Abstract
Context
Observational studies have suggested associations between adipokines and cardiovascular disease (CVD), but the roles of certain adipokines remain controversial, and these associations have not yet been ascertained causally.
Objective
To investigate whether circulating adipokines causally affect the risk of CVD using 2-sample Mendelian randomization (MR).
Methods
Independent genetic variants strongly associated with adiponectin, resistin, chemerin, and retinol binding protein 4 (RBP4) were selected from public genome-wide association studies. Summary-level statistics for CVD, including coronary artery disease (CAD), myocardial infarction, atrial fibrillation (AF), heart failure (HF), and stroke and its subtypes were collected. The inverse-variance weighted and Wald ratio methods were used for the MR estimates. The MR pleiotropy residual sum and outlier, weighted median, MR-Egger, leave-one-out analysis, MR Steiger, and colocalization analyses were used in the sensitivity analysis.
Results
Genetically predicted resistin levels were positively associated with AF risk (odds ratio [OR] 1.09; 95% confidence interval [CI], 1.04-1.13; P = 4.1 × 10-5), which was attenuated to null after adjusting for blood pressure. We observed suggestive associations between higher genetically predicted chemerin levels and an increased risk of CAD (OR 1.27; 95% CI, 1.01-1.60; P = 0.040), higher genetically predicted RBP4 levels and an increased risk of HF (OR 1.14; 95% CI, 1.02-1.27; P = 0.024). There was no causal association between genetically predicted adiponectin levels and CVD risk.
Conclusions
Our findings reveal the causal association between resistin and AF, probably acting through blood pressure, and suggest potential causal associations between chemerin and CAD, RBP4, and HF.
Keywords: adiponectin, resistin, chemerin, retinol binding protein 4, cardiovascular disease, Mendelian randomization
Cardiovascular disease (CVD) remains the leading cause of mortality globally with an increase in estimated years of life lost, though the treatment of CVD has advanced (1). The cause of CVD is not completely understood, despite substantial progress in prevention and control (2). Thus, determining the protective or causative factors in CVD remains critical.
Adipokines, a variety of bioactive compounds mainly secreted by adipose tissue, may act on the energy balance, immune responses, vascular homeostasis, angiogenesis, insulin sensitivity, and lipid metabolism and, ultimately, through these effects, directly or indirectly affect CVD (3). It is now widely believed that dysfunctional adipose tissue remodeling leads to an unbalanced production of pro- and anti-inflammatory adipokines, which contributes to a systemic pro-inflammatory state and has important adverse effects on the cardiovascular system (3, 4). However, the epidemiological conclusions remain contradictory. For instance, an earlier prospective and meta-analysis study demonstrated that adiponectin was not associated with the risk of stroke (5). But recently, a prospective study showed that adiponectin was associated with moderate-to-high stroke and indicated it could be a biomarker of poor outcome after stroke with a 31% increase in mortality risk (6). Likewise, some studies have reported that chemerin is associated with inflammatory markers and thus exhibits pro-inflammatory properties (7-9). In contrast, Yamawaki et al. indicated that chemerin showed anti-inflammatory effects (10). Such paradoxes are common in the research on adipokines. Furthermore, evidence from conventional epidemiological studies cannot resolve the confusion caused by various biases and reverse causality and is somewhat limited by small sample sizes; thus, it is still difficult to determine whether the effects of circulating adipokines on CVD risk are causal or merely shared pleiotropic factors.
In this case, Mendelian randomization (MR) provides a means of exploring the causal associations between exposures and outcomes without any potentially harmful intervention (11). A previous MR study used several coronary artery disease (CAD) datasets and adjusted a series of CAD risk factors, indicating that adiponectin had no causal relationship in the pathogenesis of CAD (12). Au Yeung et al. confirmed this finding and added that there was no reverse causal relationship between adiponectin and CAD through bidirectional MR analysis (13), although the inclusion criteria of instrumental variables in these studies are relatively loose and the causal effects on other CVDs remain unknown. To examine the role of circulating adipokines, we systematically searched genome-wide association studies (GWASs) for 4 genetically proxied circulating adipokines, namely adiponectin, resistin, chemerin, and retinol binding protein 4 (RBP4). We conducted a 2-sample MR study to clarify the potential effects of genetically predicted circulating levels of these adipokines on 9 CVDs: CAD, myocardial infarction (MI), atrial fibrillation (AF), heart failure (HF), and stroke and its subtypes. As elevated blood pressure has been demonstrated to be a risk factor for CVDs by large cohort studies, meta-analysis studies, and an MR study (14, 15), we assessed the potential mediating effects by blood pressure on the identified causal associations using multivariable Mendelian randomization (MVMR).
Materials and Methods
Study Design
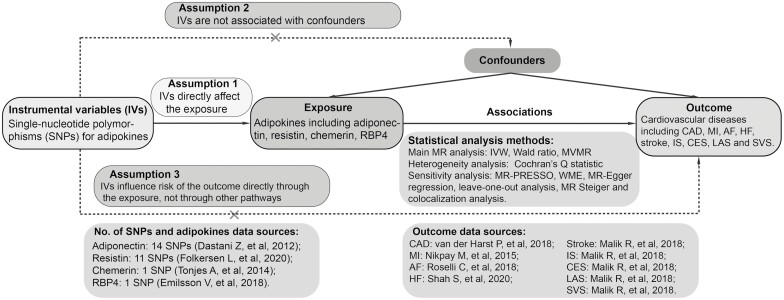
We performed a 2-sample MR study using data obtained from the publicly available GWAS catalog (https://www.ebi.ac.uk/gwas). Ethical approval and consent to participate were carried out in the original publications. An overview of the study design is shown in Fig. 1.
