−7.880), P=0.007 |
| Obermayr,
et al. (39) |
2018 | 20 | II−IV | Microfluidic ParsortixTM | RT-qPCR | EpCAM, PPIC, MAL2,
LAMB1, SERPINE2, TUSC3 |
70.0 | NR |
| Nie,
et al. (25) |
2018 | 20 | NR | Immunomagnetic
(Folic acid) |
ICC | HE4, FA | 80.0 | NR |
| Guo,
et al.(19) |
2018 | 30 | NR | Size-based microfluidic technique | ICC | EpCAM, HE4, panCK, CK7,
Vimentin |
73.3 | NR |
| Po,
et al. (40) |
2018 | 20 | III−IV | Immunomagnetic beads (EpCAM, N-cadherin) | ICC | EpCAM, N-cadherin, CK, Vecad, Vimentin | 90.0 | NR |
| Zhang,
et al. (12) |
2018 | 109 | I−IV | Immunomagnatic beads (EpCAM, HER2, MUC1) | Multiplex RT-PCR | EpCAM, HER2, MUC1, WT1, P16 and PAX8 | 90.0 (BT)
91.0 (AT) |
OS: P=0.041 |
| Kim,
et al.(20) |
2019 | 30 | I−IV | Tapered-slit membrane filters | ICC | CK-9, EpCAM | 76.7 (BT)
57.1 (AT) |
NR |
| Li N,
et al.(41) |
2019 | 30 | I−IV | Immunomagnetic
(EpCAM, FRα) |
ICC | EpCAM, FRα | 67.4 | NR |
| Banys-PM,
et al. (21) |
2020 | 43 | I−IV | Immunomagnetic
(CellSearchTM system) |
ICC (CellSearchTM system) | EpCAM, CK | 26.0 | PFS: P=0.005
OS: P=0.006 |
| Abreu M,
et al. (42) |
2020 | 38
|
I−IV | Immunomagnetic
beads (EpCAM, CK) |
RT-qPCR | GAPDH, TIMP1, CK19, MUC1 | 63.0 | NS |
Classically, the Cell Search® system is a positive enrichment method using an EpCAM targeted immunomagnetic bead to separate CTCs from peripheral blood samples (43), and is the only FDA-approved CTC detection technology that can be used for metastatic breast, colorectal and prostate cancers (11,40). However, the biggest limitation of this detection method is that EpCAM is a marker of epithelial cells and not a tumor-specific marker (44). When tumor cells undergo epithelial-mesenchymal metastasis (EMT), CTCs expressing mesenchymal-derived markers become undetectable, resulting in the loss of CTC detection (45,46). Besides, we reviewed the researches of CTC detection methods for EOC over the past 5 years, a series of new detection methods have emerged, such as streptavidin-biotin cascade amplification effect coupled with folate receptor binding method (25), invasiveness CTC subgroup labeling method (18,29) and a high-throughput microfluidic technology system designed by organically combining physical and biological characteristics (37,39). Also, based on the antigen-antibody binding enrichment methods similar to CellSearch system, studies reported the use of immunomagnetic beads coupled with two or more antibodies (including epithelial markers: EpCAM, MUC-1, CK, HER2 and ESA; interstitial markers: Vimentin and N-cadherin). The above mentioned detection methods significantly increased the detection rates by 58%−95% (Table 4 ).
