Abstract
Bone is the most frequent site of prostate cancer (PCa) metastasis; however, little is known about the role of the most common cell in bone, the osteocyte (OCy), in cancer biology. In this study we explored the crosstalk between PCa cells and OCys to determine if it contributes to PCa progression. PCa cells induced OCys to promote PCa proliferation, migration and invasion. A chemokine screen revealed that PCa cell induced OCys to produce growth-derived factor 15 (GDF15). Knockdown of GDF15 in OCys demonstrated that PCa cells conferred the ability on OCys to promote PCa proliferation, migration and invasion through GDF15. Consistent with this finding was the observation that the GDF15 receptor, GFRAL, was expressed on multiple PCa cell lines. Transcription factor array screening of PCa cells exposed to OCys with or without knockdown of GDF15 revealed that GDF15 in OCys promoted early growth response 1 (EGR1) expression in the PCa cells. Knockdown of EGR1 expression in PCa cells revealed it was required for the OCy-derived GDF15-mediated induction of in vitro PCa cell proliferation, migration and invasion. Subcutaneous co-injection of PCa cells and OCys into mice revealed that OCys promoted tumor growth in vivo, which was diminished by knockdown of GDF15 in the OCys. Knockdown of GDF15 in the tibiae diminished growth of PCa cancer cells injected into the tibiae, which was accompanied by decreased tumor cell proliferation and EGR1 expression. These results shed light on a novel mechanism through which PCa cell educate OCys to promote progression of PCa bone metastasis. They also suggest that targeting of GDF15-based and EGR1-based signaling pathways should be further explored for their potential to diminish progression of PCa bone metastasis.
Keywords: bone metastasis, prostate cancer, osteocyte, growth-derived factor, chemokine
Introduction
Prostate cancer is the most common cancer and the second leading cause of cancer-related death of men in the United States1,2. The high mortality rate is primarily due to the metastases to many tissues including bone3,4. Bone is the most common site of prostate cancer metastasis with over 80% of patients developing incurable bone metastasis5. The mechanisms through which prostate cancers favor metastasis to bone are not clearly delineated; however, they depend on a complex interplay between prostate cancer cells and the bone microenvironment6,7. Bone-derived cell types promote prostate cancer metastasis through secreting factors that directly promote the prostate cancer progression8,9.
Osteoblasts, osteoclasts and osteocytes are the major bone cells that orchestrate bone modeling and remodeling. Osteocytes, derived from osteoblasts, comprise approximately 90% of the cells found in bone tissue and coordinate the responses of osteoblasts and osteoclasts, thus playing a role as a central regulator of bone resorption and formation10,11. Osteocytes, within mineralized bone, are connected, via long cellular extensions and gap junctions, to each other and with cells on the bone surface such as osteoclasts and osteoblasts to form the osteocyte network. This network allows direct cell-to-cell contact between osteocytes and cells near bone surface and distributes osteocyte-secreted factors that regulate bone formation and resorption10. Although knowledge of the role of osteocytes and the osteocytic network in bone formation and common skeletal diseases has a solid foundation, the role of osteocytes in bone metastasis is just beginning to be defined. Osteocyte conditioned media (CM) suppressed breast cancer growth and bone metastasis12; whereas, tumor-induced pressure stimulated osteocytes to produce factor that promoted prostate cancer growth and metastasis9. These findings demonstrate that osteocytes can impact progression of bone metastasis and thus suggest that continuing to develop an understanding of the role of osteocytes in metastatic progression is warranted.
While we previously demonstrated that physical forces generated by tumors promotes prostate cancer progression through modifying osteocyte activity9, it is currently unknown if prostate cancer cells secrete factors that can modify osteocytes to impact prostate cancer progression. Accordingly, we explored if prostate cancer cells alter the bone microenvironment to stimulate osteocytes to feedback on prostate cancer cells and alter their functionality.
Results
Prostate cancer cells produce soluble factors that educate osteocytes to promote prostate cancer growth, migration and invasion.
To determine if prostate cancer cells alter osteocytes’ ability to modify prostate cancer activity, MLO-Y4 osteocytes were treated with conditioned-media (CM) from C4-2B prostate cancer cells. While neither C4-2B CM alone nor MLO-Y4 CM alone impacted C4-2B proliferation, CM from MLO-Y4 osteocytes pre-treated with C4-2B CM induced C4-2B cell proliferation (Fig. 1A). To determine if the ability of C4-2B CM to modify MLO-Y4 cells ability to promote prostate cancer growth was unique to C4-2B cells the experiments were repeated using CM from the PC3 cell line. Similar to the C4-2B CM, PC3 CM induced MLO-Y4 cells to produce CM that promoted C4-2B cell proliferation; whereas, neither PC3 CM alone nor MLO-Y4 CM alone impacted C4-2B proliferation (Fig. 1B). As prostate cancer has a predilection to metastasize to bone and osteocytes are the most abundant cell within bone, it was next explored if the prostate cancer-induced modulation of MLO-Y4 osteocytes extended to prostate cancer cell migration and invasion and thus potentially promote bone metastasis. Accordingly, C4-2B cells were treated with CM from C4-2B-CM-treated MLO-Y4 cells and their migratory and invasive abilities were measured. C4-2B CM alone had no impact on migration or invasion (Fig. 1C). In contrast, MLO-Y4 CM alone markedly increased migration and increased invasion to a lesser extent. However, CM from C4-2B CM-treated MLO-Y4 cells induced a small increase of migration; whereas, it induced a marked increase in invasion compared to the CM of untreated MLO-Y4 cells (Fig. 1C). To determine if this effect was specific to C4-2B CM it was next evaluated if a different prostate cancer cell line could induce MLO-Y4 cells to promote prostate cancer migration and invasion. Accordingly, MLO-Y4 osteocytes were incubated with PC3 CM which resulted in similar induction of migration and invasion as the C4-2B CM (Fig 1D) indicating a broad ability of prostate cancer cells to promote MLO-Y4 cells to stimulate migration and invasion. To further determine if the ability to educate MLO-Y4 cells to promote prostate cancer proliferation, migration and invasion extended to other target prostate cancer cells, the series of experiments were repeated using PC3 cells as the target cells. Similar to the previous findings on induction of C4-2B cell proliferation, migration and invasion, both PC3 CM and C4-2B CM induced MLO-Y4 cells to produce CM that promoted PC3 cell proliferation, migration and invasion (Sup. Fig. 1). To ensure these effects were not unique to MLO-Y4 osteocytes, the experiments were repeated with primary murine osteocytes. As observed in the previous experiments, C4-2B CM induced osteocytes to promote both C4-2B proliferation, invasion and migration (Sup. Fig. 2). In contrast to the MLO-Y4 osteocytes; however, the CM from primary murine osteocytes that had not been exposed to prostate cancer cells did not promote migration or invasion, which underscores the impact of prostate cancer-mediated activation of osteocytes towards promoting pro-metastatic features. Taken together, these results demonstrate that prostate cancer cells can educate osteocytes to produce a soluble factor that promotes prostate cancer growth, migration and invasion.
Figure 1. Prostate cancer educates MLO-Y4 osteocytes to promote C4-2B prostate cancer cell proliferation, migration and invasion.
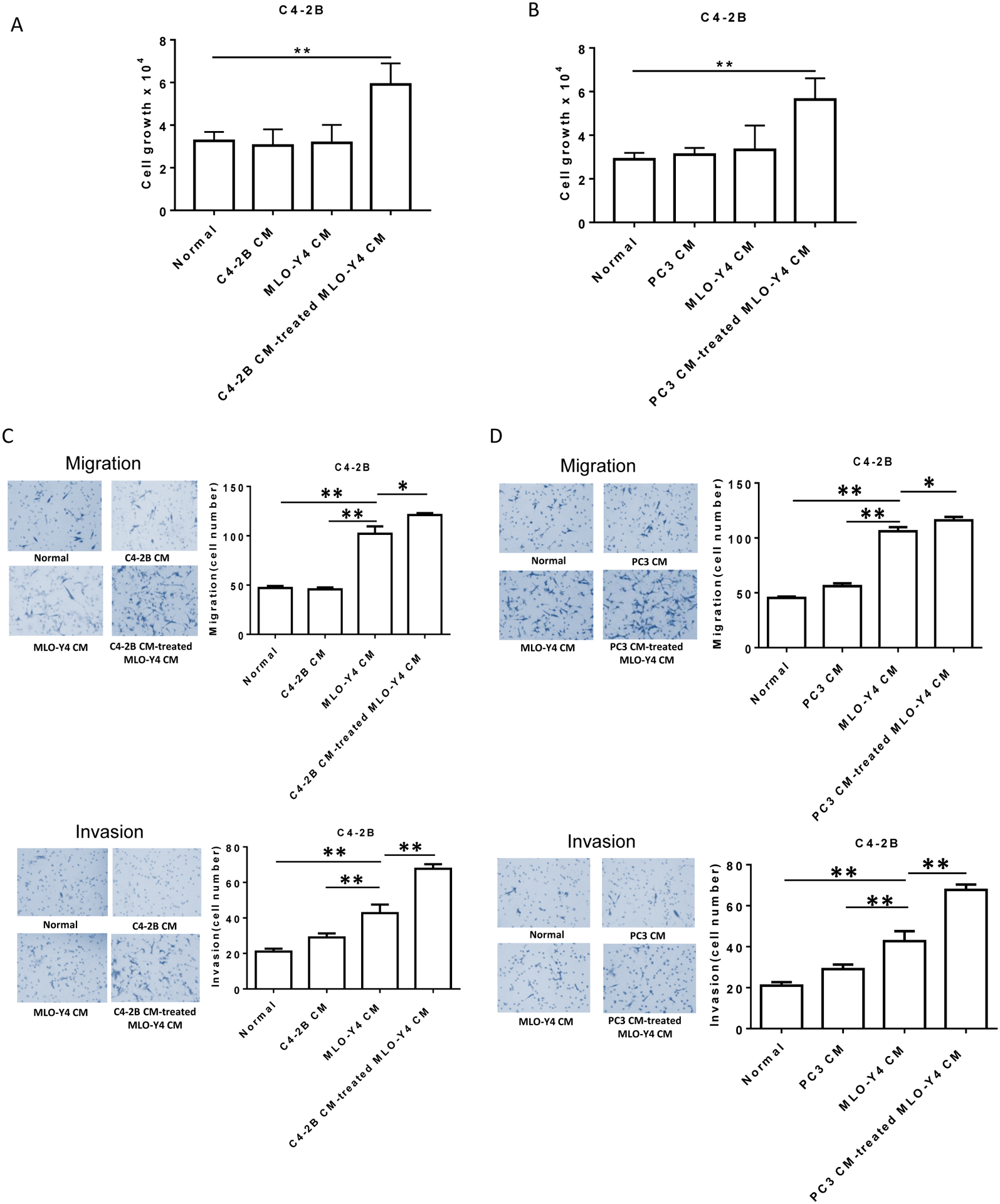
Conditioned-media (CM) from both C4-2B and PC3 prostate cancer cells was made as described in methods. CM was added as described to a final concentration of 50%. (A). Plain media, C4-2B CM, MLO-Y4 CM and C4-2B CM-treated MLO-Y4 cell CM were added to C4-2B cells (1.5×104 per well). After 48 hours, cell numbers were counted using a hemocytometer. (B). Plain media, PC3 CM, MLO-Y4 CM and PC3 CM-treated MLO-Y4 cell CM were added to C4-2B cells (1.5×104 per well). After 48 hours, cell numbers were counted using a hemocytometer. (C). Migration (upper figures) and invasion (lower figures) were assessed using a transwell assay. C4-2B cells (1.5×105 per cells) were treated with plain media, C4-2B CM, MLO-Y4 CM and C4-2B CM-treated MLO-Y4 cell CM for 24 hours. The membrane was stained using differential Quick staining kit and photographed under light microscopy (20x). The numbers of migrating (no Matrigel on membrane) and invading (Matrigel present on membrane) cells were counted in five random fields for each insert. (D). Migration (upper figures) and invasion (lower figures) were assessed using a transwell assay. C4-2B cells (1.5×105 per cells) were treated with plain media, PC3 CM, MLO-Y4 CM and PC3 CM-treated MLO-Y4 cell CM for 24 hours. The membrane was stained using differential Quick staining kit and photographed under light microscopy (20x). The numbers of migrating (no Matrigel on membrane) and invading (Matrigel present on membrane) cells were counted in five random fields for each insert. Data are shown as the mean±SD of 3 independent experiments. * P < 0.05; ** P < 0.01.
