Abstract
The development of AlphaFold2 marked a paradigm-shift in the structural biology community. Herein, we assess the ability of AlphaFold2 to predict disordered regions against traditional sequence-based disorder predictors. We find that AlphaFold2 performs well at discriminating disordered regions, but also note that the disorder predictor one constructs from an AlphaFold2 structure determines accuracy. In particular, a naïve, but non-trivial assumption that residues assigned to helices, strands, and H-bond stabilized turns are likely ordered and all other residues are disordered results in a dramatic overestimation in disorder; conversely, the predicted local distance difference test (pLDDT) provides an excellent measure of residue-wise disorder. Furthermore, by employing molecular dynamics (MD) simulations, we note an interesting relationship between the pLDDT and secondary structure, that may explain our observations and suggests a broader application of the pLDDT for characterizing the local dynamics of intrinsically disordered proteins and regions (IDPs/IDRs).
Keywords: AlphaFold2, disordered proteins, IDPs/IDRs, machine-learning, biophysics, structural bioinformatics, molecular dynamics, simulation
1. Introduction
Predicting the three-dimensional structure of a protein from its primary amino acid sequence is a grand challenge in molecular structural biology dating back to the late 1950’s [1,2]. About a year and a half ago, AlphaFold2 (AF2), a deep-learning program, provided a paradigm-shift in this problem [3]. Not only did it outperform all other groups at the 14th Critical Assessment of protein Structure Prediction (CASP14) [3], but it did so with astonishing accuracy and a large margin. Consequently, this breakthrough has caused enthusiasm in several related fields, including drug development [4].
The full problem of protein folding is, however, multi-faceted, and despite AlphaFold’s stellar success, many problems and open questions remain. As has already been pointed out by several authors [5,6,7,8,9], dynamics of protein folding remains a formidable problem; prediction of the folding pathways, effects of mutations, the solution environment, aggregation and, as a very particular category, intrinsically disordered proteins and regions (IDPs/IDRs).
IDPs remain a major challenge since they are almost entirely devoid of native structure and because they function primarily as conformational ensembles [10,11,12,13,14,15,16,17] with folding free energy landscapes that are relatively flat [18,19,20]. This is a direct consequence of their amino acid sequences [21,22,23], in particular the enrichment of disorder-promoting residues over and above order-promoting ones [24,25,26,27]. The application of AF2 to the prediction of IDRs and IDPs has only briefly been discussed in the literature [6,7,8,28], and its performance against multiple traditional predictor methods is currently absent.
In light of the recent publication of the critical assessment of protein intrinsic disorder (CAID) benchmark [29], detailing the performance of over three dozen sequence-based disorder predictors and their datasets, we saw an excellent opportunity to benchmark AF2. Herein, we compare the performance of AF2 to the top performing sequence-based disorder predictors as determined at CAID. Importantly, while we find AlphaFold2 to perform exceptionally well on disorder identification; we also note that the disorder predictor one constructs from an AlphaFold2 structure determines accuracy. Specifically, a naïve, non-trivial assumption that the structure assignment provided by DSSP [30], the primary method for assigning secondary structure based on protein geometry, can be used for the determination of disordered regions, leads to a dramatic overestimation in disorder content and represents a potential pitfall for researchers who are less familiar with IDPs and structural prediction methods.
The predicted local distance difference test (pLDDT), which is correlated to the confidence of the structure prediction, provides a better metric for identifying ordered and disordered regions. Furthermore, we find that traditional predictors are capable of outperforming AF2 in disorder prediction even when the pLDDT is used. We also show how secondary structure and pLDDT scores are interestingly related, providing a potential explanation for the observed performance discrepancy and highlight a link between local protein dynamics and the pLDDT using a well characterized IDP and MD simulations.
2. Methodology
2.1. Dataset Generation
Two datasets were used in this work, DisProt and DisProt-PDB, derived from the DisProt database [31]. Both reference sets are based on the CAID benchmark dataset and are composed of 475 targets, annotated between June and November 2018 (DisProt release 2018_11). Note that this is less than the 646 targets used at CAID because AF2 predicted structures do not exist for some sequences. In the DisProt reference set, all residues not labeled as disordered (1) are labeled as ordered (0). We would like to note that such a definition has significant limitations and the conclusions we draw herein are principally based on the DisProt-PDB dataset. Figures and tables based on the DisProt set are found in Supplemental Information and care should be taken when drawing conclusions from them. Our decision to include them here is simply for completeness. The DisProt-PDB reference set, on the other hand, only annotates residues for which some experimental data are available; either a PDB structure that suggests a residue to be ordered or experimental findings, catalogued in DisProt, which suggest a residue to be disordered. Note that if a conflict arises between a DisProt entry suggesting disorder and a PDB structure suggesting order, a disordered assignment is made. All residues not covered by PDB structures or DisProt annotations are masked and were excluded from analysis. As a result, the DisProt-PDB dataset contains no ’uncertain’ residues. All residues considered in this set have either a DisProt annotation, based on prior literature, or belong to a PDB structure. We note that the EMBL/AF2 database contains some structures that are present in the dataset. The degree to which this improves the performance of AF2 is not easily measured; however, it is our belief the impact to be small. Additional details pertaining to dataset construction are provided in Supplementary Information and the full list of proteins, structures, and combined disorder data are available at https://github.com/SoftSimu/AlphaFoldDisorderData (accessed on 21 September 2021).
AF2 structures were downloaded from the EMBL database (https://alphafold.ebi.ac.uk/, accessed on 21 September 2021) and run using DSSP [30] to assign secondary structure. We assume that residues belonging to helices, strands, or H-bond stabilized turns are ordered (0) and all other residues are disordered (1). We refer to this as the näive DSSP predictor or DSSPp for short.
We also collected pLDDT values for each structure. Every residue in an AF2 structure is assigned a value, scaled between 0 and 100, which predicts the C local distance difference test (lDDT) [3,28,32] score of a model; in short, this metric captures the residue-wise confidence of an AF2 model. We transform this value according to the equation,
| (1) |
as suggested by Tunyasuvunakool et al. [28], giving us a pLDDT-based predictor of disorder, where 1 is disordered and 0 is ordered. We refer to this prediction method as the transformed pLDDT or tpLD for short.
