Abstract
Chest X-ray becomes one of the most common medical diagnoses due to its noninvasiveness. The number of chest X-ray images has skyrocketed, but reading chest X-rays still has been manually performed by radiologists, which creates huge burnouts and delays. Traditionally, radiomics, as a subfield of radiology that can extract a large number of quantitative features from medical images, demonstrates its potential to facilitate medical imaging diagnosis before the deep learning era. With the rise of deep learning, the explainability of deep neural networks on chest X-ray diagnosis remains opaque. In this study, we proposed a novel framework that leverages radiomics features and contrastive learning to detect pneumonia in chest X-ray. Experiments on the RSNA Pneumonia Detection Challenge dataset show that our model achieves superior results to several state-of-the-art models (> 10% in F1-score) and increases the model’s interpretability.
Keywords: radiomics, medical imaging, CNN, chest X-ray, neural networks, interpretability
1. INTRODUCTION
Pneumonia is the leading cause of people hospitalized in the US [1]. It requires timely and accurate diagnosis for immediate treatment. As one of the most ubiquitous diagnostic imaging tests in medical practice, chest X-ray plays a crucial role in pneumonia diagnosis in clinical care and epidemiological studies [2]. However, rapid pneumonia detection in chest X-rays is not always available, particularly in the low-resource settings where there are not enough trained radiologists to interpret chest X-rays. There is, therefore, a critical need to develop an automated, fast, and reliable method to detect pneumonia on chest X-rays.
With the great success of deep learning in various fields, deep neural networks (DNNs) have proven to be powerful tools that can detect pneumonia to augment radiologists [3, 4, 5, 6]. However, most of the DNNs lacks explainability due to their black-box nature. Thus researchers still have a limited understanding of DNNs’ decision-making process.
One method of increasing the explainability of DNNs in chest radiographs is to leverage radiomics. Radiomics is a novel feature transformation method for detecting clinically relevant features from radiological imaging data that are difficult for the human eye to perceive. It has proven to be a highly explainable and robust technique because it is related to a specific region of interest (ROI) of the chest X-rays [7]. However, directly combining radiomic features and medical image hidden features provides only marginal benefits, a result mostly due to the lack of correlations at a “mid-level”; it can be challenging to relate raw pixels to radiomic features. In efforts to make more efficient use of multimodal data, several recent studies have shown promising results from contrastive representation learning [8, 9]. But, to the best of our knowledge, no studies have exploited the naturally occurring pairing of images and radiomic data.
In this study, we proposed a framework that leverages radiomic features and contrastive learning to detect pneumonia in chest X-ray. Our framework improves chest x-ray representations by maximizing the agreement between true image-radiomics pairs versus random pairs via a bidirectional contrastive objective between the image and human-crafted radiomic features. Experiments on the RSNA Pneumonia Detection Challenge dataset [10] show that our methods can fully utilize unlabeled data, provide a more accurate pneumonia diagnosis, and remedy the black-box’s transparency.
Our contribution in this work is three-fold: (1) We introduce a framework for pneumonia detection that combines the expert radiographic knowledge (radiomic features) with deep learning. (2) We improve chest X-ray representations by exploring the use of contrastive learning. Our model thus has the advantages of utilizing the paired radiomic features requiring no additional radiologist input. (3) We find that our models significantly outperform baselines in pneumonia detection with improved model explainability.
2. RELATED WORK
Pneumonia detection is a binary classification task which requires to classify a chest radiology image into either pneumonia or normal. Popular pneumonia detection dataset includes RSNA Pneumonia Detection Challenge [10] and pediatric pneumonia diagnosis1.
Traditionally, non-image features (e.g., patient age, gender, and body temperature) and radiomic features [11] are used for automatic chest disease classification. In recent years, many studies explored deep neural networks (DNNs) for this task [12, 13, 14]. For instance, Rajpurkar et al. introduced the CheXNet, a deep CNN trained to predict 14 diseases on chest X-ray [13]. Liang and Zheng used the Residual Neural Network (ResNet-18) [15] pre-trained on the NIH ChestX-ray 14 dataset and fine-tuned on the child’s chest X-rays dataset for pediatric pneumonia diagnosis [14].
