Abstract
Background
Inherited retinal dystrophies (IRD) and optic neuropathies (ION) are the two major causes world-wide of early visual impairment, frequently leading to legal blindness. These two groups of pathologies are highly heterogeneous and require combined clinical and molecular diagnoses to be securely identified. Exact epidemiological studies are lacking in North Africa, and genetic studies of IRD and ION individuals are often limited to case reports or to some families that migrated to the rest of the world. In order to improve the knowledge of their clinical and genetic spectrums in North Africa, we reviewed published data, to illustrate the most prevalent pathologies, genes and mutations encountered in this geographical region, extending from Morocco to Egypt, comprising 200 million inhabitants.
Main body
We compiled data from 413 families with IRD or ION together with their available molecular diagnosis. The proportion of IRD represents 82.8% of index cases, while ION accounted for 17.8%. Non-syndromic IRD were more frequent than syndromic ones, with photoreceptor alterations being the main cause of non-syndromic IRD, represented by retinitis pigmentosa, Leber congenital amaurosis, and cone-rod dystrophies, while ciliopathies constitute the major part of syndromic-IRD, in which the Usher and Bardet Biedl syndromes occupy 41.2% and 31.1%, respectively. We identified 71 ION families, 84.5% with a syndromic presentation, while surprisingly, non-syndromic ION are scarcely reported, with only 11 families with autosomal recessive optic atrophies related to OPA7 and OPA10 variants, or with the mitochondrial related Leber ION. Overall, consanguinity is a major cause of these diseases within North African countries, as 76.1% of IRD and 78.8% of ION investigated families were consanguineous, explaining the high rate of autosomal recessive inheritance pattern compared to the dominant one. In addition, we identified many founder mutations in small endogamous communities.
Short conclusion
As both IRD and ION diseases constitute a real public health burden, their under-diagnosis in North Africa due to the absence of physicians trained to the identification of inherited ophthalmologic presentations, together with the scarcity of tools for the molecular diagnosis represent major political, economic and health challenges for the future, to first establish accurate clinical diagnoses and then treat patients with the emergent therapies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-022-02340-7.
Keywords: Inherited retinal dystrophies, Inherited optic neuropathies, Molecular diagnosis, North Africa, Consanguinity, Phenotypic spectrum, Genetic spectrum
Background
North Africa encompasses five countries, Morocco, Algeria, Tunisia, Libya and Egypt, which historically and geographically were at the crossroad between Africa, Europe and Asia. This area is delimited by the Mediterranean Sea in the north, and separated from other African countries by the Sahara in the south. The North African population, today numbering 202 million people [1], is mainly occupied by two ethnic groups which are Arab Muslims and Berbers, and a minority of Jews and Christians.
Historically, since the earliest times, the Berber people, also called Amazigh have populated North Africa, before this area became a targeted land of several colonization and demographic migratory movements, first by Phoenicians, then by Romans, Vandals, Byzantines, Arabs, Ottomans, and finally by Europeans during the eighteenth and nineteenth centuries [2].
The interactions between all these populations have made the genetic pool of the North African population very complex and heterogeneous, even among populations living in close geographical areas, or within the same ethnic groups, which have been described by several molecular studies involving mitochondrial DNA, Y chromosome and autosomal markers [3–9]. Jews have appeared in North Africa since the Phoenician influence. Their communities have grown as a result of conversion and admixture with the local Berber population at that time, in addition to several migrations from the middle East and Europe, mainly by the Jews of the Iberian peninsula, after their expulsion from Spain and Portugal between 1492–1497 [10], and were named Sephardic. In 1950s, Jews of North Africa have migrated to the current Israel [10]. Genome-wide studies defined North African Jews as a distinct group, with a strong link to the European and Middle Eastern Jews, and with European non-Jews, rather than North African non-Jews. In addition, they show a high degree of identity of descent segment haplotypes, witnessing the high level of endogamy within the Jewish diaspora [11] that led to high frequency of founder mutations [12].
North African countries show a high rate of endogamy and consanguinity, ranging from 20 to 40% [13, 14], and up to some 60% in some remote villages [15]. This marriage practice is also conserved within North African immigrants who live in other parts of the world, mainly in Europe [16]. Consanguinity increases the chance of bi-allelic associations of a single mutant allele in an individual, and as a consequence, a predominance of diseases related to autosomal recessive transmission. In this respect, an epidemiological study of genetic diseases in the North African population identified 532 pathologies, of which 60% were inherited in an autosomal recessive manner [17].
Vision is the most important of the five senses, and visual impairment has major impacts on the psychological, educational and socioeconomic conditions of affected individuals. Worldwide, 19 million children are estimated to be visually impaired [18]. In developed countries, retinal and optic nerve problems are the major causes of visual impairments, [19–21], up to 24% and 23% respectively, in the UK [21]. Exact epidemiological studies on these disorders are lacking in North Africa, as other vision problems, like refractive errors and cataract are the most common causes of visual impairment in these countries, as in many other regions of the world [22]. Inherited disorders affecting the retina and the optic nerve encompass two heterogeneous groups of diseases that lead to visual impairment, and in some cases, to legal blindness. They are caused by many genes, with autosomal, X-linked and mitochondrial patterns of inheritance. Large cohort studies of inherited retinal dystrophies (IRD) or optic neuropathies (ION) with the spectrum of causal genes and mutations are absent in North African individuals, except for two studies performed on Tunisian patients with IRD [23, 24], or some studies conducted on specific syndromes [25–28]. The majority of genetically characterized individuals are reported in a single family or in few families, or as a part of cohorts located in other countries, mainly in Europe. Recently, a review on the genetics of non-syndromic rod-cone dystrophies in Arab countries disclosed the spectrum of genes and mutations identified in 26 North African Arab families, highlighting the major contribution of variants in MERTK, RLBP1, RPE65 and PDE6B genes [29]. However, in order to access the phenotypic and genetic spectrum of all non-syndromic and syndromic IRD and ION within North African families, we performed a review of the data published in the literature using PubMed and Scopus databases. Different search terms were used (Additional file 1) in combination with the names of North African countries (Morocco OR Algeria OR Tunisia OR Libya OR Egypt OR North Africa OR Maghreb), or terms that refer to patients’ origin (Moroccan OR Algerian OR Tunisian OR Libyan OR Egyptian OR North African OR Maghrebian). Abstracts, full-length and additional materials of articles in English and French published between 1994 and December 2021 and their references’ listing, where both clinical and genetic findings are reported, were examined. Genes and mutations found in families from North Africa were incorporated and are discussed in this review.
Inherited retinal dystrophies (IRD)
IRD describe a large heterogeneous group of disorders characterized by the dysfunction of the neural retina or retinal pigment epithelium cells [30]. There are at least 280 genes involved in one or more IRD disorders [31]. Indeed, variants in the same gene may lead to different clinical outcomes between and within families. These highly heterogeneous genetic findings gave rise to different clinical manifestations that can be classified on their mode of inheritance, the course of the disease (progressive or stationary), the predominant phenotype (rod-dominant, cone-dominant or macular) and the occurrence of additional systemic symptoms (syndromic IRD) or exclusively eye related symptoms (non-syndromic IRD). More than 80 syndromic forms are described [32], among which, the predominant Usher syndrome, accounting for 20 to 40% of the recessive diseases affecting both the visual and hearing capacities [33].
Large cohort studies across the world on patients with non-syndromic IRD (NS-IRD) showed that retinitis pigmentosa is the most frequent phenotype, followed by Stargardt diseases, Leber congenital amaurosis, and cone-rod dystrophies/cone dystrophies, whereas phenotypes like choroideremia, achromatopsia and Best diseases remained at very low frequencies [30, 34–36]. ABCA4, USH2A, and EYS are the most frequently mutated IRD genes, while their mutation frequencies may vary according to the studied populations, their clinical presentations, the recruited cohorts, and their pattern of inheritance [30, 34–36].
Non-syndromic IRD in North African individuals
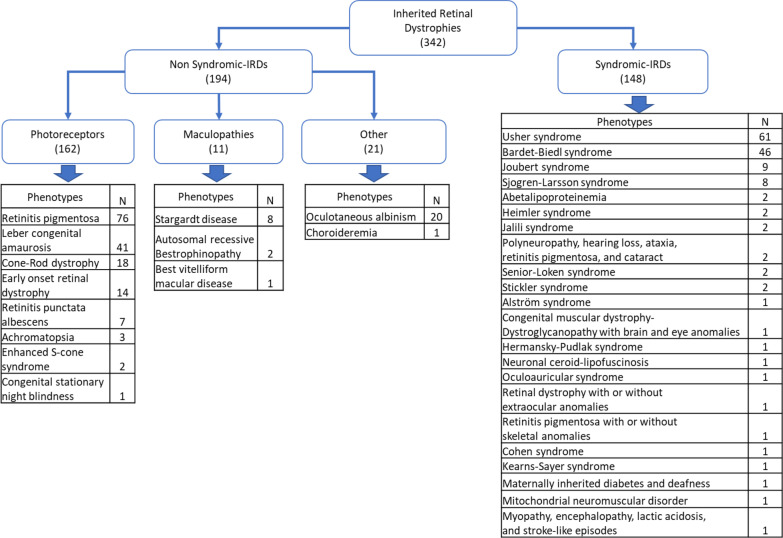
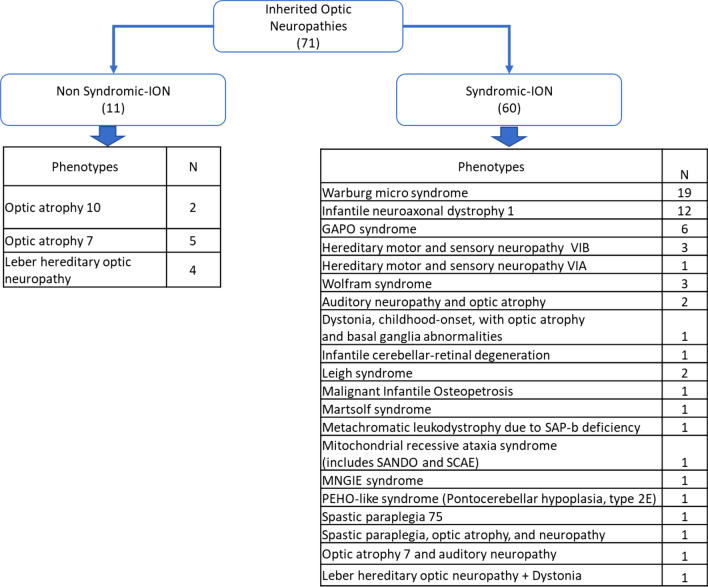
One hundred ninety-four families originating from North Africa with NS-IRD phenotypes have been characterized and reported in the literature, together with the causal genes and mutations. Their phenotypes can be classified into three categories; photoreceptors’ diseases, maculopathies and others (Fig. 1).
Fig. 1.
Classification of inherited retinal dystrophies (IRD) and number of affected families (N)
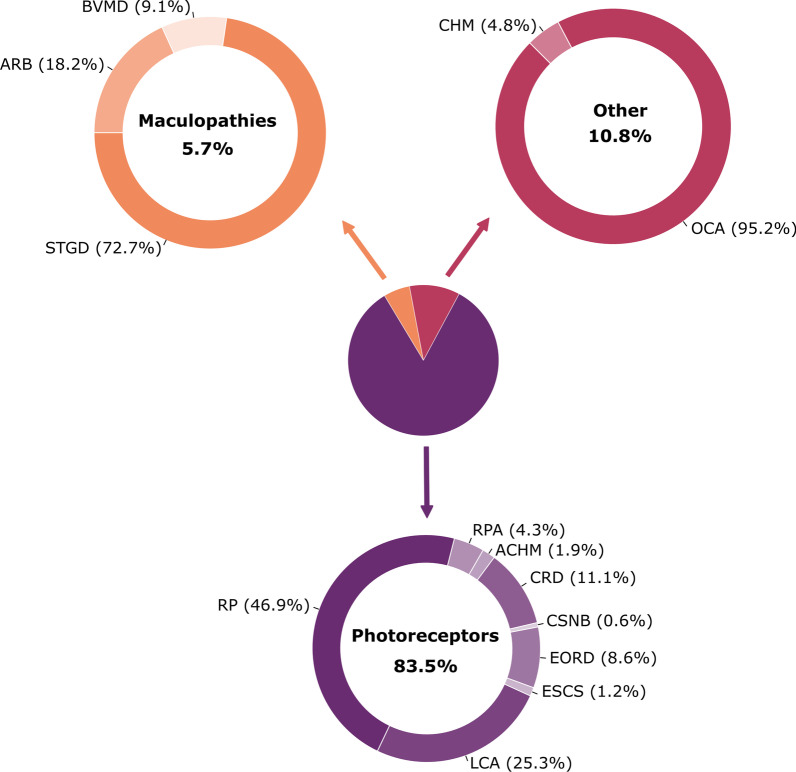
The first group characterized by photoreceptor dysfunctions is the most frequent (83.5%). It includes retinitis pigmentosa (46.9%), Leber congenital amaurosis (25.3%), cone and cone-rod dystrophies (11.1%), early onset retinal dystrophies (8.6%), retinitis punctata albescens (4.3%), achromatopsia (1.9%), enhanced S-cone syndrome (1.2%) and finally congenital stationary night blindness (0.6%). The second group, consisting in maculopathies, represents 5.7% of the reported families, Stargardt disease being the most frequent (72.7%), autosomal recessive bestrophinopathy and best vitelliform macular dystrophy representing 18.2% and 9.1%, respectively. The third group accounting for 10.8% includes two phenotypes: oculocutaneous albinism which has been described in twenty families and choroideremia in a single family (Fig. 2).
Fig. 2.
Distribution of the phenotypes of 194 North African families with non-syndromic IRD. According to disease categories in the center chart, and within each category in the outer charts. ACHM: achromatopsia, ARB: autosomal recessive bestrophinopathy, BVMD: Best vitelliform macular dystrophy, CHM: choroideremia, CRD: cone-rod dystrophy, CNSB: congenital stationary night blindness, EORD: early onset retinal dystrophy, ESCD: enhanced S-cone syndrome, LCA: Leber congenital amaurosis, OCA: oculocutaneous albinism, RP: retinitis pigmentosa, RPA: retinitis punctata albescens, STGD: Stargardt disease
Consanguinity has a major impact among North African families with NS-IRD. 71.4% of the families for whom the relation between parents has been investigated were consanguineous, reflecting the predominance of autosomal recessive (AR) pattern of inheritance. Indeed, 93.3% of these families disclosed mutations on both alleles, while autosomal dominant (AD) transmission has been identified in only three families, with variants in PRPH2, NR2E3 and PRPF31, respectively. An X-linked pattern was found in only one family with a CHM variant.
Retinitis pigmentosa (RP, OMIM: 268,000) is one of the most frequent IRD leading to legal blindness, with a prevalence estimated at 1:4.000 affected individuals worldwide [37]. RP is characterized by a progressive loss of photoreceptors, predominantly the rods associated to the scotopic system, rather than the cones associated to the photopic system. Night blindness or nyctalopia is the earliest symptom in RP patients, followed by a gradual loss of the peripheral visual field leading to a tunnel vision. In addition, pigmentary deposits in the peripheral retina, attenuation of retinal vessels and a waxy pallor of the optic disc represent RP hallmarks [38, 39].
With 76 families, RP is the most common IRD reported in patients originating from North Africa, among which consanguinity rate reaches 63.9%. Almost all RP families (96.1%) present an AR pattern of inheritance, with only three families presenting an AD pattern, and no mutation in an X-linked gene was yet reported.