We can discretize this pLDDT predictor by classifying a residue with a pLDDT score ≥n as ordered (0) and disordered (1) otherwise; we use pLDDT (or pLD for short), to indicate this binary predictor. Thresholds for n were chosen based on the Matthews correlation coefficient (MCC), which has been documented to be an excellent metric for assessing the accuracy of binary classifiers [33] and was the approach used at CAID [29]. Notice this gives us two predictors: (1) a continuous predictor (tpLDDT) where a residue’s degree of disorderedness is captured, and (2) a discrete predictor (pLD) where a residue is either disordered or ordered depending on the pLDDT and chosen threshold (n). The CAID dataset contains predictions made by three dozen predictors. We selected the top 10 performing on the DisProt and DisProt-PDB giving a combined non-redundant set of 11 (fIDPnn [34], SPOT-Disorder2 [35], RawMSA [36], fIDPlr [34], PreDisorder [37], AUCpreD [38], SPOT-Disorder1 [39], SPOT-Disorder-Single (SPOT-Disorder-S) [40], DisoMine [41], AUCpreD-np [38] and ESpritz-D [42]). The sequence predictors provide a score between 0 and 1, inclusive, as well as a binary disorder/order assignment. No modification to the classification thresholds for these predictors was attempted. Descriptions of disorder prediction methods are provided in the Supplementary Information of the original CAID paper [29]. For two vectors, v and w, we compute the RMSD as
| (2) |
where m is the number of elements (residues) in each vector (protein), v and w. Given binary vectors, a random predictor has an RMSD of on a uniform dataset. Receiver operating characteristic (ROC), area under the curve (AUC), precision–recall, F-score, and correlation analysis were all performed using scikit-learn [43], and kernel density estimate (KDE) analysis was performed in seaborn [44]. Descriptions of statistical methods are provided in Supplementary Information.
2.2. Nrf2 Structure Generation
We used ColabFold [45] to generate both Neh4 and Neh5 structures, our model IDP systems. Two approaches were used: the first was to consider the peptide sequences used in our previous work [46,47], specifically 111SDALYFDDCMQLLAQTFPFVDDN133 and 180MQQDIEQVWEELLSIPELQCLNIENDKLVE209. These are the Neh4 and Neh5 domains, respectively. The second approach was to consider the more realistic construct that includes the linker 106AHIPKSDALYFDDCMQLLAQTFPFVDDNEVSSATFQSLVPDIPGHIESPVFIATNQAQSPETSVAQVAPVDLDGMQQDIEQVWEELLSIPELQCLNIENDKLVETTMVP214 and extract the local structures comprising the domains. ColabFold generates five ranked structures per sequence giving rise to three pools of structures. Alignment of the structures within the Neh4 and Neh5 pools showed excellent agreement and we opted to simply consider the top-ranked structures in each pool, denoted Neh4 (P) and Neh5 (P). Alignment of these peptide structures to the longer construct suggests good agreement; however, there were some constructs with structural differences. We consider the construct that was the most heterogeneous with respect to the smaller peptides and extracted the local Neh4 and Neh5 structures, denoted Neh4 (C) and Neh5 (C).
2.3. Molecular Dynamics Simulations
The MD simulation protocols for the two force fields were almost identical, the primary difference was that the simulations using the Amber-99SB*-ILDNP [48,49,50] force field were performed at 310 K with the TIP3P water model [51] while the Amber99SB-disp [52] simulations were performed at 298.15 K with the TIP4P-disp water model [52]. Note that the Amber-99SB*-ILDNP simulations were taken from our previous work [46] while the Amber-99SB*-disp runs were new to the work discussed herein. In both cases, the steepest descent algorithm was utilized for energy minimization, temperature was maintained using the Parrinello–Donadio–Bussi velocity rescaling method [53] with a 1.0 ps coupling time and pressure were maintained using the Parrinello–Rahman barostat [54] at 1 bar with a coupling time of 5.0 ps. The simulation time step was 2.0 fs. Long-range electrostatic interactions were calculated using the particle-mesh Ewald (PME) method [55] with a Fourier spacing of 0.12 nm and a real-space cut-off of 1.0 nm; the Lennard–Jones interactions were computed with a 1.2 nm cut-off. H-bonds were constrained using the LINear Constraint Solver (P-LINCS) [56]. K or Cl ions were added to neutralize excess charge, i.e., overall charge neutrality was always preserved. Each simulation was performed in quadruplicate for 3 μs, totalling 12 μs of simulation time for each force field–protein combination.
3. Results
3.1. pLDDT Performs Better Than Conventional Predictors and a Näive Use of DSSP for Disorder Identification
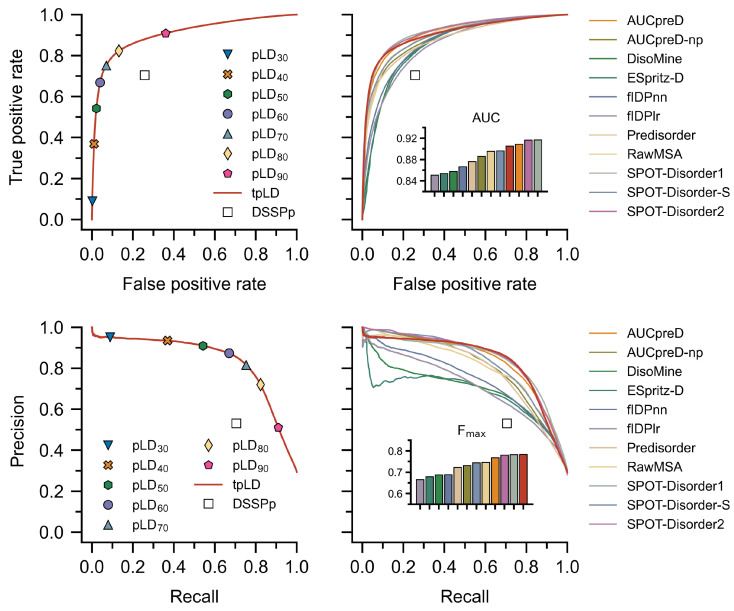
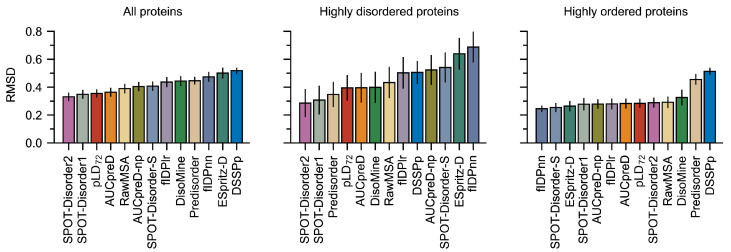
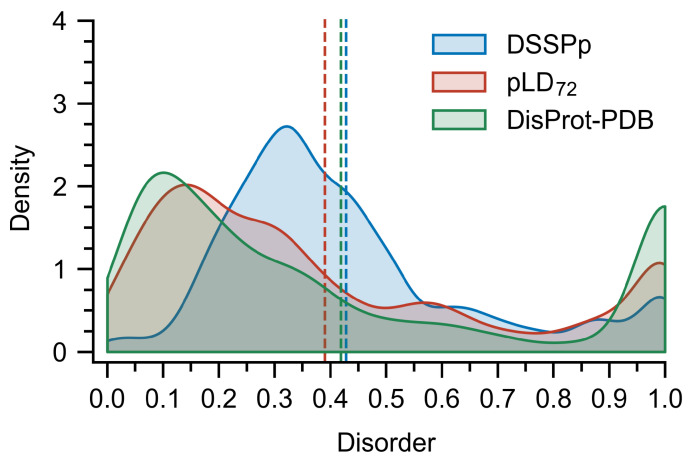
