Abstract
Immunotherapy using anti-programmed death 1 ligand 1 (PD-L1) antibodies has shown clinical efficacy against hepatocellular carcinoma (HCC) and is recognized as the first-line treatment for unresectable HCC. PD-L1 expression is affected by various cytokines produced by immune cells in the tumor microenvironment; however, there is limited information about the effects of cytokine interactions on PD-L1 expression. In this study, we examined how cytokines induce PD-L1 expression in HCC cells. Both interferon gamma (IFN-γ) and interleukin 1 beta (IL-1β) induced PD-L1 expression, and the two cytokines enhanced PD-L1 expression in combination compared to that when administered alone. The Janus kinase/signal transducer and activator of transcription signaling pathway activated by IFN-γ is the major pathway of PD-L1 expression. The increase in interferon regulatory factor 1 expression and IFN-γ receptor expression induced by IL-1β was associated with the synergistic effect of IFN-γ and IL-1β on PD-L1 expression. These findings strongly indicate that IFN-γ and IL-1β affect the mechanism underlying immune resistance in HCC cells.
Keywords: Synergistic effect, PD-L1, IFN-γ, IL-1β, Hepatocellular carcinoma
Highlights
-
•
IFN-γ and IL-1β synergistically increase the expression of PD-L1 in HCC cells.
-
•
IFN-γ enhances PD-L1 expression via STAT1 signaling.
-
•
IL-1β enhances PD-L1 expression via the NF-κB and the p38 MAPK pathways.
-
•
IRF-1 and IFNGR also contribute to the synergistic effect of IFN-γ and IL-1β in HCC.
Abbreviations
- CTL
cytotoxic T cell
- DMSO
dimethyl sulfoxide
- HCC
hepatocellular carcinoma
- IFN-γ
interferon gamma
- IFNGR
interferon gamma receptor
- IHC
immunohistochemistry
- IL
interleukin
- IL-1β
interleukin 1beta
- IRF-1
interferon regulatory factor 1
- JAK
Janus kinase
- JNK
Jun amino terminal kinase
- MAPK
Mitogen-activated protein kinase
- NF-κB
Nuclear factor-kappa B
- PD-1
programmed death 1
- PD-L1
programmed death-1 ligand 1
- qRT-PCR
Quantitative reverse-transcription - polymerase chain reaction
- TSS
transcription start site
- SAPK
Stress-activated protein kinase
- STAT
Signal transducer and activator of transcription
- TNF-α
tumor necrosis factor-alpha; WB, Western blotting
- WB
Western blotting
1. Introduction
Liver cancer is the fourth leading cause of cancer-related deaths worldwide. According to the 2018 statistics of the International Agency for Research on Cancer, the worldwide estimated age-standardized mortality rates of liver cancer are 10.2% and 5.6% per 100,000 patients per year for men and women, respectively, whereas the mortality rate by site is the second and sixth highest, respectively [1].
Hepatocellular carcinoma (HCC) is the most common type of liver cancer. Previously, the first-line treatment for unresectable HCC was a multi-targeted tyrosine kinase inhibitor, which exerts antitumor effects by suppressing angiogenesis and tumor cell growth [2]. Recently, immune checkpoint inhibitors, such as anti-programmed death 1 ligand 1 (PD-L1) antibodies, have shown excellent clinical efficacy in some solid tumors [3]; therefore, anti-PD-L1 antibodies are currently considered the first-line treatment for unresectable HCC.
PD-L1 binds to programmed death 1 (PD-1), a receptor expressed on cytotoxic T cells (CTLs), and negatively regulates the antitumor effects of CTLs [4]. PD-L1 is upregulated in tumor cells [5]; therefore, tumor cells escape the attack by CTLs and evade the immune response [6]. Anti-PD-1/PD-L1 antibodies block this pathway and prevent the suppression of CTL activity in tumor cells [7].
PD-L1 expression in tumor cells is primarily regulated by cytokines, which are synthesized owing to immune responses within the tumor microenvironment [8,9]. With advancements in computer technology, molecular dynamics-based PD-L1 and cytokine dynamics prediction studies are becoming increasingly useful to elucidate the molecular recognition of PD-1/PD-L1 structures, identify drug efficacy, and develop new drugs with higher efficacy [10,11]. Therefore, it is very important to understand the dynamics of cytokines and PD-L1, the effects of cytokines on PD-L1, and the regulatory mechanisms of PD-L1 expression.
Interferon gamma (IFN-γ) is a primary cytokine that affects PD-L1 expression [9]. IFN-γ receptor (IFNGR) is composed of two subunits; IFNGR1 and IFNGR2. Binding of IFN-γ to the extracellular domain of the IFNGR1 receptor subunit leads to engagement of the IFNGR2 subunit [12]. Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling is subsequently activated, and PD-L1 expression is induced [13,14]. Interferon regulatory factor 1 (IRF-1) is a transcription factor activated by the JAK/STAT signaling pathway [15] and activates numerous downstream genes [12]. Moreover IRF-1 inhibits antitumor immunity by upregulating PD-L1 expression [16].
With respect to HCC, in which inflammatory processes are involved in carcinogenesis [17], several pro-inflammatory cytokines, such as IFN-γ and tumor necrosis factor-alpha (TNF-α), are known to increase PD-L1 expression [18]. Additionally, a previous report also demonstrated the synergistic effect of IFN-γ and TNF-α on PD-L1 expression [19], but only a few studies have reported the effects of other combinations.
