Abstract
The pathogenesis of atopic dermatitis (AD) results from complex interactions between environmental factors, barrier defects, and immune dysregulation resulting in systemic inflammation. Therefore, we sought to characterize circulating inflammatory profiles in pediatric AD patients and identify potential signaling nodes which drive disease heterogeneity and progression. We analyzed a sample set of 87 infants that were at high risk for atopic disease based on AD diagnoses. Clinical parameters, serum, and peripheral blood mononuclear cells (PBMCs) were collected upon entry, and at 1 and 4 years later. Within patient serum, 126 unique analytes were measured using a combination of multiplex platforms and ultrasensitive immunoassays. We assessed the correlation of inflammatory analytes with AD severity (SCORAD). Key biomarkers, such as IL-13 (rmcorr = 0.47) and TARC/CCL17 (rmcorr = 0.37), among other inflammatory signals, significantly correlated with SCORAD across all timepoints in the study. Flow cytometry and pathway analysis of these analytes implies that CD4 T-cell involvement in type 2 immune responses was enhanced at the earliest time point (year 1) relative to the end of study collection (year 5). Importantly, forward selection modeling identified 18 analytes in infant serum at study entry which could be used to predict change in SCORAD 4 years later. We have identified a pediatric AD biomarker signature linked to disease severity which will have predictive value in determining AD persistence in youth and provide utility in defining core systemic inflammatory signals linked to pathogenesis of atopic disease.
Keywords: atopic dermatitis, IL-13, pediatric, SCORAD, TARC/CCL17
Introduction
Atopic dermatitis (AD) is a highly complex, heterogeneous, chronic inflammatory skin condition characterized by a pruritic, erythematous rash whose immune pathophysiology remains unclear. While AD has been shown to affect up to 25% of children and 7–10% of adults worldwide, incidence rates has been increasing 2- to 3-fold over the last several decades [1]. Although many pediatric cases are transient in nature, AD is often the first step of the atopic march and active disease throughout life with up to 75% of adults with AD reporting onset during childhood [2]. An early diagnosis of AD has been linked with the development of additional atopic comorbidities, such as allergic rhinitis, food allergy, and asthma [3]. This link may be the result of similar underlying mechanisms of disease development involving the circulating inflammatory immune milieu in AD patients since type 2 cytokines, barrier function proteins, and allergen-specific IgE responses have been implicated in both disease onset and persistence [4, 5].
The comorbid nature of AD, as well as its development in early childhood, makes it an attractive target for predictive studies of disease progression. In turn, this highlights the growing importance of characterizing these early stages of pathogenesis. The systemic nature of AD has only recently been explored within younger patients demonstrating the presence and persistence of cell populations that produce biomarkers like IL-22, IL-17a, and IFNγ, and strong correlations to cells capable of producing IL-13 were shown to increase from childhood to adulthood [6, 7]. Because AD is a complex disease involving multiple cell types, chemokines, and cytokines, further longitudinal studies identifying additional biomarkers and correlating them to disease manifestations are needed to truly characterize the complex underlying immune signals of AD pathogenesis that drive barrier disruption and atopy development. Therefore, using high-throughput and ultrasensitive protein quantitation methods, we evaluated 126 secreted proteins from the serum of 87 children diagnosed with AD in their first year of life and correlated levels of inflammatory proteins to disease progression over the course of this 5-year study.
Our findings provide further insight into the systemic inflammatory serum protein and cell population profile present in the circulation of pediatric AD patients. Previous studies utilizing this cohort of patients revealed connections between atopic and non-AD, food allergy, and asthma [8, 9]. Several biomarkers indicative of active type 2 immune responses were correlated with both progression of disease and cell phenotypes which have been linked to comorbidities associated with AD, though not directly connected, such as childhood wheezing [10]. These correlations in early life demonstrate the potential for development of targeted therapeutics for younger patients with mild to moderate disease. In this manuscript, we highlight key insights into the pathology of AD, outlining the correlations with analytes previously identified in adult moderate to severe disease and their role in disease development even in this mild to moderate cohort of pediatric subjects.
Methods
Patients and samples
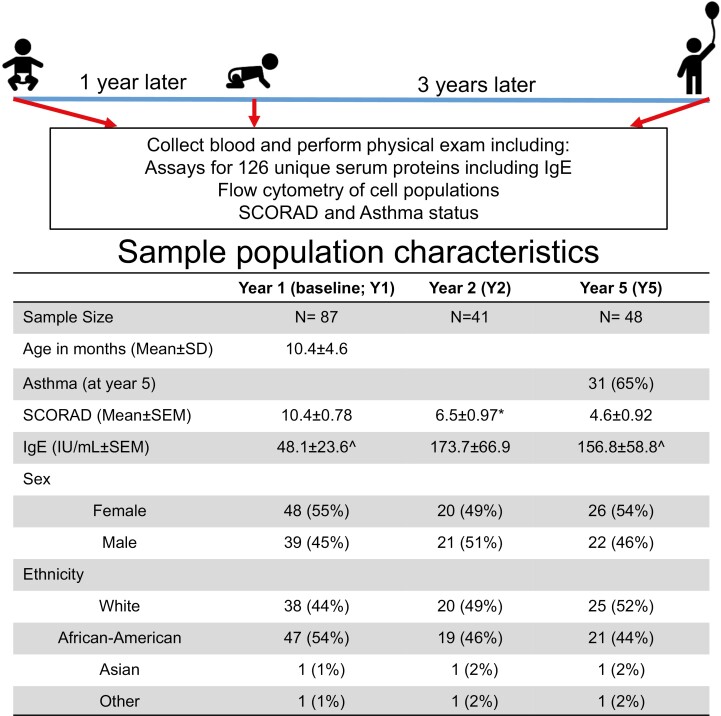
