Abstract
In subgingival plaque biofilms, Fusobacterium nucleatum is closely related to the occurrence and development of periodontitis. Streptococcus gordonii, as an accessory pathogen, can coaggregate with periodontal pathogens, facilitating the subgingival colonization of periodontal pathogens. Studies have shown that F. nucleatum can coaggregate with S. gordonii and colonize the subgingival plaque. However, most studies have focused on monocultures or coinfection of species and the potential impact of coaggregation between the two species on periodontal interactions to human gingival epithelial cells (hGECs) remains poorly understood. The present study explored the effect of coaggregation between F. nucleatum and S. gordonii on subgingival synergistic virulence to hGECs. The results showed that coaggregation inhibited the adhesion and invasion of F. nucleatum to hGECs compared with that in the F. nucleatum monoculture and coinfection group. Coaggregation and coinfection with F. nucleatum both enhanced S. gordonii adhesion to hGECs, but neither of the two groups affected S. gordonii invasion to hGECs compared with S. gordonii monoculture. The gene expression levels of TLR2 and TLR4 in hGECs in the coaggregation group were higher than those in the monoculture groups but lower than those in the coinfection group. Compared with coinfection, the coaggregation inhibited apoptosis of hGECs and promoted the secretion of the proinflammatory cytokines TNF-α and IL-6 by hGECs, showed a synergistic inflammatory effect, while coaggregation inhibited the secretion of the anti-inflammatory cytokine TGF-β1. Coaggregation enhanced the phosphorylation of p65, p38, and JNK proteins and therefore activated the NF-κB and MAPK signaling pathways. Pretreatment with a pathway antagonist/inhibitor decreased the phosphorylation levels of proteins and the secretion of TNF-α and IL-6. In conclusion, coaggregation inhibited the adhesion and invasion of F. nucleatum to hGECs. However, it enhanced the adhesion of S. gordonii to hGECs. Compared with coinfection, coaggregation inhibited the apoptosis of hGECs. The coaggregation coordinately promoted the secretion of TNF-α and IL-6 by hGECs through the TLR/NF-κB and TLR/MAPK signaling pathways while inhibiting the secretion of TGF-β1, thus aggravating the inflammatory response of hGECs.
Keywords: coaggregation, Fusobacterium nucleatum, Streptococcus gordonii, synergistic virulence, human gingival epithelial cells
Introduction
The oral microbiome is comprised of more than 700 prevalent taxa at the species level (Dewhirst et al., 2010; Gao et al., 2018). The physical and metabolic interactions between bacteria, as well as bacteria and their hosts, promote the dynamic development of microbial communities and form dental plaque biofilms. Periodontitis is a common oral disease in which dental plaque biofilms are the main pathogenic factor (Frencken et al., 2017; Peres et al., 2019). In the process of dental plaque formation, different types of bacterial species recognize and bind to each other through coaggregation (Kolenbrander et al., 2010; Guo et al., 2014). The gram-negative bacterium, Fusobacterium nucleatum, is closely related to the occurrence and development of periodontitis, which could coaggregate with early and late colonizers (Kolenbrander et al., 2010; Okuda et al., 2012; Park et al., 2017; Wu et al., 2021). Streptococcus gordonii is commonly considered an early colonizer in the formation of dental plaque biofilms (Jakubovics and Kolenbrander, 2010; Nobbs et al., 2011; Jakubovics et al., 2021). With accumulating evidence showing that S. gordonii can coaggregate with periodontal pathogens, it has been newly recognized as an accessory pathogen for facilitating the subgingival colonization of periodontal pathogens (Daep et al., 2011; Hendrickson et al., 2017; Kuboniwa et al., 2017; Brown et al., 2018). Studies have shown that F. nucleatum can adhere to S. gordonii by the outer membrane proteins RadD and CmpA, which help F. nucleatum colonize the subgingival plaque (Kaplan et al., 2009; Lima et al., 2017).
The first defense barrier of periodontal tissue against microorganisms is gingival epithelial cells (GECs), which not only form an attachment to the tooth surface, but also form a physical and chemical barrier against infection (Kantrong et al., 2019). GECs can bind to bacteria through special receptors on the cell surface to release antimicrobial peptides such as human β-defensins (hBDs), cytokines, or proteases to resist the invasion of external risk factors and maintain epithelial microecological balance (Handfield et al., 2008; Lee and Yilmaz, 2021). As an opportunistic pathogen, F. nucleatum can not only adhere to and invade GECs (Han et al., 2000; Gursoy et al., 2008; Stathopoulou et al., 2010; Jung et al., 2017; Hung et al., 2018), but also promote the invasion of the nonperiodontal pathogen Streptococcus cristatus into GECs (Edwards et al., 2006). This suggests that in the subgingival environment, F. nucleatum, which is located in the same ecological locus as S. gordonii, may also influence the adhesion or invasion of S. gordonii into GECs.
Studies have shown that compared with S. gordonii, Porphyromonas gingivalis, and Aggregatibacter actinomycetemcomitans, F. nucleatum can effectively induce the inflammatory response of GECs and trigger high levels of interleukin (IL)-1β, IL-6, and IL-8, while S. gordonii shows the lowest ability to induce inflammation (Stathopoulou et al., 2010; Peyyala et al., 2012). Expression microarrays revealed that the biological pathways in GECs significantly impacted by F. nucleatum and S. gordonii included toll-like receptors (TLRs) and mitogen-activated protein kinase (MAPK) signaling pathways (Hasegawa et al., 2007). TLRs are innate immune pattern recognition receptors (PRRs) that can identify the proteins, nucleic acids, lipids of pathogenic microorganisms, and intermediate products and metabolites synthesized in the reaction process, such as the lipopolysaccharide (LPS) of gram-negative bacteria (Kantrong et al., 2019) and the lipoteichoic acid (LTA) of the cell wall of gram-positive bacteria (Saito et al., 2020). The downstream NF-κB and MAPK signaling pathways could be activated through MyD88-dependent pathways, inducing the expression of proinflammatory cytokines (IL-1β, IL-6, IL-8, tumor necrosis factor [TNF]-α) and anti-inflammatory cytokines (IL-10, transforming growth factor [TGF]-β1), which play an important role in inflammation, immune regulation, cell survival, and proliferation (Tartey and Takeuchi, 2017).
Previous studies explored the inflammatory effect of bacteria on GECs in monoculture or coinfection states. Coinfection is only a physical mixture of bacteria that cannot truly reflect the biological functions of bacteria in the flora. Interspecies physical attachment initiates signal transduction cascades that trigger important physical changes in partner species, which could not be observed by monospecies or coinfected species experiments. There is now strong evidence that cell-cell interactions could lead to phenotypic adaptations that affect physiological and pathological functions, such as adhesion, cooperation in substrate utilization, environmental adaptation, and virulence (Jakubovics et al., 2008a; Jakubovics et al., 2008b; Ramsey et al., 2011; Meuric et al., 2013). In recent years, RNA-Seq has been gradually applied to the analysis of transcriptional regulation stimulated by interactions between bacteria. The transcriptional responses of S. gordonii and F. nucleatum subsp. nucleatum to coaggregation had been reported (Mutha et al., 2018). Among the five subspecies of F. nucleatum, subsp. nucleatum and polymorphum are both associated with apical periodontitis and periodontitis (Han, 2015). But F. nucleatum subsp. polymorphum showed the greatest ability to increase phagocytic capacity of neutrophils and to block superoxide generation (Kurgan et al., 2017). Our previous study, for the first time, reported that coaggregation between F. nucleatum subsp. polymorphum, and S. gordonii altered bacterial transcriptional profiling and attenuated the immune responses of macrophages (Liu et al., 2021), which may provide some insights into the present study.
In a subgingival plaque, F. nucleatum and S. gordonii coexist in a limited ecological site through coaggregation. However, it is still unclear how coaggregation between F. nucleatum and S. gordonii influences the subgingival synergistic virulence to GECs. This study built coaggregation model of F. nucleatum subsp. polymorphum and S. gordonii to explore the effects of coaggregation on subgingival synergistic virulence to hGECs and analyze the relevant mechanisms. We aimed to deepen the understanding of coaggregation regulation between F. nucleatum and accessory pathogen, providing a new experimental basis for the inhibition of dental plaque biofilm formation and the prevention or treatment of periodontal disease.
Materials and Methods
Bacterial Strains and Growth Conditions
F. nucleatum subsp. polymorphum ATCC 10953 was grown in brain heart infusion (BHI) broth (Difco, USA) supplemented with 5 μg/ml hemin (Sigma-Aldrich, USA), 1 μg/ml vitamin K (Sigma-Aldrich, USA), and 0.5% yeast extract (Difco, USA). S. gordonii DL1 was grown in BHI broth. Both bacterial strains were grown under anaerobic conditions (N2 90%, CO2 5%, H2 5%) at 37°C.
Coaggregation of F. nucleatum subsp. polymorphum and S. gordonii
Coaggregation assays were performed in modified coaggregation buffer (CAB) containing 150 mM NaCl, 1 mM Tris HCl pH 8, 0.1 mM CaCl2, and 0.1 mM MgCl2 as previously described (Kaplan et al., 2009; Kaplan et al., 2014; Lima et al., 2017). The bacterial cells were collected at the late exponential phase of growth. The optical density at 600nm (OD600nm) of F. nucleatum subsp. polymorphum was measured to be 0.80 (~109 CFU/mL), and the OD600nm of S. gordonii was around 0.65 (~109 CFU/mL). The colony-forming units (CFUs) of bacteria was quantified by incubating F. nucleatum subsp. polymorphum on 5% sheep blood agar plates and incubating S. gordonii on BHI agar plates in serial dilutions under anaerobic conditions. Bacterial cells were cleaned and resuspended in CAB to a final concentration of ~2×109 CFU/mL. Equal numbers of bacterial cells from each species were added together and vortexed for 10 seconds in a new reaction tube. The suspensions were settled at room temperature for 10 min to allow the bacteria to coaggregate with each other. The reaction tube was centrifuged at low speed (100×g) for 1 min to pellet coaggregated bacterial cells while leaving the nonaggregated cells in the supernatant. The supernatant was collected carefully for OD600nm measurement. The coaggregation index (C.I.) was calculated as follows (Kaplan et al., 2009; Kaplan et al., 2014): C.I. = (OD600nm(Fnp) + OD600nm (Sg)-OD600nm (Fnp-Sg))/[OD600nm (Fnp)+OD600nm (Sg)]. In this formula, OD600nm(Fnp) and OD600nm (Sg) were the optical density of F. nucleatum subsp. polymorphum and S. gordonii respectively, while OD600nm (Fnp-Sg) was the optical density of the supernatant after coaggregation. Because saliva is the common coaggregation buffer in the oral cavity, the coaggregation index of F. nucleatum subsp. polymorphum and S. gordonii in different concentrations of artificial saliva (Phygene, China) was also calculated. The coaggregation and autoaggregation of the two bacterial species in CAB at different time points were also evaluated and observed with phase contrast microscopy. The autoaggregation index was calculated as follows (Nagaoka et al., 2008; Karched et al., 2015; Toh et al., 2019): (OD600nm (time zero value)- OD600nm (sample value))/(OD600nm (time zero value).
Confocal Laser Scanning Microscopy Identification of Coaggregation of F. nucleatum subsp. polymorphum and S. gordonii
F. nucleatum subsp. polymorphum and S. gordonii were cultured to the late-exponential phase. Bacterial cells were washed three times and resuspended in sterile PBS. For visualization, F. nucleatum subsp. polymorphum was stained green with 5-(and-6)-carboxyfluorescein succinimidyl ester (CFSE) (Thermo Fisher, USA), while S. gordonii was stained red with hexidium iodide (Thermo Fisher, USA) according to the manufacturer’s instructions. Samples were incubated for 15 min in darkness at room temperature. Fluorescently stained bacteria were washed three times with sterile PBS and resuspended in CAB. The coaggregated F. nucleatum subsp. polymorphum and S. gordonii (Fnp-Sg) were obtained as described above. Coculture of the two species (Fnp+Sg) in PBS, where they did not coaggregate with each other but only mixed physically, were used as controls. After coaggregation reactions, 10 μL of coaggregated Fnp-Sg was transferred to a glass slide and covered with a cover glass. The coaggregation and coculture samples were visualized by an Olympus confocal microscope (FV3000, Olympus, Japan) using excitation (Ex) at 492 nm and emission (Em) at 517 nm for CFSE and Ex/Em = 518 nm/600 nm for hexidium iodide.
