Abstract
Background and aims
The albumin-to-globulin ratio (AGR) has been used to predict severity and mortality in infectious diseases. The aim of this study is to evaluate the prognostic value of the AGR in COVID-19 patients.
Methods
A systematic review and meta-analysis were conducted. We included observational studies assessing the association between the AGR values upon hospital admission and severity or all-cause mortality in COVID-19 patients. In the meta-analyses we used random effect models. Risk of bias was assessed using the Newcastle-Ottawa Scale (NOS). The effect measures were expressed as mean difference (MD) and their 95% confidence intervals (CI). We performed Egger's test and funnel plots to assess the publication bias.
Results
The included studies had a total of 11356 patients corresponding to 31 cohort studies. Severe COVID-19 patients had lower AGR values than non-severe COVID-19 patients (mean difference (MD), −0.27; 95% IC, −0.32 to −0.22; p < 0.001; I2 = 88%). Non-survivor patients with COVID-19 had lower AGR values than survivor patients (MD, −0.29; 95% IC, −0.35 to −0.24; p < 0.001; I2 = 79%). In the sensitivity analysis, we only included studies with low risk of bias, which decreased the heterogeneity for both outcomes (severity, I2 = 20%; mortality, I2 = 5%).
Conclusions
Low AGR values upon hospital admission were found in COVID-19 patients with a worse prognosis.
Keywords: Albumin, COVID-19, Globulin, Prognosis
Albumin, COVID-19, Globulin, Prognosis.
1. Introduction
At present, the coronavirus disease 2019 (COVID-19) pandemic, which is responsible for 4.5 million deaths at the time of writing [1], has been considered one of the most critical issues of global health. Due to the high number of COVID-19 cases in medical centers, tools are needed to help stratify patients and avoid the overloading and collapse of hospitals. In this context, the biomarkers are essential tools used when assessing a patient's prognosis with COVID-19 [2]. Several biomarkers that can predict the severity and risk of mortality in patients with SARS-COV-2 infection have been studied. Some examples of these are the neutrophil/lymphocyte ratio, the C-reactive protein, and the fibrinogen/albumin ratio, among others [3, 4, 5, 6, 7, 8].
The prognostic function of the albumin-to-globulin ratio (AGR) has been studied in several other pathologies, such as those of small cell lung cancer, hepatocellular carcinoma, heart failure, and infectious diseases [9, 10, 11, 12, 13]. In COVID-19 infection, the prognostic value of the AGR is based on the role that these serum proteins play in the process of systemic inflammation.
Increased globulin levels have been observed in patients with chronic inflammatory processes, such as those with hepatocellular carcinoma [14]. On the other hand, albumin levels have been shown to decrease in patients with altered nutritional status, systemic inflammation, and organ dysfunction [15, 16]. Moreover, serum albumin levels have been shown to be prognostic marker in COVID-19 patients [17, 18]. In this way, the AGR was postulated as a potential independent biomarker of both severity and mortality for COVID-19.
The aim of the present study is to evaluate and synthesize the published evidence regarding the ability of the AGR to predict the severity and mortality in patients diagnosed with SARS-COV-2 infection.
2. Methods
2.1. Report and register
The Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [19] statement was used for reporting this systematic review and meta-analysis. Moreover, a short protocol version was registered in the International Prospective Register of Systematic Review (PROSPERO) [CRD42021275262].
2.2. Data sources and searches
Our search strategy was constructed in accordance with the Peer Review of Electronic Search Strategies (PRESS) Guidelines [20]. Studies evaluating the association between AGR and severity of COVID-19 patients were searched on September 20, 2021, in the following databases: PubMed, LILACS, Web of Science, Ovid MEDLINE, Embase, Scopus, and the WHO COVID-19 Global Research Database. Additionally, a manual search was performed in the CNKI databases, Wanfang Database, Research Square, medRixv, and SciELO Preprints (Table 1).
Table 1.
Search strategy.
|
PUBMED |
|
Albumin (#1) Albumin [MH] OR Albumins [MH] OR Serum Albumin [MH] OR Albumin, Serum [MH] OR Plasma Albumin [MH] OR Albumin [TIAB] OR Albumins [TIAB] OR “Serum Albumin” [TIAB] OR “Plasma Albumin” [TIAB] OR Albumin [OT] OR Albumins [OT] OR “Serum Albumin” [OT] OR “Plasma Albumin” [OT] Globulin (#2) Globulin [MH] OR Globulins [MH] OR Serum Globulins [MH] OR Globulins, Serum [MH] OR Pseudoglobulins [MH] OR Euglobulins [MH] OR Globulin [TIAB] OR Globulins [TIAB] OR “Serum Globulins” [TIAB] OR Pseudoglobulins [TIAB] OR Euglobulins [TIAB] OR Globulin [OT] OR Globulins [OT] OR “Serum Globulins” [OT] OR Pseudoglobulins [OT] OR Euglobulins [OT] Albumin/Globulin ratio (#3) “Albumin/globulin ratio” [OT] OR “Albumin/globulin index” [OT] OR “Ratio albumin/globulin” [OT] OR “Index albumin/globulin” [OT] OR “Albumin to globulin ratio” [OT] OR “Albumin-to globulin ratio” [OT] OR “Albumin to-globulin ratio” [OT] OR “Albumin-to-globulin ratio” [OT] OR “Albumin to globulin index” [OT] OR “Albumin-to globulin index” [OT] OR “Albumin to-globulin index” [OT] OR “Albumin-to-globulin index” [OT] OR “Ratio Albumin to globulin” [OT] OR “Ratio Albumin-to globulin” [OT] OR “Ratio Albumin to-globulin” [OT] OR “Ratio Albumin-to-globulin” [OT] OR “Index Albumin to globulin” [OT] OR “Index Albumin-to globulin” [OT] OR “Index Albumin to-globulin” [OT] OR “Index Albumin-to-globulin” [OT] Covid-19 (#4) COVID 19 [MH] OR COVID-19 Virus Disease [MH] OR COVID 19 Virus Disease [MH] OR COVID-19 Virus Diseases [MH] OR Disease, COVID-19 Virus [MH] OR Virus Disease, COVID-19 [MH] OR COVID-19 Virus Infection [MH] OR COVID 19 Virus Infection [MH] OR COVID-19 Virus Infections [MH] OR Infection, COVID-19 Virus [MH] OR Virus Infection, COVID-19 [MH] OR 2019-nCoV Infection [MH] OR 2019 nCoV Infection [MH] OR 2019-nCoV Infections [MH] OR Infection, 2019-nCoV [MH] OR Coronavirus Disease-19 [MH] OR Coronavirus Disease 19 [MH] OR 2019 Novel Coronavirus Disease [MH] OR 2019 Novel Coronavirus Infection [MH] OR 2019-nCoV Disease [MH] OR 2019 nCoV Disease [MH] OR 2019-nCoV Diseases [MH] OR Disease, 2019-nCoV [MH] OR COVID19 [MH] OR Coronavirus Disease 2019 [MH] OR Disease 2019, Coronavirus [MH] OR SARS Coronavirus 2 Infection [MH] OR SARS-CoV-2 Infection [MH] OR Infection, SARS-CoV-2 [MH] OR SARS CoV 2 Infection [MH] OR SARS-CoV-2 Infections [MH] OR COVID-19 Pandemic [MH] OR COVID 19 Pandemic [MH] OR COVID-19 Pandemics [MH] OR Pandemic, COVID-19 [MH] OR "COVID 19" [TIAB] OR "COVID-19 Virus Disease" [TIAB] OR "COVID 19 Virus Disease" [TIAB] OR "COVID-19 Virus Diseases" [TIAB] OR "COVID-19 Virus Infection" [TIAB] OR "COVID 19 Virus Infection" [TIAB] OR "COVID-19 Virus Infections" [TIAB] OR "Infection, COVID-19 Virus" [TIAB] OR "2019-nCoV Infection" [TIAB] OR "2019 nCoV Infection" [TIAB] OR "2019-nCoV Infections" [TIAB] OR "Coronavirus Disease-19" [TIAB] OR "Coronavirus Disease 19" [TIAB] OR "2019 Novel Coronavirus Disease" [TIAB] OR "2019 Novel Coronavirus Infection" [TIAB] OR "2019-nCoV Disease" [TIAB] OR "2019 nCoV Disease" [TIAB] OR "2019-nCoV Diseases" [TIAB] OR "COVID19" [TIAB] OR "Coronavirus Disease 2019" [TIAB] OR "Disease 2019, Coronavirus" [TIAB] OR "SARS Coronavirus 2 Infection" [TIAB] OR "SARS-CoV-2 Infection" [TIAB] OR "Infection, SARS-CoV-2" [TIAB] OR "SARS-CoV-2 Infections" [TIAB] OR "COVID-19 Pandemic" [TIAB] OR "COVID 19 Pandemic" [TIAB] OR "COVID-19 Pandemics" [TIAB] OR "COVID 19" [OT] OR "COVID-19 Virus Disease" [OT] OR "COVID 19 Virus Disease" [OT] OR "COVID-19 Virus Diseases" [OT] OR "COVID-19 Virus Infection" [OT] OR "COVID 19 Virus Infection" [OT] OR "COVID-19 Virus Infections" [OT] OR "Infection, COVID-19 Virus" [OT] OR "2019-nCoV Infection" [OT] OR "2019 nCoV Infection" [OT] OR "2019-nCoV Infections" [OT] OR "Coronavirus Disease-19" [OT] OR "Coronavirus Disease 19" [OT] OR "2019 Novel Coronavirus Disease" [OT] OR "2019 Novel Coronavirus Infection" [OT] OR "2019-nCoV Disease" [OT] OR "2019 nCoV Disease" [OT] OR "2019-nCoV Diseases" [OT] OR "COVID19" [OT] OR "Coronavirus Disease 2019" [OT] OR "Disease 2019, Coronavirus" [OT] OR "SARS Coronavirus 2 Infection" [OT] OR "SARS-CoV-2 Infection" [OT] OR "Infection, SARS-CoV-2" [OT] OR "SARS-CoV-2 Infections" [OT] OR "COVID-19 Pandemic" [OT] OR "COVID 19 Pandemic" [OT] OR "COVID-19 Pandemics" [OT] Search Formula: ((#1 AND #2) OR #3) AND #4 |