Our study also used an antigen-antibody binding method to screen and separate CTCs with immunomagnetic beads, which was then combined with multiple RT-PCR to detect CTCs in the peripheral blood of EOC patients. To compensate for the decrease in detection rate caused by weak expression of a single target antigen, three molecules (EpCAM, MUC1 and WT1), which are significantly more abundant in ovarian cancer cells, based on a literature review, were selected as gene markers to establish an optimized CTC detection model. Early in 2018, our laboratory successfully used this method to detect CTCs in the peripheral blood of EOC patients (12). If PCR concentration of any of the six marker (EpCAM, HER2, MUC1, WT1, P16 and PAX8) was greater than or equal to 0.3 ng/μL, it was defined as positive for CTCs. Based on this detection method, we detected CTCs in 90% (98/109) of the newly diagnosed EOC patients, demonstrating the feasibility of this detection method. However, our previous study only enrolled healthy females in the control group which inevitably becomes a limitation, as benign ovarian tumor is the first need to be differentiated from EOC. Our pre-experiment showed that, when compared with benign ovarian tumor patients, only EpCAM, MUC1 and WT1 transcripts were significantly more abundant in EOC patients (P<0.05). This result suggests that the other three marker (HER2, P16 and PAX8) fail to differentiate benign ovarian tumor from EOC. Therefore, to establish optimized detection model, we expanded the sample sizes and focused on EpCAM, MUC1 and WT1 to achieve optimal differentiation between the 160 EOC and 90 non-cancerous patients.
Similar to our results, relevant studies demonstrated that EpCAM, MUC1 and WT1 are significantly more abundant in ovarian cancer (32,47-50). Among them, EpCAM is a type I transmembrane glycoprotein located on epithelial cells (51) and is a tumor-associated antigen that is highly expressed in ovarian cancer and is also the most widely used marker for ovarian cancer CTC detection technology (47,48). Zhao et al. (52) used the Gene Expression Omnibus (GEO) database to analyze differentially expressed genes in ovarian cancer and found that the abundance of EpCAM transcripts in ovarian cancer differs significantly from normal tissues (P=0.0048), and especially is high in ovarian cancers with recurrence, metastasis, invasiveness and chemotherapy resistance (53). This supports EpCAM role in the occurrence and metastasis of ovarian cancer. Mucin 1 (MUC1) gene encodes a membrane-bound O-glycosylated protein that plays an essential role in forming protective mucous barriers on epithelial surfaces (54), which is overexpressed in 90%−100% of serous carcinomas (55-57). The role of MUC1 has been verified in mouse models where MUC1 expression is necessary for the occurrence of ovarian cancer (56,57). The study by Hou et al. (49) confirmed that the expression level of MUC1 in malignant EOC tissues was significantly higher than that in borderline, benign and normal ovarian tissues (P<0.05).WT1 (Wilms tumor gene) is located in the nuclei and encodes a tumor-associated antigen (58). In epithelial ovarian tumors, WT1 expression has been detected, and studies showed that ovarian cancers with high expression of WT1 are more aggressive (50) and suggestive of poor prognosis (59,60).
Based on single indicators for CTC detection, our results showed that the specificity of each gene marker was high (92.1%−95.5%), but their sensitivity was relatively low (56.1%−62.6%), which suggests that we need to combine multiple markers to construct an optimized model to increase the sensitivity of detection while ensuring a high specificity. However, the combined application of these three markers for the detection of CTCs in ovarian cancer has been rarely reported. We used a binary logistic regression analysis to set up the regression equation of the optimized CTC detection model. When the threshold was set as 0.617 (Y≥0.617 as positive for CTCs), the sensitivity was 79.4%, and the specificity was 92.2%. Compared with the application of a single index, the sensitivity of the optimized detection model was improved without any evident decrease in specificity. Similar to our results, Abreu et al. (42) published results from their collection of peripheral blood CTCs from 38 patients with advanced ovarian high-grade serous cancer and also established an optimized gene panel for CTC RT-qPCR detection. Their model was composed of GAPDH, TIMP1, CK19 and MUC1 (AUC: 0.938, P<0.01; 95% CI: 0.868−1), and its detection sensitivity was 63%, which is lower than our optimized model, but had a better specificity of 100%.