Culture and Infection of Human Gingival Epithelial Cells In Vitro
Human gingival epithelial cells (hGECs) were obtained from the American Type Culture Collection (ATCC CRL-3397) and incubated in DMEM containing 10% fetal bovine serum (FBS) (Gibco, USA) at 37°C in the presence of 5% CO2 (Huang et al., 2020). Cells were seeded at 3.5 × 105 cells per well in 6-well cell culture plates (Corning, USA). hGECs were infected with F. nucleatum subsp. polymorphum monoculture (Fnp), S. gordonii monoculture (Sg), coinfection of F. nucleatum subsp. polymorphum and S. gordonii (Fnp+Sg), and coaggregation of F. nucleatum subsp. polymorphum and S. gordonii (Fnp-Sg) at an MOI of 100, respectively. The coinfection of F. nucleatum subsp. polymorphum and S. gordonii (Fnp+Sg) was only a physical mixture of bacteria in PBS where they did not coaggregate with each other. To ensure the number of bacterial cells in coaggregates was similar with monocultures, the coaggregates were resuspended in PBS, vigorously vortexed and disrupted until no visible pellet existed with validation under a microscope (Liu et al., 2021). The CFU of the coaggregates were determined by incubating the resuspension solution on 5% sheep blood agar plates and BHI agar plates in serial dilutions. The volumes of bacterial cells used in the coaggregation group were adjusted to ensure the number of bacterial cells in coaggregates were similar with monoculture groups. After incubation at 37°C in 5% CO2 for 4 hours, the culture medium containing bacteria was removed, and the bacteria were washed with PBS three times to remove planktonic bacteria. Cells in each well were added to 2 mL of DMEM containing 10% FBS, 200 μg/mL metronidazole (Solarbio, China) and 300 μg/mL of gentamicin (Solarbio, China) and incubated at 37°C in 5% CO2 for 60 min, 90 min, and 120 min to test the antibiotic treatment time for completely killing of extracellular bacteria in all groups. In detail, hGECs infected with Fnp, Sg, Fnp+Sg, and Fnp-Sg of the same antibiotic treatment time were digested and mixed together. The effect of killing extracellular bacteria was confirmed by incubating the digested cells mixture on a plate containing 10% sterile sheep’s blood at 37°C with 90% N2 + 5% CO2 + 5% H2 for 2-3 days. If bacterial colonies grew on the plate, it meant not all groups achieved a complete killing of extracellular bacteria. After killing the extracellular bacteria, cells were washed with PBS three times and incubated at 37°C in 5% CO2 for different time points. The experiment was performed three times.
Confocal Laser Scanning Microscopy (CLSM) Evaluation of hGEC Infection by F. nucleatum subsp. polymorphum and S. gordonii In Vitro
To examine bacterial infection, CFSE-labeled F. nucleatum subsp. polymorphum and S. gordonii were cocultivated with hGECs for 4 hours on cell slides. After infection, hGECs were washed 3 times with PBS to remove planktonic bacteria. The cells were fixed with 4% paraformaldehyde for 15 min and treated with 0.1% Triton X-100 (Beyotime, China) for 10 min. The cytoskeleton was stained with phalloidin (Thermo Fisher, USA) for 30 min, and the nucleus was stained with DAPI (ZSGB-BIO, China). All CLSM images were obtained by an Olympus confocal microscope using Ex/Em = 492 nm/517 nm for CFSE, Ex/Em = 540 nm/565 nm for phalloidin and Ex/Em = 340 nm/488 nm for DAPI.
Adhesion and Invasion Assay
hGECs were infected with Fnp, Sg, Fnp+Sg, and Fnp-Sg at a MOI of 100. After 4 hours of infection, the cells were washed 3 times with PBS to remove the planktonic bacteria and lysed in sterile water for 90 min to release intracellular bacteria. The total number of F. nucleatum subsp. polymorphum and S. gordonii adhering to and invading hGECs was counted by serial dilution and plating on BHI agar supplemented with yeast extract, hemin, and vitamin K. The agar plates were incubated anaerobically at 37°C with 90% N2 + 5% CO2 + 5% H2 for 2-3 days. For the invasion assay, after 4 hours of infection, the cells were washed 3 times with PBS to remove planktonic bacteria and treated with fresh DMEM supplemented with 10% FBS, 200 μg/mL metronidazole, and 300 μg/mL gentamicin for 120 min to kill extracellular bacteria. Cells were lysed in sterile water for 90 min and the number of intracellular bacteria was determined by serial dilution and plating as described above.
Cell Viability of hGECs Infected by F. nucleatum subsp. polymorphum and S. gordonii
hGECs were inoculated into a 96-well plate (200 μL/well) at a density of 1.0×104 cells per well. Fnp, Sg, Fnp+Sg and Fnp-Sg were added to cells at an MOI of 100. hGECs without any bacterial stimuli were used as the blank control group (control). After 4 hours, samples were treated with DMEM supplemented with 10% FBS, 200 μg/mL metronidazole, and 300 μg/mL gentamicin for 120 min and fresh DMEM supplemented with 10% FBS was added. The proliferation activity of hGECs was determined by the Cell Counting Kit-8 (CCK8 kit, Dojindo, Japan). After the addition of 10 μL of CCK8 solution to each well, the plate was incubated at 37°C in 5% CO2 for 1-4 h. The absorbance at 450 nm (OD450nm) was detected by a microplate reader. The effect of antibiotics alone on the proliferation activity of hGECs was also determined.
Cell Apoptosis of hGECs Infected by F. nucleatum subsp. polymorphum and S. gordonii
hGECs were inoculated into a 6-well plate at 3.5×105 cells per well. Bacterial stimuli were added as described above and hGECs without any bacterial stimuli were used as blank control. After antibiotic treatment, cells were cultured at 37°C in 5% CO2 and digested with trypsin without EDTA at different time points. Cells were washed with PBS twice and collected in flow cytometry tubes with 1~5×105 cells by centrifugation at 1000 rpm for 5 min. An Annexin V-FITC Apoptosis Kit (BD, USA) was used to detect the apoptosis of hGECs according to the manufacturer’s instructions. After 500 μL of binding buffer was used to resuspend the cells, 5 μL of Annexin V-FITC was added and mixed gently. Samples were placed on ice for 15 min, mixed with 5 μL of propodium iodide (PI) and then detected by Beckman Coulter CytoFLEX immediately using Ex/Em = 488 nm/530 nm. Cells without Annexin V-FITC and PI were used as negative controls.
RT–qPCR of the mRNA Expression Levels of TLR2 and TLR4 in hGECs Infected by F. nucleatum subsp. polymorphum and S. gordonii
The expression levels of TLR2 and TLR4 in hGECs were determined by quantitative reverse transcription PCR (RT–qPCR). Total RNA was isolated from hGECs using RNAzol according to the manufacturer’s protocol (Sigma-Aldrich, USA). The concentrations of RNA samples were determined by a NanoDrop 2000C Spectrophotometer (Thermo Fisher, USA). cDNA was synthesized using PrimeScript RT Master Mix (Takara, Japan). RT–qPCR analysis was performed in a 20-μL reaction mixture containing 10-μL of master mix (Hieff qPCR SYBR Green Master Mix, Yeasen) using a Light Cycler 480 (Roche Applied Science, Germany). The reaction product was quantified by the standard curve method. Levels of GAPDH mRNA served as internal controls. The primer sequences were as follows (F/R): TLR2 (ATCAGGCTTCTCTGTCTTGTG/TCTGTAGGTCACTGTTGCTAATG); TLR4 (GGAAGGAGCAGAATCAGGATATG/CTCCATTCACTCCACTAACCAC); and GAPDH (AATCCCATCACCATCTTCCAG/AAATGAGCCCCAGCCTTC).
Cytokine Detection
The hGECs were stimulated with Fnp, Sg, Fnp+Sg and Fnp-Sg as described above. Cell-free supernatants were harvested and stored at -80°C for cytokine assays. Cytokine levels (TNF-α, IL-6, -8, -10 and TGF-β1) in the culture supernatants were measured by ELISA kits (Neobioscience, Shenzhen, China) according to the manufacturer’s instructions.
The Activation of the NF-κB and MAPK Signaling Pathways in hGECs Infected With F. nucleatum subsp. polymorphum and S. gordonii
After removing the supernatant, the cells were washed twice with PBS at 4°C. RIPA lysis buffer (Beyotime, China) containing 1% protease inhibitor (Sigma–Aldrich, USA) and 1% serine protease inhibitor (Sigma–Aldrich, USA) was added for 30 min to lyse cells and extract proteins from each sample. The concentrations of total proteins were detected by a BCA protein assay kit (Beyotime, China) according to the manufacturer’s instructions. Protein samples were mixed with 5× loading buffer (ThermoFisher, USA) at a ratio of 4:1 and boiled at 99°C for 10 min. Samples were loaded and run on SDS–PAGE gels (CWBIO, China) and transferred onto PVDF membranes (Millipore, USA). Membranes were blocked with 2% skim milk (BD, USA) for 1 h at room temperature and then incubated with anti-IKKα, anti-IKKβ, anti-pIKKα/β, anti-p65, anti-pp65, anti-p38, anti-pp38, anti-SAPK/JNK, anti-pSAPK/JNK, and anti-GAPDH primary antibodies (Abcam, UK) overnight at 4°C. After primary incubation, blots were washed and incubated with secondary goat anti-rabbit or goat anti-mouse HRP (Abcam, UK) for 1 hour. Membranes were washed and exposed to chemiluminescent HRP substrate (Millipore, USA). Images were obtained using the GeneGnome XRQ system (Syngene, USA) and analyzed using ImageJ software.
The Inhibition of the NF-κB and MAPK Signaling Pathways in hGECs Infected With F. nucleatum subsp. polymorphum and S. gordonii
Before bacterial stimuli, hGECs were pretreated the TLR2/4 signaling pathway antagonist OxPAPC (In vivoGen, USA) at 30 μg/mL, 400 nM p38 inhibitor SB 239063 (MCE, USA), 2 μM JNK inhibitor SP600125 (MCE, USA), and 4 μM NF-κB signaling pathway inhibitor BAY 11-7082 (MCE, USA) for 1 h. hGECs treated without any inhibitors and bacterial stimuli were used as blank controls, hGECs treated with inhibitors but without any bacterial stimuli were used as negative controls, and hGECs treated with Fnp-Sg infection but without any inhibitors were used as positive controls. Total proteins were extracted at 24 h and 48 h after incubation. The phosphorylation of NF-κB pathway protein p65, MAPK pathway protein p38 and JNK was detected by western blot as described above. The supernatant was collected and used to detect the changes in the secretion of inflammatory cytokines by ELISA.
Statistical Analysis
The bacterial counts data were log (10) transformed for subsequent analysis. All data were presented as the mean ± standard deviation and assessed for normality by Kolmogorov-Smirnov test. The results showed that the data fitted a normal distribution. Differences between two groups were analyzed by Student’s t tests. Differences in the quantitative data between multiple groups were evaluated by one-way ANOVA combined with Bonferroni’s post hoc test. P values less than 0.05 were designated as significant differences. Statistical analyses were conducted by SPSS Statistics v.20 software (IBM, Inc., Chicago, IL, USA) and GraphPad Prism 9 software (GraphPad Software, Inc., San Diego, CA, USA).
