Abstract
Background
Segmentation of medical image volumes is a time‐consuming manual task. Automatic tools are often tailored toward specific patient cohorts, and it is unclear how they behave in other clinical settings.
Purpose
To evaluate the performance of the open‐source Multi‐Planar U‐Net (MPUnet), the validated Knee Imaging Quantification (KIQ) framework, and a state‐of‐the‐art two‐dimensional (2D) U‐Net architecture on three clinical cohorts without extensive adaptation of the algorithms.
Study Type
Retrospective cohort study.
Subjects
A total of 253 subjects (146 females, 107 males, ages 57 ± 12 years) from three knee osteoarthritis (OA) studies (Center for Clinical and Basic Research [CCBR], Osteoarthritis Initiative [OAI], and Prevention of OA in Overweight Females [PROOF]) with varying demographics and OA severity (64/37/24/53/2 scans of Kellgren and Lawrence [KL] grades 0–4).
Field Strength/Sequence
0.18 T, 1.0 T/1.5 T, and 3 T sagittal three‐dimensional fast‐spin echo T1w and dual‐echo steady‐state sequences.
Assessment
All models were fit without tuning to knee magnetic resonance imaging (MRI) scans with manual segmentations from three clinical cohorts. All models were evaluated across KL grades.
Statistical Tests
Segmentation performance differences as measured by Dice coefficients were tested with paired, two‐sided Wilcoxon signed‐rank statistics with significance threshold α = 0.05.
Results
The MPUnet performed superior or equal to KIQ and 2D U‐Net on all compartments across three cohorts. Mean Dice overlap was significantly higher for MPUnet compared to KIQ and U‐Net on CCBR ( vs. and ), significantly higher than KIQ and U‐Net OAI ( vs. and , and not significantly different from KIQ while significantly higher than 2D U‐Net on PROOF ( vs. , , and . The MPUnet performed significantly better on KL grade 3 CCBR scans with vs. for KIQ and for 2D U‐Net.
Data Conclusion
The MPUnet matched or exceeded the performance of state‐of‐the‐art knee MRI segmentation models across cohorts of variable sequences and patient demographics. The MPUnet required no manual tuning making it both accurate and easy‐to‐use.
Level of Evidence
3
Technical Efficacy
Stage 2
Keywords: knee segmentation, deep learning, magnetic resonance imaging, open‐source software
Recent advances in machine learning have pushed automatic segmentation tools close to human performance for medical image analysis. 1 , 2 This includes the automatic quantification of cartilage compartments from magnetic resonance imaging (MRI) scans, which facilitates robust, large‐scale quantitative studies of osteoarthritis (OA). 3 Until recently, most validated automatic segmentation software, such as the Knee Imaging Quantification (KIQ) framework, were specialized and relied at least partially on task‐specific knowledge. 4 Gan et al review a range of successful classical approaches based on, for example, random forests, deformable models, graph‐based algorithms, and atlas registration. 3 , 5 , 6 , 7 , 8 With advances in deep learning it is now possible to create automatic segmentation models given sufficient training examples alone. 9 Numerous deep learning based approaches have been suggested in recent years alone. The majority consider models from the family of fully convolutional networks (FCNs) in the popular encoder‐decoder architecture, typically inspired by the U‐Net. 10 , 11 , 12 , 13 The FCN‐centered methods for knee MRI segmentation vary in complexity, ranging from using a single‐stage U‐Net to combining several U‐nets (eg, both two‐dimensional [2D] and three‐dimensional [3D]) and shape model refinement steps (eg, a 2D U‐Net followed by shape model refinement used to identify regions of interest which are then segmented by a 3D U‐Net followed by another shape model refinement step). 11 , 13 , 14 , 15 , 16 Different strategies have been employed to render deep learning on 3D data efficient and to cope with limited training data. To increase the efficiency of 3D FCNs, it has been suggested to operate on overlapping patches or on down‐sampled scans. 17 Another strategy is to employ a 2D FCN to segment each scan slice independently. 11 , 12 , 13 , 14 The 2D approach has been extended in different ways including considering multiple planes or 3D surface model optimization schemes. 14 , 15 , 16 , 18 , 19 , 20
Despite a vast number of existing deep learning based methods for OA segmentation (often shown to perform accurately compared to human annotators), no method has seen widespread clinical adaptation. While such adaptation is complex due to practical, ethical, and legal factors, central research problems related to the models themselves also remain. For instance, it is largely unclear how different models and methods compare even on a single cohort. Which type of model should be perused for clinical validation for a given task? Second, it is even less clear if one model designed to work well on a single dataset can also be expected to work well in other clinical scenarios, for example, on data from new patient cohorts, scanner sequences, or scanner manufactures.
The 2019 OA MRI segmentation challenge made one attempt toward addressing the former problem. 21 A range of deep‐learning‐based methods were compared and evaluated for knee MRI segmentation on a single cohort. Multiple methods were found to perform at clinically applicable levels. Surprisingly, the challenge demonstrated that even the simple 2D U‐Net baseline model was highly competitive. 22 This result indicates that many deep learning based approaches are viable when tuned to a specific dataset, and that the method need not be very complex.
As for the second problem, however, it is unclear how the 2D U‐Net and other challenge methods (often using more complex setups with, eg, cascaded models or multiple postprocessing steps) would perform when trained on new clinical cohorts (eg, a smaller set of annotated scans or knees with different levels of OA severity) without re‐tuning of the hyperparameters, or when trained across multiple cohorts at once. The robustness of a model under such cross‐cohort scenarios is crucial when adapting it for clinical practice, as tuning a neural network model for new data typically requires both compute resources and access to technical experts.
The purpose of this study was to investigate the cross‐cohort performance and robustness of state‐of‐the‐art (classical as well as deep learning based) automatic knee segmentation methods. Its primary focus was on the recently proposed Multi‐Planar U‐Net (MPUnet) model. The MPUnet extends the popular 2D U‐Net with a unique data‐resampling technique, and has been found able to output accurate segmentations across clinical cohorts (and different segmentation tasks) without hyperparameter‐tuning. 23 , 24 It scored a top‐position in the 2019 OA MRI segmentation challenge and a top‐5 position in the 2018 Medical Segmentation Decathlon. 21 , 24 The MPUnet is hyperparameter search free in the sense that the default settings have proven to give good results on variable medical image segmentation tasks, so no machine learning expertise is required to train the MPUnet on new data. These findings indicated that the MPUnet could serve as an accurate, yet easy‐to‐use tool for robust cross‐cohort knee MRI segmentation also in clinics with limited access to technical experts.
To test this hypothesis, this study investigated the performance and robustness of the MPUnet as compared to other state‐of‐the‐art models for OA segmentation when applied across cohorts without manual adaptation of model‐ or optimization hyperparameters. A total of four OA segmentation models were considered:
The default MPUnet. 23 The MPUnet relies on a single 2D U‐Net (fully convolutional neural network) model fit to 2D image slices sampled isotropically along V = 6 viewing planes through the image volume. The amount of training data increases V times, but the different views of a volume are not independent of each other. In this way the extension of the training data resembles data augmentation. 25 Random elastic deformations are applied to a subset of the sampled images to further augment the training dataset, see Fig. S1 in the Supplemental Material. 26 During optimization, images from all planes are fed to the (a priori plane‐agnostic) model without additional information about the corresponding image plane, see Figure 1. This training setup forces the model to learn to segment the medical target of interest as seen from multiple views. When segmenting a new scan, the model first predicts along each plane in the isotropic scanner space creating a set of V full segmentation volumes for each input scan. The V segmentation suggestions are combined into one final output using a learned fusion model. The single neural network model thus plays the role of V experts in an ensemble‐like method. The approach is illustrated in Figure 2. The output of the MPUnet is considered the final segmentation with no postprocessing steps applied. The MPUnet and its optimization is described in further detail in the Supplemental Material. Additional technical details are given by Perslev et al. 23
The MPUnet using only a single view (the sagittal view). This corresponds to training a simple 2D U‐Net but using the augmentation strategy (ie, random elastic deformations) and training pipeline of the MPUnet. This ablatation study tests the effectiveness of including additional views.
