Abstract
Background
The association between vitamin E and cancer risk has been widely investigated by observational studies, but the findings remain inconclusive. Here, we aimed to evaluate the causal effect of circulating vitamin E on the risk of ten common cancers, including bladder, breast, colorectal, esophagus, lung, oral and pharynx, ovarian, pancreatic, prostate, and kidney cancer.
Methods
A Mendelian randomization (MR) analytic framework was applied to data from a cancer-specific genome-wide association study (GWAS) comprising a total of 297,699 cancer cases and 304,736 controls of European ancestry. Three genetic instrumental variables associated with circulating vitamin E were selected. Summary statistic-based methods of inverse variance weighting (IVW) and likelihood-based approach, as well as the individual genotyping-based method of genetic risk score (GRS) were used. Multivariable IVW analysis was further performed to control for potential confounding effects. Furthermore, the UK Biobank cohort was used as external validation, supporting 355,543 European participants (incident cases ranged from 437 for ovarian cancer to 4882 for prostate cancer) for GRS-based estimation of circulating vitamin E, accompanied by a one-sample MR analysis of dietary vitamin E intake underlying the time-to-event analytic framework.
Results
Specific to cancer GWAS, we found that circulating vitamin E was significantly associated with increased bladder cancer risk (odds ratios [OR]IVW = 6.23, PIVW = 3.05×10-3) but decreased breast cancer risk (ORIVW = 0.68, PIVW = 8.19×10-3); however, the significance of breast cancer was dampened (Pmultivariable IVW > 0.05) in the subsequent multivariable MR analysis. In the validation stage of the UK Biobank cohort, we did not replicate convincing causal effects of genetically predicted circulating vitamin E concentrations and dietary vitamin E intake on the risk of ten cancers.
Conclusions
This large-scale population study upon data from cancer-specific GWAS and a longitudinal biobank cohort indicates plausible non-causal associations between circulating vitamin E and ten common cancers in the European populations. Further studies regarding ancestral diversity are warranted to validate such causal associations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-022-02366-5.
Keywords: Circulating vitamin E, Cancer risk, Mendelian randomization, GWAS, UK Biobank
Background
Vitamin E is a group of fat-soluble antioxidant nutrients consisting of tocopherols and tocotrienols. Tocopherol, a major isoform of vitamin E, has been found to eliminate reactive oxygen species, inhibit carcinogenesis and tumor growth, and stimulate cancer cell apoptosis [1, 2].
Albeit the associations between vitamin E and cancer risk have been explored by several epidemiological studies, their findings remain inconsistent [3]. For instance, the Selenium and Vitamin E Cancer Prevention Trial (SELECT) found that supplementation with vitamin E was associated with an increased risk of prostate cancer among 34,887 men [4], but this was not confirmed in the Physicians’ Health Study II randomized trial following 14,641 men [5]. Although randomized trials are commonly recognized as the gold standard for making causal inferences, they are usually not widely available due to high cost and long duration. Nevertheless, even randomized trials are likely to be underpowered given the low incidence of endpoint phenotypes such as rare cancers [6].
Mendelian randomization (MR), a novel statistical approach that uses genetic variants associated with exposure of interest as instruments, can be applied to estimate a causal relationship between exposure and outcome [7]. MR is designed based on the fact that genetic variants are randomly allocated during gamete formation and conception, therefore independent of confounding factors. Results from MR designs are thus less susceptible to reverse causality and confounding bias [8]. In this study, we leveraged large-scale genome-wide genetic data and UK Biobank cohort of European ancestry to apply an MR framework, to estimate a putative causal association of circulating vitamin E with the risk of ten common cancers (Additional file 1: Fig. S1).
Methods
Study subjects
Cancer-specific case-control genome-wide association studies (GWASs)
The current MR analysis was comprehensively performed by leveraging information from ten GWASs totaling 602,435 participants of European ancestry, including 297,699 cancer cases and 304,736 controls across the bladder, breast, colorectal, esophagus, lung, oral and pharynx, ovarian, pancreatic, prostate, and kidney cancer. The characteristics of each cancer-specific GWAS including sample sizes and data sources are illustrated in Additional file 1: Table S1.
Briefly, as outcomes of interest, we collected available GWAS data across ten cancers. For summary-level GWAS data of 4 cancers (i.e., breast, ovarian, prostate, and lung cancer), quality control procedures and population details have been described elsewhere [9–12]. For six cancers (bladder, colorectal, esophagus, oral and pharynx, pancreatic, and kidney cancer) which we had access to individual-level genotyping data [13–23], we performed stringent quality control procedures of population via removing unexpected duplicates or probable relatives based on pairwise identity by descent, guaranteeing all individuals to be of European ancestry.
UK Biobank cohort data
The UK Biobank cohort was a prospective population-based study that recruited 502,528 adults aged 40–69 years from the general population between April 2006 and December 2010. The study protocol and information about data access are available online (http://www.ukbiobank.ac.uk/), and more details of the recruitment and study design have been published in previous studies [24]. The UK Biobank resource used by this study was under Application #45611.
After the quality control of the following population: (i) excluded individuals with prevalent cancer (except non-melanoma skin cancer, based on the International Classification of Diseases, 10th revision [ICD-10, C44]) at baseline; (ii) excluded individuals of sex discordance; (iii) excluded outliers for genotype missingness or excess heterozygosity; (iv) retained unrelated participants; (v) restricted to “white British” individuals of European ancestry; and (vi) removed individuals who decided not to participate in this program, a total of 355,543 participants remained for analysis. Moreover, we defined the ten cancers using the ICD-10 codes (Additional file 1: Table S2). The follow-up time was calculated from baseline assessment to the first diagnosis of cancer, loss to follow-up, death, or last follow-up (December 14, 2016), whichever occurred first.