|
WEB OF SCIENCE |
| Query #1 Albumin OR Albumins OR Serum Albumin OR Albumin, Serum OR Plasma Albumin OR Albumin OR Albumins OR “Serum Albumin” OR “Plasma Albumin” OR Albumin OR Albumins OR “Serum Albumin” OR “Plasma Albumin” (All Fields) Query #2 ALL=(Globulin OR Globulins OR Serum Globulins OR Globulins, Serum OR Pseudoglobulins OR Euglobulins OR Globulin OR Globulins OR “Serum Globulins” OR Pseudoglobulins OR Euglobulins OR Globulin OR Globulins OR “Serum Globulins” OR Pseudoglobulins OR Euglobulins) Query #3 ALL=(“Albumin/globulin ratio” OR “Albumin/globulin index” OR “Ratio albumin/globulin” OR “Index albumin/globulin” OR “Albumin to globulin ratio” OR “Albumin-to globulin ratio” OR “Albumin to-globulin ratio” OR “Albumin-to-globulin ratio”OR “Albumin to globulin index” OR “Albumin-to globulin index” OR “Albumin to-globulin index” OR “Albumin-to-globulin index” OR “Ratio Albumin to globulin” OR “Ratio Albumin-to globulin” OR “Ratio Albumin to-globulin” OR “Ratio Albumin-to-globulin” OR “Index Albumin to globulin” OR “Index Albumin-to globulin” OR “Index Albumin to-globulin” OR “Index Albumin-to-globulin”) Query #4 ALL=(“COVID-19” OR “COVID 19” OR “COVID-19 Virus Disease” OR “COVID 19 Virus Disease” OR “COVID-19 Virus Diseases” OR “Disease, COVID-19 Virus” OR “Virus Disease, COVID-19” OR “COVID-19 Virus Infection” OR “COVID 19 Virus Infection” OR “COVID-19 Virus Infections” OR “Infection, COVID-19 Virus” OR “Virus Infection, COVID-19” OR “2019-nCoV Infection” OR “2019 nCoV Infection” OR “2019-nCoV Infections” OR “Infection, 2019-nCoV” OR “Coronavirus Disease-19” OR “Coronavirus Disease 19” OR “2019 Novel Coronavirus Disease” OR “2019 Novel Coronavirus Infection” OR “2019-nCoV Disease” OR “2019 nCoV Disease” OR “2019-nCoV Diseases” OR “Disease, 2019-nCoV” OR “COVID19” OR “Coronavirus Disease 2019” OR “Disease 2019, Coronavirus” OR “SARS Coronavirus 2 Infection” OR “SARS-CoV-2 Infection” OR “Infection, SARS-CoV-2” OR “SARS CoV 2 Infection” OR “SARS-CoV-2 Infections” OR “COVID-19 Pandemic” OR “COVID 19 Pandemic” OR “COVID-19 Pandemics” OR “Pandemic, COVID-19”) Search Formula: (((#1) AND #2) OR #3) AND #4 |
|
OVIDMEDLINE |
|
|
WHO’S GLOBAL RESEARCH ON CORONAVIRUS DISEASE (COVID-19) DATABASE |
| tw:(Albumin/globulin ratio) AND collection:("01-internacional" OR "04-international_org" OR "09-preprints") |
|
SCOPUS |
| (( ( ALL ( albumin OR albumins OR serum AND albumin OR albumin, AND serum OR plasma AND albumin OR albumin OR albumins OR "Serum Albumin" OR "Plasma Albumin" OR albumin OR albumins OR "Serum Albumin" OR "Plasma Albumin") ) AND (ALL ( globulin OR globulins OR serum AND globulins OR globulins, AND serum OR pseudoglobulins OR euglobulins OR globulin OR globulins OR "Serum Globulins" OR pseudoglobulins OR euglobulins OR globulin OR globulins OR "Serum Globulins" OR pseudoglobulins OR euglobulins) )) OR (ALL ( "Albumin/globulin ratio" OR "Albumin/globulin index" OR "Ratio albumin/globulin" OR "Index albumin/globulin" OR "Albumin to globulin ratio" OR "Albumin-to globulin ratio" OR "Albumin to-globulin ratio" OR "Albumin-to-globulin ratio" OR "Albumin to globulin index" OR "Albumin-to globulin index" OR "Albumin to-globulin index" OR "Albumin-to-globulin index" OR "Ratio Albumin to globulin" OR "Ratio Albumin-to globulin" OR "Ratio Albumin to-globulin" OR "Ratio Albumin-to-globulin" OR "Index Albumin to globulin" OR "Index Albumin-to globulin" OR "Index Albumin to-globulin" OR "Index Albumin-to-globulin") )) AND (TITLE-ABS-KEY ( "COVID-19" OR "COVID 19" OR "COVID-19 Virus Disease" OR "COVID 19 Virus Disease" OR "COVID-19 Virus Diseases" OR "Disease, COVID-19 Virus" OR "Virus Disease, COVID-19" OR "COVID-19 Virus Infection" OR "COVID 19 Virus Infection" OR "COVID-19 Virus Infections" OR "Infection, COVID-19 Virus" OR "Virus Infection, COVID-19" OR "2019-nCoV Infection" OR "2019 nCoV Infection" OR "2019-nCoV Infections" OR "Infection, 2019-nCoV" OR "Coronavirus Disease-19" OR "Coronavirus Disease 19" OR "2019 Novel Coronavirus Disease" OR "2019 Novel Coronavirus Infection" OR "2019-nCoV Disease" OR "2019 nCoV Disease" OR "2019-nCoV Diseases" OR "Disease, 2019-nCoV" OR "COVID19" OR "Coronavirus Disease 2019" OR "Disease 2019, Coronavirus" OR "SARS Coronavirus 2 Infection" OR "SARS-CoV-2 Infection" OR "Infection, SARS-CoV-2" OR "SARS CoV 2 Infection" OR "SARS-CoV-2 Infections" OR "COVID-19 Pandemic" OR "COVID 19 Pandemic" OR "COVID-19 Pandemics" OR "Pandemic, COVID-19") ) |
|
LILACS |
| Albumin [Palabras] and Globulin [Palabras] and Covid [Palabras] |
|
EMBASE |
| Query Results #5 #3 AND #4 #4 #1 OR #2 #3 'covid-19':ab,ti OR 'covid 19':ab,ti OR 'covid-19 virus disease':ab,ti OR 'covid 19 virus disease':ab,ti OR 'covid-19 virus diseases':ab,ti OR 'disease, covid-19 virus':ab,ti OR 'virus disease, covid-19':ab,ti OR 'covid-19 virus infection':ab,ti OR 'covid 19 virus infection':ab,ti OR 'covid-19 virus infections':ab,ti OR 'infection, covid-19 virus':ab,ti OR 'virus infection, covid-19':ab,ti OR '2019-ncov infection':ab,ti OR '2019 ncov infection':ab,ti OR '2019-ncov infections':ab,ti OR 'infection, 2019-ncov':ab,ti OR 'coronavirus disease-19':ab,ti OR 'coronavirus disease 19':ab,ti OR '2019 novel coronavirus disease':ab,ti OR '2019 novel coronavirus infection':ab,ti OR '2019-ncov disease':ab,ti OR '2019 ncov disease':ab,ti OR '2019-ncov diseases':ab,ti OR 'disease, 2019-ncov':ab,ti OR 'covid19':ab,ti OR 'coronavirus disease 2019':ab,ti OR 'disease 2019, coronavirus':ab,ti OR 'sars coronavirus 2 infection':ab,ti OR 'sars-cov-2 infection':ab,ti OR 'infection, sars-cov-2':ab,ti OR 'sars cov 2 infection':ab,ti OR 'sars-cov-2 infections':ab,ti OR 'covid-19 pandemic':ab,ti OR 'covid 19 pandemic':ab,ti OR 'covid-19 pandemics':ab,ti OR 'pandemic, covid-19':ab,ti #2 'albumin/globulin ratio' OR 'albumin/globulin index' OR 'ratio albumin/globulin' OR 'index albumin/globulin' OR 'albumin to globulin ratio'/exp OR 'albumin to globulin ratio' OR 'albumin-to globulin ratio' OR 'albumin to-globulin ratio' OR 'albumin-to-globulin ratio' OR 'albumin to globulin index' OR 'albumin-to globulin index' OR 'albumin to-globulin index' OR 'albumin-to-globulin index' OR 'ratio albumin to globulin' OR 'ratio albumin-to globulin' OR 'ratio albumin to-globulin' OR 'ratio albumin-to-globulin' OR 'index albumin to globulin' OR 'index albumin-to globulin' OR 'index albumin to-globulin' OR 'index albumin-to-globulin' #1 ('albumin, serum':ab,ti OR albumin:ab,ti OR albumins:ab,ti OR 'serum albumin':ab,ti OR 'plasma albumin':ab,ti) AND ('globulins, serum':ab,ti OR globulin:ab,ti OR globulins:ab,ti OR 'serum globulins':ab,ti OR pseudoglobulins:ab,ti OR euglobulins:ab,t |
2.3. Selection process and data extraction
All retrieved articles that complied with the following criteria were included: (i) case-control and cohort studies, (ii) studies on adult patients (≥18 years old) diagnosed with COVID-19, and (iii) studies assessing the relationship between the AGR values and severity or mortality in COVID-19 patients. Duplicates and studies that did not comply with the entirety of the eligibility criteria were excluded. COVID-19 severity was our primary outcome, defined as complying with at least one of these criteria: ICU admission, shortness of breath, respiration rate of ≥30 times per minute, blood oxygen saturation of ≤93% at rest, or PaO2/FiO2 ratio of ≤300 mmHg. However, definitions for severity vary widely among studies. Moreover, mortality was considered as a secondary outcome.
