Abstract
Actinomycetes are a rich source of bioactive natural products important for novel drug leads. Recent genome mining approaches have revealed an enormous number of secondary metabolite biosynthetic gene clusters (smBGCs) in actinomycetes. However, under standard laboratory culture conditions, many smBGCs are silent or cryptic. To activate these dormant smBGCs, several approaches, including culture-based or genetic engineering-based strategies, have been developed. Above all, coculture is a promising approach to induce novel secondary metabolite production from actinomycetes by mimicking an ecological habitat where cryptic smBGCs may be activated. In this review, we introduce coculture studies that aim to expand the chemical diversity of actinomycetes, by categorizing the cases by the type of coculture partner. Furthermore, we discuss the current challenges that need to be overcome to support the elicitation of novel bioactive compounds from actinomycetes.
Keywords: Actinomycetes, Streptomyces, Coculture, Secondary metabolite
Introduction
Natural products are organic compounds produced by living organisms mainly in the form of secondary metabolites, most of which have therapeutic bioactivity, including antimicrobial, antifungal, and anticancer (Harvey, 2008). The representative sources of these bioactive secondary metabolites are Gram-positive soil-living bacteria actinomycetes, particularly Streptomyces, whose products comprise approximately 70% of commercially available antibiotics (Nett et al., 2009). From the 1950s to 1970s, the golden period of antibiotic discovery, a number of compounds produced by Streptomyces strains were explored and utilized to deal with infectious diseases (Aminov, 2010; Procopio et al., 2012). However, after two decades of success, antibiotic discovery became depressed owing to the continuously increasing rediscovery rate of known chemical entities, while pathogenic microbes gradually cultivated antimicrobial resistance to the latest generation of antibiotics (Koehn & Carter, 2005; Ventola, 2015). Even worse, currently, the emergence of multidrug-resistant pathogens such as “ESKAPEE” (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter species, and Escherichia coli) has triggered an urgent need for new and improved antimicrobial drugs (Boucher et al., 2009; Pendleton et al., 2013; Rice, 2008; Tacconelli et al., 2018).
Recent advances in high-throughput genome sequencing techniques and in silico genome mining tools have elucidated that actinomycetes, especially Streptomyces, possess a tremendous number of unexplored secondary metabolite biosynthetic gene clusters (smBGCs), indicating that the biosynthetic capability of Streptomyces has been underestimated (Craney et al., 2013; Lee et al., 2020b). For example, the genome mining of Streptomyces griseus, a well-known producer of the first aminoglycoside antibiotic streptomycin, identified 34 smBGCs in the genome, which include 28 putative smBGCs in addition to the previously characterized 6 smBGCs (Ohnishi et al., 2008). Considering that 1,110 Streptomyces strains possess approximately 40 smBGCs on average (Belknap et al., 2020) and that other actinomycete families such as Pseudonocardiales, Streptosporanqineae, Micromonosporaceae, and Corynebacteriales have 19.8, 15.0, 13.3, and 8.4 smBGCs per genome, respectively (Doroghazi et al., 2014), the genetic potential of actinomycetes has not been fully utilized because most of the smBGCs are apparently silent (cryptic) under laboratory pure culture conditions. Secondary metabolites are involved in inter- or intraspecies interactions in the natural habitat of the producer, but they are not essential for cell growth. Moreover, secondary metabolites are assembled by mega-enzyme complexes, the expression of which requires a large amount of energy and resources. Thus, the expression of smBGCs is inhibited until the action of specific environmental stimuli, such as microbial competition and physical stresses from the natural habitat.
To overcome this limitation, a variety of strategies have been developed and applied to activate the silent or poorly expressed smBGCs of actinomycetes. These approaches also provided useful information for understanding the regulatory mechanisms related to secondary metabolism. The culture-based method “OSMAC” (one strain many compounds) is one of the basic and simple ways to activate silent smBGCs (Bode et al., 2002). By changing culture conditions, including media composition (e.g., nutrient contents and chemical elicitors) (Chen et al., 2000; Kawai et al., 2007; Pettit, 2011; Tanaka et al., 2010) or physical parameters (e.g., temperature, pH, osmotic stress, and salinity) (Bode et al., 2002), a single strain can be induced to produce various molecules. Genetic engineering-based smBGC activation methods categorized into targeted (e.g., promoter exchange, heterologous expression, and cluster-situated regulator engineering) (Laureti et al., 2011; Luo et al., 2013; Zhang et al., 2017) and non-targeted (e.g., ribosome engineering and global regulator engineering) approaches are also widely used to induce substantial changes in the secondary metabolism of actinomycetes (Gao et al., 2012; Hosaka et al., 2009).
In addition to the aforementioned conventional strategies, coculture of different species is also effective in awakening silent smBGCs. Compared to the conventional strategies, coculture has the advantage of simplicity in that there is no need of prior knowledge of smBGCs or genetic engineering tools (Reen et al., 2015; Romano et al., 2018). Furthermore, coculture not only mimics ecological stresses like nutrient depletion during interspecies competition (Patin et al., 2018; van Bergeijk et al., 2020), but also enables real-time monitoring of secondary metabolite bioactivities toward the participants of coculture via the analysis of morphological changes or cell density (Wu et al., 2010). Under these conditions, several ideal combinations of the producer and partner (inducer) have been identified, which efficiently induce production of novel secondary metabolites, including antibiotics, antifungals, anticancers, and siderophores. However, owing to the chemical and molecular complexity of microbial interaction, the precise underlying mechanisms of the interaction are remarkably unexplored.
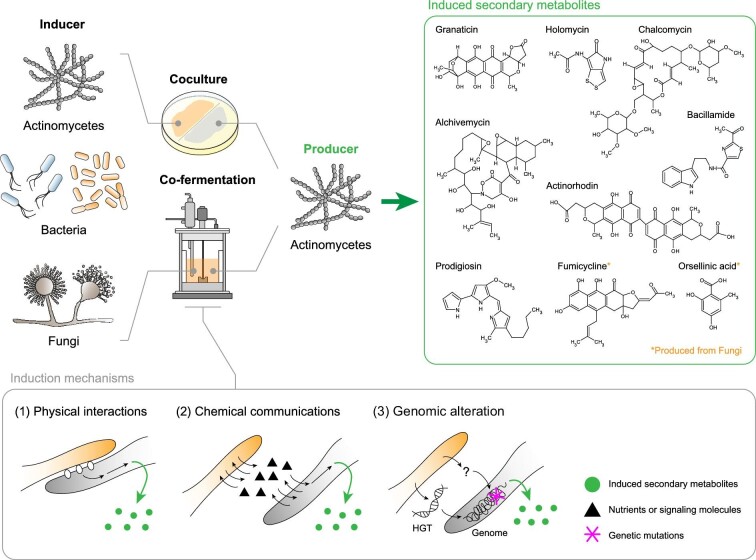
In this review, we briefly introduce the conventional strategies to awaken the silent smBGCs, and thereafter, focus on the coculture approach for unlocking the secondary metabolite production potential of actinomycetes. Coculture approaches are categorized into three sections depending on the coculture partners (Fig. 1): (i) actinomycetes–actinomycetes, (ii) actinomycetes–non-actinomycetes bacteria, and (iii) actinomycetes–fungi. The difference between bacteria and fungi as participants in coculture is presented from the perspective of induced secondary metabolites, bioactivity of secondary metabolites, and producer–inducer relationship. Finally, we highlight the future challenges of increasing the chemical diversity of actinomycetes using coculture.
Fig. 1.
Overview of expanding chemical diversity of actinomycetes via coculture. HGT, horizontal gene transfer.
Conventional Strategies for Awakening Silent smBGCs
Genome sequencing and genome mining approaches have revealed numerous potential smBGCs from actinomycetes. However, most of them are inactive under laboratory culture conditions and only subsets of these smBGCs are produced. To activate these silent smBGCs, various strategies have been developed that could be categorized into (i) culture-based strategies and (ii) genetic engineering-based strategies.
In the ecological habitat of secondary metabolite producers, biotic stresses (e.g., nutrient competition with nearby microbes) and abiotic stresses (e.g., acidity, drought, temperature, and salinity) are prevalent, which stimulate the production of various secondary metabolites (Cihak et al., 2017). In this respect, altering the culture conditions of actinomycetes is a simple and basic approach for unlocking cryptic smBGCs, which has been labeled the OSMAC approach (Bode et al., 2002). Secondary metabolite production is usually initiated when the cell growth slows down, indicating that exhaustion of a nutrient is a major key for awakening the silent smBGCs (Bibb, 2005). Therefore, changing nutrient regimes like carbon, nitrogen, sulfur, phosphorus, or trace element sources has been implemented for the secondary metabolite production from actinomycetes. Carbon source, in particular, is one of the main factors that controls secondary metabolite production (Sanchez et al., 2010). Rapidly used or preferred carbon sources, such as glucose, are known to repress the biosynthesis of various secondary metabolites in actinomycetes (i.e., carbon catabolite repression) (Bhatnagar et al., 1988; Sankaran & Pogell, 1975); thus, decreasing or altering the repressing carbon source could increase or induce inactivated secondary metabolite production. For example, actinorhodin production by Streptomyces lividans is inhibited when glucose is used as a carbon source, whereas inhibition is relieved when glucose is replaced with glycerol (Kim et al., 2001). In addition, modifying physical culture conditions, including temperature, salt concentration, or pH, also has a dramatic effect on the hierarchical regulatory network of actinomycetes and induces the production of novel secondary metabolites. For example, recently 18 types of thermotolerant actinomycetes were cultured between 30 and 45°C, and secondary metabolite production was compared. As a result, it was found that 131 secondary metabolites were produced when the actinomycetes were cultured at high temperature (Saito et al., 2020). Production of several secondary metabolites was induced in order to deal with the changed physical culture condition, as in the case of Nocardiopsis gilva YIM 90087 that accumulates ectoine and hydroxyectoine under salt stress conditions in order to regulate osmotic pressure (Han et al., 2018).