The validated KIQ automatic segmentation method. 4 , 8 The KIQ method was developed and extensively validated over many years and is partly based on task‐specific knowledge on cartilage segmentation. The framework first aligns the considered scan to a reference knee MRI model using rigid multi‐atlas registration. Gaussian derivative features are then computed within regions of interest for each segmentation compartment individually. The computed features support voxel‐wise classifications using compartment specific classifiers, and largest connected component analysis is used to select final segmentation volumes for each compartment.
A 2D U‐Net as implemented by Panfilov et al which represents state‐of‐the‐art performance on the Osteoarthritis Initiative (OAI) dataset, 13 see Materials and Methods section. The optimization hyperparameters, including loss function, learning rate, weight decay, batch size, number of epochs, and so on, have been tuned for the OAI dataset, and this comparison thus allows to study how the popular 2D U‐Net transfers to other datasets without re‐tuning of its hyperparameters. The Panfilov 2D U‐Net performs slice‐wise segmentation in the sagittal view without postprocessing of the obtained masks. Random augmentations, such as gamma corrections, scaling and bilateral filtering, are applied during training.
This study aimed to investigate how each of these models perform when applied with default hyperparameters across three distinct OA cohorts to measure their robustness to scanner‐ and patient demographic variations. Average model performance across all scans in a cohort and as a function of Kellgren and Lawrence (KL) grades were compared. 27 Finally, the ability of the MPUnet to learn segmentations across multiple cohorts at once was investigated.
FIGURE 1.
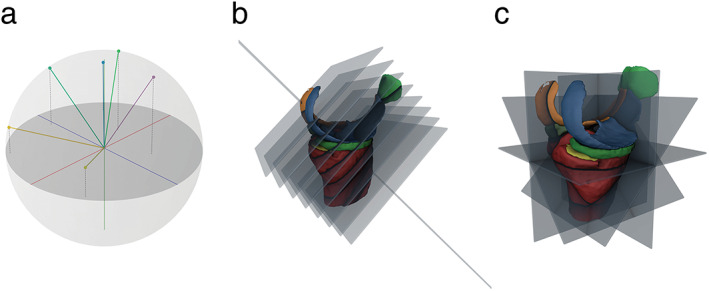
(a) Visualization of a set of sampled view axis unit vectors. (b) Illustration of images sampled along one view. (c) Illustration of multiple images sampled along multiple unique views. Adapted from Perslev et al. 23
FIGURE 2.
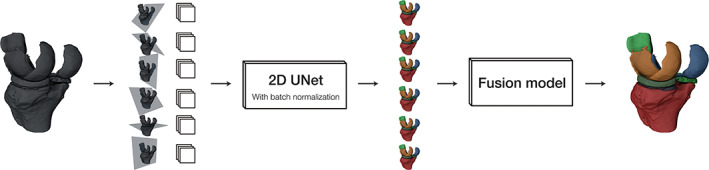
Model overview. In the inference phase, the input volume (left) is sampled on 2D isotropic grids along multiple view axes. The model predicts a full volume along each axis and maps the predictions into the original image space. A fusion model combines the proposed segmentation volumes into a single final segmentation. Adapted from Perslev et al. 23
Materials and Methods
Cohorts
The performance of all segmentation models were evaluated on three distinct cohorts of MR knee scans:
OAI cohort subset consisting of 88 baseline scans and 88 follow‐up scans with approximately 1‐year interval. Scans were acquired using a Siemens 3 T Trio (Erlangen, Germany) scanner and a sagittal 3D dual‐echo steady‐state (DESS) with water excitation sequence. The cohort consists of 45 males and 43 females of ages years and body mass indexes (BMIs) . All enrolled participants either had or were at increased risk of developing OA. OA severity was assessed for 44 baseline scans with 0/2/10/30/2 scans of KL grades 0–4.
Center for Clinical and Basic Research (CCBR) consisting of 140 scans from 140 subjects. 28 Scans were acquired using a 0.18 T Esaote C‐Span scanner (Genova, Italy) and a Turbo 3D T1w sequence. The cohort consists of 78 females and 62 males of ages and BMIs . Enrolled participants had both healthy knees and varying degrees of OA with 50/24/13/22/0 scans of KL grades 0–4.
Prevention of OA in Overweight Females (PROOF) consisting of 25 knees imaged with 1.5 T Simens Symphony (Erlangen, Germany), 1.5 T Siemens Magnetom Essenza (Erlangen, Germany), and 1.0 T Phillips Intera (Eindhoven, Netherlands) scanners using a 3D sagittal DESS sequence with water excitation. 29 Women aged 50–60 years with BMI 27 and free of knee OA (according to clinical American College of Rheumatology criteria) were included in the original study. The sub‐cohort considered here consists of 25 females of ages and BMIs and 12/11/1/1/0 scans of KL grades 0–4.
Cohort statistics are summarized in Table 1. MRI sequence details are given in Table 2. All MRIs of right knees were mirrored to resemble left knees. Informed consent was given by all participants for inclusion into any of the original study cohorts. All data considered in this study were handled and processed in accordance with the relevant data sharing agreements for each study.
TABLE 1.
Overview of Study Populations
| No. of Scans | No. of Subjects | No. of Compartments | Age (years), mean ± SD | BMI, mean ± SD | Sex (M/F) (%) | |
|---|---|---|---|---|---|---|
| OAI | 176 | 88 | 6/8 a | 61 ± 10 | 31.1 ± 4.6 | 51/49 |
| CCBR | 140 | 140 | 2 | 55 ± 15 | 25.8 ± 4.0 | 44/56 |
| PROOF | 25 | 25 | 6 | 56 ± 3 | 32.2 ± 4.1 | 0/100 |
Statistics were computed over 88, 140, and 25 subjects for the OAI, CCBR, and PROOF cohorts, respectively.
OAI = Osteoarthritis Initiative; CCBR = Center for Clinical and Basic Research; PROOF = Prevention of OA in Overweight Females.
The Tibia bone was only annotated in the 88 baseline scans. In the baseline scans, the Medial & Lateral Femoral Cartilages were annotated separately, whereas in the 88 follow‐up scans the Femoral Cartilage was annotated as a single compartment.
TABLE 2.
Overview of Cohort MRI Sequences
| Cohort | OAI | CCBR | PROOF |
|---|---|---|---|
| Scanner | Siemens Trio | Esaote C‐Span |
Siemens Symphony Siemens Magnetom Essenza Phillips Intera |
| Vendor location | Erlangen, Germany | Genoa, Italy |
Erlangen, Germany Erlangen, Germany Eindhoven, Netherlands |
| Scan | 3D DESS | Turbo 3D T1w | 3D DESS |
| Field strength (T) | 3.0 | 0.18 |
1.5 1.5 1.0 |
| Acquisition time (min) | 10 | 10 | 5–10 |
| Plane | Sagittal | Sagittal | Sagittal |
| Fat suppression | Water Excitation | None | Water excitation |
| Field of view (mm) | 140 | 180 | 160 |
| Number of slices | 160 | 110 | 50–62 |
| Voxel size (mm3) | 0.700 × 0.365 × 0.365 | 0.781 a × 0.703 × 0.703 |
1.500 × 0.420 × 0.420 1.500 × 0.500/0.625 × 0.500/0.625 1.500 × 0.310 × 0.310 |
| Flip angle (°) | 25 | 40 | 25 |
| Bit depth | 12 | 8 | 12 |
| Echo/Repetition time (msec/msec) | 4.7/16.3 | 16/50 |
6.0/19.5 8.0/21.4 11.3/22.3 b |
Variable slice thicknesses in 0.703–0.938 mm, typically 0.781 mm.
Minor variations in echo/repetition times in 11.1–11.4 msec/22.2–22.6 msec.