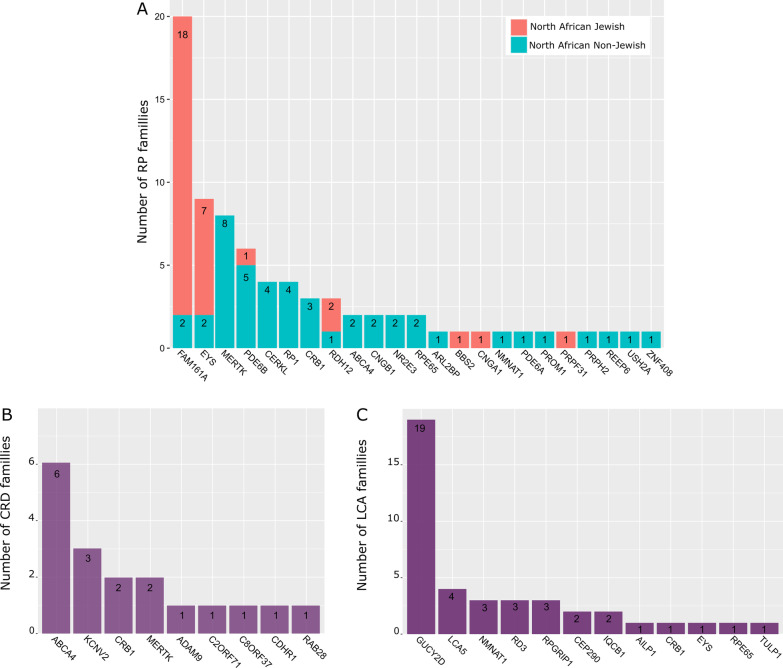
RP families can be divided in two groups, with different ethnic backgrounds. The first group corresponds to non-Jewish families, characterized by a high genetic heterogeneity, in which variants in twenty different genes were identified in 45 families (Fig. 3a). Among the 20 genes, 5 of them are responsible for 53.3% of the observed phenotypes. MERTK is the most frequent gene mutated within 8/45 families, followed by PDE6B with 5/45 families and CERKL and RP1 with 4/45 families, respectively. CRB1 variants have been identified in 3 families, while the other genes were reported only once or twice. A single MERTK splice site mutation, c.2189 + 1G > T, is responsible for 4 out of the 8 families reported (Additional file 2), suggesting a founder effect in the North African population [40]. A second frequent mutation c.1133 + 3_1133 + 6delAAGT deletion in CERKL seems also to result from a founder effect, as it was reported in 4 unrelated Tunisian families from the same geographical area, sharing a 5.7 cM homozygous region (Additional file 2) [23]. The second group corresponds to a Jewish ethnical background, with 31 reported families originating from North Africa, and only seven genes affected (Fig. 3a), among which FAM161A and EYS are mutated in 80.6% of cases. Two founder variants in this community were predominant, the FAM161A p.Thr452SerfsTer3 found at homozygous state in 14 out of the 18 families, and the EYS p.Thr135LeufsTer7 found at homozygous state in 5 of the 7 families (Additional file 2). Both founder variants were not identified in other ethnical groups [41, 42].
Fig. 3.
Distribution of the number of families with non-syndromic IRD per causal genes. A Families with retinitis pigmentosa. B Families with cone-rod dystrophy. C Families with Leber congenital amaurosis
Overall, the relative contributions of RP genes in other studies including populations with different ethnical backgrounds [30, 34, 35, 39, 43], disclosed that USH2A variants are very often encountered in the world, whereas only one family form Tunisia disclosed heterozygous compound USH2A mutations, indicating that USH2A variants are not frequently encountered in RP from North Africa.
Retinitis punctata albescens (RPA, OMIM:136,880): is a progressive hereditary disease considered to be a subtype of RP, since both presentations share various common ophthalmological symptoms, among them the predominant alteration of rod photoreceptors and night blindness, occurring in early childhood. In addition, absence or rarity of pigmentary deposits in the peripheral retina, moderate narrowing of retinal vessels and frequent macular atrophy are characterizing RPA [44, 45].
A single mutation, corresponding to a 7.36-kb deletion of three exons [7, 8 and 9], within RLBP1 was found homozygous in 7 families, 6 of them from Morocco and one from Algeria (Additional file 2). Interestingly, a clinical heterogeneity was found in the last family, with a phenotype in between cone-rod dystrophy and RP. This RLBP1 mutation was found only in individuals from a close geographical origin, and most probably results from a founder effect [45].
Cone dystrophy (CD)/Cone-Rod dystrophy (CRD) (OMIM:120,970) describes a heterogeneous group of IRD, characterized by a predominant progressive loss of cone photoreceptors and pigmentary deposits, frequently encountered in the macular area [46]. The prevalence has been estimated between 1:30.000 and 1:40.000 [47]. The earliest symptoms are a decreased of visual acuity and photophobia, a central vision loss marked by a central scotoma in the visual field, and color vision defects occurring in childhood or early adulthood [48]. Additional abnormalities may occur later in most cases, including alterations of rod photoreceptors in CRD and nyctalopia [46].
Eighteen families with a CRD phenotype have been reported with the causal gene in North Africa, 11 with a documented consanguinity (Additional file 2). Fifteen homozygous mutations, and two heterozygous compound mutations in 9 different genes were found. ABCA4 and KCNV2 were affected in 9 families, which represent 50% of North African CRD families (Fig. 3b), in accordance with other studies reporting that the majority of autosomal recessive CD/CRD are related to these two genes [48].
Leber congenital amaurosis (LCA, OMIM:204,000) is the earliest and most severe retinal dystrophy with a prevalence of 1:80.000, manifesting at birth or during the first 2 years of life [49]. The main clinical LCA features are early blindness or severe visual loss, undetectable ERG responses, nystagmus and amaurotic pupils [50, 51]. LCA patients may have additional clinical features like keratoconus, hypermetropia and cataract [52].
Forty-one families were identified with a LCA phenotype, 36 having a homozygous variant and 5 heterozygous compound variants (Additional file 2). Thirty-one different variants in 12 genes were reported: GUCY2D mutations were found in 19 families and represents 46.3% of all cases, followed by LCA5 in 4 families, RPGRIP1, RD3 and NMNAT1 in 3 families, CEP290 and IQCB1 in 2 families, and AILP1, EYS, RPE65, CRB1 and TULP1, each in a single family (Fig. 3c).
GUCY2D variants on chromosome 17p13.1 were first identified in LCA individuals from 8 North African consanguineous families [53, 54]. Interestingly, studies conducted on a large French LCA cohort disclosed that the majority of LCA families harboring GUYCY2D mutations are originating from North Africa [55, 56]. In this respect, the c.389delC, p.Pro130LeufsTer36 mutation identified in North Africa, is responsible for 42.1% of cases with a GUCY2D mutation and 19.5% of LCA families. This variant was homozygous in 7 consanguineous families and related to a founder effect (Additional file 2), most probably originating from a Jewish ancestry [56]. Another GUCY2D variant, the p.Phe565Ser, was found homozygous in three Algerian families, in addition to a family of Moroccan and Belgian mixed origins (Additional file 2).
Early onset retinal dystrophy (EORD), together with LCA represent the most severe form of retinal dystrophies. It is also considered as a moderate and/or delayed form of LCA, constituting a single clinical entity with very subtle differences [57]. Indeed, LCA patients develop symptoms in the first 3 months of life until 1 year of age, with unrecordable or severely diminished ERG responses [58], while EORD patients disclose similar symptoms, but appearing around 5 years of age [59].
Fourteen families were reported with an EORD phenotype, 13 being consanguineous with homozygous variants (Additional file 2). RPE65 is the most frequently affected gene, representing 50% of EORD families. The predominant c.271C > T (p.Arg91Trp) missense variant was found in 6 families, and is widely described in several studies in individuals from different ethnical backgrounds [60–63]. RDH12 variants come in the second place with 35.7% of all EORD families with two mutations: the p.Arg65Ter found in 2 Tunisian families from the same region and the p.Leu99Ile found in 3 Jewish North African families. The last one is known for its founder effect, and was already described among Caucasian patients [64, 65].
Stargardt disease (STGD, OMIM:248,200) is the most frequent macular dystrophy characterized by central visual loss in childhood, with a prevalence ranging from 1:8.000 to 1:10.000 [66]. Clinical features include beaten-bronze appearance or bull’s eye maculopathy, progressive atrophy of the macula and yellowish flecks in the posterior pole of the retina [67]. Ninety-five % of STGD cases are inherited as a recessive trait and are caused by ABCA4 variants. The remaining 5% cases are caused by dominant variants in ELOVL4 and PROM1, and are responsible for STGD3 (OMIM:600,110) and STGD4 (OMIM:603,786), respectively [66, 68].
Eight Tunisian recessive families were characterized with STGD phenotype, and consanguinity was reported among 7 of them, with homozygous ABCA4 variants. 62.5% of cases are explained by two variants, the p.Arg681Ter mutation found in three families, twice in a homozygous state and once in compound heterozygous state, and the p.Glu1087Lys mutation which was found homozygous in 2 families (Additional file 2).
Two additional maculopathies were characterized within 3 consanguineous families with homozygous BEST1 variants, 2 families with autosomal recessive Bestrophinopathy (OMIM:611,809) and one with Best vitelliform macular dystrophy (BVMD) (OMIM:153,700) (Additional file 2). BVMD is the second cause of maculopathies after STGD [69], and is often inherited in an AD mode, although the AR mode was also reported [70, 71].
Other NS-IRD have been also reported, and are mainly congenital stationary IRD. Achromatopsia (OMIM:216,900), also called rod monochromatism, characterized by dysfunction of the cone photoreceptors was observed in 3 consanguineous families displaying mutations in CNGA3, CNGB3 and GNAT2, respectively (Additional file 2). The Enhanced S-cone syndrome (OMIM:268,100), in which biallelic variants in NR2E3, and more rarely in NRL lead to increased number of S-type cone photoreceptors and degeneration of L/M-type cones and rod photoreceptors. This phenotype disclosed by electroretinography measurements [72], was described in 2 families with homozygous and heterozygous compound NRL variants (Additional file 2). Congenital stationary night blindness (OMIM:613,216) and Choroideremia (OMIM:303,100) were also described in 2 consanguineous families, with a homozygous TRPM variant and a hemizygote CHM variant, respectively (Additional file 2). Oculocutaneous albinism (OMIM:203,100) is also frequent, in particular among Jewish of Moroccan origin. Twenty families were reported, one with SLC45A2 and 19 with TYR variants, among which 9 with the p.Gly47Asp variant in a homozygous state and 4 in a heterozygous state (Additional file 2).
Syndromic IRD in North African population
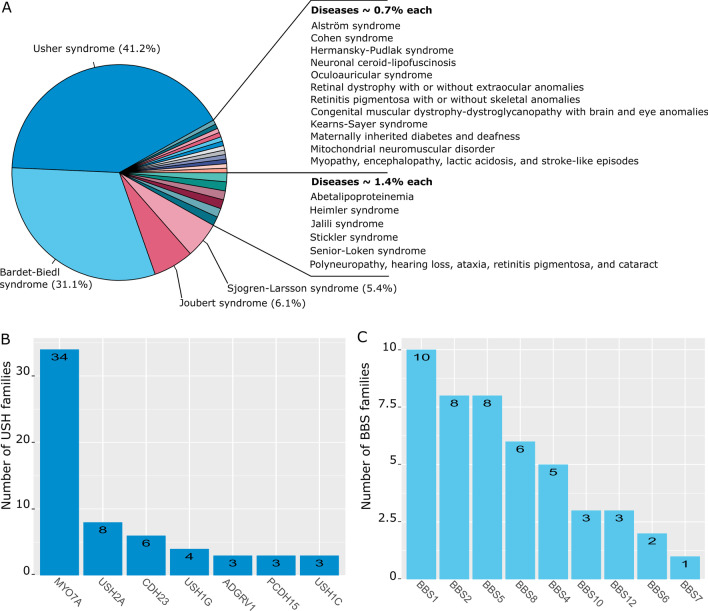
At least, 24 different disorders associated to syndromic inherited retinal dystrophies (S-IRD) were reported and genetically characterized in individuals originating from North Africa. These disorders consist in ciliopathies, in which the Usher and Bardet Biedl syndromes occupy 41.2% and 31.1% of all cases, respectively (Fig. 4a). In addition, diseases related to inborn errors of metabolism, mitochondrial defects related to mitochondrial DNA variants, and associated to cerebellar ataxia were reported.
Fig. 4.
Genetic and phenotypic spectrum of North African families with syndromic IRD. A Distribution of the phenotypes of 143 families with syndromic IRD. B distribution of families with Usher syndrome phenotype per causal gene. C Distribution of families with Bardet-Biedl syndrome phenotype per causal gene
Usher syndrome (USH) is the most common syndromic IRD that associates RP to hearing impairment. It accounts for 10–20% of all RP cases and up to 50% of deaf-blind patients [37, 73]. Three clinical subtypes (USH1-3) are described based on the age of onset, degree of hearing loss (profound, severe or moderate), presence or absence of vestibular dysfunction and age of RP onset. USH1 (OMIM:276,900) is the most severe form, characterized by a profound congenital hearing loss, vestibular dysfunction and prepubescent RP onset. USH2 (OMIM:276,901) consists in a congenital and less severe deafness, without vestibular dysfunction, and a postpubescent RP onset. USH3 (OMIM:276,902), is characterized by a progressive and mild deafness, variable presentation of vestibular dysfunction and variable age of RP onset [74, 75]. Variants in USH genes are characterized by high clinical heterogeneity that can cause either USH, non-syndromic RP and non-syndromic deafness. Many studies including North African individuals with non-syndromic deafness have evidenced USH variants, but only few associated to RP [76–82].
In North Africa, 61 families have been clinically and genetically characterized with Usher syndrome. This number might be underestimated because individuals have not systematically access to both ophthalmological and audiological examinations to confirm the USH diagnosis. The consanguinity rate within USH families is 77.3%, and 96.7% of them have variants identified on both alleles. Distribution of USH phenotypes is predominated by USH1 (82%). MYO7A is the most prevalent gene, with variants identified in 34 families, followed by USH2A in 8 families, CDH23 in 6 families, USH1G in 4 families, ADGRV1, PCDH15 and USH1C in 3 families each (Fig. 4b).
Five variants were found routinely, four in MYO7A and one in USH2A genes (Additional file 3), with different occurrences among non-Jewish and Jewish families. Two MYO7A splice site mutations are involved in 10 non-Jewish families: one, the c.2283-1G > T mutation was found homozygous in 4 families and heterozygous in 2 families, and is present in Moroccan, Algerian and Tunisian individuals [26, 82–84]. The second MYO7A variant, c.470 + 1G > A was found homozygous in 4 families of Algerian and Tunisian origins [26, 81, 85, 86]. Together, these 2 common variants might reflect founder effects in close geographic areas.
Among Jewish families, the p.Ala826Thr MYO7A variant was described in 8 families of Moroccan and Algerian origins, while a large MYO7A deletion p.Gln2119_Lys2215del was observed in 3 Tunisian families [85, 87, 88]. In addition, the p.Arg334Trp USH2A variant was found in several Moroccan and Tunisian Jewish families with a possible founder effect [87, 89, 90].
Bardet-Biedl syndrome (BBS, OMIM:209,900), is a multisystemic disorder characterized by symptoms in several organs that represents 5–6% of syndromic RP cases [37]. The wide heterogeneity of symptoms is divided into 2 categories based on clinical features and frequencies, encountered in patients. Primary features present at high frequencies are RP, polydactyly, obesity, genital and renal anomalies and learning difficulties. Secondary less frequent features are speech and/or development delay, diabetes mellitus, dental anomalies, congenital heart diseases and additional peculiar symptoms [91, 92]. The combination of 3 to 4 primary features associated to 2 secondary features are required to establish a BBS clinical diagnosis [91].