Genetic engineering-based strategies are promising for activating either (i) targeted or (ii) non-targeted smBGCs, if genome sequences and genetic manipulation tools for target actinomycetes are available. First, in the case of targeted smBGC activation, by altering genetic components, such as promoters of smBGC-encoded genes, expression of silent smBGCs could be stimulated. Recently, CRISPR/Cas9 systems have been applied to several Streptomyces species, enabling insertion of a strong and constitutive promoter in the upstream of the core biosynthetic genes or positive regulatory genes encoded in the target smBGC (Cobb et al., 2015; Huang et al., 2015; Zhang et al., 2017). For example, activation of pentangular type II polyketide BGC of Streptomyces viridochromogenes via CRISPR/Cas9-mediated promoter exchange of the main biosynthetic operon resulted in the production of a novel pigmented compound (Zhang et al., 2017). Meanwhile, targeted smBGC awakening in native hosts is often hampered by endogenous complex regulatory systems; thus, in many cases, smBGCs of interest are expressed in heterologous hosts to bypass the original regulatory systems. For instance, a PKS–NRPS-type BGC of S. griseus containing nine domains of biosynthetic mega-enzyme was reconstructed and heterologously expressed in S. lividans, resulting in the production of three novel tetramic acid-containing macrolactams (Luo et al., 2013). Additionally, non-targeted smBGC activation relies on reshaping the global transcriptome or translatome via genetic engineering, followed by analyzing the change in produced secondary metabolite pools. A representative method involves altering the expression of pleiotropic transcriptional regulators. For instance, overexpression of cyclic AMP receptor protein (Crp), which is a transcription regulator involved in diverse cellular processes, enhanced secondary metabolite production ability of various Streptomyces species, including S. coelicolor (Gao et al., 2012). In addition, introducing mutations in RNA polymerase or ribosomal proteins to change transcriptional or translational activity, respectively, led 66 strains out of 353 soil-isolated actinomycetes to acquire an antibacterial-producing ability (Hosaka et al., 2009).
Coculture of Actinomycetes
Coculture is another effective culture-based strategy for discovering novel bioactive secondary metabolites from microorganisms by mimicking the environmental habitat where microbes continuously interact with nearby residents. It is defined as “coculture or co-cultivation” when performed on solid media, such as Petri dishes or a solid support system, and called “mixed fermentation” when performed in liquid media, such as co-fermentation, transwell, microfluidic, or droplet culture systems (Tan et al., 2019). Compared to conventional strategies, coculture offers complex and unpredictable stimuli over the sole nutrient or physical condition changes, allowing microbes to produce various novel secondary metabolites, which are not observed in pure culture conditions (Abdelmohsen et al., 2015). Also, coculture enables the real-time bioactivity screening of newly induced secondary metabolites when producers are cocultured with target pathogens. Furthermore, the coculture method is beneficial not only for awakening novel secondary metabolites but also for comprehending microbial interactions related to complex regulations of secondary metabolite production. In this context, coculture methods have been intensively applied to bacteria and fungi, especially to actinomycetes (Abdelmohsen et al., 2015; Yu et al., 2019). In this section, various actinomycete coculture studies are classified into three categories, depending on the type of coculture partner, as follows: (i) actinomycetes–actinomycetes, (ii) actinomycetes–non-actinomycetes bacteria, and (iii) actinomycetes–fungi.
Actinomycetes Coculture With Actinomycetes
Streptomyces coculture with Streptomyces
More than 3,000 species of Streptomyces reside together in their ecological habitats and numerous interspecies interactions exist within them (Christova et al., 1995); therefore, many attempts have been made to coculture different Streptomyces species to expand the chemical diversity of Streptomyces (Table 1). For example, coculture of 76 Streptomyces species revealed that production of various antibiotics or sporulation was induced in 72 combinations (Ueda et al., 2000). Interspecies interaction mediated by diffusible substrates (e.g., γ-butyrolactones [GBLs] and secondary metabolites themselves) is regarded as a general factor triggering the secondary metabolism during Streptomyces–Streptomyces coculture. Especially, GBLs (e.g., A-factor, virginiae butanolides, and IM-2) are well-known and widely distributed signaling molecules involved in communications of Streptomyces species (Niu et al., 2016). GBLs produced from various Streptomyces species including S. viridochromogenes, S. bikiniensis, and S. cyaneofuscatus induced antibiotic production, cellular differentiation, and aerial mycelium formation of S. griseus, as A-factor, the GBL of S. griseus, did (Grafe et al., 1983; Hara & Beppu, 1982; Horinouchi & Beppu, 1992; Khokhlov et al., 1973; Yamada et al., 1987).
Table 1.
Actinomycetes and Actinomycetes Coculture
| Producer | Inducer | Induced compounds and bioactivity | Category (producer–inducer) | References |
|---|---|---|---|---|
| Streptomyces 76 species | Streptomyces 76 species | Various antibiotics from 72 combinations | S–S | Ueda et al. (2000) |
| Streptomyces 33 isolates | Streptomyces 33 isolates | Various antibiotics from 31 combinations | S–S | Ueda et al. (2000) |
| Streptomyces griseus st-21-2 | Streptomyces tanashiensis IAM0016 | Desferrioxamine E (siderophore) | S–S | Yamanaka et al. (2005) |
| Streptomyces tanashiensis IAM0016 | Streptomyces griseus st-21-2 | Unknown antibiotics | S–S | Yamanaka et al. (2005) |
| Streptomyces coelicolor M145 | Streptomyces sp. E14 | Four acyl-desferrioxamine derivatives (siderophore) | S–S | Traxler et al. (2013) |
| Streptomyces coelicolor M145 | Streptomyces sp. SPB74 | γ-Actinorhodin (antibiotics)Prodiginine (antibiotics)Acyl-desferrioxamines (siderophore) | S–S | Traxler et al. (2013) |
| Streptomyces coelicolor M145 | Streptomyces viridochromogenes | Actinorhodin (antibiotics)Prodiginine (antibiotics)Coelichelin (siderophore) Desferrioxamine E (siderophore) | S–S | Traxler et al. (2013) |
| Streptomyces coelicolor M145 | Streptomyces albus J1074 | Actinorhodin (antibiotics)Prodiginine (antibiotics) Desferrioxamines B and E (siderophore) | S–S | Traxler et al. (2013) |
| Streptomyces strains 574, 001, 023, and 555 | Streptomyces strain 153 | Unknown antibiotics | S–S | Amano et al. (2010) |
| Streptomyces lividans TK-23 | Tsukamurella pulmonis TP-B0596Rhodococcus sp.Corynebacterium sp.Nocardia sp.Dietzia sp.Gordonia sp.Mycobacterium sp.Williamsia sp. | Actinorhodin (antibiotics)Undecylprodigiosin (antibiotics) | S– MACB | Onaka et al. (2011) |
| Streptomyces endus S-522 | Tsukamurella pulmonis TP-B0596Corynebacterium glutamicum | Alchivemycins A and B (antibiotics) | S–MACB | Onaka et al. (2011) |
| Streptomyces cinnamoneus NBRC 13823 | Tsukamurella pulmonis TP-B0596 | BE-13793C (cytotoxicity)Arcyriaflavin E (cytotoxicity)Arcyriaflavin A | S–MACB | Hoshino et al. (2015c) |
| Streptomyces sp. CJ-5 | Tsukamurella pulmonis TP-B0596 | Chojalactones A–C (cytotoxicity) | S–MACB | Hoshino et al. (2015b) |
| Streptomyces sp. NZ-6 | Tsukamurella pulmonis TP-B0596 | Niizalactams A–C | S–MACB | Hoshino et al. (2015a) |
| Streptomyces lividans TK-23 | Tsukamurella pulmonis | Prodiginine (antibiotics) | S–MACB | Onaka et al. (2015) |
| Streptomyces nigrescens HEK616 | Tsukamurella pulmonis TP-B0596Corynebacterium glutamicum | 5-Alkyl-1,2,3,4-tetrahydroquinoline (antifungal) | S–MACB | Sugiyama et al. (2015) |
| Streptomyces nigrescens HEK616 | Tsukamurella pulmonis TP-B0596 | Streptoaminals (antifungal) | S–MACB | Sugiyama et al. (2016) |
| Streptomyces tendae KMC006 | Gordonia sp. KMC005 | Gordonic acid (antibiotics) | S–MACB | Park et al. (2017) |
| Nocardiopsis sp. FU40 | Rhodococcus wratislaviensis | Ciromicins A and B (cytotoxicity) | A–MACB | Derewacz et al. (2015) |
| 12 Micromonosporaceae | Mycobacterium sp. WMMA-183Rhodococcus sp. WMMA-185 | Unknown antibiotics | A–MACB | Adnani et al. (2015) |
| Micromonospora sp. WMMB235 | Rhodococcus sp. WMMA185 | Keyicin (antibiotics) | A–MACB | Adnani et al. (2017) |
| Micromonospora wenchangensis HEK797 | Tsukamurella pulmonis TP-B0596 | Dracolactams A and B | A–MACB | Hoshino et al. (2017) |
| Catenuloplanes sp. RD067331 | Tsukamurella pulmonis TP-B0596 | Catenulobactin ACatenulobactin B (siderophore and cytotoxicity) | A–MACB | Hoshino et al. (2018a) |
| Actinosynnema mirum NBRC 14064 | Tsukamurella pulmonis TP-B0596 | Mirilactams C–E | A–MACB | Hoshino et al. (2018b) |
| Pseudonocardiales Umezawaea sp. RD066910 | Tsukamurella pulmonis TP-B0596 | Umezawamides A and B (cytotoxicity) | A–MACB | Hoshino et al. (2018c) |
| Nocardiopsis sp. RV163 (producer unknown) | Actinokineospora sp. EG49 | N-(2-Hydroxyphenyl)-acetamide1,6-Dihydroxyphenazine5a,6,11a,12-Tetrahydro-5a,11a-dimethyl[1,4]benzoxazino[3,2b][1,4]benzoxazine (antibiotics and antitrypanosomal) | A–A | Dashti et al. (2014) |
| Streptomyces coelicolor M145 | Amycolatopsis sp. AA4 | γ-Actinorhodin (antibiotics)Prodiginine (antibiotics)Four acyl-desferrioxamine derivatives (siderophore)Amychelin from S. coelicolor (siderophore) | S–A | Traxler et al. (2012, 2013) |
| Rhodococcus fascians 307CO | Streptomyces padanus | Rhodostreptomycins A and B (antibiotics) | MACB–S | Kurosawa et al. (2008) |
S: Streptomyces; MACB: mycolic acid-containing bacteria; A: actinomycetes.