This study included a cohort of 87 American pediatric patients recruited in infancy from pediatric allergy and dermatology clinics associated with IU Health and Riley Children’s Hospital (Fig. 1). Infants and toddlers with dermatitis were recruited to the study following an initial qualifying assessment in an allergy or dermatology clinic using SCORAD to define severity. Of note, 8 patients (9%) had a SCORAD of 0 at the baseline research visit (Y1) despite having clinically diagnosed dermatitis at the initial qualifying assessment; 18 (21%) between Y1 and Y2 and 22 (25%) between Y2 and Y5 resolved from a positive SCORAD to a SCORAD of 0. Only 7 (8%) had their SCORAD value drop from >0 at Y1 to 0 for both Y2 and Y5 visits; however, this numerical change does not fully indicate resolution of AD. Subjects were excluded for a history of prior wheezing, lower respiratory tract illness, treatment with asthma medications, or congenital heart disease. The majority of the population (85%) had a family history of asthma or allergy, which was used as the criteria for high risk. Asthma was physician diagnosed based on parameters including persistent wheezing, the requirement for asthma-controlling medications, respiratory function, and response to methacholine challenge [11]. Serum samples were collected from children diagnosed with AD as previously described but only patients with baseline (Y1) sample were included in this study [9]. Demographic information (i.e. ethnicity and sex) was collected upon entry by the parent/guardian based on selecting all that apply: Caucasian (White), African American, Asian, Native American, Hispanic, or Other. Sex was selected by parent/guardian upon enrollment as male or female. These demographics are used as potential covariates for inter-subject variability. The subset of patients analyzed here was not significantly different from the entire population except for an enrichment in White and male subjects (Fig. 1). Flow cytometry analysis of peripheral blood mononuclear cells (PBMCs) collected throughout the course of the study was previously described [12]. The primary outcome of this cohort was to analyze airway function in a high-risk population of infants [9, 12, 13]. Total IgE concentration was determined by ImmuneTech and the assay had linear detection between 2 and 2000 IU/ml. All studies with human subjects and samples were performed with approval from the Indiana University Institutional Review Board and required informed consent from a parent or guardian.
Fig. 1.
Sample population characteristics. This study included a cohort of 87 patients with serum sample collection at infancy (Y1), 1-year follow-up (Y2), and 4 years later (Y5). Samples collected from children diagnosed with dermatitis for this study have been previously described and assessed for clinical features including asthma [9]. Samples missing from groups were denoted as such: ^n = 1, ∗n = 2.
ELISA and multiplex immunoassays
Olink multiplex assay
Serum samples were analyzed with the Olink Inflammation I Proseek multiplex assay (95302) according to the manufacturer’s specifications. The levels of analyte-specific DNA amplicons for each patient were quantified by Fluidigm Biomark HD-generated Ct values and represented on log 2 scale as NPX (Normalized Protein Expression) values.
Luminex bead-based multiplex immunoassay
Luminex bead-based sandwich immunoassay was used to assess serum protein levels quantitatively and simultaneously on the Millipore Luminex 200 Bead Reader System. Two MILLIPLEX MAP Human Cytokine/Chemokine 41-plex Magnetic Bead Panels (HCYTOMAG-60K-PX41 and HCP2MAG-62K-PX23) were assayed on Curiox Biosystems DA-Bead plates and plates were washed on a DropArray LT210 washing station following the Curiox low-volume protocol.
ELISA
For additional quantitative assessment of important analytes conventional sandwich enzyme-linked immunoassays (ELISA) were utilized including Periostin (DY3548B) and sST2 (DST200) from R&D Systems. Levels of IL-22 and TARC/CCL17 were measured using MesoScale Discovery (MSD)-based sandwich ELISA. The IL-22 assay, consisting of proprietary antibodies and recombinant protein, utilizes a Small Spot SA–MSD plate (L45SA-1) and in-house diluents. Following incubations, wells were washed with 1× TBST using a Bio-TEK ELx405 and analyzed on the Meso Quick Plex SQ 120 plate reader. TARC/CCL17 (K151NTD) was measured according to the manufacturer’s specifications. Levels of IL-13 (102732) and IL-17A (101599) were assessed by Quanterix single molecule array (Simoa) bead-based 2.0 assays on the Simoa HD-1 analyzer as per the manufacturer’s protocols.
Statistical analyses
Correlation analysis of AD severity
Repeated measures correlation (R package: rmcorr) was applied to SCORAD and log2-transformed serum protein concentration or NPX values across all years to account the dependent structure of the same patient over time for each marker and Benjamini-Hochberg multiple comparison adjustment across markers to report the correlation values and adjusted P-values [14]. Volcano plots were used to identify significant correlations and, due to the repeated nature of these measurements, rmcorr values of 0.3 or better are highlighted to outline the mild-to-moderate correlations in this data set (i.e. adjusted P-values <0.05 and repeated measures correlation >0.3 or <−0.3) [15].
Serum protein changes over time
For each analyte, a mixed-effect model with year, sex, and ethnicity as fixed covariates was applied to the log2-transformed analyte concentration or NPX values. Mixed-effect model with repeated measurements was used to account for the dependent relationship over time for each pediatric AD patient and utilize all pediatric AD patients, including patients that dropped out from the study prior to Y5. For analytes with expression levels lower than detectable range, we imputed a value set to half of the lower limit of detection of the assay using the Beal Method [16]. Volcano plots were generated and used to identify changes from Y5 to Y1 and from Y2 to Y1 with criteria (P-values <0.05 and fold change >1.5× or <−1.5×) set to account for the proportion of missing values. Comparisons of each analyte at each time point (Y1 to Y2, Y1 to Y5, and Y2 to Y5) were made and significance assessed using the Tukey Multiple Comparison Test.
