Abstract
Background
The potential pathogenic role of Stenotrophomonas maltophilia in lung disease and in particular in cystic fibrosis is unclear. To develop further understanding of the biology of this taxa, the taxonomic position, antibiotic resistance and virulence factors of S. maltophilia isolates from patients with chronic lung disease were studied.
Results
A total of 111 isolates recovered between 2003 and 2016 from respiratory samples from patients in five different countries were included. Based on a cut-off of 95%, analysis of average nucleotide identity by BLAST (ANIb) showed that the 111 isolates identified as S. maltophilia by Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF/MS) belonged to S. maltophilia (n = 65), S. pavanii (n = 6) and 13 putative novel species (n = 40), which each included 1–5 isolates; these groupings coincided with the results of the 16S rDNA analysis, and the L1 and L2 ß-lactamase Neighbor-Joining phylogeny. Chromosomally encoded aminoglycoside resistance was identified in all S. maltophilia and S. pavani isolates, while acquired antibiotic resistance genes were present in only a few isolates. Nevertheless, phenotypic resistance levels against commonly used antibiotics, determined by standard broth microbroth dilution, were high. Although putative virulence genes were present in all isolates, the percentage of positive isolates varied. The Xps II secretion system responsible for the secretion of the StmPr1–3 proteases was mainly limited to isolates identified as S. maltophilia based on ANIb, but no correlation with phenotypic expression of protease activity was found. The RPF two-component quorum sensing system involved in virulence and antibiotic resistance expression has two main variants with one variant lacking 190 amino acids in the sensing region.
Conclusions
The putative novel Stenotrophomonas species recovered from patient samples and identified by MALDI-TOF/MS as S. maltophilia, differed from S. maltophilia in resistance and virulence genes, and therefore possibly in pathogenicity. Revision of the Stenotrophomonas taxonomy is needed in order to reliably identify strains within the genus and elucidate the role of the different species in disease.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-022-02466-5.
Keywords: Stenotrophomonas, Cystic fibrosis, Respiratory infection, Taxonomy, Antibiotic resistance, Virulence
Background
Stenotrophomonas species are non-fermenting gram-negative rods. Currently sixteen species are recognized within the genus: S. acidaminiphila, S. bentonitica, S. chelatiphaga, S. daejeonensis, S. ginsengisoli, S. humi, S. indicatrix, S. koreensis, S. lactitubi, S. maltophilia, S. nitrireducens, S. pavanii, S. pictorum, S. rhizophila, S. terrae, S. tumulicola. To note that S. africana is no longer recognized as a species and has been added to S. maltophilia [1]. However, the taxonomy of the genus is not completely resolved. S. maltophilia forms a complex of which the isolates show considerable heterogeneity in genetic and phenotypic characteristics. Based on amplified fragment length polymorphism and DNA hybridizations 10 genogroups (#1–10) were defined [2]. This was expanded by the use of Multi-Locus Sequence Typing (MLST) with a representative of each genogroup and additional strains, which confirmed the ten genogroups and found five additional groups (A-E) [3]. Another MLST-based study added lineage (F) and genospecies Smc 1–4 and, e.g., genogroup #8 was shown to cluster with the S. rhizophila type strain [4]. In this scheme, S. pavanii was included within S. maltophilia.
Soils are an important reservoir for Stenotrophomonas species, but some species found in the environment may be opportunistic human pathogens, in particular S. maltophilia. Although S. maltophilia is considered a low-grade pathogen, it carries a large number of (putative) virulence factors [5]. The (putative) virulence factors include proteases (StmPr1/alkaline serine protease, StmPr2, StmPr3, StmPr4, gelatinase, elastase, and fibrinolysin/streptokinase), nucleases (DNase and 2 RNases), lipases (lipase, and phospholipase C and D) adhesion factors (SMF-1, TadE-like, giant cable pilus-like afimbrial adhesin, and type IV pilus machinery), siderophores, biofilm formation factors (phosphoglucomutase/phosphomannose bifunctional protein, glucose-1-phosphate thymylyltransferase, biofilm and swimming motility regulator, and Ax21 outer membrane protein), and a range of other factors (polysaccharide lyase, nitrate reductase, ankyrin repeat domain-containing protein, regulators RpfC and RpfF, hemagglutinin, esterase, hyaluronidase, heparinase, hemolysin, cytotoxin(s), and Xps type II secretion system). However, it cannot be excluded that some of these factors are part of the normal house-keeping processes.
S. maltophilia isolates usually exhibit high levels of phenotypic antibiotic resistance, due to an arsenal of intrinsic resistance genes and mutationally acquired resistance. S. maltophilia encodes several efflux pumps, of which at least six have been shown to expel a range of different antibiotics. Furthermore, S. maltophilia encodes the metallo-ß-lactamase L1 and the extended-spectrum ß-lactamase L2. L1 hydrolyzes carbapenems and other ß-lactam antibiotics with the exception of monobactams, whereas L2 is a serine hydrolase that acts as a cephalosporinase [6–9]. A chromosomally encoded qnr gene (smqnr) confers low-level fluoroquinolone resistance. Finally, acquisition of sul1, sul2, and sul3 has been also described, resulting in sulfonamide resistance and contributing to co-trimoxazole resistance. Adegoke et al. recently published a review of S. maltophilia antibiotic resistance determinants [10].
Infections by Stenotrophomonas species are usually caused by species identified as S. maltophilia, and mostly involve the airways [11]. However, due to their high intrinsic antibiotic resistance, Stenotrophomonas species may be recovered as a colonizers in patients treated with long-term broad spectrum antibiotic therapy, which obscures their role in chronic lung disease [12]. This is further complicated by the heterogeneity within the S. maltophilia complex, which is not distinguished by current microbiological diagnostics.
The aim of the present study was to determine the taxonomic position of S. maltophilia isolates from persons with cystic fibrosis (CF) and patients with other chronic respiratory infections, and to characterize their antibiotic resistance genes and virulence factors.
Results
Genome characteristics
Sequencing and assembly of the whole genomes of the isolates resulted in an average of 181 contigs larger than 1000 bp, an average coverage of 81x, and a genome size of approximately 4.70 MB, indicating high quality of the sequences. The isolates belonged to 55 different Sequence Types (ST) including 16 novel types. Forty-three novel alleles were found: five for atpD, four for gapA, thirteen for guaA, seven for mutM, three for nuoD, six for ppsA, and five for recA. Twenty-one isolates belonged to ST5, nine to ST4 and to ST31, seven to the novel ST535, five to ST162, and four to ST26 (Table 1). Of other STs three isolates or less were found.
Table 1.