As for the study selection, four authors (JRUB, EAAB, MDMR, and EAHB) screened titles and abstracts independently using the data management software Rayyan QCRI (Rayyan Systems Inc©) [21]. Afterwards, the same four authors independently screened the remaining articles by full text against the eligibility criteria. Discrepancies were resolved through discussion until a consensus was reached. Finally, two authors (PHA and MDMR) extracted the data from selected studies in a standardized data extraction sheet in Microsoft Excel 2018 (Microsoft Corporation©). The extracted data included the following: first author, publication date, study title, study design, study location, population characteristics (number of participants, age, sex, comorbidities, and stratified sample data), exposures (mean or median AGR of the whole sample according to sample stratification) and outcomes types (severity or mortality).
2.4. Evaluation of study quality and publication bias
Two authors (EAAB, VB-Z) critically assessed the included studies using the Newcastle-Ottawa Scale (NOS) [22]. Risk of bias was categorized as low (scores ≥6), moderate (scores ranging 4–5), and high (scores <4). Moreover, funnel plots and Egger's tests were conducted to assess the publication bias. P-values of >0.1 were considered indicative of no publication bias.
2.5. Data synthesis and analysis
The effect size of each study was calculated using the means and standard deviations (SD) of the AGR values for severe vs. non-severe patients and survivor vs. non-survivor patients and pooled as mean difference (MD) with 95% confidence intervals (CI). Continuous data was presented as median and interquartile range (IQR) which was converted into relative means and SD by Hozo et al. [23]. The software Review Manager 5.4 (RevMan 5.4) (The Cochrane Collaboration, Copenhagen, Denmark) was used for statistical analysis. Statistical heterogeneity was determined using I2 statistics and categorized as severe (≥60%) and non-severe (<60%). Cochran's Q test was also conducted, and a p-value of <0.05 was considered statistically significant. A random-effects model was conducted as we anticipated heterogeneity among the studies. Subgroup analyses by study country (Chinese vs. non-Chinese studies) were performed, and the interaction between the p-value and each subgroup was investigated. Furthermore, we performed sensitivity analyses using studies with a low risk of bias.
3. Results
3.1. Study selection
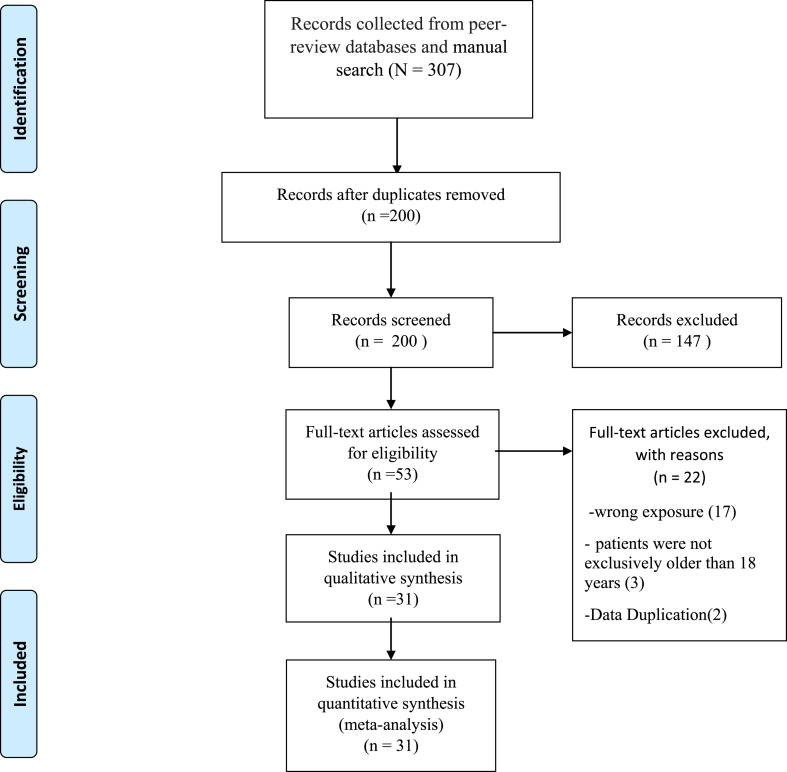
The systematic search retrieved 307 records, and after removing 107 duplicates, 200 records remained. Screening by titles and abstracts left 53 studies for full text review. A careful and rigorous assessment of full texts found 31 studies that complied with all eligibility criteria. This process is documented in a PRISMA flow diagram (Figure 1).
Figure 1.
PRISMA Flow Diagram.
3.2. Study characteristics
All 31 included studies were cohort studies, and their characteristics are outlined in Table 2 and Table 3 [24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54]. Six studies compared the AGR values upon hospital admission of survival and non-survival COVID-19 patients, and the remaining 25 studies compared the AGR values upon hospital admission between severe and non-severe COVID-19 patients. Moreover, 4 studies were conducted in India, 1 in Turkey, 1 in Algeria, 1 in Japan, and the remaining 24 in China.
Table 2.
Characteristics of the included studies comparing severe and non-severe COVID-19 patients.