Secondary metabolites themselves also play crucial role in promoting production of various secondary metabolites between Streptomyces–Streptomyces interactions. Among the secondary metabolites, iron-chelating compound, siderophore, is a type of secondary metabolite that stimulates secondary metabolism, such as antibiotic production or development of another nearby species (Challis & Hopwood, 2003). Desferrioxamine E, which is a siderophore produced by S. griseus, stimulated growth and antibiotic production of Streptomyces tanashiensis (Yamanaka et al., 2005). In addition, siderophores made by four different Streptomyces species and Amycolatopsis sp. AA4 induced production of γ-actinorhodin, prodiginine, or 12 different desferrioxamines from S. coelicolor (Traxler et al., 2013). While iron competition with neighboring strains is suspected to be the reason for increased secondary metabolite production of S. coelicolor, the underlying mechanism inducing the other secondary metabolites remains to be elucidated. Meanwhile, non-siderophore secondary metabolites were also involved in Streptomyces interspecies communications. For example, polyether antibiotic promomycin, produced by Streptomyces stain 153, induced the production of unknown antibiotics from other Streptomyces species. Polyether antibiotics act as ionophore, which increases K+ ion efflux through cell membrane by forming pores; thus, it is supposed to inhibit bacterial growth and induce the production of antibiotics. Indeed, other polyether antibiotics including salinomycin, monensin, and nigericin all promoted the antibiotic production of Streptomyces strain 574 (Amano et al., 2010). Taken together, Streptomyces–Streptomyces coculture examples pointed out that signaling molecules involved in interspecies interactions between Streptomyces species triggered production of cryptic secondary metabolites and other interaction-mediating chemicals have the potential to be utilized as cues for increasing the chemical diversity of Streptomyces.
Streptomyces coculture with non-Streptomyces actinomycetes
In addition to Streptomyces–Streptomyces coculture, intergenus interactions between Streptomyces and non-Streptomyces actinomycetes have also been exploited to activate dormant smBGCs of Streptomyces (Table 1). Coculturing S. lividans with 400 different bacteria discovered that Tsukamurella pulmonis, a rare actinomycete, is an effective coculture partner that activated prodiginine production by S. lividans (Onaka et al., 2011). In addition, several Tsukamurella-related actinomycetes such as Rhodococcus, Corynebacterium, Nocardia, Dietzia, Gordonia, Mycobacterium, and Williamsia showed the same effect (Onaka et al., 2011). Common characteristic of these close actinomycetes is the presence of mycolic acid in the outer layer of the cells, so they are called mycolic acid-containing bacteria (MACB). These MACB have been widely cocultured with various Streptomyces species, and consequently induced production of numerous secondary metabolites with a variety of bioactivity including antibacterial (e.g., alchivemycin, prodiginine, streptoaminals, and gordonic acid) (Onaka et al., 2011, 2015; Park et al., 2017; Sugiyama et al., 2016), antifungal (e.g., 5a-THQ and streptoaminals) (Sugiyama et al., 2015, 2016), and cytotoxic (e.g., BE-13793C, arcyriaflavin E, and chojalactones A–C) (Hoshino et al., 2015b). Coculturing MACB with non-Streptomyces actinomycetes also successfully awakened several cryptic smBGCs. For example, Mycobacterium sp. and Rhodococcus sp. induced production of several secondary metabolites from 12 out of 65 marine invertebrate-associated Micromonosporaceae (Adnani et al., 2015).
However, the underlying mechanism of MACB coculture is still ambiguous. Most of MACB coculture studies argued that physical cell-to-cell contact between actinomycetes and live MACB cells is required for inducing secondary metabolite production of actinomycetes, because both MACB culture extract treatment and dead MACB cell coculture were not able to induce secondary metabolite production (Onaka et al., 2011). On the contrary, keyicin production of Micromonospora sp. WMMB235 was still observed when only the chemical substance from MACB was treated, indicating that physical contact is not required (Adnani et al., 2017). Moreover, there is a report that horizontal gene transfer between MACB and actinomycetes induces the production of a novel secondary metabolite called rhodostreptomycin, although MACB is the producer and its partner actinomycete is the inducer in this case (Kurosawa et al., 2008). Overall, silent or poorly expressed smBGCs of actinomycetes could be induced by coculture between actinomycetes (Table 1). Interspecific signaling molecules between Streptomyces species, including siderophore, and intergenus communications between MACB and actinomycetes triggered production of numerous bioactive compounds. Further mechanical studies on the microbial interactions that trigger the secondary metabolism will provide valuable information to understand the regulatory network of secondary metabolism and to increase the chemical diversity of actinomycetes.
Actinomycetes Coculture With Non-Actinomycetes Bacteria
Actinomycetes coculture with predatory bacteria
As actinomycetes dwell in various habitats with diverse species, they have long evolved while interacting with many coexisting bacteria (Baltz, 2008; Jose & Jebakumar, 2012; Quillet et al., 1995). Among these bacteria, several predatory groups, which feed on nearby bacterial cells in the environmental habitat, are attractive coculture partners to stimulate protective response of the actinomycetes. For example, when motile predator bacteria Myxococcus xanthus was cocultured with S. coelicolor, M. xanthus secreted lytic enzymes, which triggered abnormal hyphae formation of S. coelicolor, and S. coelicolor produced actinorhodin to repel the intrusion of the M. xanthus (Perez et al., 2011). Although other bacteria, including several Bacillus species (B. megaterium, B. subtilis, and B. thuringiensis) and Serratia sp., slightly induced the production of actinorhodin from S. coelicolor, M. xanthus was the strongest inducer, representing the potential of predatory bacteria as coculture partner (Perez et al., 2011) (Table 2).
Table 2.
Actinomycetes and Non-Actinomycetes Bacteria
| Producer | Inducer | Induced compounds and bioactivity | Category (producer–inducer) | References |
|---|---|---|---|---|
| Streptomyces coelicolor M145 | Myxococcus xanthusBacillus megateriumBacillus subtilisBacillus thuringiensisSerratia sp. | Actinorhodin (antibiotics) | S–PRB | Perez et al. (2011) |
| Streptomyces coelicolor M145 | Myxococcus xanthus | Actinorhodin (antibiotics)Myxochelin from M. xanthus (siderophore) | S–PRB | Lee et al. (2020a) |
| Streptomyces sp. PTY087I2 | Methicillin-sensitive Staphylococcus aureus Methicillin-resistant Staphylococcus aureusPseudomonas aeruginosa | Granatomycin D (antibiotics)Granaticin (antibiotics)Dihydrogranaticin B | S–PAB | Sung et al. (2017) |
| Streptomyces albogriseolus B24 | Bacillus cereus | Dentigerumycin E (anticancer) | S–PAB | Shin et al. (2018) |
| Streptomyces clavuligerus (adapted by ALE) | Staphylococcus aureus N315 | Holomycin (antibiotics) | S–PAB | Charusanti et al. (2012) |
| Streptomyces coelicolor M145 | Staphylococcus aureus (heat-killed cell) | Undecylprodigiosin (antibiotics, immunosuppressive, and anticancer) | S–PAB | Luti and Mavituna (2011) |
| Streptomyces sp. PTY087I2 | Bacillus subtilis | Granatomycin D (antibiotics)Granaticin (antibiotics)Dihydrogranaticin B | S–NB | Sung et al. (2017) |
| Streptomyces coelicolor | Bacillus subtilis (bacillaene-deficient) | Undecylprodigiosin (antibiotics, immunosuppressive, and anticancer) | S–NB | Straight et al. (2007) |
| Streptomyces coelicolor M145 | Bacillus subtilis (heat-killed cell) | Undecylprodigiosin (antibiotics, immunosuppressive, and anticancer) | S–NB | Luti and Mavituna (2011) |
| Streptomyces sp. Mg1 | Bacillus subtilis 3610 | Chalcomycin A (antibiotics) | S–NB | Barger et al. (2012) |
| Streptomyces lividans | Bacillus subtilis (bacillaene pks operon deletion) | Undecylprodigiosin (antibiotics, immunosuppressive, and anticancer) | S–NB | Vargas-Bautista et al. (2014) |
| Streptomyces sp. | Bacillus mycoides | Bacillamides A–C (algicidal)N-Acetyltryptamine (algicidal)N-Propanoyltryptamine (algicidal) | S–NB | Yu et al. (2015) |
| Streptomyces coelicolor M145 | Escherichia coli | Undecylprodigiosin (antibiotics, immunosuppressive, and anticancer) | S–NB | Mavituna et al. (2016) |
| Streptomyces coelicolor M145 | Corallococcus coralloides B035 | Undecylprodigiosin (antibiotics, immunosuppressive, and anticancer) | S–NB | Schaberle et al. (2014) |
| Streptomyces venezuelae (methyltransferase from S. avermitilis) | Engineered Escherichia coli | O-Methylated phenylpropanoids (antibiotics)Multimethylated phenylpropanoids (antibiotics) | S–NB | Cui et al. (2019) |
| Streptomyces griseorubiginosus 43708 | Pseudomonas maltophilia 1928 | Biphenomycins A and C (antibiotics) | S–NB | Ezaki et al. (1992, 1993) |
| Streptomyces tenjamariensis SS-939 ATCC31603 | 12 unidentified bacteria | Istamycins A and B (antibiotics) | S–NB | Slattery et al. (2001) |
| Streptomyces sp. B033 | Brucella neotomae ATCC 23459Burkholderia vietnamiensis ATCC BAA-248Yersinia pestis A1122Xanthomonas axonopodis ATCC 8718 | Resistomycin (antibiotics) | S–NB | Carlson et al. (2015) |
| Streptomyces cinnabarinus PK209 | Alteromonas sp. KNS-16 | Lobocompactol (antifouling, antioxidant, and anticancer) | S–NB | Cho and Kim (2012) |
S: Streptomyces; PRB: predatory bacteria; PAB: pathogenic bacteria; NB: non-actinomycetes bacteria.
Recently, transcriptome analysis on both M. xanthus and S. coelicolor during coculture revealed that iron competition between them, not physical contact, triggered actinorhodin production of S. coelicolor (Lee et al., 2020a). During coculture, S. coelicolor actively absorbed the extracellular iron, causing M. xanthus to face an iron-reduced environment. To respond to the iron-depletion condition, M. xanthus upregulated biosynthesis of siderophore, myxochelin, and myxochelin-mediated iron uptake systems, leading M. xanthus to dominate iron scavenging. Consequently, S. coelicolor experienced an iron-restricted condition and activated actinorhodin production along with upregulating branched amino acid catabolism, which implies the potential to produce precursors of actinorhodin. Based on these results, seven Streptomyces species (i.e., S. subrutilus, S. kanamyceticus, S. coeruleorubidus, S. cinereoruber, S. roseosporus, S. rimosus, and S. venezuelae) were cultured in iron-restricted conditions, resulting in upregulation of 21 smBGCs out of a total of 260 smBGCs in seven species’ genomes. Among secondary metabolites expected to be produced from upregulated smBGCs, several secondary metabolites, including actinorhodin, cosmomycin D, and chloramphenicol, possess putative iron-interacting sites, implying that these secondary metabolites might have both antibiotic and iron-chelating functions, which would be highly advantageous during iron competition with nearby microbes (Lee et al., 2020a).
