Abstract
Despite SARS-CoV-2 being a “novel” virus, early detection of anti-spike IgG in severe COVID-19 patients may be caused by the amplification of humoral memory responses against seasonal coronaviruses. Here, we examine this phenomenon by characterizing anti-spike IgG responses in non-hospitalized convalescent individuals across a spectrum of COVID-19 severity. We observe that disease severity positively correlates with anti-spike IgG levels, IgG cross-reactivity against other betacoronaviruses (β-CoVs), and FcγR activation. Analysis of IgG targeting β-CoV-conserved and non-conserved immunodominant epitopes within the SARS-CoV-2 spike protein revealed epitope-specific relationships: IgG targeting the conserved heptad repeat (HR) 2 region significantly correlates with milder disease, while targeting the conserved S2′FP region correlates with more severe disease. Furthermore, a lower HR2-to-S2′FP IgG-binding ratio correlates with greater disease severity, with ICU-hospitalized COVID-19 patients showing the lowest HR2/S2′FP ratios. These findings suggest that HR2/S2′FP IgG profiles may predict disease severity and offer insight into protective versus deleterious humoral recall responses.
Keywords: COVID-19, SARS-CoV-2, Fc-gamma receptor activation, heptad repeat region, fusion protein region, disease severity, antibody-mediated effector responses, ADE, SARS1, OC43, human betacoronavirus
Graphical abstract
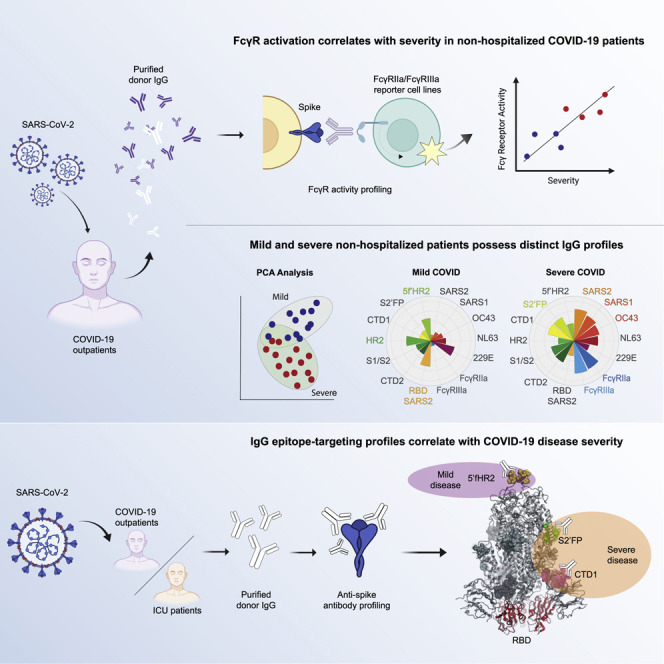
Garrido et al. find that humoral memory responses against seasonal coronaviruses contribute to COVID-19 disease severity, conferring either protection or risk, depending on epitope targeting. These data suggest an explanatory mechanism underlying the atypical bimodality of COVID-19 disease severity, the observation of which was previously obscured by aggregate epitope analysis.
Introduction
During the first waves of the coronavirus disease 2019 (COVID-19) pandemic, approximately 80% of individuals experienced asymptomatic or mild disease, 15% experienced moderately severe pneumonia, and 5% required hospitalization with severe acute respiratory distress syndrome (ARDS) (Wu and McGoogan, 2020). Studies conducted in patients with severe COVID-19 commonly observed a hyperimmune activation state (Del Valle et al., 2020; Tang et al., 2020; Ye et al., 2020), which coincided with the onset of adaptive immunity. In particular, several studies correlated early detection and higher anti-spike protein immunoglobulin G (IgG) titers with more severe disease (Long et al., 2020; Qu et al., 2020; Young et al., 2020; Zhang et al., 2020; Zhao et al., 2020), suggesting that humoral immunity may exacerbate COVID-19. Altogether, these findings have led several researchers to hypothesize that adaptive antibody (Ab) immunity may exacerbate disease severity through Ab-dependent enhancement (ADE) of disease (Iwasaki and Yang, 2020; Larsen et al., 2020; Lee et al., 2020; Ricke, 2021).
As a category, ADE refers to the processes by which pathogen-specific Abs increase virus replication (Ab-dependent enhancement of infection) and/or proinflammatory mediators (Ab-dependent immune enhancement [ADI]), both of which can enhance the severity of disease (Iwasaki and Yang, 2020). The effects of ADE have been observed in the context of several viral infections, including dengue, respiratory syncytial virus (RSV), and even other coronaviruses (Bournazos et al., 2020; Halstead, 2014; Smatti et al., 2018). Studies examining dengue-induced ADE have identified IgG-FcγR interactions as one of the primary factors governing increased disease severity (Mohsin et al., 2015; Thulin et al., 2020; Wang et al., 2017). Fcγ receptors (FcγRs) are a family of IgG-binding receptors that trigger a diverse array of non-neutralizing effector functions, such as Ab-dependent cellular cytotoxicity (ADCC), Ab-dependent cellular phagocytosis (ADCP), dendritic cell (DC) maturation and antigen presentation, and effector cytokine production (Forthal and Moog, 2009; Pincetic et al., 2014; Vogelpoel et al., 2015). Though FcγR activation is of critical importance in controlling viral infections in vivo, in the context of pathogens that induce ADE, high IgG-mediated FcγR activation has been shown to be detrimental. Notably, several studies have reported ADE/ADI effects during severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) infections in vitro and in vivo (Jaume et al., 2011; Liu et al., 2019; Wang et al., 2014; Yip et al., 2014). In contrast, vaccine studies in hamsters demonstrated that despite FcγR-IgG-mediated cellular uptake of virus, vaccinated animals were protected in viral challenge experiments (Kam et al., 2007). Therefore, the contribution of FcγR-IgG in the resolution of severe coronavirus infection remains unclear.
In most instances of virus-induced ADE, immunological memory responses from a previous infection become activated and mount inefficient and detrimental responses to a similar but distinct heterologous strain. While SARS-CoV-2 has now evolved variant strains that could act like heterologous strains, these strains arose only several months after the initial outbreak and cannot account for any ADE-like effects observed early in the pandemic. However, seasonal human coronaviruses (shCoVs), which are ubiquitous (Gorse et al., 2010), share some regions of high sequence identity with the SARS-CoV-2 spike protein, the primary viral antigen targeted by neutralizing Abs. In particular, the S2 subunit of the SARS-CoV-2 spike contains regions of high sequence conservation with shCoVs. In contrast, the receptor-binding domain (RBD)-containing S1 subunit, which interacts with host angiotensin-converting enzyme 2 (ACE2) receptor, bears far less sequence similarity to shCoVs. Importantly, several studies have shown that Abs targeting the RBD region mediate potent neutralization of SARS-CoV-2 in vitro and in vivo (Rogers et al., 2020; Wang et al., 2020b; Wec et al., 2020; Zost et al., 2020). Indeed, recent studies have found that pre-pandemic and SARS-CoV-2-naive samples possess IgG directed against the S2, but not the S1, subunit of the spike protein (Anderson et al., 2021; Ng et al., 2020; Nguyen-Contant et al., 2020). Notably, these sera are non-neutralizing and display high reactivity against the shCoV, OC43 (Anderson et al., 2021). In addition, other studies have observed that cross-reactive Ab responses against OC43 are elevated after SARS-CoV-2 infections (Shrock et al., 2020; Wang et al., 2020c). However, the contribution of shCoV cross-reactive IgG to the humoral response against SARS-CoV-2 remains unclear, with different studies showing both positive and negative correlations with severity (Anderson et al., 2021; Shrock et al., 2020; Wang et al., 2020c).
Beyond the characterization of IgG S1- versus S2-binding, other studies have conducted peptide walks to identify the linear epitopes targeted by humoral responses against the spike protein. These studies have identified several immunodominant regions that overlap or flank several functional features, such as the S1/S2 furin cleavage site (S1/S2), the S2′ fusion peptide region (S2′FP), and the heptad repeat (HR) 1 region and HR2 sites (Li et al., 2021; Mishra et al., 2021; Wang et al., 2020a; Zamecnik et al., 2020). Interestingly, some of these immunodominant regions possess high sequence identity with shCoVs (Ladner et al., 2021; Shrock et al., 2020). These findings—coupled with IgG recognition of the S2 region in naive individuals and the elevated levels of OC43 cross-reactive IgG in convalescent individuals—suggest that preexisting recall responses against shCoVs likely contribute to the humoral response against SARS-CoV-2. However, the contribution of these cross-reactive Abs to the evolution of effective humoral immunity, disease severity, and outcomes remains unclear. In this study, we examined how anti-spike IgG responses correlate with disease severity, FcγR activation, and epitope targeting in a cohort of non-hospitalized SARS-CoV-2 convalescent donors, as well as in intensive care unit (ICU)-hospitalized COVID-19 patients.
Results
Non-hospitalized SARS-CoV-2 convalescent individuals display a spectrum of COVID-19 symptom severity
To obtain a better understanding of the humoral responses generated against SARS-CoV-2, a total of 48 blood donors were recruited during the first wave of the COVID-19 pandemic in the spring of 2020. The cohort was separated into two groups based on the history of a positive or negative SARS-CoV-2 test (PCR or serology): convalescent donors (positive test; convalescent) and negative controls (negative test; naive). The negative control group was composed of age- and sex-matched SARS-CoV-2-naive donors (Table S1). Importantly, in convalescent donors, the median time from the onset of symptoms to the time of blood draw was 43 days, ensuring that anti-SARS-CoV-2 IgG levels were sufficiently high for pathogen-specific IgG analysis.
To assess the relative severity of disease, we surveyed the symptom history of each convalescent donor, including the intensity and duration of each symptom. This information was used to calculate both an average severity score for each symptom (Table S2) and a composite symptom severity score for every convalescent donor. In doing so, we observed that the convalescent group presented a wide range of symptoms and severity ranging from mild to more severe COVID-19. Moreover, we observed two categories of convalescent donors: those who experienced milder symptoms (n = 13), with composite severity scores below 45, and those with more severe disease (n = 15), with scores above 45. For simplicity, these two groups are henceforth referred to as mild and severe, respectively, as they reflect these two ends of the non-hospitalized COVID-19 spectrum, with the acknowledged caveat that all of the donors represent non-hospitalized, non-fatal COVID-19 cases. Despite not being hospitalized, individuals with the highest severity scores (>45) commonly experienced high fevers (>37.8°C) for more than 1 week, severe myalgia, headaches, and difficulty breathing. In addition, two severe patients experienced weight loss of >15% of body weight.
