To the Editor:
The gene encoding the anti-apoptotic BCL-2 family member MCL-1 is frequently amplified (~10%) in diverse human malignancies [1] and the finding that its inducible deletion impairs the growth of several types of tumours in vivo makes it an attractive target for anti-cancer therapy [2]. Lymphomas driven by c-MYC are particularly dependent on MCL-1, with loss of only one Mcl-1 allele preventing expansion of such murine lymphomas in vivo [2]. Accordingly, MYC-driven lymphomas are highly sensitive to MCL-1 inhibitors [3] and deletion of one Mcl-1 allele greatly delays c-MYC-driven lymphomagenesis in mice [4]. However, some c-MYC-driven lymphomas can tolerate the loss of one Mcl-1 allele, which reduces MCL-1 protein levels by ~40% [5]. These lymphomas uniformly display mutations of the tumour suppressor Trp53 [2]. Wild-type (wt) TRP53 function is required for optimal responses to BH3-mimetic drugs [6], and loss of Trp53 overcomes the delayed lymphomagenesis seen in Eμ-Myc;Mcl1+/− mice [4]. These studies suggest that loss or mutation of Trp53 diminishes the dependence of c-MYC-driven lymphomas on MCL-1. To explore this further, we conditionally deleted Mcl-1 in TRP53 deficient Eμ-Myc mouse lymphomas in vivo, to determine whether the loss of TRP53 allows them to tolerate the complete absence of MCL-1. This is an important question given that several BH3-mimetic drugs targeting MCL-1 have entered clinical trials in haematological cancers, where defects in TRP53 function frequently underlie resistance to standard therapies.
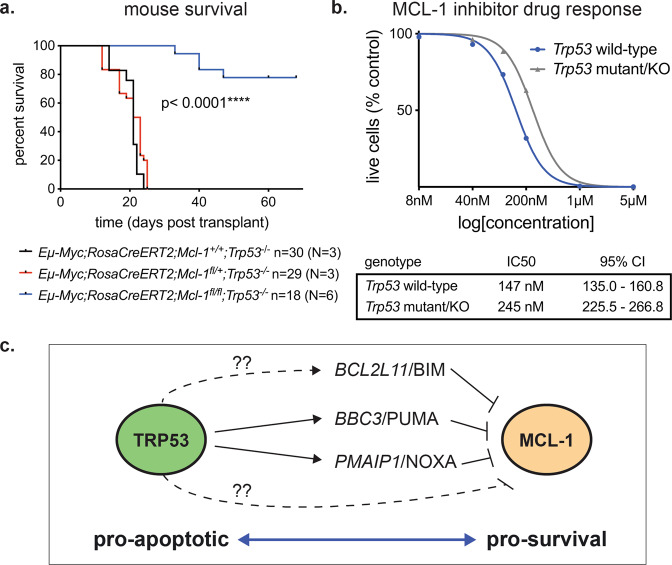
We utilised Eμ-Myc transgenic mice carrying conditional loxP flanked (floxed) Mcl-1 alleles [2] and a constitutively expressed but conditional tamoxifen-inducible Cre-recombinase (Rosa-CreERT2) (Supplementary Fig. S1). We engineered Eμ-Myc mice with germline loss of one Trp53 allele and one or two floxed Mcl-1 alleles plus the CreERT2 transgene (Supplementary Fig. S1). Eμ-Myc;Trp53+/−;Mcl1fl/+;RosaCreERT2 and Eμ-Myc;Trp53+/−;Mcl1fl/fl;RosaCreERT2 mice rapidly developed pre-B/B lymphomas (median survival ~30 days), and all selected for loss of the wt Trp53 allele, rendering them TRP53 deficient (Supplementary Fig. S2d). Ly5.2+ Eμ-Myc;Trp53−/−;Mcl1fl/+;RosaCreERT2 (N = 3; n = 29), Eμ-Myc;Trp53−/−;Mcl1fl/fl;RosaCreERT2 (N = 6; n = 18) and control Eμ-Myc;Trp53−/−;Mcl1+/+ RosaCreERT2 (N = 1; n = 3) lymphomas were transplanted into immune-competent C57BL/6-Ly5.1 recipients that were treated with either vehicle or tamoxifen on days 5 and 6 post-transplantation to induce Mcl-1fl deletion in the malignant cells in vivo (Supplementary Fig. S1b). Recipients bearing Eμ-Myc;Trp53−/−;Mcl1+/+;CreERT2 control lymphomas exhibited a marginal survival advantage (3 days) following tamoxifen treatment (Supplementary Fig. S2e). In the absence of TRP53, removal of one Mcl-1 allele in Eμ-Myc;Trp53−/−;Mcl1fl/+;RosaCreERT2 lymphomas