Taxonomy, ST, isolate source, country of origin and resistance genes of the isolates
| Order nr | ANIb | ST | Isolate source | Country | Resistance genes |
|---|---|---|---|---|---|
| 543,227 | A | 89 | CF | Northern Ireland | – |
| 533,506 | A | 215 | CF | the Netherlands | – |
| 534,845 | A | 549 | CF | Spain | – |
| 534,836 | B | 130 | CF | Spain | – |
| 546,334 | C | 133 | other | Northern Ireland | – |
| 543,210 | D | 536 | CF | Northern Ireland | – |
| 533,529 | D | 337 | CF | the Netherlands | – |
| 534,834 | D | 337 | CF | Spain | – |
| 533,533 | E | 39 | CF | the Netherlands | – |
| 534,812 | E | 39 | CF | Spain | – |
| 534,818 | E | 39 | CF | Spain | aac(6′)-Iz |
| 542,535 | E | 39 | CF | Northern Ireland | – |
| 543,206 | E | 39 | CF | Northern Ireland | – |
| 543,208 | E | 39 | CF | Northern Ireland | – |
| 543,215 | E | 39 | CF | Northern Ireland | – |
| 543,217 | E | 39 | CF | Northern Ireland | – |
| 546,336 | E | 39 | other | Northern Ireland | – |
| 543,216 | E | 180 | CF | Northern Ireland | – |
| 542,523 | F | 535 | CF | Northern Ireland | |
| 542,525 | F | 535 | CF | Northern Ireland | |
| 542,527 | F | 535 | CF | Northern Ireland | |
| 542,530 | F | 535 | CF | Northern Ireland | |
| 543,225 | F | 535 | CF | Northern Ireland | |
| 547,141 | F | 535 | CF | Northern Ireland | |
| 548,978 | F | 535 | other | Northern Ireland | |
| 534,846 | G | 537 | CF | Spain | – |
| 543,222 | H | 79 | CF | Northern Ireland | – |
| 533,514 | H | 545 | CF | the Netherlands | – |
| 548,948 | I | 34 | other | the Netherlands | – |
| 534,842 | J | 548 | CF | Spain | – |
| 533,515 | K | 29 | CF | the Netherlands | – |
| 533,513 | K | 77 | CF | the Netherlands | – |
| 543,213 | K | 77 | CF | Northern Ireland | – |
| 548,945 | K | 219 | other | Northern Ireland | – |
| 533,531 | K | 224 | CF | the Netherlands | – |
| 534,799 | K | 538 | CF | Spain | – |
| 534,811 | L | 15 | CF | Spain | – |
| 542,542 | L | 15 | CF | Northern Ireland | – |
| 534,821 | M | 533 | CF | Spain | aac(6′)-Ib3, ant(2″)-Ia, aac(6′)-Ib-cr |
| 534,822 | M | 534 | CF | Spain | aac(6′)-Ib3, ant(2″)-Ia, aac(6′)-Ib-cr |
| 533,501 | S. maltophilia | 4 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 533,507 | S. maltophilia | 4 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 533,524 | S. maltophilia | 4 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 533,532 | S. maltophilia | 4 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 533,534 | S. maltophilia | 4 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 534,831 | S. maltophilia | 4 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,838 | S. maltophilia | 4 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 542,522 | S. maltophilia | 4 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 548,946 | S. maltophilia | 4 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 533,502 | S. maltophilia | 5 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz, aph(3″)-Ib, aph(6)-Id |
| 533,518 | S. maltophilia | 5 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 534,802 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,804 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz, aph(6)-Id, aph(4)-Ia, aac(6′)-Ib-cr, aph(3″)-Ib, aac(3)-IV, aph(3′)-Ia, aac(6′)-Ib-Hangzhou, tet(G), sul1, cmx |
| 534,807 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,823 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,825 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,829 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,839 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,843 | S. maltophilia | 5 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 540,831 | S. maltophilia | 5 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz, aph(6)-Id, aac(6′)-Ib-cr, aph(3″)-Ib, aac(6′)-Ib3, ant(2″)-Ia, sul1 |
| 542,536 | S. maltophilia | 5 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 542,538 | S. maltophilia | 5 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 542,539 | S. maltophilia | 5 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 543,203 | S. maltophilia | 5 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 543,219 | S. maltophilia | 5 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 546,340 | S. maltophilia | 5 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 546,343 | S. maltophilia | 5 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 547,144 | S. maltophilia | 5 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 547,147 | S. maltophilia | 5 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz, aph(3″)-Ib, aph(6)-Id |
| 548,947 | S. maltophilia | 5 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 533,517 | S. maltophilia | 8 | CF | the Netherlands | aph(3′)-IIc |
| 534,844 | S. maltophilia | 26 | CF | Spain | aph(3′)-IIc |
| 542,532 | S. maltophilia | 26 | CF | Northern Ireland | aph(3′)-IIc |
| 542,534 | S. maltophilia | 26 | CF | Northern Ireland | aph(3′)-IIc |
| 543,223 | S. maltophilia | 26 | CF | Northern Ireland | aph(3′)-IIc |
| 533,525 | S. maltophilia | 27 | CF | the Netherlands | aph(3′)-IIc |
| 533,509 | S. maltophilia | 31 | CF | the Netherlands | aph(3′)-IIc |
| 533,530 | S. maltophilia | 31 | CF | the Netherlands | aph(3′)-IIc |
| 546,338 | S. maltophilia | 31 | CF | Northern Ireland | aph(3′)-IIc |
| 534,806 | S. maltophilia | 84 | CF | Spain | aph(3′)-Iic, aac(3)-IV, aph(3″)-Ib, aph(6)-Id, aph(4)-Ia, drfA15, tet(A), sul1 |
| 534,817 | S. maltophilia | 92 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,840 | S. maltophilia | 146 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,835 | S. maltophilia | 162 | CF | Spain | aph(3′)-IIc |
| 543,221 | S. maltophilia | 162 | CF | Northern Ireland | aph(3′)-IIc |
| 546,339 | S. maltophilia | 162 | CF | the Netherlands | aph(3′)-IIc |
| 547,143 | S. maltophilia | 162 | CF | Northern Ireland | aph(3′)-IIc |
| 548,949 | S. maltophilia | 162 | CF | the Netherlands | aph(3′)-IIc |
| 548,950 | S. maltophilia | 199 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 533,512 | S. maltophilia | 246 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 534,832 | S. maltophilia | 246 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 543,220 | S. maltophilia | 531 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 534,809 | S. maltophilia | 532 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 533,510 | S. maltophilia | 539 | CF | the Netherlands | aph(3′)-IIc |
| 533,527 | S. maltophilia | 540 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 533,528 | S. maltophilia | 541 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 539,959 | S. maltophilia | 541 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 534,828 | S. maltophilia | 542 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,808 | S. maltophilia | 543 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 534,801 | S. maltophilia | 544 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 543,211 | S. maltophilia | 546 | CF | Northern Ireland | aph(3′)-IIc |
| 534,805 | S. maltophilia | 547 | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 546,337 | S. maltophilia | 550 | CF | Northern Ireland | aph(3′)-IIc, aac(6′)-Iz |
| 546,342 | S. maltophilia | 551 | CF | the Netherlands | aph(3′)-IIc, aac(6′)-Iz |
| 547,148 | S. maltophilia | 539 | other | Northern Ireland | aph(3′)-IIc |
| 534,819 | S. maltophilia | xxa | CF | Spain | aph(3′)-IIc, aac(6′)-Iz |
| 547,145 | S. pavanii | 23 | other | Northern Ireland | aac(6′)-Iak |
| 534,815 | S. pavanii | 24 | CF | Spain | aac(6′)-Iak |
| 545,270 | S. pavanii | 220 | other | USA | aac(6′)-Iak |
| 534,800 | S. pavanii | 233 | CF | Spain | aac(6′)-Iak |
| 542,603 | S. pavanii | 233 | CF | Australia | aac(6′)-Iak |
| 533,516 | S. pavanii | 306 | CF | the Netherlands | aac(6′)-Iak |
axx no ST available because allele mutM is partially deleted
Taxonomy, ST, isolate source, country of origin and resistance genes of the isolates used in this study
Stenotrophomonas taxonomy
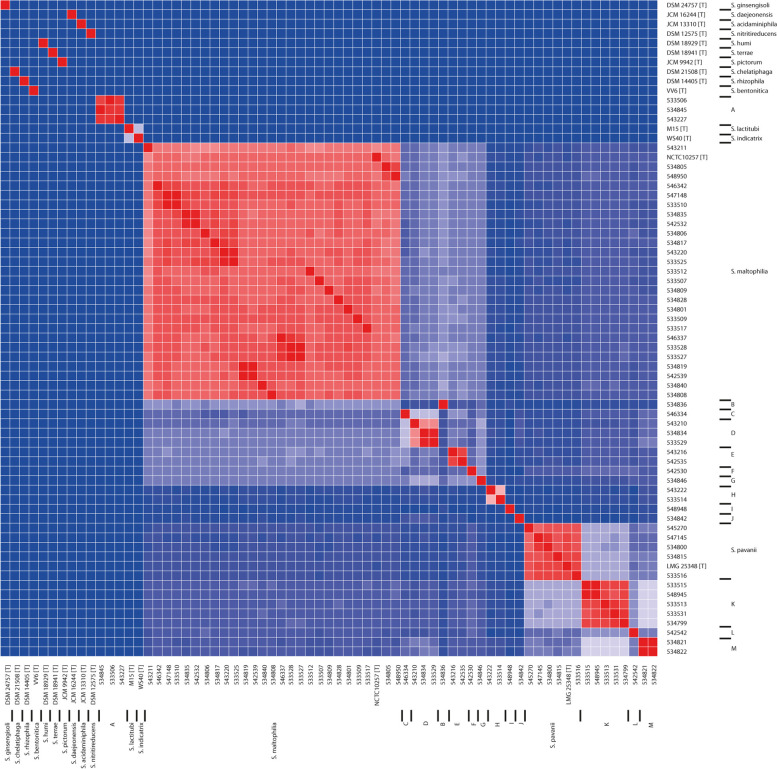
To assess the taxonomic position of the Stenotrophomonas isolates an average nucleotide identity by BLAST (ANIb) was performed (Fig. 1 and Supplementary Fig. 1). Based on a cut-off of 95% identity for the boundary between species, the type strains of the different species were separated. However, also 13 groups of 1–5 isolates (indicated by a capital letter) may be considered as separate species when applying this cut-off. S. maltophilia, S. pavanii, and putative species B-M appeared to be more closely related to each other (identity = > 90%) than to other species within the genus (identity < 90%). Based on the ANIb analysis, only 65 of the 111 isolates (58.6%) initially identified as S. maltophilia by MALDI-TOF/MS could be confirmed as belonging to this species. The 46 other isolates would either be S. pavani (n = 6) or belong to the putative novel species B-M (n = 40).
Fig. 1.
