Summary
Humans harbor numerous species of colonic bacteria that digest fiber polysaccharides in commonly consumed terrestrial plants. More recently in history, regional populations have consumed edible macroalgae seaweeds containing unique polysaccharides. It remains unclear how extensively gut bacteria have adapted to digest these nutrients. Here, we show that the ability of gut bacteria to digest seaweed polysaccharides is more pervasive than previously appreciated. Enrichment-cultured Bacteroides harbor previously discovered genes for seaweed degradation, which have mobilized into several members of this genus. Additionally, other examples of marine bacteria-derived genes, and their mobile DNA elements, are involved in gut microbial degradation of seaweed polysaccharides, including genes in gut-resident Firmicutes. Collectively, these results uncover multiple separate events that have mobilized the genes encoding seaweed degrading-enzymes into gut bacteria. This work further underscores the metabolic plasticity of the human gut microbiome and global exchange of genes in the context of dietary selective pressures.
eTOC blurb
Humans in several geographic regions have historically consumed seaweeds in forms like sushi, introducing novel dietary fiber polysaccharides to the gut. Pudlo, Pereira et al. show that a variety of mobile DNA elements have participated in transferring genes from marine bacteria, enabling gut bacteria to digest seaweed polysaccharides.
Introduction
More than any other animal on Earth, as humans have extended into new territory and developed civilization, we have expanded our nutritional repertoire out of necessity, curiosity or the pleasure of cuisine. One example is the consumption of edible red and brown macroalgae seaweeds, originally by populations living near coasts, which introduced a variety of novel dietary fiber polysaccharides such as laminarin, alginate, carrageenan, agarose and porphyran (Figure 1A). Compared to commonly consumed fruits, vegetables, grains and nuts, the polysaccharides in seaweeds have different chemical structures that require specific degradative enzymes (Hehemann et al., 2014; Porter and Martens, 2017). As with nearly all dietary fibers, humans lack these enzymes and therefore rely on colonic bacteria for the ability to digest them. Many gut bacteria are equipped with the ability to digest some dietary polysaccharides, although bacteria studied to date typically degrade only a subset of all fibers and some species are specialized for just one or a few substrates. Among the major bacterial phyla that are present in the gut, the Firmicutes, Actinobacteria and Bacteroidetes all include members associated with dietary fiber degradation (Cockburn and Koropatkin, 2016; Glowacki and Martens, 2020).
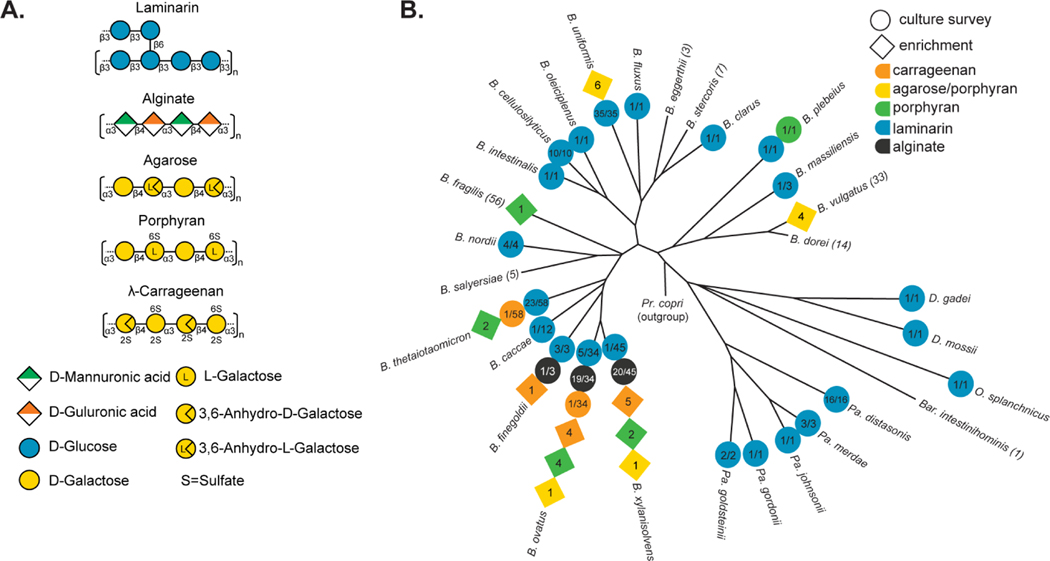
Figure 1. Seaweed polysaccharide structures and their utilization by human gut Bacteroidetes.
(A) Structures of the 5 different seaweed-derived polysaccharides used in this study. Note that carrageenan has multiple isomers and only l-carrageenan, which supports growth of gut Bacteroides strains identified here is shown for simplicity. (B) A phylogeny of human gut Bacteroidetes based on core gene alignment (Larsbrink et al., 2014) and illustration of seaweed polysaccharide-degrading abilities present in members of each species. Degradative abilities that were observed in isolates cultured without targeted enrichment are represented as circles and the numbers indicate the number of positive strains over the total number tested. Isolates recovered by enrichment on agarose, porphyran or carrageenan are shown in diamonds and the number indicates the total number of isolates recovered for that species from different donors. B.= Bacteroides, Pa.= Parabacteroides, D.= Dysgonomonas, O.= Odoribacter, Bar.= Barnesiella.
Previous studies have documented a few examples in which symbiotic bacteria belonging to Bacteroides—a dominant Bacteroidetes genus in humans—possess genes for degrading seaweed-derived porphyran (Hehemann et al., 2010; Shepherd et al., 2018), agarose (Pluvinage et al., 2018), alginate (Mathieu et al., 2018; Thomas et al., 2012) and laminarin (Déjean et al., 2020). In the first three cases, the genes involved often have closest homologs in marine Bacteroidetes, which are physiologically different from gut Bacteroides but share similar mechanisms for polysaccharide degradation (Grondin et al., 2017). In the case of porphyran degradation, a mobile DNA element was identified that presumably carried the required genes into the human microbiome, integrating in a site-specific manner into the genomes of at least two species (Hehemann et al., 2012a; Shepherd et al., 2018). Selection for this lateral gene transfer (LGT) event was presumably driven by historic consumption of porphyran in various forms (e.g., sushi) in parts of Asia. However, it remains unknown how pervasively genes involved in seaweed degradation have permeated the human gut microbiome, the sources and taxonomic range of these “genetic upgrades” and the DNA transfer mechanisms involved.
Here we used culture-based approaches to characterize several dozen isolates of human gut bacteria with the ability to digest various seaweed-derived polysaccharides, identify the genes involved in degradation, the corresponding LGT elements and characterize key enzymes. We show that known genes for porphyran degradation have mobilized into at least 5 different species and identify a second LGT event associated with porphyran utilization by human gut Bacteroides. We also identify mobile DNA involved in transfer of agarose utilization, demonstrating that this mechanism is still active in some strains, identify mobile DNA that confers carrageenan degradation, and report several human gut Firmicutes with the ability to degrade porphyran and agarose. Given the emerging interest in leveraging these comparatively unique nutrients to engineer “orthogonal niches”—which can potentially be filled by recombinant bacteria that are programmed not just to use these nutrients but also execute functions to treat diseases—our findings provide insight into both the adaptability of gut microbes to utilize new nutrients and novel tools to engineer them.
Results
Distribution of algal polysaccharide degradation in human gut Bacteroidetes
To determine the extent to which gut bacteria have acquired seaweed degrading abilities, we surveyed a collection of human and animal gut Bacteroidetes representing 30 different species (354 total isolates, Table S1A) on four of the five algal polysaccharides mentioned above (all except agarose due to solubility problems in high-throughput). The ability to use the β-linked glucan laminarin from Laminaria digitata, for which the corresponding genes were recently identified in strains of Bacteroides uniformis and Bacteroides thetaiotaomicron (Déjean et al., 2020), was broadly represented and present in members of 22 different species (Figure 1B). This may reflect the ubiquity of structurally related, but non seaweed-derived, β-linked glucans in many different plants and fungi, albeit with subtle variations in structure that can also influence their utilization.
The remaining seaweed polysaccharides were used by far fewer species. Alginate was the second most prevalent trait but was confined to strains belonging to just three related species (Figure 1B). Among strains that grew on alginate, all for which a sequenced genome was available contained known alginate utilization genes (Mathieu et al., 2018; Thomas et al., 2012) and loci from representatives in each species were upregulated >100-fold in response to growth on alginate (Figure S1A,B). Growth on the remaining polysaccharides was even less prevalent, with only the single, previously known isolate of Bacteroides plebeius from a Japanese adult showing growth on porphyran (Hehemann et al., 2012a) and individual isolates belonging to two different species, B. thetaiotaomicron and B. ovatus, growing on carrageenan (a mixture of κ, λ isomers). Since carrageenan utilization has not been previously associated with human gut bacteria, we focused on the genetic basis of this phenotype.
Carrageenan utilization is associated with mobile DNA
Unlike agarose and porphyran, which have been traditionally consumed in Asian cuisine, the red algae that contain carrageenan (CGN) have been associated with both European and Chinese consumption as far back as 400 BC and CGN is now a widely used food additive (Loureiro RR et al., 2017). To determine the genes involved in CGN utilization we grew B. thetaiotaomicron strain 3731 (herein, Bt3731) on λ-CGN and performed RNA-seq-based transcriptional profiling and proteomics to identify upregulated functions compared to growth on galactose. RNA-seq revealed a total of 343 genes were upregulated based on the criteria used (Table S2A) and 56 of these genes were contained in two regions of a polysaccharide utilization locus (PUL), a hallmark of Bacteroidetes nutrient assimilation (Figure 2A,B) (Grondin et al., 2017). More than half of these genes had nearest homologs in Gram-negative marine bacteria, suggesting transfer from marine species (Table S3A), which are known to harbor genes for CGN utilization (Ficko-Blean et al., 2017). Concurrent proteomics analysis revealed strong concordance with the transcriptome, demonstrating that Bt3731 dedicates substantial resources to production of 44 different proteins encoded in the candidate CGN-PUL that are increased greater than 0.01% of normalized spectral abundance factor (NSAF) during growth in CGN (Figure 2C, Table S4A-B). A similar PUL was identified in the genome of CGN-degrading B. ovatus CL02T12C04 (herein, Bo12C04), the second strain in our survey that grew on this seaweed polysaccharide (Figures 2B, S2A).
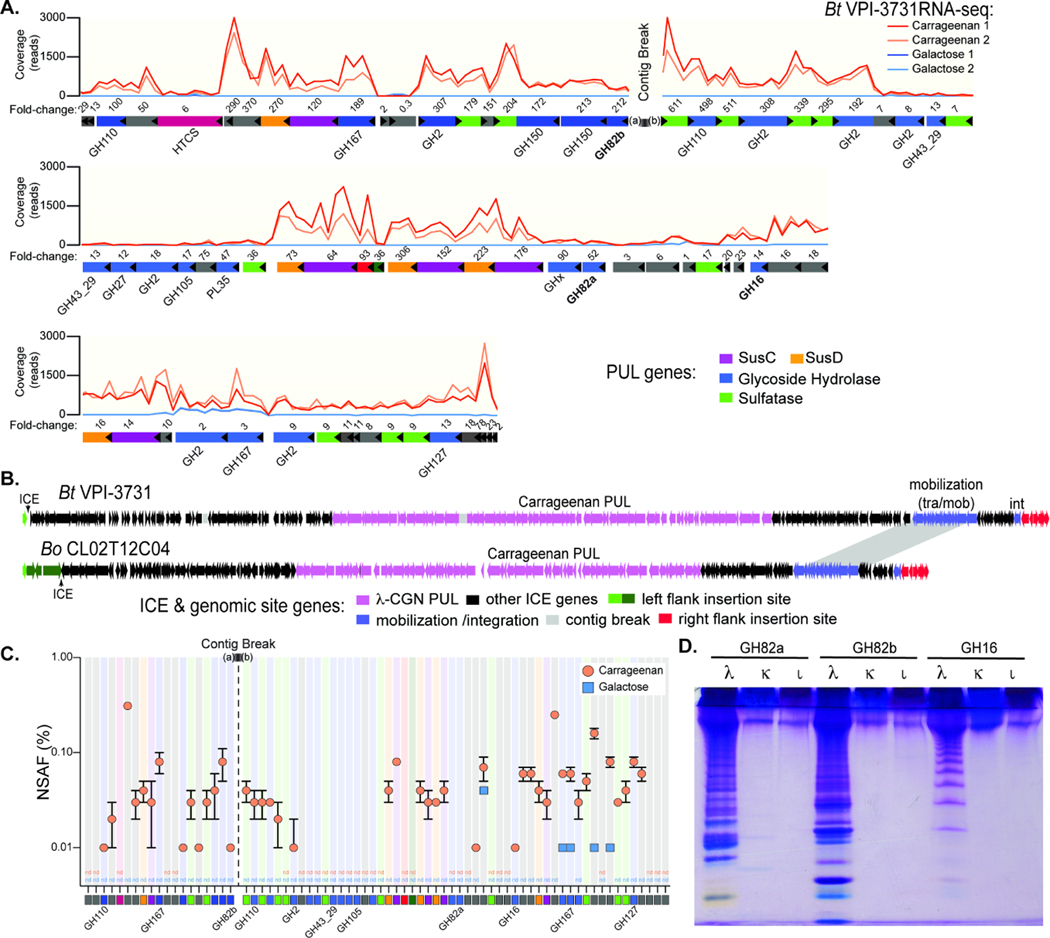
Figure 2. Bacteroides genes and enzyme involved in carrageenan degradation.