Prostate cancer cells induce MLO-Y4 cells to secrete growth differentiation factor 15 (GDF15).
A variety of cytokines can modulate prostate cancer activity. To determine if prostate cancer cells modify the ability of osteocytes to produce cytokines, MLO-Y4 cells were treated with CM from C4-2B cells or vehicle control then after 24 hours total protein from both the MLO-Y4 cells themselves and from their CM were harvested and subjected to analysis on a cytokine PCR array. In order to evaluate for a consistent induction of cytokine production by different prostate cancer cells, the experiment was repeated using VCaP prostate cancer cell line CM as the stimulus for MLO-Y4 cells. Both C4-2B CM and VCaP CM prostate cancer CMs induced a variety of cytokines from the MLO-Y4 osteocytes; however, Endoglin and GDF15 were the only two cytokines that were consistently produced by MLO-Y4 cells stimulated by both C4-2B and VCaP prostate CMs (Fig. 2A–C) which led us to focus on these two cytokines. To validate these findings, MLO-Y4 cells were incubated with several different prostate cancer CMs and the Endoglin and GDF15 production by the MLO-Y4 cells was measured. The prostate cancer CM had no impact on Endoglin mRNA production by the MLO-Y4 cells (Fig 2D). Unfortunately, a reliable antibody to detect murine Endoglin could not be identified, thus Endoglin protein was not measured. In contrast to no effect on Endoglin mRNA expression, prostate cancer CM induced GDF15 protein and mRNA production by the MLO-Y4 cells (Fig. 2E left upper immunoblot and left lower graph, respectively). This appeared as an overall increase in both the pro-GDF15 and mature GDF15 forms. Furthermore, to determine if the increase of GDF15 production resulted in increased GDF15 release from the MLO-Y4 cells, we performed an immunoblot and ELISA of the CM. PCa CM induced MLO-Y4 cells to produce an increase of the mature form, but not pro-form of GDF15 in the supernatant (Fig. 2E right upper immunoblot). The levels of GDF15 varied among the PCa cell lines used to simulate the MLO-Y4 cells, but were all above 1.2 ng/ml (Fig. 2E right lower graph). Based on the lack of Endoglin mRNA induction; and the positive induction of GDF15 mRNA and protein expression, the remaining portion of the study was focused on GDF15.
Figure 2. Prostate cancer induces GDF15 expression in osteocytes.
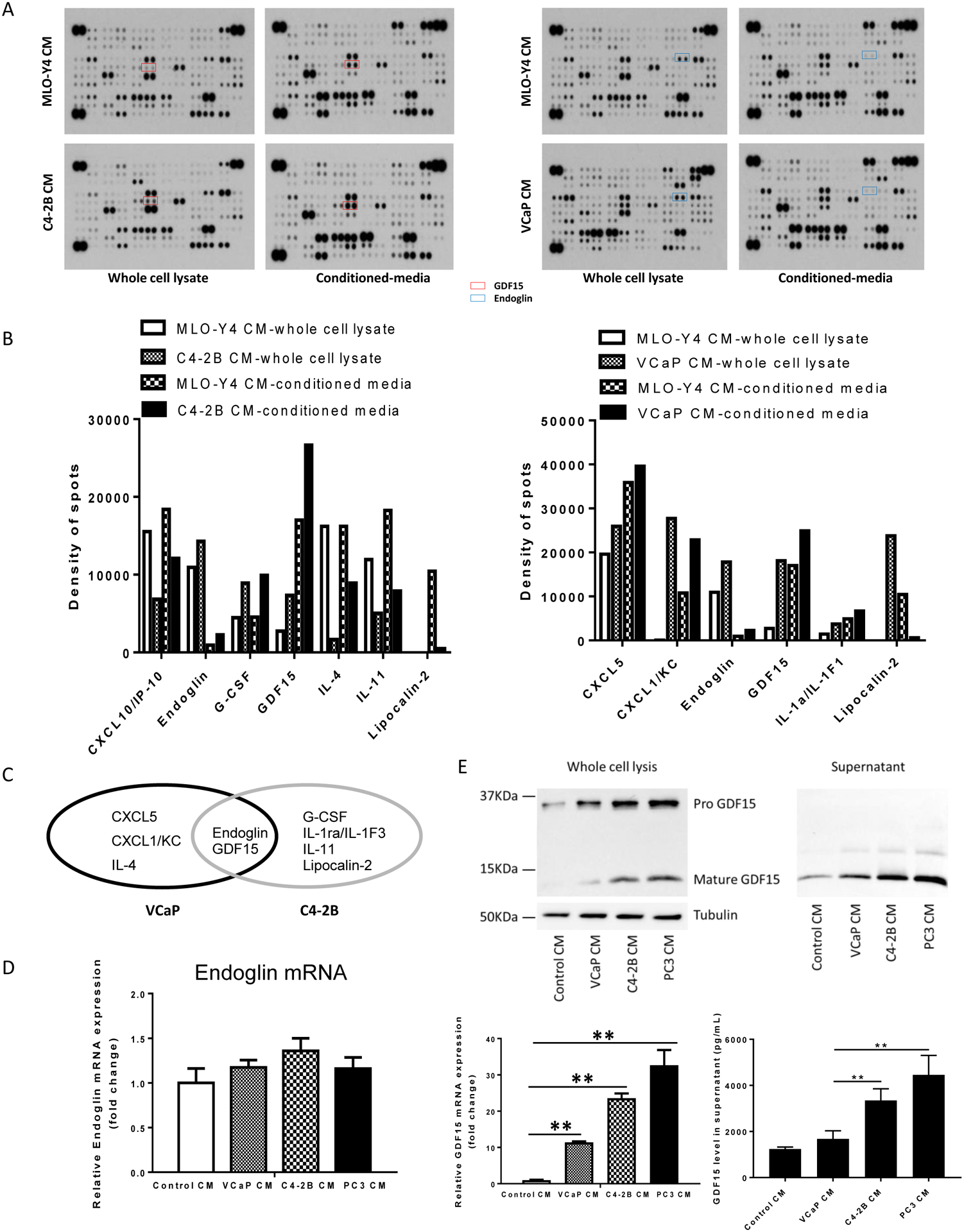
(A). MLO-Y4 cells were treated with C4-2B conditioned-media (CM) (Left), VCaP CM (Right) or MLO-Y4 CM as control for 24 hours. CM and whole cell lysates were collected and subjected to cytokine array. (B). Spots were subjected to densitometry using Image J. Results are shown as change in optical density of the cells subjected to prostate cancer CM relative to Control CM. (C). Ovals indicate the differentially regulated proteins. The overlap indicates the cytokines regulated similarly in both VCaP and C4-2B cells. (D). MLO-Y4 cells treated with MLO-Y4 CM, VCaP CM, C4-2B CM or PC3 CM. After 24 hours, total RNA was collected and subjected to RT-PCR for endoglin mRNA. (E). MLO-Y4 cells treated with MLO-Y4 CM, VCaP CM, C4-2B CM or PC3 CM. After 24 hours, total RNA and proteins was collected and subjected to immunoblot for GDF15 protein (upper left figure) and RT-PCR for GDF15 mRNA (lower left graph). After 24 hours of cell culture, supernatants were collected and subjected to immunoblot (upper right figure) and ELISA (lower right graph) for GDF15 protein. Data are shown as the mean±SD of 3 independent experiments. ** P < 0.01.
GDF15 promotes prostate cancer migration and invasion.
To evaluate the ability of GDF15 to recapitulate the ability of prostate cancer-CM-induced activation of MLO-Y4 cells to promote prostate cancer proliferation, migration and invasion recombinant GDF15 (rGDF15) was evaluated on its ability to promote these activities in prostate cancer cells. For these experiments, we used rGDF15 in the range of 100pg/ml to 4000pg/ml to capture the range of low normal levels of GDF1513 and the higher pathological levels of GDF15 we observed in the MLO-Y4 supernatants (Fig. 2E, right lower graph). GDF15 induced migration and invasion of both C4-2B and PC3 prostate cancer cells (Fig. 3A and Sup. Fig 3A, respectively). It also stimulated cell growth of both C4-2B and PC3 cells (Fig. 3B and Sup Fig. 3B, respectively). This result was consistent with the increased GDF15 in the MLO-Y4 CM as contributing to prostate cancer migration and invasion. To further determine if education of MLO-Y4 cells mediated its prostate cancer stimulatory ability through GDF15, shRNA targeting GDF15 was used to knockdown GDF15 in the MLO-Y4 cells and a scrambled shRNA was used as control. The GDF15 shRNA reduced GDF15 protein and mRNA expression (Fig. 3C left immunoblot and middle graph, respectively) which translated into a >80% decline of GDF15 in the MLO-Y4 supernatant (Fig. 3C right graph). The impact of GDF15 knockdown in MLO-Y4 was evaluated on the ability of prostate cancer CM to simulate MLO-Y4 to promote prostate cancer proliferation (as observed in Fig. 1A and B). Both C4-2B CM (Fig. 3D upper) and PC3 CM (Suppl. Fig 4 upper) stimulated shControl MLO-Y4 osteocytes to produce CM that promoted C4-2B proliferation. The results were similar for stimulating proliferation of PC3 cells (Sup. Fig. 3C). However, knockdown of GDF15 in the MLO-Y4 (MLO-Y4-shGDF15) abrogated the ability of C4-2B CM and PC3 CM to activate MLO-Y4 osteocytes to induce C4-2B proliferation (Fig. 3D upper and Sup. Fig. 4 upper) and PC3 proliferation (Sup. Fig. 3C). Furthermore, addition of rGDF-15 rescued the ability of the prostate cancer CM-treated MLO-Y4-shGDF15 CM to stimulate C4-2B proliferation (Fig. 3D upper and Sup. Fig. 4 upper) and PC3 proliferation (Sup. Fig. 3C). To determine if these results were due to GDF in the CM produced by MLO-Y4 cells, we treated the prostate cancer-treated MLO-Y4-derived CM with anti-GDF15 antibody prior to treating the prostate cancer cells. The antibody inhibited the ability of both C4-2B-stimulated MLO-Y4 CM (Fig. 3D lower) and PC-3-stimulated MLO-Y4 CM (Sup. Fig. 4 lower) to stimulate C4-2B growth. These results indicate that prostate cancer-induced osteocyte-derived GDF 15 expression contributes to prostate cancer proliferation..
Figure 3. GDF15 promotes proliferation, migration and invasion of C4-2B cells.