Information on dietary vitamin E intake in UK Biobank participants was retrieved from data field #100025 (description: vitamin E; category: estimated nutrients yesterday—diet by a 24-h recall—online follow-up). Measurements were performed at baseline (2006–2010) and/or subsequent follow-up visits. In the present study, we included 49,579 individuals (23,107 males and 26,472 females) with baseline vitamin E measurements.
Two-sample MR analysis and sensitivity analysis of cancer-specific GWAS
Based on cancer-specific GWAS databases, depends on the availability of data, we applied a summary statistics-based approach to all cancers, and additionally, a genetic risk score (GRS)-based approach to some of the cancers (bladder, colorectal, esophagus, oral and pharynx, pancreatic, and kidney cancer), followed by sensitivity analysis.
Instrumental variable (IV) selection
Circulating vitamin E was the main exposure of interest. We collected 3 independent GWAS-identified circulating vitamin E-associated single-nucleotide polymorphisms (SNPs; rs964184, rs11057830, and rs2108622) from a large GWAS available to date [25], which met the following criteria as instruments for MR analysis: (i) reported P-value < 5.00×10-8, (ii) minor allele frequency (MAF) ≥ 0.05, (iii) call rate ≥ 95%, and (iv) Hardy-Weinberg equilibrium (HWE) P-value in controls ≥ 1×10-6 (Additional file 1: Table S3). The online web tool mRnd (https://cnsgenomics.shinyapps.io/mRnd/) was used to estimate statistical power [26]. To calculate the minimum detectable effect size, we set 80.0% statistical power and 5.0% alpha level and used the proportion of circulating vitamin E variance (R2, i.e., 1.7% estimated by Major et al.) explained by the 3 IVs as calculated in the previous GWAS [25, 27]. We further quantified the strength of IVs by F-statistics, where F-statistics > 10 provided good evidence for the IV being a strong instrument [28].
Summary statistic-based method
The summary statistics-based methods, including an inverse variance weighting (IVW) method and a likelihood-based method, were primarily used to infer causal associations. The formula of IVW method was as follows: ; , where i is the ith SNP, βXi, and σXi are the estimate and standard error of genetic association with the exposure that were derived from IVs, and βYi and σYi are the estimate and standard error of genetic association with the outcome that were derived from cancer-specific GWAS. In addition, we adopted the likelihood-based method, which can be used to obtain appropriately sized confidence intervals when there is considerable imprecision in the estimates.
GRS-based method
We further constructed a weighted GRS to integrate the genetic effects of candidate SNPs on the exposure of interest for available individual-level genotyping data. We summed three circulating vitamin E-associated SNPs weighted by corresponding effect sizes using the formula: , where n is the number of SNPs, SNPi is the number of risk alleles (0, 1, 2) carried by the ith SNP, and βi is the previously published regression coefficient for ith SNP. We then evaluated the association of circulating vitamin E-GRS with cancer risk through the logistic regression model with adjustment for sex, age, and the first ten principal components when appropriate.
Multiple testing correction was performed by false discovery rate (FDR) method using the “p.adjust” function in R software.
Sensitivity analysis
Estimates from MR can only be reliably interpreted when three model assumptions are valid, including (i) the IVs are associated with exposure variables, (ii) the IVs are not related to other confounding factors, and (iii) the IVs only influence outcome variables through their effects on exposure variables. Therefore, we performed heterogeneity analysis and MR-Egger regression analysis to evaluate the potential violation to the second and third assumptions. The heterogeneity test was used to assess whether a genetic variant’s effect on cancer risk was proportional to its effect on circulating vitamin E. MR-Egger regression (MR-Egger intercept test) was fitted to evaluate the presence of horizontal pleiotropy. We additionally conducted a leave-one-out analysis where we excluded one SNP at a time and performed IVW analysis on the remaining two SNPs to evaluate the robustness of our results.
Furthermore, to control for the effects of potential confounding factors on significant associations of univariable MR analyses, we also conducted multivariable IVW analysis using the effect size retrieved from the Gene ATLAS database (http://geneatlas.roslin.ed.ac.uk/) [29].
Validation in the UK Biobank cohort
Circulating vitamin E based GRS analysis
We used Cox proportional hazards models to calculate hazard ratios (HRs) and 95% confidence intervals (CIs) for the associations between circulating vitamin E-GRS and the risk of ten cancers, with the adjustment of sex, age, study centers, body mass index (BMI), smoking status, drinking status, and first ten principal components when appropriate.
One-sample MR analysis for dietary vitamin E intake
One-sample MR analysis was used to evaluate the association between dietary vitamin E intake at baseline and the cancer risk. The genetic IVs for one sample MR were extracted from the UK Biobank imputation dataset, which followed the extensive quality control of SNPs, including (i) imputation confidence score (info score) ≥ 0.3, (ii) MAF ≥ 0.05, (iii) call rate ≥ 95%, and (iv) HWE P-value ≥ 1×10-6. Then, we performed linear regression analysis between each variant and log-transformed dietary vitamin E measurements, to provide independent (linkage disequilibrium r2 < 0.1) dietary vitamin E-associated IVs under different significance thresholds (i.e., P-value ≤ 5.00×10-7, P-value ≤ 5.00×10-6, P-value ≤ 5.00×10-5). These IVs with different significance thresholds were further used to construct weighted GRS, as well as unweighted GRS to avoid potential over-fitting. In addition, we also annotated the dietary vitamin E-associated lead loci with functional activity (with HaploReg v4.1, https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) [30] and expression quantitative trait loci (eQTL) analysis (with eQTLGen consortium of 31,684 blood samples, https://www.eqtlgen.org/cis-eqtls.html) [31].
Briefly, a two-stage method was implemented for one-sample MR analysis: (i) the first-stage model consisted of a linear regression of the log-transformed dietary vitamin E measurements on the weighted and unweighted GRSs and (ii) the second-stage model consisted of a Cox regression of the cancer risk on the fitted values from the first-stage optimal regression model (with the strongest correlation with observed dietary vitamin E level). The covariates included sex, age, study centers, BMI, smoking status, drinking status, and the first ten principal components when appropriate.