(A) Transcriptional responses of genes within the 130.5kbp candidate λ-CGN PUL in Bt3731 during growth on λ-CGN compared to a galactose reference. Numbers above genes refer to fold change induction by CGN relative to galactose (Table S2A). Two replicate experiments were conducted for each treatment condition. (B) A comparison of the surrounding Bt3731 and Bo12C04 ICE architecture showing the genes at the flanking chromosomal insertion gene in green and red (see Figure S2B for higher resolution view of insertion site). (C) Relative abundances (in % of normalized spectral abundance factor, NSAF) of PUL-encoded proteins detected in the total protein fraction of cultures grown on carrageenan (orange) and galactose (blue). Error bars are the standard error of the mean (SEM). Proteins that could be detected in at least two out of three independent biological replicates of each substrate condition are shown and color coded squares are at bottom to match genetic architecture in (A). Three replicate experiments were conducted for each treatment condition. (D) C-PAGE gel showing digestion of three different CGN isomer types (λ, lambda; κ, kappa; Ɩ, iota) with recombinant GH16 and GH82 enzymes that are highly expressed within the Bt3731 PUL. Shorter CGN oligosaccharides migrate toward the bottom of the gel.
To further test the role of the genes that were identified through transcriptomic and proteomic approaches, we performed transposon sequencing (TnSeq) on the Bo12C04 strain. A library containing transposon insertions in 4,307 different genes (69.6% of all genes annotated in this strain) was separately passaged in minimal medium with either λ-CGN or glucose as the sole carbon source and the differences in the final abundance of transposon mutants in each gene was measured between the two growth conditions (Goodman et al. 2009) (Figure S2A). A total of 170 genes with transposon insertions were found to be significantly more abundant (≥10-fold, p < 0.01) after passaging in glucose compared to CGN, suggesting that they provide a growth advantage during CGN utilization (Table S5). Sixteen of the disrupted genes were located within the Bo12C04 CGN PUL for which 56/69 genes were disrupted by transposons in the library. Notable within the list of 16 disrupted functions were a hybrid two-component system sensor regulator, several glycoside hydrolases (GH16, GH110, GH127), one of 4 SusC/SusD-like pairs encoded in this PUL and several sulfatases. Most of these functions were encoded in a contiguous stretch of 17 genes located at the “right edge” of the locus, a region that had homologs in Bt3731 of all but one gene (Figure S2A). This finding suggests that one or more enzymes or transport functions encoded in this region are critical to λ-CGN utilization, while much of the remaining locus is either less important or functionally redundant. Among the list of genes that apparently provide fitness advantage during growth on CGN but are unlinked to the CGN PUL, a gene encoding anaerobic sulfatase maturing enzyme (anSME), which is essential for use of sulfated glycans like CGN (Benjdia et al. 2011), was depleted in CGN by 1,069-fold as expected. Aside from an annotated galactokinase, another sugar kinase and a component of an ABC transporter (reduced by 576-, 1,488-, and 1,146-fold respectively in CGN; Table S5), which may have been involved in galactose metabolism, there was little additional indication of what roles the other non PUL-linked genes may play in CGN metabolism.
The genes immediately adjacent to the CGN PULs are homologous in Bt3731 and Bo12C04 and suggest that lateral gene transfer may have played a role in introducing them into their respective genomes. In particular we observed a set of mobilization genes near one side of the PUL (Figure 2B), suggesting that this region is part of an integrative conjugative element (ICE). To test this further, we performed comparative genomics with several B. thetaiotaomicron and B. ovatus strains that do not degrade CGN, revealing the presence of identical 16 bp direct repeat sequences at the ends of each putative ICE. Both flanking genomic regions were syntenic between CGN-degrading and non-degrading isolates with only a single copy of the 16 bp repeat in non-degraders that was downstream of the housekeeping gene dnaK (Figure S2B). PCR and sequence analysis of this region demonstrated the ability of this ICE to circularize from the Bt3731 genome (Figure S2B-D), supporting the idea that Bacteroides carrageenan utilization was conferred by acquisition of a 245–265 kbp ICE.
Finally, analysis of recombinant Bt3731 enzymes from the putative CGN PUL supported its role in CGN degradation. A recombinant GH16 enzyme, and two GH82 enzymes, revealed that they are each endo-acting carrageenases, but not endo-processive, with specificity for λ-CGN (Figures 2D, S2E-G). Bt3731 that was actively growing on λ-CGN released oligosaccharides in a similar size range as those produced by recombinant endo-acting enzymes (Figure S2H). This is notable because degraded forms of CGN, so called “poligeenan”, have been associated with intestinal inflammation and colorectal cancer development in animal models (Onderdonk, 1985), leading to the exclusion of low molecular weight dietary CGN in many countries (Tobacman, 2001). Thus, it is possible that some humans harbor CGN-degrading Bacteroides that might convert the non-harmful high molecular weight form of this polysaccharide into potentially harmful oligosaccharides during catabolism.
Enrichment culture to isolate seaweed degrading strains
As an additional approach to identify gut bacteria that degrade seaweed polysaccharides, we surveyed stool samples from 240 different healthy volunteers using an enrichment strategy (see Materials and Methods) that was previously successful in isolating an agarose-degrading strain of B. uniformis (Hehemann et al., 2012a). We isolated 33 different strains that were enriched based on the ability to grow on CGN, porphyran or agarose (Table S1B, Figure S3). 16S rDNA gene sequencing revealed that these isolates belong to 7 different species (Figure 1B). CGN utilization was confined to members of four closely related species, including the two identified above and extending to B. xylanisolvens and B. finegoldii. Consistent with the recombinant enzyme data above, growth on CGN was specific in all strains for λ-CGN and not the Ɩ or κ isomers (Figure S3). All of the isolates that grew on agarose were also capable of some growth on the porphyran preparation used, which is expected since porphyran from Porphyra yezoensis is a heteropolysaccharide with agarose segments containing 3,6 anhydro-Lgalactose instead of L6-S-galactose (Hehemann et al., 2010). However, growth on porphyran was sometimes better than agarose (e.g., B. xylanisolvens AO201–2, Figure S3) making it possible that the sensory and/or enzymatic machinery in some strains is optimized towards porphyran. Such combined agarose/porphyran utilization was previously observed in B. uniformis (Pluvinage et al., 2018) and also observed here in three more species, B. vulgatus, B. ovatus and B. xylanisolvens. The ability to utilize just porphyran was broadly distributed across the Bacteroides genus and, including previously identified isolates (Hehemann et al., 2010; Shepherd et al., 2018), is now known to be present in five different species.
To determine if the seaweed polysaccharide-degrading isolates harbor genes for seaweed utilization that are related to the previously identified PULs and associated mobile elements, we performed shallow-draft genome sequencing on each isolate and assembled the reads using the known gene clusters as scaffolds. All of the identified CGN-degrading strains had regions with high coverage (62–100% of PUL length) and nearly identical homology (>99% nt identity) when mapped to the known Bo12C04 PUL, the closest match for all isolates (Figure S4A). This suggests that the additional CGN degrading strains isolated here all contain close variants of the genes identified above.
We previously used phylogenetic analysis of a common Bacteroides conjugation protein (TraJ) to identify groups of ICEs that are associated with various genetic cargo (including PULs), examine their relationship to one another and identify integration site preferences (Martens et al., 2014). This analysis revealed that the original B. plebeius ICE that confers porphyran utilization and integrates into tRNALys(TTT) is closely related in both sequence and architecture to a family of ICEs that integrate into tRNAPhe(GAA) and carry variable gene cargo, including a B. thetaiotaomicron PUL for fungal α-mannan degradation (Cuskin et al., 2015). To determine if the transfer functions associated with Bacteroides CGN ICEs are related to the previously characterized ICEs, we created a phylogeny of 2,255 TraJ sequences from currently sequenced gut, marine and environmental Bacteroidetes (Figure S4B). The TraJ proteins associated with the CGN ICEs are only distantly related to those involved in porphyran transfer to B. plebeius, sharing 22% amino acid identity and supporting the conclusion that distinct mobile elements have captured their respective genetic cargo for degrading these seaweed polysaccharides.
All of the agarose/porphyran utilizing strains contained sequences that were nearly identical (>99% nt homology) to the B. uniformis PUL that was previously demonstrated to be involved predominantly in agarose degradation (Figure S5A). However, the surrounding genome region could not be resolved for this PUL in the initial strain that was sequenced (Pluvinage et al., 2018). Interestingly, a recent B. uniformis isolate that was recovered from a healthy Japanese adult and sequenced using long-read technology, also contains this known agarose PUL, which was resolved as part of a 124 kbp circular plasmid. Using this plasmid template, we mapped sequences from all 11 strains isolated here, finding that each contains sequences that match to between 46.2–94.2 % of this plasmid, with all of the B. uniformis and one B. xylanisolvens strain containing most of the episome (Figure 3A). A previously identified plasmid containing agarolytic genes was isolated from a marine sediment Bacteroidetes strain but found to be non-conjugal and lack mobilization machinery (Zhong et al., 2001). A potentially self-replicating plasmid that transfers polysaccharide utilization functions among Bacteroidetes has not yet been described and raises new possibilities for biotechnology applications. The mobilization genes that were contained on this plasmid were also divergent from the examples discussed above and lack TraJ homologs that would allow them to be placed in our phylogeny. However, they underscore the diversity of mechanisms that have been coopted to capture useful genetic cargo and transfer it between related bacteria in different environments.
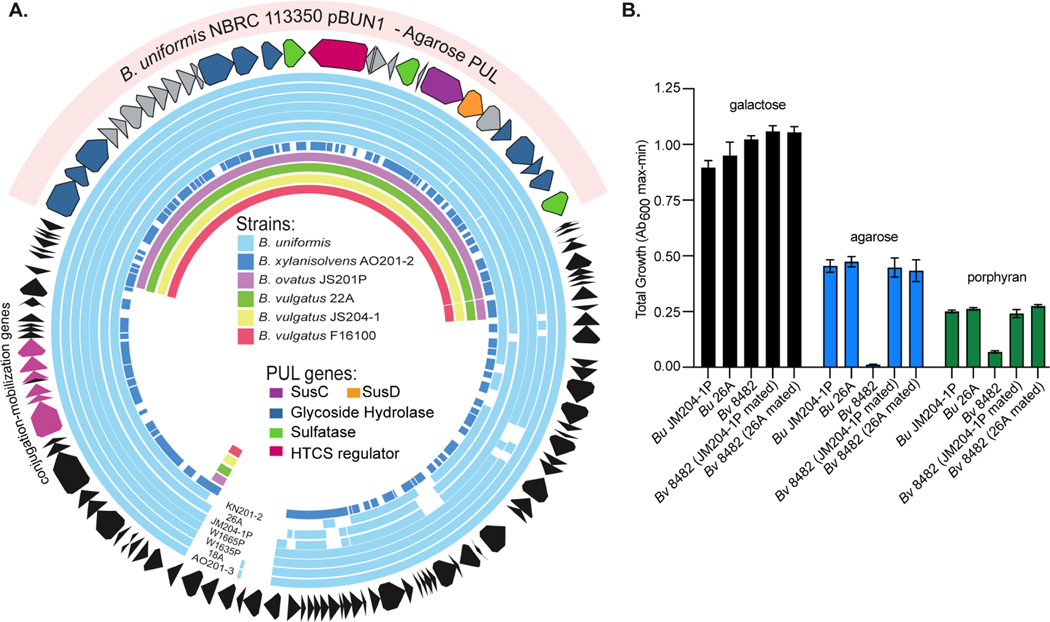
Figure 3. B. uniformis contains a mobile plasmid that confers agarose utilization.
(A) Circular map of the B. uniformis strain NBRC 113350 plasmid pBUN1 that contains an agarose utilization PUL. Strain designations for B. uniformis isolates in this study are located at the bottom. Sequence reads from each of the additional isolates in this study were mapped based on a minimum of 97% nucleotide identity to this scaffold, demonstrating that B. uniformis and B. xylanisolvens strains contain most of the episome. (B) Total growths on glucose, agarose and porphyran by B. uniformis plasmid donors (JM204–1P and 26A), B. vulgatus ATCC 8482 type strain and recipient B. vulgatus strains after mating with the indicated B. uniformis donor (Bv 8482 JM204–1P and 26A mated) showing acquisition of agarose degrading phenotypes. Error bars indicate standard deviation (SD) of the mean. Three replicate experiments were conducted for each treatment condition.
To test the ability of this agarolytic plasmid to mobilize between species, we performed conjugations between agarose-degrading B. uniformis strains and 5 genetically manipulatable Bacteroides type strains, B. thetaiotaomicronVPI−5482, B. ovatusATCC 8483, B. fragilisNCTC 9343, B. cellulosilyticusDSM14838 and B. vulgatusATCC8482. We engineered each of these strains to contain 2–3 antibiotic resistance traits to enable selection from the B. uniformis, which were susceptible to these drugs (Materials and Methods). After individually mating each B. uniformis donor with the Bacteroides type strains and plating on selective media, we found B. vulgatusATCC8482 to be the only strain capable of receiving the plasmid and gaining the ability to grow on agarose (Figure 3B). However, only 2 of the 5 donor strains (B. uniformisJM204−1P and B. uniformis26A) exhibited successful conjugation, suggesting transfer is limited both between recipients and donors.