For the medical image classification task, semi-supervised or unsupervised learning methods have benefited this task hugely because preparing annotated corpora is generally time-consuming and expensive. It also requires domain expertise and significant effort to ensure accuracy and consistency. To relieve this problem, one method is to utilize unlabeled image data. For example, Tang et al., [16] introduces the task-oriented unsupervised adversarial network, which consists of a cyclic I2I translation framework for RSNA Pneumonia Detection Challenge and a pediatric pneumonia diagnosis dataset.
Another popular trend, especially in recent years, is the contrastive representation learning [17, 18, 9]. Nevertheless, it may not be beneficial to directly apply these visual contrastive learning methods to medical images than pre-training models on ImageNet and fine-tuning them on medical images, mainly because the medical images have high interclass similarity [8]. Thus, Zhang et al. [8] proposed to use contrastive learning to learn visual representations from radiology images and text reports by maximizing the agreement between image-text representation pairs. Different from these works, we studied the contrastive learning between radiomics and convolutional neural networks (CNN) features to obtain medical visual representations. Therefore, our model does not require radiology text reports which are usually not publicly available. To this end, we deem that our framework is simple yet scalable when coupled with large-scale medical image datasets.
3. PROPOSED METHOD
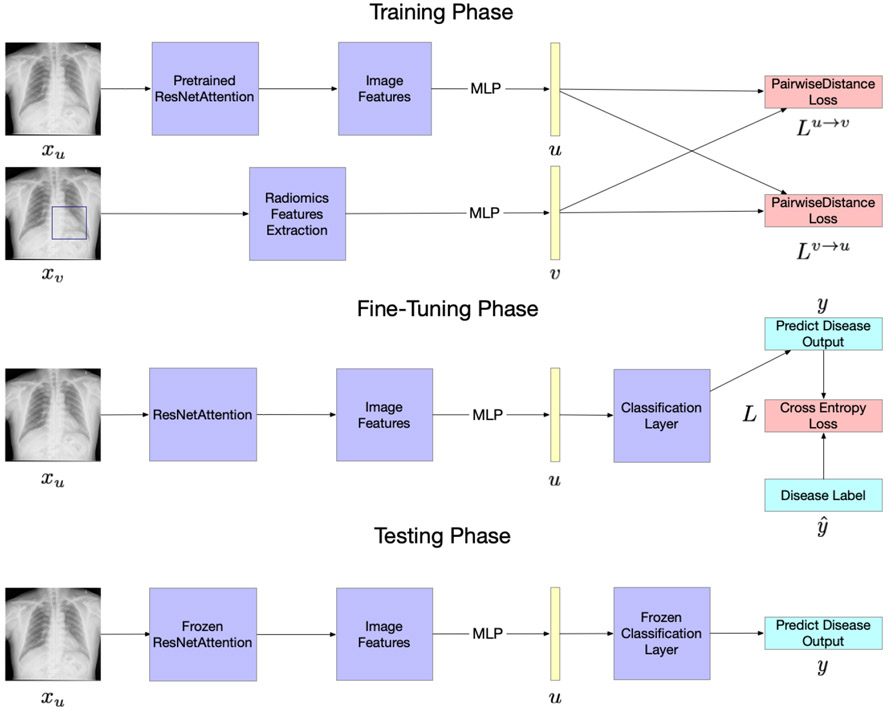
Inspired by recent contrastive learning algorithms [8], our model learns representations by maximizing agreement between radiomics features related to pneumonia ROI of the chest X-rays and the image features extracted by the attention-based convolutional neural network (CNN) model, via a contrastive loss in the latent space. Since radiomics can be considered as the quantified prior knowledge of radiologists, we deem that our model is more interpretable than others. As illustrated in Figure 1, our framework consists of three phases: contrastive training, supervised fine-tuning, and testing.
Fig. 1.
An overview of the proposed model.
Contrastive training.
