SUMMARY
Neurons must function for decades of life, but how these non-dividing cells are preserved is poorly understood. Using mouse serotonin (5-HT) neurons as a model, we report an adult-stage transcriptional program specialized to ensure the preservation of neuronal connectivity. We uncover a switch in Lmx1b and Pet1 transcription factor function from controlling embryonic axonal growth to sustaining a transcriptomic signature of 5-HT connectivity comprising functionally diverse synaptic and axonal genes. Adult-stage deficiency of Lmx1b and Pet1 causes slowly progressing degeneration of 5-HT synapses and axons, increased susceptibility of 5-HT axons to neurotoxic injury, and abnormal stress responses. Axon degeneration occurs in a die back pattern and is accompanied by accumulation of α-synuclein and amyloid precursor protein in spheroids and mitochondrial fragmentation without cell body loss. Our findings suggest that neuronal connectivity is transcriptionally protected by maintenance of connectivity transcriptomes; progressive decay of such transcriptomes may contribute to age-related diseases of brain circuitry.
Graphical abstract
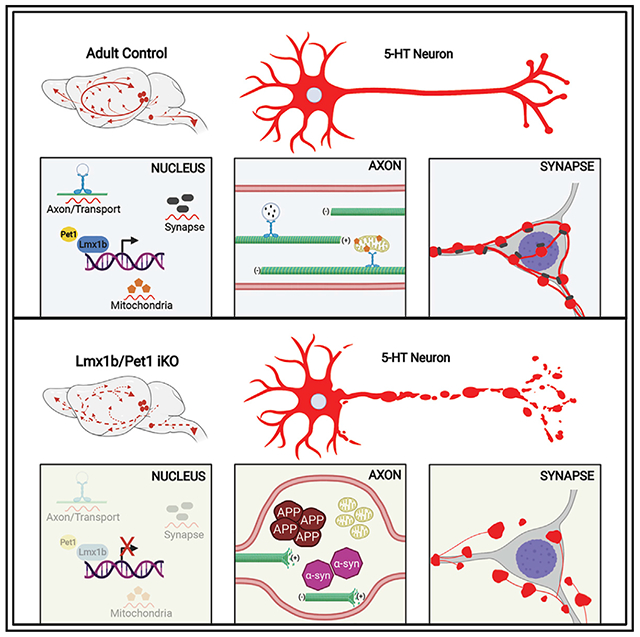
In brief
Kitt et al. show that adult serotonergic connectivity is protected from expansive degeneration by an Lmx1b/Pet1-regulated transcriptional program. Adult-stage deficiency of these transcription factors causes progressive breakdown of 5-HT synapses and axons accompanied by accumulation of α-synuclein and amyloid precursor protein in spheroids and mitochondrial fragmentation without cell body loss.
INTRODUCTION
Neural circuitry in the brain relies on the functional integrity of constituent neurons and their stable connectivity. Post-mitotic neurons must function for up to more than a century of life to preserve cognition, perception, and movement (Heckman and Doe, 2021; Lin and Koleske, 2010). Life-long function of the brain depends on the stability of neuronal gene expression to enable continuous homeostatic replacement of materials in various cellular compartments including long distance axons and synapses, which are particularly vulnerable to pathology during aging and in many neurodegenerative disorders (De Vos et al., 2008; Guedes-Dias and Holzbaur, 2019). The intrinsic regulatory programs operating to generate different neuron types during development are relatively well understood (Deneris and Gaspar, 2018; Fishell and Kepecs, 2020; Hobert and Kratsios, 2019; Lodato and Arlotta, 2015). Whether neurons possess adult-stage intrinsic regulatory programs dedicated to safeguarding their post-developmental health against breakdown of neural circuit connectivity has received little attention and is thus largely obscure (Deneris and Hobert, 2014). This represents a critical gap in knowledge, as it seems likely that failure to maintain neuronal gene expression may precipitate post-developmental neurologic disease through transcriptomic decay and consequent failure to compensate for constant turnover of molecules that are continually needed to preserve neuronal connectivity and function (Coleman and Hoke, 2020).
The LIM homeodomain protein Lmx1b and the ETS domain protein Pet1 are terminal-selector-type transcription factors required for the development of 5-HT neurons (Deneris and Wyler, 2012; Hobert, 2011; Spencer and Deneris, 2017). They function, initially, in 5-HT postmitotic precursors to coordinately activate 5-HT neurotransmission genes encoding serotonin (5-HT) synthesis (Tryptophan hydroxylase 2 [Tph2], Dopa decarboxylase [Ddc], GTP cyclohydrolase 1 [Gch1], GTP cyclohydrolase 1 feedback regulator [Gchfr]), reuptake (serotonin transporters, Sert [Slc6a4], Oct3 [Slc22a3]), autoregulatory receptors [Htr1a, Htr1b], vesicular transport (vesicular monoamine transporter 2, VMAT2 [Slc18a2]), and enzymatic degradation (monoamine oxidases A and B, [Maoa, Maob]) (Hendricks et al., 2003; Wyler et al., 2016; Zhao et al., 2006). In addition, they function at different embryonic to early postnatal stages of 5-HT neuron maturation to control long-distance growth and arborization of 5-HT axons, hence enabling expansive serotonergic neuromodulation in the brain and spinal cord (Donovan et al., 2019). Pet1 and Lmx1b retain robust expression in all adult 5-HT neurons (Dai et al., 2008; Hendricks et al., 1999). Targeting of Lmx1b and Pet1 in adult 5-HT neurons revealed that they are required to maintain normal levels of expression of Tph2, Slc6a4, and Slc18a2 (Liu et al., 2010; Song et al., 2011). Yet, it remains unknown whether Lmx1b and Pet1 function in adulthood is limited to maintaining these small numbers of terminal effector genes or whether they are needed more broadly to safeguard the health of 5-HT neurons.
Here, we comprehensively investigated the long-term consequences of adult-stage Lmx1b and Pet1 deficiency in 5-HT neurons. Our results show that these transcription factors govern an adult-stage intrinsic program specialized to maintain a transcriptomic signature of serotonergic connectivity. Loss of adult-stage Lmx1b and Pet1 led to a progressive degeneration of 5-HT synapses, breakdown of ascending and descending 5-HT axon architectures in the absence of 5-HT cell body loss, increased susceptibility of 5-HT axons to amphetamine-induced injury, and abnormal stress responses. Axon degeneration was accompanied by mitochondrial fragmentation and accumulation of α-synuclein (α-syn), amyloid precursor protein (APP), and phosphorylated neurofilament (p-NF) in abnormally swollen varicosities and spheroids. Our findings suggest that neuronal connectivity is transcriptionally protected against degeneration and injury by maintenance of a transcriptomic signature of connectivity; impaired preservation of connectivity transcriptomes may therefore contribute to aging- and disease-related degeneration of adult-brain circuitry.
RESULTS
To investigate adult-stage Lmx1b and Pet1 functions, we developed a tamoxifen (TAM)-inducible strategy with the Tph2-CreER transgenic line to target Pet1 (iPKO: Pet1fl/fl; Tph2-CreER; Ai9), or Lmx1b (iLKO: Lmx1bfl/fl; Tph2-CreER; Ai9), or both (iDKO: Pet1fl/fl/Lmx1bfl/fl; Tph2-CreER; Ai9) in adult 5-HT neurons while simultaneously genetically marking targeted neurons with Tdtomato using the Cre-dependent Ai9 allele. In 2- to 3-month-old control mice (iCON: +/+; Tph2-CreER; Ai9) treated with TAM for 5 consecutive days, 98% of Tph2+ neurons were TdTomato+, and 96% of TdTomato+ neurons were Tph2+, indicating consistent and robust TAM-inducible targeting efficiency and specificity, respectively (Figure S1A). In situ hybridization (ISH) analyses in the dorsal raphe nucleus (DRN) of iPKO mice indicated near elimination of Pet1 expression (Figure S1B).
Lmx1b and Pet1 sustain a broad transcriptomic signature of 5-HT connectivity
We performed RNA sequencing with flow-sorted adult 5-HT neurons to investigate the whole-genome functions of Lmx1b and Pet1 in sustaining 5-HT neuron transcriptomes. Comparable numbers of flow-sorted TdTomato+ cells were obtained at 10 days and 1 month post-TAM (Figure S1C). RNA-seq reads mapped to the largest Lmx1b-recombined exon (exon 4) were substantially decreased in iLKO versus iCON at 1 month post-TAM, indicating highly reproducible targeting of Lmx1b (Figure S1D). The total number of iLKO differentially expressed genes (DEGs) was small at 10 days post-TAM but showed decreased expression of Tph2 and Slc6a4, demonstrating that Lmx1b-dependent gene expression was effectively targeted (Figures S1E and S1F). At 1 month post-TAM, 571 DEGs including many 5-HT neurotransmission genes and Slc17a8 encoding the vesicular glutamate transporter, VGlut3, were significantly altered in iLKO 5-HT neurons (Figures 1A and 1B). Loss of Pet1 expression in iLKO mice reveals that Lmx1b is required to maintain Pet1 expression in adult 5-HT neurons similar to its dependence on Lmx1b at embryonic stages (Donovan et al., 2019). Over 98% of the iLKO DEGs resulted from decreased expression, suggesting that Lmx1b in adult 5-HT neurons functions largely as a transcriptional activator to guard against serotonergic transcriptomic decay. In contrast to iLKO, far fewer DEGs were detected in iPKO 5-HT neurons (Figures 1A, S1G, and S1H). Yet, a greater number of DEGs, including all 5-HT neurotransmission genes, were found in iDKO versus iLKO 5-HT neurons (Figures 1A and 1B). The direction of change was highly concordant between iLKO and iDKO DEGs, but the magnitude of expression changes was generally greater in iDKO than in iLKO, further suggesting a requirement for Pet1 in sustaining 5-HT transcriptomes (Figure 1C). As independent verification, AAV-Cre was stereotaxically injected into the DRN of adult Pet1fl/fl (vPKO), Lmx1bfl/fl (vLKO), Pet1fl/fl/Lmx1bfl/fl (vDKO), or +/+ (vCON) mice, which virtually eliminated Pet1 and Lmx1b expression (Figure S1I). Expression deficits comparable to those found after TAM-inducible targeting of Pet1 and Lmx1b were found for 5-HT neurotransmission genes at 1 month post-injection in vPKO, vLKO, and vDKO mice. Analyses at 6 months post-injection revealed further decreases in the levels of expression of these genes (Figures S1J and S1K). Likewise, we found a greater decrease in 5-HT immunofluorescence at 6 months post-TAM (Figure 1D). Thus, decreased expression of endogenous 5-HT neuron markers became evident as early as 10 days after targeting and worsened to include loss of expression of all 5-HT markers at 6 months post-targeting, providing evidence in support of a progressive decay of 5-HT neuron transcriptomes.
Figure 1. An adult-stage transcriptomic signature of 5-HT connectivity controlled by Lmx1b and Pet1.
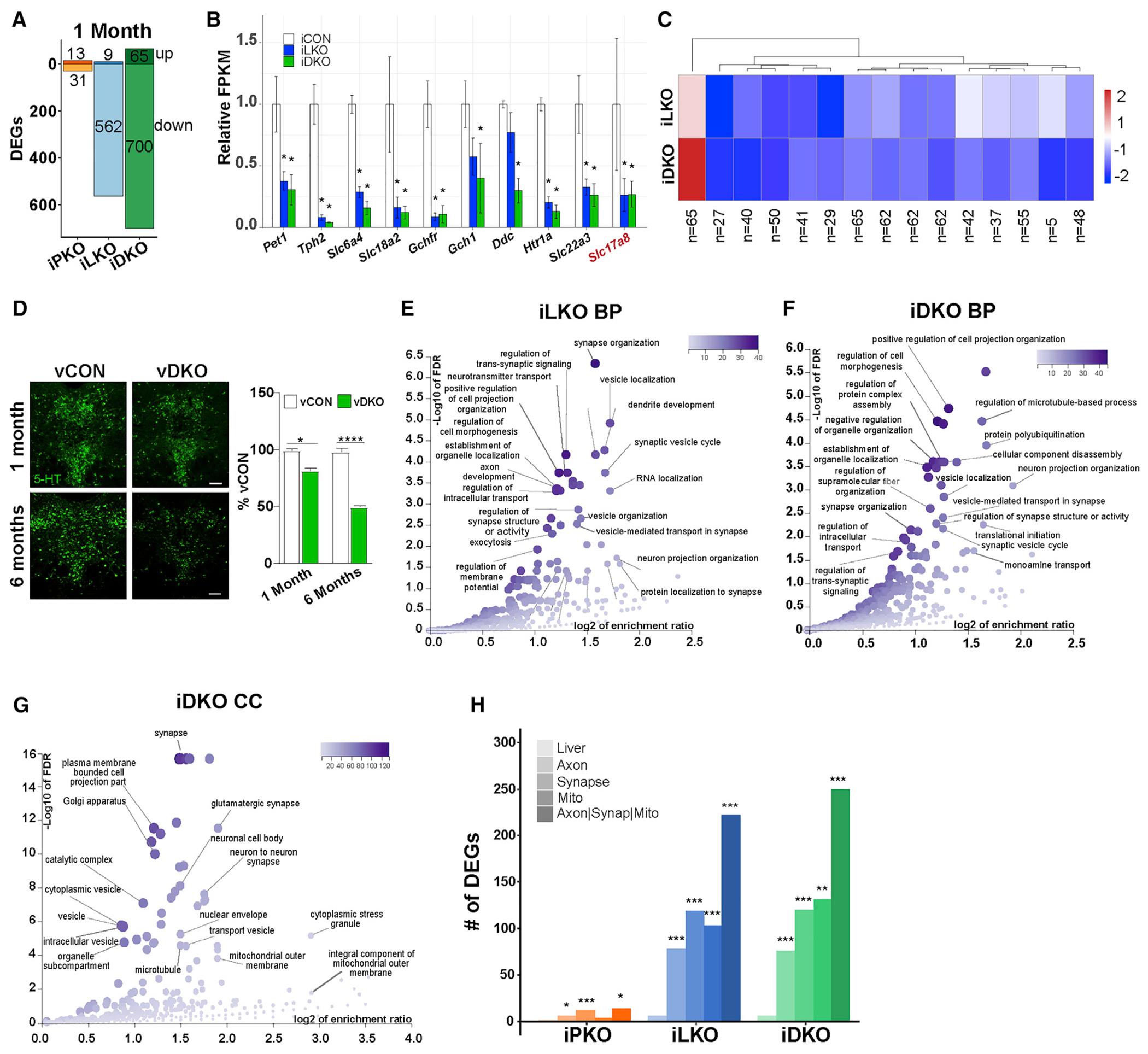
(A) iPKO, iLKO, and iDKO DEGs at 1 month post-TAM; FDR ≤ 0.05. Up or down DEG numbers are indicated for each genotype.
(B) Relative expression levels of 5-HT and glutamatergic (Slc17a8) neurotransmission genes at 1 month post-TAM.
(C) Heatmap of 1 month post-TAM iDKO and iLKO DEG expression levels; k = 15, k-means clustering. Each box represents the scaled mean expression level of the group of genes for each cluster and genotype. The dendrogram indicates the degree of similarity between each k-means cluster. The number of genes in each cluster is shown below each cluster. Red and blue indicate up- and down-regulated direction, respectively.
(D) Representative 5-HT immunostaining 1 and 6 months after AAV-Cre injection; ±SEM; n = 1,000–1,500 neurons analyzed in three sections per animal per genotype; 1-way ANOVA. Scale bar, 100 μm.
(E and F) Biological process (BP) term enrichment of 1 month post-TAM iLKO (E) and iDKO (F) down DEGs.
(G) Cellular component (CC) term enrichment of 1 month post-TAM iDKO down DEGs.
(H) DEG counts for indicated GO terms. Hypergeometric test of DEG counts for each GO term versus counts of all protein-coding genes annotated with the GO term. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S1.
To gain insight into the function of genes controlled by Lmx1b and Pet1 in adult 5-HT neurons, we performed gene ontology (GO) enrichment analyses. GO analyses of iLKO datasets at 10 days post-TAM using FDR ≤ 0.05 revealed an enrichment of terms related to synapse and actin cytoskeleton (Figure S1L). At 1 month post-TAM, analysis of iLKO DEGs revealed a striking increase in the enrichment of terms related to synapse- and axon-related processes (Figure 1E). Genes supporting enrichment for these terms encode proteins that possess a range of functions, including ones involved in actin and microtubule cytoskeletal structure and signaling, microfilament motor activity, anterograde and retrograde transport machinery, lipid homeostasis enzymes, synapse assembly, and mitochondrional function. GO enriched terms in 1-month post-TAM iDKO datasets were either identical or highly related to the 1 month post-TAM iLKO-enriched GO terms (Figures 1F and 1G). We found that 40% (222/562, p = 6.6 X 10−20 hypergeometric test) of iLKO-down DEGs and 36% (250/700, p = 8.6 × 10−16 hypergeometric test) of iDKO-down DEGs were annotated with at least one of these classification terms, indicating that a large portion of the serotonergic transcriptome maintained by Lmx1b and Pet1 is associated with synapse, axon, or mitochondrial processes (Figure 1H).
Progressive loss of serotonergic pericellular baskets
Given the significant enrichment of GO terms related to synapses in iLKO and iDKO DEGs, we investigated whether the adult-stage decay of the serotonergic transcriptomic connectivity signature impacted the integrity of 5-HT neuron synapses. Although many 5-HT axon terminals do not form junctional synapses and therefore appear to engage in diffuse transmission, junctional serotonergic synaptic appositions are abundant in many areas of the brain (Descarries and Mechawar, 2000; Papadopoulos et al., 1987). An excellent example is found in the dorsolateral septum (dLS) where 5-HT axon fibers and boutons conspicuously innervate target cells to form what are called “pericellular baskets” (Gall and Moore, 1984; Senft and Dymecki, 2021). Pericellular baskets comprise one or more converging 5-HT axon terminals whose boutons envelop the postsynaptic target cell somas and proximal dendritic shafts to form symmetrical or asymmetrical synapses. Notably, pericellular basket synapses were remarkably discernable throughout the rostrocaudal axis of the dLS (Figure 2A) in iCON mice, as TdTomato efficiently filled approaching axons and presynaptic boutons that tightly wrapped DAPI-labeled cell somas (Figure S2A).
Figure 2. Lmx1b and Pet1 act in adult 5-HT neurons to ensure survival of pericellular baskets.
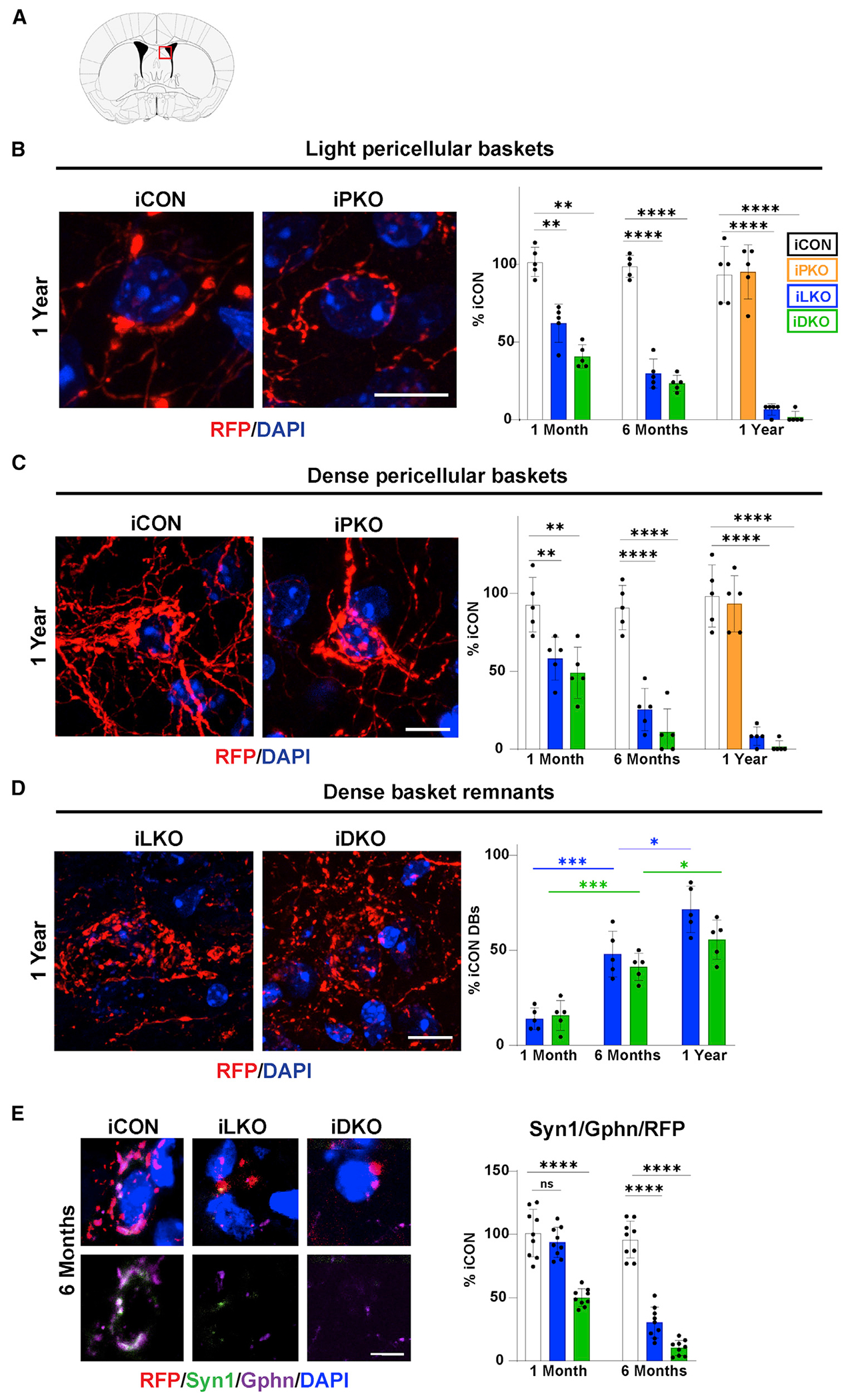
(A) Coronal schematic highlighting location (red box) of pericellular baskets in the dorsolateral septum (dLS).