In the present study, we investigated cytokines that induce PD-L1 expression in HCC cells and determined whether these cytokines synergistically affect PD-L1 expression. We found that IFN-γ and interleukin (IL) 1 beta (IL-1β) increased PD-L1 expression in HCC cells, and their synergistic effect further promoted PD-L1 expression. The JAK/STAT1 signaling pathway activated by IFN-γ is the major pathway for PD-L1 expression, and the increase in IRF-1 and IFNGR expression induced by IL-1β affects the synergistic effect of PD-L1 expression. Our findings suggest that the inhibition of both IFN-γ and IL-1β signaling could contribute to the reduction of PD-L1 expression in HCC cells, which could aid the development of therapeutic strategies in future.
2. Materials and methods
Detailed methods are available in the supplementary information.
2.1. Cell culture and chemicals
In the present study, we used two representative human HCC cell lines: HLF (JCRB0405) and Hep 3B (ECACC 86062703). HLF cells were purchased from the Laboratory of the National Institutes of Biomedical Innovation, Health and Nutrition, JCRB Cell Bank (Osaka, Japan), and Hep3B cells were purchased from the European Collection of Authenticated Cell Cultures.
We treated HLF and Hep3B cells with human recombinant cytokines, such as IFN-γ, IL-1β, IL-4, IL-6, IL-9, IL-10, IL-13, IL-17, IL-33, and TNF-α. Initially, we conducted experiments with the concentration of all cytokines set at 20 ng/mL. Subsequently, the concentrations of IFN-γ and IL-1β were fixed at 20 and 10 ng/mL, respectively. We harvested the cells treated with cytokines for mRNA expression analysis after 6 h based on the results of our examination (Supplementary Fig. S1), and for protein expression analysis after 24 h based on previous reports [19,20].
2.2. Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the HCC cells, and complementary DNA was synthesized. qRT-PCR was conducted on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, MA, USA), using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, MA, USA). The primer sequences used in this study are listed in the Supplementary Information. The gene expression levels of the target molecules were normalized to those of β-actin. The 2−ΔΔCt method was used to calculate the relative expression levels of each gene.
2.3. Western blotting (WB)
Cells were lysed in the presence of a protease inhibitor. Extracted proteins were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were probed with primary antibodies and a secondary antibody. Next, the membranes were incubated with a luminescence detection reagent and visualized for luminescence. The γ-actin protein served as an internal control for the normalization of the expression levels of other proteins.
The intensity of each band was quantitated by densitometry, analyzed using the Image J software, and normalized to the protein level of γ-actin.
2.4. Inhibitor experiment
After 1 h of pre-treatment with signal inhibitors, the HCC cells were stimulated with IFN-γ, IL-1β, or both for the indicated time period, and were then harvested for qRT-PCR assays.
2.5. Construction of luciferase gene plasmids
The candidate PD-L1 promoter region was amplified from the genomic DNA purified from THP-1 (RCB1189) cells using PCR. The reason for using the reporter vector with the PD-L1 promoter sequence generated from the genomic DNA derived from THP-1 cells in this study is that it had already been constructed and used for other experiments.
pPDL1-Luc is a pGL4.10 basic luciferase plasmid (Promega, WI, USA) with the PCR product inserted upstream of the luciferase gene. pPDL1-Luc-delSTAT1 is pPDL1-Luc with a deleted signal transducer and activator of transcription 1 (STAT1) binding site (Fig. 2B). The cloned DNA fragments were confirmed by sequencing prior to the assay.
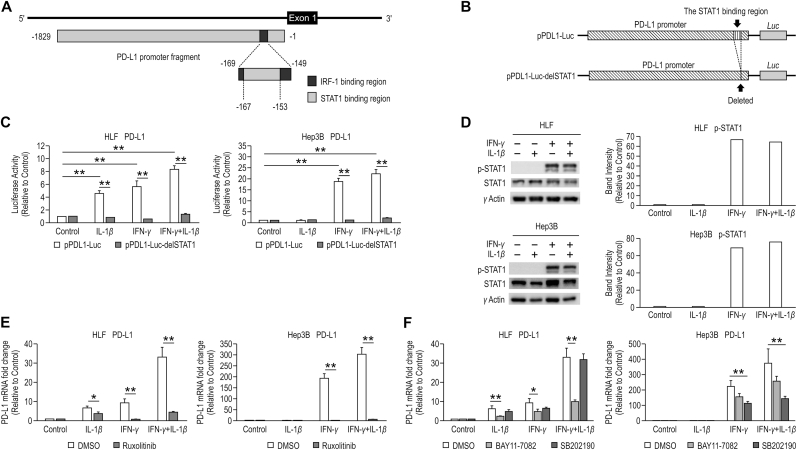
Fig. 2.
STAT1, NF-κB, and p38 MAPK are involved with the synergistic effect of IFN-γ and IL-1β on PD-L1 expression. (A) Diagrammatic representation of the PD-L1 regulatory element. The numbers annotated below the fragments are positions relative to the PD-L1 transcription start site. (B) Diagram of luciferase vectors. The GL4.17 vector with the PD-L1 promoter cloned upstream of the luciferase gene (pPDL1-Luc), and the PD-L1 promoter excluding the STAT1 binding site cloned upstream of the luciferase gene (pPDL1-Luc-delSTAT1). (C) PD-L1 mRNA expression was examined using reporter assays in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 24 h. Firefly luciferase activity was normalized by Renilla luciferase activity. (D) Activation of STAT1 was analyzed by western blotting (WB) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 15 min. γ-actin is used as a loading control in WB. The results were expressed as relative ratios of band intensity between pSTAT1 and γ-actin. (E) PD-L1 mRNA expression was examined using quantitative reverse transcription polymerase chain reaction (qRT-PCR) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β in the presence of dimethyl sulfoxide (DMSO) and Ruxolitinib (1 μM). The samples treated without IFN-γ or IL-1β were used as controls. (F) PD-L1 mRNA expression was examined using qRT-PCR in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β in the presence of DMSO, BAY11-70825 (5 μM), and SB202190 (5 μM). The samples treated without IFN-γ or IL-1β were used as controls. *P < 0.05, **P < 0.01.