Forty-six families originating from North Africa were reported, among which 28 in the Tunisian population (Additional file 3). Homozygous variants were found in the majority (93,5%) of these families. Thirty-four variants in 9 genes were reported; BBS1 in 10 families, BBS2 and BBS5 in 8 families each, BBS8 in 6 families, BBS4 in 5 families, BBS10 and BBS12 in 3 families each, while variants in BBS6 and BBS7 were only associated twice and once to BBS phenotypes, respectively (Fig. 4c). BBS1 is the most frequently mutated gene, accounting for 23.2–33.6% of all BBS families, according to different studies performed across the world in multiethnic cohorts [93–95]. In contrast, BBS2, BBS5 and BBS8 appear more frequently mutated in the North African population than in the rest of the world [94–97].
Two recurrent variants are found frequently in Caucasian BBS families, the p.Met390Arg contributing to 73.3–82.6% of BBS1 families [93, 95, 98], and the p.Cys91fsX95 contributing to 48.3% to 88.8% of BBS10 families [95, 96]. The first was found only once in a Tunisian family in a homozygous state, while the second one was never reported in North Africa. Conversely, some BBS variants were frequent in Tunisian families with a common haplotype [28], the c.459 + 1G > A in BBS8 and the p.Arg189Ter in BBS2, the latter also encountered in Algeria, witnessing a migration between these 2 countries. In addition, the c.1473 + 4A > G and c.448C > T in BBS1, and ex4-5-6del in BBS4 variants were evidenced in Algerian and Tunisian families, whereas the c.149T > G and c.123delA in BBS5 variants also frequent, were restricted to Tunisia (Additional file 3).
Senior-Loken syndrome (SLS, OMIM:266,900) and Joubert syndrome (JBTS, OMIM:213,300) are 2 inherited nephronophthisis related ciliopathies, representing a heterogeneous group of disorders characterized by autosomal recessive cystic kidney and renal anomalies. SLS with a prevalence of 1:1.000.000 is defined by the combination of retinal degeneration ranging from RP to LCA associated to nephronophthisis [99]. JBTS is defined by primary features including hypotonia/ataxia, developmental delay, oculomotor apraxia, mental retardation and breathing anomalies [100], and often associated to retinal degeneration [101–103].
Only two SLS families with mutations in NPHP4 and IQCB1 have yet been reported. Conversely, individuals with JBTS phenotype originating from North Africa, especially from Egypt, were well characterized in several studies [104–109], although few of them had a retinal dystrophy. Pathological variants (Additional file 3) responsible for JBTS associated with retinal dystrophies involve AHI1 in 4 families, followed by CEP290 and INPP5E in 2 families each, and NPHP1 in one family. CEP290 is the most frequent JBTS causative gene, but variants in AHI1 and INPP5E are the most frequently associated to retinal dystrophies [102–104, 108]. Finally, only 10% of variants in NPHP1, were reported to cause RP [101]. In addition, Alstom and Cohen syndromes, 2 other ciliopathies have been reported, but only in one family each, harboring mutations in ALMS1 and VPS13B, respectively.
Sjogren-Larsson syndrome (SJLS, OMIM:270,200) is caused by mutations in ALDH3A2, encoding the fatty aldehyde dehydrogenase (FALDH) enzyme, implicated in the metabolism of fatty alcohol [110]. SJLS consists in 3 main symptoms, ichthyosis, mental retardation and spastic diplegia [110], although, additional abnormalities including retinal manifestations are widely described [25, 111–113]. A single study conducted in 25 Egyptian SJLS families has evidenced 8 families with retinal abnormalities consisting in yellowish retinal dots [25]. Three recurrent variants, p.Ser365Leu, p.Arg9ter and p.Gly400Arg contributes for 25%, 12% and 12% of all cases, respectively. Patients harboring the latter 2 variants shared a common haplotype, supporting a founder effect. Interestingly, families harboring the same variant exhibited different retinal manifestations, most probably related to a genetic modifier effect. Additional North African families without retinal defect were further reported [114–118].
Abetalipoproteinemia (ABL, OMIM:200,100) is a systemic disease caused by errors in lipid metabolism that manifests by the absence of apolipoproteins b, very low density of lipoproteins and low density lipoproteins in the plasma [119], in addition to diarrhea caused by fat mal absorption, acanthocytosis and retinal dystrophy [112, 119]. Five ABL families of Tunisian origin were reported, 2 of them presenting retinal degeneration and homozygous MTTP variants (Additional file 3).
Heimler syndrome (HS) is an autosomal recessive peroxisomal biogenesis disorder (PBD) characterized by sensorineural deafness, enamel hypoplasia, nail abnormalities and occasionally retinal pigmentation [120]. PBD represent a group of disorders characterized by anomalies in peroxisome assembly and/or biochemical functions. Peroxisomes are including over 70 enzymes that ensure several metabolic pathways critical for normal cell functioning [121]. HS, the mildest form of the PBD spectrum, is caused by hypomorphic PEX1 variants defining the Heimler syndrome type 1 (OMIM:234,580) or by PEX6 variants defining the Heimler syndrome type 2 (OMIM:616,617) [120]. Variants in both genes have been identified in 2 consanguineous families from Moroccan and Egyptian origins, respectively (Additional file 3).
Familial isolated vitamin E deficiency (AVED, OMIM:277,460) is an autosomal recessive cerebellar ataxia characterized by low vitamin E level in the serum. Clinical symptoms are resembling those of Friedreich ataxia (FA) with dysarthria, progressive limb and gait ataxia, absent tendon reflexes and skeletal abnormalities [122]. Absence of diabetes and cardiomyopathy are the main characteristics that differentiate AVED from FA [123]. Since the first description by Burk et al. [124], several North African families were described, allowing the identification of the causative locus at 8q and later of variants in the TTPA gene, as responsible for the phenotypes [125–128]. A specific TTPA frameshift corresponding to an adenine deletion at position 744 (c.744delA) was found in the majority of AVED patients from Mediterranean countries, which correlates with an ancient founder effect [122, 126, 128–134]. Association between AVED and RP has been first described in patients with the p.His101Gln variant causing a less severe and a latter onset presentation [135, 136]. Further studies of North African cohorts revealed that individuals with the c.744delA variant may also present RP, with an incidence varying from 0% in Algeria, 4.4% in Tunisia to 27% in Morocco [122, 131–133]. Other variants have been also reported in AVED North African individuals, but no retinal abnormality was associated [122, 137].
Spinocerebellar ataxia type 7 (SCA7, OMIM:164,500) refers to an entity of progressive cerebellar ataxia associated with a cone-rod macular dystrophy, and also known as autosomal dominant cerebellar ataxia type II. SCA7 is caused by a CAG triplet expansion in ATXN7 at the 3p14.1 locus. The normal allele contains 7 to 17 repeats, and up to 35 repeats in asymptomatic patients, whereas symptomatic patients presents 36 or more CAG triplets [138–141]. SCA7 has been described in several North African families, with extended CAG repeats [139, 142–144].
Other occasional syndromic IRD were identified in 9 consanguineous families with 7 different syndromes, 2 families with Stickler syndrome (OMIM:614,134) harboring a single COL9A1 variant, 2 families with Jalili syndrome (OMIM:217,080) with 2 different CNNM4 variants, one family with congenital muscular dystrophy-dystroglycanopathy with brain and eye anomalies (OMIM:616,538), Hermansky-Pudlak syndrome (OMIM:614,072), oculoauricular syndrome (OMIM:612,109), retinal dystrophy with or without extraocular anomalies (OMIM:617,175), retinitis pigmentosa with or without skeletal anomalies (OMIM:250,410), harboring variants in DAG1, HPS3, HMX1, RCBTB1 and CWC27, respectively (Additional file 3).
Finally, retinal alterations like pigmentary retinopathy and maculopathy can be further associated to disorders related to mitochondrial genome mutations. Pigmentary retinopathy is frequently associated to the Kearns-Sayer syndrome (KSS), the neuropathy, ataxia and retinitis pigmentosa (NARP), the mitochondrial encephalomyopathy, lactic acidosis, and stroke like episodes (MELAS) and the maternally inherited deafness and diabetes (MIDD) [145, 146]. Studies performed on North African individuals affected by mitochondrial diseases and retinal manifestations are rare, and only 4 families were reported with KSS, MIDD, MELAS and a mitochondrial neuromuscular disorder, with mtDNA specific mutations or large deletions (Additional file 6).
Inherited optic neuropathies
Inherited optic neuropathies (ION) define a group of diseases characterized by the dysfunction of the optic nerve as a result of the loss of retinal ganglion cells and their axons [147]. Familial expression of the disease can help its diagnosis, although genetic analysis can also reveal the etiology of the disease, even in absence of familial history. Typically, ION manifest as bilateral symmetric central visual loss with a central scotoma, resulting from the injury of the papillomacular nerve fibers. In this context, its always crucial to rule out IRD that can interfere with primary IONs, especially the CRD affecting the central visual field, in which a pallor of the optic nerve head can also be found [148]. Optic atrophy can be the only manifestation of the IONs in non-syndromic ION (NS-ION), or associated to additional various symptoms, defining syndromic ION (S-ION) [149, 150]. All patterns of inheritance were observed in ION: autosomal dominant or recessive, X-linked and maternally transmitted by the mitochondrial genome. In general NS-ION are less heterogeneous compared to IRD, as only 12 loci have been mapped, 6 being dominant, 5 recessive and 1 X-linked, with 4 genes still to be identified.
Leber hereditary optic neuropathy (LHON;OMIM:535,000) and dominant optic atrophy (DOA;OMIM:165,500) are the 2 most common optic neuropathies worldwide, with a prevalence ranging around 1/15.000 to 1/50.000 for each, depending on the geographic area [147, 149, 151, 152]. LHON was first described by Leber [153], with a maternal mode of inheritance associated to a mitochondrial DNA (mtDNA) variant [154], then to many additional variants of the mtDNA [155]. DOA or Kjer’s optic atrophy was described first in 1959 [156], as an infantile optic atrophy with autosomal dominant mode of inheritance, characterized by a progressive loss of visual acuity, tritanopia, a pallor of the nerve optic heads, mainly in the temporal area, and a centrocoecal scotoma [147, 150, 157]. Genetic studies associated DOA to chromosome 3q28 [158], then to OPA1, encoding a mitochondrial dynamin-like GTPase essential for fusion of the mitochondrial network, the production of ATP and the control of apoptosis [157, 159–161]. OPA1 is mutated in more than 70% of DOA individuals [162], followed by WFS1 and ACO2 genes. Recessive loci were also described, as OPA6, OPA7, OPA9, OPA10 and OPA11 [163–167].
Interestingly, all DOA genes discovered so far, encode proteins involved in mitochondrial physiology, with functions involved in their biogenesis, network structure, calcium homeostasis and respiration [168–171], thus emphasizing the frailty of the optic nerve to mitochondrial dysfunctions.
In the following section, we present the phenotypic and the genotypic spectrums associated with ION in 71 North African families (Figs. 5 and 6).
Fig. 5.
Classification of inherited optic neuropathies (ION) and number of affected families (N)
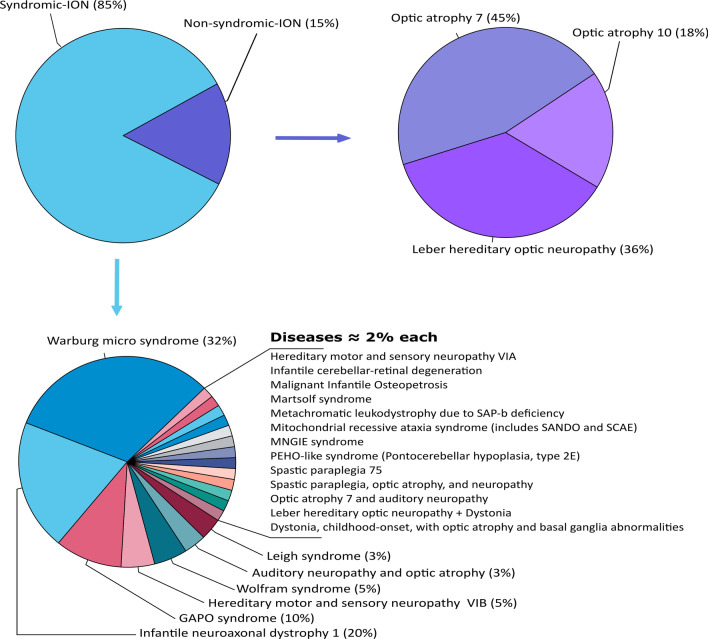
Fig. 6.
Distribution of the phenotypes of the 71 North African families with ION. According to the disease presentation (syndromic or non-syndromic) in the center chart, and within each category in the outer charts
Non syndromic inherited optic neuropathies in North African families
Leber hereditary optic neuropathy (LHON, OMIM:535,000): is the first disease which was associated to a mtDNA mutation, and now the most frequent among mitochondrial diseases [154, 172]. LHON is a non-syndromic optic neuropathy affecting predominantly men, characterized by a subacute, rapid and painless central visual loss in one eye, then of the fellow eye within weeks or months in the vast majority of patients. Ophthalmological examination discloses a centrocaecal scotoma in the visual field, temporal pallor of the optic nerve head and frequent dyschromatopsia [172–174]. 90–95% of patients with LHON phenotype harbor one of the 3 common mutations m.11778G > A, m.14484T > C, and m.3460G > A affecting the ND4, ND6 and ND1 genes, respectively [154, 174–176]. Only, the first 2 mutations were identified in individuals from 4 families originating from North Africa, 3 with the m.11778G > A and one with the m.14484T > C variant [177–179] (Additional file 6).
Other clinical symptoms, such as ataxia, dystonia, and encephalopathy can be associated with LHON, namely LHON plus phenotype [180], which was reported associated to the m.G14459A variant found in an Algerian family presenting LHON associated to dystonia [181, 182].
Other reported NS-ION cases are related to nuclear DNA variants (Additional file 4). Although the majority of ION individuals reported worldwide present an AD pattern of inheritance, with OPA1 as the most frequently mutated gene, no ION with a dominant transmission has yet been reported in North Africa. Indeed, 8 consanguineous families with an AR pattern of inheritance were reported with variants in TMEM126A and RTN4IP1 genes. TMEM126A was the first gene identified in a large consanguineous Algerian family, harboring a homozygous nonsense mutation p.Arg55Ter. The same mutation was found in two additional Moroccan and one Tunisian families [164]. Genotype analyses revealed a common haplotype within these families, witnessing a founder effect that most probably appeared 80 generations ago. The same TMEM126A nonsense variant was further identified in an Algerian consanguineous family with optic atrophy and auditory neuropathy [183] and a Moroccan consanguineous family with isolated optic atrophy [184]. Two additional families presented RTN4IP1 homozygous variants: the p.Arg103His and the p.Ile362Phe, respectively [163, 185]. Although RTN4IP1 variants can cause very severe syndromic diseases, North African families presented only a NS-ION phenotype.
Syndromic inherited optic neuropathies in North African families
Wolfram syndrome (WFS, OMIM:222,300) describes a syndromic neurodegenerative disease characterized by optic atrophy, diabetes mellitus, diabetes insipidus and deafness, also called (DIDMOAD) [186]. Additional neurological, endocrinological and other peculiar symptoms can be associated [186, 187]. Variants in 2 nuclear genes WFS1 and CISD2, in addition to heteroplasmic deletions of mtDNA were reported to cause this phenotype [188–191]. Three North African families were reported so far with a WFS phenotype, 2 of them are consanguineous and carry a homozygous mutation in WFS1 and CISD2 (Additional file 5). The third family, which presents WFS associated to a cardiomyopathy, was reported with multiple mtDNA deletions in addition to the homoplasmic m.3337G > A mitochondrial variant (Additional file 6). Additional WFS families were also described, but without molecular diagnosis [192–194].