Improved performance with tpLD (Equation (1)) over and against conventional predictors and a näive application of DSSPp is evidenced by the ROC curves and AUC values (Figure 1 and Figure S1), as well as the precision–recall (PR) curves and F values (Figure 1 and Figure S1) on both the DisProt-PDB and DisProt datasets (Tables S1 and S2). Thresholds for the binary pLD predictor were selected based on the Matthews correlation coefficients, which gave values of 76 and 68 for the DisProt and DisProt-PDB datasets respectively (Tables S3 and S4). We refer to these discrete predictors as pLD and pLD. Unsurprisingly, these values agree with the minimum distance from the ROC curve to the top left of the plot (i.e., (0, 1)) (Figure 1). The difference between these two values undoubtedly stems from the nature of the underlying datasets: while DisProt-PDB contains no uncertain residues, DisProt does. For analysis purposes, we opted to use a combined pLDDT metric, denoted pLD, which is the mean of these two. Data using multiple pLDDT values are provided in Tables S1 and S2. RMSD (Equation (2)) calculations comparing DSSPp and pLD demonstrate improved performance for all protein classes, including highly disordered (i.e., >95%) and highly ordered (i.e., <10%), irrespective of dataset (Figure 2 and Figure S2). We note that overall RMSD values are on average lower for the DisProt-PDB dataset, again likely a result of it lacking “uncertain” residues—residues for which no PDB or experimental data exists. Shifts towards lower RMSD irrespective of dataset, or protein length and disorder content, are also evident for pLD (Figures S4 and S5). A regression analysis revealed stronger correlations between pLD and the traditional disorder predictors with respect to residue-wise disorder RMSD when compared with DSSPp (Figures S6–S9). Considering global disorder content prediction, we find that on the DisProt dataset pLD shows slightly better performance than DSSPp with a lower mean and a more accurate distribution; however, we note that both methods significantly overestimate disorder content (Figure 3 and Figure S3). On the DisProt-PDB dataset, closer agreement between pLD and DSSPp is evident based on the mean with both methods returning values similar to experiment. The two distributions are, however, notably different. While that produced by pLD has a peak around 0.15, in close agreement with the experiment, the peak in the distribution produced by DSSPp is larger and shifted to a higher value around 0.3. This is all to say that a näive application of DSSP for the prediction of disordered and ordered regions for AF2 structures, specifically the assumption that helical and strand regions are ordered, and coiled regions are unstructured, leads to poorer prediction (i.e., higher RMSD, lower AUC, and higher F) of disordered regions and an overestimation in disorder content.
Figure 1.
Receiver operating characteristic (ROC) curves (top) and precision–recall (bottom) are depicted for various predictors calculated per residue on the DisProt-PDB dataset. Note that a ROC curve captures the probability of true and false positives at all thresholds, where an ideal predictor will have an area under the curve (AUC) equal to 1. Further note that a precision–recall curve captures the trade-off between precision and recall; again, in the ideal case the harmonic mean of the precision and recall (F) will be equal to 1; bar colors correspond to the legend, red denotes tpLD. In all cases the tpLD (Equation (1)) and various discrete pLD predictors are indicated alongside DSSPp. The tpLD predictor resulted in one of the highest AUC values and the highest F on the DisProt-PDB dataset. pLDDT is abbreviated as pLD for plotting purposes.
Figure 2.
Average RMSD (Equation (2)) values calculated for the DisProt-PDB datasets using various prediction methods calculated per protein. Proteins were assigned to classes (highly disordered i.e., >90% disorder and highly ordered i.e., <10% disorder) based on datasets. Bootstrapping—that is, sampling with replacement—was used to compute averages and estimate errors with 10,000 samples of size 60. pLD resulted in lower RMSD values on the DisProt-PDB dataset compared to DSSPp. pLDDT is abbreviated as pLD for plotting purposes.
Figure 3.
Distribution of disorder content per protein in the DisProt-PDB dataset depicted alongside the distributions predicted by pLD and DSSPp. Bin-widths were set at 0.5 and bootstrapping that is, sampling with replacement, was used to compute the distributions and average values (vertical dashed lines) with 10,000 samples of size 60. Close agreement between the experiment and pLD is evident, conversely, DSSPp predicted a higher disorder content. pLDDT is abbreviated pLD for plotting purposes.
3.2. Sequence Predictors Can Still Outperform AlphaFold2 on Disorder Prediction
Comparing the pLDDT-based and DSSPp predictors to various sequence-based predictors revealed performance differences amongst the methods. Notably, tpLD (Equation (1)) performed exceptionally well on the DisProt-PDB dataset posting the largest F (0.784) and one of the largest AUC (0.905) values of the methods considered (Figure 1, Tables S1 and S3). This was also evidenced by pLD, which had the highest MCC (0.701) (Table S1) and one of the lowest RMSD values (Figure 2) on the DisProt-PDB dataset. Unsurprisingly, on the DisProt dataset, both tpLD (Equation (1)) and DSSPp performed significantly worse and were readily outperformed by the other predictor methods, in particular fIDPnn (F: 0.357 (DSSPp), 0.429 (tpLD), 0.457 (fIDPnn); AUC: 0.635 (DSSPp), 0.731 (tpLD), 0.794 (fIDPnn)), which outperformed all other predictors, as evidenced by the ROC, PR, and RMSD analyses. We note that with respect to MCC, pLD still performed well on both the DisProt and DisProt-PDB datasets achieving scores of 0.310 and 0.697, respectively (Tables S1 and S2). In agreement with the CAID results, we found that SPOT-Disorder2, fIDPnn, RawMSA, and AUCpreD all performed exceptionally well (Figure 1 and Figure S1, Tables S3 and S4) [29].
