Abstract
Objective
The effect of cardiac arrest (CA) on cerebral transcriptomics and metabolomics is unknown. We previously demonstrated hemodynamic-directed CPR (HD-CPR) improves survival with favorable neurologic outcomes versus standard CPR (Std-CPR). We hypothesized HD-CPR would preserve the cerebral transcriptome and metabolome compared to Std-CPR.
Design
Randomized pre-clinical animal trial.
Setting
Large animal resuscitation laboratory at an academic children’s hospital.
Subjects
Four-week-old female piglets (8–11 kg).
Interventions
Pigs (1-month-old), three groups: 1) HD-CPR (compression depth to systolic BP 90 mmHg, vasopressors to coronary perfusion pressure 20 mmHg); 2) Std-CPR and 3) shams (no CPR). HD-CPR and Std-CPR underwent asphyxia, induced ventricular fibrillation, 10–20 min of CPR and post-resuscitation care. Primary outcomes at 24 h in cerebral cortex: 1) transcriptomic analysis (n = 4 per treatment arm, n = 8 sham) of 1727 genes using differential gene expression and 2) metabolomic analysis (n = 5 per group) of 27 metabolites using one-way ANOVA, post-hoc Tukey HSD.
Measurements and main results
65 genes were differentially expressed between HD-CPR and Std-CPR and 72 genes between Std-CPR and sham, but only five differed between HD-CPR and sham. Std-CPR increased the concentration of five AA compared to HD-CPR and sham, including the branched chain amino acids (BCAA), but zero metabolites differed between HD-CPR and sham.
Conclusions
In cerebral cortex 24 h post CA, Std-CPR resulted in a different transcriptome and metabolome compared with either HD-CPR or sham. HD-CPR preserves the transcriptome and metabolome, and is neuroprotective. Global molecular analyses may be a novel method to assess efficacy of clinical interventions and identify therapeutic targets.
Institutional protocol number
IAC 16-001023.
Keywords: Cardiac arrest, Cardiopulmonary resuscitation, Metabolomics, Transcriptome, RNA sequence
Introduction:
Pediatric in-hospital cardiac arrests (CA) affect thousands of children each year, and permanent brain injury is common amongst those that survive to hospital discharge.1., 2. Very little is known about alterations in RNA or metabolite profiles in cerebral cortex after CA. Describing the effect of distinct cardiopulmonary resuscitation (CPR) strategies on RNA and metabolite profiles may serve as an additional way of evaluating CPR efficacy and uncover novel pathways for future study.
Our group has previously shown that in a pediatric porcine model of asphyxia-associated CA, a hemodynamic-directed CPR strategy (HD-CPR) resulted in improved cerebral mitochondrial bioenergetics and greater 24-hour survival with favorable neurologic outcome compared to a guideline-based CPR strategy (Std-CPR).3., 4., 5., 6. Among survivors, we have also previously shown improved intra-arrest hemodynamics and neurologic scoring 24 h post ROSC after HD-CPR compared to Std-CPR.7 To understand the effect of HD-CPR and Std-CPR on transcriptomics and metabolomics in the immature brain, we performed a quantitative RNA and metabolite analysis of cerebral cortex 24 hours after return of spontaneous circulation (ROSC) following asphyxia-associated ventricular fibrillation in a pediatric porcine model of CA with these two distinct CPR strategies. We hypothesized that among survivors of CA, HD-CPR would better preserve the cerebral cortical transcriptome and metabolome than Std-CPR when compared to sham animals that did not undergo CPR.
Materials and methods
Experimental protocol
Cohort description
These experiments were performed as a component of a previously reported pre-clinical trial in which animals were randomized to Std-CPR versus HD-CPR and complied with ARRIVE guidelines.4 In that study, 7 of 10 animals treated with HD-CPR and 5 of 12 animals treated with Std-CPR survived to 24 hours. Available specimens were used from these survivors without knowledge of pre-arrest and intra-arrest characteristics or neurologic outcomes.7 Tissue samples available and sufficient for analysis were chosen, which yielded five samples in each treatment arm for metabolomic analysis and four samples in each treatment arm for transcriptomic analysis. Additional sham animals were available from concurrent studies with identical sham groups. Investigators performing the downstream analyses remained blinded to the characteristics and neurologic outcomes of the animals.