A second lateral transfer event for porphyran utilization into Bacteroides
All but one of the identified porphyran-degrading strains had sequences with high coverage (75–100%) and nucleotide identity (>96%) to the known ICE originally identified in B. plebeius (Hehemann et al., 2012a) (Figure S5B). Thus, variants of this mobile DNA have successfully integrated into at least 5 species (based on ≥97% near full length 16S rRNA gene homology to named species in new or existing isolates). Interestingly, one isolated B. xylanisolvens strain (Bx18−002P) lacked extensive sequence homology to the previously identified porphyran ICE (Figure 4A), suggesting that its growth on porphyran is attributable to an alternative set of genes. To explore this, we generated a high-quality, nearly closed genome for this isolate and performed RNA-seq on cells grown on porphyran compared to a galactose reference. A notably large set of 96 genes within two closely linked chromosomal regions showed strong responses (Table S2B). These genes also have close relatives in marine Bacteroidetes (Table S3B) and encode Bacteroides PUL functions, including enzymes predicted to degrade porphyran, such as families GH16, GH86 and GH117, sulfatases and many other enzymes (Figure 4B). Comparison of these gene clusters to another recently cultured and sequenced B. xylanisolvens (BxBIOML-A58) revealed that this second strain—which was presumably not isolated based on its ability to grow on porphyran (Poyet et al., 2019)—also contained the same genes that were porphyran-inducible in Bx18−002P. However, this strain lacked a region present in Bx18−002P that separates the two blocks of porphyran-responsive genes and contains several mobilization genes. In addition, both strains have syntenic and homologous genes on both sides of the porphyran-inducible PUL, with additional mobilization genes contained on one side (Figure 4C). Based on this, we are unable to conclude which, if any, of the mobilization genes might be associated with carrying these porphyran-degrading functions into Bx18−002P and Bx BIOML-A58. However, the simplest scenario is that the common DNA shared between these strains represents an ancestral ICE architecture and strain Bx18−002P harbors a second, unrelated ICE that integrated into the middle of a large porphyran PUL without disrupting its function. As with the CGN-degrading strains, comparison to other Bx strains that lack the ability to grow on porphyran suggested that an ancestral genomic region, in this case encoding a nearby tRNALys(TTT) and thymidine kinase (tdk) gene was the site of ICE integration via a 21 bp direct repeat, which interestingly targets the same tRNA as the first porphyran ICE but via a different sequence (Figure S6A). This idea was supported by PCR and sequence analysis of these regions, revealing that this ICE is capable of excising to restore the predicted ancestral genome architecture (Figure S6B).
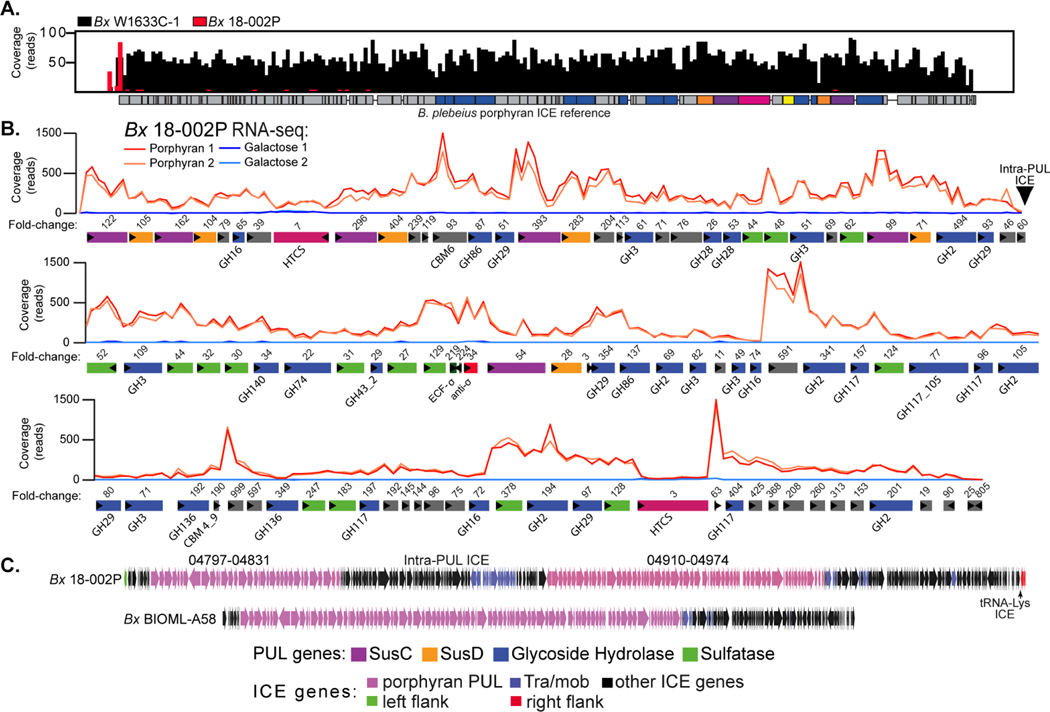
Figure 4. Identification of a second mobile element and PUL that is associated with porphyran degradation by human gut Bacteroides.
(A) Histogram of reference-guided mapping results to the B. plebeius porphyran ICE for two different strains: BxW1633C−1 (black) and Bx18−002P (red). BxW1633C−1 has full coverage across the entire Bp ICE, while sequences for Bx18−002P do not align to the known B. plebeius genes, except for a small region at the far left. (B) RNA-seq based transcriptional responses of genes within a 168.6kbp PUL in Bx18−002P during growth on porphyran compared to galactose. Numbers of above genes refer to fold change induction by prophyran relative to galactose (Table S2B). Two replicate experiments were conducted for each treatment condition. (C) Lower resolution comparison of the ICEs between the Bx18−002P and BxBIOML-A58 strains, highlighting the presence of extra genes, possibly the result of an additional ICE insertion into the middle of the Bx18−002P PUL without disrupting its function.
The Bx18−002P PUL also encodes additional GH families that have been shown to degrade other substrates, suggesting that it encodes broader glycan degrading ability. Using SACCHARIS (Jones et al., 2018), putative enzymes within the Bx18−002P locus were aligned with carbohydrate active enzymes with known functions. In addition to red algal galactans, this locus may target a diversity of substrates, possibly including pectins (GH28s, GH140), ulvan (GH105), fucoidan (GH29), and β-mannans (GH2s). The presence of a β-galactofuranosidase (GH117), α-L-arabinofuranosidase (GH43), β-glucuronidase (GH2), and β-xylosidases (GH3s) suggests that this pathway may consume both red and green seaweed polysaccharides (Table S6). Homo- and hetero-xylans are found in red and green seaweeds (Viana et al., 2011), and complex xylogalacto(furano)arabinan is a primary component of edible green seaweed Cladophora falklandica (Arata et al., 2016). Notably, however, there is a lack of polysaccharide lyases typically associated with the depolymerization of polyuronic acids found in these species. The collection of this diverse group of activities indicates that the degradative pathway(s) encoded in this complex gene cluster, which also encodes multiple regulators and SusC/D-like transporter systems, is tailored to hydrolyze structurally complex substrates found in edible seaweeds. While many of the additional seaweed polysaccharides that are candidate substrates for this system are not readily available and vary between algal sources, Bx18−002P could not grow on ulvan polysaccharide from Ulva sp. or fucoidan from Laminaria digitata when presented as the sole carbon source (Figure S6C).
The carrageenan- (Bt3731, Bo12C04) and porphyran-degrading (Bx18−002P, Bp17135) strains encode several GH16 family enzymes characterized in the CAZy database as carrageenases, agarases and porphyranases, amongst others. Therefore, we used a GH16 family phylogeny as a proxy to explore GH family function into subfamilies, further demonstrating evolutionary trajectories towards substrate specificity. While the Bt3731 and Bo12C04 proteins remain tightly clustered in a separate clade, the Bx18−002P and Bp17135 GH16 enzymes display broader subfamily assignments including subfamilies 12, 15 and 16 (Figure S6D). This is further exemplified by the various oligosaccharides produced from different subfamilies of GH16 enzymes that allow for efficient depolymerization of seaweed glycans (Figure S6E). These evolutionary adaptations towards glycan specificity undoubtedly play a critical role in proficient carbon utilization by these microbes but also provide an advantage for colonizing the human GI tract.
Human gut Firmicutes with genetic upgrades to degrade seaweed polysaccharides
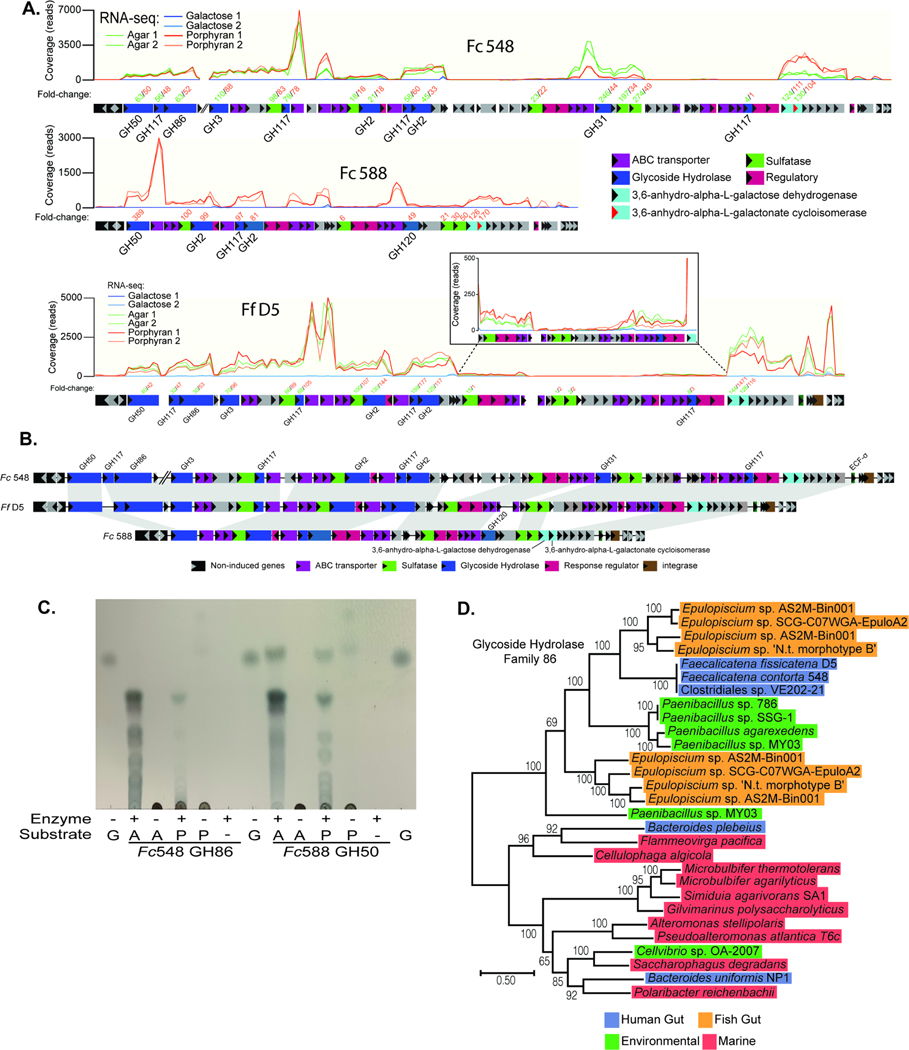
To date, all of the examples of seaweed-degrading human gut bacteria, including the isolates described above, have been Gram-negative Bacteroidetes. The Gram-positive Firmicutes that thrive in the human gut are typically more abundant, taxonomically more diverse and are also proficient fiber degraders. This raises the question of whether or not members of this phylum have received similar “genetic upgrades”. During studies with the same cohort of healthy human volunteers used to isolate seaweed-degrading Bacteroides, we fortuitously observed that some spore-forming gut bacteria (isolated by ethanol selection and re-germination (Browne et al., 2016)) had the property of pitting the agar plates on which they were cultivated. This phenotype is exhibited by the agarose and porphyran-degrading Bacteroides and suggested that these bacteria might possess the ability to degrade these substrates. From two separate people, we obtained isolates belonging to the same spore-forming species, Faecalicatena contorta, which have varying abilities to grow on agarose and/or porphyran (Figure S3). We generated high-quality, nearly closed genomes for each of these isolates (herein, Fc548 and Fc588) and performed RNA-seq during growth on porphyran or agarose relative to a galactose reference for each strain. Each strain’s transcriptional response during growth on seaweed polysaccharides was characterized by strong upregulation of a single gene cluster that was homologous between the two isolates (Figure 5A,B; Tables S2C-D). Based on homology to the genes identified, a third, related strain of Faecalicatena fissicatena (strain D5; herein, Ff D5) that harbors the same locus was located in the public database. Growth and RNA-seq analysis of this strain on porphyran and agarose, confirmed the prediction that it possesses the ability to grow on these polysaccharides with similar upregulation of the genes identified in the other two strains (Figure 5A,B; Table S2E).
Figure 5. Identification of genes in human gut Firmicutes associated with agarose and porphyran utilization.
(A) RNA-seq based transcriptional responses of the homologous loci in Fc548, Fc588 and Ff D5 during growth on agarose and/or porphyran compared to galactose. The inset box shows a closer view of a section of genes that are induced less compared to their surrounding neighbors, but still show a positive response compared to galactose. Numbers of above genes refer to fold change induction by agarose or porphyran (according to color code when applicable) relative to galactose (Tables S2C-E). Two replicate experiments were conducted for each treatment condition. (B) A genomic comparison of the three Faecalicatena spp. locus maps showing synteny and variation among the genes responsible for degrading agarose and/or porphyran. Note that the first three enzymes in Fc548 are separated genomically from the rest of the genes in the loci by 110 genes (∼87.9 kbp). (C) Thin-layer chromatography of Fc548 GH86 and Fc588 GH50 enzymes on high molecular weight agarose (A) and porphyran (P) with galactose (G) serving as a monosaccharide standard. (D) Phylogenetic position of Faecalicatena GH86 enzymes compared to other gut and environmental enzymes in this family, highlighting the similarity to enzymes found in Epulopiscium and Paenibacillus spp. Clostridiales sp. VE-202–2 is a Faecalicatena contorta isolate from a Japanese adult that has a copy of this locus and likely degrades these polysaccharides.
