Abstract
Epidemiological studies and work in animal models indicate that immune activation may be a risk factor for autism spectrum disorders (ASD). We measured levels of 60 cytokines and growth factors in 869 maternal mid-gestational (MMG) and 807 child cord blood (CB) plasma samples from 457 ASD (385 boys, 72 girls) and 497 control children (418 boys, 79 girls) from the Norwegian Autism Birth Cohort. We analyzed associations first using sex-stratified unadjusted and adjusted logistic regression models, and then employed machine learning strategies (LASSO + interactions, Random Forests, XGBoost classifiers) with cross-validation and randomly sampled test set evaluation to assess the utility of immune signatures as ASD biomarkers. We found prominent case-control differences in both boys and girls with alterations in a wide range of analytes in MMG and CB plasma including but not limited to IL1RA, TNFα, Serpin E1, VCAM1, VEGFD, EGF, CSF1 and CSF2. MMG findings were most striking, with particularly strong effect sizes in girls. Models did not change appreciably upon adjustment for maternal conditions, medication use or emotional distress ratings. Findings were corroborated using machine learning approaches, with area under the receiver operating characteristic curve values in the test sets ranging from 0.771 to 0.965. Our results are consistent with gestational immunopathology in ASD, may provide insights into sex-specific differences, and have the potential to lead to biomarkers for early diagnosis.
INTRODUCTION
Autism spectrum disorders (ASD) comprise a set of pervasive neurodevelopmental conditions characterized by restricted and repetitive behavior patterns and impairments in social interaction and communication.1 ASD diagnosis is based on clinical criteria and requires specialized expertise.2 Although caregivers may detect behavioral abnormalities at earlier time points, the mean age for diagnosis is age 4 to 5 years.3 ASDs have a profound impact on public health. The Autism and Developmental Disabilities Monitoring Network, comprising 11 sites in the United States, reported a prevalence of 1 in 54 in 2016.4 World Health Organization estimates are lower at 1 in 270. This may represent a true difference in prevalence or in the efficiency of case ascertainment. ASDs are four times more common in boys than in girls. One proffered explanation for sex bias is a multiple-threshold multifactorial liability model wherein the minimum genetic liability sufficient to cause ASD is greater in females than in males.5
Twin studies as early as the 1970s indicate that ASD are heritable. Nonetheless extragenetic factors, including prenatal medications, toxins, nutrients, fever, and immune activation, may have an important role. Valproic acid exposure during the first trimester, for example, is associated with an increased risk of ASD.6, 7 Anatomical and behavioral outcomes of maternal immune activation in animal models vary with exposure timing8 and as a function of sex-specific differences in microglial responses.5, 9–13 Gestational exposure of rodents and non-human primates to the proinflammatory cytokines IL6 and IL17 results in structural and behavioral disturbances reminiscent of ASD.14–17 Cytokines regulate intrauterine immune responses, neurogenesis, neuronal migration and synaptogenesis, and have the capacity to signal through cognate receptors on microglial cells and other neural components distributed throughout brain circuitry.18–22 Immune molecules, including VEGF, serpin E1, VCAM1, and TNFa, may be produced by neural progenitors during fetal brain development, encourage angiogenesis, and contribute to the sculpting of the brain during development; disruptions in these immune molecules may contribute to the abnormalities in angiogenesis in ASD.23
There is only sparse literature on prenatal cytokine surveys in ASD. Most studies assayed dried neonatal blood spots instead of umbilical cord blood (CB).24–28 Few have described large population-based samples or prospective designs with extended longitudinal follow-up to ensure capture of ASD cases or subsets that elude earlier diagnosis. Furthermore, none have had access to the serial samples needed to examine exposures at different time points during brain development. To investigate potential associations between immune activation during gestation and ASD risk, we characterized cytokine profiles early in gestation (maternal mid-gestation or MMG) and at birth CB in cases and controls from the Autism Birth Cohort study (ABC Study), a case-control study nested within a population-based pregnancy cohort.29
METHODS
Study design, participants, and specimen collection.
The Norwegian Mother, Father and Child Cohort Study (MoBa).
The MoBa is a population-based pregnancy cohort administered by the Norwegian Institute of Public Health (NIPH),30 comprising 114,473 children born in 1999–2009 and more than 95,000 mothers and 75,000 fathers. Pregnant women attending routine ultrasound examinations at approximately 18 weeks of gestation were recruited between 1999 and 2008 throughout Norway, with an overall participation rate of 41%.31 Biological samples collected included maternal samples at MMG and birth, paternal samples, and child umbilical CB.
The ABC Study.
The ABC Study is a sub-study nested within the MoBa cohort and is comprised of cases of ASD and a random sample of the cohort selected as controls.32
ASD cases.
The ABC Study protocol defined cases according to DSM-IV-TR criteria for any ASD (Autistic Disorder, Asperger’s Disorder, Pervasive Developmental Disorder-Not Otherwise Specified). ASD cases in the ABC cohort were ascertained by multiple methods, including MoBa questionnaire screening (child ages 3, 5, and 7 years), referrals (parental or professional), and annual linkages to the Norwegian Patient Register (NPR). The ABC methods for diagnosis and confirmation are outlined in Stoltenberg, et al.32 Of the cases included in cytokine analyses reported here, 146 children were diagnosed with ASD in the ABC Study Clinic by trained clinical psychologists and child psychiatrists. The assessments included the ADI-R and the ADOS, medical history, neurological examination and tests of intellectual and adaptive functioning and language capacity. Via the NPR, 309 children were diagnosed with ASD by a physician or a psychologist using ICD-10 F84 criteria corresponding to DSM-IV-TR criteria. Patient records included developmental, medical, and neurological exams, the ADI-R, the ADOS, language assessments, adaptive functioning, and direct observation in a nursery, school, or clinic. Cases were additionally classified according to comorbidity with intellectual disability (ID) or attention deficit hyperactivity disorder (ADHD), combining data from ABC clinic assessments, record review, and the NPR.12 A record review study was conducted to determine the validity of NPR-registered ASD diagnoses. Suren et al.33 found that 95% of NPR-sourced ASD diagnoses were consistent with the ABC Study case definitions, both for the subset of children examined at the ABC Study Clinic and for those undergoing record review.
ABC controls.
Approximately 1.63% of those reaching 37.5 months of age in a given week were randomly-selected from the MoBa cohort to serve as a pool of eligible controls, principally for laboratory studies (original n=1811).32 Controls assigned an ASD diagnosis at the ABC clinic or ascertained with an ASD by the NPR were reclassified as cases (n=8). MMG plasma was collected at 17–21 weeks gestation. CB plasma was collected on the day of birth.
Study sample selection for cytokine analyses.
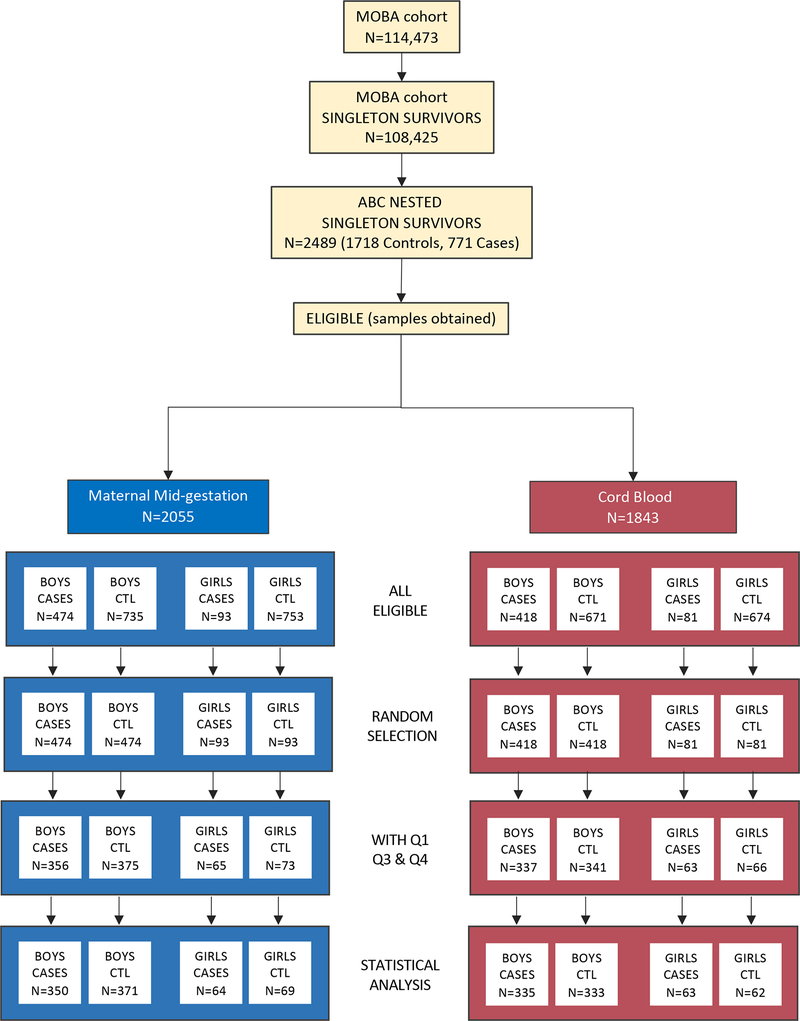
The sample derives from ASD cases ascertained through 2015 and ABC controls meeting inclusion criteria (singleton birth, continued participation in the cohort and survival to at least age three years, availability of 120 μl or more of MMG and/or umbilical CB plasma, and subjects with data available from MoBa questionnaires containing covariates relevant to both MMG and CB analyses (completed by mothers at gestational week 17 and 30, and 6 months post-partum). MMG and CB study samples were selected based on ASD case availability for each sample type, stratified on sex. Subjects with both MMG and CB samples were prioritized for the current analyses. Male and female ABC control samples were then randomly selected from among the pool of eligible male and female ABC controls in numbers equal to that of each sex- and sample type-stratified ASD case group. Demographic details of the subjects are shown in Table 1. The derivation of the final analytic study sample appears in Figure 1.
Table 1.
Subject characteristics.
| MMG Plasma (n=854) | |||||||||
| Subject Characteristics | Sex |
ASD Cases n=414 64 F/350 M |
Controls n=440 69 F/371M |
Total MMG 133 F/721M |
p-value 1 | ||||
| n | % | n | % | n | % | ||||
| Maternal Characteristics | |||||||||
| Maternal age | <30 years | F | 27 | 42.2 | 23 | 33.3 | 50 | 37.6 | 0.292 |
| M | 177 | 50.6 | 144 | 38.8 | 321 | 44.5 | 0.001 | ||
| Parental education2 | <12 years | F | 9 | 14.1 | 2 | 2.9 | 11 | 8.3 | 0.070 |
| M | 17 | 4.9 | 11 | 3.0 | 28 | 3.9 | 0.098 | ||
| 12 years | F | 13 | 20.3 | 12 | 17.6 | 25 | 18.9 | ||
| M | 102 | 29.3 | 89 | 24.1 | 191 | 26.6 | |||
| 13–16 years | F | 21 | 32.8 | 21 | 30.9 | 42 | 31.8 | ||
| M | 126 | 36.2 | 134 | 36.2 | 260 | 36.2 | |||
| >/=17 years | F | 21 | 32.8 | 33 | 48.5 | 54 | 40.9 | ||
| M | 103 | 29.6 | 136 | 36.8 | 239 | 33.3 | |||
| Maternal exposures any time in pregnancy | Non-NSAID medications | F | 22 | 34.4 | 29 | 42.0 | 51 | 38.3 | 0.364 |
| M | 138 | 39.4 | 128 | 34.5 | 266 | 36.9 | 0.171 | ||
| Fever | F | 5 | 7.8 | 3 | 4.3 | 8 | 6.0 | 0.401 | |
| M | 22 | 6.3 | 17 | 4.6 | 39 | 5.4 | 0.312 | ||
| Infection | F | 40 | 62.5 | 42 | 60.9 | 82 | 61.7 | 0.847 | |
| M | 231 | 66.0 | 218 | 58.8 | 449 | 62.3 | 0.045 | ||
| Autoimmune/ allergic disorders | F | 16 | 25.0 | 18 | 26.1 | 34 | 25.6 | 0.886 | |
| M | 97 | 27.7 | 100 | 27.0 | 197 | 27.3 | 0.819 | ||
| Obstetrical/Perinatal Factors | |||||||||
| Mode of delivery | Caesarean section | F | 5 | 7.8 | 5 | 7.2 | 10 | 7.5 | 0.902 |
| M | 27 | 7.7 | 25 | 6.7 | 52 | 7.2 | 0.613 | ||
| Child Characteristics | |||||||||
| Birth year | 2000 | F | 4 | 6.3 | 2 | 2.9 | 6 | 4.5 | 0.021 |