Radiological Assessment and Segmentation
OAI
The tibial medial and tibial lateral cartilages (TMC and TLC), femoral medial and femoral lateral cartilages (FMC and FLC), medial and lateral menisci (MM and LM), and patellar cartilage (PC) were manually segmented in all 176 scans by iMorphics (Manchester, UK). The tibia bone (TB) was further annotated in the 88 baseline scans. In the baseline scans FMC and FLC were annotated separately, whereas in the 88 follow‐up scans the femoral cartilage was annotated as a single compartment. KL grades were assessed for all scans by trained radiologists from the David Felson Lab, School of Medicine, Boston University.
CCBR
TMC and FMC were manually segmented in all scans by trained radiologist Paola C Pettersen (PCP) (Denmark). KL grades were assessed by PCP for 109 out of the total 140 scans.
PROOF
TMC, TLC, FMC, FLC, PC, and TB were segmented in all scans by clinical epidemiologist and trained physiotherapist Dieuwke Schiphof (DS) of Erasmus Medical Center, Rotterdam University. KL grades were assessed by DS for all scans.
Segmentation results on TB are reported in Results section but not further discussed, because the compartment is easily segmented by all considered methods.
Segmentation Models
Four segmentation models were evaluated on each of the three MRI cohorts: 1) A default MPUnet model using planar views (see the Supplemental Material for details) 23 ; 2) A MPUnet using only the sagittal view to test the effect of using multiple views; 3) the KIQ automatic segmentation framework 4 , 8 ; and 4) a 2D U‐Net as implemented by Panfilov et al 13 marking the state‐of‐the‐art in deep learning for knee MRI segmentation. The MPUnet and KIQ framework were applied with default settings across all cohorts. The 2D U‐Net was applied with optimization hyperparameters as in Panfilov et al 13 using the codebase (https://github.com/MIPT-Oulu/RobustCartilageSegmentation) provided by the authors with the following exceptions: 1) the input image sizes were modified from the default 300 × 300 on the OAI dataset to 256 × 256 on CCBR (to match the size of those scans) and 336 × 336 on PROOF (bilinear resampling was used to down‐sample PROOF images from their original variable sizes of 320 × 320, 384 × 384, or 512 × 512 depending on scan; the resampled pixel size was set to 0.47 × 0.47 mm). The size of the images input to the 2D U‐Net matched those of the MPUnet on corresponding datasets. 2) A common batch‐size of 32 was used across the datasets (down from 64) to allow the larger 336 × 336 PROOF images to fit in our GPU memory. 3) The learning rate was reduced to 0.0005 (down from 0.001 on OAI) and the number of training epochs increased to 150 (up from 50 on OAI) when training on the PROOF dataset due to severe overfitting observed using the default parameters on this small dataset. 4) Random horizontal flips were disabled in the augmentation pipeline (leaving random gamma corrections, scaling and bilateral filtering) as all MRI images considered here were mirrored to resemble right knees as described earlier.
Experiments and Statistical Analysis
All models were trained and evaluated on each of the three study cohorts individually. The MPUnet was further evaluated in a cross‐cohort setup. The trained models were applied to a subset of the data held out during training to test their generalization properties.
Single Cohort Setup
On CCBR and OAI, all models were trained and evaluated on a fixed dataset split. On PROOF, all models were trained and evaluated in a leave‐one‐out (LOO) cross‐validation (25‐fold CV) setup (training 25 model instances each evaluated on a single, held‐out testing scan). We considered fixed training/testing splits and cross‐validation strategies for each dataset as Dam et al. 4 The CCBR dataset was split into 30 training and 110 evaluation images and the OAI dataset was split into 44 training and 44 evaluation images. The MPUnet was further evaluated using larger training datasets facilitated by either a cross‐validation setup with more folds (for CCBR and OAI) or through training on additionally images taken from a different dataset (for PROOF). Specifically, we included 88 images taken from the OAI dataset and added them to the training dataset of PROOF to investigate if the publicly available OAI dataset could reduce the need for new manual segmentations when applying the MPUnet on a new cohort.
Cross‐Cohort Setup
A single instance of the MPUnet model was trained on MRIs from the OAI, CCBR and PROOF datasets simultaneously. For OAI and CCBR, we used the same fixed dataset splits defined above in the single‐cohort setup. We also included all 25 PROOF images into the training set to expose the model to as many and variable images as possible. The model was evaluated on the test‐set images of CCBR and OAI. We did not evaluate on PROOF images as no fixed dataset split is available for this small dataset. The cross‐cohort model segments only the tibial‐ and femoral medial cartilages, as those are the only two annotated compartments of the CCBR cohort.
Evaluation
Model performances were compared using the Dice‐Sørensen coefficient (Dice), 30 , 31 which ranges from 0 to 1 with values close to 1 indicating a perfect segmentation overlap between the predicted and ground truth masks. Dice coefficients were computed for each compartment and for each patient scan separately and reported as summary statistics across patients. Specifically, for each segmentation class the mean, standard deviation, and minimum observed Dice coefficients across subjects were considered. Similar statistics were computed for all scans sub‐divided by KL grade classification scores to investigate the effect of OA on model performances.
Statistical Tests
Statistical tests were conducted to assess for differences in the observed median performance scores on each individual compartment of each dataset (CCBR, OAI, and PROOF) between:
The MPUnet and KIQ/2D U‐Net for models trained in the single‐cohort setup with limited data (ie, when the MPUnet and KIQ/2D U‐Net were trained and evaluated on identical datasets).
The MPUnet trained in the single‐cohort setup with limited and with additional training data.
The MPUnet trained in the single‐cohort setup with limited data and the MPUnet trained under the cross‐cohort setup.
All reported P‐values were computed from paired, two‐sided Wilcoxon signed‐rank statistics unless explicitly stated otherwise. The Wilcoxon test is a nonparametric test and suitable for comparing Dice scores, which are not normally distributed. Performance differences were considered statistically significant at P‐value threshold . In all cross‐validation experiments, each scan in a dataset appears in the test‐set of a single fold, and the entire dataset is predicted once and used for computation of evaluation metrics and subsequent statistical tests. In CV the individual hold‐out datasets are not statistically independent of each other, because the hold‐out data in one fold is in the training data of all other folds. This has to be taken into account in when interpreting the statistical results (eg, see the recent work by Bates et al 32 ).
Results
Single‐Cohort Experiments
Table 3 summarizes the segmentation performance of the MPUnet, KIQ, and 2D U‐Net methods on all three study cohorts (see Tables 1 and 2). When trained on the same number of samples, the MPUnet performed significantly better in terms of the mean macro Dice scores (mean across compartments and patients) on the OAI dataset compared to KIQ ( vs. , ), the 2D U‐Net ( vs. , ), and the single‐view MPUnet ( vs. , . The MPunet performed significantly better on the CCBR dataset compared to KIQ ( vs. , ), the 2D U‐Net ( vs. , ), and the single‐view MPunet ( vs. , ). The MPUnet performed significantly better on the PROOF dataset compared to the 2D U‐Net ( vs. , ) and the single‐view MPunet ( vs. , ) and indifferent from KIQ ( vs. , ).
TABLE 3.