COVID-19 severity correlates with higher anti-spike Ab titers in SARS-CoV-2 convalescent individuals
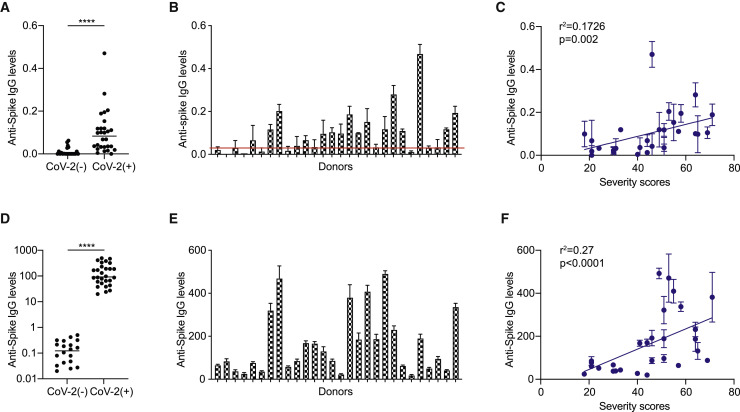
Next, we quantified the levels of IgG directed against the SARS-CoV-2 spike protein in convalescent donors by ELISA (Okba et al., 2020). We detected significantly higher levels of anti-spike IgG in the convalescent versus SARS-CoV-2-naive donors (Figure 1 A). Among the convalescent donors, we observed a range of anti-spike IgG levels, which differed by approximately 30-fold between the lowest and highest samples (Figure 1B). Interestingly, 2 donors who tested positive for SARS-CoV-2 infection by PCR possessed no detectable anti-spike titers and 8 additional convalescent donors possessed titers that were less than 3-fold above background, which was comparable to the anti-spike levels observed in some SARS-CoV-2-naive donors. In comparing anti-spike IgG levels against convalescent donor severity scores, we observed a significant positive correlation (Figure 1C). In line with these findings, several studies have found similar correlations between anti-spike IgG and COVID-19 severity in hospitalized patients (Liu et al., 2020; Qu et al., 2020; Young et al., 2020; Zhang et al., 2020).
Figure 1.
Anti-spike IgG levels in convalescent donors correlate with COVID-19 severity
(A) Anti-spike IgG titers in SARS-CoV-2 convalescent (CoV-2+) and naive (CoV-2−) donors, as quantified by ELISA (n = 28 and 20, respectively).
(B) Levels of anti-spike IgG in convalescent (CoV-2+) donors, as quantified by ELISA. Red line represents 3-fold above the mean anti-spike levels of naive (CoV-2−) donors, as quantified by ELISA.
(C) Comparison of ELISA-based anti-spike IgG titers versus CoV-2+ composite severity scores.
(D–F) Levels of anti-spike IgG in naive (CoV-2−) and CoV-2+ donors, as quantified using a cell-based assay (CBA).
(D) Levels of anti-spike IgG titers in CoV-2− and CoV-2+ donors.
(E) Levels of anti-spike titers in CoV-2+ donors, as quantified by CBA.
(F) Comparison of CBA anti-spike IgG titers versus CoV-2+ composite severity scores. The SEMs of N = 3 experiments are shown.
In addition to neutralization, non-neutralizing effector IgG responses against virus-infected host cells involve the recognition of viral antigens expressed on the surface of host cells (Forthal and Moog, 2009). Therefore, we quantified the levels of anti-spike IgG binding to cell surface-expressed forms of the SARS-CoV-2 spike in 293T endothelial cells transfected to express the SARS-CoV-2 spike. In this system, the levels of anti-spike IgG are quantified using a binding index that accounts for both the percentage and median fluorescence intensity (MFI) of IgG bound to spike-expressing cells. We have previously used this method to discern viral antigen-specific IgG levels in convalescent donors (Alvarez et al., 2014, 2017; Garrido et al., 2018). The benefit of this composite metric is that both the frequency and density of IgG-antigen binding are captured, as both parameters contribute to the activation of non-neutralizing effector functions. In addition, cell surface-expressed spike protein likely retains a tertiary structure and glycosylation that are consistent with natural infection, potentially capturing a greater range of paratopes (i.e., higher sensitivity).
Using this cell-based assay (CBA), we detected an approximately 1.5-log difference in anti-spike IgG titers between naive and SARS-CoV-2 convalescent donors, and a further 2-log difference among the convalescent donors (Figures 1D and 1E). Interestingly, the two PCR-positive SARS-CoV-2 donors whose anti-spike IgG levels were undetectable by ELISA had detectable, albeit low, anti-spike IgG levels in the cell-based assay. These levels were 1 log above the values obtained with IgG from naive donors, demonstrating greater resolution of the cell-based assay compared to ELISA. In addition, anti-spike IgG levels were readily detectable above negative control (naive) IgG levels in the eight convalescent donors who possessed low-to-negative anti-spike IgG levels using the ELISA assay (Figure S1A). Other studies have similarly observed higher sensitivity with cell-based detection assays (Fafi-Kremer et al., 2020; Grzelak et al., 2020), which may be attributable to the display of conformational epitopes that are dependent on the tertiary or quaternary structure of the natively folded trimeric spike protein. Altogether, we observed a strong positive correlation between anti-spike IgG levels and disease severity (R2 = 0.27, p < 0.0001) (Figure 1F), with severe donors possessing significantly higher anti-spike IgG levels (Figures S1B and S1C).
Higher levels of anti-spike IgG-mediated FcγR activation correlate with COVID-19 severity
Elevated levels of anti-spike IgG and proinflammatory cytokines are detected in severe hospitalized COVID-19 patients, suggesting that IgG may exacerbate disease severity via FcγR-mediated ADI effects. Therefore, we next examined the levels of Fc-γ receptor IIa and IIIa signaling (FcγRIIa, FcγRIIIa), since both of these FcγRs activate several important cellular effector functions, such as ADCC and ADCP, and are capable of activating an array of proinflammatory cytokines (Nimmerjahn and Ravetch, 2010; Vogelpoel et al., 2015).
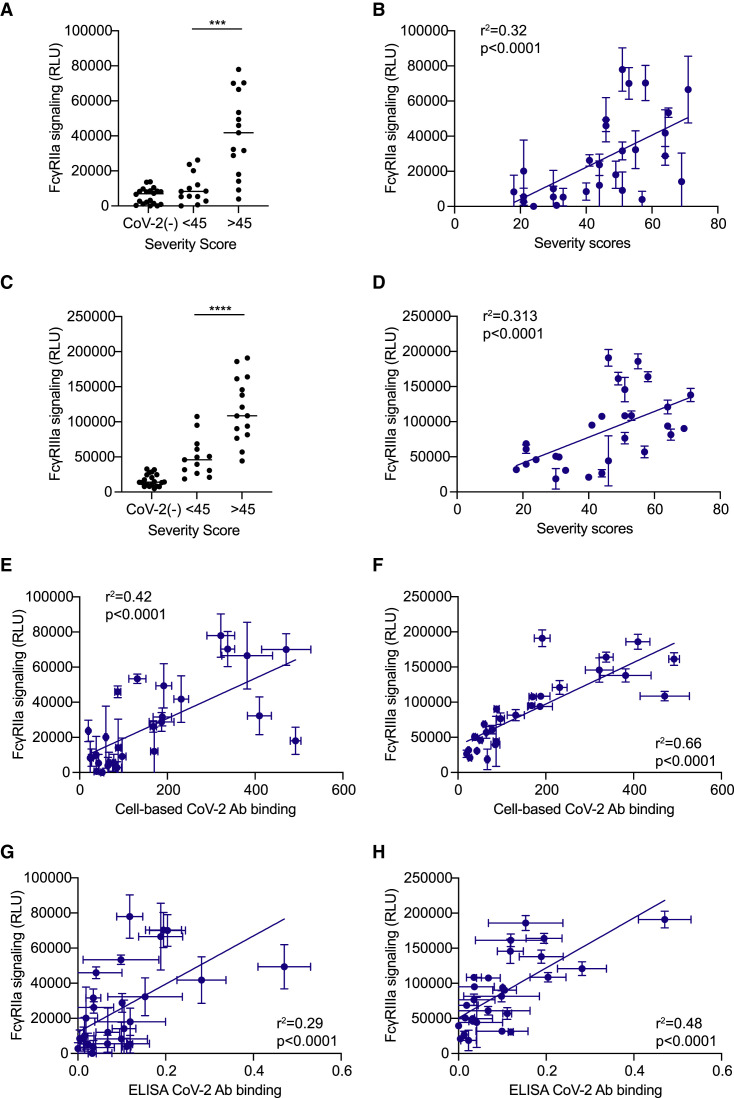
To quantify FcγR activation, we transfected 293T cells to express the SARS-CoV-2 spike and co-cultured these cells with FcγRIIa or FcγRIIIa reporter cell lines in the presence of various concentrations of purified donor IgG (Alvarez et al., 2014, 2017). Using this system, we observed a strong positive correlation between severity scores and both FcγRIIa and FcγRIIIa activation across convalescent donors (Figures 2 B, 2D, S2C, and S2D), with higher overall FcγR signaling in severe versus mild donors (Figures 2A and 2C). Only background levels of FcγR activation were detected in response to SARS-CoV-2-naive donor IgG (Figure 2A). Interestingly, the majority of IgG from mild donors did not induce greater FcγRIIa activation than SARS-CoV-2 naive IgG (Figure 2A). In contrast, both mild and severe convalescent donors induced FcγRIIIa signaling levels that were at least 2-fold above naive control IgG (Figure 2C).
Figure 2.
FcγR activation correlates with COVID-19 severity and anti-spike titers
The figure depicts the levels of FcγR-signaling induced by purified IgG from CoV-2+ donors in response to SARS-CoV-2 spike protein expressed on the surface of 293T cells.
(A–D) Graphs show the levels of (A and B) FcγRIIa and (C and D) FcγRIIIa signaling using 25 μg/mL IgG by (B and D) composite symptom severity scores or by (A and C) CoV−/+ status and severity scores.
(E–H) The panels compare the anti-spike IgG titers as quantified by CBA or ELISA assays versus the levels of FcγR signaling. (E and F) CBA anti-spike IgG versus (E) FcγRIIa and (F) FcγRIIIa signaling using 25 μg/mL IgG. (G and H) ELISA anti-spike IgG titers versus (G) FcγRIIa and (H) FcγRIIIa signaling using 25 μg/mL IgG. All of the FcγR results are the SEMs of N = 3 experiments.
Next, we evaluated the relationship between FcγR activation and the levels of anti-spike IgG. We observed a significant positive correlation with both FcγRIIa and FcγRIIIa signaling and the levels of anti-spike IgG in all convalescent donors, using both ELISA and CBA anti-spike IgG-binding assays (Figures 2E–2H and S2E–S2H). However, anti-spike IgG levels as quantified by CBA were much more highly correlated with the levels of FcγRIIIa signaling (R2 = 0.66) (Figure 2F) compared to anti-spike titers obtained by ELISA (R2 = 0.48) (Figure 2H). Altogether, these data show a strong positive correlation between anti-spike IgG titers, the levels of FcγR activation, and COVID-19 severity in non-hospitalized individuals.
