Abstract
The l-DOPA dioxygenase of Amanita muscaria (AmDODA) participates in the biosynthesis of betalain- and hygroaurin-type natural pigments. AmDODA is encoded by the dodA gene, whose DNA sequence was inferred from cDNA and gDNA libraries almost 30 years ago. However, reports on its heterologous expression rely on either the original 5′-truncated cDNA plasmid or artificial gene synthesis. We provide unequivocal evidence that the heterologous expression of AmDODA from A. muscaria specimens is not possible by using the coding sequence previously inferred for dodA. Here, we rectify and reannotate the full-length coding sequence for AmDODA and express a 205-aa His-tagged active enzyme, which was used to produce the l-DOPA hygroaurin, a rare fungal pigment. Moreover, AmDODA and other isozymes from bacteria were submitted to de novo folding using deep learning algorithms, and their putative active sites were inferred and compared. The wide catalytic pocket of AmDODA and the presence of the His-His-His and His-His-Asp motifs can provide insight into the dual cleavage of l-DOPA at positions 2,3 and 4,5 as per the mechanism proposed for nonheme dioxygenases.
1. Introduction
Molecular studies on the biosynthesis of secondary metabolites of basidiomycete fungi have been resurfacing because of modern heterologous gene expression platforms and multiomic technologies.1 Among these, the fly agaric Amanita muscaria (L.) Lam., with its white-dotted red pileus, is possibly the most iconic mushroom-forming species.2 Apart from its cultural and symbolic importance, the study of the chemical constituents of fly agaric provides us with the opportunity to explore the evolutionary connection between plants and fungi, to establish structure–bioactivity relationships of its metabolites, and to unravel the biochemical pathways leading to bright natural colors.
The biological and ecological functions of pigments in plants and fungi are more complex than meets the eye.3 The fly agaric and other basidiomycetes of the genus Amanita are pigmented by betalains and hygroaurins, whose biosynthesis requires the enzymatic extradiol insertion of molecular oxygen into the catechol ring of l-DOPA (Figure 1).4−6 Betalains also impart bright red-violet and orange colors and occasionally green fluorescence7,8 to most plants of the order Caryophyllales and have gained considerable attention because of their broad applicability,9−14 prompt availability,15 and importance as molecular markers in phylogenetic studies.16,17 In contrast, hygroaurins occur exclusively in some species of Amanita, Hygrocybe,18 and Hygrophorus fungi,19 and information on their occurrence, synthesis, and application is virtually nonexistent.20
Figure 1.
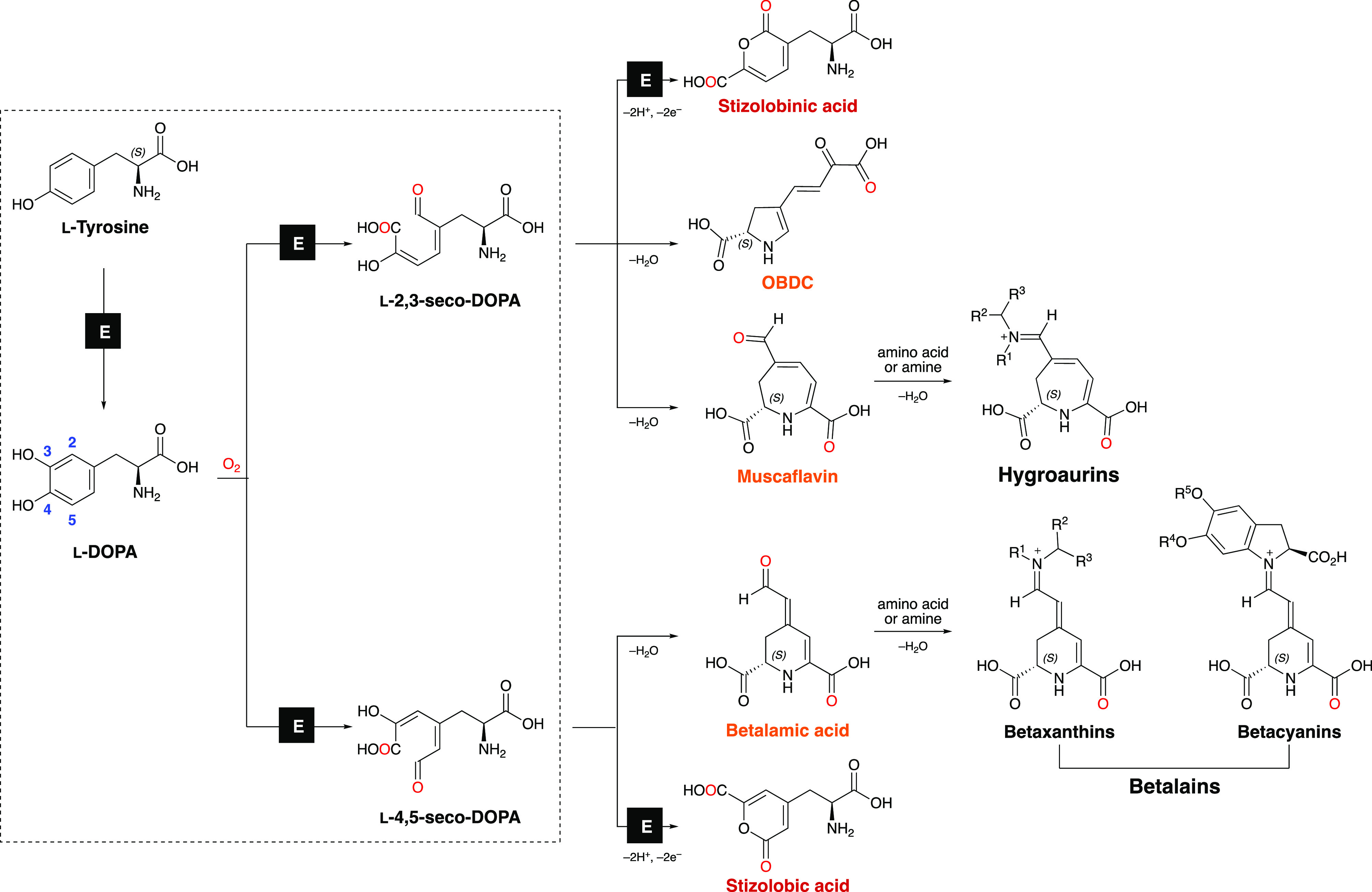
Biosynthesis of betalains, hygroaurins, and related compounds. Betacyanins and betaxanthins, the two classes of betalains, are produced by the spontaneous coupling between betalamic acid with cyclo-DOPA derivatives or amino acids/amines, respectively. The reaction of muscaflavin with amines or amino acids produces hygroaurins. The dehydrative cyclization of l-4,5-seco-DOPA is spontaneous under acidic aqueous conditions.32 Matching colored names (red/orange) indicate isomers. “E” designates enzymatic transformations. Amino and carboxyl groups are presented in the uncharged forms for clarity. Adapted with permission from Bastos, E. L.; Schliemann, W. Betalains as Antioxidants. In Plant Antioxidants and Health, Ekiert, H. M., Ramawat, K. G., Arora, J., Eds.; Springer International Publishing: Cham, 2021; pp 1–44. Copyright 2021 Springer Nature.