Single‐Cohort Experiments: Segmentation Performance Across Subjects for the MPUnet, Single‐View MPUnet, 2D U‐Net, and KIQ Methods on the OAI, CCBR, and PROOF Cohorts
| Dataset | Method | Eval. Type | Eval. Images | Tibia Bone a | Tibial Medial Cartilage | Tibial Lateral Cartilage | Femoral Medial Cartilage | Femoral Lateral Cartilage | Patellar Cartilage | Medial Meniscus | Lateral Meniscus | Macro Dice |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCBR | KIQ | Fixed split | 110 | — | 0.83 ± 0.06 | — | 0.79 ± 0.06 | — | — | — | — | 0.81 ± 0.06 |
| 0.47 | 0.52 | 0.57 | ||||||||||
| P < 0.05 | P < 0.05 | P < 0.05 | ||||||||||
| 2D U‐Net | Fixed split | 110 | — | 0.83 ± 0.06 | — | 0.81 ± 0.05 | — | — | — | — | 0.82 ± 0.05 | |
| 0.57 | 0.64 | 0.64 | ||||||||||
| P = 0.18 | P < 0.05 | P < 0.05 | ||||||||||
| MP (V = 1) | Fixed split | 110 | — | 0.82 ± 0.06 | — | 0.80 ± 0.06 | — | — | — | — | 0.81 ± 0.06 | |
| 0.60 | 0.57 | 0.59 | ||||||||||
| P < 0.05 | P < 0.05 | P < 0.05 | ||||||||||
| MP (V = 6) | Fixed split | 110 | — | 0.84 ± 0.04 | — | 0.82 ± 0.05 | — | — | — | — | 0.83 ± 0.04 | |
| 0.68 | 0.65 | 0.69 | ||||||||||
| MP (V = 6) | 5‐CV | 140 | — | 0.85 ± 0.04 | — | 0.83 ± 0.04 | — | — | — | — | 0.84 ± 0.04 | |
| 0.65 | 0.68 | 0.68 | ||||||||||
| OAI | KIQ | Fixed split | 44 | 0.98 ± 0.00 | 0.84 ± 0.05 | 0.89 ± 0.04 | 0.83 ± 0.05 | 0.86 ± 0.04 | 0.78 ± 0.11 | 0.80 ± 0.10 | 0.86 ± 0.04 | 0.84 ± 0.04 |
| 0.98 | 0.69 | 0.73 | 0.68 | 0.73 | 0.40 | 0.34 | 0.75 | 0.72 | ||||
| P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | ||||
| 2D U‐Net | Fixed split | 44 | 0.89 ± 0.01 | 0.85 ± 0.05 | 0.89 ± 0.03 | 0.85 ± 0.05 | 0.88 ± 0.04 | 0.81 ± 0.12 | 0.82 ± 0.07 | 0.87 ± 0.03 | 0.85 ± 0.03 | |
| 0.87 | 0.71 | 0.80 | 0.65 | 0.74 | 0.33 | 0.57 | 0.79 | 0.77 | ||||
| P < 0.05 | P = 0.16 | P = 0.06 | P < 0.05 | P = 0.09 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | ||||
| MP (V = 1) | Fixed split | 44 | 0.98 ± 0.0 | 0.84 ± 0.05 | 0.89 ± 0.04 | 0.84 ± 0.05 | 0.86 ± 0.05 | 0.82 ± 0.10 | 0.82 ± 0.07 | 0.88 ± 0.04 | 0.85 ± 0.03 | |
| 0.98 | 0.68 | 0.75 | 0.61 | 0.70 | 0.51 | 0.60 | 0.74 | 0.76 | ||||
| P < 0.05 | P < 0.05 | P = 0.06 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | ||||
| MP (V = 6) | Fixed split | 44 | 0.98 ± 0.0 | 0.85 ± 0.05 | 0.90 ± 0.04 | 0.86 ± 0.05 | 0.88 ± 0.04 | 0.83 ± 0.11 | 0.83 ± 0.06 | 0.89 ± 0.03 | 0.86 ± 0.03 | |
| 0.98 | 0.72 | 0.79 | 0.66 | 0.73 | 0.26 | 0.66 | 0.82 | 0.75 | ||||
| MP (V = 6) | 5‐CV | 176 (174) | — | 0.85 ± 0.05 | 0.89 ± 0.03 | 0.88 ± 0.03 | 0.81 ± 0.10 | 0.82 ± 0.07 | 0.87 ± 0.03 | 0.85 ± 0.03 | ||
| 0.67 | 0.76 | 0.71 | 0.26 | 0.55 | 0.66 | 0.70 | ||||||
| PROOF | KIQ | 25‐CV | 25 | 0.96 ± 0.02 | 0.79 ± 0.06 | 0.76 ± 0.09 | 0.77 ± 0.10 | 0.80 ± 0.05 | 0.72 ± 0.11 | — | — | 0.77 ± 0.07 |
| 0.91 | 0.61 | 0.41 | 0.44 | 0.64 | 0.36 | 0.52 | ||||||
| P < 0.05 | P = 0.97 | P = 0.17 | P = 0.09 | P < 0.05 | P < 0.05 | P = 0.10 | ||||||
| 2D U‐Net b | 25‐CV | 25 | 0.97 ± 0.01 | 0.73 ± 0.09 | 0.67 ± 0.11 | 0.73 ± 0.08 | 0.75 ± 0.07 | 0.76 ± 0.07 | — | — | 0.73 ± 0.07 | |
| 0.94 | 0.48 | 0.39 | 0.52 | 0.59 | 0.60 | 0.54 | ||||||
| P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | ||||||
| MP (V = 1) | 25‐CV | 25 | 0.95 ± 0.05 | 0.76 ± 0.09 | 0.69 ± 0.15 | 0.75 ± 0.09 | 0.77 ± 0.09 | 0.78 ± 0.06 | — | — | 0.75 ± 0.08 | |
| 0.75 | 0.41 | 0.20 | 0.48 | 0.38 | 0.60 | 0.50 | ||||||
| P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | P = 0.19 | P < 0.05 | ||||||
| MP (V = 6) | 25‐CV | 25 | 0.96 ± 0.02 | 0.79 ± 0.06 | 0.72 ± 0.13 | 0.78 ± 0.08 | 0.80 ± 0.07 | 0.79 ± 0.07 | — | — | 0.78 ± 0.07 | |
| 0.89 | 0.63 | 0.29 | 0.50 | 0.47 | 0.59 | 0.56 | ||||||
| MP (V = 6) | 25‐CV + 88 OAI | 25 | — | 0.78 ± 0.08 | 0.73 ± 0.13 | 0.79 ± 0.09 | 0.83 ± 0.04 | 0.81 ± 0.04 | — | — | 0.79 ± 0.06 | |
| 0.53 | 0.26 | 0.45 | 0.67 | 0.70 | 0.60 | |||||||
| P = 0.58 | P = 0.44 | P = 0.09 | P < 0.05 | P < 0.05 | P < 0.05 | |||||||
Individual scores where the other models score better than the MPUnet are marked in bold. Accuracy is given as the Dice volume overlap showing mean ± SD and minimum values. P‐values for the paired, two‐sided Wilcoxon signed‐rank statistic are shown for all compartments comparing the MPUnet performance against itself when trained on additional data, and the KIQ method, the single‐view MPUnet and the 2D U‐Net when evaluated on identical dataset.
CV = cross validation; LOO = leave one out (number of CV folds identical to the number of evaluation images); OAI = Osteoarthritis Initiative; CCBR = Center for Clinical and Basic Research; PROOF = Prevention of OA in Overweight Females.
Tibia bone excluded from computation of Macro Dice scores.
Lower LR, higher epochs compared to Panfilov et al, 2019 13 .
Table 3 also details the performance of all methods on each individual compartment across the three datasets and shows the minimal Dice scores observed for the compartment across all subjects in the cohort. Across a total of 14 segmentation compartments (tibia bone excluded as it is easily segmented by all methods), the MPUnet performed significantly better than the KIQ model on 11 compartments (TMC, TLC, FMC, FLC, PC, MM, and LM on OAI; TMC and FMC on CCBR; FLC and PC on PROOF; for all) and with no significant difference on the remaining 3 (TMC, , TLC, , and FMC, , all on PROOF). The MPUnet performed significantly better than the Paniflov 2D U‐Net on 10 compartments (FMC, PC, MM, and LM on OAI; FMC on CCBR; TMC, TLC, FMC, FLC, and PC on PROOF; for all) and with no significant difference on the remaining 4 (TMC, , TLC, , FLC, on OAI; TMC, on CCBR). The MPUnet performed significantly better than its single‐view counterpart on 12 compartments (TMC, FMC, FLC, PC, MM, and LM on OAI; TMC and FMC on CCBR; TMC, TLC, FMC, and FLC on PROOF; for all) and with no significant difference on the remaining 2 (TLC, on OAI; PC, on PROOF). None of the other models performed significantly better than the MPUnet on any compartment.