Heatmap based on percentage of ANIb for strains belonging to the different STs that were analyzed in this study (one isolate per ST was included) as well as available type strain sequences. The color bar shows the percentage of ANIb between any two strains starting from blue (0.8 or 80%) to red (1 or 100%); the blue/red cutoff is 0.94/0.95 (see Methods section for more details). Type strains are indicated by (T) behind the strain identification. The putative species (see text) are indicated at the right. For details about strains and the similarity between strains see Supplementary Fig. 1
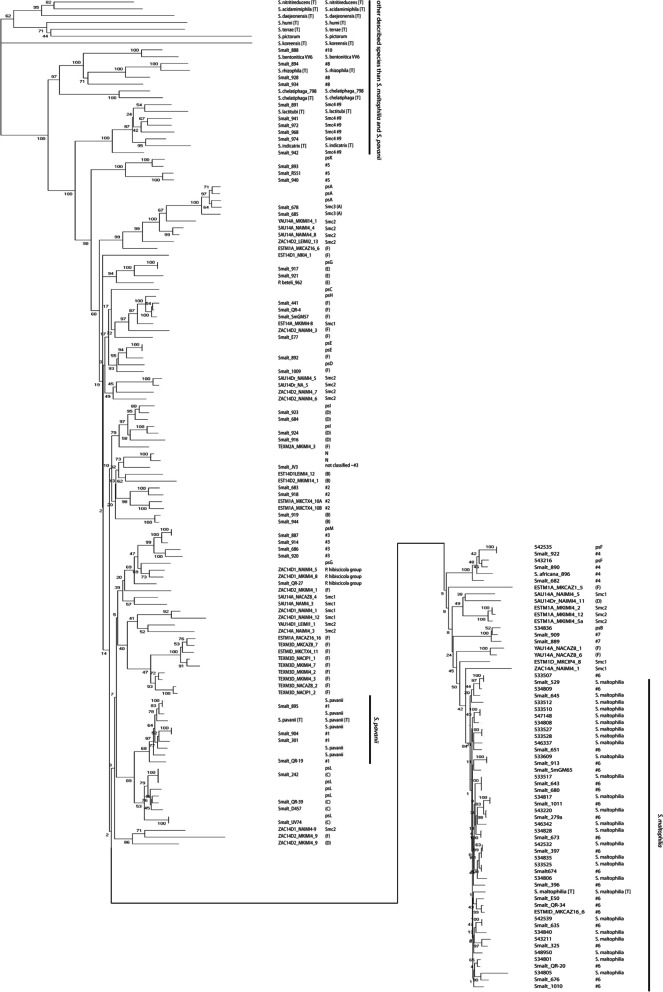
To assess the relationship of our isolates with the data of Ochoa-Sánchez and Vinuesa, a Neighbor-Joining tree of the concatenated MLST alleles was created with the STs in our collection, the dataset described by Ochoa-Sánchez and Vinuesa, and the available type strains [4]. Results showed that isolates previously identified based on MLST as lineages #1-#10 by Hauben et al., lineages (A)-(E) by Kaiser et al., and lineage (F) and genospecies Smc 1–4 by Ochoa-Sánchez and Vinuesa, as well as isolates that belonged to previously defined species other than S. maltophilia and S. pavanii, clustered in well-defined branches (Fig. 2) [2–4]. Lineage #8 isolates clustered with the S. rhizophila type strain and the lineage #10 isolate with the S. bentonitica type strain. The lineage #9 isolates clustered with both the S. indicatrix and the S. lactitubi type strains. Lineages #1 and #6 corresponded with S. pavanii and S. maltophilia, respectively (Fig. 2). Lineage #3 isolates clustered with the putative species L isolate; lineage #4 isolates with putative species E isolates; lineage #5 isolates with the putative species J isolate; lineage #7 isolates with the putative species B isolate. From the other lineages that have been defined, cluster lineage (C) isolates with putative species K isolates and Smc3 isolates clustered with putative species A isolates. Lineage (E) isolates cluster with the putative species I isolate. One of the (E) isolates has previously been named Pseudomonas beteli, which is now considered synonymous with S. maltophilia. Similarly, isolates previously named Pseudomonas hibiscicola, which is also considered synonymous with S. maltophilia, clustered with putative species F [1]. Isolates from some other lineages, e.g. #2 and (B) also clustered, but not with any of the known or putative species as determined by ANIb. However, the isolates previously grouped as (D), (F), Smc1 or Smc2 did not group together in the ST Neighbor-Joining phylogeny.
Fig. 2.
Evolutionary history of Stenotrophomonas strains based on concatenated alleles from the S. maltophilia MLST scheme (pubmlst.org). The figure combines data from Ochoa-Sánchez and Vinuesa and our study (Ochoa-Sánchez and Vinuesa, 2017). Lineages #1-#10 were defined by Hauben et al. (Hauben et al., 1999). Lineages (A) to (F) were defined by Kaiser et al. (Kaiser et al., 2009) and Ochoa-Sanchez et al. (Ochoa-Sanchez 2017) expanded this further. The putative species A-N from the ANIb analysis are indicated by ps and the letter. The evolutionary history was inferred using the Neighbor-Joining method (Saitou, et al., 1987). The optimal tree with the sum of branch length = 1.82770971 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 190 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 3710 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). The left column denotes the isolates and strain identification. The right column the grouping. Type strains are indicated by (T) behind the name.
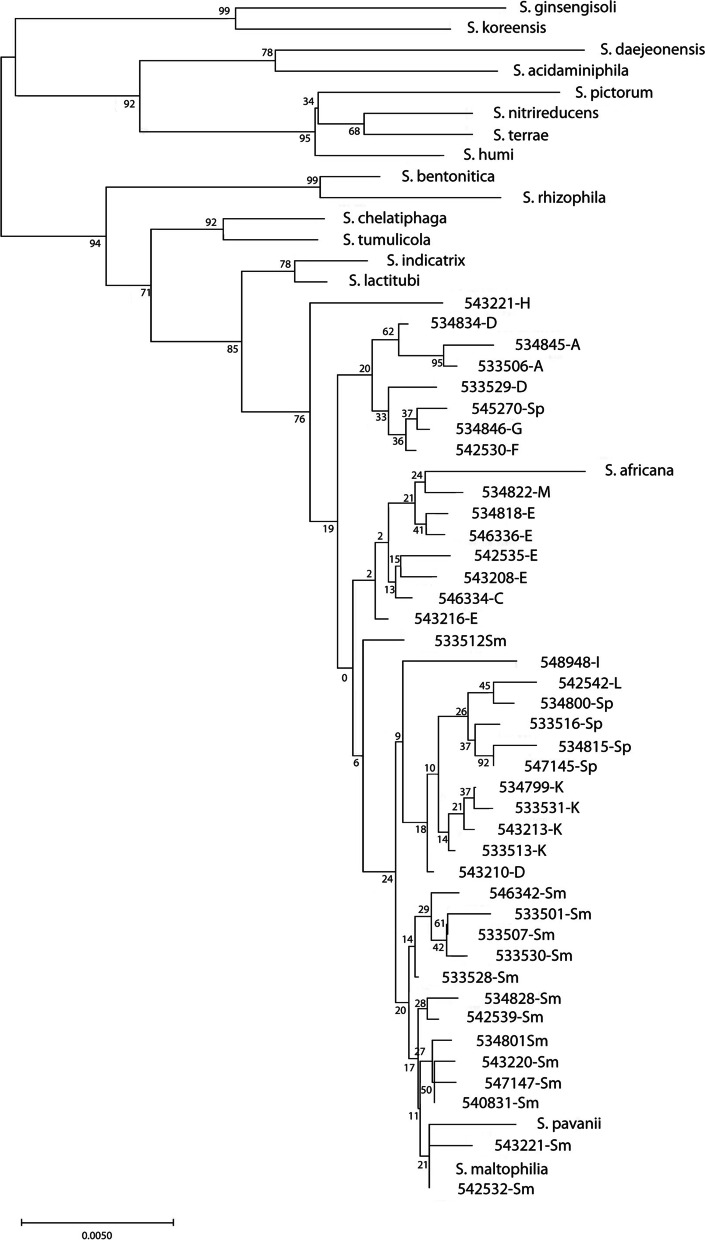
A neighbor-joining tree of our isolates based on unique 16S rRNA sequences for each species combined with the 16S rDNA sequences of the type strains showed that the type strains clustered separately, with the exception of S. maltophilia and S. pavanii, which clustered together. Sequences of all isolates identified as S. maltophilia by MALDI-TOF/MS clustered together. However, they generally clustered by (putative) species with a number of exceptions, e.g., S. maltophilia isolate 533,512 and S. pavanii isolate 545,270 and the type strain. S. africana, which is not recognized as a separate species anymore but considered as synonymous with S. maltophilia, also clustered with the isolates that were initially identified as S. maltophilia (Fig. 3).
Fig. 3.