Hygroaurins and betalains are produced by the spontaneous coupling of nitrogen nucleophiles, such as amino acids and amines, with either muscaflavin or its isomer betalamic acid. The DOPA dioxygenase of fly agaric (AmDODA, EC:1.14.99.-) catalyzes the conversion of l-DOPA into l-2,3- and l-4,5-seco-DOPA, that spontaneously cyclize to muscaflavin and betalamic acid (Figure 1).21,22 However, the nonheme DOPA 4,5-extradiol dioxygenases of betalain-producing plants only catalyze the formation of only l-4,5-seco-DOPA (Figure 1).7
OBDC (l-4-(2-oxo-3-butenoic-acid)-4,5-dihydropyrrole-2-carboxylic acid),23 as well as stizolobinic and stizolobic acids24,25 are other secondary metabolites of applied interest, whose biosynthesis is strictly related to the catalytic properties of DOPA extradiol dioxygenases and the metabolism of seco-DOPAs. Discerning betalamic acid, muscaflavin, and OBDC requires the use of standards and modern mass spectrometry techniques, as these isomers have a mass-to-charge ratio (m/z) of 212 and produce a main daughter ion of m/z 166.26
The dodA gene (GenBank Y12886, 1629-bp) encodes the AmDODA protein (Uniprot P87064), and its description by Hinz and coauthors was the starting point to unravel the genetic basis of betalain biosynthesis.27 By screening a cDNA library from the pileus of A. muscaria using anti-DOPA dioxygenase antibodies, 20 positive clones were identified, all having a 612-bp open reading frame (ORF).27 The recombinant expression of these cDNA clones resulted in a fully functional enzyme as active as the native enzyme, and an extended coding sequence (687-bp) for this 228-aa AmDODA was predicted from an upstream ATG found in genomic clones.28,29 However, to date AmDODA recombinant expression was demonstrated using either the original cDNA plasmid28,30 or artificial gene synthesis.31
In this work, we rectify and reannotate the dodA gene and provide insights into the structures of AmDODA and other DODAs from muscaflavin-producing organisms. Combining the DNA sequencing of amplicons for dodA gene obtained from A. muscaria complementary and genomic DNA (cDNA and gDNA) samples, the heterologous expression of AmDODA, and in silico protein structure analysis, we provide the genetic basis for the heterologous expression of AmDODA from natural resources and contribute to the understanding of the catalytic properties and natural occurrence of DODAs.
2. Results and Discussion
2.1. Attempt to Express the Recombinant AmDODA from the Coding DNA Sequence (CDS) Isolated from A. muscaria Specimens
To obtain the DNA template for cloning the CDS for AmDODA, RNA was extracted from A. muscaria specimens collected in São Paulo, Brazil and used to synthesize cDNA. PCR amplification of cDNA samples using primers designed from the 687-bp CDS27 produced a 784-bp long DNA amplicon that is 97-bp longer than expected. The AmDODA CDS was inferred by Hinz and coauthors using a partial genomic library constructed from A. muscaria mushrooms collected in the Jorat forest near Lausanne, Switzerland.27,28 Sequencing of the internal transcribed spacer elements (ITS1 and 2) of the nuclear rDNA cluster of our specimens confirmed they correspond to the Eurasian fly agaric (GenBank: AB080779 and AB015700, Figure S1), eliminating any inconsistencies caused by the study of a different species.33−37
A multiple alignment between the DNA sequences of the 784-bp amplicon and dodA gene revealed that the exceeding 97-bp segment in the amplicon corresponds to the retention of the first intron of dodA, probably due to a GA sequence replacing the canonical AG signal at the 3′ splice site of this putative intron of dodA (positions 748–749, Figure 2).27 This result was confirmed by DNA sequencing of dodA amplicons from A. muscaria gDNA (Figure 2a), which also revealed that all other introns exhibited the conserved GT-AG at the 5′ and 3′ splice sites, respectively. In silico translation of the 784-bp amplicon produced a truncated 35-aa polypeptide encoded by a 105-bp upstream open reading frame (uORF) beginning at the same start codon inferred for the CDS for AmDODA but ending earlier due to a stop codon (TAG) located at the putative first intron (Figure 2b).
Figure 2.
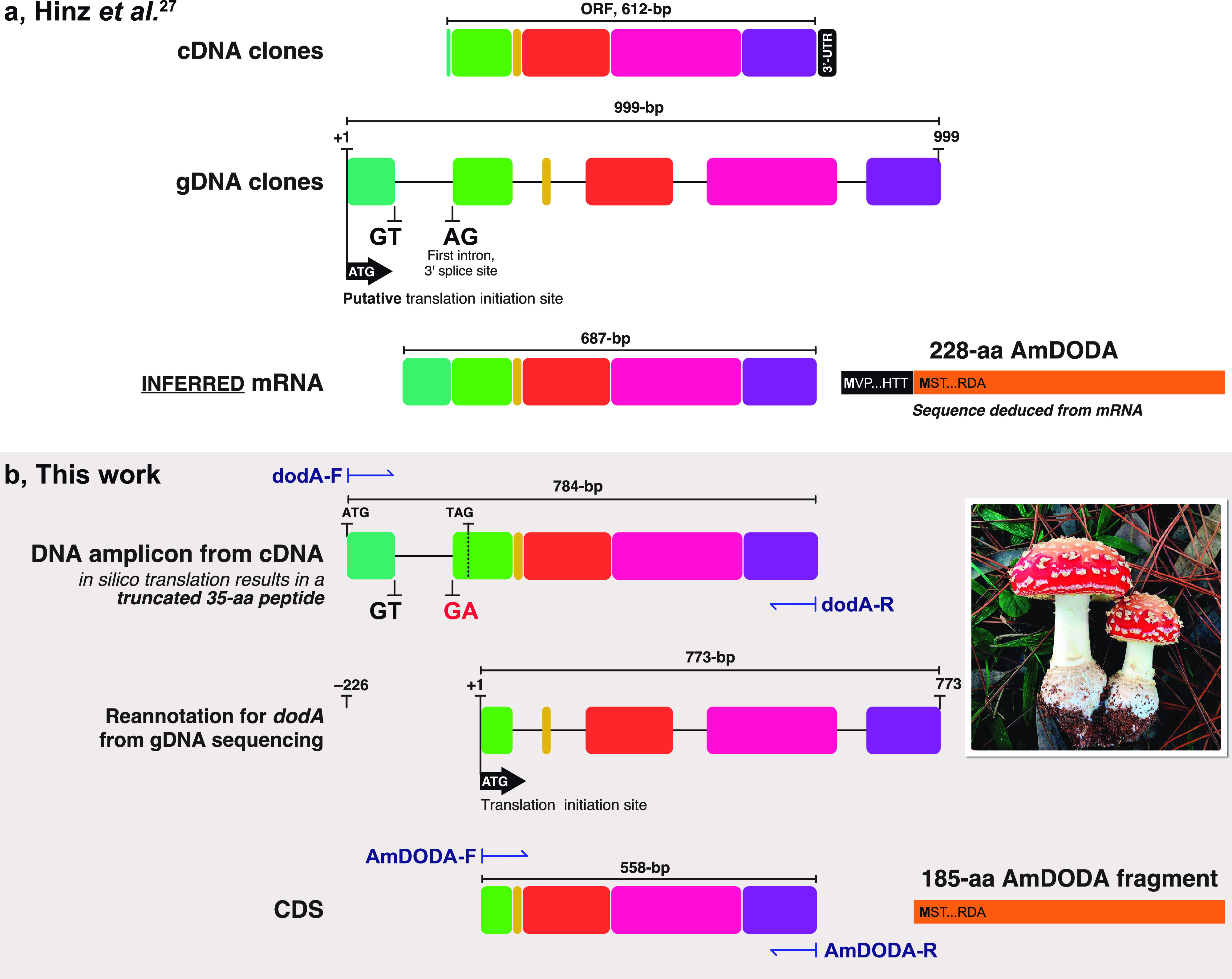
Comparison of the study reported by Hinz and coauthors27 and this work. (a) Structures of cDNA and gDNA clones isolated by Hinz and coauthors and the inferred 687-bp mRNA. Introns are presented as lines and exons as colored boxes; this schematic representation was created from the sequences reported in ref (27). The 228-aa AmDODA is represented by the black and orange bar showing the initial and final amino acid triads. The sequence was deduced by Hinz and coauthors from the mRNA and is presented in Figure S3. (b) DNA amplicon obtained by PCR amplification of the CDS for AmDODA from cDNA samples of Eurasian A. muscaria specimens collected in São Paulo, Brazil (inset photo; credit: Prof. C. V. Stevani). The “GA” replacing the canonical “AG” reported by Hinz and coathors leads to the retention of the first intron. A TAG stop codon indicates the 3′-end of the ORF used for the in silico translation of the 784-bp DNA fragment, which would result in a 35-aa truncated polypeptide. The structure of the dodA gene, as determined from DNA sequencing of gDNA amplicons, is shown and the transcription initiation site is indicated. The 558-bp CDS encoding the 185-aa AmDODA described in this work is presented. PCR primer sites are indicated using dark blue harpoon arrows. The complete sequence of the dodA gene is presented in Figure S2.
