SUMMARY
Older individuals are at increased risk of developing severe respiratory infections. However, our understanding of the impact of aging on the respiratory tract remains limited as samples from healthy humans are challenging to obtain and results can be confounded by variables such as smoking and diet. Here, we carry out a comprehensive cross-sectional study (n = 34 adult, n = 49 aged) to define the consequences of aging on the lung using the rhesus macaque model. Pulmonary function testing establishes similar age and sex differences as humans. Additionally, we report increased abundance of alveolar and infiltrating macrophages and a concomitant decrease in T cells were in aged animals. scRNAseq reveals shifts from GRZMB to IFN expressing CD8+ T cells in the lungs. These data provide insight into age-related changes in the lungs’ functional, microbial, and immunological landscape that explain increased prevalence and severity of respiratory diseases in the elderly.
Graphical abstract
In brief
Rhoades et al. describe age-associated functional, microbial, and immunological changes in the lung using the rhesus macaque model. These data will support further studies aimed at designing and testing interventions to mitigate the impact of age-associated shifts in the lung environment to reduce age-related pulmonary disease in the elderly.
INTRODUCTION
It is estimated that, by 2050, 16% of the world population will be older than 65 years and that the proportion of those older than 85 years of age will triple (Organization, 2019). This shift in demographics presents significant challenges to the health care system; older individuals are more susceptible to microbial infections and more likely to develop severe disease necessitating hospitalizations and admission to the intensive care unit (Stupka et al., 2009; Esme et al., 2019). Indeed, chronic lower respiratory disease are the third leading cause of death for aged individuals (Prevention, 2019). Moreover, aging is associated with severe respiratory bacterial, fungal, and viral infections (Hernandez-Vargas et al., 2014; Li et al., 2015; Kothe et al., 2008). For instance, influenza and pneumonia remain among the leading causes of mortality in the elderly (Stupka et al., 2009; Woolf et al., 2021). This pattern can be at least partially attributed to physiological and structural changes in the lungs, such as impaired mucociliary clearance and cough strength (Freitas et al., 2010; Lowery et al., 2013; Svartengren et al., 2005). Aging also leads to decreased expiratory flows, volumes, and overall gas exchange capability (Sharma and Goodwin, 2006; Garcia-Rio et al., 2004; Coffman et al., 2017; Vaz Fragoso et al., 2020; Vaz Fragoso and Gill, 2012; Stanojevic et al., 2008).
Aging leads to drastic shifts in systemic immunity, notably the accumulation of terminally differentiated T cells and the development of low-grade chronic inflammation, referred to as inflammaging (Oh et al., 2019; Onder et al., 2020; Lopez-Otin et al., 2013). These changes lead to reduced antimicrobial defenses and responses to vaccination (Ferrucci et al., 2010; Li, 2013; Wong et al., 2017). In contrast to peripheral blood, changes at mucosal sites such as the lung remain poorly understood. Most studies characterizing the impact of aging on lung immunity have used mouse models or clinical samples procured from individuals with chronic diseases (Schneider et al., 2021). These studies indicate functional deficits in alveolar macrophages (AM), including a refractory response to interferon (IFN)-γ, decreased phagocytosis, and increased basal cytokine production (Albright et al., 2016; Wong et al., 2017), as well as the presence of dysfunctional CD8+ tissue resident T cells (Trm) unable to adequately respond to viral challenges (Goplen et al., 2020). However, data from rodent studies are confounded by their specific pathogen-free status. Clinical studies have reported an increased proportion of neutrophils and alterations in AM abundance in bronchoalveolar lavage (BAL) with age (Meyer et al., 1996; Wallace et al., 1993).
The lung harbors a diverse microbial community that is primarily composed of oral microbes such as Prevotella, Fusobacterium, Streptococcus, and Corynebacterium (Dickson et al., 2015, 2017). The lung microbial community is modulated by environmental exposures (Fillion-Bertrand et al., 2019), changes in pulmonary function (Lee et al., 2019), and local immune responses (Segal et al., 2016). Changes in the lung microbiome have also been reported with multiple pulmonary disease states including asthma, idiopathic pulmonary fibrosis, and chronic obstructive pulmonary disease (COPD) (Molyneaux et al., 2014; O’Dwyer et al., 2019; Wang et al., 2016; Sverrild et al., 2017). For example, asthma is associated with an increased abundance of Haemophilius, Neisseria, and occasionally Pseudomonas and Klebsiella (Simpson et al., 2016; Loverdos et al., 2019; Durack et al., 2017; Li et al., 2017). Similarly, the lung microbiome of patients with COPD experiencing an exacerbation shows increased abundance of Haemophilius and Acinetobacter (Huang et al., 2014; Wang et al., 2016). However, shifts in the lung microbiome with aging alone remain poorly defined. A recent study that used sputum reported an increase in Firmicutes and a decrease in Proteobacteria with age (Lee et al., 2019). Additional studies have shown a loss of compartmentalization of upper respiratory tract communities with advanced age (Whelan et al., 2014; Odamaki et al., 2016; Claesson et al., 2011).
The mechanisms underlying the increased susceptibility to respiratory infections are multifactorial. Using clinical specimens is challenging owing to the difficulties associated with collecting respiratory specimens such as BAL samples from healthy humans and the challenges of disentangling the contribution of other confounding factors, such as diet and smoking. In contrast, rhesus macaque are the gold standard model for the study of multiple human respiratory pathogens such as tuberculosis, high pathogenic influenza, and severe acute respiratory syndrome coronavirus 2 (Miller et al., 2017; Munster et al., 2020; Darrah et al., 2020), given their significant genetic and physiological similarities to humans. Indeed, aged rhesus macaques recapitulate the hallmarks of immune senescence, including the depletion of circulating naive T cells, accumulation of memory T cells, lymphoid fibrosis, and chronic low-grade inflammation (Jankovic et al., 2003; Chiou et al., 2020; Haberthur et al., 2010; Thompson et al., 2019; Asquith et al., 2012). Moreover, outdoor-housed rhesus macaques experience a life-long exposure to environmental and microbial antigens and have been used to study microbiome development and stability at multiple sites (Rhoades et al., 2019, 2021b; Cadena et al., 2018; Morris et al., 2016).
Here we carried out a comprehensive analysis of the impact of age on the immunological landscape, microbial community, and pulmonary function using adults (4.2–9.9 years old) and aged (18–29 years) rhesus macaques. We report that the microbiome of aged animals becomes more heterogeneous across multiple sites. Additionally, as previously reported, a Tropheryma-dominated lung microbiome is common (approximately 50%) in rhesus macaques, especially adult animals. Since Tropheryma is known to infect AM, we stratified our immunological analysis by both host age and Tropheryma status. Aging led to an increase in the frequency of myeloid cell populations at the expense of T cells. Transcriptionally, we found that aging led to a shift from GRZMB+ to IFNG+ CD8+ T cells. Finally, IL1B transcripts in AM decreased with age, while those of the senescence gene FN1 increased. These findings improve our understanding the impact of aging on the lung and provide clues to increased respiratory infection severity with age.
RESULTS
Rhesus macaques display the circulating hallmarks of immunological aging
We profiled circulating immune cell populations for known hallmarks of aging. Frequencies of CD4+ and CD8+ T cells were comparable between age-groups, while those of B cells and natural killer (NK) cells decreased, and those of dendritic cells (DCs) and monocytes increased with age (Figures S1A and S1B). As expected, the significant loss of naive CD4+ and CD8+ T cells that was accompanied by an increase in both the central and effector memory (EM) cells in CD4+ T cells and EM cells in CD8+ T cells was observed with age (Figures S1C and S1D). Interestingly, the frequency of naive and memory B cells was increased while that of marginal zone B cells decreased with age in this cohort (Figure S1E). No differences in the frequency of DC cell subsets, non-classical CD16+ monocytes, or CD16+ NK cells were observed with age (Figures S1F-S1H). Functionally, frequency of tumor necrosis factor (TNF)α- producing monocytes and T cells was significantly increased with age in response to stimulation with bacterial and viral toll-like receptor (TLR) agonists (Figures S1I-S1K. These data indicate that our study population exhibited the canonical hallmarks of immune senescence and inflammaging (Oh et al., 2019; Onder et al., 2020; Lopez-Otin et al., 2013).
Aging leads to reduced lung function
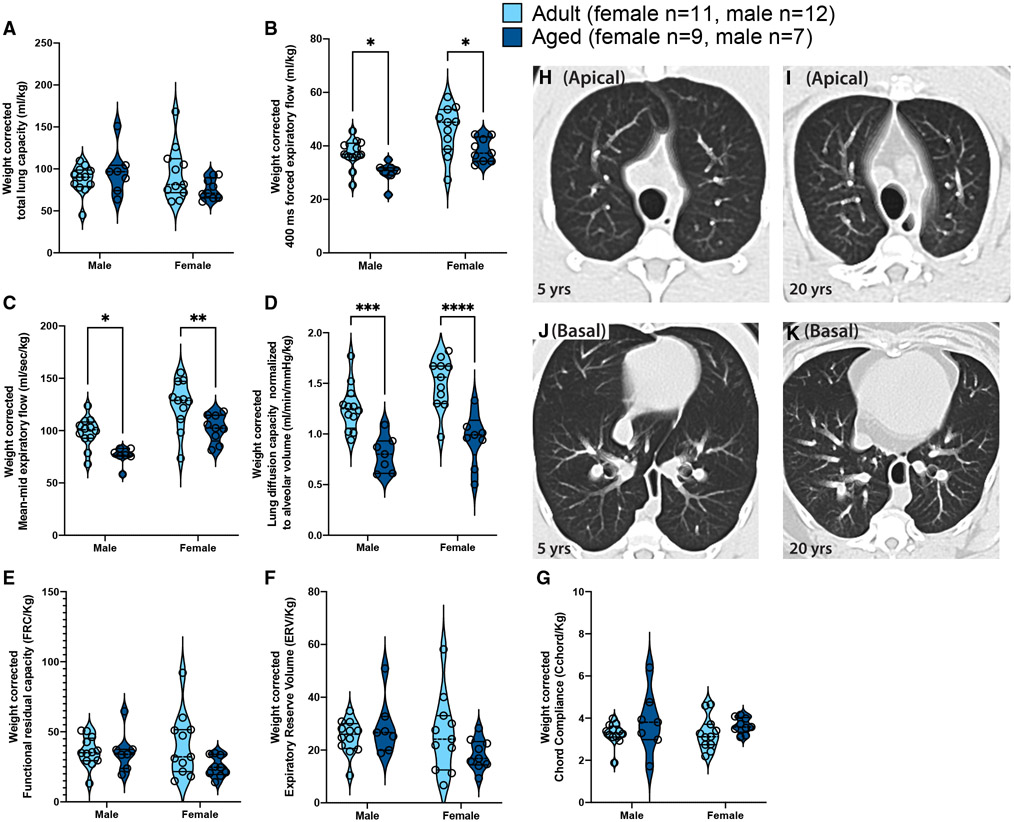
We next examined the effects of aging on lung function using a battery of lung function tests that measured lung volumes, resistance and compliance, forced expiratory flows, and volumes and gas exchange (Sekhon et al., 2001; Proskocil et al., 2005). Most measurements of lung function are highly related to height (Stanojevic et al., 2008; Beydon et al., 2007), and, for rhesus monkeys, weight is typically used as a surrogate for height, so all data are presented as absolute values and normalized for weight (Figure 1 and Table S2). The total lung capacity increased with age in males, which had significantly greater lung capacity than females (Table S2); however, when corrected for weight, no significant differences were noted with age or sex (Figure 1A). Forced expiratory flows, mean mid-expiratory flow, and gas exchange capacity were comparable between adult and aged animals, regardless of sex (Table S2); however, when corrected for weight, forced expiratory flows (Figures 1B and 1C) and gas exchange (Figure 1D) were all significantly decreased with age in males and females. No changes with aging were observed for functional residual capacity, expiratory reserve volume, or measures of static compliance (Figures 1E-1G). Thoracic computed tomography (CT) scans with controlled lung inflation in a subset of animals showed no emphysematous or fibrotic differences between adult (n = 5) versus old (n = 6) animals (Figure 1H-K). These findings recapitulate changes in human pulmonary function, supporting the use of this model.
Figure 1. Age- and sex-related changes in pulmonary function.
Pulmonary function tests were carried out in adult and aged rhesus monkeys and measures were normalized to animal weight.
(A) Total lung capacity (TLC).
(B) Forced expiratory flow in first 400 msec (FEV400) equivalent to the clinical FEV1, measure.
(C) Mean mid-expiratory flow (MMEF, average expiratory flow rate between 25 and 75% of forced vital capacity).
(D) Lung diffusion capacity normalized to alveolar volume (DLCO/VA).
(E) Functional residual capacity (FRC).
(F) Expiratory reserve volume (ERV).
(G) Chord compliance (Cchord). Impact of age and sex and the interaction of these two factors on pulmonary function was determined by two-way ANOVA with Sidak post hoc tests. Full ANOVA results can be found in Table S2 and significant (p < 0.05) post hoc comparisons are displayed in each panel (*p < 0.05, **p < 0.01, *“p < 0.001, *“*p < 0.0001).