Prediction model of SCORAD change
The prediction model of the change in SCORAD at Y5 from Y1 was built with JMP software using all the baseline (Y1) analytes concentrations [17]. With forward selection, the data set started with a null (empty) model and added individual analytes one at a time based on most predicted measure (e.g. smallest P-values) until the addition of further analytes provided no increase in predictive power. The stopping criteria are based on minimum corrected Akaike Information Criterion (AICC) [18]. To evaluate the performance of prediction model, we calculate the R squares and Root Mean Square Error (RMSE) between predicted values and observed values for all patients and RMSE. Furthermore, the cross-validation on the proposed prediction model with selected markers was applied with 100 iterations [19]. At each iteration, the entire baseline patient set was split into training set (75%) and testing test (25%). The training set data were used to build the model and estimate model parameters. The model was then used to predict the testing set data. The R squares and RMSE between predicted values and observed values were calculated for each iteration to assess the robustness of prediction model.
Pathway analysis
Pathway analysis was conducted with MetaCore using Fisher’s exact test to define relevant pathways based on differentially expressed serum protein signatures between Y5 and Y1 samples [20].
Results
Sample cohort description
This study sought to understand the interplay among analytes, relationships to disease severity, and to delineate patterns of dysregulation through the first 5 years of life. Serum samples were collected from a total of 87 pediatric AD patients upon entry, 1, and 4 years later [9]. The mean age at study entry was 10.4 months (SD 4.6 months) and the population was evenly distributed both by sex and between White and African American subjects (Fig. 1). Incidence of asthma at Y5 was observed in 65% of patients. AD severity, measured by SCORAD, was mild at time of enrollment (range 1–28) averaging 10.4 across the study cohort, although a proportion of the population demonstrated improved SCORAD over time (Figs 1 and 2).
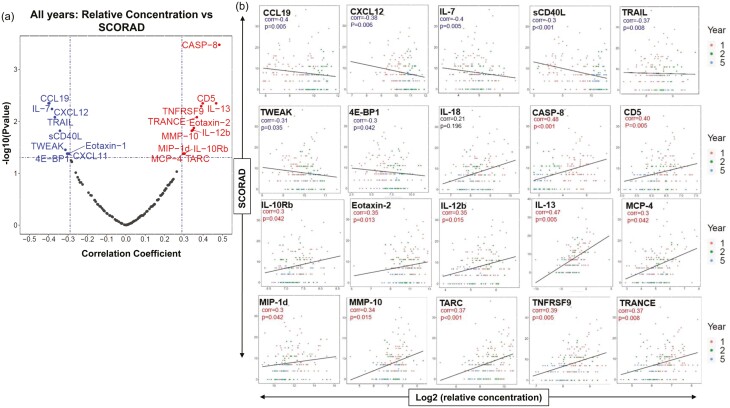
Fig. 2.
Correlations to SCORAD. (a) Repeated measurement correlation of SCORAD to the relative concentrations of all analytes across all study years. (b) Log2 protein expression of serum protein concentrations correlated to SCORAD (rmcorr) for key analytes of interest. Eight patients had a SCORAD = 0 at Y1, 18 at Y2, and 22 at Y5. 1 = year 1; 2 = year 2; 5 = year 5.
Serum protein analytes correlate to SCORAD over time
A total of 126 serum proteins, including IgE, were analyzed across different platforms to identify relevant connections between various immunological markers and disease progression, using SCORAD, across each time point of data collections. An important measure of atopy, circulating IgE, was measured on average to be 29.6 IU/ml at year 1 (Y1; baseline), 32.7 IU/ml at year 2 (Y2; 1 year after baseline), and 29.9 IU/ml at year 5 (Y5; 4 years after baseline). On average, total IgE (IU/ml) was 2-fold higher in African American than in White children though these levels did not correlate to higher SCORAD and there were no other observable differences in serum or cell phenotype profiles based on ethnicity alone (data not shown). IgE was only weakly correlated to SCORAD (rmcorr = −0.22) based on combined analysis across all patients and all timepoints of the study; therefore, we shifted focus to other key inflammatory serum markers possessing positive correlation with clinical severity across all three visits. The strongest positive correlation to SCORAD across all visits were CASP-8 (rmcorr = 0.48) and IL-13 (rmcorr = 0.47) (Fig. 2). Additional proteins positively correlated with SCORAD in these pediatric AD patients that have been previously linked to severity in adult AD included TARC/CCL17 (rmcorr = 0.37) and MCP-4/CCL13 (rmcorr = 0.3) (Fig. 2) [21]. Interestingly, we observed proteins shown to be elevated in the circulation of adult AD patients to be negatively correlated with SCORAD across all three timepoints taken in the first 5 years of life from these pediatric AD patients, such as sCD40L (rmcorr = −0.3), Eotaxin-1 (rmcorr = −0.3), and IL-7 (rmcorr = −0.4) (Fig. 2, Supplementary Table 1) [21].