(B–D) Left, representative confocal images (63×) of TdTomato+ (anti-RFP) light pericellular baskets (B), dense baskets (C), and dense basket remnants (D) at 1 year post-TAM. Right, quantification of basket or remnant numbers relative iCON numbers; ±SEM; n = 3 mice per genotype; 2–3 sections per animal were used for counting; two-way ANOVA with Welch’s correction; unpaired t test with Welch’s correction for iCON versus iPKO. Scale bars, 10 μm.
(E) Top, immunohistochemical analysis of synaptic triads in dLS at 6 months post-TAM, identified with RFP, presynaptic marker, synapsin 1 (Syn1), postsynaptic marker gephyrin (Gphn), and DAPI. Bottom, highlights overlap of pre- and post-synaptic makers. Right, relative numbers of triads; ±SEM; n = 3 mice with three sections analyzed per animal; one-way ANOVA. Scale bar, 5 μm. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant. See also Figure S2.
We identified two types of pericellular baskets in the dLS of iCON mice based on the density and extent of circumferential wrapping of TdTomato+ fibers around DAPI+ cell somas. Baskets designated “light baskets” (LBs) were defined as those arising from 1–2 TdTomato+ fibers and their associated boutons that wrap up to about 50% of the DAPI+ cell soma circumference; LBs were identifiable only at high magnification (Figure 2B). A morphologically more robust basket designated “dense baskets” (DBs) were found that comprise multiple TdTomato+ fibers whose boutons almost always entirely encased target DAPI+ cell somas; these were readily identified at low magnification (Figures 2C, S2A, and S2B). High-power confocal microscopy at 1 month post-TAM revealed a large and significant decrease in the number of LBs in iLKO and iDKO brains compared with their numbers in iCON brains (Figure 2B). At 6 months post-TAM, a further loss of LBs was found, and by 1 year post-TAM there were very few LBs in the dLS of iLKO and virtually none in iDKO brains (Figure 2B). These findings suggest that LBs fail to survive in iLKO and iDKO mice. Similarly, DBs were significantly reduced at 1 month post-TAM in iLKO and iDKO brains (Figure 2C). Examination at later time points indicated that DB numbers progressively decreased in abundance in iLKO and iDKO brains, so that by 1 year post-TAM few DBs could be identified in iLKO brains, and nearly none were present in the iDKO brain (Figure 2C).
Confocal imaging revealed that, instead of the presence of DBs, there were many abnormal clusters of TdTomato+ swollen boutons loosely arranged and uncoupled from DAPI+ cell somas in iLKO and iDKO brains; typically, these were either connected to severely thinned axons or appeared to be completely detached from incoming and surrounding fibers (Figures 2D, S2C, and S2D). Reasoning that these abnormal clusters of fibers and boutons were likely DBs prior to tamoxifen treatment, we designated these fiber/bouton structures “dense basket remnants” (DBRs). Counts at all three time points after tamoxifen revealed a progressive increase in the numbers of DBRs (Figure 2D). However, when compared with the numbers of DBs in iCON dLS, which we found to be stable across all time points examined (Figure S2E), the overall numbers of DBRs in iLKO and iDKO brains did not reach the numbers of DBs in iCON, suggesting that a substantial number of DBRs did not survive and were completely lost in the iLKO and iDKO brains. In contrast to these findings, we did not find a reduction in the numbers of LBs or DBs in iPKO brains even 1 year after tamoxifen gavage (Figures 2B, 2C, and S2B).
Serotonergic boutons have been shown to contact GABAergic synapses thus forming 5-HT-inhibitory synaptic triads (Figure S2F) (Belmer et al., 2017). To determine whether 5-HT pericellular baskets form such triads in the dLS, we used antibodies against TdTomato (RFP) and synapsin1 (Syn1) to distinguish 5-HT (RFP+/Syn1+) presynaptic elements from non-serotonergic (RFP−/Syn1+) presynaptic elements. Antibodies against gephyrin (Gphn) were included to identify GABAergic post-synaptic elements in apposition to RFP+/Syn1+ pre-synaptic elements. Syn1 was chosen as a pre-synaptic marker because its RNA expression in 5-HT neurons was not altered after Lmx1b targeting. In iCON brains, we found numerous examples of RFP+/Syn1+ double-stained pericellular baskets in close apposition to or even overlapping with Gphn+ cell somas, indicating that 5-HT boutons do indeed appear to form triads with inhibitory synapses in the dLS. However, in iLKO and iDKO brains there was a progressive decrease in the numbers of such triads in remaining terminals, which suggests that the degeneration of pericellular baskets led to the breakdown of post-synaptic GABAergic elements (Figures 2E and S2G).
Lmx1b and Pet1 protect adult 5-HT axon architectures against progressive degeneration
We next investigated 5-HT axon densities by comparing fluorescent pixel intensities of TdTomato+ axons in the brains of iPKO, iLKO, and iDKO mice. In iLKO and iDKO mice, small but significant decreases in TdTomato+ pixel intensities were evident at 1 month post-TAM in some regions, such as the cortex and hippocampus (Figures S3A and S3B). Axon densities in more proximal regions, thus possessing shorter axon lengths, such as the paraventricular nucleus of the thalamus (PVT), were unchanged at this time point (Figure S3C). At 6 months post-TAM, significant losses of TdTomato+ fiber densities were evident in all of the brain regions we examined. including the PVT in iLKO and iDKO mice, which worsened at 1 year post-TAM (Figures S3A–S3C). The progressive deficit in TdTomato+ axons in iLKO and iDKO mice was particularly noteworthy in the subventricular zone (SVZ), which receives dense 5-HT innervation. These axons terminate in the ventricular wall as a serotonergic ependymal plexus (SEP) enveloping the various SVZ cell types and extending past ependymal cells to the ventricular wall (Chan-Palay, 1976; Hawthorne et al., 2011; Jahanshahi et al., 2011; Kast et al., 2017). TdTomato labeling highlighted remarkably well the dense criss-crossing meshwork of 5-HT+ axons forming the SEP throughout the lateral ventricles of iCON mice (Figures 3A and 3B). In iLKO and iDKO mice, there were no significant axon density deficits at 1 month post-TAM in the SEP. However, at 6 months post-TAM, a breakdown of the SEP became evident, and at 1 year post-TAM, only remnants of the plexus were visible (Figure 3B). Similarly, we found a progressive breakdown of TdTomato+ axons in the supracallossal stria fiber tract (Figures 3A and 3C). In the prelimbic cortex, vertically oriented fibers in layer 1 and the irregular radially oriented TdTomato+ axons in layers 2–5 were largely missing (Figures 3A and 3D). Remnants of the TdTomato+ axonal plexus in the lateral ventricles showed reduced β3-tubulin immunostaining, suggesting disruption of the microtubule network and further corroborating a breakdown in 5-HT axon integrity (Figure S3D). In contrast to iLKO and iDKO, no such deficits were detected in iPKO mice at any of the three post-TAM time points. However, at all time points post-TAM and in all brain regions where differences were detected, there was a statistically significant greater deficit in TdTomato+ fiber intensities in iDKO compared with iLKO (Figures 3 and S3).
Figure 3. Progressive breakdown of 5-HT axon architectures.
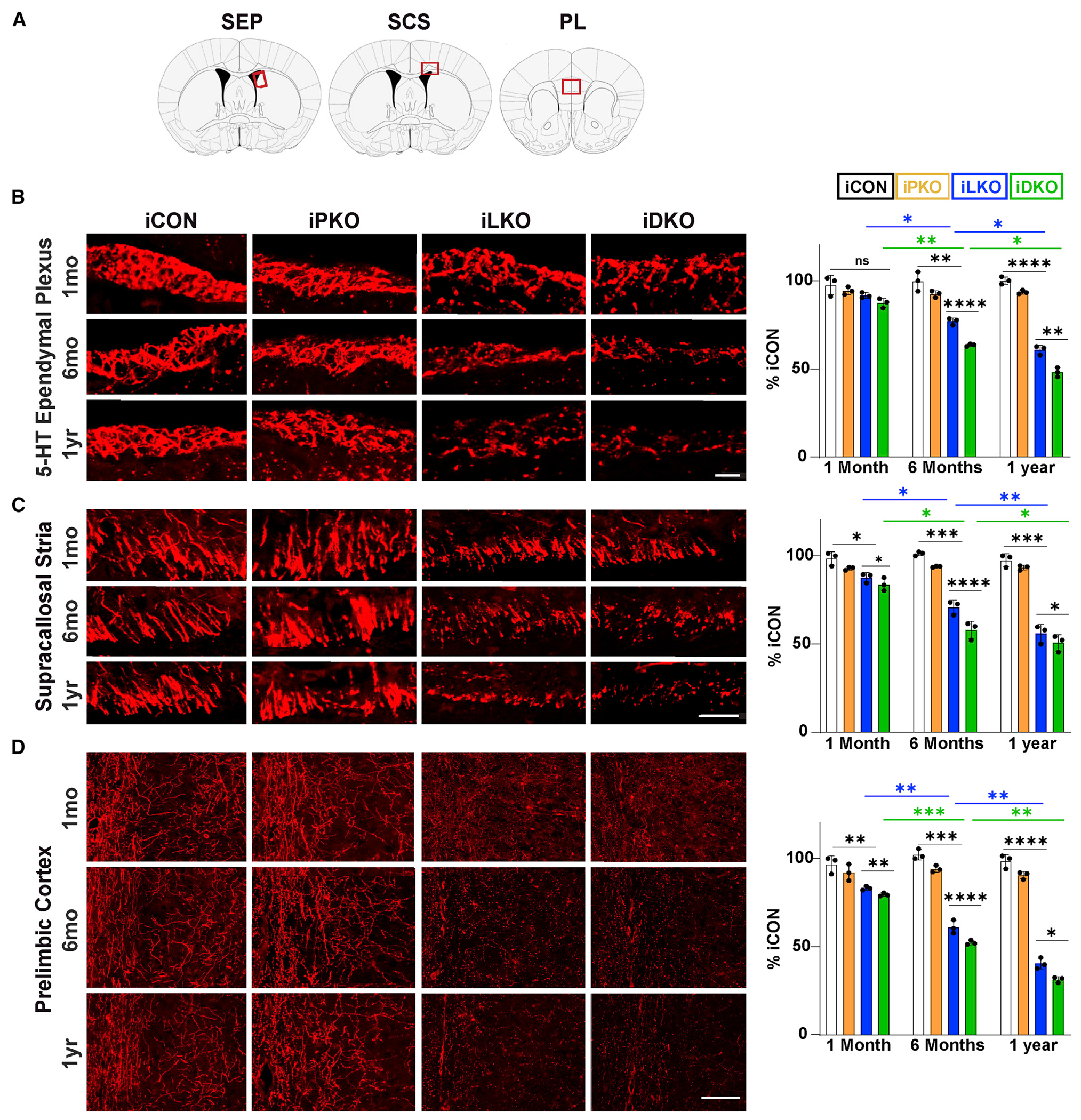
(A) Coronal schematics highlighting location (red rectangles) of analyzed regions: serotonergic ependymal plexus (SEP), supracallosal stria (SCS), prelimbic cortex (PL).
(B–D) Left, representative confocal images (63×) of TdTomato+ (anti-RFP) axons at indicated times post-TAM. Right, Quantification of relative RFP fluorescence intensity; ±SEM; n = 3 mice per genotype; two sections per animal; two-way ANOVA with Welch’s correction. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant. Scale bars, 50 μm in (B) and (C), 100 μm in (D). See also Figure S3.
Axon fragmentation, swollen varicosities, and spheroid formation
The deterioration of TdTomato+ fluorescence intensities in the SEP and supracallosal stria of iLKO and iDKO mice likely resulted from a complete loss of axon segments. In other areas of the brain, a dystrophic fiber morphology was evident in the iLKO and iDKO mice beginning at 6 months post-TAM and progressively worsening at 1 year post-TAM (Figures 4A and 4B). Using high-power confocal imaging, we characterized the dystrophic morphology and found that, whereas 80% of TdTomato+ fiber diameters in iCON mice were in the range 0.3–0.8 μm at all time points after TAM, less than 30% of TdTomato+ fibers in iLKO and iDKO mice at 6 months and 1 year post-TAM were in this range (Figure 4B). Instead, there was a greater frequency of thinner fibers with diameters in the 0.1–0.3 μm range in iLKO and iDKO brains at 6 months and 1 year post-TAM (Figure 4B). The abnormally thinned fibers were regularly interspersed with abnormally swollen TdTomato+ varicosities ranging in diameters of 0.8–1.0 μm and intermittently with dramatically larger abnormal structures (>5.0 μm in diameter) designated as spheroids (Figures 4A–4C). While normal-size single varicosities are regularly distributed throughout iCON axonal segments, the swollen varicosities found in iLKO and iDKO axons could sometimes be found clustered as tandems (Figure 4C, bottom). An additional feature of dystrophic axons in iLKO and iDKO brains was abnormal fiber clusters similar to those found in neurotoxin-induced degeneration and in the aging brain (Figure 4D) (van Luijtelaar et al., 1992). Further examination at 6 months post-TAM revealed that the constellation of dystrophic morphological features was pervasive, as a similar morphology was evident in long-distance descending fibers extending to the sacral spinal cord and even in 5-HT axons more proximal to 5-HT cell bodies such as those in the hypothalamus and inferior colliculus (Figure S4A). At 1 year post-TAM, virtually no area of the central nervous system was spared from severe 5-HT axon thinning and spheroid formation (Figure S4A).
Figure 4. Adult-stage progressive axon fragmentation and spheroid formation.
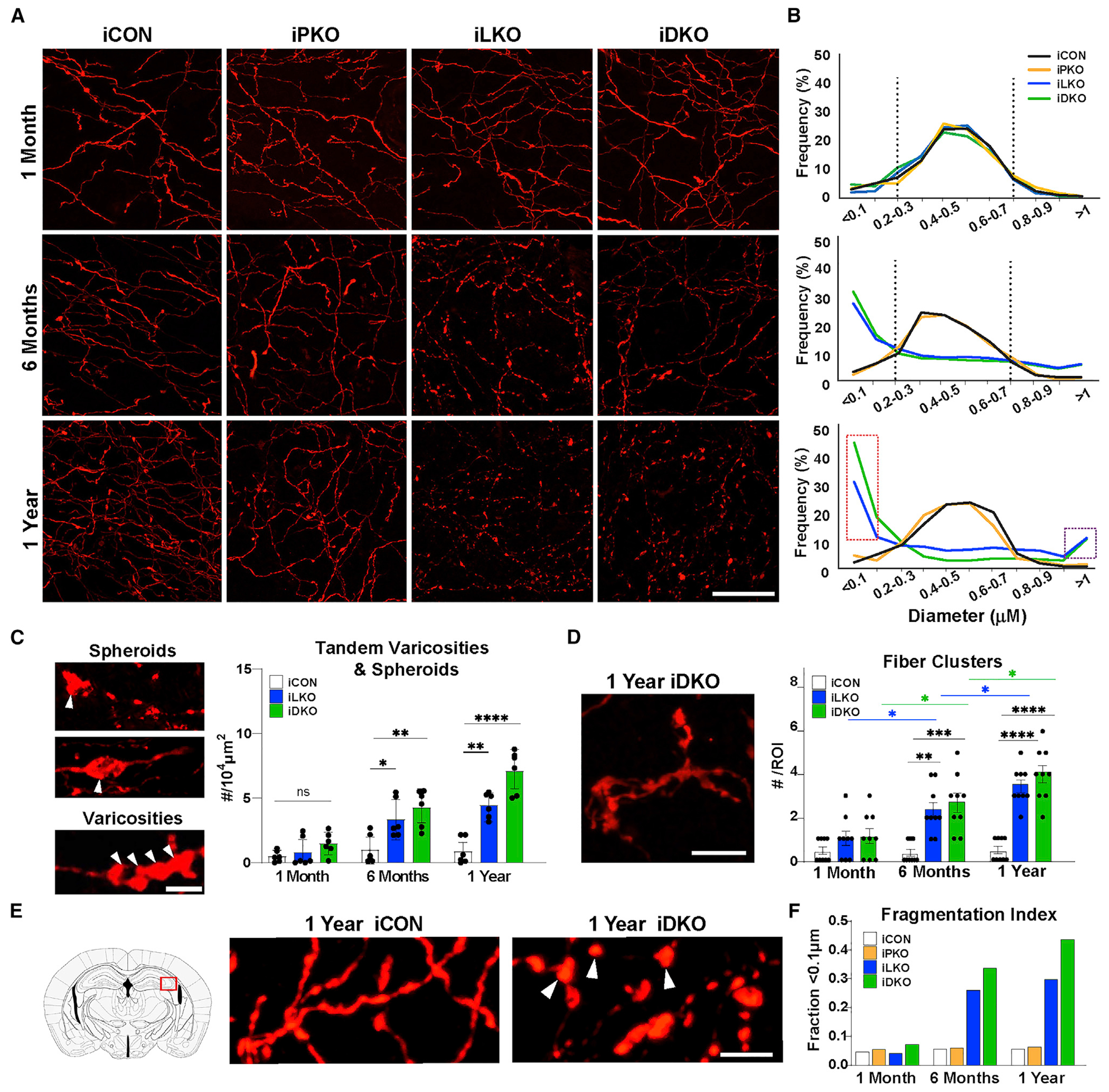
(A) Representative images of TdTomato+ (anti-RFP) axons in hippocampal CA2. Scale bar, 20 μm.
(B) Imaris quantification of binned (0.1 μm) axon diameter frequencies. Vertical dotted lines mark peak frequencies of iCON axon diameters. Dotted red rectangle highlights increased frequency of smaller fiber diameters in iLKO and iDKO. Dotted purple rectangle highlights increased frequency of large-diameter elements in iLKO and iDKO designated as swollen varicosities and spheroids; n = 2 images (63×) from two mice per genotype.
(C) Left, representative images of spheroids (arrowheads, top) and tandem swollen varicosities (arrowheads, bottom) in iDKO axons. Right, quantification of tandem swollen varicosities and spheroid numbers at indicated times post-TAM: ±SEM; n = 3 mice pergenotype. Two sections per animal were analyzed; one-way ANOVA at each time point.
(D) Left, representative image of abnormal fibers clusters in iDKO mice. Right, quantification of cluster numbers: ±SEM; n = 3 mice per genotype. Three sections per animal were analyzed; two-way ANOVA.
(E) High magnification image of iCON versus iDKO axons highlighting isolated swollen varicosities (arrowheads) in iDKO hippocampal CA2.
(F) Fragmentation index based on proportion of fiber diameters below the detection limit of <0.1 μm. (n = 2 mice per genotype with two images (63×) from each animal). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant. Scale bars, 5 μm. See also Figures S4–S7.