| Author | Year | Country | Participants (male) |
Median/mean age (IQR/SD) | Comorbidities (n) |
AGR mean in severe patients | AGR mean in non-severe patients | Cut-off | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Diabetes mellitus | Hypertension | Obesity | ||||||||
| Fu L et al. | 2020 | China | 350 (190) | NR | 145 | 125 | NR | 1.37 (0.37) | 1.37 (0.22) | NR |
| Guiling L et al. | 2020 | China | 107 (53) | 65 (60–70) | 12 | 44 | NR | 1.32 (0.22) | 1.47 (0.22) | NR |
| Gemcioglu E et al. | 2021 | Turkey | 301 (161) | 45 (24) | 107 | 169 | 4 | 1.46 (0.59) | 1.87 (0.39) | 1.52 |
| Cao Z et al. | 2020 | China | 80 (38) | 53 (20) | 6 | 20 | NR | 0.8 (0.2) | 1.1 (0.2) | NR |
| Shi S et al. | 2021 | China | 87 (49) | 60 (22–88) | NR | NR | NR | 0.91 (0.25) | 1.24 (0.23) | 1 |
| Yang R et al. | 2020 | China | 495 (235) | 55 (40–67) | 47 | 137 | NR | 1.21 (0.24) | 1.50 (0.31) | NR |
| Tsui E et al. | 2020 | China | 535 (264) | 37.8 (18) | 33 | 87 | NR | 1.01 (0.30) | 1.33 (0.30) | NR |
| Bennouar S et al. | 2020 | Algeria | 330 (206) | 66.6 (9) | NR | NR | NR | 1.0 (0.18) | 1.43 (0.3) | 1.15 |
| Huang Jiana et al. | 2021 | China | 98 (46) | 44 (33–62) | 7 | 19 | NR | 1.23 (0.22) | 1.39 (0.24) | NR |
| Mishra C et al. | 2021 | India | 500 (308) | 60 (16.3) | 126 | 259 | NR | 0.94 (0.23) | 1.16 (0.3) | NR |
| Wang Changzheng et al. | 2020 | China | 45 (23) | 39 (18–62) | 4 | 4 | NR | 1.23 (0.23) | 1.71 (4.02) | NR |
| Fang Z et al. | 2020 | China | 239 (118) | 54.87 (14) | 14 | 28 | NR | 1.32 (0.28) | 1.54 (0.3) | NR |
| Wang Menghan et al. | 2021 | China | 151 (64) | 63 (14) | 32 | 60 | NR | 0.96 (6.84) | 1.32 (0.29) | NR |
| Zhao C et al. | 2021 | China | 172 (82) | 65 (57–71) | 27 | 63 | NR | 1.09 (0.28) | 1.42 (0.37) | NR |
| Kalal CR et al. | 2021 | India | 134 (89) | 45.5 (18–86) | 31 | 40 | NR | 1.07 (0.22) | 1.4 (0.29) | NR |
| Huang Juan et al. | 2020 | China | 1187 (537) | 55 (32–76) | NR | 309 | NR | 1.36 (0.33) | 1.52 (0.37) | NR |
| Shang H et al. | 2020 | China | 514 (271) | 54 (48–68) | 99 | 222 | NR | 0.9 (0.29) | 1.02 (0.22) | NR |
| Chen Xu et al. | 2020 | China | 291 (145) | 46 (34–59) | 22 | 39 | NR | 1.19 (0.3) | 1.33 (0.52) | 1.5 |
| Yamamoto A et al. | 2021 | Japon | 152 (74) | 53.5 (38–70) | 11 | 17 | NR | 1.02 (0.14) | 1.42 (0.22) | 1.1 |
| Barya P et al. | 2021 | India | 75 (43) | 47.51 (20–90) | 16 | 4 | NR | 0.89 (0.28) | 1.43 (0.28) | 1.1 |
| Fang Hu et al. | 2020 | China | 91 (40) | 47.53 (15) | 9 | 17 | NR | 1.24 (0.18) | 1.38 (0.22) | NR |
| Qi J et. al | 2020 | China | 104 (47) | 42 (33–56) | NR | NR | NR | 1.1 (0.14) | 1.32 (0.24) | 1.2 |
| Xu F et al. | 2020 | China | 251 (132) | 60 (16) | NR | NR | NR | 0.82 (0.22) | 1.1 (0.29) | NR |
| Dai Wanfa et al. | 2020 | China | 61 (40) | 50 (17) | 6 | 16 | NR | 1.12 (0.23) | 1.43 (0.51) | NR |
| Bing H et al. | 2020 | China | 53 (28) | 50 (27–68) | NR | NR | NR | 1.1 (0.29) | 1.75 (0.44) | NR |
NR: NOT REPORTED.
Table 3.
Characteristics of the included studies comparing survivor and non-survivor COVID-19 patients.
| Author | Year | Country | Participants (male) |
Median/mean age (IQR/SD) | Comorbidities (n) |
AGR mean in non-survivor patients | AGR mean in survivor patients | Cut-off | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Diabetes Mellitus | Hypertension | Obesity | ||||||||
| Wei Y et al. | 2020 | China | 112 (73) | 61 (14.9) | 21 | 40 | NR | 1.2 (0.2) | 1.5 (0.3) | NR |
| Wang Xue et al. | 2020 | China | 131 (56) | 64 (56–71) | 28 | 52 | NR | 0.9 (0.17) | 1.27 (0.25) | NR |
| Caillon A et al. | 2021 | China | 157 (75) | 64 (46–76) | 24 | 55 | NR | 1.09 (0.22) | 1.34 (0.26) | NR |
| Wang Kun et al. | 2020 | China | 296 (140) | 47.32 (14.95) | 30 | 42 | NR | 1.2 (0.3) | 1.6 (0.4) | NR |
| Elavarasi A et al. | 2021 | India | 2017 (1320) | 47.4 (17.6) | 437 | 457 | NR | 1.3 (0.29) | 1.52 (0.22) | NR |
| Huang Wei et al. | 2020 | China | 2240 (1136) | 64 (52–71) | NR | NR | NR | 0.85 (0.17) | 1.16 (0.31) | NR |
NR: NOT REPORTED.
A total of 11356 patients were evaluated, of which 53.56% were men. In 15 studies, the days elapsed for the development of severity from the day of admission were reported, ranging between 5 and 17 days. Only one study did not report the age of the patients. Regarding the comorbidities of the included patients, a total of 1294 (11.4%) diabetes patients, 2325 (20.47%) hypertensive patients, and 4 (0.03%) overweight patients were recorded.
Seven studies evaluated the optimal AGR cut-off values for severity that ranged from 1 to 1.52. Likewise, no studies have evaluated the AGR cut-off values for mortality. Fourteen studies had low risk of bias in the study quality appraisal, while the remaining 17 had a moderate to high risk of bias (Table 4).
Table 4.
Newcastle - Ottawa quality assessment scale for included studies.
| Newcastle - Ottawa Quality Assessment Scale for Cohort Studies | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study | Selection |
Comparability |
Outcome |
Score | Evidence quality | |||||
| Representativeness of the exposed cohort | Selection of the non-exposed cohort | Ascertainment of exposure | Demonstration that outcome of interest was not present at start of study | Comparability of Cohorts on the Basis of the Design or Analysis Maximum: ☆☆ | Assessment of outcome | Was follow-up long enough for outcomes to occur | Adequacy of follow up of cohorts | |||
| Fu L et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Guiling L et al. | ☆ | ☆ | ☆ | ☆ | ☆ | 5 | Moderate risk of bias | |||
| Gemcioglu E et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Cao Z et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 7 | Low Risk of bias | |
| Shi S et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Yang R et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Tsui E et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Huang Jiana et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Mishra C et al. | ☆ | ☆ | ☆ | ☆ | ☆ | 5 | Moderate risk of bias | |||
| Wang Changzheng et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Fang Z et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Wang Menghan et al. | ☆ | ☆ | ☆ | 3 | High Risk of bias | |||||
| Zhao C et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 7 | Low Risk of bias | |
| Kalal CR et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Huang Juan et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Shang H et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Chen Xu et al. | ☆ | ☆ | ☆ | ☆ | ☆ | 5 | Moderate risk of bias | |||
| Yamamoto A et al. | ☆ | ☆ | ☆ | ☆ | ☆ | 5 | Moderate risk of bias | |||
| Barya P et al. | ☆ | ☆ | ☆ | 3 | High Risk of bias | |||||
| Fang Hu et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Qi J et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Xu F et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Wei Y et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 6 | Low Risk of bias | ||
| Wang Xue et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 8 | Low Risk of bias |
| Caillon A et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 7 | Low Risk of bias | |
| Wang Kun et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Elavarasi A et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Huang Wei et al. | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 8 | Low Risk of bias |
| Dai Wanfa et al. | ☆ | ☆ | ☆ | ☆ | 4 | Moderate risk of bias | ||||
| Bing H et al. | ☆ | ☆ | ☆ | ☆ | ☆ | 5 | Moderate risk of bias | |||
3.3. AGR and COVID-19 severity
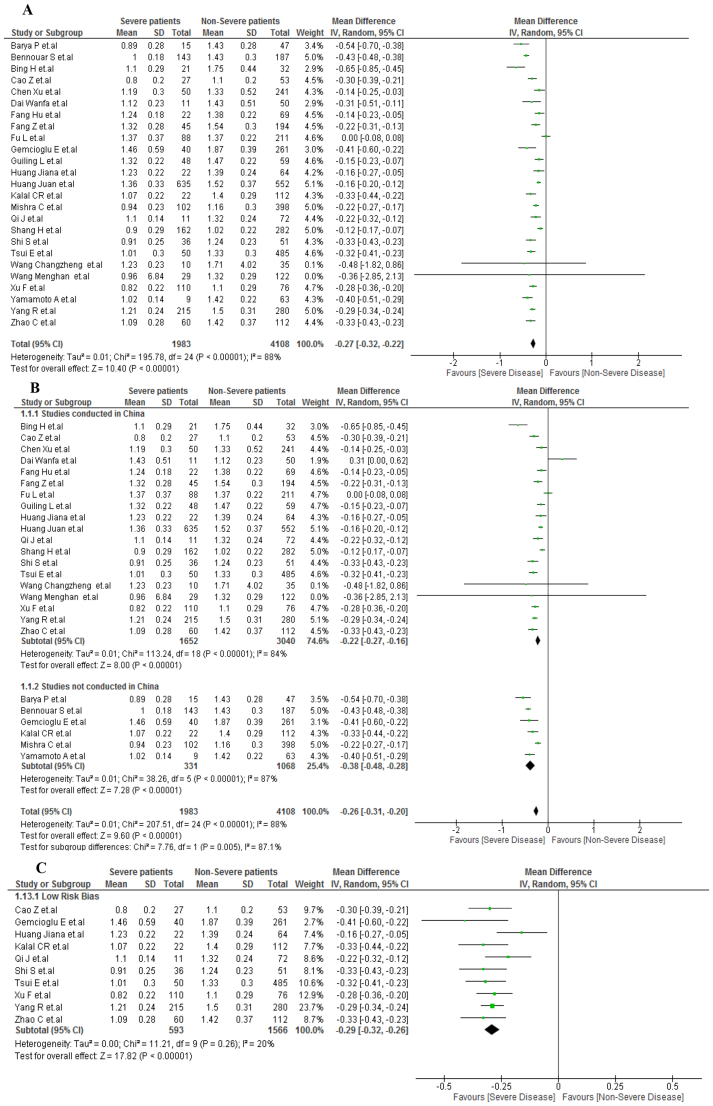
A total of 6081 patients were evaluated in 25 included articles, with 1983 patients developing severe COVID-19. Patients with severe COVID-19 had lower AGR values than non-severe COVID-19 patients (MD, −0.27; 95% IC, −0.32 to −0.22; p < 0.001) (Figure 2A). However, due to severe heterogeneity (I2 = 88%), a subgroup analysis was performed by study location. No significant differences were observed between the Chinese studies (MD, −0.22; 95% CI, −0.27 to −0.16; p < 0.001) and non-Chinese studies (MD, −0.38; 95% CI, −0.48 to −0,28; p < 0,001), with an interaction test p-value of <0.002 and severe heterogeneity in both subgroups (I2 > 85%) (Figure 2B). The sensitivity analysis included only low risk of bias studies and showed the same result: low AGR values for severe COVID-19 patients (MD, −0.29; 95% CI, −0.32 to −0.26; p < 0.001). Nonetheless, heterogeneity decreased significantly (I2 = 20%) (Figure 2C).