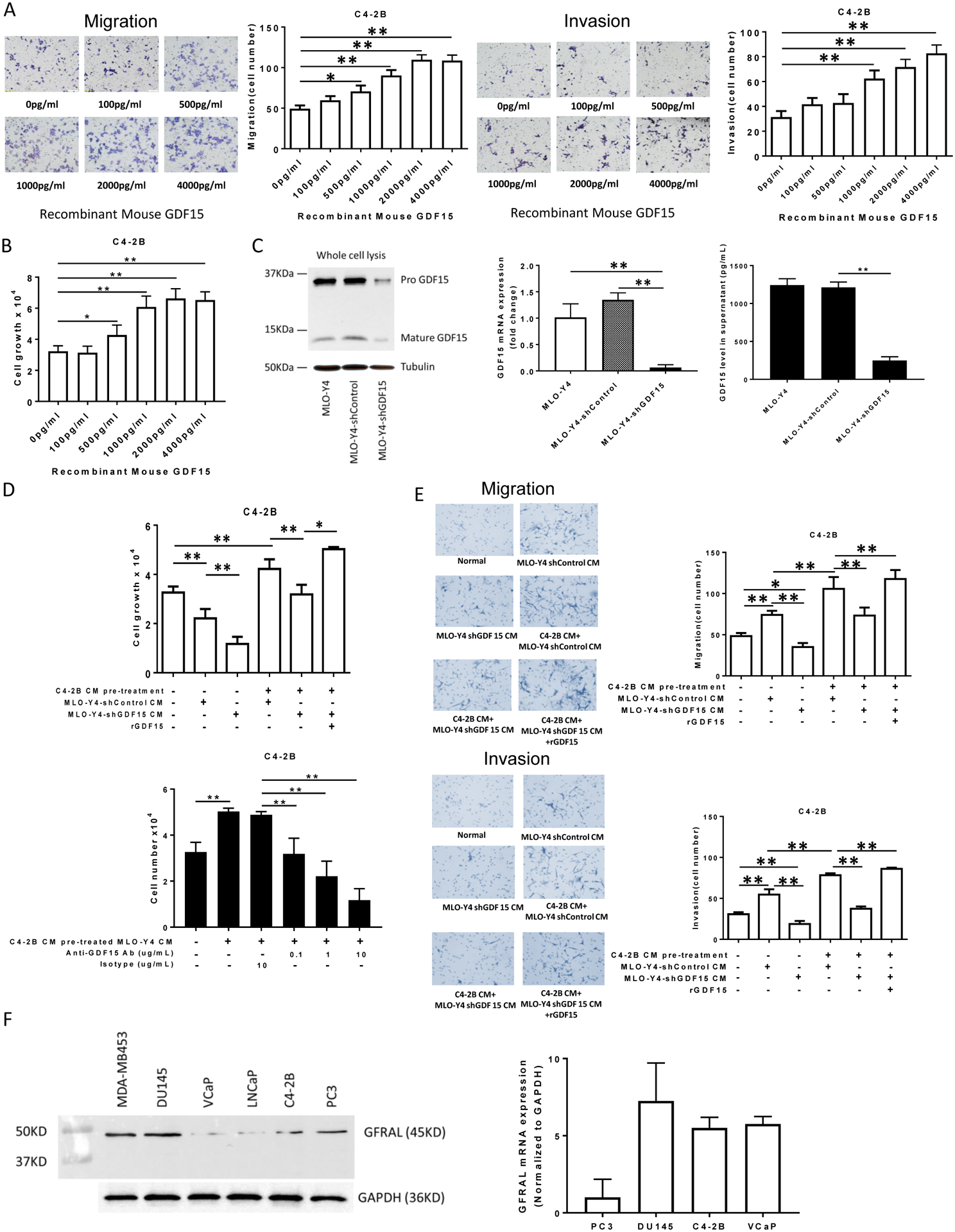
(A). Migration (left figure) and invasion (right figure) were assessed using a transwell assay. C4-2B cells (1.5×105 per cells) were treated with the indicated concentrations of rGDF15 for 24 hours. The membrane was stained using differential Quick staining kit and photographed under light microscopy (20x). The numbers of migrating (no Matrigel on membrane) and invading (Matrigel present on membrane) cells were counted in five random fields for each insert. (B). C4-2B cells (1.5×104 per well) were treated with the indicated concentrations of rGDF15 for 48 hours. Cell numbers were quantified using a hemocytometer. (C). Validation of GDF15 knockdown in MLO-Y4 cells: Total cell protein and RNA were collected and subject to immunoblot (left figure) and real-time PCR (middle graph), respectively. Supernatant was collected from the indicated cells after 24 hours of incubation and subjected to ELISA for GDF15 (right graph). (D). Upper Figure: C4-2B cells (1.5×104 per well) were treated with the indicated CM from the genetically-modified MLO-Y4 cells. In some cases, rGDF15 (500 pg/ml) was added as indicated. After 48 hours, cell numbers were quantified using a hemocytometer. Lower Figure: C4-2B cells (1.5×104 per well) were treated with the indicated CM with the addition of either anti-GDF15 antibody or isotype antibody from the genetically-modified MLO-Y4 cells. After 48 hours, cell numbers were quantified using a hemocytometer. (E). Migration (upper figures) and invasion (lower figures) were assessed using a transwell assay. C4-2B cells (1.5×105 per cells) were treated with CM from untreated or C4-2B CM pre-treated MLO-Y4 cells with knockdown (or control) of GDF15. In some cases, rGDF15 was added to the CM as indicated. After 24 hours, the membrane was stained using differential Quick staining kit and photographed under light microscopy (20x). The numbers of migrating (no Matrigel on membrane) and invading (Matrigel present on membrane) cells were counted in five random fields for each insert. Data are shown as the mean±SD of 3 independent experiments. * P < 0.05; ** P < 0.01. (F). Total protein and RNA from the indicated cell lines were subjected to immunoblot (left figure) and real time PCR (right graph) for GFRAL. The PCR data are shown as the mean±SD of 3 independent experiments.
To explore if prostate cancer’s ability to induce osteocytes to promote prostate cancer migration and invasion requires GDF15, C4-2B CM was used to activate MLO-Y4 cells with GDF15 knockdown (or scrambled control). In the absence of C4-2B CM pre-treatment, MLO-Y4-shControl cell CM induced migration and invasion, which was abrogated by GDF15 knockdown (MLO-Y4-shGDF15 CM) (Fig. 3E). In the presence of C4-2B CM pre-treatment, MLO-Y4-shControl CM induced a further increase in migration and invasion then untreated MLO-Y4-shControl cell CM which is consistent with the ability of prostate cancer cell pretreatment to enhance osteocyte’s ability to promote prostate cancer migration and invasion. However, this activity was diminished by knockdown of GDF15 (MLO-Y4-shGDF15 CM) (Fig. 3E). Addition of rGDF15 rescued the GDF15 knockdown loss of migration and invasion (Fig. 3E). Similar findings were observed when PC3 CM was used to activate the MLO-Y4 cells (Sup. Fig. 5). Furthermore, similar results were observed when PC3 cells were used as the target cells for evaluation of knockdown of GDF15 in MLO-Y4 cells (Sup. Figs. 3D–E). Specifically, knockdown of GDF15 in MLO-Y4 cells reduced the ability of PC3 CM and C4-2B CM to activated MLO-Y4 cells to stimulated PC3 migration and invasion. Similarly, exogenous GDF15 rescued both invasion and migration of PC3 cells. To provide further evidence that appropriate receptor for GDF15 was present prostate cancer cells, we evaluated for expression of the GDF receptor, GDNF Family Receptor α-like (GFRAL)13,14, expression in the prostate cancer cells. All cell lines tested expressed both GFRAL protein and mRNA expression (Fig. 3F).Taken together, these results indicate that prostate cancer cells induce a positive feedback cycle that promotes their own invasion and migration in the bone, in part, through induction of GDF15 production from osteocytes.
GDF15 upregulates EGR1 expression in prostate cancer cells.
To determine downstream targets of osteocyte-derived GDF15 on prostate cancer cells, PC3 and C4-2B cells were incubated with either C4-2B CM-treated MLO-Y4-shControl CM or C4-2B CM-treated MLO-Y4-shGDF15 CM and then the prostate cancer cells’ total protein and RNA was evaluated on a transcription factor antibody array (Fig. 4A). Knockdown of GDF15 in the MLO-Y4 cells resulted in their CM altering a variety of transcription factors in each prostate cancer cell line compared to CM from MLO-Y4 cells with intact GDF15 (Fig. 4A). However, when comparing both PC3 and C4-2B prostate cancer cell line, knockdown of GDF15 in MLO-Y4 resulted in decreased early growth response 1 (EGR1) and increased peroxisome proliferator activated receptor (PPAR) and activating transcription factor 2/3/4 (ATF2/3/4) expression in both prostate cancer cell lines (Fig. 4A) suggesting a consistent effect among prostate cancers. These results indicated that MLO-Y4-derived GDF15 promotes EGR1 expression and inhibits PPAR and ATF2/3/4 expression in prostate cancer cells. To validate these results, C4-2B and PC3 cells were incubated with C4-2B-treated MLO-Y4-shControl and ML-Y4-shGDF CMs, followed by measurement of target mRNA and protein. The changes in ATF2/3/4 and PPAR were not consistent (not shown); however, knockdown of GDF15 in MLO-Y4 cells resulted in them producing a CM that decreased EGR1 mRNA and protein expression in both prostate cancer cells lines compared to CM from MLO-Y4 cells with wildtype levels of GDF15 (Fig. 4B). These results validate the array findings and confirm that PC3 CM-treated MLO-Y4 CM requires GDF15 to induce EGR1 expression in prostate cancer cells.
Figure 4. GDF15 up-regulates EGR1 in prostate cancer cells.
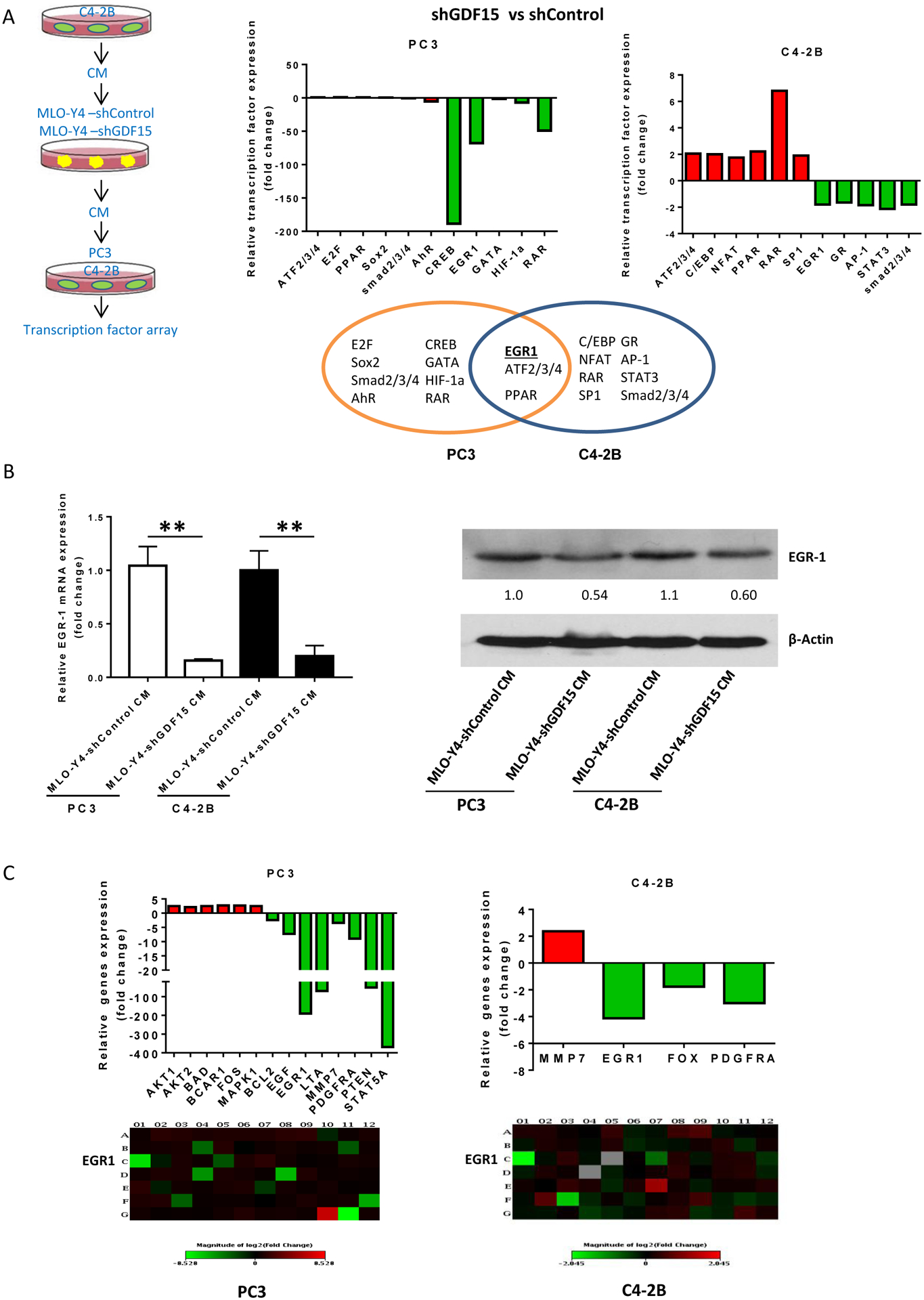
(A). PC3 and C4-2B cells (4×104 per well) cells were plated onto a Cignal transcription factor array that contains 45 different transcription factor reporter genes and reverse transfected with the reporters. 24 hours post-transfections, the media was changed and replaced with MLO-Y4-shControl CM or MLO-Y4-shGDF15 CM and 12 hours later reporter activity was measured. Results are expressed as fold change of expression in MLO-Y4-shGDF15 compared to that of MLO-Y4-shControl. Venn diagram shows the transcription factors differentially expressed in each cell line and the overlap indicates the genes differentially expressed in both PC3 and C4-2B cells. (B). Validation of EGR1 mRNA (left) and protein expression (right) in PC3 and C4-2B cells treated with MLO-Y4-shControl CM or MLO-Y4-shGDF15 CM. Numbers below the EGR-1 immunoblot indicate the EGR1 band density relative to MLO-Y4-shControl CM-treated cells. (C). Cells were treated as in describe in (A) and total RNA was collected and subject to analysis using the RT2 Profiler™ PCR Array Human EGF / PDGF Signaling Pathway Kit. Results are shown as mRNA level MLO-Y4-shGDF15 CM-treated relative to MLO-Y4-shControl CM-treated PC3 (left) or C4-2B (right) cells. A map that indicates what signaling molecule is present in each row and column of the array is presented in Supplemental Figure 6.