Experimental protocol
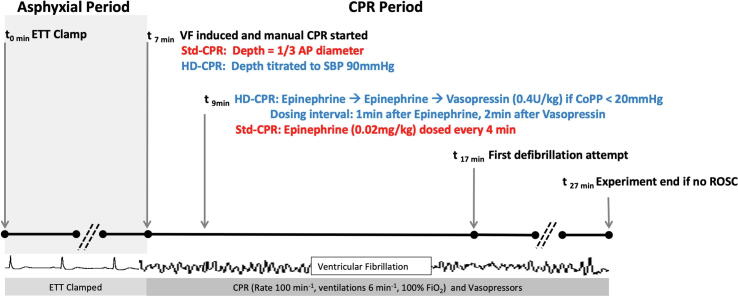
All animal procedures were approved by the Institutional Animal Care and Use Committee at the Children’s Hospital of Philadelphia (CHOP) and were conducted in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals. Detailed description can be found in prior publications.4., 5., 8. Briefly, one-month-old female swine, Sus scrofa domesticus, were anesthetized with isoflurane and mechanically ventilated and underwent seven minutes of asphyxia by endotracheal tube clamping during which fentanyl was administered. Ventricular fibrillation (VF) was induced to ensure a standard 10-minute period of CPR that would only be terminated by defibrillation (Fig. 1). Animals were randomized to receive HD-CPR or Std-CPR. In HD-CPR, force of chest compressions was adjusted to target systolic blood pressure of 90 mm Hg and vasopressors were titrated to maintain coronary perfusion pressure (CoPP) greater than 20 mm Hg using epinephrine (0.02 mg/kg) and vasopressin (0.4 U/kg) as previously described. In Std-CPR, target depth was one-third of the anterior-posterior chest diameter based on real-time feedback from a quality monitoring defibrillator. Epinephrine (0.02 mg/kg) was delivered at two minutes of CPR and every four minutes thereafter. Ten minutes after the start of CPR, defibrillation was attempted every two minutes as necessary in both treatment arms. If ROSC was attained, animals received standardized post-arrest intensive care. Sham animals underwent similar anesthetic duration to CA animals, but did not undergo CA or CPR. Following emergence, animals were extubated and monitored for 24 hours. At 24 hours post-CA, animals were re-anesthetized, a craniectomy was performed, and cortical tissue was rapidly extracted and snap frozen in liquid nitrogen and then stored at −80 degrees Celsius. Animals were humanely euthanized while under general anesthesia. Frontoparietal cortex was used for downstream outcome metrics by members of the scientific team blinded to treatment status.
Fig. 1.
Schematic of Experimental Protocol. Definitions of abbreviations: CPR cardiopulmonary resuscitation; ETT = endotracheal tube; VF = ventricular fibrillation; Std = Standard, CPR = cardiopulmonary resuscitation; HD = hemodynamic-directed; SBP = systolic blood pressure; AP = anterior–posterior; CoPP = coronary perfusion pressure; ROSC = return of spontaneous circulation.
RNA sequencing
RNA sequencing was performed for 16 animals (HD-CPR n = 4, Std-CPR n = 4, sham n = 8). RNA was extracted using a QIAsymphony® (Qiagen, Hilden, Germany) automated RNA extraction robot and reverse transcribed using a high-capacity cDNA reverse transcription kit with RNase inhibitor (Cat. 4387406 Life Technologies, NY. Ribozero rRNA Removal Kit, Human/Mouse/Rat probe) (Illumina, San Diego, CA, USA) was used for rRNA depletion, followed by library preparation using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA). Sequencing was performed using an Illumina HiSeq instrument for sequencing in 2 × 150 paired end configuration.
Reads were aligned to the SusScrofa11.1 reference genome using STAR.9 We used featureCounts from the subreads package to quantify transcripts at the gene level.10 DESeq2 v1.26.0 and R package v3.6.3 were used for downstream analyses.11 A false discovery rate (FDR) of 0.05 and 2-fold change was used to identify differentially expressed genes.
