ABSTRACT
Fostering a “balanced” gut microbiome through the administration of beneficial microbes that can competitively exclude pathogens has gained a lot of attention and use in human and animal medicine. However, little is known about how microbes affect the horizontal gene transfer of antimicrobial resistance (AMR). To shed more light on this question, we challenged neonatal broiler chicks raised on reused broiler chicken litter—a complex environment made up of decomposing pine shavings, feces, uric acid, feathers, and feed—with Salmonella enterica serovar Heidelberg (S. Heidelberg), a model pathogen. Neonatal chicks challenged with S. Heidelberg and raised on reused litter were more resistant to S. Heidelberg cecal colonization than chicks grown on fresh litter. Furthermore, chicks grown on reused litter were at a lower risk of colonization with S. Heidelberg strains that encoded AMR on IncI1 plasmids. We used 16S rRNA gene sequencing and shotgun metagenomics to show that the major difference between chicks grown on fresh litter and those grown on reused litter was the microbiome harbored in the litter and ceca. The microbiome of reused litter samples was more uniform and enriched in functional pathways related to the biosynthesis of organic and antimicrobial molecules than that in fresh litter samples. We found that Escherichia coli was the main reservoir of plasmids encoding AMR and that the IncI1 plasmid was maintained at a significantly lower copy per cell in reused litter compared to fresh litter. These findings support the notion that commensal bacteria play an integral role in the horizontal transfer of plasmids encoding AMR to pathogens like Salmonella.
IMPORTANCE Antimicrobial resistance spread is a worldwide health challenge, stemming in large part from the ability of microorganisms to share their genetic material through horizontal gene transfer. To address this issue, many countries and international organizations have adopted a One Health approach to curtail the proliferation of antimicrobial-resistant bacteria. This includes the removal and reduction of antibiotics used in food animal production and the development of alternatives to antibiotics. However, there is still a significant knowledge gap in our understanding of how resistance spreads in the absence of antibiotic selection and the role commensal bacteria play in reducing antibiotic resistance transfer. In this study, we show that commensal bacteria play a key role in reducing the horizontal gene transfer of antibiotic resistance to Salmonella, provide the identity of the bacterial species that potentially perform this function in broiler chickens, and also postulate the mechanism involved.
KEYWORDS: AMR, broiler chickens, commensal bacteria, HGT, Salmonella
INTRODUCTION
Horizontal gene transfer (HGT) is recognized as the main mechanism by which bacteria acquire antimicrobial resistance (AMR), and exposure to antibiotics has been shown to drive antimicrobial resistance gene (ARG) transfer (1). Consequently, the rise in AMR in bacteria from hospital settings has been linked to the overuse of antibiotics in humans and their use in food animal production (2, 3). These public health concerns have led to a reduction in antibiotics used for raising food animals (4, 5), including a ban on antibiotic use in Europe (6). We previously showed that neonatal broiler chicks challenged with a nalidixic acid-resistant (nalR) Salmonella enterica serovar Heidelberg (S. Heidelberg) strain and raised antibiotic free on fresh litter composed of pine shavings were colonized at a high rate with S. Heidelberg strains that harbored IncI1 plasmids encoding AMR (7). We selected S. Heidelberg as the model pathogen because of its promiscuity to plasmids carrying AMR (8, 9).
There is limited research on if and how commensal bacteria reduce AMR in foodborne pathogens (10–16), such as Salmonella enterica, and the role it plays in AMR reduction and transfer. Therefore, the goal of this study was to determine the role commensal bacteria play in limiting Salmonella enterica from acquiring AMR and to provide information on the bacterial species and mechanism involved. To do this, we compared the dynamics of AMR transfer in neonatal broiler chicks raised on fresh litter to that of chicks raised on reused litter. We chose reused litter because it is a complex environment made up of decomposing plant-based bedding (e.g., wood shavings, sawdust, and rice or peanut hulls) mixed with chicken feces, uric acid, feathers, feed, insects, and other broiler-sourced materials. Therefore, reused litter carries a unique and complex population of bacteria, fungi, and viruses (17–19) interacting with various forms of eukaryotes, making it a suitable environment to study competitive exclusion. Broiler chickens are commonly raised on litter, but how litter is managed differs between countries, producers, and farmers. For instance, the practice of reusing litter over multiple flocks of broiler chickens is a widespread practice in the United States and Brazil, while Canada (20) and Europe (21) recommend fresh litter bedding for every flock. One argument against litter reuse is that it harbors pathogenic bacteria that can be transferred to the next flock (22). Contrastingly, proponents of litter reuse argue that it confers competitive exclusion against pathogens when effectively managed and it is cost-effective (23–26). Beyond broiler chicken production, competitive exclusion by commensal bacteria has received enormous attention in the 20th century, resulting in the bloom of commercially marketed probiotics (27) and the underlying theory behind the application of fecal microbiota transplantation in human medicine (28).
RESULTS
S. Heidelberg abundance and prevalence in ceca and litter.
We confirmed that neonatal chicks were Salmonella free by testing the chick pads used for transportation from the hatchery for Salmonella. None of the chick pads was positive for Salmonella. The reused litter for this study was confirmed to be Salmonella free in earlier studies (29, 30). To determine if the microbiome offers protection against Salmonella colonization, we challenged neonatal broiler chicks with a nalidixic acid-resistant (nalR) strain of S. Heidelberg and raised the chicks on either new bedding made up of fresh pine shavings (here referred to as fresh litter) (n = 75) or reused litter (litter previously used to raise three flocks of broiler chickens) (n = 75). Chicks were challenged either through oral gavage (n = 25), cloacal inoculation (n = 25), or by the seeder method (i.e., a few chicks [n = 5] were challenged orally and comingled with unchallenged chicks [n = 20]) (7). Unchallenged chicks on fresh (n = 25) and reused litter (n = 25) were used as controls. Broiler chicks on fresh and reused litter were housed separately for 14 days (four separate pens for each treatment in a house), and chicks were not administered any medication or antibiotics for the duration of the study.
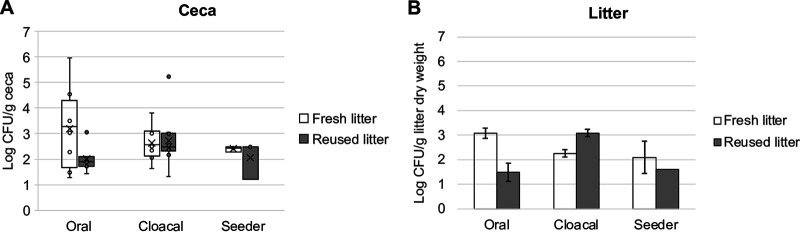
Afterwards, we determined the concentration of nalR S. Heidelberg in the ceca and litter of chicks 14 days after they were challenged. The average nalR S. Heidelberg concentrations in the ceca (n = 10 for oral and cloacal and n = 15 for seeder) were not significantly different between chicks raised on fresh litter compared to those raised on reused litter (shown here as W statistic, P value: oral = 41, 0.15; cloacal = 41, 0.96; seeder = 5, 1.00) (Fig. 1A). However, the ceca of chicks raised on fresh litter were more likely to be positive for nalR S. Heidelberg (χ2 = 16.07, degree of freedom [df] = 2, P = 0.0003), than chicks on reused litter. The percentages of cecal samples positive for nalR S. Heidelberg were 100%, 100%, and 50%, respectively, for oral, cloacal, and seeder chicks raised on fresh litter, compared to 80%, 100%, and 33.3%, respectively, for chicks raised on reused litter. For the seeder treatment on fresh litter, one seeder was lost due to premature mortality and three of the four seeders were positive for nalR S. Heidelberg, while the ceca of four of the 10 uninoculated contact chicks were positive. In contrast, the ceca of the five orally challenged seeders raised on reused litter were positive for nalR S. Heidelberg, but all contact chicks were negative (n = 10).
FIG 1.
S. Heidelberg concentration in the ceca and litter of broiler chicks. Box plot of nalR S. Heidelberg concentration in the (A) ceca (n = 10 for oral and cloacal, n = 15 for seeder) (B) and litter (n = 2 subsamples from pooled litter) of chicks raised on fresh or reused litter (P > 0.05 by Wilcoxon signed-rank test). Cecal and litter samples were collected 14 days after challenging chicks with nalR S. Heidelberg through oral gavage, cloacal inoculation, or the seeder method. The concentrations of nalR S. Heidelberg in chicks on fresh litter have been described previously (7).
For the litter, there was no significant difference in nalR S. Heidelberg concentration in fresh litter compared to that in reused litter (P > 0.05); however, oral and seeder treatments carried higher levels of nalR S. Heidelberg in fresh litter compared to reused litter, while cloacally inoculated chicks had higher levels in reused litter compared to fresh litter (Fig. 1B). All litter samples from challenged chicks were positive for nalR S. Heidelberg by direct culture or after enrichment in buffered peptone water (BPW), except for one reused litter sample from the seeder treatment that was negative. For unchallenged chicks used as controls, one litter sample each from fresh and reused litter tested positive after enrichment in BPW. These data suggest that litter age/type did not have a significant effect on the abundance of nalR S. Heidelberg in the ceca, but chicks raised on reused litter (66%) had a lower Salmonella positivity rate compared to chicks on fresh litter (79%) (P = 0.0003).
Broiler litter age/type affected the horizontal transfer of AMR.
Next, we questioned if the litter plays a role in the HGT of AMR. First, we performed antibiotic susceptibility testing (AST) on S. Heidelberg isolates recovered from the ceca and litter of broiler chicks raised on fresh litter (n = 158) and reused litter (n = 141). On average, five nalR S. Heidelberg isolates (range, 1 to 7) were randomly selected from the ceca of each challenged chick that was Salmonella positive (fresh litter, n = 25 chicks; reused litter, n = 23 chicks) and used for AST. For AST of litter isolates, an average of 11 nalR S. Heidelberg isolates (range, 3 to 17) were randomly selected per pen for fresh and reused litter (n = 3 pens each for challenged chicks on fresh or reused litter).