2.2. Reannotation of the dodA Gene Structure
Thirteen possible ATG codons were found in the 784-bp CDS amplicon. The ATG start codon at the beginning of exon 2 (position 227–229, Figure 2b) is embedded in a consensus Kozak sequence for basidiomycetes (ACUACCAUGU)38 and remains in the same reading frame as the original CDS for AmDODA. In silico translation of the resulting 558-bp sequence leads to a 185-aa polypeptide lacking the first 43 N-terminal amino acid residues of the published 228-aa AmDODA (Figures 2b and S3).27 All other start codons produce truncated polypeptides.
The 558-bp DNA sequence was amplified from A. muscaria cDNA samples and cloned into a pET28b-AmDODA plasmid, which encodes for 20 additional N-terminal amino acid residues, including a 6×His-tag. After the transformation of E. coli BL21(DE3) cells with the plasmid, the 205-aa His-tagged recombinant AmDODA expression was successfully induced, and its activity to promote the oxidation of l-DOPA was fully characterized, as described in the following section. These results confirm the correct CDS for AmDODA.
Hinz and coauthors used genomic clones to infer the structure of the dodA gene and the CDS for AmDODA.27 Even though their cDNA clones contain a 612-bp ORF encoding for functional AmDODA, which includes the 558-bp DNA sequence reported in this work, there are no further reports on the cloning of the predicted 687-bp CDS and its heterologous expression from A. muscaria specimens. The engineered production of betalains in stable transgenic non-Caryophyllales plants, namely, potato (Solanum tuberosum L.) and snapdragon (Antirrhinum majus L.),30 was performed using the original cDNA clones obtained by Hinz and coauthors.27 Likewise, the production of fluorescent betaxanthins in mammalian cells was reported by the expression of a plasmid containing a synthetic codon optimized CDS for AmDODA.31 Therefore, the rectification and reannotation of the dodA gene reported here is important for the description of the gene and protein sequences and enables its cloning and the heterologous expression of an active AmDODA enzyme from A. muscaria specimens. Moreover, AmDODA has the potential application as a genetically encodable reporter for noninvasive monitoring gene expression.39,40 The structure proposed for dodA comprises five exons interrupted by four introns (Figure 2b) and the complete CDS sequence was deposited at the GenBank (accession number MK922469).
2.3. Catalytic Activity of AmDODA and Chemoenzymatic Synthesis of l-DOPA-Hygroaurin
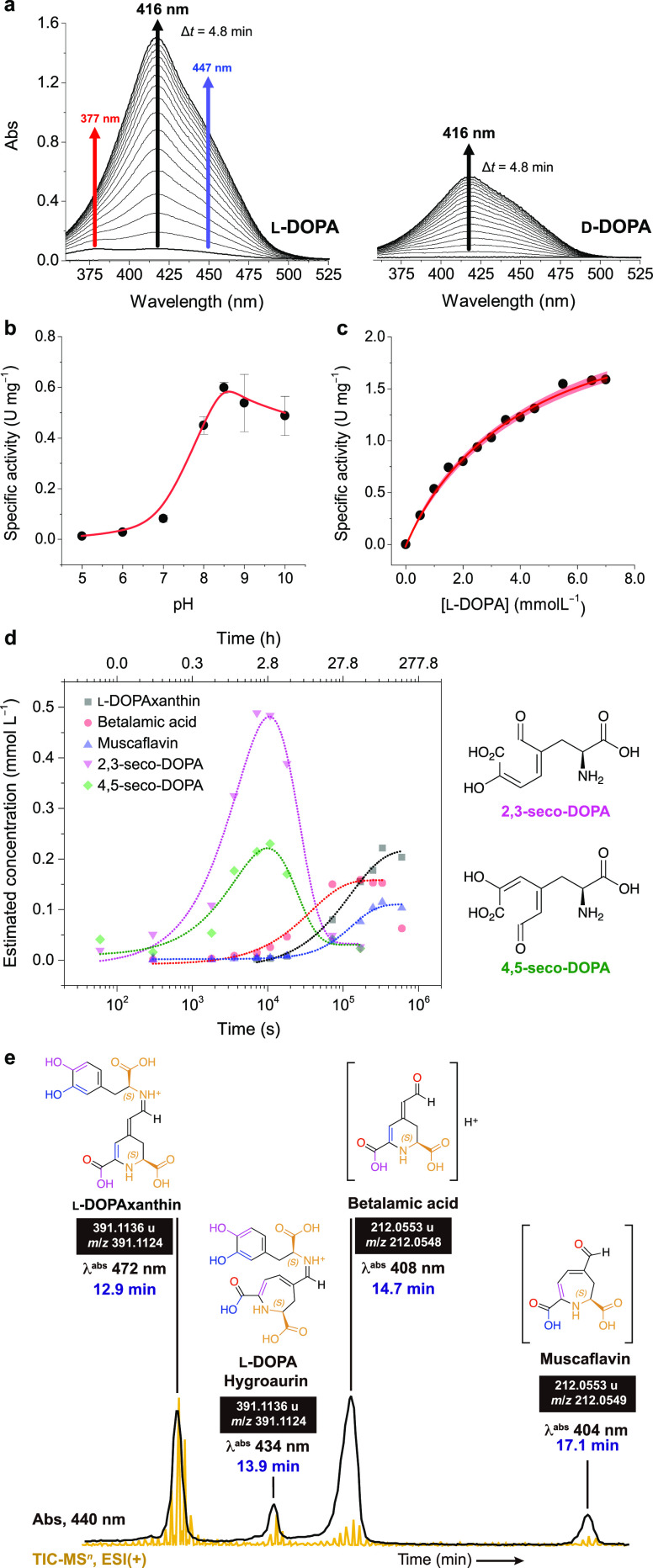
The 205-aa recombinant His-tagged AmDODA catalyzed the oxidation of l-DOPA into two different seco-DOPAs under slightly alkaline conditions at 25 °C. Ascorbic acid was used as an antioxidant to prevent the auto-oxidation of l-DOPA that is known to produce browning substances (Figures S4 and S5).41 The addition of l-DOPA to the biocatalytic system promptly turned the reaction yellow and was accompanied by the appearance of a broad absorption band with a maximum at 414 nm and shoulders at approximately 380 and 450 nm (Figure 3a). HPLC/PDA analysis (Figure S6) and comparison with literature data23 supports the formation of betalamic acid (λabs = 405 nm), muscaflavin (λabs = 400 nm), 2,3- and 4,5-seco-DOPA (λabs = 385 and 383 nm, respectively), and l-DOPAxanthin (λabs 466 nm). d-DOPA is converted into the respective stereoisomers and condensation products thereof, that is, isobetalamic acid and isomuscaflavin, at a much lower rate (Figure 3a). The maximum specific activity was reached at pH 8.5 (Figure 3b), as reported for the native enzyme,42 and the activity of AmDODA toward l-DOPA (0.5 U mg–1) was higher than d-DOPA (0.3 U mg–1).
Figure 3.
Enzyme activity. (a) Change in the UV–vis spectra during the reaction of d- and l-DOPA with oxygen in the presence of AmDODA and AscH. Effect of pH (b) and l-DOPA concentration (c) on the specific activity of AmDODA. The red shaded region shows the 95% confidence band for the nonlinear fit of the data to the Michaelis–Menten equation. (d) Kinetic traces of the reaction products and intermediates formed by the oxidation of l-DOPA in the presence of AmDODA. The reaction was monitored for up to 7 d. The structures of the seco-DOPAs are shown for clarity. Chromatograms, peak retention time, absorption spectra, and reaction conditions are shown in Figure S6 and were obtained using the chromatographic condition 1. (e) Chromatogram of products formed after 2 h of reaction at room temperature and a freezing-thawing cycle (chromatographic condition 2). Figure S8 shows the MS2 spectra and ion fragments of muscaflavin and betalamic acid. Experimental conditions: [AmDODA] = 1 μM, [AscH] = 10 mM, [l-DOPA] = 2.5 mM, sodium phosphate buffer (50 mM, pH 8.5).