Results
The Coaggregation Between F. nucleatum subsp. polymorphum and S. gordonii
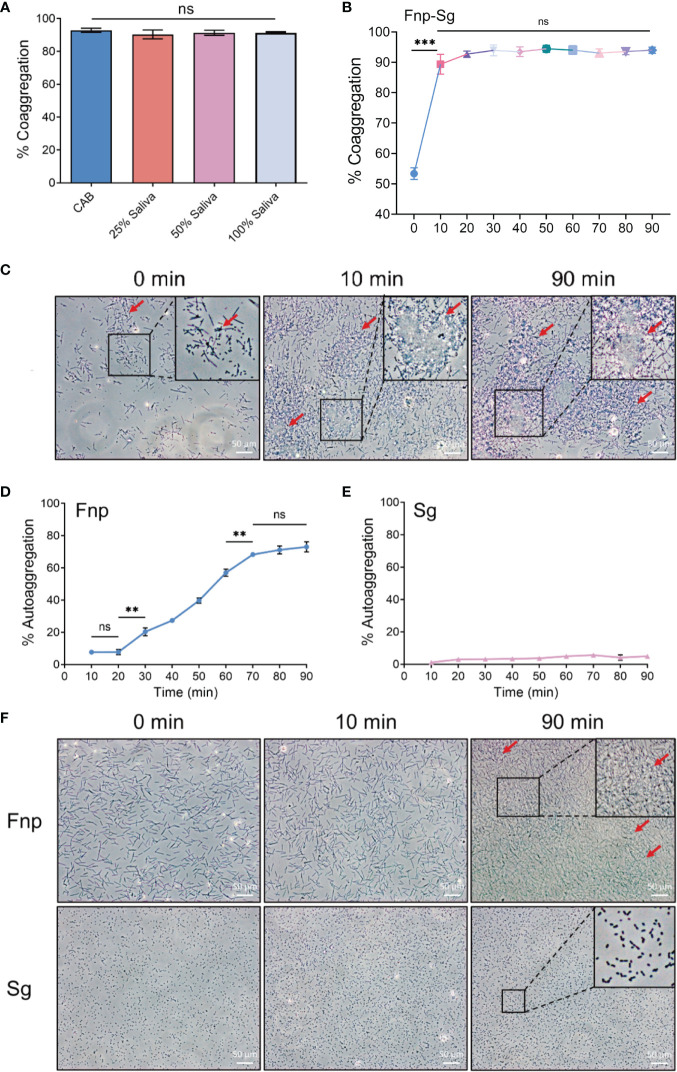
Studies have confirmed that different bacterial strains coaggregate adequately with each other in CAB (Kaplan et al., 2009; Kaplan et al., 2014). Saliva was also used as a coaggregation buffer, of which the composition was complex and included various enzymes, immunoglobulins, and mucins (Heller et al., 2017; Carpenter, 2020). In the present study, F. nucleatum subsp. polymorphum coaggregated strongly with S. gordonii in 10 min with large numbers of coaggregation clumps formed at the bottom of the centrifuge tube and a clear upper suspension. The coaggregation index (C.I.) was 89.370% ± 3.269% (mean ± standard deviation). There was no significant difference between coaggregation indices in CAB and different concentrations of artificial saliva ( Figure 1A ). The coaggregation was stable in 10-90 min with a range of coaggregation indices from 89.370% ± 3.269% to 94.450% ± 1.161% ( Figures 1B, C ). Thus, CAB was used in the present study to exclude the influence of saliva components on the results. Autoaggregation is the adhesion of bacteria of the same strain, which is common with oral bacteria (Khemaleelakul et al., 2006; Merritt et al., 2009). It is mediated by autoagglutinins which is related to surface proteins in general or related to carbohydrates, particularly exopolysaccharides in some cases (Trunk et al., 2018; Yakovlieva and Walvoort, 2020). Although F. nucleatum had been extensively studied on coaggregation, little was known about its autoaggregation. Previous studies showed the autoaggregation of F. nucleatum was strain-dependent and occurs via both saliva-dependent and -independent mechanisms (Merritt et al., 2009; Karched et al., 2015). There were few studies focused on the autoaggregation of S. gordonii. A previous study showed no autoaggregation of S. gordonii (Levin-Sparenberg et al., 2016), while another study showed an increased autoaggregation of S. gordonii depending on the concentration of composite resin containing surface reaction-type pre-reacted glass ionomer eluate used (Shimazu et al., 2016). However, the specific mechanisms of autoaggregation in F. nucleatum and S. gordonii still needed further investigations. In the present study, the autoaggregation of F. nucleatum subsp. polymorphum significantly increased at 20 min ( Figure 1D ) which was consistent with a previous study (Karched et al., 2015). We chose 10 min as the coaggregation time in the present study when there was little autoaggregation of F. nucleatum subsp. polymorphum and S. gordonii ( Figures 1E, F ).
Figure 1.
(A) Quantitative coaggregation assays between F. nucleatum subsp. polymorphum and S. gordonii in CAB and different concentrations of artificial saliva. (B) The stability of coaggregation between F. nucleatum subsp. polymorphum and S. gordonii in CAB in 90 min. (C) Phase contrast microscopy images of coaggregation between F. nucleatum subsp. polymorphum and S. gordonii in CAB at 0, 10 and 90 min (red arrows: coaggregates). (D) The autoaggregation of F. nucleatum subsp. polymorphum at 90 min. (E) The autoaggregation of S. gordonii at 90 min. (F) Phase contrast microscopy images of autoaggregation of F. nucleatum subsp. polymorphum and S. gordonii in CAB at 0, 10 and 90 min (red arrows: autoaggregates) (**p <0.01, ***p <0.001, ns: not statistically significant).
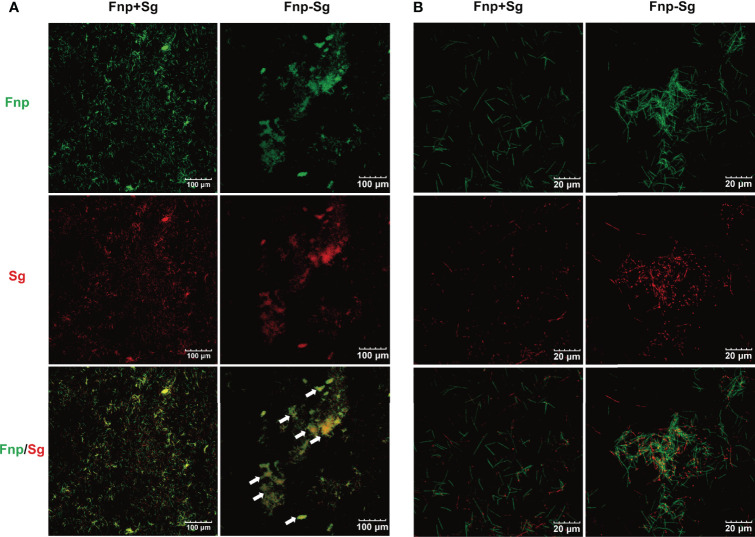
CLSM images at low magnification showed coaggregation between F. nucleatum subsp. polymorphum and S. gordonii in the form of a large number of clumps (white arrows), while the coculture F. nucleatum subsp. polymorphum and S. gordonii were noncoaggregated and distributed separately ( Figure 2A ). Under high magnification, the coaggregation group showed that F. nucleatum subsp. polymorphum and S. gordonii cells adhered to each other tightly and were distributed quite evenly throughout coaggregates ( Figure 2B ).
Figure 2.
CLSM images of the coculture (Fnp+Sg) and coaggregation (Fnp-Sg) of F. nucleatum subsp. polymorphum (green) and S. gordonii (red) at low magnification (A) and high magnification (B). The white arrows show the coaggregates.
The Effect of Antibiotics on Killing Extracellular Bacteria and Cell Proliferation of hGECs
The results showed that the number of extracellular bacteria decreased with prolonged antibiotic treatment time (** p <0.01, *** p <0.001). After 120 min, no visible bacterial colonies grew, indicating the complete killing of extracellular bacteria among all the groups with bacterial stimuli ( Figure S1A ). The CCK-8 results showed that antibiotic treatment had no significant effect on the proliferation of hGECs ( Figure S1B ). Therefore, the extracellular bacteria were killed by antibiotic treatment for 120 min in the present study.
The Infection of hGECs by F. nucleatum subsp. polymorphum and S. gordonii
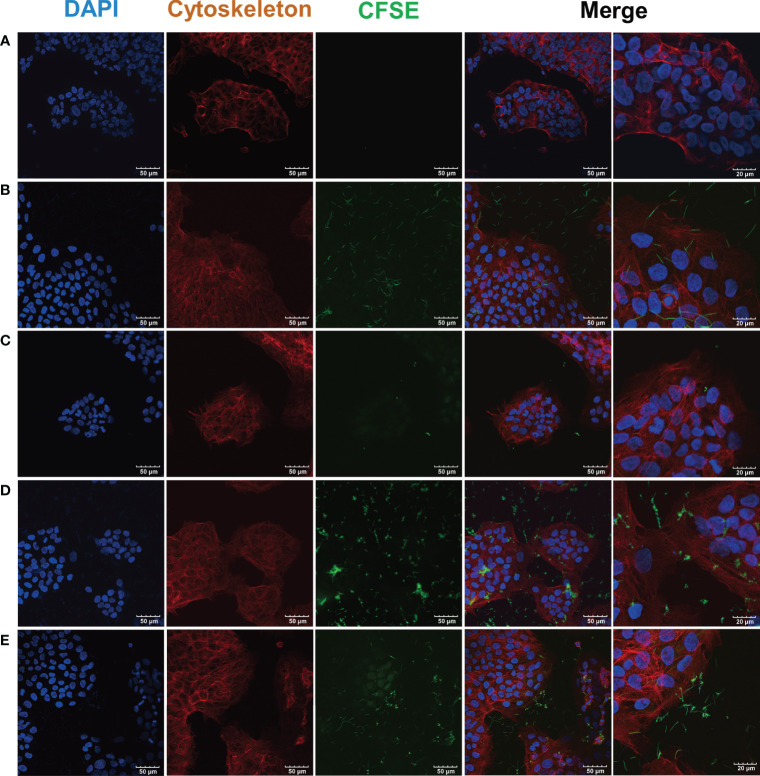
As the CLSM images showed, cell cytoskeleton was stained red with phalloidin and the nucleus was stained blue with DAPI ( Figure 3A ). There was a large number of F. nucleatum subsp. polymorphum infected- hGECs ( Figure 3B ), while S. gordonii hardly infected hGECs ( Figure 3C ). Compared with the coinfection group ( Figure 3D ), the number of F. nucleatum subsp. polymorphum and S. gordonii that infected hGECs seemed reduced in the coaggregation group ( Figure 3E ).
Figure 3.
CLSM images of F. nucleatum subsp. polymorphum and S. gordonii infection of hGECs for 4 h F. nucleatum subsp. polymorphum and S. gordonii were stained green by CFSE. The cytoskeleton was stained red by phalloidin, and the nucleus was stained blue by DAPI. (A) Blank control group. (B) F. nucleatum subsp. polymorphum monoculture group (Fnp). (C) S. gordonii monoculture group (Sg). (D) F. nucleatum subsp. polymorphum and S. gordonii coinfection group (Fnp+Sg). (E) F. nucleatum subsp. polymorphum and S. gordonii coaggregation group (Fnp-Sg).