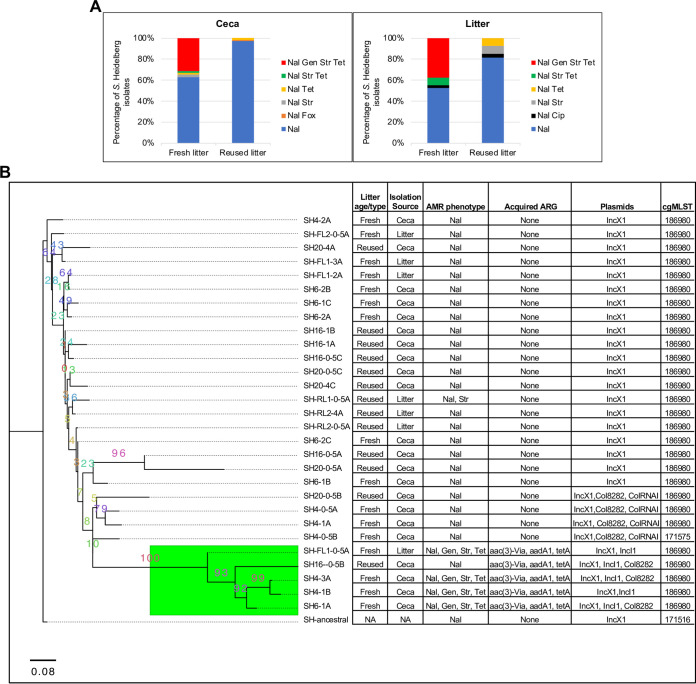
As expected, all S. Heidelberg isolates were resistant to nalidixic acid (Fig. 2). S. Heidelberg isolates from the ceca and litter of chicks on fresh litter were more likely to be resistant to one or more antibiotics compared to isolates from chicks on reused litter (ceca, χ2 = 138, df = 2, P < 2.2e−16; litter, χ2 = 11.65, df = 2, P = 0.003). Thirty-one percent of isolates from the ceca (37/118) and 37.5% of those from the litter (15/40) of chicks on fresh litter acquired resistance to gentamicin, tetracycline, and streptomycin (7) (Fig. 2A). Contrastingly, no S. Heidelberg isolate from the ceca or litter of chicks on reused litter acquired resistance to gentamicin, tetracycline, and streptomycin. Three percent of S. Heidelberg isolates from the ceca of chicks on reused litter acquired resistance to either cefoxitin (1/114) or tetracycline (2/114), while ∼18% of isolates from reused litter acquired resistance to either ciprofloxacin (1/27), tetracycline (2/27), or streptomycin (2/27) (Fig. 2A). The percentage of S. Heidelberg isolates that acquired AMR in fresh litter differed by the route of inoculation (7). For instance, 40% of isolates from orally challenged chicks acquired AMR compared to 24% for cloacal isolates (7).
FIG 2.
Broiler chicks raised on reused litter carried lower levels of antimicrobial-resistant S. Heidelberg. (A) Distribution of antibiotic resistance acquired by nalR S. Heidelberg isolates from the ceca and litter of chicks on fresh (n = 158) and reused litter (n = 141). (B) Maximum likelihood tree constructed using accessory genes present in S. Heidelberg isolates (n = 30) recovered from the ceca and litter of cloacally inoculated chicks raised on fresh litter and reused litter. S. Heidelberg isolates with strain IDs SH4 and SH6 were recovered from the ceca of two chicks on fresh litter, while IDs SH16 and SH20 are from two chicks raised on reused litter. The GTR model of nucleotide substitution and the GAMMA model of rate heterogeneity were used for sequence evolution prediction. Numbers shown next to the branches represent the percentage of replicate trees where associated isolates cluster together based on ∼100 bootstrap replicates. All S. Heidelberg strains were assembled using Illumina short reads, except SH4-3A and SH-ancestral, which were assembled by combining Illumina short reads with PacBio or MinION long reads. The tree was rooted with the ancestral S. Heidelberg strain (SH-ancestral; GenBank accession no: CP066851). The genomes of S. Heidelberg strains from fresh litter have been previously published (7). The green subclade shows strains that harbored IncI1-pST26. All isolates are expected to be resistant to nalidixic acid. Nal, nalidixic acid; Gen, gentamicin; Str, streptomycin; Tet, tetracycline; Fox, cefoxitin; Cip, ciprofloxacin.
We previously demonstrated that S. Heidelberg isolates from fresh litter that acquired resistance to gentamicin, tetracycline, and streptomycin harbored IncI1 plasmids (plasmid MLST 26; here referred to as IncI1-pST26) (7). The IncI1-pST26 plasmid carries ARGs for aminoglycoside [aadA1, aac(3′)-Via], tetracycline (tetA), and mercury (mer operon) resistance (see Fig. S1 in the supplemental material). Here, we investigated if S. Heidelberg isolates recovered from chicks raised on reused litter differed in their core genome or in plasmids harbored, compared to isolates from fresh litter. To answer this question, we performed whole-genome sequence (WGS) analysis on S. Heidelberg isolates (n = 29) recovered from the ceca of two cloacally challenged chicks from fresh litter (SH4 and SH6) and reused litter (SH16 and SH20), as well as the litter samples collected from the floor pen. We chose cloacal samples because only this route of challenge resulted in 100% colonization of the ceca of chicks raised on fresh litter and reused litter.
There was no difference in the core genome multilocus sequence types (cgMLSTs) of evolved S. Heidelberg isolates from fresh versus reused litter; however, 28 of the 29 evolved isolates had a different cgMLST from the ancestral nalR S. Heidelberg (Fig. 2B). We used the genomes sequenced to construct a maximum likelihood (ML) tree based on the pangenome and mutations of S. Heidelberg strains recovered. The core genome (genes present in ≥95% of the strains) and accessory genome (genes present in <95% of the strains) were composed of 4,373 and 356 genes, respectively. After removing 10 isolates with identical mutations, a total of 118 informative sites (single nucleotide polymorphisms [SNPs] and indels) was used for ML SNP tree construction.
The three constructed ML trees did not group isolates by litter age/type (fresh versus reused) (Fig. 2B; see Fig. S2A and B in the supplemental material), but S. Heidelberg isolates that harbored IncI1-pST26 (n = 5) formed a separate clade on the accessory genome tree (Fig. 2B). Isolates from the ceca (n = 3) and litter (n = 1) of chicks on fresh litter that harbored IncI1-pST26 displayed the antibiotic resistance phenotype predicted by WGS (Fig. 2B). Contrastingly, one isolate from reused litter carried IncI1-pST26 but was found to be susceptible to all antibiotics tested, except for the ancestral nalidixic resistance. These results indicated that chicks grown on reused litter in this study were less likely to carry S. Heidelberg isolates harboring AMR compared to chicks on fresh litter.
The microbiomes of neonatal chicks differed for fresh and reused litter.
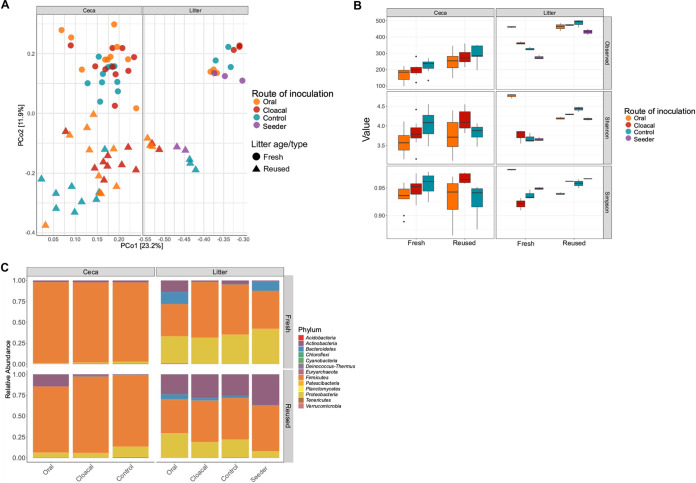
We used 16S rRNA gene sequencing to examine the differences in the microbiome between neonatal chicks raised on fresh litter and those raised on reused litter. The beta diversities of the cecal (n = 59) and litter (n = 22) microbiome of chicks were significantly different between fresh litter and reused litter (ceca, P = 0.0002; litter, P = 0.0002) (Fig. 3A). Furthermore, the route of inoculation used for S. Heidelberg affected the beta diversities of the ceca and litter for fresh litter and reused litter (P < 0.05). For example, chicks challenged orally (n = 10) or through the cloaca (n = 10) and raised on fresh litter harbored a significantly different beta diversity than uninoculated chicks (n = 10) in the ceca (P < 0.002) (Fig. 3A). Likewise, the beta diversity of the ceca of chicks challenged orally (n = 10) differed from those of cloacal (n = 9) and uninoculated controls (n = 10) for reused litter (P < 0.05). In the litter, pairwise comparisons between routes of inoculation were not significant (P > 0.05) for beta diversity. Nevertheless, fresh litter samples (n = 3) from orally challenged chicks clustered next to litter samples (n = 3) from seeder treatments, while litter samples (n = 2) from cloacally inoculated chicks were different from those of oral and seeder treatments (Fig. 3A). For reused litter samples, litter from orally (n = 3) and cloacally (n = 3) challenged chicks clustered together, while litter (n = 2) from seeder treatments was similar to litter samples (n = 3) from control chicks. Finally, the bacterial community structure (assessed using beta dispersion) of reused litter was less variable than that of fresh litter (P = 0.001); however, there was no difference in variability for the ceca (P = 0.15). Moreover, the route of challenge affected the bacterial community beta dispersion in the ceca and litter of chicks on fresh litter and reused litter (P = 0.001).
FIG 3.
Reused litter samples harbored a more uniform and diverse microbiome than fresh litter samples. (A) Principal-coordinate analysis of Bray-Curtis distances based on 16S rRNA gene libraries obtained from ceca and litter samples. Points represent the values from individual libraries, with colors denoting the route used for S. Heidelberg challenge for respective samples. (B) Average alpha diversity indices of rarefied ceca and litter samples grouped by the litter age/type and the route of S. Heidelberg challenge. Boxes indicate the interquartile range (75th to 25th) of the data. Whiskers extend to the most extreme value within 1.5× interquartile range, and dots represent outliers beyond that range. The black bar represents the median. (C) Phylum‐level classification of 16S rRNA gene sequence reads in each ceca and litter sample grouped by the route of S. Heidelberg challenge. 16S rRNA gene sequencing was performed on the ceca (59 chicks from fresh litter [n = 30] and reused litter [n = 29]) and litter (n = 22 [11 samples each from fresh and reused litter]). The microbiome data of chicks on fresh litter have been described previously (7).
The alpha diversity (i.e., the number of amplicon sequence variants [ASVs]) of the microbiome present in the ceca and litter of chicks on reused litter was higher for the observed species measure of diversity (ceca, P < 0.001; litter, P = 0.001), but not for Shannon (ceca, P = 0.25; litter, P = 0.05) and Simpson (ceca, P = 0.97; litter, P = 0.28) indices. The route of S. Heidelberg challenge affected the alpha diversity of the cecal and litter microbiomes (P < 0.05) (Fig. 3B). The alpha diversity of the ceca of challenged and control chicks raised on reused litter was higher than that of chicks grown on fresh litter for the observed species measure of diversity (P < 0.01). Cloacally challenged chicks raised on reused litter had higher alpha diversity in their ceca compared to chicks on fresh litter for the Shannon and Simpson indices (P < 0.05). Uninoculated control chicks on fresh litter had higher alpha diversity in their ceca than control chicks on reused litter for the Simpson index (Fig. 3B). The litter of cloacal, seeder, and control chicks on reused litter had higher alpha diversity than the litter of chicks on fresh litter for the Shannon measure of diversity (P < 0.05) (Fig. 3B). In contrast, the litter of orally challenged chicks on fresh litter had higher alpha diversity than the litter of gavaged chicks on reused litter for the Shannon index (P = 0.0001). There was no significant difference in alpha diversity between fresh litter and reused litter for the observed and Simpson indices (P > 0.05).