3.3. Secondary Structure Codons (SSC) Suggests Relationships between the pLDDT and Secondary Structure
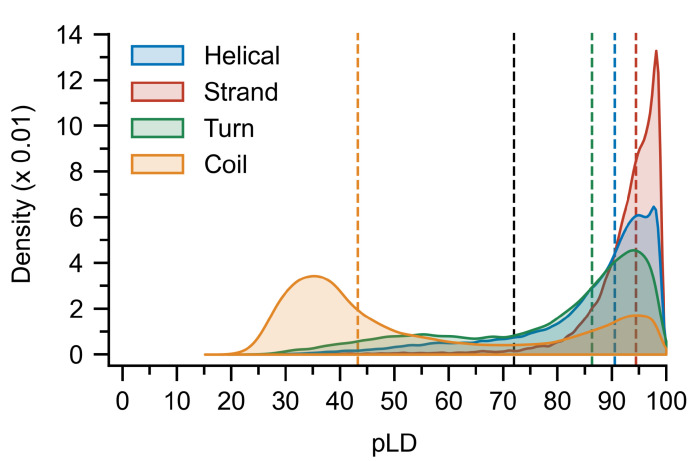
In order to explain the discrepancy between the pLDDT-based and DSSP predictors with respect to local and global disorder prediction, we considered how pLDDT values were assigned to the secondary structures. Kernel density estimates (KDE) of the distribution of pLDDT values sampled over all residues revealed a strong left-skew for all but the coil secondary structure, which exhibited a right-skewed bimodal distribution with peaks around 94 and 35 (Figure 4). Residues assigned to -strand and -bridge structures are the most likely to be assigned to large pLDDT values, followed by helical and H-bond stabilized turns. To provide a more detailed picture of the distributions, we introduce the concept of a secondary structure codon (SSC), a triplet describing the local secondary structure at a given residue. Analysis of the distributions of pLDDT values for each SSC revealed that residues predicted to belong to both the ends (HHC/CHH/HHT/THH) and middle (HHH) of helices can have pLDDT values (Figure S10), this was not observed for residues belonging to the middle (EEE) and ends of -strands (EEC/CEE/EET/TEE) (Figure S11). For highly coiled residues (CCC/CCT/TCC) and several turn residues (CTT/TTC), both high () and low () pLDDT values were observed (Figures S12 and S13).
Figure 4.
Distribution of pLDDT values per residue calculated for each secondary structure class. Bin-widths were set at 0.5 and bootstrapping, that is, sampling with replacement, was used to compute the distributions and mean values (colored vertical dashed lines; black dashed line represents pLD) with 10,000 samples of size 500. A bimodal distribution is evident for the coil structures, and while strand, helical, and turn regions are on average assigned to high pLDDT values, residues belonging to each can sample much lower values. pLDDT is abbreviated pLD for plotting purposes.
3.4. Nrf2: A Case Study
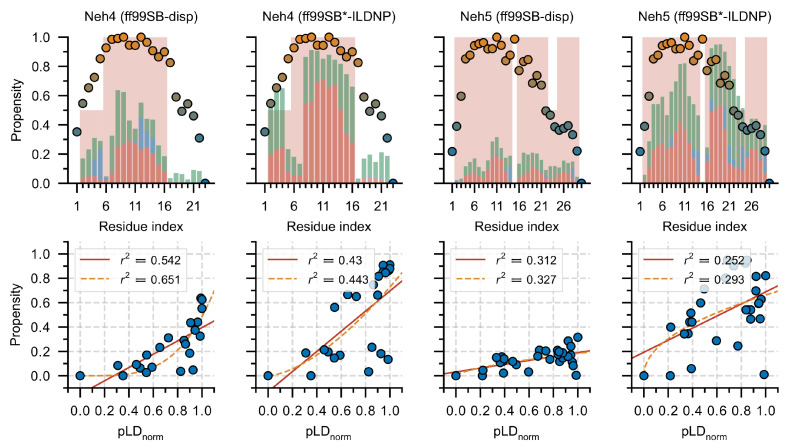
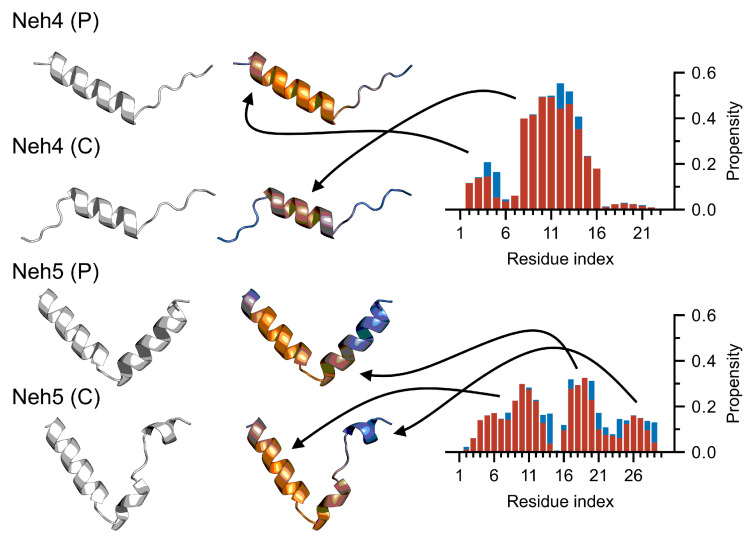
Nrf2 (nuclear factor erythroid 2-related factor 2) is a partially disordered transcription factor [47,57] and is the master regulator of the cellular anti-oxidative response. Within the multi-domain Nrf2 protein, two transactivation domains, namely Neh4 and Neh5, are responsible for binding the transcriptional adaptor zinc-binding domains, TAZ1 and TAZ2, of CBP; references [58,59]; previous work has elucidated the free-state ensembles of Neh4 and Neh5 using both MD simulations and circular dichroism [46]. We consider the AF2 predicted structures of the Neh4 and Neh5 peptides (Neh4 (P) and Neh5 (P)) and the structures predicted for Neh4/5 within a larger construct (Neh4 (C) and Neh5 (C)). Comparison of the secondary structures determined from the AF2 predictions and simulated ensembles suggested relatively good agreement; regions of low helical propensity in the ensemble corresponded to lower helical propensity in the AF2 structures, and the converse was also true (Figure 5). There also appeared to be some agreement between pLDDT and secondary structure; however, these correlations were weak (Figure 5) and depended strongly on the system considered (Neh4 vs. Neh5). We also overlaid the pLDDT with the predicted structures seeking to assess the potential for additional insights. Immediately evident was the heterogeneity in the predicted structures when considering the peptide and the larger construct. Notably, the differences in the structure occurred precisely where the pLDDT was lower (e.g., the N-terminal of the Neh4 (P) that was not present in the Neh4 (C) and the C-terminal helix in Neh5 (P) that was split in Neh 5 (C)). The pLDDT and heterogeneity of the structures in particular with Neh5, agreed closely with the observed secondary structure from the ensembles (Figure 5 and Figure 6); specifically, the triple helix, with a hard break at I14-P15 and a transient break from N22–E24. These structural dynamics—that is the exchange between a large and a small helix in the C-termini of Neh5—appeared to be captured explicitly by the pLDDT and implicitly by the heterogeneity of the AF2 structures.