EGR1 is induced in response to serum stimulation of quiescent or growing cells15,16 and acts as a tumor suppressor in multiple cancers; however, paradoxically in prostate cancer it is oncogenic17–21. Thus, the role of EGR1 in osteocyte-mediated activation of prostate cancer cell progression was further explored. Many genes that EGR1 targets belong to the epidermal growth factor receptor EGFR signaling pathway22,23. Accordingly, since prostate cancer cells induce osteocytes to produce GDF15 that induces EGR1 in prostate cancer cells, it was explored if the prostate cancer cells stimulate osteocytes to also induce EGFR targets in prostate cancer cells through GDF15. To accomplish this, an EGF signaling pathway-focused transcription factor reporter array, consisting of a library of various response elements driving luciferase was used (A map of the array is presented in Sup. Fig. 6). PC3 and C4-2B cells were transfected with the library of reporters and treated with PC3 CM-treated MLO-Y4-shGDF15 CM or MLO-Y4-shControl and the reporter activity was compared. Consistent with the previous findings, knockdown of GDF15 resulted in diminished EGR1 expression in both PC3 and C4-2B cells (Fig. 4C). Additionally, while several targets were differentially expressed (≥2-fold) in the two prostate cancer cell lines due to knockdown of GDF15 in the MLO-Y4 cells, only one, PDGFRA, was decreased in both cell lines secondary to knockdown of GDF15 in the MLO-Y4 cells. Taken together, these findings indicate that PDFGRA is a consistent change among GDF15-mediated activity in prostate cancer cells, but also indicates that there are differential effects among different prostate cancer cell lines.
Osteocytes promote prostate cancer progression through EGR1 in prostate cancer cells.
To determine if EGR1 functionally contributes to the ability of osteocytes to promote prostate cancer proliferation, migration and invasion, EGR1 was knocked down in C4-2B prostate cancer cells (Fig. 5A) and then the cells were evaluated for their responses to prostate cancer CM-activated osteocytes (Note that the prostate cancer cells to stimulate the MLO-Y4 osteocytes has intact EGR1; only the target prostate cancer cells had knockdown of EGR1). Decreased EGR1 expression in the prostate cancer cells resulted in diminished proliferation (Fig. 5B) and migration and invasion (Fig. 5C) of C4-2B cells in response to C4-2B cell CM-treated MLO-Y4 CM. The results were similar for proliferation (Fig. 5B), migration and invasion (Fig. 5D) when MLO-Y4 cells were treated with PC3 CM. To determine if these results extended to an additional prostate cancer cell lines, we repeated the experiments using PC-3 cells as targets of the osteocytes. Similar results were observed when EGR1 was knocked down in PC3 prostate cancer cells (Sup. Fig. 7A). Specifically, knockdown of EGR1 in PC3 cells diminished their proliferative, migratory and invasive response to both C4-2B CM-treated and PC3 CM-treated MLO-Y4 cells (Sup. Fig. 7B–D). To determine if this result extended beyond MLO-Y4 osteocytes, primary murine osteocytes were treated with C4-2B CM and the treated murine osteocyte CM was evaluated for its ability to modulate C4-2B cells with knockdown of EGR1. Similar to the MLO-Y4 CM, C4-2B CM-treated primary murine osteocyte CM stimulated proliferation, migration and invasion of siControl-treated C4-2B cells; however, the C4-2B CM-treated primary murine osteocyte CM was not able to stimulate these activities in C4-2B cells with knockdown of EGR1 (Sup. Fig. 8). Taken together, these results demonstrate that prostate cancer cells stimulate osteocytes to produce soluble factors, including GDF15, that promote prostate cancer proliferation, migration and invasion through EGR1 in the target prostate cancer cells.
Figure 5. Prostate cancer-activated MLO-Y4 cells promote C4-2B prostate cancer cell proliferation, migration and invasion through EGR1 in the prostate cancer cells.
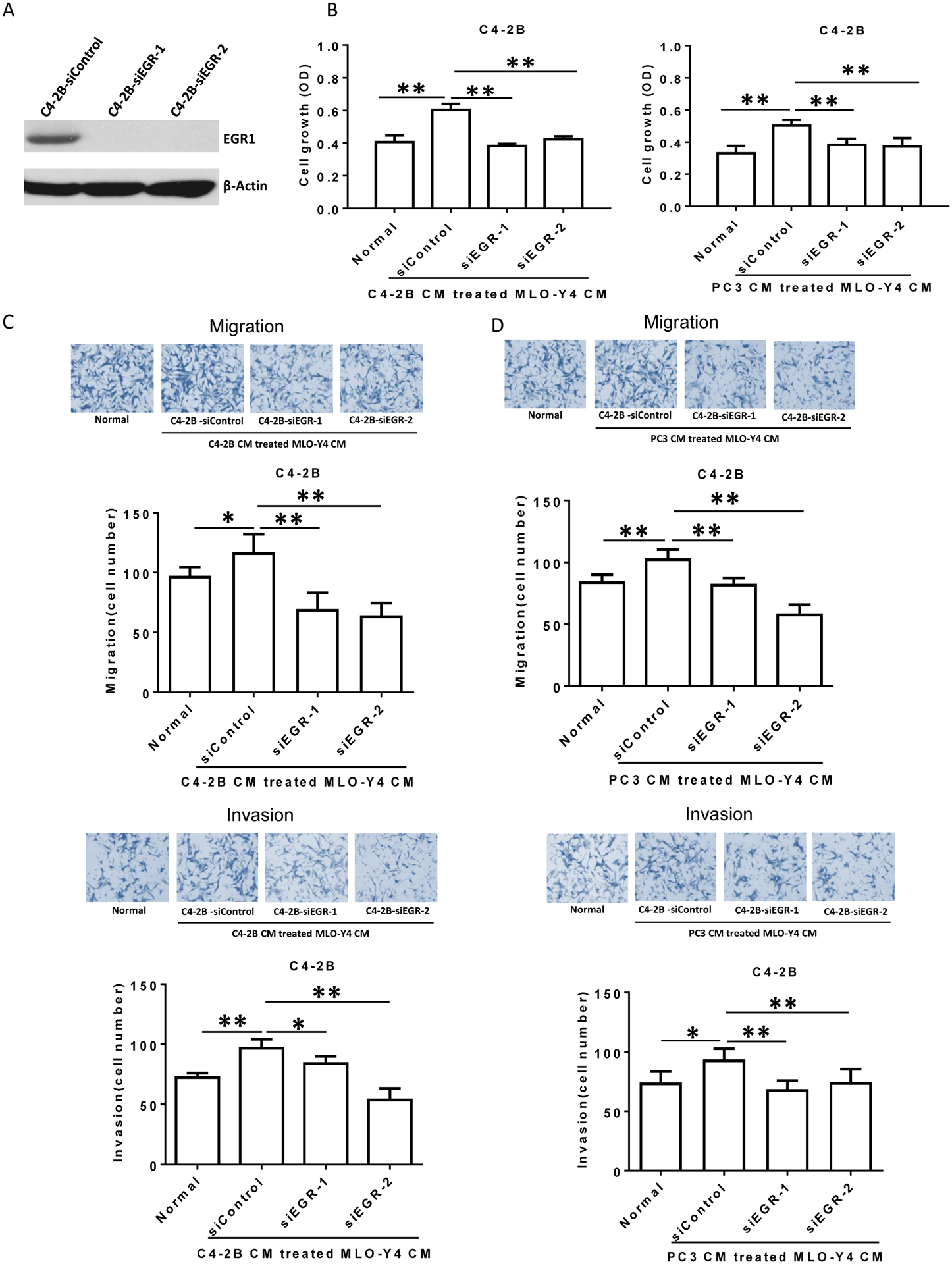
(A). Confirmation of siRNA-mediated EGR1 protein expression knockdown in C4-2B cells. Total cell protein lysate was subjected to immunoblot for EGR1. (B). C4-2B cells (1.5×104 per well) were treated with siControl or siEGR followed by C4-2B (left) or PC3 (right) CM-treated MLO-Y4 cell CM (or plain media as control). After 48 hours, cell numbers were counted using a hemocytometer. (C and D). Migration (upper figures) and invasion (lower figures) were assessed using a transwell assay. Both siControl-treated and siEGR-treated C4-2B cells (2×105 per cells) were treated with C4-2B (C) or PC3 (D) CM-treated MLO-Y4 cell CM, as indicated, for 24 hours. The membrane was stained using differential Quick staining kit and photographed under light microscopy (20x). The numbers of migrating (no Matrigel on membrane) and invading (Matrigel present on membrane) cells were counted in five random fields for each insert. Data are shown as the mean±SD of 3 independent experiments. * P < 0.05; ** P < 0.01.
Osteocytes promote prostate tumor growth through production of GDF15 that is associated with increased EGR1 expression and prostate cancer cell proliferation in vivo.
To explore if osteocytes’ ability to promote prostate cancer cell proliferation through GDF15 occurs in vivo, GDF15 expression was knocked down in primary murine osteocytes (Fig. 6A) then control or GDF knockdown murine osteocytes were mixed with C4-2B prostate cancer cells (10:1 ratio of C42B cells:osteocytes) and the cell mixtures or C4-2B cells alone were injected subcutaneously into mice. (Primary osteocytes injected alone did not grow in the subcutaneous site (data not shown)). The presence of sicontrol osteocytes mixed with C4-2B cells induced larger tumors than C4-2B cells injected alone; whereas, knockdown of GDF15 in the osteocytes abrogated this effect (Fig. 6B). These results indicate that osteocytes can increase tumor growth in vivo and that this pro-growth effect is dependent, in part, on expression of GDF15 in the osteocytes. To explore if increased proliferation and/or decreased apoptosis contributed to the increased tumor growth, we subjected tumor tissues to staining for Ki67 for proliferation and Caspase 3 for apoptosis. The presence of sicontrol osteocytes increased the percentage of proliferative calls, and this was abrogated by knockdown of GDF15; whereas, osteocytes had no impact on apoptosis (Fig. 6C and 6D). To explore if osteocytes increased EGR1 expression in the tumors as observed in the in vitro studies, we subjected the tumor tissues to immunohistochemistry and immunoblot for EGR1. The presence of sicontrol osteocytes at the time of cell injection was associated with increased EGR1 expression in the tumor cells; whereas, knockdown of GDF15 partially diminished this increase (Fig. 6C and D, lower row; and Fig. 6E). These results demonstrate that osteocytes can promote prostate cancer tumor growth through GDF15. Furthermore, they demonstrate the increased tumor growth is due, in part, to increased cell proliferation and is associated with increased EGR1 expression.