On the taxonomic level, uninoculated control chicks on fresh litter had higher relative abundance of Actinobacteria and Firmicutes in the ceca compared to control chicks on reused litter (P = 0.004), while the abundance of Proteobacteria and Bacteroidetes was higher in the ceca of control chicks on reused litter compared to that of control chicks on fresh litter (P < 0.05) (Fig. 3C). Orally challenged chicks on reused litter carried a higher abundance of Actinobacteria in the ceca compared to chicks on fresh litter (P = 0.003), while the abundance of Proteobacteria was higher in the ceca of oral and cloacal chicks on reused litter compared to chicks on fresh litter (P < 0.01). Firmicutes had higher relative abundance in the ceca of chicks on fresh litter compared to reused litter for all challenged chicks (P < 0.01), while the abundance of Bacteroidetes was higher in the ceca of cloacal chicks on reused litter compared to chicks on fresh litter (P < 0.05) (Fig. 3C).
For the litter, the abundance of Actinobacteria was higher in reused litter compared to fresh litter (P < 0.05), while the abundances of Proteobacteria and Bacteroidetes were higher in fresh litter compared to reused litter (P < 0.05). There was no significant difference in phylum abundance between routes of inoculation (P > 0.05) for the litter. These results shows that the microbiome of chicks raised on fresh litter differed from that of chicks grown on reused litter, and reused litter cecal and litter samples of challenged chicks were more uniform than fresh litter samples.
S. Heidelberg challenge modulated the gut microbiome.
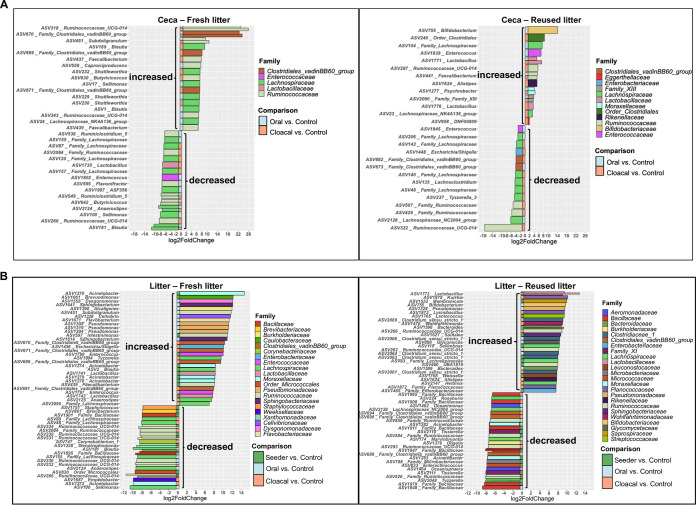
We determined that the core microbiome (i.e., ASVs of >0.1% relative abundance and present in at least 80% of samples) of fresh litter differs from that of reused litter (see Table S1 in the supplemental material) and found that S. Heidelberg challenge perturbed the cecal and litter microbiomes of broiler chicks (Fig. 4). For example, Enterobacteriaceae (Klebsiella pneumoniae and Proteus mirabilis) were part of the core microbiome in fresh litter, while they were not part of the core microbiome of reused litter (Table S1). Contrastingly, Actinobacteria (23 ASVs) were part of the core microbiome of reused litter, while only one ASV classified as Actinobacteria was determined to be a core member of fresh litter (Table S1). Unsurprisingly, the bacterial ASVs that increased or decreased in abundance after S. Heidelberg challenge differed between chicks on fresh litter and reused litter (Fig. 4A and B).
FIG 4.
S. Heidelberg challenge modulated the microbiome of the ceca and litter of broiler chicks. Plots of amplicon sequence variants (ASVs) that were significantly differentially abundant (adjusted P value [Padj] of <0.05) in the (A) ceca and (B) litter of challenged chicks on fresh and reused litter compared to uninoculated controls. Significant ASVs are plotted individually and colored according to their family-level classification. The route of S. Heidelberg challenge is shown on the edge of each plot as colored squares.
To determine the ASVs that were modulated by S. Heidelberg challenge (i.e., ASVs that increased or decreased after challenge), we compared the relative abundances of ASVs in the ceca and litter of challenged chicks to those of uninoculated control chicks (Fig. 4). In the ceca of chicks on fresh litter, the ASVs significantly modulated were assigned to five bacterial families (Fig. 4A). Three ASVs matching family Clostridiales vadinBB6 group, nine ASVs classified as Ruminococcaceae, and seven ASVs classified as Lachnospiraceae increased in abundance in challenged chicks compared to control chicks. Contrastingly, ASVs classified as Lactobacillaceae, Enterococcaceae, Ruminococcaceae (n = 8), and Lachnospiraceae (n = 10) decreased in abundance in challenged chicks compared to control chicks (Fig. 4A).
For the ceca of chicks on reused litter, the ASVs modulated were assigned to 12 bacterial families. Amplicon sequence variants matching Bifidobacteriaceae, order Clostridiales, Moraxellaceae, Rikenellaceae, Eggerthellaceae, Enterococcaceae, family XIII, Lactobacillaceae (n = 2), Ruminococcaceae (n = 2), and Lachnospiraceae (n = 2) increased in abundance in challenged chicks compared to control chicks. In contrast, ASVs classified as Enterobacteriaceae, Enterococcaceae, Clostridiales vadinBB6 group (n = 2), Ruminococcaceae (n = 3), and Lachnospiraceae (n = 7) decreased in abundance in challenged chicks compared to control chicks (Fig. 4A).
For the litter, the ASVs that were significantly modulated in fresh litter were assigned to 21 bacterial families (Fig. 4B). The ASVs that increased in abundance in fresh litter of challenged chicks belonged to Caulobacteraceae, Dysgonomonadaceae, Burkholderiaceae, Flavobacteriaceae, Cellvibrionaceae, Enterobacteriaceae, Enterococcaceae (n = 2), Lactobacillaceae (n = 2), Sphingobacteriaceae (n = 2), Pseudomonadaceae (n = 3), Ruminococcaceae (n = 3), Moraxellaceae (n = 4), Clostridiales vadinBB6 group (n = 4), and Lachnospiraceae (n = 4). Conversely, ASVs assigned to Staphylococcaceae, Brevibacteriaceae, order Micrococcales, Corynebacteriaceae, Xanthomonadaceae, Weeksellaceae, Bacillaceae (n = 2), Ruminococcaceae (n = 6), and Lachnospiraceae (n = 6) decreased in abundance in the litter of challenged chicks compared to control chicks (Fig. 4B).
Amplicon sequence variants modulated in reused litter after challenge were grouped into 24 bacterial families. The ASVs that increased in abundance in reused litter of challenged chicks were assigned to Lactobacillaceae, Saprospiraceae, Bifidobacteriaceae, Rikenellaceae, Pseudomonadaceae, Sphingobacteriaceae, Leuconostocaceae, Glycomycetaceae, Streptococcaceae, Enterobacteriaceae, Wohlfahrtiimonadaceae, Bacteroides (n = 2), Ruminococcaceae (n = 3), Planococcaceae (n = 3), Lachnospiraceae (n = 3), and Clostridiaceae 1 (n = 5). The ASVs that decreased in the reused litter of challenged chicks were classified as Burkholderiaceae, Microbacteriaceae, Micrococcaceae, Moraxellaceae (n = 2), Wohlfahrtiimonadaceae (n = 2), Aeromonadaceae (n = 3), Clostridiales vadinBB6 group (n = 3), Ruminococcaceae (n = 4), Lachnospiraceae (n = 4), and Bacillaceae (n = 6).
Not all bacterial families were modulated by the cloacal route of S. Heidelberg challenge. For instance, Enterococcaceae was only modulated by oral challenge in the ceca of chicks grown on fresh and reused litter (Fig. 4A). Similarly, Pseudomonadaceae was only modulated in the litter of chicks that were orally challenged (Fig. 4B). Together, these results show that the litter microbiomes modulated by S. Heidelberg challenge differed between chicks on fresh litter and those on reused litter, and the route of challenge affected the ASVs that increased or decreased in abundance.
The microbiomes of chicks raised on reused litter were enriched in functional pathways for the biosynthesis of antimicrobials.
A functional protein pathway analysis was performed using Hi-C metagenome-assembled genomes (MAGs) of two cecal samples each from cloacally challenged chicks on fresh and reused litter. The average assembly size and number of contigs for the four cecal samples were 1,238,395,273 ± 167,820 bp and 1,797,423 ± 281,253, respectively. The number of MAGs found in cecal samples from fresh and reused litter was 456 ± 422. Pathways related to the biosynthesis of organic and antimicrobial molecules were enriched to a greater degree in the ceca of chicks on reused litter than those in chicks on fresh litter (see Fig. S3 in the supplemental material). Of note was the enrichment of biosynthetic pathways for antimicrobials and secondary metabolites in reused litter (e.g., macrolides, tetracyclines, carbapenems, vancomycin, and polyketides), suggesting that reused litter harbored a higher abundance of microbial species with antimicrobial properties than fresh litter. Contrastingly, pathways associated with the biosynthesis of secondary metabolites of plants, including flavonoids, indole-alkaloids, and stilbenoid, diarylheptanoid, and gingerol were enriched in samples from fresh litter (Fig. S3).
Enterobacteriaceae and Clostridiales were the major bacterial hosts for AMR.
We also used Hi-C MAGs to explore the bacterial reservoir of AMR in the cecal microbiome. We previously used this approach to identify Escherichia coli as the main bacterial reservoir of transferable plasmids in fresh litter (7). Here, we extended our analysis to cecal samples from reused litter (n = 2). We searched the MAGs for sequences matching plasmid incompatibility (inc) groups and replicons available on the PlasmidFinder database. The inc groups found matched common Enterobacteriaceae plasmids, including IncF, IncI, IncX, IncH, IncY, IncB/O/K/Z, p0111, and Col-like plasmids (Table 1). Three plasmid replicons were found for Gram-positive bacteria, including rep2, rep18b, and repA (Table 1). The low representation of Gram-positive plasmids is expected, since the PlasmidFinder database is much more comprehensive for Enterobacteriaceae than other bacterial species, and within the well-studied Proteobacteria, most plasmids cannot be typed (31).
TABLE 1.
Plasmid incompatibility groups and replicons found in cecal metagenome-assembled genomes using PlasmidFindera
| Bacterial group | Incompatibility group(s) or replicon(s) in: |
|||
|---|---|---|---|---|
| Hi-C-FL1 | Hi-C-FL2 | Hi-C-RL1 | Hi-C-RL2 | |
| Gram negative | Col(MG828), Col156, ColpVC, ColRNAI, IncFIB, IncFIC(FII), IncFII(pRSB107), IncHI1B(pNDM-CIT), IncI1-I(Alpha), IncI2(Delta), IncX1, IncX2, p0111 | Col(MG828), ColpVC, ColRNAI, IncB/O/K/Z, IncFIB, IncFII(pSFO), IncI2, IncX1, IncX2 | Col(MG828), Col(pHAD28), Col156, ColRNAI, IncFIB, IncFII(pSFO), IncI1-I(Alpha), IncI2, IncX1, IncX2, IncY | Col(MG828), Col156, IncFIB, IncFII(pCoo), IncHI1B(pNDM-CIT), IncI1-I(Alpha) |
| Gram positive | rep2, repA(pB82) | None | rep18b | repA(pB82) |
Sample IDs: FL, fresh litter; RL, reused litter.