Figure 2.
A. AGR values in severe vs non-severe COVID-19 patients. B. Subgroup analysis according to country of origin between severe vs nonsevere COVID-19 patients. C. Sensitivity analysis according to the risk of bias between severe vs nonsevere COVID-19 patients.
3.4. AGR and COVID-19 mortality
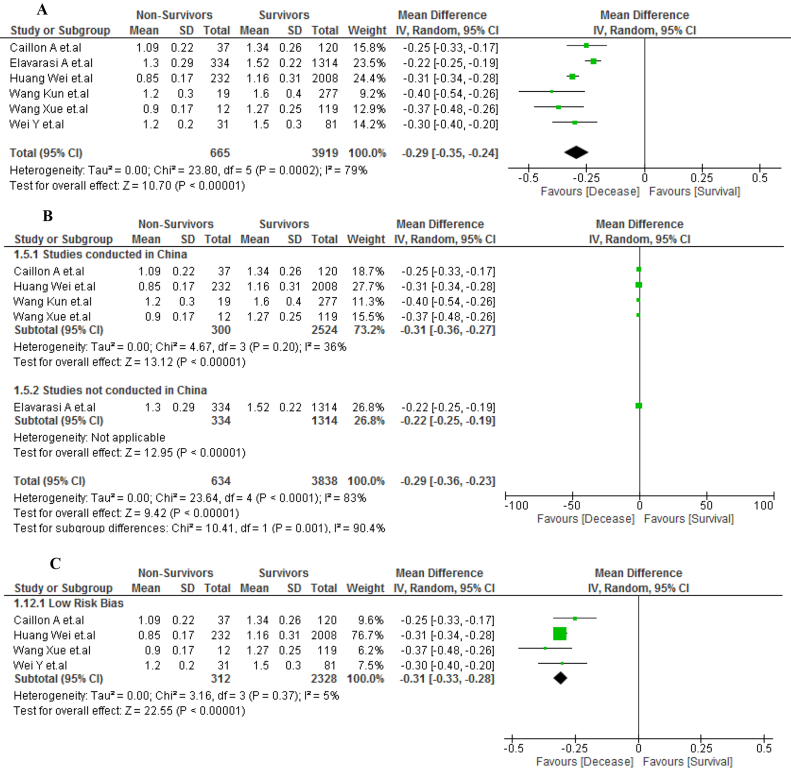
A total of 4584 patients were evaluated in six included articles, of which 665 patients diagnosed with COVID-19 died after hospitalization. The non-survivor patients with COVID-19 had lower AGR values than survivor patients (MD, −0.29; 95% IC, −0.35 to −0.24; p < 0.001) (Figure 3A). However, due to severe heterogeneity (I2 = 79%), a subgroup analysis was performed by study location. A mild heterogeneity (I2 = 36%) was obtained from the Chinese studies (MD, −0.31; 95% CI, −0.36 to −0.27; p < 0.001) (Figure 3B). The sensitivity analysis included only low risk of bias studies and showed the same result: low AGR values for non-survivor patients of COVID-19 (MD, −0.31; 95% CI, −0.33 to −0.28; p < 0.001). However, heterogeneity decreased significantly (I2 = 5%) (Figure 3C).
Figure 3.
A. AGR values in survivors vs non-survivors COVID-19 patients. B. Subgroup analysis according to country of origin between survivors vs non-survivors COVID-19 patients. C. Sensitivity analysis according to the risk of bias between survivors vs non-survivors COVID-19 patients.
3.5. Publication bias
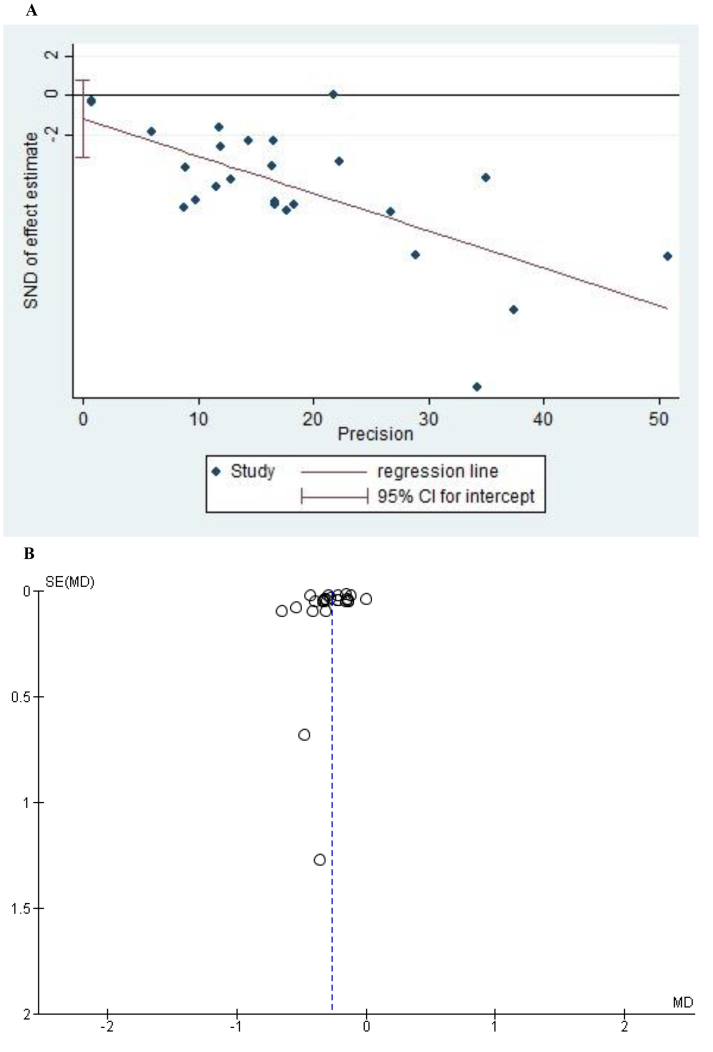
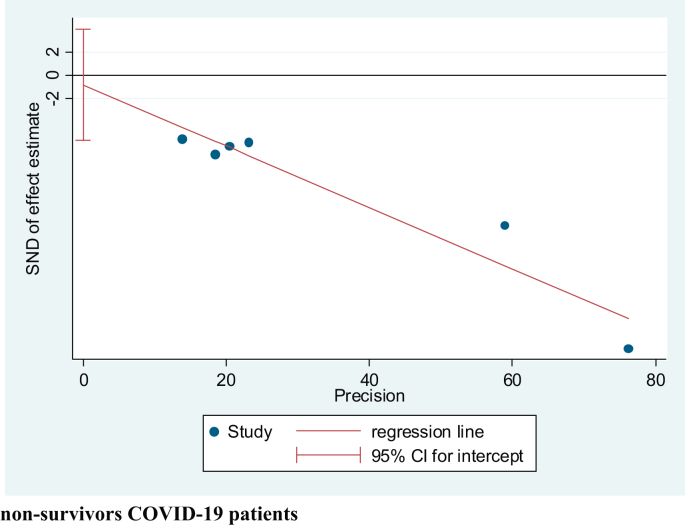
The Egger's tests performed to evaluate the publication bias of the studies (Figures 4A and 5) assessing the mortality and severity of the disease showed no indication of small-study effects (p = 0.658 and p = 0.219, respectively). Moreover, the funnel plot for the studies assessing severity showed a symmetrical pattern (Figure 4B).