(H–K) Apical and basal five -slice projections (from 64 slice scans) from representative breath-hold CT scans from a 20-year-old and a 5-year-old rhesus monkey.
The lung microbiome of rhesus macaques is often dominated by a single microbe
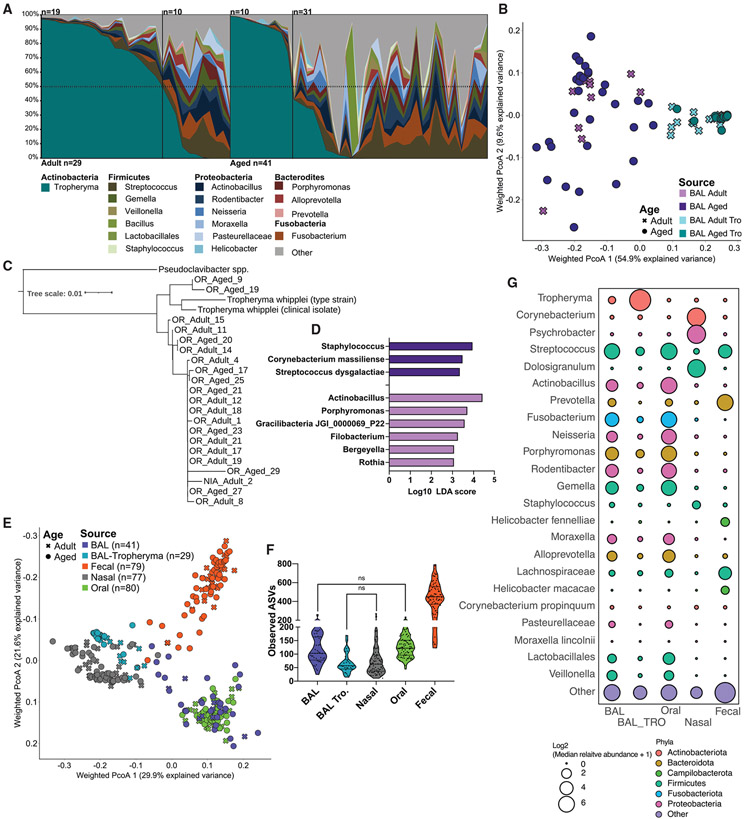
Next, we profiled the BAL microbiome using 16S rRNA gene amplicon sequencing. In total 16 amplicon sequencing variants (ASVs) were found in extraction (DNA extracted from phosphate-buffered saline [PBS]) and polymerase chain reaction (PCR)-negative controls (Figure S2A). Many of these ASVs were environmental contaminants (higher in negative controls than in samples) and were most assigned to Comamonadaceae, Stenotrophomonas, Roseobacter, and Alcaligenaceae, all of which were found in low abundance within BAL samples (Figure S2A). Additionally, some ASVs in negative controls were highly prevalent in our biological samples, suggesting cross-contamination between PCR reactions from biological samples and negative controls such as Streptococcus ASV_535, Prevotella ASV_912, and Faecalibacterium ASV_950 (Figure S2A).
Next, we determined the most abundant members of the lung microbial community. Surprisingly, we found that a single genus (Tropheryma) dominated a large subset of samples making up more than 50% of the overall community in approximately two-thirds of adult animals and approximately one-fourth of aged animals (Figure 2A). In animals without Tropheryma-dominated community we observed a high relative abundance of the genera Streptococcus, Fusobacterium, and Actinobacillus (Figure 2A). Interrogation of overall community composition using principal coordinate analysis of weighted UniFrac dissimilarity showed greater intra-group variability in non-Tropheryma-dominated communities from aged animals compared with their adult counterparts (Figure 2B and S2C). To better classify the Tropheryma, we sequenced the full-length 16S rRNA gene from a subset of samples. The majority of Tropheryma sp. sequenced from the rhesus macaques BAL samples were highly similar and on average approximately 97% similar to the T. whipplei type strain (Figure 2C). Next, we determined which bacterial taxa were differentially abundant between adult and aged animals. Only species (level 7 taxonomy) detected in at least 10% of samples at greater than 0.1% relative abundance within the sampled site were included in this analysis. No differentially abundant taxa were observed in Tropheryma-dominated animals; however, several differentially abundant species were detected in non-Tropheryma-dominated BAL communities (Figure 2D). Specifically, aging led to an increased abundance of Staphylococcus, Streptococcus dysgalactiae, and Corynebacterium massiliense and reduced abundance of Actinobacillus, Porphyromonas, and Rothia in non-Tropheryma-dominated BAL samples (Figure 2D). Of note, there was no correlation of pulmonary function with Tropheryma colonization.
Figure 2. Tropheryma colonization and composition of the rhesus macaque lung microbiome.
(A) Lung microbiome composition across all animals at the genus level. Taxa found at <1% average abundance across samples were grouped into the “other” category.
(B) Principal coordinate analysis of weighted UniFrac distance from BAL samples, colored by host age and Tropheryma status.
(C) Tropheryma phylogenetic tree generated from the alignment of full length 16S rRNA gene sequences.
(D) Differentially abundant bacterial taxa between age groups determined using LEFsE (Log10 LDA score >2) of L7 (species level) taxonomy.
(E) Principal coordinate analysis of weighted UniFrac distance generated from 16S amplicon microbiome data, colored by sampling site and shaped based on host age.
(F) Violin plots of observed ASVs across sampling sights. All comparisons not denoted as NS on the figure were significant (p < 0.05).
(G) Bubble plot of the most abundant genera of bacteria across sampling sites. The size of each bubble represents the mean relative abundance and color denotes Phyla.
The lung microbiome in humans is believed to be primarily composed of bacteria descending from the oral cavity (Dickson et al., 2015, 2017). Moreover, studies have shown a loss of tight compartmentalization with age in the upper respiratory tract (Whelan et al., 2014; Odamaki et al., 2016; Claesson et al., 2011). Therefore, we sequenced paired nasal, oral, and fecal samples from all animals. The overall composition of the microbiome at these three sites were dissimilar owing to a combination of distinct bacterial genera and differences in alpha diversity at each site (Figures 2E-2G). As expected, the number of ASVs was significantly higher in fecal samples compared with the BAL, nasal, and oral communities (Figure S2B). The oral bacterial community was dominated by Streptococcus and Fusobacterium, while the nasal microbiome was dominated by Corynebacterium, Psychrobacter, and Dolosigranulum that were largely absent at all other sites (Figure 2G). Finally, fecal samples had a high abundance of Prevotella, Helicobacter, and Streptococcus (Figure 2G).
We next determined how the microbial community of the lung related to other sampled sites. The Tropheryma-dominated lung microbiomes were phylogenetically most similar to nasal microbiomes owing to high abundance of Actinobacteria (albeit different species) and low alpha diversity rather than shared bacterial genera (Figures 2E and S2B). Conversely the non-Tropheryma lung and oral microbiomes shared several genera (Streptococcus, Actinobacillus, and Fusobacterium) (Figure 2E). We next examined which ASVs were shared and unique between lungs and other body sites (Figures S3A-S3C). Only ASVs detected within at least 10% of samples at more than 0.1 % relative abundance at the indicated sites were included in this analysis. Surprisingly, a large number of ASVs were found to be unique to both Tropheryma-dominated and non-Tropheryma BAL microbiomes on average accounting for 16.5% and 82% of the total community, respectively (Figures S3A-S3C). As expected, Tropheryma (ASV_4722, ASV_4310) was the most abundant lung-specific microbe (Figures S3B and S3C). Additional abundant lung-specific microbes included Porphyomonas canoris ASV_1309, Prevotella ASV_2044, Acinetobacter ASV_4580, and Comamonadaceae ASV_3409 (Figures S3B and S3C). A large portion of ASVs identified in BAL samples were also identified in the oral cavity (Figures S3A). However, the abundance of these shared microbes varied widely between Tropheryma-dominated and non-Tropheryma animals (Figures S2B-S2D). A large portion of the BAL microbiome of non-Tropheryma animals was composed of ASVs shared primarily with the oral microbiome, notably, Gemella ASV_3506, Porphyromonas ASV_761, and Fusobacterium ASV_1429 (Figures S3A-S3C). In contrast, a small number of ASVs were shared between the BAL and nasal or fecal microbiomes and these ASVs were found at low abundance (Figures S3A-S3C). Additionally, some ASVs were shared between the BAL and oral microbiomes, as well as at least one other site, such as Neisseria ASV_4242 and Moraxella ASV_338 (BAL, oral nasal) along with Actinobacillus ASV_1196 and Streptococcus ASV_535 (BAL, oral, and fecal) (Figures S3A). Finally, three ASVs were shared across all sites, with Streptococcus ASV_4404 being the most abundant (Figures S3A-S3C). Therefore, while a large percentage of the BAL microbiome is shared with the oral cavity, it also harbors a significant unique microbial community.
Aging did not lead to changes in observed ASVs, a measure of microbiome richness at the other sites (Figure S2B). However, aging was associated with a higher within-site dissimilarity in the nasal, fecal, and non-Tropheryma BAL samples and a decrease with age in Tropheryma BAL (Figure S2C). Next, we determined which bacterial taxa were differentially abundant between adult and aged animals at each site. Only species (level 7 taxonomy) detected in at least 10% of samples at greater than 0.1% relative abundance within the sampled site were included in this analysis (Figures S2D-S2F). An abundance of pathobiont Proteobacteria such as Escherichia-Shigella and Campylobacter corcagiensis increased in the fecal samples of aged animals, while that of Lactobacillus and Treponema was decreased (Figure S2D and Table S3). Interestingly, some species were differentially abundant in aged animals across multiple sites, including Streptococcus dysgalactiae, Corynebacterium massiliense, and Corynebacterium propinquum (Figures 2D and S2D-S2F).
Impact of aging on the pulmonary immunological landscape
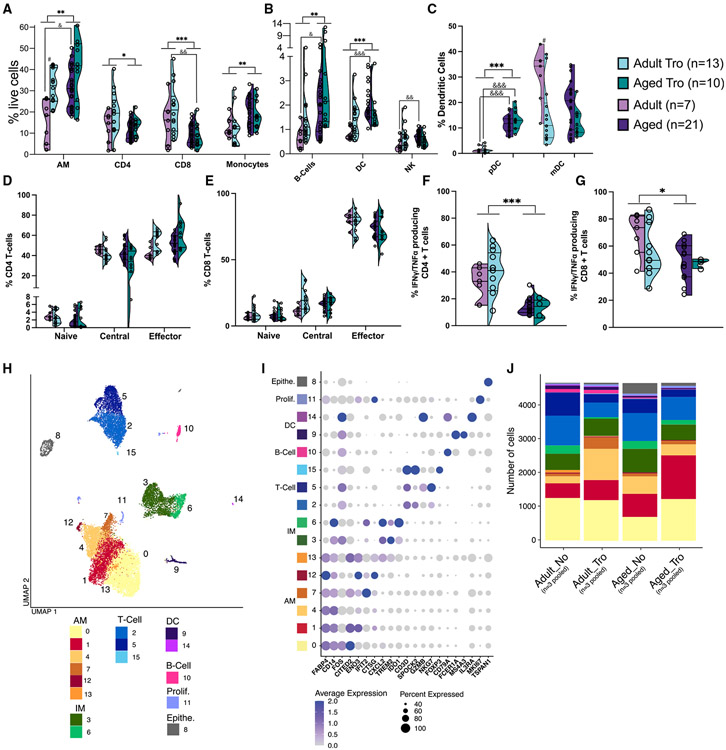
Significant differences in frequencies of AM, infiltrating macrophage (IM), T cells, B cells, NK cells, and DCs with both age and Tropheryma status were noted (Figures 3A and 3B). Specifically, the frequencies of AM, IM, DC, and B cells increased, while those of CD4+ and CD8+ T cells decreased with age regardless of Tropheryma status (Figures 3A and 3B). The increased frequency of DCs was mediated by an increase of plasmacytoid DCs (pDCs) in aged animals, while the presence of Tropheryma-dominated community was associated with decreased frequency of mDC in adult animals (Figure 3C). No differences were observed in the breakdown of CD4+ or CD8+ T cells memory subsets (Figures 3D and 3E). The frequency of INFγ/TNFα-producing CD4+ and CD8+ T cells in response to phorbol 12-myristate 13-acetate plus ionomycin (PMAi) was reduced in aged animals regardless of Tropheryma status (Figures 3F and 3G). In summary, aging results in a remodeling of the immunological landscape with a reduction in both the frequency and functional capacity of T cells while myeloid cells became dominant.
Figure 3. Age-related differences in lung immune cell frequency and function.
(A–E) Violin plots of BAL immune cell frequency measured by flow cytometry (A) major immune cell subsets, (B) CD16+ monocytes, (C) plasmacytoid and myeloid DCs, (D) CD4+ T cell subsets, and (E) CD8+ T cell subsets.