Figure 1.
Study flow diagram. Dashed lines indicate potential pleiotropic or direct causal effects between variables that may violate MR assumptions. Abbreviations: AF, atrial fibrillation; CAD, coronary artery disease; CES, cardioembolic stroke; HF, heart failure; IS, ischemic stroke; IV, instrumental variable; IVW, multiplicative random effects inverse-variance weighted; LAS, large-artery atherosclerotic stroke; MI, myocardial infarction; MR, Mendelian randomization; MR-PRESSO, MR pleiotropy residual sum and outlier; MVMR, multivariable Mendelian randomization; RBP4, retinol binding protein 4; SVS, small-vessel stroke; WME, weighted-median estimator.
Selection of Genetic Instrumental Variables
The GWAS catalog was searched to extract single-nucleotide polymorphisms (SNPs) as genetic instrumental variables. This study collected SNPs that were at genome-wide significance (5 × 10-8) and independently (linkage disequilibrium [LD], r2 ≤ 0.001) predicted circulating adipokines in populations of European ancestry. When multiple studies existed for a single adipokine, only the largest one with replication was selected. When there were SNPs with highly linkage disequilibrium, only the one with the minimum P value was selected.
For adiponectin, 14 SNPs were obtained from a large-scale GWAS with 45 891 participants (including 29 347 European ancestry individuals in the discovery phase and multiethnic individuals in the follow-up phase) that were adjusted for age, sex, body mass index (BMI), and principal components (PCs) (16). For resistin, 11 SNPs were identified from the recent genome-wide meta-analysis of 30 931 individuals (including 21 758 individuals in the discovery cohort and 9173 individuals in the replication cohort) in 15 studies that adjusted for age, sex, and PCs (17). For chemerin, 1 SNP was obtained from the GWAS with 2791 European participants in the discovery cohort and 967 participants in the replication cohort, which were adjusted for age, sex, and BMI (18). For RBP4, 1 SNP was identified from a public GWAS containing 5457 Icelanders that was adjusted for age (19). For adiponectin and resistin, considering the availability of full summary-level statistics, genetic instruments were extracted from the discovery cohorts. To prevent potential pleiotropy, the SNPs were further searched using PhenoScanner V2 (http://www.phenoscanner.medschl.cam.ac.uk/) to evaluate whether the instrumental variables were potentially associated with confounding or risk factors for CVD (20, 21). We used 3 sets of instrumental variables in our study (except chemerin and RBP4, with only 1 SNP in the corresponding gene locus):
Conservative analysis, considering both the number of SNPs for sensitivity analysis and the statistical power of results, potentially pleiotropic variables were discarded.
Liberal analysis, covering all identified SNPs to test our results and further detect the confounding or mediating effects of those potentially pleiotropic variables.
Gene locus analysis, restricting MR estimate to SNPs at corresponding adipokine gene locus because such variants might be less biased by horizontal pleiotropy and more associated with the perturbation of drug targets (22).
Outcome Data Sources
Summary data for CAD were obtained from a large-scale GWAS that meta-analyzed 122 733 cases and 424 528 controls from UK Biobank and the CARDIoGRAMplusC4D consortium (23). Genetic associations with MI were also derived from the CARDIoGRAMplusC4D consortium, which conducted a GWAS comprising 43 676 MI cases and 128 199 controls across 48 studies, 77% of whom were of European ancestry (24). Summary-level data for AF were extracted from a large-scale meta-analysis of GWAS, including 537 409 subjects of European ancestry (55 114 cases and 482 295 controls) (25). Summary statistics for HF came from the HERMES consortium, which comprised 47 309 cases and 930 014 controls of European ancestry from 26 studies (26). Summary statistics for stroke were extracted from the MEGASTROKE consortium, including 446 696 subjects of European ancestry (40 585 cases and 406 111 controls) (27). Among these stroke cases, 34 217 were defined as ischemic stroke, which was further divided into 3 subtypes, namely cardioembolic stroke (7193 cases), large-artery atherosclerotic stroke (4373 cases), and small-vessel stroke (5386 cases). There were no overlapping populations between the exposures and outcomes GWASs.
Statistical Power and Instrument Strength
Variance (R2) in MR studies represents the proportion of the variability of the exposure explained by each genetic instrument. The R2 for the adipokines was calculated in accordance with what has been described previously using the following formula: , where EAF means effect allele frequency, beta and se means the estimated effect and its standard error of SNP on certain adipokine, and Ν means the sample size of the GWAS for the SNP-adipokine association (28, 29). Subsequently, power calculations were performed using an online tool available at http://cnsgenomics.com/shiny/mRnd/ (30). To avoid potential weak instrument bias, we calculated F-statistics to test the strength of each instrument using the following formula: , where N refers to the sample size of the GWAS for the selected SNP (31).