Sensitivity analysis
Several sensitivity analyses were also performed in the UK Biobank cohort, including (i) re-analyzed the association using logistic regression model with incident and prevalent cancer cases in a case-control design and (ii) additionally adjusted for socioeconomic (i.e., education and employment status) and chronic disease status (i.e., coronary artery disease, stroke, hypertension, and type 2 diabetes).
All statistical analyses were performed using R version 3.6.1, and a two-sided P-value less than 0.05 was considered as strong evidence for a causal association.
Results
Power analysis and genetic effect estimation
For each cancer-specific GWAS, the F-statistics of the 3 IVs are summarized in Table 1. The smallest F-statistic was 72.48 (larger than 10), indicating a strong instrumental strength. In general, our MR analyses obtained sufficient power—we had 80% power to detect moderate effect sizes, with odds ratios (ORs) ranging from 0.44 (kidney cancer) to 0.91 (breast cancer) per standard deviation (SD) increase in circulating vitamin E levels.
Table 1.
Statistical power in Mendelian randomization (MR) study of circulating vitamin E and cancer risk in cancer-specific GWAS
| Cancer type | Sample size (N = 602,435) | F-statistics | Minimum detectable ORa | |
|---|---|---|---|---|
| Cases (N = 297,699) | Controls (N = 304,736) | |||
| Bladder cancer | 5930 | 5468 | 198.12 | 0.67/1.49 |
| Breast cancer | 122,977 | 105,974 | 3960.48 | 0.91/1.09 |
| Colorectal cancer | 24,476 | 23,073 | 823.31 | 0.82/1.22 |
| Esophagus cancer | 2268 | 1865 | 72.48 | 0.53/1.97 |
| Lung cancer | 29,266 | 56,450 | 1483.37 | 0.85/1.16 |
| Oral and pharynx cancer | 4950 | 2907 | 136.88 | 0.63/1.70 |
| Ovarian cancer | 22,406 | 40,941 | 1096.52 | 0.83/1.19 |
| Pancreatic cancer | 4970 | 3532 | 148.03 | 0.64/1.63 |
| Prostate cancer | 79,148 | 61,106 | 2426.55 | 0.89/1.12 |
| Kidney cancer | 1308 | 3420 | 82.77 | 0.44/1.81 |
aMinimum detectable OR (per 1 SD of vitamin E) was calculated based on 80% power, 5% alpha level, and 1.7% of vitamin E variance (R2) explained by 3 SNPs used in this study
We next evaluated the genetic effects of each circulating vitamin E-associated SNP on cancer risk and observed that no SNPs were significantly associated with any cancer risk (Additional file 1: Table S4), except for a marginal risk effect of rs964184 on breast cancer (OR = 0.98, P = 0.043); as well as rs11057830 (OR = 1.10, P = 0.015) and rs2108622 (OR = 1.08, P = 0.013) on bladder cancer.
Causal association between circulating vitamin E and cancer risk
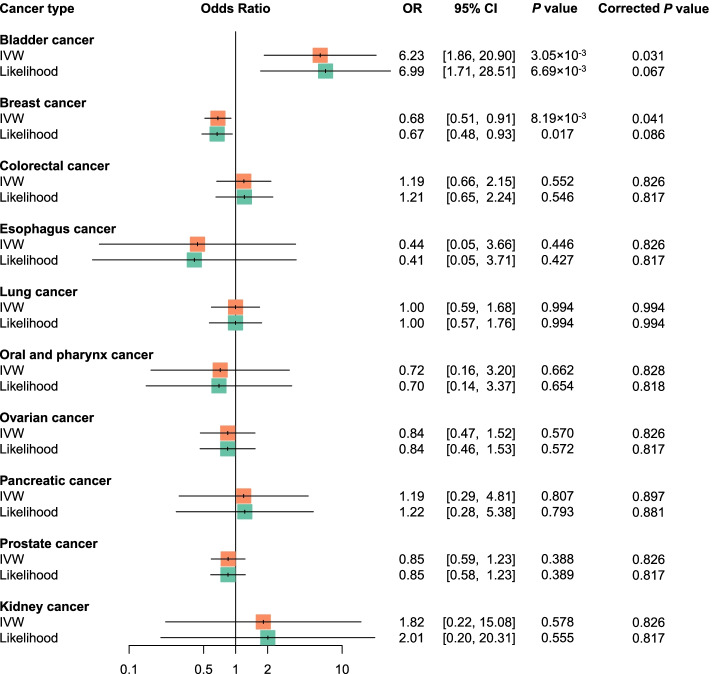
Figure S2 shows MR estimates of circulating vitamin E and each cancer risk. For the univariable MR analyses shown in Fig. 1, circulating vitamin E was not associated with risk of eight cancers, including colorectal, esophagus, lung, oral and pharynx, ovarian, pancreatic, prostate, and kidney cancer, where all P-values were above 0.05 (PIVW > 0.05, PLikelihood > 0.05, PGRS > 0.05, Additional file 1: Table S5). Notably, circulating vitamin E was causally associated with an increased risk of bladder cancer (ORIVW = 6.23, PIVW = 3.05×10-3; ORLikelihood = 6.99, PLikelihood = 6.69×10-3; ORGRS = 7.34, PGRS = 1.57×10-3), but a decreased risk of breast cancer (ORIVW = 0.68, PIVW = 8.19×10-3; ORLikelihood = 0.67, PLikelihood = 0.017). These associations remained borderline significant after accounting for multiple comparisons across ten cancers (bladder cancer: PIVW = 0.031, PLikelihood = 0.067; breast cancer: PIVW = 0.041, PLikelihood = 0.086).
Fig. 1.