Then we compared the diagnostic value of our CTC detection model with serum CA125 level. CA125 is widely used as a clinical serological marker, which is commonly found in the serum of EOC patients (61), so it can assist the diagnosis of EOC and can be used to monitor the progress of the disease (62). However, the sensitivity of CA125 in early-staged EOC is quite low, resulting in an early diagnosis rate of only 50% (3). Also, the specificity of CA125 is rather poor, as inflammation, endometriosis and other benign diseases can also present with elevated serum CA125 levels (4). Therefore, relying solely on serum CA125 levels to assist the diagnosis of ovarian cancer, monitor treatment effects and recurrence has limitations. The detection of CTCs can to some extents make up for the limitations of CA125. Studies (12,19,35) have shown that, compared with CA125, CTCs have a higher detection rate for early diagnosis. Our study also confirms this point of view. In early-stage EOC (stage I and II), the detection rate of our CTCs detection model was significantly higher than the positive rate of CA125 (74.5%vs. 58.2%, P=0.069), and also showed a higher specificity than that of CA125 (92.2% vs. 82.2%, P<0.05). This suggests the advantage of our CTC detection model in the early diagnosis of EOC and the feasibility to combine CTCs and CA125 to improve the diagnostic rate of EOC. Moreover, univariate analysis showed that the detection rate of CTCs was significantly higher in patients with ascitic volume ≥500 mL and in patients with suboptimal cytoreductive surgery. The large amount of ascites and the incomplete cytoreductive surgery all indicate the presence of a large tumor burden and advanced stage, which suggests that the detection of CTCs can reflect the severity of the disease.
A large number of studies have confirmed that CTCs can indirectly reflect the tumor proliferation and chemo-sensitivity through dynamically monitoring the changes in CTCs counts during chemotherapy, indicating that CTCs can be used as a predictive marker to assess the effect of chemotherapy (63-65). However, results from relevant studies regarding this question have been controversial. Some studies concluded that there was no statistically significant association between CTC detection and chemo-resistance (26,66). Although the results of our study showed no statistical difference between CTCs detection and chemo-resistance. In clinical practice, we found that patients with elevated serum CA125 levels after more than 2 courses of postoperative chemotherapy often had poor outcomes and usually ended up with a high incidence of chemoresistance or uncontrolled tumor progression. Our study also found that the detection rate of CTCs in this group of patients was significantly higher, suggesting a correlation between CTCs and chemo-resistance. Besides, an in-depth analysis of the abundance of the different marker genes suggested that CTCs characterized by positive expression of EpCAM or MUC1 have a predictive value for chemoresistance in EOC patients.
Recent studies have been devoted to exploring the predictive value of CTCs as tumor markers for prognosis of ovarian cancer, but the result is yet undetermined (12,18,21,34,36,38,39). We reviewed 20 articles published in the past 10 years, and more than half of the studies concluded that the detection of CTCs in peripheral blood is closely related to the shortening of PFS and OS (Table 4 ). Three published meta-analysis articles summarize relevant studies that CTCs could predict the prognosis of ovarian cancer patients (67-69), but there are also a few studies that concluded that CTCs are not significantly related to PFS and OS (17,26,70). Unfortunately, our study showed that there was no significant correlation between the CTC detection model and the prognosis. However, we found that patients with CTCs characterized by positive expression of MUC1 (CTCMUC1+) had a significantly shorter OS, and a trend for shorter PFS than patients without, which suggests that the detection of CTCMUC1+ may indicate poor prognosis. This finding indicates a correlation between different CTCs’ molecular phenotypes and clinical features. The rapid development of single-cell sequencing has paved the way for the molecular characterization of CTCs (71). Our following studies are needed to resolve the roles of the molecular phenotypic characteristics of CTCs, and explore the mechanisms for ovarian cancer metastasis, recurrence and chemotherapy resistance, and potentially guide individualized treatment of ovarian cancer patients.
There are some limitations of our study that should be improved in subsequent studies. The detection method of our study was not to capture intact CTCs from the blood, but instead to obtain mRNA after cell lysis from enriched cells and to evaluate expression of marker genes by RT-PCR. In addition, since the follow-up period of most patients was short, future studies should extend the follow-up period and expand sample size to validate the above findings.