At pH 8.5 and 25 °C, the KM and Vmax of AmDODA were found to be 4.2 ± 0.4 mM and 2.6 ± 0.1 mM min–1 (Figure 3c), respectively, corresponding to a turnover number (kcat) of 54.6 ± 0.1 min–1 (0.9 s–1), and a specificity constant (kcat/KM) of 13.0 ± 0.1 min–1 mM–1 (2.2 × 10–4 s–1 M–1). The KM of AmDODA was nearly identical to that of the native enzyme (3.9 mM)42 and Zrÿd’s recombinant enzyme (4.5 mM).28 Incubation of AmDODA with Chelex 100 to produce the apoenzyme precludes catalysis (Figure S7), indicating the requirement of a metal cofactor. Attempts to restore enzyme activity by adding different metal cations, including iron(II),21 were unsuccessful, possibly due to enzyme denaturation. Further, the participation of other metal cations such as Zn(II) and Mn(II) in AmDODA catalysis could not be ruled out by existing experimental evidence.
Although temporal changes in absorption spectra have been used to investigate the catalytic activity of AmDODA and other l-DOPA dioxygenases,28,42−45 spectral overlap makes the identification of intermediates and products challenging.46 Hence, the reaction kinetics were monitored by HPLC-PDA-ESI-qTOF-MS analysis (Figures 3d, S6, and S8), and the area of each chromatographic peak was converted into an estimated concentration using the kinetic model presented in Figure S9.
The concentration of the 2,3- and 4,5-seco DOPA intermediates reached their maxima after approximately 170 min of reaction (Figure 3d). Accordingly, change in the concentration of betalamic and muscaflavin over time showed sigmoid profiles, although betalamic acid was produced faster and in slightly higher (about 40%) amounts. DOPA-betaxanthin, but not OBDC and DOPA-hygroaurin, were detected under these experimental conditions. The use of a freezing–thawing cycle promotes the regeneration of hydrolyzed betalains47,48 and could favor the coupling of muscaflavin and l-DOPA. To test this hypothesis, we repeated the reaction by subjecting the mixture to a freezing–thawing cycle after 2 h at 25 °C, which led to the formation of l-DOPA-hygroaurin (Figure 3e). No evidence of hygroaurin formation was obtained when the reaction was performed at 25 °C possibly because of its labile character.
2.4. Phylogenetic Relationships and Insights into the Structure of AmDODA
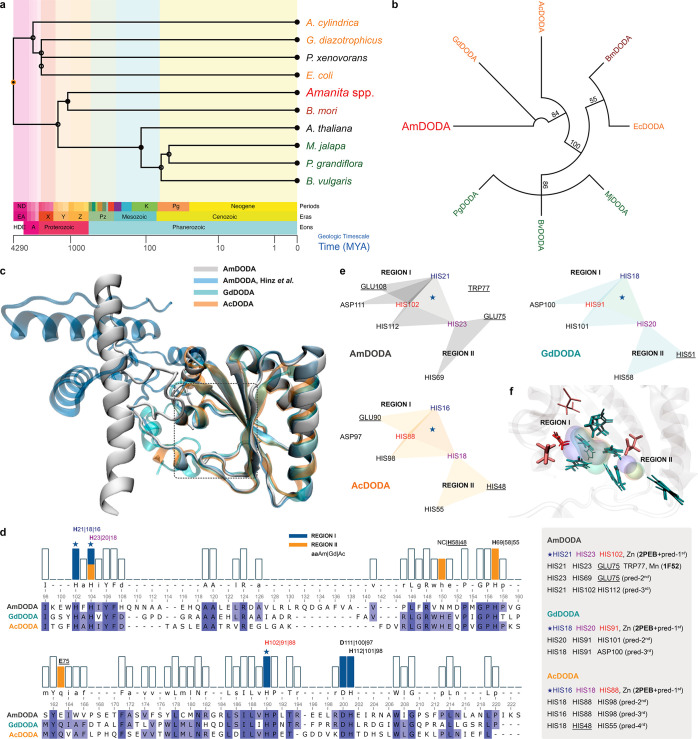
DODAs participating in muscaflavin biosynthesis were found in fungi (A. muscaria, AmDODA),27,28,42 bacteria [(G. diazotrophicus, GdDODA),43A. cylindrica, AcDODA,49 and E. coli, EcDODA],49 and insect (B. mori, BmDODA)50 but not in plants [M. jalapa, MjDODA,51B. vulgaris, BvDODAs44 and P. grandiflora, PgDODA5]. A time tree from these taxa, P. xenovorans, whose 2NYH protein shows high similarity with the fungal DODA,43 and the anthocyanin-pigmented A. thaliana was constructed to estimate the timing of species divergence (Figure 4a).52
Figure 4.
Phylogenetic relations and structural models of DODAs. (a) Divergence times for selected species producing dioxygenases. (b) Neighbor-Joining consensus tree inferred from protein sequences of functionally characterized l-DOPA extradiol dioxygenases (DODAs). Different groups of organisms are presented in the following colors: fungi (red), bacteria (orange), insects (brown), and plants (green). Branch support values >50% are indicated. Except for AmDODA, whose amino acid sequence was generated from the translation of the 558-bp CDS, protein sequences were obtained from the Uniprot database. (c) Structural alignment of AmDODA, GdDODA, and AcDODA. Enzymes were treated as monomeric units for simplicity, and the cutoff distance for the α-Cs was set to 140 pm (picometer). (d) Sequence alignment according to the structural alignment, and the results of the metal site prediction and homology search. The two regions of the putative catalytic pocket were assigned as I (blue) and II (orange). The star indicates the highest score motif for all three sequences. (e) Amino acid residues that are involved in complexation. Triangles show triads of amino acids that were predicted to bind metal cations in AmDODA (gray), GdDODA (green), and AcDODA (orange); underlined aa are not conserved in all three enzymes. (f) Amino acids at the putative catalytic pocket of the three DODAs. His (H), Glu (E), and Asp (D) residues are colored in cyan, pink, and red, respectively. Thick residues are from AmDODA. Colored volumes show the most probable region of metal cation binding; AmDODA (blue), GdDODA (green), and AcDODA (orange).
Hygroaurin-producing organisms diverged earlier than plants pigmented by either betalains or anthocyanins, an observation that is confirmed by the phylogenetic relationships between these species obtained comparing the amino acid sequences of their DODAs (Figures 4b and S10). Although AmDODA and plant DODAs share no common evolutive origin, AmDODA shows a pairwise identity of 31.7% with the bacterial AcDODA and GdDODA and all three enzymes show a significant identity with putative dioxygenases from the proteobacterium P. xenovorans strain LB400 (PDB Databank: 2P8I_D)53 and the filamentous cyanobacterium Nostoc punctiforme strain PCC 73102 (PDB Databank: 2PEB_A) (Table S1), whose structure has been elucidated. Therefore, to gain further insight into the structure of AmDODA, we used de novo folding based on deep learning algorithms to create models of DODAs from betalain- and hygroaurin-producing organisms. The resulting protein structures were subjected to conformational refinement, scanning for metal binding sites, and structural alignment (Figure 4c).
Structural differences in the N-terminal region of the 185-aa AmDODA fragment and the 228-aa protein described by Hinz and coauthors did not affect the putative active site, whose location is loosely indicated by a rectangle in Figure 4c and magnified in Figure S11. The active site of AmDODA, AcDODA, and GdDODA was predicted considering the most likely binding sites for metal cations, as well as by homology with metalloenzymes (Figure 4d–f and Table S2). The initial portion of AmDODA was not conserved in other fungal dioxygenases (Figure S12), as well as in AcDODA and GdDODA, in agreement with previous reports which suggest that the lack of about 30 N-terminal residues does not affect the overall stability and activity of the fungal enzyme.27,28 These findings also corroborate our observation that the lack of the first 43 amino acids in the 185-aa AmDODA compared to the inferred 228-aa AmDODA does not affect its catalytic properties.