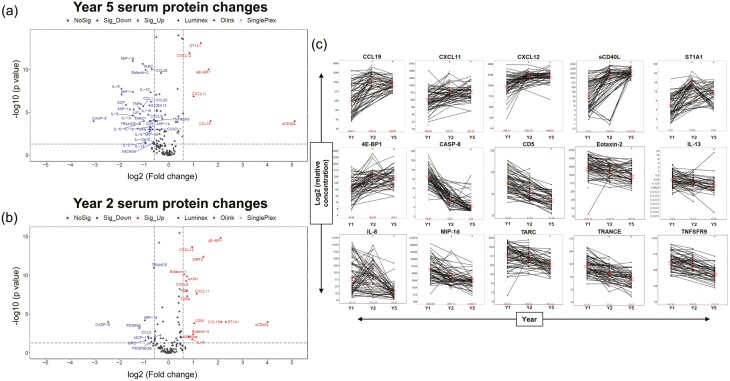
Understanding the dramatic developmental changes that occur in early childhood, we utilized volcano plots to visualize differentially expressed proteins between Y1 and both Y2 and Y5 (Fig. 3a and b). Proteins which had the highest positive correlations with disease severity, such as IL-13, CASP-8, and TARC/CCL17 (Fig. 2a) were observed to have the highest concentrations early at Y1 relative to Y5 (Fig. 3a). We also observed decreasing serum concentrations of Th2 markers (TARC/CCL17, IL-13, and MCP-4/CCL13) from Y1 to Y5, perhaps indicative of the dramatic elevation at early stages of life for these patients (Fig. 3c, Supplementary Fig. 1). The Th22 marker, IL-22, a highly expressed cytokine in adult AD was not observed to change over the course of the study, suggesting the absence of a significant role for IL-22 in the first 5 years of pediatric disease within this cohort (Supplementary Fig. 1) [22]. Similarly, many Th1/Th17 markers, including CXCL10, IL-17A, and IFNγ shown to be upregulated in adult AD were unchanged in our study [6]; however, elevated serum concentration of the related marker, CXCL11, was observed as early as Y2 (Supplementary Fig. 1).
Fig. 3.
Analyte patterns in the population. (a, b) Volcano plot showing serum protein change from Y1. Proteins expressed at Y1 that decrease in Y5 or Y2 are colored blue. Proteins expressed at Y1 that increase at Y5 or Y2 are colored red. (c) Log2 relative protein expression of serum protein concentrations for key analytes of interest. ∗Significant change from Y1, P-value < 0.05; Y1 = year 1; Y2 = year 2; Y5 = year 5.
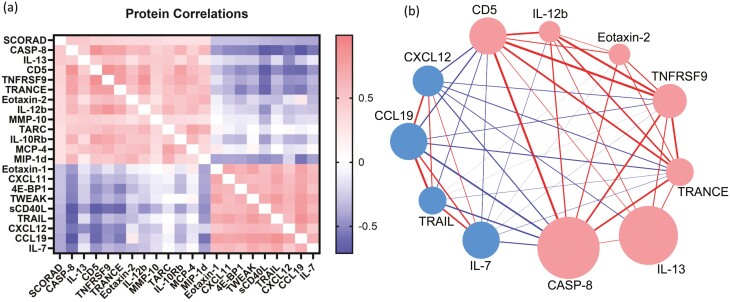
Given the correlation of inflammatory markers with SCORAD, we wanted to determine how these serum proteins associated with each other. Thus, we examined correlations among SCORAD-associated analytes across all time points of the study. We observed strong interconnectivity within two groups of analytes: (i) CD5, CASP8, IL-12B, TRANCE, and TNFRSF9 and (ii) CXCL12, CCL19, TRAIL, and IL-7 (Fig. 4). Surprisingly, IL-13, which emerged as one of the most significant correlates with SCORAD showed only moderate associations with other protein concentrations across all timepoints (Fig. 4, Supplementary Table 1). This observed lack of correlation to other analytes by IL-13, coupled with our finding that many inflammatory markers decrease in concentration over the course of the study, as patients age and utilize standard of care therapies, initiated further assessment of pathway nodes connecting these markers to disease severity.
Fig. 4.
Correlations among cytokines. (a) Heatmap representation of cytokine correlations to one another shown with additional proteins of interest across the course of the study. (b) Visual representation of cytokine correlations to each other (thickness of connecting line) and to SCORAD (size of circle) across all time points of the study. Red color denotes correlations ≥0.35. Blue color denotes correlations ≤0.35.
Serum protein analytes correlate to cell populations in peripheral blood
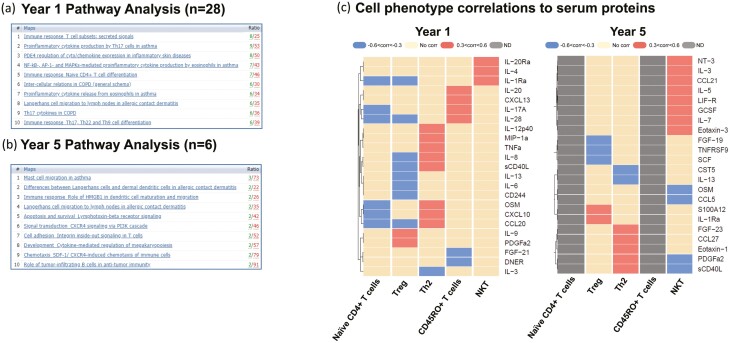
Molecular processes linked to the analytes with higher concentrations observed at Y1 relative to Y5 were significantly associated with pathways related to T-cell activation and cytokine secretion profiles including Immune response pathways in T-cell differentiation and cytokine secretion (Figs 3a and 5a). Juxtaposed with the Y1 pathway analysis of T-cell-driven response mechanisms in early AD development, Y5 pathway analysis highlights the shift from T-cell-driven disease to include innate immunity. In Y5, peripheral cell profiling indicates the involvement of Langerhans and dendritic cells (DCs) in AD, and the rise of asthma-related mast cell mechanisms over time (Fig. 5a and b). Proteins increased in concentration at subsequent visits (Y2 and Y5) relative to Y1 were sCD40L, ST1A1, 4E-BP1, CXCL12, CXCL11, CCL19, and pathway analysis connected these to innate mechanisms (Figs 3b and 5b). Most notably, a 34-fold magnitude increase was observed in the circulating levels of sCD40L at Y5 as compared to Y1 (Fig. 3a). Molecular processes observed to be upregulated at Y2 from Y1 were also affiliated with innate immunity (Fig. 3b) and tracked with similar innate cell-influenced mechanisms observed at Y5 (Fig. 5b) further suggesting the role on innate cells in disease progression. Several of these markers overlapped between Y2 and Y5 and all markers having significantly higher concentrations in Y5 (from Y1) were also seen in Y2 (Fig. 3a and b).