Statistical Analysis
Our 2-sample MR approach harmonized the effect of adipokines and CVD datasets, which comprised integrated information on SNPs, specifically effect allele, beta-coefficient, standard error, P value, and sample size. Proxy SNPs (r2 > 0.8) were applied to specific adipokine-associated SNPs that were absent in outcome datasets, and SNPs without suitable proxies were excluded from the analyses. The primary MR analysis was performed by employing the multiplicative random effects inverse variance-weighted (IVW) method that meta-analyzed the SNP-specific Wald estimates with the assumption of balanced pleiotropy (32). The Wald ratio method was applied if the MR estimate contained a single SNP. MVMR was performed to assess the potential mediating effect of blood pressure using GWAS summary statistics obtained from the International Consortium of Blood Pressure (33). Results are presented as odds ratios (ORs) and 95% confidence intervals (CIs) on CVD risk for per-unit change in circulating concentrations of adipokines on inverse-normal transformed levels (resistin, ng/mL) or natural log transformed levels (adiponectin, µg/mL; chemerin, ng/mL; RBP4, µg/mL).
Sensitivity Analysis
Several statistical tests were performed to examine the existence of horizontal pleiotropy that violated the main MR assumptions. We calculated the Cochran Q statistic that quantifies the heterogeneity in effect sizes resulting from the selected genetic instrumental variables. An MR pleiotropy residual sum and outlier (MR-PRESSO) analysis was applied to detect and adjust for horizontal pleiotropy through removing outliers (34). We also performed an MR-Egger regression and weighted-median estimator (WME) in the sensitivity analyses (35). Horizontal pleiotropy was evaluated by estimating the deviation of the MR-Egger intercept, with the difference from 0 indicating potential bias in the MR estimates (36). The WME method was complemented to generate a robust and consistent estimate of the effect, even if up to 50% of the weight came from invalid instrumental variables (37). A leave-one-out analysis was also applied to check for any pleiotropy affected by a single SNP. We performed the MR Steiger test to estimate the potential reverse causal impact of CVD on adipokines (38). In addition, we conducted colocalization analysis using the commonly applied Bayesian model to investigate whether adipokines and CVD share a common causal variant in a given region (39). For adipokine gene locus in which there was evidence to support a causal relationship on CVD (P < 0.05), variables within 200 kb of the corresponding instrumental SNP were extracted and sent to calculate the posterior probability (PP). As a convention, a PP.H4 of 0.80 or higher was considered evidence of colocalization.
The P values in this study were 2-sided, and values < 0.05 were deemed as suggestive significance, whereas the highly reliable findings were those survivals with a Bonferroni-corrected threshold of 0.001 (0.05/36). All these analyses were implemented using the “TwoSampleMR” and “coloc” packages in R Version 4.1.1.
Results
Genetic Instruments for Adipokines
Genetic variants used in the conservative analysis are listed in Table 1. After searching in Phenoscanner and the GWAS catalog, rs1108842, rs1597466, rs2980879, rs601339, rs2927324, rs731839, rs2239619, and rs445 were removed for being associated with known confounders (body traits, hypertension, and lipid levels) of CVD or directly affecting CAD and AF (Supplementary Table 1) (40). The summary characteristics of the final instrumental variables for adipokines and CVD are shown in Supplementary Table 2 (40). The collectively explained variance for adiponectin, resistin, chemerin, and RBP4 was 4.6%, 3.5%, 1.5%, and 1.1%, respectively. The F values of the selected variables ranged from 33 to 315, suggesting there was no strong evidence for weak instrument bias. In the present study, because of the relatively large sample size of the outcome databases, given a type I error of 5% and a statistical power of 0.80, the minimum detectable ORs for the 9 CVDs ranged from 1.04 to 1.41 (Supplementary Table 3) (40).
Table 1.
Characteristics of instrumental variables for circulating adipokines in the conservative analysis