COVID-19 disease severity is correlated with higher anti-spike IgG cross-reactivity against other betacoronaviruses (β-CoVs)
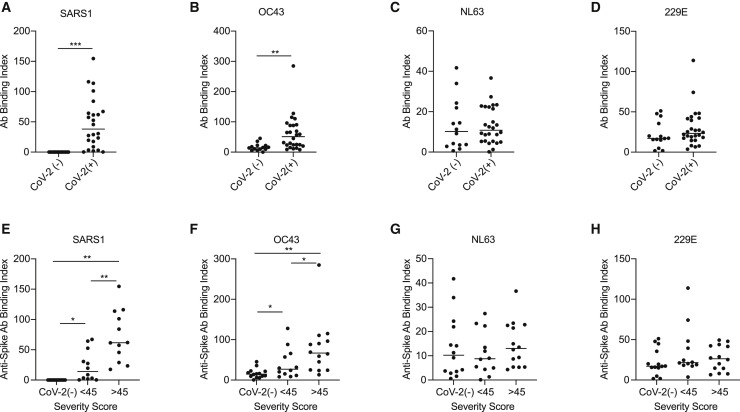
To determine whether humoral recall responses against shCoVs may contribute to SARS-CoV-2 anti-spike IgG responses, we measured the levels of anti-spike IgG binding against the spike of a seasonal β-CoV, OC43, as well as two alphacoronaviruses (-CoVs), NL63 and 229E. In addition, we examined the level of cross-reactivity of SARS-CoV-2 convalescent IgG against the spike protein of SARS-CoV-1 (SARS1). Since we observed more sensitive detection of anti-spike IgG binding using CBA versus ELISA, we quantified the levels of hCoV anti-spike IgG binding using CBA. We observed that the majority of SARS-CoV-2 convalescent donors possessed IgG, which recognized the SARS1 spike protein (Figure 3 A). Moreover, IgG recognition of the OC43 spike was significantly elevated in SARS-CoV-2 convalescent donors as compared to SARS-CoV-2-naive donors (Figure 3B). In contrast, IgG binding to -CoV spike proteins NL63 and 229E was not significantly different among SARS-CoV-2-convalescent and -naive donors (Figures 3C and 3D). This finding suggests that IgG responses against SARS-CoV-2 did not boost IgG responses against -CoVs. When further explored in relation to disease severity, IgG from severe donors possessed higher cross-reactivity to the spike protein of SARS1 and OC43 as compared to mild donors (Figures 3E and 3F). These findings show that infection with SARS-CoV-2 induces IgG that are cross-reactive with other β-CoVs, and higher levels of cross-reactive IgG correlate with more severe disease.
Figure 3.
Higher β-coronavirus cross-reactive IgG titers are correlated with COVID-19 severity
(A–H) Graphs compare the level of IgG binding to spike proteins of (A and E) SARS1, (B and F) OC43, (C and G) NL63, and (D and H) 229E coronavirus as assessed by CBA and detected by flow cytometry. Graphs (A–D) compare the level of anti-spike IgG in SARS-CoV-2-naive versus -convalescent donors. Graphs (E–H) compare the levels of anti-spike IgG among donor groups separated by SARS-CoV-2 status and COVID-19 severity scores, n = 13 mild, 15 severe. The SEMs of N = 3 experiments are shown.
Immunodominant regions with high OC43 sequence identity differentially correlate with COVID-19 severity
Recently, several groups have conducted SARS-CoV-2 peptide walk experiments using Abs from hospitalized COVID-19 patients to identify immunodominant epitopes that correlate with severe COVID-19 (Li et al., 2021; Mishra et al., 2020; Wang et al., 2020a; Zamecnik et al., 2020). Notably, some of the immunodominant epitopes share high sequence identity with shCoVs (Ladner et al., 2021; Shrock et al., 2020) and overlap with functional regions within the SARS-CoV-2 spike protein (i.e. S1/S2; S2′FP, and HR regions).
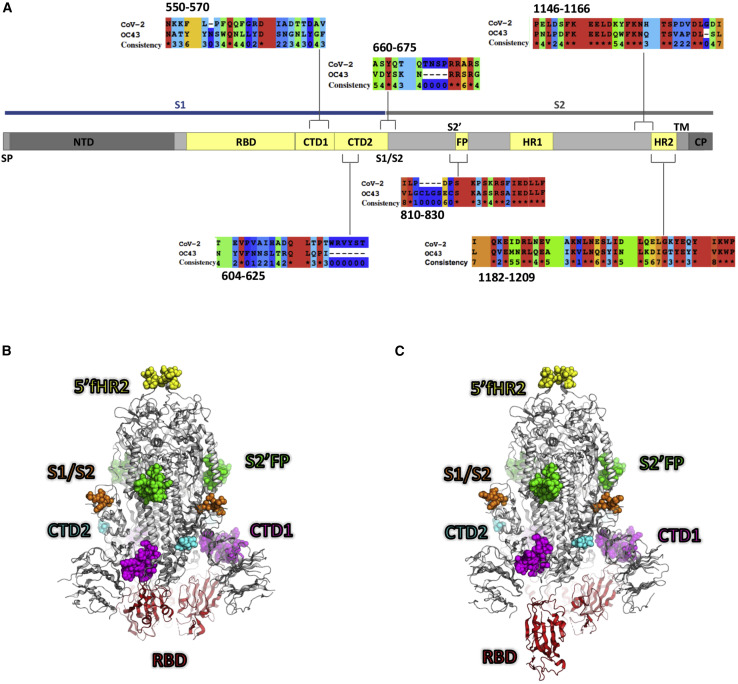
To explore how epitope targeting relates to infection severity in non-hospitalized individuals, we screened our cohort against a panel of immunodominant SARS-CoV-2 peptides that possessed high identity with shCoVs (Table S3), in particular β-CoV OC43 (Figure 4 ). To contrast these conserved regions, we also screened the levels of IgG-targeting immunodominant regions that possessed little identity with shCoVs. In addition, we quantified the levels of IgG targeting the RBD region, since Abs targeting this region mediate potent SARS-CoV-2 neutralization in vitro and in vivo (Rogers et al., 2020; Wang et al., 2020b; Wec et al., 2020; Zost et al., 2020).
Figure 4.
Localization of SARS-CoV-2 spike immunodominant regions
(A) Diagram depicts SARS-CoV-2 spike protein subdomains, which include the N-terminal domain (NTD), the receptor-binding domain (RBD), S1-C terminus domains 1 and 2 (CTD1 and -2), the furin cleavage site (S1/S2); the S2′ cleavage site and fusion protein domain (S2′FP), the heptad repeat domains 1 and 2 (HR1 and -2), the transmembrane domain (TM), and the cytosolic domain (CP). Immunodominant regions with either high or low sequence identity with β-coronavirus OC43 are shown; asterisk indicates 100% sequence identity (red) and a consistency score of zero (blue) indicates no conservation of amino acid characteristics. (B and C) The homotrimeric SARS-CoV-2 spike protein is shown in the (B) closed and (C) open state (PDB: 6VXX and 6VYB, respectively). In each protomer of the spike, the protein mainchain is illustrated in white, except for the RBD, which is red. The atoms in the 6 peptide epitopes that were tested are shown as space-filling models, colored according to peptide number. There are regions of missing density in the models, presumably due to conformational flexibility, and these regions are omitted here; CTD1 (magenta) and S2′FP (green) are fully resolved; CTD2 (cyan), S1/S2 (orange), and 5′fHR2 (yellow) are partially resolved; and HR2 is completely absent in the structure.
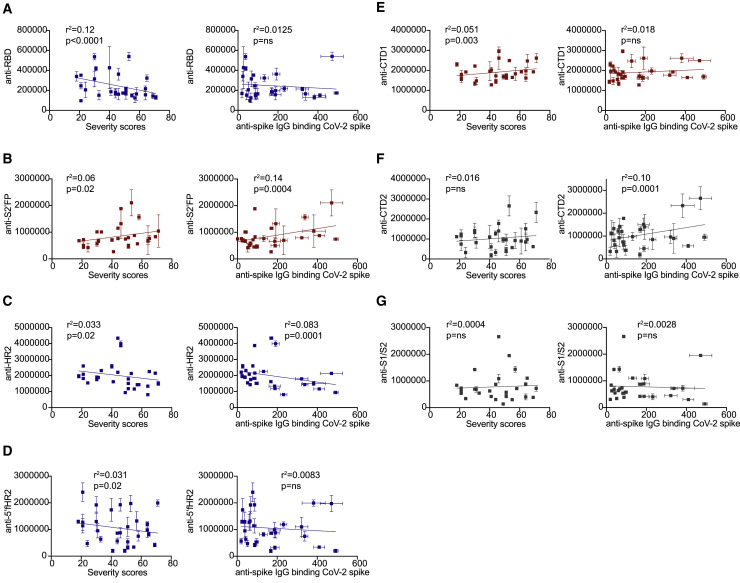
To quantify the levels of IgG targeting these regions, we developed a luciferase-based ELISA to allow for the sensitive detection of IgG binding. Using this assay, we observed a significant inverse correlation between the levels of anti-RBD IgG and overall severity scores among convalescent donors (p < 0.0001) (Figure 5 A). In comparing donors with mild versus severe disease, we observed significantly higher levels of anti-RBD IgG targeting in donors with mild disease (Figure S3A). Next, we quantified the levels of IgG targeting of three representative immunodominant peptides with high sequence identity to shCoVs. We observed a significant positive correlation between S2′FP region IgG-binding levels and severity (Figure 5B).
Figure 5.
SARS-CoV-2 convalescent IgG from mild and severe donors differentially target seasonal CoV-conserved and non-conserved SARS-CoV-2 spike protein immunodominant epitopes
(A–G) Graphs compare the levels of IgG-binding to (A) RBD, (B) S2′FP, (C) HR2, (D) 5′fHR2, (E) CTD1, (F) CTD2, and (G) S1/S2 regions in CoV-2+ convalescent donors as detected by luminescent ELISA versus severity scores (left) or total CBA anti-spike IgG levels (right). Inverse, positive, or no correlation is indicated by blue, red, or gray, respectively. n = 28; the SEMs of N = 3 experiments are graphed.
In contrast, we observed a significant inverse correlation with the levels of IgG targeting the HR2 region and severity (Figure 5C), with mild donors possessing higher IgG targeting the HR2 region, as compared to severe donors (Figure S3E). We also observed that IgG targeting of the HR2 region was inversely correlated with overall anti-spike IgG titers (Figure 5C). Lastly, we examined IgG targeting of a conserved region located just upstream of the HR2 domain, also known as the 5′ flank HR2 (5′fHR2). Similar to direct targeting of the HR2 region, the levels of IgG binding in the 5′fHR2 region were also inversely correlated with severity (Figure 5D); however, IgG levels were not inversely correlated with overall anti-spike IgG titers (Figure 5D).
Next, we quantified the levels of IgG-targeting immunodominant regions that were not highly conserved with OC43 or other shCoV, which include the novel furin cleavage site at the S1/S2 junction and two other regions located within the C-terminal domain (CTD) just downstream of the RBD region (Figure 4). In screening these regions, we observed a significant positive correlation with the levels of CTD1 targeting and severity (Figure 5E), but observed no correlation between IgG targeting of the CTD2 or S1/S2 regions and severity (Figures 5F and 5G). In comparing binding to these OC43 non-conserved regions and overall anti-spike levels, we detected a significant correlation between IgG targeting of CTD2 and higher anti-spike IgG levels (Figure 5F), but not with any other regions (Figures 5E and 5G).