The gene clusters identified in the three Faecalicatena isolates encode predicted enzymes that belong to some of the same GH families as in agarose- and porphyran-degrading Bacteroides. To determine if these enzymes degrade these polysaccharides, we produced recombinant forms of 5 proteins from 3 different families (GH50, GH86, GH117). We observed activity with the GH50 (Fc588) and GH86 (Fc548) enzymes, which produced oligosaccharides from agarose and porphyran suggestive of endo-acting function (Figure 5C). However, three different GH117 enzymes— homologs of which are known to remove 3,6-anhydro-L-galactose from neoagarooligosaccharides (Hehemann et al., 2012b; Sugano et al., 1994)—displayed little detectable activity against either native substrates or GH86/GH50-derived oligosaccharides, suggesting that their activity is likely similar to known homologs and requires prior activity of other enzymes encoded in this pathway (Figure S7A,B).
The seaweed-degrading enzymes found in human gut Bacteroides have close relatives in marine Bacteroidetes, which are plentiful in the ocean and critical for turnover of algal biomass (Becker et al., 2020; Kappelmann et al., 2019). Interestingly, the enzymes contained in human gut Faecalicatena spp. share close relatives with marine Gram-positive bacteria, including Paenibacillus and Epulopiscium spp. (Figures 5D, S7D, Tables S3C-E), the latter being abundant colonizers of marine surgeonfish (Ngugi et al., 2017). Sequence-based surveys of surgeonfish gut microbiomes have detected enzymes in these same families in metagenome- and single cell isolation-based genome sequences belonging to Epulopiscium spp. Thus, it appears that the selective pressure imposed by human consumption of seaweed has been sufficient to not only select for multiple, independent transfers of agarose, carrageenan and porphyran utilization functions into gut Bacteroides, but also into human gut Firmicutes from a different genetic reservoir. Interestingly, each of the Faecalicatena agarose/porphyran loci was flanked by an integrase, which may have been involved in its mobilization (Figure 5B). However, despite identifying and sequencing isolates of Faecalicatena spp. that do not possess the ability to grow on agarose or porphyran and performing comparative genomics we were unable to convincingly determine an ancestral genome state or identify a discrete mobile DNA signature.
To investigate some of the possible metabolites produced by the Faecalicatena spp., we screened stationary phase cultures grown in galactose, agarose and porphyran for short- and branch-chain fatty acids (SCFAs, BCFAs) via high performance liquid chromatography (Figure S7E). Although no butyrate, isobutyrate or isovalerate were detected in any sample, acetate was detected to appreciable levels in all three strains regardless of substrate. However, formate appears to be the primary metabolite produced during growth on galactose, whereas succinate, propionate and valerate are more prevalent in porphyran supernatants. Interestingly, Fc548 seems to produce more of nearly all detectable SCFAs measured when grown on porphyran.
Porphyran-mediated engraftment of a seaweed-degrading community in humanized mice
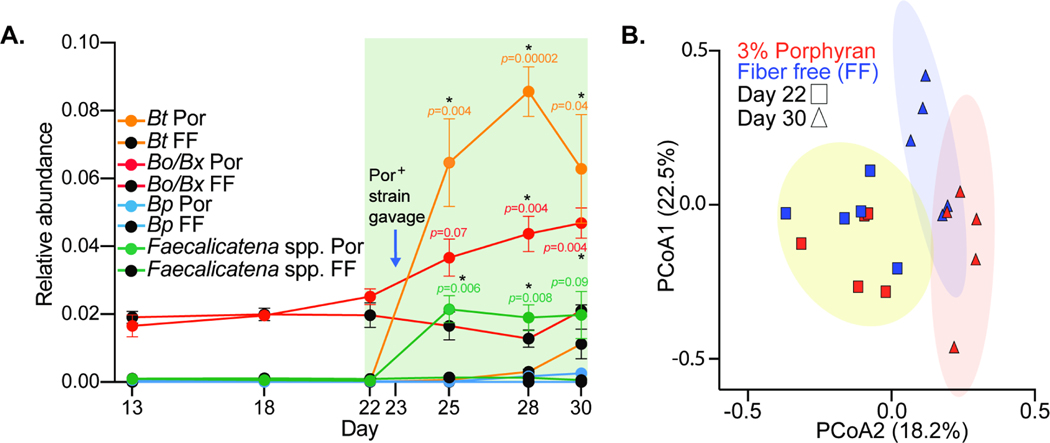
The presence of human gut Bacteroides with the ability to utilize polysaccharides like porphyran has catalyzed interest in leveraging these relatively rare fiber-degrading abilities to engineer “orthogonal niches” in the gut microbiome (Kearney et al., 2018), which may be useful in implanting exogenous gut bacteria with engineered metabolism (Shepherd et al., 2018). To determine if our expanded collection of human gut Bacteroides and Firmicutes could successfully colonize the gut in a dietary porphyran-dependent fashion, we colonized germfree mice (n=10 total) with feces from a single healthy human donor from which we repeatedly failed to enrich for indigenous agarose or porphyran-degrading bacteria. After 22 days of colonization with the human microbiota, we switched one group of 5 mice to a low fiber diet that was supplemented with 3% Porphyra yezoensis seaweed from food-grade unroasted nori sheets, while the other group was maintained on a nearly identical diet that lacked porphyran. On day 23 we gavaged both groups of mice with a community containing eight strains with varying abilities to utilize porphyran (B. plebeius17135, B. thetaiotaomicronPUR2.2, B. xylanisolvens18−002P, B. xylanisolvensW1633C−1, B. ovatusPUR1.1, F. contorta548, F. contorta588, F. fissicatenaD5) and maintained each group on their respective diets for another week (Day 30). Compared to the control group not fed P. yezoensis, mice fed this culinary seaweed showed significant increases in the relative abundances of operational taxonomic units (OTUs) corresponding to B. thetaiotaomicron (4.6-fold higher) and Faecalicatena spp. (36.1-fold higher), which were two of the porphyran-degrading species that we introduced and did not have corresponding OTUs in the donor microbiome (Figure 6A). In addition, P. yezoensis fed mice also showed an increase in an OTU corresponding to B. ovatus/B xylanisolvens (1.2-fold higher). Members of this OTU group were among the porphyran-degraders added to the engraftment community and separately contained both types of PULs involved in porphyran degradation. OTUs belonging to this group were already present in the transplanted human microbiome and remained ∼2% before the intervention. After switching to the low fiber diet, or the matching diet containing P. yezoensis, B. ovatus/B. xylanisolvens decreased in abundance in mice fed the reference diet, which is consistent with the response of this fiber-degrading group in humanized mice fed low fiber diets (Desai et al., 2016). However, in mice consuming P. yezoensis, B. ovatus/B. xylanisolvens showed the opposite trend and steadily increased over the one-week time period. Overall, feeding the P. yezoensis enriched diet, in concert with implanting seaweed-degrading bacteria, resulted in a significantly altered microbiome (Figure 6B).
Figure 6. Dietary porphyran promotes engraftment of Bacteroides and Faecalicatena strains that can degrade it as a nutrient.
(A) Engraftment of porphyran-degrading strains into gnotobiotic mice with a non porphyran-degrading human microbiota. Significant differences were observed in OTUs corresponding to Bt, Bo/Bx and Faecalicatena species when comparing the 3% porphyran (Por) diet to a fiber free (FF) control diet lacking porphyran (n=5 mice per group). Error bars are the standard error of the mean (S.E.M.) and unpaired t-tests were run using the Holm-Sidak correction to determine significance for days 25, 28 and 30 with significant p-values (<0.05, *) noted. (B) PCoA plot of individual mice shown in C before and after porphyran intervention in the presence of porphyran-degrading bacteria.
Geographic distribution of seaweed degrading genes in human gut microbiomes
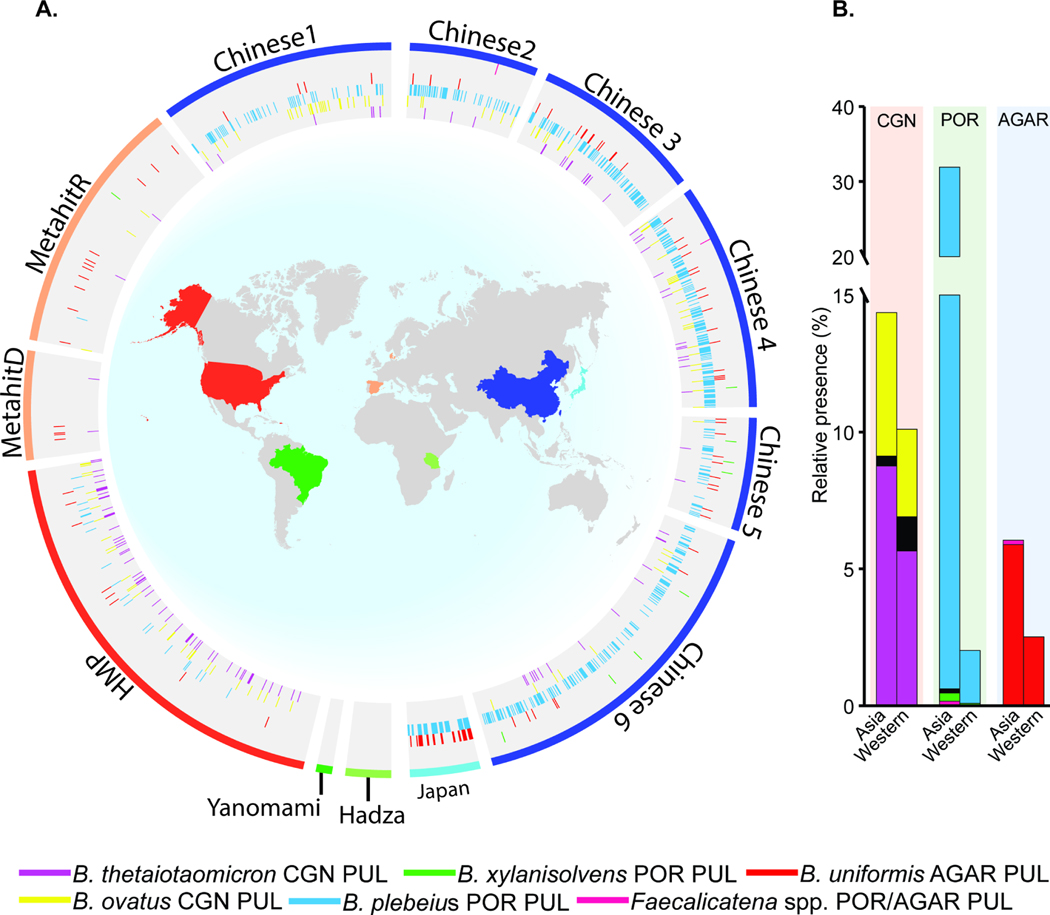
The close coupling of the gut microbiome to diet has made it possible for indigenous human gut bacteria to acquire new genes that enable them to compete for novel nutrients like seaweed polysaccharides. Since the selection for these genes is likely driven by regional dietary habits (e.g., consumption of Porphyra based seaweed products as Japanese sushi, Korean kimbap, Welsh laver bread and other dishes), we measured the presence of the known genes for seaweed degradation in metagenomic surveys of human fecal samples taken in North and South America, Africa, Europe and Asia (a total of 2,440 individuals). Consistent with several types of seaweed, including those that contain CGN, being consumed in parts of Asia for several millennia, the prevalence of these genes was substantially higher in metagenomic surveys of Chinese and Japanese subjects (Figure 7A,B). The B. plebeius-type porphyran PUL was the most abundant, while the B. uniformis-type agarose PUL appeared to be enriched in the limited Japanese samples, often cooccurring with the B. plebeius-type PUL. The λ-CGN PUL also showed increased prevalence in the metagenomic samples from the United States, consistent with its use as a food additive and historical consumption in European populations. Prevalence of CGN utilization was noticeably low in European samples, which could be attributable to the available data deriving only from individuals from Denmark and Spain (Qin et al., 2012) and not from Ireland or Scotland, the regions in which CGN has been used for food throughout history. While sampling has not yet been extended as deeply to populations living more traditional hunter-gatherer lifestyles, it is notable that none of these genes were detected in such populations from South America or Africa. Finally, the prevalence of the B. xylanisolvens-type porphyran PUL and gene clusters from Gram-positive Faecalicatena spp. were sparse in all samples. While detection of these genes will be influenced by sequencing depth, this did not hinder detection of the B. plebeius-type PUL, suggesting that species containing the latter genes are either more successful or more abundant.
Figure 7. Global distribution of known seaweed degrading gene clusters in existing metagenomic sequencing surveys.
(A) Existing global metagenomic data shown by country/region of origin. Colored wedges correspond to the presence of indicated PULs and outer band colors relate to country of project/data origin. Please note that the colored lines surrounding the perimeter of the each figure represent the country/region of origin and the radial marks in the inner circles correspond to the presence of PULs, which may be present in the species listed in the legend at the bottom or others if LGT has occurred. (B) Summary of CGN, porphyran and agarose degrading gene cluster presence in Western and Asian datasets based on the data shown in A.