Figure 6. Primary osteocytes promote C4-2B tumor growth through GDF15, which is associated with up-regulation of EGR1 in prostate cancer tumors in vivo.
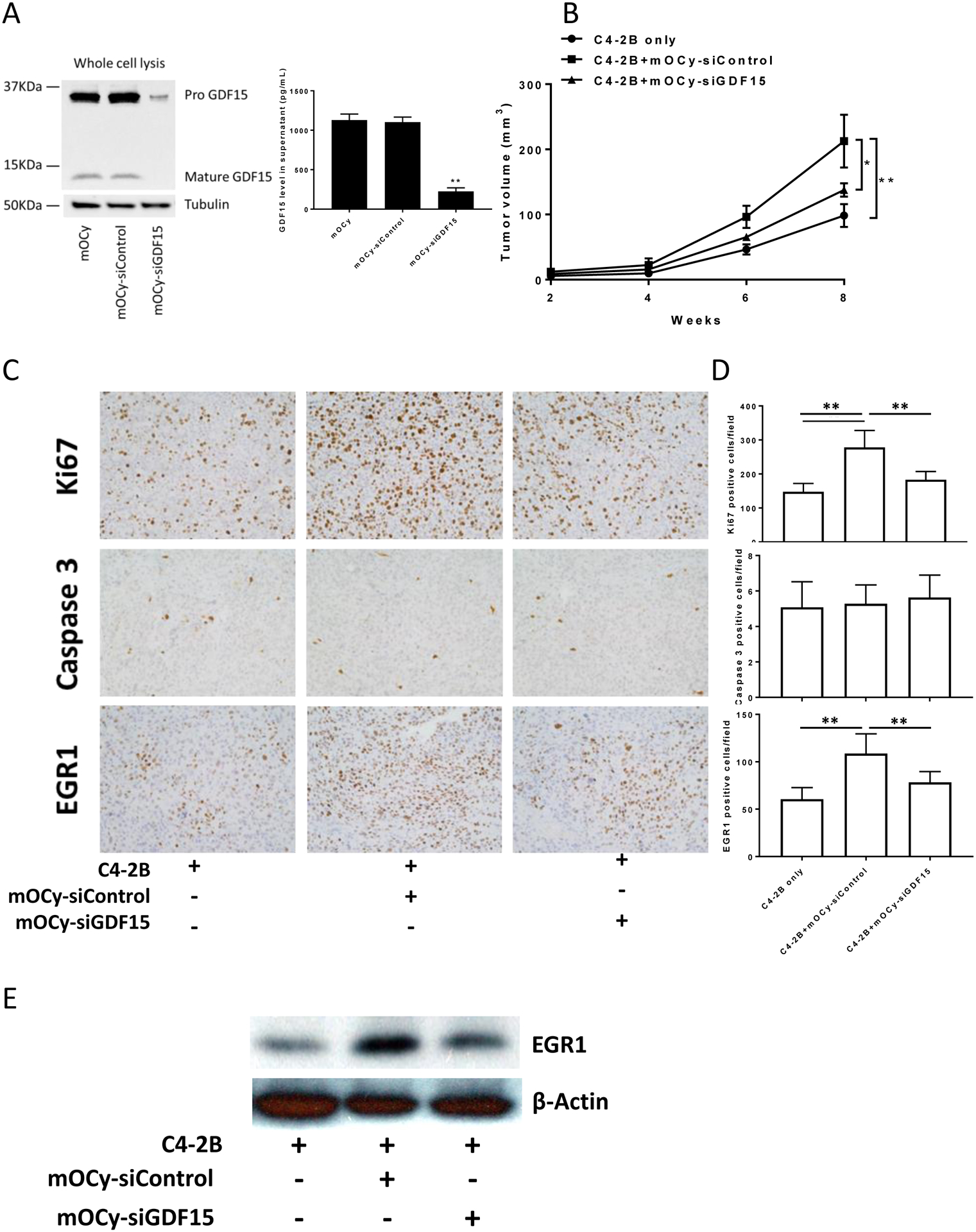
(A). Confirmation of siRNA-mediated GDF15 protein expression knockdown in mouse primary osteocytes cells by immunoblot (left figure) and ELISA (right graph). (B). SCID mice were subcutaneously injected with a C4-2B cells alone or a mixture of C4-2B cells and mouse primary osteocytes that had been pre-treated with siGDF15 or siControl. The ratio of cells injected was 10 C4-2B cells to 1 osteocyte. Subcutaneous tumor volume was measured using caliper. Results are reported as mean±SEM. (C). Subcutaneous tumors from SCID mice implanted with C4-2B cells and mouse primary osteocytes pre-treated with siGDF15 or siControl were stained using Ki67, Caspase 3 and EGR1. (D). Quantification of Ki67, Caspase 3 and EGR1 in subcutaneous tumors from SCID mice implanted with C4-2B cells and mouse primary osteocytes pre-treated with siGDF15 or siControl. ** P < 0.01. (E). EGR1 protein expression in subcutaneous tumors from SCID mice implanted with C4-2B cells and mouse primary osteocytes pre-treated with siGDF15 or siControl.
Bone microenvironment-derived GDF15 promotes prostate cancer cell proliferation in the bone.
To confirm that the GDF15 contributes to prostate cancer progression in the bone, we knocked down GDF15 expression in the tibial bone microenvironment using siRNA as previously described24 and injected PC3 or C4-2B prostate cancer cells into the tibia of mice. Since siRNA typically have an effective duration of approximately 7 days, an additional intratibial injection of siRNA was performed on day 7 after the initial injection to extend the duration of GDF15 knockdown. Tumor growth was monitored using bioluminescence every two weeks. Knockdown of GDF15 in the bone microenvironment was associated with decreased tumor growth and tumor-induced skeletal remodeling for both PC3 and C4-2B tumors (Figs. 7A and B). To explore if decreased proliferation and/or increased apoptosis contributed to the decreased tumor growth, we wanted to assess this at the time when siRNA would still be active. Thus, we had parallel groups of siRNA-treated tumor-bearing mice from which the tibiae were harvested at day 14 post-initial injection (these mice received siRNA on day 1 and day 7). The tumor tissues were subjected to staining for Ki67 for proliferation and Caspase 3 for apoptosis. Treatment of the bone microenvironment with siGDF reduced the percentage of Ki67 positive cells; whereas, it had no impact on apoptosis (Fig. 7C). To explore if knockdown of GDF15 expression impacted EGR1 expression in the tumors as observed in the in vitro studies, we subjected tumor tissues to immunoblot for EGR1. Pretreatment of the bone microenvironment with siGDF was associated with decreased EGR1 expression in the tumor cells (Fig. 7C and D). Finally, to confirm that GDF15 expression was decreased by the use of the siRNA, the tibiae of a parallel set of siRNA-treated mice were collected at day 14 post-initial injection and processed for total RNA procurement. The total RNA was subjected to qPCR for murine-specific GDF15 expression which was found decreased in the bones pre-treated with GDF15 siRNA (Fig. 7D). Furthermore, using human specific EGR1 primers, we confirmed that EGR1 expression was downregulated in the tumor tissue. Taken together, these results indicate that GDF15 expression contributes to prostate cancer progression in the bone microenvironment.
Figure 7. GDF15 inhibition reduces prostate cancer growth in bone and is associated with down-regulation of EGR1 in prostate cancer tumors.
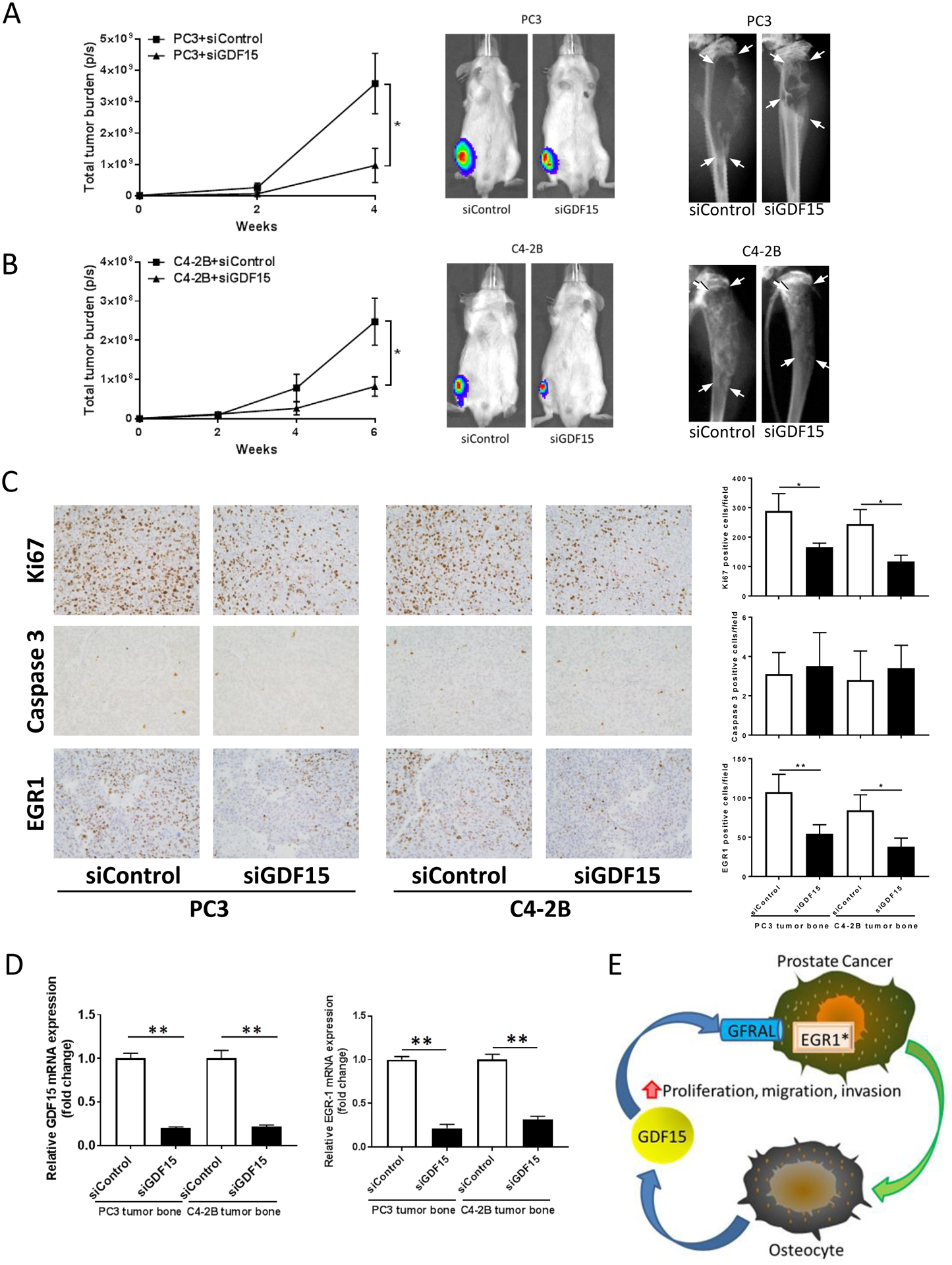
(A). SCID mice injected with mixture of prostate cancer PC3-luc cells and siRNA (siGDF15 or siControl) into right tibia followed by an additional intratibial injection of siRNA at day 7. Tumor burden was evaluated using bioluminescent imaging (left graph and middle figure) and radiography (right figure). The tumor-laden tissue area is delineated by all 4 arrows within each radiograph. Results are shown as mean±SEM. * P < 0.05. (B). SCID mice injected with mixture of prostate cancer C4-2B-luc cells and siRNA (siGDF15 or siControl) into right tibia followed by an additional intratibial injection of siRNA at day 7. Tumor burden was evaluated using bioluminescent imaging (left and middle) and radiography (right). Results are shown as mean±SEM. * P < 0.05. (C). One set of mice for each tumor type and treatmen (n=5 per tumor and treatment) were sacrificed at day 14-post initial injection and the intratibial tumors were subjected to immunohistochemistry for Ki67, Caspase 3 and EGR1. Images (left) and quantification (right) of Ki67, Caspase 3 and EGR1 staining. * P < 0.05, ** P < 0.01. (D). One set of mice (n=5) were sacrificed at day 14 -post initial injection, the tibiae flash frozen, total RNA collected and subjected to qPCR for murine GDF15 and human EGR1 mRNA expression. ** P < 0.01. (E). A proposed model system for secreted GDF15 promoting prostate cancer progression. Prostate cancer cells disseminate to bone, interact with osteocytes to induce production and release of GDF15 into the bone microenvironment which enhances prostate cancer proliferation, migration and invasion resulting in progression of bone metastasis.. Furthermore, our finding of the GDF15 receptor, GFRAL, on the prostate cancer cells, suggests that GDF15 mediates its activity through GFRAL in the prostate cancer cells. Finally, GDF15 induced EGR1 signaling in the prostate cancer cells, suggesting that EGR1 may contribute to the pro-metastatic effects of GDF15. *The data for EGR1 activation was supported by our in vitro findings and consistent with our in vivo studies; however, it has not been evaluated critically in vivo as of yet.