With the continuous optimization of CTCs detection technology, with increased sensitivity and specificity, it may become possible to isolate viable and intact CTCs. Development of CTCs in vitro cell culture technology, the construction of biomimetic models, and the combined application of genetic assessment and bioinformatics analysis should help reveal the molecular biological characteristics of primary ovarian cancer, metastases and recurrences. This should lead to improve understanding of the mechanisms of ovarian cancer metastasis and chemotherapy resistance, identify new therapeutic targets, and guide the individualized treatment of ovarian cancer patients.
Conclusions
This study used immunomagnetic beads targeting EpCAM and MUC1 for CTC separation, then quantified the expression of EpCAM, MUC1 and WT1 in peripheral blood for CTC detection, and further used these three markers to establish an optimized detection model for CTCs. Compared with a single marker gene, the optimized detection model had higher sensitivity and specificity, and had better diagnostic value for early-staged ovarian cancer than CA125. However, for predicting drug resistance and prognosis, positive CTCs characterized by a single marker appeared more advantageous than the optimized detection model. Detection of CTCEpCAM+ and CTCMUC1+ has predictive value for chemotherapy resistance and the detection of CTC MUC1+ may indicate poor prognosis.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Figure S4.
Comparison of PFS and OS curves between CTCEpCAM+ and CTCEpCAM− patients. (A) PFS of CTCEpCAM+ and CTCEpCAM− patients was not statistically different (HR=1.049; 95% CI: 0.659−1.670; P=0.841); (B) OS of CTCEpCAM+ and CTCEpCAM− patients was not statistically different (HR=0.960; 95% CI: 0.505−1.823; P=0.899). PFS, progression-free survival; OS, overall survival; CTC, circulating tumor cell; HR, hazard ratio; 95% CI, 95% confidence interval.
Figure S5.
Comparison of PFS and OS curves between CTCWT1+ and CTCWT1− patients. (A) Median PFS of CTCWT1+ and CTCWT1− patients was 42.0 and 35.0 months, respectively, but there was no statistical difference (HR=0.869; 95% CI: 0.546−1.384; P=0.308); (B) OS of CTCWT1+ and CTCWT1− patients was not statistically different (HR=0.735; 95% CI: 0.391−1.381; P=0.201). PFS, progression-free survival; OS, overall survival; CTC, circulating tumor cell; HR, hazard ratio; 95% CI, 95% confidence interval.
Acknowledgements
None.
Contributor Information
David M. Irwin, Email: david.irwin@utoronto.ca.
Huanran Tan, Email: tanlab@bjmu.edu.cn.
Hongyan Guo, Email: bysyghy@163.com.
References
- 1.Siegel RL, Miller KD, Jemal A Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Stewart C, Ralyea C, Lockwood S Ovarian cancer: An integrated review. Semin Oncol Nurs. 2019;35:151–6. doi: 10.1016/j.soncn.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Sölétormos G, Duffy MJ, Othman AHS, et al Clinical use of cancer biomarkers in epithelial ovarian cancer: Updated guidelines from the European group on tumor markers. Int J Gynecol Cancer. 2016;26:43–51. doi: 10.1097/IGC.0000000000000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stabile G, Zinicola G, Romano F, et al Pelvic mass, ascites, hydrothorax: a malignant or benign condition. Meigs syndrome with high levels of CA 125. Prz Menopauzalny. 2021;20:103–7. doi: 10.5114/pm.2021.106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bono JS, Scher HI, Montgomery RB, et al Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Jiang J, Breast Disease Center, Southwest Hospital, Third Military Medical University Progress in biological characteristics of circulating breast-tumor cells. Zhonghua Zhong Liu Fang Zhi Za Zhi. 2012;16:1272–5. doi: 10.16073/j.cnki.cjcpt.2012.16.004. [DOI] [Google Scholar]