(F and G) Percent abundance of IFNγ and/or TNFα producing CD4+ and CD8+T cells after 16-h PMAi stimulation. Cytokine producing cells were measured using intra-cellular cytokine staining and flow cytometry. Comparisons were made using an unpaired t test between age-groups for each cell subset independent of Tropheryma (Tro.) colonization (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Additional comparisons were made between Adult vs. Adult Tro. (#p < 0.05), Aged vs. Aged Tro. (#p < 0.05), Adult vs. Aged (&p < 0.05), and Adult Tro. Vs. Aged Tro. (&p < 0.05).
(H) UMAP projection of all 18,712 BAL cells, colored by cluster with major immune cell subsets are annotated.
(I) Bubble plot of genes highly expressed in each cluster identified using the FindMarkers function in Seurat. Normalized expression is shown using a color scale ranging from low (white) to high (purple). Size of the bubble represents the fraction of cells within each cluster expressing the marker.
(J) Stacked bar plot depicting the absolute abundance of cell clusters within each group. Prolif. = Proliferating cell, Epithe. = epithelial cell.
To gain a deeper understanding of the impact of aging on the immunological landscape, BAL cells from 12 animals (n = 3/group with each group pooled into a single library) were profiled using scRNA-Seq. Although this pooling strategy ensured biological diversity, it precluded statistical analyses of changes in subset distribution. Dead cells and doublets were removed; then, each group was down-sampled to 4678 cells before downstream analysis. Principal component analysis and dimensional reduction using uniform manifold approximation and projection (UMAP) revealed distinct clustering into major myeloid and lymphocyte subsets (Figure 3H). Clusters were annotated based on highly expressed marker genes using Seurat’s FindMarkers function (Figures 3I and Table S4). This approach revealed changes in proportions of AM and IM, as well as major T and B cell subsets with age and Tropheryma status (Figure 3J).
Aging leads to functional shifts in BAL T cells
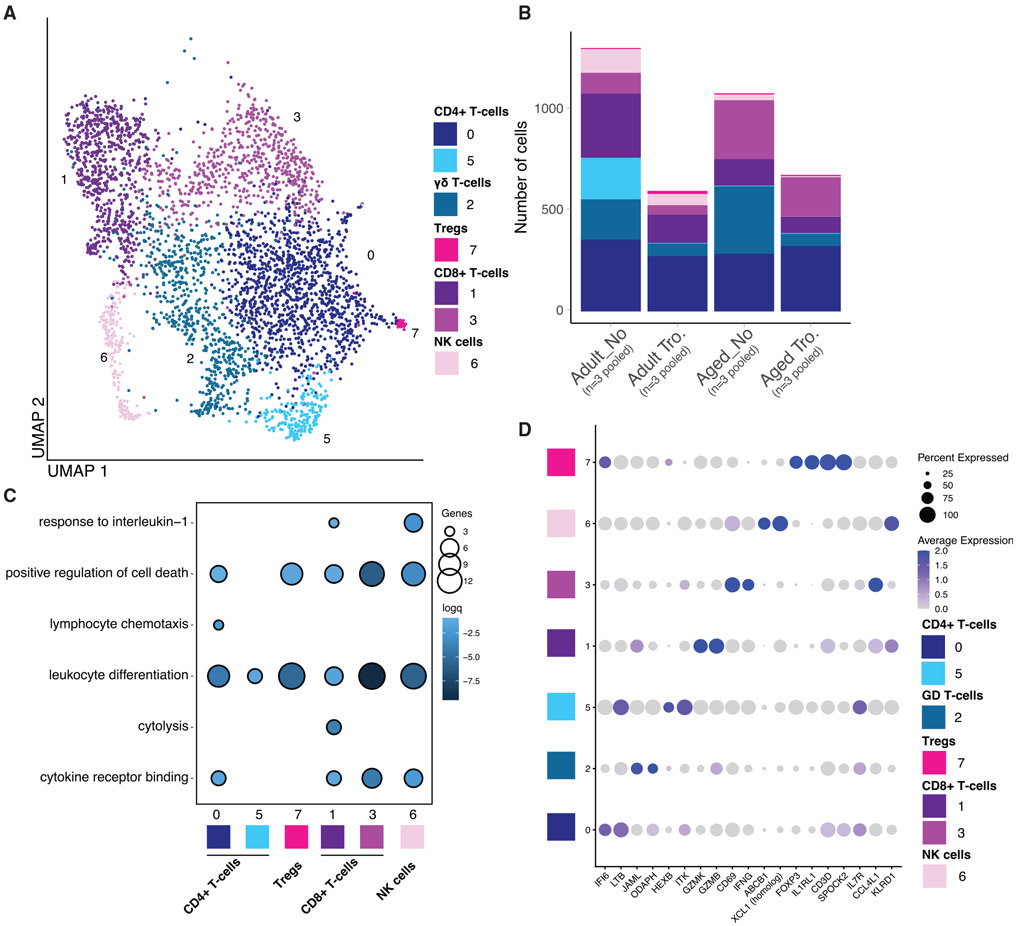
T cell subsets were re-clustered to identify changes in cell states with higher resolution (Figure 4A). This led to the identification of 7 clusters that can be partitioned into CD8+T cells (clusters 1 and 3), CD4+ T cells (clusters 0 and 5), gamma-delta T cells (γδ T cells; cluster 2), regulatory T cells (Tregs; cluster 7), and NK cells (cluster 6) (Figures 4A and 4D). Both aging and Tropheryma colonization led to a decrease in the total T and NK cells (Figure 4B). In the absence of Tropheryma colonization, this decrease was driven by a loss of ITK/IL7R CD4 T cells (cluster 5) (Figures 4B and 4D), and expansion of γδ T cells (cluster 2) and CD8 T cells (clusters 1 and 3), independent of age (Figures 4B and 4D).
Figure 4. Aging leads to a reduction of GZMK/GZMB expressing and increase of IFNG/CCL4L1 expressing CD8+ T cells.
(A) UMAP projection of T cells and NK cells re-clustered from the main UMAP. Major cell types are labeled.
(B) Stacked bar plot depicting the absolute abundance of T cells and NK cell clusters withing each group.
(C) Functional enrichment of genes differentially expressed each T cells and NK cell cluster when compared with all other clusters. Color and size of the bubble represents statistical significance and number of genes respectively. Differentially expressed genes determined using the Seurat FindMarkers function.
(D) Bubble plot identifying genes highly expressed in AM and IM cell clusters determined using the FindMarkers function in Seurat. Normalized expression is shown using a color scale ranging from low (white) to high (purple). Size of the bubble represents the fraction of cells within each cluster expressing the marker.
We next carried out functional enrichment of the marker genes that defined each cell cluster to elucidate the functional potential of each cell cluster (Figures 4C and Table S5). Marker genes that defined all CD8 T cell clusters enriched to the gene ontology (GO) term leukocyte differentiation (e.g., FOS, TNF, CD83), and this enrichment was strongest for IFNG expressing CD8 T cells (clusters) (Figure 4C). Interestingly, all gene markers except for those defining cluster 5 (which was primarily found in non-Tropheryma adult animals) enriched to the GO term positive regulation of cell death (e.g., CTLA4, CD40LG, and NTRK1) (Figure 4C). Genes that defined cluster 1 CD8 T cells, which decreased with age, enriched to both cytolysis (e.g., GZMH, GZMA, GZMB, and GZMM) and response to IL-1 (e.g., IL1B, CCL5, CCL4L1), while markers of cluster 3 CD8 T cells, which increased with age, enriched to cytokine receptor binding (e.g., CSF1, IFNG, and IL21) (Figure 4C), suggesting a potential shift in functional capacity of CD8 T cells with age.
We next determined how aging impacts the transcriptional profiles of CD4, CDS, and γδ T cells using pseudobulk analysis with DEGs between adult and aged animals within each group (based on Tropheryma status) defined as those with a log2FC of more than 0.25 and a false discovery rate p < 0.05 (Figures S4A-S4E). Within BAL samples obtained from animals without a Tropheryma-dominated community, CD4 T cell DEGs enriched predominantly to GO terms associated with chemotaxis (e.g., CCR6 and CCL4L1) and apoptosis (e.g., GZMB), while γδ T cells DEGs enriched predominantly to GO terms associated with migration (e.g., CCL5) and cytokine signaling (e.g., IFNG and LTB). CD4 T cell DEGs from Tropheryma-dominated BAL samples enriched to activation (e.g., CXCL13 and CD40LG), apoptosis (e.g., CTSD and EGLN3), response to bacterium (e.g., LYZ and IL1B), and type I IFN signaling pathway (e.g., IRF1 and ISG15) (Figures S4A and S4B). The γδ T cells DEGs from Tropheryma-dominated BAL samples enriched to GO terms associated with metabolism (e.g., SLC2A1, SLC2A3, and NR4A3), cell adhesion, and signaling (e.g., TMIGD2 and ITGA6) (Figures S4A and S4C).
Within CD8 T cells, DEGs enriched predominantly to GO terms immune activation, antiviral immunity, and apoptosis (Figures S4D). Overall, the genes associated with immune activation were significantly upregulated with age, regardless of Tropheryma status, notably IFNG, TNF, and NFKBID (Figures S4E). Genes important for the response to oxidative stress such as SOD2 and ID3 were downregulated with age (Figures S4E). These data suggest that, independent of Tropheryma colonization, aging leads to shifts in BAL CD8 T cell functional potential, while Tropheryma colonization has a bigger impact on gene expression within CD4 and γδ T cells.
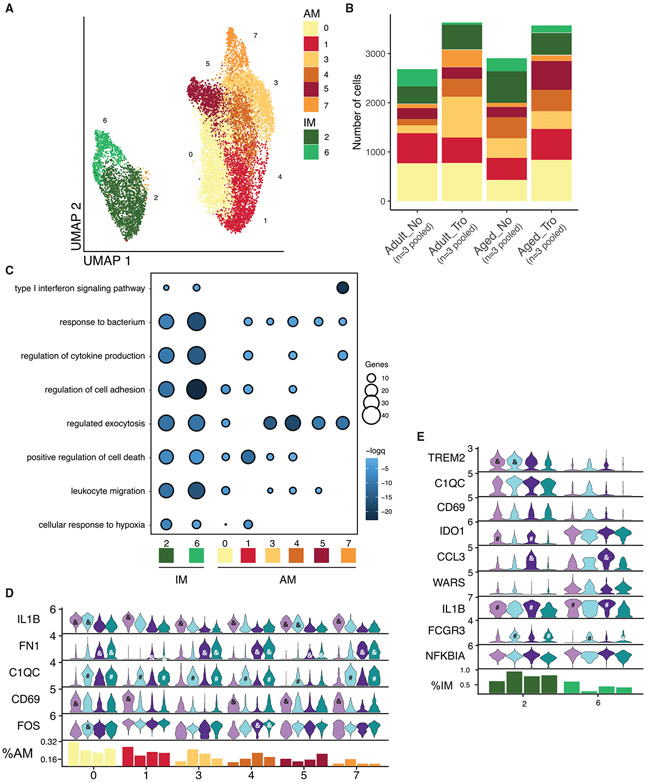
Aging and Tropheryma status impact the transcriptional profile of macrophages
In both humans and rhesus macaque, AM and IM are the most abundant immune cells in the lung. Re-clustering of the myeloid cells (Figures 5A and 5B) revealed 6 cell states for AM (clusters 0, 1, 3, 4, 5, and 7 expressing high levels of FABP4 and MARCO) (Figure 5A and S5A). Within the AM subsets, clusters 0, 1, and 4 express markers consistent with a self-renewing regulatory population (RGCC, SOCS1, and ENPP1) (Figure S5A). In contrast, AM cluster 7 expressed high levels of IFN-stimulated genes (ISGs) IFIT2 and MX1 suggesting this subset may play a role in anti-viral defense (Figure S5A). Tropheryma colonization led to increased abundance of AM, especially clusters 3 and 4 in adult animals and cluster 5 in aged animals (Figure 5B). Additionally, we observed age- and Tropheryma-dependent transcriptional patterns across clusters. For example, the expression of the inflammatory mediator IL1B was decreased with age across all clusters while that of the pro-fibrotic gene FN1 was increased (Figure 5D). Moreover, the expression of the complement factor C1QC was higher in AM from Tropheryma-colonized animals across clusters (Figure 5D). Finally, the expression of CD69, a marker of early activation, was highest in adult animals that lack a Tropheryma-dominated community across all clusters, while FOS, a suppressor of the inflammatory response was upregulated with age and Tropheryma status (Figure 5D). Within IM, cluster 2 was most abundant across all groups and expressed high levels of the M2-like marker TREM2, complement gene C1QC, and CPVL (a gene that plays a critical role in major histocompatibility complex I antigen presentation). In contrast, cluster 6 expressed high levels of M1-like markers ID01 and CCL3 (Figures 5B, 5E, and S5A). An abundance of cells in cluster 6 was reduced with aging and Tropheryma colonization (Figure 5B), while FCGR3 (CD16) was upregulated in both Tropheryma colonized groups in both IM clusters (Figure 5E).