Table 4 details the performance of each model on the CCBR, OAI, and PROOF datasets grouped by KL grade assessments of each scan. Figure 3 shows box‐plot Dice score distributions for each compartment of the CCBR dataset as segmented by the MPUnet, KIQ, and 2D U‐Net models similarly grouped by KL grades. Box‐plot figures for the OAI and PROOF datasets are shown in Figs. S2 and S3 in the Supplemental Material.
TABLE 4.
Single‐Cohort Experiments — KL Groups: Segmentation Performance Across Subjects for the MPUnet, Single‐View MPUnet, 2D U‐Net, and KIQ Methods on the OAI, CCBR, and PROOF cohorts on KL Subgroups
| Dataset | Method | Eval. Type | Eval. Images | KL 0 | KL 1 | KL 2 | KL 3 | KL 4 |
|---|---|---|---|---|---|---|---|---|
| CCBR | KIQ | Fixed split | 50/24/13/22/0 | 0.84 ± 0.03 | 0.82 ± 0.03 | 0.78 ± 0.04 | 0.75 ± 0.08 | — |
| 0.73 | 0.72 | 0.68 | 0.57 | |||||
| P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | |||||
| 2D U‐Net | Fixed split | 50/24/13/22/0 | 0.84 ± 0.03 | 0.83 ± 0.03 | 0.80 ± 0.04 | 0.76 ± 0.06 | — | |
| 0.77 | 0.75 | 0.72 | 0.64 | |||||
| P < 0.05 | P = 0.03 | P = 0.74 | P < 0.05 | |||||
| MP (V = 1) | Fixed split | 50/24/13/22/0 | 0.84 ± 0.02 | 0.83 ± 0.03 | 0.78 ± 0.04 | 0.73 ± 0.06 | — | |
| 0.79 | 0.77 | 0.68 | 0.59 | |||||
| P < 0.05 | P < 0.05 | P < 0.05 | P < 0.05 | |||||
| MP (V = 6) | Fixed split | 50/24/13/22/0 | 0.85 ± 0.03 | 0.84 ± 0.03 | 0.81 ± 0.02 | 0.78 ± 0.06 | — | |
| 0.80 | 0.77 | 0.77 | 0.69 | |||||
| OAI | KIQ | Fixed split | 0/2/10/30/2 | — | 0.88 ± 0.03 | 0.84 ± 0.04 | 0.83 ± 0.04 | 0.83 ± 0.02 |
| 0.86 | 0.76 | 0.72 | 0.82 | |||||
| P = NA | P = 0.23 | P < 0.05 | P = NA | |||||
| 2D U‐Net | Fixed split | 0/2/10/30/2 | — | 0.87 ± 0.03 | 0.85 ± 0.04 | 0.85 ± 0.03 | 0.86 ± 0.02 | |
| 0.85 | 0.78 | 0.77 | 0.85 | |||||
| P = N/A | P = 0.16 | P < 0.05 | P = NA | |||||
| MP (V = 1) | Fixed split | 0/2/10/30/2 | — | 0.87 ± 0.02 | 0.84 ± 0.03 | 0.85 ± 0.03 | 0.86 ± 0.01 | |
| 0.85 | 0.77 | 0.76 | 0.85 | |||||
| P = NA | P < 0.05 | P < 0.05 | P = NA | |||||
| MP (V = 6) | Fixed split | 0/2/10/30/2 | — | 0.88 ± 0.03 | 0.86 ± 0.03 | 0.86 ± 0.04 | 0.87 ± 0.01 | |
| 0.86 | 0.78 | 0.75 | 0.87 | |||||
| PROOF | KIQ | 25‐CV | 12/11/1/1/0 | 0.76 ± 0.06 | 0.77 ± 0.09 | 0.81 ± 0.00 | 0.80 ± 0.00 | — |
| 0.63 | 0.52 | 0.81 | 0.80 | |||||
| P = 0.08 | P = 0.41 | P = N/A | P = N/A | |||||
| 2D U‐Net a | 25‐CV | 12/11/1/1/0 | 0.74 ± 0.06 | 0.72 ± 0.08 | 0.76 ± 0.00 | 0.71 ± 0.00 | — | |
| 0.60 | 0.54 | 0.76 | 0.71 | |||||
| P < 0.05 | P < 0.05 | P = NA | P = NA | |||||
| MP (V = 1) | 25‐CV | 12/11/1/1/0 | 0.74 ± 0.08 | 0.76 ± 0.08 | 0.77 ± 0.00 | 0.77 ± 0.00 | — | |
| 0.50 | 0.52 | 0.77 | 0.77 | |||||
| P < 0.05 | P = 0.07 | P = NA | P = NA | |||||
| MP (V = 6) | 25‐CV | 12/11/1/1/0 | 0.77 ± 0.07 | 0.78 ± 0.07 | 0.82 ± 0.00 | 0.79 ± 0.00 | — | |
| 0.56 | 0.59 | 0.82 | 0.79 |
Individual scores where the other models score better than the MPUnet are marked in bold.
OAI = Osteoarthritis Initiative; CCBR = Center for Clinical and Basic Research; PROOF = Prevention of OA in Overweight Females.
Lower LR, higher epochs compared to Panfilov et al. 13
FIGURE 3.
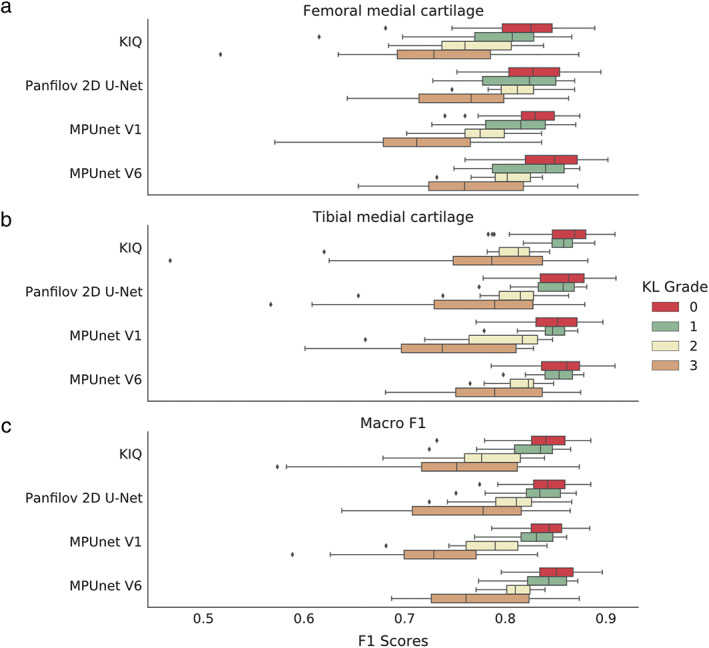
Box‐plots showing the distribution of Dice scores for the MPUnet, KIQ, and the 2D U‐Net on the CCBR dataset grouped according to the KL‐grade score of the individual MRIs. (a) Dice scores on the Femoral Medial Cartilage. (b) Dice scores on the Tibial Medial Cartilage. (c) Macro Dice scores.
On the CCBR dataset, all models had decreasing average performance for increasing KL grades with mean Dice scores across KL‐0 grade scans and KL‐3 grade scans dropping from to for KIQ, from to for the 2D U‐Net, from to for the single‐view MPUnet, and from to for the MPUnet ( for all, Mann–Whitney U test). The MPUnet had significantly higher average performance on CCBR KL‐3 grade scans compared to both KIQ, 2D U‐Net and the single view MPUnet ( for all).
On the OAI dataset, for all models there was a nonsignificant difference between their performances on KL‐2 () and KL‐3 () grade scans (KIQ: and , ; 2D U‐Net: and , ; MPUnet (): and , ; MPUnet (): and , ; Mann–Whitney U tests). The MPUnet had significantly higher average performance compared to all other models on the KL‐3 group scans with vs. for KIQ, for the 2D U‐Net and for the single‐view MPUnet ( for all). On KL‐2 scans the MPUnet performed significantly better than the single‐view MPUnet ( vs. , ) and indifferent from both KIQ ( vs. , ) and 2D U‐Net ( vs. , ). No statistics were computed for KL‐1 or KL‐4 scans as the sample sizes of were too small.