The resulting structural alignment of AmDODA, AcDODA, and GdDODA reveals that these proteins have more than 99-aa overlapping residues (Table S1) and that a conserved His-His-His motif in the region I of the catalytic pocket is the most likely metal cation binding site (Figure 4e). This prediction is supported by the fact that this motif includes the conserved His residue found from the multiple alignment of all DODAs (Figure S10 and Figure 4d; AmDODA/H102, Pg/H177, Bv/H175, Mj/H168, Ec/H177, Bm/H175, Ac/H88, and Gd/H91), which has been considered essential for the catalytic activity of extradiol dioxygenase class III enzymes,5 and by the high homology with the Zn(II) binding site of the putative dioxygenase from N. punctiforme PCC 73102. Notably, an Asp residue near the 3His motif is conserved in these DODAs, and proximal His-His-His and His-His-Asp motifs are involved in Fe(II) complexation at the active site of several extradiol dioxygenases.21,54 Although the His21-His23-Glu75-Trp77 motif of AmDODA has high homology with the binding site for Mn(II) in glutamine synthetase from Salmonella typhimurium (1F52), it is not conserved in AcDODA and GdDODA, and hence it is unlikely to participate in the oxidation of l-DOPA. Region II has two conserved His residues and the Glu75 residue for AmDODA or the His48 and His51 residues for GdDODA and AcDODA, respectively, making its participation in catalysis unlikely as well. However, the combined regions I and II make the catalytic pocket wide enough to accommodate l-DOPA, and perhaps to favor the cyclization of the 2,3- and 4,5-seco-DOPAs, as proposed for the EcDODA protein.55
3. Conclusion
The complete CDS of the fly agaric l-DOPA dioxygenase was rectified and reannotated, enabling its heterologous expression from A. muscaria cDNA samples to produce muscaflavin and hygroaurins. The recombinant AmDODA produced is 43-aa residues shorter than previously reported but shows the same catalytic performance of both the native and larger recombinant enzymes, confirming the cloning and expression of the full-length cDNA. Hygroaurins can be conveniently obtained using the freezing–thawing protocol described here, making it possible to study this rare class of natural pigments. The dual cleavage of l-DOPA by DODAs participating in muscaflavin biosynthesis could be explained by their conserved His-His-His and His-His-Asp motifs in the putative catalytic pocket, which is wide enough to accommodate large substrates.
4. Material and Methods
4.1. Molecular Identification of A. muscaria
A. muscaria mushrooms were collected in Santana de Parnaíba, São Paulo, Brazil (23°28′18.8″ S, 46°51′50.2″ W) on June 20, 2018. The internal transcribed spacer (ITS) region of its nuclear rRNA gene cluster was analyzed for taxonomic purposes. Initially, 100 mg of the red-colored cap tissue was disrupted using a mortar and pestle in the presence of liquid nitrogen. Genomic DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN), following the manufacturer’s instructions. PCR reactions were carried out in a final volume of 25 μL with the enzyme Platinum Taq DNA Polymerase (Invitrogen) using 50 ng of gDNA and the conserved primers flanking the rDNA region containing the ITS sequences: ITS5F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4R (5′-TCCTCCGCTTATTGATATGC-3′). The reactions were incubated in the SimpliAmp thermal cycler (Applied Biosystems) at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, primer annealing at 50 °C for 30 s, and DNA extension at 72 °C for 1 min. PCR products were separated by electrophoresis on a 1.5% agarose gel for 35 min at 130 V. DNA bands (∼700 bp) were excised and purified using the GenElute gel extraction kit (Sigma-Aldrich). Purified PCR amplicons were cloned into pGEM-T Easy vectors (Promega) using a molar ratio of 5:1 (insert/vector), following the manufacturer’s instructions. Chemocompetent Escherichia coli Stellar cells were transformed by ligation reactions. Reactions were plated in selective LB media containing ampicillin and incubated at 37 °C for 16 h. Positive clones were confirmed by plasmid DNA extraction, followed by EcoRI digestion and electrophoresis on a 1.5% agarose gel. DNA sequencing reactions were performed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) using 5 μL of plasmid DNA (100 ng/μL) and 2.5 μL of 5 μM sequencing primers M13F (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) or M13R (5′-CAGGAAACAGCTATGAC-3′). Two positive clones were sequenced in duplicate using the Sanger method on an ABI 3730 DNA analyzer (Applied Biosystems) at the Centro de Pesquisa sobre o Genoma Humano e Células-Tronco da Universidade de São Paulo (CEGH-USP, Brazil). DNA sequences were analyzed using the software Geneious Prime 2020.2.4 (Biomatters), and the consensus sequences produced were aligned with the nucleotide database of the National Center for Biotechnology Information (NCBI) using the online tool Blastn.
4.2. Cloning of the Coding Sequence of dodA
DNA and mRNA samples were extracted from the red-pigmented pileus using DNeasy and RNeasy kits (QIAGEN). cDNA was synthesized from mRNA (1 μg) using SuperScript III reverse transcriptase (Thermo Fisher Scientific). Primer design was carried out using the DNA sequence of A. muscaria dodA (GenBank Y12886).27 The coding sequence of dodA was PCR amplified from DNA and cDNA samples using the primers dodA-F (CACCATGGTGCCAAGCTTCGTTGT) and dodA-R (CTATGCATCTCGATGGGGCGCTCT). PCR was carried out under standard conditions using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific). Samples were kept at 98 °C for 30 s (1 cycle), 98 °C for 10 s (30 cycles), 62 °C for 30 s (1 cycle), and 72 °C for 1 min (1 cycle), followed by a final extension phase at 72 °C for 7 min. Products were cloned into pENTR/SD/d-TOPO (Thermo Fisher Scientific), and transformed into DH5α-competent E. coli (Thermo Fisher Scientific), according to the manufacturer’s instructions. The sequence was checked by DNA sequencing of the positive colonies using the primers M13 forward (5′-GTAAAACGACGGCCAG-3′) and M13 reverse (5′-CAGGAAACAGCTATGAC-3′). On the basis of the results obtained, a new CDS was proposed for dodA, which was deposited in the NCBI database (GenBank accession number MK922469). cDNA samples from A. muscaria were used for PCR gene amplification using the AmDODA-F (5′-ACTTTAAGAAGGAGATATACATGTCCACCAAGCCAGAG-3′) and AmDODA-R (5′-GTCGACGGAGCTCGAATTCGGTGCATCTCGATGGGGCG-3′) primers. PCR was performed under standard conditions using Q5 High-Fidelity DNA Polymerase (New England Biolabs). Samples were kept at 98 °C for 30 s (1 cycle), 98 °C for 10 s (30 cycles), 60 °C for 30 s (1 cycle), and 72 °C for 1 min (1 cycle), followed by a final extension phase at 72 °C for 2 min. PCR products were cloned into the pET28b vector (Novagen) linearized with BamHI and NotI, following the sequence and ligation-independent cloning (SLIC) method.56 The recombinant plasmid pET28b-AmDODA was confirmed by DNA sequencing. CDS amplification from A. muscaria cDNA samples was repeated in triplicate to eliminate any technical issues, and the DNA sequence of the 784-bp amplicon was determined subsequently.
4.3. Expression and Purification of AmDODA
E. coli strain BL21(DE3) (New England Biolabs) was transformed with the recombinant plasmid pET28b-AmDODA to express the C-terminal His-tagged AmDODA. 2-YT (tryptone/yeast extract) medium (3 mL) supplemented with kanamycin (50 μg mL–1) was inoculated with E. coli BL21(DE3) pET28b-AmDODA and maintained overnight at 37 °C in an orbital shaker operated at 200 rpm. The cultures were transferred to 1.0 L baffled Erlenmeyer flasks containing 250 mL 2-YT/kanamycin medium and shaken at 200 rpm and 37 °C for approximately 2 h to a final optical density of 0.4 to 0.6 at 600 nm. Isopropyl β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 0.5 mM to induce AmDODA expression, and the flasks were incubated for 16 h at 30 °C. Cells were harvested by centrifugation (8000×g, 4 °C, 30 min) and resuspended in sodium phosphate lysis buffer (Supporting Information). Cell lysis was performed in a French Press Cell G-M (Thermo Fisher Scientific), and the recombinant protein was purified by gravity-flow chromatography using a nickel-charged resin Ni-NTA Agarose (QIAGEN) equilibrated with 10 mM imidazole in lysis buffer. An elution buffer (linear gradient of imidazole from 100 mM to 500 mM) was used to elute AmDODA. Fractions containing the enzyme were identified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)57 using 15% polyacrylamide gels and stained using the standard Coomassie Blue method. Pure fractions were pooled and desalted by dialysis in a sodium phosphate buffer (50 mM, pH 7.4). Protein concentrations were determined by the dye-binding method of the Bradford assay (Bio-Rad); bovine serum albumin was used as the calibration standard. The revised nucleotide sequence was deposited in the GenBank database (accession number MK922469).58
4.4. Enzyme Catalysis
Oxidation of l-DOPA in the presence of AmDODA was monitored every 18 s for 5 min by UV–vis absorption spectroscopy (300–600 nm, scan rate: 2400 nm min–1) using a Varian Cary 50 Bio spectrophotometer, equipped with a cell holder thermostatic at 25 °C. The reaction was initiated by adding 1 μM of the enzyme to a solution of l-DOPA (1 mM) and ascorbic acid (AscH, 10 mM) in sodium phosphate buffer (50 mM, pH 8.5) unless otherwise stated. The pH was corrected using a base after the addition of AscH. Product formation was monitored by the increase in absorption at 414 nm over time, and the initial rate of product formation (in μM min–1) was calculated by linear regression, assuming the molar absorptivity coefficient (ε) at 424 nm to be 24 000 M–1 cm–1 for both products.43,59 The specific activity of the enzyme was calculated by dividing the initial rate of product formation (in μM min–1) by the initial enzyme concentration (in mg L–1) and thus corresponding to the molar quantity of product converted each minute per unit mass of enzyme, that is, μmol min–1 mg–1 or U mg–1.60 The specific activity at each substrate concentration (l-DOPA, 0.5 to 7 mM range) was plotted, and the values of KM and Vmax were calculated by nonlinear fitting of the data to the Michaelis–Menten equation without considering substrate inhibition. All regression analyses were performed using Origin 2016 software (OriginLab).