Next, we used proximity ligation to find the bacterial host of ARGs and to determine if they are encoded on plasmids or the chromosome. One hundred ARGs were found in the MAGs, and the majority (∼50%) were harbored by family Enterobacteriaceae (n = 3 MAGs) (Fig. 5). Escherichia coli was the only member of the Enterobacteriaceae found to be the putative host for these ARGs in fresh and reused litter. Members of the order Clostridiales were the putative hosts for 27% of ARGs, while unclassified bacterial species (n = 6) and members of the phyla Firmicutes (n = 9), Bacteriodetes (n = 3), and Actinobacteria (n = 1) were the hosts for the remaining ARGs. Fifty-four percent of the ARGs were found to be carried on the chromosome, while 46% were found on plasmids. One cecal sample from fresh litter (Fig. 5) harbored E. coli MAGs that contained silver and copper resistance genes (silABCFRS and pcoABCDRS) on the chromosome, while virulence genes (iroBCDEN) were found on plasmids. A cecal sample from a chick raised on reused litter harbored E. coli MAGs that carried ampicillin (blaTEM-1B), tetracycline (tetA), and mercury resistance (merCPRT) genes on plasmids (Fig. 5). This result suggests that the microbiomes of chicks on fresh litter and reused litter harbored AMR, and Enterobacteriaceae and Clostridiales were the putative bacterial hosts of the ARGs.
FIG 5.
Enterobacteriaceae and Clostridiales were the bacterial hosts of AMR in broiler chicks. AMR gene hosts found by Hi-C contacts. AMR genes (horizontal axis of heat map) associated with metagenome-assembled genomes (MAGs) present in cecal samples from chicks on fresh litter (Hi-C-FL1 and Hi-C-FL2) and reused litter (Hi-C-RL1 and Hi-C-RL2). MAGs and AMR gene sources were derived from Hi-C-based deconvolution of the metagenome assembly and placed into a bacterial phylogeny using Mash and CheckM by the ProxiMeta platform. The MAG assigned to family Enterobacteriaceae in each sample matches most closely to an Escherichia coli genome. Legend: gold, AMR genes that are associated with plasmid sequences on the same contigs; cyan, AMR genes that are likely to be genomically integrated; black, no contact above statistical thresholds.
E. coli isolates harbored AMR on plasmids and genomic islands.
In our earlier work, we found that E. coli MAGs were the primary hosts of IncI1 by retrospectively screening the cecal contents of chicks on fresh litter for E. coli isolates (7). Using this approach, we confirmed that the IncI1-pST26 plasmid acquired by S. Heidelberg was identical to the IncI1-pST26 plasmid present in E. coli strains (Fig. S1). Here, we performed AST and WGS on additional E. coli isolates recovered from the ceca of chicks used for Hi-C metagenomics (see Table S2 in the supplemental material), and a pangenome analysis was done on 19 of them. The isolates were randomly selected from CHROMagar plates supplemented with or without gentamicin and tetracycline.
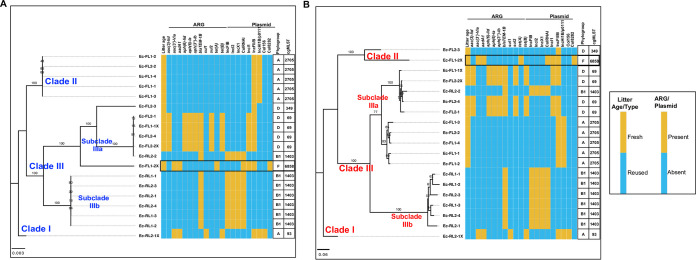
The inferred phylogeny of the E. coli strains by using their core (n = 3,385) and accessory (n = 6,024) genes grouped the isolates by the litter used for raising the chicken host, except for one isolate (Ec-RL2-2) from reused litter that clustered with E. coli strains from fresh litter (Fig. 6). The ML trees grouped the isolates into three main clades (clades I to III). One E. coli isolate from the ceca of chicks on reused litter made up clade I on the core and accessory gene tree, while the numbers of isolates in clade II and III strains differed between the core (Fig. 6A) and accessory (Fig. 6B) gene trees. Clade II isolates were clonal and recovered from chicks on fresh litter. Clade III was divided into two subclades represented by isolates from fresh litter (subclade IIIa) and reused litter (subclade IIIb), and subclade IIIb isolates were more clonal compared to subclade IIIa isolates. E. coli isolates classified as phylogroups A, D, and F were more likely to be found in chicks on fresh litter, while phylogroup B1 was found in the ceca of chicks from reused litter (Fig. 6; Table S2). Likewise, cgMLST classification showed that the sequence types ST69, ST2705, and ST6858 were associated with E. coli strains originating from chicks on fresh litter, while ST1403 was found in reused litter. These results suggest that the litter type/age used for raising the chicks in this study affected the pangenome of E. coli strains recovered from the ceca.
FIG 6.
Escherichia coli was the main reservoir of plasmids and AMR in broiler chicks. Pangenome analysis was performed on E. coli strains (n = 19) recovered from the ceca of broiler chicks challenged with S. Heidelberg and raised on fresh litter (FL) (n = 2) or reused litter (RL) (n = 2). The maximum likelihood tree was constructed using the core (A) and accessory (B) genes of the E. coli strains. Illumina short reads were combined with either PacBio or MinION long reads to assemble the genomes of E. coli strains Ec-FL1-1X, Ec-FL1-2X, Ec-FL1-3, Ec-RL2-1, and Ec-RL2-1X. The GTR model of nucleotide substitution and GAMMA model of rate heterogeneity were used for sequence evolution prediction. Numbers shown next to the branches represent the percentage of replicate trees where associated isolates cluster together based on ∼100 bootstrap replicates. Clade numbers were assigned arbitrarily to ease discussion on the differences in phylogeny between isolates. The tree was rooted with Ec-RL2-1X. cgMLST, core genome multilocus sequence type. The black rectangular box shows the E. coli strain carrying IncI1-pST26 identical to that of S. Heidelberg.
Some E. coli strains from fresh litter harbored ARGs on the chromosome and plasmids. E. coli strains classified as the pandemic lineage of extraintestinal pathogenic ST69 from fresh litter harbored an ∼128-kb genomic island containing virulence genes (pyelonephritis-associated pilus [pap] operon) and ARGs for aminoglycoside [strAB and aph(3′)-Ia] tetracycline (tetB), sulfonamide (sul2), and metal resistance (silver [sil] and copper [pco] operons) (see Table S2 and Fig. S4 in the supplemental material). The presence of a genomic island encoding AMR suggests that the ARGs are mobile. We found DNA regions carrying identical metal resistance genes (10 to 40 kb), as seen in the genomic island in p0111/IncH1B plasmids harbored by E. coli strains from this study and in E. coli genomes found in the NCBI database (Fig. S4). In addition, ST69 E. coli strains harbored a multireplicon IncF plasmid carrying blaTEM-1B, aac(3′)-Ild, and virulence genes (colicin M and catecholate siderophore uptake system [iroBCDEN]), as well as an unknown IncI1 plasmid (see Data Set S1). Data Sets S1 to S5 are available in the Dryad Repository at https://doi.org/10.5061/dryad.c866t1g6c.
As previously reported (7), E. coli ST6858 from fresh litter carried an identical S. Heidelberg IncI1-pST26 plasmid and harbored IncI2, IncF, and cryptic Col-type plasmids (see Data Set S2 in the Dryad Repository). Antibiotic-susceptible ST2705 E. coli isolates from fresh litter harbored no plasmid containing antibiotic resistance genes but carried a phage-like plasmid classified as p0111 and a multireplicon IncF plasmid containing metal resistance and virulence genes (see Data Set S3 in the Dryad Repository). The common ARG carried by ST1403 E. coli strains from reused litter was blaTEM-1B. The gene was carried on an IncX1 plasmid (see Data Set S4 in the Dryad Repository), and it conferred ampicillin resistance. Additionally, ST1403 E. coli strains harbored IncFIB, IncI2, and ColRNAi plasmids. One E. coli isolate (ST93) from reused litter harbored an IncH1B/p0111 plasmid encoding ARGs for aminoglycoside [aadA1, aac(3)-Vla], tetracycline (tetB), sulfonamide (sul1), and mercury resistance (mer operon) and an IncF plasmid containing colicin M and iroBCDEN genes (see Data Set S5 in the Dryad Repository).
Bacterial hosts from reused litter maintained IncI1 plasmid at low copies.
The low rate of AMR acquisition by S. Heidelberg isolates from reused litter made us hypothesize that IncI1-pST26 plasmids are present at lower copies in reused litter. To test this hypothesis, we used quantitative PCR (qPCR) assays targeting the region upstream of the repA of IncI1 plasmid, including incRNAI (small antisense RNA essential for control of IncI plasmids replication), to determine their copy number in the ceca of chicks on fresh and reused litter. We normalized the abundance of IncI1 against glyceraldehyde-3-phosphate dehydrogenase A (gapA) housekeeping gene carried on the chromosome of several enteric bacteria (32) including Salmonella and E. coli. We targeted Enterobacteriaceae with gapA because E. coli was determined to be the host for AMR by metagenomics and WGS in this study. The relative abundance of the Enterobacteriaceae population carrying IncI1 was higher in the ceca of orally challenged chicks on fresh litter compared to chicks on reused litter (W = 85, P = 0.00041) and was not significantly different for cloacally inoculated (W = 23, P = 0.24) and uninoculated control (W = 63, P = 0.16) chicks (Fig. 7).
FIG 7.
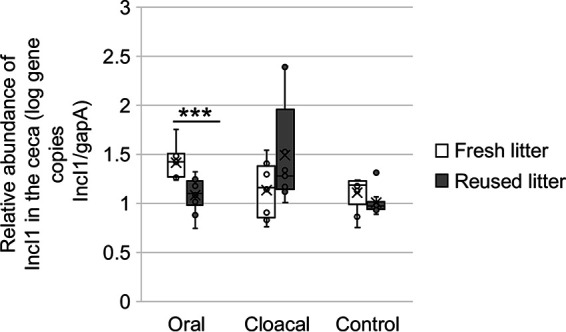
Relative abundance of IncI1 in Enterobacteriaceae bacterial population of the ceca. Box plot of the ratio of IncI1 gene copies/gram of ceca to gene copies of gapA/gram of ceca for chicks raised on fresh liter (oral, n = 9; cloacal, n = 8; control, n = 9) and reused litter (oral, n = 10; cloacal, n = 9; control, n = 10). ***, P < 0.001 by Wilcoxon signed-rank test.
To further explore the effect of low-copy IncI1 plasmids on AMR transfer, we selected an S. Heidelberg isolate from reused litter harboring IncI1-pST26 that was unexpectedly susceptible to antibiotics predicted by ARGs carried on the plasmid. The isolate (SH-16-0-5B) was confirmed twice by broth microdilution to be susceptible to gentamicin, streptomycin, and tetracycline and once on Mueller-Hinton agar supplemented with various concentrations (0, 2, 4, 8, and 16 mg/L) of gentamicin (see methods in the supplemental material). To ensure that the plasmid contig identified as IncI1-pST26 in this strain was not an artifact of short-read sequencing, we performed qPCR on plasmid DNA and targeted four coding DNA sequences on IncI1-pST26 including its incRNAI, class 1 integron (intI1), aminoglycoside (aadA1), and tetracycline resistance (tetA) genes (see Table S3 in the supplemental material). This confirmed that the strain carried IncI1-pST26. (The qPCR cycle threshold ranged from 20.2 to 27.9 for the four targets.) Afterwards, we used WGS (depth of coverage of IncI1-pST26 contig/depth of coverage of the largest chromosome contig) to compare the copy number of IncI1-pST26 in SH-16-0-5B to the copy number of IncI1-pST26 in four S. Heidelberg isolates (here referred to as SH-IncI1-FL) recovered from the ceca of cloacally inoculated chicks on fresh litter. This revealed that SH-16-0-5B maintained IncI1-pST26 at ∼0.2 copy/cell (i.e., 1 of 5 cells harbored IncI1) compared to 2.8 ± 0.2 copies/cell in SH-IncI1-FL from fresh litter.