Discussion
Our results provide insight into regulation and function of osteocytes in promoting prostate cancer bone metastasis. In the current study, we demonstrate that prostate cancer cells induce a positive feedback pro-metastatic loop through promoting osteocytes to produce GDF15, which then stimulates prostate cancer cell growth, migration and invasion (Fig. 7E). Furthermore, we identified that the GDF15 receptor, GFRAL is present on the prostate cancer cell lines. Finally, this activity is associated with stimulation of EGR1 expression in the prostate cancer cells. Overall, this activity can promote bone metastasis progression through enhancement of both growth and invasion of prostate cancer into bone.
The tumor microenvironment plays an important role in cancer establishment and progression. It may achieve this through a combination of a direct effect on cancer cells, such as release of cytokines that stimulate tumor growth, and indirectly through modifying other tumor-active cells (e.g. through down-regulation of anti-tumor immune cell activity). In terms of bone metastases, multiple studies have identified that osteoblasts produce a variety of factors that directly promote prostate cancer growth6,25–28. In addition to osteoblasts, many reports indicate that osteoclasts promote bone metastasis, indirectly, through resorption of mineralized bone matrix and subsequent release of tumor-promoting growth factors from the matrix26,29. While extensive research activity has focused on mechanisms through which osteoblasts and osteoclasts contribute to bone metastasis, there is a paucity of information regarding the role of osteocytes in bone metastasis. Considering the observation that osteocytes are the most common cell in bone tissue30,31 and the key role they play in coordinating osteoblast and osteocyte activity30,32, it follows that a greater understanding of the role of osteocytes in bone metastasis is warranted.
Our observation that PCa cells modify osteocytes to promote metastasis is consistent with previous reports that cancers can modify the tumor and metastatic microenvironment. For example, exosomes derived from primary melanomas travel through circulation to bone and educate the bone marrow to promote melanoma metastasis through activation of MET33. In the current study, we focused on the mechanism through which osteocytes enhance the pro-metastatic phenotype, thus we did not explore for the mechanism through which the prostate cancer cells modified the osteocytes. It is unlikely that there is direct contact, at least early on with osteocytes and prostate cancer cells. Our observation that conditioned media from prostate cancer cells was able to activate the osteocytes is consistent with soluble factors, such as prostate-produced exosomes34,35 or soluble proteins, stimulate the osteocyte. Dissecting the factors through which prostate cancer directly modulates osteocytes to promote metastasis is a promising area for future studies.
In our study, we demonstrated that education of osteocytes by prostate cancer cells resulted in enhancing osteocytes’ ability to promote both proliferation, migration and invasion of prostate cancer cells. This finding adds to our previous demonstration that tumor-induced pressure modifies osteocytes to promote prostate cancer growth and invasion9. A common theme of the previous study and the current study is that the tumor modifies osteocytes to further enhance bone metastasis in a positive feedback cycle. However, differences include both the mediator of osteocyte education; i.e., in the previous study it was the tumor-induced physical force of pressure sensed by osteocytes, which are key mechanotransducing cells in bone; whereas, in the current study, the mediator was prostate cancer-produced soluble factor(s). Furthermore, the osteocyte-produced effectors were different among these mediators of osteocyte education; specifically, pressure induced release of CCL5 and matrix metalloproteinases from the osteocyte as key mediators of pro-metastatic activity9; whereas, in the current study, the prostate cancer-produced soluble factor(s) induced release of GDF15. At this point, it is unknown if these different mechanisms would work together in clinical disease. However, it is plausible that early in metastatic development, prior to development of gross tumor, there would be minimal pressure changes, and thus the prostate cancer-produced soluble factors could initiate the process of educating the osteocytes, and as the tumor develops the pressure-induced changes on the osteocytes either synergize with the soluble factor effects or eclipse these effects. Either way, the current study adds an additional and novel path through which prostate cancers induce a positive feedback loop through modulating osteocytes.
Osteocytes have been shown to have both anti- and pro- tumor effects that appear to be dependent on tumor type. In the context of breast cancer, bisphosphonate treatment of osteocytes induces them to open Cx43 hemichannels and release ATP which inhibited breast cancer cell migration and tumor development12,36. In contrast, conditioned media from untreated osteocytes has been previously shown to promote proliferation of a variety of breast cancer and prostate cancer cell lines; albeit, results were inconsistent among cell lines37. In the case of multiple myeloma, direct contact between osteocytes and multiple myeloma cells reciprocally activates Notch signaling and increases Notch receptor expression that stimulates multiple myeloma cell growth38. Our results are overall consistent with these previous studies, but moreover demonstrate for the first time that a soluble factor can activate cancer cell-mediated education of osteocytes that promotes prostate cancer cell growth and invasion.
The current study revealed that osteocytes mediate their pro-metastatic activity, in part, through secreting GDF15 (also known as macrophage inhibitory cytokine-1 (MIC1)). GDF15 is a member of the bone morphogenetic protein subfamily of the transforming growth factor-beta (TGFβ) super family of proteins39. The production of GDF15 expression by MLO-Y4 cells is consistent with a previous report that demonstrated hypoxia induced GDF15 expression in osteocytes40. GDF15 is typically expressed in the range of 0.1–1.2 ng/ml13. However, in our study we observed that the MLO-Y4 cells could produce GDF15 at levels > 4.0 ng/ml. This finding is consistent with high levels found under various pathological conditions. For example Li et al. demonstrated normal serum GDF15 levels (measured as MIC-1) in the range of 0.53 to 1.33 ng/ml; whereas, for all prostate cancer patients, the range was 0.63–2.76 ng/ml41. In advanced prostate cancer, Welsh et al., demonstrated serum levels up to approximately 12.5 ng/ml and for colorectal cancer patients, the levels were higher than 15 ng/ml42. This finding was further supported by the observation that men with relapse of prostate cancer that developed into bone metastases had higher levels (a mean of 5.59 ng/ml) than those without bone metastases43.
It was recently identified that GDF15 mediates its activity through the GDNF family receptor alpha-like (GFRAL)14,44–46. GFRAL expression was evaluated in many tissues and shown to be restricted to the hindbrain neurons in healthy mice and in human brain tissue14,44,46. However, to the best of our knowledge, evaluation of GFRAL expression in prostate tissue and cancers has not been reported. Thus, our results that GFRAL was expressed in prostate cancer cell lines do not conflict with the previous reports and is consistent with multiple previous reports that GDF15 is functional in prostate cancer cells47.
Several lines of evidence suggest that GDF15 plays a role in prostate cancer progression. It has been shown to be an independent serum biomarker for the presence of prostate cancer in high-grade prostate cancer48. In one study, GDF15 appeared to limit local prostate cancer growth, but increased tumor aggressiveness resulting in increased metastatic burden in the TRAMP prostate cancer mouse model49. This finding is consistent with the observation that a specific GDF15 gene polymorphism (H6D C-to-G) was associated with decreased risk of prostate cancer occurrence, but increased risk of prostate-cancer-specific death50, which suggests the presence of increased tumor aggressiveness. The observation that GDF15 in the context of osteocyte-conditioned media promoted prostate cancer proliferation and invasion is are consistent with the observation that overexpression of GDF15 in stromal cells increased prostate cancer proliferation and invasion51.
Our finding that prostate cancer-educated osteocyte-conditioned media promoted prostate cancer cell proliferation and invasion in the prostate cancer cells through inducing EGR1 expression is consistent with previously reported observations that EGR1 promotes cancer proliferation and invasion through several mechanisms including promoting expression of proteases such as matrix metalloproteinase-852,53, protease-activated receptor 1 (PAR-1) expression54, and cytokine-mediated proliferation and invasion55,56. This result is particularly striking because in most tissues, EGR1 is considered a tumor-suppressor57; however, in the context of prostate cancer it is considered to promote tumorigenesis and aggression15,58,59. While we demonstrated that induction of EGR1 was dependent on expression of GDF15 from the osteocytes, the mechanism through which GDF15 promotes EGR1 expression was not defined.
The in vivo findings that osteocytes promoted tumor growth through GDF15 and was associated with increased EGR1 expression in the tumors is consistent with our in vitro findings. Furthermore, based on the immunohistochemistry of tumor tissues, the pro-tumorigenic effect appeared to be due primarily to increased tumor cell proliferation as opposed to inhibition of apoptosis. A weakness of the intratibial tumor experimental design was that the knockdown of intratibial GDF15 was most likely not specifically targeted to osteocytes, thus we cannot conclude from the in vivo study that the osteocyte is the main mediator of the increased tumor growth. Regardless the intratibial tumor results do support that GDF15 plays a role in bone metastasis progression and taken together with the subcutaneous tumor studies in which knockout of GDF15 in primary murine osteocytes diminished the pro-tumor effect of the osteocytes strongly indicates that osteocytes promote bone metastasis.
Overall, our model (Fig. 7E), posits that OCy-produce GDF15 directly effects prostate cancer cells. However, our studies do not rule out other possible mechanisms of GDF15 activity that may also contribute to the overall pro-metastatic activity. Specifically, it is possible that intracellular or autocrine functions of GDF15 may stimulate OCys to produce other factors that can promote pro-metastatic activity on prostate cancer cells.
In summary, our study offers a novel paradigm that contributes to the high incidence of bone metastasis in men with prostate cancer. Through a positive feedback loop, prostate cancer cells promote osteocytes to produce GDF15, which in turn targets prostate cancer cells to enhance their EGR1 expression and growth and invasive properties. These findings underscore the importance of osteocytes in contributing to the progression of prostate cancer bone metastases and identify both microenvironmental and innate tumor molecular targets to diminish bone metastasis progression.
Materials and Methods
Cell lines
The murine osteocyte cell line MLO-Y4 (a kind gift of Dr. Lynda Bonewald; University of Missouri, Kansas City, MO) was maintained α-MEM containing 5% fetal bovine serum (FBS), 5% bovine calf serum (BCS) and 1.0% penicillin/streptomycin as previously described60. Human prostate cancer cell line PC3, DU145 and LNCaP, human breast cancer cell line MDA-MB453 were obtained from the American Type Culture Collection (ATCC; Rockville, MD) and cultured in RPMI 1640 with 10% FBS and 1% penicillin/streptomycin. The human prostate cancer cell line VCaP was previously described61 and cultured in DMEM/F12 with 10% FBS and 1% penicillin/streptomycin. The C4-2B human prostate cancer cell line (kindly provided by Dr. Leland Chung, Cedars Sinai, Hollywood, CA), are LNCaP sublines selected to grow in bone in vivo62 and were grown in T medium. All cells were serially passage using trypsinization and maintained at 37°C and 5% CO2 in a humidified atmosphere. Cells are evaluated for mycoplasma monthly and authenticated by single tandem repeat analysis every 6 months.