Figure 5.
Secondary structure of ensembles versus AF2. Top: Secondary structure was computed from molecular simulation (red = -helix, -helix or -helix; blue = -strand or -bridge; and green = turn). The red background color depicts the AF2 predicted secondary structure propensities, no strand/turn content was predicted. Bottom: Min–max normalized pLDDT values () are plotted (circles) with colors ranging from 0 to 1 (orange implies and blue implies ). We plot correlations between the total secondary structure propensity computed from MD simulations and the , and fit the data to a line (red) or a power law (orange). pLDDT is abbreviated pLD for plotting purposes.
Figure 6.
AF2 predicted structures correlate with simulated secondary structure. We consider the peptide (i.e., Neh4/5 (P)) and construct (i.e., Neh4/5 (C)) structures predicted from AF2, without a colormap and with a pLDDT colormap scaled between 70 and 100 (i.e., blue implies and orange implies ). Note how the coloring of the structures provides non-trivial insights that are undetectable without it. These are depicted alongside the average secondary structure computed using both the ff99SB*-ILDNP and ff99SB-disp simulations (red = -helix, -helix or -helix; blue = -strand or -bridge). Note that arrows indicate corresponding regions between AF2 structures (left) and structural propensities computed from MD simulations (right).
4. Discussion
AF2 has been a paradigm-shift in structural biology, providing a tentative solution to the protein folding problem that has persisted over half a century [1]. Since the time that problem was posed by Perutz and Kendrew, a new class of proteins, intrinsically disordered proteins, has been discovered and IDPs have become the focus of much study [10,11,13,14,60,61]. Over the past two decades, much effort has been devoted to developing methods for identifying disordered regions given the primary sequence of a protein [29,62,63,64,65,66]. Herein, we assess the applicability of AF2 to this problem.
We find (and strongly stress) that simply inferring a residue in an AF2 structure assigned by DSSP to a helical, strand, or H-bond stabilized turn is ordered, and otherwise is disordered, results in an overestimation of disorder content and a poor prediction of disordered regions. While this may seem like a trivial observation, the abundance of AF2 structures generated for disordered proteins has made such a pitfall increasingly likely for researchers who are less familiar with IDPs and structural prediction methods. Instead, employing the pLDDT, a measure of the expected position error at a given residue and originally purposed to assess the residue-wise structural confidence, provides a much more accurate metric for determining global and local disorder content. Using the pLDDT as a disorder predictor metric, we observe impressive performance on the DisProt-PDB dataset when compared to conventional disorder predictors (Figure 1). We here note the work by Akdel et al. [8], who found that, in addition to the pLDDT, the solvent accessible surface area of an AF2 structure provides another strong predictor of disorder. Similar to our 2021 benchmark published in bioRxiv [67], this was recently extended by Piovesan et al. [68], wherein a combined RSA-pLDDT metric for assessing IDP binding was considered.
Secondary structure and global disorder analyses point to a potential root of the prediction discrepancy between pLDDT and DSSP; simply put, for AF2, not all secondary structures are created equal. AF2 will readily assign a coiled geometry and a high pLDDT value to the same residue, and conversely assign low pLDDT values to structured regions (Figure 4). While a näive DSSP predictor assumes that coils and bends are disordered while helices, strands, and turns are ordered, a pLDDT predictor captures the biophysical reality that a coil may be more "ordered" and a helix more "disordered" for certain residues in certain proteins. It is this former case that likely results in the improved performance observed for pLDDT and underscores the importance of the nuance provided by this metric for disordered protein prediction. It also opens the door to another interesting question: is the conclusion to be drawn from two helices A and B of comparable geometry with significantly different average pLDDT () simply that A is less likely to be “real”, or is it that both helices exist, however, A exists transiently?
The above question alludes to a second problem associated with IDP prediction, namely predicting the structural dynamics and transitions (i.e., order-to-disorder, disorder-to-order, disorder-to-disorder) that an IDP may undergo [62,69]. In light of the secondary structure analysis, the pLDDT may be just such a means for extracting this information, namely the transientness of secondary structures, their potential for transition upon binding and their functional importance. A helix with a low pLDDT may be more transient (i.e., existing frequently in a disordered, unfolded state) than a helix with a high pLDDT and conversely, a coiled region with a high pLDDT, may suggest a disorder–order transition and/or its conserved role in some biophysical interaction. The strength of AF2 as a predictor is that both a pLDDT score and a three-dimensional structure are provided, allowing for more comprehensive insights into an IDPs structure and dynamics. This is anecdotally evidenced by Nrf2, where considering the structure alone presents an incomplete story, that is quite literally colored in by the pLDDT, revealing something about the transientness of the C-terminal helix of Neh5. This hypothesis, pertaining to the relationship between the pLDDT and the structural transitions of IDRs, originally proposed in our 2021 pre-print [67], has been further substantiated by the findings of an impressive study by Alderson et al. [70] that systematically compared both NMR and AF2 data.
While the significance of this insight is buffeted by the unrealistically high helical content predicted by AF2, it appears to suggest that continued research into the pLDDT and heterogeneity of AF2 predicted structures may provide novel insights. We reiterate that, by their very nature, IDPs exhibit a high degree of conformational flexibility, allowing them to interact with multiple binding partners in a variety of ways [71,72,73,74,75,76,77,78]. While it is the case that a single, static, AF2 structure cannot adequately describe the totality of an often large conformational ensembles [13,14,15], the ability of the program to predict with relatively high accuracy the location of disordered regions is nonetheless impressive, and refinement of the training set to account for more accurate disordered structures could further improve performance. In addition, thorough analysis of the pLDDT score as it relates to structural transientness, as well as the local function and dynamics of IDP motifs, may further enhance the utility of AF2 to the IDP community.
While experimental NMR [47,79,80,81,82,83,84,85,86,87], and high-quality molecular simulations [46,88,89,90,91,92,93,94,95,96,97,98] are some of the most accurate methods for determining the (dis)ordered nature and dynamics of proteins, fast and computationally efficient methods play an important role. Unlike conventional predictors however, AF2 supplies both a pLDDT score, that can provide an accurate prediction of protein disorder, in addition to a three-dimensional structure, and when taken in tandem, these appear to provide insights into the underlying local dynamics (i.e., disorder–order transition) of disordered protein regions.