Leigh syndrome (LS, OMIM:256,000): is a syndromic neurodegenerative disease characterized by bilateral necrotic lesions in brainstem, basal ganglia, thalamus, cerebellum and spinal cord [195]. LS is a very heterogeneous disease, with more than 35 nuclear and mitochondrial genes involved, thus inherited as AR and maternal mitochondrial or X-linked patterns [196]. Most LS genes are implicated in mitochondrial energy production, encoding proteins involved in the formation of the respiratory chain complexes or their assembly. Ophthalmological abnormalities are frequently described in LS (79% to 82%), including refraction errors, ptosis, strabismus, nystagmus, optic atrophy and pigmentary retinopathy. However the last two abnormalities occur only in 17 to 22.5% and 15 to 22.5% of all individuals, respectively [197, 198]. Several studies described North African LS individuals [195, 199–202], but only 2 families had optic atrophy with either a SURF1 variant c.516_517delAG (Additional file 5), or a heteroplasmic variant m.9478T > C in the mitochondrial genome (Additional file 6).
Hereditary motor and sensory neuropathy type VI (OMIM:601,152) is a neurological disorder characterized by distal neuropathy and optic neuropathy [203]. Mutations in 2 genes, MFN2 and SLC25A46, were reported with an AD and AR inheritance pattern. They encode outer mitochondrial membrane proteins acting on mitochondrial dynamics [204, 205]. Four North African families were reported so far, 3 consanguineous families with SLC25A46 homozygous variants and one with a dominant MFN2 variant (Additional file 5).
Five additional families with 4 different phenotypes were reported with variants in genes encoding proteins that ensure mitochondrial physiology (Additional file 5). They affect FDXR, which encodes a ferredoxin reductase implicated in Fe-S cluster formation, [206, 207], MECR, which encodes a mitochondrial trans-2-enoyl-Coa reductase enzyme essential for fatty acid synthesis and respiratory competence [208], POLG, which encodes the mitochondrial polymerase gamma ensuring mtDNA replication and repair [209], and ACO2, encoding the mitochondrial aconitase converting citrate to isocitrate in the Krebs cycle [166].
GAPO syndrome (OMIM:230,740) is a AR disorder characterized by growth retardation, alopecia, pseudo-anodontia and frequent optic atrophy, caused by ANTXR1 variants [210]. Six consanguineous Egyptian GAPO families were reported in addition to one family without optic atrophy [211], all presenting homozygous ANTXR1 variants (Additional file 5).
Infantile neuroaxonal dystrophy (INAD, OMIM:256,600) is an early onset AR neurodegenerative disorder appearing in the first 2 years of age, characterized by axial hypotonia, psychomotor regression, ataxia, in addition to several ophthalmological features like, nystagmus, strabismus and a frequent optic atrophy [212, 213]. Twelve North African INAD families with optic atrophy have been reported so far with PLA2G6 variants, of which 8 were homozygous and 4 compound heterozygous (Additional file 5). A particular mutation p.Val691del, previously reported in other Mediterranean families, was found in 5 Tunisian and one Libyan families, who shared a common haplotype, witnessing a founder effect that occurred 12 generations ago [214].
Warburg Micro syndrome (OMIM:600,118) is an autosomal recessive disorder combining neurological symptoms as microcephaly, mental retardation, hypotonia, and eye abnormalities with severe visual impairment with microphthalmia, microcornea, cataract and frequent optic atrophy, in addition to hypogenitalism [215]. More than 32 North African families, mainly from Egypt were reported by several studies [216–219]. Among them, optic atrophy was clearly established in 19 families, with 79% of these families presenting RAB3GAP1 variants, 2 families with RAB3GAP2 variants, and 2 others with RAB18 and TBC1D20 variants, respectively (Additional file 5).
Discussion
The data presented in this review provide an extensive overview of IRD and ION phenotypic and genotypic spectrums reported in the literature, regarding 413 North African families. The respective contributions of these 2 groups of diseases show a high predominance of IRD (82.8%) compared to ION (17.2%); fitting with earlier studies from Western countries, suggesting that the origins of blindness are more frequently associated to retinal dysfunctions than to optic nerve degeneration [220].
Nevertheless, by contrast with Europe, North African countries show high rate of consanguinity, as marriage practices are maintained by religion, cultural and socio-economic conditions, in addition to their geographical isolation delimited by the Mediterranean Sea in the North and the Sahara in the South. In this respect, first cousin mating is the most frequent consanguineous marriage in North Africa [13], and progeny of individuals who shared common ancestors are the most likely to be autozygous for recessive variants. This is the case for 71.4% of NS-IRD families for whom investigations of the relationships between parents disclosed a consanguinity, with 82.5% presenting homozygous mutations in recessive genes. For S-IRD, 84.4% were consanguineous, among which 88.2% presented homozygous variants. The same observations were found in ION families, with 84.3% presenting homozygous mutations in nuclear genes.
This high level of consanguinity has further drastic consequences on small endogamous communities, alike the North African Jewish ones, leading to the identification of founder effects associated to variants unusually present at high frequencies in specific genes. For example, although FAM161A is not a major cause of RP [39], 18 out of 31 North African Jewish families present a mutation in this gene, among which 14 disclosed with the same homozygous mutation c.1355_1356delCA, 13 living in Morocco. Similarly, 5 out of the 7 Jewish Moroccan RP families presented the same homozygous variant in EYS. These peculiarities were also found in non-Jewish North African families, with 4 out of 8 RP families related to the homozygous c.2189 + 1G > T MERTK mutation; and virtually all RP families with CERKL variants presented the unique c.1133 + 3_1133 + 6delAAGT mutation at a homozygous state, all being of Tunisian ancestry. Similarly, all families reported with RPA were related to the same homozygous 7.36 kb deletion in RLBP1. This was also true for all autosomal NS-ION families related to the OPA7 locus, which presented the same TMEM126A recessive variant, and for those related to the OPA10 locus, presenting the same RTN4IP1 recessive variants, both being now encountered respectively in North African and Gipsy families living in Europe, respectively [163, 164, 183–185].
Other biases associated to the clinical spectrum of North African IRD and ION consist in the significant differences in the time course and severity of the visual impairments. Indeed, while S-IRD and S-ION are easily early characterized by severe visual impairment associated to other symptoms, diseases with milder effects on the visual acuity, often occurring later in the life-time, like some NS-IRD and most NS-ION, might be far less clinically diagnosed, as patients can be unaware of their visual difficulties.
This might explain why the proportion of each IRD among the North African population that we report here, is so different compared to what was observed in Western countries. Indeed, RP and STGD diseases, which are the less severe presentations are under-represented, whereas LCA, USH and BBS diseases, which have the worse visual prognosis are significantly over-represented (Table 1).
Table 1.
Frequencies of IRD subtypes across different countries
| Diseases | USA | UK | France | North Africa |
|---|---|---|---|---|
| Occurrence and frequencies | ||||
| RP | 341 (34.1%) | 311 (43.1%) | 922 (47.1%) | 76 (22.2%) |
| STGD | 189 (18.9%) | 45 (6.2%) | 118 (6%) | 8 (2.3%) |
| USH | 81 (8.1%) | 37 (5.1%) | 207 (10.6%) | 61 (17.8%) |
| CD/CRD | 45 (4.5%) | 74 (10.2%) | 140 (7.2%) | 18 (5.3%) |
| Other macular dystrophies | 93 (9.3%) | 37 (5.1%) | 231 (11.8%) | 3 (0.9%) |
| LCA | 14 (1.4%) | 18 (2.5%) | 52 (2.7%) | 41 (12%) |
| BBS | 25 (2.5%) | 7 (1%) | 23 (1.2%) | 46 (13.5%) |
USA data are from Stone et al., 2017 [221], based on the study of 1000 families
UK data are from Carss et al., 2017 [30], based on 722 individuals
French data are from Bocquet et al., 2013 [220], based on 1957 individuals
North Africa IRD data are from this review, based on 342 families
Data are provided as the number of families or individuals with the disease and their relative frequency in-between parenthesis within the cohort
This has obvious consequences on the respective frequency of each gene identified in IRD individuals among the North African population, with a lower contribution of variants in the ABCA4, USH2A and PRPH2 genes and a higher contribution of variants in the MYO7A, FAM161A, GUCY2D and MERTK genes (Table 2).
Table 2.
Frequencies of IRD genes across different countries
| Genes | USA | UK | France | North Africa |
|---|---|---|---|---|
| Occurrence and frequencies | ||||
| ABCA4 (AR) | 173 (22.8%) | 73 (18.1%) | 72 (17.3%) | 16 (4.7%) |
| USH2A (AR) | 76 (10%) | 61 (15.1%) | 59 (14.1%) | 9 (2.6%) |
| RPGR (X-L) | 48 (6.3%) | 13 (3.2%) | 23 (5.5%) | – |
| RHO (AD/AR) | 34 (4.5%) | 7 (1.7%) | 15 (3.6%) | – |
| PRPH2 (AD) | 32 (4.2%) | 6 (1.5%) | 20 (4.8%) | 1 (0.3%) |
| EYS (AR) | 6 (0.8%) | 16 (4%) | – | 10 (2.9%) |
| MYO7A (AR) | 8 (1.1%) | 8 (2%) | 26 (6.2%) | 34 (9.9%) |
| FAM161A (AR) | 9 (1.2%) | 2 (0.5%) | – | 20 (5.8%) |
| GUCY2D (AR) | 4 (0.5%) | 4 (1%) | 1 (0.2%) | 19 (5.6%) |
| MERTK (AR) | 3 (0.4%) | 4 (1%) | 1 (0.2%) | 10 (2.9%) |
USA data are from Stone et al., 2017 [221], based on 760 families
UK data are from Carss et al., 2017 [30], based on 404 individuals
French data are from Bocquet et al., 2013 [220], based on 417 individuals
North Africa data are from this review, based on 342 IRD families
Data are provided as the number of families or individuals and with autosomal dominant (AD) or recessive (AR) variants in the gene considered, and their frequency in-between parenthesis within the cohort with a molecular diagnosis
The same reasons explain why S-ION are proportionally so frequent (88.7%) in this region of the world, while NS-ION are rare (11.3%), and in particular, why no case of isolated Dominant Optic Atrophy has yet been reported in North Africa, although AD ION are at least ten times more frequent than the recessive ones in Western countries [220, 222]. Consequently, these peculiarities explain why the most prevalent ION among the North African families are recessive and syndromic, with the Warburg micro syndrome and the GAPO syndrome predominating in Egypt, and the Infantile neuroaxonal dystrophy 1 predominating in Tunisia, while a single family with dominant S-ION has been reported, with a hereditary motor and sensory neuropathy type-VIA, associated to a heterozygous ACO2 variant.
Thus, we provided here the first epidemiological data regarding IRD and ION in North Africa, combining clinical and molecular data. Our bibliographical review suggests that the North African populations are far from been fully characterized, because these two groups of diseases are today under-diagnosed, most probably due to the remote location of many patients far away from an Ophthalmologist, the restricted training of Ophthalmologists to rare inherited eye diseases, the difficulties to access local high throughput next generation sequencing, and the limited resources of the population. We also believe that the actual IRD and ION clinical and molecular spectrums in the North African countries are fragmented, as most reports of such information are limited to case reports, or as a part of patient cohorts included in studies from other countries.
Nevertheless, the structural peculiarities of the North African population, with the high rate of consanguinity and the presence of informative large pedigrees with many affected generations, offer outstanding opportunities to identify novel clinical presentations and genes responsible for these inherited eye diseases. With these perspectives in mind, research priorities should now focus on the training of local ophthalmo-geneticists dedicated to the clinical and molecular diagnoses of inherited blinding diseases.
Conclusion
Inherited retinal dystrophies and optic neuropathies are major causes of early visual impairment that often lead to legal blindness, which are individually rare, but collectively frequent. With a population of some 200 million inhabitants in North Africa, we can expect that some 100.000 individuals (1/2.000 persons) are affected by one of these diseases, a population nowadays far under-diagnosed with respect to the 413 North African families yet reported in medical databases, and compiled here. As both IRD and ION constitute real public health challenges, with major impacts on the educational, autonomy and socioeconomic conditions of affected individuals, future politics should invest in the training of physicians and biologists to improve the diagnosis of inherited ophthalmologic presentations and in equipment dedicated to their molecular diagnosis, to first establish accurate clinical diagnoses, and second constitute well clinically characterized cohorts of individuals eligible to treatments with the emergent therapies for these eye diseases.
Supplementary Information
Additional file 1: Keywords used in the medical Pubmed and Scopus databases.
Additional file 2: Phenotypic and mutational spectrum of 194 North African families with non-syndromic inherited retinal dystrophies.
Additional file 3: Phenotypic and mutational spectrum of 143 North African families with syndromic inherited retinal dystrophies.
Additional file 4: Mutational spectrum of North African families with non-syndromic inherited optic neuropathies.
Additional file 5: Phenotypic and mutational spectrum of North African families with syndromic inherited optic neuropathies.
Additional file 6: Mutational spectrum of North African families with hereditary mitochondrial diseases with retinal or optic nerve manifestations.
Acknowledgements
This project was supported by Ministry of Europe and Foreign Affairs, the Ministry of Higher Education, Research and Innovation, and the French Institute of Rabat and the French-Morocco bilateral program PHC TOUBKAL 2019 Grant Number: 39005ZL. We are indebted to the support of the Université d’Angers, CHU d’Angers, the Région Pays de la Loire, Angers Loire Métropole, the Fondation Maladies Rares, Retina France, Ouvrir Les Yeux, UNADEV, Kjer-France, Fondation de France, and Fondation VISIO.
Abbreviations
- IRD
Inherited retinal dystrophy
- ION
Inherited optic neuropathy
- NS-IRD
Non-syndromic inherited retinal dystrophy
- NS-ION
Non-syndromic inherited optic neuropathy
- S-IRD
Syndromic inherited retinal dystrophy
- S-ION
Syndromic inherited optic neuropathy
- AR
Autosomal recessive
- AD
Autosomal dominant
- RP
Retinitis pigmentosa
- RPA
Retinitis punctata albescens
- CD
Cone dystrophy
- CRD
Cone-rod dystrophy
- LCA
Leber congenital amaurosis
- EORD
Early onset retinal dystrophy
- STGD
Stargardt disease
- BMVD
Best vitelliform macular dystrophy
- USH
Usher syndrome
- USH1
Usher syndrome type 1
- USH2
Usher syndrome type 2
- USH3
Usher syndrome. Type 3
- BBS
Bardet-Biedl syndrome
- SLS
Senior-Loken syndrome
- JBTS
Joubert syndrome
- SJLS
Sjogren-Larsson syndrome
- ABL
Abetalipoproteinemia
- HS
Heimler syndrome
- PBD
Peroxisomal biogenesis disorder
- AVED
Familial isolated vitamin E deficiency
- FA
Friedreich ataxia
- SCA7
Spinocerebellar ataxia type 7
- KSS
Kearns-Sayer syndrome
- MELAS
Mitochondrial encephalomyopathy, lactic acidosis, and stroke like episodes
- MIDD
Maternally inherited deafness and diabetes
- LHON
Leber hereditary optic neuropathy
- DOA
Dominant optic atrophy
- mtDNA
Mitochondrial DNA
- WFS
Wolfram syndrome
- LS
Leigh syndrome
- INAD
Infantile neuroaxonal dystrophy
Author contributions
AB: Collecting and compiling of data, writing the manuscript and production of figures. HC: Design and production of figures. GA: Compilation of data and correction of the manuscript. MC: Conception and the correction of the manuscript. MK: Conception and support in manuscript writing. ABa: Conception and support in manuscript writing. GL: Conception, writing and surveillance of the manuscript and figures. All authors read and approved the final manuscript.