- 7.Antonarakis ES, Lu C, Luber B, et al Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol. 2017;35:2149–56. doi: 10.1200/JCO.2016.70.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorente D, Olmos D, Mateo J, et al Decline in circulating tumor cell count and treatment outcome in advanced prostate cancer. Eur Urol. 2016;70:985–92. doi: 10.1016/j.eururo.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SJ, Punt CJ, Iannotti N, et al Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 10.Heller G, McCormack R, Kheoh T, et al Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: A comparison with prostate-specific antigen across five randomized phase III clinical trials. J Clin Oncol. 2018;36:572–80. doi: 10.1200/JCO.2017.75.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Gao P, Song Y, et al Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch System in colorectal cancer. BMC Cancer. 2015;15:202. doi: 10.1186/s12885-015-1218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Li H, Yu X, et al Analysis of circulating tumor cells in ovarian cancer and their clinical value as a biomarker. Cell Physiol Biochem. 2018;48:1983–94. doi: 10.1159/000492521. [DOI] [PubMed] [Google Scholar]
- 13.Wang T, Yao Y, Li Y, et al. Research progress of circulating tumor cells in ovarian epithelial carcinoma. Zhongguo Fu Chan Ke Lin Chuang Za Zhi (in Chinese) 2019;20:280-2.
- 14.Marth C, Kisic J, Kaern J, et al Circulating tumor cells in the peripheral blood and bone marrow of patients with ovarian carcinoma do not predict prognosis. Cancer. 2002;94:707–12. doi: 10.1002/cncr.10250. [DOI] [PubMed] [Google Scholar]
- 15.Judson PL, Geller MA, Bliss RL, et al Preoperative detection of peripherally circulating cancer cells and its prognostic significance in ovarian cancer. Gynecol Oncol. 2003;91:389–94. doi: 10.1016/j.ygyno.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu JF, Kindelberger D, Doyle C, et al Predictive value of circulating tumor cells (CTCs) in newly-diagnosed and recurrent ovarian cancer patients. Gynecol Oncol. 2013;131:352–6. doi: 10.1016/j.ygyno.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Ning N, Zhan T, Zhang Y, et al Improvement of specific detection of circulating tumor cells using combined CD45 staining and fluorescence in situ hybridization. Clin Chim Acta. 2014;433:69–75. doi: 10.1016/j.cca.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Pearl ML, Dong H, Tulley S, et al Treatment monitoring of patients with epithelial ovarian cancer using invasive circulating tumor cells (iCTCs) Gynecol Oncol. 2015;137:229–38. doi: 10.1016/j.ygyno.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo YX, Neoh KH, Chang XH, et al Diagnostic value of HE4+ circulating tumor cells in patients with suspicious ovarian cancer. Oncotarget. 2018;9:7522–33. doi: 10.18632/oncotarget.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim M, Suh DH, Choi JY, et al Post-debulking circulating tumor cell as a poor prognostic marker in advanced stage ovarian cancer: A prospective observational study. Medicine (Baltimore) 2019;98:e15354. doi: 10.1097/MD.0000000000015354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banys-Paluchowski M, Fehm T, Neubauer H, et al Clinical relevance of circulating tumor cells in ovarian, fallopian tube and peritoneal cancer. Arch Gynecol Obstet. 2020;301:1027–35. doi: 10.1007/s00404-020-05477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng H, Wang S, Luan W, et al Combined detection and subclass characteristics analysis of CTCs and CTECs by SE-iFISH in ovarian cancer. Chin J Cancer Res. 2021;33:256–70. doi: 10.21147/j.issn.1000-9604.2021.02.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daly MB, Pal T, Berry MP, et al. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;19:77-102.