Murine studies
7–8 week-old male NOD.CB17-Prkdcscid/NCrCrl (NOD/SCID) mice (Charles River, Wilmington, MA) were housed under pathogen-free conditions. All experimental protocols were approved by the University of Michigan Institutional Animal Care and Use Committee. To model tumors, either 1×106 C4-2B cells alone or a 10:1 mixture of 9.1 × 105 C4-2B cells plus 9 × 104 murine osteocytes (either with shControl or shGDF15 knockdown) (total of 1 × 106 cells) were injected subcutaneously. Tumors were measured using calipers and tumor volume was calculated following the formula: tumor volume V = a ×b2×0.52 where a=length of the tumor, b=width of the tumor. The experimental protocols did not require animal randomization. Investigators were not blinded to the experimental groups. In some cases, mice were excluded if they had unexpected deaths.
Primary mouse osteocyte isolation and culture
Osteocytes were isolated from mouse long bones utilizing a modification of a previously reported protocol63. Briefly, long bones (femora, tibia, and humeri) were aseptically dissected from skeletally mature 3-month-old C57BL/6 mice (Charles River Laboratories, Wilmington, MA, USA) and diced into 1 to 2 mm2 particles using a scalpel. The bone particles were processed using serial digestion with 8 ml warmed collagenase type-IA (300 active units/mL) and 8 ml warmed EDTA (5 mM) for 25 min at 37°C in water bath. Following each sequential digestion, the digest solution of last step with suspended cells was collected and kept for cell plating. The bone particles were then rinsed with 5 ml Hank’s balanced salt solution (HBSS) for three times and the rinse solutions were saved. The digest solution and rinse solutions were combined and centrifuged at 200g for 5 min and the supernatant was removed from the cell pellet. The cells and bone particles were resuspended respectively in α-MEM supplemented with 5% FBS, 5% BCS, and 1% penicillin and streptomycin and cultured on type-I rat tail collagen-coated six-well plates and maintained at 37°C and 5% CO2 in a humidified incubator.
Conditioned medium
Conditioned medium (CM) for treating MLO-Y4 cells and primary mouse osteocytes was prepared by incubating C4-2B, PC3, VCaP and MLO-Y4 cells in 10 ml dishes for 24 hours with alpha-MEM containing 0.5% FBS, 0.5% BCS and 1.0% penicillin/streptomycin. Supernatant was removed and centrifuged at 1000g for 3 minutes to remove the cell debris. Cells were counted and CMs were normalized by adding media to a final volume that would represent 1×106 cells per ml. The CM for treating prostate cancer cells was prepared by incubating primary mouse osteocytes, MLO-Y4 cells, MLO-Y4-scramble cells and MLO-Y4–GDF15 KD cells on 10ml dishes with prostate cancer CM for 24 hours, the cells were washed and then cell culture medium appropriate for the target prostate cancer cell containing 0.5% FBS and 1.0% penicillin/streptomycin was added. After 24 hours of incubation, CM was collected and centrifuged at 1000 g for 3 minutes to remove the cell debris. Primary mouse osteocytes and MLO-Y4 cells were counted and CM was normalized by adding media to a final volume that would represent 1×106 cells per ml.
ELISA assays
Mouse GDF15 level in supernatants of MLO-Y4 cells and mouse primary osteocytes, and mouse TGF-β level in recombinant mouse GDF15 were analyzed by antibody sandwich ELISA (R&D Systems, Minneapolis, MN) following manufacturer’s instructions.
Cytokine array
Identification of MLO-Y4 secreted cytokines was achieved by using a Mouse XL Cytokine Array Kit (Cat#.ARY028, R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions. Protein concentration was normalized before starting the assay. Pixel density of each spot was measured using Image J system. Spots were averaged, background subtracted and average values reported for each cytokine.
Pathway finder reporter array
Identification of transcription factor activity was achieved by using a Cignal Finder Signal Transduction 45-pathway Reporter Array (Cat.CCA-901L-2, Qiagen, Frederick, MD) according to the manufacturer’s instructions. Briefly, PC3 and C4-2B cells were reverse transfected by plating cells onto the Cignal array plate in which wells contain an array of 45 different pathway reporter vectors. Medium was replaced 24 hours of transfection with the indicated MLO-Y4 CM and 12 hours later the dual-luciferase assay was performed. The activity of a pathway-specific firefly luciferase reporter was detected using the Dual-Luciferase® Assay System (Promega, Madison, WI) allowing for simultaneous evaluation of transfection efficiency and promoter activation. Results are expressed as fold change of CM-treated vs. control-treated cells.
Real time RT-PCR analysis of Epidermal Growth Factor (EGF) signaling genes
Measurement of changes in mRNA levels of 84 EGF signaling related genes were achieved with the RT2 Profiler™ PCR Array Human EGF / PDGF Signaling Pathway Kit (Cat.PAHS-040Z, Qiagen). Total RNA was extracted using RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Then cDNA was synthesized from 2.0 μg mRNA using Superscript III first strand synthesis system (Thermo Fisher Scientific, Waltham, MA). SYBR green (Qiagen) was used to amplify cDNA and then cDNA was subjected to quantitative real time PCR, using SYBR green labeling, as directed by the manufacturer. Expression of EGF regulated transcripts was compared between the groups.
Creation of gene knockdown cells
For stable knockdown studies, MLO-Y4 cells and mouse primary osteocytes were seeded in 12-well plates and allowed to attach overnight to achieve 50% confluency. Cells were then transduced with GDF15 shRNA lentiviral particles (Cat. sc-39799-V, Santa Cruz Biotechnology, Santa Cruz, CA) or shRNA control lentiviral particles (Cat. sc-108080, Santa Cruz Biotechnology). Selection of stably transduced cells was initiated 48h after transduction using 8 μg/ml puromycin for 14 days. For siRNA silencing studies, PC3 and C4-2B cells were seeded in 6-well plates at 30% confluence on the day before the transfection, then transfected with siRNA EGR1(Cat. 4392420, 4390824, Thermo Fisher Scientific,Waltham, MA) and Negative Control siRNA(Cat.4390843, Thermo Fisher Scientific,) using siRNA Transfection Reagent (Thermo Fisher Scientific) as recommended by the manufacturer.
Transwell migration and Matrigel invasion assays
Migration and invasion results were performed as previously described64 using transwell assays with Matrigel (invasion) and without Matrigel (migration) according to the manufacturer’s directions (Corning Inc., Corning, NY). Prostate cancer cells (PC3=0.5–1×105; C4-2B=1–2×105) were plated in the top well in no FBS RPMI 1640 or T medium. In the bottom well, control media, CM or different concentrations of recombinant GDF15 (Cat.8944-GD-025, R&D Systems Minneapolis, MN) and 10% FBS were added as a chemoattractant. Cells underneath the inserts were analyzed using light microscopy at 20x magnification and photographed. The numbers of cells were counted in five random fields for each insert. Each data point represents the average number from three wells.
In vitro cell growth
Prostate cancer cells (PC3=2000, C4-2B=3000) were plated in 96-well plates, and appropriate test CM or control, anti-GDF15 antibody or isotype antibody were added and then cells incubated for 24 hours at which time10μl cell proliferation reagent WST-1 (Clontech, Mountain View, CA) was added into 100 μl medium and incubated at 37°C and 5% CO2 for 4 hours respectively. Absorbance was then measured at 440 nm with a plate reader (Multi-Mode Microplate Reader, SpectraMax M5, Molecular Devices MDS Analytical Technologies, Sunnyvale, CA).
Quantitative Real-time PCR
Cells and soft tissues were processed using standard RNA isolation methods. For tibiae, the bone was flash frozen and ground to powder which was then processed using standard RNA isolation methods. Briefly, total RNA was extracted using Trizol reagent (Thermo Fisher Scientific), Two micrograms of total RNA were reversed-transcribed in a final 20 μl volume, Quantitative real time PCR was performed in triplicate using SYBR Green qPCR MasterMix (Qiagen) in a 10 μl reaction volume on a Roche LightCycler 480 (Roche, Pleasanton, CA). The PCR conditions were: 30 at 95°C; followed by 40 cycles of 95°C for 30 s; 65°C for 20 s. Mouse GDF15 primers (Cat. no. PPM04436C-200), mouse Endoglin primers (Cat. no. PPM03790C-200) and human EGR1 primers (Cat. no. PPH00139A-200) were purchased from SABiosciences (Qiagen). All analyses were performed in triplicate in three independent experiments. Measurements from triplicate Ct values were normalized to GAPDH, averaged, and reported.
Western blot analysis
Cell lysates were prepared in RIPA buffer containing protease and phosphatase inhibitors. The protein concentration of tumor extracts was determined using BCA Protein Assay Kit (Thermo Fisher). Protein lysates (50 μg) were resolved by electrophoresis on 10% SDS-PAGE, and proteins were transferred to PVDF membrane. After blocking in 5% non-fat milk in 1×TBST/0.1% Tween-20 at room temperature for 1 hour, the membranes were incubated at 4°C overnight with primary antibodies including mouse GDF15 (Cat. ABIN3020659, Antibodies-Online, Atlanta, GA), human EGR1 (Cat. 4153S, Cell Signaling Technology, Danvers, MA), human GFRAL (Cat. ab235111, abcam, Cambridge, MA) and β-actin (Cat. A5441, Sigma-Aldrich), The antibodies were diluted as recommended by the manufacturers.
Subcutaneous injection of C4-2B cells and murine osteocytes.
Pre-designed siRNA for mouse GDF-15 (Origene, Rockville, MD) were used to knockdown the target genes in mouse primary osteocytes followed the manufacturer’s directions. Briefly, the cells were transfected with siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific, USA) for 24 h followed by validation of expected change in expression of the target protein using immunoblot. To inject cells, 0.5ml Corning™ Matrigel™ Membrane Matrix (Thermo Fisher Scientific, USA) was mixed with C4-2B (2.5×106) cells alone or a combination of C4-2B cells and and siRNA control-treated or siRNA GDF-15-treated primary murine osteocytes (ration of 10 C4-2B to 1 osteocyte) and the mixture was injected subcutaneously into flank of 8 weeks old SCID mouse (5 mice/each group). The tumor sizes were measured using caliper every two weeks. The subcutaneous tumors were collected at 8 weeks for immunohistochemistry and western blot.
Intratibial injection of PC3 or C4-2B cells with siRNA
The siGDF-15 or siControl transfection complex was prepared and injected intratibially as previously described24. In brief, 20 μg of siRNA was dissolved in 20 μl of RNase-free water, followed by the addition of 10 μl of EntransterTM-in vivo (Engreen, Beijing, China), the mixture was then incubated for 15 min at room temperature. Finally, 30 μl of Poloxamer 407 (Sigma-Aldrich) was added to the solution with sufficient mixing. 20 μg of siGDF-15 or siControl in 50 μl of transfection medium was mixed with C4-2B (2.5×106) or PC3 (5×105) cells and was injected into right tibia of 8 week-old SCID mouse (5 mice/each group plus an additional 10 mice for early harvesting). Either siControl or siGDF-15 was reinjected intratibially 7 days post the initial injection. At day 14 post-initial injection, the 10 additional mice from each group were euthanized and tibiae from 5 of the mice from each group were processed for evaluation of tumor tissue and tibiae from 5 of the mice from each group was processed for total RNA isolation for evaluation of GDF-15 and EGR1 expression. In the remaining mice, the tumor growth was evaluated weekly using bioluminescent imaging (BLI) and radiographs.