Forest plots of univariable Mendelian randomization (MR) estimates between circulating vitamin E and cancer risk in cancer-specific GWAS. The odds ratio (OR) was estimated using inverse variance weighting (IVW) and likelihood-based methods. The corrected P-value was calculated with false discovery rate (FDR) method
Sensitivity analysis for causal estimation across each cancer
There was no heterogeneity or directional pleiotropy for each causal estimation (Pheterogeneity > 0.05; Additional file 1: Fig. S2; PMR-Egger intercept > 0.05; Additional file 1: Table S5). Besides, leave-one-out analysis did not identify any outlying instruments (Additional file 1: Table S6). When profiling the association of each IV and multiple traits, we found that rs964184 and rs11057830 were associated with a total of 24 traits under P < 5.00×10-8 (Additional file 1: Table S7). Therefore, we performed multivariable MR analysis to adjust for the influence of each confounding trait, that is, the effect acting in particular through these traits. The association of circulating vitamin E with breast cancer risk attenuated to non-significant, indicating that the effect was most likely mediated by lipid-related traits such as cholesterol and lipoprotein. However, bladder cancer retained a robust, potentially causal relationship with circulating vitamin E (almost all adjusted P < 0.05; Table 2).
Table 2.
Multivariable Mendelian randomization (MR) analysis for the associations of circulating vitamin E with the risk of bladder cancer and breast cancer
| Corrected trait | Bladder cancer | Breast cancer | ||||
|---|---|---|---|---|---|---|
| Betaa | 95% CIa | Pa | Betaa | 95% CIa | Pa | |
| E70-E90 metabolic disorders | 3.03 | 0.69, 5.37 | 0.011 | −0.18 | −0.73, 0.36 | 0.507 |
| E78 disorders of lipoprotein metabolism and other lipidemias | 2.96 | 0.91, 5.01 | 4.70×10-3 | −0.23 | −0.70, 0.25 | 0.349 |
| Eosinophill count | 2.61 | 1.15, 4.07 | 4.53×10-4 | −0.36 | −0.71, −0.01 | 0.043 |
| Eosinophill percentage | 2.34 | 1.02, 3.66 | 5.28×10-4 | −0.37 | −0.69, −0.05 | 0.022 |
| High cholesterol | 2.99 | 1.16, 4.83 | 1.38×10-3 | −0.28 | −0.71, 0.15 | 0.204 |
| High light scatter reticulocyte count | 3.07 | 1.26, 4.89 | 8.93×10-4 | −0.30 | −0.73, 0.13 | 0.165 |
| High light scatter reticulocyte percentage | 2.94 | 1.22, 4.65 | 7.85×10-4 | −0.31 | −0.71, 0.10 | 0.137 |
| I20-I25 ischemic heart diseases | 1.58 | −0.15, 3.32 | 0.073 | −0.26 | −0.66, 0.15 | 0.211 |
| I25 chronic ischemic heart disease | 1.62 | −0.28, 3.51 | 0.095 | −0.23 | −0.67, 0.21 | 0.308 |
| Lymphocyte count | 1.29 | −0.15, 2.74 | 0.080 | −0.34 | −0.67, −0.01 | 0.046 |
| Mean corpuscular hemoglobin | 1.13 | −0.70, 2.95 | 0.225 | −0.28 | −0.70, 0.15 | 0.202 |
| Mean corpuscular hemoglobin concentration | 2.67 | 1.17, 4.17 | 4.67×10-4 | −0.35 | −0.71, 0.01 | 0.057 |
| Mean corpuscular volume | 2.83 | 0.34, 5.32 | 0.026 | −0.15 | −0.73, 0.43 | 0.607 |
| Mean platelet (thrombocyte) volume | 2.66 | 1.05, 4.27 | 1.18×10-3 | −0.29 | −0.67, 0.08 | 0.126 |
| Mean reticulocyte volume | 3.09 | 1.19, 4.98 | 1.42×10-3 | −0.28 | −0.72, 0.17 | 0.223 |
| Mean sphered cell volume | 3.14 | 1.04, 5.23 | 3.29×10-3 | −0.24 | −0.72, 0.25 | 0.341 |
| Monocyte count | 1.81 | 0.60, 3.02 | 3.40×10-3 | −0.38 | −0.67, −0.10 | 0.008 |
| Monocyte percentage | 1.47 | 0.19, 2.74 | 0.025 | −0.38 | −0.67, −0.08 | 0.012 |
| Platelet count | 2.74 | 1.14, 4.34 | 7.81×10-4 | −0.31 | −0.69, 0.07 | 0.109 |
| Platelet crit | 2.71 | 1.17, 4.25 | 5.70×10-4 | −0.33 | −0.69, 0.04 | 0.080 |
| Platelet distribution width | 2.81 | 1.18, 4.43 | 7.21×10-4 | −0.31 | −0.70, 0.07 | 0.112 |
| Red blood cell (erythrocyte) distribution width | 2.51 | 1.09, 3.94 | 5.43×10-4 | −0.34 | −0.67, 0.00 | 0.051 |
| Reticulocyte count | 3.15 | 1.25, 5.05 | 1.17×10-3 | −0.29 | −0.74, 0.16 | 0.207 |
| Reticulocyte percentage | 3.01 | 1.23, 4.79 | 9.32×10-4 | −0.30 | −0.72, 0.12 | 0.164 |
aMultivariable inverse variance weighting (IVW) method
Validation in the UK Biobank cohort
In the validation stage with the UK Biobank cohort, there was no evidence to support the associations between genetically predicted circulating vitamin E and the risk of ten cancers (all PGRS > 0.05; Table 3). In particular, the positive association between circulating vitamin E and bladder cancer observed in GWAS was not replicated in this cohort (HR = 0.86, P = 0.918). Further random effects meta-analysis combining the GRS results for bladder cancer from GWAS and UK Biobank cohort still yielded a non-significant result (I2 = 45.8%, Pmeta = 0.186). In the sensitivity analysis with incident and prevalent cancer cases, the association of circulating vitamin E with the risk of esophagus and kidney cancer became significant (P < 0.05), but they failed to survive the FDR correction (PFDR > 0.05; Additional file 1: Table S8). In addition, the random effects meta-analysis combing the GRS results from GWAS and UK Biobank cohort yielded non-significant results for the two cancers (esophagus: I2 = 73.3%, Pmeta = 0.607; kidney: I2 = 81.8%, Pmeta = 0.540).