Fig. 5.
Pathway analysis. (a) Top 10 pathway maps for highly expressed proteins at year 1 that decrease expression by year 5. The 28 specific proteins used for this analysis are outlined in Fig. 3a. (b) Top 10 pathways of most highly expressed proteins at year 5 that were low or absent at year 1. The six specific proteins used for this analysis are outlined in Fig. 3a. (c) Heat map correlations of cell phenotypes previously assessed by flow cytometry [12] to serum protein analysis from same time point.
Utilizing heatmap visualizations, we next assessed the correlations of serum protein analytes to T-cell phenotypes in PBMCs from the same individuals. These protein correlations track with changes in T-cell development from Y1 to Y5 (Fig. 5c) [12]. We observed distinct profiles at Y1 between circulating Th2 and Treg cells and epithelial-derived chemokines, including two proteins most significantly upregulated at Y1 to Y5; MIP-1a/CCL3, sCD40L, CCL20, and IL-8 all showed positive correlations with Th2 populations and were unchanged or negatively correlated with Treg populations indicating these signaling mediators promote T-cell-mediated immune responses very early in pediatric development (Fig. 5c). sCD40L showed strong correlations at both Y1 and Y5 with Th2 cells and did not correlate with any other cell types (Fig. 5c). Th2 cytokines, IL-4 and IL-5, strongly correlated with NKT cell populations at Y1 and Y5, respectively, and IL-9 and PDFGa2 positively correlated with Treg levels in Y1, but not at Y5 (Fig. 5c).
Predictive model defines change in SCORAD from Y1 to Y5
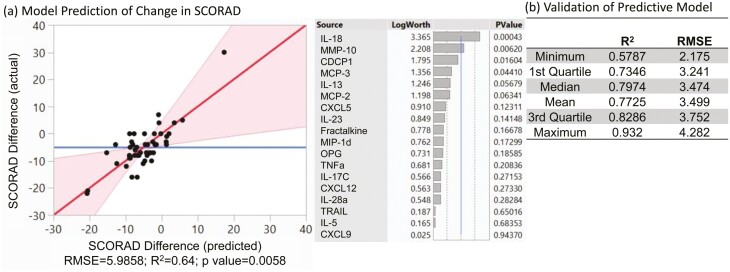
Notwithstanding the complexity of immune responses observed in these samples, the longitudinal nature of this study suggested that we may be able to identify predictive biomarkers related to the course of disease. We utilized the forward selection modeling approach with a corrected AICC stopping rule to predict the change of SCORAD from Y1 to Y5 using the Y1 serum protein concentrations. A biomarker panel of 18 analytes was selected in the prediction model to predict progression of severity from infancy to Y5 (Fig. 6a, R2 = 0.64, P = 0.0058). To further illustrate the robustness of prediction, a cross-validation with 100 iterations on the proposed 18 analytes predicted model resulted in the mean of the R squares and RMSE of 0.77 and 3.5 for the 100 iterations, respectively (Fig. 6b). The highlighted serum proteins, which could be involved in persistence or resolution of AD, include IL-18, MMP-10, and IL-13 (Fig. 6). These analyses highlight the objective and predictive nature of correlated analytes present in the serum of infants for subsequent disease severity.
Fig.6.
Predictive model. (a) Using forward modeling with AICC, we use the listed 18 serum analytes to predict change in SCORAD over time. The fit of these 18 analytes within this model are defined by RMSE and R square adjusted (R2). (b) Samples were split into either test or training sets to assess the reproducibility of the model.
Discussion
Minimally invasive biomarkers, such as serum proteins, are widely sought after for their value in defining disease states and outcomes. In adolescent and adult AD patients, a high total serum IgE level was a predictive factor for poor disease prognosis [23]. Interestingly, in these pediatric patients, many inflammatory serum biomarkers demonstrated stronger correlation to disease severity better than IgE alone [24]. Thus, the focus of our study was to highlight key biomarkers in early AD that correlate with disease and potentially predict the persistence or resolution of disease.
Repeated measurement correlations across all time points
Multiple analytes correlated strongly with pediatric AD severity and, most significantly, known disease markers, IL-13, MCP-4/CCL13, and TARC/CCL17, correlated with SCORAD across all time points from infancy to 5 years of age, recapitulating the importance of type 2 immune response early in AD pathogenesis. Correlations with chemokines and monocyte chemoattractant proteins underscores the role of eosinophils and monocytes in early AD, while TARC/CCL17 and IL-13 are both mediators of Th2 inflammation and implicated in pediatric and adult AD, emphasizing that these analytes could represent critical nodes for better understanding AD heterogeneity and disease progression [7, 25, 26]. Although standard of care treatments lead to a decreased concentration of these biomarkers, the observations and changes observed in our study demonstrate the strong relationship these markers have with the disease pathology in pediatric patients. Importantly, this study included ultrasensitive measurements of numerous cytokines for which quantitative assessment has only recently been possible, including IL-13, which is implicated as a driver of AD based on clinical responses to lebrikizumab and dupilumab, further implicating IL-13 in disease severity even in infancy [5, 27, 28].
