Abstract
The circadian clock refers to the intrinsic biological rhythms of physiological functions and behaviours. It synergises with the solar cycle and has profound effects on normal metabolism and organismal fitness. Recent studies have suggested that the circadian clock exerts great influence on the differentiation of stem cells. Here, we focus on the close relationship between the circadian clock and mesenchymal stem cell fate decisions in the skeletal system. The underlying mechanisms include hormone signals and the activation and repression of different transcription factors under circadian regulation. Additionally, the clock interacts with epigenetic modifiers and non-coding RNAs and is even involved in chromatin remodelling. Although the specificity and safety of circadian therapy need to be further studied, the circadian regulation of stem cells can be regarded as a promising candidate for health improvement and disease prevention.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-022-02878-0.
Keywords: Circadian clock, Mesenchymal stem cells, Cell fate decision, Transcription factors, Epigenetics
Background
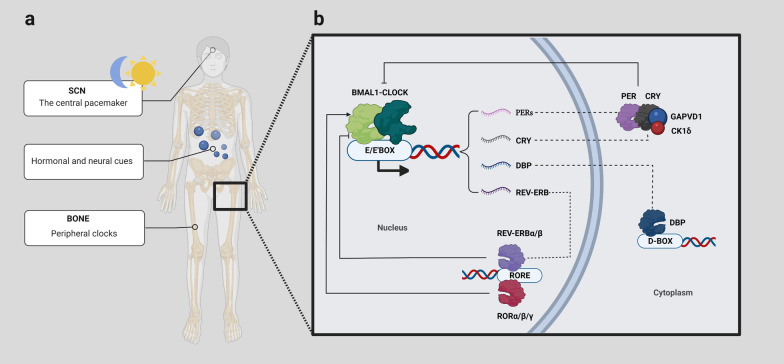
Circadian rhythm, also known as the circadian clock, directs and maintains nearly 24-h periodic changes in behaviour and physiology [1]. The central pacemaker of endogenous circadian clocks lies in the SCN (suprachiasmatic nuclei) of the anterior hypothalamus, which is sensitive to environmental cues (called zeitgebers) and orchestrates peripheral synchronicity through hormonal and neural mechanisms [2, 3]. Light input is one of the essential entraining daily oscillations and triggers the sympathetic release of norepinephrine that synchronises the peripheral clocks (Fig. 1a) [4]. The molecular clock is essentially a multilevel transcription–translation feedback loop that integrates a number of regulatory cues, including Bmal1 (brain and muscle ARNT-like protein 1/aryl hydrocarbon receptor nuclear translocator-like protein 1), Clock, Per (period), Cry (cryptochrome), RORα/β/γ (retinoid-related orphan receptor α/β/γ), and Rev–erbα/β (Fig. 1b) [5].
Fig. 1.
a The circadian clock is distributed throughout the body. The retina perceives light information, which will be relayed to the SCN. In turn, the SCN entrains peripheral system clocks via neural, humoral, and metabolic cues [6]. b The multilevel transcription–translation feedback loop of circadian rhythm. The central loop is based on reciprocal regulation between the BMAL1-CLOCK complex and the PER complex. BMAL1 and CLOCK bind to E-boxes in the promoters of a subset of clock-controlled genes (CCGs) in a heterodimer form, promoting the expression of Per, Cry, Rev-erb, etc. Once PERs and CRY in the cytoplasm accumulate to a certain extent, they will enter the nucleus under GAPVD1 modulation and suppress the transcriptional activity of the BMAL1-CLOCK complex [7]. Casein kinases 1δ/ε phosphorylate PERs, which affects their nuclear translocation and stability [8]. In the second transcription loop, nuclear receptors RORα/β/γ and REV-ERBα/β compete for the ROR element of BMAL1 [9]
The circadian clock has a great influence on the differentiation process in stem cells derived from different sources [10]. Bmal1 knockdown changes the stem cells’ differentiation potential with an aberrant induction of differential gene expression in vitro, and it is essential for transcriptional programmes in cell differentiation [11]. However, how mesenchymal stem cell (MSC) differentiation responds to rhythmic changes timely is a fundamental question to be answered in bone metabolism. Vast amounts of research continue to emerge and are yet to be summarised. This review links new developments to earlier findings, aiming to clarify how circadian clock genes regulate stem cell differentiation fate in the skeletal system and hoping to exploit the MSC differentiation potential by targeting the circadian clock. This review opens up the possibility of a new strategy for MSC regulation and will potentially improve MSC differentiation efficiency for future clinical applications.
Stem cells and the circadian clock
Prior to the late 1970s, researchers sought to interrogate how stem cells controlled by the circadian clock perform tissue homeostasis and regenerative functions. Today, rhythmicity has been shown to be instrumental in stem cells’ self-renewal and activation in the event of injury and mammalian development [12]. Multiple studies have confirmed that circadian components exist in undifferentiated ESCs (embryonic stem cells), even before zygotic genome activation [13, 14]. However, ESCs show non-oscillating clock gene expression and are thought to lack clock function. A self-sustained oscillator occurs during postnatal development with the gradual appearance of CLOCK, which is controlled by Dicer/Dgcr8-mediated post-transcriptional mechanisms [15, 16]. Interestingly, Paulose et al. [17] identified the rhythmicity of glucose utilisation and glucose transporter mRNA in ESCs in the absence of clock gene oscillation. This meant that circadian variations in some physiological activities are independent of the core clock, such as peroxiredoxin hyperoxidation [18]. Additionally, there may be other clock regulatory mechanisms in ESCs, and core clock genes may have a non-clock function in ESCs. The deletion of Clock reduces proliferation and increases apoptosis in ESCs, while Bmal1 has been demonstrated to be essential for a proper lineage differentiation of ESCs. Depletion of Bmal1 adversely affects the subsequent differentiation, leading to an imbalance of lineage-specific or selective activation of genes with increased pluripotency transcription factor Nanog, which may be closely related to the change of PI3K, MAPK, and Wnt pathways [19].
Although induced pluripotent stem cells (iPSCs) do not display oscillating clock genes, expression rhythms arise spontaneously in their directed differentiation process [16], which can also be reversed if iPSCs are reprogrammed to pluripotency. This indicates that clock function may be a common feature of differentiated and mature cells [20].
Multipotent stem cells from different tissues show differential clock gene expression profiles with different amplitude ranges. For example, compared with bone marrow stem cells (BMSCs) and adipose tissue-derived mesenchymal stem cells (ADSCs), dental pulp stem cells (DPSCs) have higher Bmal1 expression. After circadian synchronisation by dexamethasone, PER2 peaked at 32 h in BMSCs, with a nadir at 16 h in ADSCs [10]. The circadian clock and daily changes in light and darkness operating at the RNA and protein levels control the proliferation and differentiation of various multipotent cells such as cardiac progenitor cells, stem cells from the epidermis, hair follicle, nervous system, and haematopoietic tissue [4, 12]. For example, the rhythmic Per and Cycle underlie fate decisions and regeneration of intestinal epithelial stem cells [21].
-
2.
MSC differentiation exhibits a circadian pattern in the skeletal system
The circadian regulation of the skeletal system and the differentiation of MSCs have been widely recognised [22]. Luciferase reporter assays detect the 24-h rhythm of OCN mRNA, an osteogenesis marker [23]. BMP2 and RUNX2 change with the light cycle and the level of melatonin [24]. Serum bone turnover markers also show diurnal fluctuations [25], and the irregular light–dark cycle experienced by pregnant mice will affect the offspring’s circadian rhythm and lead to decreased bone perimeter, endocortical volume, and bone strength [26]. The dynamic balance of bone formation and resorption under strict time control ensures bone mass integrity and bone structure and function [27]. The fate change in MSCs resulting from a disrupted circadian clock is implicated in osteoporosis, elevated fracture risk, and osteoarthritis. Continuous exposure to artificial light significantly increases osteoporosis risk in humans and animals [28], and shift workers have a higher fracture risk [29]. Bioinformatic analysis of osteoarthritis patients’ transcriptome shows that clock imbalance may be the leading cause for these complications [30].
The intrinsic molecular clock underlies MSC differentiation at the genetic level. Clock gene (Bmal1, Pers, etc.) knockout and knockdown lead to the phase change of MSC differentiation. Disruption of Clock and Per2 results in a significant reduction in adipogenic differentiation without apparent changes in osteogenic differentiation [31]. BMAL1 oscillates in skeletal tissue and seems to promote osteogenesis and inhibit adipogenesis in a model-dependent manner. Bmal1−/− mice show low bone mass phenotypes with decreased bone trabeculae, bone density, and a thinner cortical bone, resulting from osteogenic MSC differentiation defects [32, 33], and the BMP2 signal may be a key target for BMAL1 [34]. Per−/− and Cry−/− mice show a significant increase in bone mass in both vertebrae and long bones [35]. This high bone phenotype worsens over time [36]. REV-ERBα, a negative regulator of BMAL1, gradually decreases with MSC osteogenesis, and its overexpression reverses the process [37]. However, the results are contradictory. RORα, a positive regulator of BMAL1, increases with MSC osteogenesis and late adipogenesis [38]. Bmal1 interference promotes adipogenesis with Wnt pathway down-regulation in preadipocytes. A high expression pattern of Bmal1 has been testified in mature adipocytes [39]. The knockdown of Bmal1 by RNA interference in mature 3T3-L1 cells only leads to a small amount of lipid droplet formation [40]. These findings suggest the distinct roles of the circadian clock at different differentiation stages that require further research.