Actinomycetes coculture with pathogenic bacteria
Human pathogenic bacteria such as Staphylococcus aureus have been tried to coculture with actinomycetes due to the advantage in real-time screening of induced secondary metabolites’ bioactivity against pathogenic bacteria (Table 2). For example, marine Streptomyces sp. PTY08712 was isolated from a complex tunicate community and cocultured with antibiotic-resistant human pathogens, including methicillin-sensitive S. aureus, methicillin-resistant S. aureus (MRSA), and P. aeruginosa. As a result, coculture extracts showed increased bioactivity against human pathogens, except P. aeruginosa, which results from enhanced production of three secondary metabolites: granatomycin D (antibacterial), granaticin (strong antibacterial), and dihydrogranaticin B (not known) (Sung et al., 2017). As with coculturing actinomycetes with stressors, coculturing with antibiotic-resistant pathogens could stimulate production of novel secondary metabolites effective to them. So far, why and how actinomycetes produce bioactive compounds against pathogenic bacteria have not been fully revealed, but from the example of S. coelicolor cultured with heat-killed pathogenic bacteria S. aureus, antibiotic producer S. coelicolor might recognize some proteins like receptors on the surface of pathogenic bacteria via physical cell-to-cell contact (Luti & Mavituna, 2011). Still, further revelation of mechanism is needed for elucidation of novel bioactive secondary metabolites from actinomycetes–pathogenic bacteria coculture.
Sometimes, actinomycetes require long-term microbial interaction to acquire the ability to produce antibiotics against nearby microbes. Continuous adaptive laboratory evolution (ALE) of Streptomyces clavuligerus implementing coculture with MRSA as a driving force resulted in S. clavuligerus acquiring the ability to constitutively produce a pyrrothine class of antibiotic, holomycin, which inhibits growth of MRSA. Competition between the two microbes led to genomic mutations of S. clavuligerus, including loss of megaplasmid and five single-nucleotide polymorphisms, which might affect the secondary metabolism (Charusanti et al., 2012). These results indicate that long-term coculture can activate silent smBGC by inducing genetic mutations, which keep silent under short-term coculture.
Actinomycetes coculture with other bacteria
Well-characterized model bacteria such as B. subtilis and E. coli have also been utilized as coculture partner with actinomycetes. For example, when Streptomyces sp. Mg1 was cocultured with competitor B. subtilis, chalcomycin A, which inhibits growth and even lyses B. subtilis, was produced by Streptomyces sp. Mg1 (Barger et al., 2012). Also, both live and heat-killed B. subtilis activated the undecylprodigiosin production of S. coelicolor and S. lividans (Luti & Mavituna, 2011; Straight et al., 2007; Vargas-Bautista et al., 2014). In addition, coculturing other Bacillus species such as B. mycoides and B. cereus with Streptomyces species activated production of bioactive secondary metabolites including algicides (e.g., bacillamide and tryptamines) (Yu et al., 2015) and antibiotics (e.g., dentigerumycin E), which are protective against B. subtilis (Shin et al., 2018). Meanwhile, in case of E. coli, when S. coelicolor was cocultured with live E. coli cell, undecylprodigiosin production of S. coelicolor was 3.5-fold increased, whereas actinorhodin production was 15-fold decreased. This secondary metabolism change was proven to be induced from the chemical compound in cell-free supernatant of E. coli (Mavituna et al., 2016).
Taken together, a broad range of bacteria has been utilized to trigger the production of numerous secondary metabolites from actinomycetes (Table 2). Predatory microbes and competitive participants, including human pathogens and model bacteria, have been cocultured with actinomycetes to induce production of defensive or inhibitory secondary metabolites, which have the potential for the development of antibiotics. Nevertheless, many of underlying principles of secondary metabolite production have not been clearly elucidated, which hinders further understanding of communications between actinomycetes and bacteria.
Actinomycetes Cocultured With Fungi
Actinomycetes as a producer
Fungal species have been revealed to possess about 50 cryptic smBGCs per genome like actinomycetes (Nierman et al., 2005; Pel et al., 2007; Wortman et al., 2009) and many fungi coexist with actinomycetes in various ecological habitats, implying inter-kingdom interactions between them are commonly present (Frey-Klett et al., 2011; Hibbing et al., 2010; Kroiss et al., 2010). Indeed, interaction between actinomycetes and fungi activated the secondary metabolism of actinomycetes (Table 3). For example, when S. lividans was cocultured with Verticillium dahlia, the production of the antibiotic undecylprodigiosin was upregulated. Undecylprodigiosin strongly reduced the microsclerotia formation of V. dahlia, possibly by interfering with the signal transduction pathway (Meschke et al., 2012). Another example is that coculturing Streptomyces leeuwenhoekii C58 with Aspergillus fumigatus MR2012 in various culture media induced the production of nocardamine, pentalenic acid, and chaxapeptin by S. leeuwenhoekii C58, but none of these metabolites were proved to have antifungal bioactivity (Wakefield et al., 2017).
Table 3.
Actinomycetes and Fungi Coculture
| Producer | Inducer | Induced compounds and bioactivity | Category (producer–inducer) | References |
|---|---|---|---|---|
| Streptomyces lividans | Verticillium dahliae | Undecylprodigiosin (antifungal) | S–F | Meschke et al. (2012) |
| Streptomyces leeuwenhoekii C58 | Aspergillus fumigatus MR2012 | Chaxapeptin | S–F | Wakefield et al. (2017) |
| Streptomyces rochei MB037 | Rhinocladiella similis 35 | Borrelidin J (antibiotics)Borrelidins K and F 7-Methoxy-2,3-dimethylchromone-4-one | S–F | Yu et al. (2019) |
| Aspergillus nidulans | Streptomyces rapamycinicus | Orsellinic acidLecanoric acid (inhibit ATP synthesis)Cathepsin K inhibitors F-9775A and F-9775B (antiosteoporosis) | F–S | Fischer et al. (2018), Nutzmann et al. (2011), and Schroeckh et al. (2009) |
| Aspergillus fumigatus | Streptomyces rapamycinicus | Fumicyclines A and B (antibiotics) | F–S | Konig et al. (2013) |
| Aspergillus fumigatus | Streptomyces rapamycinicusStreptomyces iranensisStreptomyces coelicolorStreptomyces lividans | Fumigermin (antibiotics) | F–S | Stroe et al. (2020) |
| Aspergillus fumigatus | Streptomyces peucetius ATCC 29050 | FumiformamideN,N′-[(1Z,3Z)-1,4-Bis(4-methoxyphenyl)buta-1,3-diene-2,3-diyl]diformamide (cytotoxicity)N-Formyl derivatives (cytotoxicity)BU-4704Xanthocillin X monoetherXanthocillin X dietherXanthocillin dimethyl etherXanthoascin | F–S | Zuck et al. (2011) |
| Aspergillus fumigatus MBC-F1-10 | Streptomyces bullii | ErgosterolBrevianamide F (cytotoxicity)Spirotryprostatin A (antibiotics)6-Methoxyspirotryprostatin B (leishmanicidal and cytotoxicity)Fumitremorgin C (antitrypanosomal and leishmanicidal)12,13-Dihydroxyfumitremorgin C (antitrypanosomal and leishmanicidal)Fumitremorgin B (antitrypanosomal and leishmanicidal)Verruculogen (antitrypanosomal and leishmanicidal)11-O-Methylpseurotin A (cytotoxicity)11-O-Methylpseurotin A2 (antitrypanosomal and leishmanicidal)Emestrins A and B (induced by quorum-sensing molecule) | F–S | Rateb et al. (2013) |
| Aspergillus fumigatus MR2012 | Streptomyces leeuwenhoekii C34 | Luteoride DPseurotin GTerezine D11-O-Methylpseurotin A Chaxapeptin | F–S | Wakefield et al. (2017) |
| Aspergillus niger | Streptomyces coelicolor | cyclo-(Phe–Phe)cyclo-(Phe–Tyr)Phenylacetic acid2-HydroxyphenylaceticFuran-2-carboxylic acid | F–S | Wu et al. (2015) |
| Aspergillus flavipes | Streptomyces sp. | RosellichalasinAspochalasins E, P, H, and M19,20-Dihydro-aspochalasin D | F–S | Yu et al. (2016) |
| Aspergillus austroafricanus | Streptomyces lividans | AustramideViolaceols I and II (antibiotics)Diorcinol (antibiotics) | F–S | Ebrahim et al. (2016) |
| Aspergillus sp. | Streptomyces sp. | Heronapyrrole B from Streptomyces (antifungal)Debromomarinonecyclo-(l-Phe-trans-4-hydroxy-l-Pro) from Aspergillus (antibiotics) | F–S | Khalil et al. (2019) |
| Fusarium tricinctum | Streptomyces lividans | Fusatricinones A–DDihydrolateropyroneLateropyrone (antibiotics)Zearalenone(−)-CitreoisocoumarinMacrocarpon C7-Hydroxy-2-(2-hydroxypropyl)-5-methylchromoneDepsipeptide enniatins A1, B, and B1 (antibiotics)Lipopeptide fusaristatin A | F–S | Moussa et al. (2019) |
| Bionectria sp. | Streptomyces lividans TK24 | Bionectriamines A and BTris(2,4-di-tert-butylphenyl) phosphate6,8-Dihydroxyisocoumarin-3-carboxylic acid | F–S | Kamdem et al. (2018) |
| Penicillium sp. WC-29-5 | Streptomyces fradiae 007 | DeoxyfuniconeAlternariolVermistatin(9R,14S)-Epoxy-11-deoxyfunicone (cytotoxicity)(9S,14R)-Epoxy-11-deoxyfunicone (cytotoxicity) | F–S | Wang et al. (2014) |
| Heterobasidion abietinum 331 | Streptomyces AcH 505 | 5-Formylsalicylic acid (virulence factor and siderophore) | F–S | Keilhofer et al. (2018) |
| Emericella sp. CNL-878 | Salinispora arenicola sp. CNH-665 | Emericellamides A and B (anticancer) | F–S | Oh et al. (2007) |
S: Streptomyces; F: fungi.