| M | 7 | 2.0 | 4 | 1.1 | 11 | 1.5 | <0.001 | ||
| 2001 | F | 3 | 4.7 | 5 | 7.2 | 8 | 6.0 | ||
| M | 26 | 7.4 | 15 | 4.0 | 41 | 5.7 | |||
| 2002 | F | 11 | 17.2 | 3 | 4.3 | 14 | 10.5 | ||
| M | 52 | 14.9 | 30 | 8.1 | 82 | 11.4 | |||
| 2003 | F | 17 | 26.6 | 10 | 14.5 | 27 | 20.3 | ||
| M | 62 | 17.7 | 44 | 11.9 | 106 | 14.7 | |||
| 2004 | F | 7 | 10.9 | 9 | 13.0 | 16 | 12.0 | ||
| M | 56 | 16.0 | 52 | 14.0 | 108 | 15.0 | |||
| 2005 | F | 9 | 14.1 | 9 | 13.0 | 18 | 13.5 | ||
| M | 50 | 14.3 | 51 | 13.7 | 101 | 14.0 | |||
| 2006 | F | 5 | 7.8 | 11 | 15.9 | 16 | 12.0 | ||
| M | 44 | 12.6 | 69 | 18.6 | 113 | 15.7 | |||
| 2007 | F | 7 | 10.9 | 9 | 13.0 | 16 | 12.0 | ||
| M | 30 | 8.6 | 53 | 14.3 | 83 | 11.5 | |||
| 2008 | F | 0 | 0.0 | 9 | 13.0 | 9 | 6.8 | ||
| M | 22 | 6.3 | 45 | 12.1 | 67 | 9.3 | |||
| 2009 | F | 1 | 1.6 | 2 | 2.9 | 3 | 2.3 | ||
| M | 1 | 0.3 | 8 | 2.2 | 9 | 1.2 | |||
| Birth season | Winter (Dec-Feb) | F | 13 | 20.3 | 18 | 26.1 | 31 | 23.3 | 0.347 |
| M | 87 | 24.9 | 76 | 20.5 | 163 | 22.6 | 0.093 | ||
| Spring (Mar-May) |
F | 27 | 42.2 | 19 | 27.5 | 46 | 34.6 | ||
| M | 93 | 26.6 | 113 | 30.5 | 206 | 28.6 | |||
| Summer (Jun-Aug) |
F | 14 | 21.9 | 17 | 24.6 | 31 | 23.3 | ||
| M | 74 | 21.1 | 96 | 25.9 | 170 | 23.6 | |||
| Fall (Sept-Nov) |
F | 10 | 15.6 | 15 | 21.7 | 25 | 18.8 | ||
| M | 96 | 27.4 | 86 | 23.2 | 182 | 25.2 | |||
| GA3 | <37 weeks | F | 4 | 6.3 | 5 | 7.2 | 9 | 6.8 | 0.812 |
| M | 27 | 7.7 | 16 | 4.3 | 43 | 6.0 | 0.041 | ||
| 37 to <42 weeks | F | 56 | 88.9 | 59 | 85.5 | 115 | 87.1 | ||
| M | 276 | 79.1 | 318 | 85.9 | 594 | 82.6 | |||
| >/=42 weeks | F | 3 | 4.8 | 5 | 7.2 | 8 | 6.1 | ||
| M | 46 | 13.2 | 36 | 9.7 | 82 | 11.4 | |||
| Birth weight4 | <1500 g | F | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0.597 |
| M | 4 | 1.1 | 2 | 0.5 | 6 | 0.8 | 0.363 | ||
| 1500 to <2500 g | F | 0 | 0.0 | 1 | 1.5 | 1 | 0.8 | ||
| M | 13 | 3.7 | 8 | 2.2 | 21 | 2.9 | |||
| 2500 to <4000 g | F | 51 | 79.7 | 52 | 76.5 | 103 | 78.0 | ||
| M | 233 | 66.8 | 242 | 65.2 | 475 | 66.0 | |||
| >/=4 000 g | F | 13 | 20.3 | 15 | 22.1 | 28 | 21.2 | ||
| M | 99 | 28.4 | 119 | 32.1 | 218 | 30.3 | |||
| CB Plasma (n=793) | |||||||||
| Subject Characteristics | Sex |
ASD Cases n=398 63 F/335 M |
Controls n=395 62 F/333 M |
Total MMG 125 F/668 M |
p-value 1 | ||||
| n | % | n | % | n | % | ||||
| Maternal Characteristics | |||||||||
| Maternal age | <30 years | F | 31 | 49.2 | 21 | 33.9 | 52 | 41.6 | 0.082 |
| M | 171 | 51.0 | 128 | 38.4 | 299 | 44.8 | 0.001 | ||
| Parental education2 | <12 years | F | 8 | 12.7 | 2 | 3.2 | 10 | 8.0 | 0.140 |
| M | 19 | 5.7 | 12 | 3.6 | 31 | 4.7 | 0.059 | ||
| 12 years | F | 11 | 17.5 | 9 | 14.5 | 20 | 16.0 | ||
| M | 99 | 29.6 | 73 | 22.1 | 172 | 25.9 | |||
| 13–16 years | F | 23 | 36.5 | 21 | 33.9 | 44 | 35.2 | ||
| M | 114 | 34.1 | 129 | 29.0 | 243 | 36.5 | |||
| >/=17 years | F | 21 | 33.3 | 30 | 48.4 | 51 | 40.8 | ||
| M | 102 | 30.5 | 117 | 35.3 | 291 | 32.9 | |||
| Maternal exposures any time in pregnancy | Non-NSAID medications | F | 29 | 46.0 | 28 | 45.2 | 57 | 45.6 | 0.922 |
| M | 167 | 49.9 | 162 | 48.6 | 329 | 49.3 | 0.756 | ||
| Fever | F | 14 | 22.2 | 6 | 9.7 | 20 | 16.0 | 0.056 | |
| M | 76 | 22.7 | 66 | 19.8 | 142 | 21.3 | 0.365 | ||
| Infection | F | 53 | 84.1 | 49 | 79.0 | 102 | 81.6 | 0.462 | |
| M | 273 | 81.5 | 278 | 83.5 | 551 | 82.5 | 0.498 | ||
| Autoimmune/ allergic disorders | F | 17 | 27.0 | 14 | 22.6 | 31 | 24.8 | 0.569 | |
| M | 117 | 34.9 | 115 | 34.5 | 232 | 34.7 | 0.916 | ||
| Obstetrical/Perinatal Factors | |||||||||
| Mode of delivery | Caesarean section | F | 6 | 9.5 | 2 | 3.2 | 8 | 6.4 | 0.150 |
| M | 19 | 5.7 | 15 | 4.5 | 34 | 5.1 | 0.493 | ||
| Child Characteristics | |||||||||
| Birth year | 2000 | F | 2 | 3.2 | 2 | 3.2 | 4 | 3.2 | 0.058 |
| M | 8 | 2.4 | 3 | 0.9 | 11 | 1.6 | <0.001 | ||
| 2001 | F | 3 | 4.8 | 5 | 8.1 | 8 | 6.4 | ||
| M | 27 | 8.1 | 15 | 4.5 | 42 | 6.3 | |||
| 2002 | F | 11 | 17.5 | 3 | 4.8 | 14 | 11.2 | ||
| M | 53 | 15.8 | 27 | 8.1 | 80 | 12.0 | |||
| 2003 | F | 18 | 28.6 | 11 | 17.7 | 29 | 23.2 | ||
| M | 63 | 18.8 | 45 | 13.5 | 108 | 16.2 | |||
| 2004 | F | 10 | 15.9 | 8 | 12.9 | 18 | 14.4 | ||
| M | 51 | 15.2 | 44 | 13.2 | 95 | 14.2 | |||
| 2005 | F | 9 | 14.3 | 8 | 12.9 | 17 | 13.6 | ||
| M | 46 | 13.7 | 47 | 14.1 | 93 | 13.9 | |||
| 2006 | F | 3 | 4.8 | 10 | 16.1 | 13 | 10.4 | ||
| M | 38 | 11.3 | 60 | 18.0 | 98 | 14.7 | |||
| 2007 | F | 6 | 9.5 | 7 | 11.3 | 13 | 10.4 | ||
| M | 30 | 9.0 | 43 | 12.9 | 73 | 10.9 | |||
| 2008 | F | 1 | 1.6 | 6 | 9.7 | 7 | 5.6 | ||
| M | 19 | 5.7 | 39 | 11.7 | 58 | 8.7 | |||
| 2009 | F | 0 | 0.0 | 2 | 3.2 | 2 | 1.6 | ||
| M | 0 | 0.0 | 10 | 3.0 | 10 | 1.5 | |||
| Birth season | Winter (Dec-Feb) |
F | 12 | 19.0 | 16 | 25.8 | 28 | 22.4 | 0.490 |
| M | 79 | 23.6 | 69 | 20.7 | 148 | 22.2 | 0.042 | ||
| Spring (Mar-May) |
F | 24 | 38.1 | 18 | 29.0 | 42 | 33.6 | ||
| M | 89 | 26.6 | 103 | 30.9 | 192 | 28.7 | |||
| Summer (Jun-Aug) |
F | 18 | 28.6 | 15 | 24.2 | 33 | 26.4 | ||
| M | 69 | 20.6 | 89 | 26.7 | 158 | 23.7 | |||
| Fall (Sept-Nov) |
F | 9 | 14.3 | 13 | 21.0 | 22 | 17.6 | ||
| M | 98 | 29.3 | 72 | 21.6 | 170 | 25.4 | |||
| GA3 | <37 weeks | F | 4 | 6.5 | 3 | 4.8 | 7 | 5.6 | 0.927 |
| M | 22 | 6.6 | 9 | 2.7 | 31 | 4.7 | 0.005 | ||
| 37 to <42 weeks | F | 54 | 87.1 | 55 | 88.7 | 109 | 87.9 | ||
| M | 265 | 79.3 | 293 | 88.3 | 558 | 83.8 | |||
| >/=42 weeks | F | 4 | 6.5 | 4 | 6.5 | 8 | 6.5 | ||
| M | 47 | 14.1 | 30 | 9.0 | 77 | 11.6 | |||
| Birth weight4 | <1500 g | F | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0.474 |
| M | 1 | 0.3 | 0 | 0.0 | 1 | 0.1 | 0.229 | ||
| 1500 to <2500 g | F | 1 | 1.6 | 0 | 0.0 | 1 | 0.8 | ||
| M | 9 | 2.7 | 3 | 0.9 | 12 | 1.8 | |||
| 2500 to <4000 g | F | 51 | 81.0 | 47 | 77.0 | 98 | 79.0 | ||
| M | 224 | 67.1 | 222 | 66.7 | 446 | 66.9 | |||
| >/=4000 g | F | 11 | 17.5 | 14 | 23.0 | 25 | 20.2 | ||
| M | 100 | 29.9 | 108 | 32.4 | 208 | 31.2 | |||
chi-square, 2-sided p-value.
Parental education missing: MMG, n=3M, 1F; CB, n=3M.
Gestational age missing: MMG, n=2M; CB, n=2M, 1F.
Birth weight missing: MMG, n=1M, 1F; CB, n=1M, 1F. ASD autism spectrum disorders, CB cord blood, F female, GA gestational age at birth, M male, MMG maternal mid-gestation.
Figure 1.
Pipeline for sample selection.
Covariates.
Maternal illness and medications consumed during pregnancy have the potential to influence both the risk of having altered immune marker concentrations in MMG or CB plasma, and the risk of ASD. Accordingly, we extracted data from MoBa questionnaires covering the time period prior to the maternal mid-gestational blood draw (MMG analyses), or for all of pregnancy up until birth (CB analyses) including maternal report of fever, respiratory infection, other infection, autoimmune/allergic disorders, emotional distress ratings (Hopkins Symptom Check List) (SCL-5)34, and use of acetaminophen and related non-NSAID-type antipyretic drugs (ATC codes N02BE01, N02BA01, N02BA51, N02BB51). We also included maternal age (dichotomized as <30 years or >/=30 years). MMG analyses additionally included gestational age (in days) at maternal mid-gestational blood draw. For MMG sensitivity analyses, to examine the possibility that some questionnaire data was provided beyond +/− 28 days from the timing of the acquisition of the maternal blood sample, we also extracted the dates of return of the MoBa questionnaires containing data regarding the period preceding the MMG blood draw, calculated the difference in days between the MMG blood draw and the return of the relevant MoBa questionnaire, and categorized subjects as within or outside the 28-day window between questionnaire completion and blood sample collection. For CB sensitivity analyses, we extracted data regarding preeclampsia/eclampsia, mode of delivery (Caesarean section or not) and gestational age at birth (categorized as <37 weeks; >/=37 weeks and <42 weeks; >/=42 weeks).
Human subjects.
Studies were approved by the Regional Committee for Medical and Health Research Ethics for Southeastern Norway and the Columbia University Medical Center Institutional Review Board (protocol number AAA2258). All samples were obtained using informed consent from mothers for both MMG plasma and CB.
MMG and CB samples.
MMG and umbilical CB samples were collected via syringe into K2 EDTA tubes, processed and stored at −80°C,31 with quality assurance procedures as previously described.35 MMG and CB plasma samples were shipped from the MoBa Biobank to Columbia on dry ice, stored at −80°C until aliquoting and returned to −80°C storage until use.
Immune profiling assays.
We assayed a wide range of cytokines, chemokines, cellular and growth factors reflecting key processes relating to systemic activation of inflammatory/immune signaling pathways involved in autoimmunity and anti-inflammatory responses as well as others implicated in CNS inflammation, neurovascular disruption and neurogenesis. Immune molecules within this panel are also found to be dysregulated during infection with certain pathogens, including those that trigger autoimmunity,36 as well as in some studies in ASD27,28,37 (Supplementary Table 1). The fluorescent intensity levels of the following immune molecules were determined using a bead-based, 60-plex immunoassay: interleukin (IL)1 superfamily, IL1α, IL1β, IL18, IL1RA; IL2 family, IL2, IL4, IL7, IL9, IL15, IL21; IL6 (gp130) family, IL6, IL31, LIF; IL12 family, IL12p40, IL12p70, IL23, IL27; IL17 family, IL17A, IL17F, IL22; Th2 type, IL5, IL10, IL13; tumor necrosis factor (TNF) superfamily, TNFα (TNFSF2), TNFβ (TNFSF1), sFasL (TNFSF6), TRAIL (TNFSF10); type I interferons (IFN), IFNα2, IFNβ; type II IFN, IFNγ; CC chemokines, CCL2 (MCP1), CCL3 (MIP1α), CCL4 (MIP1β), CCL5 (RANTES), CCL7 (MCP3), CCL11 (eotaxin); CXC chemokines, CXCL1 (GROα), CXCL8 (IL8), CXCL9 (MIG), CXCL10 (IP10), CXCL12a (SDF1); Other growth factors, βNGF, EGF, HGF, TGFα, TGFβ, FGFb; PDGF family/VEGF subfamily, PDGFBB, VEGFA, VEGFD; Serine protease inhibitor, PAI1 (serpin E1); Cell adhesion molecules, sICAM1 (CD54), VCAM1 (CD106); Neurotrophic/stimulating factors, BDNF, CSF1 (MCSF), CSF2 (GMCSF), CSF3 (GCSF), SCF; Adipose-derived hormones, leptin, resistin (customized Procarta immunoassay, Affymetrix/eBioscience, Santa Clara, CA, USA, Thermo Fisher Scientific).