Finally, to determine if close copies of the abundant B. plebeius-type porphyran PUL and the Bo12C04 CGN PUL exist in bacterial genomes from more diverse environments, we searched a database consisting of 122,126 metagenome assembled genome (MAG) bins from 14,557 metagenomic surveys that are publicly available in the Integrated Microbial Genomes database (Chen et al., 2021). Using a relatively stringent cutoff for percent nucleotide identity, which was required based on manual inspection of potential hits to avoid false positives (Materials and Methods), we surprisingly found only 6 high-confidence copies of the Bo12C04 CGN PUL, while the porphyran PUL was not detected
Discussion
As human beings have spread around the planet, we have varied and altered our diets more than any other mammal. The regional fruits, vegetables, seeds, gums and other foods that some humans have introduced to their diets often contain polysaccharides with unique or variant structures that necessitate gut bacteria possess requisite degradative enzymes if they are to utilize them. While many novel diet introductions may be too infrequent to create the selective pressure for gut bacteria to acquire such functions, consumption of seaweeds has clearly selected for multiple lateral gene transfer events into human gut bacteria. A key aspect of this process is the presence of an environmental reservoir of appropriate genes and gene transfer mechanisms to enable these events. The existence of diverse marine Bacteroidetes, which use biochemically similar, PUL-based systems to degrade the algal polysaccharides that are abundant in their environment may help accelerate these events (Ficko-Blean et al., 2017; Hehemann et al., 2014; Kappelmann et al., 2019; Thomas et al., 2012). Our observation of Gram-positive Firmicutes that have acquired the ability to degrade porphyran and agarose suggests that there are additional environmental sources of these genes for human gut bacteria. While marine Bacteroidetes are abundant as planktonic and biomass associated bacteria involved in carbon cycling (Reintjes et al., 2019), the possibility that human gut Firmicutes derived their genes for agarose and porphyran degradation from organisms like Epulopiscium is intriguing. These bacteria are themselves intestinal symbionts of fishes and metagenomic surveys have revealed that they harbor CAZymes for degrading seaweed polysaccharides (Ngugi et al., 2017). Thus, it is intuitive that there are compatibility requirements for recipient gut bacteria based on their cellular organization (Grampositive or -negative) as well as genes expression signals and LGT mechanisms that determine if these genes can move between environments.
The precise paths through which genes that presumably originated in marine bacteria were transferred into the human gut microbiome remain unclear. The simplest scenario is that mobile elements containing genes for degrading seaweed polysaccharides already exist in marine bacteria and the bacteria that contain these genes were consumed along with the seaweed itself, leading to transfer into gut bacteria. This model is complicated by the requirement that conjugative transfer mechanisms, which the Bacteroides ICEs discussed here appear to rely on, require the donor species to be viable to initiate transfer. While it is not impossible, the requirement for aerobic marine Bacteroidetes to both survive seaweed processing (e.g., nori is pressed into sheets, dried and often roasted) as well as passage through the upper gastrointestinal tract to allow them to transfer genes in the colon present additional barriers that may limit the probability of this model. In a previous phylogenetic analysis of the ICE that harbors the B. plebeius porphyran PUL, we showed that this mobile element has closest relatives that exist in other mammalian gut bacteria and that many other variant ICEs contain different cargo genes located at the exact same site at which the porphyran PUL is located (Martens et al., 2014). Thus, a more complicated model could involve gut resident ICEs that have evolved the ability to acquire new DNA cargo from exogenous sources. Since the gut bacterial proteins involved in seaweed degradation often display distant homology to their marine counterparts and neither environmental intermediates or reservoirs of genes that are closely related to gut examples (Table S7) have been found, it is possible that these events occurred a long time ago. This scenario is probable given the long history of human seaweed consumption and the respective marine and human gut PULs may have undergone diversification after LGT into gut bacteria. Indeed, transfer might have even occurred into other mammalian gut bacteria prior to humans, although some level of selection based on seaweed consumption would presumably be needed as selection. Towards this latter point, it is interesting to note that a PUL with nearly identical organization to the B. plebeius porphyran PUL is present in the genome of a B. sartori strain (formerly B. chinchillae), which is a Bacteroides species typically associated with rodents (Kitahara et al., 2011). This strain was isolated from a pet Chinchilla in Japan. While it was not reported if this pet was ever provided P. yezoensis for food, it is apparent that the known genes for degrading this polysaccharide entered the genome of a species not commonly associated with the human gut.
Most of the algal polysaccharides investigated here have fundamentally different structures compared to those present in terrestrial plants, enabling our enrichment culture-based approach. However, there is undoubtedly a spectrum of gut microbial adaptations to the botanical source-specific variations in our foods and some of these may be quite subtle. One such example is gut bacterial adaptation to xyloglucan polysaccharides commonly found in fruits and vegetables. In solanaceous plants (eggplant, tomato) these polysaccharides have sidechains that terminate in α-linked arabinose residues, while in leafy vegetables (lettuce) they terminate in α-linked fucose. The human gut Bacteroides species that have so far been shown to degrade these polysaccharides commonly have enzymes to remove arabinose (Larsbrink et al., 2014). However, only some Bacteroides harbor PULs with the required fucosidases to fully degrade the fibers present in vegetables like lettuce (Dejean et al., 2019; Larsbrink et al., 2014), highlighting that adaptation to a different polysaccharide can involve a change as simple as the presence of one extra gene.
Much additional research is required to investigate the biochemistry of novel foods, especially those from local plants, fungi or fermentations, that are consumed by regional populations around the world. This knowledge will be required to fully understand the corresponding adaptations by members of the gut microbiome. As metagenomic sequencing methods become more sophisticated and applied more broadly to global populations, parallel exploration of the novel genes that have permeated the human gut microbiome as a function of dietary selective pressure will not only expand our knowledge of the microbiome’s adaptability but also reveal strategies for developing engineered probiotics that are coupled to short- or longterm term diet supplementation. Exploitation of these “orthogonal niches” holds powerful potential for engineering functions into the microbiome and implanting new microorganisms (Kearney et al., 2018; Shepherd et al., 2018). However, as evidenced by the unequal competition of the agarose/porphyran-degrading species that were implanted into humanized gnotobiotic mice, some of which had very similar genes, there is likely to be much left to understand about how these different gene clusters will function in different bacterial backgrounds and microbiomes. For example, B. thetaiotaomicronPUR2.2 was one of three strains with very similar porphyran PULs, but it was the dominant competitor when implanted into gnotobiotic mice. B. thetaiotaomicron and B. ovatus are polysaccharide generalists, each with the ability to degrade over 12 different glycans (Martens et al., 2011) and expression of the PULs involved is subject to catabolite repression (Rogers et al., 2012, Tuncil et al., 2017). Thus, it will be interesting to both determine how new PULs that are acquired through LGT are positioned in each host strain’s broader physiological hierarchy and how these phenomena influence engraftment and competition. Indeed, the alternative substrates that a strain can use, the actual substrates that are present in the host diet at the time of engraftment and the amount to which the presence of these nutrients influences expression of the newly acquired PUL may all be key determinants of success in filling orthogonal niches and therefore need to be investigated.
Star Methods
Resource availability
Lead contact:
Further information may be obtained from the Lead Contact Eric C. Martens (Email: emartens@umich.edu; address: University of Michigan Medical School, Ann Arbor, Michigan 48109, USA).
Materials availability:
All resources related to this study, including bacterial strains, are available by contacting the Lead Author.
Data and Code availability:
Shallow draft genome data of all isolates can be accessed using BioProject PRJNA625151 and corresponding BioSamples SAMN14593814–45. Bx18–002P, Fc548 and Fc588 WGS data can be accessed using Bioproject accession number PRJNA625151. All RNA-sequencing data can be accessed using GEO accession project number GSE149357 and corresponding samples GSM4498559–82. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD019149.
No new code was developed in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Subject Details
Animals
Germfree mice were from the C57BL/6NCrl strain and were reared, gavaged and maintained during the course of the experiment by the University of Michigan Germ-Free Mouse Facility. All experiments involving animals, including euthanasia via carbon dioxide asphyxiation, were approved by the University Committee on Use and Care of Animals at the University of Michigan (NIH Office of Laboratory Animal Welfare number A3114–01) and overseen by a veterinarian. Each experimental group contained 3 males and 2 females housed in groups according to the same sex.
Humans
All human fecal sample collections were approved by the Institutional Review Board of the University of Michigan Medical School (HUM00094242 and HUM00118951) and were conducted in compliance with the Helsinki Declaration. Study participants were recruited through the Authentic Research Sections of the introductory biology laboratory course at the University of Michigan (BIO173). individuals who had taken antibiotics withing the last 6 months or with self-reported history of inflammatory bowel syndrome, inflammatory bowel disease, or colorectal cancer were excluded from the study. All participants gave written, informed consent prior to participating in the study. Participants under the age of 18 were granted permission by a parent or legal guardian. Participants ranged in age from 17 to 29 years old, with a median age of 19 years old.
Method Details
Culture collection bacterial strains and growth conditions
A total of 354 human and animal gut Bacteroidetes were included in our initial survey. A complete list is provided in Table S1A, along with species designation based on 16S rDNA and associated meta-data. Species classifications were made based on BLAST alignment of each strain’s near full length 16S rRNA gene sequence to a database containing the type strains of the >40 named human gut Bacteroidetes species. Isolates with >98% 16 rDNA gene sequence identity to the type strain of a named species were labeled with that species designation. This classification strategy included all except 3 of the strains examined, which were all related to B. uniformis and had slightly more divergent 16S sequences (∼96% identity to the type strain). Because of the small number of strains that did not satisfy our 98% cutoff, we grouped these unclassified strains with their nearest relative and labeled them as more divergent in Table S1A. All seaweed-degrading strains are listed in Table S1B. All Bacteroidetes strains were routinely grown in an anaerobic chamber (Coy Lab Products, Grass Lake, MI) at 37°C under an atmosphere of 10% H2, 5% CO2, and 85% N2 on brain-heart infusion (BHI, Beckton Dickinson) agar that included 10% defibrinated horse blood (Colorado Serum Co.). Faecalicatena strains were grown in the same atmosphere on solid YCFA medium containing 0.1% taurocholic acid. For Bacteroidetes growth analysis, a single colony was picked into custom Chopped Meat Broth (CMB) and then sub-cultured into minimal medium (MM); For Faecalicatena strain growth analysis, CMB was used followed by a depleted medium (DM) (all media recipes can be found in Table S1C).
Human subjects and enrichment for seaweed degrading strains
Study participants were recruited through the Authentic Research Sections of the introductory biology laboratory course at the University of Michigan (BIO173). All participants gave written, informed consent prior to participating in the study. Participants under the age of 18 were granted permission by a parent or legal guardian. Participants ranged in age from 17 to 29, with a median age of 19. Individuals with self-reported history of inflammatory bowel syndrome, inflammatory bowel disease, or colorectal cancer were excluded from the study. This study was approved by the Institutional Review Board of the University of Michigan Medical School (HUM00094242 and HUM00118951) and was conducted in compliance with the Helsinki Declaration.
Human fecal samples were collected from healthy donors and grown in custom Chopped Meat Broth (CMB) in an anaerobic chamber (Coy Labs, Grass Lake, MI) with an 85% N2, 10% H2, 5% CO2 atmosphere. To isolate seaweed-degrading species, we transferred the CMB cultures (1:50) into minimal medium containing only one seaweed polysaccharide and incubated for ≥24 hrs. If growth was detected (defined as >0.1 OD600 above baseline, which was the same culture grown in minimal medium without added carbohydrates), cultures were streaked onto BHI plates containing 10% defibrinated horse blood (Quad Five, Ryegate, MT) and incubated for ≥48 hrs. Isolated colonies were sequentially purified by streaking 2–3x onto BHI-blood plates, then picked into fresh CMB and incubated for 24 hrs. Cultures were then grown in minimal media (MM) containing monosaccharides (MM-M) and incubated 24 hours prior to pelleting bacteria, washing twice in MM-no carbon (MM-NC) and transferring (1:100) into MM containing phenotypic respective seaweed polysaccharide. If isolates were still positive (i.e., based on the same >0.1 OD600 cutoff used above), isolates were stocked at −80ºC and pelleted for DNA extractions. Kinetic growth assays were performed using 100ul 2X concentrated MM-NC mixed with 100 μL, 1% carbohydrates (200 μL total culture volume containing 1:100 diluted and washed bacteria in 0.5% final carbohydrate) in 96-well plates (Costar) using a plate stacker coupled to a spectrophotometer (Biotek, Winooski, VT) as previously described(Martens et al., 2011). The above isolation methods resulted in all Bacteroides species isolated in this study except the previously isolated Bt3731 and Bo12C04, which were part of existing collections and the former was isolated in the 1960s-1970s. However, Faecalicatena contorta 548 and 588 strains were fortuitously discovered by noticing a characteristic agarolytic “pitting” around colonies on YCFA agar plates. Elevated background growth in liquid YCFA-no carbohydrate led us to formulate a depleted medium (DM, described above) used for all Faecalicatena species analysis.
Low-melt agarose (Lonza, 50101) and carrageenan (Sigma, 22049, C1138 and 22048) were used for all experiments. Ulvan (Elicityl) and fucoidan (V-Labs) were used for Bx18002-P growths. Porphyran was prepared by the following method: Unroasted nori was purchased locally, ground in a blender and autoclaved for 3hrs at 50g/L in water. The solution was cooled to room temperature with stirring and crude filtered to remove debris. Porphyran was precipitated with 80% final volume ethanol overnight at 4ºC. The precipitate was pelleted, supernatant removed and dissolved in water. The solution was centrifuged again to remove excess debris and co-extracted/pelleted polysaccharides (xylans). The porphyran containing supernatant was retained, lyophilized to reduce volume and extensively dialyzed against a 12–14 KDa dialysis membrane. Finally, the porphyran was lyophilized ad prepared at a 1% (10 mg/mL) autoclaved solution in water.