The circadian clock regulates MSC differentiation by targeting hormones
Melatonin, synthesised in the pineal gland, is mainly stimulated by darkness and inhibited by light [41]. It keeps the bodily rhythms synchronised with the outside environment and has a positive effect on bone health [42, 43]. Decreased melatonin levels caused by ageing will increase the probability of adverse skeletal events [44], and this can be rescued by its timed administration. Melatonin’s capacity to reduce MSC oxidative stress-induced toxicity and stress-induced cellular senescence by increasing SIRT1 (sirtuin1) [45, 46] provides further evidence on its positive role in promoting bone health [47, 48]. It drives MSCs into an osteoblast lineage by inhibiting miRNA sponge circ-0003865 [49] and by taking part in osteogenesis signalling cascades, such as p38, BMP2, JNK, and Wnt-βcatenin pathways [50, 51]. Melatonin also increases chondrocyte cell volume and enhances chondrogenic differentiation through the Wnt pathway [52].
Parathyroid hormone (PTH) resets the skeletal clock by up-regulating Per mRNA expression [53], and its concentration in bodily fluids exhibits a circadian rhythm. PTH has been widely used in cartilage tissue engineering in recent years, as it inhibits chondrogenic differentiation but mitigates cartilage degeneration. Intermittent PTH protects cartilage by attenuating chondrocyte hypertrophy in 3D-printed tissue engineering scaffolds [54], although it reduces MSC cartilage terminal differentiation markers [55]. The deletion of PTH receptors in MSCs leads to decreased bone formation and increased bone marrow adipose tissue (BMAT), and intermittent PTH administration significantly reduced BMAT in control mice, also found in male patients with osteoporosis [56]. Research identifies that the number of bone marrow adipocytes in men with idiopathic osteoporosis decreased by 27% after 18 months of PTH treatment [56].
Glucocorticoids (GCs) mediate the transcription of Per1 and reset of a peripheral circadian rhythm [57]. SCN controls GC levels in plasma via the hypothalamic–pituitary–adrenal axis, and Per2 may be critical for this process [58]. Oscillation amplitudes of GCs are a crucial determinant of their biological function. The periodic GC-activated GC receptor directly influences the differentiation and functional integration of stem cells in hippocampi. GCs are directly or indirectly tied to osteoporosis by regulating PER2 [59]. Besides, GCs change the methylation profile of cell cycle regulation and Wnt signalling pathway genes [60].
The molecular clock influences MSC multilineage differentiation by regulating transcription factors
Various transcription factors spatiotemporally control the MSC differentiation in response to different stimuli. They directly interpret the genome, and their binding to it is the first step in DNA decoding. In bone metabolism, osteogenic factors, like RUNX2, guide MSCs to the osteogenic lineage; PPARγ promotes adipogenic differentiation; SOX9 participates in chondrogenesis. These transcription factors form a transcription network that plays a crucial role in MSC fate decisions. Network imbalance may lead to skeletal malformations and bone mass disorders. On the other hand, the mechanisms underlying these factors may provide solutions for the treatment of bone diseases.
Regulation and role of transcription factors in osteogenesis
RUNX2 is the ‘master switch’ of osteogenic differentiation, with a highly conserved DNA-binding runt domain. Runx −/− mice show a lack of osteogenic markers and complete blockage of intramembranous and endochondral ossification [61]. Cleidocranial dysplasia resulting from the mutation of RUNX2 is characterised by skeletal dysplasia, such as clavicle hypoplasia and abnormal clavicle [62]. Surprisingly, the overexpression of RUNX2 also shows bone defects in mice, including a dramatic reduction in osteoblast numbers with immature phenotypes. Therefore, RUNX2 plays a major role in MSCs’ early lineage commitment to osteoblasts and maintains the latter at an earlier stage to provide a large number of immature osteoblasts.
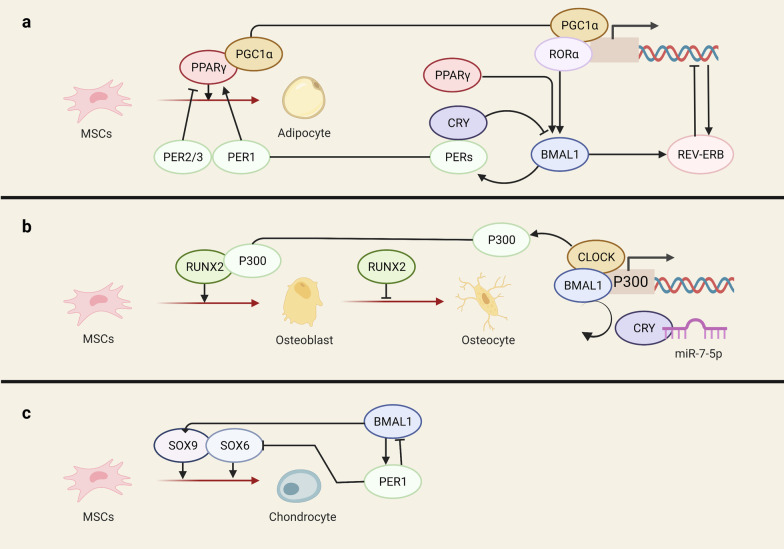
RUNX2 shows expression rhythmicity in various tissues, including the SCN and bone [63], and is involved in connecting the circadian clock and MSC fate decision. RUNX2 influences enamel formation in MSCs under the control of circadian pacemaker [64]. Some researchers suppose that RUNX2 relies on the activity of BMAL1, as the rhythm of RUNX2 mRNA disappears in mouse SCN without Bmal1 [63]. Still, some believe REV-ERBα is the upstream regulator of RUNX2 and disturbs the recruitment of RUNX2 to the targeted promoter. Knockdown of REV-ERBα enhances the differentiation and function of pre-osteoblasts [65].
miRNA is also one of the primary mechanisms underlying the rhythm of RUNX2. miR-7-5p disentangles CRY2’s inhibition on the BMAL1-CLOCK complex. The released BMAL1-CLOCK stimulates P300 transcription, promoting histone H3 acetylation and forming a transcription complex with RUNX2 to enhance bone formation (Fig. 2) [66]. Another miRNA, miR-433, shows robust circadian rhythmicity with a peak after dark and targets RUNX2 3’-UTR causing its degradation [67]. In miR-433-inhibited mice skulls, the mRNA levels of RUNX2 and its downstream osteocalcin increased significantly [68].
Fig. 2.
a Both PPARγ and its cofactor PGC-1α serve as master transcription factors in adipogenic differentiation and are tightly regulated by the circadian clock. b miR-7-5p liberates CRY from the BMAL1-CLOCK complex to promote the transcription of P300. P300 and RUNX2 act simultaneously on the targeted promoter. c BMAL1 promotes the expression of SOX9 and PER1 inhibits SOX6 expression
Besides acting as a transcription factor, RUNX2 also indirectly regulates the transcription of clock genes and/or clock-controlled genes (CCGs) by working as a scaffold to recruit growth factors and cytokines [63, 69], including P300, CCAAT/enhancer-binding protein, and histone deacetylase [70], with its protein–protein interacting domains.
To our knowledge, Osterix, cAMP-dependent transcription factor 4 (ATF4), Forkhead box O (FOXO), and other transcription factors in osteogenic differentiation have rarely been described in circadian rhythm studies. The role of these transcription factors in circadian rhythm remains to be identified.
Adipogenic master regulator PPARγ
Peroxisome proliferator-activated receptor γ (PPARγ) belongs to the nuclear hormone receptor superfamily with the highest expression in adipose tissue. It plays a vital role in the balance between osteogenic and adipogenic differentiation of MSCs [71–73]. PPARγ up-regulation promotes adipogenic differentiation and inhibits the generation of osteoblasts and chondrocytes [74], with its down-regulation having opposite effects [75]. Besides, the PPARγ agonist thiazolidinedione (TZD) is associated with bone loss and fracture [76]. Clinical experience and animal studies suggest that the fate change of MSCs associated with PPARγ may be the main reason for this.
PPARγ and its cofactor PGC-1α display a clear circadian expression pattern in many tissues, and PPARγ even acts as a crucial peripheral clock activator of the cardiovascular system and metabolism [77, 78]. Multiple randomised clinical trials have shown that the clock hormone melatonin up-regulates PPARγ [79, 80]. And the inhibition of Clock or Per2 changes the expression of osteocalcin in MSCs, in association with extraordinary increases in PPARγ [31]. BMAL1 and REV-ERBα are induced by PPARγ and PGC-1α [77, 81, 82]. PPARγ is deeply involved in the regulatory networks between circadian rhythms and MSC fate decisions (Fig. 2), with the mechanisms described hereafter.
Nocturnin−/− mice show deregulated circadian patterns in PPARγ gene expression and increased bone mass [83]. Nocturnin adapts to daily environmental changes by changing the organism’s metabolic state [84, 85]. It works as a pivotal mediator of mitochondrial function by dephosphorylating nicotinamide adenine dinucleotide phosphate (NADP+/NADPH) [86]. Nocturnin is highly expressed in MSCs, and its overexpression enhances the lipogenesis of preadipocytes 3T3-L1. It enhances PPARγ transcription activities and guides PPARγ nuclear translocation.
Per1 and Per2 have antagonistic influences on PPARγ activity. PER1 enhances PPARγ activity operating as its natural modulator, while PER2 directly and specifically inhibits the recruitment of PPARγ to the target promoter [87]. However, the PER2 regulation of adipogenesis has significant cell/tissue specificity [87], as it is regulated by a complicated network and works with other core clock products, such as PER1, BMAL1, and REV-ERBα. For example, a substantial reduction in PPARγ is found in per1/2−/− mice [88]. However, Per3 is believed to be a nonessential clock component with modest effects on circadian behaviours [89, 90], and it does inhibit adipogenesis in vivo in a clock manner [91]. The deletion of Per3 promotes adipogenesis by clock outputs [92]. PER3 binds to PPARγ and inhibits its activity [91, 93]. Increasing PPAR is observed in Per3−/− mice, which means that the differentiation arrest resulting from Per3 happens during the early stages.