5. Conclusions
In this study, we assessed the ability of AF2 to predict disordered protein regions. We benchmark the program on two datasets developed for CAID [29], and find it to perform quite well, exceeding the performance of 11 traditional predictors on the DisProt-PDB dataset. Furthermore, we observe that the pLDDT score assigned to each residue by AF2 provides an impressive metric for assessing disorder, far surpassing a näive, but by no means, non-trivial application of DSSP for researchers who are less familiar with IDPs and structural prediction methods. Our analysis, in particular that of Nrf2, also suggests a novel link between secondary structure transience and the pLDDT score, intimating that continued research into this metric may reveal a connection to the local dynamics of disordered proteins.
Acknowledgments
The authors thank SharcNet and Compute Canada for computational resources.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23094591/s1.
Author Contributions
Conceptualization: all authors; methodology: all authors; simulations and analysis: C.J.W.; writing—original draft: C.J.W.; writing—review and editing: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
All authors were supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). M.K. also acknowledges support from the Canada Research Chairs Program. Computational resources were provided by Compute Canada.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Additional details pertaining to dataset construction are provided in Supplementary Information and the full list of proteins, structures and combined disorder data are available at https://github.com/SoftSimu/AlphaFoldDisorderData (accessed on 21 September 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dill K.A., MacCallum J.L. The Protein-Folding Problem, 50 Years On. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 2.Nassar R., Dignon G.L., Razban R.M., Dill K.A. The Protein Folding Problem: The Role of Theory. J. Mol. Biol. 2021;433:167126. doi: 10.1016/j.jmb.2021.167126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullard A. What does AlphaFold mean for drug discovery? Nat. Rev. Drug Discov. 2021;20:725–727. doi: 10.1038/d41573-021-00161-0. [DOI] [PubMed] [Google Scholar]
- 5.Serpell L.C., Radford S.E., Otzen D.E. AlphaFold: A Special Issue and A Special Time for Protein Science. J. Mol. Biol. 2021;433:167231. doi: 10.1016/j.jmb.2021.167231. [DOI] [PubMed] [Google Scholar]
- 6.Strodel B. Energy Landscapes of Protein Aggregation and Conformation Switching in Intrinsically Disordered Proteins. J. Mol. Biol. 2021;433:167182. doi: 10.1016/j.jmb.2021.167182. [DOI] [PubMed] [Google Scholar]
- 7.Ruff K.M., Pappu R.V. AlphaFold and Implications for Intrinsically Disordered Proteins. J. Mol. Biol. 2021;433:167208. doi: 10.1016/j.jmb.2021.167208. [DOI] [PubMed] [Google Scholar]
- 8.Akdel M., Pires D.E.V., Pardo E.P., Jänes J., Zalevsky A.O., Mészáros B., Bryant P., Good L.L., Laskowski R.A., Pozzati G., et al. A structural biology community assessment of AlphaFold 2 applications. bioRxiv. 2021 doi: 10.1101/2021.09.26.461876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buel G.R., Walters K.J. Can AlphaFold2 predict the impact of missense mutations on structure? Nat. Struct. Mol. Biol. 2022;29:1–2. doi: 10.1038/s41594-021-00714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright P.E., Dyson H. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 11.Dunker A., Lawson J., Brown C.J., Williams R.M., Romero P., Oh J.S., Oldfield C.J., Campen A.M., Ratliff C.M., Hipps K.W., et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 12.Dunker A.K., Brown C.J., Lawson J.D., Iakoucheva L.M., Obradović Z. Intrinsic Disorder and Protein Function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 13.Uversky V.N. Intrinsically Disordered Proteins and Their “Mysterious” (Meta)Physics. Front. Phys. 2019;7:10. doi: 10.3389/fphy.2019.00010. [DOI] [Google Scholar]
- 14.DeForte S., Uversky V.N. Intrinsically Disordered Proteins in PubMed: What can the tip of the iceberg tell us about what lies below? RSC Adv. 2016;6:11513–11521. doi: 10.1039/C5RA24866C. [DOI] [Google Scholar]
- 15.Lyle N., Das R.K., Pappu R.V. A quantitative measure for protein conformational heterogeneity. J. Chem. Phys. 2013;139:121907. doi: 10.1063/1.4812791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi U.B., Sanabria H., Smirnova T., Bowen M.E., Weninger K.R. Spontaneous Switching among Conformational Ensembles in Intrinsically Disordered Proteins. Biomolecules. 2019;9:114. doi: 10.3390/biom9030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salem A., Wilson C.J., Rutledge B.S., Dilliott A., Farhan S., Choy W.Y., Duennwald M.L. Matrin3: Disorder and ALS Pathogenesis. Front. Mol. Biosci. 2022;8:794646. doi: 10.3389/fmolb.2021.794646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turoverov K.K., Kuznetsova I.M., Uversky V.N. The protein kingdom extended: Ordered and Intrinsically Disordered Proteins, their folding, supramolecular complex formation, and aggregation. Prog. Biophys. Mol. Biol. 2010;102:73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uversky V.N. Unusual biophysics of Intrinsically Disordered Proteins. Biochim. Biophys. Acta Proteins Proteom. 2013;1834:932–951. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Fisher C.K., Stultz C.M. Constructing ensembles for Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2011;21:426–431. doi: 10.1016/j.sbi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R.K., Ruff K.M., Pappu R.V. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das R.K., Pappu R.V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao A.H., Crick S.L., Vitalis A., Chicoine C.L., Pappu R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K. Sequence complexity of disordered protein. Proteins Struct. Funct. Bioinf. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Radivojac P., Iakoucheva L.M., Oldfield C.J., Obradovic Z., Uversky V.N., Dunker A.K. Intrinsic Disorder and Functional Proteomics. Biophys. J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theillet F.X., Kalmar L., Tompa P., Han K.H., Selenko P., Dunker A.K., Daughdrill G.W., Uversky V.N. The alphabet of intrinsic disorder. Intrinsically Disord. Proteins. 2013;1:e24360. doi: 10.4161/idp.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky V.N. The alphabet of intrinsic disorder. Intrinsically Disord. Proteins. 2013;1:e24684. doi: 10.4161/idp.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Necci M., Piovesan D., CAID Predictors. DisProt Curators. Tosatto S.C.E. Critical assessment of protein intrinsic disorder prediction. Nat. Methods. 