| SNP | Trait | chr:position | Nearest gene | EA | OA | EAF | Beta | SE | P value | Sample size | R2 | F statistic |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2062632 | Adiponectin | chr3:186461181 | KNG1 | C | T | 0.686 | -0.055 | 0.006 | 2.52E-19 | 29 028 | 0.003 | 86 |
| rs17366568 | Adiponectin | chr3:186570453 | ADIPOQ | A | G | 0.908 | -0.154 | 0.009 | 1.00E-200 | 24 865 | 0.013 | 315 |
| rs6810075 | adiponectin | chr3:186830776 | ADIPOQ | C | T | 0.633 | -0.066 | 0.005 | 1.00E-200 | 29 140 | 0.007 | 192 |
| rs7615090 | Adiponectin | chr3:186591003 | ADIPOQ | G | T | 0.883 | -0.058 | 0.008 | 2.81E-11 | 21 869 | 0.002 | 47 |
| rs7955516 | Adiponectin | chr12:20498036 | PDE3A | C | A | 0.442 | 0.026 | 0.005 | 2.43E-08 | 29 178 | 0.001 | 33 |
| rs7964945 | Adiponectin | chr12:124437668 | CCDC92 | A | T | 0.808 | 0.037 | 0.006 | 2.61E-08 | 29 252 | 0.001 | 33 |
| rs8042532 | Adiponectin | chr15:74255230 | LOXL1 | G | T | 0.992 | -0.340 | 0.055 | 2.86E-09 | 7850 | 0.005 | 38 |
| rs12051272 | Adiponectin | chr16:82663288 | CDH13 | T | G | 0.020 | -0.277 | 0.018 | 1.00E-200 | 15 593 | 0.015 | 233 |
| rs17405635 | Resistin | chr2:43355763 | ZFP36L2 | A | G | 0.260 | 0.080 | 0.011 | 6.60E-14 | 20 793 | 0.003 | 53 |
| rs7589428 | Resistin | chr2:43561771 | THADA | A | G | 0.510 | 0.063 | 0.010 | 4.60E-11 | 21 747 | 0.002 | 44 |
| rs6775731 | Resistin | chr3:128306894 | RPN1 | T | C | 0.300 | -0.063 | 0.011 | 3.20E-09 | 21 747 | 0.002 | 33 |
| rs73008259 | Resistin | chr6:144411338 | SF3B5 | A | G | 0.053 | 0.190 | 0.020 | 3.70E-21 | 16 199 | 0.006 | 90 |
| rs77691416 | Resistin | chr6:144354119 | PLAGL1 | A | C | 0.910 | 0.120 | 0.016 | 7.90E-14 | 18 353 | 0.003 | 56 |
| rs10103048 | Resistin | chr8:130602281 | GSDMC | A | C | 0.420 | 0.060 | 0.010 | 5.20E-10 | 21 747 | 0.002 | 39 |
| rs3087852 | Resistin | chr17:38137033 | PSMD3 | A | G | 0.460 | -0.086 | 0.009 | 6.60E-20 | 21 749 | 0.004 | 84 |
| rs10401670 | Resistin | chr19:7742802 | MCEMP1 | T | C | 0.430 | 0.150 | 0.012 | 9.00E-37 | 16 221 | 0.01 | 156 |
| rs3745367 | Resistin | chr19:7734511 | RETN | A | G | 0.240 | 0.120 | 0.012 | 7.50E-23 | 18 353 | 0.005 | 100 |
| rs7806429 | Chemerin | chr7:150013393 | ACTR3C | T | C | 0.721 | -0.067 | 0.009 | 7.79E-14 | 3758 | 0.015 | 56 |
| rs36014035 | RBP4 | chr10:95348182 | RBP4 | A | C | 0.643 | 0.147 | 0.025 | 7.20E-09 | 3200 | 0.011 | 35 |
Abbreviations: EA, effect allele; EAF, effect allele frequency; OA, other allele; SE, standard error; RBP4, retinol binding protein 4.
Causal Effects of Adipokines on CVD
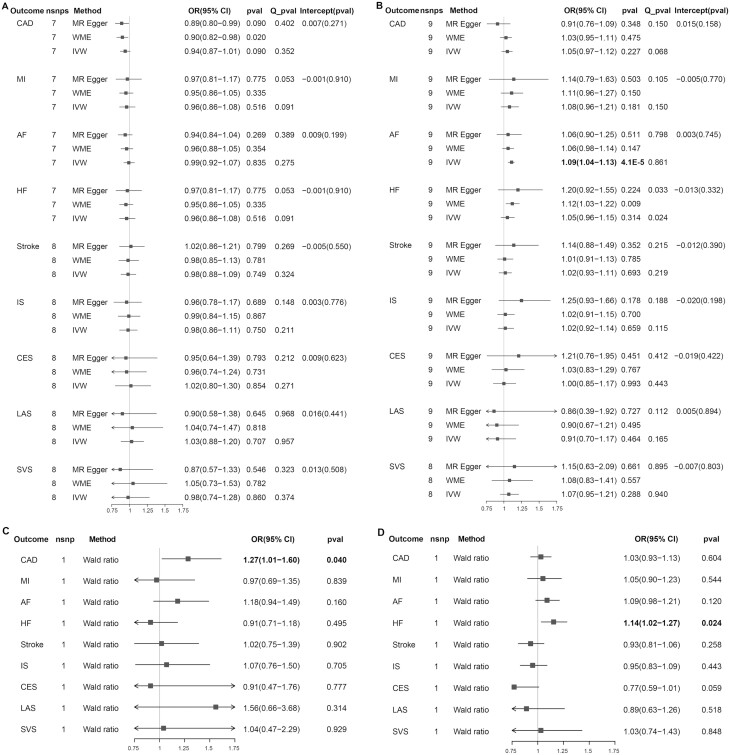
The results of the conservative analysis are shown in Fig. 2. We found evidence that higher genetically predicted circulating resistin levels were associated with an increased risk of AF (OR per unit increase, 1.09; 95% CI, 1.04-1.13; P = 4.1 × 10-5) but were not associated with the other 8 CVDs. We found a suggestive causal effect of genetically predicted circulating chemerin on CAD; for the per unit increase in chemerin levels, the OR was 1.27 for CAD (95% CI, 1.01-1.60; P = 0.040), whereas it was not significant in the other CVDs. We observed a suggestive association between higher genetically predicted circulating RBP4 levels and increased HF risk (OR per unit increase, 1.14; 95% CI, 1.02-1.27; P = 0.024), whereas the associations with the other CVDs were not significant. In addition, genetically determined circulating adiponectin levels were not associated with any CVD. The results of the liberal analysis of adiponectin and resistin were consistent with those obtained through conservative analysis, with no causal effect across the CVD types except circulating resistin on AF (OR per unit increase, 1.11; 95% CI, 1.04-1.20; P = 0.003) (Supplementary Table 4) (40). In the gene locus analysis, the SNPs for chemerin and RBP4 were located in the corresponding gene region and the results were the same as those obtained through conservative analysis. The genetically determined circulating adiponectin levels played no causal role in the nine CVDs. Moreover, the genetically determined resistin levels were suggestively associated with increased risk of AF (OR per unit increase, 1.16; 95% CI, 1.01-1.34; P = 0.040) and MI (OR per unit increase, 1.28; 95% CI, 1.03-1.59; P = 0.027) (Supplementary Table 5) (40).