The varicose appearance of 5-HT fibers in iLKO and iDKO mice seemed similar to the transiently varicose or beaded morphology of developing 5-HT axons (Maddaloni et al., 2017). However, we found that developing 5-HT axons did not exhibit swollen varicosities or spheroids (Figure S4B). We also considered the possibility that the abnormal morphological features of axons in iLKO and iDKO mice were indicative of a compensatory regenerative reaction to axonal dystrophy. To address this, we treated iCON mice with p-chloroamphetamine (PCA) that potently destroys select 5-HT axon terminals (Mamounas et al., 1991) and examined 5-HT axon morphology 1 month later when 5-HT axons were actively regenerating (Jin et al., 2016). Although regenerating 5-HT axons took on a beaded morphology, we did not find swollen varicosities or spheroids (Figure S4B). Furthermore, regenerating axons exhibited an accumulation of the pre-synapse marker Syn1 in varicosities, suggestive of new pre-synaptic element formation, but spheroids in iDKO 5-HT neurons did not (Figure S4C). Together, these findings argue against iLKO and iDKO 5-HT axons mounting a regenerative response or reversion to an immature developmental state.
Many sequentially ordered swollen varicosities and spheroids in iLKO and iDKO mice were seemingly physically free of interconnecting TdTomato+ fiber segments suggesting fragmentation (Figures 4A and 4E). We sought to quantitate the extent of fragmentation using semiautomatic 3-D reconstruction and axon diameter analysis (Figure S5) (Maddaloni et al., 2017). The lower limit of TdTomato+ fiber detection was 0.1 μm, below which fibers could not be detected and were seemingly missing. We first counted the number of TdTomato+ fiber segments every 0.2 μm along the length of an axon that possessed diameters ≥0.1 μm. We then analyzed cases in which sequentially ordered swollen varicosities were devoid of TdTomato+ interconnecting segments (<0.1 μm diameter) but which we could reasonably surmise were once interconnected with axon segments of normal diameters. In these cases, we counted TdTomato-negative segments every 0.2 μm between swollen varicosities to obtain a tally of the number of undetectable axon segments between isolated varicosities. This analysis suggested a progressive increase in undetectable axon segments iLKO and iDKO brains (Figure 4F).
To determine whether progressive 5-HT synapse and axon degeneration elicited glial activation, as occurs in many chronic neurodegenerative conditions, we investigated the status of glial fibrillary acidic protein-positive (GFAP+) astrocytes and Iba1+ microglia in iLKO and iLKO cortex. GFAP immunofluorescence significantly increased in iLKO and iDKO brains compared with iCON brain at all examined ages (Figures S6A and S6C). In iCON brains at all examined ages there was a low level of Iba1 immunostaining in cells featuring small cell bodies and thin elongated processes suggesting a resting state (Figure S6A). However, in iLKO and iDKO brains at 6 months post-TAM there was a striking increase in the intensity of Iba1 immunofluorescence; Iba1+ cells in iLKO and iDKO had larger cell bodies and thicker processes, suggesting microglial activation (Figures S6A–S6C). CD68 is a transmembrane glycoprotein often used to assess the activated phagocytic state of microglia. We found a significant increase in CD68 immunofluorescence intensity in iLKO and iDKO compared with iCON (Figures S6A and S6C). These findings suggest that 5-HT synapse and axon decay in iLKO and iDKO brains elicited a robust inflammatory response characterized by microglia activation and astrogliosis.
5-HT deficiency does not contribute to degeneration or dystrophic morphology
Previous studies have shown that depletion of 5-HT after adult-stage Tph2 targeting resulted in increased 5-HT fiber densities in the hippocampus and decreased 5-HT fiber densities in the PVT at 1 month post-TAM (Pratelli et al., 2017). The severe loss of Tph2 expression and 5-HT immunostaining in the iLKO and iDKO brains (Figure 1), prompted us to investigate whether decreased 5-HT contributed to the degeneration of pericellular baskets and loss of 5-HT axon density in iLKO and iDKO mice. We used a similar adult-stage tamoxifen-inducible strategy to target Tph2 in Tph2fl/fl; Tph2-CreER; Ai9 (iTKO) adult mice. Adult-stage targeting of Tph2 led to complete loss of Tph2 and 5-HT immunostaining in the iTKO adult brain (Figure S7A). At 6 months post-TAM, we found no loss of LBs or DBs, no DBRs, and no altered TdTomato+ fiber densities or swollen varicosities or spheroids in iTKO brains (Figures S7B–S7D). Furthermore, Syn1 immunostaining was readily detected in TdTomato+ iTKO varicosities (Figure S7E). Interestingly, iTKO fibers at 6 months post-TAM exhibited a highly beaded morphology consisting of normal-size varicosities, which was quite distinct from the morphology of iCON, iLKO, and iDKO fibers. Together, these findings suggest that 5-HT loss does not contribute to 5-HT synapse and axon degeneration in iLKO and iDKO mice.
Lmx1b and Pet1 protect adult 5-HT axons against neurotoxin-induced injury
We next sought to determine whether Lmx1b and Pet1 might also protect axons against environmental insults. To investigate this, we tested the susceptibility of Lmx1b- and Pet1-deficient 5-HT axons to PCA-induced damage. PCA (20 mg/kg) was injected intraperitoneally twice per day for four consecutive days at either 10 days or 1 or 3 months after the last tamoxifen gavage (Figure 5A). Acute susceptibility to neurotoxic damage was assessed 1 day after the last PCA injection. At 10 days post-TAM, there was, as expected, a substantial loss of 5-HT fibers in the somatosensory cortex of iCON, iLKO, and iDKO mice but no significant differences among the genotypes. However, at 1 month post-TAM, we found significantly greater deficits in TdTomato+ fibers in iLKO and iDKO mice 1 day after PCA compared with iCON mice (Figures 5B and 5C). Susceptibility to PCA injury progressively worsened, as evidenced by the significant differences in TdTomato+ fiber deficits at 1 month versus 3 months post-TAM in iLKO and iDKO mice and was most severe in iDKO mice (Figure 5C). The progressive fiber pathology was accompanied by progressive increases in Iba1 and GFAP immunofluorescence, suggesting activated microglia and astrogliosis (Figures 5D–5G). These findings suggest that Lmx1b and Pet1 not only control an intrinsic adult-stage survival program for 5-HT synapses and axons but also provide protection against extrinsically induced injury.
Figure 5. Adult-stage targeting of Lmx1b and Pet1 causes increased susceptibility of 5-HT axons to neurotoxic injury.
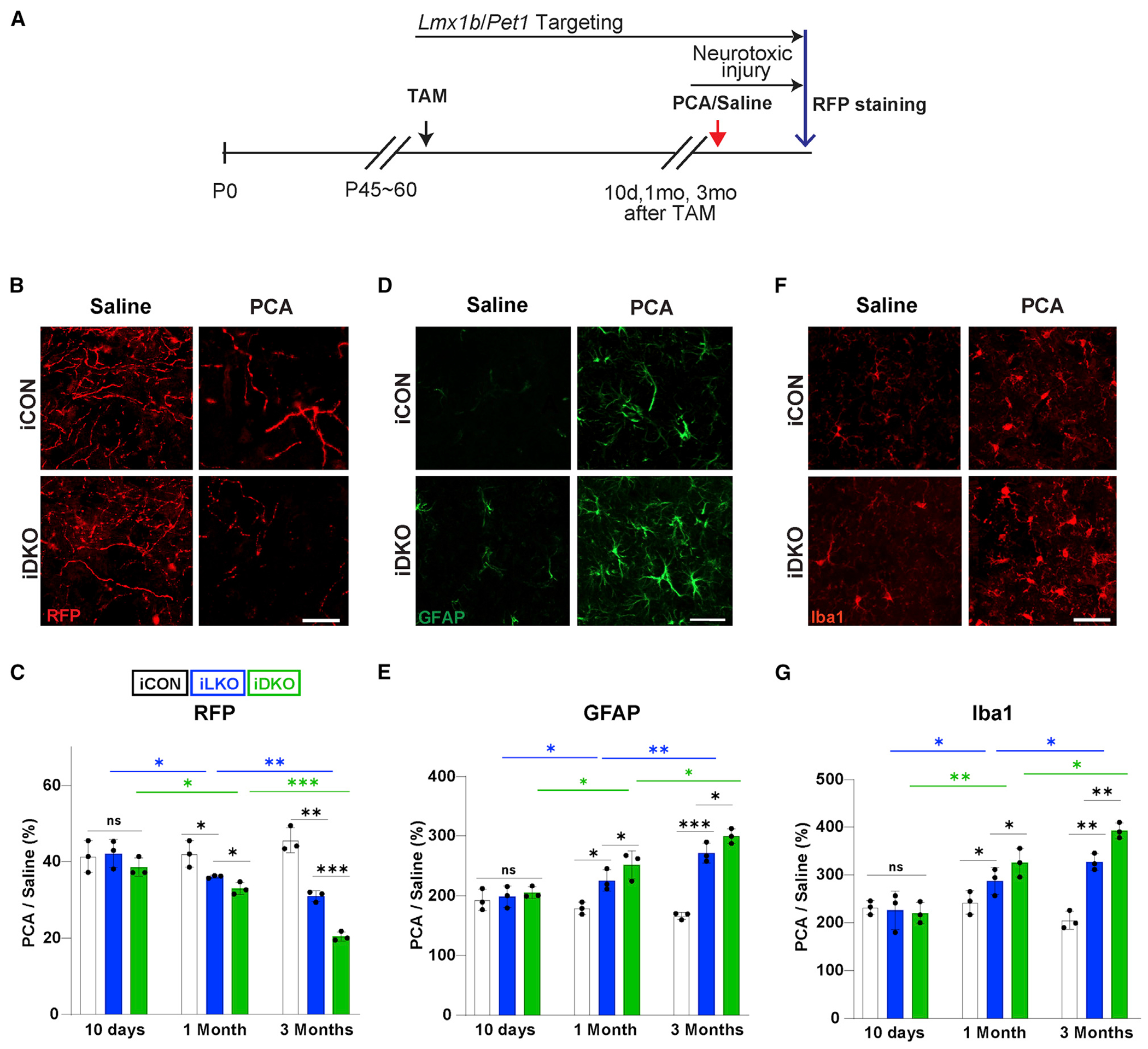
(A) Experimental design.
(B and C) Representative images at 1 month post-TAM (B) and quantification (C) of anti-RFP fluorescence intensities at indicated times post-TAM shows greater loss of TdTomato+ axons in somatosensory cortex of iLKO and iDKO versus iCON mice treated with PCA. Presented as ratio of TdTomato+ fiber intensities in PCA versus saline treated mice.
(D and E) Representative images of GFAP immunopositive cells (D) and quantification of immunofluorescence intensities (E) in somatosensory cortex at 1 month post-TAM.
(F and G) Representative images of Iba1 immunopositive cells (F) and quantification of immunofluorescence intensities (G) in somatosensory cortex at 1 month post-TAM. two-way ANOVA; ±SEM; n = 3 mice per genotype. *p < 0.05, **p < 0.01, ***p < 0.001. ns, not significant. Scale bars, 5μm.
5-HT neuron cell bodies retain long term survivability
We used whole-cell patch-clamp recordings to investigate the functional integrity of Lmx1b- and Pet1-deficient 5-HT cell bodies. Adult 5-HT neuron passive membrane properties, such as tau time constant, input resistance, membrane resistance, and voltage threshold, were not altered in iLKO and iDKO 5-HT neurons compared with iCON 5-HT neurons at 1 month post-TAM (Figure S8A). However, there was a significant increase in the firing rate of 5-HT neurons in iLKO or iDKO slices at 1 month post-TAM, which worsened at 3 months post-TAM (Figures 6A and 6B). Activation of somatodendritic 5-HT1a autoreceptors can inhibit 5-HT neuron firing (Andrade et al., 2015; Courtney and Ford, 2016). Given the significant loss of Htr1a expression in iLKO and iDKO 5-HT neurons (Figure 1B), we investigated 5-HT-inhibitory autoreceptor responses at 1 month post-TAM. In slices taken from 1 month post-TAM iCON mice, application of the 5-HT1a receptor agonist 8-OH-DPAT elicited inwardly rectifying currents. In contrast, no such changes in inward currents were elicited with 8-OH-DPAT in iLKO or iDKO slices (Figures 6C and 6D). Along with the reduced levels of 5-HT in the adult brain (Figure 1D) these findings suggest the loss of autoreceptor-mediated inhibition of 5-HT neuron firing may contribute to the increased firing rate of iLKO and iDKO 5-HT neurons.
Figure 6. Adult 5-HT neuron cell bodies exhibit long-term survivability after Lmx1b and Pet1 targeting.
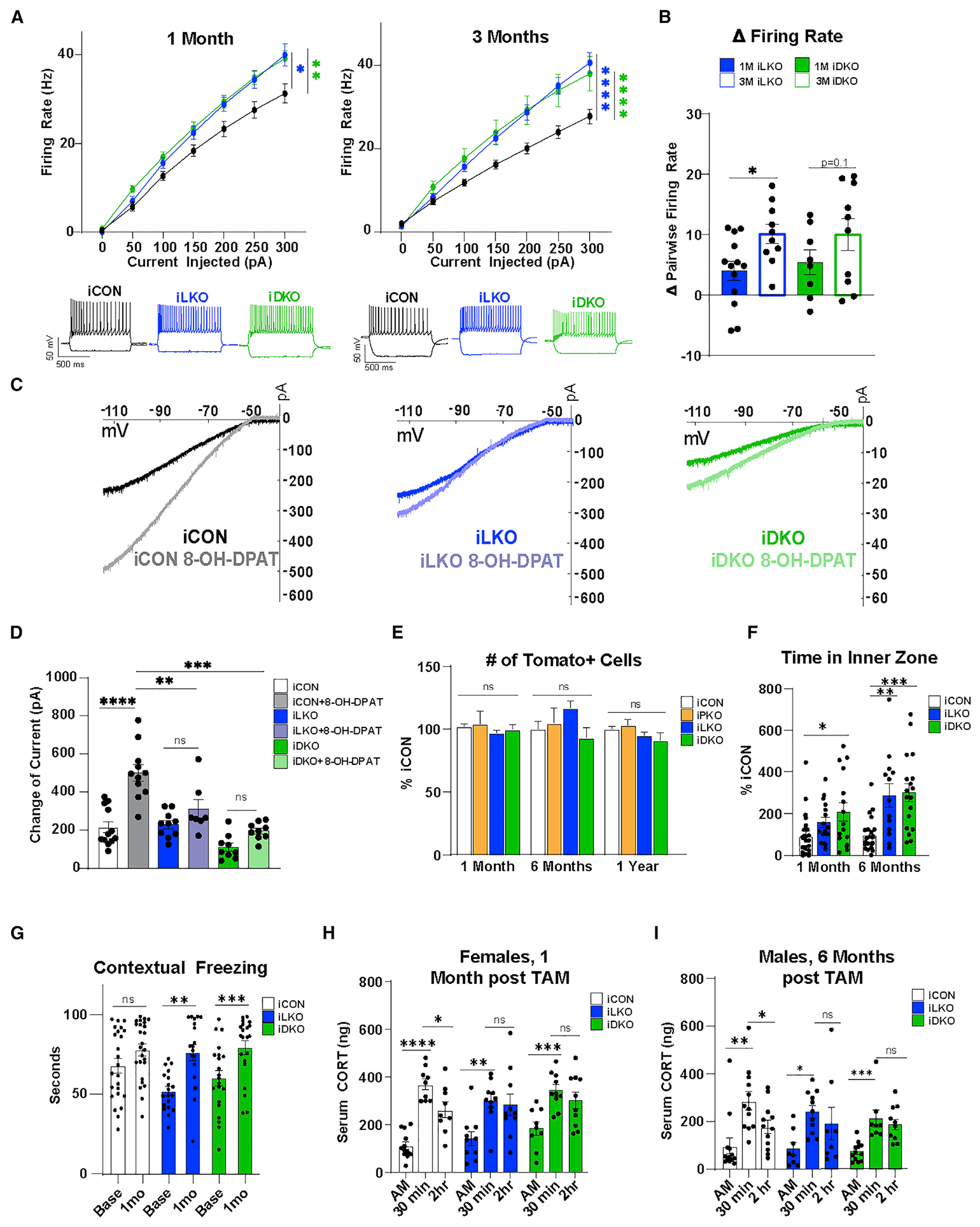
(A) iLKO and iDKO 5-HT neurons exhibit increased firing rates after indicated times post-TAM relative to iCON; ±SEM; one-way ANOVA.
(B) Excitability progressively increased in iLKO and trends in iDKO 5-HT neurons; ±SEM; pairwise t test.
(C) 8-OH-DPAT failed to elicit significant changes in inward-rectifying currents in 1-month post-TAM iLKO and iDKO 5-HT neurons.
(D) Quantification of change in 8-OH-DPAT elicited current responses in the indicated genotype at 1 month post-TAM; ±SEM; two-way ANOVA.
(E) TdTomato+ (anti-RFP) cell body counts at indicated times post-TAM; ±SEM; n = 3 mice per genotype; one-way ANOVA.
(F) Open field. Time in inner zone at indicated times post-TAM; ±SEM; n = 20–28 mice per genotype; two-way repeated-measures ANOVA.
(G) Contextual fear memory at baseline versus 1 month post-TAM; ±SEM; n = 20–28 mice per genotype; two-way repeated-measures ANOVA.
(H and I) Serum CORT levels measured 30 min before and 2 h after 30-min restraint stress in females (H) and males (I); ±SEM; n = 8–12 mice per genotype; two-way repeated-measures ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant. See also Figure S8.
We next assessed the physical integrity of 5-HT neurons to determine whether cell body degeneration accompanied altered firing properties and severe synapse and axon pathology. Measurement of TdTomato+ cell bodies revealed decreased cell body volumes beginning at 6 months post-TAM in iLKO and iDKO mice, which persisted at 1 year post-TAM. In contrast, volumes of iPKO TdTomato+ cell bodies were not different from those in iCON at any age post-TAM examined (Figure S8B). We counted TdTomato+ cell bodies throughout the rostrocaudal extent of the DRN and the median raphe nucleus (MRN) and found comparable numbers of TdTomato+ neurons across all genotypes up to 1 year post-TAM (Figure 6E). Long-term viability was further supported by the substantial numbers of flow-sorted TdTomato+ neurons obtained from iLKO and iDKO at 6 months post-TAM (Figure S8C). Thus, adult-stage Lmx1b and Pet1 are not required to sustain 5-HT cell body numbers up to 1 year post-TAM, and the early-stage increased firing rate of iLKO and iDKO 5-HT neurons did not impair their survival.
Altered stress-related behaviors accompany connectivity breakdown
We investigated the behavioral consequences of loss of adult-stage Lmx1b and Pet1. In the open field, total distance traveled by iLKO and iDKO was similar to that of controls prior to and after gene targeting (Figure S8D). However, iLKO and iDKO mice spent progressively more time in the inner zone of the open field compared with iCON mice after but not before targeting, indicating altered anxiety behaviors (Figures 6F and S8E). In assays of contextual fear memory, we found significantly increased freezing in iLKO and iDKO mice at 1 month post-TAM relative to baseline freezing measured prior to tamoxifen (Figure 6G). 5-HT function has been implicated in hypothalamic-pituitary-adrenal (HPA) axis function and stress responses, and Nr3c1 encoding the glucocorticoid receptor was a DEG in iLKO and iDKO at 1 month post-TAM (Figure S8F). Therefore, we assessed stress responses by measuring serum corticosterone (CORT) levels following 30-min restraint stress. At 1 and 6 months post-TAM, all control mice exhibited significant increases in serum CORT 30 min after restraint compared with baseline morning levels, which significantly declined 2 h following stress. We observed sex-dependent progressive alterations in stress responses in iLKO and iDKO mice. At 1 month post-TAM, iLKO and iDKO females exhibited early susceptibility to altered stress responses as their restraint-induced increase in CORT levels did not normalize when measured 2 h later (Figure 6H). Stress-induced serum CORT increases and recoveries in iLKO and iDKO males were similar to those in iCON mice at 1-month post-TAM (Figure S8G), but by 6 months post-TAM iLKO and iDKO males also failed to exhibit a normal return to baseline CORT levels, suggesting a late-stage onset of altered stress behaviors (Figure 6I).
An adult-stage transcriptional switch for preservation of connectivity
To probe the transcriptional mechanisms through which Lmx1b and Pet1 control an adult-stage 5-HT connectivity transcriptome, we next compared 1 month-post-TAM iLKO DEGs with E17.5 Lmx1bcKO DEGs (Donovan et al., 2019) and found that the majority of iLKO DEGs were adult stage-specific, with only 25% overlapping with genes that Lmx1b controls during 5-HT axon development (Figure S9A). GO analysis revealed that these adult-stage-specific DEGs retained the robust enrichment of terms related to synapse and axon as was found for all iLKO and iDKO DEGs (Figure S9B). GO analysis of the genes continuously regulated by Lmx1b revealed enrichment of terms related to serotonin neurotransmitter-related processes as well as synapse- and axon-related processes (Figure S9C).