Severe non-hospitalized COVID-19 correlates with high FcγR activation and IgG targeting of the spike protein S2′’ fusion protein site
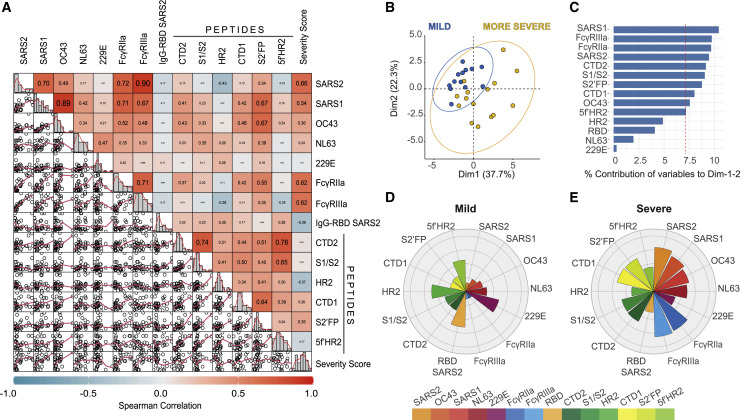
To gain insight into the associations between all of the Ab features tested and disease severity, we performed a Spearman’s chart correlation on all of the variables analyzed (Figure 6 A). Through this correlation analysis, we observed that individuals with more severe COVID-19 were characterized by higher anti-SARS-CoV-2 spike IgG (R = 0.66), higher IgG cross-reactivity to β-CoVs (SARS1 R = 0.54; OC43 R = 0.36), and higher proinflammatory FcγR activation (R = 0.62), along with higher levels of IgG targeting the CTD1 (R = 0.26) and S2’FP regions (R = 0.35), as compared to mild donors (Figure 6A). These results reaffirm the significance of all the previous findings in this study (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5). Notably, higher severity scores were inversely correlated with the levels of IgG targeting the RBD and HR2 regions, suggesting that in severe individuals, there is a relative lack of IgG targeting these previously described SARS-CoV-2 neutralizing regions. Interestingly, while we observed that both FcγRIIa and FcγRIIIa activation were positively correlated with β-CoV cross-reactivity, the levels of FcγRIIa signaling were more highly correlated with IgG targeting the CTD1 (R = 0.42) and S2′FP (R2 = 0.55) regions, as compared to FcγRIIIa signaling (CTD1 R = 0.14; S2′FP R = 0.35).
Figure 6.
Multivariable analysis identifies distinct humoral immune profiles in mild versus severe COVID-19
(A) Scatter matrix chart summarizes the Spearman’s correlation (upper) and the scatterplots (lower) between all analyzed variables using the entire cohort (n = 28). The Spearman’s r values are shown inside the colored squares, and the scale of blue-to-red color indicates a negative-to-positive correlation. The small bar graphs (diagonal) represent the distribution of data for each variable.
(B) Biplot shows the principal component analysis (PCA) depicting the mild-scored (n = 13) and more severe-scored (n = 15) COVID-19 patients, according to their severity scores.
(C) The contribution of each variable to PCA for dimension 1 and 2 is represented by bars, and its threshold is indicated as a red dotted line.
(D and E) Polar plots show the different profiles of humoral response for mild and more severe groups. Each bar in the plot represents the mean of Z scores for each variable.
To further explore the contribution of different IgG features, we performed a principal-component analysis (PCA) using all convalescent donors (Figures 6B and 6C). The results of this analysis showed that two dimensions explain the 60% of variance, and the severe individuals are clearly separated according to dimension 1 (37.7% of variance), with a response focused on both FcγRIIa and FcγRIIIa signaling, β-CoV anti-spike IgG (including SARS-CoV-2), and IgG targeting of CTD1, CTD2, and S2′FP regions. Interestingly, the individuals with more severe COVID-19 were highly represented on this dimension, suggesting that cross-reactive IgG responses against β-CoVs, which included the induction of high FcγRIIa and FcγRIIIa activation that targeted CTD1, CTD2, and S2′FP regions, are predictive of severe disease. In contrast, mild individuals possessed different IgG responses, with lower levels on the features within dimension 1, coupled with higher representation of dimension 2 (22.3% of variance), which was primarily represented by IgG targeting of the RBD, S1/S2 furin site, HR2, and 5′fHR2 regions. This analysis suggests a differential course of disease when the IgG response is directed against different regions of the SARS-CoV-2 spike.
Next, we conducted a Spearman’s chart correlation using either mild or severe donor groups (Figures S4A and S4C). We observed that FcγRIIa activation was highly correlated with targeting of CTD1, CTD2, and S2′FP regions in severe individuals (Figure S4C), but no correlation between FcγRIIa activation and IgG targeting of immunodominant regions in mild individuals (Figure S4A). In only examining the anti-spike region IgG-targeting profiles, we observed that individuals with milder infection possessed anti-spike IgG levels that were inversely correlated with the levels of S2′FP (R = −0.27) and CTD1 (R = −0.41) region IgG targeting (Figure S4B). The converse was observed in severe profiles, in which total anti-spike IgG levels were inversely correlated with IgG targeting the S1/S2 (R = −0.39) and HR2 (R = −0.42) regions and positively correlated with IgG targeting the S2′FP (R = 0.37) region (Figure S4D). Interestingly, we did not observe a relationship between anti-RBD levels and HR2 region targeting (R = −0.077) in mild individuals.
To further elucidate the differences between the overall profiles of mild and severe individuals, the mean of Z score values for each Ab feature was calculated and represented on a polar plot graph (Figures 6D and 6E). In comparing the relative targeting of α- versus β-CoVs, we observed that mild individuals expressed higher Z scores for IgG targeting α-CoVs, with the inverse being true for severe profiles. In support of the PCA analysis, the polar plot shows that individuals with mild disease had lower FcγRIIa and FcγRIIIa enrichment scores (Figure 6D), which was coupled with relative lower enrichment of IgG targeting the CTD1 and S2′FP regions, both of which were significantly correlated with elevated FcγR activity in severe individuals (Figure 6D). Crucially, despite severe donors possessing higher overall levels of anti-spike IgG, the enrichment scores for RBD IgG-targeting were higher in mild donors (Figures 6D and 6E), suggesting an enrichment in neutralizing IgG. In severe individuals, the inverse was observed: IgG targeting CTD1 and S2′FP regions was enriched, and HR2 region and 5’fHR2 targeting was decreased, relative to total anti-spike IgG (Figure 6E). Taken together, these data suggest that the efficacy of IgG responses and corresponding disease severity is highly dependent on the specific epitopes targeted in the SARS-CoV-2 spike protein, which in turn may be influenced by prior shCoV exposure.
The ratio of HR2/S2′FP IgG targeting correlates with disease severity in non-hospitalized convalescent and ICU-hospitalized patients
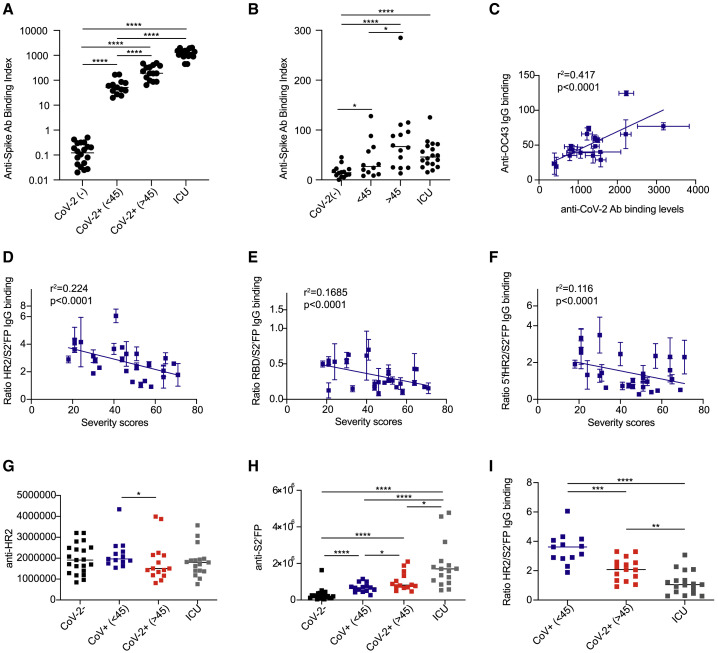
Having established that IgG targeting of the SARS-CoV-2 spike protein was predictive of mild versus severe disease in non-hospitalized patients (Figure 6), we examined whether this Ab-targeting profile was recapitulated in ICU-hospitalized COVID-19 patients. For this analysis, we obtained a cohort of 17 patients hospitalized in the ICU with COVID-19 during the first waves of the COVID-19 pandemic in 2020. We quantified the levels of IgG directed against the SARS-CoV-2 spike protein in ICU-hospitalized individuals using our cell-based assay. We observed a range of anti-spike IgG levels that were significantly higher than the anti-spike IgG levels detected in non-hospitalized donors (Figure 7 A). Next, we examined the levels of anti-OC43 IgG in ICU patients. Interestingly, while the anti-OC43 IgG levels were significantly higher in ICU patients as compared to SARS-CoV-2-naive donors, ICU patient anti-OC43 IgG levels were in the same range as those detected in non-hospitalized convalescent patients (Figure 7B). In comparing the levels of SARS-CoV-2 versus OC43 IgG-binding levels in ICU patients, we detected a positive correlation between total anti-SARS-CoV-2 and anti-OC43 spike titers (Figure 7C).
Figure 7.
The ratio of IgG targeting the HR2/S2′FP regions of the SARS-CoV-2 spike protein correlates with COVID-19 severity in convalescent and ICU-hospitalized patients
(A and B) (A) Anti-SARS-CoV-2 or (B) anti-OC43 spike IgG titers as detected by CBA in SARS-CoV-2-naive (CoV-2−), non-hospitalized convalescent (CoV-2+), and ICU-hospitalized COVID-19 patients (n = 20 naive, 28 convalescent, and 17 ICU).
(C) Comparison of SARS-CoV-2 versus OC43 anti-spike IgG titers in ICU-hospitalized COVID-19 patients.
(D–F) The ratio of (D) HR2/S2′FP, (E) RBD/S2′FP, and (F) 5′fHR2/S2′FP in convalescent non-hospitalized donors.
(G and H) The levels of IgG binding to (G) HR2 and (H) S2′FP regions as detected by luminescent ELISA in SARS-CoV-2-naive (CoV-2−), convalescent non-hospitalized donors split by mild (<45), more severe (>45), and ICU-hospitalized COVID-19 patients.
(I) The ratio of HR2/S2′FP IgG-binding levels in SARS-CoV-2-convalescent donors versus ICU-hospitalized COVID-19 patients. The SEMs of N = 3 experiments are shown.