Fungi as a producer
Unlike the above examples, in most cases of actinomycetes–fungi interactions, fungi, especially Aspergillus species, act as producers while actinomycetes induce the secondary metabolism of fungi (Table 3). Coculturing Aspergillus nidulans with a collection of 58 soil-dwelling actinomycetes is a representative example of activating silent fungal smBGCs by coculture with actinomycetes. As a result, four secondary metabolites (orsellinic acid [OA], lecanoric acid [LA], F-9775A, and F-9775B) were produced from A. nidulans only when cocultured with Streptomyces hygroscopicus (renamed as Streptomyces rapamycinicus). Interestingly, further analysis revealed that physical interaction between A. nidulans and S. rapamycinicus is required for inducing the secondary metabolism of A. nidulans (Schroeckh et al., 2009). It was discovered that physical contact between the two organisms triggered histone acetylation of the OA-encoding ors gene of A. nidulans by histone acetyltransferase Saga/Ada complex, ultimately inducing OA and LA production (Nutzmann et al., 2011). The latest study elucidated that transcriptional factor BasR acts as a central “node” for linking external signals from physical interaction with actinomycetes and secondary metabolic regulation, including OA production (Fischer et al., 2018). In addition, the fungal species A. fumigatus was cocultured with the inducer S. rapamycinicus, resulting in the production of fungal secondary metabolites fumicyclines A and B and fumigermin (Konig et al., 2013; Stroe et al., 2020). In the case of fumicyclines A and B, the same principle of histone modifications was working when S. rapamycinicus was cocultured with A. fumigatus, whereas in the case of fumigermin, it was not clarified whether elicitation was via histone modification or not (Konig et al., 2013; Stroe et al., 2020). As for the bioactivity of induced compounds, fumicyclines A and B showed antibacterial effect to S. rapamycinicus and fumigermin inhibited germination of S. rapamycinicus, indicating that compounds induced during coculture with S. rapamycinicus are considered as fungal defensive systems.
We categorized actinomycetes–fungi coculture into two sections: (i) actinomycetes as a producer and (ii) fungi as a producer. Despite this, chemical and physical interactions between the two kingdoms often cause complex metabolic shifts of both organisms to produce various secondary metabolites as a defensive response (Table 3). Considering that most of the aforementioned cases have been focused on analyzing a few induced secondary metabolites, it is expected that there may have been more diverse alterations in secondary metabolite production than reported. For example, Aspergillus sp. CMB-StM0423 produces a bacteriostatic compound, diketopiperazine, when cocultured with Streptomyces sp. CMB-StM0423 (Khalil et al., 2019). Actually, diketopiperazines are common secondary metabolites and are known to be overproduced by Aspergillus when cocultured with Streptomyces (Wakefield et al., 2017; Wu et al., 2015). Transcriptome analysis revealed that diketopiperazine stimulated Streptomyces to repress nitric oxide (NO) dioxygenase, which reduced the level of NO gas in the cell, resulting in a high intracellular concentration of NO gas. As a result, a high concentration of NO-activated novel smBGCs and antifungal compound, heronapyrrole B, was produced by Streptomyces (Khalil et al., 2019). In addition, when marine-derived Streptomyces rochei MB037 was cultured with the fungi Rhinocladiella similis, two novel antibacterial borrelidins, J and K, were produced by S. rochei and one antibacterial chromone was produced by R. similis (Yu et al., 2019).
Overall, actinomycetes act as both inducer and producer when cocultured with various fungal species (Table 3). In some instances, actinomycetes trigger epigenetic modification of fungi, resulting in complex secondary metabolism changes, and sometimes fungi produce certain secondary metabolites, which alter the secondary metabolism of actinomycetes. The interaction between fungi and actinomycetes is mainly attack and defense, so if coculturing pathogenic actinomycetes or pathogenic fungi, it seems likely novel secondary metabolites that can kill each other will be discovered.
Conclusion
To date, numerous bioactive secondary metabolites have been elicited through coculture of actinomycetes with various bacteria or fungi. Coculture provides complex stimuli, which dramatically affect secondary metabolism of actinomycetes, and allows the real-time bioactivity screening of newly induced secondary metabolites; thus, it is highly advantageous to the discovery of novel bioactive secondary metabolites with triggering mechanisms. However, the coculture method is often irreproducible and inappropriate for large-scale culture to produce target secondary metabolites abundantly. Yet, the secondary metabolite induction stimuli elucidated from coculture study can be exploited in industrial applications for secondary metabolite production by single culture. Thus, a precise and comprehensive understanding of the underlying coculture mechanism is a top priority (Lee et al., 2020a).
After examining the previous reports in an effort to discover the underlying principles of coculture, induction mechanisms can be categorized into three scenarios (Fig. 1): (i) physical interactions, (ii) chemical communications (e.g., nutrient competition and quorum sensing), and (iii) genomic alteration (e.g., horizontal gene transfer and genomic mutation by ALE). However, still only a few in-depth studies about the genetic regulatory network linked with those inducing signals. For example, physical cell-to-cell interactions between fungi and Streptomyces triggered chromosome acetylation of fungi, which implies not just physical interaction itself but also a further underlying mechanism to bring out the secondary metabolism changes (Nutzmann et al., 2011). In recent years, various tools have been developed and applied for elucidating these inducing mechanisms during coculture. In particular, transcriptomic analysis enables the examination of the genetic responses of each coculture participant. Functional analysis of differently expressed genes during coculture allows tracing the triggering factors and responses of producer and inducer. In addition, comparative proteomic and metabolic analysis between axenic culture and coculture enables the clarification of the dynamics of proteins and molecules related to secondary metabolism. Multi-omics technology-based mechanical studies on the coculture will improve our understanding of the secondary metabolic regulation of actinomycetes.
Moreover, previous cocultures of actinomycetes were limited in range of culture partner, which may have restricted the range of secondary metabolism involved; therefore, coculture with more diverse partners, such as amoeba or phages, is needed (Klapper et al., 2016; Kronheim et al., 2018). For instance, coculturing actinomycetes with double-stranded DNA phages unveiled a secondary metabolism of Streptomyces involved in defense against phage infection (Kronheim et al., 2018). Accumulation of diverse microbial coculture studies will help us to understand the relationship between coculture conditions (e.g., coculture partner, culture media, and culture type) and type of induced secondary metabolites. Indeed, recent comprehensive analysis demonstrating the induction of 259 compounds via coculture revealed that production of “linear polyketides, oxylipins, and fatty acids” and “cyclic peptides, diketopiperazines, and related compounds” seems to occur mostly during liquid fermentation compared to solid coculture, independent of the type of coculture participants (Arora et al., 2020). As pointed out in the study, lack of information provided by previous coculture studies is the main hurdle to comprehensive understanding; thus, general guidelines are needed for the coculture studies to provide accurate and sufficient information.
In conclusion, numerous coculture studies have successfully discovered novel secondary metabolites from actinomycetes to date, but even so, the precise mechanisms of interaction are rarely understood. Broader and deeper identification of the inducing mechanisms during coculture is required to understand complex secondary metabolic regulation and to set directions to genetic engineering-based strategies for inducing or increasing production of target secondary metabolites.
Contributor Information
Ji Hun Kim, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea.
Namil Lee, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea.
Soonkyu Hwang, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea.
Woori Kim, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea.
Yongjae Lee, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea.
Suhyung Cho, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea.
Bernhard O Palsson, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA; Department of Pediatrics, University of California, San Diego, La Jolla, CA 92093, USA; Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, 2800 Lyngby, Denmark.
Byung-Kwan Cho, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea; Intelligent Synthetic Biology Center, Daejeon 34141, Republic of Korea.
Funding
This work was supported by the Bio & Medical Technology Development Program (2018M3A9F3079664 to B.-K.C.) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT). This work was also supported by a grant from the Novo Nordisk Foundation (NNF10CC1016517 to B.O.P).
Conflict of Interest
The authors declare no conflict of interest.