Table 3.
Genetic risk score (GRS) analysis for the associations of vitamin E with cancer risk in the UK Biobank cohort
| Cancer type | Cases | Methoda | HRb | 95% CIb | Pb | Corrected Pc |
|---|---|---|---|---|---|---|
| Bladder cancer | 526 | Circulating vitamin E-based GRS | 0.86 | 0.05, 14.64 | 0.918 | 0.983 |
| One-sample weighted GRS | 0.70 | 0.28, 1.75 | 0.444 | 0.989 | ||
| One-sample unweighted GRS | 0.75 | 0.29, 1.93 | 0.551 | 0.988 | ||
| Breast cancer | 4350 | Circulating vitamin E-based GRS | 2.30 | 0.86, 6.14 | 0.096 | 0.337 |
| One-sample weighted GRS | 1.06 | 0.82, 1.36 | 0.674 | 0.989 | ||
| One-sample unweighted GRS | 1.06 | 0.82, 1.38 | 0.664 | 0.988 | ||
| Colorectal cancer | 2621 | Circulating vitamin E-based GRS | 0.43 | 0.12, 1.54 | 0.194 | 0.400 |
| One-sample weighted GRS | 1.19 | 0.79, 1.80 | 0.404 | 0.989 | ||
| One-sample unweighted GRS | 1.22 | 0.80, 1.87 | 0.348 | 0.988 | ||
| Esophagus cancer | 460 | Circulating vitamin E-based GRS | 14.06 | 0.72, 275.16 | 0.082 | 0.337 |
| One-sample weighted GRS | 1.01 | 0.38, 2.69 | 0.989 | 0.989 | ||
| One-sample unweighted GRS | 0.99 | 0.36, 2.72 | 0.988 | 0.988 | ||
| Lung cancer | 1700 | Circulating vitamin E-based GRS | 0.79 | 0.16, 3.89 | 0.772 | 0.983 |
| One-sample weighted GRS | 1.17 | 0.70, 1.96 | 0.542 | 0.989 | ||
| One-sample unweighted GRS | 1.16 | 0.69, 1.97 | 0.573 | 0.988 | ||
| Oral and pharynx cancer | 458 | Circulating vitamin E-based GRS | 1.03 | 0.05, 21.74 | 0.983 | 0.983 |
| One-sample weighted GRS | 0.93 | 0.35, 2.50 | 0.891 | 0.989 | ||
| One-sample unweighted GRS | 0.87 | 0.32, 2.39 | 0.792 | 0.988 | ||
| Ovarian cancer | 437 | Circulating vitamin E-based GRS | 2.09 | 0.09, 47.08 | 0.644 | 0.983 |
| One-sample weighted GRS | 0.87 | 0.39, 1.92 | 0.724 | 0.989 | ||
| One-sample unweighted GRS | 0.86 | 0.38, 1.96 | 0.722 | 0.988 | ||
| Pancreatic cancer | 506 | Circulating vitamin E-based GRS | 1.08 | 0.06, 20.08 | 0.960 | 0.983 |
| One-sample weighted GRS | 0.94 | 0.37, 2.41 | 0.898 | 0.989 | ||
| One-sample unweighted GRS | 0.96 | 0.37, 2.52 | 0.934 | 0.988 | ||
| Prostate cancer | 4882 | Circulating vitamin E-based GRS | 0.54 | 0.21, 1.39 | 0.200 | 0.400 |
| One-sample weighted GRS | 0.93 | 0.72, 1.19 | 0.572 | 0.989 | ||
| One-sample unweighted GRS | 0.97 | 0.75, 1.26 | 0.840 | 0.988 | ||
| Kidney cancer | 649 | Circulating vitamin E-based GRS | 0.11 | 0.01, 1.54 | 0.101 | 0.337 |
| One-sample weighted GRS | 0.68 | 0.30, 1.57 | 0.370 | 0.989 | ||
| One-sample unweighted GRS | 0.73 | 0.31, 1.70 | 0.466 | 0.988 |
aCirculating vitamin E-based GRS, derived from three circulating vitamin E-SNPs; one-sample weighted and unweighted GRS, derived from dietary vitamin E-SNPs (P-value ≤ 5×10-5)
bAdjusted for sex, age, study centers, body mass index (BMI), smoking status, drinking status, and first ten principal components when appropriate
cThe corrected P-value was calculated with false discovery rate (FDR) method
Subsequently, we performed the genome-wide analysis to identify variants associated with dietary vitamin E intake, but no SNPs were found beyond genome-wide significance threshold (P ≤ 5×10-8; Additional file 1: Fig. S3). Based on the suggestive significance threshold (P ≤ 5×10-7), we identified three significant variants (rs11889555 on 2q32.2, beta = 0.02, P = 7.59×10-8; rs139695510 on 13q32.1, beta = -0.03, P = 1.29×10-7; and rs12165526 on 22q13.31, beta = 0.03, P = 2.79×10-7) in all population, one significant variant (rs11889555 on 2q32.2, beta = 0.03, P = 4.33×10-7) in males, and one significant variant (rs201524387 on 13q21.1, beta = 0.03, P = 1.96×10-7; Additional file 1: Table S9) in females. Further, we annotated these loci with functional activity and cis-eQTL analysis. Interestingly, rs11889555 had a high function score and significantly affected the expression of multiple nearby genes in blood samples (Additional file 1: Table S10).
For the one-sample MR analysis of dietary vitamin E intake, in the first-stage model, the weighed and unweighted vitamin E associated GRSs with a threshold of P-value ≤ 5.00×10-5 showed the strongest correlation with observed vitamin E level and were then used for predicting dietary vitamin E in the second-stage model (Additional file 1: Table S11). We found that the genetically predicted dietary vitamin E intake was not significantly associated with the risk of all ten cancers (PFDR of weighted and unweighted GRS > 0.05; Table 3), consistent with findings of sensitivity analysis (Additional file 1: Table S8).