2.6. Luciferase reporter assay
The PD-L1 promoter was inserted into pGL4.10 plasmids containing luciferase sequences (Promega). These plasmids, along with the pGL4.74 hRluc/TK (Promega) plasmids containing Renilla luciferase sequences, were transfected into HLF and Hep3B cells. Luciferase activity was measured, and Renilla luciferase activity was used as an internal control.
2.7. Immunohistochemistry (IHC)
HLF and Hep3B cells were collected and embedded in iPGell (GenoStaff, Tokyo, Japan) and fixed with 4% paraformaldehyde. The gels containing the cells were embedded in paraffin and sectioned at 5 μm. IHC of the sections was performed according to the protocol described in the Supplementary Information. The intensity of IHC was quantified using Fiji ImageJ2 software (Fiji, RRID:SCR_002285), according to the protocol described in the Supplementary Information.
2.8. Statistical analysis
All qRT-PCR data are presented in terms of the relative level of gene expression level for the replicate group computed using normalized quantities ± standard deviations. Statistical significance of the difference between any two experimental groups was analyzed using Student's t-test. Statistical significance was set at P < 0.05.
3. Results
3.1. IFN-γ and IL-1β synergistically increase PD-L1 expression in HCC cells
To investigate the cytokines that enhance PD-L1 expression in HCC cells, we examined the effect of several cytokines such as IFN-γ, IL-1β, IL-4, IL-6, IL-9, IL-10, IL-13, IL-17, IL-33, and TNF-α on PD-L1 expression in HLF and Hep3B cells. IFN-γ, IL-1β, and TNF-α increased the expression of PD-L1 (Fig. 1A). However, we decided to examine the effects of IFN-γ and IL-1β on PD-L1 expression in HCC cells because (1) IFN-γ greatly increased PD-L1 expression in HLF and Hep3B cells; (2) IL-1β and TNF-α induced a similar increase in PD-L1 expression in Hep3B cells, but IL-1β increased PD-L1 expression more than TNF-α did in HLF cells; and (3) the effect of IFN-γ and IL-1β on PD-L1 expression in HCC cells has not been previously reported.
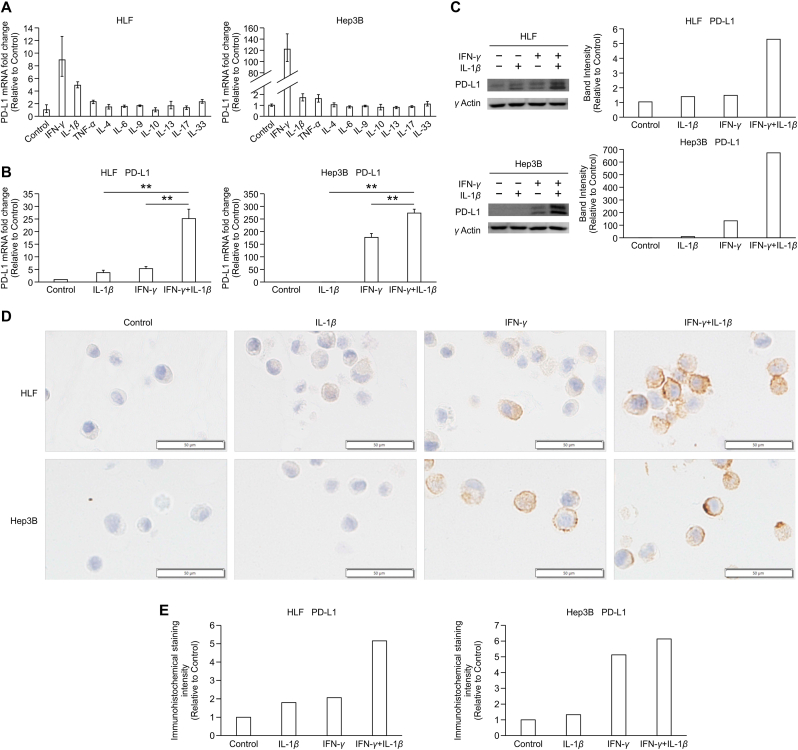
Fig. 1.
IFN-γ and IL-1β synergistically increase the expression of PD-L1 in HCC cells. (A) PD-L1 expression was measured using quantitative reverse transcription polymerase chain reaction (qRT-PCR) in HLF and Hep3B cells treated with cytokines IFN-γ, IL-1β, IL-4, IL-6, IL-9, IL-10, IL-13, IL-17, IL-33, and TNF-α (all at 20 ng/mL). Data are presented as normalized quantities ±95% confidence level (n = 1). (B) PD-L1 mRNA expression was examined using qRT-PCR in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 6 h. In both cells, IL-1β or IFN-γ significantly increased PD-L1 expression compared with control. PD-L1 expression induced by the combination of IFN-γ and IL-1β was significantly higher than that induced by IFN-γ alone (p < 0.01, n = 3) or IL-1β alone (p < 0.01, n = 3). (C) PD-L1 protein expression was analyzed by western blotting (WB) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 24 h. γ-actin is used as a loading control in WB. (D) Immunohistochemistry (IHC) of PD-L1 in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 24 h (original magnification, 200×). Scale bars: 50 μm (E) Quantitative analysis of PD-L1 IHC staining intensity. The samples treated without IFN-γ or IL-1β were used as controls.