We next used CUT&RUN (Skene and Henikoff, 2017) data obtained with Lmx1b and Pet1 antibodies to generate a map of Lmx1b and Pet1 genomic occupancy in flow-sorted 5-HT neurons (Zhang et al. https://doi.org/10.1101/2021.10.13.463782). We intersected the occupancy maps with iLKO and iPKO DEGs to determine the potential for direct control of the adult-stage connectivity signature. We found that 292/562 iLKO-down DEGs and 353/700 iDKO-down DEGs contained Lmx1b occupancy within 5 kb of gene transcriptional start sites (TSSs) (Figure S9D). Moreover, 95% of Lmx1b-occupied sites also showed Pet1 occupancy within 5 kb of gene TSSs, suggesting that Lmx1b and Pet1 directly sustain the transcriptomic signature of serotonergic connectivity (Figure S9D). Co-occupancy was often found at DEG TSSs and H3K4me3 and H3K27ac-enriched active promoters (Figure 7A).
Figure 7. An adult-stage transcriptional switch to regulation of a connectivity survival program.
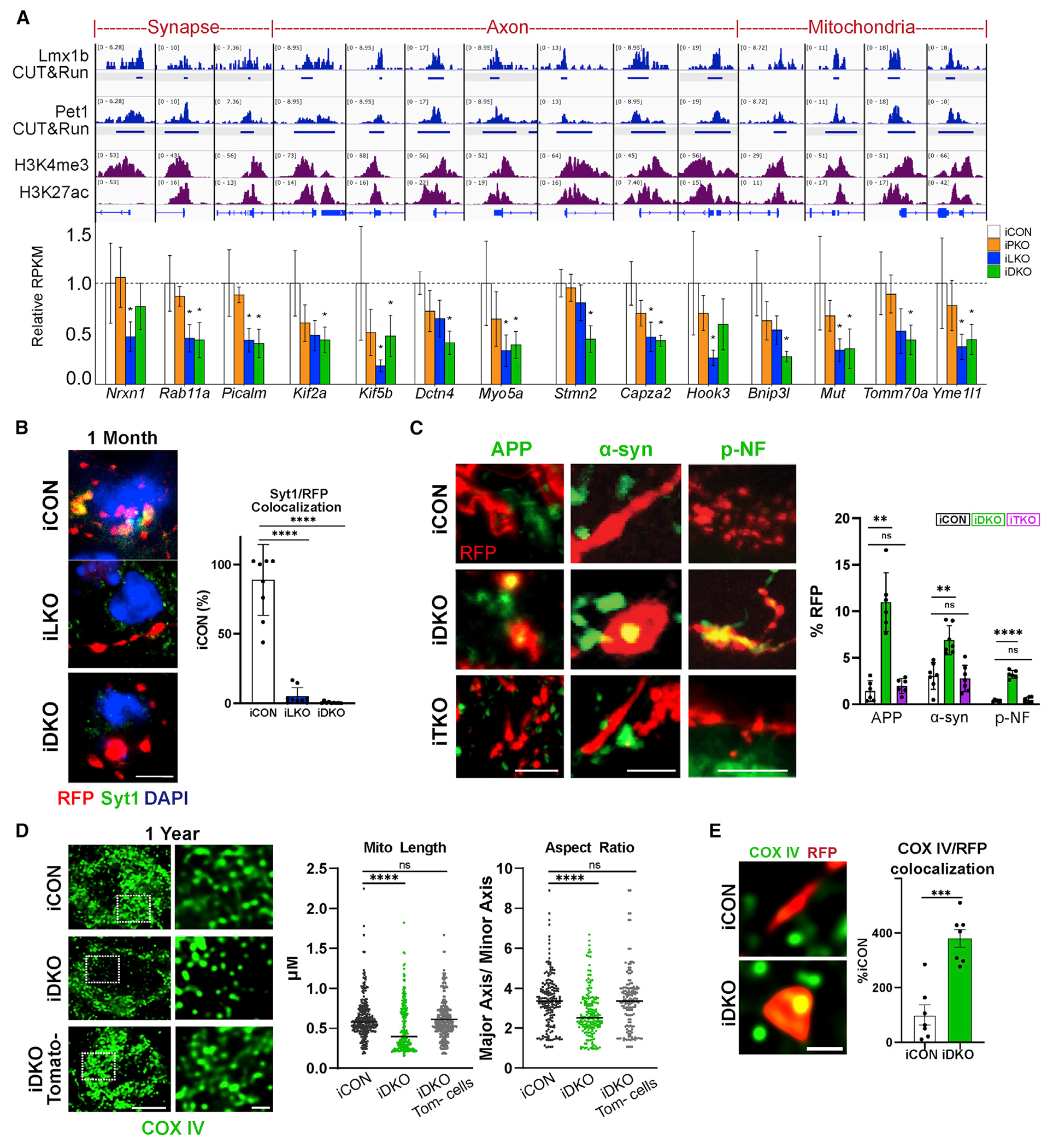
(A) Top, genome browser views of Lmx1b and Pet1 co-occupancy (reads in blue) at adult-stage-specific iLKO and iDKO DEG promoters, defined by enrichment of H3K4me3 and H3K27ac (reads in purple). Bottom, bar plots of adult-stage-specific DEGs; *FDR ≤ 0.05.
(B) Left, representative images of RFP and anti-Syt1 co-immunostaining in pericellular baskets. Right, quantification of Syt1/RFP colocalization; ±SEM; n = 3; one-way ANOVA. Scale bar, 5 μm.
(C) Left, axonal RFP co-immunostaining with APP, α-syn, and pNF. Scale bars from left, 2, 5, and 10 μm. Right, quantification of APP, α-syn, and p-NF accumulation in iDKO or iTKO at 6 months post-TAM; ±SEM; n = 3 mice per genotype; one-way ANOVA.
(D) Left, mitochondria labeled with COX IV exhibit tubular morphology in iCON mice and appear fragmented in iDKO mice at 1 year post-TAM. Right, quantification of mitochondrial length and aspect ratio; ±SEM; n = 200–300 mitochondria from two to three mice per genotype; unpaired t test with Welch’s correction. Scale bar, 1 μm.
(E) COX IV-labeled mitochondria in iDKO spheroids; ±SEM; n = 7 samples from three mice per genotype; unpaired t test with Welch’s correction. Scale bar, 1 μm. **p < 0.01, ***p < 0.001, ****p < 0.0001 in (B–E). See also Figure S9.
Fifty-two percent of adult-stage-specific DEGs exhibited Lmx1b occupancy, with 49% of these showing Lmx1b/Pet1 co-occupancy within 5 kb of gene TSSs. Adult-stage-specific genes with Lmx1b/Pet1 co-occupancy at TSSs include those encoding synapse components Nrxn1, Rab11, Pclo, and Picalm; axon structure and transport proteins Capza2, Hook3, Dctn4, Kif2a, and Kif5b, unconventional actin motor proteins Myo5a and Myo9a; Stmn2, involved in microtubule dynamics; and several nuclear genes encoding proteins with diverse mitochondrial functions, including methylmalonyl-CoA mutase, Mut, for production of key intermediates in the citric acid cycle; Tomm70a, a translocase of the outer mitochondrial membrane; Trak2, an adaptor that links kinesin and dynein motors for mitochondrial transport (Fenton et al., 2021); Yme1l1, an i-AAA protease component whose deficiency leads to mitochondrial fragmentation and spinal axon degeneration (MacVicar et al., 2019; Sprenger et al., 2019); and Bnip3l, a mitochondrial outer-membrane protein, which functions in PINK1/PARK2 signaling to mediate mitophagy and protects against stress-induced synaptic defects (Gao et al., 2015) (Figure 7A).
Fifty-one percent of continuously regulated DEGs exhibited Lmx1b occupancy, with 48% showing Lmx1b/Pet1 co-occupancy within 5 kb of gene TSSs. These include genes encoding synaptic components, synaptotagmin 1 (Syt1), Liprin-a2 (Ppfia2), and Rab3c; dynein motor component LIS1 (Pafah1b1); Trim2, which ubiquitinates neurofilament light-chain; axon transport protein Kif21a; Tau tubulin kinase 2 (Ttbk2); axon elongation protein Raph1; microtubule end-binding protein Mapre1; axon growth protein Rufy3; and nuclear encoded mitochondrial transport protein Armcx1 (Figure S9E). Comparing expression levels of iLKO DEGs in control 5-HT neurons from E17.5 to adulthood indicated that adult-stage-specific genes were strongly upregulated, continuously regulated genes slightly increased in expression, and embryonic-specific DEGs decreased in adulthood (Figure S9F). These findings suggest that Lmx1b switches to a new set of targets while also continuing to regulate a subset of embryonic genes for survival of 5-HT connectivity.
In support of failed maintenance of synapses, we found that pericellular basket remnants were rapidly depleted of Syt1 protein at 1 month post-TAM (Figure 7B). The lack of Syn1 protein in iLKO and iDKO pericellular basket remnants suggested impaired axonal transport (Figure S2G). To further assess this, we performed immunohistochemistry for APP, α-syn, and p-NF. Although expression of the genes encoding these endogenous markers of neurodegeneration were not altered at 1 month post-TAM, we found robust accumulation of all three markers specifically in iDKO swollen varicosities and spheroids but not in iCON or iTKO TdTomato+ fibers (Figure 7C). Finally, immunohistochemical analysis of mitochondria with a COX IV antibody revealed circular, fragmented mitochondria characterized by decreased length and aspect ratio at 1 year post-TAM in iDKO cell bodies (Figures 7D and S9G) and accumulation of mitochondria in spheroids (Figure 7E), further indicating altered mitochondrial dynamics and transport deficits, respectively.
DISCUSSION
Using mouse serotonin neurons in vivo as a model, we discovered an adult-stage transcriptional program that is specialized to ensure lifelong survival of neuronal connectivity and function. The adult-stage program we have uncovered is distinct from other programs that promote neuronal survival either during development or in mature neurons by suppressing programmed cell death (Huang and Reichardt, 2001; Lin et al., 2020). Indeed, the absence of overt 5-HT cell body loss suggests that the adult-stage program governed by Lmx1b and Pet1 is geared expressly to protect 5-HT synapses and axons. A key element of the connectivity survival program we have discovered is the preservation of 5-HT neurotransmission, yet we found a surprisingly broader, but highly interrelated, role for this adult-stage program in which it acts to ensure the health of 5-HT synapses and axons by safeguarding against their degeneration. GO analyses of adult-stage DEGs after targeting of Lmx1b and Pet1 revealed that these terminal selectors preserve an extensive transcriptomic signature of connectivity comprising functionally diverse synaptic, axonal, and mitochondrial genes. A portion of the connectivity genes controlled in adult 5-HT neurons are ones that Lmx1b also regulates during its embryonic control of long-distance 5-HT axon growth and arborization (Donovan et al., 2019). However, the majority comprise a new set of upregulated adult-stage-specific synaptic and axonal genes including ones specifically involved in axonal transport and diverse mitochondrial functions. Interestingly, loss of function of several of these genes has been implicated in axon fragmentation and axon neuropathy, respectively (Balastik et al., 2008; Klim et al., 2019; Shin et al., 2012; Ylikallio et al., 2013). These findings suggest that Lmx1b, with some contribution by Pet1, switches, in part, to new targets in adulthood to ensure a continuous supply of materials needed for long-term survival, rather than formation, of 5-HT connectivity.
Concomitant with the decay of the transcriptomic connectivity signature, we found a dramatic loss of 5-HT pericellular basket synapses and widespread 5-HT axon degeneration throughout the CNS. Axon degeneration comprised slowly progressive breakdown of distal axon tracts and termini together with the formation of swollen varicosities and spheroids. Accumulation of α-syn, APP, and p-NF in swollen varicosities and spheroids and a lack of the presynaptic marker protein Syn1, which is not transcriptionally controlled by Lmx1b or Pet1, in pericellular baskets remnants and spheroids, provide evidence in support of defective axon transport. In addition to the progressive degeneration that occurred in iLKO and iDKO mice, we found that Lmx1b deficiency resulted in greater amphetamine-induced loss of 5-HT axons, suggesting adult-stage transcriptional protection of axons against neurotoxic injury. Consistent with the major role of Lmx1b in maintaining gene expression, adult-stage degeneration of 5-HT connectivity largely resulted from the lack of Lmx1b, although dual deficiency of Lmx1b and Pet1 resulted in accelerated progressive breakdown of 5-HT synapses and axons.
Our analysis of iTKO mice indicated that adult-stage loss of 5-HT did not recapitulate any of the abnormal morphological features found in iLKO and iDKO 5-HT pericellular baskets and axons. The highly beaded morphology of iTKO 5-HT fibers resulting from increased numbers of normal-size varicosities was distinct from the dystrophic morphology of iLKO and iDKO fibers and may result from a long-term compensatory feedback mechanism that builds more release sites as transmitter levels gradually fall during the 6-month post-TAM period. In contrast to Pratelli et al., who observed increased 5-HT fiber densities in the hippocampus 1 month after adult-stage targeting of Tph2 (Pratelli et al., 2017), we never observed increased fiber densities in iLKO or iDKO mice at 1 month, 6 months, or 1 year post-TAM. In further contrast to our findings, 5-HT fiber morphology was not altered (Pratelli et al., 2017). Consistent with our findings, Pratelli et al. also found decreased 5-HT fiber densities in the PVT at 1 month after Tph2 targeting. However, at 1 month post-TAM, PVT fiber densities were not altered in iLKO and iDKO mice.
Axon degeneration can result from physical injury or exposure to neurotoxins and is an early feature of many age-related neurological and neurodegenerative disorders (Raff et al., 2002; Salvadores et al., 2017; Vickers et al., 2009). Axon degeneration often occurs in a die-back pattern in which degeneration initiates at axon termini and progressively propagates retrogradely toward the cell somas, which subsequently can result in cell soma death (Burke and O’Malley, 2013; Coleman and Hoke, 2020; Kneynsberg et al., 2017). Dying-back degeneration has been discussed as resulting from a failure to nourish distal regions of the neuron with new materials for renewal of axonal and synapse structures, axon transport machinery, and energy production (Cavanagh, 1984; Conforti et al., 2014). Consistent with this idea is that long-range axons are particularly susceptible to die-back pathology (Cavanagh, 1984). The small numbers of 5-HT neurons situated in the midbrain, pons, and ventral medulla extend extremely arborized, mostly unmyelinated, axons to nearly all areas of the brain and spinal cord, with estimated mean axon lengths per neuron of more than 170 m in humans (Wu et al., 2014). This expansive morphological feature likely places extraordinary demand for continual synthesis and long-distance anterograde delivery of new gene products to maintain labyrinthine axon membranes, cytoskeletal integrity, synaptic function, and energy production. Dystrophic morphological changes and loss of 5-HT axon densities occur during aging and in neurodegenerative diseases such as Parkinson’s disease (Azmitia and Nixon, 2008; van Luijtelaar et al., 1992), suggesting that breakdown of the connectivity survival program described here could be involved. Lmx1b may perform similar protective roles in other neurons: targeting of Lmx1b and its closely related paralog Lmx1a in adult midbrain dopamine (DA) neurons resulted in failed maintenance of expression of DA neuron marker tyrosine hydroxylase and enlarged DA nerve terminals in the striatum, which exhibited increased presence of autophagic vesicles and α-syn (Doucet-Beaupre et al., 2016; Laguna et al., 2015).
The spatiotemporal sequence of 5-HT connectivity breakdown reported here exhibited features of die-back degeneration. First, we found dramatic losses of TdTomato+ pericellular baskets in iLKO and iDKO beginning at 1 month post-TAM, which progressively worsened at 6 months and 1 year post-TAM. In contrast, at 1 month post-TAM, distal axon fiber densities were either mildly reduced in some regions of the brain or were not significantly different from axon densities in iCON brain until 6 months post-TAM. Second, more proximal axons such as those in the PVT did not exhibit axon density deficits until 6 months post-TAM. Third, deficient Syn1 immunostaining in distal iLKO and iDKO pericellular baskets and remnants was an early feature of the degeneration. Fourth, although iLKO and iDKO cell bodies had smaller diameters, their numbers were not reduced for at least up to 1 year after adult-stage targeting despite their increased firing rate, suggesting the degeneration of 5-HT connectivity does not arise from a dying forward pathology originating in 5-HT cell bodies.
Wallerian degeneration (WD) is a well-recognized form of programmed axon degeneration (Gerdts et al., 2016). WD is thought to result from an initial deficit in axons distal to transection in the levels of the NAD+ biosynthetic enzyme NMNAT2, and activation of sterile-α and Toll/interleukin-1 receptor (TIR) motif-containing protein 1 (Sarm1), resulting in cytoskeletal disintegration, mitochondrial swelling, and axon fragmentation (Figley et al., 2021; Gilley et al., 2019). We examined our RNA-seq datasets to determine whether adult-stage loss of Lmx1b and Pet1 altered the expression of NMNAT2 or SARM1 and did not find significant changes in their expression. This does not preclude the possibility that newly synthesized NMNAT2 fails to reach distal 5-HT axons as a result of axon transport impairment and thus contributes to the 5-HT axon degeneration that we report here. Yet, considering the breadth of transcriptome decay in iLKO and iDKO 5-HT neurons, it seems likely that the accompanying progressive 5-HT synapse and axon degeneration results from a more generalized failure of targeted 5-HT neurons to produce and transport cargoes to distal axons and synapses rather than a specific deficiency of a single labile maintenance factor. Thus, we propose that a progressive decay of connectivity transcriptomes brought about by impairment of adult-stage connectivity survival programs may represent a previously unrecognized path to aging- and disease-related degeneration of adult brain circuitry.
Limitations of the study
Our RNA-seq analyses provide evidence in support of an embryonic- to adult-stage switch in Lmx1b and Pet1 targets. One limitation is that we do not know how this switch is accomplished or when it occurs between embryonic and adult stages. A second limitation is that our CUT&RUN findings were obtained using embryonic 5-HT neurons and showed that Lmx1b and Pet1 occupy the promoter regions of many of their continuously controlled and adult–stage-specific targets at least before birth. Our efforts to obtain a CUT&RUN dataset with adult flow-sorted 5-HT neurons were not successful. Further work is required to unequivocally identify continuously controlled and adult-stage-specific direct targets of Lmx1b and Pet1. However, we believe that the simplest interpretation of our findings is that Lmx1b and Pet1 directly control at least many of the adult–stage-specific genes that we have identified by RNA-seq.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Evan Deneris (esd@case.edu).
Materials availability
Mouse lines used for this study are available at The Jackson Laboratory (Tph2-CreERT2 Strain #016584, Tph2fl Strain #027590, Lmx1bfl Strain #031287, Ai9 Strain #007909, Pet1-Cre Strain #012712) or upon request (Pet1fl).
Data and code availability
All data for our RNA-seq analyses have been deposited in the NCBI GEO (Gene Expression Omnibus) and are publicly available as of the date of publication. The accession number for the datasets is listed in the key resources table.
This paper analyzes existing, publicly available data. The accession number for the datasets is listed in the key resources table.
This paper does not generate original code.