Evolutionary history of Stenotrophomonas strains belonging to unknown species based on concatenated alleles of the MLST scheme using 16S rRNA sequences was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 0.18050158 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1050 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 57 nucleotide sequences. There were a total of 1392 positions in the final dataset
Antibiotic resistance
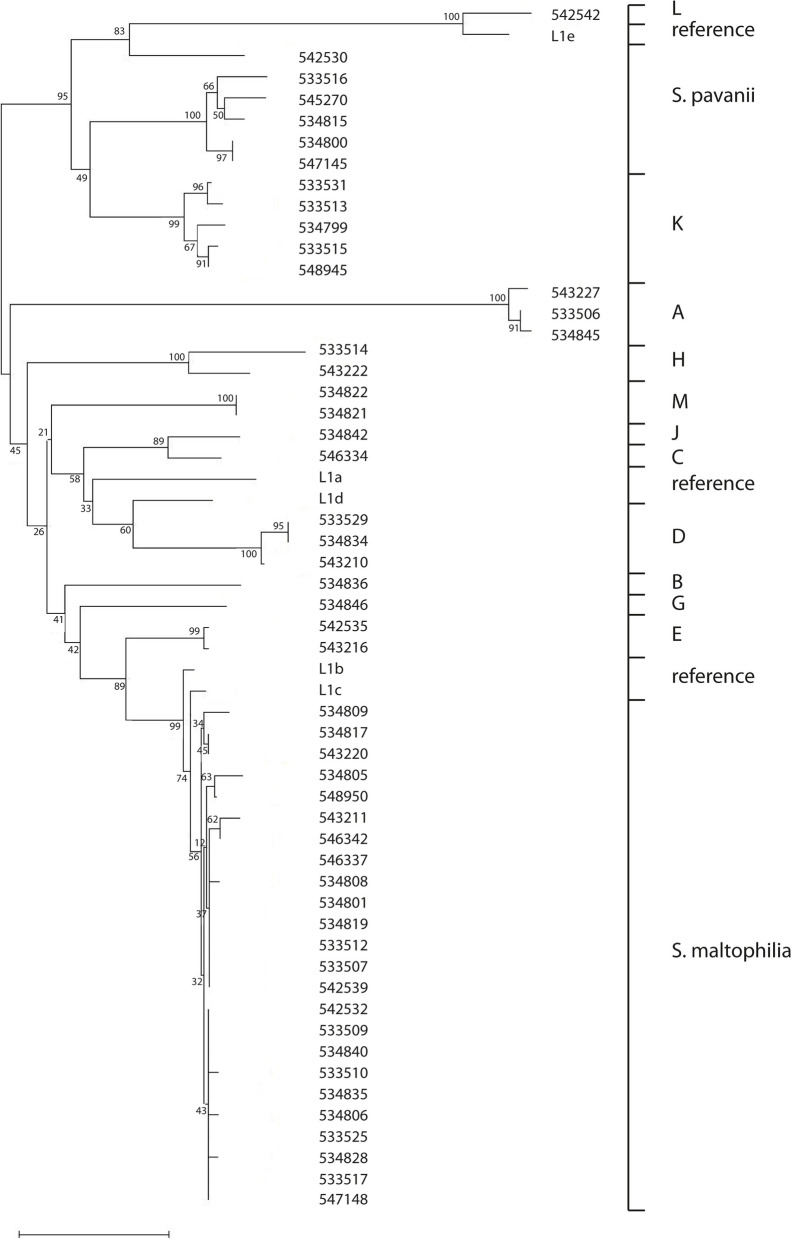
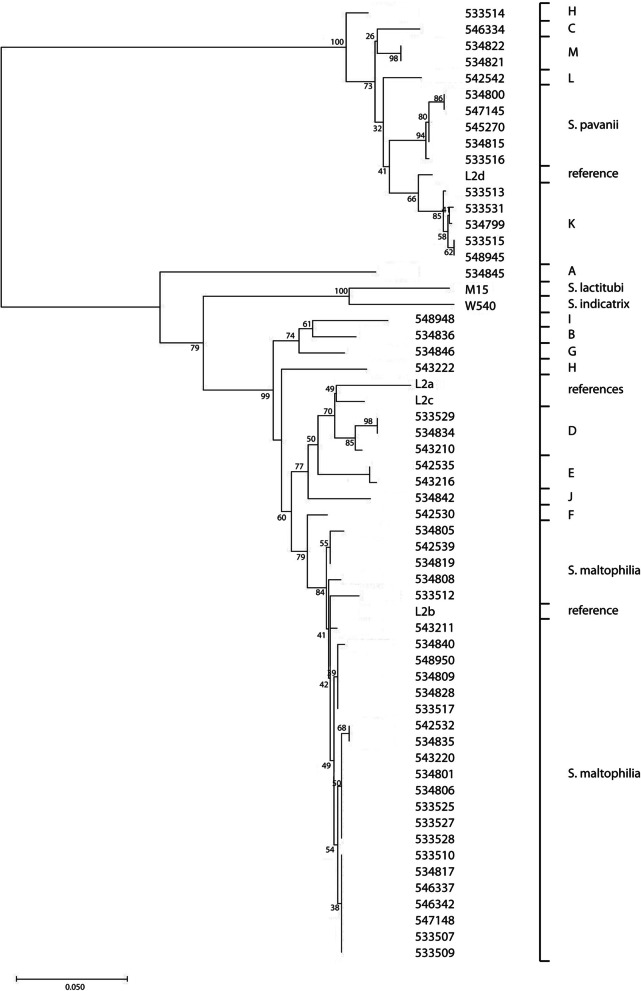
In S. maltophilia the L1 and L2 ß-lactamases have been described [6–8]. Most isolates in our study encoded a L1 family ß-lactamase, but in four the gene could not be detected: 3 S. maltophilia isolates and the isolate of the putative species I. L2 was present in all isolates collected for this study, although only partial sequences were available for four isolates (two incomplete sequences and two truncated sequences with 24-132x sequence coverage). The type strains for S. maltophilia and S. pavanii contained the genes for both ß-lactamases. S. indicatrix and S. lactitubi harbored only the L2 gene, but the type strains of the other species lacked the genes for the L1 and L2 ß-lactamase. A Neighbor-Joining tree of the amino acid sequences of both ß-lactamases showed that these cluster according to the putative species A-M, S. maltophilia, and S. pavanii as defined by ANIb (Figs. 4 and 5). The two isolates from putative species H were an exception. These showed clear differences between the ß-lactamases, in particular for the L2 ß-lactamases.
Fig. 4.
The evolutionary history of L1 ß-lactamases was inferred using the Neighbor-Joining method. One amino acid sequence per ST was included. L1a-L1d were used as reference sequences from literature (Walsh et al., 1994; Crowder et al., 1998). The optimal tree with the sum of branch length = 1.25626229 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1050 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 57 amino acid sequences. There were a total of 295 positions in the final dataset. No L1 sequences were found for the ST34, STH, and STI isolates. The letter behind the strain number indicate the presumptive species (Sm = Stenotrophomonas maltophilia, Sp = S. pavani) or putative novel species (A-M) as based on the analysis of the ANIb
Fig. 5.
The evolutionary history of L2 ß-lactamases was inferred using the Neighbor-Joining method. One amino acid sequence per ST was included. L2a-L2d were used as reference sequences from literature (Walsh et al., 1997). Strains M15 and W540 are the type strains for S. lactitubi and S. indicatrix, respectively. The optimal tree with the sum of branch length = 1.04210294 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1050 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 59 amino acid sequences. There were a total of 305 positions in the final dataset. There were a total of 304 positions in the final dataset. No L2 ß-lactamase sequences for ST89 and ST215 isolates were found
All S. pavanii isolates contained an aac (6′)-Iak aminoglycoside resistance gene and all isolates that were S. maltophilia based on the ANIb had an aph (3′)-IIc aminoglycoside resistance gene. Based on other genes present on the same contig, these resistance genes were located chromosomally, which is consistent with their presence in all isolates (Table 1). Many S. maltophilia isolates (72%) also contained an aac (6′)-Iz aminoglycoside resistance gene, which appeared to be present on the chromosome. The data suggest that the presence or absence of aac (6′)-Iz is ST-dependent (Table 1). The putative species M encoded an aac (6′)-Ib3 and ant (2″)-Ia aminoglycoside resistance gene; both genes appeared to be chromosomally located. The other putative species lacked aminoglycoside resistance genes (Table 1); these generally displayed lower tobramycin MICs, although some were also > 128 mg/L. Seven S. maltophilia isolates (11%) also had acquired aminoglycoside resistance genes. Although the majority of the isolates lacked acquired aminoglycoside resistance genes, isolate 534,804 encoded 7 of these genes (Table 1). This isolate also harbored cmx, tet(G), and sul1, encoding resistance against phenicols, tetracyclines, and sulfonamides respectively. Resistance genes other than those encoding aminoglycoside resistance were observed in isolate 534,806. These were dfrA15, tet(A), and sul1 encoding trimethoprim, tetracycline and sulfonamide resistance (Table 1). The sul1 gene was associated with a class 1 integron, but the integrase encoding genes could not be identified.