References
- Abdelmohsen U. R., Grkovic T., Balasubramanian S., Kamel M. S., Quinn R. J., Hentschel U. (2015). Elicitation of secondary metabolism in actinomycetes. Biotechnology Advances, 33, 798–811. 10.1016/j.biotechadv.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Adnani N., Chevrette M. G., Adibhatla S. N., Zhang F., Yu Q., Braun D. R., Nelson J., Simpkins S. W., McDonald B. R., Myers C. L., Piotrowski J. S., Thompson C. J., Currie C. R., Li L., Rajski S. R., Bugni T. S. (2017). Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chemical Biology, 12, 3093–3102. 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnani N., Vazquez-Rivera E., Adibhatla S. N., Ellis G. A., Braun D. R., Bugni T. S. (2015). Investigation of interspecies interactions within marine Micromonosporaceae using an improved co-culture approach. Marine Drugs, 13, 6082–6098. 10.3390/md13106082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano S., Morota T., Kano Y. K., Narita H., Hashidzume T., Yamamoto S., Mizutani K., Sakuda S., Furihata K., Takano-Shiratori H., Takano H., Beppu T., Ueda K. (2010). Promomycin, a polyether promoting antibiotic production in Streptomyces spp. Journal of Antibiotics, 63, 486–491. 10.1038/ja.2010.68 [DOI] [PubMed] [Google Scholar]
- Aminov R. I. (2010). A brief history of the antibiotic era: Lessons learned and challenges for the future. Frontiers in Microbiology, 1, 134. 10.3389/fmicb.2010.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora D., Gupta P., Jaglan S., Roullier C., Grovel O., Bertrand S. (2020). Expanding the chemical diversity through microorganisms co-culture: Current status and outlook. Biotechnology Advances, 40, 107521. 10.1016/j.biotechadv.2020.107521 [DOI] [PubMed] [Google Scholar]
- Baltz R. H. (2008). Renaissance in antibacterial discovery from actinomycetes. Current Opinion in Pharmacology, 8, 557–563. 10.1016/j.coph.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Barger S. R., Hoefler B. C., Cubillos-Ruiz A., Russell W. K., Russell D. H., Straight P. D. (2012). Imaging secondary metabolism of Streptomyces sp. Mg1 during cellular lysis and colony degradation of competing Bacillus subtilis. Antonie Van Leeuwenhoek, 102, 435–445. 10.1007/s10482-012-9769-0 [DOI] [PubMed] [Google Scholar]
- Belknap K. C., Park C. J., Barth B. M., Andam C. P. (2020). Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Scientific Reports, 10, 2003. 10.1038/s41598-020-58904-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar R. K., Doull J. L., Vining L. C. (1988). Role of the carbon source in regulating chloramphenicol production by Streptomyces venezuelae: Studies in batch and continuous cultures. Canadian Journal of Microbiology, 34, 1217–1223. 10.1139/m88-214 [DOI] [PubMed] [Google Scholar]
- Bibb M. J. (2005). Regulation of secondary metabolism in Streptomycetes. Current Opinion in Microbiology, 8, 208–215. 10.1016/j.mib.2005.02.016 [DOI] [PubMed] [Google Scholar]
- Bode H. B., Bethe B., Hofs R., Zeeck A. (2002). Big effects from small changes: Possible ways to explore nature's chemical diversity. ChemBioChem, 3, 619–627. [DOI] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., Scheld M., Spellberg B., Bartlett J. (2009). Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clinical Infectious Diseases, 48, 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Carlson S., Tanouye U., Omarsdottir S., Murphy B. T. (2015). Phylum-specific regulation of resistomycin production in a Streptomyces sp. via microbial coculture. Journal of Natural Products, 78, 381–387. 10.1021/np500767u [DOI] [PubMed] [Google Scholar]
- Challis G. L., Hopwood D. A. (2003). Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proceedings of the National Academy of Sciences of the USA, 100, 14555–14561. 10.1073/pnas.1934677100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charusanti P., Fong N. L., Nagarajan H., Pereira A. R., Li H. J., Abate E. A., Su Y., Gerwick W. H., Palsson B. O. (2012). Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction. PLoS One, 7, e33727. 10.1371/journal.pone.0033727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wang G. Y., Li X., Waters B., Davies J. (2000). Enhanced production of microbial metabolites in the presence of dimethyl sulfoxide. Journal of Antibiotics, 53, 1145–1153. 10.7164/antibiotics.53.1145 [DOI] [PubMed] [Google Scholar]
- Cho J. Y., Kim M. S. (2012). Induction of antifouling diterpene production by Streptomyces cinnabarinus PK209 in co-culture with marine-derived Alteromonas sp. KNS-16. Bioscience, Biotechnology, and Biochemistry, 76, 1849–1854. 10.1271/bbb.120221 [DOI] [PubMed] [Google Scholar]
- Christova K., Sholeva Z., Chipeva V. (1995). Application of molecular biological methods in taxonomy of genus Streptomyces. Journal of Culture Collections, 1, 3–10. [Google Scholar]
- Cihak M., Kamenik Z., Smidova K., Bergman N., Benada O., Kofronova O., Petrickova K., Bobek J. (2017). Secondary metabolites produced during the germination of Streptomyces coelicolor. Frontiers in Microbiology, 8, 2495. 10.3389/fmicb.2017.02495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb R. E., Wang Y., Zhao H. (2015). High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synthetic Biology, 4, 723–728. 10.1021/sb500351f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craney A., Ahmed S., Nodwell J. (2013). Towards a new science of secondary metabolism. Journal of Antibiotics, 66, 387–400. 10.1038/ja.2013.25 [DOI] [PubMed] [Google Scholar]
- Cui H., Song M. C., Ban Y. H., Jun S. Y., Kwon A. S., Lee J. Y., Yoon Y. J. (2019). High-yield production of multiple O-methylated phenylpropanoids by the engineered Escherichia coli–Streptomyces cocultivation system. Microbial Cell Factories, 18, 67. 10.1186/s12934-019-1118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti Y., Grkovic T., Abdelmohsen U. R., Hentschel U., Quinn R. J. (2014). Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Marine Drugs, 12, 3046–3059. 10.3390/md12053046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewacz D. K., Covington B. C., McLean J. A., Bachmann B. O. (2015). Mapping microbial response metabolomes for induced natural product discovery. ACS Chemical Biology, 10, 1998–2006. 10.1021/acschembio.5b00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi J. R., Albright J. C., Goering A. W., Ju K. S., Haines R. R., Tchalukov K. A., Labeda D. P., Kelleher N. L., Metcalf W. W. (2014). A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nature Chemical Biology, 10, 963–968. 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim W., El-Neketi M., Lewald L. I., Orfali R. S., Lin W. H., Rehberg N., Kalscheuer R., Daletos G., Proksch P. (2016). Metabolites from the fungal endophyte Aspergillus austroafricanus in axenic culture and in fungal-bacterial mixed cultures. Journal of Natural Products, 79, 914–922. 10.1021/acs.jnatprod.5b00975 [DOI] [PubMed] [Google Scholar]
- Ezaki M., Iwami M., Yamashita M., Komori T., Umehara K., Imanaka H. (1992). Biphenomycin A production by a mixed culture. Applied and Environmental Microbiology, 58, 3879–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki M., Shigematsu N., Yamashita M., Komori T., Umehara K., Imanaka H. (1993). Biphenomycin C, a precursor of biphenomycin A in mixed culture. Journal of Antibiotics, 46, 135–140. 10.7164/antibiotics.46.135 [DOI] [PubMed] [Google Scholar]
- Fischer J., Muller S. Y., Netzker T., Jager N., Gacek-Matthews A., Scherlach K., Stroe M. C., Garcia-Altares M., Pezzini F., Schoeler H., Reichelt M., Gershenzon J., Krespach M. K., Shelest E., Schroeckh V., Valiante V., Heinzel T., Hertweck C., Strauss J., Brakhage A. A. (2018). Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites. eLife, 7, e40969. 10.7554/eLife.40969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey-Klett P., Burlinson P., Deveau A., Barret M., Tarkka M., Sarniguet A. (2011). Bacterial–fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiology and Molecular Biology Reviews, 75, 583–609. 10.1128/Mmbr.00020-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Hindra, Mulder D., Yin C., Elliot M. A. (2012). Crp is a global regulator of antibiotic production in Streptomyces. mBio, 3, e00407–12. 10.1128/mBio.00407-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafe U., Reinhardt G., Schade W., Eritt I., Fleck W. F., Radics L. (1983). Interspecific inducers of cytodifferentiation and anthracycline biosynthesis from Streptomyces bikinensis and Streptomyces cyaneofuscatus. Biotechnology Letters, 5, 591–596. 10.1007/Bf00130838 [DOI] [Google Scholar]
- Han J., Gao Q. X., Zhang Y. G., Li L., Mohamad O. A. A., Rao M. P. N., Xiao M., Hozzein W. N., Alkhalifah D. H. M., Tao Y., Li W. J. (2018). Transcriptomic and ectoine analysis of halotolerant Nocardiopsis gilva YIM 90087(T) under salt stress. Frontiers in Microbiology, 9, 618. 10.3389/fmicb.2018.00618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara O., Beppu T. (1982). Induction of streptomycin-inactivating enzyme by A-factor in Streptomyces griseus. Journal of Antibiotics, 35, 1208–1215. 10.7164/antibiotics.35.1208 [DOI] [PubMed] [Google Scholar]
- Harvey A. L. (2008). Natural products in drug discovery. Drug Discovery Today, 13, 894–901. 10.1016/j.drudis.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Hibbing M. E., Fuqua C., Parsek M. R., Peterson S. B. (2010). Bacterial competition: Surviving and thriving in the microbial jungle. Nature Reviews Microbiology, 8, 15–25. 10.1038/nrmicro2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. (1992). Autoregulatory factors and communication in actinomycetes. Annual Review of Microbiology, 46, 377–398. 10.1146/annurev.mi.46.100192.002113 [DOI] [PubMed] [Google Scholar]