DNA sequencing
Bacteroides DNA was isolated directly using the DNeasy Blood and Tissue Kit (Qiagen). For Faecalicatena isolates, a standard phenol-chloroform (1:1) method with bead-beating was employed. The 16S rDNA sequencing was performed at the University of Michigan DNA Sequencing Core with universal primers 8F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’). Shallow draft genome sequencing of Bacteroides isolates was performed at the University of Michigan through the Host Microbiome Initiative Microbiome Core. However, strains with unknown genetic architectures, Bx18−002P, Fc548 and Fc588, were sequenced by MicrobesNG (Birmingham, UK) to obtain deeper, near-closed genome sequences. Reference based genome assemblies were generated using a 90% identity threshold in SeqMan NGen (DNASTAR) to determine coverage and generate corresponding read-mapping histograms. Unassembled short-read sequences for each sequenced Bacteroides isolate were aligned against the known PUL or ICE sequences from B. uniformis NP1 (agarose), B. plebeius DSM17135 (porphyran), B. thetaiotaomicron VPI-3731 (carrageenan) and/or B. ovatus CL02T12C04 (carrageenan) loci involved in seaweed polysaccharide degradation. A corresponding reference guided variant analysis was also performed in SeqMan NGen to calculate percent nucleotide identity over covered regions to the respective ICE or PULS using 90% identity threshold, ≥50% depth variance and at least 3X sequencing coverage as cutoffs.
RNA sequencing
Total RNA was extracted using a modified phenol-chloroform procedure to reduce seaweed polysaccharide carryover, namely for agarose and carrageenan, but not porphyran, which interfere with molecular biology applications by binding to RNA purification columns or forming solids. All strains were grown to near mid-log phase in 50 mL conical tubes and centrifuged to pellet bacteria. Cell pellets were washed with PBS to remove as much media as possible prior to resuspension in 3 mL RNA Protect Bacteria Reagent (Qiagen). After incubating for 5 min at room temperature (RT), cells were centrifuged at 7000xg for 10 min at RT, supernatants decanted and stored at −80ºC until RNA extractions. Cell pellets were thawed on ice, resuspended in 200 μL TE (10mM Tris, 1mM EDTA, pH8.0) buffer with lysozyme (1mg/mL) and incubated at RT for 2 min prior to adding 700 μL RLT Buffer (Qiagen) with 10ul/mL B-mercaptoethanol (Sigma). After mixing thoroughly, 500ul phenol:chloroform:isoamyl alcohol (125:24:1; pH 4.5) was added and mixed by inversion. Following an 18,000xg centrifugation for 3 min at 4ºC, the aqueous phase was removed into a new RNase free tube and another 500 μL phenol:chloroform:IAA was added, mixed and centrifuged as above. The aqueous phase was removed, 0.1 volume of 3M sodium acetate (pH5.5) and 600 μL cold 100% isopropanol were added and mixed by inversion. Importantly, tubes were rapidly centrifuged at 18,000xg for 1 min at RT to pellet precipitated polysaccharides. Some RNA may be lost at this stage but is required to remove residual seaweed polysaccharides (agarose and carrageenan) that interferes with downstream applications. Larger initial bacterial culture volumes (i.e., 25–100 mL) were required to overcome this loss. The RNA containing supernatant was retained and RNA was precipitated at −80ºC for 20 min before centrifuging at 18,000xg for 20 min at 4ºC. Supernatants were discarded and RNA pellets were washed with 70% isopropanol prior to centrifuging at 18,000xg for 5 min at 4ºC. Supernatants were discarded and pellets were dried at RT before resuspending in 100 μL nuclease free water. RNA was subjected to DNaseI treatment (NEB) prior to re-precipitating and pelleting of RNA as above, including the polysaccharide removal centrifugation step. Finally, RNA pellets were washed with 1.3 mL 70% isopropanol, prior to drying and resuspension in 50 μL RNase free water. Total RNA was quantified using a Qubit 2.0 (Invitrogen).
To ensure a DNA-free RNA extraction was successful, quantitative polymerase chain reaction (qPCR) was performed on all RNA samples using a standard curve of genomic DNA and 16S rDNA primers for respective strains. All samples had <0.01ng/μL of 16S rDNA and proceeded to rRNA depletion using the MICROBExpress Bacterial mRNA Enrichment Kit (Thermo Fisher) two times to sufficiently remove rRNA. Residual mRNA was quality controlled and converted to sequencing libraries at the University of Michigan Sequencing Core with an Illumina HiSeq-4000. Barcoded data were demultiplexed and analyzed using Arraystar software (DNASTAR, Inc.) using RPKM normalization with default parameters. Gene expression during growth on seaweed polysaccharides was determined by comparison to a galactose reference. Genes with significant up-or down-regulation were determined by the following criteria: genes with an average fold-change >10-fold and both replicates with a normalized expression level >1% of the overall average RPKM expression level.
Proteome analysis
For proteome analysis, Bt3731 was cultured in triplicate in MM with either 0.1% carrageenan from Cladosiphon okamurans (IEX purified) or 0.2% galactose, using previously described culture conditions (Martens et al., 2008) Pre-cultures with the same carbon source were used to set the starting OD600 of the main culture to 0.05. Cells were harvested in mid-exponential phase (∼0.25 OD600) by centrifugation at 4,000 x g for 10 min at 4°C and stored at −80°C until analysis. The soluble proteome (all soluble proteins), the secretome (all secreted proteins) and the membrane proteome (all proteins attached to membranes) were selectively enriched for further analysis. The soluble total proteome was extracted from cell pellets of carrageenan- and galactose-grown Bt3731 by sonication in TE buffer as previously described (Heinz et al., 2012). The secretome of carrageenan-grown cells was enriched from carrageenan culture supernatants using StrataClean beads (Otto et al., 2017) and the membrane proteome was enriched from pellets of carrageenan-grown cells using the trypsin shaving approach (Reisky et al., 2018). Soluble proteome and secretome samples were further analyzed in a gel-based proteomic approach, i.e. protein extracts were separated on a 10% polyacrylamide sodium dodecyl sulfate mini gel as previously described (Heinz et al., 2012). After CBB staining, entire gel lanes were dissected into 10 equal pieces, which were destained and overnight-digested with trypsin (1 μg/mL, sequencing grade, Promega), before MS analysis of the peptide mixes. Bt3731’s secretome samples were subjected to gel-free digestion (Otto et al., 2017) before MS analysis. Peptides were separated on a C18 column and subjected to reversed-phase chromatography on a nano-ACQUITY-UPLC (Waters Corporation). Mass spectrometry (MS) and tandem mass spectrometry (MS/MS) data were acquired using an online-coupled LTQ-Orbitrap mass spectrometer (Thermo Fisher) (Reisky et al., 2018). MS spectra were searched against a target-decoy protein sequence database, which included all predicted proteins of Bt3731 in forward and reverse directions (decoys) and a set of common laboratory contaminants. Validation of MS/MS-based peptide and protein identifications was performed with Scaffold v4 (Proteome Software Inc., Portland, OR, USA) using a maximum false discovery rate of 0.01 (1%) on the peptide level and 0.01 (1%) on the protein level. Only proteins that could be detected in at least two out of three biological replicates were considered identified. Normalized spectral abundance factors (%NSAF) were calculated for each protein by normalizing Scaffold’s ‘total spectrum counts’ against protein size and against the sum of all proteins in the same. %NSAF values give a proteins percentage relative to total protein abundance in a given sample, thus allowing for comparisons between individual samples (Zybailov et al., 2006).
Carbohydrate-polyacrylamide gel electrophoresis (C-PAGE) of carrageenases
The genes encoding for carrageenases GH82a, GH82b and GH16 were amplified by PCR using gene-specific primers and cloned into a pET-28a(+) vector. In the resulting plasmids, GH82a was fused to an N-terminal His6 tag and GH82b, GH16 to C-terminal His6 tags. The signal peptide and few N-terminal sequences were removed to increase solubility. All the constructs were transformed into E.coli BL21 (DE3) cells. A single colony from each of these constructs was used to inoculate an LB culture containing 50 μg/mL kanamycin at 37°C overnight with shaking at 150 rpm, respectively. These starter cultures of GH82a and GH16 were used to inoculate 1L of auto-induction ZYP-5052 medium supplemented with 50 μg/mL kanamycin and incubated at 20°C for 4 days with shaking at 150 rpm. 1L LB containing 50 μg/mL kanamycin was inoculated with GH82b starter culture at 37°C until an OD600 of 0.8 was reached. IPTG at a final concentration of 0.5 mM was added to the LB culture to induce protein production at 16°C with overnight shaking at 150 rpm. Cells were harvested and stored at −80°C.
To purify enzymes, stored cells were first thawed and then lysed using chemical lysis. Tris(2-carboxyethyl)phosphine (TCEP) was added at a final concentration of 1 mM to prevent disulphide bridge formation during lysis. A soluble lysate containing expressed protein was obtained by centrifugation at 16,000 x g at 4°C for 45 minutes. The enzymes were purified by immobilized metal ion affinity chromatography (IMAC) using a Ni2+ loaded HiTrap HP column. Protein elution was achieved using 20 mM Tris-HCl pH 8 and 500mM NaCl buffer with an increasing imidazole gradient up to 500 mM concentration. Fractions were verified for successful protein expression using SDS-PAGE and dialysed overnight in 20 mM Tris-HCl pH 8, 1 mM DTT and 250 mM NaCl. Samples were concentrated (Amicon) using a 10 KDa cut-off membrane and stored at 4°C.
Carrageenase activity screening was performed by incubating enzymes GH82a, GH82b and GH16 with 3 different types of carrageenan: λ, κ and ɩ-carrageenan at a final concentration of 0.3% at 37°C overnight. The enzymatic degradation products were analysed by carbohydratepolyacrylamide gel electrophoresis (C-PAGE) and visualized using 0.005% Stains-All in 50% ethanol solution followed by destaining with 10% ethanol.
Mariner Transposon Sequencing (Tn-Seq) of B. ovatus CL02T12C04
The B. ovatus CL02T12C04 was used for Tn-Seq because the Bt VPI-3731 isolate is both erythromycin and tetracycline resistant, while the former strain was only found to be resistant to erythromycin. To perform TnSeq using the pSAM mariner transposon and tetracycline resistance, the tetQ gene was incorporated into the plasmid using restriction sites flanking the originally included ermG gene (Goodman et al., 2009). The tetQ gene and promoter from the pKNOCK-bla-tetQ plasmid (Martens et al. 2008) was PCR-amplified using primers containing XhoI and XbaI restriction enzyme cut sites. The original pSAM_Bt (bla + ermG) was digested using XhoI and XbaI to remove ermG, purified vector fragment ligated with the tetQ PCR product creating a pSAM_Bt_tet (bla + tetQ) vector. Conjugations were performed using 10ml of B. ovatus CL02T12C04 grown in TYG broth and 10ml E. coli + pSAM_Bt_tet grown in LB broth containing ampicillin (300μg/ml), cells were pelleted and washed to remove ampicillin before pooling into 1ml that was plated onto BHI agar plates containing 10% defibrinated horse blood (Quad Five). This was done in parallel for a total of 15 conjugations. Plates were incubated agar side down at 37°C overnight in aerobic conditions. At 18h the conjugation mixtures were resuspended in 75ml TYG broth prior to plating each of 85 BHI-blood + gentamycin (200μg/ml) + tetracycline (20μg/ml) with 250μl of the cell resuspension. Plates were grown for 2d anaerobically (85% N2, 10% CO2, 5% H2) at 37°C resulting in ∼300 colonies/plate for a total of ∼25,000 transposon mutants. Bioinfomatics analysis revealed that at least 4,307 of the 6,186 (69.6%) of the annotated genes in this strain were disrupted. Insertions in intergenic regions were not considered. All colonies were scraped from plates and resuspended into 15ml TYG broth. This cell suspension was initially inoculated at a 1:10 dilution rate (to decrease risk of bottleneck) into 10ml of minimal media (MM) containing either glucose or carrageenan as the sole carbon source (n=3 replicate cultures per treatment). After 24 hours of anaerobic growth at 37°C, cultures were passaged into a fresh culture using the respective media and a dilution rate of 1:20. This was repeated a third time at a 1:50 dilution for a total of 3 passages per medium condition. Due to the interfering properties of negatively-charged carrageenan on nucleic acid purification, especially on cationic purification columns, both glucose and carrageenan triplicates were transferred one last time to 50ml MM-glucose to remove contaminating carrageenan from the culture that had previously been passaged in this substrate (attempts to isolate sufficient DNA directly from MM-CGN, even after growth was exhausted, were unsuccessful). These final cultures were grown for 48h cells, cells harvested and stored at −20°C prior to DNA extraction.