Metabolite analysis
Metabolomic analysis was performed for 15 animals (HD-CPR n = 5, Std-CPR n = 5, sham n = 5). Analysis prioritized pathways that feed into the TCA cycle and may impact cerebral cortex bioenergetics, as oxidative phosphorylation dysfunction after CA in the immature brain may contribute to post-ROSC neurologic injury.3., 4. Amino acid concentrations were determined with an Agilent 1260 Infinity HPLC system, utilizing pre-column derivatization with O-phthalaldehyde.12., 13. Lactate levels were determined enzymatically.12., 13. Organic acid concentrations were determined by isotope dilution approach using GC-MS system.14 Briefly, an aliquot of the brain sample (100 ul) was spiked with a mixture of 13C-labeled organic acids of known concentrations. Then, GC-MS measurement of 13C isotopic abundance in each sample was performed and concentrations of organic acids in the sample were calculated. Protein level was determined by Coomassie blue (ThermoFisher Scientific).12., 13.
Processing and analysis of metabolite data was done using the MetaboAnalystR (PMID: 29955821) package in R.15 Metabolite data was normalized using median norm row-normalization, log-norm transformation, and Pareto scaling, which yielded a probability distribution closest to Gaussian. Individual metabolite effect was determined by one-way ANOVA corrected by FDR with post-hoc Tukey honestly significant difference testing. Treatment arms were compared using Pearson correlation of sample medians to measure similarity between groups.16
Porcine cerebral performance category
Porcine Cerebral Performance Category (CPC) was independently determined 24 h after ROSC by two blinded, trained investigators. The scoring system is as follows: 1 is normal (no difficulty standing, walking, etc.); 2 is mild disability (can stand, but unsteady, slow to respond, drinking, not eating); 3 is severe disability (awake but not responding, cannot stand, walk, eat, drink); 4 is comatose; and 5 is dead.17., 18.
Data analysis
A priori planned analyses were to compare metabolome and transcriptome data among the 2 treatment groups (HD-CPR versus Std-CPR) and shams to address potential benefits of HD-CPR on outcomes and the potential mechanism of the outcome benefits in translational models before extending to clinical trials.
Results
RNA sequencing analysis
RNA expression in cerebral cortex at 24 h post-CA demonstrated little separation between HD-CPR and sham treatment arms (Fig. 2a). Of 1727 genes analyzed, only five were differentially expressed between HD-CPR and sham animals, while 65 genes differed between the Std-CPR and sham groups and 72 between Std-CPR and HD-CPR (Fig. 2b). Std-CPR increased expression of multiple genes related to immune function when compared to either of the other two groups, including adhesion molecules (ICAM1), chemokines and cytokines (CCL2, PLEK, TNFSF18, CCL3L1), genes critical to neutrophil function (CSF2RA, NCF2), regulatory molecules important in preventing autoimmunity (CD274) and transcription factors integral to innate immunity (ELF4) (Fig. 2c, 2d). HD-CPR increased expression of ASPA, which hydrolyzes N-acetylaspartate to maintain white matter compared to both Std-CPR and sham (Fig. 2e).
Fig. 2.
Transcriptome of HD-CPR following asphyxia CA global RNA expression is similar to sham animals that did not undergo cardiac arrest, while Std-CPR animals exhibited distinct RNA expression. A. PCA loading plot for the first two principal components of the RNA expression indicate HD-CPR and sham animals are similar to one another, but distinct from Std-CPR animals. B. Venn Diagram illustrating RNA transcripts differentially expressed between treatment arms. Of those analyzed, there are five genes differentially expressed between HD-CPR and sham, 72 between Std-CPR and sham, and 65 between Std-CPR and HD-CPR. Three of the five genes differentially expressed between HD-CPR and sham were also different between Std-CPR and HD-CPR. Similarly, 14 genes differentially expressed between Std-CPR and HD-CPR were also different between Std-CPR and sham. C. Volcano plot of RNA transcripts between Std-CPR and HD-CPR. Red values indicate transcripts with increased expression in Std-CPR versus HD-CPR. Blue values indicate decreased expression in Std-CPR versus HD-CPR. D. Volcano plot of RNA transcripts between Std-CPR and sham. Red values indicate transcripts with increased expression in Std-CPR versus sham. Blue values indicate decreased expression in Std-CPR versus sham. E. Volcano plot of RNA transcripts between HD-CPR and sham. Red values indicate transcripts with increased expression in HD-CPR versus sham. Blue values indicate decreased expression in HD-CPR versus sham.