D-box binding protein (DBP) and E4 promoter-binding protein 4 (E4BP4), contributors to circadian regulatory loops, interact with the active site of the PPARγ promoter to regulate its rhythmic expression and are enhanced by histone deacetylase inhibitor (HDACi) [94]. Decreased H3K9 acetylation of the DBP promoter leads to a lower level of DBP and a diminished binding between DBP and the PPARγ promoter, which gives rise to a lower level of PPARγ [95].
Chondrogenic factors
The circadian clock is critical to early chondrogenesis and endochondral ossification [96, 97]. Indian Hedgehog (Ihh) is key in governing chondrogenic differentiation, and its activity is regulated by PER1 and BMAL1 in the growth plate [97]. Bmal1 deficiency leads to the deregulated rhythmic expression of Ihh and Per [98]. Our previous studies show that Prg4 and related chondrogenic genes are downstream targets of Bmal1 in cartilage formation and chondrogenic differentiation [99]. Cartilage tissue engineering, which combines MSCs with other factors, such as collagen, is a promising therapy for degenerated temporomandibular joint and osteoarthritis. Sry-related HMG box 9 (SOX9), a key transcription regulator of chondrocyte differentiation [100], controls the synthesis of collagen type II, the hallmark of chondrogenic commitment [101]. Impaired circadian rhythm induces osteoarthritis resulting from SOX9 inhibition in articular chondrocytes, and the deletion of Bmal1 also leads to reduced SOX9 [102] (Table 1). Two additional SOX family members, SOX6 and SOX5, are also involved in chondrogenesis regulation. SOX6 is negatively regulated by its transactivator PER1 (Fig. 2). It shows a long-term decrease after exposure to 10 nM PTH for 1 h in chondrocyte-like ATDC5 cells with no distinct changes in level SOX5 and RUNX2 levels [103].
Table 1.
Epigenetic modifying enzymes shared both in the circadian clock and in the MSC fate decisions
The circadian clock determines MSC fate through epigenetic events
Approximately 43% of mammalian genes oscillate in a circadian fashion, but no more than 30% of circadian mRNAs are driven by de novo transcription, indicating that post-transcriptional mechanisms mainly regulate the rhythmic expression of genes in a cell-/tissue-specific fashion [109, 110]. Epigenetics is an essential component of post-transcriptional regulation and has rapidly developed into various fields and disciplines in the twenty-first century, enhancing our understanding of gene regulation in health and disease.
The term ‘epigenetics’ refers to heritable alterations in gene expression without directly altering the nucleotide sequence [111]. The epigenome mediates a crosstalk between cells and the environment through covalent modifications in chromatin (also called epigenetic code) and epigenetic regulators, which include DNA methylation, histone modification, non-coding RNAs, etc. [112]. The epigenome opens or closes chromatin fibre around genes to control subsequent transcription factor binding, exerting great influence on MSC differentiation [113]. The circadian clock has also been implicated in stem cell differentiation, which far exceeds its daily timekeeping roles [114]. Interestingly, accumulating research shows the close relationship between the epigenetic mechanism and circadian rhythm. The rhythmic transcriptome is assisted by a specific epigenetic mechanism which cooperates with the clock to orchestrate daily oscillations [115]. Therefore, we postulate that epigenetic regulators function in the crosstalk between the circadian clock and stem cell fate decisions to determine how and when transcription oscillations occur. The coordinated mechanisms include the clock proteins and epigenetic modifying enzymes constituting complexes to regulate transcription and the collaborative interaction between the clock and non-coding RNAs or chromatin remodellers.
Circadian proteins form complexes with epigenetic modifying enzymes
Histone-modifying enzymes
Genome-wide expression analysis reveals the rhythm of global histone labels that, depending on an intact clock, are under tight regulation by enzymatic feedback loops [109]. The clock recruits epigenetic enzymes to the promoter of important transcription factors and key signalling molecules involved in differentiation. Besides, CLOCK, functioning as an epigenetic modifier, was subsequently found to have a histone acetyltransferase (HAT) activity greatly amplified by its combination with BMAL1 [116]. Additionally, clock-controlled SIRTs and EZH2 provide possibility in elucidating how the clock is linked to epigenetic mechanism in MSCs differentiation.
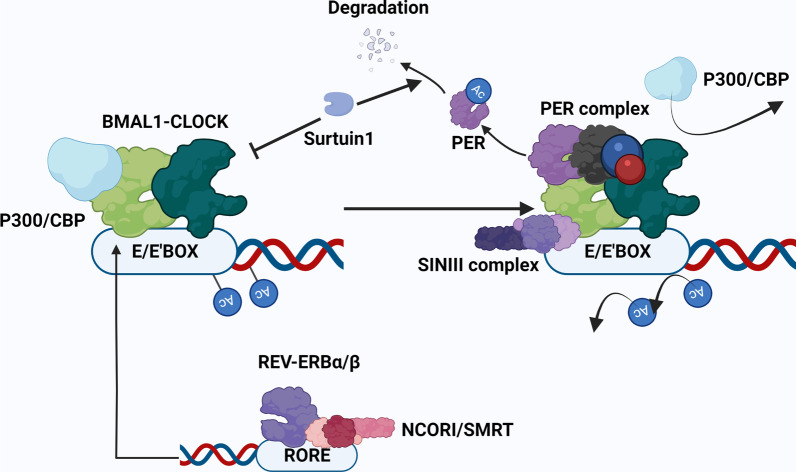
At the stage of transcriptional activation, the BMAL1-CLOCK complex acetylates the RUNX2 promoter by Clock itself and by recruiting acetyltransferases P300/CBP, thus promoting osteogenesis in MSCs [66]. At the repression stage, CRY (a constitutive component of the PER complex) occupies the C-terminal transactivation domain of BMAL1, leaving P300/CBP dissociated from its binding site [117]. Once CRY is loaded to the BMAL1-CLOCK complex, it directly recruits the SIN3 complex that comprises HDAC1 and HDAC2 to remove the acetylation tag of promoters (Fig. 3). Thus, the PER-SIN-HDACs pathway targeting histones H3 and H4 controls gene expression precisely and dynamically [118]. In addition, REV-ERBα and RORα exert downstream effects on the expression of NF-kB (which inhibits osteoblast differentiation) by recruiting HDACs [104]. REV-ERBα calls up co-repressors NCOR1/SMRT to ROR element. HDAC3, serving as the catalytic component of NCOR1/SMRT, rhythmically modulates histone deacetylation of the ROR element [119, 120].
Fig. 3.
The PER complex, competing with P300/CBP, binds the BMAL1-CLOCK complex. SIRT1 and REV-ERBα/β precisely regulate this process: SIRT1 prompts PER complex dissociation from the BMAL1-CLOCK complex and promotes PER2 degradation. It also plays a negative role in CLOCK HAT activity. REV-ERBα/β recruits the co-repressors NCOR1/SMRT to ROR element to promote P300/CBP binding with the BMAL1-CLOCK complex
Sirtuins (SIRTs) are class III NAD + dependent deacetylases that participate in many biological processes, including the circadian clock and cell differentiation [121–123]. This family has seven members (SIRT1–7) distributed across the cytoplasm, nucleus, and mitochondria. SIRT1 binds BMAL1-CLOCK in a circadian manner and counteracts the HAT activity of CLOCK [124]. Its deacetylation ability extends to BMAL1 and PER2 and relieves BMAL1-CLOCK from CRY and promotes PER2 degradation [125, 126]. SIRT1 requires the cofactors NAD + and NAMPT to function, which are both under tight circadian control [86, 127]. Thus, the deacetylase activity of SIRT1 exhibits a circadian pattern.
The MSC-specific overexpression of SIRT1 increased alveolar bone mass in vivo and promoted MSCs’ proliferation and osteogenic differentiation [128]. Aged mice with a MSC-specific knockout of SIRT1 show reduced subcutaneous fat and a progressive loss of trabecular and cortical bone volume [129]. SIRT1 exerts its effects mainly by deacetylating important transcription factors and signalling pathways, influencing their activity and localisation. For example, SIRT1 directly or indirectly deacetylates RUNX2and PPARγ [130, 131] and deacetylates β-catenin, facilitating its accumulation in the nucleus [132]. Along these lines, SIRT1, serving as a circadian hallmarker, causes dynamic changes in the epigenome to generate transcriptional responses controlling MSC differentiation.
Methylation is also a widespread histone modification in mammalian cells and is linked to transcriptional repression. EZH2, mediating gene silencing via trimethylation of H3K27, is directly regulated by the circadian clock via the E-BOX and RORE motifs in fish species [133]. EZH2 is a repressor of differentiation-associated genes in vitro. It inhibits osteogenesis by inhibiting BMP2 and inhibits chondrogenic differentiation by inhibiting ATOH8 [106, 107] (Table 1). However, in in-vivo animal studies, the additive effects of VDR‐mediated EZH2 transcriptional increase show increased osteogenesis, as EZH2 also inhibits MSC senescence by repressing p16 transcription [134]. MSCs are cells of mesodermal origin which derived from ESCs. The level of EZH2 in MSCs is significantly lower than that in ESCs, and a specific EZH2 inhibitor, GSK126, dramatically drives ESCs differentiation into MSCs by reducing H3K27me3 [135], suggesting that EZH2 levels in specific cell types are crucial for the balance of pluripotency maintenance, cell differentiation, and cell senescence.