Figure 5. Aging leads to increased fibronectin 1 expression in AM and shift in expression of IL1B from AM to IM.
(A) UMAP projection of AM and IM cells re-clustered from the main UMAP. Major cell types are labeled.
(B) Stacked bar plot depicting the absolute abundance of AM and IM cell clusters within each group.
(C) Functional enrichment of genes differentially expressed by each AM and IM cell cluster when compared with all other clusters. Size and color of the bubble represents statistical significance and number of genes respectively. Differentially expressed genes determined using the Seurat FindMarkers function.
(D and E) Violin plots depicting the expression of key genes between groups across AM (D) and IM (E) cell clusters. Bar graph at the bottom of the figure represents the relative abundance of each cluster with the indicated group as a percentage of the total AM (D) and IM (E) cells. For (D) and (E), significance was determined for each gene within each cell clusters using the non-parametric Kruskal–Wallis ANOVA with Dunn’s multiple comparison test for post hoc pairwise comparison when the initial ANOVA was significant. Post hoc significance between Adult vs. Adult Tro. (#p < 0.05), Aged vs. Aged Tro. (#p < 0.05), Adult vs. Aged (&p < 0.05), and Adult Tro. Vs. Aged Tro. (&p < 0.05) are show in each panel.
The functional enrichment of the marker genes was carried out to infer differences in functional potential between the various cell states (Figures 5C and Table S6). Genes that define IM enriched more significantly to most GO terms (e.g., response to bacterium, regulation of cytokine production, and leukocyte migration) (Figure 5C). Marker genes of AM Cluster 1 enriched more significantly to GO term positive regulation of cell death, while clusters 3–7 enriched more significantly to regulated exocytosis, and cluster 7 genes to type 1 IFN signaling pathway (Figure 5C).
From a transcriptional standpoint, aging in the absence of Tropheryma colonization resulted in a more profound shift (Figure S5B). IM DEGs enriched to GO terms, such as leukocyte activation involved in immune response and response to bacterium (Figure S5B). Specifically, inflammatory genes such as TNF, S100A8, IL1B, RGS1, and NFKBIA were significantly upregulated with age, while genes that encode anti-inflammatory proteins (CHIT1 and FTL) and play a role in endocytosis (CTSB, CTSD, and SPP1) were downregulated (Figure S5C). Interestingly, expression of CXCL1, CD63, and CCL17 within IM showed opposite but significant age-related expression changes based on the Tropheryma status (Figure S5C).
AM DEGs enriched to GO terms such as leukocyte degranulation and response to bacterium (Figure S5B). Multiple pro-phagocytic and inflammatory genes (e.g., IL1B, CXCL1, CCL4L1, IDO1, CCL2, and SOD2) were downregulated, while genes important for tissue homeostasis (e.g., LGALS3, GRN, LCN2, MRC1, and GPNMB) were upregulated with aging (Figure S5D). In AM from animals with a Tropheryma-dominated BAL community, genes associated with apoptotic signaling (e.g., BAG3 and ATF3) were upregulated, coupled with a downregulation of ISGs (e.g., IFI27, IGHM and ISG15) (Figure S5D). Finally, expression of alarmins S100A8, and S100A9 was increased across all BAL macrophages with age (Figure S5C and S5D). Together, these data suggest that aging leads to the upregulation of inflammatory and pro-fibrotic genes, and downregulation of genes important for tissue homeostasis and antigen uptake and processing.
DISCUSSION
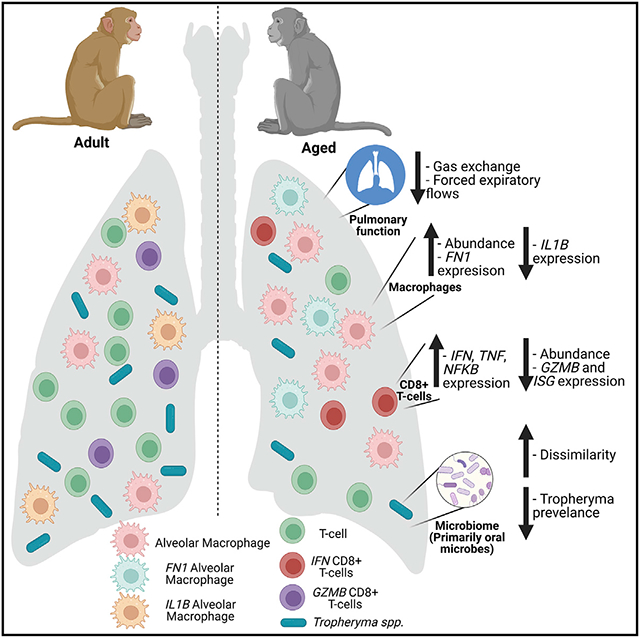
While systemic markers of immunosenescence and inflammaging have been well explored, there is a paucity of studies exploring patterns of immunological aging in the lungs owing to challenges associated with collecting samples from healthy humans. In this study we sought to determine the impact of healthy aging on the functional, microbial, and immunological landscapes of the lung using the rhesus macaque model, a highly reliable pre-clinical model, using BAL samples from adult and aged macaques and a multi-pronged approach. Rhesus macaques recapitulate the hallmarks of systemic immune senescence described in humans (Asquith et al., 2012; Jankovic et al., 2003) and suffer from more severe respiratory disease, including from severe acute respiratory syndrome coronavirus 2 and influenza viruses (Josset et al., 2012; Urano et al., 2021). We report similar declines as seen in humans (Sharma and Goodwin, 2006; Garcia-Rio et al., 2004; Coffman et al., 2017; Vaz Fragoso et al., 2020; Vaz Fragoso and Gill, 2012; Stanojevic et al., 2008), including hallmark decreases in forced expiratory volumes, forced expiratory flows, and gas exchange. These differences do not seem to derive from emphysematous changes or increased fibrosis; no such changes were seen on CT scans. This is not, however, definitive; no histologic analysis was performed and emphysematous changes have previously been reported in aged rhesus macaques (Uno, 1997). This decrease in respiratory capacity and alveolar function, when combined with infection, could increase the likelihood of clinically significant disease owing to poor clearance of pathogens and warrants further investigation. The decrease in alveolar gas exchange capacity increases the likelihood of clinically significant decreases in blood oxygen levels with respiratory infection, leading to increased tissue damage.
As described in earlier studies (Cadena et al., 2018; Morris et al., 2016), several animals had a Tropheryma spp.-dominated BAL microbial community. An earlier longitudinal analysis showed a majority of the animals had Tropheryma-dominated lung microbiome prior and up to 1-year after simian HIV infection, suggesting that Tropheryma is a common member of the healthy captive macaque lung microbiome (Morris et al., 2016). In humans, Tropheryma is most associated with Whipple’s disease, an infection in which T. whipplei infects macrophages of the small intestine (Petrides et al., 1998; Mahnel and Marth, 2004). However, Tropheryma spp. species can also be found in the human lung without any clear clinical symptoms, and their abundance in increased in HIV+ individuals (Lozupone et al., 2013; Qin et al., 2016). Since the lung is one of the least sampled human tissues for microbiome composition, the prevalence of Tropheryma may be higher than currently estimated (Dickson et al., 2016). Additionally, gut and vaginal microbiomes of rhesus macaques are more similar to those of humans in the developing world than Western countries (Rhoades et al., 2019, 2021a; Yatsunenko et al., 2012; Vargas-Robles et al., 2020). It is possible that Tropheryma lung colonization among humans could also vary in this manner.
Most animals in our study (41/70) had diverse lung microbial communities with little or no Tropheryma and were dominated by microbes typically found in the oral cavity, which has been well established as the primary source of the lung microbio me in humans (Bassis et al., 2015; Dickson et al., 2017). While it was initially hypothesized that the presence of oral microbes in the lungs was due to contamination during the bronchoscopy procedure, more in-depth studies have shown that this contamination is negligible and that oral microbes infiltrate the lungs primarily by micro-aspiration (Dickson et al., 2015). Most microbes that colonize the lungs of rhesus macaques (i.e., Streptococcus, Neisseria, Prevoteiia, Fusobacterium) are also found in humans (Bassis et al., 2015; Dickson et al., 2015, 2017), with some exceptions such as Rodentibacter, which is commonly found in the oral microbiome of rodents, but not humans (Matejkova et al., 2020).
Additionally, we show that aging leads to increased intragroup heterogeneity of microbial communities across multiple sites (fecal, nasal, and non-Tropheryma BAL) as previously observed in the gut microbiome of humans (Wilmanski et al., 2021). The fact that this pattern is seen in aged macaques in the absence of significant differences in diet and exposure to environmental factors strongly suggest other age-mediated changes are important (e.g., changes in immunological control and tissue elasticity). In agreement with human studies (Claesson et al., 2011; Odamaki et al., 2016; Kong et al., 2019), we also observed an increased relative abundance of proteobacteria pathobionts (Escherichia-Shigella and Campylobacter corcagiensis) in the gut microbiome with aging. Multiple age-related differentially abundant bacterial taxa within the lungs, including Streptococcus dysgalatiae, Corynebacterium massiliense, and Staphylococcus, were identified in aged animals. Conversely, the lung microbiome of non-Tropheryma adult animals had an increased abundance of Actinobacillus, Rothia, and Porphyromonas, among others. This observation suggests that aging leads to an environment that is more permissive to colonization by specific microbes.
Despite the role of AM as the primary orchestrators of the pulmonary immune system in both health and disease (Baharom et al., 2017; Hashimoto et al., 2013), little is known about the impact of healthy aging on these cells. The loss or impaired function of AM can lead to increased pathology and mortality after viral infection in mice (Hussell and Bell, 2014; Purnama et al., 2014). In contrast with data from inbred mice (Wong et al., 2017), aging led to an increased abundance of AM in rhesus macaques. A recent study in humans showed no changes in the abundance of airway macrophages (Byrne et al., 2020), possibly owing to differences in sampling procedures. Previous studies have found that pulmonary neutrophils increase with age and are a significant contributor to age-related pulmonary inflammation (Meyer et al., 1998); however, we were unable to assess changes in neutrophil frequencies as we used cryopreserved BAL samples.
We observed an increased expression of the complement factor C1QC across all AM clusters obtained from animals colonized by Tropheryma. Complement plays an important role in linking the innate and adaptive immune systems in pulmonary disease (Pandya and Wilkes, 2014); however, it is unclear whether Tropheryma infects AM and if complement is a mechanism by which this bacterium is controlled. Independent of Tropheryma colonization, AM from aged individuals had heightened expression of Fibronectin 1 (FN1). The expression of FN1 expression and other extracellular matrix proteins that play a role in tissue repair is tightly regulated in the lungs and under homeostatic conditions (Balestrini and Niklason, 2015). Overexpression of these factors may lead to the development of idiopathic pulmonary fibrosis (Herrera et al., 2018; Genovese and Karsdal, 2016) and acts as a predictor of resistance to chemotherapies in some cancers (Jerhammar et al., 2010; Yang et al., 2013). Transcripts of IL1B were decreased within AM with age, which may indicate reduced immunosurveillance by AM in aged animals. In contrast with AM, expression of IL1B, CCL3, and TNF in IM increased with age in non-Tropheryma animals. The lack of TNF induction in IM from Tropheryma-dominated samples is in line with reports of reduced TNF expression by bystander macrophages in T. whipplei-infected gut samples (Ben Azzouz et al., 2018).
The increased abundance of AM and IM was accompanied by a corresponding decrease in CD4+ and CD8+ T cells in the BAL. Moreover, the frequency of cytokine-producing T cells in response to polyclonal stimulation decreased with age. Transcriptionally, CD8+ T cells in aged animals expressed lower levels of granzymes (GZMK and GZMB), suggesting a reduced cytolytic capacity, a key factor for clearing viral pathogens (Kohlmeier et al., 2010; Salti et al., 2011). In contrast, CD8+ T cells in aged animals expressed high levels of IFNG and their marker genes are predicted to play a role in differentiation and apoptosis. IFNγ does not play a prominent role in CD8+ Trmmediated respiratory viral clearance (Durbin et al., 2002; Rutigliano and Graham, 2004; Wiley et al., 2001), but may play a role in immunopathology (Wiley et al., 2001). Indeed, previous studies of influenza in mice have found that viral infection causes a distinct accumulation of CD8+ Trm in aged animals (Goplen et al., 2021). However, these cells lack key effector functions (Goplen et al., 2020). Our findings are consistent with a transcriptional shift indicative of reduced anti-microbial function with aging.