Imaging
For BLI, mice were injected intraperitoneally with 100μl luciferin (40 mg/ml in PBS), anesthetized with 1.5% isoflurane and imaged 15 minutes post-luciferin injection on the IVIS BLI system (Perkin Elmer, Waltham, MA) as previously described65,66. Signal intensity was quantified as the sum of all detected photons within the region of interest during a 1-minute luminescent integration time. Radiographs were taken using a digital Faxitron MX-20 (Wheeling, IN) at 4x magnification.
Immunohistochemistry
Subcutaneous tumors were fixed in 10% neutral-buffered formalin and paraffin-embedded. Intratibial tumors were decalcified using Cal-Ex II (Fisher Scientific) then fixed in 10% neutral-buffered formalin and paraffin-embedded. Five-micron (5μM) sections were used for immunohistochemistry. Ki67 (1:500, Santa Cruz Biotechnology), cleaved-caspase 3 (1:300, Cell Signaling Biotechnology) and human EGR1 (1:250, Cell Signaling Technology) were stained using standard methodology. To quantify, five 20x random fields each well were selected for positive staining cells counting. Results are reported as positive staining cells counted per field.
Statistical analysis
Statistical analysis was performed using Prism5 (Graph Software). Sample size was selected based on the ability to detect a difference of greater than 30% change at a power of 80% with p<0.05. Sample size for animal models was based on ability to detect a difference of greater than 20% with a power of 80% at p<0.05. Student’s t-test was used for analysis and p <0.05 was considered as statistically significant. All experiments were repeated independently a minimum of two times.
Supplementary Material
Acknowledgements
This work was supported by NIH R01-CA190554, NIH P01-CA093900, NSFC Key Project 81130046, NSFC 81773146 and JCYJ20170412154619484.
Footnotes
Conflicts of Interests
The authors declare that they have no conflict of interest.
References
- 1.Siegel RL, Miller KD & Jemal A Cancer Statistics, 2017. CA Cancer J Clin 67, 7–30 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD & Jemal A Cancer statistics, 2016. CA Cancer J Clin 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Gartrell BA et al. Metastatic Prostate Cancer and the Bone: Significance and Therapeutic Options. Eur Urol 68, 850–858 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Koutsilieris M Osteoblastic metastasis in advanced prostate cancer. Anticancer Res 13, 443–449 (1993). [PubMed] [Google Scholar]
- 5.Shah RB et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res 64, 9209–9216 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Logothetis CJ & Lin SH Osteoblasts in prostate cancer metastasis to bone. Nat Rev Cancer 5, 21–28 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Turner CJ & Edwards CM The Role of the Microenvironment in Prostate Cancer-Associated Bone Disease. Curr Osteoporos Rep 14, 170–177 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Miftakhova R et al. Cyclin A1 and P450 Aromatase Promote Metastatic Homing and Growth of Stem-like Prostate Cancer Cells in the Bone Marrow. Cancer Res 76, 2453–2464 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Sottnik JL, Dai J, Zhang H, Campbell B & Keller ET Tumor-induced pressure in the bone microenvironment causes osteocytes to promote the growth of prostate cancer bone metastases. Cancer Res 75, 2151–2158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalajzic I et al. In vitro and in vivo approaches to study osteocyte biology. Bone 54, 296–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Compton JT & Lee FY A review of osteocyte function and the emerging importance of sclerostin. J Bone Joint Surg Am 96, 1659–1668 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou JZ et al. Osteocytic connexin hemichannels suppress breast cancer growth and bone metastasis. Oncogene 35, 5597–5607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai VWW, Husaini Y, Sainsbury A, Brown DA & Breit SN The MIC-1/GDF15-GFRAL Pathway in Energy Homeostasis: Implications for Obesity, Cachexia, and Other Associated Diseases. Cell Metab 28, 353–368 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Mullican SE et al. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat Med 23, 1150–1157 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Adamson ED & Mercola D Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumour Biol 23, 93–102 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Yi JH, Park SW, Kapadia R & Vemuganti R Role of transcription factors in mediating post-ischemic cerebral inflammation and brain damage. Neurochem Int 50, 1014–1027 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y, Han CC, Huang Q, Sun WY & Wei W GRK2 overexpression inhibits IGF1-induced proliferation and migration of human hepatocellular carcinoma cells by downregulating EGR1. Oncol Rep 35, 3068–3074 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Parra E, Gutierrez L & Ferreira J Increased expression of p21Waf1/Cip1 and JNK with costimulation of prostate cancer cell activation by an siRNA Egr-1 inhibitor. Oncol Rep 30, 911–916 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Han MH, Kim GY, Yoo YH & Choi YH Sanguinarine induces apoptosis in human colorectal cancer HCT-116 cells through ROS-mediated Egr-1 activation and mitochondrial dysfunction. Toxicol Lett 220, 157–166 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Baron V, Adamson ED, Calogero A, Ragona G & Mercola D The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther 13, 115–124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baron V et al. Inhibition of Egr-1 expression reverses transformation of prostate cancer cells in vitro and in vivo. Oncogene 22, 4194–4204 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Arora S et al. Egr1 regulates the coordinated expression of numerous EGF receptor target genes as identified by ChIP-on-chip. Genome Biol 9, R166 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florkowska M et al. EGF activates TTP expression by activation of ELK-1 and EGR-1 transcription factors. BMC Mol Biol 13, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W et al. A Convenient In Vivo Model Using Small Interfering RNA Silencing to Rapidly Assess Skeletal Gene Function. PLoS One 11, e0167222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng X et al. Recent advances in bone-targeted therapies of metastatic prostate cancer. Cancer Treat Rev 40, 730–738 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hensel J & Thalmann GN Biology of Bone Metastases in Prostate Cancer. Urology 92, 6–13 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Hall CL & Keller ET The role of Wnts in bone metastases. Cancer Metastasis Rev 25, 551–558 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Keller ET et al. Prostate carcinoma skeletal metastases: cross-talk between tumor and bone. Cancer Metastasis Rev 20, 333–349 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Ortiz A & Lin SH Osteolytic and osteoblastic bone metastases: two extremes of the same spectrum? Recent Results Cancer Res 192, 225–233 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Dallas SL, Prideaux M & Bonewald LF The osteocyte: an endocrine cell … and more. Endocr Rev 34, 658–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonewald LF The amazing osteocyte. J Bone Miner Res 26, 229–238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonewald LF & Johnson ML Osteocytes, mechanosensing and Wnt signaling. Bone 42, 606–615 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peinado H et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18, 883–891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishn SR et al. Prostate cancer sheds the alphavbeta3 integrin in vivo through exosomes. Matrix Biol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Languino LR et al. Exosome-mediated transfer from the tumor microenvironment increases TGFbeta signaling in squamous cell carcinoma. Am J Transl Res 8, 2432–2437 (2016). [PMC free article] [PubMed] [Google Scholar]
- 36.Bultynck G The anti-metastatic micro-environment of the bone: Importance of osteocyte Cx43 hemichannels. Biochim Biophys Acta 1866, 121–127 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Cui YX, Evans BA & Jiang WG New Roles of Osteocytes in Proliferation, Migration and Invasion of Breast and Prostate Cancer Cells. Anticancer Res 36, 1193–1201 (2016). [PubMed] [Google Scholar]
- 38.Delgado-Calle J et al. Bidirectional Notch Signaling and Osteocyte-Derived Factors in the Bone Marrow Microenvironment Promote Tumor Cell Proliferation and Bone Destruction in Multiple Myeloma. Cancer Res 76, 1089–1100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paralkar VM et al. Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J Biol Chem 273, 13760–13767 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Hinoi E et al. Positive regulation of osteoclastic differentiation by growth differentiation factor 15 upregulated in osteocytic cells under hypoxia. J Bone Miner Res 27, 938–949 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Li J, Veltri RW, Yuan Z, Christudass CS & Mandecki W Macrophage inhibitory cytokine 1 biomarker serum immunoassay in combination with PSA is a more specific diagnostic tool for detection of prostate cancer. PLoS One 10, e0122249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welsh JB et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc Natl Acad Sci U S A 100, 3410–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selander KS et al. Serum macrophage inhibitory cytokine-1 concentrations correlate with the presence of prostate cancer bone metastases. Cancer Epidemiol Biomarkers Prev 16, 532–537 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Yang L et al. GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat Med 23, 1158–1166 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Emmerson PJ et al. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat Med 23, 1215–1219 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Hsu JY et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 550, 255–259 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Vanhara P, Hampl A, Kozubik A & Soucek K Growth/differentiation factor-15: prostate cancer suppressor or promoter? Prostate Cancer Prostatic Dis 15, 320–328 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Brown DA et al. Measurement of serum levels of macrophage inhibitory cytokine 1 combined with prostate-specific antigen improves prostate cancer diagnosis. Clin Cancer Res 12, 89–96 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Husaini Y et al. Macrophage inhibitory cytokine-1 (MIC-1/GDF15) slows cancer development but increases metastases in TRAMP prostate cancer prone mice. PLoS One 7, e43833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes VM et al. Macrophage inhibitory cytokine-1 H6D polymorphism, prostate cancer risk, and survival. Cancer Epidemiol Biomarkers Prev 15, 1223–1225 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Bruzzese F et al. Local and systemic protumorigenic effects of cancer-associated fibroblast-derived GDF15. Cancer Res 74, 3408–3417 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Shin SY, Kim JH, Baker A, Lim Y & Lee YH Transcription factor Egr-1 is essential for maximal matrix metalloproteinase-9 transcription by tumor necrosis factor alpha. Mol Cancer Res 8, 507–519 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Ozen E et al. Heparin inhibits Hepatocyte Growth Factor induced motility and invasion of hepatocellular carcinoma cells through early growth response protein 1. PLoS One 7, e42717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salah Z, Maoz M, Pizov G & Bar-Shavit R Transcriptional regulation of human protease-activated receptor 1: a role for the early growth response-1 protein in prostate cancer. Cancer Res 67, 9835–9843 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Ma J et al. Targeted knockdown of EGR-1 inhibits IL-8 production and IL-8-mediated invasion of prostate cancer cells through suppressing EGR-1/NF-kappaB synergy. J Biol Chem 284, 34600–34606 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun T, Tian H, Feng YG, Zhu YQ & Zhang WQ Egr-1 promotes cell proliferation and invasion by increasing beta-catenin expression in gastric cancer. Dig Dis Sci 58, 423–430 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Liu C, Calogero A, Ragona G, Adamson E & Mercola D EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit Rev Oncog 7, 101–125 (1996). [PubMed] [Google Scholar]
- 58.Adamson E et al. Egr1 signaling in prostate cancer. Cancer Biol Ther 2, 617–622 (2003). [PubMed] [Google Scholar]
- 59.Gitenay D & Baron VT Is EGR1 a potential target for prostate cancer therapy? Future Oncol 5, 993–1003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato Y, Windle JJ, Koop BA, Mundy GR & Bonewald LF Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res 12, 2014–2023 (1997). [DOI] [PubMed] [Google Scholar]
- 61.Korenchuk S et al. VCaP, a cell-based model system of human prostate cancer. In Vivo 15, 163–168 (2001). [PubMed] [Google Scholar]
- 62.Wu TT et al. Establishing human prostate cancer cell xenografts in bone: induction of osteoblastic reaction by prostate-specific antigen-producing tumors in athymic and SCID/bg mice using LNCaP and lineage-derived metastatic sublines. Int J Cancer 77, 887–894 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Stern AR et al. Isolation and culture of primary osteocytes from the long bones of skeletally mature and aged mice. Biotechniques 52, 361–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai J et al. Cabozantinib inhibits prostate cancer growth and prevents tumor-induced bone lesions. Clin Cancer Res 20, 617–630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai J et al. Reversal of chemotherapy-induced leukopenia using granulocyte macrophage colony-stimulating factor promotes bone metastasis that can be blocked with osteoclast inhibitors. Cancer Res 70, 5014–5023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang M et al. Bone Microenvironment Changes in Latexin Expression Promote Chemoresistance. Mol Cancer Res 15, 457–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