-
(2)
DNA-modifying enzymes
Tissue-specific DNA methylation patterns play central roles in establishing cell identity during differentiation. Generally, DNA methylation is considered one of the hallmark mechanisms of gene silencing. For example, DNMT inhibitors, such as 5-aza-2'-deoxycytidine, induce gene activation and cell differentiation [136]. However, the genome-wide DNA methylation profiling of MSCs shows an increasing methylation level of the global genome and CpG sites in the process of osteogenic differentiation [137]. These results appear to be incompatible with the gene-silencing role of DNA methylation. Recent studies suggest that the function of DNA methylation varies depending on the methylation location. The methylation of the gene body, and not the promoter, promotes genome stability and even transcription [138], suggesting that promoter segments may not be the DNA methyltransferase target sites during MSCs differentiation.
In recent years, growing evidence in chronobiology hints at a close connection between the circadian rhythm and DNA methylation. DNMTs oscillate in a clock pattern in the mouse liver and brain [139], and excessive exposure to artificial light leads to tissue-specific changes in DNMTs expression [140]. Taken together, this evidence suggests that DNMT may exert great influence during clock-controlled differentiation.
Reciprocal regulation between the circadian clock and non-coding RNAs
miRNAs are a class of short non-coding RNAs with a length of about 22 nucleotides. The circadian clock regulates miRNAs expression and activity, and the clock itself is regulated by miRNAs at post-transcriptional levels [141]. And miRNAs taking part in clock regulation have been extensively reviewed [142, 143]. In healthy young males, some circulating miRNAs display 24-h rhythms with two expression patterns: one peaks at night and the other peaks during the daytime [144, 145]. There are 104 miRNAs differently expressed in CLOCK-mutant mice compared with the controlled group, and they are linked to transcription regulation and PI3K-AKT and MAPK signalling pathways [146], which are closely related to MSC differentiation.
However, some researchers argue that mature miRNAs do not oscillate in a circadian pattern. Studies show at least 57 pri-miRNAs oscillated in the mouse liver, with only four mature miRNAs showing the same rhythm as their primary transcript [147]. Interestingly, the rhythmic expression of Dicer (the essential enzyme for miRNA maturation) is found in various tissues [148]. Studies are contradictory and may be model-dependent. The inconsistent findings among these studies can be partly explained by variations in the study samples, methods of assessment, and, most importantly, tissue specificity, which may hint at the role of circadian miRNA in establishing cell identity.
Mounting research has unveiled the importance of miRNAs in the regulation of MSC differentiation. For example, the expression profile in patients with fractures or osteoporosis differs from that in healthy individuals. Hackl et al. summarized the different expression of miRNAs in patients with bone disease and the control group. They rasied the possibilility that circulating miRNAs are used as biomarkers for bone disease [149]. Furthermore, many miRNAs are shared between the circadian clock and MSCs fate decisions. A candidate miRNA set potentially involved in circadian control has been established by various means, including microarrays [150], CHIP [142], and in silico prediction analysis of databases [151]. We found that some of them also function in the differentiation process of MSCs, as summarised in the Additional file 1.
Long non-coding RNAs (lncRNAs) are non-coding RNAs with 2000–100,000 nucleotides in length. A mouse transcriptome analysis indicated that more than 1000 lncRNAs show a diurnal rhythm [152]. The expression of a substantial portion of rhythmic lncRNAs is pronounced in regions of enhancer clusters and regulates gene expression, showing high similarity in rhythm phase and expression patterns with genes in their proximity. For instance, lnc-CROT is highly expressed at super-enhancer regions and modulates long-range circadian gene regulation [153]. Accumulating research has shown that lncRNAs, such as lnc-MEG3 and lnc-XIST, take part in osteogenic differentiation [154, 155]. Unfortunately, there are few reports on the role of lncRNAs in the clock-controlled differentiation process, and follow-up bioinformatics studies should be carried out.
Circadian proteins influencing chromatin architecture
Different kinds of epigenetic mechanisms show a pile-up effect on gene expression levels by influencing chromatin architecture and accessibility, which influence the binding of transcription factors to the genome. The molecular clock, working with various epigenetic regulators, contributes to spatiotemporal balance and the oscillation of chromatin accessibility [109, 156]. For example, RORα/β, BMAL1, and other circadian proteins promote global chromatin decondensation during the activation phase via the SWI/SNF complex (one of the epigenetic remodelling complexes). In turn, this promotes the load of REV-ERB, which rapidly responds to structure changes and inhibits circadian protein expression [157]. Furthermore, the chromatin landscape shaped by circadian regulation is critical for MSCs self-renewal and multilineage differentiation [158].
Future perspective
Circadian dysfunction is associated with various diseases in mammals [159], such as myocardial infarctions (MIs) and bone diseases [160]. Interrupted light at night causes depressive-like behaviours in mice by the circadian ipRGCs-dpHb-NAc (retinal ganglion cells–the dorsal perihabenular nucleus–the nucleus accumbens) pathway [161]. Besides, the symptom complexity and the onset of various diseases show marked circadian rhythm. For example, the incidence of MIs at 9 a.m. is three times greater than that at 11 p.m., periods associated with the blood pressure rhythm [162]. Thus, manipulating and maintaining the circadian rhythm is important for disease prevention. The circadian rhythm of disease onset and chrono-drug discovery should be taken into account when deciding treatment times [163].
Luckily, the circadian rhythm can be remodelled by environmental cues. Light, temperature, eating frequency, and regular exercises are proved to be daily circadian temporal cues [164, 165]. Regular exercise rescued the disrupted circadian rhythm by increasing Per2 expression amplitude [166]. Time-restricted feeding increasing circadian amplitude prevents metabolic diseases and obesity in mice fed a high-fat diet [167]. Short-term caloric restriction is associated with cardio-protection [168]. This suggests that we may alter MSC function and restore bone function and structure by rectifying eating frequency and the sleep–wake pattern and reducing night-time exposure to artificial light to reset the circadian clock.
In addition to behavioural improvements, mechanical stress, biotic factors, and small molecule modifiers are promising interventions for a robust rhythm. Mechanical stress regulates the clock of skeletal muscle cells (C2C12 cells) by decreasing Per/Cry expression and increasing Bmal1 expression [169]. The Per1 inhibition of BMSC by artificial materials can be rescued by vitamin D supplements and osteogenic media [170]. Melatonin supplements, overcoming rhythm disruption, exhibit excellent treatment efficacy and safety as ideal candidates for preventing bone loss. GCs like dexamethasone are widely used to generate synchronised clocks in vitro. Some small molecule modifiers of circadian clocks alter the circadian period length, phase, and amplitude with favourable benefits. For example, Rev-erb activation by SR9009 is beneficial for the reparative process after ischaemia–reperfusion injury by attenuating NLRP3 inflammasome activation and immune infiltration, being also essential for protection against the development of cardiac hypertrophy and heart failure in mice [171, 172]. Furthermore, the proliferation and differentiation of neural stem cells are promoted by SR9009 in a dose-dependent manner through targeting Rev-erb [173]. However, clock is found in almost all tissues in mammals, and systematic influences on clock activity may interrupt normal physiological processes in healthy tissues. Thus, the specificity and safety of those modifiers should be fully reviewed before it is widely applied in clinical practice.
Additionally, different stem cells are sensitive to specific entraining cues with different circadian patterns. In DPSCs, mechanosensitive synchronisation is more accessible than dexamethasone synchronisation. BMSCs are more responsive to chemical stimulation than dexamethasone, and stem cells show different responses to entraining cues at certain times of the day [10, 162]. This hints at specific entraining choices for the circadian rhythm of different cells at specific times.
Our study has several limitations, since the interplay between the circadian rhythm and cell differentiation is a complicated network rather than a unidirectional hierarchical structure. How chromatin modifiers [174], systemic conditions, signalling pathways, kinases and phosphatases [175], RNA-binding factors [176], and other factors are involved in the whole regulatory network remains to be identified. However, the development of high-throughput techniques and novel sequencing technologies makes it possible to depict this underlying network, which is beneficial for a deeper understanding of MSC [1]. We may potentially improve MSC differentiation efficiency for future clinical applications.
Conclusions
The connections between the circadian rhythm and cell differentiation have been intensively researched in recent years. The underlying associated mechanisms remain to be summarised. This review elaborates on key coupling mechanisms, which include the following: (1) The circadian clock affects hormone secretion to regulate cell differentiation; (2) molecular clock core products directly modulate the activity and expression of key transcription factors; and (3) histone modifications, DNA modification, non-coding RNAs, and large multi-subunit chromatin remodelling complexes and other epigenetic modifications are critical nodes in the circadian regulation of cell fate decision. We offer an in-depth insight into the interplay between the circadian clock and cell differentiation in the skeletal system. Additionally, we suggest that incorporating circadian regulation (ranging from chemical, biological, and physical cues) may have profound benefits in optimising tissue engineering approaches and stem cell therapies.
Supplementary Information
Additional file 1. Predicted clock-targeted miRNAs in mice and human which also working in the MSC differentiation
Acknowledgements
Figures were created with BioRender.com.