Plasma samples from male and female ASD cases and ABC controls were coded, randomized and assayed in duplicate. Median fluorescence intensities (MFI) of each analyte-specific immunoassay bead set were detected by the flow- and fluorescence-based Luminex 200™ detection platform (Luminex Corporation, Austin, TX, USA).38 Data were processed using a quality control (QC) algorithm that calibrates performance of an expanded set of serial standard curves and in-house plasma controls included on every plate, and monitors intra- and inter-plate coefficient of variation (CV) and bead counts. Only plates with mean intra-assay %CV <15% were accepted. Samples failing to meet QC criteria were designated for re-run when feasible (CVs >25%, bead counts <30). MFI values exceeding machine limits of reliable detection (>25,000) were excluded. Because interpolated concentrations can introduce bias39–42 for samples with very low or high values in relation to the serial standard curves, we based our analyses on MFI rather than concentration estimates. Analyte concentrations in Luminex assays are derivatives of MFI values. As noted by Breen et al., low abundance analytes are frequently not in the linear range of the standard curve.39 Accordingly, accurate measurements of analyte concentration would require individualized, calibrated standard curves for each of the 60 analytes on every plate. Moreover, MFI has a lower inter-assay CV.40 Thus, analyses based on MFI are more reliable than analyses based on estimates of analyte concentrations. Averaged MFI values meeting quality control criteria for all 60 cytokines were used in the final statistical analyses. The final data set (test samples) had mean intra-assay %CV of 5.5% (SD 5.2%) and 0.73% of all possible intra-assay %CV values were >25%, across all cytokines. All per-cytokine intra-assay %CV values were similarly <10%, with the exception of CCL2, for which the mean intra-assay %CV was 10.5%. The medians and ranges of the MFI values of all 60 cytokines in MMG and CB are reported in Supplementary Table 2.
Statistical analysis.
Immune data processing: missing data, imputation, outliers, and transformations.
Missing data and imputation.
Five data points were lacking in MMG immunoassay data due to insufficient bead counts (<0.01% of 98,820 potential values derived from the MMG and CB immunoassays). These included one MMG boy lacking both CCL2 and IL22, one MMG boy lacking CCL2, and one girl lacking both CCL2 and IL22. No CB immunoassay values were missing. For logistic regression analyses, procedures were pursued using all 60 analytes, excluding subjects with these missing data; for the predictive modeling through machine learning, the missing data points were imputed using the mean value of the corresponding analyte.
Outliers and data transformations.
Outliers were identified through principal component analysis (PCA).43 After eliminating samples identified by PCA as outliers (visual inspection, MMG female n=5, MMG male n=10, CB female n=4, CB male n=10; for these samples, data from all 60 analytes were excluded from analyses), levels of each analyte were natural log-transformed and divided by the standard deviation of that analyte within the control group.
Data analyses.
We used logistic regression models separately for boys and girls, and within each sample type to test for an association between each analyte and ASD risk. Two models were explored for MMG samples: one unadjusted, and another adjusting for the questionnaire- and MBRN-derived covariate data reflecting maternal age, illnesses (fever, infection, inflammatory, autoimmune, allergic disorders), emotional distress scores (SCL-5) and use of non-NSAID antipyretic medications (e.g., acetaminophen) in pregnancy up until sample acquisition, as well as gestational age at MMG blood sample collection. Two models were applied for CB samples: one unadjusted, and another adjusting for the questionnaire-derived covariate data reflecting maternal age, maternal illnesses during pregnancy (fever, infection, inflammatory/autoimmune/allergic disorders), maternal emotional distress scores and use of non-NSAID antipyretic medications in pregnancy. We imputed missing values within the maternal variables including gestational age at MMG blood draw (female n=1, male n=2), and gestational age at birth, (female n=1, male n=2). Missing items for maternal emotional distress scores (SCL-5) were addressed by mean imputation (n=10 missing all SCL-5 items on the gestational week 17 questionnaire; n=6 subjects missing all SCL-5 items on the gestational week 30 questionnaire). The standard errors reduced by mean/mode imputations are negligible given the low prevalence of missing information. For MMG samples, we repeated the adjusted models in one sensitivity analysis restricting the study population to those whose mid-gestational sample collection times were within +/− 28-day window from the dates of return of the MoBa questionnaires. Three sensitivity analyses were explored for CB samples: one restricting the study population to those not born by Caesarean section, one restricting to those whose mothers did not experience preeclampsia or eclampsia during the pregnancy, and one restricting to those with gestational ages between weeks 37 and 41 of gestation.
Multiple comparisons over the 60-plex immunoassay panel were corrected using the Benjamini-Hochberg procedure,44 controlling the overall false discovery rate (FDR) at the level of 0.05. Odds ratios (ORs) and their associated 95% confidence intervals (CIs) were calculated. To explore the utility of the multiplex panel as a biomarker tool for ASD, we employed three machine learning algorithms: LASSO (least absolute shrinkage and selection operator),45 Random Forests (RF)46 and XGBoost.47 For LASSO, we fitted the 60 immune signature analytes, with and without their two-way interaction terms, as predictors in 2 separate models. Models were built and evaluated within each sample type and sex, separately. The models were first trained in the 80% randomly selected training set using 10-fold cross-validation, and the remaining 20% of the study population was used as the independent test set to validate model performance. We also applied the Bayesian Model Averaging method that combines the predictions of multiple models using weighted averages in which the weights are Bayesian posterior probabilities that the given model is the true model, conditional on the training data.48 The predictive performance of the 5 models (LASSO without interaction terms, Lasso with interaction terms, RF, XGBoost, and Model Average) in the test set was evaluated using Area under the Receiver Operating Characteristic curve (AUROC) values and Receiver Operating Characteristic (ROC) curves. Data analysis was implemented using MATLAB and Statistics Toolbox release 2013a (MathWorks, Inc., Natick, MA), R version 3.6.3 (RStudio, Inc., Boston, MA) and IBM SPSS Statistics for Windows, version 24.0. All p-values were 2-tailed.
RESULTS
Subject characteristics.
Table 1 summarizes the maternal and child characteristics for the MMG and CB study samples. In the MMG and CB analyses, mothers of ASD boys were younger than mothers of control boys (p=0.001 for both MMG and CB analyses). Birth year was differently proportioned between ASD boys and control boys in both MMG and CB analyses (p<0.001). Distribution of birth year was also different between ASD girls and control girls in MMG analysis (p=0.021) and showed a nonsignificant trend in CB (p=0.058). Gestational age at birth for ASD boys was more likely to be outside of the 37–41 gestational week window as compared with control boys (p=0.041 for MMG boys, p=0.005 for CB boys). These parameters did not differ for girls in either MMG or CB analyses.
ASD is associated with altered cytokine profiles.
We used unadjusted and adjusted logistic regression models to test for ASD association with levels of cytokines in MMG and CB samples (Table 2). We considered a cytokine to be significantly associated with risk of ASD if it satisfied the following criteria with respect to control samples: 1) adjusted odds ratio (aOR) >1.5 or <0.667 and 2) FDR adjusted p-value <0.05. An odds ratio of 1.5 (or the reciprocal ~0.667) is roughly equivalent to a Cohen’s effect size d=0.224,49 and Cohen’s d=0.2 was proposed as an indicator of a small effect size.50 Table 2A and 2B report the sex-stratified unadjusted odds ratio (OR) and aOR of each cytokine, together with their associated 95% CI, crude p-value and FDR adjusted p-value, in MMG and CB samples, respectively. We also rebuilt the adjusted logistic models with further adjustments for the year of birth and found no significant changes in the results (Supplementary Table 3).
Table 2.
Estimates from logistics regression models testing for ASD association with each analyte. We considered a cytokine to be significantly associated with risk of Autism Spectrum Disorder (ASD) if it satisfied adjusted odds ratio (aOR) >1.5 or <0.667 and false discovery rate (FDR) adjusted p-value <0.05. Green denotes levels positively associated with ASD. Blue denotes levels negatively associated with ASD. Maternal mid-gestation (MMG) n=854. Cord blood (CB) n=793.
| A | MMG: Boys (n=350 ASD, 371 controls) | ||||||||||
| Immune family | Immune molecule | Unadjusted model | Adjusted model 1 | ||||||||
| OR | 95% CI | p-value | Adj p 3 | aOR | 95% CI | p-value | Adj p 3 | ||||