Fluoroquinolone resistance is associated with over expression of the efflux pumps SmQnr, SmeDEF or SmeVWX. For SmeVWX a number of changes in the regulator SmeRv are associated with overexpression [13–16], and the following mutations with amino acid changes were detected in our isolates: G266D in isolates 553,507 and 543,203; G266S in 533,509; C310Y in 533,517 and 533,533; and C310W in 534,828. The MICs for ciprofloxacin ranged from 2- > 32 μg/mL. Isolates 533,532, 534,846, 545,270, and 547,144 were truncated at positions 305, 305, 207, and 173, respectively; the range of ciprofloxacin MICs in these isolates was 2–16 μg/mL.
The only susceptibility breakpoint that EUCAST has currently defined for S. maltophilia is to co-trimoxazole; strains with a MIC ≤4 mg/L are susceptible with increased exposure (i.e. using a dose of 1440 mg q12h) [17]. Twenty-five of the 65 S. maltophilia isolates were resistant (39%). Applying the S. maltophilia co-trimoxazole breakpoint to S. pavanii, two of six isolates would be resistant (Supplementary Table 1). Based on this breakpoint, 14 of the 40 isolates (35%) with undefined taxonomic status would be resistant to co-trimoxazole.
Putative virulence genes and expression of protease and esterase activity
The genes for the proteases StmPr1, StmPr2, and StmP3, as well as the DNase, phospholipase C and D, esterase, fimbriae, TadE-like protein, the RpfC regulator, glucose-1-phosphate thymylyltransferase, phosphoglucomutase/phosphomannose bifunctional protein, and Ax21 outer membrane protein were present in nearly all isolates, independent of their taxonomic status; only six isolates lacked one or a few of these genes (Table 2, Supplementary Table 2). The genes for the type IV pilus machinery, Xps type II secretion system, and hemolysin were only found in S. maltophilia and in two isolates in the group of putative species (B and J) (Table 2, Supplementary Table 2). The genes for the other virulence factors showed a more mixed picture (Table 2, Supplementary Table 2), although generally the virulence factors were more frequently present in the S. maltophilia than in the putative novel species.
Table 2.
Percentage isolates positive in each species group
| Function | Genetic identifier | Total (n/%) | S. maltophilia (n/%) | Others (n/%) |
|---|---|---|---|---|
| alkaline serum protease stmPr1 | stmPr1 | 111/100.0 | 65/100.0 | 46/100.0 |
| protease | stmPr2 | 111/100.0 | 65/100.0 | 46/100.0 |
| protease | stmPr3 | 110/99.1 | 64/98.5 | 46/100.0 |
| Xps type II secretion system | smlt0687–0697 | 62/55.8 | 60/92.3 | 2/4.3 |
| DNase | smlt3212 | 111/100.0 | 65/100.0 | 46/100.0 |
| phospholipase C | smlt1755 | 110/99.1 | 65/100.0 | 45/97.8 |
| phospholipase D | smlt3521 | 110/99.1 | 65/100.0 | 45/97.8 |
| esterase | smlt3773 | 111/100.0 | 65/100.0 | 46/100.0 |
| polysaccharide lyase | smlt1473 | 85/76.6 | 55/84.6 | 30/65.2 |
| nitrate reductase | narG | 53/47.7 | 41/63.1 | 12/26.1 |
| fimbriae | smf-1 | 109/98.2 | 64/98.5 | 45/97.8 |
| TadE-like protein | smlt2869 achter | 108/97.3 | 64/98.5 | 44/8.7 |
| giant cable pilus-like protein | smlt3830voor | 93/83.8 | 64/98.5 | 29/63.0 |
| afimbrial adhesin | smlt4423voor | 56/50.5 | 39/60.0 | 17/37.0 |
| type IV pilus machinery | smlt1621–1627 | 61/55.0 | 59/90.8 | 2/4.3 |
| ankyrin repeat domain-containing protein | smlt3054 | 27/24.3 | 16/24.6 | 11/23.9 |
| filamentous hemagglutinin | fha | 61/55.0 | 55/84.6 | 6/13.0 |
| hemolysin | hly | 14/12.6 | 14/21.5 | 0/0.0 |
| regulator RpfC | rpfC | 111/100.0 | 65/100.0 | 46/100.0 |
| regulator RpfD | rpfF | 84/75.7 | 56/86.2 | 28/60.9 |
| glucose-1-phosphate thymdylyltransferase | rmlA | 110/99.1 | 65/100.0 | 45/97.8 |
| phosphoglucomutase/phosphomannose bifunctional protein | spgM | 110/99.1 | 65/100.0 | 45/97.8 |
| Ax21 outer membrane protein | ax1 | 110/99.1 | 65/100.0 | 45/97.8 |
Extensive sequence variation was observed for the type IV pilin adhesion precursor gene (Supplementary Table 2). Seven isolates exhibited only partial genes for filamentous hemagglutinin. DNase also showed some variation. Two isolates (545,270 and 533,529) had a truncated esterase. The glucose-1-phosphate thymylyltransferase was present in all strains and some variation was observed. Variation occurred also within STs (ST5, ST15, ST39, ST162). The Ax21 outer membrane protein was nearly identical within S. maltophilia and S. pavanii with a variation in only one amino acid position, but it was more variable within the isolates of putative species B-M (variations in 31 positions). Ax21, DNase, phospholipase D sequences in S. maltophilia cluster together as do the S. pavanii sequences. The sequences of the putative species form several clusters (data not shown).
The RPF (Regulation of Pathogenicity Factors) cluster of genes encodes a two-component system with a sensor and phosphorylation protein, as well as enoyl-CoA hydratase and a long chain-fatty-acid-CoA-ligase; together this composes a quorum sensing system with the fatty acid cis-11-methyl-2-dodecenoic acid (DSF) as the signal [18, 19]. The RPF cluster showed considerable sequence variation, especially in the sensor protein, with four main groups. (Supplementary Fig. 2).
The phenotypic expression of protease and esterase activity was also assessed: only 71.6% of the isolates exhibited protease activity and 40.4% exhibited esterase activity. Both the protease and the esterase activities showed associations with (putative) species or STs.
Discussion
The taxonomy of the genus Stenotrophomonas is complex and requires revision [4]. The individual Stenotrophomonas species recognized by the List of Prokaryotic names with Standing in Nomenclature may be distinguished from each other by using the conservative cut-off for species of 0.95 in the ANIb (Fig. 1, Supplementary Fig. 1) [1, 20]. Using this same criterion, 65 of 111 isolates collected for this study and identified by MALDI-TOF/MS as S. maltophilia would belong to this species. Six isolates were S. pavanii; the other isolates would represent a total of 13 different novel species, which we here indicate as putative species A-M. Isolates that cluster as putative species B-M cluster more closely to S. maltophilia than to the type strains of other Stenotrophomonas species, suggesting that either the divergence of these putative species is more recent or that genetic exchange between these isolates has been more common.
A neighbor-joining tree of the concatenated alleles of the S. maltophila MLST scheme used to compare the ANIb results with those of Ochoa-Sánchez and Vinuesa showed that most groups of isolates or new species grouped comparably (Fig. 2) [4]. Group #1 isolates clustered with the S. pavanii type strain, and also our pairwise clustering suggests that these isolates are S. pavanii. The previously identified group #6 clustered with the S. maltophilia type strain as well as with a number of our isolates. Group #9 was previously proposed as a novel species (Smc4), but, based on their clustering with the type strain in the MLST based phylogeny, the isolates in these group are most likely either S. indicatrix or S. lactitubi. Several previously proposed lineages clustered with the putative species in the ANIb analysis, e.g., lineage #3, #4, #5, #7 with putative species L, E, J, and B, respectively. Some of the putative species from the ANIb clustered with isolates that previously belonged to a different species now considered synonymous with S. maltophilia, such as P. beteli. Our data suggest that these isolates differ significantly from S. maltophilia and may indeed represent different species.
Some groups ((D), (F), Smc1, and Smc2) did not group in our analysis. This may be due to a different algorithm to generate the trees, but another explanation is that concatenated STs are not optimal for defining species [3, 4]. However, it should be noted that the bootstrap values for some branches are low, indicating low confidence for the branching.
To further elucidate the taxonomic position of our isolates, a 16S rDNA sequence analysis was performed. This analysis was basically in agreement with the ANIb analysis. Remarkably, the 16S rDNA sequence of the S. pavanii type strain clustered closely with that of the S. maltophilia type strain and with the S. maltophilia isolates collected for this study, whereas our other S. pavanii isolates clustered elsewhere (Fig. 3). As already suggested by the ANIb, the 16S rDNA data suggest that S. maltophilia, S. pavanii and the putative new species are still closely related. Based on 16S analysis, S. africana, which is currently considered synonymous with S. maltophilia, should be considered as a separate species.