Figure 2.
Associations and sensitivity analysis for the effect of per unit increase in inverse-normal transformed resistin (ng/mL) values, and in natural log-transformed adiponectin (µg/mL), chemerin (ng/mL), and RBP4 (µg/mL) values on cardiovascular diseases. The Cochran’s Q statistic was used to assess the heterogeneity among estimates for selected SNPs. The P value for the intercept in the MR-Egger regression was used to present the pleiotropy (P < 0.05). (A) adiponectin; (B) resistin; (C) chemerin; (D) RBP4. Abbreviations: CI, confidence interval; nsnp, number of single nucleotide polymorphism; OR, odds ratio; Q_pval, Cochran’s Q statistic P value. See Fig. 1 for other abbreviations.
We conducted MVMR to adjust for CVD-related traits, reassess the causal associations observed in our primary analysis, and explore the potential mediators. Because of the limited full summary-level statistics for chemerin and RBP4, and the positive causal association of resistin with AF in our 3 analyses, we further explored this association with MVMR. As shown in Supplementary Table 7 and Supplementary Figure 1, the association between genetically predicted circulating resistin and AF was attenuated to null after adjusting for systolic blood pressure (OR per unit increase, 1.04; 95% CI, 0.97-1.12; P = 0.270), diastolic blood pressure (OR per unit increase, 1.04; 95% CI, 0.97-1.12; P = 0.308), or both (OR per unit increase, 1.02; 95% CI, 0.96-1.09; P = 0.509), suggesting that resistin might affect AF risk through blood pressure (40).
Sensitivity Analyses
In the Cochran’s Q statistic and the MR-PRESSO analysis, heterogeneity or outliers were detected mainly in the liberal analysis. After correcting the outliers, the analysis indicated a potential causal association between adiponectin and CAD or MI, which could not be replicated in the other 2 analyses, suggesting that these potentially pleiotropic variables biased our results instead of acting as mediators (Fig. 2 and Supplementary Table 4-6) (40). The results were consistent using the WME method compared with those achieved with the IVW method. There was no evidence of horizontal pleiotropy across the analyses in the MR-Egger regression (P for intercept > 0.05). The combined MR-PRESSO, MR Egger, and WME results showed that our MR estimates were robust. In the leave-one-out analysis, there was no significant change in the risk estimations for genetically predicted resistin levels and AF risk after removing 1 SNP at a time, demonstrating that the causal association was not driven by specific SNPs (Supplementary Figures 2 and 3) (40). We found no evidence of reverse causality across the analyses in the MR Steiger test (Supplementary Table 8) (40). Moreover, there was no evidence that genomic test regions contained a shared causal variant for resistin and AF, resistin, and MI (Supplementary Table 9) (40).
Discussion
The association between circulating adipokines and CVD has been extensively investigated in recent studies, most of which have focused on atherosclerosis and CAD (41-44). However, evidence from current studies is limited to showing observational correlations, and reverse causality may exist. The purpose of this study is to clarify the causal effect of a specified adipokine on a certain CVD. In the present study, based on the available GWAS, we investigated the relationship between 4 adipokines and the risk of 9 CVDs using MR. Our findings indicate that genetically predicted higher resistin levels are significantly associated with increased AF risk, probably acting through blood pressure. There is suggestive evidence that genetically predicted chemerin causally affects the risk of CAD and genetically predicted RBP4 causally affects the risk of HF. We failed to find evidence to support a causal association between genetically predicted adiponectin and the risk of CVD. Also, our results do not support the causal role of the 4 adipokines in stroke and its subtypes.
To examine the reliability of our findings, we conducted a series of sensitivity analyses. For adiponectin and resistin, 3 sets of SNPs were used and pleiotropic SNPs were tested in the liberal analysis. We found that these pleiotropic SNPs were confounders rather than mediators. The results were robust when MR-PRESSO, MR Egger, WME, and leave-one-out analysis were used for the sensitivity analyses. A causal association between genetically predicted resistin and AF was found using either set of SNPs. In the gene locus analysis, we also found a causal association between genetically predicted resistin and MI risk, whereas we failed to find this association in the other 2 analyses, which reassessed the results using various statistical methods; thus, this association tended to be vulnerable and might have resulted from LD with other genetic variants. For chemerin and RBP4, given that we obtained only 1 SNP for each adipokine, methodological sensitivity analysis was not applicable. We searched in PhenoScanner and the GWAS catalog and confirmed no evidence of pleiotropy, indicating that our results were plausible. The MR Steiger test further showed that there was no evidence of reverse causality in our study. In the colocalization analysis, there was little evidence that 2 traits showing evidence of a causal association in the MR analysis were colocalized, which suggested that the SNPs in the gene locus might be LD with proxied variants and might not be suitable for perturbing drug targets, or that multiple causal variants might exist within the given region, which was not suitable for colocalization analysis (22). Meanwhile, the value of PP.H1 was relatively high, indicating variants in the region were significantly associated with resistin and were unlikely to affect the outcome, thus obeying the MR main assumption and further supporting our MR estimates (45). Because of the limited summary-level statistics for chemerin and RBP4, colocalization analysis was not performed on the causal associations between chemerin and CAD, RBP4 and HF.