Funding
This project was financially supported by Ministry of Europe and Foreign Affairs, the Ministry of Higher Education, Research and Innovation, and the French Institute of Rabat and the French-Morocco bilateral program PHC TOUBKAL 2019 Grant Number: 39005ZL.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary files.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Worldometers, African Countries by population. 2021. https://www.worldometers.info/population/countries-in-africa-by-population/
- 2.Newman JL. The peopling of Africa: a geographic interpretation. New Haven: Yale University Press; 1995. p. 235. [Google Scholar]
- 3.Arauna LR, Mendoza-Revilla J, Mas-Sandoval A, Izaabel H, Bekada A, Benhamamouch S, et al. Recent historical migrations have shaped the gene pool of arabs and berbers in North Africa. Mol Biol Evol. 2017;34(2):318–329. doi: 10.1093/molbev/msw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekada A, Arauna LR, Deba T, Calafell F, Benhamamouch S, Comas D. Genetic heterogeneity in Algerian human populations. PLoS ONE. 2015;10(9):e0138453. doi: 10.1371/journal.pone.0138453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J. High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet. 2001;68(4):1019–1029. doi: 10.1086/319521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coudray C, Olivieri A, Achilli A, Pala M, Melhaoui M, Cherkaoui M, et al. The complex and diversified mitochondrial gene pool of Berber populations. Ann Hum Genet. 2009;73(2):196–214. doi: 10.1111/j.1469-1809.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- 7.Font-Porterias N, Solé-Morata N, Serra-Vidal G, Bekada A, Fadhlaoui-Zid K, Zalloua P, et al. The genetic landscape of Mediterranean North African populations through complete mtDNA sequences. Ann Hum Biol. 2018;45(1):98–104. doi: 10.1080/03014460.2017.1413133. [DOI] [PubMed] [Google Scholar]
- 8.Kefi R. Anthropology and population heterogeneity in North Africa. In: Kumar D, editor. Genomics and health in the developing world. Oxford University Press, Oxford, 2012.
- 9.Plaza S, Calafell F, Helal A, Bouzerna N, Lefranc G, Bertranpetit J, et al. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann Hum Genet. 2003;67(Pt 4):312–328. doi: 10.1046/j.1469-1809.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- 10.Chouraqui A. Between East and West: a history of the Jews of North Africa. Skokie, Ill.: Varda Books; 2001. http://search.ebscohost.com/login.aspx?direct=true&scope=site&db=nlebk&db=nlabk&AN=106851
- 11.Campbell CL, Palamara PF, Dubrovsky M, Botigue LR, Fellous M, Atzmon G, et al. North African Jewish and non-Jewish populations form distinctive, orthogonal clusters. Proc Natl Acad Sci. 2012;109(34):13865–13870. doi: 10.1073/pnas.1204840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behar DM, Metspalu E, Kivisild T, Rosset S, Tzur S, Hadid Y, et al. Counting the founders: the matrilineal genetic ancestry of the Jewish Diaspora. PLoS ONE. 2008;3(4):e2062. doi: 10.1371/journal.pone.0002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romdhane L, Mezzi N, Hamdi Y, El-Kamah G, Barakat A, Abdelhak S. Consanguinity and inbreeding in health and disease in North African populations. Annu Rev Genom Hum Genet. 2019;20(1):155–179. doi: 10.1146/annurev-genom-083118-014954. [DOI] [PubMed] [Google Scholar]
- 14.Tadmouri GO, Nair P, Obeid T, Al Ali MT, Al Khaja N, Hamamy HA. Consanguinity and reproductive health among Arabs. Reprod Health. 2009;8(6):17. doi: 10.1186/1742-4755-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badr FM. Genetic studies of Egyptian Nubian populations. I. Frequency and types of consanguineous marriages. Hum Hered. 1972;22(4):387–98. doi: 10.1159/000152515. [DOI] [PubMed] [Google Scholar]
- 16.Anwar WA, Khyatti M, Hemminki K. Consanguinity and genetic diseases in North Africa and immigrants to Europe. Eur J Public Health. 2014;24(suppl 1):57–63. doi: 10.1093/eurpub/cku104. [DOI] [PubMed] [Google Scholar]
- 17.Romdhane L, Abdelhak S. . Genetic disorders in North African Populations. In: Kumar D, editor. Genomics and health in the developing world. Oxford University Press, Oxford; 2012. http://oxfordmedicine.com/view/10.1093/med/9780195374759.001.0001/med-9780195374759-chapter-34
- 18.Solebo AL, Rahi J. Epidemiology, aetiology and management of visual impairment in children. Arch Dis Child. 2014;99(4):375–379. doi: 10.1136/archdischild-2012-303002. [DOI] [PubMed] [Google Scholar]
- 19.Alagaratnam J, Sharma TK, Lim CS, Fleck BW. A survey of visual impairment in children attending the Royal Blind School, Edinburgh using the WHO childhood visual impairment database. Eye. 2002;16(5):557–561. doi: 10.1038/sj.eye.6700149. [DOI] [PubMed] [Google Scholar]
- 20.Kong L, Fry M, Al-Samarraie M, Gilbert C, Steinkuller PG. An update on progress and the changing epidemiology of causes of childhood blindness worldwide. J Am Assoc Pediat Ophthalmol Strabismus. 2012;16(6):501–507. doi: 10.1016/j.jaapos.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Rahi JS, Cable N, British Childhood Visual Impairment Study Group. Severe visual impairment and blindness in children in the UK. Lancet. 2003;362(9393):1359–65. [DOI] [PubMed]
- 22.Kahloun R, Khairallah M, Resnikoff S, Cicinelli MV, Flaxman SR, Das A, et al. Prevalence and causes of vision loss in North Africa and Middle East in 2015: magnitude, temporal trends and projections. Br J Ophthalmol. 2019;103(7):863–870. doi: 10.1136/bjophthalmol-2018-312068. [DOI] [PubMed] [Google Scholar]
- 23.Habibi I, Chebil A, Falfoul Y, Allaman-Pillet N, Kort F, Schorderet DF, et al. Identifying mutations in Tunisian families with retinal dystrophy. Sci Rep. 2016;6(1):37455. doi: 10.1038/srep37455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habibi I, Falfoul Y, Turki A, Hassairi A, El Matri K, Chebil A, et al. Genetic spectrum of retinal dystrophies in Tunisia. Sci Rep. 2020;10(1):11199. doi: 10.1038/s41598-020-67792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel-Hamid MS, Issa MY, Elbendary HM, Abdel-Ghafar SF, Rafaat K, Hosny H, et al. Phenotypic and mutational spectrum of thirty-five patients with Sjögren-Larsson syndrome: identification of eleven novel ALDH3A2 mutations and founder effects. J Hum Genet. 2019;64(9):859–865. doi: 10.1038/s10038-019-0637-x. [DOI] [PubMed] [Google Scholar]
- 26.Abdi S, Bahloul A, Behlouli A, Hardelin J-P, Makrelouf M, Boudjelida K, et al. Diversity of the genes implicated in algerian patients affected by usher syndrome Del Bene F, editor. PLoS ONE. 2016;11(9):e0161893. doi: 10.1371/journal.pone.0161893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M’hamdi O, Redin C, Stoetzel C, Ouertani I, Chaabouni M, Maazoul F, et al. Clinical and genetic characterization of Bardet-Biedl syndrome in Tunisia: defining a strategy for molecular diagnosis: Clinical and genetic characterization of BBS. Clin Genet. 2014;85(2):172–7. [DOI] [PubMed]
- 28.Smaoui N, Chaabouni M, Sergeev YV, Kallel H, Li S, Mahfoudh N, et al. Screening of the eight BBS genes in tunisian families: no evidence of Triallelism. Invest Ophthalmol Vis Sci. 2006;47(8):3487. doi: 10.1167/iovs.05-1334. [DOI] [PubMed] [Google Scholar]
- 29.Jaffal L, Joumaa H, Mrad Z, Zeitz C, Audo I, El Shamieh S. The genetics of rod-cone dystrophy in Arab countries: a systematic review. Eur J Hum Genet. 2021;29(6):897–910. doi: 10.1038/s41431-020-00754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017;100(1):75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Retinal Information Network [Internet]. 2021. https://sph.uth.edu/retnet/
- 32.Tatour Y, Ben-Yosef T. Syndromic inherited retinal diseases: genetic, clinical and diagnostic aspects. Diagnostics (Basel). 2020 Oct 2;10(10). [DOI] [PMC free article] [PubMed]
- 33.Boughman JA, Vernon M, Shaver KA. Usher syndrome: definition and estimate of prevalence from two high-risk populations. J Chronic Dis. 1983;36(8):595–603. doi: 10.1016/0021-9681(83)90147-9. [DOI] [PubMed] [Google Scholar]
- 34.Bernardis I, Chiesi L, Tenedini E, Artuso L, Percesepe A, Artusi V, et al. Unravelling the complexity of inherited retinal dystrophies molecular testing: added value of targeted next-generation sequencing. Biomed Res Int. 2016;2016:1–14. doi: 10.1155/2016/6341870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez-Muñoz A, Aller E, Jaijo T, González-García E, Cabrera-Peset A, Gallego-Pinazo R, et al. Expanding the clinical and molecular heterogeneity of nonsyndromic inherited retinal dystrophies. J Mol Diagn. 2020;22(4):532–543. doi: 10.1016/j.jmoldx.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Tiwari A, Bahr A, Bähr L, Fleischhauer J, Zinkernagel MS, Winkler N, et al. Next generation sequencing based identification of disease-associated mutations in Swiss patients with retinal dystrophies. Sci Rep. 2016;6(1):28755. doi: 10.1038/srep28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 38.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;11(1):40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, et al. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018;66:157–186. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 40.Audo I, Mohand-Said S, Boulanger-Scemama E, Zanlonghi X, Condroyer C, Démontant V, et al. MERTK mutation update in inherited retinal diseases. Hum Mutat. 2018;39(7):887–913. doi: 10.1002/humu.23431. [DOI] [PubMed] [Google Scholar]
- 41.Bandah-Rozenfeld D, Mizrahi-Meissonnier L, Farhy C, Obolensky A, Chowers I, Pe’er J, et al. Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2010;87(3):382–91. [DOI] [PMC free article] [PubMed]
- 42.Beryozkin A, Shevah E, Kimchi A, Mizrahi-Meissonnier L, Khateb S, Ratnapriya R, et al. Whole exome sequencing reveals mutations in known retinal disease genes in 33 out of 68 Israeli families with inherited retinopathies. Sci Rep. 2015;5(1):13187. doi: 10.1038/srep13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birtel J, Gliem M, Mangold E, Müller PL, Holz FG, Neuhaus C, et al. Next-generation sequencing identifies unexpected genotype-phenotype correlations in patients with retinitis pigmentosa. Lewin AS, editor. PLoS ONE. 2018;13(12):e0207958. doi: 10.1371/journal.pone.0207958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dessalces E, Bocquet B, Bourien J, Zanlonghi X, Verdet R, Meunier I, et al. Early-onset foveal involvement in retinitis punctata albescens with mutations in RLBP1. JAMA Ophthalmol. 2013;131(10):1314. doi: 10.1001/jamaophthalmol.2013.4476. [DOI] [PubMed] [Google Scholar]
- 45.Humbert G, Delettre C, Se´ne´chal A, Bazalgette C, Barakat A, Bazalgette C, et al. Homozygous deletion related to alu repeats in RLBP1 causes retinitis Punctata Albescens. Invest Ophthalmol Vis Sci. 2006;47(11):4719. [DOI] [PubMed]
- 46.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2(1):7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang SH, Sharma T. Progressive Cone Dystrophy and Cone-Rod Dystrophy (XL, AD, and AR). In: Tsang SH, Sharma T, editors. Atlas of inherited retinal diseases [Internet]. Cham: Springer International Publishing; 2018 [cited 2020 Nov 17]. p. 53–60. (Advances in Experimental Medicine and Biology; vol. 1085). http://link.springer.com/10.1007/978-3-319-95046-4_12 [DOI] [PubMed]
- 48.Gill JS, Georgiou M, Kalitzeos A, Moore AT, Michaelides M. Progressive cone and cone-rod dystrophies: clinical features, molecular genetics and prospects for therapy. Br J Ophthalmol. 2019 Jan 24; [DOI] [PMC free article] [PubMed]
- 49.Tsang SH, Sharma T. Leber congenital amaurosis. Adv Exp Med Biol. 2018;1085:131–137. doi: 10.1007/978-3-319-95046-4_26. [DOI] [PubMed] [Google Scholar]
- 50.den Hollander AI, Roepman R, Koenekoop RK, Cremers FPM. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27(4):391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Koenekoop RK. An overview of Leber congenital amaurosis: a model to understand human retinal development. Surv Ophthalmol. 2004;49(4):379–398. doi: 10.1016/j.survophthal.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Chung DC, Traboulsi EI. Leber congenital amaurosis: clinical correlations with genotypes, gene therapy trials update, and future directions. J AAPOS. 2009;13(6):587–592. doi: 10.1016/j.jaapos.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Camuzat A, Rozet J-M, Dollfus H, Gerber S, Perrault I, Munnich A, et al. Evidence of genetic heterogeneity of Leber’s congenital amaurosis (LCA) and mapping of LCA1 to chromosome 17p13. Hum Genet. 1996;97(6):798–801. doi: 10.1007/BF02346192. [DOI] [PubMed] [Google Scholar]
- 54.Camuzat A, Dollfus H, Rozet J-M, Gerber S, Bonneau D, Bonnemaison M, et al. A gene for Leber’s congenital amaurosis maps to chromosome 17p. Hum Mol Genet. 1995;4(8):1447–1452. doi: 10.1093/hmg/4.8.1447. [DOI] [PubMed] [Google Scholar]
- 55.Hanein S, Perrault I, Gerber S, Tanguy G, Barbet F, Ducroq D, et al. Leber congenital amaurosis: Comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum Mutat. 2004;23(4):306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 56.Perrault I, Rozet J-M, Gerber S, Ghazi I, Ducroq D, Souied E, et al. Spectrum of retGC1 mutations in Leber’s congenital amaurosis. Eur J Hum Genet. 2000;8(8):578–582. doi: 10.1038/sj.ejhg.5200503. [DOI] [PubMed] [Google Scholar]
- 57.Kuniyoshi K, Ikeo K, Sakuramoto H, Furuno M, Yoshitake K, Hatsukawa Y, et al. Novel nonsense and splice site mutations in CRB1 gene in two Japanese patients with early-onset retinal dystrophy. Doc Ophthalmol. 2015;130(1):49–55. doi: 10.1007/s10633-014-9464-8. [DOI] [PubMed] [Google Scholar]
- 58.Mataftsi A, Schorderet DF, Chachoua L, Boussalah M, Nouri MT, Barthelmes D, et al. Novel TULP1 mutation causing leber congenital amaurosis or early onset retinal degeneration. Invest Ophthalmol Vis Sci. 2007;48(11):5160. doi: 10.1167/iovs.06-1013. [DOI] [PubMed] [Google Scholar]