Any additional information required to reanalyze the data included in this report is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| chicken anti-RFP 1:200 | Rockland | Cat# 600-901-379S, RRID:AB_10703148 |
| mouse anti-RFP 1:200 | Rockland | Cat# 200-301-379S, RRID:AB_2611064 |
| rabbit anti-RFP 1:200 | Rockland | Cat# 600-401-379S, RRID:AB_11182807 |
| rabbit anti-5-HT 1:200 | Immunostar | Cat# 20080, RRID:AB_572263 |
| rabbit anti-Tph2 1:500 | Millipore | Cat# ABN60, RRID:AB_11212793 |
| mouse anti-Tph2 1:500 | Millipore | Cat# MAB5278, RRID:AB_11214093 |
| rabbit anti-AADC 1:200 | Millipore | Cat# AB1569, RRID:AB_90789 |
| mouse anti-GFAP 1:500 | Sigma | Cat# G3893, RRID:AB_477010 |
| rabbit anti-Iba1 1:500 | FUJIFILM | Cat# 019-19741, RRID:AB_839504 |
| rabbit anti-COX4 1:500 | Thermo Fischer | Cat# PA5-29992, RRID:AB_2547466 |
| chicken anti-beta 3 tubulin 1:500 | Novus Biologicals | Cat# NB100-1612, RRID:AB_10000548 |
| rabbit anti-Sert 1:400 | Immunostar | Cat# 24330, RRID:AB_572209 |
| guinea pig anti-Synapsin1 1:500 | Synaptic Systems | Cat# 106 104, RRID:AB_2721082 |
| mouse anti-Synaptotagmin1 1:500 | Sigma | Cat# SAB1404433, RRID:AB_10759362 |
| mouse anti-Gephyrin 1:500 | Synaptic Systems | Cat# 147 011, RRID:AB_887717 |
| rabbit anti-α-Synuclein 1:500 | Cell Signaling | Cat# 2642, RRID:AB_2192679 |
| rabbit anti-APP 1:500 | Sigma | Cat# SAB4300464, RRID:AB_10621836 |
| mouse anti-P-Neurofilament 1:500 | BioLegend | Cat# SMI-31P-100, RRID:AB_2028812 |
| rat anti-CD68 1:500 | BIO RAD | Cat# MCA1957, RRID:AB_322219 |
| Goat anti-Mouse, Alexa 647 1:500 | Thermo Fisher Scientific | Cat# A-21240, RRID:AB_2535809 |
| Goat anti-Rabbit, Alexa 647 1:500 | Thermo Fisher Scientific | Cat# A-21246, RRID:AB_2535814 |
| Goat anti-Rabbit, Alexa 594 1:500 | Thermo Fisher Scientific | Cat# A-11012, RRID:AB_2534079 |
| Goat anti-Mouse, Alexa 594 1:500 | Thermo Fisher Scientific | Cat# A-21155, RRID:AB_2535785 |
| Goat anti-Chicken, Alexa 594 1:500 | Thermo Fisher Scientific | Cat# A32759, RRID:AB_2762829 |
| Goat anti-Rabbit, Alexa 488 1:500 | Thermo Fisher Scientific | Cat# A-11070, RRID:AB_2534114 |
| Goat anti-Mouse, Alexa 488 1:500 | Thermo Fisher Scientific | Cat# A-21131, RRID:AB_2535771 |
| VECTASTAIN ABC-Peroxidase Kit | Vector Laboratories | Cat# PK-4001, RRID:AB_2336810 Cat# PK-4004,RRID:AB_2336813 Cat# PK-4002, RRID:AB_2336811 Cat# PK-4007, RRID:AB_2336816 |
| Anti-Digoxigenin-AP Fab Fragments | Roche | Cat# 11093274910, RRID:AB_2734716 |
| Bacterial and virus strains | ||
| pENN.AAV.CMVs.PI.Cre.rBG | PennVector/ADDGENE | RRID:Addgene_105537 |
| Chemicals, peptides, and recombinant proteins | ||
| Protease | Sigma | Cat# P8811 |
| bupivacaine HCL | Hospira | Cat# C0409-3613-01 |
| Meloxicam | Pfizer | Cat# 71125-38-7 |
| Tamoxifen | Sigma | Cat# T5648 |
| PCA | Sigma | Cat# 274763 |
| 8-OH-DPAT | Tocris | Cat# 87394874 |
| D-AP5 | Tocris | Cat# 0106 |
| DNQX | Tocris | Cat# 0189 |
| TTX | Abcam | Cat# ab120054 |
| TRIZOL LS | Ambion | Cat# 10296010 |
| Chloroform | Fisher Scientific | Cat# C298500 |
| NBT | Roche | Cat# 11383213001 |
| BCIP | Roche | Cat# 11383221001 |
| Formimide | Roche | Cat# 11814320001 |
| Critical commercial assays | ||
| PerfeCTa PreAmp Supermix | QuantaBio | Cat# 89409170 |
| PerfeCTa FastMix II ROX Mastermix | QuantaBio | Cat# 97065990 |
| RNA Clean and Concentrator kit | ZYMO | Cat# R1013 |
| Ovation® SoLo RNA-Seq Library Preparation Kit | Tecan (Nugen Inc.) | Cat# 040732 |
| CORT ELISA kit | ENZO Life Sciences | Cat# 101106156 |
| Transcriptor First Strand cDNA Kit | Roche | Cat# 04896866001 |
| Deposited data | ||
| Lmx1b & Pet1 CUT&RUN and histone mark ChIPmentation | https://doi.org/10.1101/2021.10.13.463782 | GSE185737 |
| Raw and analyzed data | This paper | GSE189052 |
| Experimental models: Organisms/strains | ||
| Mouse: Tph2-CreERT2 | Jackson Laboratory | RRID:IMSR_JAX:016584 |
| Mouse: Lmx1bflox | Jackson Laboratory; PMID:17151281 | Stock # 031287; RRID:MGI:3810753 |
| Mouse: Pet1flox | PMID:20818386 | RRID:MGI:4837213 |
| Mouse: Tph2flox | Jackson Laboratory; PMID:24972638 | RRID:IMSR_JAX:027590 |
| Mouse: Pet1-Cre | Jackson Laboratory; PMID:16251278 | Stock #012712; RRID:MGI:4837211 |
| Mouse: Ai9 RosaTdTom | Jackson Laboratory | Stock #: 007909; RRID:MGI:4436851 |
| Oligonucleotides | ||
| Tph2 Probe | TaqMan | Cat# Mm00557722 |
| Pet1 Probe | TaqMan | Cat# Mm00462220 |
| Ddc Probe | TaqMan | Cat# Mm00516688 |
| Slc6a4 Probe | TaqMan | Cat# Mm00439391 |
| Software and algorithms | ||
| MATLAB | https://www.mathworks.com/products/matlab.html | RRID:SCR_001622 |
| Fiji/ImageJ | https://imagej.net/Fiji | RRID:SCR_002285 |
| FASTQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 |
| Trimmomatic | Bolger et al., 2014 | RRID:SCR_011848 |
| Hisat2 (v2.1.0) | Kim et al., 2019 | RRID:SCR_015530 |
| Cufflinks v2.2.2 | Trapnell et al., 2010 | RRID:SCR_014597 |
| Webgestalt | http://www.webgestalt.org | RRID:SCR_006786 |
| Imaris 7.4.2 | http://www.bitplane.com/ | RRID:SCR_007370 |
| Filament Analysis 19 Beta | Matthew J. Gastinger | https://github.com/Ironhorse1618/Python3.7-Imaris-XTenstions/blob/master/XT_MJG_Filament_Analysis19_beta.py |
| PClamp | Molecular Devices | RRID:SCR_011323 |
| ANY-maze | Stoelting | RRID:SCR_014289 |
| Video Freeze software | Med Associates | RRID:SCR_014574 |
| Integrated Genome Viewer v2.9.3 | https://software.broadinstitute.org/software/igv/ | RRID:SCR_011793 |
| bamCoverage function v3.5.0 | https://deeptools.readthedocs.io/en/develop/ | RRID:SCR_016366 |
| Prism 8.0 | https://www.graphpad.com/scientific-software/prism/ | RRID:SCR_002798 |
| Huygens Deconvolution | Scientific Volume Imaging http://svi.nl | RRID:SCR_014237 |
| ggpubr | https://rdocumentation.org/packages/ggpubr/versions/0.4.0 | RRID:SCR_021139 |
| pheatmap | https://www.rdocumentation.org/packages/pheatmap/versions/0.2/topics/pheatmap | RRID:SCR_016418 |
| DEXSeq | http://bioconductor.org/packages/release/bioc/html/DEXSeq.html | RRID:SCR_012823 |
| ENCODE | https://www.encodeproject.org/ | RRID:SCR_015482 |
| Bowtie 2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | RRID:SCR_016368 |
| samtools | http://htslib.org/ | RRID:SCR_002105 |
| SEACR | https://seacr.fredhutch.org/ | Meers et al., 2019 |
| BEDTools | https://github.com/arq5x/bedtools2 | RRID:SCR_006646 |
| GREAT | http://great.stanford.edu/public/html/splash.php | RRID:SCR_005807 |
| MEME suite | http://meme-suite.org/ | RRID:SCR_001783 |
| MACS2 | https://github.com/macs3-project/MACS | RRID:SCR_013291 |
| ENCODE ChIP-seq processing pipeline | https://github.com/ENCODE-DCC/chip-seq-pipeline2/ | RRID:SCR_021323 |
| ChIPSeqSpikeInFree | https://github.com/stjude/ChIPseqSpikeInFree | https://doi.org/10.1093/bioinformatics/btz720 |
| Other | ||
| Microvette | Sarstedt | Cat# 201278100 |
| Plastic Feeding Tubes | Instech | Cat# FTP2225 |
| Artificial Tears | CWRU Animal Resource Center Henry Schein |
Cat# 11695-68321 |
| FBS | Gibco | Cat# 10439024 |
| Corn Oil | Sigma | Cat# C8267 |
| VetBond (webglue) | CWRU Animal Resource Center Patterson Veterinary |
Cat# 078910065 |
| OCT solution | Fisher Scientific | Cat# 23730571 |
| MAS-GP Adhesive Glass Slides | Matsunami | Cat# 10748176 |
| Betadine | Emerson Healthcare Llc. | Cat# 19027132 |
| 20X SSC | Fischer BioReagents | Cat# BP13251 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mouse husbandry procedures were approved by the Institutional Animal Care and Use Committees (IACUC) of Case Western Reserve University. Experiments were performed with adult (2-3 months old) male and female mice of mixed background using age-matched and sex-matched controls. Mice were generated using the following alleles: Pet1fl (Liu et al., 2010), Lmx1bfl (Zhao et al., 2006), or Tph2fl (Kim et al., 2014), Tph2-CreERT2 (Jackson labs), Ai9, (RosaTom; Jackson lab). Control mice were either: +/+ (vCON) or +/+; Tph2-CreER; Ai9, (iCON). 5-HT axon varicosities at postnatal day zero were imaged in Pet1-Cre; Ai9 animals (Scott et al., 2005). Tail or ear genomic DNA was used to determine genotypes. Mice were housed in ventilated cages on a 12 hr light/dark cycle with free access to food and water with 2–5 mice per cage.
Mice treatments
Mice 2-3 months of age, were subjected to surgery or administered tamoxifen and rested for 1, 3, 6, or 12 months before being sacrificed for tissue collection. An experimental group was treated with para-chloroamphetamine (PCA) after tamoxifen administration prior to tissue collection. Additional cohorts were subjected to a battery of behavioral tests prior to tamoxifen administration for baseline monitoring including Open Field and Contextual Fear Conditioning, and then subjected to behaviors again 1 and 6 months following tamoxifen treatment. Restraint stress was performed 1 and 6 months following tamoxifen. All procedures were approved by CWRU IACUC in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
METHOD DETAILS
Tamoxifen gavage
Mice were orally gavaged with a 250mg/kg dose of tamoxifen dissolved in corn oil using 22ga X 25mm Plastic Feeding Tubes 1x/day for consecutive 5 days.
PCA administration
PCA dissolved in saline, (20 mg/kg) or saline alone was delivered intraperitoneally to mice 10 days, 1 month or 3 months following tamoxifen. Mice were injected 2 times a day 6 hours apart, for 4 consecutive days. Intervals between days was 16 hours and cages were maintained at 25°C-26°C (Jin et al., 2016).
Cell sorting
Adult 5-HT neuron dissociation was performed on male and female mouse brains at 10 days or 1 month after tamoxifen administration. Mice were sacrificed and brains collected individually into 15mL conical tubes containing 10mL ice cold artificial cerebrospinal fluid (aCSF) solution (3.5 mM KCl, 126 mM NaCl, 20 mM NaHCO3, 20 mM Dextrose, 1.25 mM NaH2PO4, 2 mM CaCl2, 2 mM MgCl2, 50 μm AP-V, 20 μm DNQX, and 100 nM TTX) for 5min. Brains were then sectioned at 400 μm on a vibratory microtome (Pelco easiSlicer) in continuously bubbling (95% O2; 5% CO2) aCSF. Sections containing dorsal and median raphe 5-HT neurons were incubated in 1 mg/mL Protease from Streptomyces griseus in bubbling aCSF solution for 75 min at room temperature (RT). Sections were then incubated for 15 min in bubbling aCSF alone at RT. Brain regions containing TdTomato+ neurons were microdissected from sections in cold aCSF/10%FBS solution. Samples were slowly triturated 30-100X with fire-polished Pasteur pipettes of decreasing bore size until a single cell suspension was achieved. Samples were then filtered, and flow sorted on a BD FACS Aria SORP.
qPCR
RNA samples were obtained from TdTomato+flow sorted cells (2 mice pooled per sample (n), n=3-5 per genotype) and collected into Trizol LS followed by chloroform extraction and concentrated with RNA Clean and Concentrator-5 kit. cDNA was then synthesized using equal input RNA with the Transcriptor First Strand cDNA Synthesis Kit (Roche). cDNA was then amplified (10 PCR cycles) using PerfeCTa PreAmp SuperMix. RT-qPCR was performed using PerfeCTa FastMix II ROX mastermix with TaqMan probes.
RNA sequencing
Flow-sorted neurons were collected into Trizol LS and isolated using chloroform extraction and purification with RNA Clean and Concentrator-5 kit. Total concentration and quality assessment were performed using Quantifluor RNA system (Promega) and Fragment analyzer (Advanced Analytics). Samples (2 mice pooled per sample defined a single replicate (n), with n=3-4 per genotype and time point) were converted to cDNA, depleted of rRNA transcripts, and amplified using the Ovation® SoLo RNA-Seq Library Preparation Kit for mouse (Nugen Inc). Single-end sequencing was performed on a Nextseq 550 (Illumina) for 76 cycles. Read quality was assessed using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and adapters were trimmed using Trimmomatic (Bolger et al., 2014). Filtered and trimmed reads were aligned to the mouse genome (mm10, UCSC) using Hisat2 (v2.1.0) (Kim et al., 2019).
AAV-Cre injections
Mice were anesthetized with Isoflurane and stereotaxically injected unilaterally at two sites with 1.5 μl and 1 μl pENN.AAV.CMVs.PI.Cre.rBG at X = 0.6 mm, Y = −4.2 mm, Z = −3.2 mm and X = 0.6 mm, Y = −4.2 mm, Z = −4.5 mm relative to Bregma respectively. Procedure was performed in an approved surgical suite. Animals were anesthetized using an Isoflurane chamber, treated with a local anesthetic (bupivacaine HCL) administered subcutaneously prior to surgery and with an analgesic (Meloxicam 5 mg/kg) for 3 days post-surgery. Once anesthetized, mouse scalps were shaved and cleaned with betadine and ethanol. Mice were placed in stereotaxic apparatus on a warming pad. Sterile ophthalmic ointment was applied to lubricate the eyes. A midline incision was made down the top of the scalp, holes were drilled through the skull to expose brain and a Hamilton syringe was slowly lowered to desired coordinates. An infusion rate of 0.1 μl/min was used with 10 min after each injection to allow diffusion of virus, after which the needle was slowly retracted. Incisions were closed using Vetbond and sutures. Animals were recovered in a clean chamber warmed with a heat pad. Mice were then returned to group housing monitored for 5 days and sacrificed at either 1 month or 6 months post-surgery.
In situ hybridization
In situ hybridization was performed using a standard protocol using a digoxigenin-11-UTP labeled antisense RNA probes (Roche diagnostics) as described elsewhere (Wyler et al., 2016). Briefly, under RNAse free conditions, slide mounted tissue sections (18μm thick) were fixed in 4% PFA for 10 min, followed by 11 min of proteinase K treatment, 5 min PFA, acetylated in TEA buffer for 10 min, incubated in pre-hybridization buffer for 2 hours at RT and hybridized with probes O/N at 65°C. 3x5 min washes in PBS followed each PFA incubation, and the acetylation step. The next day slides were washed in preheated 5x SSC/50% formamide solution for 5 minutes at then 2x60 min washes in 2x SSC/50% formamide at 65°C and 1xSSC solution for 15 min at 37°C. Slides were then equilibrated in 1x B1 buffer (0.1 M Trizma, 0.15 M NaCl, pH 7.5) for 10 min, blocked at RT in 10% heat inactivated goat serum in B1 buffer for 60 min and incubated in anti-DIG-AP antibody in 5% serum O/N at 4°C. On the last day slides were washed 5x5 min in B1 buffer, equilibrated in buffer NTMT for 10 min and developed in NTMT/NBT/BCIP developing solution. Probes were generated using the following primer sets:
Pet1:F:5′-CCAGTGACCAATCCCATCCTC-3′ and R:5′-AGAATTAACCCTCACTAAAGGGTTAATGGGGCTGAAAGGGATA-3′.
Tph2:F:5′-AGGATCCCTACACGCAGAGCaTTGAA-3′ and R:5′-GAATTAACCCTCACTAAAGGGATCCATCCCAACTGCTGTGT-3′.
Slc6a4:F:5′-TGAGATGAGGAACGAAGACGTGTC-3′ and R:5′-AGAATTAACCCTCACTAAAGGGTAGCCCAAGATGATACTCCAGTG-3′.
Ddc:F:5′-GTTTTTAGAATGTACGGAGTCAAGGG-3′ and R:5′-AGAATTAACCCTCACTAAAGGGATACAGAAAATATTTCTTCATTGATAAGTCACC-3′.
Immunofluorescence
Adult mice were anesthetized with Avertin (44 mM tribromoethanol, 2.5% tert-amyl alcohol, 0.02 ml/g body weight) and transcardially perfused for 2–3 min with ice cold PBS followed by 20 min cold 4% paraformaldehyde (PFA) in PBS. Brains and spinal cords were removed and post-fixed in 4% PFA for 2 hr prior to placement in 30% sucrose/PBS overnight (O/N) for cryoprotection. For processing, tissue samples were frozen in Optimal Cutting Temperature (OCT) solution and sectioned on a cryostat at 20μm (DRN/MRN) or 25-30μm (forebrain and spinal cord). Tissue sections were mounted on MAS-GP Adhesive Glass Slides and vacuum dried. Sections were then permeabilized in 0.3% Triton 100X-PBS (PBS-T)for 15 min followed by antigen retrieval in Sodium Citrate buffer for 5 min in a microwave at low power. Sections were blocked with 10% NGS in PBS-T for 1 hr followed by incubation in primary antibody at 4°C O/N. Primary antibodies used were rabbit, mouse or chicken anti-RFP, rabbit anti-5-HT, rabbit or mouse anti-Tph2, rabbit anti-AADC, mouse anti-GFAP, rabbit anti-Iba1, rabbit anti-COXIV, and chicken anti-beta 3 tubulin. Secondary antibodies (goat anti-rabbit, mouse or chicken, Alexa Fluor 488, 594, or 647) were used at room temperature for 1 hour. Primary antibodies used with the Avidin/Biotin Blocking kit and VECTASTAIN ABC kit were: rabbit anti-Sert, guinea pig anti-Synapsin1, mouse anti-Synaptotagmin1, mouse anti-Gephyrin, rabbit anti-α-Synuclein, rabbit anti-APP, mouse anti-P-Neurofilament, and rat anti-CD68.
Microscopy
Images of in situ hybridizations were captured with an Olympus Optical BX51 microscope. Immunofluorescent images were captured with an LSM800 confocal microscope or Axio Imager M2 microscope (Carl Zeiss). Mitochondria were imaged using a Leica TCS SP8 gated STED. The same brightness/contrast settings and background subtraction were applied for all image analyses except where noted below.
Quantification of signal intensities
For cell body and Sert+ fiber immunofluorescence intensities and in situ hybridization signals 1000-1500 individual cells in 3 sections from each of three mice per genotype were analyzed. For Sert+ axon signals, 3 representative images from 3 mice per genotype were analyzed. Signal intensities were measured using FIJI cell counter and Measure analysis.
Pericellular baskets
Manual counting of TdTomato+ LBs, DBs and DBRs in the dLS was performed using 2-3 25μm thick coronal sections taken from each of 3 animals for each genotype and each timepoint. For each section, 16-20 non-overlapping images were captured with a Zeiss LSM800 with 63x magnification and stitched to uniform size using Zeiss 2.3 software. LBs, DBs, and DBRs were then counted separately. Baskets numbers were normalized to numbers in iCON. Two-way ANOVA with Welch’s correction analysis was performed with Prism 8.4.3. Swollen varicosities and spheroids were counted in a similar manner.
TdTomato+ axon densities
TdTomato+ (anti-RFP) fluorescent pixel intensities were compared using two 25μm thick coronal sections taken from each of 3 animals for each genotype and each timepoint to quantitate axon densities. For each section, 16-20 non-overlapping images from comparable regions of the brain were captured with a Zeiss LSM800 with 20x magnification and stitched to uniform size using Zeiss 2.3 software. TdTomato+ intensities were determined in each stitched image using ImageJ-Fiji and the six values within each genotype were summed to obtain a mean ± SEM axon density value for that genotype. GFAP, Iba1, and CD68 intensities were analyzed in a similar manner.