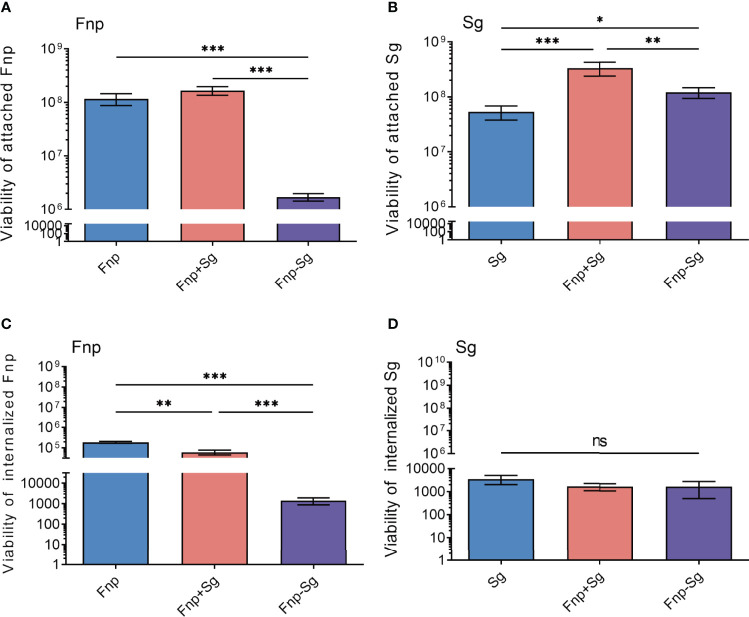
To further explore the infection of F. nucleatum subsp. polymorphum and S. gordonii to hGECs, we quantified the bacteria that adhered to and invaded hGECs by serial dilution and plating, respectively. The results showed that coaggregation significantly inhibited F. nucleatum subsp. polymorphum adhesion and invasion of hGECs (** p <0.01, *** p <0.001) ( Figures 4A, C ). For S. gordonii, coaggregation and coinfection both enhanced the adhesion of hGECs, with the coinfection group showing a stronger effect than the coaggregation group (* p <0.05, ** p <0.01, *** p <0.001) ( Figure 4B ). Nevertheless, the invasion ability of S. gordonii in hGECs was weak among all groups and neither coinfection nor coaggregation influenced the invasion ability of S. gordonii ( Figure 4D ).
Figure 4.
The numbers of attached F. nucleatum subsp. polymorphum (A) and S. gordonii (B), internalized F. nucleatum subsp. polymorphum (C) and S. gordonii (D) to hGECs with an MOI of 100 after 4 h of infection (*p <0.05, **p <0.01, ***p <0.001, ns: not statistically significant).
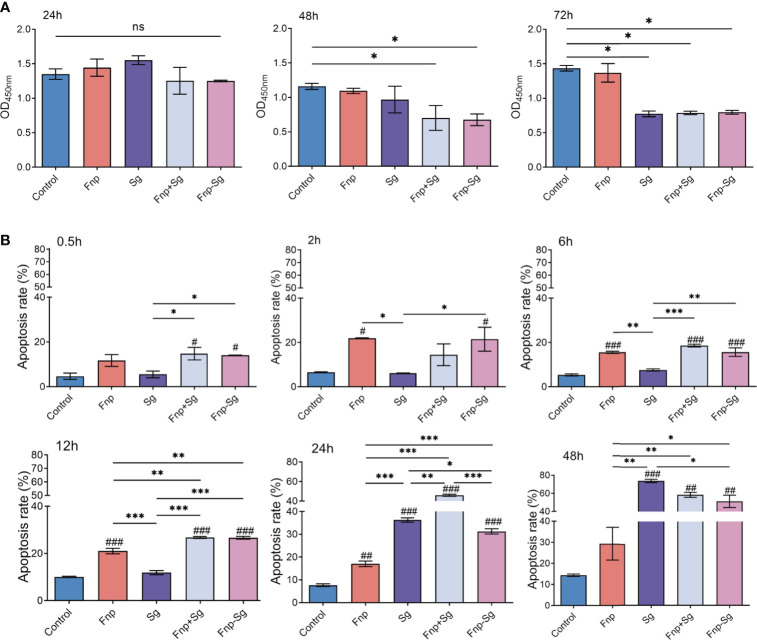
The Effect of Coaggregation Between F. nucleatum subsp. polymorphum and S. gordonii on the Proliferation Activity and Apoptosis of hGECs
The results showed that the monocultures, coinfection, and coaggregation of F. nucleatum subsp. polymorphum and S. gordonii had no significant effect on the proliferation activity of hGECs after 24 h of infection ( Figure 5A ). After 48 h of infection, the proliferation activity of hGECs was significantly reduced in both the coinfection and coaggregation groups with no significant difference between the two groups (* p <0.05). The monoculture of S. gordonii inhibited the proliferation activity of hGECs after 72 h of infection (* p <0.05), while F. nucleatum subsp. polymorphum monoculture had no significant influence on the proliferation activity of hGECs at various time points.
Figure 5.
(A) The proliferation activity of hGECs infected by Fnp, Sg, Fnp+Sg and Fnp-Sg after 24, 48 and 72 h of infection (*p <0.05, ns: not statistically significant). (B) The apoptosis rate of hGECs infected with Fnp, Sg, Fnp+Sg and Fnp-Sg after various time points (*p < 0.05, **p < 0.01, ***p < 0.001; #p < 0.05, ##p < 0.01, ###p < 0.001, compared with the control group).
As shown in Figure 5B , F. nucleatum subsp. polymorphum showed a relatively weak ability to promote cell apoptosis after 2 h of infection and sustained through 24 h. After 12 h of infection, the coaggregation and coinfection of F. nucleatum subsp. polymorphum and S. gordonii significantly promoted hGECs apoptosis compared with the other groups (### p <0.001, ** p <0.01, *** p <0.001, #compared with the control group). There was no significant difference between the coinfection and coaggregation groups within 12 h of infection, however, the coinfection group significantly promoted cell apoptosis at 24 h compared with the coaggregation group (*** p <0.001). After 24 h of infection, the ability of S. gordonii to promote cell apoptosis was enhanced and was the strongest after 48 h. The images of flow cytometry were shown in Figure S2 .
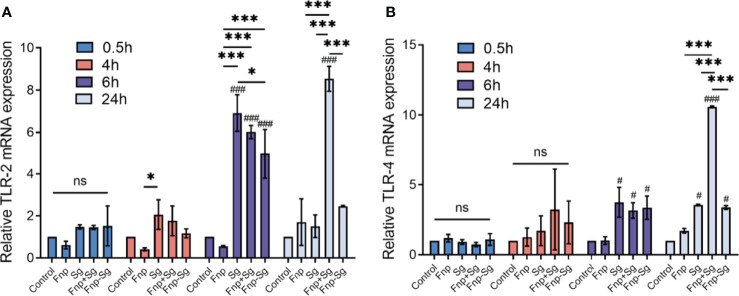
The Effect of Coaggregation Between F. nucleatum subsp. polymorphum and S. gordonii on TLR2 and TLR4 mRNA Expression Levels in hGECs
RT–qPCR was used to detect the effect of coaggregation between F. nucleatum subsp. polymorphum and S. gordonii on the TLR2 and TLR4 mRNA expression levels in hGECs ( Figure 6 ). The results showed that after 6 h of infection, the TLR2 and TLR4 mRNA expression levels were significantly increased in the S. gordonii monoculture, coinfection, and coaggregation groups compared with the control group (# p < 0.05, ### p < 0.001). After 24 h of infection, the TLR2 and TLR4 mRNA expression levels were significantly decreased in the coaggregation group compared with the coinfection group (***p <0.001). In the present study, S. gordonii monoculture increased both TLR2 and TLR4 mRNA expression levels in hGECs, while F. nucleatum subsp. polymorphum monoculture had no significant influence on the expression levels.
Figure 6.
TLR2 (A) and TLR4 (B) mRNA expression levels in hGECs infected with Fnp, Sg, Fnp+Sg and Fnp-Sg (*p < 0.05, ***p < 0.001, #compared with the control group, # p < 0.05, ### p < 0.001, ns: not statistically significant).
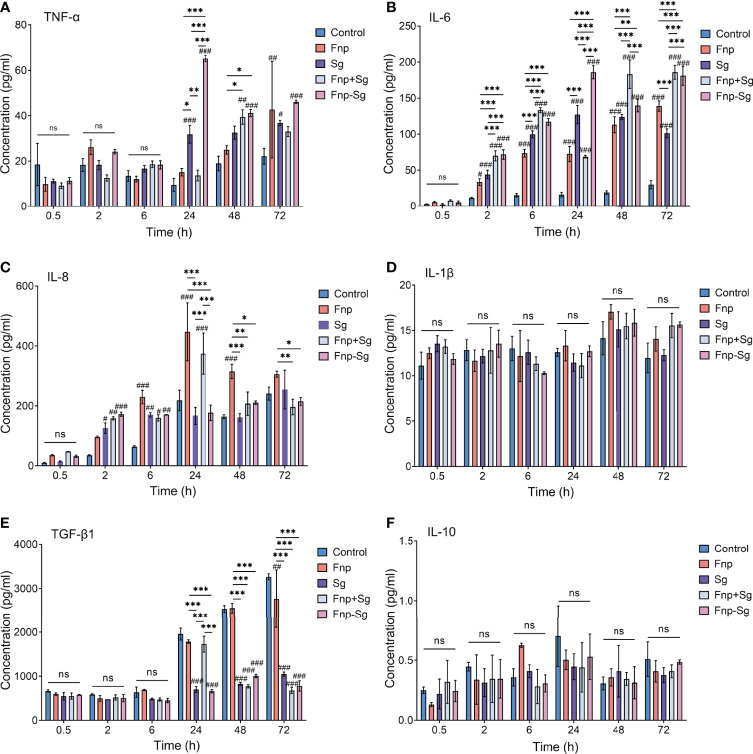
The Effect of Coaggregation Between F. nucleatum subsp. polymorphum and S. gordonii on the Secretion of Inflammatory Cytokines by hGECs
ELISA results showed that the secretion level of TNF-α by hGECs in the coaggregation group was significantly higher than that in other groups at 24 h of infection (***p <0.001, ### p <0.001, #compared with the control group), while no difference was found between groups at 0.5 h, 2 h, and 6 h. Afterwards, the secretion level of TNF-α in the coaggregation group decreased with no difference compared with the coinfection group ( Figure 7A ). The secretion level of IL-6 steadily reached the highest in the coaggregation group at 24 h of infection, showing a significantly higher level than that in other groups (***p <0.001, ### p <0.001, #compared with the control group). The secretion level of IL-6 in the coinfection group increased within 6 h and decreased at 24 h of infection. However, the secretion level of IL-6 in the coinfection group restored and was dramatically higher than that in the coaggregation group at 48 h of infection ( Figure 7B ). As for IL-8, F. nucleatum subsp. polymorphum monoculture and coinfection groups resulted in high secretion levels at 24 h of infection and then decreased. The coaggregation group did not significantly promote IL-8 secretion at various time points ( Figure 7C ). The secretion levels of TGF-β1 were higher in the control and F. nucleatum subsp. polymorphum monoculture groups than that in other groups at 48 h and 72 h of infection (***p <0.001, ### p <0.001, #compared with the control group). However, the secretion level of TGF-β1 in the coinfection group increased significantly at 24 h with no difference compared with the control and F. nucleatum subsp. polymorphum monoculture groups and decreased afterwards ( Figure 7E ). No significant difference was found in the secretion of IL-1β and IL-10 among the groups ( Figures 7D, F ).
Figure 7.
The production of TNF-α, IL-6, IL-8, IL-1β, TGF-β1 and IL-10 in hGECs infected with Fnp, Sg, Fnp+Sg and Fnp-Sg, as assessed by ELISA (*p < 0.05, **p < 0.01, ***p < 0.001; # p < 0.05, ## p < 0.01, ### p < 0.001, #compared with the control group, ns: not statistically significant).