Metabolite analysis
Metabolite quantities in cerebral cortex at 24 h post-CA demonstrated little separation between HD-CPR and sham treatment arms (Fig. 3a). Of 27 metabolites studied, there were no differences between HD-CPR and sham animals. Std-CPR increased expression of 5 amino acids compared to HD-CPR and sham, including branch chain amino acids (BCAA) isoleucine (p = 0.005), leucine (p = 0.001), valine (p = 0.008) as well as glycine (p = 0.005) and phenylalanine (p = 0.003) (Fig. 3b, 3c). Overall metabolite expression in HD-CPR was more similar to sham (r = 0.12, p = 0.56) than either HD-CPR compared to Std-CPR (r = -0.51, p = 0.006) or between Std-CPR and sham (r = -0.47, p = 0.014). After normalization, positive Pearson correlation indicates similarity and negative correlation indicates differences between groups.
Fig. 3.
Cerebral cortex metabolite profiles of animals receiving HD-CPR are similar to sham animals that did not undergo cardiac arrest, while Std-CPR animals exhibited a distinct metabolite profile. A. PCA loadings plot for the first two principal components for all metabolite expression indicate HD-CPR and sham are similar to one another, but distinct from Std-CPR animals. B. Assembly of major metabolic pathways in the brain with differentially expressed metabolites shaded. Branched chain amino acid metabolic pathway is altered in Std-CPR animals. C. Std-CPR animals exhibited an increase in all branched chain amino acids compared to both HD-CPR and sham animals in cerebral cortex at 24 h after CA.
Porcine cerebral performance category
As previously described, at 24 h post ROSC all HD-CPR survivors with metabolome or transcriptome data attained CPC 1 (neurologically normal), whereas 4/5 Std-CPR survivors had CPC 3 (severe disability) and 1/5 had CPC 1.7
Discussion
Our group has previously shown that in a pediatric asphyxia-associated cardiac arrest swine model, HD-CPR leads to improved cerebral mitochondrial respiration and higher rates of 24 h survival with favorable neurologic outcome compared to Std-CPR.3., 4. Among survivors, we have also previously demonstrated improved intra-arrest hemodynamics and neurologic scoring 24 h post ROSC after HD-CPR compared to Std-CPR.7 We hypothesized that HD-CPR animals better preserve the cerebral transcriptome and metabolome than Std-CPR in comparison to sham animals that did not undergo CPR. We now show that 24 hours after CA, cerebral cortex from animals that received HD-CPR had a similar transcriptome and metabolome compared to sham animals that did not undergo CA, and both groups were distinct from animals that received Std-CPR. These results, in concert with already established clinical and mitochondrial benefits, establish the benefit of HD-CPR on global molecular changes in the developing brain at 24 hours after CA. Our findings are the first to document alterations in cerebral transcriptomics and metabolomics after distinct cardiopulmonary resuscitation strategies in a highly relevant, pediatric large animal model.
There is no published data on the effect of CA on the transcriptome. Std-CPR results in differential RNA expression of numerous transcripts, many of which are pro-inflammatory compared to HD-CPR and shams. Neuroinflammation mediated by microglial activation plays a pivotal role in post CA brain injury, and immunomodulation has successively been neuroprotective in rodent studies.19., 20. Trials of immunomodulation in large animals are needed to identify the link between neuroinflammation and poorer neurologic outcomes after CA treated with Std-CPR.4
We describe BCAA increase after Std-CPR compared to both HD-CPR and sham animals. BCAA are important for protein and neurotransmitter synthesis, though excess is neurotoxic.21 In adult rats undergoing CA, numerous cerebral amino acids, including BCAAs, are elevated after 30 minutes of reperfusion.22 Following Std-CPR, BCAA elevation may be a direct mediator of neurologic injury or a neuroprotective response to severe injury. Our findings warrant further mechanistic study of cerebral BCAA after CA associated brain injury.