To investigate if the copy number difference determined using WGS exists under relevant environmental conditions, we first acclimatized the isolates in prereduced cecal extract (pH 6.5) for 2 h before exposing them to prereduced cecal extract at pH 2.5 under microaerophilic conditions. We reasoned that the low pH of the upper gastrointestinal tract (GIT) of the chicken host will interfere with the copy number of plasmids kept in a bacterial cell, which would have a direct effect on the rate of HGT. The copy number of IncI1-pST26 was significantly lower (0.00005 to 0.21 copy/per cell [i.e., 1 in 20,000 to 1 in 5 cells carried IncI1]) in SH-16-0-5B compared to SH-IncI1-FL (4.94 to 6.65 copies/per cell) at pH 6.5 (W = 9, P = 0.1) and pH 2.5 (W = 81, P = 4.114e−05) (Table 2). These results suggest that Enterobacteriaceae populations in reused litter harbored lower copies of IncI1-pST26 than the populations in fresh litter.
TABLE 2.
IncI1 copy number in S. Heidelberg isolates recovered from fresh and reused litter
| S. Heidelberg strain ID | Time (h) | pH of cecal extract | IncI1 copy no./cell (mean ± SD)a | Coefficient of variation (%) |
|---|---|---|---|---|
| SH-16-0-5B-RL | 2 | 6.52 | 0.001 ± 0.001 | 155.8 |
| SH-IncI1-FL | 2 | 6.52 | 5.594 ± 0.582 | 10.4 |
| SH-16-0-5B-RL | 0.5 | 2.52 | 0.001 ± 0.0003 | 47.9 |
| 4 | 2.52 | 0.141 ± 0.064 | 46.0 | |
| 24 | 2.52 | 0.084 ± 0.038 | 46.4 | |
| SH-IncI1-FL | 0.5 | 2.52 | 6.304 ± 0.165 | 2.6 |
| 4 | 2.52 | 6.301 ± 0.343 | 5.4 | |
| 24 | 2.52 | 5.892 ± 0.378 | 6.4 | |
Three individual S. Heidelberg isolates from fresh litter (FL) were used to determine the copy number of IncI1 for SH-IncI1-FL, while the IncI1 copy number in S. Heidelberg strain SH-16-0-5B from reused litter (RL) was determined using three replicates.
DISCUSSION
Litter is commonly used as a bedding material for raising poultry, and it is ingested by chickens during pecking activities. Therefore, the microbiome of the litter is one of the first inocula that colonize the GIT of broiler chicks. Litter can be managed as single use (i.e., complete removal of litter from a broiler house after each flock), or it can be reused over multiple flocks. The benefits of litter reuse include lower cost for growing chickens and the sustainable management of litter waste (33). It has been shown that chickens raised on reused litter harbor a different microbiome than chicks grown on fresh litter (17, 34) and that reused litter stimulates higher humoral and cell-mediated immune responses than fresh litter in chickens (35). Muniz et al. (23) and Roll et al. (26) showed that the number of litter samples positive for Salmonella significantly decreases as the number of litter reuses increased compared with the first use of the litter.
In this study, we corroborate these findings by showing that neonatal chicks challenged with S. Heidelberg and raised on reused litter were more resistant to S. Heidelberg colonization (i.e., had a lower positivity rate compared to chicks on fresh litter). In addition, we found that chicks raised on reused litter were less likely to be colonized with S. Heidelberg isolates harboring plasmids encoding AMR compared to chicks on fresh litter. We determined that the major difference between chicks grown on fresh and reused litter was the bacterial community harbored in the litter and ceca.
Pathogen-induced gut microbiome modulation can create an imbalance in the abundance of bacterial species (i.e., some species outgrow known resident bacteria), leading to a dysbiosis of the gut microbiome and successful Salmonella colonization and AMR transfer (36). However, gut conditions that allow the maintenance of a diverse and beneficial microbiome are likely to succeed in establishing homeostasis and limit the transfer of AMR (37, 38). In this study, we found that the microbiome of reused litter was associated with a lower HGT of AMR to S. Heidelberg populations in neonatal chicks. The microbiome data led us to draw one main conclusion: that the litter used for growing chicks and the route of Salmonella challenge affected which ASVs increased or decreased in abundance in the ceca and litter.
This was the case for the ASV assigned to Bifidobacterium that increased in the ceca and litter of orally challenged chicks raised on reused litter but was not modulated in cloacally inoculated chicks or in chicks on fresh litter. Consequently, Bifidobacterium was not part of the core microbiome of the ceca or litter in this study (Table S1) and most likely originated with neonatal chicks. Bifidobacterium species are strict anaerobic Actinobacteria and are among the first microbes to colonize the GIT of vertebrates (39). Additionally, Bifidobacterium has been shown to affect the abundance of AMR and virulence genes in the GIT of infants (40–42). Casaburi et al. (42) reported that infants fed a probiotic strain of Bifidobacterium longum harbored lower levels of ARG compared to nonfed controls. Alignment of shotgun metagenomic reads generated from the ceca of broiler chicks (n = 4) to the Kraken database (43) revealed that 21% and 11% of the reads mapping to the genus Bifidobacterium, were aligned to B. longum and Bifidobacterium catenulatum, respectively (see Fig. S5 in the supplemental material). Their increase in abundance for only orally challenged chicks on reused litter suggests that the route S. Heidelberg used for gaining entry into the GIT and the litter microbiome affected how Bifidobacterium was perturbed. Therefore, it is conceivable that the reused litter microbiome created conditions that allowed Bifidobacterium to flourish and proliferate, while the microbiome in fresh litter led to a reduction or elimination of Bifidobacterium species.
We used metagenomics and WGS to show that E. coli was the main reservoir of AMR in the cecal microbiomes of fresh and reused litter. Importantly, we found the identical IncI1-pST26 plasmid, acquired by S. Heidelberg, to be present in E. coli, suggesting that the plasmid was transferred in vivo within chicks raised on fresh litter. The possible sources of the E. coli strains in this study include the chicken host and the broiler house environment. Although we did not establish if the meconium harbored E. coli, it is plausible that post-hatch chicks were colonized with E. coli from the hatchery (44). The evolution of these ancestral E. coli strains would be shaped by selective pressures, such as exposure to antibiotics and metals, and competition with resident microbiota in the environment. For chicks placed on fresh litter, the bacterial community of the first fecal droppings will compete with mostly a pine shaving/plant-associated microbiome. In contrast, chicks placed on reused litter will compete with microbes and metabolites deposited from earlier flocks, including resident E. coli and Salmonella strains that are well adapted to the broiler house environment.
Also, the physiochemical properties of the litter will affect the litter structure, the native microbiome, and the survival of the invading pathogen. The difference in litter physiochemical parameters in this study was inferred using pH and moisture. The pH of reused litter was higher (7.12 ± 0.26) than that of fresh litter (6.54 ± 0.17) (P = 0.028), while fresh litter had higher moisture (20.83% ± 3.54%) than reused litter (14.65% ± 3.51%) (P = 0.11). Litter moisture and pH are known factors that affect pathogen survival and microbial diversity (45, 46). We did not administer antibiotics to chicks in this study and have shown that metals did not significantly affect the metabolism of S. Heidelberg (7). Hence, the litter microbiome and physiochemical properties are the major selection pressure that could explain the evolutionary trajectory of E. coli in this study. This hypothesis was further supported after a functional protein pathway analysis was performed on the cecal metagenome of chicks raised on fresh versus reused litter. Although we measured only two cross-sectional time points for reused litter and fresh litter, the relative abundance of plant metabolic-related pathways in fresh litter compared to the enrichment of antimicrobial molecules in reused litter suggests that a selective pressure gradient exists over time for the colonizing microbiome in reused litter. Thus, the microbiome of reused litter has the antimicrobial capability to competitively exclude invading pathogens, including strains carrying AMR-encoding plasmids.
Plasmid carriage is expected to impose a fitness cost on the host, but plasmid-bearing microbes are pervasive in nature (47). We showed previously that carriage of IncI1-pST26 by S. Heidelberg isolates from fresh litter presented a variable fitness cost to the host (7). We found here that Enterobacteriaceae populations in the ceca of chicks on reused litter harbored lower copies of the IncI1 plasmid per cell compared to chicks on fresh litter. In addition, we did not obtain any E. coli isolate carrying IncI1 from the ceca of chicks raised on reused litter. The limited number of members of the Enterobacteriaceae IncI1 donor population would affect the rate of transfer to S. Heidelberg recipients as direct contact is a requirement for conjugation (48). Nevertheless, we found one S. Heidelberg isolate from the ceca of one chick on reused litter that harbored IncI1-pST26; however, only a fraction of its population harbored the IncI1 plasmid. This isolate did not exhibit the resistance phenotype predicted by the ARGs carried on the IncI1-pST26 plasmid. These results suggest that IncI1 plasmid carriage in reused litter posed a fitness cost on the bacterial host, but this cost was ameliorated by maintaining IncI1 at low copies in our study.
It is important to mention that the litter used for this study is not representative of all broiler litter. Litter compositions may differ between farms, and there are multiple physiochemical factors that can affect the litter microbiome: e.g., moisture and pH, antibiotic usage, and feed additives. Furthermore, the age of the litter (i.e., numbers of flocks that have been grown on the litter) and the length of the downtime between flocks are examples of management practices that may influence microbiome succession. Nonetheless, our study showed that the litter microbiome significantly affects the bacterial diversity of broiler chicks and the HGT of AMR.
MATERIALS AND METHODS
Details of methods used for preparing the S. Heidelberg inocula, challenging neonatal chicks, bacterial and DNA analyses, whole-genome and metagenome sequencing, and bioinformatics have been described before for neonatal chicks raised on fresh litter (7). We briefly redescribe these methods and present others below.
Determining if post-hatch chicks were Salmonella free.
Chick pads that conveyed chicks from the hatchery were preenriched in 500 mL of buffered peptone water (BPW) for 18 to 24 h at 37°C. Two different enrichment broths were used to isolate Salmonella from the BPW preenrichment broths: tetrathionate (TT) broth (Becton, Dickinson, Sparks, MD) and Rappaport-Vassiliadis (RV) medium (Becton, Dickinson). After overnight incubation at 42°C in both enrichment broths, 10-μL aliquots from each enrichment broth were spread on two different differential media—Brilliant green sulfa (BGS) agar (Becton, Dickinson) and xylose lysine tergitol-4 agar (XLT-4) (Becton, Dickinson)—and incubated for 18 to 24 h at 37°C. Isolated colonies characteristic of Salmonella were stabbed and streaked onto triple sugar iron agar (TSI) (Becton, Dickinson) and lysine iron agar fermentation (LIA) (Becton, Dickinson) and incubated for 18 to 24 h at 37°C for biochemical confirmation.