The model is given two inputs, xu and xv. xu is the original chest X-rays without a corresponding paired bounding box. xv is the original chest X-rays with an additional paired bounding box. For normal chest X-rays, we take the whole image as a bounding box.
For xu, we utilize the pre-trained attention-based CNN models, Residual Attention Network (ResNet-18Attention) [19] pre-trained on CIFAR-10 [20], as the backbone of the network. We replace the last fully-connected layer with a multilayer perceptron (MLP) to generate a 128-dimensional image features vector u. For xv, we apply the PyRadiomics2 to extract 102-dimensional quantitative features, and [21] showed the details of these quantitative features and extraction process. We then use an MLP to map the features to a 128-dimensional radiomics feature v.
At each epoch of training, we sample a mini-batch of N input pairs (Xu, Xv) from the training data, and calculate their image features and radiomics features pairs (U, V). We use (ui, vi) to denote the ith pair. The training loss function will be divided into two parts. The first part is a contrastive image-to-radiomics loss:
| (1) |
where < ui, vi > represents the pairwise distance, i.e. and p represents the norm degree, e.g., p = 1 and p = 2 represent the Taxicab norm and Euclidean norm, respectively; and represents a temperature parameter. In our model, we set p to 2 and τ to 0.1. Like previous work [8], which uses a contrastive loss between inputs of the different modalities, our image-to-radiomics contrastive loss is also asymmetric for each input modality. We thus define a similar radiomics-to-image contrastive loss as:
| (2) |
Our final loss is then computed as a weighted combination of the two losses averaged over all pairs in each minibatch where λ ∈ [0, 1] is a scalar weight
| (3) |
Supervised fine-tuning.
We follow the work of Zhang et al. [8] by fine-tuning both the CNN weights and the MLP blocks together, which closely resembles how the pre-trained CNN weights are used in practical applications. In this process, the loss function is the cross-entropy loss where and y represent the true and predicted disease label, respectively.:
| (4) |
Testing.
The model is only given one input, the original chest X-rays xu without a corresponding paired bounding box. Image features are extracted then mapped into the 128-dimensional feature representation u. Finally, the predicted output is calculated based on u.
4. EXPERIMENTS AND DISCUSSION
4.1. Dataset and Experimental Settings
To evaluate the performance of our proposed model, we conducted experiments on a public Kaggle dataset: RSNA Pneumonia Detection Challenge3. It contains 30,227 frontal-view images, out of which 9,783 images has pneumonia with a corresponding bounding box. We used 75% imaged for training and fine-tuning and 25% for testing.
We used SGD as our optimizer and set the initial learning rate as 0.1. We iterated the training and fine-tuning process for 200 epochs with batch size 64 and early stooped if the loss did not decrease. We reported accuracy, F1 score, and the area under the receiver operating characteristic curve (AUC).
4.2. Results
We compared four models: (1) ResNet-18, (2) ResNet-18 with radiomics features (ResNet-18Radi), (3) ResNet-18 with the attention mechanism (ResNet-18Att), and (4) ResNet-18Attention with radiomics features (ResNet-18AttRadi).
Experimental results are shown in Table 1. Compared with the baseline models (ResNet-18 and ResNet-18Att), our radiomics-based models (ResNet-18Radi and ResNet-18AttRadi) achieved better performance on the pneumonia/normal binary classification task. It suggests that radiomic features can provide additional strengths over the image features extracted by the CNN model. Compared ResNet-18Att with ResNet-18 and ResNet-18AttRadi with ResNet-18Radi, we observed that the attention mechanism could effectively boost the classification accuracy. It proves our hypothesis that pneumonia is often related to some specific ROI of chest X-rays. Hence, the attention mechanism makes it easier for the CNN model to focus on those regions.
Table 1.