We observed the most pronounced age-related shift in the transcriptional profiles of BAL CD4+T cells in Tropheryma-dominated animals, including the upregulation of genes associated with anti-bacterial responses (LYZ and CXCL13). Since Tropheryma colonization was not associated with clinical disease, these observations suggest that CD4 T cells may play a role in maintaining this homeostasis. Indeed, colonization of the lung by Tropheryma in humans is most common in HIV+ individuals who have reduced and dysfunctional CD4+ T cells (Lozupone et al., 2013), and Whipple’s disease (an infection of the small intestine by T. whipplei) is associated with a reduced CD4+ Th1 response (Moos et al., 2006). CD4+ T cells from the BAL non-Tropheryma animals showed limited age-related transcriptional changes with the exception of increased expression of CCL4L1. CCL4L1 is subsequently translated into macrophage inflammatory protein 1β (MIP-1β) a key chemoattractant of monocytes. In the absence of infection, increased T cell production of MIP-1β increases with age may be associated with aberrant inflammation and accumulation of myeloid cells (Chen et al., 2003). Finally, we identified limited age-related transcriptional changes in BAL γδ T cells that was depleted in both adult and aged Tropheryma-dominated animals. Given the cross-sectional nature of our study it is not possible to determine if this depletion is a cause or consequence of Tropheryma colonization.
Key findings from this study indicate that aging leads to a more inflammatory environment with compromised anti-microbial function. This change combined with the decrease in pulmonary function and gas exchange could increase the risk of clinically significant respiratory infections with aging. Independent of age, Tropheryma is a prevalent and dominant colonizer of the rhesus macaque lung and had a significant impact on pulmonary immune composition and function, despite the absence of respiratory symptoms. Given these findings, future studies should determine how differences in the lung microbiome may impact the host response to infection. Specifically in macaques, which serve as a gold standard model for multiple respiratory pathogens, the impact of Tropheryma colonization should be assessed. These data pave the way for studies aimed at designing and testing interventions to reduce or reverse age-associated shifts in the lung environment to reduce age-related pulmonary disease in the elderly.
Limitations of the study
The current study has multiple limitations, most notably the use of pooled samples for single cell RNA sequencing, precluding us from assessing the statistical significance of these findings. Future studies that use cell hashing technologies cab overcome this pitfall. Additionally, our analysis is limited to BAL. Observations reported in this manuscript could be validated in future terminal studies using samples from throughout the lung to better understand the impact of healthy aging.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ilhem Messaoudi (Ilhem.messaoudi@uky.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The 16S rRNA gene amplicon, and host transcriptional data have been deposited at the NCBI Sequence Read Archive and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 510™ anti-human CD20 Antibody | Biolegend | Cat#302340; RRID:AB_2561941; Lot# 13327426; Clone: 2H7 |
| CD8β-ECD | Beckman Coulter | Cat#6607123; RRID: N/A; Clone: 2ST8.5H7 |
| PerCP-Cyanine5.5 Anti-Human CD4 | Ton bo biosciences | Cat#65-0048-CM01; RRID:AB_2621878; Lot#C004807121365C; Clone: OKT4 |
| Brilliant Violet 711™ anti-human CD27 Antibody | Biolegend | Cat#356430; RRID:AB_2650751; Lot#B307472; Clone: M-T271 |
| PE Anti-Human CD28 | Ton bo biosciences | Cat#50-0289-CM01; RRID:AB_2687623; Lot#C027906251320C; Clone: CD28.2 |
| APC anti-human CD95 (Fas) Antibody | Biolegend | Cat#305612; RRID:AB_314550; Lot#B266759; Clone: DX2 |
| FITC Mouse Anti-Human CD3ε | BD Biosciences | Cat#556611; RRID:AB_396484; Lot#0286772; Clone: SP34 |
| FITC anti-human CD20 Antibody | Biolegend | Cat#302304; RRID: RRID:AB_314252; Lot#B289592; Clone: 2H7 |
| APC/Cyanine7 anti-human HLA-DR Antibody | Biolegend | Cat#307618; RRID:AB_493586; Lot#B295253; Clone: L243 |
| APC anti-human CD14 Antibody | Biolegend | Cat#301808; RRID:AB_314190; Lot#B259538; Clone M5E2 |
| PE/Cyanine7 anti-human CD8a Antibody | Biolegend | Cat#301012; RRID:AB_314130; Lot#B267334; Clone: RPA-T8 |
| CD11c Monoclonal Antibody PE-eFluor 610 | Invitrogen | Cat#61-0116-42; RRID:AB_2574532; Lot#2331068; Clone: 3.9 |
| Pacific Blue™ anti-human CD16 Antibody | Biolegend | Cat#302032; RRID:AB_2104003; Lot#B288858; Clone: 3G8 |
| PE Mouse Anti-Human CD206 | BD biosciences | Cat#555954; RRID:AB_396250; Lot#0065122; Clone: 19.2 |
| PerCP/Cyanine5.5 anti-human CD123 Antibody | Biolegend | Cat#306016; RRID:AB_2264693; Lot#0065122; Clone: 6H6 |
| Brilliant Violet 711™ anti-human CD28 Antibody | Biolegend | Cat#302948; RRID: AB_2616857; Lot#B314478; Clone: 28.2 |
| Pacific Blue™ anti-human CD20 Antibody | Biolegend | Cat#302328; RRID: AB_1595435; Lot#B285815; Clone: 2H7 |
| Alexa Fluor® 700 anti-human CD14 Antibody | Biolegend | Cat#301822; RRID: AB_493747; Lot#B315064; M5E2 |
| PE-Cyanine7 IFN gamma Monoclonal Antibody | eBioscience | Cat#25-7319-82; RRID:AB_469682; Lot# 21141213; Clone: 4S.B3 |
| APC TNF alpha Monoclonal Antibody | eBioscience | Cat#17-7349-82; RRID: AB_469512; Lot#2101117; Clone: MAb11 |
| PE Rat Anti-Human IL-6 | BD Biosciences | Cat#551473; RRID: AB_2127751; Lot#5292975; Clone: MQ2-6a3 |
| Ghost Dye™ Violet 510 | Tonbo Biosciences | Cat#13-0870-T100; RRID: N/A; Lot#D0870040521133 |
| Ghost Dye™ Violet 450 | Tonbo Biosciences | Cat#13-0863-T100; RRID: N/A; Lot#D0863071019133 |
| Ghost Dye™ Violet 710 | Tonbo Biosciences | Cat#13-0871-T100; RRID: N/A; Lot#D0871083018133 |
| Goat Anti-Human IgD-BIOT | Southern Biotech | Cat#2030-08; RRID: N/A; Lot#I1010-N071B |
| Streptavidin, Alexa Fluor™ 700 conjugate | Invitrogen | Cat#S21383; RRID: N/A; Lot#2126789 |
| Fc Receptor Binding Inhibitor Polyclonal Antibody | Invitrogen | Cat#14-9165-42; RRID: N/A; Lot#2251850 |
| True-Stain Monocyte Blocker™ | Bioiegend | Cat#426103; RRID: N/A |
| Biological samples | ||
| Rhesus macaque biological fluids (whole blood, whole BAL) | ONPRC | N/A |
| Rhesus macaque cells (PBMCs, BAL cells) | ONPRC | N/A |
| Rhesus macaque microbiome swabs (nasal, oral, fecal) | ONPRC | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Phorbol 12-myristate 13-acetate (PMA) | Sigma Aldrich | Cat#P8139-1 MG |
| Ionomycin | Sigma Aldrich | Cat#I9657-1MG |
| brefeldin A | Biolegend | Cat#420601 |
| Phosphate Buffered Saline (PBS) | Global Life | Cat#SH30028.02 |
| Fetal Bovine Serum (FBS) | Omega Scientific | Cat#FB-02; Lot#881991 |
| Bovine Serum Albumine (BSA) | Gemini | Cat#700-106P; Lot#C82H81J |
| Dimethyl sulfoxide (DMSO) | Sigma Aldrich | Cat#d4540 |
| GoTaq master mix | Promega | Cat#M71333; Lot#000428913 |
| nuclease-free H20 | Promega | Cat#P1193 |
| ODN 2216 - TLR9 ligand | InvivoGen | Cat#tlrl-2216-1; Lot#3603-39T |
| ssRNA40/LyoVec™ | InvivoGen | Cat#tlrl-lrna40; Lot#A40-39-03 |
| Imiquimod (R837) | InvivoGen | Cat#tlrl-imq; Lot#IMQ-36-02A |
| FSL-1 | InvivoGen | Cat#tlrl-fsl; Lot#FSL-37-04 |
| Pam3CSK4 | InvivoGen | Cat#tlrl-pms; Lot#PMS-35-03 |
| LPS | InvivoGen | Cat#tlrl-b5lps |
| Flow Cytometry Perm Buffer (10x) | Tonbo Biosciences | Cat#TNB-1213-L150; Lot#B1213062817TN |
| Foxp3/Transcription Factor Fix/Perm Diluent (1x) | Ton bo Biosciences | Cat#TNB-1022-L160; Lot#B1022062415 |
| Foxp3/Transcription Factor Fix/Perm Concentrate (4x) | Tonbo Biosciences | Cat#TNB-1020-L050; Lot#B10200322420TN |
| Fixation Buffer | Biolegend | Cat#420801; Lot#B331895 |
| Intracellular Staining Perm Wash Buffer (10x) | Biolegend | Cat#421002; Lot#B32433 |
| Critical commercial assays | ||
| DNeasy Powersoil Pro kit | Qiagen | Cat#47016; Lot#169022930 |
| MinElute 96 UF PCR Purification Kit | Qiagen | Cat#28053; Lot#154032872 |
| MiSeq Reagent Kit v3 600 cycles | Illumina | Cat#MS-102-3003 |
| Chromium Single Cell 3′ Feature Barcoding Library Kit v3.1 chemistry | 10X Genomics | Cat#1000079 |
| Deposited data | ||
| Raw sequencing data | This Study | Sequence Read Archive- BioProject SRA: PRJNA800766 (https://www.ncbi.nlm.nih.gov) |
| Oligonucleotides | ||
| 16S rRNA gene 515F forward primer 5′-GTGYCAGCMGCCGCGGTAA-3′ | IDT | (Parada et al., 2016) |
| 16S rRNA gene 806R reverse primer 5′-GGACTACNVGGGTWTCTAAT-3′ | IDT | (Parada et al., 2016) |
| 16S rRNA gene B8F forward primer | IDT | (Edwards et al., 1989) |
| 16S rRNA gene 1492R reverse primer | IDT | (Edwards et al., 1989) |
| Software and algorithms | ||
| Quantitative Insights Into Microbial Ecology 2 (QIIME2)(2019.10) Built in tools: Dada2, mafft, sklearn, FastTree 2 | (Bolyen et al., 2019) | https://qiime2.org |
| DNAsubway | CyVerse and DNA Learning Center | https://dnasubway.cyverse.org |
| iTOL (Interactive Tree of Life) | (Letunic and Bork, 2021) | https://itol.embl.de/ |
| FlowJo | BD Life Sciences | https://www.flowjo.com/ |
| Cell Ranger Single-Cell Software Suite v4.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Seurat v4.0 | (Hao et al., 2021) | https://satijalab.org/seurat/ |
| R software (v4.0.2) R packages: vegan (v2.5-7), SystemPipeR (v1.26.3), edgeR (V3.34.0), ggplot2 (v3.3.3) | R Foundation | https://www.r-project.org |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Samples were obtained from a total of 82 rhesus macaques and a full breakdown of animal data and experiment can be found in Table S1. All rhesus macaque studies were overseen and approved by the Oregon Health & Science University/Oregon National Primate Research Center (ONPRC) and the NIA Intramural Institutional Animal Care and Use Committees per the National Institutes of Health guide for the care and use of laboratory animals. Animals were housed per the standards established by the US Federal Animal Welfare Act and The Guide for the Care and Use of Laboratory Animals. Animals observed at ONPRC were pre-screened to establish their suitability for experimental procedures. All animals 18 years and older receive physical examinations every 6 months and are also tested for normality in complete blood count, blood chemistry, and a fecal occult test. An electronic animal records system documents an animals long-term health record and identifies any criteria that invalidates its use. Whole blood samples were collected in EDTA vacutainer tubes. Peripheral blood mononuclear cells (PBMC) were obtained by standard density gradient centrifugation over the blood separation polymer Ficoll in Sepmate tubes. BAL cells were isolated from total BAL by centrifugation at 1200 rcf for 15 min. PBMC and BAL cells were cryopreserved using 10% DMSO/fetal bovine serum (FBS) and Mr. Frosty Freezing containers (ThermoFisher Scientific) at −80°C and then transferred to a cryogenic unit until analysis. Whole BAL for microbiome analysis was collected before the isolation of BAL cells. All swabs (nasal, oral, and fecal) were collected at the time of BAL.
METHOD DETAILS
Pulmonary function testing
For pulmonary function testing, rhesus macaques were anesthetized with ketamine and dexmedetomidine, and then intubated. Pulmonary function tests were performed using the Buxco pulmonary function apparatus from DSI/Harvard Bioscience as previously described (Sekhon et al., 2001; Proskocil et al., 2005). Gas exchange (lung diffusion capacity) and alveolar volume were measured using carbon monoxide (CO) diffusion after the methods of Castillo et al. (2006) and Fallica et al. (2011). The reference gas containing 0.5% CO, 0.5% neon, 21% O2, and 78% N2 was injected into the lungs, withdrawn after 4 s and gas levels measured by gas chromatography. All tests were performed a minimum of three times. After testing was complete, animals were extubated, monitored and returned to their home cages. Thoracic CT scanning with controlled lung inflation was performed in a subset of animals (5 young, 6 old) on a GE Optima CT 660 with a 64-slice detector and analyzed by a cardiothoracic radiologist.