4.5. Phylogenetic Analysis and In Silico Enzyme Modeling
The amino acid sequences of DOPA extradiol dioxygenases of Anabaena cylindrica Lemmermann 1896 (AcDODA, GenBank ID WP_015213489), A. muscaria (AmDODA, GenBank ID MK922469), Beta vulgaris L. (BvDODA1, GenBank ID HQ656027), Bombyx mori strain Dazao (BmDODA1, GenBank ID MG882761), E. coli (EcDODA, GenBank ID WP_000188362), Gluconacetobacter diazotrophicus (GdDODA, GenBank ID WP_012222467), Mirabilis jalapa L. (MjDODA, GenBank ID B6F0W8), and Portulaca grandiflora (PgDODA, GenBank ID AJ580598) were aligned using the MUSCLE (Multiple Sequence Comparison by Log-Expectation) tool in the Geneious Prime software (Biomatters, v. 2020.2.4). Pairwise distances obtained from this multiple sequence alignment were used to produce a neighbor-joining consensus tree with 100 000 bootstrap replicates. All of the amino acid sequences were also subjected to de novo folding with restraints, using both deep residual neural networks and homologous templates using the trRosetta server.61 The resulting structures were refined using the GalaxyRefine2 service at the GalaxyWEB server,62 scanned for metal binding sites using the BioMetAll software,63 and submitted to a homology search for metalloenzymes using the COACH-D server.64 Structural alignment was performed considering a maximum distance between residues of 140 pm and restrained to a minimum number of matching residues of 50. The timing of species divergence was estimated by constructing a time tree from the species listed above, Paraburkholderia xenovorans, and the anthocyanin-pigmented Arabidopsis thaliana using the TimeTree online server.52
4.6. Chromatographic Analysis
The reaction of l-DOPA (2.5 mM) and oxygen in the presence of AscH (10 mM) and AmDODA (1.0 μM) in sodium phosphate buffer (50 mM, pH 8.5) at 25 °C was monitored over time by HPLC-PDA using a Shimadzu Prominence liquid chromatograph equipped with an Ascentis C18 column (5 μm, 250 × 4.6 mm, Supelco) and an SPD-M20A detector. The reaction was analyzed at 1.0 mL min–1 at 25 °C under a linear gradient from 2% to 60% B over 20 min (solvent A, water; solvent B, acetonitrile, both containing 0.05% v/v formic acid) (condition 1), or isocratic 5% B for 5 min, followed by a linear gradient from 5% to 25% B over 15 min (condition 2). After equilibrium was reached, the reaction mixture was subjected to HPLC-HRMS analysis using a Shimadzu Prominence liquid chromatograph equipped with a Luna C18 column (3 μm, 150 × 2 mm, Phenomenex), and coupled to a Bruker Daltonics microTOF-QII mass spectrometer fitted with an electrospray source operated in positive mode. The reaction mixture analyzed at 0.2 mL min–1 at 30 °C under a linear gradient from 5% to 95% B over 15 min (solvent A, 0.05% v/v formic acid in water; solvent B, 0.05% v/v formic acid in acetonitrile). l- and d-DOPA, betalamic acid, and dopaxanthin were used as standards (see Supporting Information). Betalamic acid was quantified by absorption spectroscopy using a molar absorption coefficient of 24 000 M–1 cm–1 at 424 nm.43
Acknowledgments
We thank Mr. Enrico Florence Stevani for collecting the A. muscaria specimens.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c01365.
Additional methods, complete amino acid and DNA sequences, sequence alignments, kinetic and HPLC/MS data (PDF)
Author Contributions
E.L.B. and C.T.H. conceived the study. C.O.M, F.A.D., E.P., and L.C.P.G. performed chromatographic analyses. L.C.E. prepared l-DOPAxanthin. C.V.S. provided the A. muscaria fungi. L.C.P.G. and D.M.M.S. performed kinetic studies. D.M.M.S. and F.M.M.A. cloned the dodA gene and expressed AmDODA. C.T.H., D.M.M.S., and C.C.O. conceived the molecular biology experiments. E.L.B. conceived and performed in silico analysis and kinetic modeling. E.L.B., C.T.H., D.M.M.S., and L.C.P.G. wrote the paper. All authors discussed the results and revised the manuscript.
This research was funded by the São Paulo Research Foundation, FAPESP (DMMS, 2019/12605-0, ELB, 2019/06391-8 and 2019/15412-9; COM, 2015/24760-0; LCPG, 2018/25842-8; CVS 2017/22501-2; EP 2013/07914-8), the Brazilian National Council for Scientific and Technological Development, CNPq (ELB, 301623/2019-8), University of São Paulo (PIPAE 21.1.10424.1.9), and Coordination for the Improvement of Higher Education Personnel (CAPES, Finance code 001).
The authors declare no competing financial interest.