| IL1 superfamily | IL1α | 1.31 | 1.13 | 1.54 | <0.001 | 0.001 | 1.34 | 1.14 | 1.56 | <0.001 | <0.001 |
| IL1β | 1.58 | 1.35 | 1.85 | <0.0001 | <0.0001 | 1.58 | 1.35 | 1.85 | <0.0001 | <0.0001 | |
| IL18 | 1.14 | 0.98 | 1.33 | 0.086 | 0.105 | 1.17 | 1.00 | 1.36 | 0.051 | 0.068 | |
| IL1RA | 2.35 | 1.93 | 2.86 | <0.0001 | <0.0001 | 2.33 | 1.91 | 2.84 | <0.0001 | <0.0001 | |
| IL2 family | IL2 | 1.84 | 1.56 | 2.16 | <0.0001 | <0.0001 | 1.83 | 1.55 | 2.15 | <0.0001 | <0.0001 |
| IL7 | 1.48 | 1.27 | 1.72 | <0.0001 | <0.0001 | 1.47 | 1.26 | 1.71 | <0.0001 | <0.0001 | |
| IL9 | 1.47 | 1.24 | 1.74 | <0.0001 | <0.0001 | 1.47 | 1.24 | 1.75 | <0.0001 | <0.0001 | |
| IL12 family | IL27 | 1.35 | 1.15 | 1.59 | <0.001 | <0.001 | 1.37 | 1.17 | 1.62 | <0.001 | <0.001 |
| TNF superfamily | TNFα | 2.58 | 2.14 | 3.10 | <0.0001 | <0.0001 | 2.63 | 2.18 | 3.18 | <0.0001 | <0.0001 |
| sFasL | 1.20 | 1.02 | 1.41 | 0.026 | 0.039 | 1.19 | 1.01 | 1.40 | 0.039 | 0.056 | |
| IL6 (gp130) cytokine family | IL6 | 0.96 | 0.82 | 1.12 | 0.585 | 0.627 | 0.96 | 0.83 | 1.13 | 0.644 | 0.678 |
| IL31 | 1.18 | 1.01 | 1.39 | 0.035 | 0.049 | 1.17 | 1.00 | 1.38 | 0.051 | 0.068 | |
| Th2 type | IL5 | 1.69 | 1.43 | 1.99 | 0.0000 | <0.0001 | 1.68 | 1.42 | 1.99 | <0.0001 | <0.0001 |
| IL13 | 1.76 | 1.47 | 2.09 | 0.0000 | <0.0001 | 1.74 | 1.46 | 2.08 | <0.0001 | <0.0001 | |
| IL10 | 1.27 | 1.09 | 1.48 | 0.002 | 0.004 | 1.27 | 1.09 | 1.49 | 0.003 | 0.005 | |
| CC chemokines | CCL5 (RANTES) | 1.76 | 1.49 | 2.06 | <0.0001 | <0.0001 | 1.77 | 1.50 | 2.09 | <0.0001 | <0.0001 |
| CCL3 (MIP1a) | 1.35 | 1.15 | 1.59 | <0.001 | <0.001 | 1.35 | 1.15 | 1.59 | <0.001 | <0.001 | |
| CCL4 (MIP1b) | 1.38 | 1.18 | 1.61 | <0.0001 | <0.001 | 1.39 | 1.19 | 1.63 | <0.0001 | 0.0001 | |
| CCL11 (eotaxin) | 1.24 | 1.06 | 1.44 | 0.008 | 0.012 | 1.30 | 1.11 | 1.53 | 0.001 | 0.002 | |
| CXC chemokines | CXCL8 (IL8) | 1.61 | 1.35 | 1.93 | <0.0001 | <0.0001 | 1.60 | 1.33 | 1.92 | <0.0001 | <0.0001 |
| CXCL9 (MIG) | 1.11 | 0.96 | 1.29 | 0.152 | 0.172 | 1.15 | 0.99 | 1.34 | 0.075 | 0.091 | |
| CXCL10 (IP10) | 1.63 | 1.38 | 1.93 | <0.0001 | <0.0001 | 1.68 | 1.42 | 1.99 | <0.0001 | <0.0001 | |
| CXCL12a (SDF1) | 1.30 | 1.11 | 1.51 | <0.001 | 0.002 | 1.33 | 1.14 | 1.55 | <0.001 | <0.001 | |
| Neurotrophic/stimulating factors | CSF3 (G-CSF) | 1.90 | 1.61 | 2.25 | <0.0001 | <0.0001 | 1.90 | 1.61 | 2.25 | <0.0001 | <0.0001 |
| SCF (KITLG) | 1.27 | 1.08 | 1.48 | 0.003 | 0.005 | 1.31 | 1.12 | 1.54 | <0.001 | 0.001 | |
| Serine protease inhibitors | Serpin E1 (PAI-1) | 2.41 | 2.01 | 2.90 | <0.0001 | <0.0001 | 2.44 | 2.03 | 2.93 | <0.0001 | <0.0001 |
| Cell adhesion molecules | VCAM1 (CD106) | 2.56 | 2.13 | 3.06 | <0.0001 | <0.0001 | 2.58 | 2.15 | 3.10 | <0.0001 | <0.0001 |
| sICAM1 (CD54) | 1.15 | 0.99 | 1.34 | 0.074 | 0.092 | 1.19 | 1.02 | 1.40 | 0.027 | 0.040 | |
| PDGF family/VEGF subfamily | VEGFD | 1.53 | 1.32 | 1.78 | <0.0001 | <0.0001 | 1.59 | 1.36 | 1.86 | <0.0001 | <0.0001 |
| VEGFA | 1.40 | 1.17 | 1.66 | <0.001 | <0.001 | 1.41 | 1.18 | 1.68 | <0.001 | <0.001 | |
| PDGFBB | 1.20 | 1.03 | 1.39 | 0.016 | 0.025 | 1.19 | 1.02 | 1.38 | 0.025 | 0.039 | |
| Other growth factors | EGF | 1.54 | 1.33 | 1.79 | <0.0001 | <0.0001 | 1.51 | 1.30 | 1.75 | <0.0001 | <0.0001 |
| FGFb | 1.37 | 1.16 | 1.61 | <0.001 | <0.001 | 1.36 | 1.15 | 1.60 | <0.001 | <0.001 | |
| βNGF | 1.22 | 1.02 | 1.46 | 0.034 | 0.048 | 1.21 | 1.01 | 1.45 | 0.042 | 0.059 | |
| TGFα | 1.12 | 0.96 | 1.31 | 0.149 | 0.172 | 1.15 | 0.98 | 1.35 | 0.083 | 0.099 | |
| TGFβ | 1.12 | 0.97 | 1.31 | 0.132 | 0.156 | 1.13 | 0.97 | 1.32 | 0.116 | 0.134 | |
| Neurotrophic/stimulating factors | BDNF | 1.11 | 0.96 | 1.29 | 0.174 | 0.193 | 1.09 | 0.93 | 1.26 | 0.292 | 0.325 |
| Adipose-derived hormones | Resistin | 1.16 | 1.00 | 1.35 | 0.050 | 0.066 | 1.19 | 1.02 | 1.38 | 0.028 | 0.041 |
| Type I IFN | IFNβ | 1.13 | 0.97 | 1.32 | 0.118 | 0.141 | 1.17 | 1.00 | 1.37 | 0.053 | 0.069 |
| IL2 family | IL4 | 0.58 | 0.49 | 0.68 | <0.0001 | <0.0001 | 0.59 | 0.50 | 0.70 | <0.0001 | <0.0001 |
| IL15 | 0.85 | 0.73 | 1.00 | 0.044 | 0.060 | 0.86 | 0.74 | 1.01 | 0.068 | 0.085 | |
| IL21 | 0.97 | 0.83 | 1.13 | 0.668 | 0.691 | 0.97 | 0.83 | 1.14 | 0.750 | 0.776 | |
| IL6 (gp130) cytokine family | LIF | 0.86 | 0.74 | 1.00 | 0.056 | 0.072 | 0.87 | 0.75 | 1.02 | 0.087 | 0.103 |
| IL12 family | IL12p40 | 0.54 | 0.46 | 0.63 | <0.0001 | <0.0001 | 0.54 | 0.46 | 0.64 | <0.0001 | <0.0001 |
| IL12p70 | 0.83 | 0.71 | 0.98 | 0.027 | 0.039 | 0.85 | 0.72 | 1.00 | 0.055 | 0.070 | |
| IL23 | 0.79 | 0.67 | 0.93 | 0.005 | 0.008 | 0.80 | 0.68 | 0.95 | 0.009 | 0.015 | |
| IL17 family | IL17A | 0.77 | 0.66 | 0.90 | 0.001 | 0.002 | 0.78 | 0.67 | 0.92 | 0.002 | 0.004 |
| IL17F | 1.00 | 0.85 | 1.16 | 0.964 | 0.964 | 1.02 | 0.87 | 1.19 | 0.805 | 0.805 | |
| IL22 | 0.43 | 0.35 | 0.52 | <0.0001 | <0.0001 | 0.43 | 0.35 | 0.53 | <0.0001 | <0.0001 | |
| Type I IFN | IFNα2 | 0.86 | 0.74 | 1.00 | 0.047 | 0.062 | 0.89 | 0.76 | 1.04 | 0.139 | 0.157 |
| Type II IFN | IFNγ | 0.47 | 0.39 | 0.56 | <0.0001 | <0.0001 | 0.48 | 0.40 | 0.57 | <0.0001 | <0.0001 |
| CC chemokines | CCL2 (MCP1) | 0.44 | 0.37 | 0.53 | <0.0001 | <0.0001 | 0.45 | 0.37 | 0.53 | <0.0001 | <0.0001 |
| CCL7 (MCP3) | 1.04 | 0.90 | 1.21 | 0.566 | 0.618 | 1.06 | 0.91 | 1.23 | 0.463 | 0.506 | |
| CXC chemokines | CXCL1 (GROa) | 0.76 | 0.64 | 0.90 | 0.001 | 0.003 | 0.75 | 0.63 | 0.89 | <0.001 | 0.002 |
| Neurotrophic/stimulating factors | CSF1 (M-CSF) | 0.42 | 0.35 | 0.50 | <0.0001 | <0.0001 | 0.42 | 0.36 | 0.51 | <0.0001 | <0.0001 |
| CSF2 (GM-CSF) | 0.51 | 0.43 | 0.60 | <0.0001 | <0.0001 | 0.51 | 0.43 | 0.60 | <0.0001 | <0.0001 | |
| Other growth factors | HGF | 0.63 | 0.53 | 0.74 | <0.0001 | <0.0001 | 0.64 | 0.54 | 0.76 | <0.0001 | <0.0001 |
| TNF superfamily | TNFβ (LTA) | 0.78 | 0.67 | 0.92 | 0.003 | 0.005 | 0.79 | 0.67 | 0.92 | 0.004 | 0.006 |
| TRAIL | 1.03 | 0.89 | 1.20 | 0.658 | 0.691 | 1.05 | 0.91 | 1.22 | 0.498 | 0.533 | |
| Adipose-derived hormones | Leptin | 1.02 | 0.87 | 1.19 | 0.805 | 0.819 | 1.02 | 0.87 | 1.20 | 0.792 | 0.805 |
| A | MMG: Girls (n=64 ASD, 69 controls) | ||||||||||
| Immune family | Immune molecule | Unadjusted model | Adjusted model 1 | ||||||||
| OR | 95% CI | p-value | Adj p 3 | aOR | 95% CI | p-value | Adj p 3 | ||||
| IL1 superfamily | IL1α | 2.21 | 1.48 | 3.31 | <0.001 | <0.001 | 2.21 | 1.46 | 3.33 | <0.001 | <0.001 |
| IL1β | 4.33 | 2.62 | 7.17 | <0.0001 | <0.0001 | 5.00 | 2.84 | 8.80 | <0.0001 | <0.0001 | |
| IL18 | 1.28 | 0.95 | 1.73 | 0.103 | 0.126 | 1.26 | 0.93 | 1.70 | 0.134 | 0.164 | |
| IL1RA | 5.48 | 3.14 | 9.58 | <0.0001 | <0.0001 | 6.14 | 3.37 | 11.18 | <0.0001 | <0.0001 | |
| IL2 family | IL2 | 4.37 | 2.68 | 7.14 | <0.0001 | <0.0001 | 4.70 | 2.78 | 7.93 | <0.0001 | <0.0001 |
| IL7 | 3.04 | 1.95 | 4.74 | <0.0001 | <0.0001 | 3.56 | 2.18 | 5.79 | <0.0001 | <0.0001 | |
| IL9 | 2.91 | 1.77 | 4.80 | <0.0001 | <0.0001 | 3.07 | 1.84 | 5.14 | <0.0001 | <0.0001 | |
| IL12 family | IL27 | 2.51 | 1.57 | 4.03 | <0.001 | <0.001 | 2.54 | 1.56 | 4.13 | <0.001 | <0.001 |
| TNF superfamily | TNFα | 7.45 | 3.78 | 14.68 | <0.0001 | <0.0001 | 8.42 | 4.03 | 17.59 | <0.0001 | <0.0001 |
| sFasL | 2.09 | 1.30 | 3.34 | 0.002 | 0.004 | 2.11 | 1.30 | 3.42 | 0.002 | 0.004 | |
| IL6 (gp130) cytokine family | IL6 | 1.15 | 0.82 | 1.61 | 0.417 | 0.455 | 1.11 | 0.79 | 1.57 | 0.547 | 0.597 |
| IL31 | 2.38 | 1.54 | 3.67 | <0.0001 | <0.001 | 2.43 | 1.56 | 3.80 | <0.0001 | <0.001 | |
| Th2 type | IL5 | 4.41 | 2.63 | 7.39 | <0.0001 | <0.0001 | 4.67 | 2.72 | 8.04 | <0.0001 | <0.0001 |
| IL13 | 4.34 | 2.61 | 7.22 | <0.0001 | <0.0001 | 4.79 | 2.77 | 8.30 | <0.0001 | <0.0001 | |
| IL10 | 2.19 | 1.42 | 3.40 | <0.001 | <0.001 | 2.22 | 1.41 | 3.48 | <0.001 | <0.001 | |
| CC chemokines | CCL5 (RANTES) | 2.47 | 1.67 | 3.63 | <0.0001 | <0.0001 | 2.64 | 1.76 | 3.95 | <0.0001 | <0.0001 |
| CCL3 (MIP1a) | 2.95 | 1.88 | 4.62 | <0.0001 | <0.0001 | 2.97 | 1.88 | 4.70 | <0.0001 | <0.0001 | |
| CCL4 (MIP1b) | 2.64 | 1.75 | 3.99 | <0.0001 | <0.0001 | 2.69 | 1.77 | 4.09 | <0.0001 | <0.0001 | |
| CCL11 (eotaxin) | 1.95 | 1.33 | 2.85 | <0.001 | 0.001 | 1.97 | 1.34 | 2.90 | <0.001 | <0.001 | |
| CXC chemokines | CXCL8 (IL8) | 3.37 | 2.03 | 5.57 | <0.0001 | <0.0001 | 3.65 | 2.15 | 6.21 | <0.0001 | <0.0001 |
| CXCL9 (MIG) | 1.54 | 1.05 | 2.27 | 0.026 | 0.036 | 1.56 | 1.05 | 2.32 | 0.029 | 0.039 | |
| CXCL10 (IP10) | 2.86 | 1.82 | 4.48 | <0.0001 | <0.0001 | 3.19 | 1.97 | 5.18 | <0.0001 | <0.0001 | |
| CXCL12a (SDF1) | 2.61 | 1.70 | 3.99 | <0.0001 | <0.0001 | 2.63 | 1.70 | 4.08 | <0.0001 | <0.0001 | |
| Neurotrophic/stimulating factors | CSF3 (G-CSF) | 6.51 | 3.40 | 12.45 | <0.0001 | <0.0001 | 7.82 | 3.76 | 16.25 | <0.0001 | <0.0001 |
| SCF (KITLG) | 3.33 | 2.00 | 5.52 | <0.0001 | <0.0001 | 3.46 | 2.03 | 5.89 | <0.0001 | <0.0001 | |
| Serine protease inhibitors | Serpin E1 (PAI-1) | 5.73 | 3.30 | 9.96 | <0.0001 | <0.0001 | 7.64 | 3.90 | 14.96 | <0.0001 | <0.0001 |
| Cell adhesion molecules | VCAM1 (CD106) | 5.37 | 3.15 | 9.16 | <0.0001 | <0.0001 | 5.84 | 3.30 | 10.35 | <0.0001 | <0.0001 |
| sICAM1 (CD54) | 1.50 | 0.97 | 2.32 | 0.067 | 0.084 | 1.65 | 1.03 | 2.64 | 0.038 | 0.049 | |
| PDGF family/VEGF subfamily | VEGFD | 2.35 | 1.64 | 3.37 | <0.0001 | <0.0001 | 2.53 | 1.72 | 3.72 | <0.0001 | <0.0001 |
| VEGFA | 3.62 | 2.21 | 5.93 | <0.0001 | <0.0001 | 3.87 | 2.30 | 6.50 | <0.0001 | <0.0001 | |
| PDGFBB | 1.87 | 1.27 | 2.76 | 0.002 | 0.003 | 2.10 | 1.38 | 3.22 | <0.001 | 0.001 | |