Discussion
In this large-scale genetic association study, we evaluated the causal relationship of circulating vitamin E with the risk of ten common cancers capitalizing on the largest available cancer-specific GWAS data and UK Biobank cohort of European ancestry. Our current MR study, despite its largely augmented sample size and strong instruments, did not reveal convincing evidence to support causal associations of genetically predicted circulating vitamin E and dietary vitamin E intake with the risk of ten cancers.
Previous observational epidemiological studies have reported associations between vitamin E intake and the risk of these cancers [4, 32–42], and part of our results were supported by these reports. A previous meta-analysis including 24 studies suggested an inverse association between plasma α-tocopherol and breast cancer risk, but the association was not significant in the European population [41]. de Munter et al found that intake of dietary vitamin E did not support a protective association with oral and pharynx cancer risk using Netherlands cohort study data with 120,852 participants [33]. A systematic review including prospective cohort studies with over 200 ovarian cancer cases (n = 24) did not find a significant association between vitamin E concentrations and the risk of ovarian cancer [34]. Another cohort study including 10 studies in North America and Europe with 501,857 women also indicated that intakes of vitamins A, C, and E were not significantly associated with ovarian cancer risk [35]. Besides, the association of vitamin E supplementation with the risk of prostate cancer was not found in the Physicians’ Health Study II randomized trial with 14,641 men [5].
In addition, multiple observational studies have reported significant associations between vitamin E and a decreased risk of esophagus, colorectal, lung, pancreatic, kidney, and bladder cancer [36–40, 43]. For instance, a meta-analysis with 6431 subjects found that colorectal cancer patients had lower concentrations of serum vitamin E compared to healthy controls, especially in European populations [36]. A recent prospective study with 22,781 Finnish male smokers reported a 24% significant reduction in the risk of lung cancer for the fifth quintile compared with the bottom quartile of baseline α-tocopherol concentration [37]. A meta-analysis of 10 studies with 2976 patients and 254,393 controls observed a 13% lower risk of pancreatic cancer for the highest compared with the lowest level of vitamin E intake among European populations [38]. Cui et al. found an inverse relationship between dietary vitamin E intake and the risk of esophagus cancer among European and non-European populations using meta-analysis including 14 studies with 3013 cases and 11,384 non-cases [39]. Shang et al. reported a significant reduction in the risk of renal cell carcinoma for the highest intake compared with the lowest intakes of vitamin E concentrations among European populations using meta-analysis including 7 studies with 5789 cases and 14,866 controls [40]. A recent meta-analysis with 575,601 participants from the USA and Europe indicated that vitamin E consumption was inversely associated with the risk of bladder cancer; Chen et al. also found that α-tocopherol, the main isoform of vitamin E, was associated with a decreased risk of bladder cancer [42, 43]. However, our MR analysis with sufficient power did not support the associations between circulating vitamin E and the risk of above six cancers, indicating that the results of observational studies may need to be validated in further studies.
Vitamin E is a group of fat-soluble antioxidant nutrients consisting of eight natural isoforms. All isoforms scavenge reactive oxygen species through the presence of phenolic hydrogen in their chromanol ring [44]. Oxidative stress has been demonstrated to be involved in the pathogenesis of multiple diseases, especially for cancer. Oxidative stress can lead to free radical chain reaction causing lipid peroxidation, but vitamin E plays a vital role in breaking the free radical chain reaction, preventing lipid peroxidation, and protecting biological membrane [45, 46]. Therefore, the anticancer activity of vitamin E has been studied extensively. However, our findings of this MR study indicated that increasing circulating vitamin E concentrations or vitamin E intake was unlikely to result in clinical benefit for reducing cancer risk, which provided an important public health message that vitamin E supplementation may not be useful for cancer prevention.
Our study has several strengths. This was the first large-scale MR analysis that systemically evaluated a causal association between circulating vitamin E and the risk of multiple cancers, leveraging cancer-specific GWAS data of 602,435 solid cancer cases and controls, and a validation in UK Biobank cohort of 355,543 individuals, the largest study of its kind. In addition, this MR analysis was performed with no signs of violation to MR assumptions, as tested by MR-Egger and median-based approaches. We performed multiple MR analyses based on individuals of European descent, largely reducing population stratification.
Several limitations also need to be acknowledged. The main challenge with this study is the limited availability of genetic instruments for circulating vitamin E concentrations, with only 3 genetic variants explaining 1.7% of variation. This has implications for the detection of pleiotropy using MR Egger—although none of our pleiotropy tests reveals statistically significant violations, these diagnostic analyses are likely to be underpowered; therefore, more IVs related to circulating vitamin E and dietary vitamin E need to be identified. In addition, the 3 IVs were only associated with α-tocopherol levels, and we need to consider the effects of other isoforms of tocopherol and tocotrienol (e.g., γ- and δ-tocopherols) on cancer risk.
Conclusions
In summary, this is the first largest MR study making causal inferences between circulating vitamin E concentrations and the risk of multiple cancers among European population. Our MR does not convincingly support a causal effect of vitamin E on the risk of cancer development, which delivers an important public health message that administration of vitamin E supplementation may not be necessary for prevention of cancers. Nevertheless, further studies are warranted to validate such findings as well as to demonstrate causal associations across ancestries.