This model is limited in that the RNA and metabolite profiles are only available in survivors at 24 hours after CA. The transcriptome and metabolome may have an evolving time-course, which should be investigated. This study does not evaluate proteomics, which would bridge the gap between the transcriptome and metabolome. Additionally, due to the small number of genes studied, it was not possible to conduct a gene-ontology enrichment analysis.
In the cerebral cortex 24 h after CA, Std-CPR resulted in a different transcriptome and metabolome compared with both HD-CPR and shams. Notably, HD-CPR and sham animals were not substantially different from each other, suggesting that HD-CPR better preserved the native transcriptome and metabolome and appears to be a potent neuroprotective intervention. The effect of CA on immune function and the role of BCAA following CA are promising future directions. Global molecular analyses may be a novel method to assess efficacy of clinical interventions and identify potential therapeutic targets.
Reprints ordered: No.
Financial support for this study: Children’s Hospital of Philadelphia Department of Anesthesiology and Critical Care Medicine. Mitochondrial RAG Institutional Core Grant 26026‐7260260617-03 (Dr Senthil), NIH National Heart, Lung and Blood Institute K23HL148541 (Dr Morgan), NIH National Heart, Lung and Blood Institute R01HL141386 (Dr Kilbaugh).
CRediT authorship contribution statement
Kumaran Senthil: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Visualization, Funding acquisition. Marco M. Hefti: Formal analysis, Writing – original draft, Data curation, Visualization. Larry N. Singh: Formal analysis, Writing – original draft, Data curation, Visualization. Ryan W. Morgan: Conceptualization, Methodology, Formal analysis, Investigation, Writing – review & editing, Supervision, Project administration. Constantine D. Mavroudis: Methodology, Investigation, Writing – review & editing. Tiffany Ko: Conceptualization, Methodology, Investigation, Writing – review & editing. Hunter Gaudio: Visualization. Vinay M. Nadkarni: Conceptualization, Methodology, Investigation, Resources, Writing – review & editing, Supervision. Johannes Ehinger: Conceptualization, Methodology, Writing – review & editing. Robert A. Berg: Conceptualization, Methodology, Investigation, Resources, Writing – review & editing, Supervision, Project administration. Robert M. Sutton: Conceptualization, Methodology, Investigation, Resources, Writing – review & editing, Supervision, Project administration. Francis X. McGowan: Conceptualization, Methodology, Supervision. Todd J. Kilbaugh: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgments
Acknowledgments
We thank E. Daikhin, O. Horyn, Itzhak Nissim and Ilana Nissim for performing the analysis of metabolites in the Metabolomics Core Facility, The Children’s Hospital of Philadelphia.
We thank the Mitochondria Research Affinity Group through the Children’s Hospital of Philadelphia Research Institute for awarding our team the institutional core grant that supported this project.