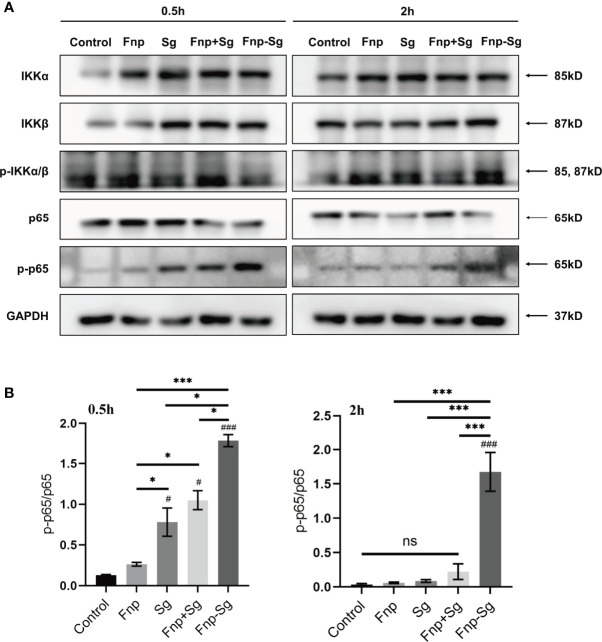
Activation of the NF-κB and MAPK Signaling Pathways in hGECs Infected by Coaggregation Between F. nucleatum subsp. polymorphum and S. gordonii
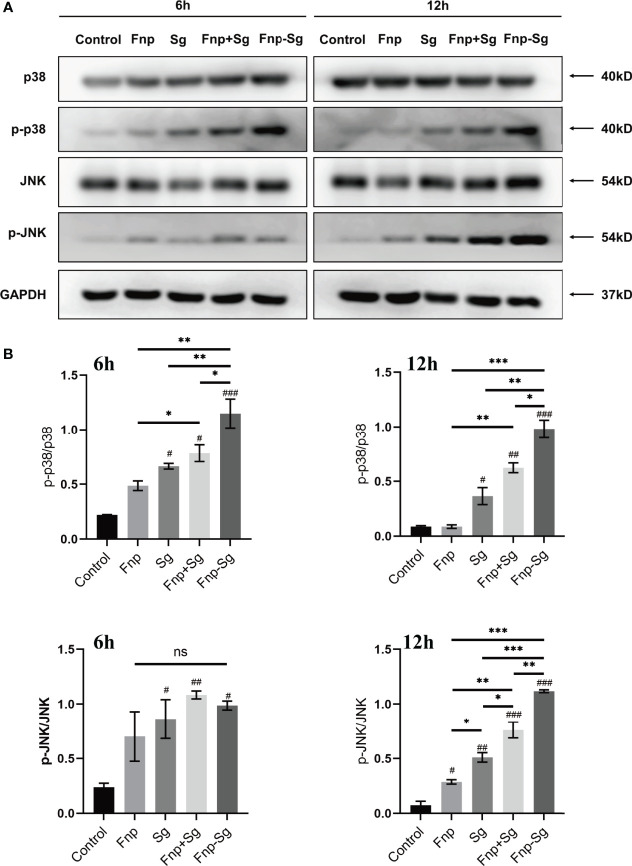
The western blot and semiquantitative analysis results showed that the phosphorylation level of p65 (p-p65) protein in the coaggregation group was higher than that of the other groups after 0.5 h and 2 h of infection (* p <0.05, ** p <0.01, *** p <0.001, #p <0.05, ###p <0.001, #compared with the control group) ( Figure 8 ). Moreover, the phosphorylation level of p38 (p-p38) protein in the coaggregation group was higher than that of the other groups after 6 h and 12 h of infection (* p <0.05, ** p <0.01, *** p <0.001, #p <0.05, ##p <0.01, ###p <0.001, #compared with the control group) ( Figure 9 ). After 12 h of infection, the phosphorylation level of JNK (p-JNK) protein in the coaggregation group was higher than that in the other groups (* p <0.05, ** p <0.01, *** p <0.001, #p <0.05, ##p <0.01, ###p <0.001, #compared with the control group) ( Figure 9 ).
Figure 8.
The phosphorylation levels (A) and semiquantitative analysis (B) of proteins in NF-κB signaling pathways in hGECs infected with Fnp, Sg, Fnp+Sg and Fnp-Sg (*p < 0.05, ***p < 0.001; #p < 0.05, ###p < 0.001, # compared with the control group; ns: not statistically significant).
Figure 9.
The phosphorylation levels (A) and semiquantitative analysis (B) of proteins in MAPK signaling pathways in hGECs infected with Fnp, Sg, Fnp+Sg and Fnp-Sg (*p < 0.05, **p < 0.01, ***p < 0.001; #p < 0.05, ##p < 0.01, ###p < 0.001, # compared with the control group; ns: not statistically significant).
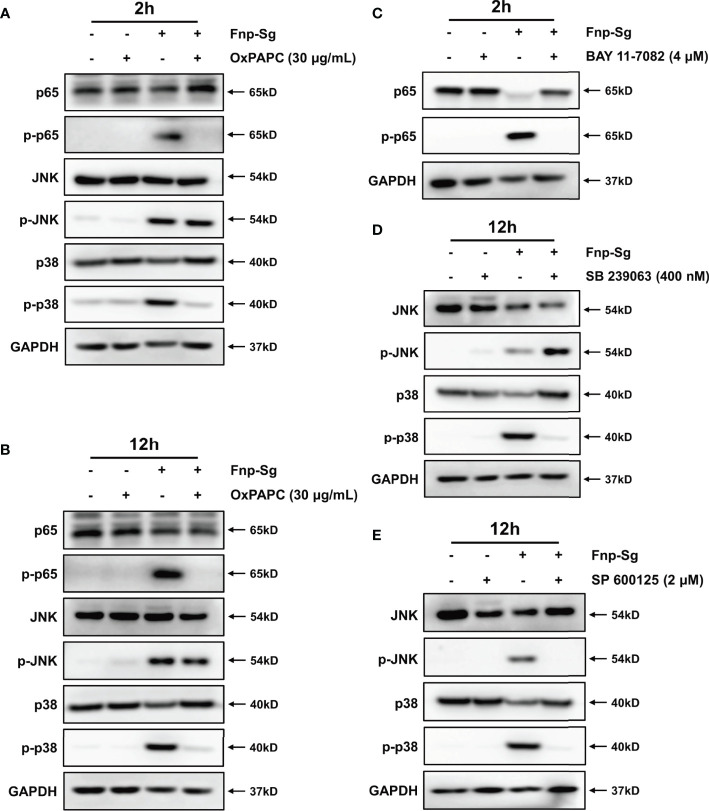
The Regulation of the NF-κB and MAPK Signaling Pathways in hGECs by Coaggregation Between F. nucleatum subsp. polymorphum and S. gordonii
By pretreatment of hGECs with the TLR2/4 antagonist OxPAPC for 1 h, the protein expression of p-p65, p-p38, and p-JNK in the coaggregation group was decreased ( Figures 10A, B ). After pretreatment with the NF-κB inhibitor BAY 11-7082, the protein expression of p-p65 decreased significantly ( Figure 10C ). Pretreatment with the p38 MAPK inhibitor SB 239063 decreased the p-p38 protein level; thus, the p-JNK protein level showed a compensatory increase ( Figure 10D ). Pretreatment with the JNK MAPK inhibitor SP 600125 significantly decreased the p-JNK protein level. Because the targets of the inhibitor SP 600125 also included the upstream kinases MKK3, MKK4, and MKK6 in the p38 MAPK signaling pathway, the p-p38 protein level was decreased significantly ( Figure 10E ).
Figure 10.
The regulations of the NF-κB and MAPK signaling pathways in hGECs by coaggregation between F. nucleatum subsp. polymorphum and S. gordonii by western blot. hGECs were pretreated with the TLR2/4 antagonist OxPAPC for 2 h (A) and 12 h (B), the NF-κB inhibitor BAY 11-7082 (B) for 2 h (C), the p38 MAPK inhibitor SB 239063 for 12 h (D) and the JNK MAPK inhibitor SP 600125 for 12 h (E).
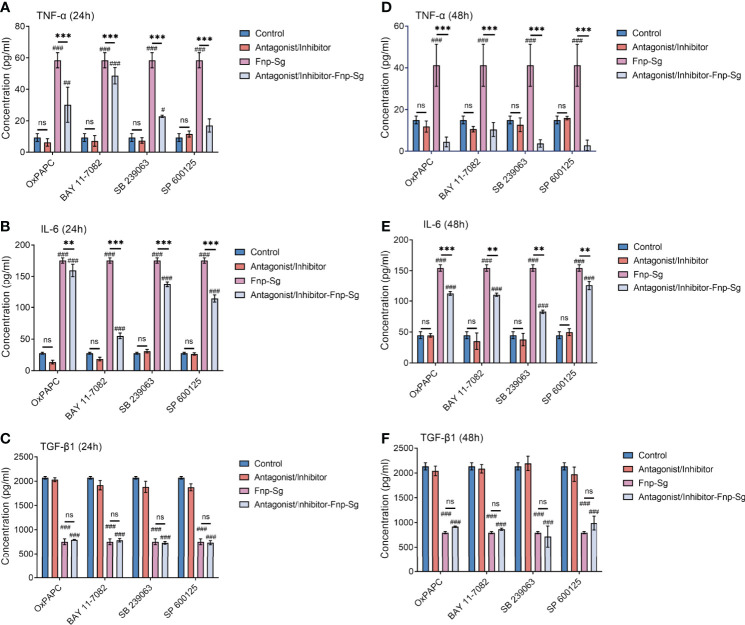
The changes in the secretion of inflammatory cytokines by hGECs were also evaluated by ELISA. By pretreatment of hGECs with antagonist or inhibitors, the levels of TNF-α and IL-6 decreased in the coaggregation group after 24 h and 48 h of infection (** p < 0.01, *** p < 0.001) ( Figures 11A, B, D, E ), but no significant difference was detected in the secretion of TGF-β1 ( Figures 11C, F ).
Figure 11.
The secretion levels of inflammatory cytokines by hGECs pretreated with antagonist or inhibitors at 24 h (A–C) and 48 h of infection (D–F) (**p < 0.01, ***p < 0.001, ## p < 0.01, ### p < 0.001, #compared with the control group, ns: not statistically significant).
Discussions
Coaggregation with early colonizers is important for the colonization of F. nucleatum in the oral flora (Guo et al., 2014). Although S. gordonii are generally considered early colonizers and commensal organisms, increasing evidence shows that they are becoming recognized as important associated pathogens during the development of periodontal disease (Croft et al., 2018). The metabolite of S. gordonii, 4-aminobenzoate/p-aminobenzoic acid (pABA), can increase the subgingival colonization and intracellular survival of P. gingivalis but decrease its pathogenicity (Kuboniwa et al., 2017). Therefore, the interactions between bacterial species may affect bacterial colonization and pathogenicity. Although studies have proven that F. nucleatum can use the outer membrane proteins RadD and CmpA to adhere to S. gordonii and colonize the same ecological locus of the subgingival plaque (Kaplan et al., 2009; Lima et al., 2017), this largely remains to be investigated. Here, we evaluated the effects of coaggregation between F. nucleatum subsp. polymorphum and S. gordonii on the subgingival synergistic interactions to hGECs and analyzed the potential mechanisms in the development of periodontal disease.