On the PROOF dataset, the MPUnet performed indifferent from KIQ on both KL‐0 scans ( vs. , ) and KL‐1 scans ( vs. , ) and significantly better than the 2D U‐Net ( vs. , ) and single‐view MPunet ( vs. , ) on KL‐0 scans and significantly better than 2D U‐Net ( vs. , ) and indifferent from the single‐view MPUnet ( vs. , ) on KL‐1 scans. No statistics were computed for the KL‐2 or KL‐3 scans as the sample sizes were too small.
Figure 4 displays a surface model fit to the manual and MPUnet predicted segmentation masks on a single subject of the OAI cohort. The output was generated by the MPUnet trained in the fixed‐split setup (model trained with less data) and having the mean Dice on this image closest to the mean performance over the OAI cohort. Thus, the figure shows the typical performance of the model. An animation showing a rotation of the predicted segmentation compartments is also available in the Supplemental Material (Video S1).
FIGURE 4.
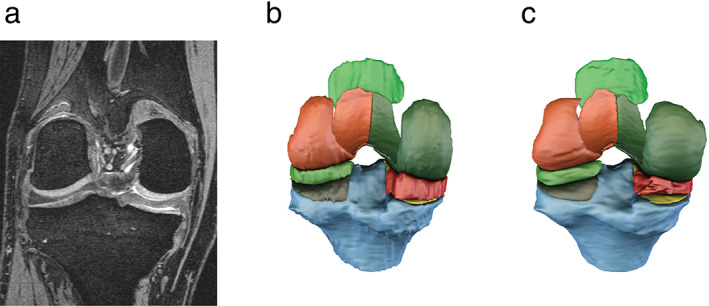
Surface models visually comparing the expert annotated segmentation (b) to the annotations of MPUnet (c) on an average performing sample of the OAI dataset. (a) Shows a reference coronal slice from the MRI volume with KL grade = 3. See also the Supplemental Material for a rotating animation of the predicted segmentation compartments.
Single‐Cohort Experiments: Training with Additional Data
Table 3 also summarizes the performance of the MPUnet model when trained on larger versions of the CCBR, OAI and PROOF datasets. On CCBR, the average Dice scores improved slightly from to on TMC and from to on FMC with the inclusion of additional training data, while the worst‐case performance decreased on TMC and increased on FMC. On the OAI dataset, the 5‐CV models obtained slightly lower Dice scores than the single‐split model on average ( vs. ). However, for both CCBR and OAI, direct statistical comparisons were not made, because the evaluation datasets differ.
On the PROOF dataset, the addition of 88 OAI scans (significantly different in both resolution, noise level and contrast compared to the scans of PROOF) to the training set significantly improved average Dice scores on FLC (from to , ) and PC (from to , ), nonsignificantly increased average Dice scores on TLC (from to , ) and FMC (from to , ) and nonsignificantly decreased performance on TMC (from to , ). The mean macro Dice scores were significantly improved from to (.
Cross‐Cohort Experiment
Table 5 summarizes the performance sores of an MPUnet model trained on images from all the OAI, CCBR and PROOF datasets simultaneously and evaluated on test‐set images from OAI and CCBR. The cross‐cohort model matched the performance of its specialized counterpart on the CCBR dataset (mean macro Dice scores of and , respectively, ) while the cross‐cohort MPUnet model showed significantly decreased, but still high, performance compared to its specialized counterpart on the OAI dataset ( and , respectively, ).
TABLE 5.
Cross‐Cohort Experiment: Segmentation Performance Across Subjects in the Test‐Splits of OAI and CCBR of a Single MPUnet Model Instance Trained on MRIs From All of the CCBR, OAI, and PROOF Cohorts
| Method | MP | |
|---|---|---|
| Training images | 30 CCBR + 44 OAI + 25 PROOF | |
| Evaluation images | 110 CCBR | 44 OAI |
| Tibial medial cartilage | 0.84 ± 0.05 | 0.83 ± 0.06 |
| 0.59 | 0.59 | |
| P = 0.49 | P < 0.05 | |
| Femoral medial cartilage | 0.82 ± 0.05 | 0.85 ± 0.05 |
| 0.68 | 0.66 | |
| P = 0.07 | P < 0.05 | |
| Macro Dice | 0.83 ± 0.04 | 0.84 ± 0.04 |
| 0.66 | 0.72 | |
| P = 0.71 | P < 0.05 | |
Accuracy is given as the Dice volume overlap showing mean ± SD and minimum values. P‐values compare the per‐compartment mean Dice scores of the cross‐cohort model to the MPUnet trained and evaluated on the individual cohorts.
Individual scores where the cross‐cohort model scores better than the respective specialized MPUnet model are marked in bold.
OAI = Osteoarthritis Initiative; CCBR = Center for Clinical and Basic Research; PROOF = Prevention of OA in Overweight Females.
Discussion
In this study, three models for automatic MRI knee segmentation were evaluated across three clinical cohorts. Each model was applied as‐is without prior tuning of its hyperparameters to simulate a clinical scenario in which the model is to be applied in a new setting (eg, in a new clinic, for a new scanner or for a new segmentation task), but where manual tuning of the model's hyperparameters is not feasible (eg, due to lack of technical experts, computational resources or time). The MPUnet was hypothesized to perform well under these restrictions, because it was designed for participation in the 2018 Medical Segmentation Decathlon. 24 Participating models were tasked to solve highly variable medical segmentation tasks without (manual) task‐specific modifications. The MPUnet ranked 5th without expensive hyperparameter tuning 24 (see in the Supplemental Material for details). In addition, the MPUnet later scored a top position in the 2019 OA MRI segmentation challenge using the same set of hyperparameters. 21
Here, the MPUnet was compared to the validated KIQ method as well as a state‐of‐the‐art 2D U‐Net implementation for knee MRI segmentation by Panfilov et al 13 on the OAI, CCBR and PROOF datasets. The considered cohorts varied in both patient demographics, size and scanner sequences, see Tables 1 and 2. All three models were able to reach high performance on both the CCBR and OAI datasets. However, the MPUnet reached a significantly higher mean macro Dice score on the OAI and CCBR datasets compared to both KIQ and the 2D U‐Net. None of the comparison models reached significantly higher Dice scores on any individual compartment across the datasets. The performance scores of the MPUnet were only slightly below the best of models submitted to the 2019 OA MRI segmentation challenge (a set of models which included the MPUnet itself) on the same dataset. There, models achieved mean Dice scores in the range of 0.86–0.88 but on fewer segmentation compartments than considered in this study (ie, the 2019 OA MRI segmentation challenge task was simpler). 21 In the challenge, only four compartments were segmented while eight were segmented here. The MPUnet even performed slightly better than the 2D U‐Net model, which was tuned specifically for the OAI dataset, and has reported the highest (to our knowledge) mean Dice scores so far with on femoral cartilage, on tibial cartilage, on patellar cartilage and on menisci. 13 It is important to note that the re‐trained version of the 2D U‐Net model applied to the OAI dataset in this work scores significantly lower on average. Again, this is due to the different number of segmentation compartments considered (four and eight, respectively). For instance, the models trained here must separate the femoral cartilage into both a medial and lateral sub‐compartment which is a harder task with uncertainty even in the ground‐truth labelling.