All our isolates harbored a chromosomally encoded L1 and L2 ß-lactamase. However, the (type) strains of species other than S. maltophilia and S. pavanii lacked these ß-lactamases, except for S. indicatrix and S. lactitubi, which encoded the L2 gene. A Neighbor-joining analysis of the amino acid sequences was in agreement with the results of the ANIb and concatenated STs (Fig. 4, Fig. 5): isolates identified as S. maltophilia in the ANIb clustered together with limited sequence variability; the other (putative) species displayed more diversity. The L1 ß-lactamase hydrolyzes carbapenems and other ß-lactam antibiotics (but not monobactams), whereas L2 ß-lactamase is a serine hydrolase that acts as a cephalosporinase. Stenotrophomonas MICs for imipenem and meropenem are generally high; more variation is seen for ceftazidime. The considerable sequence variation for both ß-lactamases, which was already described when the first genes were sequenced, may at least in part explain differences between isolates [6–9].
All S. pavanii isolates contained a chromosomally encoded aac (6′)-Iak aminoglycoside resistance gene and all isolates that were S. maltophilia based on the ANIb had an aph (3′)-IIc aminoglycoside resistance gene, whereas the other isolates lacked chromosomally encoded aminoglycoside resistance genes. S. maltophilia and S. pavanii tended to have higher MICs for tobramycin than the putative species (Supplementary Table 1). However, this correlation was not perfect, suggesting that other factors play a role in expression and thereby resistance, including regulation of aminoglycoside resistance genes. In addition to aph (3′)-IIc some S. maltophilia had aac (6′)-Iz, a chromosomally encoded aminoglycoside resistance gene (Table 1).
Only a few isolates possessed acquired resistance genes (Table 1), but one isolate encoded 7 different ones, including a sul1 resistance gene. Sul1 is associated with class 1 integrons [21], but presence of an integron could not be confirmed. Nevertheless, this suggests that S. maltophilia can acquire plasmids or transposons with class 1 integrons, as has been previously described [21, 22]. Differences between the (presence of) chromosomally encoded antibiotic resistance genes further supports the hypothesis that our isolates included different species (S. maltophilia, S. pavanii and a set of putative novel species). Although the chromosomally encoded ß-lactamases and aminoglycoside resistance genes may be clinically relevant, their function is probably to aid in competition with other micro-organisms in their natural niche.
The extensive resistance pattern of S. maltophilia severely limits the antibiotic treatment options for infections, and only co-trimoxazole is considered a reliable treatment option. Described alternatives include fluoroquinolones and tetracyclines [10], but the MICs may vary considerably. Currently little is known about the genetic factors determining resistance to these antibiotics with the exception of fluoroquinolone resistance, which is associated with overexpression of efflux pumps. These are either SmQnr, SmeDEF or SmeVWX [14–16]. Overexpression of SmeVWX is associated with amino acid changes in its repressor, SmeRv [15, 16]. Five isolates had previously described amino acid substitutions; however, their ciprofloxacin MICs ranged from 2 to > 32 mg/L. For the newly described variants (a C310W in 534,828 and truncated sequences) the range was 2 to 16 mg/L. No mutations in the quinolone resistance determining regions of gyrAB and parCE were observed [13].
The presence of the genes for the proteases StmPr1, StmPr2, and StmP3, as well as the DNase, phospholipase C and D, esterase, fimbriae, TadE-like protein, and the RpfC regulator genes in all or nearly all isolates (Table 2, Supplementary Table 2). However, somewhat lower percentages have been reported in literature. StmPr1, StmPr2, esterase, and fimbriae were present in more than 90% of the isolates in a collection from CF patients attending a pediatric hospital in Rome, Italy [23]. Possibly, the collections differed in the distribution of different (putative) species, and that these genes do not belong to the core genome of all (putative) species.
Differences in the type IV pilus adhesion precursor should likely be sought in the different adherence properties that may lead to advantages in different natural niches.
StmPr1, − 2, and − 3 degrade a wide variety of extracellular matrix components. StmPr1 degrades collagen type I and IL-8, and can kill A549 lung epithelial cells; StmP3 degrades fibronectin, fibrinogen, and IL-8, and contributes to cell rounding and detachment in vitro; the activities of StmPr2 have not exactly been defined, but this protease contributes to degradation of extracellular matrix proteins and cell rounding. The three proteases are secreted in Xps-dependent manner [24, 25]. However, the genes encoding Xps type II secretion system could only be identified in 90% of the S. maltophilia isolates and in putative species B and J andabsent in the other (putative) species. Since phenotypic protease activity did not correlate with species or STs, proteases may likely also be secreted by an alternative mechanism. All isolates contained protease genes and an esterase gene, but phenotypic expression of protease and esterase activity was found in only 71.6 and 40.4% of the isolates respectively. This suggests that the regulation of expression of these activities is complex. Alternatively, additional proteases may be present which are secreted by a different mechanism.
Polysaccharide lyase degrades alginate, poly-ß-D-glucuronic acid and hyaluronic acid [26]. These first two compounds are found widely in nature and hyaluronic acid is an important constituent of human skin, but it is also found elsewhere in the human body. The polysaccharide lyase was present in more than three quarters of the isolates, and its absence/presence did not follow the (putative) species boundaries. The absence of the gene in many isolates obtained from CF and lung infections indicate that it is not essential for colonization or infection of the human lung.
The secreted ankyrin-repeat protein, which interacts with actin, was present in only approximately a quarter of the isolates [27]. This interaction is thought to alter the cytoskeletal structure. However, StmPr1 and StmPr2 also contribute to this process [25].
The RPF quorum sensing system showed considerable sequence variability, in particular for the sensor protein (Supplementary Fig. 2). This quorum sensing system has been reported to regulate motility, biofilm development, antibiotic resistance and virulence in S. maltophilia, but the implications for (CF) lung infections are not immediately clear [18, 19, 28]. It has been reported that the signal molecule DSF generated by the expression of the RPF system influences Pseudomonas aeruginosa biofilm formation and antibiotic resistance [29–31]. Some isolates lack a 190 amino acid sequence in the sensor domain region in the middle of the protein. Huedo et al showed sequence variation in the RPF cluster of S. maltophilia and designated two variants: rpf-1 and rpf-2 [32]. The first is more similar to the system found in Xanthomonas, whereas the latter is more similar to the system in Pseudoxanthomonas, Arenimonas, and Lysobacter. In a rpf-2 system, with a shorter sensor domain, it appears that some quorum sensing signal DSF needs to be present for activation of DSF synthesis. It has been speculated that under some conditions the rpf-2 system saves energy, and that its activation is dependent on the presence of other bacterial isolates or species [32].
The majority of S. maltophilia isolates are capable of biofilm production, but marked differences have been observed. On average more biofilm formation on polystyrene was observed by non-CF isolates than by CF isolates, which in turn displayed more biofilm formation than environmental isolates, but large variation among isolates within the groups was present [33]. Variable biofilm formation was also observed on IB3–1 bronchial cells in vitro [34].
The spgM, and rmlA genes, encoding aphosphoglucomutase/phosphomannose bifunctional protein and glucose-1-phosphate thymylyltransferase, respectively, have been implicated in biofilm formation and were reported in 83.3 and 87.5% of 37 isolates tested [35]. However, all our isolates contained these genes; therefore, either there were differences in the populations tested, or the PCR used in prior studies did not detect all genes due to sequence variation in the primer region(s). Although the RPF system has been reported to influence biofilm formation, no correlation with the detection of rpfF by PCR was found [35]. The nitrate reductase has also been associated with biofilm by enabling growth in micro-oxic conditions [36]. Nearly half of our isolates harbored the narG gene encoding the nitrate reductase; this was in agreement with a previous study that reported 37/63 positive isolates [36]. The Ax21 outer membrane protein is also involved in biofilm formation, as well as motility, reduced tolerance to tobramycin, and virulence in an insect model, but its regulation and mode action of action have not been resolved [37].
Although these data help to identify putative virulence their role should ultimately be proven in vivo.
Conclusions
This study has shown that either S. maltophilia is highly heterogeneous and S. pavanii should be included in this species, or that S. maltophilia is more limited and many of our isolates belong to currently undescribed and unnamed species. The putative novel Stenotrophomonas species recovered from patient samples and identified by MALDI-TOF/MS as S. maltophilia, differed from S. maltophilia in resistance and virulence genes, and, as a consequence, possibly in pathogenicity. Revision of the Stenotrophomonas taxonomy is needed to reliably identify strains within the genus and elucidate the role of the different species in disease.
Methods
The study
The strains were collected for the iABC-project, in which novel antibiotics for CF and bronchiectasis are being developed. A total of 111 isolates were included in the study. These isolates were recovered from respiratory samples of CF (n = 103) and diverse pulmonary infections (n = 8) between 2003 and 2016 from five different countries: Australia (n = 1), United Kingdom (n = 41), Spain (n = 35), the Netherlands (n = 33), and USA (n = 1) (Table 1). Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF/MS, Bruker) identified all isolates as S. maltophilia.