did not impact lymphoma growth or survival of recipient mice (Fig. 1a). Deletion of the floxed Mcl-1 gene and encoded protein were confirmed in these lymphoma cells following treatment of mice with tamoxifen (Supplementary Fig. S2a-c). Remarkably, biallelic Mcl-1 deletion in Eμ-Myc;Trp53−/−;Mcl1fl/fl;RosaCreERT2 (N = 6; n = 18) lymphomas, resulting in complete removal of MCL-1, significantly prolonged tumour-free survival, with 14/18 (77.8%) of recipients cured (Fig. 1a). The four relapsed lymphomas may have selected against Mcl-1fl recombination and hence loss of MCL-1 protein or acquired changes that compensated for MCL-1 loss.
Fig. 1. Eμ-Myc lymphoma cells with loss of TRP53 remain dependent on anti-apoptotic MCL-1.
a Kaplan–Meier survival curve for mice transplanted with Eμ-MycT/+;CreERT2KI/+;Trp53−/− lymphomas bearing either wt Mcl-1, a single floxed Mcl-1 allele or two floxed Mcl-1 alleles and treated with tamoxifen on days 5 and 6 to delete the floxed Mcl-1 alelle(s) in the lymphomas. p value determined by Log-rank (Mantel–Cox) test. N indicates number of independent lymphomas of a given genotype tested; n indicates the number of recipient mice transplanted with the indicated lymphomas examined. b Response of Eμ-Myc lymphoma-derived cell lines to the MCL-1 inhibitor, S63845, comparing a panel of cell lines with deficient vs wild-type TRP53 function; N = 9 of each. Data represent non-linear regression of the means with IC50 and 95% confidence interval (CI) indicated. c Schematic depicting the intersecting and opposing roles of TRP53 and MCL-1 in the regulation of apoptosis.
We further assessed the impact of Trp53 loss on MCL-1 dependency in Eμ-Myc lymphomas by using the MCL-1 inhibitor S63845. Eμ-Myc lymphoma-derived cell lines with spontaneous Trp53 loss (N = 5) or mutation (N = 4) were compared with control Eμ-Myc lymphoma lines with intact TRP53 function (N = 9). Loss of TRP53 function conferred only minor resistance to MCL-1 inhibition (increase in IC50 value), with higher doses of MCL-1 inhibitor effectively killing Trp53-deficient lymphoma cells (Fig. 1b). In the human diffuse large B cell lymphoma (DLBCL) line DoHH2, CRISPR-mediated loss of TP53, validated by resistance to the MDM2 inhibitor Nutlin-3a (Supplementary Fig. S3a), did not impact the response to S63845 in short-term killing assays (Supplementary Fig. S3b). However, the TP53 deficient DoHH2 cells had a competitive advantage over their wt TP53 counterparts when cultured together in sub-optimal (~IC25) doses of S63845 for 14 days (Supplementary Fig. S3c), consistent with previous observations [6].
Considering the potential underlying mechanism, there is a direct relationship between TRP53 and MCL-1 in their opposing functions in regulating apoptosis (Fig. 1c). TRP53 can induce apoptosis by increasing the expression of the pro-apoptotic BH3-only proteins, PUMA/Bbc3, NOXA/Pmaip1 and BIM/Bcl2l11 [7].