Interestingly, the KIQ and the MPUnet performed equally good on the small and variable () PROOF dataset without requiring modifications (for reference, the menisci have previously been automatically segmented with a mean Dice of 0.75 on the same dataset, but a direct comparison is not possible as the menisci were not segmented here 33 ). The 2D U‐Net model, however, experienced significant overfitting when trained using its default parameters. Overfitting was decreased by lowering the learning rate and increasing the number of training epochs, but still the obtained 2D U‐Net model performed significantly worse than both KIQ and the MPUnet. This result illustrates the premise of this paper, namely that adapting automatic segmentation models in practice is challenging. While the 2D U‐Net model of Panfilov et al. is one of the best models fit so far on the OAI dataset for the segmentation of the four considered compartments, that result alone does not provide a guarantee that the model will work well on other, for example, smaller, datasets or even the same dataset with a different number of segmentation compartments. With systematic hyperparameter tuning, the 2D U‐Net model could likely be brought to a high performance also on the PROOF dataset, but such a process may not be feasible in many clinical settings. The KIQ model, although slightly inferior on average to the 2D U‐Net on the CCBR and OAI datasets, does not suffer from this limitation when transferred to the small PROOF dataset. This is likely because the framework builds on expert knowledge of knee segmentation, which acts as a strong prior when learning a new dataset. Therefore, the KIQ framework requires less data compared to the 2D U‐Net, which must learn from scratch how to segment the 25 new MRIs. Interestingly, the MPUnet, which is also a deep‐learning model based on the 2D U‐Net and accordingly must also learn from scratch on the small PROOF dataset, did surprisingly well and even outperforms the KIQ framework as measured by the average Dice scores. The MPUnet's robustness and ability to learn from small datasets may result from its unique multi‐planar data augmentation strategy. This is supported by the observation that the single‐view MPUnet model performs significantly worse than the normal (6 viewed) MPUnet model on all datasets, and often below both KIQ and the 2D U‐Net.
The average performance of automatic knee MRI segmentation models are likely to drop for knees of increasing KL grades as increased OA severity may cause the target compartments to vary abnormally in both shape and volume. Consequently, the robustness and clinical relevance of any automatic model is reflected above all in its performance on high KL‐grade scans. This study systematically investigated the performance of all models as a function of KL grades 0 to 3. The considered cohorts contained too few scans of KL grade 4 for statistical analysis. On both the OAI and CCBR cohorts, the MPUnet had a significantly higher average performance on knees with moderate OA (KL‐3) as compared to all other models. None of the other models performed significantly better than the MPUnet on any individual KL‐grade group across the datasets.
On the CCBR dataset, all considered models dropped in performance as a function of KL grade. On the OAI and PROOF datasets, however, the picture is less clear. For instance, the MPUnet performed similar on KL‐2 () and KL‐3 () OAI scans, but with only KL‐1 and KL‐4 grade scans available it could not be concluded if there is an overall decreasing trend or not. Similarly, a decreasing trend could not be concluded on the PROOF data due to the limited number of available scans of KL grades 2 and 3.
As the performance of deep learning models generally improves with increasing amounts of training data, the potential for further improvement of the MPUnet performance was tested by training separate instances of the model on larger training datasets. As expected, increasing the size of the PROOF training dataset (by using more folds in CV) increased performance as measured by most metrics on all compartments. On the OAI dataset, the performance instead dropped slightly. In both cases, however, a direct comparison is difficult because the evaluation sets differ (evaluation on a fixed test‐set vs. CV). Interestingly, including 88 MRIs from the OAI dataset into the PROOF training set significantly improved the macro Dice performance of the MPUnet. The cross‐cohort experiment further showed that a single instance of the MPUnet can learn to segment knee MRIs from two different scanner sequences and patient cohorts with high performance on both. These results suggest that a great potential exists to obtain robust and clinically applicable models by training on larger, merged knee MRI datasets even if they differ with regards to, for example, scanner sequences, clinical site, and cohort demographics. This strategy of mixing even highly variable training datasets has recently led to the development of robust & clinically applicable models in the field of automated sleep analysis. 34 Given the demonstrated high performance of the MPUnet across clinical cohorts, MRI sequences and KL grades, such a model, if trained on enough and variable data, is perhaps achievable also for knee MRI segmentation and could ultimately serve as a ready‐to‐use, robust model for general knee MRI segmentation.
The pre‐trained MPUnet models are made available. These models may be used directly or serve as initializations for training new models. This transfer learning can help building well generalizing models for new data even if the new dataset is very small.
Limitations
This study considered mean Dice scores as a direct proxy for general knee MRI segmentation performance. Further studies should be made to address if the presented observations hold also for other clinically relevant metrics such as surface distances, volumes, and so on. Ultimately, future studies should address if the segmentation masks obtained by deep learning allow for accurate assessments of pathologies such as OA associated cartilages. In addition, this study did not include data from all major producers of MRI scanners (eg, GE Healthcare). Finally, it is a limitation of the study that data selection was done retrospectively.
Conclusion
This study found that the MPUnet improves on the state‐of‐the‐art in knee MRI segmentations across cohorts without the need for manual adaptations. It was found accurate even on high KL‐grade scans and could learn across multiple cohorts at once. This robustness of the MPUnet makes it practical and applicable also for research groups with limited specialist knowledge of deep learning, because the framework may be easily adapted to new data or even applied directly using one of the pre‐trained models that were made available.
Author Contributions
Mathias Perslev developed the MPUnet framework and conducted all experiments related to this system. Akshay Pai, Christian Igel, and Erik B. Dam all made substantial contributions to the conceptual and theoretical development of the method, the experimental design, and result interpretations. All authors took active part in writing and revising the manuscript. All authors have accepted the final manuscript.
Conflict of Interest
Erik B. Dam is a shareholder of Biomediq. Biomediq holds the intellectual property rights to the KIQ framework used for comparison in this study.
Supporting information
Fig. S1 Visualization of the effect of random elastic deformations. (a) Input image. (b) Augmented image.
Fig. S2. Box‐plots showing the distribution of dice scores for the MPUnet, KIQ and the Panfilov 2D U‐Net on the OAI dataset grouped according to the KL‐grade score of the individual MRIs.
Fig. S3. Box‐plots showing the distribution of dice scores for the MPUnet, KIQ and the Panfilov 2D U‐Net on the PROOF dataset grouped according to the KL‐grade score of the individual MRIs.
Video S1. Animation showing a rotation of segmentation compartments as predicted by the MPUnet on a typical example from the OAI cohort.
Acknowledgments
Mathias Perslev and Christian Igel gratefully acknowledge support from the Independent Research Fund Denmark through the project “U‐Sleep” (project number 9131‐00099B). Akshay Pai and Christian Igel gratefully acknowledge support from The Danish Industry Foundation as part of the initiative AI Denmark. The OAI collection was provided by the OAI. The OAI is a public–private partnership comprised of five contracts (N01‐AR‐2‐2258, N01‐AR‐2‐2259, N01‐AR‐2‐2260, N01‐AR‐2‐2261, and N01‐ AR‐2‐2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use dataset and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Data Availability Statement
Pre‐trained MPUnet models for cohorts OAI, CCBR, and PROOF are available at https://doi.org/10.17894/ucph.b3963d9a-78f6-4b89-b257-dc043486cd83. The multi‐planar convolutional neural network method is available as open‐source software. The software can be used without prior knowledge of deep learning, is open sourced under the MIT license and is available along with tutorials at https://github.com/perslev/MultiPlanarUNet. To fit the model to new MRI sequences, a set of manually annotated segmentation masks are required. With these at hand, the included Python scripts will perform training, evaluation, and predictions on future images with launching the script on properly organized data folders being the only involved human action. The software requires just a single GPU but can utilize additional GPUs if available. For most applications, 12GB GPU memory is required for optimal performance. On our system with a single GPU segmenting a new scan takes 2–6 minutes.