- 24.Rustin GJ, Vergote I, Eisenhauer E, et al Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1. 1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG). Int J Gynecol Cancer. 2011;21:419–23. doi: 10.1097/IGC.0b013e3182070f17. [DOI] [PubMed] [Google Scholar]
- 25.Nie L, Li F, Huang X, et al Folic acid targeting for efficient isolation and detection of ovarian cancer CTCs from human whole blood based on two-step binding strategy. ACS Appl Mater Interfaces. 2018;10:14055–62. doi: 10.1021/acsami.8b02583. [DOI] [PubMed] [Google Scholar]
- 26.Fan T, Zhao Q, Chen JJ, et al Clinical significance of circulating tumor cells detected by an invasion assay in peripheral blood of patients with ovarian cancer. Gynecol Oncol. 2009;112:185–91. doi: 10.1016/j.ygyno.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aktas B, Kasimir-Bauer S, Heubner M, et al Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int J Gynecol Cancer. 2011;21:822–30. doi: 10.1097/IGC.0b013e318216cb91. [DOI] [PubMed] [Google Scholar]
- 28.Sang M, Wu X, Fan X, et al Multiple MAGE-A genes as surveillance marker for the detection of circulating tumor cells in patients with ovarian cancer. Biomarkers. 2014;19:34–42. doi: 10.3109/1354750X.2013.865275. [DOI] [PubMed] [Google Scholar]
- 29.Pearl ML, Zhao Q, Yang J, et al Prognostic analysis of invasive circulating tumor cells (iCTCs) in epithelial ovarian cancer. Gynecol Oncol. 2014;134:581–90. doi: 10.1016/j.ygyno.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhlmann JD, Wimberger P, Bankfalvi A, et al ERCC1-positive circulating tumor cells in the blood of ovarian cancer patients as a predictive biomarker for platinum resistance. Clin Chem. 2014;60:1282–9. doi: 10.1373/clinchem.2014.224808. [DOI] [PubMed] [Google Scholar]
- 31.Blassl C, Kuhlmann JD, Webers A, et al Gene expression profiling of single circulating tumor cells in ovarian cancer - Establishment of a multi-marker gene panel. Mol Oncol. 2016;10:1030–42. doi: 10.1016/j.molonc.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolostova K, Pinkas M, Jakabova A, et al Molecular characterization of circulating tumor cells in ovarian cancer. Am J Cancer Res. 2016;6:973–80. [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Guo W, Zhang D, et al Circulating tumor cell detection in hepatocellular carcinoma based on karyoplasmic ratios using imaging flow cytometry. Sci Rep. 2016;6:39808. doi: 10.1038/srep39808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chebouti I, Kasimir-Bauer S, Buderath P, et al EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget. 2017;8:48820–31. doi: 10.18632/oncotarget.16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suh DH, Kim M, Choi JY, et al Circulating tumor cells in the differential diagnosis of adnexal masses. Oncotarget. 2017;8:77195–206. doi: 10.18632/oncotarget.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee M, Kim EJ, Cho Y, et al Predictive value of circulating tumor cells (CTCs) captured by microfluidic device in patients with epithelial ovarian cancer. Gynecol Oncol. 2017;145:361–5. doi: 10.1016/j.ygyno.2017.02.042. [DOI] [PubMed] [Google Scholar]
- 37.Rao Q, Zhang Q, Zheng C, et al Detection of circulating tumour cells in patients with epithelial ovarian cancer by a microfluidic system. Int J Clin Exp Pathol. 2017;10:9599–606. [PMC free article] [PubMed] [Google Scholar]
- 38.Obermayr E, Bednarz-Knoll N, Orsetti B, et al Circulating tumor cells: potential markers of minimal residual disease in ovarian cancer. a study of the OVCAD consortium. Oncotarget. 2017;8:106415–28. doi: 10.18632/oncotarget.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obermayr E, Maritschnegg E, Agreiter C, et al Efficient leukocyte depletion by a novel microfluidic platform enables the molecular detection and characterization of circulating tumor cells. Oncotarget. 2017;9:812–23. doi: 10.18632/oncotarget.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Po JW, Roohullah A, Lynch D, et al. Improved ovarian cancer EMT-CTC isolation by immunomagnetic targeting of epithelial EpCAM and mesenchymal N-cadherin. J Circ Biomark 2018;7: 1849454418782617.