2021;18:472–481. doi: 10.1038/s41592-021-01117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W., Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 31.Hatos A., Hajdu-Soltész B., Monzon A.M., Palopoli N., Álvarez L., Aykac-Fas B., Bassot C., Benítez G.I., Bevilacqua M., Chasapi A., et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2019;48:D269–D276. doi: 10.1093/nar/gkz975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani V., Biasini M., Barbato A., Schwede T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29:2722–2728. doi: 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chicco D., Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020;21:6. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G., Katuwawala A., Wang K., Wu Z., Ghadermarzi S., Gao J., Kurgan L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021;12:4438. doi: 10.1038/s41467-021-24773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson J., Paliwal K.K., Litfin T., Zhou Y. SPOT-Disorder2: Improved Protein Intrinsic Disorder Prediction by Ensembled Deep Learning. Genom. Proteom. Bioinform. 2019;17:645–656. doi: 10.1016/j.gpb.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirabello C., Wallner B. rawMSA: End-to-end Deep Learning using raw Multiple Sequence Alignments. PLoS ONE. 2019;14:e0220182. doi: 10.1371/journal.pone.0220182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng X., Eickholt J., Cheng J. PreDisorder: ab initio sequence-based prediction of protein disordered regions. BMC Bioinform. 2009;10:436. doi: 10.1186/1471-2105-10-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S., Ma J., Xu J. AUCpreD: Proteome-level protein disorder prediction by AUC-maximized deep convolutional neural fields. Bioinformatics. 2016;32:i672–i679. doi: 10.1093/bioinformatics/btw446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanson J., Yang Y., Paliwal K., Zhou Y. Improving protein disorder prediction by deep bidirectional long short-term memory recurrent neural networks. Bioinformatics. 2016;33:685–692. doi: 10.1093/bioinformatics/btw678. [DOI] [PubMed] [Google Scholar]
- 40.Hanson J., Paliwal K., Zhou Y. Accurate Single-Sequence Prediction of Protein Intrinsic Disorder by an Ensemble of Deep Recurrent and Convolutional Architectures. J. Chem. Inf. Model. 2018;58:2369–2376. doi: 10.1021/acs.jcim.8b00636. [DOI] [PubMed] [Google Scholar]
- 41.Orlando G., Raimondi D., Codice F., Tabaro F., Vranken W. Prediction of disordered regions in proteins with recurrent Neural Networks and protein dynamics. bioRxiv. 2020 doi: 10.1101/2020.05.25.115253. [DOI] [PubMed] [Google Scholar]
- 42.Walsh I., Martin A.J.M., Domenico T.D., Tosatto S.C.E. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics. 2011;28:503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- 43.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 44.Waskom M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021;6:3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 45.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold—Making protein folding accessible to all. bioRxiv. 2021 doi: 10.1101/2021.08.15.456425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang M., Wilson C.J., Karunatilleke N.C., Moselhy M.H., Karttunen M., Choy W.Y. Exploring the Conformational Landscape of the Neh4 and Neh5 Domains of Nrf2 Using Two Different Force Fields and Circular Dichroism. J. Chem. Theory Comput. 2021;17:3145–3156. doi: 10.1021/acs.jctc.0c01243. [DOI] [PubMed] [Google Scholar]
- 47.Karunatilleke N.C., Fast C.S., Ngo V., Brickenden A., Duennwald M.L., Konermann L., Choy W.Y. Nrf2, the Major Regulator of the Cellular Oxidative Stress Response, is Partially Disordered. Int. J. Mol. Sci. 2021;22:7434. doi: 10.3390/ijms22147434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aliev A.E., Kulke M., Khaneja H.S., Chudasama V., Sheppard T.D., Lanigan R.M. Motional timescale predictions by molecular dynamics simulations: Case study using proline and hydroxyproline sidechain dynamics. Proteins. 2014;82:195–215. doi: 10.1002/prot.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Best R.B., Hummer G. Optimized Molecular Dynamics Force Fields Applied to the Helix-Coil Transition of Polypeptides. J. Phys. Chem. B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 52.Robustelli P., Piana S., Shaw D.E. Developing a molecular dynamics force field for both folded and disordered protein states. Proc. Natl. Acad. Sci. USA. 2018;115:E4758–E4766. doi: 10.1073/pnas.1800690115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 54.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 55.Darden T., York D., Pedersen L. Particle mesh Ewald: An Nlog (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 56.Hess B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 57.Moi P., Chan K., Asunis I., Cao A., Kan Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang J., Hosoya T., Maruyama A., Nishikawa K., Maher J.M., Ohta T., Motohashi H., Fukamizu A., Shibahara S., Itoh K., et al. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem. J. 2007;404:459–466. doi: 10.1042/BJ20061611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T., et al. Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uversky V.N. Recent Developments in the Field of Intrinsically Disordered Proteins: Intrinsic Disorder–Based Emergence in Cellular Biology in Light of the Physiological and Pathological Liquid–Liquid Phase Transitions. Annu. Rev. Biophys. 2021;50:135–156. doi: 10.1146/annurev-biophys-062920-063704. [DOI] [PubMed] [Google Scholar]
- 62.Miskei M., Horvath A., Vendruscolo M., Fuxreiter M. Sequence-Based Prediction of Fuzzy Protein Interactions. J. Mol. Biol. 2020;432:2289–2303. doi: 10.1016/j.jmb.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 63.Peng Z., Mizianty M.J., Kurgan L. Genome-scale prediction of proteins with long intrinsically disordered regions. Proteins. 2013;82:145–158. doi: 10.1002/prot.24348. [DOI] [PubMed] [Google Scholar]
- 64.Ward J., Sodhi J., McGuffin L., Buxton B., Jones D. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Piovesan D., Necci M., Escobedo N., Monzon A.M., Hatos A., Mičetić I., Quaglia F., Paladin L., Ramasamy P., Dosztányi Z., et al. MobiDB: Intrinsically disordered proteins in 2021. Nucleic Acids Res. 2020;49:D361–D367. doi: 10.1093/nar/gkaa1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y., Wang X., Liu B. A comprehensive review and comparison of existing computational methods for intrinsically disordered protein and region prediction. Brief. Bioinform. 2017;20:330–346. doi: 10.1093/bib/bbx126. [DOI] [PubMed] [Google Scholar]
- 67.Wilson C.J., Choy W.Y., Karttunen M. AlphaFold2: A role for disordered protein prediction? bioRxiv. 2021 doi: 10.1101/2021.09.27.461910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piovesan D., Monzon A.M., Tosatto S.C. Intrinsic Protein Disorder, Conditional Folding and AlphaFold2. bioRxiv. 2022 doi: 10.1101/2022.03.03.482768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindorff-Larsen K., Kragelund B.B. On the Potential of Machine Learning to Examine the Relationship Between Sequence, Structure, Dynamics and Function of Intrinsically Disordered Proteins. J. Mol. Biol. 2021;433:167196. doi: 10.1016/j.jmb.2021.167196. [DOI] [PubMed] [Google Scholar]