Axon morphology analysis
Axon tracing was performed using semiautomatic Filament Tracer module in Imaris 7.4.2 (Bitplane, Zurich, Switzerland). Individual fibers were traced to generate a cylindrical filament object. At 6 months and 1 year iLKO and iDKO axons had many instances where TdTomato+ signal between sequential varicosities or axon segments was below the detection threshold, manually determined to be 0.1μm. In these cases, a “below threshold” line was automatically generated to connect the two discrete TdTomato+ points along the shortest possible path. Volume threshold was automatically applied using set parameters assigned by the user for each animal. Filaments were then converted to dot objects and further analyzed with Filament Analysis 19 Beta module, https://github.com/Ironhorse1618/Python3.7-Imaris-XTenstions/blob/master/XT_MJG_Filament_Analysis19_beta.py. Dot objects contained individual points of reference equally distributed along the generated cylindrical filament object with each dot representing a single diameter value. Dots assigned along the “below threshold” lines were assigned a diameter value of <0.1μm. The frequency of dot diameters was plotted using 0.1μm bins. A Fragmentation Index was calculated as the sum total of the number of dots with <0.1μm diameters divided by the sum total of all dots (dots with values ≥0.1μm + dots of value <0.1μm).
Mitochondria fragmentation analysis
Mitochondria were stained using a COX IV antibody. Single neurons were then imaged at 100x using a Leica TCS SP8 gated STED. Images were then deconvoluted using Huygens Deconvolution. Length and width of mitochondria were measured and used to generate aspect ratio (longest length/shortest length). iDKO images in Figures 7D and S9G were brightened to enhance visualization of mitochondrial fragments.
Cell body counts
Every 4th section through the entire rostro-caudal extent of the DRN/MRN 5-HT system was counted. RFP and Tph2 doubly stained sections were used to calculate RFP+/Tph2+ and Tph2+/RFP+ ratios to determine Tph2-CreER efficiency and specificity, respectively. TdTomato+ cell bodies were counted as described and presented as a percent of iCON for each time point to assess survivability of cell bodies.
Cell body volume analysis
Sections were prepared as described above and cell bodies were imaged with a 40x oil objective on an LSM800 confocal microscope. Unstacked images were then used to generate a reconstructed 3D rendering using the Surfaces tool in Imaris x64 9.6.0. One-way ANOVA statistical analysis was performed and values were expressed as a percent of iCON for each time point.
Electrophysiology
Due to heterogeneity of electrophysiological properties in 5-HT neurons, recordings were performed in TdTomato+ neurons located in similar locations of the lateral wing of the DRN. 2-3 month old mice were gavaged with tamoxifen. At 1 or 3 months post-TAM mice were sacrificed for cell recordings.
Slice preparation
Mice were anesthetized with isoflurane and transcardially perfused with ice-cold oxygenated (95% O2 and 5% CO2) aCSF cutting solution. Brains were quickly removed and placed into ice-cold oxygenated aCSF cutting solution, which contained (in mM): 110 choline Cl, 2.5 KCl, 0.5 CaCl2, 7 MgCl2, 1.3 NaH2PO4, 25 NaHCO3 and 20 glucose. Coronal brain slices (300 μm thick) containing the DRN were cut on a vibratome (VT1200, Leica) and allowed to recover at 34°C for 20 minutes before recording. Recovery and recording aCSF contained (in mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 1.3 MgCl2, 1.3 NaH2PO4, 25 NaHCO3 and 10 glucose. Individual slices were transferred to a recording chamber submerged with aCSF and continuously perfused with aCSF (3.0 mL/min) at 32°C. Slices were visualized under a microscope (BX51WI, Olympus) using infrared video microscopy and differential interference contrast optics.
Recordings
Recordings were done using a MultiClamp700B amplifier and PClamp software (Molecular Devices, USA). The data was low pass filtered at 2 kHz and digitized at 10 kHZ with Digidata 1550B (Molecular Devices, USA). Patched pipettes were pulled (3-6 MΩ resistance) from borosilicate glass capillaries (B-120-69-15, Sutter Instruments) using micropipette puller (P1000, Sutter instruments), then filled with internal solution containing (in mM): 125 K-gluconate, 15 KCl, 10 HEPES, 4 MgCl2, 4 Na2ATP, 0.4 Na3GTP, 10 Tris-phosphocreatine and 0.2 EGTA. Intrinsic membrane properties were measured using whole-cell recordings. Action potentials and voltage threshold were recording in current-clamp mode. Action potentials were measured with current injections (−100 to 300 pA at 50 pA steps). Voltage thresholds were done by injecting a ramp current where the voltage threshold was the voltage where the derivative equaled 10 mV/ms. Input resistance and inwardly rectifying potassium channels were assessed by voltage-clamp mode. Input resistance was assessed with a hyperpolarizing voltage step (−10 mV, 500 ms), where the resistance was solved for using Ohms Law. Inwardly rectifying potassium channels were assessed by measuring currents elicited from a voltage ramp to −120 mV from a holding potential of −40 mV (0.1 V/s). Serotonin autoreceptor function was assessed using Htr1a agonist (+)-8-Hydroxy-2-dipro-pylaminotetralin hydrobromide (8-OH-DPAT). Analysis was done using a combination of Clampfit (Molecular Devices, USA) and custom written scripts in MATLAB (Mathworks, USA). For firing rates at 1 month post-TAM we used 19 neurons from 7 iCON mice, 2-3 neurons per animal; 13 neurons from 5 iLKO mice, 2-3 neurons per animal; 11 neurons from 5 iDKO mice, 2-3 neurons per animal. For firing rates at 3 months post-TAM we used 10 neurons from 2 iCON mice, 5 neurons per animal; 8 neurons from 2 iLKO mice, 4 neurons per animal; 10 neurons from 3 iDKO mice, 3-4 neurons per animal. For control autoreceptor responses, we used 12 neurons from 7 iCON mice, 1-2 neurons per animal; 10 neurons from 5 iLKO mice, 2 neurons per animal; 9 neurons from 5 iDKO mice, 1-2 neurons per animal. For 8-OH-DPAT elicited responses, we used 11 neurons from 7 iCON mice, 1-2 per animal; 7 neurons from 5 iLKO mice, 1-2 per animal; 9 neurons from 5 iDKO mice, 1-2 neurons per animal.
Behavior
Mice were assessed for baseline performance prior to tamoxifen treatment. They were then rested for 3 days prior to tamoxifen gavage and assessed in the same behaviors 1 month following the last tamoxifen gavage and again at 6 months after the last gavage. Male and female mice were tested separately after being acclimated to the testing room for at least 30 min. Assays were performed during the light phase and experimenters were blinded to genotype.
Open field test
Mice were placed in the center of a 40 cm × 40 cm box in a dimly lit room and allowed to freely explore for 10 min. ANY-maze video-tracking software was used to track animal movement in digitally subdivided center and outer areas. During this period, total distance traveled and time spent in inner zone were measured.
Contextual fear conditioning
During training, mice were placed in a conditioning chamber (Med Associates) for 3min to acclimate and then presented with a single tone (30sec at 90dB and 5000 Hz) co-terminating with a 2 sec foot shock at 0.5 mA. Mice were then removed 1 min after the tone/shock was presented and returned to home cage. About 24 hrs later mice were returned to the same chamber and allowed 5 min of free exploration without auditory cues or foot shock during which freezing behavior was scored using Video Freeze software.
Restraint stress & corticosterone measurements
Mice were habituated overnight to the room where stress tests and blood collection were performed. For AM and PM blood collection mice were placed into a ventilated 50mL conical tube, during which tails were numbed in ice cold ethanol for 30 sec before nicking of tail tips with a sterile razor blade (AM collections) or gently rubbed with a clean wipe to remove blood clot from prior cut (PM collections). Approximately 2μl of blood was collected into sterile microvettes between 9am and 10am and between 5pm and 6pm. Mice were rested for a day before undergoing stress tests at which point they were restrained in ventilated 50mL conical tubes for 30 min between 9am and 10am and immediately returned to home cage. 30 min and 2 hr following stress, blood was collected using similar methods. Blood samples were spun at 4°C for 20 min at 12,000xg and serum was collected for use in CORT ELISA performed according to manufacturer’s instructions.
Genomic visualization
Histone ChIPmentation and CUT&RUN data from https://doi.org/10.1101/2021.10.13.463782 were visualized using the Integrated Genome Viewer (IGV, v2.9.3). All datasets were normalized to 1X genome coverage using the bamCoverage function in deeptools (v3.5.0) (Ramirez et al., 2016).
QUANTIFICATION AND STATISTICAL ANALYSIS
One-way ANOVA was performed for all analyses involving 3 or more genotypes assessed one time point using Welch’s correction where noted in the figure legend. Exceptions include the Hypergeometric test for the number of DEGs with GO terms in Figure 1H and Wilcoxon rank sum test for stage specific gene expression in Figure S9F. Two-way ANOVA with Welch’s correction where noted was performed for analyses involving more than 2 genotypes across 2 or more time points. Comparisons within the same genotype across time points are designated by color matched bars and stars. In some cases, only 1 comparison was made between 2 genotypes, in which case an unpaired t-test was performed. A pairwise comparison t-test was performed for Figure 6B. For behavior experiments two-way repeated measures ANOVA was performed. Graphpad Prism 8 was used for analysis and graphs using mean ± SEM unless otherwise noted. RNA-seq and Hypergeometric bar graphs were generated using ggplot2 in R. Violin plot of embryonic vs adult genes were generated using ggpubr in R (v4.0.4) (Wickham et al., 2016). The heatmap of all iLKO and iDKO DEGs was clustered using k-means clustering with pheatmap (v1.0.12) in R (v4.0.4) with k = 15. Specific details regarding n and significance parameters are specified in figure legends.
Gene expression and genomics
Gene expression quantification and differential expression were analyzed using Cufflinks v2.2.2 (Trapnell et al., 2010). Differentially expressed genes (DEGs) were defined as fold-change ≥1.5 and false discovery rate (FDR) ≤0.05. Gene Ontology analysis was performed using Webgestalt (http://www.webgestalt.org), requiring five genes per category and FDR ≤ 5% (Liao et al., 2019). Differential exon expression analysis was performed to test for differences in floxed Lmx1b exon 4 expression with DEXSeq (Anders et al., 2012). RNA-seq reads were mapped to the Mus musculus genome (Ensembl, v. 96) and reads falling within annotated exons were counted using featureCounts (Rsubread). DEXSeq default settings were used and significant exon expression differences were determined at FDR ≤ 5%. CUT&RUN and ChIPmentation datasets were normalized to 1X genome coverage using the bamCoverage function in deeptools (v3.5.0) (Ramirez et al., 2016).
CUT&RUN data processing and analysis
Details of CUT&RUN data generation are available elsewhere (https://doi.org/10.1101/2021.10.13.463782). The standard ENCODE ChIP-seq software pipeline from the ENCODE Consortium was used to process CUT&RUN data. Sequencing reads were mapped to the UCSC mm10 by Bowtie v2.3.4.3 (Langmead and Salzberg, 2012). Removal of PCR duplicates was done using samtools v1.9 (Li et al., 2009). Unique mapped reads were used to call peaks comparing TF-bound chromatin with an IgG control with SEACR using both relaxed and stringent threshold settings (Meers et al., 2019). The relaxed threshold uses the total signal of the “knee” to the peak of the total signal curve and the stringent threshold uses the peak of the total signal curve. Both threshold settings have been shown to have high specificity and sensitivity (Meers et al., 2019). Shared intersections between identified peaks (1bp minimum overlap) were identified using BEDTools (Quinlan and Hall, 2010). CUT&RUN data for Lmx1b and Pet1 were normalized together for visualization using the ChIPseqSpikeInFree method to determine the scaling factor for adjusting signal coverage across the genome (Jin et al., 2020). Gene annotation of CUT&RUN peaks was performed using the Genomic Regions Enrichment of Annotations Tool (GREAT), with each peak assigned to the nearest gene (McLean et al., 2010). Motif matching was performed with fimo from the MEME suite (Bailey et al., 2015) with a p-value threshold of 0.001.
ChIPmentation data processing and analysis
Details of ChIPmentation data generation are available elsewhere (https://doi.org/10.1101/2021.10.13.463782). The standard ENCODE ChIP-seq software pipeline from the ENCODE Consortium was used to process ChIPmentation data. Sequencing reads were mapped to the UCSC mm10 by Bowtie v2.3.4.3. Removal of PCR duplicates was done using samtools v1.9 (Li et al., 2009). The unique mapped reads were used to call peaks comparing immunoprecipitated chromatin with input chromatin with MACS2 v2.2.4 with FDR adjusted p-value cutoff of 0.01 (Zhang et al., 2008).
Supplementary Material
Highlights.
Lmx1b and Pet1 sustain an adult-stage connectivity transcriptome in 5-HT neurons
Adult-stage loss of Lmx1b and Pet1 causes degeneration of 5-HT synapses and axons
APP and α-syn accumulate in spheroids and mitochondria fragment as fibers degenerate
Connectivity decay is accompanied by physiological and behavioral alterations
Grant support:
T32GM008056 to M.M.K., F30 MH122173 and T32 GM007250 to X.L.Z., and the Uehara Memorial Foundation Fellowship (201940009) and JSPS Overseas Research Fellowship to N.T., R01MH062723 to E.S.D., and a NIH Shared Instrumentation Grant S10-OD016164. Graphical abstract was created with BioRender.com.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110711.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anders S, Reyes A, and Huber W (2012). Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade R, Huereca D, Lyons JG, Andrade EM, and McGregor KM (2015). 5-HT1A receptor-mediated autoinhibition and the control of serotonergic cell firing. ACS Chem. Neurosci 6, 1110–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmitia EC, and Nixon R (2008). Dystrophic serotonergic axons in neurodegenerative diseases. Brain Res. 1217, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, and Noble WS (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balastik M, Ferraguti F, Pires-da Silva A, Lee TH, Alvarez-Bolado G, Lu KP, and Gruss P (2008). Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc. Natl. Acad. Sci. U S A 105, 12016–12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmer A, Klenowski PM, Patkar OL, and Bartlett SE (2017). Mapping the connectivity of serotonin transporter immunoreactive axons to excitatory and inhibitory neurochemical synapses in the mouse limbic brain. Brain Struct. Funct 222, 1297–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RE, and O’Malley K (2013). Axon degeneration in Parkinson’s disease. Exp. Neurol 246, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh JB (1984). The problems of neurons with long axons. Lancet 1, 1284–1287. [DOI] [PubMed] [Google Scholar]
- Chan-Palay V (1976). Serotonin axons in the supra- and subependymal plexuses and in the leptomeninges; their roles in local alterations of cerebrospinal fluid and vasomotor activity. Brain Res. 102, 103–130. [DOI] [PubMed] [Google Scholar]
- Coleman MP, and Hoke A (2020). Programmed axon degeneration: from mouse to mechanism to medicine. Nat. Rev. Neurosci 21, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti L, Gilley J, and Coleman MP (2014). Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci 15, 394–409. [DOI] [PubMed] [Google Scholar]
- Courtney NA, and Ford CP (2016). Mechanisms of 5-HT1A receptor-mediated transmission in dorsal raphe serotonin neurons. J. Physiol 594, 953–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai JX, Hu ZL, Shi M, Guo C, and Ding YQ (2008). Postnatal ontogeny of the transcription factor Lmx1b in the mouse central nervous system. J. Comp. Neurol 509, 341–355. [DOI] [PubMed] [Google Scholar]
- De Vos KJ, Grierson AJ, Ackerley S, and Miller CC (2008). Role of axonal transport in neurodegenerative diseases. Annu. Rev. Neurosci 31, 151–173. [DOI] [PubMed] [Google Scholar]
- Deneris E, and Gaspar P (2018). Serotonin neuron development: shaping molecular and structural identities. Wiley Interdiscip. Rev. Dev. Biol 7, e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneris ES, and Hobert O (2014). Maintenance of postmitotic neuronal cell identity. Nat. Neurosci 17, 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneris ES, and Wyler SC (2012). Serotonergic transcriptional networks and potential importance to mental health. Nat. Neurosci 15, 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descarries L, and Mechawar N (2000). Ultrastructural evidence for diffuse transmission by monoamine and acetylcholine neurons of the central nervous system. Prog. Brain Res 125, 27–47. [DOI] [PubMed] [Google Scholar]
- Donovan LJ, Spencer WC, Kitt MM, Eastman BA, Lobur KJ, Jiao K, Silver J, and Deneris ES (2019). Lmx1b is required at multiple stages to build expansive serotonergic axon architectures. eLife 8, e48788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Beaupre H, Gilbert C, Profes MS, Chabrat A, Pacelli C, Giguere N, Rioux V, Charest J, Deng Q, Laguna A, et al. (2016). Lmx1a and Lmx1b regulate mitochondrial functions and survival of adult midbrain dopaminergic neurons. Proc. Natl. Acad. Sci. U S A 113, E4387–E4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AR, Jongens TA, and Holzbaur ELF (2021). Mitochondrial adaptor TRAK2 activates and functionally links opposing kinesin and dynein motors. Nat. Commun 12, 4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley MD, Gu W, Nanson JD, Shi Y, Sasaki Y, Cunnea K, Malde AK, Jia X, Luo Z, Saikot FK, et al. (2021). SARM1 is a metabolic sensor activated by an increased NMN/NAD(+) ratio to trigger axon degeneration. Neuron 109, 1118–1136.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, and Kepecs A (2020). Interneuron types as attractors and controllers. Annu. Rev. Neurosci 43, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall C, and Moore RY (1984). Distribution of enkephalin, substance P, tyrosine hydroxylase, and 5-hydroxytryptamine immunoreactivity in the septal region of the rat. J. Comp. Neurol 225, 212–227. [DOI] [PubMed] [Google Scholar]
- Gao F, Chen D, Si J, Hu Q, Qin Z, Fang M, and Wang G (2015). The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum. Mol. Genet 24, 2528–2538. [DOI] [PubMed] [Google Scholar]
- Gerdts J, Summers DW, Milbrandt J, and DiAntonio A (2016). Axon self-destruction: new links among SARM1, MAPKs, and NAD+ metabolism. Neuron 89, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Mayer PR, Yu G, and Coleman MP (2019). Low levels of NMNAT2 compromise axon development and survival. Hum. Mol. Genet 28, 448–458. [DOI] [PubMed] [Google Scholar]
- Guedes-Dias P, and Holzbaur ELF (2019). Axonal transport: driving synaptic function. Science 366, eaaw9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne AL, Hu H, Kundu B, Steinmetz MP, Wylie CJ, Deneris ES, and Silver J (2011). The unusual response of serotonergic neurons after CNS Injury: lack of axonal dieback and enhanced sprouting within the inhibitory environment of the glial scar. J. Neurosci 31, 5605–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman EL, and Doe CQ (2021). Establishment and maintenance of neural circuit architecture. J. Neurosci 41, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, and Deneris E (1999). The ETS domain factor Pet-1 is an early and precise marker of central 5-HT neurons and interacts with a conserved element in serotonergic genes. J. Neurosci 19, 10348–10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, and Deneris ES (2003). Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron 37, 233–247. [DOI] [PubMed] [Google Scholar]