To define the IgG-targeting profile that best segregated mild versus severe disease, we individually examined the ratios of the three epitopes (RBD, HR2, 5′fHR2) that were associated with mild disease over the epitope (S2′FP) that was most highly associated with severe disease in non-hospitalized donors. Calculating the IgG-targeting ratio of each donor normalizes against the higher anti-SARS-CoV-2 IgG titers detected in more severe patients. In plotting the ratio of RBD/S2′FP, HR2/S2′FP, and 5′fHR2/S2′FP versus severity scores, we observed that all of these IgG-targeting ratios inversely correlated with higher severity scores (Figures 7D–7F); however, the ratio of HR2/S2′FP displayed the highest R2 values (R2 = 0.2239), compared to either RBD/S2′FP (R2 = 0.1685) or 5′fHR2/S2′FP (R2 = 0.1158). We next examined the levels of IgG targeting the HR2 and S2′FP regions using IgG from the ICU-hospitalized COVID-19 cohort. We observed that ICU patients possess similar levels of IgG targeting the HR2 region as compared to non-hospitalized convalescent donors (Figure 7G). In contrast, we detected significantly higher levels of IgG targeting the S2′FP region in ICU-hospitalized patients, as compared to non-hospitalized individuals (Figure 7H). In calculating HR2/S2′FP IgG-binding ratios, we observed significantly lower ratios in ICU patients as compared to non-hospitalized individuals (Figure 7I).
Discussion
Early in the pandemic, it was largely assumed that the adaptive immune system treated SARS-CoV-2 as a novel pathogen, despite the significant sequence overlap of the virus with shCoVs. Although subsequent studies examined seasonal coronavirus immune recall, immunodominant epitopes, and COVID-19 severity individually, none looked at all three factors together. Accordingly, the role of memory Abs in disease pathogenesis has been unclear and clinically disregarded. Researchers have noted an unusual aspect of COVID-19 disease course; namely, some patients show IgG within days of contracting SARS-CoV-2 (Liu et al., 2020; Zhang et al., 2020; Zhao et al., 2020). This is in contrast to the 7–14 days that is usually required for naive B cells to become activated, class switch, and begin producing IgG (Murphy et al., 2012). Notably, these early anti-spike IgG responses are associated with disease severity, not protection. Higher anti-spike IgG titers, along with elevated levels of proinflammatory cytokines, correlate with more severe disease in hospitalized individuals (Young et al., 2020; Zhang et al., 2020). In this study, we detected a significant positive correlation between anti-SARS-CoV-2 spike IgG levels and disease severity in a cohort of non-hospitalized convalescent individuals with mild to severe COVID-19, as well as even higher anti-spike IgG titers in ICU patients. Our findings substantiate previous studies that detected high levels of anti-spike IgG in severe hospitalized COVID-19 cases, and further extend this correlation to non-hospitalized cases.
Along with higher anti-spike IgG levels, we detected that IgG cross-reactivity to other β-CoVs (SARS1 and OC43) is positively correlated with disease severity. Several studies have now observed similar phenomena (Anderson et al., 2021; Shrock et al., 2020; Wang et al., 2020c), suggesting that anti-spike IgG responses in severe COVID-19 patients target regions of high sequence and structural identity with other β-CoVs. Although the time course is atypical for IgG, theoretically, these responses could arise from rapid de novo evolution of IgG targeting these conserved and immunodominant regions. However, inconsistent with rapid evolution, in acute severe hospitalized COVID-19 patients, Unterman et al. (2022) observed a population of expanded persistent B cell clones with stable somatic hypermutation (SHM) that are instead more likely memory B cell lineages. Furthermore, studies examining pre-pandemic and SARS-CoV-2-naive plasma samples observed that some sera were able to recognize the SARS-CoV-2 spike (Anderson et al., 2021; Ng et al., 2020; Nguyen-Contant et al., 2020). Notably, samples predominantly bound the S2 subunit, which contains the regions of high identity with shCoVs (Shah et al., 2021). In general, sera containing high S2-targeting IgG has been shown to be highly cross-reactive against the shCoV OC43 (Anderson et al., 2021; Nguyen-Contant et al., 2020). Considering the ubiquitous seroprevalence of shCoVs (Gorse et al., 2010)—4%−27% of the population tests positive for any given shCoV at any given time (Khan et al., 2021)—it is a realistic possibility that early, high levels of cross-reactive anti-spike IgG arise from recall memory responses against shCoVs, such as OC43. The role of these cross-reactive recall Abs has been unclear, however, and studies conflict as to whether they possess neutralizing activity against SARS-CoV-2.
In addition to higher levels of anti-spike IgG and cross-reactive IgG, we also detected higher levels of anti-spike IgG-mediated FcγRIIa and FcγRIIIa signaling in severe patients in the non-hospitalized cohort. Other studies have shown similar findings, as well as a correlation between severity and the afucosylated glycoform of anti-spike IgG, which is inherently more FcγR activating (Chakraborty et al., 2021; Hoepel et al., 2020; Larsen et al., 2020). These afucosylated anti-spike IgG were shown to activate cytokine expression from macrophages (Chakraborty et al., 2021; Hoepel et al., 2020), suggesting anti-spike IgG can contribute to elevated proinflammatory cytokine profiles in severe COVID-19 (Del Valle et al., 2020; Tang et al., 2020; Ye et al., 2020).
We initially hypothesized that any shCoV recall responses amplified during acute SARS-CoV-2 infections would favor S2—and not the novel RBD-containing S1—subunit targeting inefficient non-neutralizing Ab responses that would lead to uncontrolled virus replication (similar to original antigenic sin), elevated inflammation, and worse outcomes. However, we detected no uniform association between high sequence identity and severity. Instead, we observed that IgG targeting of two highly conserved regions (HR2 region, 5′fHR2) was significantly correlated with milder infections, while targeting of another conserved region (S2′FP) was correlated with more severe infections. All three of the immunodominant conserved regions studied were correlated with disease severity, whether inverse or positive. In contrast, only one of the three non-homologous regions had a significant relationship with disease course. HR2 and S2′FP have been previously shown to be two of the most immunodominant regions in the SARS-CoV-2 spike (Ladner et al., 2021; Shrock et al., 2020). Notably, SARS-CoV-2-naive individuals can have Abs that react to these regions (Ladner et al., 2021), demonstrating that prior exposure to shCoVs could give rise to cross-reactive memory responses against the HR2 and S2′FP regions. Kaplonek et al. (2021) found that OC43 cross-reactive responses were expanded in individuals who survived infection and experienced milder infections, suggesting that some proportion of patients leverage shCoV immunity to control SARS-CoV-2 infection. Similarly, we observed that IgG targeting of two OC43-conserved regions (HR2 and 5′fHR2) correlated with milder infection. It is possible that cross-reactive IgG responses targeting these regions represent effective/efficient cross-reactive responses, possibly due to their neutralizing activity, allowing for the early and potent control of SARS-CoV-2 infections. In contrast, McNaughton et al. (2021) detected that fatal COVID-19 outcomes are associated with Ab targeting betacoronavirus-conserved S2 regions in the SARS-CoV-2 spike. This later study is consistent with our findings that higher levels of overall OC43 cross-reactive IgG correlate with more severe disease in non-hospitalized individuals. It is possible that high S2′FP-targeting, with a lack of RBD and/or HR2 targeting, is an inefficient cross-reactive memory response. In this scenario, this inefficient recall response could abrogate or divert the humoral immune response away from more effective de novo SARS-CoV-2 responses. In addition, these S2′FP IgGs may contribute to the actively deleterious ADE/ADI responses recently observed in COVID-19 (Junqueira et al., 2022). Thus, these findings highlight that the potential contribution of recall Abs to COVID-19 disease severity isnuanced, and that the preexisting shCoV IgG repertoire of an individual before becoming infected with SARS-CoV-2 may contribute to the efficiency of Ab-mediated immune responses and COVID-19 severity in beneficial or deleterious ways.
Beyond defining the spike protein-immunodominant regions based on their similarity to β-CoVs (OC43), these regions also represent known functional regions within the spike protein. Examining the IgG profiles of convalescent individuals in the context of functional regions, we observed that IgG targeting of RBD, HR2, and HR flanking regions significantly correlated with lower severity scores and, in the case of HR2, also correlated with lower levels of anti-spike IgG. This latter finding suggests that humoral responses with a higher proportion of IgG targeting the HR2 region may be highly efficient at controlling infection, since lower anti-spike titers were independently associated with milder infections in the present study and others (Long et al., 2020; Young et al., 2020; Zhang et al., 2020; Zhao et al., 2020). The HR2 region functions to mediate viral fusion and entry into host cells through the formation of a six-helix bundle in conjunction with the HR1 domain (Walls et al., 2017). In previous studies examining Ab targeting of the RBD and HR2 regions, targeting both of these regions have been shown to mediate virus neutralization (Li et al., 2020; Rogers et al., 2020; Wec et al., 2020; Zost et al., 2020). In addition, Ab targeting of both the HR2 region and 5′HR2 flanking regions can mediate neutralization of both SARS-CoV-1 and -2 in vitro (Keng et al., 2005; Lai et al., 2005; Li et al., 2020; Lip et al., 2006; Tripet et al., 2006). In fact, peptide-based SARS-CoV-2 fusion inhibitors targeting the HR1-HR2 fusion complex within regions that are highly conserved with OC43 were also shown to potently inhibit virus replication (Xia et al., 2020). When we examined the levels of anti-RBD IgG targeting in mild donors, we observed a positive correlation with the S1/S2 furin site and 5′fHR2 IgG targeting, but not with HR2 region targeting alone. Therefore, it is possible that targeting 5′fHR2 sufficiently disrupts the interaction between HR2 and HR1, thereby preventing viral fusion. Altogether, these data show that IgG targeting in and around the HR2 region may induce potent neutralization of SARS-CoV-2. Furthermore, in comparing the levels of RBD and immunodominant epitope targeting in mild donors, we observed no correlation between RBD and HR2 region IgG targeting, despite both being correlated with mild disease. This raises the possibility of at least two distinct neutralization profiles, possibly driven by whether humoral responses are predominately recall or de novo.