The inducible expression of the PD-L1 gene was upregulated by IFN-γ in a dose-dependent manner, unlike the effect exerted by IL-1β (Supplementary Fig. S2). Based on the results of the experiments, we decided to use IFN-γ at the concentration of 20 ng/mL and IL-1β at 10 ng/mL in this study for their effective concentration in increasing PD-L1 expression in HCC cells.
To explore the contributions of IFN-γ and IL-1β to PD-L1 expression in HCC cells, we stimulated HLF and Hep3B cells with IL-1β, IFN-γ, or both (Fig. 1B). The combination of IFN-γ and IL-1β augmented PD-L1 expression in HLF cells compared to stimulation with IL-1β or IFN-γ alone. In Hep 3B cells, IFN-γ increased PD-L1 expression compared with the control. Similar to HLF cells, the combination of IFN-γ and IL-1β augmented PD-L1 expression in Hep3B cells compared with that in Hep3B cells stimulated with IL-1β or IFN-γ alone.
WB and IHC also demonstrated that the combination of IFN-γ and IL-1β elicited a marked synergistic response on PD-L1 expression in HCC cells (Fig. 1C, D, E).
3.2. STAT1, nuclear factor kappa B (NF-κB), and p38 mitogen-activated protein kinase (MAPK) are involved in the synergistic effect of IFN-γ and IL-1β on PD-L1 expression
To understand the mechanism underlying the regulation of PD-L1 expression, we examined the activity of the PD-L1 promoter regions using JASPAR (http://jaspar.genereg.net/), an open-access database of transcription factor binding profiles. The candidate binding sites for IRF-1 (−169 to −149 from the PD-L1 transcription start site (TSS)) and STAT1 (−167 to −153 from the PD-L1 TSS) were found upstream of the PD-L1 gene. The IRF-1 binding site included the STAT1 binding site (Fig. 2A).
We constructed the luciferase gene plasmids, pPDL1-Luc and pPDL1-Luc-delSTAT1 (Fig. 2B), and examined PD-L1 expression with them using luciferase reporter assays (Fig. 2C). The results demonstrated that stimulation with IL-1β, IFN-γ, and both IFN-γ and IL-1β increased the PD-L1 promoter-driver luciferase reporter activity in HLF cells, and stimulation with IFN-γ and both IFN-γ and IL-1β significantly increased the same in Hep3B cells (p < 0.01, n = 3). In contrast, stimulation with these cytokines did not increase the activity of a luciferase reporter gene in either cell lines transfected with a plasmid containing a promoter with a deleted STAT1 responsive element.
WB also showed that unlike IL-1β stimulation, IFN-γ stimulation enhanced STAT1 phosphorylation (Fig. 2D).
In the presence of ruxolitinib, a Janus kinase (JAK) inhibitor, PD-L1 expression was significantly reduced in HLF and Hep3B cells stimulated with IFN-γ compared to that in the control (p < 0.01, n = 3) (Fig. 2E).
WB also showed that IL-1β stimulation enhanced NF-κB and p38 MAPK phosphorylation in both HLF and Hep3B cells. (Supplementary Fig. S3). BAY11-7082 (NF-кB inhibitor) significantly reduced the PD-L1 expression induced by IL-1β stimulation alone to 38% (p < 0.01, n = 3) and by the combination of IFN-γ and IL-1β to 30% in HLF cells (p < 0.01, n = 3). However, BAY11-7082 did not significantly reduce the PD-L1 expression induced by IL-1β stimulation alone, IFN-γ stimulation alone, or a combination of the two in Hep3B cells. SB202190 (p38 MAPK inhibitor) significantly reduced PD-L1 expression induced by IFN-γ stimulation alone to 51% (p < 0.01, n = 3) and by the combination of IFN-γ and IL-1β to 39% in Hep3B cells (p < 0.01, n = 3), while it did not inhibit the same in HLF cells (Fig. 2F).
3.3. IRF-1 is a major factor of the synergistic effect of IFN-γ and IL-1β on PD-L1 expression
According to the results of the experiment conducted using JASPAR, IRF-1 may be involved in PD-L1 expression (Fig. 2A). We examined the expression of IRF-1 in HLF and Hep3B cells treated with IL-1β, IFN-γ, or both, using qRT-PCR (Fig. 3A) and WB (Fig. 3B). An additive effect was observed in HLF and Hp3B cells in IRF-1 expression by combined stimulation with IFN-γ and IL-1β.