Abbreviations
- SCN
Suprachiasmatic nuclei
- Bmal1
Brain and muscle ARNT-like protein 1
- Per
Period
- Cry
Cryptochrome
- RORα/β/γ
Retinoid-related orphan receptor α/β/γ
- MSCs
Mesenchymal stem cells
- ESCs
Embryonic stem cells
- iPSCs
Induced pluripotent stem cells
- BMSCs
Bone marrow stem cells
- ADSCs
Adipose tissue-derived mesenchymal stem cells
- DPSCs
Dental pulp stem cells
- PTH
Parathyroid hormone
- BMAT
Bone marrow adipose tissue
- GCs
Glucocorticoids
- SOX9
Sry-related HMG box 9
- PPAR γ
Peroxisome proliferator-activated receptor γ
- TZD
Thiazolidinedione
- CCGs
Clock-controlled genes
- ATF4
CAMP-dependent transcription factor 4
- FOXO
Forkhead box O
- NADP + /NADPH
Nicotinamide adenine dinucleotide phosphate
- DBP
D-box binding protein
- E4BP4
E4 promoter-binding protein 4
- CBP
CREB-binding protein
- HDACs
Histone deacetylases
- SIRTs
Sirtuins
- EZH2
Histone-lysine N-methyltransferase
- DNMTs
DNA methyltransferases
- CpG
Cytosine-phosphate-guanine
- lncRNAs
Long non-coding RNAs
- MIs
Myocardial infarctions
- ipRGCs-dpHb-NAc
Retinal ganglion cells–the dorsal perihabenular nucleus–the nucleus accumbens
Author contributions
WZG wrote the initial manuscript draft; RL and MLY revised the manuscript draft; LXZ and JWZ revised the manuscript to make it clearer; XYW and YQY reviewed and corrected grammar mistakes; QZ edited and approved the final manuscript for publication. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81870745, 82171003).
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing financial or professional interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chirico N, Van Laake LW, Sluijter JPG, et al. Cardiac circadian rhythms in time and space: the future is in 4D. Curr Opin Pharmacol. 2021;57:49–59. doi: 10.1016/j.coph.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Silver R, LeSauter J, Tresco PA, et al. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382(6594):810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 3.Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science (New York, NY) 2000;288(5466):682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 4.Fan SMY, Chang YT, Chen CL, et al. External light activates hair follicle stem cells through eyes via an ipRGC-SCN-sympathetic neural pathway. Proc Natl Acad Sci USA. 2018;115(29):E6880–E6889. doi: 10.1073/pnas.1719548115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96(2):271–290. doi: 10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 6.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 7.Aryal RP, Kwak PB, Tamayo AG, et al. Macromolecular assemblies of the mammalian circadian clock. Mol Cell. 2017;67(5):770–782. doi: 10.1016/j.molcel.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narasimamurthy R, Hunt SR, Lu Y, et al. CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci USA. 2018;115(23):5986–5991. doi: 10.1073/pnas.1721076115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Fang B, Emmett MJ, et al. Gene regulation. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science (New York, NY) 2015;348(6242):1488–1492. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers EH, Fawcett SA, Pekovic-Vaughan V, et al. Comparing circadian dynamics in primary derived stem cells from different sources of human adult tissue. Stem Cells Int. 2017;2017:2057168. doi: 10.1155/2017/2057168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ameneiro C, Moreira T, Fuentes-Iglesias A, et al. BMAL1 coordinates energy metabolism and differentiation of pluripotent stem cells. Life Sci Alliance. 2020;3(5). [DOI] [PMC free article] [PubMed]
- 12.Janich P, Pascual G, Merlos-Suárez A, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480(7376):209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 13.Hamatani T, Carter MG, Sharov AA, et al. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6(1):117–131. doi: 10.1016/S1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 14.Delaunay F, Thisse C, Thisse B, et al. Differential regulation of Period 2 and Period 3 expression during development of the zebrafish circadian clock. Gene Expr Patterns. 2003;3(3):319–324. doi: 10.1016/S1567-133X(03)00050-4. [DOI] [PubMed] [Google Scholar]
- 15.Umemura Y, Koike N, Ohashi M, et al. Involvement of posttranscriptional regulation of in the emergence of circadian clock oscillation during mouse development. Proc Natl Acad Sci USA. 2017;114(36):E7479–E7488. doi: 10.1073/pnas.1703170114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dierickx P, Vermunt MW, Muraro MJ, et al. Circadian networks in human embryonic stem cell-derived cardiomyocytes. EMBO Rep. 2017;18(7):1199–1212. doi: 10.15252/embr.201743897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulose JK, Rucker EB, Cassone VM. Toward the beginning of time: circadian rhythms in metabolism precede rhythms in clock gene expression in mouse embryonic stem cells. PLoS ONE. 2012;7(11):e49555. doi: 10.1371/journal.pone.0049555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putker M, O'Neill JS. Reciprocal control of the circadian clock and cellular redox state—a critical appraisal. Mol Cells. 2016;39(1):6. doi: 10.14348/molcells.2016.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallardo A, Molina A, Asenjo HG, et al. The molecular clock protein Bmal1 regulates cell differentiation in mouse embryonic stem cells. Life Sci Alliance. 2020;3(5):e201900535 . doi: 10.26508/lsa.201900535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umemura Y, Yoshida J, Wada M, et al. An in vitro ES cell-based clock recapitulation assay model identifies CK2α as an endogenous clock regulator. PLoS ONE. 2013;8(6):e67241. doi: 10.1371/journal.pone.0067241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karpowicz P, Zhang Y, Hogenesch JB, et al. The circadian clock gates the intestinal stem cell regenerative state. Cell Rep. 2013;3(4):996–1004. doi: 10.1016/j.celrep.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudek M, Meng Q-J. Running on time: the role of circadian clocks in the musculoskeletal system. Biochem J. 2014;463(1):1–8. doi: 10.1042/BJ20140700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gafni Y, Ptitsyn AA, Zilberman Y, et al. Circadian rhythm of osteocalcin in the maxillomandibular complex. J Dent Res. 2009;88(1):45–50. doi: 10.1177/0022034508328012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witt-Enderby PA, Slater JP, Johnson NA, et al. Effects on bone by the light/dark cycle and chronic treatment with melatonin and/or hormone replacement therapy in intact female mice. J Pineal Res. 2012;53(4):374–384. doi: 10.1111/j.1600-079X.2012.01007.x. [DOI] [PubMed] [Google Scholar]
- 25.Tsang K, Liu H, Yang Y, et al. Defective circadian control in mesenchymal cells reduces adult bone mass in mice by promoting osteoclast function. Bone. 2019;121:172–180. doi: 10.1016/j.bone.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaves I, van der Eerden B, Boers R, et al. Gestational jet lag predisposes to later-life skeletal and cardiac disease. Chronobiol Int. 2019;36(5):657–671. doi: 10.1080/07420528.2019.1579734. [DOI] [PubMed] [Google Scholar]
- 27.Feng X, McDonald JM. Disorders of bone remodeling. Annu Rev Pathol. 2011;6:121–145. doi: 10.1146/annurev-pathol-011110-130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucassen EA, Coomans CP, van Putten M, et al. Environmental 24-hr cycles are essential for health. Curr Biol CB. 2016;26(14):1843–1853. doi: 10.1016/j.cub.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 29.Feskanich D, Hankinson SE, Schernhammer ES. Nightshift work and fracture risk: the Nurses' Health Study. Osteoporos Int J Establ Result Coop Between Eur Found Osteoporos Natl Osteoporos Found USA. 2009;20(4):537–542. doi: 10.1007/s00198-008-0729-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soul J, Dunn SL, Anand S, et al. Stratification of knee osteoarthritis: two major patient subgroups identified by genome-wide expression analysis of articular cartilage. Ann Rheum Dis. 2018;77(3):423. doi: 10.1136/annrheumdis-2017-212603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boucher H, Vanneaux V, Domet T, et al. Circadian clock genes modulate human bone marrow mesenchymal stem cell differentiation, migration and cell cycle. PLoS ONE. 2016;11(1):e0146674. doi: 10.1371/journal.pone.0146674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieben L. Bone: The circadian clock controls bone remodelling. Nat Rev Rheumatol. 2016;12(3):132. doi: 10.1038/nrrheum.2016.10. [DOI] [PubMed] [Google Scholar]
- 33.Min H-Y, Kim K-M, Wee G, et al. Bmal1 induces osteoblast differentiation via regulation of BMP2 expression in MC3T3-E1 cells. Life Sci. 2016;162:41–46. doi: 10.1016/j.lfs.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Huang Z, Wei H, Wang X, et al. Icariin promotes osteogenic differentiation of BMSCs by upregulating BMAL1 expression via BMP signaling. Mol Med Rep. 2020;21(3):1590–1596. doi: 10.3892/mmr.2020.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maronde E, Schilling AF, Seitz S, et al. The clock genes Period 2 and Cryptochrome 2 differentially balance bone formation. PLoS ONE. 2010;5(7):e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu L, Patel MS, Bradley A, et al. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122(5):803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 37.He Y, Lin F, Chen Y, et al. Overexpression of the circadian clock gene Rev-erbα affects murine bone mesenchymal stem cell proliferation and osteogenesis. Stem Cells Dev. 2015;24(10):1194–1204. doi: 10.1089/scd.2014.0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer T, Kneissel M, Mariani J, et al. In vitro and in vivo evidence for orphan nuclear receptor RORalpha function in bone metabolism. Proc Natl Acad Sci USA. 2000;97(16):9197–9202. doi: 10.1073/pnas.150246097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tal Y, Chapnik N, Froy O. Non-obesogenic doses of palmitate disrupt circadian metabolism in adipocytes. Adipocyte. 2019;8(1):392–400. doi: 10.1080/21623945.2019.1698791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimba S, Ishii N, Ohta Y, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102(34):12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stehle JH, Saade A, Rawashdeh O, et al. A survey of molecular details in the human pineal gland in the light of phylogeny, structure, function and chronobiological diseases. J Pineal Res. 2011;51(1):17–43. doi: 10.1111/j.1600-079X.2011.00856.x. [DOI] [PubMed] [Google Scholar]