Experimental results
| Model | Accuracy | F1 score | AUC |
|---|---|---|---|
| ResNet-18 | 0.763 | 0.782 | 0.795 |
| ResNet-18Att | 0.815 | 0.826 | 0.848 |
| ResNet-18Radi | 0.851 | 0.901 | 0.898 |
| ResNet-18AttRadi | 0.886 | 0.927 | 0.923 |
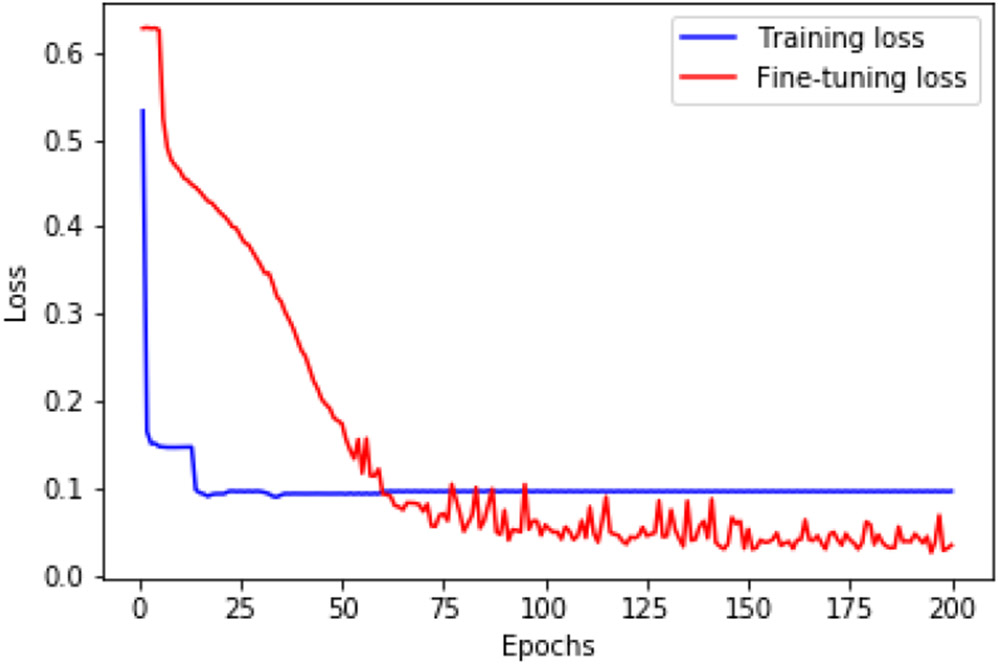
Figure 2 shows the training and fine-tuning loss convergence for the ResNet-18AttRadi model on the training set. We find that the loss drops rapidly during the pre-training stage within just a few epochs, revealing that contrastive learning makes the model learn to extract image features fast and effectively.
Fig. 2.
The training and fine-tuning loss convergence for the ResNet-18AttRadi model.
To fairly evaluate the impact of radiomics features on ROI, we conducted additional experiments using the whole image as a bounding box to extract the radiomics features, denoted as ResNet-18FairRadi and ResNet-18AttFairRadi. Table 2 shows that even if without ROI, the radiomics features could improve the performance of the deep learning model by 5% in F1 score. This observation further demonstrates that combining radiomics features with a deep learning model for reading chest X-rays is necessary.
Table 2.
Experimental Results Without Using Bounding Box
| Model | Accuracy | F1 score | AUC |
|---|---|---|---|
| ResNet-18 | 0.763 | 0.782 | 0.795 |
| ResNet-18FairRadi | 0.821 | 0.841 | 0.864 |
| ResNet-18Att | 0.815 | 0.826 | 0.848 |
| ResNet-18AttFairRadi | 0.854 | 0.884 | 0.877 |
4.3. Visualization of the deep learning model
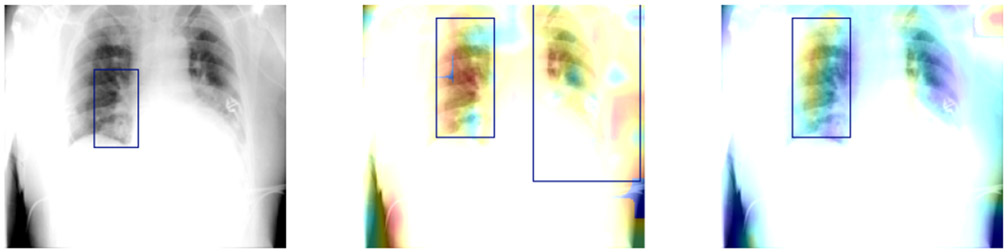
To demonstrate the interpretability of our model, we show some selected examples of model visualization, i.e., attention maps of ResNet-18Att and ResNet-18AttRadi. Figure 3 shows the original chest X-ray with a bounding box, attention map of the final attention layer of the ResNet-18Att and ResNet-18AttRadi, respectively. These examples suggest that our ResNet-18AttRadi model can focus on a more accurate area of the chest X-ray while ResNet-18Att attends to almost the whole image and contains plenty of attention noise. This illustrates that contrastive learning can help the model learn from radiomics features related to certain ROIs and thus attend more to the correct regions. And more examples of the attention maps can be found in the supplemental material.