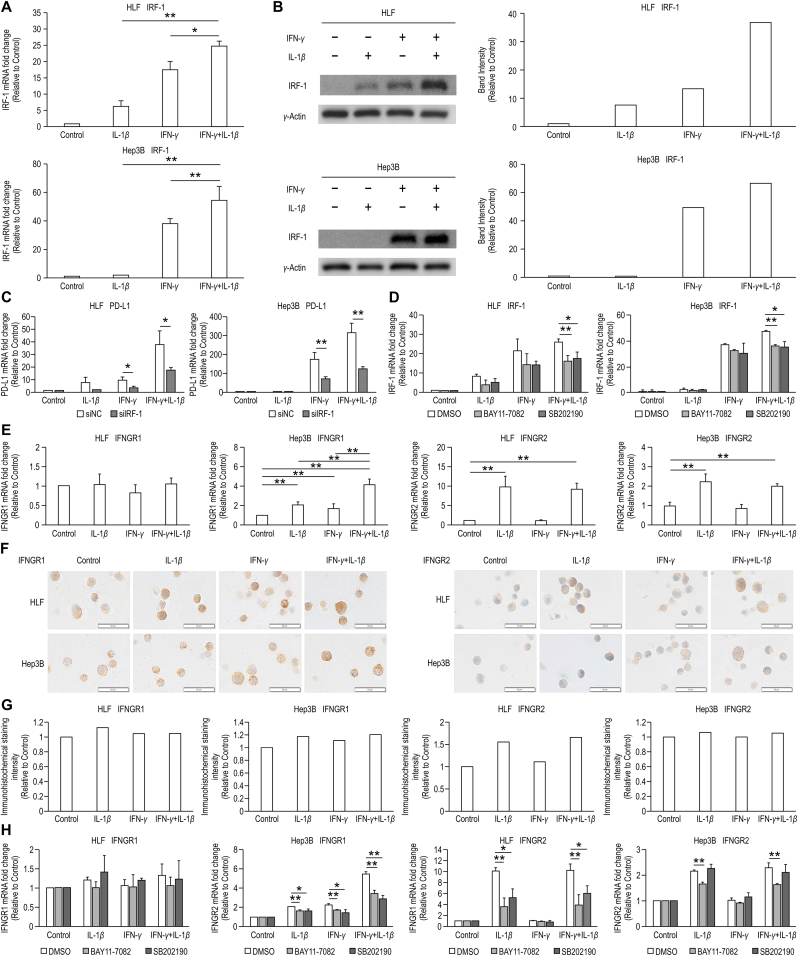
Fig. 3.
IRF-1 is a major and IFNGRs is a contributing factor of the synergistic effect of IFN-γ and IL-1β on PD-L1 expression. (A) IRF-1 protein expression was analyzed by western blotting (WB) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β. The results were expressed as relative ratios of band intensity between pSTAT1 and γ-actin. (B) IRF-1 mRNA expression was examined using quantitative reverse transcription polymerase chain reaction (qRT-PCR) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 6 h. The samples treated without IFN-γ or IL-1β were used as controls. (C) PD-L1 mRNA expression was examined using qRT-PCR in HLF and Hep3B cells transfected with siRNA of negative control (siNC) or IRF-1 (siIRF-1) and subsequently treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 6 h. The samples treated without IFN-γ or IL-1β were used as controls. (D) IRF-1 mRNA expression was examined using quantitative reverse transcription polymerase chain reaction in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β in the presence of DMSO, BAY11-7082 (5 μM), and SB202190 (5 μM). The samples treated without IFN-γ or IL-1β were used as controls. (E) Interferon gamma receptor (IFNGR)1 and IFNGR2 mRNA expression was examined using quantitative reverse transcription polymerase chain reaction (qRT-PCR) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β. (F) Immunohistochemistry (IHC) of IFNGR1 and IFNGR2 in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 24 h (original magnification, 200×). Scale bars: 50 μm (G) Quantitative analysis of IFNGR1 and IFNGR2 IHC staining intensity. The samples treated without IFN-γ or IL-1β were used as controls. (H) IFNGR1 and IFNGR2 mRNA expression was examined using qRT-PCR in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β in the presence of DMSO, BAY11-7082 (5 μM), and SB202190 (5 μM). The samples treated without IFN-γ or IL-1β were used as controls.*P < 0.05,**P < 0.01.
We investigated PD-L1 expression in HLF and Hep3B cells after transfection with siRNA for IRF-1 and stimulation with IL-1β, IFN-γ, or both using qRT-PCR (Fig. 3C). We found that IRF-1 knockdown in HLF and Hep3B cells reduced PD-L1 expression. qRT-PCR revealed that BAY11-7082 significantly reduced IRF-1 expression induced upon combined stimulation with IFN-γ and IL-1β in HLF and Hep3B cells (p < 0.01, n = 3). Similar results were observed in the presence of SB202190 (p < 0.05, n = 3) (Fig. 3D).
3.4. IFNGRs are contributing factors of the synergistic effect of IFN-γ and IL-1β on PD-L1 expression
Because IFNGR reportedly affects the synergistic effect of IFN-γ and TNF-α on PD-L1 expression in HCC cells, we examined the expression of IFNGR1 and IFNGR2 in HLF and Hep3B cells using qRT-PCR after stimulation with IL-1β, IFN-γ, or both (Fig. 3E). IFNGR1 expression in response to cytokine stimulation differed between HLF and Hep3B cells. Stimulation with neither IFN-γ nor IL-1β affected IFNGR1 expression in HLF cells. In contrast, IFN-γ and IL-1β stimulation significantly increased IFNGR1 expression compared with that in the control (p < 0.01, n = 3), and combined IFN-γ and IL-1β treatment significantly increased the expression compared with IFN-γ or IL-1β stimulation alone in Hep3B cells (p < 0.01, n = 3). IFNGR2 expression was upregulated by IL-1β stimulation but not by IFN-γ stimulation in HLF and Hep3B cells. This observation was confirmed using IHC for IFNGR1 and IFNGR2 (Fig. 3F, G).
BAY11-7082 and SB202190 did not affect IFNGR1 expression in HLF cells stimulated with cytokines; however, both inhibitors suppressed the synergistic enhancement of IFNGR2. Both inhibitors reduced IFNGR1 expression in Hep3B cells stimulated with IL-1β, IFN-γ, and both IFN-γ and IL-1β. BAY11-7082 reduced IFNGR2 expression in Hep3B cells stimulated with IL-1β and with both IFN-γ and IL-1β. (p < 0.01, n = 3) (Fig. 3H).