References
- 1. Shen D, Wu G, Il SH. Deep learning in medical image analysis. Annu Rev Biomed Eng 2017;19:221‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60‐88. [DOI] [PubMed] [Google Scholar]
- 3. Gan HS, Ramlee MH, Wahab AA, Lee YS, Shimizu A. From classical to deep learning: Review on cartilage and bone segmentation techniques in knee osteoarthritis research. Artif Intell Rev 2020;54:2445‐2494. [Google Scholar]
- 4. Dam EB, Runhaar J, Bierma‐Zienstra S, Karsdal M. Cartilage cavity—An MRI marker of cartilage lesions in knee OA with data from CCBR, OAI, and PROOF. Magn Reson Med 2018;80:1219‐1232. [DOI] [PubMed] [Google Scholar]
- 5. Kashyap S, Zhang H, Rao K, Sonka M. Learning‐based cost functions for 3‐D and 4‐D multi‐surface multi‐object segmentation of knee MRI: Data from the osteoarthritis initiative. IEEE Trans Med Imaging 2018;37:1103‐1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seim H, Kainmüller D, Lamecker H, Bindernagel M, Malinowski J, Zachow S. Model‐based auto‐segmentation of knee bones and cartilage in MRI data. Proceedings ‐ Medical Image Analysis for the Clinic: a Grand Challenge in Conjunction with MICCAI. Lecture Notes in Computer Science, 6361 Cham, Switzerland: Springer; 2010;215‐223. [Google Scholar]
- 7. Ababneh SY, Prescott JW, Gurcan MN. Automatic graph‐cut based segmentation of bones from knee magnetic resonance images for osteoarthritis research. Med Image Anal 2011;15:438‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dam EB, Lillholm M, Marques J, Nielsen M. Automatic segmentation of high‐ and low‐field knee MRIs using knee image quantification with data from the osteoarthritis initiative. J Med Imaging 2015;2:024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lecun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436‐444. [DOI] [PubMed] [Google Scholar]
- 10. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:640‐651. [DOI] [PubMed] [Google Scholar]
- 11. Norman B, Pedoia V, Majumdar S. Use of 2D U‐net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 2018;288:177‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wirth W, Eckstein F, Kemnitz J, et al. Accuracy and longitudinal reproducibility of quantitative femorotibial cartilage measures derived from automated U‐Net‐based segmentation of two different MRI contrasts: Data from the osteoarthritis initiative healthy reference cohort. Magn Reson Mater Phys Biol Med 2021;34:337‐354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Panfilov E, Tiulpin A, Klein S, Nieminen MT, Saarakkala S. Improving robustness of deep learning based knee mri segmentation: Mixup and adversarial domain adaptation. Proceedings ‐ IEEE/CVF International Conference on Computer Vision Workshop (ICCVW); International Conference on Computer Vision, New Jersey, USA: The Institute of Electrical and Electronics Engineers; 2019;450‐459. [Google Scholar]
- 14. Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 2018;80:2759‐2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tack A, Mukhopadhyay A, Zachow S. Knee menisci segmentation using convolutional neural networks: Data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2018;26:680‐688. [DOI] [PubMed] [Google Scholar]
- 16. Ambellan F, Tack A, Ehlke M, Zachow S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative. Med Image Anal 2019;52:109‐118. [DOI] [PubMed] [Google Scholar]
- 17. Raj A, Vishwanathan S, Ajani B, Krishnan K, Agarwal H. Automatic knee cartilage segmentation using fully volumetric convolutional neural networks for evaluation of osteoarthritis. Proceedings ‐ IEEE 15th International Symposium on Biomedical Imaging (ISBI); International Symposium on Biomedical Imaging, New Jersey, USA: The Institute of Electrical and Electronics Engineers; 2018. p 851‐854. [Google Scholar]
- 18. Lee H, Hong H, Kim J. BCD‐NET: A novel method for cartilage segmentation of knee MRI via deep segmentation networks with bone‐cartilage‐complex modeling. Proceedings ‐ IEEE 15th International Symposium on Biomedical Imaging (ISBI); International Symposium on Biomedical Imaging, New Jersey, USA: The Institute of Electrical and Electronics Engineers; 2018. p 1538‐1541. [Google Scholar]
- 19. Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical image computing and computer assisted intervention. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. p 246‐253. [DOI] [PubMed] [Google Scholar]
- 20. Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 2018;79:2379‐2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Desai AD, Caliva F, Iriondo C, et al. The international workshop on osteoarthritis imaging knee MRI segmentation challenge: A multi‐institute evaluation and analysis framework on a standardized dataset. Radiol Artif Intell 2021;3:e200078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ronneberger O, Fischer P, Brox T. U‐net: Convolutional networks for biomedical image segmentation. Proceedings ‐ Medical Image Computing and Computer‐Assisted Intervention (MICCAI), Lecture Notes in Computer Science, Cham, Switzerland: Springer; Vol 9351;2015. p 234‐241. [Google Scholar]
- 23. Perslev M, Dam EB, Pai A, Igel C. One network to segment them all: A general, lightweight system for accurate 3D medical image segmentation. Proceedings ‐ Medical Image Computing and Computer‐Assisted Intervention (MICCAI), Lecture Notes in Computer Science, Cham, Switzerland: Springer; Vol 11765;2019. p 30‐38. [Google Scholar]
- 24. Antonelli M, Reinke A, Bakas S, et al. The medical segmentation decathlon. arXiv 2021; Preprint. [DOI] [PMC free article] [PubMed]
- 25. Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. Proceedings ‐ 32nd International Conference on Machine Learning (ICML), Proceedings of Machine Learning Research, PMLR; Vol 37;2015. p 448‐456. [Google Scholar]
- 26. Simard PY, Steinkraus D, Platt JC. Best practices for convolutional neural networks applied to visual document analysis. Proceedings ‐ International Conference on Document Analysis and Recognition (ICDAR): International Conference on Document Analysis and Recognition, 2 New Jersey, USA: The Institute of Electrical and Electronics Engineers; 2003. p 958‐963. [Google Scholar]
- 27. Kellgren JH, Lawrence JS. Radiological assessment of osteo‐arthrosis. Ann Rheum Dis 1957;16:494‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dam EB, Folkesson J, Pettersen PC, Christiansen C. Automatic morphometric cartilage quantification in the medial tibial plateau from MRI for osteoarthritis grading. Osteoarthritis Cartilage 2007;15:808‐818. [DOI] [PubMed] [Google Scholar]
- 29. Runhaar J, Van Middelkoop M, Reijman M, et al. Prevention of knee osteoarthritis in overweight females: The first preventive randomized controlled trial in osteoarthritis. Am J Med 2015;128:888‐895.e4. [DOI] [PubMed] [Google Scholar]
- 30. Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity. Kong Dansk Vidensk Selsk Biol Skr 1948;5:1‐34. [Google Scholar]
- 31. Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297‐302. [Google Scholar]
- 32. Bates S, Hastie T, Tibshirani R. Cross‐validation: What does it estimate and how well does it do it? arXiv 2021; Preprint.
- 33. Xu D, van der Voet J, Hansson NM, et al. Association between meniscal volume and development of knee osteoarthritis. Rheumatology 2020;60:1392‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perslev M, Darkner S, Kempfner L, Nikolic M, Jennum PJ, Igel C. U‐Sleep: Resilient high‐frequency sleep staging. npj Digit Med 2021;4(72):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Visualization of the effect of random elastic deformations. (a) Input image. (b) Augmented image.
Fig. S2. Box‐plots showing the distribution of dice scores for the MPUnet, KIQ and the Panfilov 2D U‐Net on the OAI dataset grouped according to the KL‐grade score of the individual MRIs.
Fig. S3. Box‐plots showing the distribution of dice scores for the MPUnet, KIQ and the Panfilov 2D U‐Net on the PROOF dataset grouped according to the KL‐grade score of the individual MRIs.
Video S1. Animation showing a rotation of segmentation compartments as predicted by the MPUnet on a typical example from the OAI cohort.
Data Availability Statement
Pre‐trained MPUnet models for cohorts OAI, CCBR, and PROOF are available at https://doi.org/10.17894/ucph.b3963d9a-78f6-4b89-b257-dc043486cd83. The multi‐planar convolutional neural network method is available as open‐source software. The software can be used without prior knowledge of deep learning, is open sourced under the MIT license and is available along with tutorials at https://github.com/perslev/MultiPlanarUNet. To fit the model to new MRI sequences, a set of manually annotated segmentation masks are required. With these at hand, the included Python scripts will perform training, evaluation, and predictions on future images with launching the script on properly organized data folders being the only involved human action. The software requires just a single GPU but can utilize additional GPUs if available. For most applications, 12GB GPU memory is required for optimal performance. On our system with a single GPU segmenting a new scan takes 2–6 minutes.