| Other growth factors | EGF | 2.67 | 1.87 | 3.82 | <0.0001 | <0.0001 | 3.10 | 2.07 | 4.65 | <0.0001 | <0.0001 |
| FGFb | 2.93 | 1.83 | 4.69 | <0.0001 | <0.0001 | 3.30 | 2.00 | 5.44 | <0.0001 | <0.0001 | |
| βNGF | 3.59 | 2.19 | 5.88 | <0.0001 | <0.0001 | 4.00 | 2.35 | 6.81 | <0.0001 | <0.0001 | |
| TGFα | 1.64 | 1.10 | 2.44 | 0.015 | 0.023 | 1.64 | 1.09 | 2.47 | 0.019 | 0.028 | |
| TGFβ | 1.90 | 1.25 | 2.90 | 0.003 | 0.005 | 1.95 | 1.26 | 3.01 | 0.003 | 0.004 | |
| Neurotrophic/stimulating factors | BDNF | 1.68 | 1.18 | 2.39 | 0.004 | 0.006 | 1.77 | 1.22 | 2.56 | 0.003 | 0.004 |
| Adipose-derived hormones | Resistin | 1.06 | 0.74 | 1.52 | 0.736 | 0.789 | 1.06 | 0.73 | 1.54 | 0.770 | 0.825 |
| Type I IFN | IFNβ | 1.58 | 1.05 | 2.36 | 0.028 | 0.037 | 1.57 | 1.03 | 2.39 | 0.034 | 0.045 |
| IL2 family | IL4 | 0.54 | 0.37 | 0.80 | 0.002 | 0.004 | 0.53 | 0.35 | 0.79 | 0.002 | 0.003 |
| IL15 | 1.25 | 0.86 | 1.82 | 0.234 | 0.265 | 1.23 | 0.84 | 1.80 | 0.294 | 0.340 | |
| IL21 | 1.48 | 1.06 | 2.06 | 0.022 | 0.031 | 1.47 | 1.04 | 2.06 | 0.027 | 0.038 | |
| IL6 (gp130) cytokine family | LIF | 1.30 | 0.88 | 1.92 | 0.187 | 0.220 | 1.27 | 0.85 | 1.89 | 0.245 | 0.288 |
| IL12 family | IL12p40 | 0.37 | 0.25 | 0.53 | <0.0001 | <0.0001 | 0.35 | 0.24 | 0.52 | <0.0001 | <0.0001 |
| IL12p70 | 1.21 | 0.81 | 1.81 | 0.343 | 0.382 | 1.18 | 0.78 | 1.77 | 0.441 | 0.490 | |
| IL23 | 0.97 | 0.65 | 1.43 | 0.860 | 0.905 | 0.98 | 0.65 | 1.46 | 0.909 | 0.940 | |
| IL17 family | IL17A | 1.01 | 0.70 | 1.47 | 0.946 | 0.979 | 0.98 | 0.67 | 1.44 | 0.936 | 0.952 |
| IL17F | 1.27 | 0.89 | 1.81 | 0.186 | 0.220 | 1.26 | 0.87 | 1.82 | 0.219 | 0.262 | |
| IL22 | 0.43 | 0.29 | 0.65 | <0.0001 | <0.0001 | 0.41 | 0.27 | 0.62 | <0.0001 | <0.0001 | |
| Type I IFN | IFNα2 | 0.68 | 0.48 | 0.96 | 0.029 | 0.038 | 0.66 | 0.46 | 0.94 | 0.023 | 0.033 |
| Type II IFN | IFNγ | 0.36 | 0.24 | 0.53 | <0.0001 | <0.0001 | 0.32 | 0.21 | 0.49 | <0.0001 | <0.0001 |
| CC chemokines | CCL2 (MCP1) | 0.22 | 0.13 | 0.38 | <0.0001 | <0.0001 | 0.22 | 0.13 | 0.37 | <0.0001 | <0.0001 |
| CCL7 (MCP3) | 1.61 | 1.07 | 2.43 | 0.022 | 0.031 | 1.59 | 1.04 | 2.41 | 0.031 | 0.041 | |
| CXC chemokines | CXCL1 (GROa) | 1.23 | 0.88 | 1.74 | 0.226 | 0.261 | 1.21 | 0.84 | 1.74 | 0.304 | 0.345 |
| Neurotrophic/stimulating factors | CSF1 (M-CSF) | 0.21 | 0.13 | 0.35 | <0.0001 | <0.0001 | 0.20 | 0.12 | 0.34 | <0.0001 | <0.0001 |
| CSF2 (GM-CSF) | 0.32 | 0.21 | 0.49 | <0.0001 | <0.0001 | 0.27 | 0.16 | 0.43 | <0.0001 | <0.0001 | |
| Other growth factors | HGF | 0.71 | 0.49 | 1.02 | 0.063 | 0.080 | 0.69 | 0.48 | 1.01 | 0.057 | 0.071 |
| TNF superfamily | TNFβ (LTA) | 1.00 | 0.68 | 1.47 | 0.997 | 0.997 | 1.00 | 0.67 | 1.49 | 0.992 | 0.992 |
| TRAIL | 1.50 | 1.08 | 2.10 | 0.016 | 0.024 | 1.49 | 1.06 | 2.09 | 0.020 | 0.030 | |
| Adipose-derived hormones | Leptin | 0.99 | 0.73 | 1.35 | 0.969 | 0.985 | 0.97 | 0.70 | 1.34 | 0.844 | 0.888 |
| B | CB: Boys (n=355 ASD, 333 controls) | ||||||||||
| Immune family | Immune molecule | Unadjusted model | Adjusted model 1 | ||||||||
| OR | 95% CI | p-value | Adj p 3 | aOR | 95% CI | p-value | Adj p 3 | ||||
| IL1 superfamily | IL1α | 1.06 | 0.92 | 1.22 | 0.440 | 0.528 | 1.05 | 0.90 | 1.21 | 0.543 | 0.651 |
| IL1β | 1.17 | 1.02 | 1.34 | 0.023 | 0.038 | 1.15 | 1.00 | 1.32 | 0.043 | 0.069 | |
| IL18 | 1.02 | 0.88 | 1.18 | 0.769 | 0.824 | 1.00 | 0.86 | 1.16 | 0.962 | 0.962 | |
| IL1RA | 1.37 | 1.17 | 1.60 | <0.0001 | <0.001 | 1.35 | 1.15 | 1.59 | <0.001 | <0.001 | |
| IL2 family | IL2 | 1.25 | 1.09 | 1.45 | 0.002 | 0.004 | 1.24 | 1.08 | 1.44 | 0.003 | 0.006 |
| IL7 | 1.70 | 1.45 | 1.99 | <0.0001 | <0.0001 | 1.68 | 1.43 | 1.97 | <0.0001 | <0.0001 | |
| IL9 | 1.12 | 0.96 | 1.31 | 0.165 | 0.230 | 1.09 | 0.93 | 1.28 | 0.276 | 0.376 | |
| IL12 family | IL27 | 0.95 | 0.82 | 1.11 | 0.525 | 0.617 | 0.96 | 0.82 | 1.12 | 0.593 | 0.679 |
| TNF superfamily | TNFα | 1.75 | 1.48 | 2.07 | <0.0001 | <0.0001 | 1.71 | 1.44 | 2.03 | <0.0001 | <0.0001 |
| sFasL | 0.96 | 0.83 | 1.11 | 0.579 | 0.668 | 0.96 | 0.83 | 1.11 | 0.600 | 0.679 | |
| IL6 (gp130) cytokine family | IL6 | 1.12 | 1.01 | 1.23 | 0.031 | 0.049 | 1.11 | 1.00 | 1.23 | 0.042 | 0.068 |
| IL31 | 0.90 | 0.77 | 1.05 | 0.174 | 0.237 | 0.88 | 0.76 | 1.03 | 0.123 | 0.184 | |
| Th2 type | IL5 | 1.38 | 1.18 | 1.62 | <0.0001 | <0.001 | 1.37 | 1.17 | 1.60 | <0.001 | <0.001 |
| IL13 | 1.24 | 1.07 | 1.43 | 0.004 | 0.006 | 1.23 | 1.07 | 1.43 | 0.005 | 0.009 | |
| IL10 | 0.86 | 0.75 | 1.00 | 0.046 | 0.070 | 0.86 | 0.75 | 1.00 | 0.048 | 0.074 | |
| CC chemokines | CCL5 (RANTES) | 1.41 | 1.21 | 1.64 | <0.0001 | <0.0001 | 1.41 | 1.21 | 1.64 | <0.0001 | <0.0001 |
| CCL3 (MIP1a) | 1.06 | 0.94 | 1.21 | 0.342 | 0.446 | 1.05 | 0.92 | 1.19 | 0.471 | 0.587 | |
| CCL4 (MIP1b) | 1.13 | 0.98 | 1.30 | 0.095 | 0.142 | 1.12 | 0.97 | 1.29 | 0.126 | 0.184 | |
| CCL11 (eotaxin) | 0.97 | 0.83 | 1.13 | 0.679 | 0.767 | 0.98 | 0.84 | 1.14 | 0.783 | 0.839 | |
| CXC chemokines | CXCL8 (IL8) | 1.09 | 0.94 | 1.26 | 0.274 | 0.365 | 1.06 | 0.91 | 1.23 | 0.458 | 0.585 |
| CXCL9 (MIG) | 0.75 | 0.64 | 0.88 | <0.001 | <0.001 | 0.75 | 0.64 | 0.88 | <0.001 | 0.001 | |
| CXCL10 (IP10) | 1.28 | 1.10 | 1.49 | 0.002 | 0.003 | 1.28 | 1.10 | 1.50 | 0.002 | 0.004 | |
| CXCL12a (SDF1) | 0.90 | 0.78 | 1.04 | 0.155 | 0.221 | 0.90 | 0.78 | 1.04 | 0.148 | 0.207 | |
| Neurotrophic/stimulating factors | CSF3 (G-CSF) | 1.19 | 1.04 | 1.37 | 0.012 | 0.019 | 1.17 | 1.02 | 1.34 | 0.028 | 0.046 |
| SCF (KITLG) | 0.75 | 0.65 | 0.88 | <0.001 | <0.001 | 0.77 | 0.66 | 0.90 | <0.001 | 0.002 | |
| Serine protease inhibitors | Serpin E1 (PAI-1) | 1.95 | 1.63 | 2.34 | <0.0001 | <0.0001 | 1.92 | 1.60 | 2.31 | <0.0001 | <0.0001 |
| Cell adhesion molecules | VCAM1 (CD106) | 1.80 | 1.53 | 2.12 | <0.0001 | <0.0001 | 1.78 | 1.51 | 2.10 | <0.0001 | <0.0001 |
| sICAM1 (CD54) | 0.97 | 0.85 | 1.12 | 0.691 | 0.767 | 0.98 | 0.85 | 1.12 | 0.735 | 0.817 | |
| PDGF family/VEGF subfamily | VEGFD | 1.37 | 1.18 | 1.58 | <0.0001 | <0.0001 | 1.38 | 1.19 | 1.59 | <0.0001 | <0.0001 |
| VEGFA | 1.07 | 0.92 | 1.24 | 0.3712 | 0.4640 | 1.06 | 0.91 | 1.23 | 0.480 | 0.587 | |
| PDGFBB | 1.13 | 0.98 | 1.31 | 0.0975 | 0.1427 | 1.12 | 0.96 | 1.30 | 0.144 | 0.205 | |
| Other growth factors | EGF | 1.75 | 1.49 | 2.04 | <0.0001 | <0.0001 | 1.71 | 1.46 | 2.00 | <0.0001 | <0.0001 |
| FGFb | 1.27 | 1.08 | 1.48 | 0.003 | 0.006 | 1.25 | 1.06 | 1.47 | 0.007 | 0.011 | |
| βNGF | 0.93 | 0.81 | 1.08 | 0.351 | 0.448 | 0.92 | 0.80 | 1.07 | 0.289 | 0.385 | |
| TGFα | 1.00 | 0.86 | 1.15 | 0.975 | 0.985 | 0.98 | 0.84 | 1.13 | 0.765 | 0.834 | |
| TGFβ | 0.78 | 0.67 | 0.91 | 0.002 | 0.004 | 0.78 | 0.67 | 0.92 | 0.002 | 0.005 | |
| Neurotrophic/stimulating factors | BDNF | 1.00 | 0.86 | 1.16 | 0.985 | 0.985 | 0.99 | 0.85 | 1.15 | 0.919 | 0.951 |
| Adipose-derived hormones | Resistin | 1.00 | 0.87 | 1.14 | 0.945 | 0.978 | 0.99 | 0.87 | 1.13 | 0.888 | 0.935 |
| Type I IFN | IFNβ | 0.74 | 0.63 | 0.87 | <0.001 | <0.001 | 0.75 | 0.63 | 0.89 | <0.001 | 0.002 |
| IL2 family | IL4 | 0.43 | 0.36 | 0.52 | <0.0001 | <0.0001 | 0.43 | 0.36 | 0.53 | <0.0001 | <0.0001 |
| IL15 | 0.63 | 0.53 | 0.75 | <0.0001 | <0.0001 | 0.63 | 0.53 | 0.76 | <0.0001 | <0.0001 | |
| IL21 | 0.63 | 0.53 | 0.75 | <0.0001 | <0.0001 | 0.63 | 0.53 | 0.75 | <0.0001 | <0.0001 | |
| IL6 (gp130) cytokine family | LIF | 0.80 | 0.69 | 0.93 | 0.004 | 0.007 | 0.81 | 0.70 | 0.94 | 0.004 | 0.008 |
| IL12 family | IL12p40 | 0.76 | 0.66 | 0.88 | <0.001 | <0.001 | 0.77 | 0.67 | 0.89 | <0.001 | 0.001 |
| IL12p70 | 0.52 | 0.43 | 0.62 | <0.0001 | <0.0001 | 0.52 | 0.43 | 0.62 | <0.0001 | <0.0001 | |
| IL23 | 0.94 | 0.81 | 1.08 | 0.383 | 0.469 | 0.93 | 0.80 | 1.08 | 0.331 | 0.432 | |
| IL17 family | IL17A | 0.54 | 0.45 | 0.64 | <0.0001 | <0.0001 | 0.54 | 0.45 | 0.64 | <0.0001 | <0.0001 |
| IL17F | 0.61 | 0.51 | 0.72 | <0.0001 | <0.0001 | 0.60 | 0.51 | 0.72 | <0.0001 | <0.0001 | |
| IL22 | 0.46 | 0.39 | 0.56 | <0.0001 | <0.0001 | 0.47 | 0.39 | 0.56 | <0.0001 | <0.0001 | |
| Type I IFN | IFNα2 | 0.78 | 0.67 | 0.92 | 0.003 | 0.005 | 0.79 | 0.67 | 0.93 | 0.004 | 0.007 |
| Type II IFN | IFNγ | 0.43 | 0.36 | 0.52 | <0.0001 | <0.0001 | 0.42 | 0.35 | 0.51 | <0.0001 | <0.0001 |
| CC chemokines | CCL2 (MCP1) | 0.79 | 0.68 | 0.92 | 0.002 | 0.004 | 0.79 | 0.68 | 0.91 | 0.002 | 0.003 |
| CCL7 (MCP3) | 0.67 | 0.57 | 0.79 | <0.0001 | <0.0001 | 0.67 | 0.57 | 0.80 | <0.0001 | <0.0001 | |
| CXC chemokines | CXCL1 (GROa) | 1.01 | 0.88 | 1.16 | 0.872 | 0.918 | 1.00 | 0.87 | 1.14 | 0.949 | 0.962 |
| Neurotrophic/stimulating factors | CSF1 (M-CSF) | 0.39 | 0.32 | 0.47 | <0.0001 | <0.0001 | 0.39 | 0.33 | 0.47 | <0.0001 | <0.0001 |
| CSF2 (GM-CSF) | 0.50 | 0.42 | 0.60 | <0.0001 | <0.0001 | 0.50 | 0.42 | 0.60 | <0.0001 | <0.0001 | |
| Other growth factors | HGF | 0.97 | 0.85 | 1.12 | 0.714 | 0.779 | 0.96 | 0.83 | 1.11 | 0.593 | 0.679 |
| TNF superfamily | TNFβ (LTA) | 0.56 | 0.47 | 0.67 | <0.0001 | <0.0001 | 0.57 | 0.47 | 0.68 | <0.0001 | <0.0001 |
| TRAIL | 0.75 | 0.64 | 0.87 | <0.001 | <0.001 | 0.75 | 0.64 | 0.88 | <0.001 | <0.001 | |
| Adipose-derived hormones | Leptin | 0.74 | 0.64 | 0.86 | <0.0001 | <0.001 | 0.73 | 0.63 | 0.85 | <0.0001 | <0.001 |
| B | CB: Girls (n=63 ASD, 62 controls) | ||||||||||
| Immune family | Immune molecule | Unadjusted model | Adjusted model 1 | ||||||||
| OR | 95% CI | p-value | Adj p 3 | aOR | 95% CI | p-value | Adj p 3 | ||||
| IL1 superfamily | IL1α | 1.15 | 0.78 | 1.71 | 0.480 | 0.610 | 1.14 | 0.75 | 1.72 | 0.541 | 0.662 |
| IL1β | 1.06 | 0.71 | 1.59 | 0.784 | 0.871 | 1.02 | 0.67 | 1.56 | 0.917 | 0.963 | |