- 59.Sallum JMF, Motta FL, Arno G, Porto FBO, Resende RG, Belfort R. Clinical and molecular findings in a cohort of 152 Brazilian severe early onset inherited retinal dystrophy patients. Am J Med Genet. 2020;184(3):728–752. doi: 10.1002/ajmg.c.31828. [DOI] [PubMed] [Google Scholar]
- 60.Lorenz B, Gyürüs P, Preising M, Bremser D, Gu S, Andrassi M, et al. Early-onset severe rod-cone dystrophy in young children with RPE65 mutations. Invest Ophthalmol Vis Sci. 2000;41(9):2735–2742. [PubMed] [Google Scholar]
- 61.Morimura H, Fishman GA, Grover SA, Fulton AB, Berson EL, Dryja TP. Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or Leber congenital amaurosis. Proc Natl Acad Sci. 1998;95(6):3088–3093. doi: 10.1073/pnas.95.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson DA, Gyürüs P, Fleischer LL, Bingham EL, McHenry CL, Apfelstedt-Sylla E, et al. Genetics and phenotypes of RPE65 mutations in inherited retinal degeneration. Invest Ophthalmol Vis Sci. 2000;41(13):4293–4299. [PubMed] [Google Scholar]
- 63.Yzer S. A Tyr368His RPE65 founder mutation is associated with variable expression and progression of early onset retinal dystrophy in 10 families of a genetically isolated population. J Med Genet. 2003;40(9):709–713. doi: 10.1136/jmg.40.9.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mackay DS, Dev Borman A, Moradi P, Henderson RH, Li Z, Wright GA, et al. RDH12 retinopathy: novel mutations and phenotypic description. Mol Vis. 2011;17:2706–2716. [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson DA, Janecke AR, Lange J, Feathers KL, Hübner CA, McHenry CL, et al. Retinal degeneration associated with RDH12 mutations results from decreased 11-cis retinal synthesis due to disruption of the visual cycle. Hum Mol Genet. 2005;14(24):3865–3875. doi: 10.1093/hmg/ddi411. [DOI] [PubMed] [Google Scholar]
- 66.Tsang SH, Sharma T. Stargardt disease. Adv Exp Med Biol. 2018;1085:139–151. doi: 10.1007/978-3-319-95046-4_27. [DOI] [PubMed] [Google Scholar]
- 67.Tanna P, Strauss RW, Fujinami K, Michaelides M. Stargardt disease: clinical features, molecular genetics, animal models and therapeutic options. Br J Ophthalmol. 2017;101(1):25–30. doi: 10.1136/bjophthalmol-2016-308823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu F, Gao F, Li J, Xu P, Wang D, Chen F, et al. Novel variants associated with Stargardt disease in Chinese patients. Gene. 2020;5(754):144890. doi: 10.1016/j.gene.2020.144890. [DOI] [PubMed] [Google Scholar]
- 69.Meunier I, Sénéchal A, Dhaenens C-M, Arndt C, Puech B, Defoort-Dhellemmes S, et al. Systematic screening of BEST1 and PRPH2 in juvenile and adult vitelliform macular dystrophies: a rationale for molecular analysis. Ophthalmology. 2011;118(6):1130–1136. doi: 10.1016/j.ophtha.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 70.Bitner H, Mizrahi-Meissonnier L, Griefner G, Erdinest I, Sharon D, Banin E. A homozygous frameshift mutation in BEST1 causes the classical form of best disease in an autosomal recessive mode. Invest Ophthalmol Vis Sci. 2011;52(8):5332. doi: 10.1167/iovs.11-7174. [DOI] [PubMed] [Google Scholar]
- 71.Chibani Z, Abid IZ, Molbaek A, Söderkvist P, Feki J, Hmani-Aifa M. Novel BEST1 gene mutations associated with two different forms of macular dystrophy in Tunisian families. Clin Experiment Ophthalmol. 2019;47(8):1063–1073. doi: 10.1111/ceo.13577. [DOI] [PubMed] [Google Scholar]
- 72.Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24(2):127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 73.Petit C. USHER SYNDROME : From Genetics to Pathogenesis. Annu Rev Genom Hum Genet. 2001;2(1):271–297. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- 74.El-Amraoui A, Petit C. The retinal phenotype of Usher syndrome: Pathophysiological insights from animal models. CR Biol. 2014;337(3):167–177. doi: 10.1016/j.crvi.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Weil D. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet. 2003;12(5):463–471. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- 76.Ammar-Khodja F, Faugère V, Baux D, Giannesini C, Léonard S, Makrelouf M, et al. Molecular screening of deafness in Algeria: High genetic heterogeneity involving DFNB1 and the Usher loci, DFNB2/USH1B, DFNB12/USH1D and DFNB23/USH1F. Eur J Med Genet. 2009;52(4):174–179. doi: 10.1016/j.ejmg.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 77.Bakhchane A, Charif M, Bousfiha A, Boulouiz R, Nahili H, Rouba H, et al. Novel compound heterozygous MYO7A mutations in Moroccan families with autosomal recessive non-syndromic hearing loss Deng H, editor. PLoS ONE. 2017;12(5):e0176516. doi: 10.1371/journal.pone.0176516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bousfiha A, Bakhchane A, Charoute H, Detsouli M, Rouba H, Charif M, et al. Novel compound heterozygous mutations in the GPR98 (USH2C) gene identified by whole exome sequencing in a Moroccan deaf family. Mol Biol Rep. 2017;44(5):429–434. doi: 10.1007/s11033-017-4129-9. [DOI] [PubMed] [Google Scholar]
- 79.Budde BS, Aly MA, Mohamed MR, Breß A, Altmüller J, Motameny S, et al. Comprehensive molecular analysis of 61 Egyptian families with hereditary nonsyndromic hearing loss. Clin Genet. 2020;98(1):32–42. doi: 10.1111/cge.13754. [DOI] [PubMed] [Google Scholar]
- 80.Neuhaus C, Eisenberger T, Decker C, Nagl S, Blank C, Pfister M, et al. Next-generation sequencing reveals the mutational landscape of clinically diagnosed Usher syndrome: copy number variations, phenocopies, a predominant target for translational read-through, and PEX26 mutated in Heimler syndrome. Mol Genet Genomic Med. 2017;5(5):531–552. doi: 10.1002/mgg3.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Talbi S, Bonnet C, Riahi Z, Boudjenah F, Dahmani M, Hardelin J-P, et al. Genetic heterogeneity of congenital hearing impairment in Algerians from the Ghardaïa province. Int J Pediatr Otorhinolaryngol. 2018;112:1–5. doi: 10.1016/j.ijporl.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 82.Riahi Z, Bonnet C, Zainine R, Lahbib S, Bouyacoub Y, Bechraoui R, et al. Whole exome sequencing identifies mutations in Usher syndrome genes in profoundly deaf Tunisian patients. PLoS ONE. 2015;10(3):e0120584. doi: 10.1371/journal.pone.0120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jaijo T, Aller E, Beneyto M, Najera C, Graziano C, Turchetti D, et al. MYO7A mutation screening in Usher syndrome type I patients from diverse origins. J Med Genet. 2006;44(3):e71–e71. doi: 10.1136/jmg.2006.045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roux A-F. Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90% J Med Genet. 2006;43(9):763–768. doi: 10.1136/jmg.2006.041954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adato A, Weil D, Kalinski H, Pel-Or Y, Ayadi H, Petit C, et al. Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am J Hum Genet. 1997;61(4):813–821. doi: 10.1086/514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ben-Rebeh I, Grati M, Bonnet C, Bouassida W, Hadjamor I, Ayadi H, et al. Genetic analysis of Tunisian families with Usher syndrome type 1: toward improving early molecular diagnosis. Mol Vis. 2016;22:827–835. [PMC free article] [PubMed] [Google Scholar]
- 87.Khalaileh A, Abu-Diab A, Ben-Yosef T, Raas-Rothschild A, Lerer I, Alswaiti Y, et al. The genetics of usher syndrome in the Israeli and Palestinian populations. Invest Ophthalmol Vis Sci. 2018;59(2):1095. doi: 10.1167/iovs.17-22817. [DOI] [PubMed] [Google Scholar]
- 88.Khateb S, Hanany M, Khalaileh A, Beryozkin A, Meyer S, Abu-Diab A, et al. Identification of genomic deletions causing inherited retinal degenerations by coverage analysis of whole exome sequencing data. J Med Genet. 2016;53(9):600–607. doi: 10.1136/jmedgenet-2016-103825. [DOI] [PubMed] [Google Scholar]
- 89.Adato A, Weston MD, Berry A, Kimberling WJ, Bonne-Tamir A. Three novel mutations and twelve polymorphisms identified in the USH2A gene in Israeli USH2 families. Hum Mutat. 2000;15(4):388. doi: 10.1002/(SICI)1098-1004(200004)15:4<388::AID-HUMU27>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 90.Auslender N, Bandah D, Rizel L, Behar DM, Shohat M, Banin E, et al. Four USH2A founder mutations underlie the majority of Usher syndrome type 2 cases among non-Ashkenazi Jews. Genet Test. 2008;12(2):289–294. doi: 10.1089/gte.2007.0107. [DOI] [PubMed] [Google Scholar]
- 91.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet. 1999;36(6):437–446. [PMC free article] [PubMed] [Google Scholar]
- 92.Forsythe E, Beales PL. Bardet-Biedl syndrome. Eur J Hum Genet. 2013;21(1):8–13. doi: 10.1038/ejhg.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-mendelian bardet-biedl syndrome. Am J Hum Genet. 2003;72(5):1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen J, Smaoui N, Hammer MBH, Jiao X, Riazuddin SA, Harper S, et al. Molecular analysis of Bardet-Biedl syndrome families: report of 21 novel mutations in 10 genes. Invest Ophthalmol Vis Sci. 2011;52(8):5317–5324. doi: 10.1167/iovs.11-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Muller J, Stoetzel C, Vincent MC, Leitch CC, Laurier V, Danse JM, et al. Identification of 28 novel mutations in the Bardet-Biedl syndrome genes: the burden of private mutations in an extensively heterogeneous disease. Hum Genet. 2010;127(5):583–593. doi: 10.1007/s00439-010-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Janssen S, Ramaswami G, Davis EE, Hurd T, Airik R, Kasanuki JM, et al. Mutation analysis in Bardet-Biedl syndrome by DNA pooling and massively parallel resequencing in 105 individuals. Hum Genet. 2011;129(1):79–90. doi: 10.1007/s00439-010-0902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stoetzel C, Laurier V, Faivre L, Mégarbané A, Perrin-Schmitt F, Verloes A, et al. BBS8 is rarely mutated in a cohort of 128 Bardet-Biedl syndrome families. J Hum Genet. 2006;51(1):81–84. doi: 10.1007/s10038-005-0320-2. [DOI] [PubMed] [Google Scholar]
- 98.Hichri H, Stoetzel C, Laurier V, Caron S, Sigaudy S, Sarda P, et al. Testing for triallelism: analysis of six BBS genes in a Bardet-Biedl syndrome family cohort. Eur J Hum Genet. 2005;13(5):607–616. doi: 10.1038/sj.ejhg.5201372. [DOI] [PubMed] [Google Scholar]
- 99.Tsang SH, Aycinena ARP, Sharma T. Ciliopathy: senior-løken syndrome. In: Tsang SH, Sharma T, editors. Atlas of Inherited Retinal Diseases [Internet]. Cham: Springer International Publishing; 2018 [cited 2020 Nov 27]. p. 175–8. (Advances in Experimental Medicine and Biology; vol. 1085). http://link.springer.com/10.1007/978-3-319-95046-4_34 [DOI] [PubMed]
- 100.Zaki MS, Abdel-Aleem A, Abdel-Salam G, Marsh SE, Silhavy JL, Barkovich AJ, et al. The molar tooth sign: A new Joubert syndrome and related cerebellar disorders classification system tested in Egyptian families. Neurology. 2008;70(7):556–565. doi: 10.1212/01.wnl.0000277644.12087.fd. [DOI] [PubMed] [Google Scholar]
- 101.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37(3):282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 102.Radha Rama Devi A, Naushad SM, Lingappa L. Clinical and Molecular Diagnosis of Joubert Syndrome and Related Disorders. Pediatr Neurol. 2020;106:43–9. [DOI] [PubMed]
- 103.Wang SF, Kowal TJ, Ning K, Koo EB, Wu AY, Mahajan VB, et al. Review of ocular manifestations of joubert syndrome. Genes (Basel). 2018;9(12). [DOI] [PMC free article] [PubMed]
- 104.Akizu N, Silhavy JL, Rosti RO, Scott E, Fenstermaker AG, Schroth J, et al. Mutations in CSPP1 lead to classical Joubert syndrome. Am J Hum Genet. 2014;94(1):80–86. doi: 10.1016/j.ajhg.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brancati F, Travaglini L, Zablocka D, Boltshauser E, Accorsi P, Montagna G, et al. RPGRIP1L mutations are mainly associated with the cerebello-renal phenotype of Joubert syndrome-related disorders. Clin Genet. 2008;74(2):164–170. doi: 10.1111/j.1399-0004.2008.01047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Mori R, Romani M, D’Arrigo S, Zaki MS, Lorefice E, Tardivo S, et al. Hypomorphic recessive variants in SUFU impair the sonic hedgehog pathway and cause Joubert syndrome with Cranio-facial and skeletal defects. Am J Hum Genet. 2017;101(4):552–563. doi: 10.1016/j.ajhg.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Latour BL, Van De Weghe JC, Rusterholz TD, Letteboer SJ, Gomez A, Shaheen R, et al. Dysfunction of the ciliary ARMC9/TOGARAM1 protein module causes Joubert syndrome. J Clin Invest. 2020;130(8):4423–4439. doi: 10.1172/JCI131656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schueler M, Halbritter J, Phelps IG, Braun DA, Otto EA, Porath JD, et al. Large-scale targeted sequencing comparison highlights extreme genetic heterogeneity in nephronophthisis-related ciliopathies. J Med Genet. 2016;53(3):208–214. doi: 10.1136/jmedgenet-2015-103304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Soliman NA, Hildebrandt F, Otto EA, Nabhan MM, Allen SJ, Badr AM, et al. Clinical characterization and NPHP1 mutations in nephronophthisis and associated ciliopathies: a single center experience. Saudi J Kidney Dis Transpl. 2012;23(5):1090–1098. doi: 10.4103/1319-2442.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rizzo WB, Craft DA, Somer T, Carney G, Trafrova J, Simon M. Abnormal fatty alcohol metabolism in cultured keratinocytes from patients with Sjögren-Larsson syndrome. J Lipid Res. 2008;49(2):410–419. doi: 10.1194/jlr.M700469-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jagell S, Polland W, Sandgren O. Specific changes in the fundus typical for the Sjögren-Larsson syndrome. An ophthalmological study of 35 patients. Acta Ophthalmol (Copenh). 1980;58(3):321–30. doi: 10.1111/j.1755-3768.1980.tb05730.x. [DOI] [PubMed] [Google Scholar]
- 112.Poll-The BT, Billette de Villemeur T, Abitbol M, Dufier JL, Saudubray JM. Metabolic pigmentary retinopathies: diagnosis and therapeutic attempts. Eur J Pediatr. 1992;151(1):2–11. doi: 10.1007/BF02073880. [DOI] [PubMed] [Google Scholar]