- 41.Li N, Zuo H, Chen L, et al Circulating tumor cell detection in epithelial ovarian cancer using dual-component antibodies targeting EpCAM and FRα. Cancer Manag Res. 2019;11:10939–48. doi: 10.2147/CMAR.S211455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abreu M, Cabezas-Sainz P, Alonso-Alconada L, et al Circulating tumor cells characterization revealed TIMP1 as a potential therapeutic target in ovarian cancer. Cells. 2020;9:1218. doi: 10.3390/cells9051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andree KC, van Dalum G, Terstappen LW Challenges in circulating tumor cell detection by the CellSearch system. Mol Oncol. 2016;10:395–407. doi: 10.1016/j.molonc.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baeuerle PA, Gires O EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96:417–23. doi: 10.1038/sj.bjc.6603494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Du F, Fujii T, Kida K, et al EpCAM-independent isolation of circulating tumor cells with epithelial-to-mesenchymal transition and cancer stem cell phenotypes using ApoStream® in patients with breast cancer treated with primary systemic therapy . PLoS One. 2020;15:e0229903. doi: 10.1371/journal.pone.0229903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou W, Ye C, Li L, et al Adipocyte-derived SFRP5 inhibits breast cancer cells migration and invasion through Wnt and epithelial-mesenchymal transition signaling pathways. Chin J Cancer Res. 2020;32:347–60. doi: 10.21147/j.issn.1000-9604.2020.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visvader JE, Lindeman GJ Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 48.Tavsan Z, Ayar K H EpCAM-claudin-tetraspanin-modulated ovarian cancer progression and drug resistance. Cell Adh Migr. 2020;14:57–68. doi: 10.1080/19336918.2020.1732761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou R, Jiang L, Liu D, et al Lewis(y) antigen promotes the progression of epithelial ovarian cancer by stimulating MUC1 expression. Int J Mol Med. 2017;40:293–302. doi: 10.3892/ijmm.2017.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Yamanouchi K, Ohtao T, et al High levels of Wilms’ tumor 1 (WT1) expression were associated with aggressive clinical features in ovarian cancer. Anticancer Res. 2014;34:2331–40. [PubMed] [Google Scholar]
- 51.Jing Y, Zhou L, Chen J, et al Quantitatively mapping the assembly pattern of epcam on cell membranes with peptide probes. Anal Chem. 2020;92:1865–73. doi: 10.1021/acs.analchem.9b03901. [DOI] [PubMed] [Google Scholar]
- 52.Zhao L, Li Y, Zhang Z, et al Meta-analysis based gene expression profiling reveals functional genes in ovarian cancer. Biosci Rep. 2020;40:BSR20202911. doi: 10.1042/BSR20202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bellone S, Siegel ER, Cocco E, et al Overexpression of epithelial cell adhesion molecule in primary, metastatic, and recurrent/chemotherapy-resistant epithelial ovarian cancer: implications for epithelial cell adhesion molecule-specific immunotherapy. Int J Gynecol Cancer. 2009;19:860–6. doi: 10.1111/IGC.0b013e3181a8331f. [DOI] [PubMed] [Google Scholar]
- 54.Williams KA, Terry KL, Tworoger SS, et al Polymorphisms of MUC16 (CA125) and MUC1 (CA15. 3) in relation to ovarian cancer risk and survival. PLoS One. 2014;9:e88334. doi: 10.1371/journal.pone.0088334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schroeder JA, Masri AA, Adriance MC, et al MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene. 2004;23:5739–47. doi: 10.1038/sj.onc.1207713. [DOI] [PubMed] [Google Scholar]
- 56.Feng H, Ghazizadeh M, Konishi H, et al Expression of MUC1 and MUC2 mucin gene products in human ovarian carcinomas. Jpn J Clin Oncol. 2002;32:525–9. doi: 10.1093/jjco/hyf111. [DOI] [PubMed] [Google Scholar]