Genomic DNA extractions were performed using phenol:chloroform extraction and bead-beating as previously described (Martens et al. 2008). DNA pellets were resuspended in 500μl water and 50μg were used in an MmeI restriction digest overnight at 37°C. Digests were heat inactivated for 20 min at 65°C prior to running at 150V/30min on a 1% agarose gel. The DNA in the gel region corresponding to 1.5–4.0kb (expected size = ∼2.8kb) was excised for each replicate and purified using the Qiagen QIAquick gel extraction kit. A dsDNA adapter was created as previously described (Goodman et al., 2009) but using new P7Seq_Lig-NN and P7Seq_Lig oligos that contain the current Illumina P7 sequencing primer. The dsDNA adapter was ligated onto the MmeI digested genomic DNA using T4 DNA ligase (NEB) as previously described (Goodman et al., 2009). Ligations were heat inactivated at 65°C for 10min prior to purification using the Qiagen MinElute PCR purification kit. A PCR approach was utilized to incorporate the current Illumina adapters. One of the primers (P7 LibAdapt_X; where X is one of the i7 indexes used in this study) is homologous on the 3’ end to the ligated dsDNA adapter molecule but also contains a 5’ tail with multiplexed P7 adapter/i7 sequences. The other primer (P5 LibAdaptUniv MmeI) contains a 22bp 3’ sequence homologous to the original pSAM_Bt backbone. This sequence contains the MmeI recognition site through the first 4bp of the obsolete P7 adapter sequence but serves as an annealing sequence for a 5’ tail that contains the current P5 adapter/i5/sequencing primer sequences. All primers are described in Star Methods. PCR was performed with SuperFi polymerase (Invitrogen) using 200ng DNA and 20μM each primer in the following conditions: 95°C/3min, 20 cycles of 95°C/10s, 55°C/30s, 72°C/30s and a final elongation at 72°C/2min. Products were run on a 1.5% agarose gel at 150V for 60 min. DNA bands corresponding to ∼185bp were excised and purified with the Qiagen MinElute PCR purification kit using 20μl buffer EB to elute. DNA was quantified by Qubit (Thermo Fisher) prior to sequencing on a 2.5% shared NovaSeq flow cell at the UM Advanced Genomics Core.
Mobilization of B. uniformis agarose plasmids
All B. uniformis agarose-degrading isolates were initially tested for erythromycin (25 μg/ml) and tetracycline (4 μg/ml) resistance. Of these, all strains were resistant to tetracycline but 5 (KN201–2, 26A, JM204–1P, W1635P and W1665P) were sensitive to erythromycin. Based on this, we created erythromycin-resistant recipient strains of B. thetaiotaomicronVPI−5482, B. ovatusATCC 8483, B. fragilisNCTC 9343, B. cellulosilyticusDSM14838 and B. vulgatusATCC8482 by integrating an empty pNBU2-bla-ermGb vector into one of the two tRNAser sites that its targets (Koropatkin et al., 2008). We then used these strains to create spontaneous rifampicin (20 μg/ml) mutants resulting in a dual selection strategy for B. fragilisNCTC 9343, B. cellulosilyticusDSM14838 and B. vulgatusATCC8482. The ermR and rifR production was performed in B. thetaiotaomicronVPI5482 and B. ovatusATCC 8483 strains already lacking the thymidine kinase (tdk) gene allowing for a third selection factor in these strains as they are resistant to the pyrimidine analog 5-fluoro-2’deoxyuridine (FUdR; 200 μg/ml). Once recipient strains were prepared, the 5 erythromycin-sensitive B. uniformis donor strains containing the agarose plasmid were mated with each of the five other Bacteroides type strains resulting in 25 individual matings. For matings, all strains were grown anaerobically in chopped meat broth at 37°C overnight, transferred to MM-glucose and incubated anaerobically at 37°C overnight. Cells were diluted 1:50 into 5 ml MM-glucose and allowed to grow for 4 hrs prior to pelleting cells 2 min at 7,800 rpm. Supernatants were discarded and cells were resuspended in 500 μl MM-agarose. Each B. uniformis donor strain was added to each possible recipient strain and added to 4 ml MM-agarose to make a 5 ml total MMagarose mating culture that was incubated anaerobically at 37°C overnight. Matings were centrifuged for 2 min at 7,800 rpm to pellet cells, supernatants discarded, and resuspended in 500 μl of chopped meat broth. The cells were plated onto MM-agarose plates with erythromycin (25 μg/ml) and rifampicin (20 μg/ml) for B. fragilisNCTC 9343, B. cellulosilyticusDSM14838 and for B. vulgatusATCC8482 matings on and erythromycin (25 μg/ml), rifampicin (20 μg/ml) and FUdR (200 μg/ml) for B. thetaiotaomicronVPI−5482 and B. ovatusATCC 8483 matings. Plates were incubated anaerobically for 4 days at 37°C. If colonies were present, they were restreaked onto BHI-blood plates and incubated anaerobically for 3 days at 37°C prior to picking colonies into chopped meat broth. Possible recipients were 16S rRNA gene sequenced to verify mobilization into new species, analyzed for agarose utilization phenotypes and subjected to PCR analysis to detect the presence of the GH86 coding gene from the donor B. uniformis strains.
Identification of closest homologs for genes involved in seaweed degradation
The amino acid sequence for each identified gene in CGN, agarose and porphyran degradation was subjected to a protein BLAST (NCBI) search against non-redundant Refseq to identify its closest relative and the corresponding environmental association of the bacteria in which that closest relative exists. We omitted results from the strains investigated in this study, and the bacterium with the highest match was investigated for its environmental source using NCBI BioProject information or PubMed literature search. Additionally, the organisms in which the closest homolog(s) resides were listed as Gram negative or positive as an additional indication of the genetic reservoir of the potentially transferred genes.
Glycoside hydrolase 16 subfamily phylogeny
The GH16 enzymes from the Bt3731, Bo12C04, Bx18−002P, and Bp17135 were identified using dbCAN, and sequences were run through the SACCHARIS pipeline (Jones et al., 2018). SACCHARIS combines user sequences with sequences from the CAZy database (Cantarel et al., 2009) and trims sequences to the catalytic domain using dbCAN2 (Zhang et al., 2018). Sequences were aligned with MUSCLE (Edgar, 2004), and a best-fit model was generated with ProtTest (Darriba et al., 2011). The final tree was generated with FastTree (Price et al., 2010) and visualized with iTOL (Letunic and Bork, 2019).
Faecalicatena spp. amino acid phylogeny
FASTA amino acid files were run through the dBCAN2 server to annotate glycoside hydrolase 50, 86 and 117 families present in the Faecalicatena genomes. A multiple sequence alignment was generated for each glycoside hydrolase family using ClustalW in MUSCLE (EMBL-EBI) from amino acid sequences found in Faecalicatena spp. genomes, CAZy database, BLAST results or Protein Data Bank. The output file was converted to a MEGA file and a Maximum-Likelihood phylogeny created using a 100-resampling bootstrap analysis in MEGA X software.
Faecalicatena spp. protein expression and activity
Putative agarases and porphyranases were analyzed for signal peptides, which were subsequently removed if present prior to PCR amplification. Purified PCR products were ligated into the pETite vector containing an N-terminal His6 tag (Lucigen) and transformed into HI-Control 10G cells. After a 1hr recovery in Luria-Bertani (LB) broth at 37ºC, cells were plated onto LB plates supplemented with kanamycin (30 μg/mL) and incubated overnight at 37ºC. The fidelity of the constructs was confirmed prior to transformation into TUNER E. coli cells and plated onto LB with kanamycin. A single colony was picked into LB broth with kanamycin and incubated at 37ºC overnight. A 1L flask of Terrific Broth (TB) was inoculated with 5 mL of overnight culture and incubated at 37ºC with shaking at 200rpm until mid-log growth (0.6 OD600) when it was placed on ice. After 1hr on ice cells were induced with 0.2 mM final isopropyl-d-1-thiogalactopyranoside (IPTG). Cells were incubated with 200rpm shaking at 16ºC overnight then pelleted and stored at −80ºC until purification.
Enzymes were purified by centrifuging thawed cells in Talon Buffer (20 mM Tris, 300 mM NaCl, pH8.0) at 8,000 x g post sonication and purified on a HisPur Cobalt Resin (Thermo Scientific). Two separate elutions (5 mL (E1) and 10 mL (E2)) were performed and run on an SDS-PAGE gel to verify expression/purification. Soluble, expressed proteins were dialyzed overnight using a 12–14KDa membrane in Talon buffer. Enzyme activity assays on agarose and porphyran were done in Talon Buffer with 1 μM of enzyme and incubated overnight at 37ºC. Assays were heat inactivated and subjected to thin-layer chromatography (TLC) to check for enzymatic activity.
Metabolite quantification
A 1 mL aliquot of stationary phase cultures of Fc548, Fc588 and FfD5 grown on galactose, agarose and porphyran (n=3/substrate) were centrifuged to pellet cells and 200 μL of supernatants were filtered through a 0.22 μm 96-well filter. Filtrates (200 μL) were transferred to 250 μL pre-slit screw cap vials (Agilent) prior to high-performance liquid chromatography (HPLC) using a Shimadzu HPLC system (Shimadzu Scientific Instruments, Columbia, MD) with LC-10AD vp pump A, LC-10AD vp pump B, degasser DGU-14A, CBM-20A, autosampler SIL-10AD HT, column heater CTO-10A(C) vp, UV detector SPD-10A(V) vp, and an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA). A 0.01 N H2SO4 mobile phase with a 0.6 mL/min total flow rate and a column oven temperature at 50 ºC was used. Sample injections were 10 μL with a 40 min elution. The concentrations were calculated using a standard curve from a cocktail of short-chain organic acid standards (acetate, butyrate, formate, isobutyrate, isovalerate, lactate, succinate, propionate, valerate) at concentrations of 40, 20, 10, 5, 2.5, 1, 0.5, 0.25, and 0.1 mM. Values were then normalized to a media controls for respective galactose and porphyran cultures (agarose media control cannot be performed) and an OD600 of 1.0 to account for optical density variabilities by strain/substrate.
Gnotobiotic mouse colonization
Ten 8-week old C57BL/6NCrl germ-free mice were gavaged with a healthy human microbiota that was repeatedly screened to be negative for the ability to enrich agarose or porphyran degrading bacteria. All mice were kept on normal chow for 13 days prior to switching to a fiber-free diet (TD.130343, Envigo). At day 22, 5 mice were switched to a diet containing 3% Porphyra yezoensis (TD.190608, Envigo), which was ground into a course powder and added to TD.190608 to replace 3% of the dextrose contained in the base diet. To investigate engraftment of the different species tested, equal volumes of B. plebeius17135, B. thetaiotaomicronPUR2.2, B. xylanisolvens18−002P, B. xylanisolvensW1633C−1, B. ovatusPUR1.1, F. contorta548, F. contorta588, F. fissicatenaD5 were gavaged into all mice at day 23. After 8 days of 3% Porphyra yezoensis diet feeding in the experimental group (n=5) and 7 days post porphyrin-degrading strain gavage (n=10), all mice were sacrificed. Feces were collected from each mouse throughout the experiment at the time points shown in Fig. 5.
For DNA extractions, fecal pellets were combined with acid-washed glass beads (212−300 μm; Sigma-Aldrich, USA), 500 μL Buffer A (200 mM NaCl, 200 mM Tris, 20 mM EDTA), 210 μL SDS (20% w/v, filter-sterilized) and 500 μL phenol:chloroform (1:1). A Mini-BeadBeater-16 (Biospec Products, USA) was used to disrupt the bacterial cells for 3 min at room temperature then centrifuged and the aqueous phase was recovered. An equal volume of phenol:chloroform (1:1) was added to the aqueous phase and was mixed with the aqueous phase by gentle inversion. After centrifugation (12,000 rpm, 4°C, 3 min), the aqueous phase was recovered and 500 μL of pure chloroform was added to the aqueous phase, mixed by inversion and the tubes were centrifuged (12,000 rpm, 4°C, 3 min). The aqueous phase was transferred into fresh tubes and 1 volume of −20°C chilled 100% ethanol and 1/10 volume 3 M sodium acetate (pH 5.2) were added to the aqueous phase. The samples were mixed by gentle inversion and incubated at −80°C for 30 min, centrifuged for 20 min (12,000 rpm, 4°C) and the supernatants were discarded. The pellets were washed in 70% ethanol, air-dried and then resuspended in nuclease-free water. The resulting DNA extracts were purified by using DNeasy Blood & Tissue Kit (Qiagen).
The V4 region of the 16S rDNA gene was sequenced at the University of Michigan Microbial Systems and Molecular Biology Laboratory using an Illumina MiSeq platform as previously described (Kozich et al., 2013). The resulting16S rRNA abundance data were processed using the Mothur software package to reduce sequencing errors and remove chimeras as previously described (Schloss et al., 2009). Sequences were aligned to the SILVA 16S rRNA sequence database (Pruesse et al., 2007). Sequences were clustered into operational taxonomic units (OTUs) using a 99% similarity cutoff. The R package ‘vegan’ was used to calculate and plot the principal coordinates analysis (PCoA) from the Bray-Curtis dissimilarity index based on phylotype classification of the community. Analysis of molecular variance (AMOVA) was used to determine significance between community structure differences of different groups of samples. The generated OTUs were analyzed by BLAST to identify the corresponding species. The average relative abundance for highly represented OTUs were generated for each treatment group and sampling date and fold-change differences calculated only at day 30. Unpaired t-tests were run in Prism (GraphPad) using the Holm-Sidak correction to determine significance of OTU changes by day.
Survey of human metagenomic data sets including metagenome assembled genome (MAG) bins
Available cohorts of human gut metagenomic sequence data (National Center for Biotechnology Information projects: PRJNA422434 (Qin et al., 2012), PRJEB10878 (Yu et al., 2017), PRJEB12123 (Liu et al., 2017), PRJEB12124 (Gu et al., 2017), PRJEB15371 (He et al., 2017), PRJEB6997 (Zhang et al., 2015), PRJDB3601 (Nishijima et al., 2016), PRJNA48479 (Lloyd-Price et al., 2017), PRJEB4336 (Le Chatelier et al., 2013), PRJEB2054 (Qin et al., 2010), PRJNA392180 (Smits et al., 2017), and PRJNA527208 (Conteville et al., 2019) were searched for the presence of CGN/POR/AGAR degrading PUL nucleotide sequences from Bt37371, Bo12C04, Bp17135, BuNP1, Bx18−002P and Faecalicatena spp. using the following workflow: Each PUL nucleotide sequence was used separately as a template and then magic-blast v1.5.0 (Boratyn et al., 2019) was used to recruit raw Illumina reads from the available metagenomic datasets with an identity cutoff of 97%. Next, the alignment files were used to generate a coverage map using bedtools v2.29.0 (Quinlan and Hall, 2010) to calculate the percentage coverage of each sample against each individual reference. We considered a metagenomic data sample to be positive for a particular PUL if it had at least 70% of the corresponding PUL nucleotide sequence covered.