16S amplicon sequencing
Total DNA was extracted from 400 μL BAL fluid or swabs (fecal, oral, or nasal) using the DNeasy Powersoil Pro Kit (Qiagen, Valencia, CA). The hypervariable V4-V5 region of the 16S rRNA gene was amplified using PCR primers (515F/806R with the forward primers including a 12-bp barcode) (Parada et al., 2016). PCR reactions were conducted in triplicates and contained 12.5 μL GoTaq master mix, 9.5 μL nuclease-free H2O, 1 μL template DNA, and 1 μL 10 μmol primer mix. Thermal cycling parameters were 94°C for 5 min, 35 cycles of 94°C for 20 s, 50°C for 20 s, and 72°C for 30 s, followed by 72°C for 5 min. PCR products were purified using a MinElute 96 UF PCR Purification Kit (Qiagen). Libraries were sequenced (2 × 300 bases) using an Illumina MiSeq.
Raw FASTQ 16S rRNA gene amplicon sequences were uploaded and processed using the QIIME2 analysis pipeline (Bolyen et al., 2019). Briefly, sequences were demultiplexed and the quality filtered using DADA2 (Callahan et al., 2016), which filters chimeric sequences and generates an amplicon sequence variant (ASV) table equivalent to an operational taxonomic unit table at 100% sequence similarity. Sequence variants were then aligned using MAFFT (Katoh and Standley, 2013) and a phylogenetic tree was constructed using FastTree2 (Price et al., 2010). Taxonomy was assigned to sequence variants using q2-feature-classifier against the SILVA database (release 138) (Quast et al., 2013). To prevent sequencing depth bias samples were rarified to 20,000 sequences per sample before alpha and beta diversity analysis. QIIME 2 was also used to generate the following alpha diversity metrics: richness (as observed ASV), Shannon evenness, and phylogenetic diversity. Beta diversity was estimated in QIIME 2 using weighted and unweighted UniFrac distances (Lozupone et al., 2011).
Full-length 16S sequencing and tree building
For Tropheryma-dominated animals, the full-length 16S rRNA gene was amplified using the B8F-1492R primers (Edwards et al., 1989). Cleaned PCR reactions were then sequenced bi-directionally using Sanger di-deoxy sequencing. Paired sequences were then merged, trimmed, and aligned against known Tropheryma 16S rRNA gene sequences using the DNAsubway pipeline (https://dnasubway.cyverse.org). The final alignment was exported as a newick file and plotted in iTOL (itol.embl.de) (Letunic and Bork, 2021).
Flow cytometry
To profile adaptive immune cell subsets 1 × 106 BAL cells or PBMC were stained using antibodies against CD4, CD8b, CD95, CD28, CD20, CD27, and IgD to delineate naïve and memory CD4 and CD8 T cells, as well as CD20 B cell subsets. T cells were then divided into naive (CD28+, CD95−; naive), central/transitional memory (CD284 CD954), and EM cells (CD28− CD95+). A second tube of 1 × 106 BAL cells or PBMC was stained using antibodies against: CD3, CD20, HLA-DR, CD14, CD11c, CD123, CD16, CD8a, and CD206 (BAL only) to delineate AMs (CD3− CD20− CD206+) IMs (CD3− CD20− CD206- CD14 + HLA-DR+), DC (CD3− CD20- CD14− HLA-DR+), and NK (CD3− CD20− CD14− HLA-DR− CD8a+) cell subsets. Macrophages were further subdivided into classical (CD16−) and non-classical macrophages (CD16+). DCs were further subdivided into myeloid DCs (CD123− CD11c+) and pDC (CD123+CD11c−). All flow cytometry samples were acquired using Attune NxT (Life Technologies, Carlsbad, CA) and analyzed using FlowJo (TreeStar, Ashland, OR).
Stimulation and ICS
BAL cells (5 × 105/well) were thawed and incubated for 16 h in the absence (unstimulated) or presence (stimulated) of 5 ng/mL PMA and 1 μg/mL ionomycin (Sigma-Aldrich, St. Louis, MO). PBMC cells (5 × 105/well) were thawed and incubated for 16 h in the absence (unstimulated) or presence (stimulated) of PMAi (5 ng/mL PMA and 1 μg/mL Ionomycin) or bacterial TLR antigen cocktail (lipopolysaccharide, FSL-1, Pam3CSK4) or viral TLR antigen cocktail (single-stranded RNA, Imiquimod, ODN2216). All cells were incubated with brefeldin A at 37°C in a humidified incubator (5% CO2). After incubation cell were stained using antibodies against CD4, CD8b, CD28, CD20, CD14, and HLA-DR to delineate total T cell and monocyte populations. Cells were next fixed and permeabilized and stained with IFNγ, TNFα, and IL-6.
Single cell RNA library preparation
Cryopreserved BAL cells from a subset of animals (n = 3/group for Adult No Tropheryma [Tro.], Adult Tro., Aged No Tro., and Aged Tro.) were thawed. Samples were washed three times in ice-cold PBS supplemented with 2% FBS and sorted on the fluorescence-activated cell sourcing Aria Fusion (BD Biosciences) with Ghost Dye Red 710 (Tonbo Biosciences) for dead cell exclusion. Live cells were counted in triplicates on a TC20 Automated Cell Counter (BioRad) and pooled in groups of three based on host age and Tropheryma status. Pooled cells were resuspended in ice-cold PBS with 0.04% BSA in a final concentration of 1200 cells/μL. Single cell suspensions were then immediately loaded on the 10X Genomics Chromium Controller with a loading target of 17,600 cells. Libraries were generated per manufacturer’s recommendation Chromium Single Cell 3′ Feature Barcoding Library Kit using the v3.1 chemistry (10X Genomics, Pleasanton, CA). Libraries were sequenced on Illumina NovaSeq with a sequencing target of 30,000 reads per cell RNA library and 2000 reads per cell hashtag-oligo barcode library.
Single cell RNA sequencing data analysis
Raw reads were aligned and quantified using the Cell Ranger Single-Cell Software Suite (version 4.0, 10X Genomics) against the Mmul_8 rhesus macaque reference genome using the STAR aligner. Downstream processing of aligned reads was performed using Seurat (version 4.0). Droplets with ambient RNA (cells fewer than 200 detected genes) and dying cells (cells with more than 25% total mitochondrial gene expression) were excluded during the initial QC. Data objects from all groups were integrated using Seurat (Hao et al., 2021). Data normalization and variance stabilization was performed on the integrated object using the SCTransform function, where a regularized negative binomial regression corrected for differential effects of mitochondrial cell cycle gene expression levels. The R package doublet finder (McGinnis et al., 2019) along with manual curation was used to remove suspected doublet cells based on the expression of multiple lineage marker. Dimensional reduction was performed using RunPCA function to obtain the first 30 principal components followed by clustering using the FindClusters function in Seurat. Clusters were visualized using the UMAP algorithm as implemented by Seurat’s runUMAPfunction. Cell types were assigned to individual clusters using FindMarkers function with a fold change cutoff of at least 0.4. Clusters from two major cell types (T/NK cells and Macrophages) were further subsetted from the total cells and re-clustered to identify minor subsets within those groups.
QUANTIFICATION AND STATISTICAL ANALYSIS
A differential expression analysis was performed using MAST using default settings in Seurat. All disease comparisons were performed relative to healthy donors from corresponding age groups. Only statistically significant genes (Fold change cutoff ≥ 1.5; adjusted p value ≤ 0.05) were included in downstream analysis. Significance of two group comparisons (i.e., all Adult vs. all Aged) were measured using the non-parametric Mann-Whitney U test. Significance of 3+ group comparison was measured using the non-parametric Kruskal-Wallis ANOVA with Dunn’s multiple comparison test for post hoc pairwise comparison when the initial ANOVA was significant. The LEfSe algorithm was used to identify differentially abundant taxa and pathways between groups with a logarithmic Linear discriminant analysis score cutoff of 2 (Segata et al., 2011).
Supplementary Material
Highlights.
Rhesus macaques display the functional hallmarks of healthy pulmonary aging
Aging increases frequency of myeloid cells at the expense of T cells in the lungs
Aging shifts from GRZMB+ to IFNG+ CD8+ T cells and fewer IL1B+ lung macrophages
Tropheryma spp. is highly prevalent in the rhesus macaque lung microbiome
ACKNOWLEDGMENTS
This work was supported, in part, by the intramural program of the National Institute on Aging, NIH (R01AI152258-02). NSR was supported by an NIH training grant (T32AI007319-32). The authors thank the ONPRC Division of Comparative Medicine and veterinary staff for their expert animal care. The authors also thank the veterinary technicians at the NIA nonhuman primate core for sample collection and support from the Division of Veterinary Resources animal care team (P51-OD011092). The graphical abstract was generated using graphics from BioRender.com.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j-celrep.2022.110725.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Albright JM, Dunn RC, Shults JA, Boe DM, Afshar M, and Kovacs EJ (2016). Advanced age alters monocyte and macrophage responses. Antioxid. Redox Signal 25, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asquith M, Haberthur K, Brown M, Engelmann F, Murphy A, Al-Mahdi Z, and Messaoudi I (2012). Age-dependent changes in innate immune phenotype and function in rhesus macaques (Macaca mulatta). Pathobiol Aging Age Relat. Dis 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharom F, Rankin G, Blomberg A, and Smed-Sorensen A (2017). Human lung mononuclear phagocytes in health and disease. Front. Immunol 8, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini JL, and Niklason LE (2015). Extracellular matrix as a driver for lung regeneration. Ann. Biomed. Eng 43, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, and Huffnagle GB (2015). Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 6, e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Azzouz E, Boumaza A, Mezouar S, Bardou M, Carlini F, Picard C, Raoult D, Mege JL, and Desnues B (2018). Tropheryma whipplei increases expression of human leukocyte antigen-G on monocytes to reduce tumor necrosis factor and promote bacterial replication. Gastroenterology 155, 1553–1563. [DOI] [PubMed] [Google Scholar]
- Beydon N, Davis SD, Lombardi E, Allen JL, Arets HG, Aurora P, Bis- gaard H, Davis GM, Ducharme FM, Eigen H, et al. (2007). an official American thoracic society/European respiratory society statement: pulmonary function testing in preschool children. Am. J. Respir. Crit. Care Med 175, 1304–1345. [DOI] [PubMed] [Google Scholar]
- Bolyen E, Rideout RJ, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander H, Aim EJ, Arumugam M,Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AJ, Powell JE, O’sullivan BJ, Ogger PP, Hoffland A, Cook J, Bonner KL, Hewitt RJ, Wolf S, Ghai P, et al. (2020). Dynamics of human monocytes and airway macrophages during healthy aging and after transplant. J. Exp. Med 3, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena AM, Ma Y, Ding T, Bryant M, Maiello P, Geber A, Lin PL, Flynn JL, and Ghedin E (2018). Profiling the airway in the macaque model of tuberculosis reveals variable microbial dysbiosis and alteration of community structure. Microbiome 6, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJ, and Holmes SP (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Llapur CJ, Martinez T, Kisling J, Williams-Nkomo T, Coates C, and Tepper RS (2006). Measurement of single breath-hold carbon monoxide diffusing capacity in healthy infants and toddlers. Pediatr. Pulmonol 41, 544–550. [DOI] [PubMed] [Google Scholar]
- Chen J, Mo R, Lescure PA, Misek DE, Hanash S, Rochford R, Hobbs M, and Yung RL (2003). Aging is associated with increased T-cell chemokine expression in C57BL/6 mice. J. Gerontol. A. Biol. Sci. Med. Sci 58, 975–983. [DOI] [PubMed] [Google Scholar]