- 113.Willemsen MA, Cruysberg JR, Rotteveel JJ, Aandekerk AL, Van Domburg PH, Deutman AF. Juvenile macular dystrophy associated with deficient activity of fatty aldehyde dehydrogenase in Sjögren-Larsson syndrome. Am J Ophthalmol. 2000;130(6):782–789. doi: 10.1016/s0002-9394(00)00576-6. [DOI] [PubMed] [Google Scholar]
- 114.Amr K, El-Bassyouni HT, Ismail S, Youness E, El-Daly SM, Ebrahim AY, et al. Genetic assessment of ten Egyptian patients with Sjögren-Larsson syndrome: expanding the clinical spectrum and reporting a novel ALDH3A2 mutation. Arch Dermatol Res. 2019;311(9):721–730. doi: 10.1007/s00403-019-01953-6. [DOI] [PubMed] [Google Scholar]
- 115.Bernardini ML, Cangiotti AM, Zamponi N, Porfiri L, Cinti S, Offidani A. Diagnosing Sjögren–Larsson syndrome in a 7-year-old Moroccan boy. J Cutan Pathol. 2007;34(3):270–275. doi: 10.1111/j.1600-0560.2006.00603.x. [DOI] [PubMed] [Google Scholar]
- 116.Gaboon NEA, Jelani M, Almramhi MM, Mohamoud HSA, Al-Aama JY. Case of Sjögren-Larsson syndrome with a large deletion in the ALDH3A2 gene confirmed by single nucleotide polymorphism array analysis. J Dermatol. 2015;42(7):706–709. doi: 10.1111/1346-8138.12861. [DOI] [PubMed] [Google Scholar]
- 117.Haruna K, Suga Y, Mizuno Y, Kourou K, Muramatsu S, Hasegawa T, et al. A Moroccan patient with Sjögren–Larsson syndrome. J Dermatol. 2007;34(2):153–155. doi: 10.1111/j.1346-8138.2006.00239.x. [DOI] [PubMed] [Google Scholar]
- 118.Rafai MA, Boulaajaj FZ, Seito A, Suga Y, Slassi I, Fadel H. Syndrome de Sjögren-Larsson en rapport avec une nouvelle mutation chez un enfant marocain. Arch Pediatr. 2008;15(11):1648–1651. doi: 10.1016/j.arcped.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 119.Zamel R, Khan R, Pollex RL, Hegele RA. Abetalipoproteinemia: two case reports and literature review. Orphanet J Rare Dis. 2008;8(3):19. doi: 10.1186/1750-1172-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ratbi I, Falkenberg KD, Sommen M, Al-Sheqaih N, Guaoua S, Vandeweyer G, et al. Heimler syndrome is caused by hypomorphic mutations in the peroxisome-biogenesis genes PEX1 and PEX6. Am J Hum Genet. 2015;97(4):535–545. doi: 10.1016/j.ajhg.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Braverman NE, Raymond GV, Rizzo WB, Moser AB, Wilkinson ME, Stone EM, et al. Peroxisome biogenesis disorders in the Zellweger spectrum: an overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab. 2016;117(3):313–321. doi: 10.1016/j.ymgme.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Euch-Fayache GE, Bouhlal Y, Amouri R, Feki M, Hentati F. Molecular, clinical and peripheral neuropathy study of Tunisian patients with ataxia with vitamin E deficiency. Brain. 2014;137(2):402–410. doi: 10.1093/brain/awt339. [DOI] [PubMed] [Google Scholar]
- 123.Koenig M. Rare forms of autosomal recessive neurodegenerative ataxia. Semin Pediat Neurol. 2003;10(3):183–192. doi: 10.1016/s1071-9091(03)00027-5. [DOI] [PubMed] [Google Scholar]
- 124.Burck U, Goebel H, Kuhlendahl H, Meier C, Goebel K. Neuromyopathy and vitamin e deficiency in man1. Neuropediatrics. 1981;12(03):267–278. doi: 10.1055/s-2008-1059657. [DOI] [PubMed] [Google Scholar]
- 125.Ben Hamida C, Doerflinger N, Belal S, Linder C, Reutenauer L, Dib C, et al. Localization of Friedreich ataxia phenotype with selective vitamin E deficiency to chromosome 8q by homozygosity mapping. Nat Genet. 1993;5(2):195–200. doi: 10.1038/ng1093-195. [DOI] [PubMed] [Google Scholar]
- 126.Cavalier L, Ouahchi K, Kayden HJ, Di Donato S, Reutenauer L, Mandel J-L, et al. Ataxia with isolated vitamin e deficiency: heterogeneity of mutations and phenotypic variability in a large number of families. Am J Hum Genet. 1998;62(2):301–310. doi: 10.1086/301699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hamida MB, Belal S, Sirugo G, Hamida CB, Panayides K, Ionannou P, et al. Friedreich’s ataxia phenotype not linked to chromosome 9 and associated with selective autosomal recessive vitamin E deficiency in two inbred Tunisian families. Neurology. 1993;43(11):2179–2179. doi: 10.1212/wnl.43.11.2179. [DOI] [PubMed] [Google Scholar]
- 128.Ouahchi K, Arita M, Kayden H, Hentati F, Hamida MB, Sokol R, et al. Ataxia with isolated vitamin E deficiency is caused by mutations in the α–tocopherol transfer protein. Nat Genet. 1995;9(2):141–145. doi: 10.1038/ng0295-141. [DOI] [PubMed] [Google Scholar]
- 129.Aparicio JM, Bélanger-Quintana A, Suárez L, Mayo D, Benítez J, Díaz M, et al. Ataxia with isolated vitamin e deficiency: case report and review of the literature. J Pediat Gastroenterol Nutrit. 2001;33(2):206–210. doi: 10.1097/00005176-200108000-00022. [DOI] [PubMed] [Google Scholar]
- 130.Bellayou H, Dehbi H, Bourezgui M, Slassi I, Nadifi S. Ataxia with vitamin E deficiency (AVED); an example of the contribution of research in molecular genetic to counselling in Morocco. Pathol Biol (Paris) 2009;57(5):425–426. doi: 10.1016/j.patbio.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 131.Benomar A, Yahyaoui M, Meggouh F, Bouhouche A, Boutchich M, Bouslam N, et al. Clinical comparison between AVED patients with 744 del A mutation and Friedreich ataxia with GAA expansion in 15 Moroccan families. J Neurol Sci. 2002;198(1–2):25–29. doi: 10.1016/s0022-510x(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 132.Bouhlal Y, Zouari M, Kefi M, Hamida CB, Hentati F, Amouri R. Autosomal recessive ataxia caused by three distinct gene defects in a single consanguineous family. J Neurogenet. 2008;22(2):139–148. doi: 10.1080/01677060802025233. [DOI] [PubMed] [Google Scholar]
- 133.Hamza W, Ali Pacha L, Hamadouche T, Muller J, Drouot N, Ferrat F, et al. Molecular and clinical study of a cohort of 110 Algerian patients with autosomal recessive ataxia. BMC Med Genet. 2015;16(1):36. doi: 10.1186/s12881-015-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mariotti C, Gellera C, Rimoldi M, Mineri R, Uziel G, Zorzi G, et al. Ataxia with isolated vitamin E deficiency: neurological phenotype, clinical follow-up and novel mutations in TTPAgene in Italian families. Neurol Sci. 2004;25(3):130–137. doi: 10.1007/s10072-004-0246-z. [DOI] [PubMed] [Google Scholar]
- 135.Yokota T, Shiojiri T, Gotoda T, Arai H. Retinitis pigmentosa and ataxia caused by a mutation in the gene for the alpha-tocopherol-transfer protein. N Engl J Med. 1996;335(23):1770–1771. doi: 10.1056/NEJM199612053352315. [DOI] [PubMed] [Google Scholar]
- 136.Yokota T, Shiojiri T, Gotoda T, Arita M, Arai H, Ohga T, et al. Friedreich-like ataxia with retinitis pigmentosa caused by the His101Gln mutation of the alpha-tocopherol transfer protein gene. Ann Neurol. 1997;41(6):826–832. doi: 10.1002/ana.410410621. [DOI] [PubMed] [Google Scholar]
- 137.Roubertie A, Biolsi B, Rivier F, Humbertclaude V, Cheminal R, Echenne B. Ataxia with vitamin E deficiency and severe dystonia: report of a case. Brain Develop. 2003;25(6):442–445. doi: 10.1016/s0387-7604(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 138.Bah MG, Rodriguez D, Cazeneuve C, Mochel F, Devos D, Suppiej A, et al. Deciphering the natural history of SCA7 in children. Eur J Neurol. 2020;27(11):2267–2276. doi: 10.1111/ene.14405. [DOI] [PubMed] [Google Scholar]
- 139.Benomar A, Krols L, Stevanin G, Cancel G, LeGuern E, David G, et al. The gene for autosomal dominant cerebellar ataxia with pigmentary macular dystrophy maps to chromosome 3p12-p21.1. Nat Genet. 1995;10(1):84–8. doi: 10.1038/ng0595-84. [DOI] [PubMed] [Google Scholar]
- 140.David G, Abbas N, Stevanin G, Dürr A, Yvert G, Cancel G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17(1):65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- 141.Harding AE. The clinical features and classification of the late onset autosomal dominant cerebellar ataxias. A study of 11 families, including descendants of the “the Drew family of Walworth”. Brain. 1982;105(Pt 1):1–28. doi: 10.1093/brain/105.1.1. [DOI] [PubMed] [Google Scholar]
- 142.Benomar A, Le Guern E, Dürr A, Ouhabi H, Stevanin G, Yahyaoui M, et al. Autosomal-dominant cerebellar ataxia with retinal degeneration (ADCA type II) is genetically different from ADCA type I. Ann Neurol. 1994;35(4):439–444. doi: 10.1002/ana.410350411. [DOI] [PubMed] [Google Scholar]
- 143.David G, Dürr A, Stevanin G, Cancel G, Abbas N, Benomar A, et al. Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7) Hum Mol Genet. 1998;7(2):165–170. doi: 10.1093/hmg/7.2.165. [DOI] [PubMed] [Google Scholar]
- 144.Stevanin G, David G, Dürr A, Giunti P, Benomar A, Abada-Bendib M, et al. Multiple origins of the spinocerebellar ataxia 7 (SCA7) mutation revealed by linkage disequilibrium studies with closely flanking markers, including an intragenic polymorphism (G3145TG/A3145TG) Eur J Hum Genet. 1999;7(8):889–896. doi: 10.1038/sj.ejhg.5200392. [DOI] [PubMed] [Google Scholar]
- 145.Finsterer J, Zarrouk-Mahjoub S, Daruich A. The eye on mitochondrial disorders. J Child Neurol. 2016;31(5):652–662. doi: 10.1177/0883073815599263. [DOI] [PubMed] [Google Scholar]
- 146.Fraser JA, Biousse V, Newman NJ. The neuro-ophthalmology of mitochondrial disease. Surv Ophthalmol. 2010;55(4):299–334. doi: 10.1016/j.survophthal.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Amati-Bonneau P, Milea D, Bonneau D, Chevrollier A, Ferré M, Guillet V, et al. OPA1-associated disorders: phenotypes and pathophysiology. Int J Biochem Cell Biol. 2009;41(10):1855–1865. doi: 10.1016/j.biocel.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 148.Biousse V, Newman NJ. Hereditary optic neuropathies. Ophthalmol Clin North Am. 2001;14(3):547–568. doi: 10.1016/s0896-1549(05)70252-2. [DOI] [PubMed] [Google Scholar]
- 149.Lenaers G, Hamel CP, Delettre C, Amati-Bonneau P, Procaccio V, Bonneau D, et al. Dominant optic atrophy. Orphanet J Rare Dis. 2012;7(1):46. doi: 10.1186/1750-1172-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Milea D, Amati-Bonneau P, Reynier P, Bonneau D. Genetically determined optic neuropathies. Curr Opin Neurol. 2010;23(1):24–28. doi: 10.1097/WCO.0b013e3283347b27. [DOI] [PubMed] [Google Scholar]
- 151.McClelland C, Meyerson C, VanStavern G. Leber hereditary optic neuropathy: current perspectives. OPTH. 2015;1165. [DOI] [PMC free article] [PubMed]
- 152.Yu-Wai-Man P, Griffiths PG, Burke A, Sellar PW, Clarke MP, Gnanaraj L, et al. The prevalence and natural history of dominant optic atrophy due to OPA1 mutations. Ophthalmology. 2010;117(8):1538–46. doi: 10.1016/j.ophtha.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leber Th. Ueber hereditäre und congenital-angelegte Sehnervenleiden. Graefe’s Arhiv für Ophthalmologie. 1871;17(2):249–291. [Google Scholar]
- 154.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242(4884):1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 155.Huoponen K, Lamminen T, Juvonen V, Aula P, Nikoskelainen E, Savontaus M-L. The spectrum of mitochondrial DNA mutations in families with Leber hereditary optic neuroretinopathy. Hum Genet. 1993;92(4):379–384. doi: 10.1007/BF01247339. [DOI] [PubMed] [Google Scholar]
- 156.Kjer P. Infantile optic atrophy with dominant mode of inheritance: a clinical and genetic study of 19 Danish families. Acta Ophthalmol Suppl. 1959;164(Supp 54):1–147. [PubMed] [Google Scholar]
- 157.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26(2):207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 158.Eiberg H, Kjer B, Kjer P, Rosenberg T. Dominant optic atrophy (OPA1) mapped to chromosome 3q region. I. Linkage analysis. Hum Mol Genet. 1994;3(6):977–980. doi: 10.1093/hmg/3.6.977. [DOI] [PubMed] [Google Scholar]
- 159.Chevrollier A, Guillet V, Loiseau D, Gueguen N, deCrescenzo M-AP, Verny C, et al. Hereditary optic neuropathies share a common mitochondrial coupling defect. Ann Neurol. 2008;63(6):794–8. [DOI] [PubMed]
- 160.Del Dotto V, Fogazza M, Lenaers G, Rugolo M, Carelli V, Zanna C. OPA1: How much do we know to approach therapy? Pharmacol Res. 2018;131:199–210. doi: 10.1016/j.phrs.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 161.Olichon A, Landes T, Arnauné-Pelloquin L, Emorine LJ, Mils V, Guichet A, et al. Effects of OPA1 mutations on mitochondrial morphology and apoptosis: relevance to ADOA pathogenesis. J Cell Physiol. 2007;211(2):423–430. doi: 10.1002/jcp.20950. [DOI] [PubMed] [Google Scholar]
- 162.Ferré M, Bonneau D, Milea D, Chevrollier A, Verny C, Dollfus H, et al. Molecular screening of 980 cases of suspected hereditary optic neuropathy with a report on 77 novel OPA1 mutations. Hum Mutat. 2009;30(7):E692–705. doi: 10.1002/humu.21025. [DOI] [PubMed] [Google Scholar]
- 163.Angebault C, Guichet P-O, Talmat-Amar Y, Charif M, Gerber S, Fares-Taie L, et al. Recessive mutations in RTN4IP1 cause isolated and syndromic optic neuropathies. Am J Hum Genet. 2015;97(5):754–760. doi: 10.1016/j.ajhg.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hanein S, Perrault I, Roche O, Gerber S, Khadom N, Rio M, et al. TMEM126A, encoding a mitochondrial protein, is mutated in autosomal-recessive nonsyndromic optic atrophy. Am J Hum Genet. 2009;84(4):493–498. doi: 10.1016/j.ajhg.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hartmann B, Wai T, Hu H, MacVicar T, Musante L, Fischer-Zirnsak B, et al. Homozygous YME1L1 mutation causes mitochondriopathy with optic atrophy and mitochondrial network fragmentation. Elife. 2016;6:5. doi: 10.7554/eLife.16078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Metodiev MD, Gerber S, Hubert L, Delahodde A, Chretien D, Gérard X, et al. Mutations in the tricarboxylic acid cycle enzyme, aconitase 2, cause either isolated or syndromic optic neuropathy with encephalopathy and cerebellar atrophy. J Med Genet. 2014;51(12):834–838. doi: 10.1136/jmedgenet-2014-102532. [DOI] [PubMed] [Google Scholar]