- Hobert O (2011). Regulation of terminal differentiation programs in the nervous system. Annu. Rev. Cell Dev. Biol 27, 681–696. [DOI] [PubMed] [Google Scholar]
- Hobert O, and Kratsios P (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr. Opin. Neurobiol 56, 97–105. [DOI] [PubMed] [Google Scholar]
- Huang EJ, and Reichardt LF (2001). Neurotrophins: roles in neuronal development and function. Annu. Rev. Neurosci 24, 677–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshahi A, Temel Y, Lim LW, Hoogland G, and Steinbusch HW (2011). Close communication between the subependymal serotonergic plexus and the neurogenic subventricular zone. J. Chem. Neuroanat 42, 297–303. [DOI] [PubMed] [Google Scholar]
- Jin Y, Dougherty SE, Wood K, Sun L, Cudmore RH, Abdalla A, Kannan G, Pletnikov M, Hashemi P, and Linden DJ (2016). Regrowth of serotonin axons in the adult mouse brain following injury. Neuron 91, 748–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kasper LH, Larson JD, Wu G, Baker SJ, Zhang J, and Fan Y (2020). ChIPseqSpikeInFree: a ChIP-seq normalization approach to reveal global changes in histone modifications without spike-in. Bioinformatics 36, 1270–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast RJ, Wu HH, Williams P, Gaspar P, and Levitt P (2017). Specific connectivity and unique molecular identity of MET receptor tyrosine kinase expressing serotonergic neurons in the caudal dorsal raphe nuclei. ACS Chem. Neurosci 8, 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol 37, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kim A, Zhao ZQ, Liu XY, and Chen ZF (2014). Postnatal maintenance of the 5-Ht1a-Pet1 autoregulatory loop by serotonin in the raphe nuclei of the brainstem. Mol. Brain 7, 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, Burberry A, Steinbaugh MJ, Gamage KK, Kirchner R, et al. (2019). ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci 22, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneynsberg A, Combs B, Christensen K, Morfini G, and Kanaan NM (2017). Axonal degeneration in tauopathies: disease relevance and underlying mechanisms. Front. Neurosci 11, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna A, Schintu N, Nobre A, Alvarsson A, Volakakis N, Jacobsen JK, Gomez-Galan M, Sopova E, Joodmardi E, Yoshitake T, et al. (2015). Dopaminergic control of autophagic-lysosomal function implicates Lmx1b in Parkinson’s disease. Nat. Neurosci 18, 826–835. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, and Zhang B (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Zhang M, Stoilov P, Chen L, and Zheng S (2020). Developmental attenuation of neuronal apoptosis by neural-specific splicing of Bak1 microexon. Neuron 107, 1180–1196.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, and Koleske AJ (2010). Mechanisms of synapse and dendrite maintenance and their disruption in psychiatric and neurodegenerative disorders. Annu. Rev. Neurosci 33, 349–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, and Deneris ES (2010). Pet-1 is required across different stages of life to regulate serotonergic function. Nat. Neurosci 13, 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, and Arlotta P (2015). Generating neuronal diversity in the mammalian cerebral cortex. Annu. Rev. Cell Dev. Biol 31, 699–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar T, Ohba Y, Nolte H, Mayer FC, Tatsuta T, Sprenger HG, Lindner B, Zhao Y, Li J, Bruns C, et al. (2019). Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature 575, 361–365. [DOI] [PubMed] [Google Scholar]
- Maddaloni G, Bertero A, Pratelli M, Barsotti N, Boonstra A, Giorgi A, Migliarini S, and Pasqualetti M (2017). Development of serotonergic fibers in the post-natal mouse brain. Front. Cell. Neurosci 11, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamounas LA, Mullen CA, O’Hearn E, and Molliver ME (1991). Dual serotoninergic projections to forebrain in the rat: morphologically distinct 5-HT axon terminals exhibit differential vulnerability to neurotoxic amphetamine derivatives. J. Comp. Neurol 314, 558–586. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Tenenbaum D, and Henikoff S (2019). Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenet. Chromatin 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GC, Parnavelas JG, and Buijs R (1987). Monoaminergic fibers form conventional synapses in the cerebral cortex. Neurosci. Lett 76, 275–279. [DOI] [PubMed] [Google Scholar]
- Pratelli M, Migliarini S, Pelosi B, Napolitano F, Usiello A, and Pasqualetti M (2017). Perturbation of serotonin homeostasis during adulthood affects serotonergic neuronal circuitry. eNeuro 4, ENEURO.0376-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Whitmore AV, and Finn JT (2002). Axonal self-destruction and neurodegeneration. Science 296, 868–871. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvadores N, Sanhueza M, Manque P, and Court FA (2017). Axonal degeneration during aging and its functional role in neurodegenerative disorders. Front. Neurosci 11, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MM, Wylie CJ, Lerch JK, Murphy R, Lobur K, Herlitze S, Jiang W, Conlon RA, Strowbridge BW, and Deneris ES (2005). A genetic approach to access serotonin neurons for in vivo and in vitro studies. Proc. Natl. Acad. Sci. U S A 102, 16472–16477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft RA, and Dymecki SM (2021). Neuronal pericellular baskets: neurotransmitter convergence and regulation of network excitability. Trends Neurosci. 44, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Miller BR, Babetto E, Cho Y, Sasaki Y, Qayum S, Russler EV, Cavalli V, Milbrandt J, and DiAntonio A (2012). SCG10 is a JNK target in the axonal degeneration pathway. Proc. Natl. Acad. Sci. U S A 109, E3696–E3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, and Henikoff S (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song NN, Xiu JB, Huang Y, Chen JY, Zhang L, Gutknecht L, Lesch KP, Li H, and Ding YQ (2011). Adult raphe-specific deletion of lmx1b leads to central serotonin deficiency. PLoS One 6, e15998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WC, and Deneris ES (2017). Regulatory mechanisms controlling maturation of serotonin neuron identity and function. Front. Cell. Neurosci 11, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger HG, Wani G, Hesseling A, Konig T, Patron M, MacVicar T, Ahola S, Wai T, Barth E, Rugarli EI, et al. (2019). Loss of the mitochondrial i-AAA protease YME1L leads to ocular dysfunction and spinal axonopathy. EMBO Mol. Med 11, e9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Luijtelaar MG, Tonnaer JA, and Steinbusch HW (1992). Aging of the serotonergic system in the rat forebrain: an immunocytochemical and neurochemical study. Neurobiol. Aging 13, 201–215. [DOI] [PubMed] [Google Scholar]
- Vickers JC, King AE, Woodhouse A, Kirkcaldie MT, Staal JA, McCormack GH, Blizzard CA, Musgrove RE, Mitew S, Liu Y, et al. (2009). Axonopathy and cytoskeletal disruption in degenerative diseases of the central nervous system. Brain Res. Bull 80, 217–223. [DOI] [PubMed] [Google Scholar]
- Wickham H, Chang W, and Wickham MH (2016). Package ‘ggplot2.’ Create Elegant Data Vis Using Gramm Graph Version (New York: Springer-Verlag; ), ISBN 978-3-319-24277-4. [Google Scholar]
- Wu H, Williams J, and Nathans J (2014). Complete morphologies of basal forebrain cholinergic neurons in the mouse. eLife 3, e02444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler SC, Spencer WC, Green NH, Rood BD, Crawford L, Craige C, Gresch P, McMahon DG, Beck SG, and Deneris E (2016). Pet-1 switches transcriptional targets postnatally to regulate maturation of serotonin neuron excitability. J. Neurosci 36, 1758–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikallio E, Poyhonen R, Zimon M, De Vriendt E, Hilander T, Paetau A, Jordanova A, Lonnqvist T, and Tyynismaa H (2013). Deficiency of the E3 ubiquitin ligase TRIM2 in early-onset axonal neuropathy. Hum. Mol. Genet 22, 2975–2983. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZQ, Scott M, Chiechio S, Wang JS, Renner KJ, Gereau R.W.t., Johnson RL, Deneris ES, and Chen ZF (2006). Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. J. Neurosci 26, 12781–12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for our RNA-seq analyses have been deposited in the NCBI GEO (Gene Expression Omnibus) and are publicly available as of the date of publication. The accession number for the datasets is listed in the key resources table.
This paper analyzes existing, publicly available data. The accession number for the datasets is listed in the key resources table.
This paper does not generate original code.
Any additional information required to reanalyze the data included in this report is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| chicken anti-RFP 1:200 | Rockland | Cat# 600-901-379S, RRID:AB_10703148 |
| mouse anti-RFP 1:200 | Rockland | Cat# 200-301-379S, RRID:AB_2611064 |
| rabbit anti-RFP 1:200 | Rockland | Cat# 600-401-379S, RRID:AB_11182807 |
| rabbit anti-5-HT 1:200 | Immunostar | Cat# 20080, RRID:AB_572263 |
| rabbit anti-Tph2 1:500 | Millipore | Cat# ABN60, RRID:AB_11212793 |
| mouse anti-Tph2 1:500 | Millipore | Cat# MAB5278, RRID:AB_11214093 |
| rabbit anti-AADC 1:200 | Millipore | Cat# AB1569, RRID:AB_90789 |
| mouse anti-GFAP 1:500 | Sigma | Cat# G3893, RRID:AB_477010 |
| rabbit anti-Iba1 1:500 | FUJIFILM | Cat# 019-19741, RRID:AB_839504 |
| rabbit anti-COX4 1:500 | Thermo Fischer | Cat# PA5-29992, RRID:AB_2547466 |
| chicken anti-beta 3 tubulin 1:500 | Novus Biologicals | Cat# NB100-1612, RRID:AB_10000548 |
| rabbit anti-Sert 1:400 | Immunostar | Cat# 24330, RRID:AB_572209 |
| guinea pig anti-Synapsin1 1:500 | Synaptic Systems | Cat# 106 104, RRID:AB_2721082 |
| mouse anti-Synaptotagmin1 1:500 | Sigma | Cat# SAB1404433, RRID:AB_10759362 |
| mouse anti-Gephyrin 1:500 | Synaptic Systems | Cat# 147 011, RRID:AB_887717 |
| rabbit anti-α-Synuclein 1:500 | Cell Signaling | Cat# 2642, RRID:AB_2192679 |
| rabbit anti-APP 1:500 | Sigma | Cat# SAB4300464, RRID:AB_10621836 |
| mouse anti-P-Neurofilament 1:500 | BioLegend | Cat# SMI-31P-100, RRID:AB_2028812 |
| rat anti-CD68 1:500 | BIO RAD | Cat# MCA1957, RRID:AB_322219 |
| Goat anti-Mouse, Alexa 647 1:500 | Thermo Fisher Scientific | Cat# A-21240, RRID:AB_2535809 |
| Goat anti-Rabbit, Alexa 647 1:500 | Thermo Fisher Scientific | Cat# A-21246, RRID:AB_2535814 |
| Goat anti-Rabbit, Alexa 594 1:500 | Thermo Fisher Scientific | Cat# A-11012, RRID:AB_2534079 |
| Goat anti-Mouse, Alexa 594 1:500 | Thermo Fisher Scientific | Cat# A-21155, RRID:AB_2535785 |
| Goat anti-Chicken, Alexa 594 1:500 | Thermo Fisher Scientific | Cat# A32759, RRID:AB_2762829 |
| Goat anti-Rabbit, Alexa 488 1:500 | Thermo Fisher Scientific | Cat# A-11070, RRID:AB_2534114 |
| Goat anti-Mouse, Alexa 488 1:500 | Thermo Fisher Scientific | Cat# A-21131, RRID:AB_2535771 |
| VECTASTAIN ABC-Peroxidase Kit | Vector Laboratories | Cat# PK-4001, RRID:AB_2336810 Cat# PK-4004,RRID:AB_2336813 Cat# PK-4002, RRID:AB_2336811 Cat# PK-4007, RRID:AB_2336816 |
| Anti-Digoxigenin-AP Fab Fragments | Roche | Cat# 11093274910, RRID:AB_2734716 |
| Bacterial and virus strains | ||
| pENN.AAV.CMVs.PI.Cre.rBG | PennVector/ADDGENE | RRID:Addgene_105537 |
| Chemicals, peptides, and recombinant proteins | ||
| Protease | Sigma | Cat# P8811 |
| bupivacaine HCL | Hospira | Cat# C0409-3613-01 |
| Meloxicam | Pfizer | Cat# 71125-38-7 |
| Tamoxifen | Sigma | Cat# T5648 |
| PCA | Sigma | Cat# 274763 |
| 8-OH-DPAT | Tocris | Cat# 87394874 |
| D-AP5 | Tocris | Cat# 0106 |
| DNQX | Tocris | Cat# 0189 |
| TTX | Abcam | Cat# ab120054 |
| TRIZOL LS | Ambion | Cat# 10296010 |
| Chloroform | Fisher Scientific | Cat# C298500 |
| NBT | Roche | Cat# 11383213001 |
| BCIP | Roche | Cat# 11383221001 |
| Formimide | Roche | Cat# 11814320001 |
| Critical commercial assays | ||
| PerfeCTa PreAmp Supermix | QuantaBio | Cat# 89409170 |
| PerfeCTa FastMix II ROX Mastermix | QuantaBio | Cat# 97065990 |
| RNA Clean and Concentrator kit | ZYMO | Cat# R1013 |
| Ovation® SoLo RNA-Seq Library Preparation Kit | Tecan (Nugen Inc.) | Cat# 040732 |
| CORT ELISA kit | ENZO Life Sciences | Cat# 101106156 |
| Transcriptor First Strand cDNA Kit | Roche | Cat# 04896866001 |
| Deposited data | ||
| Lmx1b & Pet1 CUT&RUN and histone mark ChIPmentation | https://doi.org/10.1101/2021.10.13.463782 | GSE185737 |
| Raw and analyzed data | This paper | GSE189052 |
| Experimental models: Organisms/strains | ||
| Mouse: Tph2-CreERT2 | Jackson Laboratory | RRID:IMSR_JAX:016584 |
| Mouse: Lmx1bflox | Jackson Laboratory; PMID:17151281 | Stock # 031287; RRID:MGI:3810753 |
| Mouse: Pet1flox | PMID:20818386 | RRID:MGI:4837213 |
| Mouse: Tph2flox | Jackson Laboratory; PMID:24972638 | RRID:IMSR_JAX:027590 |
| Mouse: Pet1-Cre | Jackson Laboratory; PMID:16251278 | Stock #012712; RRID:MGI:4837211 |
| Mouse: Ai9 RosaTdTom | Jackson Laboratory | Stock #: 007909; RRID:MGI:4436851 |
| Oligonucleotides | ||
| Tph2 Probe | TaqMan | Cat# Mm00557722 |
| Pet1 Probe | TaqMan | Cat# Mm00462220 |
| Ddc Probe | TaqMan | Cat# Mm00516688 |
| Slc6a4 Probe | TaqMan | Cat# Mm00439391 |
| Software and algorithms | ||
| MATLAB | https://www.mathworks.com/products/matlab.html | RRID:SCR_001622 |
| Fiji/ImageJ | https://imagej.net/Fiji | RRID:SCR_002285 |
| FASTQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 |
| Trimmomatic | Bolger et al., 2014 | RRID:SCR_011848 |
| Hisat2 (v2.1.0) | Kim et al., 2019 | RRID:SCR_015530 |
| Cufflinks v2.2.2 | Trapnell et al., 2010 | RRID:SCR_014597 |
| Webgestalt | http://www.webgestalt.org | RRID:SCR_006786 |
| Imaris 7.4.2 | http://www.bitplane.com/ | RRID:SCR_007370 |
| Filament Analysis 19 Beta | Matthew J. Gastinger | https://github.com/Ironhorse1618/Python3.7-Imaris-XTenstions/blob/master/XT_MJG_Filament_Analysis19_beta.py |
| PClamp | Molecular Devices | RRID:SCR_011323 |
| ANY-maze | Stoelting | RRID:SCR_014289 |
| Video Freeze software | Med Associates | RRID:SCR_014574 |
| Integrated Genome Viewer v2.9.3 | https://software.broadinstitute.org/software/igv/ | RRID:SCR_011793 |
| bamCoverage function v3.5.0 | https://deeptools.readthedocs.io/en/develop/ | RRID:SCR_016366 |
| Prism 8.0 | https://www.graphpad.com/scientific-software/prism/ | RRID:SCR_002798 |
| Huygens Deconvolution | Scientific Volume Imaging http://svi.nl | RRID:SCR_014237 |
| ggpubr | https://rdocumentation.org/packages/ggpubr/versions/0.4.0 | RRID:SCR_021139 |
| pheatmap | https://www.rdocumentation.org/packages/pheatmap/versions/0.2/topics/pheatmap | RRID:SCR_016418 |
| DEXSeq | http://bioconductor.org/packages/release/bioc/html/DEXSeq.html | RRID:SCR_012823 |
| ENCODE | https://www.encodeproject.org/ | RRID:SCR_015482 |
| Bowtie 2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | RRID:SCR_016368 |
| samtools | http://htslib.org/ | RRID:SCR_002105 |
| SEACR | https://seacr.fredhutch.org/ | Meers et al., 2019 |
| BEDTools | https://github.com/arq5x/bedtools2 | RRID:SCR_006646 |
| GREAT | http://great.stanford.edu/public/html/splash.php | RRID:SCR_005807 |
| MEME suite | http://meme-suite.org/ | RRID:SCR_001783 |
| MACS2 | https://github.com/macs3-project/MACS | RRID:SCR_013291 |
| ENCODE ChIP-seq processing pipeline | https://github.com/ENCODE-DCC/chip-seq-pipeline2/ | RRID:SCR_021323 |
| ChIPSeqSpikeInFree | https://github.com/stjude/ChIPseqSpikeInFree | https://doi.org/10.1093/bioinformatics/btz720 |
| Other | ||
| Microvette | Sarstedt | Cat# 201278100 |
| Plastic Feeding Tubes | Instech | Cat# FTP2225 |
| Artificial Tears | CWRU Animal Resource Center Henry Schein |
Cat# 11695-68321 |
| FBS | Gibco | Cat# 10439024 |
| Corn Oil | Sigma | Cat# C8267 |
| VetBond (webglue) | CWRU Animal Resource Center Patterson Veterinary |
Cat# 078910065 |
| OCT solution | Fisher Scientific | Cat# 23730571 |
| MAS-GP Adhesive Glass Slides | Matsunami | Cat# 10748176 |
| Betadine | Emerson Healthcare Llc. | Cat# 19027132 |
| 20X SSC | Fischer BioReagents | Cat# BP13251 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mouse husbandry procedures were approved by the Institutional Animal Care and Use Committees (IACUC) of Case Western Reserve University. Experiments were performed with adult (2-3 months old) male and female mice of mixed background using age-matched and sex-matched controls. Mice were generated using the following alleles: Pet1fl (Liu et al., 2010), Lmx1bfl (Zhao et al., 2006), or Tph2fl (Kim et al., 2014), Tph2-CreERT2 (Jackson labs), Ai9, (RosaTom; Jackson lab). Control mice were either: +/+ (vCON) or +/+; Tph2-CreER; Ai9, (iCON). 5-HT axon varicosities at postnatal day zero were imaged in Pet1-Cre; Ai9 animals (Scott et al., 2005). Tail or ear genomic DNA was used to determine genotypes. Mice were housed in ventilated cages on a 12 hr light/dark cycle with free access to food and water with 2–5 mice per cage.
Mice treatments
Mice 2-3 months of age, were subjected to surgery or administered tamoxifen and rested for 1, 3, 6, or 12 months before being sacrificed for tissue collection. An experimental group was treated with para-chloroamphetamine (PCA) after tamoxifen administration prior to tissue collection. Additional cohorts were subjected to a battery of behavioral tests prior to tamoxifen administration for baseline monitoring including Open Field and Contextual Fear Conditioning, and then subjected to behaviors again 1 and 6 months following tamoxifen treatment. Restraint stress was performed 1 and 6 months following tamoxifen. All procedures were approved by CWRU IACUC in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
METHOD DETAILS
Tamoxifen gavage
Mice were orally gavaged with a 250mg/kg dose of tamoxifen dissolved in corn oil using 22ga X 25mm Plastic Feeding Tubes 1x/day for consecutive 5 days.
PCA administration
PCA dissolved in saline, (20 mg/kg) or saline alone was delivered intraperitoneally to mice 10 days, 1 month or 3 months following tamoxifen. Mice were injected 2 times a day 6 hours apart, for 4 consecutive days. Intervals between days was 16 hours and cages were maintained at 25°C-26°C (Jin et al., 2016).
Cell sorting
Adult 5-HT neuron dissociation was performed on male and female mouse brains at 10 days or 1 month after tamoxifen administration. Mice were sacrificed and brains collected individually into 15mL conical tubes containing 10mL ice cold artificial cerebrospinal fluid (aCSF) solution (3.5 mM KCl, 126 mM NaCl, 20 mM NaHCO3, 20 mM Dextrose, 1.25 mM NaH2PO4, 2 mM CaCl2, 2 mM MgCl2, 50 μm AP-V, 20 μm DNQX, and 100 nM TTX) for 5min. Brains were then sectioned at 400 μm on a vibratory microtome (Pelco easiSlicer) in continuously bubbling (95% O2; 5% CO2) aCSF. Sections containing dorsal and median raphe 5-HT neurons were incubated in 1 mg/mL Protease from Streptomyces griseus in bubbling aCSF solution for 75 min at room temperature (RT). Sections were then incubated for 15 min in bubbling aCSF alone at RT. Brain regions containing TdTomato+ neurons were microdissected from sections in cold aCSF/10%FBS solution. Samples were slowly triturated 30-100X with fire-polished Pasteur pipettes of decreasing bore size until a single cell suspension was achieved. Samples were then filtered, and flow sorted on a BD FACS Aria SORP.
qPCR
RNA samples were obtained from TdTomato+flow sorted cells (2 mice pooled per sample (n), n=3-5 per genotype) and collected into Trizol LS followed by chloroform extraction and concentrated with RNA Clean and Concentrator-5 kit. cDNA was then synthesized using equal input RNA with the Transcriptor First Strand cDNA Synthesis Kit (Roche). cDNA was then amplified (10 PCR cycles) using PerfeCTa PreAmp SuperMix. RT-qPCR was performed using PerfeCTa FastMix II ROX mastermix with TaqMan probes.