S. Heidelberg inoculum preparation.
The S. Heidelberg strain was made resistant to 200 ppm of nalidixic acid (nalR S. Heidelberg) for selective enumeration and was grown overnight in poultry litter extract, centrifuged, and resuspended in 1× phosphate-buffered saline (PBS). The resuspended cells were used as inocula. The complete genome of the ancestor to nalR S. Heidelberg is available under GenBank accession no. CP066851.
Challenging broiler chicks with nalR S. Heidelberg.
One-day-old Cobb 500 broiler chicks were either uninoculated (n = 25), gavaged (n = 25), or (n = 25) cloacally inoculated with ∼106 CFU of nalR S. Heidelberg. We also included a seeder bird colonization method, whereby five chicks were gavaged and mingled with 20 uninoculated chicks. Afterwards, chicks were placed in floor pens at a stocking density of 0.65 m2/chick on fresh pine shavings (fresh litter, n = 100) or reused litter (n = 100). The reused litter was previously used to raise three flocks of broiler chickens antibiotic free under simulated commercial poultry production conditions and was top dressed with 0.5 cm of fresh pine shavings before the placement of each flock. Broiler chicks on fresh and reused litter were housed separately and were raised for 14 days on antibiotic-free starter diet and water. At 14 days, 45 chicks from each fresh litter and reused litter (10 chicks from the gavaged, cloacal, and uninoculated groups and 15 chickens from the seeder method [5 seeder and 10 contact chicks]) were euthanized to determine the extent of nalR S. Heidelberg colonization in ceca. Additionally, litter samples were collected as grab samples from each pen after chicks were euthanized. The experiments were performed in April 2018. The study was approved by the University of Georgia’s Office of Animal Care and Use under Animal Use Protocol A2017 04-028-A2.
Determination of nalR S. Heidelberg concentration in the ceca and litter.
Ceca were removed from the eviscera of broiler chicks and stomached for 60 s after the addition of 3× volume to the weight (vol/wt) of BPW. Litter was collected as grab samples from seven locations (4 corners of the pen and 3 locations under the waterer) in each pen after chicks were removed. The litter samples were pooled, and 30 g was subsampled in duplicates from each pen as previously described (29). Serial dilutions of the cecal and litter slurry were plated onto BGS containing 200 ppm nalidixic acid, and plates were incubated for 24 h at 37°C.
After incubation, colonies were counted and calculated per gram of ceca or litter dry weight. When no colonies appeared, preenriched cecal and litter slurry was streaked onto BGS agar supplemented with nalidixic acid and incubated overnight. These plates were then examined for the presence/absence of Salmonella colonies. Two to six single colonies were randomly selected from each plate and archived in 30% LB glycerol at −80°C. In addition, cecal slurry was saved at a 4:1 ratio in LB broth (BD Difco, MD, USA) containing 30% glycerol at −80°C, while litter samples were stored in vacuum-sealed Whirl-Pak bags at −20°C. Litter pH and moisture were determined as described previously (29).
Antibiotic resistance phenotype determination.
Antibiotic susceptibility testing was done on S. Heidelberg and E. coli isolates by following the National Antimicrobial Resistance Monitoring System (NARMS) protocol for Gram-negative bacteria. Results were interpreted according to Clinical and Laboratory Standards Institute guidelines and breakpoints established by NARMS. In addition, we used agar dilution to determine the MIC of gentamicin for one S. Heidelberg isolate (SH-16-0.5B) (see methods in the supplemental material).
Genomic, environmental, and plasmid DNA extraction.
Unless otherwise noted, DNA was extracted and purified from bacterial cultures by using FastDNA spin kit for soil (MP Biomedicals, LLC, CA, USA), while 250 mg of cecal slurry (previously saved in LB broth containing glycerol) and litter were extracted with the Qiagen DNeasy PowerSoil DNA kit (Hilden, Germany). Extracted DNA were used for qPCR, 16S rRNA gene sequencing, shotgun metagenomics, and whole-genome sequencing. Plasmid DNA was extracted from two S. Heidelberg isolates that harbored IncI1. Isolates were cultured overnight on sheep blood agar, and plasmid DNA was extracted using the Qiagen Plasmid Midi kit (Qiagen, Inc., Germantown, MD) as per the manufacturer’s instructions. Plasmid DNA was used to construct a calibration curve for IncI1 plasmid copy number determination.
Real-time quantitative PCR.
Real-time qPCR amplification was performed as described previously (45) using a CFX96 Touch real-time PCR detection system (Bio-Rad, Inc., Hercules, CA). Reaction mixtures contained 1× SsoAdvanced Universal SYBR green Supermix (Bio-Rad, Inc., Hercules, CA), 600 nM (each) primers, and 2 μL of DNA. The primers used in this study are shown in Table S3. Unless otherwise stated, primers were designed with Beacon Designer (Premier Biosoft, Palo Alto, CA) and synthesized by Integrated DNA Technologies (Coralville, IA). Calibration curves used for converting qPCR cycle threshold values to gene copies per gram of ceca were determined using the genomic DNA from an E. coli strain that harbored IncI1-pST26 plasmid. To convert qPCR cycle threshold values to IncI1 copies per cell of S. Heidelberg, plasmid DNA of a relevant S. Heidelberg strain harboring IncI1-pST26 was used for calibration curve construction (Table S3). Two primer sets specific to the gapA gene and the region upstream of the repA gene of IncI1, including incRNAI, were used to determine the gene copies of IncI1 in the ceca. gapA is a housekeeping gene carried on the chromosome of enteric bacteria, including E. coli, Klebsiella, Citrobacter, and Salmonella (32). The number of gene copies of IncI1 per cell was determined as the copy ratio of incRNAI to gapA (49).
Whole-genome sequencing and analysis.
Whole-genome sequencing libraries were prepared using either Nextera XT or Nextera DNA Flex library preparation kits (Illumina, Inc., CA, USA) following the manufacturer’s protocol. Libraries were sequenced on the Illumina MiSeq platform with 150- or 250-bp paired-end reads. Additionally, five E. coli isolates were selected for long-read sequencing using the Sequel II system (PacBio Biosciences, Inc.) or MinION device (Oxford Nanopore Technology) (Table S4). Preparation and sequencing of long-read libraries were done by sequencing core centers of University of Georgia and Colorado State University. The method used for read quality control and demultiplexing has been reported previously (7).
Genome assembly, resistome characterization, and quality assessment of assemblies were done using Reads2Resistome pipeline v.1.1.1 (50). For tools used for genome annotation and bacterial and plasmid typing, see the methods in the supplemental material. MAFFT v.1.4.0 (51), implemented in Geneious Prime v.2020.0.1, was used to align and compare sequences. Roary (52) was used for pangenome analysis, and phylogenetic trees were constructed using the maximum likelihood method implemented in RAxML-NG v.1.0.0 (53). IslandViewer (54) was used to predict genomic islands.
Identification of single nucleotide polymorphisms and indels present in S. Heidelberg isolates.
Alignment of raw FASTQ reads to S. Heidelberg (GenBank accession no. CP016573) was done using BWA (v.0.7.17) (55), and SNPs/indels were called using the Genome Analysis Toolkit (56). Variant call format (VCF) files of SNPs/indels and the script used are available in the Dryad Digital Repository (https://doi.org/10.5061/dryad.c866t1g6c).
16S rRNA gene community analysis.
The V4 hypervariable region of the 16S rRNA gene was sequenced using the paired‐end (250 × 2) method on the Illumina MiSeq platform. The cecal (n = 59; 30 and 29 chicks from fresh litter and reused litter, respectively) and litter (n = 22; 11 samples each from fresh and reused litter) samples were part of a larger sequencing run and processed together with other samples. Thus, detailed sequence processing parameters are described in reference 7. To avoid bias introduced by spurious amplicon sequence variants (ASVs) or from samples with low sequencing depth, any ASVs with less than 5 reads and samples with less than 5,000 reads were removed from the data set before further analysis.
Statistical analysis of microbial communities was performed in the R environment using the packages “phyloseq,” “Ampvis2,” and “vegan.” Alpha diversity indices were calculated with a data set rarefied to the minimum sample size (8,740 sequences) and compared using the Wilcoxon signed-rank test. Principal-coordinate analysis (PCoA) based on Bray-Curtis distances was performed with no initial data transformation and after removing ASVs present with a relative abundance of less than 0.1% in any sample. The core microbiome was determined as ASVs with 0.1% relative abundance and present in at least 80% of samples.
Cecal metagenome sequencing.
Two hundred fifty milligrams of cecal slurry from chicks cloacally challenged with S. Heidelberg and raised on fresh litter (n = 2) or reused litter (n = 2) was used to generate the shotgun library and Hi-C DNA library. For the shotgun library, cecal DNA was extracted with the Qiagen DNeasy PowerSoil DNA kit (Hilden, Germany), and the Nextera XT library preparation kit was used to make the library. The Hi-C library was made by the Phase Genomics (Seattle, WA, USA) ProxiMeta Hi-C microbiome kit following the manufacturer's instructions. Libraries were sequenced by the Novogene Corporation (Sacramento, CA, USA) on the Illumina HiSeq platform using 150-bp paired-end reads. Two libraries were sequenced per HiSeq flow cell lane, resulting in a total of ∼111 million shotgun reads, with ∼213 million Hi-C reads per sample. Quality-controlled shotgun reads were classified using Kraken2 (v.2.0.8 beta) (43) to create a count profile of the metagenome. We used cumulative sum scaling implemented in metagenomeSeq to normalize read counts (57).
Metagenome-assembled genomes and associated plasmids and ARGs.
The details of the methods used for proximity-guided metagenome assembly and deconvolution have been described before (7). Briefly, shotgun metagenomics sequence reads were filtered and trimmed for quality and normalized before assembly with metaSPAdes (58) using default options. Hi-C sequence reads were then aligned to the assembly following the ProxiMeta Hi-C kit manufacturer’s recommendations (https://phasegenomics.github.io/2019/09/19/hic-alignment-and-qc.html). Metagenome deconvolution was performed with ProxiMeta (59), resulting in the creation of putative genome and genome fragment clusters. Hi-C clusters were assessed for quality using CheckM (60) and assigned taxonomic classifications with Mash v.2.1 (61), resulting in the metagenome-assembled genomes (MAGs). We used PlasmidFinder (v.2.1.1) (62) to identify plasmids present in MAGs.
To view AMR gene connections to hosts, AMR genes in the metagenomic assembly were annotated using NCBI’s AMRFinderPlus software (version: v.3.10.5) with the –plus option specified and all other options set to defaults. The AMR assembly annotations were then used in combination with the MAG taxonomic annotations and the mobile element to host association matrices generated by ProxiMeta to annotate which AMR genes are present in each MAG, and specify whether the gene originated from a genomic, viral or plasmid contig. This was done by making an NxM matrix, C, where and , where and , where . The matrix (C) was then filled as follows:
and where , , . The matrix, C, was then filtered to remove rows and columns with only 0 values and plotted using a color-encoding system indicating whether the AMR gene was of genomic (cyan) or plasmid (gold) origin.