In conclusion, we demonstrate that loss/mutation of TRP53/TP53 renders mouse c-MYC-driven lymphomas and a human DLBCL cell line less dependent on MCL-1 for sustained survival and expansion. Importantly though, lymphomas with loss of TRP53/TP53 can still be effectively killed using either higher concentrations of the MCL-1 inhibitor or by homozygous deletion of Mcl-1. This is important for the clinical translation of MCL-1 inhibitors because it suggests that patients bearing cancers with loss/mutation of TP53 may require higher doses of MCL-1 inhibitors or combination therapies to augment the levels of pro-apoptotic BH3-only proteins [6].
Supplementary information
Acknowledgements
We thank all members of the Blood Cells and Blood Cancer Division at The Walter and Eliza Hall Institute (WEHI) for support and advice; G. Siciliano, H. Johnson, C. Gatt, and their team for animal husbandry; S. Monard and the team at the WEHI Flow Cytometry Unit; J. McManus and J. Corbin for haematology support.
Author contributions
BJA, MSB, STD, ZW, CC, MJH, GLK and AS conceptualised and planned the study, performed experiments, analysed data and wrote the paper.
Funding
This work was supported by a Leukaemia Foundation National Research Program Clinical PhD Scholarship (BJA), National Health and Medical Research Council (NHMRC) project grants 1086291 to GLK, 1145728 to MJH and 1143105 to MJH and AS, NHMRC Ideas Grants 2002618 and 2001201 to GLK and 1186575 and 1159658 to MJH, an NHMRC program grant 1016701 (AS), an NHMRC Senior Principal Research Fellowship 1020363 (AS) and an NHMRC Senior Research Fellowship 1156095 to MJH, Leukaemia and Lymphoma Society of America Specialised Center of Research (SCOR) grant 7001-13 (AS, GLK and MJH), Cancer Council Victoria grants-in-aid 1086157 and 1147328 (GLK), Victorian Cancer Agency Fellowship 17028 (GLK), a Leukaemia Foundation Australia grant (GLK and AS), a Leukaemia Foundation PhD Scholarship (MSB), Cancer Council Victoria Postdoctoral Fellowship (MSB), the Dyson Bequest and bequests from the Anthony Redstone Estate and Craig Perkins Cancer Research Foundation. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government National Health and Medical Research Council Independent Research Institutes Infrastructure Support Scheme.
Data availability
Data sharing is not applicable to this paper as no datasets were generated or analysed during the current study. Reagents are available on request.
Competing interests
AS, GLK, STD, ZW, CC and MJH are currently, and BJA and MSB have previously been, employees of the Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia, which receives Royalties and Milestone payments related to the BCL-2 inhibitor, Venetoclax/ABT-199. AS, GLK and MH have received research funding from Servier.
Ethics approval
All experiments with mice followed the guidelines of the Melbourne Directorate Animal Ethics Committee, according to The Walter and Eliza Hall Institute of Medical Research Ethics Committee.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andreas Strasser, Gemma L. Kelly.
Contributor Information
Andreas Strasser, Email: strasser@wehi.edu.au.
Gemma L. Kelly, Email: gkelly@wehi.edu.au
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00946-9.
References
- 1.Beroukhim R, Mermel C, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly GL, Grabow S, Glaser SP, Fitzsimmons L, Aubrey BJ, Okamoto T, et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 2014;28:58–70. doi: 10.1101/gad.232009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–82. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 4.Grabow S, Delbridge AR, Aubrey BJ, Vandenberg CJ, Strasser A. Loss of a Single Mcl-1 Allele Inhibits MYC-Driven Lymphomagenesis by Sensitizing Pro-B Cells to Apoptosis. Cell Rep. 2016;14:2337–47. doi: 10.1016/j.celrep.2016.02.039. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann K, Grabow S, Hyland CD, Teh CE, Alexander WS, Herold MJ, et al. The combination of reduced MCL-1 and standard chemotherapeutics is tolerable in mice. Cell Death Differ. 2017;24:2032–43. doi: 10.1038/cdd.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thijssen R, Diepstraten ST, Moujalled D, Chew E, Flensburg C, Shi MX, et al. Intact TP-53 function is essential for sustaining durable responses to BH3-mimetic drugs in leukemias. Blood. 2021;137:2721–35. doi: 10.1182/blood.2020010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–8. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable to this paper as no datasets were generated or analysed during the current study. Reagents are available on request.