Fig. 3.
An example of visualization of attention maps. The left figure is the original Pneumonia chest X-ray with a bounding box. The right two figures are the attention maps of the final attention layer ResNet-18Att and ResNet-18AttRadi, respectively.
5. CONCLUSION AND FUTURE WORK
In this work, we present a novel framework by combining radiomic features and contrastive learning to detect pneumonia from chest X-ray. Experimental results showed that our proposed models could achieve superior performance to baselines. We also observed that our model could benefit from the attention mechanism to highlight the ROI of chest X-rays.
There are two limitations to this work. First, we evaluated our framework on one deep learning model (ResNet). We plan to assess the effect of radiomic features on other DNNs in the future. Second, our model relies on bounding box annotations during the training phase. We plan to leverage weakly supervised learning to automatically generate bounding boxes on large-scale datasets to ease the expert annotating process. In addition, we will compare contrastive learning with multitask learning to further exploit the integration of radiomics with deep learning.
While our work only scratches the surface of contrastive learning using radiomics knowledge in the medical domain, we hope it will shed light on the development of explainable models that can efficiently use domain knowledge for medical image understanding.
Supplementary Material
7. ACKNOWLEDGMENTS
This project was supported by the National Library of Medicine under award number 4R00LM013001 and Amazon Machine Learning Grant. Ying Ding receives research support from Amazon. Yifan Peng is a coinventor on patents awarded and pending.
Footnotes
COMPLIANCE WITH ETHICAL STANDARDS
This research study was conducted retrospectively using human subject data made available [10]. Ethical approval was not required.
8. REFERENCES
- [1].Jain Seema, Self Wesley H, Wunderink Richard G, Fakhran Sherene, Balk Robert, Bramley Anna M, Reed Carrie, Grijalva Carlos G, Anderson Evan J, Courtney D Mark, et al. , “Community-acquired pneumonia requiring hospitalization among us adults,” New England Journal of Medicine, vol. 373, no. 5, pp. 415–427,2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tang Yu-Xing, Tang You-Bao, Peng Yifan, Yan Ke, Bagheri Mohammadhadi, Redd Bernadette A., Brandon Catherine J., Lu Zhiyong, et al. , “Automated abnormality classification of chest radiographs using deep convolutional neural networks,” NPJ digital medicine, vol. 3, pp. 70, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang Xiaosong, Peng Yifan, Lu Le, Lu Zhiyong, Bagheri Mohammadhadi, and Summers Ronald M, “Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in CVPR, 2017, pp. 2097–2106. [Google Scholar]
- [4].Jaiswal Amit Kumar, Tiwari Prayag, Kumar Sachin, Gupta Deepak, Khanna Ashish, and Rodrigues Joel JPC, “Identifying pneumonia in chest x-rays: A deep learning approach,” Measurement, vol. 145, pp. 511–518, 2019. [Google Scholar]
- [5].Wang Linda and Wong Alexander, “Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images,” arXiv:2003.09871, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kermany Daniel S, Goldbaum Michael, Cai Wenjia, Valentim Carolina CS, Liang Huiying, Baxter Sally L, McKeown Alex, Yang Ge, Wu Xiaokang, Yan Fangbing, et al. , “Identifying medical diagnoses and treatable diseases by image-based deep learning,” Cell, vol. 172, no. 5, pp. 1122–1131, 2018. [DOI] [PubMed] [Google Scholar]