| IL18 | 1.52 | 1.03 | 2.23 | 0.033 | 0.071 | 1.53 | 1.02 | 2.29 | 0.039 | 0.077 | |
| IL1RA | 1.90 | 1.26 | 2.86 | 0.002 | 0.005 | 1.79 | 1.17 | 2.74 | 0.008 | 0.019 | |
| IL2 family | IL2 | 1.79 | 1.15 | 2.78 | 0.010 | 0.025 | 1.78 | 1.13 | 2.79 | 0.012 | 0.029 |
| IL7 | 2.96 | 1.92 | 4.56 | <0.0001 | <0.0001 | 3.13 | 1.95 | 5.04 | <0.0001 | <0.0001 | |
| IL9 | 1.07 | 0.69 | 1.66 | 0.770 | 0.871 | 1.07 | 0.67 | 1.70 | 0.778 | 0.850 | |
| IL12 family | IL27 | 1.42 | 0.96 | 2.12 | 0.083 | 0.155 | 1.38 | 0.91 | 2.08 | 0.127 | 0.225 |
| TNF superfamily | TNFα | 2.94 | 1.87 | 4.64 | <0.0001 | <0.0001 | 3.24 | 1.95 | 5.36 | <0.0001 | <0.0001 |
| sFasL | 1.20 | 0.83 | 1.73 | 0.337 | 0.486 | 1.21 | 0.82 | 1.79 | 0.325 | 0.476 | |
| IL6 (gp130) cytokine family | IL6 | 0.97 | 0.68 | 1.39 | 0.886 | 0.930 | 1.01 | 0.70 | 1.46 | 0.940 | 0.963 |
| IL31 | 0.88 | 0.62 | 1.26 | 0.488 | 0.610 | 0.86 | 0.59 | 1.24 | 0.416 | 0.542 | |
| Th2 type | IL5 | 2.20 | 1.41 | 3.45 | <0.001 | 0.002 | 2.26 | 1.39 | 3.67 | 0.001 | 0.003 |
| IL13 | 2.02 | 1.41 | 2.89 | <0.001 | <0.001 | 2.28 | 1.54 | 3.39 | <0.0001 | <0.001 | |
| IL10 | 0.84 | 0.57 | 1.23 | 0.375 | 0.510 | 0.86 | 0.58 | 1.28 | 0.458 | 0.572 | |
| CC chemokines | CCL5 (RANTES) | 2.55 | 1.74 | 3.74 | <0.0001 | <0.0001 | 2.91 | 1.87 | 4.52 | <0.0001 | <0.0001 |
| CCL3 (MIP1a) | 0.94 | 0.66 | 1.33 | 0.712 | 0.838 | 0.92 | 0.63 | 1.34 | 0.650 | 0.780 | |
| CCL4 (MIP1b) | 1.28 | 0.85 | 1.90 | 0.236 | 0.372 | 1.25 | 0.83 | 1.90 | 0.290 | 0.447 | |
| CCL11 (eotaxin) | 1.02 | 0.71 | 1.47 | 0.915 | 0.931 | 0.95 | 0.64 | 1.39 | 0.779 | 0.850 | |
| CXC chemokines | CXCL8 (IL8) | 1.19 | 0.80 | 1.76 | 0.382 | 0.510 | 1.20 | 0.79 | 1.81 | 0.396 | 0.527 |
| CXCL9 (MIG) | 0.74 | 0.51 | 1.07 | 0.110 | 0.194 | 0.72 | 0.49 | 1.05 | 0.090 | 0.164 | |
| CXCL10 (IP10) | 2.07 | 1.40 | 3.07 | <0.001 | <0.001 | 2.10 | 1.38 | 3.20 | <0.001 | 0.002 | |
| CXCL12a (SDF1) | 0.87 | 0.62 | 1.22 | 0.423 | 0.552 | 0.85 | 0.60 | 1.21 | 0.367 | 0.502 | |
| Neurotrophic/stimulating factors | CSF3 (G-CSF) | 1.34 | 0.94 | 1.90 | 0.108 | 0.194 | 1.33 | 0.91 | 1.95 | 0.144 | 0.246 |
| SCF (KITLG) | 0.84 | 0.59 | 1.20 | 0.340 | 0.486 | 0.81 | 0.57 | 1.17 | 0.267 | 0.422 | |
| Serine protease inhibitors | Serpin E1 (PAI-1) | 3.04 | 1.91 | 4.83 | <0.0001 | <0.0001 | 3.40 | 2.05 | 5.65 | <0.0001 | <0.0001 |
| Cell adhesion molecules | VCAM1 (CD106) | 3.76 | 2.44 | 5.81 | <0.0001 | <0.0001 | 4.12 | 2.55 | 6.67 | <0.0001 | <0.0001 |
| sICAM1 (CD54) | 1.22 | 0.85 | 1.77 | 0.283 | 0.424 | 1.23 | 0.83 | 1.82 | 0.298 | 0.447 | |
| PDGF family/VEGF subfamily | VEGFD | 1.81 | 1.36 | 2.40 | <0.0001 | <0.001 | 2.02 | 1.45 | 2.82 | <0.0001 | <0.001 |
| VEGFA | 1.04 | 0.72 | 1.49 | 0.854 | 0.915 | 1.01 | 0.69 | 1.48 | 0.970 | 0.970 | |
| PDGFBB | 1.26 | 0.85 | 1.85 | 0.248 | 0.381 | 1.17 | 0.78 | 1.76 | 0.447 | 0.571 | |
| Other growth factors | EGF | 1.91 | 1.35 | 2.70 | <0.001 | <0.001 | 1.91 | 1.33 | 2.75 | <0.001 | 0.001 |
| FGFb | 1.59 | 1.03 | 2.47 | 0.037 | 0.077 | 1.85 | 1.13 | 3.02 | 0.014 | 0.033 | |
| βNGF | 1.02 | 0.71 | 1.45 | 0.935 | 0.935 | 1.01 | 0.69 | 1.49 | 0.947 | 0.963 | |
| TGFα | 0.96 | 0.66 | 1.39 | 0.8230 | 0.8978 | 0.93 | 0.63 | 1.36 | 0.706 | 0.830 | |
| TGFβ | 0.62 | 0.41 | 0.93 | 0.022 | 0.049 | 0.58 | 0.38 | 0.91 | 0.017 | 0.037 | |
| Neurotrophic/stimulating factors | BDNF | 0.98 | 0.69 | 1.38 | 0.899 | 0.930 | 0.94 | 0.65 | 1.36 | 0.752 | 0.850 |
| Adipose-derived hormones | Resistin | 1.06 | 0.74 | 1.51 | 0.743 | 0.858 | 1.04 | 0.72 | 1.51 | 0.825 | 0.884 |
| Type I IFN | IFNβ | 0.77 | 0.53 | 1.12 | 0.175 | 0.283 | 0.77 | 0.52 | 1.15 | 0.197 | 0.319 |
| IL2 family | IL4 | 0.28 | 0.18 | 0.44 | <0.0001 | <0.0001 | 0.26 | 0.16 | 0.42 | <0.0001 | <0.0001 |
| IL15 | 0.50 | 0.32 | 0.77 | 0.002 | 0.004 | 0.47 | 0.29 | 0.75 | 0.002 | 0.004 | |
| IL21 | 0.42 | 0.27 | 0.66 | <0.001 | <0.001 | 0.41 | 0.26 | 0.65 | <0.001 | <0.001 | |
| IL6 (gp130) cytokine family | LIF | 0.67 | 0.48 | 0.94 | 0.020 | 0.046 | 0.66 | 0.46 | 0.93 | 0.019 | 0.041 |
| IL12 family | IL12p40 | 0.71 | 0.50 | 0.99 | 0.043 | 0.085 | 0.67 | 0.47 | 0.95 | 0.024 | 0.050 |
| IL12p70 | 0.43 | 0.28 | 0.65 | <0.0001 | <0.001 | 0.39 | 0.24 | 0.62 | 0.0001 | <0.001 | |
| IL23 | 1.08 | 0.80 | 1.47 | 0.614 | 0.736 | 1.06 | 0.77 | 1.46 | 0.736 | 0.849 | |
| IL17 family | IL17A | 0.38 | 0.25 | 0.57 | <0.0001 | <0.0001 | 0.35 | 0.22 | 0.54 | <0.0001 | <0.0001 |
| IL17F | 0.36 | 0.22 | 0.58 | <0.0001 | <0.001 | 0.32 | 0.18 | 0.54 | <0.0001 | <0.001 | |
| IL22 | 0.28 | 0.18 | 0.45 | <0.0001 | <0.0001 | 0.26 | 0.15 | 0.43 | <0.0001 | <0.0001 | |
| Type I IFN | IFNα2 | 0.76 | 0.52 | 1.12 | 0.163 | 0.272 | 0.76 | 0.51 | 1.14 | 0.183 | 0.305 |
| Type II IFN | IFNγ | 0.35 | 0.22 | 0.54 | <0.0001 | <0.0001 | 0.31 | 0.19 | 0.50 | <0.0001 | <0.0001 |
| CC chemokines | CCL2 (MCP1) | 0.76 | 0.56 | 1.03 | 0.081 | 0.155 | 0.74 | 0.54 | 1.01 | 0.060 | 0.116 |
| CCL7 (MCP3) | 0.52 | 0.36 | 0.77 | <0.001 | 0.003 | 0.50 | 0.33 | 0.75 | <0.001 | 0.002 | |
| CXC chemokines | CXCL1 (GROa) | 0.72 | 0.49 | 1.08 | 0.115 | 0.198 | 0.67 | 0.44 | 1.02 | 0.064 | 0.120 |
| Neurotrophic/stimulating factors | CSF1 (M-CSF) | 0.30 | 0.21 | 0.44 | <0.0001 | <0.0001 | 0.26 | 0.16 | 0.41 | <0.0001 | <0.0001 |
| CSF2 (GM-CSF) | 0.38 | 0.24 | 0.61 | <0.0001 | <0.001 | 0.36 | 0.22 | 0.59 | <0.0001 | <0.001 | |
| Other growth factors | HGF | 1.14 | 0.78 | 1.66 | 0.502 | 0.615 | 1.20 | 0.81 | 1.80 | 0.366 | 0.502 |
| TNF superfamily | TNFβ (LTA) | 0.35 | 0.23 | 0.54 | <0.0001 | <0.0001 | 0.34 | 0.21 | 0.53 | <0.0001 | <0.0001 |
| TRAIL | 0.61 | 0.42 | 0.89 | 0.010 | 0.025 | 0.57 | 0.38 | 0.86 | 0.007 | 0.018 | |
| Adipose-derived hormones | Leptin | 0.84 | 0.58 | 1.22 | 0.359 | 0.501 | 0.84 | 0.56 | 1.24 | 0.368 | 0.502 |
Adjusted for maternal age, gestational age at MMG blood sample collection, illnesses (fever, infection, inflammatory, autoimmune, allergic disorders), emotional distress scores (SCL5) and use of non-NSAID antipyretic medications (e.g., acetaminophen) in pregnancy up until sample acquisition.
Adjusted for maternal age, maternal illnesses during pregnancy (fever, infection, inflammatory/autoimmune/allergic disorders), maternal emotional distress scores and use of non-NSAID antipyretic medications in pregnancy.
Adj p is the FDR adjusted p-value using Benjamini-Hochberg procedure controlling the FDR at 0.05 level. aOR adjusted odds ratio, CI confidence interval.
In the MMG dataset, in comparison with male controls, male ASD subjects had significantly higher levels of interleukins IL1β, IL1RA, IL2, IL5, and IL13; TNFα; CCL5; CXC chemokines CXCL8 and CXCL10; and vascular, growth and stimulating factors Serpin E1, VCAM1, VEGFD, EGF, and CSF3 (adjusted p<0.0001). Levels of interleukins IL4, IL12p40 and IL22; IFNγ; CCL2; and growth and stimulating factors HGF, CSF1 and CSF2 were significantly reduced (adjusted p<0.0001). Adjusted odds ratios ranged from 1.51 (EGF) to 2.63 (TNFα) with a mean of 1.92. The mean aOR was calculated as the natural exponential of the average absolute log-odds associated with the significant analytes (aOR >1.5 or <0.667, and FDR adjusted p-value <0.05). Compared with female controls, female ASD subjects had significantly higher levels of interleukins IL1α, IL1β, IL1RA, IL2, IL5, IL7, IL9, IL10, IL13, IL27 and IL31; TNF superfamily factors TNFα and sFasL; IFNβ; CC chemokines CCL3, CCL4, CCL5, CCL7 and CCL11; CXC chemokines CXCL8, CXCL9, CXCL10 and CXCL12a; and vascular, growth and stimulating factors Serpin E1, VCAM1, sICAM1, VEGFD, VEGFA, PDGFBB, EGF, FGFb, βNGF, TGFα, TGFβ, BDNF, CSF3 and SCF (adjusted p<0.0001 to p=0.049). Levels of IL4, IL12p40, IL22, IFNα2, IFNγ, CCL2, CSF1 and CSF2 were reduced (adjusted p<0.0001 to p=0.033). The mean effect size for a significant case-control analyte difference was aOR=3.03 in girls.
In the CB dataset, in comparison with male controls, male ASD subjects had significantly higher levels of IL7, TNFα, Serpin E1, VCAM1 and EGF (adjusted p<0.0001). Levels of interleukins IL4, IL5, IL12p70, IL17A, IL17F, IL21 and IL22; IFNγ, TNFβ, CSF1 and CSF2 were reduced (adjusted p<0.0001). Compared with female controls, female ASD subjects had significantly higher levels of interleukins IL1RA, IL2, IL5, IL7 and IL13; TNFα; CCL5; CXCL10; and vascular and other growth factors Serpin E1, VCAM1, VEGFD, EGF and FGFb (adjusted p<0.0001 to p=0.033). Levels of interleukins IL4, IL12p40, IL12p70, IL15, IL17A, IL17F, IL21, IL22 and LIF; TNF superfamily factors TNFβ and TRAIL; CCL7; IFNγ; and growth and stimulating factors TGFβ, CSF1 and CSF2 were reduced (adjusted p<0.0001 to p=0.050). As in the MMG dataset, despite having a lower sample size, female subjects had larger effect sizes (mean aOR=2.47) than male subjects (mean aOR=1.89).
Sensitivity Analyses.
We then restricted analysis in the MMG datasets to subjects who had mid-gestational sample collections within +/− 28-day window from the dates of return of the MoBa questionnaires. Three sensitivity analyses were explored for CB samples: one restricting the study population to subjects not born by Caesarean section, one restricted to subjects whose mothers did not experience preeclampsia or eclampsia during the pregnancy, and one restricted to subjects with gestational ages between week 37 and 41 of gestation. Each of these sensitivity analyses revealed similar estimations as in the main analysis (Supplementary Table 4).
Assessment of the 60-plex immune signature panel as a biomarker for ASD.