We also observed IgG targeting of the non-conserved CTD2 region and the conserved S2′FP region significantly correlated with higher severity scores in convalescent donors. In examining only severe donor profiles, we resolved that they were predominantly characterized by higher levels of IgG targeting S2′FP. Functionally, in the context of virus entry, the fusion protein region inserts into the host cell membrane, facilitating virus fusion into host cells (Harrison, 2015; Kawase et al., 2019). In contrast to HR2 and 5′fHR2, Abs targeting the S2′FP and CTD regions have been less well characterized. While we did not examine IgG neutralization activity, we did examine the capacity of IgG to induce FcγR activation, a prerequisite for inducing non-neutralizing effector functions such as proinflammatory cytokine release. In this analysis, we identify that increased FcγR signaling is highly correlated with higher IgG targeting of the CTD1 and S2′FP regions and more severe COVID-19. CTD1 had a lower R2 value than S2′FP, possibly indicating weaker association. In contrast, the levels of HR2 or 5′HR2 IgG were not correlated with FcγR signaling. These findings suggest that IgG targeting of the S2′FP and/or CTD1 region could indicate sites of enhanced FcγR activation. In only examining the severe convalescent donors, we observed that severe responses were not solely characterized by higher levels of S2′FP targeting, but also the absence of IgG targeting the S1/S2 furin site and HR2 regions. Similarly, milder IgG profiles were not only characterized by higher levels of RBD and/or HR2 targeting, but also lower proportions of CTD1 and S2′FP targeting in relation to overall anti-spike titers. These findings strongly suggest that targeting the S2′FP region, coupled with the absence of anti-RBD and/or anti-HR2 IgG targeting, is linked with severity. S2′FP targeting may be merely inefficient or it may actively cause deleterious ADE or ADI effects. Junqueira et al. (2022) recently demonstrated that, independent of neutralization, afucosylated SARS-CoV-2 anti-spike IgG facilitate viral entry into monocytes via FcγRIIIa, leading to inflammatory cell death (pyroptosis) and potent inflammation. Although the researchers did not directly examine cross-reactive epitopes, strikingly, some IgG from patients with non-COVID respiratory infections also induced Ab-dependent SARS-CoV-2 infection of monocytes, consistent with a role for shCoV Abs. It will be important to further dissect how cross-reactive shCoV Abs contribute to SARS-CoV-2 antiviral response efficiency and monocyte infection, and how FcγR activation across disease course influences outcomes.
Importantly, multivariate PCA analysis showed that profiles with higher levels of IgG targeting of HR2, 5’fHR2, and RBD regions were predictive of mild disease, while higher levels of IgG targeting the S2′FP region were predictive of severe disease in non-hospitalized donors. In these recently convalescent individuals, the ratio of IgG targeting the HR2/S2′FP regions was most tightly correlated with having mild versus severe disease, as compared to the ratios of IgG targeting 5′fHR2/S2′FP or RBD/S2′FP. Specifically, lower ratios of IgG targeting the HR2/S2′FP regions correlated with more severe disease and higher ratios correlated with mild infections. In examining this signature in ICU-hospitalized COVID-19 patients, we observed even lower ratios of IgG targeting the HR2/S2′FP regions, as compared to severe non-hospitalized donors. These data show that HR2/S2′FP IgG targeting ratios may predict COVID-19 severity. Therefore, determining the dominant epitope profile and associated predicted risk of severe disease of an individual could allow earlier interventions with treatments such as monoclonal Ab therapies and antivirals, preventing progression to ARDS.
In addition, assessing the humoral profiles induced by vaccination will be important. Evidence has emerged that SARS-CoV-2 reinfection is possible after 3–6 months, particularly with emerging variants (Abu-Raddad et al., 2020; Gupta et al., 2020; Selhorst et al., 2020; Tillett et al., 2021). This is in line with shCoVs, in which reinfection is common after 6 months (Edridge et al., 2020). Moreover, the emergence of variants able to evade current vaccines remains an ongoing threat. Both SARS-CoV-2 spike and whole-virus vaccines likely boost both novel and shCoV cross-reactive IgG responses, initially giving some level of protection, but which could lead to higher frequencies of severe breakthrough infections over time if inefficient IgG targeting remains the predominant response. Therefore, it will be important to examine the ratios of RBD to other immunodominant epitope IgG and assess the longevity of immunity and the frequency/severity of breakthrough infections over time. Altogether, epitope profiling may be essential to ensure long-term vaccine-induced protection. Even if vaccinated with current vaccines, individuals with inefficient IgG profiles (i.e., anti-S2′FP dominant) may require an RBD-specific or HR2-specific vaccine to shift their targeting profiles to induce long-lived protective humoral responses. Promisingly, HR2 offers a good target for universal hCoV treatment and vaccine design.
Overall, our study suggests that humoral memory responses contribute to COVID-19 disease severity, conferring either protection or risk, depending on epitope targeting. This may explain the atypical bimodality of COVID-19 disease severity, an observation that was previously obscured by aggregate data/epitope analysis. Thus far, it has been challenging to enumerate the underlying factors that accurately predict disease outcome, even among individuals with defined comorbidities. The findings reported here have the potential to help explain the differential correlations observed in previous studies regarding preexisting cross-reactive immunity to seasonal coronaviruses and disease severity, as well as improve the prediction accuracy of disease outcomes. This epitope-based original antigenic sin immunosurveillance, or OASiS, could be readily adapted to a clinical prognostic, offering a novel approach for the prediction of disease severity risk, as well as vaccine efficacy, on a patient-by-patient basis.
Limitations of the study
The limitations of this study include the limited number of COVID-19 donors profiled, only characterizing IgG and not other Ig classes (IgA, IgM), and only profiling the IgG targeting of SARS-CoV-2 spike linear epitopes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Human IgG (H+L) Cross-Adsorbed Secondary Antibody, HRP | ThermoFisher Scientific | A18811; RRID: AB_2535588 |
| Goat anti-Human IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | ThermoFisher Scientific | 21445; RRID: AB_2535862 |
| F(ab')2-Goat anti-Human IgG (H+L) Cross-Adsorbed Secondary Antibody, HRP | ThermoFisher Scientific | A24470; RRID: AB_2535939 |
| Bacterial and virus strains | ||
| DH5-α | ThermoFisher Scientific | 18265017 |
| Stbl2™ | ThermoFisher Scientific | 10268019 |
| Biological samples | ||
| Serum/Plasma from donors | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ficoll-Hypaque | GE Healthcare | 17-1440-03 |
| Nunc-Immuno™ MicroWell™ 96 well solid plates | MERCK | M9410 |
| Falcon® 96-well Clear Round Bottom TC-treated Cell Culture Microplate | CORNING | 353077 |
| Pierce™ Streptavidin Coated High Capacity Plates, White, 96-Well | ThermoFisher Scientific | 15502 |
| SARS-CoV-2 (2019-nCoV) Spike S1+S2 ECD-His Recombinant Protein, Biotinylated | SinoBiological | 40589-V08B1-B |
| ELISA Wash Buffer | ThermoFisher Scientific | N503 |
| BSA Blocker | ThermoFisher Scientific | 37525 |
| eBioscience™ TMB Solution | ThermoFisher Scientific | 00-4201-56 |
| SuperSignal™ ELISA Pico Chemiluminescent Substrate | ThermoFisher Scientific | 37069 |
| DMEM/High glucose without L-glutamine, sodium pyruvate | Cytiva | SH30081 |
| RPMI 1640 | CORNING | 10-040 |
| L-Glutamine | CORNING | 25-005 |
| HyClone Penicillin-Streptomycin 100X solution | Hyclone | SV30010 |
| Cosmic Calf Serum | Hyclone | SH30087 |
| Fetal Bovine Serum | Hyclone | SH30071 |
| Critical commercial assays | ||
| Zeba™ Spin Desalting Columns | ThermoFisher Scientific | 89892 |
| Easy-Titer™ Human IgG (H+L) Assay Kit | ThermoFisher Scientific | 23310 |
| Stop Solution for TMB Substrates | ThermoFisher Scientific | N600 |
| PolyJet™ In Vitro DNA Transfection Reagent | SignaGen | SL100688 |
| Luciferase Cell Culture Lysis 5X Reagent | Promega | E1531 |
| Luciferase Assay System | Promega | E1500 |
| Experimental models: Cell lines | ||
| 293T cells | ATCC | CRL-3216 |
| FcγRIIa and FcγRIIIa, CD4+ Jurkat reporter cell lines | Promega | G7010 |
| Recombinant DNA | ||
| SARS-CoV-2 (2019-nCoV) Spike Gene ORF cDNA clone expression plasmid, C-FLAG tag (Codon Optimized) | SinoBiological | VG40589-CF |
| Human SARS coronavirus (SARS-CoV) Spike glycoprotein Gene ORF cDNA clone expression plasmid (Codon Optimized), C-Flag tag | SinoBiological | VG40150-CF |
| Human coronavirus (HCoV-OC43) Spike Gene ORF cDNA clone expression plasmid(Codon Optimized), C-Flag tag | SinoBiological | VG40607-CF |
| Human coronavirus(HCoV-NL63) Spike Gene ORF cDNA clone expression plasmid(Codon Optimized), C-Flag tag | SinoBiological | VG40604-CF |
| Human coronavirus(HCoV-229E) Spike Gene ORF cDNA clone expression plasmid(Codon Optimized), C-Flag tag | SinoBiological | VG40605-CF |
| Software and algorithms | ||
| Gen5 | Agilent | https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imager-software/ |
| FlowJo | Tree Star Inc. | v10 |
| PRALINE multiple sequence alignment | IBIVU | https://www.ibi.vu.nl/programs/pralinewww/help.php |
| Adobe Illustrator | Adobe | 25.4.1 |
| GraphPad Prism | GraphPad Software | 8.4.3 |
| R | R Foundation | 4.0.4 |
| R Studio | R Foundation | 1.4.1103 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Raymond A. Alvarez (ralvarez@ichorbiologics.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects and samples
Demographic data (age, sex, COVID-19 RT-PCR status), and collection site and date of the serum/plasma donors studied herein are described in Table S1. COVID-19 convalescent donor samples (n = 28) and COVID-19 negative donor (n = 20) were collected at Ichor Biologics facility in New York City, USA. SARS-CoV-2 negative donors had no history of positive SARS-CoV-2 PCR test or serology test, and had not experienced any symptoms of infection in at least the 5 months prior to blood collection. From convalescent donors, 12 were male and 16 were female, while across naïve donors, 12 were female and 8 were male. As displayed in Table S1, the median age of convalescent and naïve groups of donors was 38 years old, and the median of days since the onset of disease to blood draw were 43 and 55.5, respectively. Blood type diversity and underlying health conditions were also presented in Table S1, showing a broad range of biological contexts among the cohort. Blood samples were collected after obtaining signed informed consent in accordance with institutionally approved IRB protocols (SSV ORD-2260). Convalescent COVID-19 patient samples were collected from donors 2–10 weeks after the onset of symptoms. A history of COVID-19 symptoms and symptom intensities, along with current medications and history of pre-existing conditions, were collected through participant questionnaires completed at the time of blood draw. The reported symptoms were summarized in Table S2, including the average severity and duration of each one. This information was used to calculate a severity score and to cluster donors into two categories: mild and more severe (n = 13, n = 15, respectively). More severe individuals, with severity scores over 45, and also mild individuals, with severity scores bellow 45, did not require hospitalization. Also, every donor recovered from disease. In addition, 17 individuals were recruited after being diagnosed with a severe course of disease which required invasive ventilation at intensive care units (ICU) from southern Chile (Hospital Base San José, Osorno and Valdivia). Their samples were obtained seven days after recruitment and signing of informed consents in accordance with IRB Servicio Salud Valdivia ORD number 226. Lithium heparin-coated tubes were used for blood collection and plasma was isolated using Ficoll-Hypaque (GE Healthcare; 17-1440-03) in accordance with manufacturer’s instructions. Polyclonal IgG was isolated from 200 μL of donor plasma using a protein A/G spin column kit, followed by desalting using Zeba spin columns according to manufacturer’s instructions (ThermoFisher Scientific; 89892). IgG yields were quantified using an Easy-Titer Human IgG Assay Kit (ThermoFisher Scientific; 23310). Remaining deidentified plasma samples were aliquoted and stored at −80C.