- [7].Chen Bojiang, Zhang Rui, Gan Yuncui, Yang Lan, and Li Weimin, “Development and clinical application of radiomics in lung cancer,” Radiation Oncology, vol. 12, no. 1, pp. 154, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang Yuhao, Jiang Hang, Miura Yasuhide, et al. , “Contrastive learning of medical visual representations from paired images and text,” arXiv:2010.00747, 2020. [Google Scholar]
- [9].Chen Ting, Kornblith Simon, Norouzi Mohammad, and Hinton Geoffrey, “A simple framework for contrastive learning of visual representations,” arXiv:2002.05709, 2020. [Google Scholar]
- [10].Shih George, Wu Carol C, Halabi Safwan S, Kohli Marc D, Prevedello Luciano M, Cook Tessa S, Sharma Arjun, Amorosa Judith K, Arteaga Veronica, Galperin-Aizenberg Maya, et al. , “Augmenting the national institutes of health chest radiograph dataset with expert annotations of possible pneumonia,” Radiology: Artificial Intelligence, vol. 1, no. 1, pp. e180041, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gillies Robert J, Kinahan Paul E, and Hricak Hedvig, “Radiomics: images are more than pictures, they are data,” Radiology, vol. 278, no. 2, pp. 563–577, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Khan Wasif, Zaki Nazar, and Ali Luqman, “Intelligent pneumonia identification from chest x-rays: A systematic literature review,” medRxiv, 2020. [Google Scholar]
- [13].Rajpurkar Pranav, Irvin Jeremy, Zhu Kaylie, Yang Brandon, Mehta Hershel, Duan Tony, Ding Daisy, Bagul Aarti, Langlotz Curtis, Shpanskaya Katie, et al. , “Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning,” arXiv:1711.05225, 2017. [Google Scholar]
- [14].Liang Gaobo and Zheng Lixin, “A transfer learning method with deep residual network for pediatric pneumonia diagnosis,” Computer methods and programs in biomedicine, vol. 187, pp. 104964, 2020. [DOI] [PubMed] [Google Scholar]
- [15].He Kaiming, Zhang Xiangyu, Ren Shaoqing, and Sun Jian, “Deep residual learning for image recognition,” in CVPR, 2016, pp. 770–778. [Google Scholar]
- [16].Tang Yuxing, Tang Youbao, Sandfort Veit, Xiao Jing, and Summers Ronald M, “Tuna-net: Task-oriented unsupervised adversarial network for disease recognition in cross-domain chest x-rays,” in MICCAI, 2019, pp. 431–440. [Google Scholar]
- [17].Hénaff Olivier J, Srinivas Aravind, De Fauw Jeffrey, Razavi Ali, Doersch Carl, Eslami SM, and van den Oord Aaron, “Data-efficient image recognition with contrastive predictive coding,” arXiv:1905.09272, 2019. [Google Scholar]
- [18].He Kaiming, Fan Haoqi, Wu Yuxin, Xie Saining, and Girshick Ross, “Momentum contrast for unsupervised visual representation learning,” in CVPR, 2020, pp. 9729–9738. [Google Scholar]
- [19].Wang Fei, Jiang Mengqing, Qian Chen, Yang Shuo, Li Cheng, Zhang Honggang, Wang Xiaogang, and Tang Xiaoou, “Residual attention network for image classification,” in CVPR, 2017, pp. 3156–3164. [Google Scholar]
- [20].Krizhevsky Alex, Hinton Geoffrey, et al. , “Learning multiple layers of features from tiny images,” 2009. [Google Scholar]
- [21].Van Griethuysen Joost JM, Fedorov Andriy, Parmar Chintan, Hosny Ahmed, Aucoin Nicole, Narayan Vivek, Beets-Tan Regina GH, Fillion-Robin Jean-Christophe, Pieper Steve, and Aerts Hugo JWL, “Computational radiomics system to decode the radiographic phenotype,” Cancer research, vol. 77, no. 21, pp. e104–e107, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.