We developed five predictive models using an 80% randomly selected training set of subject cytokine profiles: Lasso without interaction terms, Lasso with interaction terms, RF, XGBoost, and Model Average. We tested model performance in the remaining 20% of subject cytokine profiles. The test sets included 70 male cases and 70 male controls, and 12 female cases and 12 female controls.
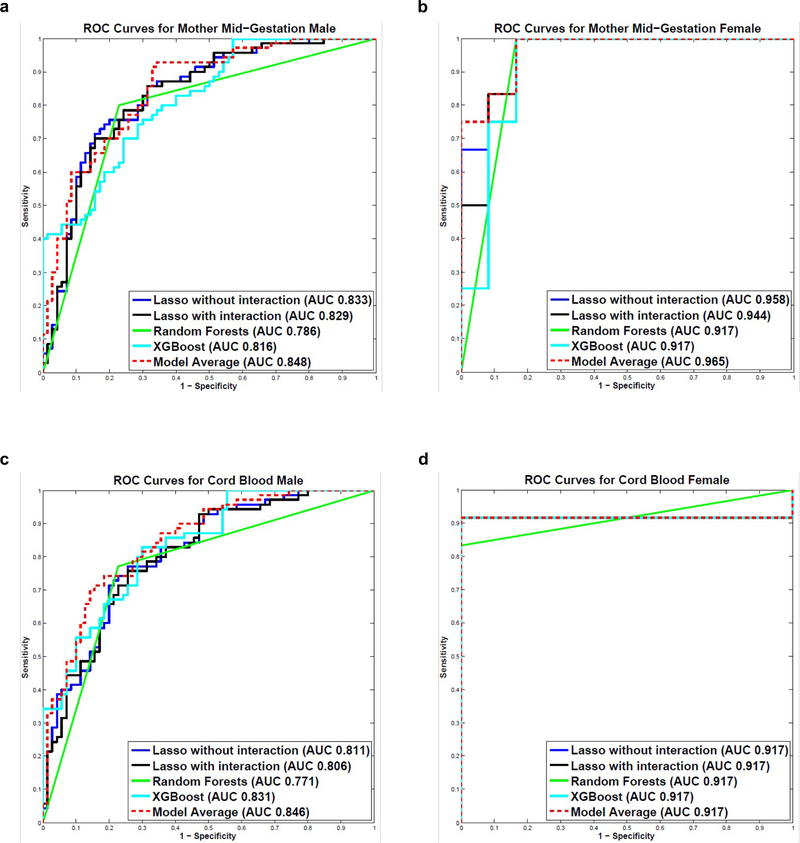
For male subjects in MMG dataset, the Model Average was the best performing classifier and significantly out-performed RF (p=0.002). All other pairwise comparisons between models were not significant. Lasso without interaction terms distinguished ASD cases from controls with an AUROC value of 0.833 (95% CI, 0.753, 0.891); Lasso with interaction terms produced an AUROC value of 0.829 (95% CI, 0.749, 0.888); RF and XGBoost yielded AUROC values of 0.786 (95% CI, 0.663, 0.855) and 0.816 (95% CI, 0.739, 0.874), respectively; Model Average separated cases from controls with an AUROC value of 0.848 (95% CI, 0.774, 0.901) (Figure 2A). The confusion matrix of the best performing classifier (Model Average) is shown in Supplementary Table 5. When the Model Average was focused on the subjects with predictive probability greater than 0.85 (n=28), the true positive rate was 89.29%; those with predictive probability greater than 0.90 (n=2) were all true positives. We also measured the importance of each cytokine in the predictive models using Bootstrapping with 1000 iterations in the training set.51 IL1RA, TNFα, CCL2, CXCL1 and Serpin E1 were ranked in top 10 in Lasso without interactions, RF and XGBoost (Supplementary Table 6).
Figure 2.
Autism Spectrum Disorders (ASD) predictive modeling. a ROC Curves for Mother Mid-Gestation Male. b ROC Curves for Mother Mid-Gestation Female. c ROC Curves for Cord Blood Male. d ROC Curves for Cord Blood Female. Five models were built to predict ASD outcome using the 60-plex immunoassay: Lasso without interaction terms, Lasso with interaction terms, Random Forests, XGBoost and Bayesian Model Averaging (Model Average). Models were built and evaluated within each sample type and sex, separately. The models were first trained in the 80% randomly-selected training set using 10-fold cross-validation, and the remaining 20% of the study population was used as the independent test set to validate model performance. The predictive performance of the 5 models in the test set was evaluated using Area under the Receiver Operating Characteristic curve (AUROC) values and Receiver Operating Characteristic (ROC) curves.
For female subjects in MMG dataset, all 5 models distinguished ASD cases from controls with AUROC values greater than 0.9. Lasso without and with interaction terms yielded AUC values of 0.958 (95% CI, 0.801, 0.992) and 0.944 (95% CI, 0.749, 0.990), respectively; both RF and XGBoost produced an AUROC value of 0.917 (RF 95% CI, 0.532, 0.986; XGBoost 95% CI, 0.666, 0.984); Model Average separated cases from controls with an AUROC value of 0.965 (95% CI, 0.823, 0.994) (Figure 2B). None of the pairwise comparisons between models were significant. The confusion matrix of the best performing classifier (Model Average) is shown in Supplementary Table 5. When we focused on the subjects with predictive probability greater than 0.80 (n=10), the true positive rate reached 100%. IL1RA, TNFα, CCL2, CSF1, and Serpin E1 were ranked in top 10 in all the models in which we measured feature importance using 1 000 iterations of Bootstrapping in the training set (Supplementary Table 6).
For male subjects in CB dataset, Lasso without interaction terms distinguished ASD cases from controls with an AUROC value of 0.811 (95% CI, 0.730, 0.872); Lasso with interaction terms produced an AUROC value of 0.806 (95% CI, 0.724, 0.868); RF and XGBoost yielded AUROC values of 0.771 (95% CI, 0.669, 0.862) and 0.831 (95% CI, 0.756, 0.887), respectively; Model Average separated cases from controls with an AUROC value of 0.846 (95% CI, 0.771, 0.899). The ROC curves of the 5 models were shown in Figure 2C. The Model Average was the best performing classifier and significantly out-performed Lasso with interaction terms (p=0.028) and RF (p<0.001). The confusion matrix of the best performing classifier (Model Average) is shown in Supplementary Table 5. When the Model Average was focused on the subjects with predictive probability greater than 0.85 (n=16), the true positive rate was 93.75%; those with predictive probability greater than 0.90 (n=4) were all true positives. IL4, CSF1 and Serpin E1 were ranked top 10 in Lasso without interactions, RF and XGBoost (Supplementary Table 6).
For female subjects in CB dataset, all 5 models distinguished ASD cases from controls with an AUROC value of 0.917. For RF, the 95% CI was (0.532, 0.986). For the other 4 models, the 95% CI was (0.565, 0.989). The ROC curves for the 5 models are shown in Figure 2D. When we focused on the subjects with predictive probability greater than 0.80 (n=10), the true positive rate reached 100%. IL4, TNFα, VEGFD, CSF1, VCAM1 and IL22 were ranked in top 10 in all the models wherein we measured feature importance (Supplementary Table 6).
DISCUSSION
We and others have shown that maternal fever and infection during pregnancy are associated with an increased risk of autism spectrum disorder (ASD).12,52,53 Research in animal models has also shown that gestational exposure to IL-6 or IL-17 or triggers of innate immunity such as double stranded nucleic acid and lipopolysaccharide cause neurodevelopmental damage reminiscent of ASD. To explore potential links between ASD and maternal immune activation during gestation, we leveraged the resources of the ABC, a nationwide birth cohort in Norway that prospectively collected questionnaire data and plasma samples. We identified subjects with ASD through the national patient registry and used MMG plasma and CB to ask three questions: (1) was systemic maternal inflammation during gestation associated with increased risk of ASD, (2) do cytokine profiles in maternal MMG plasma or CB differ by sex in ASD, and (3) could we identify biomarkers in CB that might be used to guide early identification of children at risk for ASD.
Cytokine profiles of MMG plasma, collected at 17–21 weeks gestation from mothers of both boys and girls who received a diagnosis of ASD, were consistent with systemic inflammation. In both boys and girls, we found elevations in a wide range of molecules associated with inflammation including IL1β, IL1RA, IL2, TNFα, and CCL5 (RANTES). We also found sex-specific effects despite the difference in sample sizes between boys and girls. The repertoire of elevated proinflammatory cytokines, chemokines, and adhesion molecules was larger in girls than in boys (37 versus 14). Furthermore, the effect sizes were also larger in girls than in boys (mean aOR 3.03 versus 1.92). Differences between cases and controls were less pronounced in CB with respect to the number of molecules that were elevated and the effect sizes. Levels of only five analytes were elevated in CB of ASD boys: IL7, TNFα, SerpinE1, VCAM1 and EGF. Four of the five analytes elevated in CB (TNFα, SerpinE1, VCAM1 and EGF) were also elevated in MMG plasma. Levels of thirteen analytes were elevated in girls: all five that were elevated in CB of ASD boys plus IL1RA, IL2, IL5, IL13, CCL5 (RANTES), EGF and FGFb. As in MMG analyses, effect sizes in CB were larger in girls than in boys (mean aOR 2.47 versus 1.89). Levels of IL-4, IL22, IFNγ, and growth factors CSF-1 (M-CSF) and CSF-2 (GM-CSF) were reduced in MMG and CB plasma in both ASD boys and ASD girls. A Venn diagram that indicates significant case/control alterations in cytokines present in MMG and CB samples from male and female ASD subjects is shown in Supplementary Figure 1.
Two of the proinflammatory cytokines with the largest effect sizes in MMG plasma of ASD boys and girls were IL1RA and TNFα. IL1RA is a member of the large, tightly regulated IL1 cytokine family comprised of cytokines, cytokine receptors, and modulating factors. It has potent antagonist effects with respect to IL1 signaling, achieved at least in part through the strength of its binding to IL1R1, which has affinity exceeding that of IL1.54 Although we did not find significantly elevated levels of IL-6 in MMG or CB plasma of ASD boys or girls, IL1RA may serve as a marker for elevation at an earlier time point. Infusion of IL-6 into normal human volunteers, the same molecule implicated by Patterson and colleagues in mediating maternal immune activation in the placenta and brain in rodent models of ASD, resulted in elevated levels of IL1RA. TNFα is expressed by activated monocytes/macrophages, NK and T cells, as well as endothelial cells and fibroblasts. It has myriad functions including caspase-dependent apoptotic cell death, and activation of NFκB, which in turn promotes expression of pro-inflammatory cytokines.55,56 TNFα is elevated in serum and cerebrospinal fluid of children with ASD57,58 and in neonatal blood spots of children subsequently diagnosed with ASD.59
The observation that proinflammatory cytokine expression is more pronounced in MMG than CB plasma is consistent with earlier work in the ABC wherein we found increased ASD risk with maternal exposure to fever in the first or second versus the third trimester,12 and to the presence of high titers of antibodies to herpes simplex virus type II.60 Work with this same cohort showed a trend toward increased risk in women with documented influenza infection and a prospectively collected report of influenza like illness.61 We cite these studies not to implicate a specific infection in the pathogenesis of ASD but rather to point to a period of vulnerability during gestation when exuberant immune responses may interfere with central nervous system development.
Cytokines are important as growth factors as well as mediators of inflammation. Neuropoietic cytokines, including IL6 and TGFβ (elevated in MMG ASD girls but not in MMG boys), orchestrate fate switching and differentiation of neurons, astrocytes and oligodendrocytes.62 CSF1, reduced in MMG and CB of ASD boys and girls, has been implicated in microglial and neuronal development and function.63,64 CSF1-depleted mice have reduced numbers of Purkinje cells, disordered cerebellar architecture, and deficits in both motor function and social memory.65
The use of machine learning predictive models allowed us to distinguish ASD cases from controls with high accuracies in the randomly selected test sets in both sample types (MMG and CB) and in both sexes (boys and girls). The AUROC values for MMG boys ranged from 0.786 to 0.848; for MMG girls, the AUROC values ranged from 0.917 to 0.965; the AUROC values for CB boys ranged from 0.771 to 0.846; all 5 models yielded an AUROC value of 0.917 for CB girls. The Model Average was the best performing model. When focused on subjects with predictive probabilities above 0.9, the true positive rates reached 100%. To our knowledge, there are no reported examples of comparably robust predictions where independent test sets were used for model evaluations. TNFα, Serpin E1 and CSF1 were constantly top ranked. Activation of Danger-Associated Molecular Pattern (DAMP) and Toll-like receptor (TLR) pathways could lead to increased expression of TNFα, Serpin E1, VCAM1, and IL1β. 9, 66–69 Dysregulation of these molecules may have implications for angiogenesis during brain development and have been invoked in ASD pathogenesis. 23, 69, 70
In summary, our results provide robust evidence of immune dysregulation in mothers as early as 17–21 weeks gestation and in CB of neonates later diagnosed with ASD. Future work focused on identification of genetic factors in and environmental triggers of immune activation, has the potential to lead to strategies for mitigating ASD risk.
Strengths and limitations
Among the strengths of this study are its sourcing of eligible cases and controls from a large population-based study; inclusion of multiple methods for case ascertainment and validation of ASD diagnoses; ongoing diagnostic follow up through child age 6–16 years; use of plasma samples derived from venous CB collected, handled, processed and stored under quality-controlled study procedures; methods for optimization and quality control of laboratory assays and their data; and cautious statistical approaches, including adjustment for multiple comparisons (conceptualizing the risk of Type I errors) and the use of three different machine learning algorithms (Random Forests, Lasso with and without interaction terms) to assess convergence across classifiers. Autism is less common in girls; thus, our sample size was smaller for girls than boys. A larger sample size in the female cohort might have allowed us to find more significant associations. Nonetheless, the power is similar in the female and male cohorts because the effect size is larger in the female cohort than the male cohort (Supplemental Table 7).
Supplementary Material
ACKNOWLEDGMENTS
We dedicate this paper to the memory of Sir Michael (Mike) Rutter, a dear friend and pioneer in autism and child psychiatry research, who helped build the ABC. We thank Wai Hung Wong, Nina Deoras, Parisa Zolfaghari, and Shobun Baile for laboratory analyses, Joy Ukaigwe for data preparation, Meredith Eddy for project coordination, and Kelly Magnus for assistance with manuscript preparation. We are grateful to the families in Norway participating in MoBa and the ABC study.