Supplementary Material
References
- Gressler M.; Löhr N. A.; Schäfer T.; Lawrinowitz S.; Seibold P. S.; Hoffmeister D. Mind the mushroom: natural product biosynthetic genes and enzymes of Basidiomycota. Nat. Prod Rep 2021, 38 (4), 702–722. 10.1039/D0NP00077A. [DOI] [PubMed] [Google Scholar]
- Li C.; Oberlies N. H. The most widely recognized mushroom: chemistry of the genus Amanita. Life Sci. 2005, 78 (5), 532–538. 10.1016/j.lfs.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Davies K. M.; Albert N. W.; Zhou Y.; Schwinn K. E.. Functions of flavonoid and betalain pigments in abiotic stress tolerance in plants. In Annual Plant Reviews online; Roberts J. A., Ed.; John Wiley & Sons, Ltd: Chichester, 2018; Vol. 1, pp 21–62. [Google Scholar]
- Stintzing F.; Schliemann W. Pigments of fly agaric (Amanita muscaria). Z. Naturforsch C J. Biosci 2007, 62 (11–12), 779–785. 10.1515/znc-2007-11-1201. [DOI] [PubMed] [Google Scholar]
- Christinet L.; Burdet F. X.; Zaiko M.; Hinz U.; Zryd J.-P. Characterization and functional identification of a novel plant 4,5-extradiol dioxygenase involved in betalain pigment biosynthesis in Portulaca grandiflora. Plant Physiol 2004, 134 (1), 265–274. 10.1104/pp.103.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döpp H.; Musso H. Eine chromatographische Analysenmethode für Betalainfarbstoffe in Pilzen und höheren Pflanzen. Z. Naturforsch. 1974, 29c, 640–642. 10.1515/znc-1974-9-1033. [DOI] [PubMed] [Google Scholar]
- Timoneda A.; Feng T.; Sheehan H.; Walker-Hale N.; Pucker B.; Lopez-Nieves S.; Guo R.; Brockington S. The evolution of betalain biosynthesis in Caryophyllales. New Phytol 2019, 224 (1), 71–85. 10.1111/nph.15980. [DOI] [PubMed] [Google Scholar]
- Gandía-Herrero F.; García-Carmona F.; Escribano J. Floral fluorescence effect. Nature 2005, 437 (7057), 334–334. 10.1038/437334a. [DOI] [PubMed] [Google Scholar]
- Pinheiro A. C.; Fazzi R. B.; Esteves L. C.; Machado C. O.; Dörr F. A.; Pinto E.; Hattori Y.; Sa J.; da Costa Ferreira A. M.; Bastos E. L. A bioinspired nitrone precursor to a stabilized nitroxide radical. Free Radic Biol. Med. 2021, 168, 110–116. 10.1016/j.freeradbiomed.2021.03.030. [DOI] [PubMed] [Google Scholar]
- Freitas-Dörr B. C.; Machado C. O.; Pinheiro A. C.; Fernandes A. B.; Dörr F. A.; Pinto E.; Lopes-Ferreira M.; Abdellah M.; Sa J.; Russo L. C.; Forti F. L.; Gonçalves L. C. P.; Bastos E. L. A metal-free blue chromophore derived from plant pigments. Sci. Adv. 2020, 6 (14), eaaz0421 10.1126/sciadv.aaz0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A. C. B.; Mariz I. D. A.; Macoas E. M. S.; Tonelli R. R.; Martinho J. M. G.; Quina F. H.; Bastos E. L. Bioinspired water-soluble two-photon fluorophores. Dyes Pigm 2018, 150, 105–111. 10.1016/j.dyepig.2017.11.020. [DOI] [Google Scholar]
- Gonçalves L. C. P.; Da Silva S. M.; DeRose P.; Ando R. A.; Bastos E. L. Beetroot-pigment-derived colorimetric sensor for detection of calcium dipicolinate in bacterial spores. PloS One 2013, 8 (9), e73701 10.1371/journal.pone.0073701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves L. C. P.; Tonelli R. R.; Bagnaresi P.; Mortara R. A.; Ferreira A. G.; Bastos E. L. A nature-inspired betalainic probe for live-cell imaging of Plasmodium-infected erythrocytes. PLoS One 2013, 8 (1), e53874 10.1371/journal.pone.0053874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumorkiewicz-Jamro A.; Swiergosz T.; Sutor K.; Sporna-Kucab A.; Wybraniec S. Multi-colored shades of betalains: recent advances in betacyanin chemistry. Nat. Prod Rep 2021, 38 (12), 2315–2346. 10.1039/D1NP00018G. [DOI] [PubMed] [Google Scholar]
- Quina F. H.; Bastos E. L. Chemistry inspired by the colors of fruits, flowers and wine. An Acad. Bras Cienc 2018, 90, 681–695. 10.1590/0001-3765201820170492. [DOI] [PubMed] [Google Scholar]
- Sheehan H.; Feng T.; Walker-Hale N.; Lopez-Nieves S.; Pucker B.; Guo R.; Yim W. C.; Badgami R.; Timoneda A.; Zhao L. J.; Tiley H.; Copetti D.; Sanderson M. J.; Cushman J. C.; Moore M. J.; Smith S. A.; Brockington S. F. Evolution of l-DOPA 4,5-dioxygenase activity allows for recurrent specialisation to betalain pigmentation in Caryophyllales. New Phytol 2020, 227 (3), 914–929. 10.1111/nph.16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polturak G.; Aharoni A. La vie en rose”: Biosynthesis, sources, and applications of betalain pigments. Mol. Plant 2018, 11 (1), 7–22. 10.1016/j.molp.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Bresinsky A.; Kronawitter I. Zur Kenntnis der Hygrocybenpigmente. Z. Mykol. 1986, 52 (2), 321–334. [Google Scholar]
- Gill M.; Steglich W.. Pigments of fungi (Macromycetes). In Progress in the Chemistry of Organic Natural Products; Herz W., Grisebach H., Kirby G. W., Ch T., Eds.; Springer-Verlag: New York, 1987; Vol. 51, pp 1–109. [DOI] [PubMed] [Google Scholar]
- Bastos E. L.; Schliemann W.. Betalains as Antioxidants. In Plant Antioxidants and Health; Ekiert H. M., Ramawat K. G., Arora J., Eds.; Springer International Publishing: Cham, 2021; pp 1–44. [Google Scholar]
- Vaillancourt F. H.; Bolin J. T.; Eltis L. D. The ins and outs of ring-cleaving dioxygenases. Crit Rev. Biochem Mol. Biol. 2006, 41 (4), 241–67. 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- Strack D.; Vogt T.; Schliemann W. Recent advances in betalain research. Phytochemistry 2003, 62 (3), 247–269. 10.1016/S0031-9422(02)00564-2. [DOI] [PubMed] [Google Scholar]
- Saha S.; Li W.; Gerratana B.; Rokita S. E. Identification of the dioxygenase-generated intermediate formed during biosynthesis of the dihydropyrrole moiety common to anthramycin and sibiromycin. Bioorg. Med. Chem. 2015, 23 (3), 449–54. 10.1016/j.bmc.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso H. The pigments of fly agaric, Amanita muscaria. Tetrahedron 1979, 35 (24), 2843–2853. 10.1016/S0040-4020(01)99498-0. [DOI] [Google Scholar]
- Saito K.; Komamine A. Biosynthesis of stizolobinic acid and stizolobic acid in higher plants. Eur. J. Biochem. 1978, 82 (2), 385–92. 10.1111/j.1432-1033.1978.tb12033.x. [DOI] [PubMed] [Google Scholar]
- Penna T. C.; Cervi G.; Rodrigues-Oliveira A. F.; Yamada B. D.; Lima R. Z. C.; Menegon J. J.; Bastos E. L.; Correra T. C. Development of a photoinduced fragmentation ion trap for infrared multiple photon dissociation spectroscopy. Rapid Commun. Mass Spectrom. 2020, 34, e8635 10.1002/rcm.8635. [DOI] [PubMed] [Google Scholar]
- Hinz U. G.; Fivaz J.; Girod P.-A.; Zyrd J.-P. The gene coding for the DOPA dioxygenase involved in betalain biosynthesis in Amanita muscaria and its regulation. Mol. Gen Genet 1997, 256 (1), 1–6. 10.1007/s004380050539. [DOI] [PubMed] [Google Scholar]
- Mueller L. A.; Hinz U.; Zryd J. P. The formation of betalamic acid and muscaflavin by recombinant dopa-dioxygenase from Amanita. Phytochemistry 1997, 44 (4), 567–569. 10.1016/S0031-9422(96)00625-5. [DOI] [Google Scholar]
- Mueller L. A.; Hinz U.; Uzé M.; Sautter C.; Zryd J. P. Biochemical complementation of the betalain biosynthetic pathway in Portulaca grandiflora by a fungal 3,4-dihydroxyphenylalanine dioxygenase. Planta 1997, 203 (2), 260–263. 10.1007/s004250050190. [DOI] [Google Scholar]
- Harris N. N.; Javellana J.; Davies K. M.; Lewis D. H.; Jameson P. E.; Deroles S. C.; Calcott K. E.; Gould K. S.; Schwinn K. E. Betalain production is possible in anthocyanin-producing plant species given the presence of DOPA-dioxygenase and l-DOPA. BMC Plant Biol. 2012, 12, 34. 10.1186/1471-2229-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucheli P.; Sieber S.; Fuchs D. W.; Scheller L.; Strittmatter T.; Saxena P.; Gademann K.; Fussenegger M. Genetically encoded betaxanthin-based small-molecular fluorescent reporter for mammalian cells. Nucleic Acids Res. 2020, 48 (12), e67 10.1093/nar/gkaa342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliemann W.; Kobayashi N.; Strack D. The decisive step in betaxanthin biosynthesis is a spontaneous reaction. Plant Physiol 1999, 119 (4), 1217–1232. 10.1104/pp.119.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geml J.; Laursen G. A.; O’Neill K.; Nusbaum H. C.; Taylor D. L. Beringian origins and cryptic speciation events in the fly agaric (Amanita muscaria). Mol. Ecol. 2006, 15 (1), 225–239. 10.1111/j.1365-294X.2005.02799.x. [DOI] [PubMed] [Google Scholar]