- 167.Barbet F, Gerber S, Hakiki S, Perrault I, Hanein S, Ducroq D, et al. A first locus for isolated autosomal recessive optic atrophy (ROA1) maps to chromosome 8q. Eur J Hum Genet. 2003;11(12):966–971. doi: 10.1038/sj.ejhg.5201070. [DOI] [PubMed] [Google Scholar]
- 168.Chen XJ, Wang X, Kaufman BA, Butow RA. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307(5710):714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 169.Hanein S, Garcia M, Fares-Taie L, Serre V, De Keyzer Y, Delaveau T, et al. TMEM126A is a mitochondrial located mRNA (MLR) protein of the mitochondrial inner membrane. Biochim Biophys Acta. 2013;1830(6):3719–3733. doi: 10.1016/j.bbagen.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 170.Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278(10):7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 171.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143(2):351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chinnery PF, Johnson MA, Wardell TM, Singh-Kler R, Hayes C, Brown DT, et al. The epidemiology of pathogenic mitochondrial DNA mutations. Ann Neurol. 2000;48(2):188–193. [PubMed] [Google Scholar]
- 173.Carelli V, La Morgia C, Iommarini L, Carroccia R, Mattiazzi M, Sangiorgi S, et al. Mitochondrial optic neuropathies: how two genomes may kill the same cell type? Biosci Rep. 2007;27(1–3):173–184. doi: 10.1007/s10540-007-9045-0. [DOI] [PubMed] [Google Scholar]
- 174.Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Prog Retin Eye Res. 2011;30(2):81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Howell N, Bindoff LA, McCullough DA, Kubacka I, Poulton J, Mackey D, et al. Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet. 1991;49(5):939–950. [PMC free article] [PubMed] [Google Scholar]
- 176.Johns DR, Neufeld MJ, Park RD. An ND-6 mitochondrial DNA mutation associated with Leber hereditary optic neuropathy. Biochem Biophys Res Commun. 1992;187(3):1551–1557. doi: 10.1016/0006-291x(92)90479-5. [DOI] [PubMed] [Google Scholar]
- 177.ElHefnawi M, Jeon S, Bhak Y, ElFiky A, Horaiz A, Jun J, et al. Whole genome sequencing and bioinformatics analysis of two Egyptian genomes. Gene. 2018;668:129–134. doi: 10.1016/j.gene.2018.05.048. [DOI] [PubMed] [Google Scholar]
- 178.Carelli V, Ghelli A, Bucchi L, Montagna P, De Negri A, Leuzzi V, et al. Biochemical features of mtDNA 14484 (ND6/M64V) point mutation associated with Leber’s hereditary optic neuropathy. Ann Neurol. 1999;45(3):320–328. [PubMed] [Google Scholar]
- 179.Bouzidi A, Aboussair N, Charif M, Amalou G, Goudenège D, Desquiret-Dumas V, et al. First characterization of LHON pedigrees in North Africa. Eye. 2020;34(11):2138–2139. doi: 10.1038/s41433-019-0755-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yu-Wai-Man P, Griffiths PG, Hudson G, Chinnery PF. Inherited mitochondrial optic neuropathies. J Med Genet. 2009;46(3):145–158. doi: 10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jun AS, Brown MD, Wallace DC. A mitochondrial DNA mutation at nucleotide pair 14459 of the NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc Natl Acad Sci U S A. 1994;91(13):6206–6210. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mahoui S, Belkhamsa O, Ait Kaci I, Abada Bendib M, Castelnovo G. Leber optic hereditary neuropathy plus dystonia. Revue Neurologique. 2019;175(7–8):483–484. doi: 10.1016/j.neurol.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 183.Meyer E, Michaelides M, Tee LJ, Robson AG, Rahman F, Pasha S, et al. Nonsense mutation in TMEM126A causing autosomal recessive optic atrophy and auditory neuropathy. Mol Vis. 2010;13(16):650–664. [PMC free article] [PubMed] [Google Scholar]
- 184.Désir J, Coppieters F, Van Regemorter N, De Baere E, Abramowicz M, Cordonnier M. TMEM126A mutation in a Moroccan family with autosomal recessive optic atrophy. Mol Vis. 2012;18:1849–1857. [PMC free article] [PubMed] [Google Scholar]
- 185.Charif M, Nasca A, Thompson K, Gerber S, Makowski C, Mazaheri N, et al. Neurologic phenotypes associated with mutations in RTN4IP1 (OPA10) in children and young adults. JAMA Neurol. 2018;75(1):105–113. doi: 10.1001/jamaneurol.2017.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Barrett TG, Bundey SE, Macleod AF. Neurodegeneration and diabetes: UK nationwide study of Wolfram (DIDMOAD) syndrome. Lancet. 1995;346(8988):1458–1463. doi: 10.1016/s0140-6736(95)92473-6. [DOI] [PubMed] [Google Scholar]
- 187.Ganie MA, Bhat D. Current developments in Wolfram syndrome. J Pediatr Endocrinol Metab. 2009;22(1):3–10. doi: 10.1515/jpem.2009.22.1.3. [DOI] [PubMed] [Google Scholar]
- 188.Amr S, Heisey C, Zhang M, Xia X-J, Shows KH, Ajlouni K, et al. A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am J Hum Genet. 2007;81(4):673–683. doi: 10.1086/520961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Barrientos A, Casademont J, Saiz A, Cardellach F, Volpini V, Solans A, et al. Autosomal recessive Wolfram syndrome associated with an 8.5-kb mtDNA single deletion. Am J Hum Genet. 1996;58(5):963–70. [PMC free article] [PubMed] [Google Scholar]
- 190.Rötig A, Cormier V, Chatelain P, Francois R, Saudubray JM, Rustin P, et al. Deletion of mitochondrial DNA in a case of early-onset diabetes mellitus, optic atrophy, and deafness (Wolfram syndrome, MIM 222300) J Clin Invest. 1993;91(3):1095–1098. doi: 10.1172/JCI116267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Strom TM, Hörtnagel K, Hofmann S, Gekeler F, Scharfe C, Rabl W, et al. Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet. 1998;7(13):2021–2028. doi: 10.1093/hmg/7.13.2021. [DOI] [PubMed] [Google Scholar]
- 192.Bouslama K, Naoui A, Rezgui L, Goucha S, M’Rad S, Ben Dridi M. [Wolfram syndrome. A new case report]. Tunis Med. 2002;80(11):714–7. [PubMed]
- 193.Dliga Y, Chaabouni H, Largueche S, Bennaceur Z, Ferchiou A. [The Wolfram syndrome. Apropos of a Tunisian case]. Ann Pediatr (Paris). 1985 Dec;32(10):889–92. [PubMed]
- 194.Sayouti A, Benhaddou R, Khoumiri R, Gaboune L, Guelzim H, Benfdil N, et al. Two cases of Wolfram syndrome. J Fr Ophtalmol. 2007;30(6):607–609. doi: 10.1016/s0181-5512(07)89665-3. [DOI] [PubMed] [Google Scholar]
- 195.Gerards M, Sluiter W, van den Bosch BJC, de Wit LEA, Calis CMH, Frentzen M, et al. Defective complex I assembly due to C20orf7 mutations as a new cause of Leigh syndrome. J Med Genet. 2010;47(8):507–512. doi: 10.1136/jmg.2009.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ruhoy IS, Saneto RP. The genetics of Leigh syndrome and its implications for clinical practice and risk management. Appl Clin Genet. 2014;7:221–234. doi: 10.2147/TACG.S46176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Åkebrand R, Andersson S, Seyedi Honarvar AK, Sofou K, Darin N, Tulinius M, et al. Ophthalmological characteristics in children with Leigh syndrome - a long-term follow-up. Acta Ophthalmol. 2016;94(6):609–617. doi: 10.1111/aos.12983. [DOI] [PubMed] [Google Scholar]
- 198.Han J, Lee Y-M, Kim SM, Han SY, Lee JB, Han S-H. Ophthalmological manifestations in patients with Leigh syndrome. Br J Ophthalmol. 2015;99(4):528–535. doi: 10.1136/bjophthalmol-2014-305704. [DOI] [PubMed] [Google Scholar]
- 199.Assouline Z, Jambou M, Rio M, Bole-Feysot C, deLonlay P, Barnerias C, et al. A constant and similar assembly defect of mitochondrial respiratory chain complex I allows rapid identification of NDUFS4 mutations in patients with Leigh syndrome. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2012;1822(6):1062–9. [DOI] [PubMed]
- 200.Gerards M, Kamps R, van Oevelen J, Boesten I, Jongen E, de Koning B, et al. Exome sequencing reveals a novel Moroccan founder mutation in SLC19A3 as a new cause of early-childhood fatal Leigh syndrome. Brain. 2013;136(Pt 3):882–890. doi: 10.1093/brain/awt013. [DOI] [PubMed] [Google Scholar]
- 201.Mkaouar-Rebai E, Chaari W, Younes S, Bousoffara R, Sfar MT, Fakhfakh F. Maternally Inherited Leigh Syndrome: T8993G Mutation in a Tunisian Family. Pediatr Neurol. 2009;40(6):437–442. doi: 10.1016/j.pediatrneurol.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 202.Mkaouar-Rebai E, Chamkha I, Kammoun F, Kammoun T, Aloulou H, Hachicha M, et al. Two new mutations in the MT-TW gene leading to the disruption of the secondary structure of the tRNATrp in patients with Leigh syndrome. Mol Genet Metab. 2009;97(3):179–184. doi: 10.1016/j.ymgme.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 203.Voo I, Allf BE, Udar N, Silva-Garcia R, Vance J, Small KW. Hereditary motor and sensory neuropathy type VI with optic atrophy. Am J Ophthalmol. 2003;136(4):670–677. doi: 10.1016/s0002-9394(03)00390-8. [DOI] [PubMed] [Google Scholar]
- 204.Abrams AJ, Hufnagel RB, Rebelo A, Zanna C, Patel N, Gonzalez MA, et al. Mutations in SLC25A46, encoding a UGO1-like protein, cause an optic atrophy spectrum disorder. Nat Genet. 2015;47(8):926–932. doi: 10.1038/ng.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36(5):449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 206.Paul A, Drecourt A, Petit F, Deguine DD, Vasnier C, Oufadem M, et al. FDXR mutations cause sensorial neuropathies and expand the spectrum of mitochondrial Fe-S-synthesis diseases. Am J Hum Genet. 2017;101(4):630–637. doi: 10.1016/j.ajhg.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Stehling O, Wilbrecht C, Lill R. Mitochondrial iron–sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 208.Heimer G, Kerätär JM, Riley LG, Balasubramaniam S, Eyal E, Pietikäinen LP, et al. MECR mutations cause childhood-onset dystonia and optic atrophy, a mitochondrial fatty acid synthesis disorder. Am J Hum Genet. 2016;99(6):1229–1244. doi: 10.1016/j.ajhg.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14(13):1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 210.Tipton RE, Gorlin RJ. Growth retardation, alopecia, pseudo-anodontia, and optic atrophy–the GAPO syndrome: report of a patient and review of the literature. Am J Med Genet. 1984;19(2):209–216. doi: 10.1002/ajmg.1320190202. [DOI] [PubMed] [Google Scholar]
- 211.Stránecký V, Hoischen A, Hartmannová H, Zaki MS, Chaudhary A, Zudaire E, et al. Mutations in ANTXR1 Cause GAPO syndrome. Am J Hum Genet. 2013;92(5):792–799. doi: 10.1016/j.ajhg.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gregory A, Polster BJ, Hayflick SJ. Clinical and genetic delineation of neurodegeneration with brain iron accumulation. J Med Genet. 2009;46(2):73–80. doi: 10.1136/jmg.2008.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kurian MA, Morgan NV, MacPherson L, Foster K, Peake D, Gupta R, et al. Phenotypic spectrum of neurodegeneration associated with mutations in the PLA2G6 gene (PLAN) Neurology. 2008;70(18):1623–1629. doi: 10.1212/01.wnl.0000310986.48286.8e. [DOI] [PubMed] [Google Scholar]
- 214.Romani M, Kraoua I, Micalizzi A, Klaa H, Benrhouma H, Drissi C, et al. Infantile and childhood onset PLA2G6 -associated neurodegeneration in a large North African cohort. Eur J Neurol. 2015;22(1):178–186. doi: 10.1111/ene.12552. [DOI] [PubMed] [Google Scholar]
- 215.Warburg M, Sjö O, Fledelius HC, Pedersen SA. Autosomal recessive microcephaly, microcornea, congenital cataract, mental retardation, optic atrophy, and hypogenitalism. Micro syndrome Am J Dis Child. 1993;147(12):1309–1312. doi: 10.1001/archpedi.1993.02160360051017. [DOI] [PubMed] [Google Scholar]
- 216.Abdel-Hamid MS, Abdel-Ghafar SF, Ismail SR, Desouky LM, Issa MY, Effat LK, et al. Micro and Martsolf syndromes in 34 new patients: Refining the phenotypic spectrum and further molecular insights. Clin Genet. 2020;98(5):445–456. doi: 10.1111/cge.13825. [DOI] [PubMed] [Google Scholar]
- 217.Abdel-Salam GMH, Hassan NA, Kayed HF, Aligianis IA. Phenotypic variability in Micro syndrome: report of new cases. Genet Couns. 2007;18(4):423–435. [PubMed] [Google Scholar]
- 218.Aligianis IA, Johnson CA, Gissen P, Chen D, Hampshire D, Hoffmann K, et al. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat Genet. 2005;37(3):221–223. doi: 10.1038/ng1517. [DOI] [PubMed] [Google Scholar]
- 219.Handley MT, Morris-Rosendahl DJ, Brown S, Macdonald F, Hardy C, Bem D, et al. Mutation spectrum in RAB3GAP1, RAB3GAP2, and RAB18 and genotype-phenotype correlations in warburg micro syndrome and Martsolf syndrome. Hum Mutat. 2013;34(5):686–696. doi: 10.1002/humu.22296. [DOI] [PubMed] [Google Scholar]
- 220.Bocquet B, Lacroux A, Surget M-O, Baudoin C, Marquette V, Manes G, et al. Relative frequencies of inherited retinal dystrophies and optic neuropathies in Southern France: assessment of 21-year data management. Ophthalmic Epidemiol. 2013;20(1):13–25. doi: 10.3109/09286586.2012.737890. [DOI] [PubMed] [Google Scholar]
- 221.Stone EM, Andorf JL, Whitmore SS, DeLuca AP, Giacalone JC, Streb LM, et al. Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology. 2017;124(9):1314–1331. doi: 10.1016/j.ophtha.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Charif M, Bris C, Goudenège D, Desquiret-Dumas V, Colin E, Ziegler A, et al. Use of next-generation sequencing for the molecular diagnosis of 1,102 patients with a autosomal optic Neuropathy. Front Neurol. 2021;25(12):602979. doi: 10.3389/fneur.2021.602979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Keywords used in the medical Pubmed and Scopus databases.
Additional file 2: Phenotypic and mutational spectrum of 194 North African families with non-syndromic inherited retinal dystrophies.
Additional file 3: Phenotypic and mutational spectrum of 143 North African families with syndromic inherited retinal dystrophies.
Additional file 4: Mutational spectrum of North African families with non-syndromic inherited optic neuropathies.
Additional file 5: Phenotypic and mutational spectrum of North African families with syndromic inherited optic neuropathies.
Additional file 6: Mutational spectrum of North African families with hereditary mitochondrial diseases with retinal or optic nerve manifestations.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary files.