- Hosaka T., Ohnishi-Kameyama M., Muramatsu H., Murakami K., Tsurumi Y., Kodani S., Yoshida M., Fujie A., Ochi K. (2009). Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nature Biotechnology, 27, 462–464. 10.1038/nbt.1538 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Okada M., Awakawa T., Asamizu S., Onaka H., Abe I. (2017). Mycolic acid containing bacterium stimulates tandem cyclization of polyene macrolactam in a lake sediment derived rare actinomycete. Organic Letters, 19, 4992–4995. 10.1021/acs.orglett.7b02508 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Okada M., Wakimoto T., Zhang H., Hayashi F., Onaka H., Abe I. (2015a). Niizalactams A–C, multicyclic macrolactams isolated from combined culture of Streptomyces with mycolic acid-containing bacterium. Journal of Natural Products, 78, 3011–3017. 10.1021/acs.jnatprod.5b00804 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Ozeki M., Awakawa T., Morita H., Onaka H., Abe I. (2018a). Catenulobactins A and B, heterocyclic peptides from culturing Catenuloplanes sp. with a mycolic acid-containing bacterium. Journal of Natural Products, 81, 2106–2110. 10.1021/acs.jnatprod.8b00261 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Ozeki M., Wong C. P., Zhang H., Hayashi F., Awakawa T., Morita H., Onaka H., Abe I. (2018b). Mirilactams C–E, novel polycyclic macrolactams isolated from combined-culture of Actinosynnema mirum NBRC 14064 and mycolic acid-containing bacterium. Chemical and Pharmaceutical Bulletin, 66, 660–667. 10.1248/cpb.c18-00143 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Wakimoto T., Onaka H., Abe I. (2015b). Chojalactones A–C, cytotoxic butanolides isolated from Streptomyces sp. cultivated with mycolic acid containing bacterium. Organic Letters, 17, 1501–1504. 10.1021/acs.orglett.5b00385 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Wong C. P., Ozeki M., Zhang H., Hayashi F., Awakawa T., Asamizu S., Onaka H., Abe I. (2018c). Umezawamides, new bioactive polycyclic tetramate macrolactams isolated from a combined-culture of Umezawaea sp. and mycolic acid-containing bacterium. Journal of Antibiotics, 71, 653–657. 10.1038/s41429-018-0040-4 [DOI] [PubMed] [Google Scholar]
- Hoshino S., Zhang L., Awakawa T., Wakimoto T., Onaka H., Abe I. (2015c). Arcyriaflavin E, a new cytotoxic indolocarbazole alkaloid isolated by combined-culture of mycolic acid-containing bacteria and Streptomyces cinnamoneus NBRC 13823. Journal of Antibiotics, 68, 342–344. 10.1038/ja.2014.147 [DOI] [PubMed] [Google Scholar]
- Huang H., Zheng G., Jiang W., Hu H., Lu Y. (2015). One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochimica et Biophysica Sinica, 47, 231–243. 10.1093/abbs/gmv007 [DOI] [PubMed] [Google Scholar]
- Jose P. A., Jebakumar S. R. (2012). Phylogenetic diversity of actinomycetes cultured from coastal multipond solar saltern in Tuticorin, India. Aquatic Biosystems, 8, 23. 10.1186/2046-9063-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem R. S. T., Wang H., Wafo P., Ebrahim W., Ozkaya F. C., Makhloufi G., Janiak C., Sureechatchaiyan P., Kassack M. U., Lin W. H., Liu Z., Proksch P. (2018). Induction of new metabolites from the endophytic fungus Bionectria sp. through bacterial co-culture. Fitoterapia, 124, 132–136. 10.1016/j.fitote.2017.10.021 [DOI] [PubMed] [Google Scholar]
- Kawai K., Wang G., Okamoto S., Ochi K. (2007). The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiology Letters, 274, 311–315. 10.1111/j.1574-6968.2007.00846.x [DOI] [PubMed] [Google Scholar]
- Keilhofer N., Nachtigall J., Kulik A., Ecke M., Hampp R., Sussmuth R. D., Fiedler H. P., Schrey S. D. (2018). Streptomyces AcH 505 triggers production of a salicylic acid analogue in the fungal pathogen Heterobasidion abietinum that enhances infection of Norway spruce seedlings. Antonie Van Leeuwenhoek, 111, 691–704. 10.1007/s10482-018-1017-9 [DOI] [PubMed] [Google Scholar]
- Khalil Z. G., Cruz-Morales P., Licona-Cassani C., Marcellin E., Capon R. J. (2019). Inter-kingdom beach warfare: Microbial chemical communication activates natural chemical defences. ISME Journal, 13, 147–158. 10.1038/s41396-018-0265-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlov A. S., Anisova L. N., Tovarova I. I., Kleiner E. M., Kovalenko I. V., Krasilnikova O. I., Kornitskaya E. Y., Pliner S. A. (1973). Effect of A-factor on the growth of asporogenous mutants of Streptomyces griseus, not producing this factor. Zeitschrift fur Allgemeine Mikrobiologie, 13, 647–655. 10.1002/jobm.3630130803 [DOI] [PubMed] [Google Scholar]
- Kim E. S., Hong H. J., Choi C. Y., Cohen S. N. (2001). Modulation of actinorhodin biosynthesis in Streptomyces lividans by glucose repression of afsR2 gene transcription. Journal of Bacteriology, 183, 2198–2203. 10.1128/JB.183.7.2198-2203.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper M., Gotze S., Barnett R., Willing K., Stallforth P. (2016). Bacterial alkaloids prevent amoebal predation. Angewandte Chemie, International Edition, 55, 8944–8947. 10.1002/anie.201603312 [DOI] [PubMed] [Google Scholar]
- Koehn F. E., Carter G. T. (2005). The evolving role of natural products in drug discovery. Nature Reviews Drug Discovery, 4, 206–220. 10.1038/nrd1657 [DOI] [PubMed] [Google Scholar]
- Konig C. C., Scherlach K., Schroeckh V., Horn F., Nietzsche S., Brakhage A. A., Hertweck C. (2013). Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. ChemBioChem, 14, 938–942. 10.1002/cbic.201300070 [DOI] [PubMed] [Google Scholar]
- Kroiss J., Kaltenpoth M., Schneider B., Schwinger M. G., Hertweck C., Maddula R. K., Strohm E., Svatos A. (2010). Symbiotic Streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nature Chemical Biology, 6, 261–263. 10.1038/Nchembio.331 [DOI] [PubMed] [Google Scholar]
- Kronheim S., Daniel-Ivad M., Duan Z., Hwang S., Wong A. I., Mantel I., Nodwell J. R., Maxwell K. L. (2018). A chemical defence against phage infection. Nature, 564, 283–286. 10.1038/s41586-018-0767-x [DOI] [PubMed] [Google Scholar]
- Kurosawa K., Ghiviriga I., Sambandan T. G., Lessard P. A., Barbara J. E., Rha C., Sinskey A. J. (2008). Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. Journal of the American Chemical Society, 130, 1126–1127. 10.1021/ja077821p [DOI] [PubMed] [Google Scholar]
- Laureti L., Song L., Huang S., Corre C., Leblond P., Challis G. L., Aigle B. (2011). Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proceeding of the National Academy of Sciences of the USA, 108, 6258–6263. 10.1073/pnas.1019077108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Kim W., Chung J., Lee Y., Cho S., Jang K. S., Kim S. C., Palsson B., Cho B. K. (2020a). Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus. ISME Journal, 14, 1111–1124. 10.1038/s41396-020-0594-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Kim W., Hwang S., Lee Y., Cho S., Palsson B., Cho B. K. (2020b). Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. Scientific Data, 7, 55. 10.1038/s41597-020-0395-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Huang H., Liang J., Wang M., Lu L., Shao Z., Cobb R. E., Zhao H. (2013). Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nature Communications, 4, 2894. 10.1038/ncomms3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luti K. J., Mavituna F. (2011). Elicitation of Streptomyces coelicolor with dead cells of Bacillus subtilis and Staphylococcus aureus in a bioreactor increases production of undecylprodigiosin. Applied Microbiology and Biotechnology, 90, 461–466. 10.1007/s00253-010-3032-2 [DOI] [PubMed] [Google Scholar]
- Mavituna F., Luti K. J. K., Gu L. X. (2016). In search of the E. coli compounds that change the antibiotic production pattern of Streptomyces coelicolor during inter-species interaction. Enzyme and Microbial Technology, 90, 45–52. 10.1016/j.enzmictec.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Meschke H., Walter S., Schrempf H. (2012). Characterization and localization of prodiginines from Streptomyces lividans suppressing Verticillium dahliae in the absence or presence of Arabidopsis thaliana. Environmental Microbiology, 14, 940–952. 10.1111/j.1462-2920.2011.02665.x [DOI] [PubMed] [Google Scholar]
- Moussa M., Ebrahim W., Bonus M., Gohlke H., Mandi A., Kurtan T., Hartmann R., Kalscheuer R., Lin W. H., Liu Z., Proksch P. (2019). Co-culture of the fungus Fusarium tricinctum with Streptomyces lividans induces production of cryptic naphthoquinone dimers. RSC Advances, 9, 1491–1500. 10.1039/c8ra09067j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M., Ikeda H., Moore B. S. (2009). Genomic basis for natural product biosynthetic diversity in the actinomycetes. Natural Product Reports, 26, 1362–1384. 10.1039/b817069j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S., Arroyo J., Berriman M., Abe K., Archer D. B., Bermejo C., Bennett J., Bowyer P., Chen D., Collins M., Coulsen R., Davies R., Dyer P. S., Farman M., Fedorova N., Denning D. W. (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature, 438, 1151–1156. 10.1038/nature04332 [DOI] [PubMed] [Google Scholar]
- Niu G., Chater K. F., Tian Y., Zhang J., Tan H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiology Reviews, 40, 554–573. 10.1093/femsre/fuw012 [DOI] [PubMed] [Google Scholar]
- Nutzmann H. W., Reyes-Dominguez Y., Scherlach K., Schroeckh V., Horn F., Gacek A., Schumann J., Hertweck C., Strauss J., Brakhage A. A. (2011). Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proceedings of the National Academy of Sciences of the USA, 108, 14282–14287. 10.1073/pnas.1103523108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. C., Kauffman C. A., Jensen P. R., Fenical W. (2007). Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. Journal of Natural Products, 70, 515–520. 10.1021/np060381f [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Ishikawa J., Hara H., Suzuki H., Ikenoya M., Ikeda H., Yamashita A., Hattori M., Horinouchi S. (2008). Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. Journal of Bacteriology, 190, 4050–4060. 10.1128/Jb.00204-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H., Mori Y., Igarashi Y., Furumai T. (2011). Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Applied and Environmental Microbiology, 77, 400–406. 10.1128/AEM.01337-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H., Ozaki T., Mori Y., Izawa M., Hayashi S., Asamizu S. (2015). Mycolic acid-containing bacteria activate heterologous secondary metabolite expression in Streptomyces lividans. Journal of Antibiotics, 68, 594–597. 10.1038/ja.2015.31 [DOI] [PubMed] [Google Scholar]
- Park H. B., Park J. S., Lee S. I., Shin B., Oh D. C., Kwon H. C. (2017). Gordonic acid, a polyketide glycoside derived from bacterial coculture of Streptomyces and Gordonia species. Journal of Natural Products, 80, 2542–2546. 10.1021/acs.jnatprod.7b00293 [DOI] [PubMed] [Google Scholar]
- Patin N. V., Floros D. J., Hughes C. C., Dorrestein P. C., Jensen P. R. (2018). The role of inter-species interactions in Salinispora specialized metabolism. Microbiology, 164, 946–955. 10.1099/mic.0.000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel H. J., de Winde J. H., Archer D. B., Dyer P. S., Hofmann G., Schaap P. J., Turner G., de Vries R. P., Albang R., Albermann K., Andersen M. R., Bendtsen J. D., Benen J. A., van den Berg M., Breestraat S., Caddick M. X., Contreras R., Cornell M., Coutinho P. M., Stam H. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology, 25, 221–231. 10.1038/nbt1282 [DOI] [PubMed] [Google Scholar]
- Pendleton J. N., Gorman S. P., Gilmore B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Reviews of Anti-infective Therapy, 11, 297–308. 10.1586/Eri.13.12 [DOI] [PubMed] [Google Scholar]