Analytes described as drivers of adult AD, such as IL-22 and IL-17A [7, 22], did not possess strong correlations to disease severity within these pediatric AD samples. This is likely due to the slow increase in serum concentration over time, limiting the role of IL-22 in pediatric AD as seen in previous reports [7]. Though levels of IL-17A were not correlated to SCORAD and present in very low levels, as seen in previous pediatric studies [29], Th17-related fibroblast markers, CCL19 and CCL20, were observed at various timepoints throughout our study and CCL20 positively correlated with Th2 circulating cell populations at Y1 pointing toward an intriguing immunological overlap of inflammatory fibroblasts and T cells. Pathway analysis of proteins upregulated in Y5 samples reveals the emergence of innate pathways over time and multiple gene ontology results indicate that DCs may increase with age. Increased conventional DCs were previously described as being protective for wheezing in these pediatric AD patients [10], and stimulation of these conventional DCs isolated from atopic infants produced more interferon and IL-10 than those derived from non-atopic infants at initial visit and the 1-year follow-up visit [30]. Mounting evidence demonstrates this appearance of symptoms known as the atopic march tracks with early disruption of immune development and can have lasting effect into adulthood [31]. This link between increased CCL19 and DCs further indicates a role in atopic march from AD to asthma for CCR7, the receptor for CCL19, which can coordinate T cell, B cell, and DC responses across pathologies [32].
Protein correlations support a Th2 to Th1 immune shift
Matrix remodeling proteins (MMP-10) and chemoattractant molecules (MCP-3/CCL7, MCP-2/CCL8, CXCL5) provide further insights into the complexity of disease mechanisms significantly associated with the progression of the pediatric form of AD. The connections of these markers to disease severity contextualize a shift toward Th1 immunity and myeloid involvement developing throughout the course of the pediatric AD patient journey concomitant with dominant Th2 responses throughout. The identification of CXCL5 and IL-5 as important disease severity predictive biomarkers for pediatric AD highlights the innate allergic response early in disease progression. The high levels of sCD40L that we observed in pediatric patients aligned with prior published observations that this protein is elevated in the blood of pediatric patients [6]. Additionally, sCD40L concentrations correlated throughout the study with CD4+Th2 cells which might link to the role of CD4+CD40L+ cells in the priming of other immune cell types including CD14+ monocytes and CD8+ T cells [33]. This work aids our contextualization of a shift toward Th1 immunity and myeloid involvement developing throughout the course of the pediatric AD patient journey concomitant with Th2 responses presenting initially and persisting throughout the course of disease. These differences reiterate the idea that blood samples from pediatric AD patients contain strong Th2-driven signals early, though levels decrease slightly with age as Th1 and innate-linked inflammatory markers develop.
Caspase-8 was highly correlated with SCORAD and was intriguing as a circulating analyte. Caspase-8 has been previously implicated in allergic airway inflammation and was responsible for the release of IL-1 and immune infiltration [34]. However, caspase-8 displayed opposite effects in a mouse model of dermatitis where loss is associated with disease [35]. As caspase-8 is involved broadly in cell death, it could be activated within multiple cell populations within the skin [36]. Release into the circulation could result from the physical damage of scratching lesions, or it could be a general marker of inflammatory disease. It will be interesting to discern these possibilities in future mechanistic studies.
The previously reported circulating protein profiles of pediatric AD have provided robust assessment of correlation analysis at a single timepoint [6]. Our study builds upon these important observations via the monitoring of circulating biomarkers over a development period of 4 years adding value to the concept that objective biomarkers could be used to objectively assess disease severity, as has been demonstrated in adult AD [37]. We observed subsets of inflammatory proteins that correlated with each other in a manner consistent with early disease skewing toward type 2 immune responses. Importantly, forward selection modeling with the minimum AICC stopping rule identified 18 analytes, such as IL-18, IL-13, and MMP-10, in infant serum that could be used to predict AD severity 4 years later; therefore, these analytes might represent critical nodes to better understand AD heterogeneity and disease progression. The measurement of markers identified in Fig. 6 should provide guidance for therapeutic intervention strategies at early stages of life for pediatric patients suffering from AD. However, it is important to note that the model only predicts change in SCORAD at a specific time point and a predicted negative change does not necessarily predict a durable remission. Additional testing of this model could build upon the predictive value for AD persistence past infancy and further define the pathogenesis of atopic disease. This additional testing could allow us to see ever more clearly branching nodes delineating the complex underlying immune regulation. This, in turn, may guide therapeutic intervention and the prognostic value of such serum protein measurement, outlining the need for early treatment options in pediatric patients.
Current data set outlines future goals for larger studies
There are some limitations to this data set and many overarching considerations for studies performed in pediatric populations. First our cohort has a relatively small number of patient samples (n = 87) and samples from all patients were not available for analysis at all time points. This was a result of these analyses being ancillary to the primary outcomes from this cohort of infants and the fact that not all patients had samples remaining for these studies [9, 12, 13]. Second, there was no healthy control group for comparison. The goal of the previous studies was to analyze airway function in infants at high risk for asthma, and it was not feasible to perform these analyses on healthy infants. Few other studies have been able to collect from a healthy childhood population for comparison. However, other disease-driven population studies have outlined the presence of Th1 immunity in early childhood, contrasting our study that indicates early Th2 responses [38, 39]. These studies also note changes within the biology of the skin over time and the cellular populations further highlighting the difference between children and adults and outlining the need for but lack of a relevant, age-matched control sample group [38]. Third, there could have been some bias in subject selection, in that all subjects had a positive SCORAD for entry to the study, and the majority developed sensitization to at least one common allergen [9, 12]. Finally, treatment of patients was not standardized and might contribute to variations in outcomes. Still, this population provided a novel opportunity to interrogate AD at this early stage and follow subjects to assess for AD persistence and asthma diagnosis. The large proportion of African American subjects also adds to the value of this dataset. Overall, these findings provide the basis for larger future studies.