The Bo12C04 CGN PUL (112,734 bp) and the B. plebeius porphyran PUL (60,280 bp) were separately used as queries in a large-scale search against the assembled scaffolds of metagenome assembled genomes (MAG) bins) using the Integrated Microbial Genomes & Microbiomes (IMG/M) comparative analysis system (Chen et al., 2021). Within the LAST software package, version 1066, the ‘lastal’ tool was used with default thresholds to search the 2 loci against 122,126 MAG bins from 14,557 public metagenome datasets. Metagenome bins were generated using the binning analysis method described in (Clum et al., 2021).
Quantification and Statistical Analysis
For in vivo experiments, unpaired t-tests were run in Prism (GraphPad) using the Holm-Sidak correction to determine significance of OTU changes by day. Details of statistical tests used, sample size (“n”), definition of samples and definition of error bars are provided in figure legends.
Supplementary Material
Highlights.
Human gut bacteria have acquired genetic upgrades enabling edible seaweed digestion.
Bacteroides genes for agarose degradation reside on a large, mobilizable plasmid.
Some human Firmicutes have also gained the ability to degrade seaweed polysaccharides.
At least 4 separate events mobilized porphyran and/or agarose genes into gut bacteria.
Acknowledgements
We would like to thank Laurie E. Comstock for providing B. ovatus CL02T12C04 and Emma Allen-Vercoe for providing F. fissicatena D5 and the University of Michigan Germfree Mouse facility. We thank members of the Eric Martens and Nicole Koropatkin Labs for critical feedback during this project. A grant from the Howard Hughes Medical Institute (HHMI Grant # 52008119) and support from the University of Michigan’s Host-Microbiome Initiative to TMS supported work with the human cohort. This work was supported by NIH grants (GM099513, DK118024, DK125445, DK096023 to E.C.M). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02–05CH11231. We appreciate Jana Matulla’s and Sebastian Grund’s assistance during proteome sample preparation and MS measurements, respectively. The work of SM, TS, DB and JHH was financially supported by grants (BE 3869/4–2, SCHW 595/10–2, HE 7217/2–2) of the Deutsche Forschungsgemeinschaft (DFG) in the framework of the research unit FOR 2406 “Proteogenomics of Marine Polysaccharide Utilization” (POMPU). FU was supported by a scholarship from the Institute of Marine Biotechnology e.V. An Agriculture and Agri-Food Canada grant (J-002262) supported work from DWA.
Footnotes
Competing interests: Eric C. Martens serves a consultant and Scientific Advisory Board member of Novome Biotechnologies.
Inclusion and Diversity: One or more authors self identifies as an underrepresented minority in scientific research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arata PX, Quintana I, Raffo MP, and Ciancia M (2016). Novel sulfated xylogalactoarabinans from green seaweed Cladophora falklandica: Chemical structure and action on the fibrin network. Carbohydr Polym 154, 139–150. [DOI] [PubMed] [Google Scholar]
- Becker S, Tebben J, Coffinet S, Wiltshire K, Iversen MH, Harder T, Hinrichs KU, and Hehemann JH (2020). Laminarin is a major molecule in the marine carbon cycle. Proc Natl Acad Sci 117, 6599–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjdia A, Martens EC, Gordon JI, Berteau O (2011) Sulfatases and a radical S-adenosylL-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem 286, 25973–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, & Usadel B (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, Thierry-Mieg J, Thierry-Mieg D, Busby B, and Madden TL (2019). MagicBLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinformatics 20, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, and Lawley TD (2016). Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, and Henrissat B (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nuc Acids Res 37, D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IMA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M1 , Hajek P, Ritter S, Varghese N, Seshadri R, et al. (2021) The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res 49, D751–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clum A, Huntemann M, Bushnell B, Foster B, Foster B, Roux S, Hajek PP, Varghese N, Mukherjee S, Reddy TBK, et al. (2021) DOE JGI Metagenome Workflow. mSystems 6, e00804–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn DW, and Koropatkin NM (2016). Polysaccharide Degradation by the Intestinal Microbiota and Its Influence on Human Health and Disease. J Mol Biol 428, 3230–3252. [DOI] [PubMed] [Google Scholar]
- Conteville LC, Oliveira-Ferreira J, and Vicente ACP (2019). Gut Microbiome Biomarkers and Functional Diversity Within an Amazonian Semi-Nomadic Hunter-Gatherer Group. Front Microbiol 10, 1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. (2015). Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H (2021). Twelve years of SAMtools and BCFtools. GigaScience 10, giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, and Posada D (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjean G, Tamura K, Cabrera A, Jain N, Pudlo N, Holm Viborg A, Van Petegem F, Martens E, and Brumer H (2020). Synergy between cell-surface glycosidases and glycanbinding proteins dictates the utilization of specific beta(1,3)-glucans by human gut Bacteroides. mBio 11, e00095–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean G, Tauzin AS, Bennett SW, Creagh AL, and Brumer H (2019). Adaptation of Syntenic Xyloglucan Utilization Loci of Human Gut Bacteroidetes to Polysaccharide Side Chain Diversity. Appl Environ Microbiol 85, e01491–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficko-Blean E, Prechoux A, Thomas F, Rochat T, Larocque R, Zhu Y, Stam M, Genicot S, Jam M, Calteau A, et al. (2017). Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria. Nature Comm 8, 1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacki R, and Martens E (2020). If you eat it, or secrete it, they will grow: the expanding list of nutrients utilized by human gut bacteria. J Bacteriol 203, e00481–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone C, Knight R, Gordon JI (2009) Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin JM, Tamura K, Dejean G, Abbott DW, and Brumer H (2017). Polysaccharide Utilization Loci: Fueling Microbial Communities. J Bacteriol 199, e00860–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, Zhang D, Feng Q, Xie X, Hong J, et al. (2017). Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nature Comm 8, 1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Gao Y, Jie Z, Yu X, Laursen JM, Xiao L, Li Y, Li L, Zhang F, Feng Q, et al. (2017). Two distinct metacommunities characterize the gut microbiota in Crohn’s disease patients. Gigascience 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann JH, Boraston AB, and Czjzek M (2014). A sweet new wave: structures and mechanisms of enzymes that digest polysaccharides from marine algae. Curr Opin Struct Biol 28, 77–86. [DOI] [PubMed] [Google Scholar]
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, and Michel G (2010). Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912. [DOI] [PubMed] [Google Scholar]
- Hehemann JH, Kelly AG, Pudlo NA, Martens EC, and Boraston AB (2012a). Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci USA 109, 19786–19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann JH, Smyth L, Yadav A, Vocadlo DJ, and Boraston AB (2012b). Analysis of keystone enzyme in Agar hydrolysis provides insight into the degradation (of a polysaccharide from) red seaweeds. J Biol Chem 287, 13985–13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz E, Williams TA, Nakjang S, Noel CJ, Swan DC, Goldberg AV, Harris SR, Weinmaier T, Markert S, Becher D, et al. (2012). The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS pathogens 8, e1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DR, Thomas D, Alger N, Ghavidel A, Inglis GD, and Abbott DW (2018). SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol Biofuels 11, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappelmann L, Kruger K, Hehemann JH, Harder J, Markert S, Unfried F, Becher D, Shapiro N, Schweder T, Amann RI, et al. (2019). Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J 13, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney SM, Gibbons SM, Erdman SE, and Alm EJ (2018). Orthogonal Dietary Niche Enables Reversible Engraftment of a Gut Bacterial Commensal. Cell reports 24, 1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, and Smith TJ (2008). Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara M, Tsuchida S, Kawasumi K, Amao H, Sakamoto M, Benno Y, Ohkuma M (2011) Bacteroides chinchillae sp. nov. and Bacteroides rodentium sp. nov., isolated from chinchilla (Chinchilla lanigera) faeces. Int J Syst Evol Microbiol 61, 877–881. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, and Schloss PD (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79, 51125120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg S (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, et al. (2014). A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. [DOI] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, et al. (2017). Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med 23, 859–868. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, et al. (2017). Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro RR, Cornish ML, and C N (2017). Applications of carrageenan: with special reference to iota and kappa forms as derived from the Eucheumatoid seaweeds. [Google Scholar]
- Martens EC, Chiang HC, and Gordon JI (2008). Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe 4, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS, and Brumer H (2014). The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. Journal of molecular biology 426, 3851–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. (2011). Recognition and Degradation of Plant Cell Wall Polysaccharides by Two Human Gut Symbionts. Plos Biol 9, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu S, Touvrey-Loiodice M, Poulet L, Drouillard S, Vincentelli R, Henrissat B, Skjak-Braek G, and Helbert W (2018). Ancient acquisition of “alginate utilization loci” by human gut microbiota. Scientific Reports 8, 8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi DK, Miyake S, Cahill M, Vinu M, Hackmann TJ, Blom J, Tietbohl MD, Berumen ML, and Stingl U (2017). Genomic diversification of giant enteric symbionts reflects host dietary lifestyles. Proc Natl Acad Sci USA 114, E7592–E7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima S, Suda W, Oshima K, Kim SW, Hirose Y, Morita H, and Hattori M (2016). The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res 23, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onderdonk AB (1985). The carrageenan model for experimental ulcerative colitis. Prog Clin Biol Res 186, 237–245. [PubMed] [Google Scholar]
- Otto A, Maass S, Bonn F, Buttner K, and Becher D (2017). An Easy and Fast Protocol for Affinity Bead-Based Protein Enrichment and Storage of Proteome Samples. Methods Enzymol 585, 1–13. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluvinage B, Grondin JM, Amundsen C, Klassen L, Moote PE, Xiao Y, Thomas D, Pudlo NA, Anele A, Martens EC, et al. (2018). Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont. Nat Comm 9, 1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter NT, and Martens EC (2017). The Critical Roles of Polysaccharides in Gut Microbial Ecology and Physiology. Annual review of microbiology 71, 349–369. [DOI] [PubMed] [Google Scholar]
- Poyet M, Groussin M, Gibbons SM, Avila-Pacheco J, Jiang X, Kearney SM, Perrotta AR, Berdy B, Zhao S, Lieberman TD, et al. (2019). A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat Med 25, 1442–1452. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2010). FastTree 2--approximately maximumlikelihood trees for large alignments. PLoS ONE 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, and Glockner FO (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35, 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintjes G, Arnosti C, Fuchs B, and Amann R (2019). Selfish, sharing and scavenging bacteria in the Atlantic Ocean: a biogeographical study of bacterial substrate utilisation. The ISME journal 13, 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisky L, Stanetty C, Mihovilovic MD, Schweder T, Hehemann JH, and Bornscheuer UT (2018). Biochemical characterization of an ulvan lyase from the marine flavobacterium Formosa agariphila KMM 3901(T). Appl Microbiol Biotechnol 102, 6987–6996. [DOI] [PubMed] [Google Scholar]
- Rogers TE, Pudlo NA, Koropatkin NM, Bell JSK, Balasch MM, Jasker K, Martens EC (2013) Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Micro 88, 876–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 15, 2068–9. [DOI] [PubMed] [Google Scholar]
- Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, and Sonnenburg JL (2018). An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, et al. (2017). Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano Y, Kodama H, Terada I, Yamazaki Y, and Noma M (1994). Purification and characterization of a novel enzyme, alpha-neoagarooligosaccharide hydrolase (alpha-NAOS hydrolase), from a marine bacterium, Vibrio sp. strain JT0107. J Bacteriol 176, 6812–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F, Barbeyron T, Tonon T, Genicot S, Czjzek M, and Michel G (2012). Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteria to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ Microbiol 14, 2379–2394. [DOI] [PubMed] [Google Scholar]
- Tobacman JK (2001). Review of harmful gastrointestinal effects of carrageenan in animal experiments. Environ Health Perspect 109, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncil YE, Xiao Y, Porter NT, Reuhs BL, Martens EC, Hamaker BR (2017) Reciprocal Prioritization to Dietary Glycans by Gut Bacteria in a Competitive Environment Promotes Stable Coexistence. MBio 8, e01068–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana AG, Noseda MD, Goncalves AG, Duarte ME, Yokoya N, Matulewicz MC, and Cerezo AS (2011). beta-D-(1-->4), beta-D-(1-->3) ‘mixed linkage’ xylans from red seaweeds of the order Nemaliales and Palmariales. Carbohydrate research 346, 1023–1028. [DOI] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE, (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Computational Biology 13, e1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, et al. (2017). Metagenomic analysis of faecal microbiome as a tool towards targeted noninvasive biomarkers for colorectal cancer. Gut 66, 70–78. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, and Yin Y (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46, W95–W101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 21, 895–905. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Toukdarian A, Helinski D, Knauf V, Sykes S, Wilkinson JE, O’Bryne C, Shea T, DeLoughery C, and Caspi R (2001). Sequence analysis of a 101-kilobase plasmid required for agar degradation by a Microscilla isolate. Appl Environ Microbiol 67, 5771–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, and Washburn MP (2006). Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5, 2339–2347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Shallow draft genome data of all isolates can be accessed using BioProject PRJNA625151 and corresponding BioSamples SAMN14593814–45. Bx18–002P, Fc548 and Fc588 WGS data can be accessed using Bioproject accession number PRJNA625151. All RNA-sequencing data can be accessed using GEO accession project number GSE149357 and corresponding samples GSM4498559–82. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD019149.
No new code was developed in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.