- Oda T.; Tanaka C.; Tsuda M. Molecular phylogeny and biogeography of the widely distributed Amanita species, A. muscaria and A. pant henna. Mycol Res. 2004, 108 (8), 885–896. 10.1017/S0953756204000620. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Tang L. P.; Cai Q.; Xu J. P. A review on the diversity, phylogeography and population genetics of Amanita mushrooms. Mycology 2015, 6 (2), 86–93. 10.1080/21501203.2015.1042536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T.; Yamazaki T.; Tanaka C.; Terashita T.; Taniguchi N.; Tsuda M. Amanita ibotengutake sp. nov., a poisonous fungus from Japan. Mycol Progr 2002, 1 (4), 355–365. 10.1007/s11557-006-0032-9. [DOI] [Google Scholar]
- Oda T.; Tanaka C.; Tsuda M. Molecular phylogeny of Japanese Amanita species based on nucleotide sequences of the internal transcribed spacer region of nuclear ribosomal DNA. Mycoscience 1999, 40, 57–64. 10.1007/BF02465674. [DOI] [Google Scholar]
- Hernandez G.; Osnaya V. G.; Perez-Martinez X. Conservation and variability of the AUG initiation codon context in Eukaryotes. Trends Biochem. Sci. 2019, 44 (12), 1009–1021. 10.1016/j.tibs.2019.07.001. [DOI] [PubMed] [Google Scholar]
- He Y.; Zhang T.; Sun H.; Zhan H.; Zhao Y. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic Res. 2020, 7 (1), 152. 10.1038/s41438-020-00390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoneda A.; Yunusov T.; Quan C.; Gavrin A.; Brockington S. F.; Schornack S. MycoRed: Betalain pigments enable in vivo real-time visualisation of arbuscular mycorrhizal colonisation. PLoS Biol. 2021, 19 (7), e3001326 10.1371/journal.pbio.3001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper C.; Vissers M. C. M. Ascorbate as a co-factor for Fe-and 2-oxoglutarate dependent dioxygenases: physiological activity in tumor growth and progression. Front Oncol 2014, 4, 359. 10.3389/fonc.2014.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod P. A.; Zryd J. P. Biogenesis of betalains: Purification and partial characterization of dopa 4,5-dioxygenase from Amanita muscaria. Phytochemistry 1991, 30 (1), 169–174. 10.1016/0031-9422(91)84119-D. [DOI] [Google Scholar]
- Contreras-Llano L. E.; Guerrero-Rubio M. A.; Lozada-Ramirez J. D.; Garcia-Carmona F.; Gandia-Herrero F. First betalain-producing bacteria break the exclusive presence of the pigments in the plant kingdom. mBio 2019, 10 (2), e00345 10.1128/mBio.00345-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandía-Herrero F.; García-Carmona F. Characterization of recombinant Beta vulgaris 4,5-DOPA-extradiol-dioxygenase active in the biosynthesis of betalains. Planta 2012, 236 (1), 91–100. 10.1007/s00425-012-1593-2. [DOI] [PubMed] [Google Scholar]
- Terradas F.; Wyler H. 2,3-and 4,5-Secodopa, the Biosynthetic Intermediates Generated froml-DOPA by an Enzyme System Extracted from the Fly Agaric, Amanita muscaria L., and Their Spontaneous Conversion to Muscaflavin and Betalamic Acid, Respectively, and Betalains. Helv. Chim. Acta 1991, 74 (1), 124–140. 10.1002/hlca.19910740115. [DOI] [Google Scholar]
- El Seoud O. A.; Baader W. J.; Bastos E. L.. Practical Chemical Kinetics in Solution. In Encyclopedia of Physical Organic Chemistry; Wang Z., Wille U., Juaristi E., Eds.; John Wiley & Sons, Inc., 2016; pp 1–68. [Google Scholar]
- Stintzing F. C.; Carle R. Betalains – emerging prospects for food scientists. Trends Food Sci. Technol. 2007, 18 (10), 514–525. 10.1016/j.tifs.2007.04.012. [DOI] [Google Scholar]
- Herbach K. M.; Stintzing F. C.; Carle R. Betalain Stability and Degradation-Structural and Chromatic Aspects. J. Food Sci. 2006, 71 (4), R41–R50. 10.1111/j.1750-3841.2006.00022.x. [DOI] [Google Scholar]
- Guerrero-Rubio M. A.; García-Carmona F.; Gandía-Herrero F. First description of betalains biosynthesis in an aquatic organism: characterization of 4,5-DOPA-extradiol-dioxygenase activity in the cyanobacteria Anabaena cylindrica. Microb Biotechnol 2020, 13 (6), 1948–1959. 10.1111/1751-7915.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. F.; Sun W.; Zhang Z. Functional characterization of the horizontally transferred 4,5-DOPA extradiol dioxygenase gene in the domestic silkworm, Bombyx mori. Insect Mol. Biol. 2019, 28 (3), 409–419. 10.1111/imb.12558. [DOI] [PubMed] [Google Scholar]
- Sasaki N.; Abe Y.; Goda Y.; Adachi T.; Kasahara K.; Ozeki Y. Detection of DOPA 4,5-dioxygenase (DOD) activity using recombinant protein prepared from Escherichia coli cells harboring cDNA encoding DOD from Mirabilis jalapa. Plant Cell Physiol 2009, 50 (5), 1012–6. 10.1093/pcp/pcp053. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Suleski M.; Hedges S. B. TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017, 34 (7), 1812–1819. 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Chain P. S.; Denef V. J.; Konstantinidis K. T.; Vergez L. M.; Agullo L.; Reyes V. L.; Hauser L.; Cordova M.; Gomez L.; Gonzalez M.; Land M.; Lao V.; Larimer F.; LiPuma J. J.; Mahenthiralingam E.; Malfatti S. A.; Marx C. J.; Parnell J. J.; Ramette A.; Richardson P.; Seeger M.; Smith D.; Spilker T.; Sul W. J.; Tsoi T. V.; Ulrich L. E.; Zhulin I. B.; Tiedje J. M. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (42), 15280–7. 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg T. D. H.; Ramaswamy S. Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations. Curr. Opin Chem. Biol. 2008, 12 (2), 134–140. 10.1016/j.cbpa.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Burroughs A. M.; Glasner M. E.; Barry K. P.; Taylor E. A.; Aravind L. Oxidative opening of the aromatic ring: Tracing the natural history of a large superfamily of dioxygenase domains and their relatives. J. Biol. Chem. 2019, 294 (26), 10211–10235. 10.1074/jbc.RA119.007595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.-Y.; Yim H.-S.; Ryu J.-Y.; Lee H. S.; Lee J.-H.; Seen D.-S.; Kang S. G. One-step sequence-and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 2012, 78 (15), 5440–5443. 10.1128/AEM.00844-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227 (5259), 680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Soares D. M. M.; Machado C. O.; Goncalves L. C. P.; Stevani C. V.; Oliveira C. C.; Hotta C. T.; Bastos E. L.. MK922469. Amanita muscaria DOPA extradiol dioxygenase mRNA, complete cds. In GenBank database; NCBI: 2019.
- Trezzini G.F.; Zryb J.-P. Characterization of some natural and semi-synthetic betaxanthins. Phytochemistry 1991, 30 (6), 1901–1903. 10.1016/0031-9422(91)85036-Y. [DOI] [Google Scholar]
- Harris T. K.; Keshwani M. M., Measurement of Enzyme Activity. In Methods in Enzymology; Burgess R. R., Deutscher M. P., Eds.; Academic Press: Cambridge, 2009; Vol. 463, pp 57–71. [DOI] [PubMed] [Google Scholar]
- Yang J.; Anishchenko I.; Park H.; Peng Z.; Ovchinnikov S.; Baker D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (3), 1496–1503. 10.1073/pnas.1914677117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. R.; Won J.; Heo L.; Seok C. GalaxyRefine2: simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res. 2019, 47 (W1), W451–W455. 10.1093/nar/gkz288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Aparicio J.-E.; Tiessler-Sala L.; Velasco-Carneros L.; Roldán-Martín L.; Sciortino G.; Maréchal J.-D. BioMetAll: Identifying Metal-Binding Sites in Proteins from Backbone Preorganization. J. Chem. Inf Model 2021, 61 (1), 311–323. 10.1021/acs.jcim.0c00827. [DOI] [PubMed] [Google Scholar]
- Wu Q.; Peng Z.; Zhang Y.; Yang J. COACH-D: improved protein-ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018, 46 (W1), W438–W442. 10.1093/nar/gky439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.