Our data highlight additional questions for the field regarding the progression of AD as children age through adolescence into adulthood. We did not observe elevation of IL-4 in the serum of these patients, which could be attributed to the technical limitations of assays available at time of testing, considering that IL-13 was measured on an ultra-sensitive platform. We also note that though MCP-4/CCL13 and TARC/CCL17 were moderately correlated with disease severity, they did not factor into the predictive modeling of disease progression possibly due to these proteins being more broadly involved in atopic mechanisms beyond AD [40]. Future studies with focused outcomes on other atopy measures can provide predictive insight into mechanisms of wheezing or lung function. Correlations to SCORAD allow us to minimize potential error that is introduced in a longitudinal study, especially in pediatrics as maturation and growth are heavily involved in immune system development. This cohort and data included in this manuscript enhance our knowledge regarding the systemic presentation of AD in children and potential biomarker profiles which aid in predication of disease persistence or resolution.
Supplementary data
Supplementary data are available at Clinical and Experimental Immunology online.
Supplementary Figure 1: Analyte Patterns in the Population. Log2 protein expression of serum protein concentrations for analytes of interest. Y1 = year 1; Y2 = year 2; Y5 = year 5.
Supplementary Table 1: Correlations to SCORAD and Among Cytokines. Repeated measurement correlations of SCORAD to the concentration of selected analytes and correlations of cytokine correlations to one another across all study years.
Acknowledgements
We thank Jeffrey Fill for technical assistance with the IL-13 Quanterix Simoa assay. We thank Robert J. Benschop, Ernst R. Dow, and Josh Poorbaugh for scientific input.
Glossary
Abbreviations
- AD
atopic dermatitis
- AICC
corrected Akaike information criterion
- rmcorr
repeated-measures correlation
- DC
dendritic cell
- IL
interleukin
- MCP
monocyte chemoattractant protein
- MSD
mesoscale discovery
- PBMC
peripheral blood mononuclear cell
- TARC
thymus and activation regulated chemokine
- Y1
year 1
- Y2
year 2
- Y5
year 5
Contributor Information
Sarah M Engle, Eli Lilly and Company, Indianapolis, IN, USA.
Ching-Yun Chang, Eli Lilly and Company, Indianapolis, IN, USA.
Benjamin J Ulrich, Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA; Wells Center for Pediatric Research, Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN, USA.
Allyson Satterwhite, Eli Lilly and Company, Indianapolis, IN, USA.
Tristan Hayes, Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA; Wells Center for Pediatric Research, Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN, USA.
Kim Robling, Eli Lilly and Company, Indianapolis, IN, USA.
Sean E Sissons, Eli Lilly and Company, Indianapolis, IN, USA.
Jochen Schmitz, Eli Lilly and Company, Indianapolis, IN, USA.
Robert S Tepper, Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA.
Mark H Kaplan, Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA; Wells Center for Pediatric Research, Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN, USA.
Jonathan T Sims, Eli Lilly and Company, Indianapolis, IN, USA.
Funding
This study was funded by Eli Lilly and Company and a Public Health Services grant from NIH to M.H.K. (AI095282). B.J.U. was supported by T32 AI060519 and F30 HL147515. T.H. was supported by T32 AI060519.
Conflict of interest
S.M.E., C.C., A.S., K.R., S.E.S., J.S., and J.T.S. are employees and may be shareholders of Eli Lilly and Company. B.J.U., T.H., R.S.T., and M.H.K. have no conflicts of interest related to this study.
Author contributions
S.M.E., J.T.S., and M.K. conceived the study. S.M.E., J.T.S., A.S., K.R., and S.E.S. conducted the laboratory experiments. S.M.E., J.T.S., C-Y.C., B.J.U., A.S., T.H., and M.K. designed and conducted the main analyses and interpreted the results. J.S., K.R., S.E.S., and R.S.T. also contributed to the discussion of analyses. J.T.S. and S.M.E. wrote the first draft of the manuscript. S.M.E., J.T.S., C-Y.C., and M.K. wrote the final manuscript, with contributions and review by all other authors.
Ethical approval
All studies with human subjects and samples were performed with approval from the Indiana University Institutional Review Board and required informed consent from a parent or guardian.
Data availability
Data will be made available upon reasonable request.
References
- 1. Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat Rev Dis Primers 2018, 4, 1. [DOI] [PubMed] [Google Scholar]
- 2. Silverberg JI. Adult-onset atopic dermatitis. J Allergy Clin Immunol Pract 2019, 7, 28–33. [DOI] [PubMed] [Google Scholar]
- 3. Weidinger S, Novak N. Atopic dermatitis. Lancet 2016, 387(10023), 1109–22. [DOI] [PubMed] [Google Scholar]
- 4. Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol 2003, 112(6 Suppl), S118–27. [DOI] [PubMed] [Google Scholar]
- 5. Bieber T, Traidl-Hoffmann C, Schappi G, Lauener R, Akdis C, Schmid-Grendlmeier P. Unraveling the complexity of atopic dermatitis: the CK-CARE approach toward precision medicine. Allergy 2020, 75, 2936–8. [DOI] [PubMed] [Google Scholar]
- 6. Brunner PM, He H, Pavel AB, et al. The blood proteomic signature of early-onset pediatric atopic dermatitis shows systemic inflammation and is distinct from adult long-standing disease. J Am Acad Dermatol 2019, 81, 510–9. [DOI] [PubMed] [Google Scholar]
- 7. Czarnowicki T, He H, Canter T, et al. Evolution of pathologic T-cell subsets in patients with atopic dermatitis from infancy to adulthood. J Allergy Clin Immunol 2020, 145, 215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bohme M, Wickman M, Lennart NS, Svartengren M, Wahlgren CF. Family history and risk of atopic dermatitis in children up to 4 years. Clin Exp Allergy 2003, 33, 1226–31. [DOI] [PubMed] [Google Scholar]