The aim of the present study was to determine the taxonomic position of S. maltophilia isolates from persons with CF and patients with other chronic respiratory infections, and to characterize their antibiotic resistance genes and virulence factors.
Whole genome sequencing
Bacterial DNA was purified using the Qiacube with the DNeasy Blood & Tissue kit with the enzymatic lysis protocol (Qiagen, Carlsbad, CA) and used to prepare a library for sequencing with the MiSeq or Nextseq (Illumina, San Diego, CA) platforms, using the Nextera XT library prep kit (Illumina). Contigs were assembled with SPAdes genome assembler v.3.6.2. The assembled contigs were analyzed for the presence of resistance genes by ResFinder [38, 39] from the Center for Genomic Epidemiology (DTU, Copenhagen, Denmark) [39]. Multi-locus Sequence Typing (MLST) was performed using PubMLST using the scheme for S. maltophilia [40]. Novel alleles and sequence types (ST) were submitted to the database.
Analysis of average nucleotide identity by BLAST (ANIb)
Average nucleotide identity (ANI) among the genomes was calculated using ANIb algorithm of pyani tool version 0.2.3 [41], which uses nucleotide BLAST (version 2.2.28+) alignment for whole genome alignment. Genomes were fragmented into genomic fragments of 1020 bases long. After pairwise alignments of all fragments of each genome, ANI was calculated as the percentage of nucleotide identity for matching regions of all genomes. In biclustering analysis of ANI scores, complete linkage was used as a hierarchical clustering method with the Euclidean distance metric. A heatmap of all genomes was generated using biclustering, where a color scale bar shows the pairwise ANI scores above 75% are shown in a color range starting from blue (ANI 75%) through white to red (ANI 100%). In the heatmap, each species is shown in a different color next to a leaf node of a tree. A heatmap of all genomes was generated using biclustering, with a color scale bar showing the pairwise ANI score for values above the threshold value of 0.75. A cut-off of 0.95 was used to define groups, which is also the conservative cut-off used to define species [20, 42].
Phylogenetic trees
Neighbor-joining trees of the concatenated alleles of the MLST scheme for S. malthophilia, the 16S rRNA gene, and the L1 and L2 ß-lactamase sequences were generated with WebPrank and Clustal Omega for the alignments, and with MEGA X for the generation of the tree, using 500 bootstraps [40, 43]. The trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method, in the units of the number of base substitutions per site for the 16S rDNA sequences. For the other sequences the evolutionary distances were computed using the Poisson correction method, in the units of the number of amino acid substitutions per site [44–48]. For the ß-lactamase sequences only one sequence per ST was included. KODON (Applied Maths, Belgium) was used for the whole genome alignments.
The whole genome sequences of the type strains of all species except S. tumilicola were available for analysis.
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) were determined by the standard ISO broth microdilution method with frozen panels (Trek Diagnostic Systems, Westlake, OH) using EUCAST methodology [17]. The antimicrobial agents and the concentration ranges tested were as follows: aztreonam (0.25–256 mg/L); ceftazidime (0.25–256 mg/L); ciprofloxacin (0.03–32 mg/L); colistin (0.25–16 mg/L); tobramycin (0.125–128 mg/L); imipenem (0.125–128 mg/L); meropenem (0.06–64 mg/L); and trimethoprim/sulfamethoxazole (co-trimoxazole) (0.06–32 mg/L). The susceptibility data for the isolates recovered from CF patients were previously reported [49].
Protease and esterase activity
Bacteria were grown to 1 OD600 in LB broth and 2 μl was spotted onto agar with 2% skim milk and onto plates with tributyrin-Arabic gum, and incubated overnight at 37 °C to respectively assess protease activity, and esterase activity [50, 51].
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- ANIb
Average nucleotide identity by BLAST
- CF
Cystic fibrosis
- MALDI-TOF/MS
Matrix-assisted laser desorption/ionization time of flight mass spectrometry
- MB
Megabase
- MIC
Minimum inhibitory concentrations
- MLST
Multi-Locus Sequence Typing
- RPF
Regulation of Pathogenicity Factors
- S. acidaminiphila
Stenotrophomonas acidaminiphila
- S. bentonitica
Stenotrophomonas bentonitica
- S. chelatiphaga
Stenotrophomonas chelatiphaga
- S. daejeonensis
Stenotrophomonas daejeonensis
- S. ginsengisoli
Stenotrophomonas ginsengisoli
- S. humi
Stenotrophomonas humi
- S. indicatrix
Stenotrophomonas indicatrix
- S. koreensis
Stenotrophomonas koreensis
- S. lactitubi
Stenotrophomonas lactitubi
- S. maltophilia
Stenotrophomonas maltophilia
- S. nitrireducens
Stenotrophomonas nitrireducens
- S. pavanii
Stenotrophomonas pavanii
- S. pictorum
Stenotrophomonas pictorum
- S. rhizophila
Stenotrophomonas rhizophila
- S. terrae
Stenotrophomonas terrae
- S. tumulicola
Stenotrophomonas tumulicola
- ST
Sequence type
Authors’ contributions
ACF, MA, MBE, MT, RC, SE design. BJB, JS investigation. ACF, JRB, MBE analysis. ACF, JRB, MBE, MD, MT, RC, SE writing, review-editing. All authors have read and approved the manuscript.
Funding
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement number (115721–2), resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies in kind contribution. MD-A was also partially supported by the Fundación.
Francisco Soria Melguizo (Madrid, Spain). The funders had no influence on the contents of this manuscript.
Availability of data and materials
The sequence data have been submitted to GenBank (BioProject ID: PRJNA718312 available at https://dataview.ncbi.nlm.nih.gov/object/PRJNA718312?reviewer=epjohsfos888drv6cjnimg3f8s).
Data have also been submitted to the DRYAD database. These data have a temporary link: https://datadryad.org/stash/share/keoI3aRF2r55sXSotR_oxlA6yyfdd1KMfsxG-N_-_IQ.
Declarations
Ethics approval and consent to participate
Samples and patient data were collected in compliance with the Declaration of Helsinki ICH-GCP, the Declaration of Taipei regarding Health Databases and Biobanks, and with local and European regulations for collection and handling of patient data. Since the study concerned retrospectively collected anonymized patient data and bacterial isolates, informed consent at the individual patient level was not required for this study. In addition, the Spanish and UK isolates were collected in accordance with their local ethics guidelines and described in prior studies [52–54]. In the Netherlands, use and analysis of bacterial isolates with anonymized patient data does not require approval from Institutional Review Boards/Ethics Committees [https://wetten.overheid.nl/BWBR0009408/2021-07-01].
Experimental protocols did not need approval by a named institutional and/or licensing committee because analysis of bacterial strains was not subject to approval.
Informed consent was not required by local laws because only bacterial strains were used.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.List of Prokaryotic names with Standing in Nomenclature. http://www.bacterio.net/ralstonia.html. (accessed Feb 20, 2020).