Functional pathway analysis.
Both shotgun sequences and ProxiMeta-deconvoluted genomes were also utilized to assess enrichment of functional pathways. Shotgun metagenomic sequences were aligned to the Hi-C genomic contigs by using the BWA-MEM algorithm as implemented in BWA (v.0.7.17) (55). The Hi-C genomic contigs were then annotated using Prokka (v.1.13) (63). For each annotated region on the Hi-C genomic contigs, sequences overlapping the annotated region in the sequence alignments were counted using a custom Python script to produce a count matrix of genes for all samples. Gene counts were normalized by gene length and using total sum scaling normalization to control for differences in sequencing depth (57). Enzyme commission identifiers were then mapped to KEGG functional pathways according to the KEGG ontology. For each pathway, an adjacency graph was reconstructed from the KEGG XML files, and connected components were determined using depth-first search (64). Z-scores were calculated for pathways and connected components for each sample, and statistical testing was then performed by comparing scores to the normal distribution. Details of this analysis are described in the methods in the supplemental material.
E. coli isolation from ceca.
The method used for screening frozen cecal contents has been reported previously (7). Briefly, two cecal slurries from challenged chicks raised on fresh litter and reused litter were spread plated onto CHROMagar plates supplemented with or without gentamicin (8 ppm) and tetracycline (8 ppm) or ampicillin (16 ppm) and cefoxitin (16 ppm) (Thermo Fisher, Waltham, MA). After 24 h of incubation, blue-green and blue-cream colonies were counted as presumptive E. coli, and 4 to 5 colonies from each spread plate were picked and restruck for colony purification. No colonies grew on CHROMagar plates supplemented with ampicillin and cefoxitin. Colonies were preserved in LB broth containing glycerol and frozen before they were used for inclusion of a solid agar competition experiment.
Determination of plasmid copy number of IncI1.
To compare the copy number of IncI1 harbored by S. Heidelberg strain (SH-16-0-5B) from reused litter to the copy number of IncI1 in three SH-IncI1-FL isolates from fresh litter, we exposed each isolate to acidified filter sterilized cecal extract (CE). CE was prepared from the cecal content of 2-week-old broiler chickens (see methods in the supplemental material). Cecal extracts were prereduced by covering tubes with a gas-permeable paper strip and incubating them overnight under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) at 42°C. Three single colonies of each strain were selected from overnight cultures grown on sheep blood agar and transferred to a microcentrifuge tube containing 900 μL of prereduced CE (pH 6.52): i.e., one tube for each SH-IncI1-FL strain and three tubes for SH-16-0-5B.
After transfer, tubes were vortexed, covered with a gas-permeable paper strip, and incubated at 41°C under microaerophilic conditions for 2 h. After incubation, tubes were vortexed and 100 μL of the suspension was transferred to a microcentrifuge tube containing 900 μL of CE (with the pH of CE adjusted to 2.52 using 1 M HCl [Spectrum Chemical Mfg. Corp., CA, USA] and 1 M NaOH [Fisher Chemical, NJ, USA]) Afterwards, tubes were vortexed, and incubation continued for another 24 h. To determine the copy number of IncI1, one replicate tube per strain was removed at time points 2 h for pH 6.5 and 0.5, 4, and 24 h for pH 2.5 and used for DNA extraction. DNA was extracted from 500 to 800 μL of CE samples using FastDNA Spin kit for soil. Calibration curves for gapA and incRNAI were generated using genomic and plasmid DNA extracted from ancestral S. Heidelberg (GenBank accession no. CP066851) and one SH-IncI1-FL isolate, respectively. The plasmid copy number of IncI1 was determined as the copy ratio of incRNAI to gapA (49).
Statistical analyses.
Continuous variables were log transformed before any statistical tests were performed. Moreover, continuous variables did not meet the assumption of a normal distribution; therefore, nonparametric testing for direct comparisons was performed using Wilcoxon rank sum and signed-rank tests, and the Kruskal-Wallis rank sum test was used for one-way analysis of variance tests. Additionally, a generalized linear model with a binomially distributed outcome and a log link function was performed as described earlier (65) to determine if there are significant differences in Salmonella prevalence (presence/absence) and the litter used for raising broiler chicks. The significance of the model was established using a likelihood ratio test (R function ANOVA with argument test set to “Chisq”). Multiple comparisons of means (i.e., regression coefficients for route of inoculation and litter age/type) were done using the multcomp package in R. The mcp function was used to specify linear hypotheses, and the glht function was used to make Tukey contrasts. Statistical analyses were performed using R (v.4.0.3).
Ethics statement.
All animal experiments were approved by the University of Georgia Office of Animal Care and Use under Animal Use Protocol A2017 04-028-A2.
Data availability.
All raw short and long FASTQ reads for S. Heidelberg are publicly available under NCBI accession no. PRJNA683658, while E. coli FASTQ reads are available under NCBI accession no. PRJNA684578. Shotgun and Hi-C reads from fresh litter and reused litter cecal samples are publicly available under NCBI accession no. PRJNA688069, and 16S rRNA gene sequences are available under NCBI accession no. PRJNA669215. The complete genome assemblies for E. coli strains Ec-FL1-1X and Ec-FL1-2X were previously published and are available under GenBank accession no. CP066836 and JAFCXR000000000. The complete genome of E. coli strain Ec-RL2-1X has been made available under GenBank accession no. CP066839. Variant call format (VCF) files of identified SNPs/indels and the Linux/Unix shell script used have been deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.c866t1g6c).
ACKNOWLEDGMENTS
We are grateful to Marlo Sommers, Carolina Hall, Jeromey Jackson, and Latoya Wiggins for logistical and technical help.
This work was supported by USDA Agricultural Research Service (Project no. 6040‐32000‐010‐00‐D), a Non-assistance Cooperative Agreement (58-6040-6-030) between the USDA Agricultural Research Service and University of Georgia, Research Foundation, and a Research Service Agreement (58-6040-8-035) between the USDA Agricultural Research Service and Colorado State University.
This study was supported in part by resources and technical expertise from the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia’s Office of the Vice President for Research and Office of the Vice President for Information Technology. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the USDA or the National Science Foundation, and any mention of products or trade names does not constitute recommendation for use. I.L., M.O.P., and J.R.G. are employees of Phase Genomics, Inc., and have been supported in part by NIAID grants R44AI162570 and R44AI150008. The remaining authors declare no conflict of interest.
A.O. and Z.A. designed the study. A.O., G.Z., N.A.C., and C.R. performed live broiler chicken studies. M.J.R. Jr contributed to the design of experiments performed with E. coli strains. A.O., J.L., G.Z., and D.C. performed bacteriological analyses, antibiotic susceptibility testing, DNA extraction and qPCR. G.Z. and A.O. performed Illumina whole-genome sequencing. A.O. performed cecal shotgun and Hi-C library preparation. T.L. made 16S rRNA gene libraries and sequencing. B.Z. performed 16S rRNA bacterial community analysis and interpretation. A.O., Z.A., M.O.P., I.L., J.R.G., C.W., J.C.T. IV, S.M.L., and R.W. performed bioinformatic analyses and data curation. A.O., J.L., and D.C. performed the solid agar mating experiment. A.O. and B.Z. performed statistical analyses. A.O., Z.A., N.A.C., B.Z., R.W., M.O.P., J.R.G., S.M.L., G.Z., and J.L. drafted the manuscript, which was reviewed and edited by all authors. A.O., Z.A., and S.E.A. supervised the study.
Footnotes
Supplemental material is available online only.
Contributor Information
Adelumola Oladeinde, Email: ade.oladeinde@usda.gov.
Zaid Abdo, Email: zaid.abdo@colostate.edu.
Charles M. Dozois, INRS—Institut Armand-Frappier
REFERENCES
- 1.Von Wintersdorff CJ, Penders J, Van Niekerk JM, Mills ND, Majumder S, Van Alphen LB, Savelkoul PH, Wolffs PF. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol 7:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventola C. 2015. The antibiotic resistance crisis. Part 1. Causes and threats. PT 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.Angulo FJ, Baker NL, Olsen SJ, Anderson A, Barrett TJ. 2004. Antimicrobial use in agriculture: controlling the transfer of antimicrobial resistance to humans. Semin Pediatr Infect Dis 15:78–85. 10.1053/j.spid.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, Polachek AJ, Ganshorn H, Sharma N, Kellner JD, Ghali WA. 2017. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health 1:e316–e327. 10.1016/S2542-5196(17)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer R, Porter L. 2019. Estimates of on-farm antimicrobial usage in broiler chicken and turkey production in the United States, 2013–2017. Mindwalk Consulting Group, LLC. https://www.uspoultry.org/poultry-antimicrobial-use-report/docs/USPOULTRY_Antimicrobial-Report.pdf. [DOI] [PubMed]
- 6.Kirchhelle C. 2018. Swann song: antibiotic regulation in British livestock production (1953–2006). Bull Hist Med 92:317–351. 10.1353/bhm.2018.0029. [DOI] [PubMed] [Google Scholar]
- 7.Oladeinde A, Abdo Z, Press MO, Cook K, Cox NA, Zwirzitz B, Woyda R, Lakin SM, Thomas J, Looft T, Cosby DE, Hinton A, Jr, Guard J, Line E, Rothrock MJ, Berrang ME, Herrington K, Zock G, Plumblee Lawrence J, Cudnik D, House S, Ingram K, Lariscy L, Wagner M, Aggrey SE, Chai L, Ritz C. 2021. Horizontal gene transfer is the main driver of antimicrobial resistance in broiler chicks infected with Salmonella enterica serovar Heidelberg. mSystems 6:e0072921. 10.1128/mSystems.00729-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, Rosen H, Libby T, Kissler B, Marsden-Haug N, Booth H, Kimura A, Grass J, Bicknese A, Tolar B, Defibaugh-Chavez S, Williams I, Wise M, Salmonella Heidelberg Investigation Team. 2016. National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One 11:e0162369. 10.1371/journal.pone.0162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edirmanasinghe R, Finley R, Parmley EJ, Avery BP, Carson C, Bekal S, Golding G, Mulvey MR. 2017. A whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob Agents Chemother 61:e01919-16. 10.1128/AAC.01919-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceccarelli D, van Essen-Zandbergen A, Smid B, Veldman KT, Boender GJ, Fischer EAJ, Mevius DJ, van der Goot JA. 2017. Competitive exclusion reduces transmission and excretion of extended-spectrum-beta-lactamase-producing Escherichia coli in broilers. Appl Environ Microbiol 83:e03439-16. 10.1128/AEM.03439-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chantziaras I, Smet A, Filippitzi ME, Damiaans B, Haesebrouck F, Boyen F, Dewulf J. 2018. The effect of a commercial competitive exclusion product on the selection of enrofloxacin resistance in commensal E. coli in broilers. Avian Pathol 47:443–454. 10.1080/03079457.2018.1486027. [DOI] [PubMed] [Google Scholar]
- 12.Raffatellu M. 2018. Learning from bacterial competition in the host to develop antimicrobials. Nat Med 24:1097–1103. 10.1038/s41591-018-0145-0. [DOI] [PubMed] [Google Scholar]
- 13.Achard CS, Dupouy V, Siviglia S, Arpaillange N, Cauquil L, Bousquet-Mélou A, Zemb O. 2019. Variability of the ability of complex microbial communities to exclude microbes carrying antibiotic resistance genes in rabbits. Front Microbiol 10:1503. 10.3389/fmicb.2019.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumgartner M, Bayer F, Pfrunder-Cardozo KR, Buckling A, Hall AR. 2020. Resident microbial communities inhibit growth and antibiotic-resistance evolution of Escherichia coli in human gut microbiome samples. PLoS Biol 18:e3000465. 10.1371/journal.pbio.3000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dame-Korevaar A, Fischer EA, van der Goot J, Velkers F, Ceccarelli D, Mevius D, Stegeman A. 2020. Early life supply of competitive exclusion products reduces colonization of extended spectrum beta-lactamase-producing Escherichia coli in broilers. Poult Sci 99:4052–4064. 10.1016/j.psj.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Methner U, Rösler U. 2020. Efficacy of a competitive exclusion culture against extended-spectrum β-lactamase-producing Escherichia coli strains in broilers using a seeder bird model. BMC Vet Res 16:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Lilburn M, Yu Z. 2016. Intestinal microbiota of broiler chickens as affected by litter management regimens. Front Microbiol 7:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueroa A, Derksen T, Biswas S, Nazmi A, Rejmanek D, Crossley B, Pandey P, Gallardo R. 2020. Persistence of low and highly pathogenic avian influenza virus in reused poultry litter, effects of litter amendment use, and composting temperatures. J Appl Poult Res 30:100096. 10.1016/j.japr.2020.09.011. [DOI] [Google Scholar]
- 19.Wadud S, Michaelsen A, Gallagher E, Parcsi G, Zemb O, Stuetz R, Manefield M. 2012. Bacterial and fungal community composition over time in chicken litter with high or low moisture content. Br Poult Sci 53:561–569. 10.1080/00071668.2012.723802. [DOI] [PubMed] [Google Scholar]
- 20.Council National Farm Animal Care Council. 2016. Code of practice for the care and handling of hatching eggs, breeders, chickens, and turkeys. https://www.nfacc.ca/poultry-code-of-practice.