References
- 1.Berg R.A., Sutton R.M., Holubkov R., et al. Ratio of PICU versus ward cardiopulmonary resuscitation events is increasing. Crit Care Med. 2013;41(10):2292–2297. doi: 10.1097/CCM.0b013e31828cf0c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg R.A., Nadkarni V.M., Clark A.E., et al. Incidence and outcomes of cardiopulmonary resuscitation in PICUs. Crit Care Med. 2016;44(4):798–808. doi: 10.1097/CCM.0000000000001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilbaugh TJ, Sutton RM, Karlsson M, et al. Persistently Altered Brain Mitochondrial Bioenergetics After Apparently Successful Resuscitation From Cardiac Arrest. JAmHeart Assoc. 2015;4(2047-9980 (Electronic)):1-12. doi: 10.1161/JAHA.115.002232. [DOI] [PMC free article] [PubMed]
- 4.Lautz A.J., Morgan R.W., Karlsson M., et al. Hemodynamic-directed cardiopulmonary resuscitation improves neurologic outcomes and mitochondrial function in the heart and brain. Crit Care Med. 2019;47(3):e241–e249. doi: 10.1097/CCM.0000000000003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan R.W., Kilbaugh T.J., Shoap W., et al. A hemodynamic-directed approach to pediatric cardiopulmonary resuscitation (HD-CPR) improves survival. Resuscitation. 2017;111:41–47. doi: 10.1016/j.resuscitation.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkins D.L., Berger S., Duff J.P., et al. Part 11: Pediatric basic life support and cardiopulmonary resuscitation quality: 2015 American Heart Association guidelines update for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation. 2015;132(18):S519–S525. doi: 10.1161/CIR.0000000000000265. [DOI] [PubMed] [Google Scholar]
- 7.Senthil K., Morgan R.W., Hefti M.M., et al. Haemodynamic-directed cardiopulmonary resuscitation promotes mitochondrial fusion and preservation of mitochondrial mass after successful resuscitation in a pediatric porcine model. Resusc Plus. 2020;2021(6) doi: 10.1016/j.resplu.2021.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton R.M., Friess S.H., Naim M.Y., et al. Patient-centric blood pressure-targeted cardiopulmonary resuscitation improves survival from cardiac arrest. Am J Respir Crit Care Med. 2014;190(11):1255–1262. doi: 10.1164/rccm.201407-1343OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobin A., Davis C.A., Schlesinger F., et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47(8) doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nissim I., Horyn O., Nissim I., et al. Effects of a glucokinase activator on hepatic intermediary metabolism: Study with 13C-isotopomer-based metabolomics. Biochem J. 2012;444(3):537–551. doi: 10.1042/BJ20120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissim I., Horyn O., Daikhin Y., et al. The molecular and metabolic influence of long term agmatine consumption. J Biol Chem. 2014;289(14):9710–9729. doi: 10.1074/jbc.M113.544726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberg J.M., Venkatachalam M.A., Roeser N.F., Nissim I. Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc Natl Acad Sci USA. 2000;97(6):2826–2831. doi: 10.1073/pnas.97.6.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. Published 2018. https://www.r-project.org.
- 16.Chen P.Y., Popovich P.M. Sage University; 2002. Correlation: Parametric and Nonparametric Measures. [Google Scholar]
- 17.Berg R.A., Chapman F.W., Berg M.D., et al. Attenuated adult biphasic shocks compared with weight-based monophasic shocks in a swine model of prolonged pediatric ventricular fibrillation. Resuscitation. 2004;61(2):189–197. doi: 10.1016/j.resuscitation.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 18.Berg R.A., Sanders A.B., Kern K.B., et al. Adverse hemodynamic effects of interrupting chest compressions for rescue breathing during cardiopulmonary resuscitation for ventricular fibrillation cardiac arrest. Circulation. 2001;104(20):2465–2470. doi: 10.1161/hc4501.098926. [DOI] [PubMed] [Google Scholar]
- 19.Ocak U., Ocak P.E., Huang L., et al. Inhibition of PAR-2 attenuates neuroinflammation and improves short-term neurocognitive functions via ERK1/2 signaling following asphyxia-induced cardiac arrest in rats. Shock. 2020;54(4):539–547. doi: 10.1097/SHK.0000000000001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y., Zhu J., Wang D., et al. NLRP3 inflammasome-mediated microglial pyroptosis is critically involved in the development of post-cardiac arrest brain injury. J Neuroinflammation. 2020;17(1):1–21. doi: 10.1186/s12974-020-01879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sperringer J.E., Addington A., Hutson S.M. Branched-chain amino acids and brain metabolism. Neurochem Res. 2017;42(6):1697–1709. doi: 10.1007/S11064-017-2261-5. [DOI] [PubMed] [Google Scholar]
- 22.Choi J., Shoaib M., Yin T., et al. Tissue-specific metabolic profiles after prolonged cardiac arrest reveal brain metabolome dysfunction predominantly after resuscitation. J Am Heart Assoc. 2019;8(17):1–14. doi: 10.1161/JAHA.119.012809. [DOI] [PMC free article] [PubMed] [Google Scholar]