Cell lines
293T cells: This female cell line was grown in Dulbecco’s modified Eagle’s medium (DMEM) (Cytiva) supplemented with 10% cosmic calf serum (CCS) (Hyclone), Lglutamine (Corning), and Penicillin-Streptomycin (Hyclone).
FcγRIIa and FcγRIIIa, CD4+ Jurkat reporter cell lines: This male cell line was grown in RPMI- 1640 medium (Corning) supplemented with 10% fetal bovine serum (FBS) (Hyclone), L-glutamine (Corning) and Penicillin-Streptomycin (Hyclone).
Method details
SARS-CoV-2 Spike-ELISA
High binding capacity 96-well plates (Nunc) were washed and coated with 50 μL per well of 2 μg/mL of recombinant spike protein (Sino Biological; 40589-V08B1-B), diluted in 0.1% BSA, 0.05% Tween20 TBST ELISA wash buffer (ThermoFisher Scientific; N503). Plates were coated for 2 h at room temperature while shaking at 500 rpm on a Benchmark OrbishakerTM. Plates were then washed twice with ELISA wash buffer to remove any excess unbound spike protein and blocked with 2% BSA (ThermoFisher Scientific; 37,525) in ELISA wash buffer overnight at 4°C. After overnight blocking step, plates were washed twice and incubated with 5 μg/mL of donor-derived polyclonal IgG for 1 h at room temperature. After incubation, plates were washed three times and incubated for 30 min at room temperature with cross-absorbed goat anti-human IgG-horseradish peroxidase (HRP)-conjugated secondary antibody (ThermoFisher Scientific; A18811) diluted to a 1:2500 dilution in ELISA wash buffer. After being washed again twice, 100 μL of TMB substrate solution was added to each well for 15mins and then 100 μL of 0.18M H2SO4 (ThermoFisher Scientific; N600) was added to stop the reaction. The optical density at 450 nm (OD450) was measured using a BioTek Powerwave HT plate reader using Gen5 software. Assay background was established using anti-human secondary Ab alone without donor IgG, which was subtracted from OD values of all samples tested.
Anti-spike protein IgG determination using a cell-based assay
To quantify the levels of IgG binding various coronavirus spike proteins, 293T cells were transfected with SARS-CoV-2 (Sino Biological; VG40589-CF) (Genbank: YP_009724390.1), SARS-CoV-1 (Sino Biological; VG40150-CF) (Genbank: AAP13567.1), OC43 (Sino Biological; VG40607-CF) (Genbank: AVR40344.1), NL63 (Sino Biological; VG40604-CF) (Genbank: APF29071.1), or 229E (Sino Biological; VG40605-CF) (Genbank: APT69883.1) spike protein expression vectors. For this assay, 2 × 106 293T cells were plated in 10 cm plates and incubated at 37°C overnight. The next day, 4 μg of coronavirus spike expression vectors were transfected into 293T cells using Polyjet™ transfection reagent (SignaGen; SL100688) according to manufacturer’s instructions. After 48 h, 1 × 105 293T cells were plated per well into round bottom 96-well plates. Cells were then washed and incubated with 10 μg/mL of convalescent donor-derived IgG or negative donor control IgG and incubated at 4°C for 45 min. After primary Ab incubation, IgG opsonized cells were washed and incubated with 3 μg/mL of an APC-conjugated anti-human total IgG secondary Ab (Invitrogen, catalog A21445) at 4°C for 25mins. Cells were then washed again with PBS and LIVE/DEAD™ Fixable Violet Stain (Invitrogen; L34964A) was used to stain cells for 10 min in the dark at RT. Lastly, cells were washed twice and fixed with 1.0% paraformaldehyde in PBS and analyzed by flow cytometry (BD LSRFortessa X-20). The data were quantified using Flow Jo software (Tree Star, Inc). The IgG-binding index was calculated by multiplying the percentage of anti-spike IgG positive cells by the median fluorescent intensity (MFI) of APC signal, as normalized to the average MFI of negative control IgG. To ensure that the relative differences between patient-derived IgG were maintained, all IgG were tested in parallel on the same day for each replicate.
Fc-gamma receptor signaling assay
FcγRIIa and FcγRIIIa signaling was assessed using a reporter cell co-culture system that we have previous used to assess FcγR signaling in response to viral antigens ([59, 60]). For this assay, 293T cells are transfected with SARS-CoV-2 spike expression vector and co-cultured with either a FcγRIIa, or FcγRIIIa, CD4+ Jurkat reporter cell line, which expresses firefly luciferase upon FcγR activation. For this assay, 1 × 105 SARS-CoV-2 spike-expressing 293T cells were plated in each well of a 96-well round bottom plate. The cells were then preincubated with a 5-fold dilution series of convalescent donor-derived IgG starting at a maximum concentration of 25 μg/mL. IgG opsonized 293T cells were then co-cultured with FcγRIIa or FcγRIIIa reporter cells at a 2:1 reporter-to-target cell ratio for 24 h at 37°C. After 24 h, all cells were lysed with cell lysis buffer (Promega; E1531), and the levels of firefly luciferase activity determined using a luciferase assay kit according to manufacturer’s instructions (Promega; E1500). To quantify background (i.e., IgG activation-independent) luciferase production, reporter cells were co-cultured with the spike-expressing 293T cells in the absence of any IgG. Background levels were subsequently subtracted from the signal to yield IgG-specific activation in relative light units (RLUs). Luminescence was measured on a Cytation 3 image reader using Gen5 software.
Immunodominant epitope IgG-binding assay
For this assay, N-terminus biotinylated peptides were synthesized by Genscript. N-terminal GSGS linker sequence was added to all peptide sequences. The RBD peptide contained a C-terminal avitag (GLNDIFEAQKIEWHE), for biotinylation via BirA enzyme; a Protein C tag (EDQVDPRLIDGK), and a polyhisidine tag (HHHHHHHHHH), to enable immobilized metal affinity chromatography purification. Lyophilized peptides and RBD were initially resuspended in DMSO and then used to make 5 μg/mL working dilutions in TBST ELISA wash buffer. Pierce™ white streptavidin-coated high binding capacity 96-well binding plates (ThermoFisher Scientific; 15502) were washed twice with ELISA wash buffer and coated with 5 μg/mL of biotiylated peptides at room temp. Plates were coated for 2 h while shaking at 500 rpm on a Benchmark Orbi-Shaker™. After incubation, plates were washed three times and blocked with 2% BSA blocking solution diluted in wash buffer and incubated at 4°C overnight. After incubation, plates were washed three times and incubated with 5 μg/mL of donor-derived IgG for 1 h at room temp. After primary IgG incubation, plates were washed 3 times and incubated for 25 mins with 100 μL of Invitrogen™ cross-absorbed, F(ab’)2, goat anti-human IgG secondary Ab (ThermoFisher Scientific; A24470) diluted 1:2500 in ELISA wash buffer. Plates were then washed again three times and developed using a SuperSignal™ ELISA Pico Chemiluminescent Substrate (ThermoFisher; Cat#37069) and the level of luminescence detected using a Cytation 3 image reader luminometer using Gen5 software.
Quantification and statistical analysis
Statistical and data analyses were performed using GraphPad Prism 8.4.3, R 4.0.4, and R Studio 1.4.1103. Graphs were generated in Prism and R Studio and statistical differences between two groups were calculated by Mann-Whitney U-test. Statistical significance was defined as ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Scatter plots, bar graphs, heatmaps, and polar plots were visualized with ggplot2 (v3.3.3 R Studio). Correlation analysis (in Figures 7 and S4) were performed using the R package “correlation” (v0.6.0) in R Studio. Polar plots represent the value of different variables normalized to the Z-score of data. Each variable was mean-centered and then divided by the standard deviation of the variable to ensure each variable had zero mean and unit standard deviation. Unsupervised principal components analysis (PCA) was performed in R. The completed data were scaled to unit variance using FactoMineR (v2.4 R studio). The PCA results were extracted and visualized using factoextra (v1.0.7 R Studio). Outlier exclusion was performed using Prism. n and N values are mentioned at figure captions.
Acknowledgments
We would like to thank Sheila O’Donoghue, RN, and Beth Sferrazza, RN, for conducting the blood draws for this study; Drs. Svenja Weiss and Biliana Lozanoska-Ochser for their critical review of this manuscript; Dr. Server Ertem for insights on diagnostic design; and Dean Greg Morrisett, PhD, of Cornell Tech for generous institutional support. This study was supported by NIH SBIR grant no. 1R43AI138740-01A1, awarded to Ichor Biologics and R.A.A.; Chilean National Research and Development Agency grant no. COVID0422, awarded to M.I.B.; and a Jacobs Technion-Cornell Institute Runway package, awarded to R.A.B. F.B. was supported by an ANID scholarship/PhD no.21201764 and F.F. was supported by FONDECYT grant no. 3200913. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Study design, J.L.G., R.A.A., R.A.B., C.D.B., M.C., F.F., and M.I.B. Experimental work, J.L.G., F.B., S.M., J.W.B., M.I.B., and R.A.A. Data analysis, J.L.G., M.M., S.M., C.D.B., R.A.B., and R.A.A. Reagent development, J.L.G., R.A.A., J.W.B., and C.D.B. Donor plasma acquisition, S.M., J.L.G., Y.P., and R.A.A. Writing, J.L.G., R.A.A., and R.A.B., and the final version was approved by all of the authors.