- Chiou KL, Montague MJ, Goldman EA, Watowich MM, Sams SN, Song J, Horvath JE, Sterner KN, Ruiz-Lambides AV, Martinez MI, et al. (2020). Rhesus macaques as a tractable physiological model of human ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci 375, 20190612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O’sullivan O, Greene-Diniz R, De Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U S A 108. 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman KE, Carlson AR, Miller AD, Johnson BD, and Taylor BJ (2017). The effect of aging and cardiorespiratory fitness on the lung diffusing capacity response to exercise in healthy humans. J. Appl. Physiol 122, 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah PA, Zeppa JJ, Maiello P, Hackney JA, Wadsworth MH 2nd, Hughes TK, Pokkali S, Swanson PA 2nd, Grant NL, Rodgers MA, et al. (2020). Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature 577, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Freeman CM, Mccloskey L, Beck JM, Huffnagle GB, and Curtis JL (2015). Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann. Am. Thorac. Soc 12, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Freeman CM, Mccloskey L, Falkowski NR, Huffnagle GB, and Curtis JL (2017). Bacterial topography of the healthy human lower respiratory tract. mBio 8, e02287–e02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Martinez FJ, and Hufihagle GB (2016). The microbiome and the respiratory tract. Annu. Rev. Physiol 78, 481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, Dyer AM, Israel E, Kraft M, Martin RJ, et al. (2017). Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol 140, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Johnson TR, Durbin RK, Mertz SE, Morotti RA, Peebles RS, and Graham BS (2002). The role of IFN in respiratory syncytial virus pathogenesis. J. Immunol 168, 2944–2952. [DOI] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M,and Bottger EC(1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esme M, Topeli A, Yavuz BB, and Akova M (2019). Infections in the elderly critically-ill patients. Front. Med. (Lausanne) 6, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallica J, Das S, Horton M, and Mitzner W (2011). Application of carbon monoxide diffusing capacity in the mouse lung. J. Appl. Physiol 110, 1455–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, Semba RD, Guralnik JM, Ershler WB, Bandinelli S, Patel KV, Sun K, Woodman RC, Andrews NC, Cotter RJ, et al. (2010). Proinflammatory state, hepcidin, and anemia in older persons. Blood 115, 3810–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillion-Bertrand G, Dickson RP, Boivin R, Lavoie JP, Huffnagle GB, and Leclere M (2019). Lung microbiome is influenced by the environment and asthmatic status in an equine model of asthma. Am. J. Respir. Cell Mol. Biol 60, 189–197. [DOI] [PubMed] [Google Scholar]
- Freitas FS, Ibiapina CC, Alvim CG, Britto RR, and Parreira VF (2010). Relationship between cough strength and functional level in elderly. Rev. Bras. Fisioter 14, 470–476. [PubMed] [Google Scholar]
- Garcia-Rio F,, Pino JM, Dorgham A, Alonso A, and Villamor J (2004). Spirometric reference equations for European females and males aged 65-85 yrs. Eur. Respir. J 24, 397–405. [DOI] [PubMed] [Google Scholar]
- Genovese F, and Karsdal MA (2016). Protein degradation fragments as diagnostic and prognostic biomarkers of connective tissue diseases: understanding the extracellular matrix message and implication for current and future serological biomarkers. Expert Rev. Proteomics 13, 213–225. [DOI] [PubMed] [Google Scholar]
- Goplen NP, Cheon IS, and Sun J (2021). Age-related dynamics of lung-resident memory CD8(+) T cells in the age of COVID-19. Front. Immunol 12, 636118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goplen NP, Wu Y,Son YM, Li C, Wang Z, Cheon IS, Jiang L,Zhu B, Ayasoufi K, Chini EN, et al. (2020). Tissue-resident CD8(+) T cells drive age-associated chronic lung sequelae after viral pneumonia. Sci. Immunol 5, eabc4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberthur K, Engelman F, Barron A, and Messaoudi I (2010). Immune senescence in aged nonhuman primates. Exp. Gerontol 45, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd,Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, et al. (2013). Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vargas EA, Wilk E, Canini L, Toapanta FR, Binder SC, Uvarovskii A, Ross TM, Guzman CA, Perelson AS, and Meyer-Hermann M (2014). Effects of aging on influenza virus infection dynamics. J. Virol 88, 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera J, Henke CA, and Bitterman PB (2018). Extracellular matrix as a driver of progressive fibrosis. J. Clin. Invest 128, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Sethi S, Murphy T, Nariya S, Boushey HA, and Lynch SV (2014). Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J. Clin. Microbiol 52, 2813–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T, and Bell TJ (2014). Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol 14, 81–93. [DOI] [PubMed] [Google Scholar]
- Jankovic V, Messaoudi I, and Nikoiich-Zugich J (2003). Phenotypic and functional T-cell aging in rhesus macaques (Macaca mulatta): differential behavior of CD4 and CD8 subsets. Blood 102, 3244–3251. [DOI] [PubMed] [Google Scholar]
- Jerhammar F, Ceder R, Garvin S, Grenman R, Grafstrom RC, and Ro- berg K (2010). Fibronectin 1 is a potential biomarker for radioresistance in head and neck squamous cell carcinoma. Cancer Biol. Ther 10, 1244–1251. [DOI] [PubMed] [Google Scholar]
- Josset L, Engelmann F, Haberthur K, Kelly S, Park B, Kawoaka Y, Garcia-Sastre A, Katze MG, and Messaoudi I (2012). Increased viral loads and exacerbated innate host responses in aged macaques infected with the 2009 pandemic H1N1 influenza A virus. J. Virol 86, 11115–11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier JE, Cookenham T, Roberts AD, Miller SC, and Woodland DL (2010). Type I interferons regulate cytolytic activity of memory CD8(+) T cells in the lung airways during respiratory virus challenge. Immunity 33, 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Deng F, Li Y, and Zhao J (2019). Identification of gut microbiome signatures associated with longevity provides a promising modulation target for healthy aging. Gut Microbes 10, 210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe H, Bauer T, Marre R, Suttorp N, Welte T, and Dalhoff K; Competence Network for Community-Acquired Pneumonia Study Group (2008). Outcome of community-acquired pneumonia: influence of age, residence status and antimicrobial treatment. Eur. Respir. J 32, 139–146. [DOI] [PubMed] [Google Scholar]
- Lee SY, Mac Aogain M, Fam KD, Chia KL, Binte Mohamed Ali NA, Yap MMC, Yap EPH, Chotirmall SH, and Lim CL (2019). Airway microbiome composition correlates with lung function and arterial stiffness in an age-dependent manner. PLoS One 14, e0225636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2021). Interactive tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Qiu R, Yang Z, Li J, Chung KF, Zhong N, and Zhang Q (2017). Sputum microbiota in severe asthma patients: relationship to eosinophilic inflammation. Respir. Med 131, 192–198. [DOI] [PubMed] [Google Scholar]
- Li W (2013). Phagocyte dysfunction, tissue aging and degeneration. Ageing Res. Rev 12, 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ding C, and Yin S (2015). Severe pneumonia in the elderly: a multivariate analysis of risk factors. Int. J. Clin. Exp. Med 8, 12463–12475. [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loverdos K, Bellos G, Kokolatou L, Vasileiadis I, Giamarellos E, Pecchiari M, Koulouris N, Koutsoukou A, and Rovina N (2019). Lung microbiome in asthma: current perspectives. J. Clin. Med 8, 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery EM, Brubaker AL, Kuhlmann E, and Kovacs EJ (2013). The aging lung. Clin. Interv. Aging 8, 1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, Mitreva M, Abubucker S, Martin J, Yao G, et al. (2013). Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am. J. Respir. Crit. Care Med 187, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, and Knight R (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnel R, and Marth T (2004). Progress, problems, and perspectives in diagnosis and treatment of Whipple’s disease. Clin. Exp. Med 4, 39–43. [DOI] [PubMed] [Google Scholar]
- Matejkova T, Hajkova P, Stopkova R, Stanko M, Martin JF, Kreisinger J, and Stopka P (2020). Oral and vaginal microbiota in selected field mice of the genus Apodemus: a wild population study. Sci. Rep 10, 13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KC, Ershler W, Rosenthal NS, Lu XG, and Peterson K (1996). Immune dysregulation in the aging human lung. Am. J. Respir. Crit. Care Med 153, 1072–1079. [DOI] [PubMed] [Google Scholar]
- Meyer KC, Rosenthal NS, Soergel P, and Peterson K (1998). Neutrophils and low-grade inflammation in the seemingly normal aging human lung. Mech. Ageing Dev 104, 169–181. [DOI] [PubMed] [Google Scholar]
- Miller LA, Royer CM, Pinkerton KE, and Schelegle ES (2017). Nonhuman primate models of respiratory disease: past, present, and future. ILAR J. 58, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, Murphy E, Johnston SL, Schwartz DA, Wells AU, et al. (2014). The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med 190, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos V, Kunkel D, Marth T, Feurle GE, Lascoia B, Ignatius R,Zeitz M, and Schneider T (2006). Reduced peripheral and mucosal Tropheryma whipplei-specific Th1 response in patients with Whipple’s disease. J. Immunol 177, 2015–2022. [DOI] [PubMed] [Google Scholar]
- Morris A, Paulson JN, Talukder H, Tipton L, Kling H, Cui L, Fitch A, Pop M, Norris KA, and Ghedin E (2016). Longitudinal analysis of the lung microbiota of cynomolgous macaques during long-term SHIV infection. Microbiome 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Feldmann F, Williamson BN, Van Doremalen N, Perez-Perez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, et al. (2020). Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, and Osawa R (2016). Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 16, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer DN, Ashley SL, Gurczynski SJ, Xia M, Wilke C, Falkowski NR, Norman KC, Arnold KB, Huffnagle GB, Salisbury ML, et al. (2019). Lung microbiota Contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am. J. Respir. Crit. Care Med 199, 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Lee JK, and Shin OS (2019). Aging and the immune system: the impact of immunosenescence on viral infection, immunity and vaccine immunogenicity. Immune Netw. 19, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G, Rezza G, and Brusaferro S (2020). Case-fatality rate and Characteristics of patients dying in relation to COVID-19 in Italy. JAMA 323, 1775–1776. [DOI] [PubMed] [Google Scholar]
- Organization, W.H. (2019). World Population Prospects 2019: Highlights. [Google Scholar]
- Pandya PH, and Wilkes DS (2014). Complement system in lung disease. Am. J. Respir. Cell Mol. Biol 51, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada AE, Needham DM, and Fuhrman JA (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol 18, 1403–1414. [DOI] [PubMed] [Google Scholar]
- Petrides PE, Muller-Hocker J, Fredricks DN, and Relman DA (1998). PCR analysis of T. whippelii DNA in a case of Whipple’s disease: effect of antibiotics and correlation with histology. Am. J. Gastroenterol 93, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Prevention, C.F.D.C.A. (2019). WISQARS LEADING CAUSES OF DEATH. [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proskocil BJ, Sekhon HS, Clark JA, Lupo SL, Jia Y, Hull WM, Whitsett JA, Starcher BC, and Spindel ER (2005). Vitamin C prevents the effects of prenatal nicotine on pulmonary function in newborn monkeys. Am. J. Respir. Crit. Care Med 171, 1032–1039. [DOI] [PubMed] [Google Scholar]
- Purnama C, Ng SL,Tetlak P, Setiagani YA, Kandasamy M, Baalasubramanian S, Karjalainen K, and Ruedl C (2014). Transient ablation of alveolar macrophages leads to massive pathology of influenza infection without affecting cellular adaptive immunity. Eur. J. Immunol 44, 2003–2012. [DOI] [PubMed] [Google Scholar]