Figure 4.
A: Egger Test of all the studies that evaluated AGR values in severe vs non-severe COVID-19 patients. B: Funnel Plot of the studies that evaluated AGR values in severe vs nonsevere COVID-19 patients.
Figure 5.
Egger Test of all the studies that evaluated AGR values in survivors vs non-survivors COVID-19 patients.
4. Discussion
The present systematic review and meta-analysis found that upon hospital admission, low AGR values were observed in COVID-19 patients who developed severe disease or died compared with patients who survived or did not develop severe disease.
Albumin and globulin are two primary components of serum proteins and are involved in systemic inflammation. Low albumin levels are associated with malnutrition and inflammation, whereas low globulin levels are associated with chronic inflammation and indicate cumulative exposure to various pro-inflammatory cytokines [55]. AGR is considered a more stable and reliable marker than serum albumin or globulin alone as a prognostic factor and has been associated with poor results in relation to different pathologies, such as digestive and urological cancers and solid cancers in general [56, 57, 58]. Similarly, it was associated with the prognosis of other diseases, such as heart failure and chronic kidney disease [10, 59]. In infectious diseases, AGR was associated with a worse prognosis in pediatric patients with tuberculous meningitis as a predictor of febrile urinary tract infection after ureteroscopic lithotripsy [60, 61].
In patients with COVID-19, the role of inflammation as a marker of severity and poor prognosis is suggested. A systematic review and meta-analysis of 23 studies found that in patients with severe COVID-19, high values of procalcitonin, C-reactive protein, D-dimer, and lactic dehydrogenase were found, as well as low values of albumin [62]. Similarly, a meta-analysis of 17 articles found a marked decrease in the values of lymphocytes, monocytes, eosinophils, platelets, albumin, the C-reactive protein/lymphocyte ratio, and the C-reactive protein/leukocyte ratio [63]. Likewise, the authors found high values of C-reactive protein, glomerular sedimentation rate, procalcitonin, lactate dehydrogenase, and others [63]. Thus, the inflammation in patients with COVID-19 raised the need to identify more stable markers to predict prognosis. In this way, the C-reactive protein/lymphocyte ratio, C-reactive protein/leukocyte ratio, neutrophil/monocyte ratio, and neutrophil/lymphocyte ratio demonstrated their potential as prognostic biomarkers in patients with sepsis [64, 65].
As mentioned, AGR is a better and more stable prognostic marker than albumin and globulin alone. Therefore, its association with severity and mortality in patients with COVID-19 may be related to the properties of these proteins. A meta-analysis of 67 studies had found that low albumin levels were associated with severity and worse prognosis in patients with COVID-19 [66]. Although the reasons for this association are not clear, its relationship to inflammation is believed to be one of them. A low level of albumin is related to the release of cytokines, including interleukin (IL)-6 and tumor necrosis factor (TNF)-α, and is inversely related to other markers, such as the neutrophil/lymphocyte ratio [57]. Therefore, a lower AGR value indicates more inflammation, which predicts more severity of the disease and, therefore, a greater probability of dying. Likewise, it is likely that as albumin values are influenced by other parameters independently associated with the severity and poorer prognosis of patients with COVID-19, they contribute to the prognostic value of the AGR, for example, age. In line with this, various meta-analyses had shown the association of older age with severity and poor prognosis in these patients [67]. On the other hand, various inflammation markers are associated with other sociodemographic factors of the AGR that were not evaluated and associated with worse prognosis in COVID-19 patients, which could confirm the findings of this study [68].
Although systematic reviews and meta-analyses on the role of various inflammation markers and their prognostic value in patients with COVID-19 have been published [4, 60], to our knowledge, this is the first study that focuses on the AGR in relation to COVID-19. Similarly, we performed a sensitivity analysis taking into account the biases of the studies, which gives robustness to our results. Likewise, our findings will allow us to suggest a potential low-cost prognostic marker in patients hospitalized for COVID-19 that will encourage health personnel to prioritize or individualize management strategies in patients with low AGR values.
4.1. Limitations
Our study needs to be rationally interpreted due to its limitations. First, the meta-analyses of both outcomes presented high heterogeneity, which indicates the clinical and methodological differences of the analyzed studies. However, we could explain this heterogeneity, which was led by studies that were not at low risk of bias and by differences between countries. Second, the calculated estimates come from averages without adjusting for confounding variables. Furthermore, it is necessary to conduct more epidemiological studies where an optimal cut-off point for the AGR and its prognostic value adjusted by confounding factors is defined. Third, the small sample size of some studies could cause them to be over-represented in the meta-analysis. However, the results of Egger's test did not show any statistical significance for the small-study effect. Fourth, although no language or region restriction was applied in the systematic search, most of the included studies have been published in Asia, so the assessment of other populations should be addressed in future research. Fifth, same levels of physiological parameters across countries are not directly comparable and possible levels of albumin and globulin need to have a geographical context when we say high or low. Sixth, the included studies did not consider the modulatory role played by factors, such as age, gender, and the presence or absence of comorbidities. Likewise, it is also critical to have the context of the samples included in the different studies, since it is possible that the potential role of the virus in modulating the levels of albumin and globulin has evolved. Finally, we cannot confirm the role of this marker in non-severe patients, and although the role of inflammation was suggested, the studies did not consider these markers to confirm this hypothesis.
5. Conclusion
The AGR is a biomarker that can predict the severity and mortality of COVID-19 patients. Moreover, low AGR values upon hospital admission were observed in COVID-19 patients who developed severe disease or died.
Declarations
Author contribution statement
Juan R. Ulloque-Badaracco, Melany D. Mosquera-Rojas, Enrique A Hernandez-Bustamante, Esteban A Alarcón-Braga: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Percy Herrera – Añazco and Vicente A. Benites-Zapata: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
None.
References
- 1.WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. https://covid19.who.int/ (n.d.)
- 2.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57:389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi X., Su Z., Yan H., Du J., Wang J., Chen L., Peng M., Chen S., Shen B., Li J. Prediction of severe illness due to COVID-19 based on an analysis of initial fibrinogen to albumin ratio and platelet count. Platelets. 2020;31:674–679. doi: 10.1080/09537104.2020.1760230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stringer D., Braude P., Myint P.K., Evans L., Collins J.T., Verduri A., Quinn T.J., Vilches-Moraga A., Stechman M.J., Pearce L., Moug S., McCarthy K., Hewitt J., Carter B., COPE Study Collaborators The role of C-reactive protein as a prognostic marker in COVID-19. Int. J. Epidemiol. 2021;50:420–429. doi: 10.1093/ije/dyab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonetti G., Manelli F., Patroni A., Bettinardi A., Borrelli G., Fiordalisi G., Marino A., Menolfi A., Saggini S., Volpi R., Anesi A., Lippi G. Laboratory predictors of death from coronavirus disease 2019 (COVID-19) in the area of Valcamonica, Italy. Clin. Chem. Lab. Med. 2020;58:1100–1105. doi: 10.1515/cclm-2020-0459. [DOI] [PubMed] [Google Scholar]
- 6.Piva E., Zuin J., Pelloso M., Tosato F., Fogar P., Plebani M. Monocyte distribution width (MDW) parameter as a sepsis indicator in intensive care units. Clin. Chem. Lab. Med. 2021;59:1307–1314. doi: 10.1515/cclm-2021-0192. [DOI] [PubMed] [Google Scholar]