- 9. Tepper RS, Llapur CJ, Jones MH, et al. Expired nitric oxide and airway reactivity in infants at risk for asthma. J Allergy Clin Immunol 2008, 122, 760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao W, Barbe-Tuana FM, Llapur CJ, et al. Evaluation of airway reactivity and immune characteristics as risk factors for wheezing early in life. J Allergy Clin Immunol 2010, 126, 483–8 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang D, Yao W, Tiller CJ, et al. Exhaled nitric oxide during infancy as a risk factor for asthma and airway hyperreactivity. Eur Respir J 2015, 45, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yao W, Tepper RS, Kaplan MH. Predisposition to the development of IL-9-secreting T cells in atopic infants. J Allergy Clin Immunol 2011, 128, 1357–60 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarria EE, Mattiello R, Yao W, et al. Atopy, cytokine production, and airway reactivity as predictors of pre-school asthma and airway responsiveness. Pediatr Pulmonol 2014, 49, 132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2013. Available from http://www.R-project.org/ [Google Scholar]
- 15. Akoglu H. User’s guide to correlation coefficients. Turk J Emerg Med 2018, 18, 91–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson JR, ed. Methods for Handling Concentration Values Below the Limit of Quantification in PK Studies. In: SAS Conference Proceedings: Pharmaceutical Users Software Exchange US 2018, Raleigh, North Carolina, June 3–6, 162 papers, 2018. [Google Scholar]
- 17. JMP14. Cary, NC: SAS Institute Inc., 2018. Available from https://support.sas.com/downloads/index.htm?fil=2 [Google Scholar]
- 18. Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information Theoretic Approach. 2nd ed. NY: Springer-Verlag New York, 2002. [Google Scholar]
- 19. Stone M. Cross-validatory choice and assessment of statistical predictions. J R Stat Soc Series B Stat Methodol 1974, 36, 111–33. [Google Scholar]
- 20. MetaCore: Clarivate Analytics; 2020. Available from https://clarivate.com/cortellis/solutions/early-research-intelligence-solutions/
- 21. Brunner PM, Suárez-Fariñas M, He H, et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci. Rep 2017, 7:8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nograles KE, Zaba LC, Shemer A, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol 2009, 123, 1244–52 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kiiski V, Karlsson O, Remitz A, Reitamo S. High serum total IgE predicts poor long-term outcome in atopic dermatitis. Acta Derm Venereol 2015, 95, 943–7. [DOI] [PubMed] [Google Scholar]
- 24. Hon KE, Lam MA, Leung T, et al. Are age-specific high serum IgE levels associated with worse symptomatology in children with atopic dermatitis? Int J Dermatol 2007, 46, 1258–62. [DOI] [PubMed] [Google Scholar]
- 25. Kataoka Y. Thymus and activation-regulated chemokine as a clinical biomarker in atopic dermatitis. J Dermatol 2014, 41, 221–9. [DOI] [PubMed] [Google Scholar]
- 26. Taha RA, Minshall EM, Leung DY, et al. Evidence for increased expression of eotaxin and monocyte chemotactic protein-4 in atopic dermatitis. J Allergy Clin Immunol 2000, 105, 1002–7. [DOI] [PubMed] [Google Scholar]
- 27. Guttman-Yassky E, Blauvelt A, Eichenfield LF, et al. Efficacy and safety of lebrikizumab, a high-affinity interleukin 13 inhibitor, in adults with moderate to severe atopic dermatitis: a phase 2b randomized clinical trial. JAMA Dermatol. 2020, 156, 411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bieber T. Interleukin-13: targeting an underestimated cytokine in atopic dermatitis. Allergy 2020, 75, 54–62. [DOI] [PubMed] [Google Scholar]
- 29. Black A, Bhaumik S, Kirkman RL, Weaver CT, Randolph DA. Developmental regulation of Th17-cell capacity in human neonates. Eur J Immunol 2012, 42, 311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao W, Chang J, Sehra S, et al. Altered cytokine production by dendritic cells from infants with atopic dermatitis. Clin Immunol 2010, 137, 406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davidson WF, Leung DYM, Beck LA, et al. Report from the National Institute of Allergy and Infectious Diseases workshop on “Atopic dermatitis and the atopic march: mechanisms and interventions”. J Allergy Clin Immunol 2019, 143, 894–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grinnan D, Sung SS, Dougherty JA, et al. Enhanced allergen-induced airway inflammation in paucity of lymph node T cell (plt) mutant mice. J Allergy Clin Immunol 2006, 118, 1234–41. [DOI] [PubMed] [Google Scholar]
- 33. Trella E, Raafat N, Mengus C, et al. CD40 ligand-expressing recombinant vaccinia virus promotes the generation of CD8(+) central memory T cells. Eur J Immunol 2016, 46, 420–31. [DOI] [PubMed] [Google Scholar]
- 34. Qi X, Gurung P, Malireddi RK, et al. Critical role of caspase-8-mediated IL-1 signaling in promoting Th2 responses during asthma pathogenesis. Mucosal Immunol 2017, 10, 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kovalenko A, Kim JC, Kang TB, et al. Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. J Exp Med 2009, 206, 2161–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orning P, Lien E. Multiple roles of caspase-8 in cell death, inflammation, and innate immunity. J Leukoc Biol 2021, 109, 121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thijs JL, Nierkens S, Herath A, et al. A panel of biomarkers for disease severity in atopic dermatitis. Clin Exp Allergy 2015, 45, 698–701. [DOI] [PubMed] [Google Scholar]
- 38. Georgountzou A, Papadopoulos NG. Postnatal innate immune development: from birth to adulthood. Front Immunol 2017, 8, 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siden H, Steele R, Brant R, et al. Designing and implementing a longitudinal study of children with neurological, genetic or metabolic conditions: charting the territory. BMC Pediatrics 2010, 10, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grünig G, Corry DB, Reibman J, Wills-Karp M. Interleukin 13 and the evolution of asthma therapy. Am J Clin Exp Immunol 2012, 1, 20–7. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon reasonable request.