4. Discussion
HCC is associated with chronic inflammation and mostly develops in inflamed liver cells [21]. In the tumor microenvironment in HCC, the levels of pro-inflammatory cytokines, such as IFN-γ, IL-1β, and TNF-α, produced from T cells, tumor-associated macrophages, and fibroblasts, are increased, and these are involved in the development and progression of HCC [22]. In particular, IFN-γ and TNF-α are known to increase PD-L1 expression [23,24] and exert a synergistic effect on PD-L1 expression in HCC [19].
We considered the possibility that combinations of other cytokines present in the tumor microenvironment may also exert synergistic effects on PD-L1 expression, and confirmed that IFN-γ and IL-1β exert synergistic effects on PD-L1 expression in HCC cells.
The findings of our study demonstrated that PD-L1 expression in HCC increases significantly in response to the synergistic effect of IFN-γ and IL-1β. A potential mechanism is that IFN-γ and IL-1β synergistically increase the expression of transcription factor IRF-1, which regulates PD-L1 expression. Another potential mechanism is that IL-1β enhances IFNGR expression, which consequently enhances IFN-γ signal transduction and leads to an increase in PD-L1 expression. STAT1, NF-κB, and p38 MAPK may be involved in these mechanisms. The schematic figure of what we consider the mechanism of the synergistic effect of IFN-γ and IL-1β on PD-L1 expression is shown in Fig. 4. Our data suggest that the cytokine profile in the tumor microenvironment affects PD-L1 expression in HCC cells.
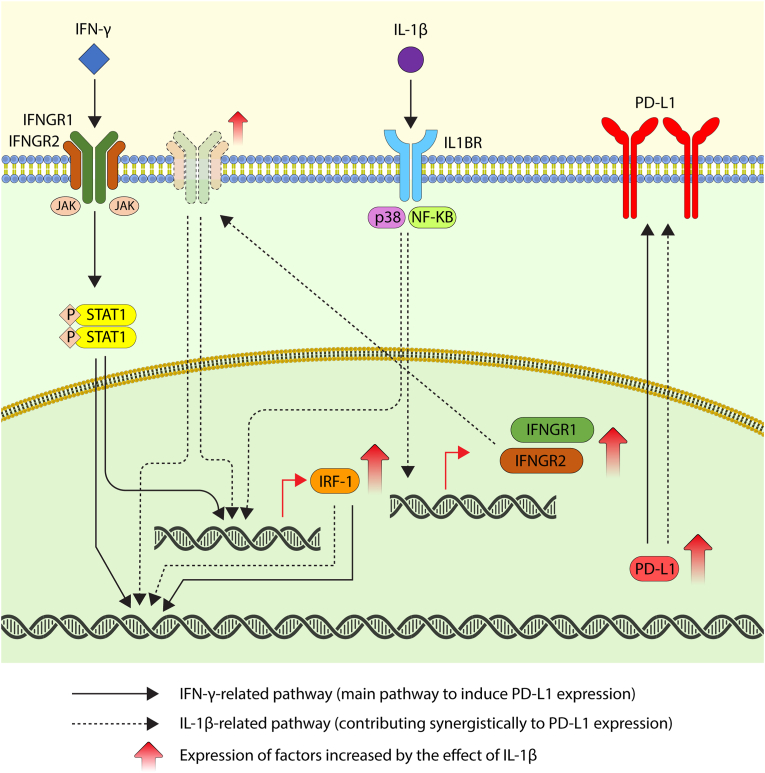
Fig. 4.
Schematic figure displaying the mechanism of the synergistic effect of IFN-γ and IL-1β on PD-L1 expression. A potential mechanism of the synergistic effect of IFN-γ and IL-1β on PD-L1 expression is that (1) IFN-γ and IL-1β synergistically increase the expression of transcription factor IRF-1, which regulates PD-L1 expression; and (2) IL-1β enhances IFNGR expression, which consequently enhances IFN-γ signal transduction and leads to an increase in PD-L1 expression. STAT1, NF-κB, and p38 MAPK may be involved in these mechanisms.
We demonstrated that IFN-γ plays an important role in PD-L1 expression via STAT signaling. We also showed that IL-1β contributes to PD-L1 expression via the NF-κB pathway in HLF cells and that NF-κB is associated with the synergistic effect of IFN-γ and IL-1β on PD-L1 expression in HLF cells and p38 in Hep3B cells.
We found that IRF-1 activity was enhanced due to the additive action of IFN-γ and IL-1β on PD-L1 expression. STAT1 and NF-κB activate IRF-1 by binding to their respective binding elements in the IRF-1 regulatory region [25]. Simultaneous stimulation with IFN-γ and TNF-α has an additive effect on increasing IRF-1 expression, because IFN-γ activates IRF-1 via STAT1 activation and TNF-α activates IRF-1 via NF-κB [26]. Since IL-1β also activates NF-κB, IRF-1 is considered to be activated by IL-1β and TNF-α, and the simultaneous stimulation of IFN-γ and IL-1β may additively enhance IRF-1 expression.
Previous reports have demonstrated that p38 MAPK promotes IRF-1 expression resulting from the activation of STAT1 [27,28]. The mechanism may involve the direct recruitment of p38 MAPK to the IRF-1 promoter, similar to that observed for NF-κB. It is speculated that IL-1β can exert a synergistic effect on PD-L1 by enhancing the activity of IRF-1 via the NF-κB and p38 MAPK signaling pathways.