RNA sequencing
Flow-sorted neurons were collected into Trizol LS and isolated using chloroform extraction and purification with RNA Clean and Concentrator-5 kit. Total concentration and quality assessment were performed using Quantifluor RNA system (Promega) and Fragment analyzer (Advanced Analytics). Samples (2 mice pooled per sample defined a single replicate (n), with n=3-4 per genotype and time point) were converted to cDNA, depleted of rRNA transcripts, and amplified using the Ovation® SoLo RNA-Seq Library Preparation Kit for mouse (Nugen Inc). Single-end sequencing was performed on a Nextseq 550 (Illumina) for 76 cycles. Read quality was assessed using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and adapters were trimmed using Trimmomatic (Bolger et al., 2014). Filtered and trimmed reads were aligned to the mouse genome (mm10, UCSC) using Hisat2 (v2.1.0) (Kim et al., 2019).
AAV-Cre injections
Mice were anesthetized with Isoflurane and stereotaxically injected unilaterally at two sites with 1.5 μl and 1 μl pENN.AAV.CMVs.PI.Cre.rBG at X = 0.6 mm, Y = −4.2 mm, Z = −3.2 mm and X = 0.6 mm, Y = −4.2 mm, Z = −4.5 mm relative to Bregma respectively. Procedure was performed in an approved surgical suite. Animals were anesthetized using an Isoflurane chamber, treated with a local anesthetic (bupivacaine HCL) administered subcutaneously prior to surgery and with an analgesic (Meloxicam 5 mg/kg) for 3 days post-surgery. Once anesthetized, mouse scalps were shaved and cleaned with betadine and ethanol. Mice were placed in stereotaxic apparatus on a warming pad. Sterile ophthalmic ointment was applied to lubricate the eyes. A midline incision was made down the top of the scalp, holes were drilled through the skull to expose brain and a Hamilton syringe was slowly lowered to desired coordinates. An infusion rate of 0.1 μl/min was used with 10 min after each injection to allow diffusion of virus, after which the needle was slowly retracted. Incisions were closed using Vetbond and sutures. Animals were recovered in a clean chamber warmed with a heat pad. Mice were then returned to group housing monitored for 5 days and sacrificed at either 1 month or 6 months post-surgery.
In situ hybridization
In situ hybridization was performed using a standard protocol using a digoxigenin-11-UTP labeled antisense RNA probes (Roche diagnostics) as described elsewhere (Wyler et al., 2016). Briefly, under RNAse free conditions, slide mounted tissue sections (18μm thick) were fixed in 4% PFA for 10 min, followed by 11 min of proteinase K treatment, 5 min PFA, acetylated in TEA buffer for 10 min, incubated in pre-hybridization buffer for 2 hours at RT and hybridized with probes O/N at 65°C. 3x5 min washes in PBS followed each PFA incubation, and the acetylation step. The next day slides were washed in preheated 5x SSC/50% formamide solution for 5 minutes at then 2x60 min washes in 2x SSC/50% formamide at 65°C and 1xSSC solution for 15 min at 37°C. Slides were then equilibrated in 1x B1 buffer (0.1 M Trizma, 0.15 M NaCl, pH 7.5) for 10 min, blocked at RT in 10% heat inactivated goat serum in B1 buffer for 60 min and incubated in anti-DIG-AP antibody in 5% serum O/N at 4°C. On the last day slides were washed 5x5 min in B1 buffer, equilibrated in buffer NTMT for 10 min and developed in NTMT/NBT/BCIP developing solution. Probes were generated using the following primer sets:
Pet1:F:5′-CCAGTGACCAATCCCATCCTC-3′ and R:5′-AGAATTAACCCTCACTAAAGGGTTAATGGGGCTGAAAGGGATA-3′.
Tph2:F:5′-AGGATCCCTACACGCAGAGCaTTGAA-3′ and R:5′-GAATTAACCCTCACTAAAGGGATCCATCCCAACTGCTGTGT-3′.
Slc6a4:F:5′-TGAGATGAGGAACGAAGACGTGTC-3′ and R:5′-AGAATTAACCCTCACTAAAGGGTAGCCCAAGATGATACTCCAGTG-3′.
Ddc:F:5′-GTTTTTAGAATGTACGGAGTCAAGGG-3′ and R:5′-AGAATTAACCCTCACTAAAGGGATACAGAAAATATTTCTTCATTGATAAGTCACC-3′.
Immunofluorescence
Adult mice were anesthetized with Avertin (44 mM tribromoethanol, 2.5% tert-amyl alcohol, 0.02 ml/g body weight) and transcardially perfused for 2–3 min with ice cold PBS followed by 20 min cold 4% paraformaldehyde (PFA) in PBS. Brains and spinal cords were removed and post-fixed in 4% PFA for 2 hr prior to placement in 30% sucrose/PBS overnight (O/N) for cryoprotection. For processing, tissue samples were frozen in Optimal Cutting Temperature (OCT) solution and sectioned on a cryostat at 20μm (DRN/MRN) or 25-30μm (forebrain and spinal cord). Tissue sections were mounted on MAS-GP Adhesive Glass Slides and vacuum dried. Sections were then permeabilized in 0.3% Triton 100X-PBS (PBS-T)for 15 min followed by antigen retrieval in Sodium Citrate buffer for 5 min in a microwave at low power. Sections were blocked with 10% NGS in PBS-T for 1 hr followed by incubation in primary antibody at 4°C O/N. Primary antibodies used were rabbit, mouse or chicken anti-RFP, rabbit anti-5-HT, rabbit or mouse anti-Tph2, rabbit anti-AADC, mouse anti-GFAP, rabbit anti-Iba1, rabbit anti-COXIV, and chicken anti-beta 3 tubulin. Secondary antibodies (goat anti-rabbit, mouse or chicken, Alexa Fluor 488, 594, or 647) were used at room temperature for 1 hour. Primary antibodies used with the Avidin/Biotin Blocking kit and VECTASTAIN ABC kit were: rabbit anti-Sert, guinea pig anti-Synapsin1, mouse anti-Synaptotagmin1, mouse anti-Gephyrin, rabbit anti-α-Synuclein, rabbit anti-APP, mouse anti-P-Neurofilament, and rat anti-CD68.
Microscopy
Images of in situ hybridizations were captured with an Olympus Optical BX51 microscope. Immunofluorescent images were captured with an LSM800 confocal microscope or Axio Imager M2 microscope (Carl Zeiss). Mitochondria were imaged using a Leica TCS SP8 gated STED. The same brightness/contrast settings and background subtraction were applied for all image analyses except where noted below.
Quantification of signal intensities
For cell body and Sert+ fiber immunofluorescence intensities and in situ hybridization signals 1000-1500 individual cells in 3 sections from each of three mice per genotype were analyzed. For Sert+ axon signals, 3 representative images from 3 mice per genotype were analyzed. Signal intensities were measured using FIJI cell counter and Measure analysis.
Pericellular baskets
Manual counting of TdTomato+ LBs, DBs and DBRs in the dLS was performed using 2-3 25μm thick coronal sections taken from each of 3 animals for each genotype and each timepoint. For each section, 16-20 non-overlapping images were captured with a Zeiss LSM800 with 63x magnification and stitched to uniform size using Zeiss 2.3 software. LBs, DBs, and DBRs were then counted separately. Baskets numbers were normalized to numbers in iCON. Two-way ANOVA with Welch’s correction analysis was performed with Prism 8.4.3. Swollen varicosities and spheroids were counted in a similar manner.
TdTomato+ axon densities
TdTomato+ (anti-RFP) fluorescent pixel intensities were compared using two 25μm thick coronal sections taken from each of 3 animals for each genotype and each timepoint to quantitate axon densities. For each section, 16-20 non-overlapping images from comparable regions of the brain were captured with a Zeiss LSM800 with 20x magnification and stitched to uniform size using Zeiss 2.3 software. TdTomato+ intensities were determined in each stitched image using ImageJ-Fiji and the six values within each genotype were summed to obtain a mean ± SEM axon density value for that genotype. GFAP, Iba1, and CD68 intensities were analyzed in a similar manner.
Axon morphology analysis
Axon tracing was performed using semiautomatic Filament Tracer module in Imaris 7.4.2 (Bitplane, Zurich, Switzerland). Individual fibers were traced to generate a cylindrical filament object. At 6 months and 1 year iLKO and iDKO axons had many instances where TdTomato+ signal between sequential varicosities or axon segments was below the detection threshold, manually determined to be 0.1μm. In these cases, a “below threshold” line was automatically generated to connect the two discrete TdTomato+ points along the shortest possible path. Volume threshold was automatically applied using set parameters assigned by the user for each animal. Filaments were then converted to dot objects and further analyzed with Filament Analysis 19 Beta module, https://github.com/Ironhorse1618/Python3.7-Imaris-XTenstions/blob/master/XT_MJG_Filament_Analysis19_beta.py. Dot objects contained individual points of reference equally distributed along the generated cylindrical filament object with each dot representing a single diameter value. Dots assigned along the “below threshold” lines were assigned a diameter value of <0.1μm. The frequency of dot diameters was plotted using 0.1μm bins. A Fragmentation Index was calculated as the sum total of the number of dots with <0.1μm diameters divided by the sum total of all dots (dots with values ≥0.1μm + dots of value <0.1μm).
Mitochondria fragmentation analysis
Mitochondria were stained using a COX IV antibody. Single neurons were then imaged at 100x using a Leica TCS SP8 gated STED. Images were then deconvoluted using Huygens Deconvolution. Length and width of mitochondria were measured and used to generate aspect ratio (longest length/shortest length). iDKO images in Figures 7D and S9G were brightened to enhance visualization of mitochondrial fragments.
Cell body counts
Every 4th section through the entire rostro-caudal extent of the DRN/MRN 5-HT system was counted. RFP and Tph2 doubly stained sections were used to calculate RFP+/Tph2+ and Tph2+/RFP+ ratios to determine Tph2-CreER efficiency and specificity, respectively. TdTomato+ cell bodies were counted as described and presented as a percent of iCON for each time point to assess survivability of cell bodies.
Cell body volume analysis
Sections were prepared as described above and cell bodies were imaged with a 40x oil objective on an LSM800 confocal microscope. Unstacked images were then used to generate a reconstructed 3D rendering using the Surfaces tool in Imaris x64 9.6.0. One-way ANOVA statistical analysis was performed and values were expressed as a percent of iCON for each time point.
Electrophysiology
Due to heterogeneity of electrophysiological properties in 5-HT neurons, recordings were performed in TdTomato+ neurons located in similar locations of the lateral wing of the DRN. 2-3 month old mice were gavaged with tamoxifen. At 1 or 3 months post-TAM mice were sacrificed for cell recordings.
Slice preparation
Mice were anesthetized with isoflurane and transcardially perfused with ice-cold oxygenated (95% O2 and 5% CO2) aCSF cutting solution. Brains were quickly removed and placed into ice-cold oxygenated aCSF cutting solution, which contained (in mM): 110 choline Cl, 2.5 KCl, 0.5 CaCl2, 7 MgCl2, 1.3 NaH2PO4, 25 NaHCO3 and 20 glucose. Coronal brain slices (300 μm thick) containing the DRN were cut on a vibratome (VT1200, Leica) and allowed to recover at 34°C for 20 minutes before recording. Recovery and recording aCSF contained (in mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 1.3 MgCl2, 1.3 NaH2PO4, 25 NaHCO3 and 10 glucose. Individual slices were transferred to a recording chamber submerged with aCSF and continuously perfused with aCSF (3.0 mL/min) at 32°C. Slices were visualized under a microscope (BX51WI, Olympus) using infrared video microscopy and differential interference contrast optics.
Recordings
Recordings were done using a MultiClamp700B amplifier and PClamp software (Molecular Devices, USA). The data was low pass filtered at 2 kHz and digitized at 10 kHZ with Digidata 1550B (Molecular Devices, USA). Patched pipettes were pulled (3-6 MΩ resistance) from borosilicate glass capillaries (B-120-69-15, Sutter Instruments) using micropipette puller (P1000, Sutter instruments), then filled with internal solution containing (in mM): 125 K-gluconate, 15 KCl, 10 HEPES, 4 MgCl2, 4 Na2ATP, 0.4 Na3GTP, 10 Tris-phosphocreatine and 0.2 EGTA. Intrinsic membrane properties were measured using whole-cell recordings. Action potentials and voltage threshold were recording in current-clamp mode. Action potentials were measured with current injections (−100 to 300 pA at 50 pA steps). Voltage thresholds were done by injecting a ramp current where the voltage threshold was the voltage where the derivative equaled 10 mV/ms. Input resistance and inwardly rectifying potassium channels were assessed by voltage-clamp mode. Input resistance was assessed with a hyperpolarizing voltage step (−10 mV, 500 ms), where the resistance was solved for using Ohms Law. Inwardly rectifying potassium channels were assessed by measuring currents elicited from a voltage ramp to −120 mV from a holding potential of −40 mV (0.1 V/s). Serotonin autoreceptor function was assessed using Htr1a agonist (+)-8-Hydroxy-2-dipro-pylaminotetralin hydrobromide (8-OH-DPAT). Analysis was done using a combination of Clampfit (Molecular Devices, USA) and custom written scripts in MATLAB (Mathworks, USA). For firing rates at 1 month post-TAM we used 19 neurons from 7 iCON mice, 2-3 neurons per animal; 13 neurons from 5 iLKO mice, 2-3 neurons per animal; 11 neurons from 5 iDKO mice, 2-3 neurons per animal. For firing rates at 3 months post-TAM we used 10 neurons from 2 iCON mice, 5 neurons per animal; 8 neurons from 2 iLKO mice, 4 neurons per animal; 10 neurons from 3 iDKO mice, 3-4 neurons per animal. For control autoreceptor responses, we used 12 neurons from 7 iCON mice, 1-2 neurons per animal; 10 neurons from 5 iLKO mice, 2 neurons per animal; 9 neurons from 5 iDKO mice, 1-2 neurons per animal. For 8-OH-DPAT elicited responses, we used 11 neurons from 7 iCON mice, 1-2 per animal; 7 neurons from 5 iLKO mice, 1-2 per animal; 9 neurons from 5 iDKO mice, 1-2 neurons per animal.
Behavior
Mice were assessed for baseline performance prior to tamoxifen treatment. They were then rested for 3 days prior to tamoxifen gavage and assessed in the same behaviors 1 month following the last tamoxifen gavage and again at 6 months after the last gavage. Male and female mice were tested separately after being acclimated to the testing room for at least 30 min. Assays were performed during the light phase and experimenters were blinded to genotype.
Open field test
Mice were placed in the center of a 40 cm × 40 cm box in a dimly lit room and allowed to freely explore for 10 min. ANY-maze video-tracking software was used to track animal movement in digitally subdivided center and outer areas. During this period, total distance traveled and time spent in inner zone were measured.
Contextual fear conditioning
During training, mice were placed in a conditioning chamber (Med Associates) for 3min to acclimate and then presented with a single tone (30sec at 90dB and 5000 Hz) co-terminating with a 2 sec foot shock at 0.5 mA. Mice were then removed 1 min after the tone/shock was presented and returned to home cage. About 24 hrs later mice were returned to the same chamber and allowed 5 min of free exploration without auditory cues or foot shock during which freezing behavior was scored using Video Freeze software.
Restraint stress & corticosterone measurements
Mice were habituated overnight to the room where stress tests and blood collection were performed. For AM and PM blood collection mice were placed into a ventilated 50mL conical tube, during which tails were numbed in ice cold ethanol for 30 sec before nicking of tail tips with a sterile razor blade (AM collections) or gently rubbed with a clean wipe to remove blood clot from prior cut (PM collections). Approximately 2μl of blood was collected into sterile microvettes between 9am and 10am and between 5pm and 6pm. Mice were rested for a day before undergoing stress tests at which point they were restrained in ventilated 50mL conical tubes for 30 min between 9am and 10am and immediately returned to home cage. 30 min and 2 hr following stress, blood was collected using similar methods. Blood samples were spun at 4°C for 20 min at 12,000xg and serum was collected for use in CORT ELISA performed according to manufacturer’s instructions.
Genomic visualization
Histone ChIPmentation and CUT&RUN data from https://doi.org/10.1101/2021.10.13.463782 were visualized using the Integrated Genome Viewer (IGV, v2.9.3). All datasets were normalized to 1X genome coverage using the bamCoverage function in deeptools (v3.5.0) (Ramirez et al., 2016).
QUANTIFICATION AND STATISTICAL ANALYSIS
One-way ANOVA was performed for all analyses involving 3 or more genotypes assessed one time point using Welch’s correction where noted in the figure legend. Exceptions include the Hypergeometric test for the number of DEGs with GO terms in Figure 1H and Wilcoxon rank sum test for stage specific gene expression in Figure S9F. Two-way ANOVA with Welch’s correction where noted was performed for analyses involving more than 2 genotypes across 2 or more time points. Comparisons within the same genotype across time points are designated by color matched bars and stars. In some cases, only 1 comparison was made between 2 genotypes, in which case an unpaired t-test was performed. A pairwise comparison t-test was performed for Figure 6B. For behavior experiments two-way repeated measures ANOVA was performed. Graphpad Prism 8 was used for analysis and graphs using mean ± SEM unless otherwise noted. RNA-seq and Hypergeometric bar graphs were generated using ggplot2 in R. Violin plot of embryonic vs adult genes were generated using ggpubr in R (v4.0.4) (Wickham et al., 2016). The heatmap of all iLKO and iDKO DEGs was clustered using k-means clustering with pheatmap (v1.0.12) in R (v4.0.4) with k = 15. Specific details regarding n and significance parameters are specified in figure legends.
Gene expression and genomics
Gene expression quantification and differential expression were analyzed using Cufflinks v2.2.2 (Trapnell et al., 2010). Differentially expressed genes (DEGs) were defined as fold-change ≥1.5 and false discovery rate (FDR) ≤0.05. Gene Ontology analysis was performed using Webgestalt (http://www.webgestalt.org), requiring five genes per category and FDR ≤ 5% (Liao et al., 2019). Differential exon expression analysis was performed to test for differences in floxed Lmx1b exon 4 expression with DEXSeq (Anders et al., 2012). RNA-seq reads were mapped to the Mus musculus genome (Ensembl, v. 96) and reads falling within annotated exons were counted using featureCounts (Rsubread). DEXSeq default settings were used and significant exon expression differences were determined at FDR ≤ 5%. CUT&RUN and ChIPmentation datasets were normalized to 1X genome coverage using the bamCoverage function in deeptools (v3.5.0) (Ramirez et al., 2016).
CUT&RUN data processing and analysis
Details of CUT&RUN data generation are available elsewhere (https://doi.org/10.1101/2021.10.13.463782). The standard ENCODE ChIP-seq software pipeline from the ENCODE Consortium was used to process CUT&RUN data. Sequencing reads were mapped to the UCSC mm10 by Bowtie v2.3.4.3 (Langmead and Salzberg, 2012). Removal of PCR duplicates was done using samtools v1.9 (Li et al., 2009). Unique mapped reads were used to call peaks comparing TF-bound chromatin with an IgG control with SEACR using both relaxed and stringent threshold settings (Meers et al., 2019). The relaxed threshold uses the total signal of the “knee” to the peak of the total signal curve and the stringent threshold uses the peak of the total signal curve. Both threshold settings have been shown to have high specificity and sensitivity (Meers et al., 2019). Shared intersections between identified peaks (1bp minimum overlap) were identified using BEDTools (Quinlan and Hall, 2010). CUT&RUN data for Lmx1b and Pet1 were normalized together for visualization using the ChIPseqSpikeInFree method to determine the scaling factor for adjusting signal coverage across the genome (Jin et al., 2020). Gene annotation of CUT&RUN peaks was performed using the Genomic Regions Enrichment of Annotations Tool (GREAT), with each peak assigned to the nearest gene (McLean et al., 2010). Motif matching was performed with fimo from the MEME suite (Bailey et al., 2015) with a p-value threshold of 0.001.
ChIPmentation data processing and analysis
Details of ChIPmentation data generation are available elsewhere (https://doi.org/10.1101/2021.10.13.463782). The standard ENCODE ChIP-seq software pipeline from the ENCODE Consortium was used to process ChIPmentation data. Sequencing reads were mapped to the UCSC mm10 by Bowtie v2.3.4.3. Removal of PCR duplicates was done using samtools v1.9 (Li et al., 2009). The unique mapped reads were used to call peaks comparing immunoprecipitated chromatin with input chromatin with MACS2 v2.2.4 with FDR adjusted p-value cutoff of 0.01 (Zhang et al., 2008).