The multilayer model of hGECs infected with P. gingivalis, A. actinomycetemcomitans, F. nucleatum or S. gordonii showed that P. gingivalis invaded intracellularly and spread cell to cell, A. actinomycetemcomitans and F. nucleatum remained extracellular and showed intercellular movement through the multilayer, while S. gordonii remained extracellular and predominantly associated with the superficial cell layer (Dickinson et al., 2011). Although the study established a multilayer structure of hGECs to stimulate the actual oral environment, oral bacteria usually did not infect hGECs in monocultures. The interactions between oral bacteria may influence adhesion to and invasion of hGECs. A previous study reported that F. nucleatum could promote noninvasive Streptococcus cristae and Streptococcus sanguinis adhesion to and invasion of hGECs (Edwards et al., 2006). Studies on the polymicrobial infections of hGECs showed that F. nucleatum improved the adhesion and invasion of periodontal pathogens P. gingivalis and A. actinomycetemcomitans in hGECs (Saito et al., 2009; Saito et al., 2012; Li et al., 2015). At the same time, P. gingivalis has been confirmed to inhibit F. nucleatum invasion of hGECs by gingipain when in a coinfection state (Jung et al., 2017). Current studies have mostly focused on monoculture infection or polymicrobial infection with periodontal pathogens, and the effects of the interactions between F. nucleatum and S. gordonii on the adhesion and invasion of hGECs remain to be investigated. In the present study, coinfection with F. nucleatum subsp. polymorphum improved S. gordonii adhesion to hGECs more significantly than coaggregation with F. nucleatum subsp. polymorphum, indicating that different mechanisms functioned during the two types of infections. As for F. nucleatum subsp. polymorphum, coaggregation with S. gordonii significantly inhibited the adhesion and invasion of hGECs. Based on a previous study reporting that F. nucleatum could survive in hGECs for no more than 12 h (Ji et al., 2010), it was speculated that coaggregation may improve the extracellular survival of F. nucleatum subsp. polymorphum by inhibiting its adhesion and invasion of hGECs.
In our study, infection time within 24 h was considered appropriate for evaluations because after 48 h of infection, the cell proliferation activity was significantly inhibited in both coaggregation and coinfection groups. Meanwhile, the cell apoptosis rate was significantly increased in S. gordonii monoculture, coaggregation, and coinfection groups after 48 h of infection, with the highest cell apoptosis rate nearly 80% in S. gordonii monoculture group. This may be related to the exhaustion of media nutrients caused by the accumulated amount of S. gordonii in these three groups, making it challenging for hGECs proliferation and survival. In the present study, F. nucleatum subsp. polymorphum significantly promoted hGECs apoptosis after 2 h and sustained through 24 h of infection (Dickinson et al., 2011), which was consistent with the previous study. However, a weak ability to induce cell apoptosis was found in S. gordonii before 12 h, possibly because of a symbiotic relationship between S. gordonii and hGECs in the early stage of infection. According to the previous study, P. gingivalis could activate the phosphoinositide 3-kinase (PI3K) signaling pathway to inhibit the apoptosis of hGECs when coinfected with F. nucleatum, facilitating the intracellular survival of P. gingivalis and F. nucleatum (Maekawa et al., 2014). This suggested that bacteria in monoculture or coinfection resulted in different regulatory mechanisms of cell apoptosis. In the present study, coaggregation of F. nucleatum subsp. polymorphum and S. gordonii showed similar effects on hGECs proliferation activity with coinfection, while coaggregation showed an inhibitory effect on cell apoptosis at 24 h of infection compared with coinfection. This suggested that, compared with coinfection of the two species, coaggregation inhibited hGECs apoptosis which may facilitate the intracellular survival of bacteria and favor a prolonged cell inflammation induction.
F. nucleatum and lipopolysaccharide can stimulate the secretion of proinflammatory cytokines and chemokines, leading to inflammation and bone resorption (Huang et al., 2004; Hasegawa et al., 2007; de Andrade et al., 2019; Kantrong et al., 2019). The outer membrane proteins FadA and Fap2 of F. nucleatum are involved in both bacterial coaggregation mediation and infection of various host cells which induce inflammatory responses (Xu et al., 2007; Ikegami et al., 2009; Fardini et al., 2011). Lipoteichoic acid and lipoprotein are the main virulence factors of S. gordonii in bacterial infection and inflammatory responses (Bi et al., 2017). Studies have shown that F. nucleatum significantly promotes the secretion of IL-6, IL-8, and IL-1β in hGECs, with no significant effect on IL-10 secretion, while S. gordonii has no significant effect on the secretion of IL-6, IL-8, IL-1β, and IL-10 at 4 h or 24 h of infection (Ji et al., 2007; Stathopoulou et al., 2010) and even inhibits IL-6 and IL-8 secretion during 8 h of infection (Hasegawa et al., 2007). Another study showed that F. nucleatum significantly promoted the secretion of TNF-α and IL-1β, while S. gordonii promoted the secretion of TNF-α, IL-6, IL-8, and IL-1β after 24 h of infection (Dickinson et al., 2011). The inconsistency of the results may be because of the different MOIs, bacterial subspecies, or epithelial cell models (monolayer or multilayers). Most studies in vitro investigated the secretion levels of inflammatory cytokines by bacterial stimuli were within 24 h of infection. In the present study, we found that the secretion levels of inflammatory cytokines in each group had a relatively consistent trend of variety from 0.5 h to 24 h ( Figure 9 ). However, not all groups maintained the original trend after 48 h, especially for IL-6 secretion level in the coinfection group, which was dramatically increased and higher than the coaggregation group after 48 h of infection. The precise reason for this fluctuation in IL-6 secretion level was not clear. However, during the experiment, we observed that the floating debris or dead cells were much more obvious in the three groups infected with S. gordonii monoculture, coinfection and coaggregation of F. nucleatum subsp. polymorphum and S. gordonii after 48 h of infection. Combined with the significant decrease in cell proliferation activity and significant increase in cell apoptosis after 48 h in the present study, these results may all be related to the exhaustion of media nutrients caused by the accumulated amount of S. gordonii in these three groups. Therefore, in the present study, we focused on analyzing and concluding the changes of secretion levels of inflammatory cytokines among the groups within 24 h of infection. Nevertheless, the results showed that compared with coinfection, coaggregation between F. nucleatum subsp. polymorphum and S. gordonii promoted hGECs to secrete the proinflammatory factors TNF-α and IL-6 at 24 h of infection, while inhibiting the secretion of the anti-inflammatory factor TGF-β1. Different from studies in vitro, animal studies usually took a long-term evaluation of host responses (Polak et al., 2009; Polak et al., 2012; de Molon et al., 2014). An animal study showed that at 42 days post-infection, coinfection with F. nucleatum and P. gingivalis synergistically promoted the loss of periodontal bone tissue and aggravated inflammatory responses in rats (Polak et al., 2009). Animal experiments with long-term evaluations could be used to explore and verify the specific mechanisms for further study.
It was reported that the NF-κB and MAPK signaling pathways were involved in IL-8 secretion by hGECs infected with F. nucleatum (Huang et al., 2004). TLR2 and TLR4 simultaneously mediated the secretion of IL-6 and TNF-α by hGECs infected by F. nucleatum, which also activated the NF-κB and MAPK signaling pathways (Park et al., 2014). Compared with hGECs infected by F. nucleatum monoculture, coinfection with P. gingivalis or A. actinomycetemcomitans significantly reduced the secretion of IL-8 and inhibited host inflammatory responses after 4 h of infection (Li et al., 2015). In the present study, western blot results verified significant activation of NF-κB in the coaggregation group at 0.5 h and 2 h, as well as significant activation of MAPK at 6 h and 12 h of infection. This indicated that both NF-κB and MAPK signaling pathways were involved in the regulatory effect of coaggregation of the two species on inflammatory responses, with NF-κB activation at an earlier stage of infection. However, an absence of an effect on IL-8 was observed based on the phosphorylation of p65. Although IL-8 is a classical downstream of the NF-κB signaling pathway, the regulatory mechanism of IL-8 seems to be complex. A previous study showed MK2 was involved in regulating the TNF-induced expression of IL-8 by p38 MAPK in human lung microvascular endothelial cells at a post-transcriptional level (Su et al., 2008). Another study showed the stimulation of synovial fibroblasts with IL-6 and TNF-α cooperatively inhibited the induction of IL-8 (Valin et al., 2020). It was speculated that a more complex mechanism in IL-8 secretion existed induced by coaggregation of F. nucleatum subsp. polymorphum and S. gordonii.
In the present study, no significant changes in TLR4 gene expression levels were observed in hGECs infected by F. nucleatum subsp. polymorphum monoculture. This may be because a relatively lower MOI was used than previous studies in which the MOI was 200 or 1000 (Ji et al., 2009; Sun et al., 2010). The MOI was limited to 100 in the present study because the number of S. gordonii was the same as that of F. nucleatum subsp. polymorphum for coaggregation. S. gordonii at a larger MOI grew exponentially and caused cell apoptosis or death rapidly because of the accelerated consumption of nutrients. At the transcriptional level, the coaggregation of F. nucleatum subsp. polymorphum and S. gordonii upregulated the expression levels of TLR2 and TLR4 in hGECs, but the expression levels were lower than those in hGECs infected by coinfection of the two species. This may indicate that the activation of the NF-κB and MAPK signaling pathways in hGECs infected by coaggregation of the two species did not occur through the upregulation of TLR2 and TLR4, but through the enhanced bacterial virulence induced by coaggregation. Moreover, compared with hGECs infected with coinfection bacteria, coaggregation inhibited the secretion of the anti-inflammatory cytokine TGF-β1, suggesting that coaggregation of F. nucleatum subsp. polymorphum and S. gordonii could aggravate the cellular inflammatory response through a two-way regulation of proinflammatory and anti-inflammatory cytokines. Interestingly, the secretion of TGF-β1 did not change with the use of related pathway antagonists/inhibitors. The underlying mechanism of the regulation of TGF-β1 secretion in hGECs induced by coaggregation still needs to be investigated.
By RNA-Seq, Mutha et al. (Mutha et al., 2018) found that by comparison with monocultures, 16 genes were regulated following coaggregation in F. nucleatum subsp. nucleatum whereas 119 genes were regulated in S. gordonii. In both species, genes involved in amino acid and carbohydrate metabolism were strongly affected by coaggregation (Mutha et al., 2018). Our previous transcriptome results indicated up-regulated genes associated with protein export systems and repressed arginine biosynthesis in S. gordonii after coaggregation might help enhance and maintain a symbiotic relationship with F. nucleatum subsp. polymorphum (Liu et al., 2021). In F. nucleatum subsp. polymorphum, genes related to LPS or peptidoglycan biosynthesis were downregulated, which might reduce the immunogenicity of F. nucleatum subsp. polymorphum and improve bacterial survival within macrophages (Liu et al., 2021). Besides, the coaggregation of F. nucleatum subsp. polymorphum and S. gordonii exhibited significantly decreased levels of propanoic acid and butyric acid than dual-species co-cultures (Liu et al., 2021). The symbiotic lifestyle and metabolic changes of F. nucleatum subsp. polymorphum and S. gordonii after dual-species coaggregation may contribute to the regulatory effect on the synergistic virulence to hGECs in the present study. In further study, bacterial mutants should be constructed for more rigorous conclusions and validations.
In contrast to previous studies that only considered S. gordonii as an early colonizer, our study revealed that the functions of S. gordonii coaggregated with F. nucleatum subsp. polymorphum in the periodontal virulence. The regulatory effect of interactions between F. nucleatum subsp. polymorphum with S. gordonii in the process of periodontal diseases may be more fully interpreted. In further studies, animal models of the colonization of coaggregated bacteria on the tooth surface or gingival sulcus are needed to investigate of the potential mechanism.
Conclusions
In summary, the coaggregation between F. nucleatum subsp. polymorphum and S. gordonii inhibited the adhesion and invasion of F. nucleatum subsp. polymorphum to hGECs but enhanced the adhesion of S. gordonii to hGECs. Coaggregation between F. nucleatum subsp. polymorphum and S. gordonii coordinately promoted the secretion of the proinflammatory cytokines TNF-α and IL-6 by hGECs through the TLR/NF-κB and TLR/MAPK signaling pathways, while inhibiting the secretion of the anti-inflammatory cytokine TGF-β1, thus aggravating the inflammatory response of hGECs.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Author Contributions
LG and RY conceived, designed, and performed experiments. XW, LG, and RY analyzed the data. RY wrote the manuscript. XW, LG, and ZL reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant number: 81670982). The funders did not play a role in manuscript design, data collection, data analysis, data interpretation, or writing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.879423/full#supplementary-material
(A) Extracellular bacteria after antibiotic treatment with 200 μg/mL metronidazole and 300 μg/mL gentamicin for 60, 90 and 120 min (**p <0.01, ***p <0.001). (B) The effect of antibiotic treatment on hGECs proliferation activity for 60, 90 and 120 min.