- 2.Hauben L, Vauterin L, Moore ER, Hoste B, Swings J. Genomic diversity of the genus Stenotrophomonas. Int J Syst Bacteriol. 1999;49:1749–1746. doi: 10.1099/00207713-49-4-1749. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser S, Biehler K, Jonas D. A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J Bacteriol. 2009;191:2934–2943. doi: 10.1128/JB.00892-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochoa-Sánchez LE, Vinuesa P. Evolutionary genetic analysis uncovers multiple species with distinct habitat preferences and antibiotic resistance phenotypes in the Stenotrophomonas maltophilia complex. Front Microbiol. 2017;8:1548. doi: 10.3389/fmicb.2017.01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trifonova A, Strateva T. Stenotrophomonas maltophilia - a low-grade pathogen with numerous virulence factors. Infect Dis (Lond) 2019;51:168–178. doi: 10.1080/23744235.2018.1531145. [DOI] [PubMed] [Google Scholar]
- 6.Walsh TR, Hall L, Assinder SJ, Nichols WW, Cartwright SJ, MacGowan AP, et al. Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 7.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1998;42:921–926. doi: 10.1128/AAC.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh TR, MacGowan AP, Bennett PM. Sequence analysis and enzyme kinetics of the L2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1460–1464. doi: 10.1128/AAC.41.7.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avison MB, Higgins CS, von Heldreich CJ, Bennett PM, Walsh TR. Plasmid location and molecular heterogeneity of the L1 and L2 beta-lactamase genes of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2001;45:413–419. doi: 10.1128/AAC.45.2.413-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adegoke AA, Stenström TA, Okoh AI. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front Microbiol. 2017;8:2276. doi: 10.3389/fmicb.2017.02276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ince N, Yekenkurul D, Danış A, Çalışkan E, Akkaş İ. An evaluation of six-year S. maltophilia infections in a university hospital. Af Health Sci. 2020;20:1118–1123. doi: 10.4314/ahs.v20i3.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berdah L, Taytard J, Leyronnas S, Clement A, Boelle PY, Corvol H. Stenotrophomonas maltophilia: a marker of lung disease severity. Pediatr Pulmonol. 2018;53:426–430. doi: 10.1002/ppul.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdezate S, Vindel A, Saéz-Nieto JA, Baquero F, Cantón R. Preservation of topoisomerase genetic sequences during in vivo and in vitro development of high-level resistance to ciprofloxacin in isogenic Stenotrophomonas maltophilia strains. J Antimicrob Chemother. 1994;56:220–223. doi: 10.1093/jac/dki182. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez MB, Martínez JL. SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2010;54:580–581. doi: 10.1128/AAC.00496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-León G, de Alegría R, Puig C, García de la Fuente C, Martínez-Martínez L, Martínez JL, Sánchez MB. High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin Microbiol Infect. 2015;21:464–467. doi: 10.1016/j.cmi.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 16.García-León G, Salgado F, Oliveros JC, Sánchez MB, Martínez JL. Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ Microbiol. 2014;16:1282–1296. doi: 10.1111/1462-2920.12408. [DOI] [PubMed] [Google Scholar]
- 17.European Committee on Antimicrobial Susceptibility Testing . Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. 2019. [Google Scholar]
- 18.Huang T-P, Lee Wong AC. Extracellular fatty acids facilitate flagella-independent translocation by Stenotrophomonas maltophilia. Res Microbiol. 2007;158:702–711. doi: 10.1016/j.resmic.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Huedo P, Yero D, Martínez-Servat S, Estibariz I, Planell R, Martínez P, et al. Two different rpf clusters distributed among a population of Stenotrophomonas maltophilia clinical strains display differential diffusible signal factor production and virulence regulation. J Bacteriol. 2014;196:2431–2442. doi: 10.1128/JB.01540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fluit AC, Schmitz F-J. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 22.Barbolla R, Catalano M, Orman BE, Famiglietti A, Vay C, Smayevsky J, et al. Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob Agents Chemother. 2004;48:666–669. doi: 10.1128/aac.48.2.666-669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicoletti M, Iacobino A, Prosseda G, Fiscarelli E, Zarrill R, De Carolis E, et al. Stenotrophomonas maltophilia strains from cystic fibrosis patients: genomic variability and molecular characterization of some virulence determinants. Int J Med Microbiol. 2011;301:34–43. doi: 10.1016/j.ijmm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 24.DuMont AL, Karaba SM, Cianciotto NP. Type II secretion-dependent degradative and cytotoxic activities mediated by Stenotrophomonas maltophilia serine proteases StmPr1 and StmPr2. Infect Immun. 2015;83:3825–3837. doi: 10.1128/IAI.00672-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DuMont AL, Cianciotto NP. Stenotrophomonas maltophilia serine protease Stmpr1 induces matrilysis, anoikis, and protease-activated receptor 2 activation in human lung epithelial cells. Infect Immun. 2017;85:e00544–e00517. doi: 10.1128/IAI.00544-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacDonald LC, Berger BW. A polysaccharide lyase from Stenotrophomonas maltophilia with a unique, pH-regulated substrate specificity. J Biol Chem. 2014;289:312–325. doi: 10.1074/jbc.M113.489195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacDonald LC, O'Keefe S, Parne M-F, MacDonald H, Stretz L, Templer SJ, et al. A secreted ankyrin-repeat protein from clinical Stenotrophomonas maltophilia isolates disrupts actin cytoskeletal structure. ACS Infect Dis. 2016;2:62–70. doi: 10.1021/acsinfecdis.5b00103. [DOI] [PubMed] [Google Scholar]
- 28.Fouhy Y, Scanlon K, Schouest K, Spillane C, Crossman L, Avison MB, et al. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol. 2007;189:4964–4968. doi: 10.1128/JB.00310-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehau K, Yang L, et al. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 30.Twomey KB, O'Connell OJ, McCarthy Y, Dow JM, O'Toole GA, Plant BJ, et al. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 2012;6:39–950. doi: 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pompilio A, De Nicola CV, S, Verginelli F, Fiscarelli E, Di Bonaventura G. Cooperative pathogenicity in cystic fibrosis: Stenotrophomonas maltophilia modulates Pseudomonas aeruginosa virulence in mixed biofilm. Front Microbiol. 2015;6:951. doi: 10.3389/fmicb.2015.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huedo P, Yero D, Martinez-Servat S, Ruyra N, Rohe À, Daura X, et al. Decoding the genetic and functional diversity of the DSF quorum-sensing system in Stenotrophomonas maltophilia. Front Microbiol. 2015;6:761. doi: 10.3389/fmicb.2015.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pompilio A, Pomponio S, Crocetta V, Gherardi G, Verginelli F, Fiscarelli E, et al. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 2011;11:159. doi: 10.1186/1471-2180-11-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pompilio A, Crocetta V, Confalone P, Nicoletti M, Petrucca A, Guarnieri S, et al. Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol. 2010;10:102. doi: 10.1186/1471-2180-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhuo C, Zhao QY, Xiao SN. The impact of spgM, rpfF, rmlA gene distribution on biofilm formation in Stenotrophomonas maltophilia. PLoS One. 2014;9:e108409. doi: 10.1371/journal.pone.0108409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcaraz E, Garcia C, Papalia M, Vay C, Friedman L, Passerini de Rossi B. Stenotrophomonas maltophilia isolated from patients exposed to invasive devices in a university hospital in Argentina: molecular typing, susceptibility and detection of potential virulence factors. J Med Microbiol. 2018;67:992–1002. doi: 10.1099/jmm.0.000764. [DOI] [PubMed] [Google Scholar]
- 37.An S, Tang J. The Ax21 protein influences virulence and biofilm formation in Stenotrophomonas maltophilia. Arch Microbiol. 2018;200:183–187. doi: 10.1007/s00203-017-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ReFinder https://cge.cbs.dtu.dk/services/ResFinder/ (accessed Oct 28, 2019).
- 39.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.PubMLST https://pubmlst.org/general.shtml (accessed Nov 10, 2020).
- 41.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 42.Schneider I, Bauernfeind A. Intrinsic carbapenem-hydrolyzing oxacillinases from members of the genus Pandoraea. Antimicrob Agents Chemother. 2015;59:7136–7141. doi: 10.1128/AAC.01112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clustal Omega https://www.ebi.ac.uk/Tools/msa/clustalo (accessed Nov 10, 2020).
- 44.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York, NY: Academic Press; 1965. pp. 97–166. [Google Scholar]
- 45.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 46.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Nat Acad Sc USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Díez-Aguilar M, Ekkelenkamp M, Morosini MI, Merino I, de Dios CJ, Jones M, et al. Antimicrobial susceptibility of non-fermenting gram-negative pathogens isolated from cystic fibrosis patients. Int J Antimicrob Agents. 2019;53:84–88. doi: 10.1016/j.ijantimicag.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Figueirêdo PMS, Furumura MT, Santos AM, Sousa ACT, Kota DJ, Levy CE, et al. Cytotoxic activity of clinical Stenotrophomonas maltophilia. Lett Appl Microbiol. 2006;43:443–449. doi: 10.1111/j.1472-765X.2006.01965.x. [DOI] [PubMed] [Google Scholar]
- 51.Wilhelm S, Gdynia A, Tielen P, Rosenau F, Jaeger KE. The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J Bacteriol. 2007;189:6695–6703. doi: 10.1128/JB.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Dios CJ, Del Campo R, Royuela A, Solé A, Máiz L, Olveira C, et al. Bronchopulmonary infection-colonization patterns in Spanish cystic fibrosis patients: results from a national multicenter study. J Cyst Fibros. 2016;15:357–365. doi: 10.1016/j.jcf.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Muhlebach MS, Hatch JE, Einarsson GG, McGrath SJ, Gilipin DF, Lavelle G, et al. Anaerobic bacteria cultured from cystic fibrosis airways correlate to milder disease: a multisite study. Eur Respi J. 2018;52:pii–1800242. doi: 10.1183/13993003.00242-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCaughey G, McKevitt M, Elborn JS, Tunney MM. Antimicrobial activity of fosfomycin and tobramycin in combination against cystic fibrosis pathogens under aerobic and anaerobic conditions. J Cyst Fibros. 2012;11:163–172. doi: 10.1016/j.jcf.2015.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data have been submitted to GenBank (BioProject ID: PRJNA718312 available at https://dataview.ncbi.nlm.nih.gov/object/PRJNA718312?reviewer=epjohsfos888drv6cjnimg3f8s).
Data have also been submitted to the DRYAD database. These data have a temporary link: https://datadryad.org/stash/share/keoI3aRF2r55sXSotR_oxlA6yyfdd1KMfsxG-N_-_IQ.