- 42.Feng Z-Y, Yang S-D, Wang T, et al. Effect of melatonin for regulating mesenchymal stromal cells and derived extracellular vesicles. Front Cell Dev Biol. 2021;9:717913. doi: 10.3389/fcell.2021.717913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu X, Yu S, Chen G, et al. Insight into the roles of melatonin in bone tissue and bone-related diseases (Review) Int J Mol Med. 2021;47(5):1–19. doi: 10.3892/ijmm.2021.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amstrup AK, Sikjaer T, Heickendorff L, et al. Melatonin improves bone mineral density at the femoral neck in postmenopausal women with osteopenia: a randomized controlled trial. J Pineal Res. 2015;59(2):221–229. doi: 10.1111/jpi.12252. [DOI] [PubMed] [Google Scholar]
- 45.Munmun F, Witt-Enderby PA. Melatonin effects on bone: implications for use as a therapy for managing bone loss. J Pineal Res. 2021;71(1):e12749. doi: 10.1111/jpi.12749. [DOI] [PubMed] [Google Scholar]
- 46.Chen W, Chen X, Chen AC, et al. Melatonin restores the osteoporosis-impaired osteogenic potential of bone marrow mesenchymal stem cells by preserving SIRT1-mediated intracellular antioxidant properties. Free Radic Biol Med. 2020;146:92–106. doi: 10.1016/j.freeradbiomed.2019.10.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu KK. Control of mesenchymal stromal cell senescence by tryptophan metabolites. Int J Mol Sci. 2021;22(2):697. doi: 10.3390/ijms22020697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao Z, Geng X, Jiang X, et al. Melatonin attenuates AlCl-induced apoptosis and osteoblastic differentiation suppression by inhibiting oxidative stress in MC3T3-E1 cells. Biol Trace Elem Res. 2020;196(1):214–222. doi: 10.1007/s12011-019-01893-2. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Chen T, Deng Z, et al. Melatonin promotes bone marrow mesenchymal stem cell osteogenic differentiation and prevents osteoporosis development through modulating circ_0003865 that sponges miR-3653-3p. Stem Cell Res Ther. 2021;12(1):150. doi: 10.1186/s13287-021-02224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Jia G, Xue P, et al. Melatonin restores osteoporosis-impaired osteogenic potential of bone marrow mesenchymal stem cells and alleviates bone loss through the HGF/PTEN/Wnt/β-catenin axis. Ther Adv Chronic Dis. 2021;12:2040622321995685. doi: 10.1177/2040622321995685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Wang C, Si J, et al. Melatonin up-regulates bone marrow mesenchymal stem cells osteogenic action but suppresses their mediated osteoclastogenesis via MT2-inactivated NF-κB pathway. Br J Pharmacol. 2020;177(9):2106–2122. doi: 10.1111/bph.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, He T, He L, et al. Melatonin contributes to the hypertrophic differentiation of mesenchymal stem cell-derived chondrocytes via activation of the Wnt/β-catenin signaling pathway: melatonin promotes MSC-derived chondrocytes hypertrophy. Stem Cell Res Ther. 2021;12(1):467. doi: 10.1186/s13287-021-02536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinoi E, Ueshima T, Hojo H, et al. Up-regulation of per mRNA expression by parathyroid hormone through a protein kinase A-CREB-dependent mechanism in chondrocytes. J Biol Chem. 2006;281(33):23632–23642. doi: 10.1074/jbc.M512362200. [DOI] [PubMed] [Google Scholar]
- 54.Music E, Futrega K, Palmer JS, et al. Intermittent parathyroid hormone (1–34) supplementation of bone marrow stromal cell cultures may inhibit hypertrophy, but at the expense of chondrogenesis. Stem Cell Res Ther. 2020;11(1):321. doi: 10.1186/s13287-020-01820-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao W, Zou T, Cui H, et al. Parathyroid hormone (1–34) promotes the effects of 3D printed scaffold-seeded bone marrow mesenchymal stem cells on meniscus regeneration. Stem Cell Res Ther. 2020;11(1):328. doi: 10.1186/s13287-020-01845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan Y, Hanai J-I, Le PT, et al. Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell Metab. 2017;25(3):661–672. doi: 10.1016/j.cmet.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balsalobre A, Brown SA, Marcacci L, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science (New York, NY) 2000;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 58.Nakamura Y, Harama D, Shimokawa N, et al. Circadian clock gene Period2 regulates a time-of-day-dependent variation in cutaneous anaphylactic reaction. J Allergy Clin Immunol. 2011;127(4):1038–1045. doi: 10.1016/j.jaci.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Abe T, Sato T, Yoda T, et al. The period circadian clock 2 gene responds to glucocorticoids and regulates osteogenic capacity. Regen Ther. 2019;11:199–206. doi: 10.1016/j.reth.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schouten M, Bielefeld P, Garcia-Corzo L, et al. Circadian glucocorticoid oscillations preserve a population of adult hippocampal neural stem cells in the aging brain. Mol Psychiatry. 2020;25(7):1382–1405. doi: 10.1038/s41380-019-0440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Komori T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem Cell Biol. 2018;149(4):313–323. doi: 10.1007/s00418-018-1640-6. [DOI] [PubMed] [Google Scholar]
- 62.Mundlos S, Otto F, Mundlos C, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89(5):773–779. doi: 10.1016/S0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 63.Reale ME, Webb IC, Wang X, et al. The transcription factor Runx2 is under circadian control in the suprachiasmatic nucleus and functions in the control of rhythmic behavior. PLoS ONE. 2013;8(1):e54317. doi: 10.1371/journal.pone.0054317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Athanassiou-Papaefthymiou M, Kim D, Harbron L, et al. Molecular and circadian controls of ameloblasts. Eur J Oral Sci. 2011;119(Suppl 1):35–40. doi: 10.1111/j.1600-0722.2011.00918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim K, Kim JH, Kim I, et al. Rev-erbα negatively regulates osteoclast and osteoblast differentiation through p38 MAPK Signaling Pathway. Mol Cells. 2020;43(1):34–47. doi: 10.14348/molcells.2019.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Z, Xu T, Li Y, et al. Inhibition of CRY2 by STAT3/miRNA-7-5p promotes osteoblast differentiation through upregulation of CLOCK/BMAL1/P300 expression. Mol Ther Nucleic Acids. 2020;19:865–876. doi: 10.1016/j.omtn.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim E-J, Kang I-H, Lee JW, et al. MiR-433 mediates ERRγ-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013;92(10):562–568. doi: 10.1016/j.lfs.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 68.Smith SS, Dole NS, Franceschetti T, et al. MicroRNA-433 dampens glucocorticoid receptor signaling, impacting circadian rhythm and osteoblastic gene expression. J Biol Chem. 2016;291(41):21717–21728. doi: 10.1074/jbc.M116.737890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lian JB, Stein JL, Stein GS, et al. Runx2/Cbfa1 functions: diverse regulation of gene transcription by chromatin remodeling and co-regulatory protein interactions. Connect Tissue Res. 2003;44(Suppl 1):141–148. doi: 10.1080/03008200390152232. [DOI] [PubMed] [Google Scholar]
- 70.Jensen ED, Schroeder TM, Bailey J, et al. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J Bone Miner Res Off J Am Soc Bone Miner Res. 2008;23(3):361–372. doi: 10.1359/jbmr.071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-X. [DOI] [PubMed] [Google Scholar]
- 72.Ren D, Collingwood TN, Rebar EJ, et al. PPARgamma knockdown by engineered transcription factors: exogenous PPARgamma2 but not PPARgamma1 reactivates adipogenesis. Genes Dev. 2002;16(1):27–32. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123(6):993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 74.Long H, Zhu Y, Lin Z, et al. miR-381 modulates human bone mesenchymal stromal cells (BMSCs) osteogenesis via suppressing Wnt signaling pathway during atrophic nonunion development. Cell Death Dis. 2019;10(7):470. doi: 10.1038/s41419-019-1693-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Fan J, Li J, Fan Q. Naringin promotes differentiation of bone marrow stem cells into osteoblasts by upregulating the expression levels of microRNA-20a and downregulating the expression levels of PPARγ. Mol Med Rep. 2015;12(3):4759–4765. doi: 10.3892/mmr.2015.3996. [DOI] [PubMed] [Google Scholar]
- 76.Riche DM, King ST. Bone loss and fracture risk associated with thiazolidinedione therapy. Pharmacotherapy. 2010;30(7):716–727. doi: 10.1592/phco.30.7.716. [DOI] [PubMed] [Google Scholar]
- 77.Wang N, Yang G, Jia Z, et al. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8(6):482–491. doi: 10.1016/j.cmet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 79.González-González A, García Nieto E, González A, et al. Melatonin modulation of radiation and chemotherapeutics-induced changes on differentiation of breast fibroblasts. Int J Mol Sci. 2019;20(16):3935. doi: 10.3390/ijms20163935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shabani A, Foroozanfard F, Kavossian E, et al. Effects of melatonin administration on mental health parameters, metabolic and genetic profiles in women with polycystic ovary syndrome: a randomized, double-blind, placebo-controlled trial. J Affect Disord. 2019;250:51–56. doi: 10.1016/j.jad.2019.02.066. [DOI] [PubMed] [Google Scholar]