Human resistin, which is functionally associated with insulin resistance and inflammatory response, can be released from epicardial adipose tissue, with increased secretion being strongly associated with ventricular dysfunction and AF risk via promoting myocardial fibrosis (46, 47). However, the role of resistin in AF is controversial. A multiethnic cohort study with 1913 participants showed that resistin was an independent risk factor for CAD, MI, and HF but not AF (48). Two studies suggested that elevated levels of circulating resistin were related to AF, whereas the impact on permanent AF yielded opposite results (49, 50). The discrepancy in results may be due to measurement error, residual confounding, or reverse causality in the observational researches. In comparison, our MR study largely excludes common confounders and theoretically eliminates reverse causality, which makes causal inferences possible. Our analysis indicates a strong causal association between genetically predicted resistin levels and AF risk; therefore, it is necessary to further investigate the role of resistin in the pathogenesis of AF.
Chemerin is involved in adipogenesis and angiogenesis and acts as a chemoattractant, thus promoting inflammation (51, 52). Higher chemerin levels have been reported to be associated with an increased risk of CAD. For instance, circulating chemerin levels were significantly elevated in CAD patients compared with healthy participants (53). Evidence from a case-control study indicated that chemerin levels were positively correlated with CAD severity (54). A GWAS study containing 2197 Taiwanese and 481 CAD cases verified the hypothesis that higher circulating chemerin levels represented a significantly poorer long-term CAD outcome (55). Our MR results are consistent with those of observational epidemiological studies and provide suggestive evidence on the causal relationship between genetically predicted chemerin levels and CAD risk. Mechanistically, chemerin may affect the development of CAD through the following mechanisms. Gu et al. revealed that increased chemerin levels were independently associated with impaired endothelial function and increased arterial stiffness (56). It was reported that chemerin was a proinflammatory chemoattractant and the change of circulating chemerin levels coincided with that of certain inflammatory factors, such as IL-6, C-reactive protein, and TNF-α (57). Furthermore, an animal experiment provided evidence that chemerin receptor 23 deficiency restricted atherosclerotic plaque formation and attenuated lesion inflammation (58). Taken together, chemerin may play a critical role in the pathogenesis of CAD.
RBP4 is secreted by the liver and the adipose tissue, acting as both hepatokine and adipokine. The relationship between RBP4 and HF remains debated. Evidence from an observational study indicated that circulating RBP4 levels were higher in advanced HF cases than in healthy controls (59). Subsequently, it was reported that RBP4 was a prognostic indicator of worse outcome in elderly patients with chronic HF (60). However, several studies failed to confirm this relationship in systolic HF patients (61) and older patients hospitalized for chronic HF (62). In the present study, we find suggestive evidence that genetically predicted RBP4 causally affects the risk of HF. Studies have suggested cardiac prohypertrophic effects of RBP4, which may lead to HF. For instance, 1 community-based cohort observational study showed that RBP4 level was positively related to the left ventricular mass index and left atrial end-systolic dimension, suggesting potential prohypertrophic effects of RBP4 (63). Also, RBP4 showed pro-inflammatory abilities, which may be the crucial pathway in the pathogenesis of CVD (64). Interestingly, Gao et al. revealed that RBP4 stimulated inflammatory and hypertrophic responses through Toll-like receptor 4 and the myeloid differentiation primary response gene 88 pathway (65). Nevertheless, the causal role of RBP4 in HF needs to be interpreted cautiously.
Adiponectin is the most abundant adipokine with multifunctional properties. Circulating adiponectin concentrations have been found to be inversely correlated with adiposity. Several meta-analysis studies have suggested that elevated adiponectin levels are implicated in the increased risk of CAD (66), AF (67), and HF (68), and are not associated with stroke risk (5, 69). However, previous MR studies have not supported a causal association between adiponectin levels and CAD pathogenesis (12, 13). Our study, using data with stricter inclusion criteria, extended the causal effects of adiponectin to MI, AF, HF, and stroke and its subtypes, and found the same negative results. This discrepancy may indicate that adiponectin is a risk marker rather than a causal risk factor for CVD. Moreover, Christen et al. demonstrated that N-terminal-pro-brain natriuretic peptide was causally related to adiponectin concentrations, supporting the existence of reverse causation in previous studies (70). Thus, further studies are needed to elucidate the internal relationship between adiponectin, as well as other adipokines with robust null results in our study, and CVD and to clarify whether there are driving factors that account for bias or confounding in previous observational studies.
Our study has several strengths. The predominant strength is the MR design, which estimates the causal effects of adipokines on 9 CVDs without interference from residual confounding or reverse causality. Before the MR analysis, we strictly screen the related SNPs using Plink clumping and Phenoscanner; we excluded the potential pleiotropic instruments in the conservative analysis to ensure the allowance of MR assumptions and further examined related instruments in the liberal analysis. There was no evidence of weak instruments in the F statistics among the selected SNPs. We chose the largest GWAS to minimize the “winner’s curse” and, although some causal estimates were relatively small, the corresponding power was enough to support the results. In addition, multiple testing adjustment was implemented using the Bonferroni method to increase the reliability of our results.