- 70.Alderson T.R., Pritišanac I., Moses A.M., Forman-Kay J.D. Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2. bioRxiv. 2022 doi: 10.1101/2022.02.18.481080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wright P.E., Dyson H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freiberger M.I., Wolynes P.G., Ferreiro D.U., Fuxreiter M. Frustration in Fuzzy Protein Complexes Leads to Interaction Versatility. J. Phys. Chem. B. 2021;125:2513–2520. doi: 10.1021/acs.jpcb.0c11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oldfield C.J., Dunker A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 74.Uversky V.N. Multitude of binding modes attainable by Intrinsically Disorder Proteins: A portrait gallery of disorder-based complexes. Chem. Soc. Rev. 2011;40:1623–1634. doi: 10.1039/C0CS00057D. [DOI] [PubMed] [Google Scholar]
- 75.Sharma R., Raduly Z., Miskei M., Fuxreiter M. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015;589:2533–2542. doi: 10.1016/j.febslet.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 76.Khan H., Cino E.A., Brickenden A., Fan J., Yang D., Choy W.Y. Fuzzy Complex Formation between the Intrinsically Disordered Prothymosin α and the Kelch Domain of Keap1 Involved in the Oxidative Stress Response. J. Mol. Biol. 2013;425:1011–1027. doi: 10.1016/j.jmb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 77.Tompa P., Fuxreiter M. Fuzzy complexes: Polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 78.Arbesú M., Iruela G., Fuentes H., Teixeira J.M.C., Pons M. Intramolecular Fuzzy Interactions Involving Intrinsically Disordered Domains. Front. Mol. Biosci. 2018;5:39. doi: 10.3389/fmolb.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Killoran R.C., Sowole M.A., Halim M.A., Konermann L., Choy W.Y. Conformational characterization of the intrinsically disordered protein Chibby: Interplay between structural elements in target recognition. Protein Sci. 2016;25:1420–1429. doi: 10.1002/pro.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gall C., Xu H., Brickenden A., Ai X., Choy W.Y. The intrinsically disordered TC-1 interacts with Chibby via regions with high helical propensity. Protein Sci. 2007;16:2510–2518. doi: 10.1110/ps.073062707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mokhtarzada S., Yu C., Brickenden A., Choy W.Y. Structural Characterization of Partially Disordered Human Chibby: Insights into Its Function in the Wnt-Signaling Pathway. Biochemistry. 2011;50:715–726. doi: 10.1021/bi101236z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zahn R., Liu A., Luhrs T., Riek R., von Schroetter C., Garcia F.L., Billeter M., Calzolai L., Wider G., Wuthrich K. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y., Fisher J.C., Mathew R., Ou L., Otieno S., Sublet J., Xiao L., Chen J., Roussel M.F., Kriwacki R.W. Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21. Nat. Chem. Biol. 2011;7:214–221. doi: 10.1038/nchembio.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong L.E., Kim T.H., Muhandiram D.R., Forman-Kay J.D., Kay L.E. NMR Experiments for Studies of Dilute and Condensed Protein Phases: Application to the Phase-Separating Protein CAPRIN1. J. Am. Chem. Soc. 2020;142:2471–2489. doi: 10.1021/jacs.9b12208. [DOI] [PubMed] [Google Scholar]
- 85.Kim D.H., Lee J., Mok K., Lee J., Han K.H. Salient Features of Monomeric Alpha-Synuclein Revealed by NMR Spectroscopy. Biomolecules. 2020;10:428. doi: 10.3390/biom10030428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kosol S., Contreras-Martos S., Cedeño C., Tompa P. Structural Characterization of Intrinsically Disordered Proteins by NMR Spectroscopy. Molecules. 2013;18:10802–10828. doi: 10.3390/molecules180910802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dyson H.J., Wright P.E. NMR illuminates intrinsic disorder. Curr. Opin. Struct. Biol. 2021;70:44–52. doi: 10.1016/j.sbi.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shaw D.E., Maragakis P., Lindorff-Larsen K., Piana S., Dror R.O., Eastwood M.P., Bank J.A., Jumper J.M., Salmon J.K., Shan Y., et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 89.Lindorff-Larsen K., Trbovic N., Maragakis P., Piana S., Shaw D.E. Structure and Dynamics of an Unfolded Protein Examined by Molecular Dynamics Simulation. J. Am. Chem. Soc. 2012;134:3787–3791. doi: 10.1021/ja209931w. [DOI] [PubMed] [Google Scholar]
- 90.Ahmed M.C., Skaanning L.K., Jussupow A., Newcombe E.A., Kragelund B.B., Camilloni C., Langkilde A.E., Lindorff-Larsen K. Refinement of α-Synuclein Ensembles Against SAXS Data: Comparison of Force Fields and Methods. Front. Mol. Biosci. 2021;8:216. doi: 10.3389/fmolb.2021.654333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilson C.J., Chang M., Karttunen M., Choy W.Y. KEAP1 Cancer Mutants: A Large-Scale Molecular Dynamics Study of Protein Stability. Int. J. Mol. Sci. 2021;22:5408. doi: 10.3390/ijms22105408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rauscher S., Gapsys V., Gajda M.J., Zweckstetter M., de Groot B.L., Grubmüller H. Structural Ensembles of Intrinsically Disordered Proteins Depend Strongly on Force Field: A Comparison to Experiment. J. Chem. Theory Comput. 2015;11:5513–5524. doi: 10.1021/acs.jctc.5b00736. [DOI] [PubMed] [Google Scholar]
- 93.Cino E.A., Choy W.Y., Karttunen M. Characterization of the Free State Ensemble of the CoRNR Box Motif by Molecular Dynamics Simulations. J. Phys. Chem. B. 2016;120:1060–1068. doi: 10.1021/acs.jpcb.5b11565. [DOI] [PubMed] [Google Scholar]
- 94.Samantray S., Yin F., Kav B., Strodel B. Different Force Fields Give Rise to Different Amyloid Aggregation Pathways in Molecular Dynamics Simulations. J. Chem. Inf. Model. 2020;60:6462–6475. doi: 10.1021/acs.jcim.0c01063. [DOI] [PubMed] [Google Scholar]
- 95.Nasica-Labouze J., Nguyen P.H., Sterpone F., Berthoumieu O., Buchete N.V., Coté S., Simone A.D., Doig A.J., Faller P., Garcia A., et al. Amyloid β Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015;115:3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Piana S., Lindorff-Larsen K., Shaw D.E. Atomic-level description of ubiquitin folding. Proc. Natl. Acad. Sci. USA. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dror R.O., Dirks R.M., Grossman J., Xu H., Shaw D.E. Biomolecular Simulation: A Computational Microscope for Molecular Biology. Annu. Rev. Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 98.Best R.B., Hummer G., Eaton W.A. Native contacts determine protein folding mechanisms in atomistic simulations. Proc. Natl. Acad. Sci. USA. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional details pertaining to dataset construction are provided in Supplementary Information and the full list of proteins, structures and combined disorder data are available at https://github.com/SoftSimu/AlphaFoldDisorderData (accessed on 21 September 2021).