Another factor contributing to the synergistic effect of IFN-γ and IL-1β on PD-L1 expression is that IL-1β increases the expression of IFN-γ receptors, primarily IFNGR2, and promotes IFN-γ activation. Our data showed that IL-1β induces IFNGR2 expression in HLF and Hep3B cells, and that the combination of IFN-γ and IL-1β enhances IFNGR1 expression in Hep3B cells, thereby enhancing the IFN-γ signaling pathway and contributing to its synergistic effect. We also showed that IL-1β upregulated IFNGR1 and IFNGR2 expression via the NF-κB and p38 MAPK signaling pathways.
Previous reports have demonstrated that IFNGR2 expression is enhanced by TNF-α [19,29]. Similarly, in this study, IFNGR2 expression was observed to be upregulated by IL-1β. These two cytokines commonly activate NF-κB [30]. IL-1β is considered to promote the expression of IFNGR2 via NF-κB or p38 MAPK, which may be more likely to form a dimer with IFNGR1 and increase IFNGR expression. Consequently, IFN-γ enhances the JAK/STAT signaling activity, which may exert a synergistic effect on PD-L1 expression.
However, our study showed discrepancies between the results of the cytokine stimulation and inhibition experiments. BAY11-7082 and SB202190 suppressed both IRF-1 and IFNGR2 expression to approximately the same levels in HLF and Hep3B cells, whereas PD-L1 expression was reduced differently in each cell type and via the action of each inhibitor. Differences in the degree of differentiation between HLF and Hep3B cells may be attributable to the differences in PD-L1 expression. HLF cells belong to a poorly differentiated HCC cell line, whereas Hep3B cells are well differentiated [31]. Compared with well differentiated cancers, poorly differentiated cancers constitutively express PD-L1 [8]. Therefore, the difference in PD-L1 expression stimulated with IFN-γ between HLF and Hep3B cells can be considered to be associated with cell differentiation. In addition, the signaling pathways and metabolic genes upregulated or downregulated in HCC cell lines differ based on the degree of differentiation [31], which may affect the expression of PD-L1.
IFNGRs could contribute to the expression of PD-L1 on the basis of the present data indicating that the expression of IFNGRs with cytokine stimulation depends on the differentiation level of HCC cell lines. Notably, IL-1β affects IFN-γ signaling by enhancing the expression of IFNGR2 in HLF cells and IFNGR1 and R2 in Hep3B cells.
These results suggest that cytokines play an important role in the development and progression of HCC and immune escape, and that cytokine regulation may be a useful therapeutic target in HCC. Additional studies are warranted to elucidate the mechanisms through which IFN-γ, IL-1β, and other cytokines, including TNF-α, affect and regulate PD-L1 expression, and to enhance the efficacy of immune checkpoint inhibitors against HCC. In recent years, through computer simulation, molecular dynamics have been used to predict the movement of molecules in complex chemical and biological systems [10,11]. Using computer simulation, it would be interesting to evaluate the extent to which IFN-γ and IL-1β around HCC synergize to increase PD-L1 expression in HCC and how this affects the functions of anti-PD-L1 antibodies.
This study had some limitations. First, our experiment regarding the association between cytokines and PD-L1 was conducted in vitro only. Therefore, additional in vivo experiments would help to understand the synergistic effect or the origin of cytokines in the tumor microenvironment. Additionally, it would be beneficial to demonstrate the correlation between PD-L1 expression and cytokines in human HCC.
In conclusion, we demonstrated the synergistic effect of IFN-γ and IL-1β on PD-L1 expression in HCC and elucidated the underlying mechanism. Our results suggest that cytokine regulation suppresses immune escape owing to PD-L1 expression in HCC and that cytokine regulation may be a useful therapeutic target in HCC therapy.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101270.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Raza A., Sood G.K. Hepatocellular carcinoma review: current treatment, and evidence-based medicine. World J. Gastroenterol. 2014;20:4115–4127. doi: 10.3748/wjg.v20.i15.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamanishi J., Mandai M., Matsumura N., Abiko K., Baba T., Konishi I. PD-1/PD-L1 blockade in cancer treatment: perspectives and issues. Int. J. Clin. Oncol. 2016;21:462–473. doi: 10.1007/s10147-016-0959-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderaro J., Rousseau B., Amaddeo G., Mercey M., Charpy C., Costentin C., Luciani A., Zafrani E.S., Laurent A., Azoulay D., Lafdil F., Pawlotsky J.M. Programmed death ligand 1 expression in hepatocellular carcinoma: relationship with clinical and pathological features. Hepatology. 2016;64:2038–2046. doi: 10.1002/hep.28710. [DOI] [PubMed] [Google Scholar]