- 7.Ulloque-Badaracco J.R., Hernandez-Bustamante E.A., Herrera-Añazco P., Benites-Zapata V.A. Prognostic value of apolipoproteins in COVID-19 patients: a systematic review and meta-analysis. Trav. Med. Infect. Dis. 2021;44:102200. doi: 10.1016/j.tmaid.2021.102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulloque-Badaracco J.R., Ivan Salas-Tello W., Al-Kassab-Córdova A., Alarcón-Braga E.A., Benites-Zapata V.A., Maguiña J.L., Hernandez A.V. Prognostic value of neutrophil-to-lymphocyte ratio in COVID-19 patients: a systematic review and meta-analysis. Int. J. Clin. Pract. 2021;75 doi: 10.1111/ijcp.14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Li S., Hu X., Wang Y., Wu Y., Li P., Che G. The prognostic value of serum albumin-globulin ratio in early-stage non-small cell lung cancer: a retrospective study. Cancer Manag. Res. 2019;11:3545–3554. doi: 10.2147/CMAR.S191288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li K., Fu W., Bo Y., Zhu Y. Effect of albumin-globulin score and albumin to globulin ratio on survival in patients with heart failure: a retrospective cohort study in China. BMJ Open. 2018;8 doi: 10.1136/bmjopen-2018-022960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng Y., Pang Q., Miao R.C., Chen W., Zhou Y.Y., Bi J.B., Liu S.S., Zhang J.Y., Qu K., Liu C. Prognostic significance of pretreatment albumin/globulin ratio in patients with hepatocellular carcinoma. OncoTargets Ther. 2016;9:5317–5328. doi: 10.2147/OTT.S109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng B., Zhou B., Zhang S., Zhu Y., Han L., Geng Y., Jin Z., Liu H., Wang D., Zhao Y., Wen Y., Cui W., Zhou Y., Gu Q., Sun C., Lu X., Wang W., Wang Y., Li C., Wang Y., Yao W., Liu P. Clinical features and factors associated with severity and fatality among patients with severe fever with thrombocytopenia syndrome bunyavirus infection in Northeast China. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Wang T., Fang Y., Wang M., Liu W., Zhao J., Wang B., Wu Z., Lv Y., Wu R. Clinical significance of serum albumin/globulin ratio in patients with pyogenic Lliver Aabscess. Front. Surg. 2021;8:1–10. doi: 10.3389/fsurg.2021.677799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W., Zhangyuan G., Wang F., Zhang H., Yu D., Wang J., Jin K., Yu W., Liu Y., Sun B. High preoperative serum globulin in hepatocellular carcinoma is a risk factor for poor survival. J. Cancer. 2019;10:3494–3500. doi: 10.7150/jca.29499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feketea G.M., Vlacha V. The diagnostic significance of usual biochemical parameters in coronavirus Disease 19 (COVID-19): albumin to globulin ratio and CRP to albumin ratio. Front. Med. 2020;7:566591. doi: 10.3389/fmed.2020.566591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S., Zhang Y., Li M., Xie C., Wu H. Serum albumin, a good indicator of persistent organ failure in acute pancreatitis. BMC Gastroenterol. 2017;17:59. doi: 10.1186/s12876-017-0615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kheir M., Saleem F., Wang C., Mann A., Chua J. Higher albumin levels on admission predict better prognosis in patients with confirmed COVID-19. PLOS One. 2021;16 doi: 10.1371/journal.pone.0248358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdeen Y., Kaako A., Ahmad Amin Z., Muhanna A., Josefine Froessl L.J., Alnabulsi M., Okeh A., Miller R.A. The prognostic effect of serum albumin level on outcomes of hospitalized COVID-19 patients. Crit. Care Res. Pract. 2021;2021:9963274. doi: 10.1155/2021/9963274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGowan J., Sampson M., Salzwedel D.M., Cogo E., Foerster V., Lefebvre C. PRESS peer review of electronic search strategies: 2015 guideline statement. J. Clin. Epidemiol. 2016;75:40–46. doi: 10.1016/j.jclinepi.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Ouzzani M., Hammady H., Fedorowicz Z., Elmagarmid A. Rayyan—a web and mobile app for systematic reviews. Syst. Rev. 2016;5:1. doi: 10.1186/s13643-016-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells G., Shea B., O’Connell D., et al. Ottawa Hospital Research Institute; 2011. The Newcastle-Ottawa scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses.http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp [Google Scholar]
- 23.Hozo S.P., Djulbegovic B., Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med. Res. Methodol. 2005;5:1. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu L., Fei J., Xu S., Xiang H.X., Xiang Y., Hu B., Li M.D., Liu F.F., Li Y., Li X.Y., Zhao H., Xu D.X. Liver dysfunction and its association with the risk of death in COVID-19 patients: a prospective cohort study. J. Clin. Transl. Hepatol. 2020;8:246–254. doi: 10.14218/JCTH.2020.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G., Du L., Cao X., Wei X., Jiang Y., Lin Y., Nguyen V., Tan W., Wang H. Follow-up study on serum cholesterol profiles and potential sequelae in recovered COVID-19 patients. BMC Infect. Dis. 2021;21:299. doi: 10.1186/s12879-021-05984-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gemcioglu E., Davutoglu M., Catalbas R., Karabuga B., Kaptan E., Aypak A., Kalem A.K., Özdemir M., Yeşilova N.Y., Kalkan E.A., Civak M., Kücüksahin O., Erden A., Ates I. Predictive values of biochemical markers as early indicators for severe COVID-19 cases in admission. Future Virol. 2021;16:353–367. [Google Scholar]
- 27.Cao Z., Li T., Liang L., Wang H., Wei F., Meng S., Cai M., Zhang Y., Xu H., Zhang J., Jin R. Clinical characteristics of coronavirus disease 2019 patients in Beijing, China. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi S., Liu X., Xiao J., Wang H., Chen L., Li J., Han K. Prediction of adverse clinical outcomes in patients with coronavirus disease 2019. J. Clin. Lab. Anal. 2021;35 doi: 10.1002/jcla.23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R., Gui X., Ke H., Gao S., Luo M., Xiong Y. The indicative role of markers for liver injury on the severity and prognosis of coronavirus disease 2019 patients. Eur. J. Gastroenterol. Hepatol. 2020 doi: 10.1097/MEG.0000000000001968. [DOI] [PubMed] [Google Scholar]
- 30.Tsui E.L.H., Lui C.S.M., Woo P.P.S., Cheung A.T.L., Lam P.K.W., Tang V.T.W., Yiu C.F., Wan C.H., Lee L.H.Y. Development of a data-driven COVID-19 prognostication tool to inform triage and step-down care for hospitalised patients in Hong Kong: a population-based cohort study. BMC Med. Inf. Decis. Making. 2020;20:323. doi: 10.1186/s12911-020-01338-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennouar S., Bachir Cherif A.B., Kessira A., Hamel H., Boudahdir A., Bouamra A., Bennouar D., Abdi S. Usefulness of biological markers in the early prediction of corona virus disease-2019 severity. Scand. J. Clin. Lab. Investig. 2020;80:611–618. doi: 10.1080/00365513.2020.1821396. [DOI] [PubMed] [Google Scholar]
- 32.Huang J., Gao J., Zhu W., Feng R., Liu Q., Chen X., Huang J., Yang Z., Lin X., Zhang Z., Lin Y. Indicators and prediction models for the severity of Covid-19. Int. J. Clin. Pract. 2021;75 doi: 10.1111/ijcp.14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra C., Yadav Y.K., Verma V., Kumar V. Association of serum albumin and lipoproteins as strong predictor of severity in patients with COVID-19. Int. J. Handheld Comput. Res. (IJHCR) 2021;4:40–42. https://www.ijhcr.com/index.php/ijhcr/article/view/1570 [Google Scholar]
- 34.Wang C., Deng R., Gou L., Fu Z., Zhang X., Shao F., Wang G., Fu W., Xiao J., Ding X., Li T., Xiao X., Li C. Preliminary study to identify severe from moderate cases of COVID-19 using combined hematology parameters. Ann. Transl. Med. 2020;8:593. doi: 10.21037/atm-20-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng F., Yao R., Liu J., Chen R., Li N. 2020. Nomogram Prediction of Severe Risk in Patients with COVID-19 Pneumonia: A Retrospective Study. [Google Scholar]
- 36.Wang M., Yu D., Shang Y., Zhang X., Yang Y., Zhao S., Su D., Liu L., Wang Q., Ren J., Li Y., Chen H. Predictive score of risk associated with progression of patients with COVID-19 pneumonia in Wuhan, China: the ALA score. Arabian J. Sci. Eng. 2021;1:1–9. doi: 10.1007/s13369-021-05808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C., Bai Y., Wang C., Zhong Y., Lu N., Tian L., Cai F., Jin R. Risk factors related to the severity of COVID-19 in Wuhan. Int. J. Med. Sci. 2021;18:120–127. doi: 10.7150/ijms.47193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalal C.R., Joshi H., Kumar V., Gopal D., Rathod D., Shukla A., Gianchandani T., Bhatt C. Clinical significance of liver function abnormality in patients with COVID-19: a single-center experience from Western India. J. Clin. Transl. Hepatol. 2021;9:878–888. doi: 10.14218/JCTH.2020.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J., Zhu L., Bai X., Jia X., Lu Y., Deng A., Li J., Jin S. Multidimensional analysis of risk factors for the severity and mortality of patients with COVID-19 and diabetes. Infect. Dis. Ther. 2020;9:981–1002. doi: 10.1007/s40121-020-00359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang H., Huang C., Chen Y., Zhang S., Yang P., Hong G., Zhang L., Hou X. 2020. Digestive Involvement in SARS-CoV-2 Infection: A Retrospective Multi-Center Study. [Google Scholar]