Flow cytometry images of hGECs infected by Fnp, Sg, Fnp+Sg and Fnp-Sg after various time points.
References
- Bi J., Koivisto L., Pang A., Li M., Jiang G., Aurora S., et al. (2017). Suppression of Alphavbeta6 Integrin Expression by Polymicrobial Oral Biofilms in Gingival Epithelial Cells. Sci. Rep. 7 (1), 4411. doi: 10.1038/s41598-017-03619-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Yates E. A., Bielecki M., Olczak T., Smalley J. W. (2018). Potential Role for Streptococcus Gordonii-Derived Hydrogen Peroxide in Heme Acquisition by Porphyromonas Gingivalis. Mol. Oral. Microbiol. 33 (4), 322–335. doi: 10.1111/omi.12229 [DOI] [PubMed] [Google Scholar]
- Carpenter G. H. (2020). Salivary Factors That Maintain the Normal Oral Commensal Microflora. J. Dent. Res. 99 (6), 644–649. doi: 10.1177/0022034520915486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft A. J., Metcalfe S., Honma K., Kay J. G. (2018). Macrophage Polarization Alters Postphagocytosis Survivability of the Commensal Streptococcus Gordonii. Infect. Immun. 86 (3), e00858–17. doi: 10.1128/IAI.00858-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daep C. A., Novak E. A., Lamont R. J., Demuth D. R. (2011). Structural Dissection and In Vivo Effectiveness of a Peptide Inhibitor of Porphyromonas Gingivalis Adherence to Streptococcus Gordonii. Infect. Immun. 79 (1), 67–74. doi: 10.1128/IAI.00361-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrade K. Q., Almeida-da-Silva C. L. C., Coutinho-Silva R. (2019). Immunological Pathways Triggered by Porphyromonas Gingivalis and Fusobacterium Nucleatum: Therapeutic Possibilities? Mediators Inflamm. 2019, 7241312. doi: 10.1155/2019/7241312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Molon R. S., de Avila E. D., Boas Nogueira A. V., Chaves de Souza J. A., Avila-Campos M. J., de Andrade C. R., et al. (2014). Evaluation of the Host Response in Various Models of Induced Periodontal Disease in Mice. J. Periodontol. 85 (3), 465–477. doi: 10.1902/jop.2013.130225 [DOI] [PubMed] [Google Scholar]
- Dewhirst F. E., Chen T., Izard J., Paster B. J., Tanner A. C., Yu W. H., et al. (2010). The Human Oral Microbiome. J. Bacteriol 192 (19), 5002–5017. doi: 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B. C., Moffatt C. E., Hagerty D., Whitmore S. E., Brown T. A., Graves D. T., et al. (2011). Interaction of Oral Bacteria With Gingival Epithelial Cell Multilayers. Mol. Oral. Microbiol. 26 (3), 210–220. doi: 10.1111/j.2041-1014.2011.00609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. M., Grossman T. J., Rudney J. D. (2006). Fusobacterium Nucleatum Transports Noninvasive Streptococcus Cristatus Into Human Epithelial Cells. Infect. Immun. 74 (1), 654–662. doi: 10.1128/IAI.74.1.654-662.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardini Y., Wang X., Temoin S., Nithianantham S., Lee D., Shoham M., et al. (2011). Fusobacterium Nucleatum Adhesin FadA Binds Vascular Endothelial Cadherin and Alters Endothelial Integrity. Mol. Microbiol. 82 (6), 1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frencken J. E., Sharma P., Stenhouse L., Green D., Laverty D., Dietrich T. (2017). Global Epidemiology of Dental Caries and Severe Periodontitis - a Comprehensive Review. J. Clin. Periodontol 44 (Suppl 18), S94–S105. doi: 10.1111/jcpe.12677 [DOI] [PubMed] [Google Scholar]
- Gao L., Xu T., Huang G., Jiang S., Gu Y., Chen F. (2018). Oral Microbiomes: More and More Importance in Oral Cavity and Whole Body. Protein Cell 9 (5), 488–500. doi: 10.1007/s13238-018-0548-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., He X., Shi W. (2014). Intercellular Communications in Multispecies Oral Microbial Communities. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursoy U. K., Kononen E., Uitto V. J. (2008). Intracellular Replication of Fusobacteria Requires New Actin Filament Formation of Epithelial Cells. APMIS 116 (12), 1063–1070. doi: 10.1111/j.1600-0463.2008.00868.x [DOI] [PubMed] [Google Scholar]
- Han Y. W. (2015). Fusobacterium Nucleatum: A Commensal-Turned Pathogen. Curr. Opin. Microbiol. 23, 141–147. doi: 10.1016/j.mib.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield M., Baker H. V., Lamont R. J. (2008). Beyond Good and Evil in the Oral Cavity: Insights Into Host-Microbe Relationships Derived From Transcriptional Profiling of Gingival Cells. J. Dent. Res. 87 (3), 203–223. doi: 10.1177/154405910808700302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. W., Shi W., Huang G. T., Kinder Haake S., Park N. H., Kuramitsu H., et al. (2000). Interactions Between Periodontal Bacteria and Human Oral Epithelial Cells: Fusobacterium Nucleatum Adheres to and Invades Epithelial Cells. Infect. Immun. 68 (6), 3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y., Mans J. J., Mao S., Lopez M. C., Baker H. V., Handfield M., et al. (2007). Gingival Epithelial Cell Transcriptional Responses to Commensal and Opportunistic Oral Microbial Species. Infect. Immun. 75 (5), 2540–2547. doi: 10.1128/IAI.01957-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller D., Helmerhorst E. J., Oppenheim F. G. (2017). Saliva and Serum Protein Exchange at the Tooth Enamel Surface. J. Dent. Res. 96 (4), 437–443. doi: 10.1177/0022034516680771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson E. L., Beck D. A., Miller D. P., Wang Q., Whiteley M., Lamont R. J., et al. (2017). Insights Into Dynamic Polymicrobial Synergy Revealed by Time-Coursed RNA-Seq. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Kuang S., Shen Z., Liang M., Lin Z. (2020). High Glucose Disrupts Autophagy Lysosomal Pathway in Gingival Epithelial Cells via ATP6V0C. J. Periodontol. 91 (5), 705–714. doi: 10.1002/JPER.19-0262 [DOI] [PubMed] [Google Scholar]
- Huang G. T., Zhang H. B., Dang H. N., Haake S. K. (2004). Differential Regulation of Cytokine Genes in Gingival Epithelial Cells Challenged by Fusobacterium Nucleatum and Porphyromonas Gingivalis. Microb. Pathog. 37 (6), 303–312. doi: 10.1016/j.micpath.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Hung S. C., Huang P. R., Almeida-da-Silva C. L. C., Atanasova K. R., Yilmaz O., Ojcius D. M. (2018). NLRX1 Modulates Differentially NLRP3 Inflammasome Activation and NF-kappaB Signaling During Fusobacterium Nucleatum Infection. Microbes Infect. 20 (9-10), 615–625. doi: 10.1016/j.micinf.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami A., Chung P., Han Y. W. (2009). Complementation of the fadA Mutation in Fusobacterium Nucleatum Demonstrates That the Surface-Exposed Adhesin Promotes Cellular Invasion and Placental Colonization. Infect. Immun. 77 (7), 3075–3079. doi: 10.1128/IAI.00209-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S., Gill S. R., Iobst S. E., Vickerman M. M., Kolenbrander P. E. (2008. a). Regulation of Gene Expression in a Mixed-Genus Community: Stabilized Arginine Biosynthesis in Streptococcus Gordonii by Coaggregation With Actinomyces Naeslundii. J. Bacteriol 190 (10), 3646–3657. doi: 10.1128/JB.00088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S., Gill S. R., Vickerman M. M., Kolenbrander P. E. (2008. b). Role of Hydrogen Peroxide in Competition and Cooperation Between Streptococcus Gordonii and Actinomyces Naeslundii. FEMS Microbiol. Ecol. 66 (3), 637–644. doi: 10.1111/j.1574-6941.2008.00585.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S., Goodman S. D., Mashburn-Warren L., Stafford G. P., Cieplik F. (2021). The Dental Plaque Biofilm Matrix. Periodontol 2000 86 (1), 32–56. doi: 10.1111/prd.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S., Kolenbrander P. E. (2010). The Road to Ruin: The Formation of Disease-Associated Oral Biofilms. Oral. Dis. 16 (8), 729–739. doi: 10.1111/j.1601-0825.2010.01701.x [DOI] [PubMed] [Google Scholar]
- Ji S., Kim Y., Min B. M., Han S. H., Choi Y. (2007). Innate Immune Responses of Gingival Epithelial Cells to Nonperiodontopathic and Periodontopathic Bacteria. J. Periodontal Res. 42 (6), 503–510. doi: 10.1111/j.1600-0765.2007.00974.x [DOI] [PubMed] [Google Scholar]
- Ji S., Shin J. E., Kim Y. C., Choi Y. (2010). Intracellular Degradation of Fusobacterium Nucleatum in Human Gingival Epithelial Cells. Mol. Cells 30 (6), 519–526. doi: 10.1007/s10059-010-0142-8 [DOI] [PubMed] [Google Scholar]
- Ji S., Shin J. E., Kim Y. S., Oh J. E., Min B. M., Choi Y. (2009). Toll-Like Receptor 2 and NALP2 Mediate Induction of Human Beta-Defensins by Fusobacterium Nucleatum in Gingival Epithelial Cells. Infect. Immun. 77 (3), 1044–1052. doi: 10.1128/iai.00449-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y. J., Jun H. K., Choi B. K. (2017). Porphyromonas Gingivalis Suppresses Invasion of Fusobacterium Nucleatum Into Gingival Epithelial Cells. J. Oral. Microbiol. 9 (1), 1320193. doi: 10.1080/20002297.2017.1320193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantrong N., To T. T., Darveau R. P. (2019). Gingival Epithelial Cell Recognition of Lipopolysaccharide. Adv. Exp. Med. Biol. 1197, 55–67. doi: 10.1007/978-3-030-28524-1_5 [DOI] [PubMed] [Google Scholar]
- Kaplan A., Kaplan C. W., He X., McHardy I., Shi W., Lux R. (2014). Characterization of Aid1, a Novel Gene Involved in Fusobacterium Nucleatum Interspecies Interactions. Microb. Ecol. 68 (2), 379–387. doi: 10.1007/s00248-014-0400-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. W., Lux R., Haake S. K., Shi W. (2009). The Fusobacterium Nucleatum Outer Membrane Protein RadD is an Arginine-Inhibitable Adhesin Required for Inter-Species Adherence and the Structured Architecture of Multispecies Biofilm. Mol. Microbiol. 71 (1), 35–47. doi: 10.1111/j.1365-2958.2008.06503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karched M., Bhardwaj R. G., Asikainen S. E. (2015). Coaggregation and Biofilm Growth of Granulicatella Spp. With Fusobacterium Nucleatum and Aggregatibacter Actinomycetemcomitans. BMC Microbiol. 15, 114. doi: 10.1186/s12866-015-0439-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemaleelakul S., Baumgartner J. C., Pruksakom S. (2006). Autoaggregation and Coaggregation of Bacteria Associated With Acute Endodontic Infections. J. Endod. 32 (4), 312–318. doi: 10.1016/j.joen.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E., Palmer R. J., Jr., Periasamy S., Jakubovics N. S. (2010). Oral Multispecies Biofilm Development and the Key Role of Cell-Cell Distance. Nat. Rev. Microbiol. 8 (7), 471–480. doi: 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- Kuboniwa M., Houser J. R., Hendrickson E. L., Wang Q., Alghamdi S. A., Sakanaka A., et al. (2017). Metabolic Crosstalk Regulates Porphyromonas Gingivalis Colonization and Virulence During Oral Polymicrobial Infection. Nat. Microbiol. 2 (11), 1493–1499. doi: 10.1038/s41564-017-0021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurgan S., Kansal S., Nguyen D., Stephens D., Koroneos Y., Hasturk H., et al. (2017). Strain-Specific Impact of Fusobacterium Nucleatum on Neutrophil Function. J. Periodontol 88 (4), 380–389. doi: 10.1902/jop.2016.160212 [DOI] [PubMed] [Google Scholar]