- Perez J., Munoz-Dorado J., Brana A. F., Shimkets L. J., Sevillano L., Santamaria R. I. (2011). Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microbial Biotechnology, 4, 175–183. 10.1111/j.1751-7915.2010.00208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit R. K. (2011). Small-molecule elicitation of microbial secondary metabolites. Microbial Biotechnology, 4, 471–478. 10.1111/j.1751-7915.2010.00196.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procopio R. E., Silva I. R., Martins M. K., Azevedo J. L., Araujo J. M. (2012). Antibiotics produced by Streptomyces. Brazilian Journal of Infectious Diseases, 16, 466–471. 10.1016/j.bjid.2012.08.014 [DOI] [PubMed] [Google Scholar]
- Quillet L., Barray S., Labedan B., Petit F., Guespin-Michel J. (1995). The gene encoding the β-1,4-endoglucanase (CelA) from Myxococcus xanthus: Evidence for independent acquisition by horizontal transfer of binding and catalytic domains from actinomycetes. Gene, 158, 23–29. 10.1016/0378-1119(95)00091-j [DOI] [PubMed] [Google Scholar]
- Rateb M. E., Hallyburton I., Houssen W. E., Bull A. T., Goodfellow M., Santhanam R., Jaspars M., Ebel R. (2013). Induction of diverse secondary metabolites in Aspergillus fumigatus by microbial co-culture. RSC Advances, 3, 14444–14450. 10.1039/c3ra42378f [DOI] [Google Scholar]
- Reen F. J., Romano S., Dobson A. D. W., O'Gara F. (2015). The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Marine Drugs, 13, 4754–4783. 10.3390/md13084754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. Journal of Infectious Diseases, 197, 1079–1081. 10.1086/533452 [DOI] [PubMed] [Google Scholar]
- Romano S., Jackson S. A., Patry S., Dobson A. D. W. (2018). Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Marine Drugs, 16, 244. 10.3390/md16070244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Kato W., Ikeda H., Katsuyama Y., Ohnishi Y., Imoto M. (2020). Discovery of “heat shock metabolites” produced by thermotolerant actinomycetes in high-temperature culture. Journal of Antibiotics, 73, 203–210. 10.1038/s41429-020-0279-4 [DOI] [PubMed] [Google Scholar]
- Sanchez S., Chavez A., Forero A., Garcia-Huante Y., Romero A., Sanchez M., Rocha D., Sanchez B., Avalos M., Guzman-Trampe S., Rodriguez-Sanoja R., Langley E., Ruiz B. (2010). Carbon source regulation of antibiotic production. Journal of Antibiotics, 63, 442–459. 10.1038/ja.2010.78 [DOI] [PubMed] [Google Scholar]
- Sankaran L., Pogell B. M. (1975). Biosynthesis of puromycin in Streptomyces alboniger: Regulation and properties of O-demethylpuromycin O-methyltransferase. Antimicrobial Agents and Chemotherapy, 8, 721–732. 10.1128/aac.8.6.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaberle T. F., Orland A., Konig G. M. (2014). Enhanced production of undecylprodigiosin in Streptomyces coelicolor by co-cultivation with the corallopyronin A-producing myxobacterium, Corallococcus coralloides. Biotechnology Letters, 36, 641–648. 10.1007/s10529-013-1406-0 [DOI] [PubMed] [Google Scholar]
- Schroeckh V., Scherlach K., Nutzmann H. W., Shelest E., Schmidt-Heck W., Schuemann J., Martin K., Hertweck C., Brakhage A. A. (2009). Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proceedings of the National Academy of Sciences of the USA, 106, 14558–14563. 10.1073/pnas.0901870106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Byun W. S., Moon K., Kwon Y., Bae M., Um S., Lee S. K., Oh D. C. (2018). Coculture of marine Streptomyces sp. with Bacillus sp. produces a new piperazic acid-bearing cyclic peptide. Frontiers in Chemistry, 6, 498. 10.3389/fchem.2018.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Rajbhandari I., Wesson K. (2001). Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microbial Ecology, 41, 90–96. [DOI] [PubMed] [Google Scholar]
- Straight P. D., Fischbach M. A., Walsh C. T., Rudner D. Z., Kolter R. (2007). A singular enzymatic megacomplex from Bacillus subtilis. Proceedings of the National Academy of Sciences of the USA, 104, 305–310. 10.1073/pnas.0609073103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroe M. C., Netzker T., Scherlach K., Kruger T., Hertweck C., Valiante V., Brakhage A. A. (2020). Targeted induction of a silent fungal gene cluster encoding the bacteria-specific germination inhibitor fumigermin. eLife, 9, e52541. 10.7554/eLife.52541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama R., Nishimura S., Ozaki T., Asamizu S., Onaka H., Kakeya H. (2015). 5-Alkyl-1,2,3,4-tetrahydroquinolines, new membrane-interacting lipophilic metabolites produced by combined culture of Streptomyces nigrescens and Tsukamurella pulmonis. Organic Letters, 17, 1918–1921. 10.1021/acs.orglett.5b00607 [DOI] [PubMed] [Google Scholar]
- Sugiyama R., Nishimura S., Ozaki T., Asamizu S., Onaka H., Kakeya H. (2016). Discovery and total synthesis of streptoaminals: Antimicrobial [5,5]-spirohemiaminals from the combined-culture of Streptomyces nigrescens and Tsukamurella pulmonis. Angewandte Chemie, International Edition, 55, 10278–10282. 10.1002/anie.201604126 [DOI] [PubMed] [Google Scholar]
- Sung A. A., Gromek S. M., Balunas M. J. (2017). Upregulation and identification of antibiotic activity of a marine-derived Streptomyces sp. via co-cultures with human pathogens. Marine Drugs, 15, 250. 10.3390/md15080250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., Ouellette M., Outterson K., Patel J., Cavaleri M., Cox E. M., Houchens C. R., Grayson M. L., Hansen P., Singh N., … WHO Pathogens Priority List Working Group . (2018). Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infectious Diseases, 18, 318–327. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Tan Z. Q., Leow H. Y., Lee D. C. W., Karisnan K., Song A. A. L., Mai C. W., Yap W. S., Lim S. H. E., Lai K. S. (2019). Co-culture systems for the production of secondary metabolites: Current and future prospects. The Open Biotechnology Journal, 13, 18–26. [Google Scholar]
- Tanaka Y., Hosaka T., Ochi K. (2010). Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2). Journal of Antibiotics, 63, 477–481. 10.1038/ja.2010.53 [DOI] [PubMed] [Google Scholar]
- Traxler M. F., Seyedsayamdost M. R., Clardy J., Kolter R. (2012). Interspecies modulation of bacterial development through iron competition and siderophore piracy. Molecular Microbiology, 86, 628–644. 10.1111/mmi.12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler M. F., Watrous J. D., Alexandrov T., Dorrestein P. C., Kolter R. (2013). Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio, 4, e00459–13. 10.1128/mBio.00459-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Kawai S., Ogawa H., Kiyama A., Kubota T., Kawanobe H., Beppu T. (2000). Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. Journal of Antibiotics, 53, 979–982. 10.7164/antibiotics.53.979 [DOI] [PubMed] [Google Scholar]
- van Bergeijk D. A., Terlouw B. R., Medema M. H., van Wezel G. P. (2020). Ecology and genomics of actinobacteria: New concepts for natural product discovery. Nature Reviews Microbiology, 18, 546–558. 10.1038/s41579-020-0379-y [DOI] [PubMed] [Google Scholar]
- Vargas-Bautista C., Rahlwes K., Straight P. (2014). Bacterial competition reveals differential regulation of the pks genes by Bacillus subtilis. Journal of Bacteriology, 196, 717–728. 10.1128/JB.01022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. (2015). The antibiotic resistance crisis: Part 1: Causes and threats. Pharmacy and Therapeutics, 40, 277–283. [PMC free article] [PubMed] [Google Scholar]
- Wakefield J., Hassan H. M., Jaspars M., Ebel R., Rateb M. E. (2017). Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Frontiers in Microbiology, 8, 1284. 10.3389/fmicb.2017.01284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang L., Zhuang Y., Kong F., Zhang C., Zhu W. (2014). Phenolic polyketides from the co-cultivation of marine-derived Penicillium sp. WC-29-5 and Streptomyces fradiae 007. Marine Drugs, 12, 2079–2088. 10.3390/md12042079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman J. R., Gilsenan J. M., Joardar V., Deegan J., Clutterbuck J., Andersen M. R., Archer D., Bencina M., Braus G., Coutinho P., von Dohren H., Doonan J., Driessen A. J., Durek P., Espeso E., Fekete E., Flipphi M., Estrada C. G., Geysens S., Turner G. (2009). The 2008 update of the Aspergillus nidulans genome annotation: A community effort. Fungal Genetics and Biology, 46(Suppl. 1), S2–S13. 10.1016/j.fgb.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Zacchetti B., Ram A. F., van Wezel G. P., Claessen D., Hae Choi Y. (2015). Expanding the chemical space for natural products by Aspergillus–Streptomyces co-cultivation and biotransformation. Scientific Reports, 5, 10868. 10.1038/srep10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. H., Huang S. B., Lee G. B. (2010). Microfluidic cell culture systems for drug research. Lab on a Chip, 10, 939–956. 10.1039/b921695b [DOI] [PubMed] [Google Scholar]
- Yamada Y., Sugamura K., Kondo K., Yanagimoto M., Okada H. (1987). The structure of inducing factors for virginiamycin production in Streptomyces virginiae. Journal of Antibiotics, 40, 496–504. 10.7164/antibiotics.40.496 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Oikawa H., Ogawa H. O., Hosono K., Shinmachi F., Takano H., Sakuda S., Beppu T., Ueda K. (2005). Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology, 151, 2899–2905. 10.1099/mic.0.28139-0 [DOI] [PubMed] [Google Scholar]
- Yu L., Ding W., Ma Z. (2016). Induced production of cytochalasans in co-culture of marine fungus Aspergillus flavipes and actinomycete Streptomyces sp. Natural Product Research, 30, 1718–1723. 10.1080/14786419.2015.1136910 [DOI] [PubMed] [Google Scholar]
- Yu L., Hu Z., Ma Z. (2015). Production of bioactive tryptamine derivatives by co-culture of marine Streptomyces with Bacillus mycoides. Natural Product Research, 29, 2087–2091. 10.1080/14786419.2015.1005619 [DOI] [PubMed] [Google Scholar]
- Yu M., Li Y., Banakar S. P., Liu L., Shao C., Li Z., Wang C. (2019). New metabolites from the co-culture of marine-derived actinomycete Streptomyces rochei MB037 and fungus Rhinocladiella similis 35. Frontiers in Microbiology, 10, 915. 10.3389/fmicb.2019.00915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. M., Wong F. T., Wang Y., Luo S., Lim Y. H., Heng E., Yeo W. L., Cobb R. E., Enghiad B., Ang E. L., Zhao H. (2017). CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nature Chemical Biology, 13, 607–609. 10.1038/nchembio.2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuck K. M., Shipley S., Newman D. J. (2011). Induced production of N-formyl alkaloids from Aspergillus fumigatus by co-culture with Streptomyces peucetius. Journal of Natural Products, 74, 1653–1657. 10.1021/np200255f [DOI] [PubMed] [Google Scholar]