Declaration of interests
J.L.G., F.B., and R.A.A. were partially supported by Ichor Biologics LLC. R.A.A. and R.A.B. are inventors on a provisional patent related to this study. The remaining authors declare no commercial or financial relationships that are potential conflicts of interest.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: May 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110904.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request, unless it is protected by law.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu-Raddad L.J., Chemaitelly H., Malek J.A., Ahmed A.A., Mohamoud Y.A., Younuskunju S., Ayoub H.H., Al Kanaani Z., Al Khal A., Al Kuwari E., et al. Assessment of the risk of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection in an intense reexposure setting. Clin. Infect. Dis. 2020;73:e1830–e1840. doi: 10.1093/cid/ciaa1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez R.A., Hamlin R.E., Monroe A., Moldt B., Hotta M.T., Rodriguez Caprio G., Fierer D.S., Simon V., Chen B.K. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J. Virol. 2014;88:6031–6046. doi: 10.1128/jvi.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez R.A., Maestre A.M., Law K., Durham N.D., Barria M.I., Ishii-Watabe A., Tada M., Kapoor M., Hotta M.T., Rodriguez-Caprio G., et al. Enhanced FCGR2A and FCGR3A signaling by HIV viremic controller IgG. JCI Insight. 2017;2 doi: 10.1172/jci.insight.88226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M., et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Fafi-Kremer S., Bruel T., Madec Y., Grant R., Tondeur L., Grzelak L., Staropoli I., Anna F., Souque P., Fernandes-Pellerin S., et al. Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. EBioMedicine. 2020;59:102915. doi: 10.1016/j.ebiom.2020.102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forthal D.N., Moog C. Fc receptor-mediated antiviral antibodies. Curr. Opin. HIV AIDS. 2009;4:388–393. doi: 10.1097/COH.0b013e32832f0a89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido J.L., Prescott J., Calvo M., Bravo F., Alvarez R., Salas A., Riquelme R., Rioseco M.L., Williamson B.N., Haddock E., et al. Two recombinant human monoclonal antibodies that protect against lethal Andes hantavirus infection in vivo. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorse G.J., Patel G.B., Vitale J.N., O'Connor T.Z. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17:1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzelak L., Temmam S., Planchais C., Demeret C., Tondeur L., Huon C., Guivel-Benhassine F., Staropoli I., Chazal M., Dufloo J., et al. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Bhoyar R.C., Jain A., Srivastava S., Upadhayay R., Imran M., Jolly B., Divakar M.K., Sharma D., Sehgal P., et al. Asymptomatic reinfection in 2 healthcare workers from India with genetically distinct severe acute respiratory syndrome coronavirus 2. Clin. Infect. Dis. 2020;73:e2823–e2825. doi: 10.1093/cid/ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479-480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepel W., Chen H.J., Allahverdiyeva S., Manz X., Aman J., Biobank A.U.C.-., Bonta P., Brouwer P., de Taeye S., Caniels T., et al. Anti-SARS-CoV-2 IgG from severely ill COVID-19 patients promotes macrophage hyper-inflammatory responses. bioRxiv. 2020 doi: 10.1101/2020.07.13.190140. Preprint at. [DOI] [Google Scholar]
- Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F., Dutry I., Callendret B., Escriou N., Altmeyer R., et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011;85:10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira C., Crespo Â., Ranjbar S., de Lacerda L.B., Lewandrowski M., Ingber J., Parry B., Ravid S., Clark S., Schrimpf M.R., et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022 doi: 10.1038/s41586-022-04702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Kien F., Roberts A., Cheung Y.C., Lamirande E.W., Vogel L., Chu S.L., Tse J., Guarner J., Zaki S.R., et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcγRII-dependent entry into B cells in vitro. Vaccine. 2007;25:729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplonek P., Wang C., Bartsch Y., Fischinger S., Gorman M.J., Bowman K., Kang J., Dayal D., Martin P., Nowak R.P., et al. Early cross-coronavirus reactive signatures of humoral immunity against COVID-19. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abj2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Kataoka M., Shirato K., Matsuyama S. Biochemical analysis of coronavirus spike glycoprotein conformational intermediates during membrane fusion. J. Virol. 2019;93:e00785–e00819. doi: 10.1128/JVI.00785-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H.P., Chou C.F., Loh C.B., Wang S., Fu J., et al. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T., Rahman M., Al Ali F., Huang S.S.Y., Ata M., Zhang Q., Bastard P., Liu Z., Jouanguy E., Béziat V., et al. Distinct antibody repertoires against endemic human coronaviruses in children and adults. JCI insight. 2021;6 doi: 10.1172/jci.insight.144499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Henson S.N., Boyle A.S., Engelbrektson A.L., Fink Z.W., Rahee F., D'Ambrozio J., Schaecher K.E., Stone M., Dong W., et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep. Med. 2021;2:100189. doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S.C., Chong P.C.S., Yeh C.T., Liu L.S., Jan J.T., Chi H.Y., Liu H.W., Chen A., Wang Y.C. Characterization of neutralizing monoclonal antibodies recognizing a 15-residues epitope on the spike protein HR2 region of severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biomed. Sci. 2005;12:711–727. doi: 10.1007/s11373-005-9004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.D., de Graaf E.L., Sonneveld M.E., Plomp H.R., Nouta J., Hoepel W., Chen H.J., Linty F., Visser R., Brinkhaus M., et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2020;371 doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- Li Y., Lai D.-y., Zhang H.-n., Jiang H.-w., Tian X., Ma M.-l., Qi H., Meng Q.-f., Guo S.-j., Wu Y., et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell. Mol. Immunol. 2020;17:1095–1097. doi: 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ma M.L., Lei Q., Wang F., Hong W., Lai D.Y., Hou H., Xu Z.W., Zhang B., Chen H., et al. Linear epitope landscape of the SARS-CoV-2 Spike protein constructed from 1,051 COVID-19 patients. Cell Rep. 2021;34:108915. doi: 10.1016/j.celrep.2021.108915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lip K.M., Shen S., Yang X., Keng C.T., Zhang A., Oh H.L.J., Li Z.H., Hwang L.A., Chou C.F., Fielding B.C., et al. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol. 2006;80:941–950. doi: 10.1128/JVI.80.2.941-950.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang J., Xu X., Liao G., Chen Y., Hu C.H. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect. 2020;9:1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- McNaughton A.L., Paton R.S., Edmans M., Youngs J., Wellens J., Phalora P., Fyfe A., Belij-Rammerstorfer S., Bolton J.S., Ball J., et al. Fatal COVID-19 outcomes are associated with an antibody response targeting epitopes shared with endemic coronaviruses. medRxiv. 2021 doi: 10.1101/2021.05.04.21256571. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Huang X., Joshi S., Guo C., Ng J., Thakkar R., Wu Y., Dong X., Li Q., Pinapati R., et al. Immunoreactive peptide maps of SARS-CoV-2 and other human coronaviruses. bioRxiv. 2020 doi: 10.1101/2020.08.13.249953. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Huang X., Joshi S., Guo C., Ng J., Thakkar R., Wu Y., Dong X., Li Q., Pinapati R.S., et al. Immunoreactive peptide maps of SARS-CoV-2. Commun. Biol. 2021;4:225. doi: 10.1038/s42003-021-01743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin S.N., Mahmood S., Amar A., Ghafoor F., Raza S.M., Saleem M. Association of FcγRIIa polymorphism with clinical outcome of dengue infection: first insight from Pakistan. Am. J. Trop. Med. Hyg. 2015;93:691–696. doi: 10.4269/ajtmh.15-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K., Travers P., Walport M., Janeway C. 8th Edition. Garland Science; 2012. Janeway's Immunobiology. [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio. 2020;11 doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F., Ravetch J.V. Antibody-mediated modulation of immune responses. Immunol. Rev. 2010;236:265–275. doi: 10.1111/j.1600-065X.2010.00910.x. [DOI] [PubMed] [Google Scholar]
- Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincetic A., Bournazos S., DiLillo D.J., Maamary J., Wang T.T., Dahan R., Fiebiger B.-M., Ravetch J.V. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 2014;15:707–716. doi: 10.1038/ni.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., Zhu Q., Liu L. Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:2255–2258. doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke D.O. Two different antibody-dependent enhancement (ADE) risks for SARS-CoV-2 antibodies. Front. Immunol. 2021;12:640093. doi: 10.3389/fimmu.2021.640093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selhorst P., van Ierssel S.H., Michiels J., Mariën J., Bartholomeeusen K., Dirinck E., Vandamme S., Jansens H., Ariën K.K. Symptomatic severe acute respiratory syndrome coronavirus 2 reinfection of a Healthcare worker in a Belgian nosocomial outbreak despite primary neutralizing antibody response. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., Canziani G.A., Carter E.P., Chaiken I. The case for S2: the potential benefits of the S2 subunit of the SARS-CoV-2 spike protein as an immunogen in fighting the COVID-19 pandemic. Front. Immunol. 2021;12:637651. doi: 10.3389/fimmu.2021.637651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. MGH COVID-19 Collection & Processing Team Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370 doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smatti M.K., Al Thani A.A., Yassine H.M. Viral-induced enhanced disease illness. Front. Microbiol. 2018;9:2991. doi: 10.3389/fmicb.2018.02991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulin N.K., Brewer R.C., Sherwood R., Bournazos S., Edwards K.G., Ramadoss N.S., Taubenberger J.K., Memoli M., Gentles A.J., Jagannathan P., et al. Maternal anti-dengue IgG fucosylation predicts susceptibility to dengue disease in infants. Cell Rep. 2020;31:107642. doi: 10.1016/j.celrep.2020.107642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Kao D.J., Jeffers S.A., Holmes K.V., Hodges R.S. Template-based coiled-coil antigens elicit neutralizing antibodies to the SARS-coronavirus. J. Struct. Biol. 2006;155:176–194. doi: 10.1016/j.jsb.2006.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterman A., Sumida T.S., Nouri N., Yan X., Zhao A.Y., Gasque V., Schupp J.C., Asashima H., Liu Y., Cosme C., Jr., et al. The Yale IMPACT Research Team Single-cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19. Nat. Commun. 2022;13:440. doi: 10.1038/s41467-021-27716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelpoel L.T.C., Baeten D.L.P., de Jong E.C., den Dunnen J. Control of cytokine production by human fc gamma receptors: implications for pathogen defense and autoimmunity. Front. Immunol. 2015;6:79. doi: 10.3389/fimmu.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U S A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., Teng F., Dai J., Duan H., Guo S., et al. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020;6:2238–2249. doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.F., Tseng S.P., Yen C.H., Yang J.Y., Tsao C.H., Shen C.W., Chen K.H., Liu F.T., Liu W.T., Chen Y.M.A., Huang J.C. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.T., Sewatanon J., Memoli M.J., Wrammert J., Bournazos S., Bhaumik S.K., Pinsky B.A., Chokephaibulkit K., Onlamoon N., Pattanapanyasat K., et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355:395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z., et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Investig. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wang B., Mao J. The pathogenesis and treatment of the 'Cytokine Storm' in COVID-19. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M.S., Leung N.H.L., Cheung C.Y., Li P.H., Lee H.H.Y., Daeron M., Peiris J.S.M., Bruzzone R., Jaume M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Ng L.F.P., Anderson D.E., Chia W.N., Chia P.Y., Ang L.W., Mak T.M., Kalimuddin S., Chai L.Y.A., et al. Singapore 2019 Novel Coronavirus Outbreak Research Team Viral dynamics and immune correlates of coronavirus disease 2019 (COVID-19) severity. Clin. Infect. Dis. 2020;73:e2932–e2942. doi: 10.1093/cid/ciaa1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamecnik C.R., Rajan J.V., Yamauchi K.A., Mann S.A., Loudermilk R.P., Sowa G.M., Zorn K.C., Alvarenga B.D., Gaebler C., Caskey M., et al. ReScan, a multiplex diagnostic pipeline, pans human sera for SARS-CoV-2 antigens. Cell Rep. Med. 2020;1:100123. doi: 10.1016/j.xcrm.2020.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhou X., Zhu C., Song Y., Feng F., Qiu Y., Feng J., Jia Q., Song Q., Zhu B., Wang J. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Front. Mol. Biosci. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request, unless it is protected by law.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.