- 41.Chen X., Zheng F., Qing Y., Ding S., Yang D., Lei C., Yin Z., Zhou X., Jiang D., Zuo Q., He J., Lv J., Chen P., Chen Y., Peng H., Li H., Xie Y., Liu J., Zhou Z., Luo H. Epidemiological and clinical features of 291 cases with coronavirus disease 2019 in areas adjacent to hubei, China: a double-center observational study. medRxiv. 2020 [Google Scholar]
- 42.Yamamoto A., Wada H., Ichikawa Y., Mizuno H., Tomida M., Masuda J., Makino K., Kodama S., Yoshida M., Fukui S., Moritani I., Inoue H., Shiraki K., Shimpo H. Evaluation of biomarkers of severity in patients with COVID-19 infection. J. Clin. Med. 2021;10:3775. doi: 10.3390/jcm10173775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barya P., Narang S., Jain V.K., Shrivastava R. Neutrophil-–lymphocytic ratio (NLR) or albumin-globulin ratio (AGR) which one is a better predictor marker in assessment of severity of COVID-19 infection. Int. J. Med. Biomed. Stud. 2021;5:188–198. [Google Scholar]
- 44.Hu F., Guo Y., Lin J., Zeng Y., Wang J., Li M., Cong L. Association of serum uric acid levels with COVID-19 severity. BMC Endocr. Disord. 2021;21:97. doi: 10.1186/s12902-021-00745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi J., He D., Yang D., Wang M., Ma W., Cui H., Ye F., Wang F., Xu J., Li Z., Liu C., Wu J., Qi K., Wu R., Huang J., Liu S., Zhu Y. Severity-associated markers and assessment model for predicting the severity of COVID-19: a retrospective study in Hangzhou, China. BMC Infect. Dis. 2021;21:1. doi: 10.1186/s12879-021-06509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai W., Ke P.F., Li Z.Z., Zhuang Q.Z., Huang W., Wang Y., Xiong Y., Huang X.Z. Establishing classifiers with Clinical Laboratory indicators to distinguish COVID-19 from community-acquired pneumonia: retrospective cohort study. J. Med. Internet Res. 2021;23 doi: 10.2196/23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He B., Wang J., Wang Y., Zhao J., Huang J., Tian Y., Yang C., Zhang H., Zhang M., Gu L., Zhou X., Zhou J. The metabolic changes and immune profiles in patients with COVID-19. Front. Immunol. 2020;11:2075. doi: 10.3389/fimmu.2020.02075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu F., Qin X., Zhang L., Wu F., Jin Y., Xu Y., Liu C., Xiong Y., Li G., Xiang X., Jiang Y., Bai T., Hou X., Song J. Clinical characteristics of gastrointestinal symptoms and liver function injury in patients with coronavirus disease 2019. Chin. J. Digest. 2020;12:249–256. [Google Scholar]
- 49.Wei Y., He J., Chen J., Zhu Y., Chen J., Ding J., Wang H., Hu Y., Huang Y., Jiang Y., Pan Z., Shen S., Zhao W., Gao W., Chen F., Lu X. 2020. Dynamic Prognosis Model for Predicting Survival in Severe and Critically Ill COVID-19 Patients Using Machine Learning. [Google Scholar]
- 50.Wang X., Li X., Shang Y., Wang J., Zhang X., Su D., Zhao S., Wang Q., Liu L., Li Y., Chen H. Ratios of neutrophil-to-lymphocyte and platelet-to-lymphocyte predict all-cause mortality in inpatients with coronavirus disease 2019 (COVID-19): a retrospective cohort study in a single medical centre. Epidemiol. Infect. 2020;148:e211. doi: 10.1017/S0950268820002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caillon A., Zhao K., Klein K.O., Greenwood C.M.T., Lu Z., Paradis P., Schiffrin E.L. High systolic blood pressure at hospital admission is an important risk factor in models predicting outcome of COVID-19 patients. Am. J. Hypertens. 2021;34:282–290. doi: 10.1093/ajh/hpaa225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K., Zuo P., Liu Y., Zhang M., Zhao X., Xie S., Zhang H., Chen X., Liu C. Clinical and laboratory predictors of in-hospital mortality in patients with coronavirus Disease-2019: a cohort study in Wuhan, China. Clin. Infect. Dis. : Off. Publ. Infect. Dis. Soc. Am. 2020;71:2079–2088. doi: 10.1093/cid/ciaa538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elavarasi A., Sagiraju H.K.R., Garg R.K., Paul S.S., Ratre B., Sirohiya P., Gupta N., Garg R., Pandit A., Vig S., Singh R., Kumar B., Meena V.P., Wig N., Mittal S., Pahuja S., Madan K., Dwivedi T., Das N., Gupta R., Vidyarthi A.J., Das A., Chaudhary R., Wundawalli L., Singh A.R., Singh S., Pandey M., Mishra A., Matharoo K.S., Kumar S., Mohan A., Guleria R., Bhatnagar S. Clinical features, demography and predictors of outcomes of SARS-CoV-2 infection in a tertiary care hospital in India-A cohort study. medRxiv. 2021 doi: 10.4103/lungindia.lungindia_493_21. 2021.08.10.21261855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang W., Li C., Wang Z., Wang H., Zhou N., Jiang J., Ni L., Zhang X.A., Wang D.-W. Decreased serum albumin level indicates poor prognosis of COVID-19 patients: hepatic injury analysis from 2,623 hospitalized cases, Science China. Life Sci. 2020;63:1678–1687. doi: 10.1007/s11427-020-1733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopal D.M., Kalogeropoulos A.P., Georgiopoulou V.V., Tang W.W., Methvin A., Smith A.L., Bauer D.C., Newman A.B., Kim L., Harris T.B., Kritchevsky S.B., Butler J., Health ABC Study Serum albumin concentration and heart failure risk the health, aging, and body composition study. Am. Heart J. 2010;160:279–285. doi: 10.1016/j.ahj.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo H.W., Yuan T.Z., Chen J.X., Zheng Y. Prognostic value of pretreatment albumin/globulin ratio in digestive system cancers: a meta-analysis. PLoS One. 2018;13 doi: 10.1371/journal.pone.0189839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y., Wang L., Lin S., Wang R. Preoperative albumin-to-globulin ratio as a significant prognostic indicator in urologic cancers: a meta-analysis. Cancer Manag. Res. 2018;10:4695–4708. doi: 10.2147/CMAR.S178271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He J., Pan H., Liang W., Xiao D., Chen X., Guo M., He J. Prognostic effect of albumin-to-globulin ratio in patients with solid tumors: a systematic review and meta-analysis. J. Cancer. 2017;8:4002–4010. doi: 10.7150/jca.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu P.-P., Hsieh Y.-P., Kor C.-T., Chiu P.-F. Association between albumin–globulin ratio and mortality in patients with chronic kidney disease. J. Clin. Med. 2019;8 doi: 10.3390/jcm8111991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li K., Tang H., Yang Y., Li Q., Zhou Y., Ren M., Long X., Shen W., Hu R., Wang X., Zeng K. Clinical features, long-term clinical outcomes, and prognostic factors of tuberculous meningitis in West China: a multivariate analysis of 154 adults. Expert Rev. Anti-infect. Ther. 2017;15:629–635. doi: 10.1080/14787210.2017.1309974. [DOI] [PubMed] [Google Scholar]
- 61.Yi S.Y., Park D.J., Min K., Chung J.W., Ha Y.S., Kim B.S., Kim H.T., Kim T.H., Yoo E.S. Significance of albumin to globulin ratio as a predictor of febrile urinary tract infection after ureteroscopic lithotripsy. Yeungnam Univ. J. Med. 2021;38:225–230. doi: 10.12701/yujm.2021.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hariyanto T.I., Japar K.V., Kwenandar F., Damay V., Siregar J.I., Lugito N.P.H., Tjiang M.M., Kurniawan A. Inflammatory and hematologic markers as predictors of severe outcomes in COVID-19 infection: a systematic review and meta-analysis. Am. J. Emerg. Med. 2021;41:110–119. doi: 10.1016/j.ajem.2020.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghahramani S., Tabrizi R., Lankarani K.B., Kashani S.M.A., Rezaei S., Zeidi N., Akbari M., Heydari S.T., Akbari H., Nowrouzi-Sohrabi P., Ahmadizar F. Laboratory features of severe vs. non-severe COVID-19 patients in Asian populations: a systematic review and meta-analysis. Eur. J. Med. Res. 2020;25:30. doi: 10.1186/s40001-020-00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kilercik M., Demirelce Ö., Serdar M.A., Mikailova P., Serteser M. A new haematocytometric index: predicting severity and mortality risk value in COVID-19 patients. PLoS One. 2021;16 doi: 10.1371/journal.pone.0254073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Z., Fu Z., Huang W., Huang K. Prognostic value of neutrophil-to-lymphocyte ratio in sepsis: a meta-analysis. Am. J. Emerg. Med. 2020;38:641–647. doi: 10.1016/j.ajem.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 66.Paliogiannis P., Mangoni A.A., Cangemi M., Fois A.G., Carru C., Zinellu A. Serum albumin concentrations are associated with disease severity and outcomes in coronavirus 19 disease (COVID-19): a systematic review and meta-analysis. Clin. Exp. Med. 2021;21:343–354. doi: 10.1007/s10238-021-00686-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J., Li Q., Jiang C., Zhou Y., Liu S., Ye C., Zhang P., Xing Y., Guo H., Tang W. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J. Infect. 2020;81:e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howard R., Scheiner A., Kanetsky P.A., Egan K.M. Sociodemographic and lifestyle factors associated with the neutrophil-to-lymphocyte ratio. Ann. Epidemiol. 2019;38:11–21. doi: 10.1016/j.annepidem.2019.07.015. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.