- Qin S, Clausen E, Lucht L, Michael H, Beck JM, Curtis JL, Freeman BM, and Morris A (2016). Presence of Tropheryma whipplei in different body sites in a cohort of healthy subjects. Am. J. Respir. Crit. Care Med 194, 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quasi C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glockner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades NS, Hendrickson SM, Gerken DR, Martinez K, Slayden OD, Slifka MK, and Messaoudi I (2021a). Longitudinal profiling of the macaque vaginal microbiome reveals similarities to diverse human vaginal communities, mSystems 6, e01322–e01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades NS, Hendrickson SM, Prongay K, Haertel A, Gill L, Edwards RA, Garzel L, Slifka MK, and Messaoudi I (2021b). Growth faltering regardless of chronic diarrhea is associated with mucosal immune dysfunction and microbial dysbiosis in the gut lumen. Mucosal Immunol. 14, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades N, Barr T, Hendrickson S, Prongay K, Haertel A, Gill L, Garzel L, Whiteson K, Slifka M, and Messaoudi I (2019). Maturation of the infant rhesus macaque gut microbiome and its role in the development of diarrheal disease. Genome Biol. 20, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutigliano JA, and Graham BS (2004). Prolonged production of TNF-alpha exacerbates illness during respiratory syncytial virus infection. J. Immunol 173, 3408–3417. [DOI] [PubMed] [Google Scholar]
- Salti SM, Hammelev EM, Grewal JL, Reddy ST, Zemple SJ, Grossman WJ, Grayson MH, and Verbsky JW (2011). Granzyme B regulates antiviral CD8+ T cell responses. J. Immunol 187, 6301–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, Rowe JH, Garcia-De-Alba C, Kim CF, Sharpe AH, and Haigis MC (2021). The aging lung: physiology, disease, and immunity. Cell 184, 1990–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal LN, Clemente JC,Tsay JC, Koralov SB, Keller BC, Wu BG, Li Y, Shen N, Ghedin E, Morris A, et al. (2016). Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol 1, 16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon HS, Keller JA, Benowitz NL, and Spindel ER (2001). Prenatal nicotine exposure alters pulmonary function in newborn rhesus monkeys. Am. J. Respir. Crit. Care Med 164, 989–994. [DOI] [PubMed] [Google Scholar]
- Sharma G, and Goodwin J (2006). Effect of aging on respiratory system physiology and immunology. Clin. Interv. Aging 1, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, Hugenholtz P, Willner D, and Gibson PG (2016). Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur. Respir. J 47, 792–800. [DOI] [PubMed] [Google Scholar]
- Stanojevic S, Wade A, Stocks J, Hankinson J, Coates AL, Pan H, Rosenthal M, Corey M, Lebecque P, and Cole TJ (2008). Reference ranges for spirometry across all ages: a new approach. Am. J. Respir. Crit. Care Med 177, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupka JE, Mortensen EM, Anzueto A, and Restrepo M.l. (2009). Community-acquired pneumonia in elderly patients. Aging health 5, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svartengren M, Falk R, and Philipson K (2005). Long-term clearance from small airways decreases with age. Eur. Respir. J 26, 609–615. [DOI] [PubMed] [Google Scholar]
- Sverrild A, Kiilerich P, Brejnrod A, Pedersen R, Porsbjerg C, Bergqvist A, Erjefalt JS, Kristiansen K, and Backer V (2017). Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J. Allergy Clin. Immunol. 140, 407–417.e11. [DOI] [PubMed] [Google Scholar]
- Thompson HL, Smithey MJ, Uhrlaub JL, Jeftic I, Jergovic M, White SE, Currier N, Lang AM, Okoye A, Park B, et al. (2019). Lymph nodes as barriers to T-cell rejuvenation in aging mice and nonhuman primates. Aging Cell 18, e12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno H (1997). Age-related pathology and biosenescent markers in captive rhesus macaques. Age (Omaha) 20, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano E, Okamura T, Ono C, Ueno S, Nagata S, Kamada H, Higuchi M, Furukawa M, Kamitani W, Matsuura Y, et al. (2021). COVID-19 cynomolgus macaque model reflecting human COVID-19 pathological conditions. Proc. Natl. Acad. Sci. U S A 118, e2104847118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Robles D, Morales N, Rodriguez I, Nieves T, Godoy-Vitorino F, Alcaraz LD, Perez ME, Ravel J, Forney LJ, and Dominguez-Bello MG (2020). Changes in the vaginal microbiota across a gradient of urbanization. Sci. Rep 10, 12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz Fragoso CA, and Gill TM (2012). Respiratory impairment and the aging lung: a novel paradigm for assessing pulmonary function. J. Gerontol. A. Biol. Sci. Med. Sci 67, 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz Fragoso CA, Rochester CL, Mcavay GJ, lannone L, and Leo-Summers LS (2020). Diffusing capacity in normal-for-age spirometry and spirometric impairments, using reference equations from the global lung function initiative. Respir. Med 170, 106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace WA, Gillooly M, and Lamb D (1993). Age related increase in the intra-alveolar macrophage population of non-smokers. Thorax 48, 668–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Bafadhel M, Haidar K, Spivak A, Mayhew D, Miller BE, Tal-Singer R, Johnston SL, Ramsheh MY, Barer MR, et al. (2016). Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J 47, 1082–1092. [DOI] [PubMed] [Google Scholar]
- Whelan FJ, Verschoor CP, Stearns JC, Rossi L, Luinstra K, Loeb M, Smieja M, Johnstone J, Surette MG, and Bowdish DM (2014). The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann. Am. Thorac. Soc 11, 513–521. [DOI] [PubMed] [Google Scholar]
- Wiley JA, Cerwenka A, Harkema JR, Dutton RW, and Harmsen AG (2001). Production of interferon-gamma by influenza hemagglutinin-specific CD8 effector T cells influences the development of pulmonary immunopathology. Am. J. Pathol 158, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski T, Diener C, Rappaport N, Patwardhan S, Wiedrick J, Lapidus J, Earls JC, Zimmer A, Glusman G, Robinson M, et al. (2021). Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab 3, 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CK, Smith CA, Sakamoto K, Kaminski N, Koff JL, and Goldstein DR (2017). Aging impairs alveolar macrophage phagocytosis and increases influenza-induced mortality in mice. J. Immunol 199, 1060–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf SH, Chapman DA, and Lee JH (2021). COVID-19 as the leading cause of death in the United States. JAMA 325, 123–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li H, Hou S, Hu B, Liu J, and Wang J (2013). The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One 8, e65309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene amplicon, and host transcriptional data have been deposited at the NCBI Sequence Read Archive and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 510™ anti-human CD20 Antibody | Biolegend | Cat#302340; RRID:AB_2561941; Lot# 13327426; Clone: 2H7 |
| CD8β-ECD | Beckman Coulter | Cat#6607123; RRID: N/A; Clone: 2ST8.5H7 |
| PerCP-Cyanine5.5 Anti-Human CD4 | Ton bo biosciences | Cat#65-0048-CM01; RRID:AB_2621878; Lot#C004807121365C; Clone: OKT4 |
| Brilliant Violet 711™ anti-human CD27 Antibody | Biolegend | Cat#356430; RRID:AB_2650751; Lot#B307472; Clone: M-T271 |
| PE Anti-Human CD28 | Ton bo biosciences | Cat#50-0289-CM01; RRID:AB_2687623; Lot#C027906251320C; Clone: CD28.2 |
| APC anti-human CD95 (Fas) Antibody | Biolegend | Cat#305612; RRID:AB_314550; Lot#B266759; Clone: DX2 |
| FITC Mouse Anti-Human CD3ε | BD Biosciences | Cat#556611; RRID:AB_396484; Lot#0286772; Clone: SP34 |
| FITC anti-human CD20 Antibody | Biolegend | Cat#302304; RRID: RRID:AB_314252; Lot#B289592; Clone: 2H7 |
| APC/Cyanine7 anti-human HLA-DR Antibody | Biolegend | Cat#307618; RRID:AB_493586; Lot#B295253; Clone: L243 |
| APC anti-human CD14 Antibody | Biolegend | Cat#301808; RRID:AB_314190; Lot#B259538; Clone M5E2 |
| PE/Cyanine7 anti-human CD8a Antibody | Biolegend | Cat#301012; RRID:AB_314130; Lot#B267334; Clone: RPA-T8 |
| CD11c Monoclonal Antibody PE-eFluor 610 | Invitrogen | Cat#61-0116-42; RRID:AB_2574532; Lot#2331068; Clone: 3.9 |
| Pacific Blue™ anti-human CD16 Antibody | Biolegend | Cat#302032; RRID:AB_2104003; Lot#B288858; Clone: 3G8 |
| PE Mouse Anti-Human CD206 | BD biosciences | Cat#555954; RRID:AB_396250; Lot#0065122; Clone: 19.2 |
| PerCP/Cyanine5.5 anti-human CD123 Antibody | Biolegend | Cat#306016; RRID:AB_2264693; Lot#0065122; Clone: 6H6 |
| Brilliant Violet 711™ anti-human CD28 Antibody | Biolegend | Cat#302948; RRID: AB_2616857; Lot#B314478; Clone: 28.2 |
| Pacific Blue™ anti-human CD20 Antibody | Biolegend | Cat#302328; RRID: AB_1595435; Lot#B285815; Clone: 2H7 |
| Alexa Fluor® 700 anti-human CD14 Antibody | Biolegend | Cat#301822; RRID: AB_493747; Lot#B315064; M5E2 |
| PE-Cyanine7 IFN gamma Monoclonal Antibody | eBioscience | Cat#25-7319-82; RRID:AB_469682; Lot# 21141213; Clone: 4S.B3 |
| APC TNF alpha Monoclonal Antibody | eBioscience | Cat#17-7349-82; RRID: AB_469512; Lot#2101117; Clone: MAb11 |
| PE Rat Anti-Human IL-6 | BD Biosciences | Cat#551473; RRID: AB_2127751; Lot#5292975; Clone: MQ2-6a3 |
| Ghost Dye™ Violet 510 | Tonbo Biosciences | Cat#13-0870-T100; RRID: N/A; Lot#D0870040521133 |
| Ghost Dye™ Violet 450 | Tonbo Biosciences | Cat#13-0863-T100; RRID: N/A; Lot#D0863071019133 |
| Ghost Dye™ Violet 710 | Tonbo Biosciences | Cat#13-0871-T100; RRID: N/A; Lot#D0871083018133 |
| Goat Anti-Human IgD-BIOT | Southern Biotech | Cat#2030-08; RRID: N/A; Lot#I1010-N071B |
| Streptavidin, Alexa Fluor™ 700 conjugate | Invitrogen | Cat#S21383; RRID: N/A; Lot#2126789 |
| Fc Receptor Binding Inhibitor Polyclonal Antibody | Invitrogen | Cat#14-9165-42; RRID: N/A; Lot#2251850 |
| True-Stain Monocyte Blocker™ | Bioiegend | Cat#426103; RRID: N/A |
| Biological samples | ||
| Rhesus macaque biological fluids (whole blood, whole BAL) | ONPRC | N/A |
| Rhesus macaque cells (PBMCs, BAL cells) | ONPRC | N/A |
| Rhesus macaque microbiome swabs (nasal, oral, fecal) | ONPRC | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Phorbol 12-myristate 13-acetate (PMA) | Sigma Aldrich | Cat#P8139-1 MG |
| Ionomycin | Sigma Aldrich | Cat#I9657-1MG |
| brefeldin A | Biolegend | Cat#420601 |
| Phosphate Buffered Saline (PBS) | Global Life | Cat#SH30028.02 |
| Fetal Bovine Serum (FBS) | Omega Scientific | Cat#FB-02; Lot#881991 |
| Bovine Serum Albumine (BSA) | Gemini | Cat#700-106P; Lot#C82H81J |
| Dimethyl sulfoxide (DMSO) | Sigma Aldrich | Cat#d4540 |
| GoTaq master mix | Promega | Cat#M71333; Lot#000428913 |
| nuclease-free H20 | Promega | Cat#P1193 |
| ODN 2216 - TLR9 ligand | InvivoGen | Cat#tlrl-2216-1; Lot#3603-39T |
| ssRNA40/LyoVec™ | InvivoGen | Cat#tlrl-lrna40; Lot#A40-39-03 |
| Imiquimod (R837) | InvivoGen | Cat#tlrl-imq; Lot#IMQ-36-02A |
| FSL-1 | InvivoGen | Cat#tlrl-fsl; Lot#FSL-37-04 |
| Pam3CSK4 | InvivoGen | Cat#tlrl-pms; Lot#PMS-35-03 |
| LPS | InvivoGen | Cat#tlrl-b5lps |
| Flow Cytometry Perm Buffer (10x) | Tonbo Biosciences | Cat#TNB-1213-L150; Lot#B1213062817TN |
| Foxp3/Transcription Factor Fix/Perm Diluent (1x) | Ton bo Biosciences | Cat#TNB-1022-L160; Lot#B1022062415 |
| Foxp3/Transcription Factor Fix/Perm Concentrate (4x) | Tonbo Biosciences | Cat#TNB-1020-L050; Lot#B10200322420TN |
| Fixation Buffer | Biolegend | Cat#420801; Lot#B331895 |
| Intracellular Staining Perm Wash Buffer (10x) | Biolegend | Cat#421002; Lot#B32433 |
| Critical commercial assays | ||
| DNeasy Powersoil Pro kit | Qiagen | Cat#47016; Lot#169022930 |
| MinElute 96 UF PCR Purification Kit | Qiagen | Cat#28053; Lot#154032872 |
| MiSeq Reagent Kit v3 600 cycles | Illumina | Cat#MS-102-3003 |
| Chromium Single Cell 3′ Feature Barcoding Library Kit v3.1 chemistry | 10X Genomics | Cat#1000079 |
| Deposited data | ||
| Raw sequencing data | This Study | Sequence Read Archive- BioProject SRA: PRJNA800766 (https://www.ncbi.nlm.nih.gov) |
| Oligonucleotides | ||
| 16S rRNA gene 515F forward primer 5′-GTGYCAGCMGCCGCGGTAA-3′ | IDT | (Parada et al., 2016) |
| 16S rRNA gene 806R reverse primer 5′-GGACTACNVGGGTWTCTAAT-3′ | IDT | (Parada et al., 2016) |
| 16S rRNA gene B8F forward primer | IDT | (Edwards et al., 1989) |
| 16S rRNA gene 1492R reverse primer | IDT | (Edwards et al., 1989) |
| Software and algorithms | ||
| Quantitative Insights Into Microbial Ecology 2 (QIIME2)(2019.10) Built in tools: Dada2, mafft, sklearn, FastTree 2 | (Bolyen et al., 2019) | https://qiime2.org |
| DNAsubway | CyVerse and DNA Learning Center | https://dnasubway.cyverse.org |
| iTOL (Interactive Tree of Life) | (Letunic and Bork, 2021) | https://itol.embl.de/ |
| FlowJo | BD Life Sciences | https://www.flowjo.com/ |
| Cell Ranger Single-Cell Software Suite v4.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Seurat v4.0 | (Hao et al., 2021) | https://satijalab.org/seurat/ |
| R software (v4.0.2) R packages: vegan (v2.5-7), SystemPipeR (v1.26.3), edgeR (V3.34.0), ggplot2 (v3.3.3) | R Foundation | https://www.r-project.org |