Supplementary Information
Additional file 1: Table S1. Summary of cancer-specific GWAS data included in Mendelian randomization (MR) analysis. Table S2. ICD-10 diagnosis codes for ten cancers in the UK Biobank cohort. Table S3. Characteristics of circulating vitamin E-associated instrumental variants (IVs). Table S4. Associations of circulating vitamin E instrumental variants (IVs) with multiple cancer risk in cancer-specific GWAS. Table S5. Mendelian randomization (MR) analysis for the associations of circulating vitamin E with cancer risk in cancer-specific GWAS. Table S6. Leave-one-out analysis for the associations between circulating vitamin E and cancer risk in cancer-specific GWAS. Table S7. Summary of 24 traits related to circulating vitamin E associated SNPs. Table S8. Sensitivity analysis of genetic risk score (GRS) analysis for the associations of vitamin E with cancer risk in the UK Biobank cohort. Table S9. Summary of dietary vitamin E intake associated SNPs derived from the UK Biobank cohort. Table S10. Functional annotation of dietary vitamin E intake associated SNPs. Table S11. Summary of dietary vitamin E associated variants in the UK Biobank cohort. Figure S1. Conceptual framework of Mendelian randomization (MR) analysis in this study. Figure S2. The effect of each variant on circulating vitamin E (exposure) and cancer risk (outcome) in cancer-specific GWAS. Figure S3. Manhattan and quantile-quantile (QQ) plots for the dietary vitamin E intake-related GWASs in the UK Biobank cohort.
Acknowledgements
We would like to thank the study staff of dbGaP, BCAC, OCAC, PRACTICAL, TRICL-ILCCO, and UK Biobank. Especially, we acknowledged the support of TRICL-ILCCO which provided the lung cancer summary-level GWAS data underlying the available access by David C. Christiani (Harvard T.H. Chan School of Public Health).
Abbreviations
- BMI
Body mass index
- CI
Confidence interval
- eQTL
Expression quantitative trait loci
- GRS
Genetic risk score
- GWAS
Genome-wide association study
- HR
Hazard ratio
- HWE
Hardy-Weinberg equilibrium
- IV
Instrumental variable
- IVW
Inverse variance weighting
- MAF
Minor allele frequency
- MR
Mendelian randomization
- OR
Odds ratio
- SD
Standard deviation
- SELECT
Selenium and Vitamin E Cancer Prevention Trial
- SNP
Single-nucleotide polymorphism
Authors’ contributions
JX, XJ, and SB contributed equally to this paper. The study was conceived and designed by JX, ZZ, MD, MW, and DC. The data were analyzed by JX, XJ, and SB. The first draft of the manuscript was prepared by JX. The data was collected and interpreted by JX, XJ, SB, QY, LS, and ZZ. All other authors provided the data and revised the manuscript critically for important intellectual content. The authors read and approved the final version of the manuscript.
Funding
This study was supported in part by the National Natural Science Foundation of China (81872697 and 82173603), the Gusu Health Talent Program (GSWS2021034) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files or available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committees of Nanjing Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
David C. Christiani is a senior author who supervised this work.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Junyi Xin, Xia Jiang and Shuai Ben contributed equally to this work.
References
- 1.Das GS, Suh N. Tocopherols in cancer: an update. Mol Nutr Food Res. 2016;60(6):1354–1363. doi: 10.1002/mnfr.201500847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Yang X, Liu A, Wang G, Bosland MC, Yang CS. delta-Tocopherol inhibits the development of prostate adenocarcinoma in prostate specific Pten-/- mice. Carcinogenesis. 2018;39(2):158–169. doi: 10.1093/carcin/bgx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang CS, Suh N, Kong AN. Does vitamin E prevent or promote cancer? Cancer Prev Res (Phila) 2012;5(5):701–705. doi: 10.1158/1940-6207.CAPR-12-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein EA, Thompson IJ, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, et al. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2011;306(14):1549–1556. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Sesso HD, Glynn RJ, Christen WG, Bubes V, Manson JE, et al. Vitamin E and C supplementation and risk of cancer in men: posttrial follow-up in the Physicians’ Health Study II randomized trial. Am J Clin Nutr. 2014;100(3):915–923. doi: 10.3945/ajcn.114.085480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan SB, Dear KB, Bruzzi P, Machin D. Strategy for randomised clinical trials in rare cancers. BMJ. 2003;327(7405):47–49. doi: 10.1136/bmj.327.7405.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey SG, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verduijn M, Siegerink B, Jager KJ, Zoccali C, Dekker FW. Mendelian randomization: use of genetics to enable causal inference in observational studies. Nephrol Dial Transplant. 2010;25(5):1394–1398. doi: 10.1093/ndt/gfq098. [DOI] [PubMed] [Google Scholar]
- 9.Schumacher FR, Al OA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar SP, Lawrenson K, Winham SJ, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49(5):680–691. doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mckay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49(7):1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42(11):978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Closas M, Ye Y, Rothman N, Figueroa JD, Malats N, Dinney CP, et al. A genome-wide association study of bladder cancer identifies a new susceptibility locus within SLC14A1, a urea transporter gene on chromosome 18q12.3. Hum Mol Genet. 2011;20(21):4282–4289. doi: 10.1093/hmg/ddr342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa JD, Ye Y, Siddiq A, Garcia-Closas M, Chatterjee N, Prokunina-Olsson L, et al. Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum Mol Genet. 2014;23(5):1387–1398. doi: 10.1093/hmg/ddt519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144(4):799–807. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51(1):76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine DM, Ek WE, Zhang R, Liu X, Onstad L, Sather C, et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett's esophagus. Nat Genet. 2013;45(12):1487–1493. doi: 10.1038/ng.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43(1):60–65. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesseur C, Diergaarde B, Olshan AF, Wunsch-Filho V, Ness AR, Liu G, et al. Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat Genet. 2016;48(12):1544–1550. doi: 10.1038/ng.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42(3):224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46(9):994–1000. doi: 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Major JM, Yu K, Wheeler W, Zhang H, Cornelis MC, Wright ME, et al. Genome-wide association study identifies common variants associated with circulating vitamin E levels. Hum Mol Genet. 2011;20(19):3876–3883. doi: 10.1093/hmg/ddr296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimitrakopoulou VI, Tsilidis KK, Haycock PC, Dimou NL, Al-Dabhani K, Martin RM, et al. Circulating vitamin D concentration and risk of seven cancers: Mendelian randomisation study. BMJ. 2017;359:j4761. doi: 10.1136/bmj.j4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 29.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50(11):1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vosa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy SJ, Anderson LA, Ferguson HR, Johnston BT, Watson PR, Mcguigan J, et al. Dietary antioxidant and mineral intake in humans is associated with reduced risk of esophageal adenocarcinoma but not reflux esophagitis or Barrett’s esophagus. J Nutr. 2010;140(10):1757–1763. doi: 10.3945/jn.110.124362. [DOI] [PubMed] [Google Scholar]
- 33.de Munter L, Maasland DH, van den Brandt PA, Kremer B, Schouten LJ. Vitamin and carotenoid intake and risk of head-neck cancer subtypes in the Netherlands Cohort Study. Am J Clin Nutr. 2015;102(2):420–432. doi: 10.3945/ajcn.114.106096. [DOI] [PubMed] [Google Scholar]
- 34.Crane TE, Khulpateea BR, Alberts DS, Basen-Engquist K, Thomson CA. Dietary intake and ovarian cancer risk: a systematic review. Cancer Epidemiol Biomarkers Prev. 2014;23(2):255–273. doi: 10.1158/1055-9965.EPI-13-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koushik A, Wang M, Anderson KE, van den Brandt P, Clendenen TV, Eliassen AH, et al. Intake of vitamins A, C, and E and folate and the risk of ovarian cancer in a pooled analysis of 10 cohort studies. Cancer Causes Control. 2015;26(9):1315–1327. doi: 10.1007/s10552-015-0626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong Y, Liu Y, Shu Y, Chen X, Hu J, Zheng R, et al. Link between risk of colorectal cancer and serum vitamin E levels: a meta-analysis of case-control studies. Medicine (Baltimore) 2017;96(27):e7470. doi: 10.1097/MD.0000000000007470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Weinstein SJ, Yu K, Mannisto S, Albanes D. A prospective study of serum vitamin E and 28-year risk of lung cancer. J Natl Cancer Inst. 2020;112:191–199. doi: 10.1093/jnci/djz077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng L, Liu X, Lu Q, Tang T, Yang Z. Vitamin E intake and pancreatic cancer risk: a meta-analysis of observational studies. Med Sci Monit. 2015;21:1249–1255. doi: 10.12659/MSM.893607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui L, Li L, Tian Y, Xu F, Qiao T. Association between dietary vitamin E intake and esophageal cancer risk: an updated meta-analysis. Nutrients. 2018;10(7):801. doi: 10.3390/nu10070801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang Y, Yi S, Cui D, Han G, Liu C. Vitamin E intake and risk of renal cell carcinoma: a meta-analysis of 7 case-control studies. J Ren Nutr. 2015;25(4):339–344. doi: 10.1053/j.jrn.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Hu F, Wu Z, Li G, Teng C, Liu Y, Wang F, et al. The plasma level of retinol, vitamins A, C and alpha-tocopherol could reduce breast cancer risk? A meta-analysis and meta-regression. J Cancer Res Clin Oncol. 2015;141(4):601–614. doi: 10.1007/s00432-014-1852-7. [DOI] [PubMed] [Google Scholar]
- 42.Lin JH, Chen SJ, Liu H, Yan Y, Zheng JH. Vitamin E consumption and the risk of bladder cancer. Int J Vitam Nutr Res. 2019;89(3–4):168–175. doi: 10.1024/0300-9831/a000553. [DOI] [PubMed] [Google Scholar]
- 43.Chen F, Li Q, Yu Y, Yang W, Shi F, Qu Y. Association of vitamin C, vitamin D, vitamin E and risk of bladder cancer: a dose-response meta-analysis. Sci Rep. 2015;5:9599. doi: 10.1038/srep09599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Q. Natural forms of vitamin E: metabolism, antioxidant, and anti-inflammatory activities and their role in disease prevention and therapy. Free Radic Biol Med. 2014;72:76–90. doi: 10.1016/j.freeradbiomed.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abraham A, Kattoor AJ, Saldeen T, Mehta JL. Vitamin E and its anticancer effects. Crit Rev Food Sci Nutr. 2019;59(17):2831–2838. doi: 10.1080/10408398.2018.1474169. [DOI] [PubMed] [Google Scholar]
- 46.Traber MG. Vitamin E regulatory mechanisms. Annu Rev Nutr. 2007;27:347–362. doi: 10.1146/annurev.nutr.27.061406.093819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Summary of cancer-specific GWAS data included in Mendelian randomization (MR) analysis. Table S2. ICD-10 diagnosis codes for ten cancers in the UK Biobank cohort. Table S3. Characteristics of circulating vitamin E-associated instrumental variants (IVs). Table S4. Associations of circulating vitamin E instrumental variants (IVs) with multiple cancer risk in cancer-specific GWAS. Table S5. Mendelian randomization (MR) analysis for the associations of circulating vitamin E with cancer risk in cancer-specific GWAS. Table S6. Leave-one-out analysis for the associations between circulating vitamin E and cancer risk in cancer-specific GWAS. Table S7. Summary of 24 traits related to circulating vitamin E associated SNPs. Table S8. Sensitivity analysis of genetic risk score (GRS) analysis for the associations of vitamin E with cancer risk in the UK Biobank cohort. Table S9. Summary of dietary vitamin E intake associated SNPs derived from the UK Biobank cohort. Table S10. Functional annotation of dietary vitamin E intake associated SNPs. Table S11. Summary of dietary vitamin E associated variants in the UK Biobank cohort. Figure S1. Conceptual framework of Mendelian randomization (MR) analysis in this study. Figure S2. The effect of each variant on circulating vitamin E (exposure) and cancer risk (outcome) in cancer-specific GWAS. Figure S3. Manhattan and quantile-quantile (QQ) plots for the dietary vitamin E intake-related GWASs in the UK Biobank cohort.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files or available from the corresponding author upon reasonable request.