- 21.ADAS, Ltd. 2016. Comparison of the regulatory framework and key practices in the poultry meat supply chain in the EU and USA: a study for the Association of Poultry Processors and Poultry Trade in the EU Countries. https://britishpoultry.org.uk/identity-cms/wp-content/uploads/2018/05/2016-ADAS-EU-US-comparison.pdf.
- 22.Dornelas KC, Henrique Mascarenhas NM, Soares Rodrigues HC, do Nascimento RT, dos Santos Lima de Brito AN, Furtado DA, do Nascimento JWB. 2020. Chicken bed: a review on reuse, treatment and influence on ambience. Poult Sci. 10.1016/j.psj.2020.09.067. [DOI] [Google Scholar]
- 23.Muniz E, Mesa D, Cuaspa R, Souza AM, Santin E. 2014. Presence of Salmonella spp. in reused broiler litter. Rev Colombiana Ciencias Pecuarias 27:12–17. [Google Scholar]
- 24.Yamak US, Sarica M, Boz MA, Ucar A. 2016. Effect of reusing litter on broiler performance, foot-pad dermatitis and litter quality in chickens with different growth rates. Kafkas Univ Vet Fak J 22:85–91. [Google Scholar]
- 25.Casey R, Fairchild B, Lacy M. 2005. Litter quality and broiler performance. UGA Extension Bulletin 1267. https://secure.caes.uga.edu/extension/publications/files/pdf/B%201267_5.PDF?msclkid=d3024dcda53f11eca99a970af80dbcfb.
- 26.Roll VF, Dai Pra MA, Roll AP. 2011. Research on Salmonella in broiler litter reused for up to 14 consecutive flocks. Poult Sci 90:2257–2262. 10.3382/ps.2011-01583. [DOI] [PubMed] [Google Scholar]
- 27.Zucko J, Starcevic A, Diminic J, Oros D, Mortazavian AM, Putnik P. 2020. Probiotic—friend or foe? Curr Opin Food Sci 32:45–49. 10.1016/j.cofs.2020.01.007. [DOI] [Google Scholar]
- 28.Gupta S, Allen-Vercoe E, Petrof EO. 2016. Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol 9:229–239. 10.1177/1756283X15607414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oladeinde A, Cook K, Orlek A, Zock G, Herrington K, Cox N, Plumblee Lawrence J, Hall C. 2018. Hotspot mutations and ColE1 plasmids contribute to the fitness of Salmonella Heidelberg in poultry litter. PLoS One 13:e0202286. 10.1371/journal.pone.0202286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bucher MG, Zwirzitz B, Oladeinde A, Cook K, Plymel C, Zock G, Lakin S, Aggrey SE, Ritz C, Looft T, Lipp E, Agga GE, Abdo Z, Sistani KR. 2020. Reused poultry litter microbiome with competitive exclusion potential against Salmonella Heidelberg. J Environ Qual 49:869–881. 10.1002/jeq2.20081. [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer E, de Sousa JAM, Touchon M, Rocha EPC. 2021. Bacteria have numerous distinctive groups of phage-plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res 49:2655–2673. 10.1093/nar/gkab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wertz JE, Goldstone C, Gordon DM, Riley MA. 2003. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J Evol Biol 16:1236–1248. 10.1046/j.1420-9101.2003.00612.x. [DOI] [PubMed] [Google Scholar]
- 33.Wiedemann S. 2015. Litter reuse: an evidence-based guide to reusing litter. Vol. project PRJ-006440, publication 14/095. Rural Industries Research and Development Corporation (RIRDC): Canberra, ACT. https://www.agrifutures.comau/product/litter-reuse-an-evidence-based-guide-to-reusing-litter/. Accessed 19 March 2019.
- 34.Cressman MD, Yu Z, Nelson MC, Moeller SJ, Lilburn MS, Zerby HN. 2010. Interrelations between the microbiotas in the litter and in the intestines of commercial broiler chickens. Appl Environ Microbiol 76:6572–6582. 10.1128/AEM.00180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K, Lillehoj H, Jang S, Lee S, Bautista D, Siragusa G. 2013. Effect of Bacillus subtilis-based direct-fed microbials on immune status in broiler chickens raised on fresh or used litter. Asian Australas J Anim Sci 26:1592–1597. 10.5713/ajas.2013.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt W-D. 2012. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci USA 109:1269–1274. 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gagliardi A, Totino V, Cacciotti F, Iebba V, Neroni B, Bonfiglio G, Trancassini M, Passariello C, Pantanella F, Schippa S. 2018. Rebuilding the gut microbiota ecosystem. Int J Environ Res Public Health 15:1679. 10.3390/ijerph15081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montassier E, Valdes-Mas R, Batard E, Zmora N, Dori-Bachash M, Suez J, Elinav E. 2021. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat Microbiol 6:1043–1054. 10.1038/s41564-021-00920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Callaghan A, Van Sinderen D. 2016. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol 7:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casaburi G, Frese SA. 2018. Colonization of breastfed infants by Bifidobacterium longum subsp. infantis EVC001 reduces virulence gene abundance. Human Microbiome J 9:7–10. 10.1016/j.humic.2018.05.001. [DOI] [Google Scholar]
- 41.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA. 2018. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere 3:e00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casaburi G, Duar RM, Vance DP, Mitchell R, Contreras L, Frese SA, Smilowitz JT, Underwood MA. 2019. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulsen LL, Thofner I, Bisgaard M, Christensen JP, Olsen RH, Christensen H. 2017. Longitudinal study of transmission of Escherichia coli from broiler breeders to broilers. Vet Microbiol 207:13–18. 10.1016/j.vetmic.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 45.de Toledo TD, Roll AAP, Rutz F, Dallmann HM, Pra MAD, Leite FPL, Roll VFB. 2020. An assessment of the impacts of litter treatments on the litter quality and broiler performance: a systematic review and meta-analysis. PLoS One 15:e0232853. 10.1371/journal.pone.0232853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joerger RD, Ganguly A, de Los Santos M, Li H. 2020. Effect of sodium bisulfate amendments on bacterial populations in broiler litter. Poult Sci 99:5560–5571. 10.1016/j.psj.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brockhurst MA, Harrison E. 2021. Ecological and evolutionary solutions to the plasmid paradox. Trends Microbiol. 10.1016/j.tim.2021.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Virolle C, Goldlust K, Djermoun S, Bigot S, Lesterlin C. 2020. Plasmid transfer by conjugation in Gram-negative bacteria: from the cellular to the community level. Genes 11:1239. 10.3390/genes11111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee C, Kim J, Shin SG, Hwang S. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol 123:273–280. 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 50.Woyda RR, Oladeinde A, Abdo Z. 2020. Reads2Resistome: an adaptable and high-throughput whole-genome sequencing pipeline for bacterial resistome characterization. bioRxiv. 10.1101/2020.05.18.102715. [DOI]
- 51.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozlov A, Darriba D, Flouri T, Morel B, Stamatakis A. 2018. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. bioRxiv. 10.1101/447110. [DOI] [PMC free article] [PubMed]
- 54.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FSL, Simon Fraser University Research Computing Group. 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paulson JN, Stine OC, Bravo HC, Pop M. 2013. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–1202. 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Press MO, Wiser AH, Kronenberg ZN, Langford KW, Shakya M, Lo C-C, Mueller KA, Sullivan ST, Chain PS, Liachko I. 2017. Hi-C deconvolution of a human gut microbiome yields high-quality draft genomes and reveals plasmid-genome interactions. bioRxiv 198713. 10.1101/198713. [DOI]
- 60.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carattoli A, Zankari E, Garcia-Fernandez A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 64.Csardi G, Nepusz T. 2006. The igraph software package for complex network research. InterJournal Complex Systems 1695:1–9. [Google Scholar]
- 65.Rothrock MJ, Guard JY, Oladeinde A. 2021. Salmonella diversity along the farm-to-fork continuum of pastured poultry flocks in the Southeastern United States. Front Anim Sci. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download aem.02517-21-s0001.xlsx, XLSX file, 0.02 MB (19.7KB, xlsx)
Supplemental materials and methods, Tables S2 and S3, and Fig. S1 to S5. Download aem.02517-21-s0002.pdf, PDF file, 1.8 MB (1.1MB, pdf)
File S1. Download aem.02517-21-s0003.xlsx, XLSX file, 0.02 MB (21.9KB, xlsx)
Data Availability Statement
All raw short and long FASTQ reads for S. Heidelberg are publicly available under NCBI accession no. PRJNA683658, while E. coli FASTQ reads are available under NCBI accession no. PRJNA684578. Shotgun and Hi-C reads from fresh litter and reused litter cecal samples are publicly available under NCBI accession no. PRJNA688069, and 16S rRNA gene sequences are available under NCBI accession no. PRJNA669215. The complete genome assemblies for E. coli strains Ec-FL1-1X and Ec-FL1-2X were previously published and are available under GenBank accession no. CP066836 and JAFCXR000000000. The complete genome of E. coli strain Ec-RL2-1X has been made available under GenBank accession no. CP066839. Variant call format (VCF) files of identified SNPs/indels and the Linux/Unix shell script used have been deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.c866t1g6c).