- 6.Jiang X., Wang J., Deng X., Xiong F., Ge J., Xiang B., Wu X., Ma J., Zhou M., Li X., Li Y., Li G., Xiong W., Guo C., Zeng Z. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer. 2019;18:10. doi: 10.1186/s12943-018-0928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng R., Zhang P., Liu W., Zeng X., Ma X., Shi L., Wang T., Yin Y., Chang W., Zhang P., Wang G., Tao K. HDAC is indispensable for IFN-gamma-induced B7-H1 expression in gastric cancer. Clin. Epigenet. 2018;10:153. doi: 10.1186/s13148-018-0589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowicki T.S., Hu-Lieskovan S., Ribas A. Mechanisms of resistance to PD-1 and PD-L1 blockade. Cancer J. 2018;24:47–53. doi: 10.1097/PPO.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X., Yan X., Zhuo W., Gu J., Zuo K., Liu W., Liang L., Gan Y., He G., Wan H., Gou X., Shi H., Hu J. PD-L1 nanobody competitively inhibits the formation of the PD-1/PD-L1 complex: comparative molecular dynamics simulations. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19071984. 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allec S.I., Sun Y., Sun J., Chang C.A., Wong B.M. Heterogeneous CPU+GPU-Enabled simulations for DFTB molecular dynamics of large chemical and biological systems. J. Chem. Theor. Comput. 2019;15:2807–2815. doi: 10.1021/acs.jctc.8b01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaidi M.R., Merlino G. The two faces of interferon-gamma in cancer. Clin. Cancer Res. 2011;17:6118–6124. doi: 10.1158/1078-0432.CCR-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon J.W., Kong S.K., Kim B.S., Kim H.J., Lim H., Noh K., Kim Y., Choi J.W., Lee J.H., Kim Y.S. IFNgamma induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci. Rep. 2017;7:17810. doi: 10.1038/s41598-017-18132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mimura K., Teh J.L., Okayama H., Shiraishi K., Kua L.F., Koh V., Smoot D.T., Ashktorab H., Oike T., Suzuki Y., Fazreen Z., Asuncion B.R., Shabbir A., Yong W.P., So J., Soong R., Kono K. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018;109:43–53. doi: 10.1111/cas.13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Diaz A., Shin D.S., Moreno B.H., Saco J., Escuin-Ordinas H., Rodriguez G.A., Zaretsky J.M., Sun L., Hugo W., Wang X., Parisi G., Saus C.P., Torrejon D.Y., Graeber T.G., Comin-Anduix B., Hu-Lieskovan S., Damoiseaux R., Lo R.S., Ribas A. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerdes I., Matikas A., Bergh J., Rassidakis G.Z., Foukakis T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene. 2018;37:4639–4661. doi: 10.1038/s41388-018-0303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Refolo M.G., Messa C., Guerra V., Carr B.I., D'Alessandro R. Inflammatory mechanisms of HCC development. Cancers. 2020;12 doi: 10.3390/cancers12030641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L., Lin Y., Kwok H.F. The function and regulation of PD-L1 in immunotherapy. ADMET and DMPK. 2017;5 doi: 10.5599/admet.5.3.442. [DOI] [Google Scholar]
- 19.Li N., Wang J., Zhang N., Zhuang M., Zong Z., Zou J., Li G., Wang X., Zhou H., Zhang L., Shi Y. Cross-talk between TNF-alpha and IFN-gamma signaling in induction of B7-H1 expression in hepatocellular carcinoma cells, Cancer Immunol. Immunotherapy. 2018;67:271–283. doi: 10.1007/s00262-017-2086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao G., Jin L.L., Liu C.Q., Wang Y.C., Meng Y.M., Zhou Z.G., Chen J., Yu X.J., Zhang Y.J., Xu J., Zheng L. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J. Immunother. Cancer. 2019;7:300. doi: 10.1186/s40425-019-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Multhoff G., Molls M., Radons J. Chronic inflammation in cancer development. Front. Immunol. 2011;2:98. doi: 10.3389/fimmu.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heim M.H., Moradpour D., Blum H.E. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 1999;73:8469–8475. doi: 10.1128/JVI.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Wang H., Yao H., Li C., Fang J.Y., Xu J. Regulation of PD-L1: emerging routes for targeting tumor immune evasion. Front. Pharmacol. 2018;9:536. doi: 10.3389/fphar.2018.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartley G., Regan D., Guth A., Dow S. Regulation of PD-L1 expression on murine tumor-associated monocytes and macrophages by locally produced TNF-alpha. Cancer Immunol. Immunother. 2017;66:523–535. doi: 10.1007/s00262-017-1955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pine R. Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 1997;25:4346–4354. doi: 10.1093/nar/25.21.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirey K.A., Jung J.Y., Maeder G.S., Carlin J.M. Upregulation of IFN-gamma receptor expression by proinflammatory cytokines influences IDO activation in epithelial cells. J. Interferon Cytokine Res. 2006;26:53–62. doi: 10.1089/jir.2006.26.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsauer K., Sadzak I., Porras A., Pilz A., Nebreda A.R., Decker T., Kovarik P. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12859–12864. doi: 10.1073/pnas.192264999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y., Antony S., Juhasz A., Lu J., Ge Y., Jiang G., Roy K., Doroshow J.H. Up-regulation and sustained activation of Stat1 are essential for interferon-gamma (IFN-gamma)-induced dual oxidase 2 (Duox2) and dual oxidase A2 (DuoxA2) expression in human pancreatic cancer cell lines. J. Biol. Chem. 2011;286:12245–12256. doi: 10.1074/jbc.M110.191031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crescioli C., Cosmi L., Borgogni E., Santarlasci V., Gelmini S., Sottili M., Sarchielli E., Mazzinghi B., Francalanci M., Pezzatini A., Perigli G., Vannelli G.B., Annunziato F., Serio M. Methimazole inhibits CXC chemokine ligand 10 secretion in human thyrocytes. J. Endocrinol. 2007;195:145–155. doi: 10.1677/JOE-07-0240. [DOI] [PubMed] [Google Scholar]
- 30.Liu T., Zhang L., Joo D., Sun S.C. NF-kappaB signaling in inflammation. Signal Transduct. Targeted Ther. 2017;2 doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nwosu Z.C., Battello N., Rothley M., Pioronska W., Sitek B., Ebert M.P., Hofmann U., Sleeman J., Wolfl S., Meyer C., Megger D.A., Dooley S. Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J. Exp. Clin. Cancer Res. 2018;37:211. doi: 10.1186/s13046-018-0872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.