- 81.Fontaine C, Dubois G, Duguay Y, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278(39):37672–37680. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 82.Liu C, Li S, Liu T, et al. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447(7143):477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 83.Le PT, Bornstein SA, Motyl KJ, et al. A novel mouse model overexpressing Nocturnin results in decreased fat mass in male mice. J Cell Physiol. 2019;234(11):20228–20239. doi: 10.1002/jcp.28623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chaturvedi P, Tyagi SC. Epigenetic mechanisms underlying cardiac degeneration and regeneration. Int J Cardiol. 2014;173(1):1–11. doi: 10.1016/j.ijcard.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kojima S, Gatfield D, Esau CC, et al. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS ONE. 2010;5(6):e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Estrella MA, Du J, Chen L, et al. The metabolites NADP and NADPH are the targets of the circadian protein Nocturnin (Curled) Nat Commun. 2019;10(1):2367. doi: 10.1038/s41467-019-10125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grimaldi B, Bellet MM, Katada S, et al. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12(5):509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adamovich Y, Rousso-Noori L, Zwighaft Z, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19(2):319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bae K, Jin X, Maywood ES, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–536. doi: 10.1016/S0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 90.Shearman LP, Jin X, Lee C, et al. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol. 2000;20(17):6269–6275. doi: 10.1128/MCB.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Costa MJ, So AYL, Kaasik K, et al. Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J Biol Chem. 2011;286(11):9063–9070. doi: 10.1074/jbc.M110.164558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aggarwal A, Costa MJ, Rivero-Gutiérrez B, et al. The circadian clock regulates adipogenesis by a Per3 crosstalk pathway to Klf15. Cell Rep. 2017;21(9):2367–2375. doi: 10.1016/j.celrep.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang T, Wang Z, Yang P, et al. PER1 prevents excessive innate immune response during endotoxin-induced liver injury through regulation of macrophage recruitment in mice. Cell Death Dis. 2016;7:e2176. doi: 10.1038/cddis.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suzuki C, Ushijima K, Ando H, et al. Induction of by a histone deacetylase inhibitor is involved in amelioration of insulin sensitivity via adipocyte differentiation in ob/ob mice. Chronobiol Int. 2019;36(7):955–968. doi: 10.1080/07420528.2019.1602841. [DOI] [PubMed] [Google Scholar]
- 95.Ushijima K, Suzuki C, Kitamura H, et al. Expression of clock gene in omental and mesenteric adipose tissue in patients with type 2 diabetes. BMJ Open Diabetes Res Care. 2020;8(1):e001465. doi: 10.1136/bmjdrc-2020-001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alagha MA, Vágó J, Katona É, et al. A synchronized circadian clock enhances early chondrogenesis. Cartilage. 2021;13(2_suppl):53S–67S. doi: 10.1177/1947603520903425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takarada T, Kodama A, Hotta S, et al. Clock genes influence gene expression in growth plate and endochondral ossification in mice. J Biol Chem. 2012;287(43):36081–36095. doi: 10.1074/jbc.M112.408963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu S, Tang Q, Xie M, et al. Circadian BMAL1 regulates mandibular condyle development by hedgehog pathway. Cell Prolif. 2020;53(1):e12727. doi: 10.1111/cpr.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cha S, Lee S-M, Wang J, et al. Enhanced circadian clock in MSCs-based cytotherapy ameliorates age-related temporomandibular joint condyle degeneration. Int J Mol Sci. 2021;22(19):10632. doi: 10.3390/ijms221910632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bi W, Deng JM, Zhang Z, et al. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 101.Hardingham TE, Oldershaw RA, Tew SR. Cartilage, SOX9 and Notch signals in chondrogenesis. J Anat. 2006;209(4):469–480. doi: 10.1111/j.1469-7580.2006.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dudek M, Gossan N, Yang N, et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J Clin Investig. 2016;126(1):365–376. doi: 10.1172/JCI82755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Le NQ, Binh NT, Takarada T, et al. Negative correlation between Per1 and Sox6 expression during chondrogenic differentiation in pre-chondrocytic ATDC5 cells. J Pharmacol Sci. 2013;122(4):318–325. doi: 10.1254/jphs.13091FP. [DOI] [PubMed] [Google Scholar]
- 104.Lee IK, Song H, Kim H, et al. RORα regulates cholesterol metabolism of CD8 T cells for anticancer immunity. Cancers. 2020;12(7):1733. doi: 10.3390/cancers12071733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qu P, Wang L, Min Y, et al. Vav1 regulates mesenchymal stem cell differentiation decision between adipocyte and chondrocyte via sirt1. Stem Cells (Dayton, Ohio) 2016;34(7):1934–1946. doi: 10.1002/stem.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dudakovic A, Samsonraj RM, Paradise CR, et al. Inhibition of the epigenetic suppressor EZH2 primes osteogenic differentiation mediated by BMP2. J Biol Chem. 2020;295(23):7877–7893. doi: 10.1074/jbc.RA119.011685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu F, Song D-Y, Huang J, et al. Long non-coding RNA CIR inhibits chondrogenic differentiation of mesenchymal stem cells by epigenetically suppressing ATOH8 via methyltransferase EZH2. Mol Med. 2021;27(1):12. doi: 10.1186/s10020-021-00272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nomura Y, Hara ES, Yoshioka Y, et al. DNA methylation-based regulation of human bone marrow-derived mesenchymal stem/progenitor cell chondrogenic differentiation. Cells Tissues Organs. 2019;207(3–4):115–126. doi: 10.1159/000502885. [DOI] [PubMed] [Google Scholar]
- 109.Koike N, Yoo S-H, Huang H-C, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science (New York, NY) 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hurley JM, Loros JJ, Dunlap JC. Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem Sci. 2016;41(10):834–846. doi: 10.1016/j.tibs.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Browne CJ, Godino A, Salery M, et al. Epigenetic mechanisms of opioid addiction. Biol Psychiatry. 2020;87(1):22–33. doi: 10.1016/j.biopsych.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Richards EJ, Elgin SCR. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108(4):489–500. doi: 10.1016/S0092-8674(02)00644-X. [DOI] [PubMed] [Google Scholar]
- 113.Fu G, Ren A, Qiu Y, et al. Epigenetic regulation of osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther. 2016;11(3):235–246. doi: 10.2174/1574888X10666150528153313. [DOI] [PubMed] [Google Scholar]
- 114.Brown SA. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development (Cambridge, England) 2014;141(16):3105–3111. doi: 10.1242/dev.104851. [DOI] [PubMed] [Google Scholar]
- 115.Orozco-Solis R, Aguilar-Arnal L. Circadian regulation of immunity through epigenetic mechanisms. Front Cell Infect Microbiol. 2020;10:96. doi: 10.3389/fcimb.2020.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 117.Xu H, Gustafson CL, Sammons PJ, et al. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol. 2015;22(6):476–484. doi: 10.1038/nsmb.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duong HA, Robles MS, Knutti D, et al. A molecular mechanism for circadian clock negative feedback. Science (New York, NY) 2011;332(6036):1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 120.Vaissière A, Berger S, Harrus D, et al. Molecular mechanisms of transcriptional control by Rev-erbα: an energetic foundation for reconciling structure and binding with biological function. Protein Sci. 2015;24(7):1129–1146. doi: 10.1002/pro.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dernie F, Adeyoju D. A matter of time: circadian clocks in osteoarthritis and the potential of chronotherapy. Exp Gerontol. 2021;143:111163. doi: 10.1016/j.exger.2020.111163. [DOI] [PubMed] [Google Scholar]
- 122.Louvet L, Leterme D, Delplace S, et al. Sirtuin 1 deficiency decreases bone mass and increases bone marrow adiposity in a mouse model of chronic energy deficiency. Bone. 2020;136:115361. doi: 10.1016/j.bone.2020.115361. [DOI] [PubMed] [Google Scholar]
- 123.Shakibaei M, Shayan P, Busch F, et al. Resveratrol mediated modulation of Sirt-1/Runx2 promotes osteogenic differentiation of mesenchymal stem cells: potential role of Runx2 deacetylation. PLoS ONE. 2012;7(4):e35712. doi: 10.1371/journal.pone.0035712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Soni SK, Basu P, Singaravel M, et al. Sirtuins and the circadian clock interplay in cardioprotection: focus on sirtuin 1. Cell Mol Life Sci CMLS. 2021;78(6):2503–2515. doi: 10.1007/s00018-020-03713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 127.Ramsey KM, Yoshino J, Brace CS, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science (New York, NY) 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Artsi H, Cohen-Kfir E, Gurt I, et al. The Sirtuin1 activator SRT3025 down-regulates sclerostin and rescues ovariectomy-induced bone loss and biomechanical deterioration in female mice. Endocrinology. 2014;155(9):3508–3515. doi: 10.1210/en.2014-1334. [DOI] [PubMed] [Google Scholar]
- 129.Simic P, Zainabadi K, Bell E, et al. SIRT1 regulates differentiation of mesenchymal stem cells by deacetylating β-catenin. EMBO Mol Med. 2013;5(3):430–440. doi: 10.1002/emmm.201201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zainabadi K, Liu CJ, Guarente L. SIRT1 is a positive regulator of the master osteoblast transcription factor, RUNX2. PLoS ONE. 2017;12(5):e0178520. doi: 10.1371/journal.pone.0178520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bäckesjö C-M, Li Y, Lindgren U, et al. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. Cells Tissues Organs. 2009;189(1–4):93–97. doi: 10.1159/000151744. [DOI] [PubMed] [Google Scholar]