Our study also has several limitations. First, the available data we used were summary-level statistics rather than individual-level statistics. We systematically searched databases and found them unable to obtain exposure data unadjusted for BMI or adjusted for PCs; therefore, it might introduce unavoidable biases to our results. Second, we could not determine the gender-specific or nonlinear causal association between circulating adipokines levels and the risk of CVD. Third, potential pleiotropy may bias our results. Either chemerin or RBP4 had only 1 SNP, which was insufficient to conduct the sensitivity analysis. We ruled out the possibility of pleiotropy by searching in PhenoScanner and the GWAS catalog, proving that our results were plausible. Sensitivity analyses for the other adipokines generated results consistent with the IVW method and the MR-Egger regression demonstrated that pleiotropy was unlikely to influence our results. Fourth, our MR study estimated the effects of lifetime exposure, which was likely to overestimate effects in the real world when considering the effectiveness of the interventions. Fifth, epigenetic phenomena, such as methylation, and the developmental compensation mechanism may also influence the associations between adipokines and CVD, whereas these are inherent defects of MR that cannot be assessed. Last, this study was confined to individuals of European origin, except for the analysis for CAD and MI, which comprised nearly 80% European individuals. We minimized the population structure bias, whereas this restriction may prevent our findings from being generalized to other populations.
In summary, our MR study supports genetic evidence that increased circulating resistin levels are causally related to higher AF risk, probably acting through blood pressure. We find suggestive evidence that increased circulating chemerin levels are causally related to higher CAD risk and RBP4 may have a positive causal effect on HF. Our results provide novel insights for exploring strategies to prevent and manage CVD.
Acknowledgments
Data used in our study were downloaded from the GWAS catalog (https://www.ebi.ac.uk/gwas/). The authors acknowledge all GWAS participants and investigators for their contributions to the summary statistics data. The authors thank all investigators for sharing these data. We thank the blogger (orange_milk_sugar, Wenyan Chen) for providing operation instructions of related data and code.
Glossary
Abbreviations
- AF
atrial fibrillation
- BMI
body mass index
- CAD
coronary artery disease
- CI
confidence interval
- CVD
cardiovascular disease
- GWAS
genome-wide association study
- HF
heart failure
- IVW
inverse variance-weighted
- LD
linkage disequilibrium
- MI
myocardial infarction
- MR
Mendelian randomization
- MR-PRESSO
MR pleiotropy residual sum and outlier
- MVMR
multivariable Mendelian randomization
- OR
odds ratio
- PC
principal component
- PP
posterior probability
- RBP4
retinol binding protein 4
- SNP
single-nucleotide polymorphism
- WME
weighted-median estimator
Financial Support
This work was supported by the National Natural Science Foundation of China (grant 82170332).
Author Contributions
D.C. and J.J. designed the study and drafted the article. Y.Z. and A.Y. conducted data acquisition, D.C., Y.Z., Y.X., Q.D., and J.J. performed data analysis and manuscript revision. All authors read and approved the final manuscript.
Disclosures
The authors declare no conflict of interest.
Data Availability
All data used in the present study are based on publicly available summary level data from the GWAS catalog. Data generated during this study are available from the corresponding author on reasonable request.
References
- 1. Diseases GBD, Injuries C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ezzati M, Obermeyer Z, Tzoulaki I, et al. Contributions of risk factors and medical care to cardiovascular mortality trends. Nat Rev Cardiol. 2015;12(9):508-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakamura K, Fuster JJ, Walsh K. Adipokines: a link between obesity and cardiovascular disease. J Cardiol. 2014;63(3-4):250-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ouchi N, Parker JL, Lugus JJ, et al. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11(2):85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arregui M, Buijsse B, Fritsche A, et al. Adiponectin and risk of stroke: prospective study and meta-analysis. Stroke. 2014;45(1):10-17. [DOI] [PubMed] [Google Scholar]
- 6. Wang Z, Li B, Wang Y, et al. The association between serum adiponectin and 3-month outcome after ischemic stroke. Cardiovasc Diabetol. 2019;18(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehrke M, Becker A, Greif M, et al. Chemerin is associated with markers of inflammation and components of the metabolic syndrome but does not predict coronary atherosclerosis. Eur J Endocrinol. 2009;161(2):339-344. [DOI] [PubMed] [Google Scholar]
- 8. Dimitriadis GK, Kaur J, Adya R, et al. Chemerin induces endothelial cell inflammation: activation of nuclear factor-kappa beta and monocyte-endothelial adhesion. Oncotarget. 2018;9(24):16678-16690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaur J, Adya R, Tan BK, et al. Identification of chemerin receptor (ChemR23) in human endothelial cells: chemerin-induced endothelial angiogenesis. Biochem Biophys Res Commun. 2010;391(4):1762-1768. [DOI] [PubMed] [Google Scholar]
- 10. Yamawaki H, Kameshima S, Usui T, et al. A novel adipocytokine, chemerin exerts anti-inflammatory roles in human vascular endothelial cells. Biochem Biophys Res Commun. 2012;423(1):152-157. [DOI] [PubMed] [Google Scholar]