- 57.Tashiro Y, Yonezawa S, Kim YS, et al Immunohistochemical study of mucin carbohydrates and core proteins in human ovarian tumors. Hum Pathol. 1994;25:364–72. doi: 10.1016/0046-8177(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 58.Cheever MA, Allison JP, Ferris AS, et al The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–37. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Netinatsunthorn W, Hanprasertpong J, Dechsukhum C, et al WT1 gene expression as a prognostic marker in advanced serous epithelial ovarian carcinoma: an immunohistochemical study. BMC Cancer. 2006;6:90. doi: 10.1186/1471-2407-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi XW, Zhang F, Wu H, et al Wilms’ tumor 1 (WT1) expression and prognosis in solid cancer patients: a systematic review and meta-analysis. Sci Rep. 2015;5:8924. doi: 10.1038/srep08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morales-Vásquez F, Pedernera E, Reynaga-Obregón J, et al High levels of pretreatment CA125 are associated to improved survival in high grade serous ovarian carcinoma. J Ovarian Res. 2016;9:41. doi: 10.1186/s13048-016-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Cheng S, Jin Y, et al Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188503. doi: 10.1016/j.bbcan.2021.188503. [DOI] [PubMed] [Google Scholar]
- 63.Wimberger P, Heubner M, Otterbach F, et al Influence of platinum-based chemotherapy on disseminated tumor cells in blood and bone marrow of patients with ovarian cancer. Gynecol Oncol. 2007;107:331–8. doi: 10.1016/j.ygyno.2007.07.073. [DOI] [PubMed] [Google Scholar]
- 64.Molnar B, Sipos F, Galamb O, et al Molecular detection of circulating cancer cells. Role in diagnosis, prognosis and follow-up of colon cancer patients. Dig Dis. 2003;21:320–5. doi: 10.1159/000075355. [DOI] [PubMed] [Google Scholar]
- 65.Gasparri ML, Savone D, Besharat RA, et al Circulating tumor cells as trigger to hematogenous spreads and potential biomarkers to predict the prognosis in ovarian cancer. Tumour Biol. 2016;37:71–5. doi: 10.1007/s13277-015-4299-9. [DOI] [PubMed] [Google Scholar]
- 66.Lou E, Vogel RI, Teoh D, et al Assessment of circulating tumor cells as a predictive biomarker of histology in women with suspected ovarian cancer. Lab Med. 2018;49:134–9. doi: 10.1093/labmed/lmx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Y, Bian B, Yuan X, et al Prognostic value of circulating tumor cells in ovarian cancer: A meta-analysis. PLoS One. 2015;10:e0130873. doi: 10.1371/journal.pone.0130873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng L, Liang X, Liu Q, et al The predictive value of circulating tumor cells in ovarian cancer: A meta analysis. Int J Gynecol Cancer. 2017;27:1109–17. doi: 10.1097/IGC.0000000000000459. [DOI] [PubMed] [Google Scholar]
- 69.Cui L, Kwong J, Wang CC Prognostic value of circulating tumor cells and disseminated tumor cells in patients with ovarian cancer: a systematic review and meta-analysis. J Ovarian Res. 2015;8:38. doi: 10.1186/s13048-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poveda A, Kaye SB, McCormack R, et al Circulating tumor cells predict progression free survival and overall survival in patients with relapsed/recurrent advanced ovarian cancer. Gynecol Oncol. 2011;122:567–72. doi: 10.1016/j.ygyno.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 71.Chen H, Su Z, Li R, et al Single-cell DNA methylome analysis of circulating tumor cells. Chin J Cancer Res. 2021;33:391–404. doi: 10.21147/j.issn.1000-9604.2021.03.10. [DOI] [PMC free article] [PubMed] [Google Scholar]