- 132.Zhou Y, Zhou Z, Zhang W, et al. SIRT1 inhibits adipogenesis and promotes myogenic differentiation in C3H10T1/2 pluripotent cells by regulating Wnt signaling. Cell Biosci. 2015;5:61. doi: 10.1186/s13578-015-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhong Y, Ye Q, Chen C, et al. Ezh2 promotes clock function and hematopoiesis independent of histone methyltransferase activity in zebrafish. Nucleic Acids Res. 2018;46(7):3382–3399. doi: 10.1093/nar/gky101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang R, Chen J, Zhang J, et al. 1,25-Dihydroxyvitamin D protects against age-related osteoporosis by a novel VDR-Ezh2-p16 signal axis. Aging Cell. 2020;19(2):e13095. doi: 10.1111/acel.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yu Y, Deng P, Yu B, et al. Inhibition of EZH2 promotes human embryonic stem cell differentiation into mesoderm by reducing H3K27me3. Stem Cell Rep. 2017;9(3):752–761. doi: 10.1016/j.stemcr.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lim HW, Iwatani M, Hattori N, et al. Resistance to 5-aza-2'-deoxycytidine in genic regions compared to non-genic repetitive sequences. J Reprod Dev. 2010;56(1):86–93. doi: 10.1262/jrd.20247. [DOI] [PubMed] [Google Scholar]
- 137.Cao Y, Yang H, Jin L, et al. Genome-wide DNA methylation analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cells Int. 2018;2018:8238496. doi: 10.1155/2018/8238496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 139.Maekawa F, Shimba S, Takumi S, et al. Diurnal expression of Dnmt3b mRNA in mouse liver is regulated by feeding and hepatic clockwork. Epigenetics. 2012;7(9):1046–1056. doi: 10.4161/epi.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Agbaria S, Haim A, Fares F, et al. Epigenetic modification in 4T1 mouse breast cancer model by artificial light at night and melatonin—the role of DNA-methyltransferase. Chronobiol Int. 2019;36(5):629–643. doi: 10.1080/07420528.2019.1574265. [DOI] [PubMed] [Google Scholar]
- 141.Feng Y-Z, Yu Y, Zhou Y-F, et al. A natural variant of miR397 mediates a feedback loop in circadian rhythm. Plant Physiol. 2020;182(1):204–214. doi: 10.1104/pp.19.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mehta N, Cheng HYM. Micro-managing the circadian clock: the role of microRNAs in biological timekeeping. J Mol Biol. 2013;425(19):3609–3624. doi: 10.1016/j.jmb.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 143.Torres M, Becquet D, Franc J-L, et al. Circadian processes in the RNA life cycle. Wiley Interdiscip Rev RNA. 2018;9(3):e1467. doi: 10.1002/wrna.1467. [DOI] [PubMed] [Google Scholar]
- 144.Assmann TS, Cuevas-Sierra A, Salas-Pérez F, et al. Crosstalk between circulating microRNAs and chronotypical features in subjects with metabolic syndrome. Chronobiol Int. 2020;37(7):1048–1058. doi: 10.1080/07420528.2020.1782419. [DOI] [PubMed] [Google Scholar]
- 145.Heegaard NHH, Carlsen AL, Lilje B, et al. Diurnal variations of human circulating cell-free micro-RNA. PLoS ONE. 2016;11(8):e0160577. doi: 10.1371/journal.pone.0160577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y, Lv K, Zhao M, et al. Analysis of miRNA expression profiles in the liver of mutant mice. PeerJ. 2019;7:e8119. doi: 10.7717/peerj.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang H, Fan Z, Zhao M, et al. Oscillating primary transcripts harbor miRNAs with circadian functions. Sci Rep. 2016;6:21598. doi: 10.1038/srep21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yan Y, Salazar TE, Dominguez JM, et al. Dicer expression exhibits a tissue-specific diurnal pattern that is lost during aging and in diabetes. PLoS ONE. 2013;8(11):e80029. doi: 10.1371/journal.pone.0080029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hackl M, Heilmeier U, Weilner S, et al. Circulating microRNAs as novel biomarkers for bone diseases—complex signatures for multifactorial diseases? Mol Cell Endocrinol. 2016;432:83–95. doi: 10.1016/j.mce.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 150.Na YJ, Sung JH, Lee SC, et al. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp Mol Med. 2009;41(9):638–647. doi: 10.3858/emm.2009.41.9.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Figueredo DDS, Barbosa MR, Gitaí DLG, et al. Predicted microRNAs for mammalian circadian rhythms. J Biol Rhythms. 2013;28(2):107–116. doi: 10.1177/0748730413476827. [DOI] [PubMed] [Google Scholar]
- 152.Zhang R, Lahens NF, Ballance HI, et al. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fan Z, Zhao M, Joshi PD, et al. A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res. 2017;45(10):5720–5738. doi: 10.1093/nar/gkx156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun H, Peng G, Wu H, et al. Long non-coding RNA MEG3 is involved in osteogenic differentiation and bone diseases (Review) Biomed Rep. 2020;13(1):15–21. doi: 10.3892/br.2020.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu J, Xiao M, Ren G. Long non-coding RNA XIST promotes osteoporosis by inhibiting the differentiation of bone marrow mesenchymal stem cell by sponging miR-29b-3p that suppresses nicotinamide N-methyltransferase. Bioengineered. 2021;12(1):6057–6069. doi: 10.1080/21655979.2021.1967711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu B, Gates LA, Stashi E, et al. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol Cell. 2015;60(5):769–783. doi: 10.1016/j.molcel.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Du W, Guo D, Du W. ATP-dependent chromatin remodeling complex in the lineage specification of mesenchymal stem cells. Stem Cells Int. 2020;2020:8839703. doi: 10.1155/2020/8839703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Deshayes N, Genty G, Berthelot F, et al. Human long-term deregulated circadian rhythm alters regenerative properties of skin and hair precursor cells. Eur J Dermatol EJD. 2018;28(4):467–475. doi: 10.1684/ejd.2018.3358. [DOI] [PubMed] [Google Scholar]
- 160.Wu Q-J, Sun H, Wen Z-Y, et al. Shift work and health outcomes: an umbrella review of systematic reviews and meta-analyses of epidemiological studies. J Clin Sleep Med JCSM Off Publ Am Acad Sleep Med. 2022;18(2):653–662. doi: 10.5664/jcsm.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.An K, Zhao H, Miao Y, et al. A circadian rhythm-gated subcortical pathway for nighttime-light-induced depressive-like behaviors in mice. Nat Neurosci. 2020;23(7):869–880. doi: 10.1038/s41593-020-0640-8. [DOI] [PubMed] [Google Scholar]
- 162.Janich P, Toufighi K, Solanas G, et al. Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell. 2013;13(6):745–753. doi: 10.1016/j.stem.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 163.Ohdo S. Chrono-drug discovery and development based on circadian rhythm of molecular, cellular and organ level. Biol Pharm Bull. 2021;44(6):747–761. doi: 10.1248/bpb.b21-00277. [DOI] [PubMed] [Google Scholar]
- 164.Parasram K, Karpowicz P. Time after time: circadian clock regulation of intestinal stem cells. Cell Mol Life Sci CMLS. 2020;77(7):1267–1288. doi: 10.1007/s00018-019-03323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mishra HK, Ying NM, Luis A, et al. Circadian rhythms in bipolar disorder patient-derived neurons predict lithium response: preliminary studies. Mol Psychiatry. 2021;26(7):3383–3394. doi: 10.1038/s41380-021-01048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Murphy BA, Wagner AL, McGlynn OF, et al. Exercise influences circadian gene expression in equine skeletal muscle. Vet J. 2014;201(1):39–45. doi: 10.1016/j.tvjl.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 167.Hatori M, Vollmers C, Zarrinpar A, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Noyan H, El-Mounayri O, Isserlin R, et al. Cardioprotective signature of short-term caloric restriction. PLoS ONE. 2015;10(6):e0130658. doi: 10.1371/journal.pone.0130658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang M, Yu D, Zheng L, et al. Mechanical stress affects circadian rhythm in skeletal muscle (C2C12 Myoblasts) by reducing Per/Cry gene expression and increasing Bmal1 gene expression. Med Sci Monit Int Med J Exp Clin Res. 2021;27:e928359. doi: 10.12659/MSM.928359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hassan N, McCarville K, Morinaga K, et al. Titanium biomaterials with complex surfaces induced aberrant peripheral circadian rhythms in bone marrow mesenchymal stromal cells. PLoS ONE. 2017;12(8):e0183359. doi: 10.1371/journal.pone.0183359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Reitz CJ, Alibhai FJ, Khatua TN, et al. SR9009 administered for one day after myocardial ischemia-reperfusion prevents heart failure in mice by targeting the cardiac inflammasome. Commun Biol. 2019;2:353. doi: 10.1038/s42003-019-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang L, Zhang R, Tien C-L, et al. REV-ERBα ameliorates heart failure through transcription repression. JCI Insight. 2017;2(17):e95177. doi: 10.1172/jci.insight.95177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shimozaki K. REV-ERB agonist SR9009 regulates the proliferation and neurite outgrowth/suppression of cultured rat adult hippocampal neural stem/progenitor cells in a concentration-dependent manner. Cell Mol Neurobiol. 2021. [DOI] [PMC free article] [PubMed]
- 174.Sahar S, Sassone-Corsi P. The epigenetic language of circadian clocks. Handb Exp Pharmacol. 2013;217:29–44. doi: 10.1007/978-3-642-25950-0_2. [DOI] [PubMed] [Google Scholar]
- 175.Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585(10):1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 176.Kowalska E, Ripperger JA, Muheim C, et al. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol Cell Biol. 2012;32(22):4585–4594. doi: 10.1128/MCB.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Predicted clock-targeted miRNAs in mice and human which also working in the MSC differentiation
Data Availability Statement
All data generated or analysed during this study are included in this published article.