This work was supported by National Institutes of Health grants NS047537 and NS086122, the Jane Botsford Johnson foundation, the Korein Foundation, the Simons Foundation Autism Research Initiative, the Norwegian Ministry of Health and Care Services, the Norwegian Ministry of Education and Research, and Research Council of Norway grants 189457, 190694, and 196452. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. None of the authors reported biomedical financial interests or potential conflicts of interest.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
SUPPLEMENTARY INFORMATION
Supplementary information is available at Molecular Psychiatry’s website.
References
- 1.Diagnostic and Statistical Manual of Mental Disorders (DSM-5(R)). American Psychiatric Publishing: Arlington, VA, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Randall M, Egberts KJ, Samtani A, Scholten RJ, Hooft L, Livingstone N et al. Diagnostic tests for autism spectrum disorder (ASD) in preschool children. Cochrane Database Syst Rev 2018; 7: CD009044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zwaigenbaum L, Penner M. Autism spectrum disorder: advances in diagnosis and evaluation. BMJ 2018; 361: k1674. [DOI] [PubMed] [Google Scholar]
- 4.Maenner MJ, Shaw KA, Baio J, Washington A, Patrick M, DiRienzo M et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill Summ 2020; 69(4): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werling DM, Geschwind DH. Sex differences in autism spectrum disorders. Curr Opin Neurol 2013; 26(2): 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen J, Grønborg TK, Sørensen MJ, Schendel D, Parner ET, Pedersen LH et al. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA 2013; 309(16): 1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiggs KK, Rickert ME, Sujan AC, Quinn PD, Larsson H, Lichtenstein P et al. Antiseizure medication use during pregnancy and risk of ASD and ADHD in children. Neurology 2020; 95(24): e3232–e3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer U, Nyffeler M, Yee BK, Knuesel I, Feldon J. Adult brain and behavioral pathological markers of prenatal immune challenge during early/middle and late fetal development in mice. Brain Behav Immun 2008; 22(4): 469–486. [DOI] [PubMed] [Google Scholar]
- 9.Bilbo SD, Block CL, Bolton JL, Hanamsagar R, Tran PK. Beyond infection - Maternal immune activation by environmental factors, microglial development, and relevance for autism spectrum disorders. Exp Neurol 2018; 299(Pt A): 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masi A, Quintana DS, Glozier N, Lloyd AR, Hickie IB, Guastella AJ. Cytokine aberrations in autism spectrum disorder: a systematic review and meta-analysis. Mol Psychiatry 2015; 20(4): 440–446. [DOI] [PubMed] [Google Scholar]
- 11.Brown AS. Epidemiologic studies of exposure to prenatal infection and risk of schizophrenia and autism. Dev Neurobiol 2012; 72(10): 1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornig M, Bresnahan MA, Che X, Schultz AF, Ukaigwe JE, Eddy ML et al. Prenatal fever and autism risk. Mol Psychiatry 2018; 23(3): 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes ML, McAllister AK. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci 2015; 16(8): 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016; 351(6276): 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci 2007; 27(40): 10695–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deverman BE, Patterson PH. Cytokines and CNS development. Neuron 2009; 64(1): 61–78. [DOI] [PubMed] [Google Scholar]
- 17.Careaga M, Murai T, Bauman MD. Maternal Immune Activation and Autism Spectrum Disorder: From Rodents to Nonhuman and Human Primates. Biol Psychiatry 2017; 81(5): 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron 1998; 21(3): 505–520. [DOI] [PubMed] [Google Scholar]
- 19.Needleman LA, McAllister AK. The major histocompatibility complex and autism spectrum disorder. Dev Neurobiol 2012; 72(10): 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shatz CJ. MHC class I: an unexpected role in neuronal plasticity. Neuron 2009; 64(1): 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpentier PA, Dingman AL, Palmer TD. Placental TNF-alpha signaling in illness-induced complications of pregnancy. Am J Pathol 2011; 178(6): 2802–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borsini A, Zunszain PA, Thuret S, Pariante CM. The role of inflammatory cytokines as key modulators of neurogenesis. Trends Neurosci 2015; 38(3): 145–157. [DOI] [PubMed] [Google Scholar]
- 23.Azmitia EC, Saccomano ZT, Alzoobaee MF, Boldrini M, Whitaker-Azmitia PM. Persistent Angiogenesis in the Autism Brain: An Immunocytochemical Study of Postmortem Cortex, Brainstem and Cerebellum. J Autism Dev Disord 2016; 46(4): 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prins JR, Eskandar S, Eggen BJL, Scherjon SA. Microglia, the missing link in maternal immune activation and fetal neurodevelopment; and a possible link in preeclampsia and disturbed neurodevelopment? J Reprod Immunol 2018; 126: 18–22. [DOI] [PubMed] [Google Scholar]
- 25.Kurzrock R, Estrov Z, Wetzler M, Gutterman JU, Talpaz M. LIF: not just a leukemia inhibitory factor. Endocr Rev 1991; 12(3): 208–217. [DOI] [PubMed] [Google Scholar]
- 26.Probert L TNF and its receptors in the CNS: The essential, the desirable and the deleterious effects. Neuroscience 2015; 302: 2–22. [DOI] [PubMed] [Google Scholar]
- 27.Abdallah MW, Larsen N, Mortensen EL, Atladottir HO, Norgaard-Pedersen B, Bonefeld-Jorgensen EC et al. Neonatal levels of cytokines and risk of autism spectrum disorders: an exploratory register-based historic birth cohort study utilizing the Danish Newborn Screening Biobank. J Neuroimmunol 2012; 252(1–2): 75–82. [DOI] [PubMed] [Google Scholar]
- 28.Abdallah MW, Larsen N, Grove J, Bonefeld-Jorgensen EC, Norgaard-Pedersen B, Hougaard DM et al. Neonatal chemokine levels and risk of autism spectrum disorders: findings from a Danish historic birth cohort follow-up study. Cytokine 2013; 61(2): 370–376. [DOI] [PubMed] [Google Scholar]
- 29.Abdallah MW, Mortensen EL, Greaves-Lord K, Larsen N, Bonefeld-Jorgensen EC, Norgaard-Pedersen B et al. Neonatal levels of neurotrophic factors and risk of autism spectrum disorders. Acta Psychiatr Scand 2013; 128(1): 61–69. [DOI] [PubMed] [Google Scholar]
- 30.Magnus P, Irgens LM, Haug K, Nystad W, Skjaerven R, Stoltenberg C et al. Cohort profile: the Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol 2006; 35(5): 1146–1150. [DOI] [PubMed] [Google Scholar]
- 31.Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK et al. Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol 2016; 45(2): 382–388. [DOI] [PubMed] [Google Scholar]
- 32.Stoltenberg C, Schjolberg S, Bresnahan M, Hornig M, Hirtz D, Dahl C et al. The Autism Birth Cohort: a paradigm for gene-environment-timing research. Mol Psychiatry 2010; 15(7): 676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suren P, Havdahl A, Oyen AS, Schjolberg S, Reichborn-Kjennerud T, Magnus P et al. Diagnosing autism spectrum disorder among children in Norway. Tidsskr Nor Laegeforen 2019; 139(14). [DOI] [PubMed] [Google Scholar]
- 34.Strand BH, Dalgard OS, Tambs K, Rognerud M. Measuring the mental health status of the Norwegian population: a comparison of the instruments SCL-25, SCL-10, SCL-5 and MHI-5 (SF-36). Nord J Psychiatry 2003; 57(2): 113–118. [DOI] [PubMed] [Google Scholar]
- 35.Ronningen KS, Paltiel L, Meltzer HM, Nordhagen R, Lie KK, Hovengen R et al. The biobank of the Norwegian Mother and Child Cohort Study: a resource for the next 100 years. Eur J Epidemiol 2006; 21(8): 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bach JF. Infections and autoimmune diseases. J Autoimmun 2005; 25 Suppl: 74–80. [DOI] [PubMed] [Google Scholar]
- 37.Goines PE, Croen LA, Braunschweig D, Yoshida CK, Grether J, Hansen R et al. Increased midgestational IFN-gamma, IL-4 and IL-5 in women bearing a child with autism: A case-control study. Mol Autism 2011; 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vignali DA. Multiplexed particle-based flow cytometric assays. J Immunol Methods 2000; 243(1–2): 243–255. [DOI] [PubMed] [Google Scholar]
- 39.Breen EJ, Tan W, Khan A. The Statistical Value of Raw Fluorescence Signal in Luminex xMAP Based Multiplex Immunoassays. Sci Rep 2016; 6: 26996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breen EJ, Polaskova V, Khan A. Bead-based multiplex immuno-assays for cytokines, chemokines, growth factors and other analytes: median fluorescence intensities versus their derived absolute concentration values for statistical analysis. Cytokine 2015; 71(2): 188–198. [DOI] [PubMed] [Google Scholar]
- 41.Helsel DR. Fabricating data: how substituting values for nondetects can ruin results, and what can be done about it. Chemosphere 2006; 65(11): 2434–2439. [DOI] [PubMed] [Google Scholar]
- 42.Antweiler RC. Evaluation of Statistical Treatments of Left-Censored Environmental Data Using Coincident Uncensored Data Sets. II. Group Comparisons. Environ Sci Technol 2015; 49(22): 13439–13446. [DOI] [PubMed] [Google Scholar]
- 43.Aggarwal CC. Outlier Analysis. 1st edn. Springer-Verlag; New York: 2013, pp XV, 446. [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995; 57(1): 289–300. [Google Scholar]
- 45.Tibshirani R Regression Shrinkage and Selection Via the Lasso. Journal of the Royal Statistical Society: Series B (Methodological) 1996; 58(1): 267–288. [Google Scholar]
- 46.Breiman L Random Forests. Machine Learning 2001; 45: 5–32. [Google Scholar]
- 47.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: San Francisco, CA, 2016, pp 785–794. [Google Scholar]
- 48.Hoeting JA, Madigan D, Raftery AE, Volinsky CT. Bayesian Model Averaging: A Tutorial. Statistical Science 1999; 14(4): 382–401. [Google Scholar]
- 49.Sanchez-Meca J, Marin-Martinez F, Chacon-Moscoso S. Effect-size indices for dichotomized outcomes in meta-analysis. Psychol Methods 2003; 8(4): 448–467. [DOI] [PubMed] [Google Scholar]
- 50.Cohen J Statistical Power Anaysis for the Behavioral Sciences. 2nd edn. Routledge: New York, 1988. [Google Scholar]
- 51.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. 1st edn. Chapman & Hall: Norwell, Massachusetts, 1993. [Google Scholar]
- 52.Brucato M, Ladd-Acosta C, Li M, Caruso D, Hong X, Kaczaniuk J et al. Prenatal exposure to fever is associated with autism spectrum disorder in the boston birth cohort. Autism Res 2017; 10(11): 1878–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zerbo O, Iosif AM, Walker C, Ozonoff S, Hansen RL, Hertz-Picciotto I. Is maternal influenza or fever during pregnancy associated with autism or developmental delays? Results from the CHARGE (CHildhood Autism Risks from Genetics and Environment) study. J Autism Dev Disord 2013; 43(1): 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity 2013; 39(6): 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atzeni F, Sarzi-Puttini P. Tumor Necrosis Factor. In: Maloy S, Kelly H (eds). Brenner’s Encyclopedia of Genetics, 2nd edn. Academic Press; 2013, pp 229–231. [Google Scholar]
- 56.Sedger LM, McDermott MF. TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants - past, present and future. Cytokine Growth Factor Rev 2014; 25(4): 453–472. [DOI] [PubMed] [Google Scholar]
- 57.Chez MG, Dowling T, Patel PB, Khanna P, Kominsky M. Elevation of tumor necrosis factor-alpha in cerebrospinal fluid of autistic children. Pediatr Neurol 2007; 36(6): 361–365. [DOI] [PubMed] [Google Scholar]
- 58.Tsilioni I, Taliou A, Francis K, Theoharides TC. Children with autism spectrum disorders, who improved with a luteolin-containing dietary formulation, show reduced serum levels of TNF and IL-6. Transl Psychiatry 2015; 5: e647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizejewski GJ, Lindau-Shepard B, Pass KA. Newborn screening for autism: in search of candidate biomarkers. Biomark Med 2013; 7(2): 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahic M, Mjaaland S, Bovelstad HM, Gunnes N, Susser E, Bresnahan M et al. Maternal Immunoreactivity to Herpes Simplex Virus 2 and Risk of Autism Spectrum Disorder in Male Offspring. mSphere 2017; 2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahic M, Che X, Susser E, Levin B, Reichborn-Kjennerud T, Magnus P et al. Epidemiological and Serological Investigation into the Role of Gestational Maternal Influenza Virus Infection and Autism Spectrum Disorders. mSphere 2017; 2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stolp HB. Neuropoietic cytokines in normal brain development and neurodevelopmental disorders. Mol Cell Neurosci 2013; 53: 63–68. [DOI] [PubMed] [Google Scholar]
- 63.Chitu V, Gokhan S, Nandi S, Mehler MF, Stanley ER. Emerging Roles for CSF-1 Receptor and its Ligands in the Nervous System. Trends Neurosci 2016; 39(6): 378–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nandi S, Gokhan S, Dai XM, Wei S, Enikolopov G, Lin H et al. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev Biol 2012; 367(2): 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kana V, Desland FA, Casanova-Acebes M, Ayata P, Badimon A, Nabel E et al. CSF-1 controls cerebellar microglia and is required for motor function and social interaction. J Exp Med 2019; 216(10): 2265–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zengeler KE, Lukens JR. Innate immunity at the crossroads of healthy brain maturation and neurodevelopmental disorders. Nat Rev Immunol 2021; 21(7): 454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han VX, Patel S, Jones HF, Dale RC. Maternal immune activation and neuroinflammation in human neurodevelopmental disorders. Nat Rev Neurol 2021; 17(9): 564–579. [DOI] [PubMed] [Google Scholar]
- 68.Costa D, Castelo R. Umbilical cord gene expression reveals the molecular architecture of the fetal inflammatory response in extremely preterm newborns. Pediatr Res 2016; 79(3): 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen L, Liu X, Zhang H, Lin J, Feng C, Iqbal J. Biomarkers in autism spectrum disorders: Current progress. Clin Chim Acta 2020; 502: 41–54. [DOI] [PubMed] [Google Scholar]
- 70.Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest 2009; 119(4): 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.