- Lee J. S., Yilmaz O. (2021). Key Elements of Gingival Epithelial Homeostasis Upon Bacterial Interaction. J. Dent. Res. 100 (4), 333–340. doi: 10.1177/0022034520973012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Sparenberg E., Shin J. M., Hastings E. M., Freeland M., Segaloff H., Rickard A. H., et al. (2016). High-Throughput Quantitative Method for Assessing Coaggregation Among Oral Bacterial Species. Lett. Appl. Microbiol. 63 (4), 274–281. doi: 10.1111/lam.12622 [DOI] [PubMed] [Google Scholar]
- Li Y., Guo H., Wang X., Lu Y., Yang C., Yang P. (2015). Coinfection With Fusobacterium Nucleatum can Enhance the Attachment and Invasion of Porphyromonas Gingivalis or Aggregatibacter Actinomycetemcomitans to Human Gingival Epithelial Cells. Arch. Oral. Biol. 60 (9), 1387–1393. doi: 10.1016/j.archoralbio.2015.06.017 [DOI] [PubMed] [Google Scholar]
- Lima B. P., Shi W., Lux R. (2017). Identification and Characterization of a Novel Fusobacterium Nucleatum Adhesin Involved in Physical Interaction and Biofilm Formation With Streptococcus Gordonii. Microbiologyopen 6 (3),e00444. doi: 10.1002/mbo3.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Yang R., Zhou J., Lu X., Yuan Z., Wei X., et al. (2021). Interactions Between Streptococcus Gordonii and Fusobacterium Nucleatum Altered Bacterial Transcriptional Profiling and Attenuated the Immune Responses of Macrophages. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.783323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Krauss J. L., Abe T., Jotwani R., Triantafilou M., Triantafilou K., et al. (2014). Porphyromonas Gingivalis Manipulates Complement and TLR Signaling to Uncouple Bacterial Clearance From Inflammation and Promote Dysbiosis. Cell Host Microbe 15 (6), 768–778. doi: 10.1016/j.chom.2014.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J., Niu G., Okinaga T., Qi F. (2009). Autoaggregation Response of Fusobacterium Nucleatum. Appl. Environ. Microbiol. 75 (24), 7725–7733. doi: 10.1128/AEM.00916-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuric V., Martin B., Guyodo H., Rouillon A., Tamanai-Shacoori Z., Barloy-Hubler F., et al. (2013). Treponema Denticola Improves Adhesive Capacities of Porphyromonas Gingivalis. Mol. Oral. Microbiol. 28 (1), 40–53. doi: 10.1111/omi.12004 [DOI] [PubMed] [Google Scholar]
- Mutha N. V. R., Mohammed W. K., Krasnogor N., Tan G. Y. A., Choo S. W., Jakubovics N. S. (2018). Transcriptional Responses of Streptococcus Gordonii and Fusobacterium Nucleatum to Coaggregation. Mol. Oral. Microbiol. 33 (6), 450–464. doi: 10.1111/omi.12248 [DOI] [PubMed] [Google Scholar]
- Nagaoka S., Hojo K., Murata S., Mori T., Ohshima T., Maeda N. (2008). Interactions Between Salivary Bifidobacterium Adolescentis and Other Oral Bacteria: In Vitro Coaggregation and Coadhesion Assays. FEMS Microbiol. Lett. 281 (2), 183–189. doi: 10.1111/j.1574-6968.2008.01092.x [DOI] [PubMed] [Google Scholar]
- Nobbs A. H., Jenkinson H. F., Jakubovics N. S. (2011). Stick to Your Gums: Mechanisms of Oral Microbial Adherence. J. Dent. Res. 90 (11), 1271–1278. doi: 10.1177/0022034511399096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T., Kokubu E., Kawana T., Saito A., Okuda K., Ishihara K. (2012). Synergy in Biofilm Formation Between Fusobacterium Nucleatum and Prevotella Species. Anaerobe 18 (1), 110–116. doi: 10.1016/j.anaerobe.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Park S. R., Kim D. J., Han S. H., Kang M. J., Lee J. Y., Jeong Y. J., et al. (2014). Diverse Toll-Like Receptors Mediate Cytokine Production by Fusobacterium Nucleatum and Aggregatibacter Actinomycetemcomitans in Macrophages. Infect. Immun. 82 (5), 1914–1920. doi: 10.1128/IAI.01226-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S., Ryu E. J., Li L., Choi B. K., Kim B. M. (2017). New Bicyclic Brominated Furanones as Potent Autoinducer-2 Quorum-Sensing Inhibitors Against Bacterial Biofilm Formation. Eur. J. Med. Chem. 137, 76–87. doi: 10.1016/j.ejmech.2017.05.037 [DOI] [PubMed] [Google Scholar]
- Peres M. A., Macpherson L. M. D., Weyant R. J., Daly B., Venturelli R., Mathur M. R., et al. (2019). Oral Diseases: A Global Public Health Challenge. Lancet 394 (10194), 249–260. doi: 10.1016/S0140-6736(19)31146-8 [DOI] [PubMed] [Google Scholar]
- Peyyala R., Kirakodu S. S., Novak K. F., Ebersole J. L. (2012). Oral Microbial Biofilm Stimulation of Epithelial Cell Responses. Cytokine 58 (1), 65–72. doi: 10.1016/j.cyto.2011.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak D., Shapira L., Weiss E. I., Houri-Haddad Y. (2012). The Role of Coaggregation Between Porphyromonas Gingivalis and Fusobacterium Nucleatum on the Host Response to Mixed Infection. J. Clin. Periodontol 39 (7), 617–625. doi: 10.1111/j.1600-051X.2012.01889.x [DOI] [PubMed] [Google Scholar]
- Polak D., Wilensky A., Shapira L., Halabi A., Goldstein D., Weiss E. I., et al. (2009). Mouse Model of Experimental Periodontitis Induced by Porphyromonas Gingivalis/Fusobacterium Nucleatum Infection: Bone Loss and Host Response. J. Clin. Periodontol. 36 (5), 406–410. doi: 10.1111/j.1600-051X.2009.01393.x [DOI] [PubMed] [Google Scholar]
- Ramsey M. M., Rumbaugh K. P., Whiteley M. (2011). Metabolite Cross-Feeding Enhances Virulence in a Model Polymicrobial Infection. PloS Pathog. 7 (3), e1002012. doi: 10.1371/journal.ppat.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Inagaki S., Ishihara K. (2009). Differential Ability of Periodontopathic Bacteria to Modulate Invasion of Human Gingival Epithelial Cells by Porphyromonas Gingivalis. Microb. Pathog. 47 (6), 329–333. doi: 10.1016/j.micpath.2009.09.012 [DOI] [PubMed] [Google Scholar]
- Saito A., Kokubu E., Inagaki S., Imamura K., Kita D., Lamont R. J., et al. (2012). Porphyromonas Gingivalis Entry Into Gingival Epithelial Cells Modulated by Fusobacterium Nucleatum is Dependent on Lipid Rafts. Microb. Pathog. 53 (5-6), 234–242. doi: 10.1016/j.micpath.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Okuno A., Cao D. Y., Peng Z., Wu H. Y., Lin S. H. (2020). Bacterial Lipoteichoic Acid Attenuates Toll-Like Receptor Dependent Dendritic Cells Activation and Inflammatory Response. Pathogens 9 (10), 825. doi: 10.3390/pathogens9100825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu K., Oguchi R., Takahashi Y., Konishi K., Karibe H. (2016). Effects of Surface Reaction-Type Pre-Reacted Glass Ionomer on Oral Biofilm Formation of Streptococcus Gordonii. Odontology 104 (3), 310–317. doi: 10.1007/s10266-015-0217-2 [DOI] [PubMed] [Google Scholar]
- Stathopoulou P. G., Benakanakere M. R., Galicia J. C., Kinane D. F. (2010). Epithelial Cell Pro-Inflammatory Cytokine Response Differs Across Dental Plaque Bacterial Species. J. Clin. Periodontol 37 (1), 24–29. doi: 10.1111/j.1600-051X.2009.01505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Ao L., Zou N., Song Y., Yang X., Cai G. Y., et al. (2008). Post-Transcriptional Regulation of TNF-Induced Expression of ICAM-1 and IL-8 in Human Lung Microvascular Endothelial Cells: An Obligatory Role for the P38 MAPK-MK2 Pathway Dissociated With HSP27. Biochim. Biophys. Acta 1783 (9), 1623–1631. doi: 10.1016/j.bbamcr.2008.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Shu R., Li C. L., Zhang M. Z. (2010). Gram-Negative Periodontal Bacteria Induce the Activation of Toll-Like Receptors 2 and 4, and Cytokine Production in Human Periodontal Ligament Cells. J. Periodontol 81 (10), 1488–1496. doi: 10.1902/jop.2010.100004 [DOI] [PubMed] [Google Scholar]
- Tartey S., Takeuchi O. (2017). Pathogen Recognition and Toll-Like Receptor Targeted Therapeutics in Innate Immune Cells. Int. Rev. Immunol. 36 (2), 57–73. doi: 10.1080/08830185.2016.1261318 [DOI] [PubMed] [Google Scholar]
- Toh Y. S., Yeoh S. L., Yap I. K. S., Teh C. S. J., Win T. T., Thong K. L., et al. (2019). Role of Coaggregation in the Pathogenicity and Prolonged Colonisation of Vibrio Cholerae. Med. Microbiol. Immunol. 208 (6), 793–809. doi: 10.1007/s00430-019-00628-3 [DOI] [PubMed] [Google Scholar]
- Trunk T., Khalil H. S., Leo J. C. (2018). Bacterial Autoaggregation. AIMS Microbiol. 4 (1), 140–164. doi: 10.3934/microbiol.2018.1.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valin A., Del Rey M. J., Municio C., Usategui A., Romero M., Fernandez-Felipe J., et al. (2020). IL6/sIL6R Regulates TNFalpha-Inflammatory Response in Synovial Fibroblasts Through Modulation of Transcriptional and Post-Transcriptional Mechanisms. BMC Mol. Cell Biol. 21 (1), 74. doi: 10.1186/s12860-020-00317-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen Y. W., Scheible M., Chang C., Wittchen M., Lee J. H., et al. (2021). Genetic and Molecular Determinants of Polymicrobial Interactions in Fusobacterium Nucleatum. Proc. Natl. Acad. Sci. USA 118 (23), e2006482118. doi: 10.1073/pnas.2006482118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Yamada M., Li M., Liu H., Chen S. G., Han Y. W. (2007). FadA From Fusobacterium Nucleatum Utilizes Both Secreted and Nonsecreted Forms for Functional Oligomerization for Attachment and Invasion of Host Cells. J. Biol. Chem. 282 (34), 25000–25009. doi: 10.1074/jbc.M611567200 [DOI] [PubMed] [Google Scholar]
- Yakovlieva L., Walvoort M. T. C. (2020). Processivity in Bacterial Glycosyltransferases. ACS Chem. Biol. 15 (1), 3–16. doi: 10.1021/acschembio.9b00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Extracellular bacteria after antibiotic treatment with 200 μg/mL metronidazole and 300 μg/mL gentamicin for 60, 90 and 120 min (**p <0.01, ***p <0.001). (B) The effect of antibiotic treatment on hGECs proliferation activity for 60, 90 and 120 min.
Flow cytometry images of hGECs infected by Fnp, Sg, Fnp+Sg and Fnp-Sg after various time points.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.