- 11. Lawlor DA, Harbord RM, Sterne JAC, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133-1163. [DOI] [PubMed] [Google Scholar]
- 12. Borges MC, Lawlor DA, de Oliveira C, et al. Role of adiponectin in coronary heart disease risk: a Mendelian randomization study. Circ Res. 2016;119(3):491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Au Yeung SL, Schooling CM. Adiponectin and coronary artery disease risk: a bi-directional Mendelian randomization study. Int J Cardiol. 2018;268:222-226. [DOI] [PubMed] [Google Scholar]
- 14. Fuchs FD, Whelton PK. High blood pressure and cardiovascular disease. Hypertension (Dallas, Tex: 1979). 2020;75(2):285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wan EYF, Fung WT, Schooling CM, et al. Blood pressure and risk of cardiovascular disease in UK Biobank: a mendelian randomization study. Hypertension (Dallas, Tex: 1979). 2021;77(2):367-375. [DOI] [PubMed] [Google Scholar]
- 16. Dastani Z, Hivert MF, Timpson N, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8(3):e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Folkersen L, Gustafsson S, Wang Q, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020;2(10):1135-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tönjes A, Scholz M, Breitfeld J, et al. Genome wide meta-analysis highlights the role of genetic variation in RARRES2 in the regulation of circulating serum chemerin. PLoS Genet. 2014;10(12):e1004854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361(6404):769-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Staley JR, Blackshaw J, Kamat MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gill D, Georgakis MK, Walker VM, et al. Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res. 2021;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122(3):433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roselli C, Chaffin MD, Weng LC, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50(9):1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shah S, Henry A, Roselli C, et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020;11(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malik R, Chauhan G, Traylor M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shim H, Chasman DI, Smith JD, et al. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS One. 2015;10(4):e0120758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Au Yeung SL, Borges MC, Lawlor DA, et al. Impact of lung function on cardiovascular diseases and cardiovascular risk factors: a two sample bidirectional Mendelian randomisation study. Thorax. 2022;77(2):164-171. [DOI] [PubMed] [Google Scholar]
- 30. Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755-764. [DOI] [PubMed] [Google Scholar]
- 32. Bowden J, Del Greco MF, Minelli C, et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36(11):1783-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burgess S, Bowden J, Fall T, et al. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28(1):30-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen D, Zhang Y, Yidilisi A, et al. Data from: Causal associations between circulating adipokines and cardiovascular disease: a Mendelian randomization study. figshare. Posted January 2, 2022; 10.6084/m9.figshare.17710916.v5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yuxiang L, Fujiu K. Human resistin and cardiovascular disease. Int Heart J. 2020;61(3):421-423. [DOI] [PubMed] [Google Scholar]
- 42. Kotnik P, Fischer-Posovszky P, Wabitsch M. RBP4: a controversial adipokine. Eur J Endocrinol. 2011;165(5):703-711. [DOI] [PubMed] [Google Scholar]
- 43. Katsiki N, Mantzoros C, Mikhailidis DP. Adiponectin, lipids and atherosclerosis. Curr Opin Lipidol. 2017;28(4):347-354. [DOI] [PubMed] [Google Scholar]
- 44. Eichelmann F, Schulze MB, Wittenbecher C, et al. Chemerin as a biomarker linking inflammation and cardiovascular diseases. J Am Coll Cardiol. 2019;73(3):378-379. [DOI] [PubMed] [Google Scholar]
- 45. Wang Y, Guo P, Liu L, et al. Mendelian randomization highlights the causal role of normal thyroid function on blood lipid profiles. Endocrinology. 2021;162(5):1-10.. [DOI] [PubMed] [Google Scholar]
- 46. Venteclef N, Guglielmi V, Balse E, et al. Human epicardial adipose tissue induces fibrosis of the atrial myocardium through the secretion of adipo-fibrokines. Eur Heart J. 2015;36(13):795-805a. [DOI] [PubMed] [Google Scholar]
- 47. Langheim S, Dreas L, Veschini L, et al. Increased expression and secretion of resistin in epicardial adipose tissue of patients with acute coronary syndrome. Am J Physiol Heart Circ Physiol. 2010;298(3):H746-H753. [DOI] [PubMed] [Google Scholar]
- 48. Muse ED, Feldman DI, Blaha MJ, et al. The association of resistin with cardiovascular disease in the Multi-Ethnic Study of Atherosclerosis. Atherosclerosis. 2015;239(1):101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samanidis G, Gkogkos A, Bousounis S, et al. Blood plasma resistin and atrial fibrillation in patients with cardiovascular disease. Cardiol Res. 2020;11(5):286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Özcan KS, Güngör B, Altay S, et al. Increased level of resistin predicts development of atrial fibrillation. J Cardiol. 2014;63(4):308-312. [DOI] [PubMed] [Google Scholar]
- 51. Goralski KB, McCarthy TC, Hanniman EA, et al. Chemerin, a novel adipokine that regulates adipogenesis and adipocyte metabolism. J Biol Chem. 2007;282(38):28175-28188. [DOI] [PubMed] [Google Scholar]
- 52. Bozaoglu K, Curran JE, Stocker CJ, et al. Chemerin, a novel adipokine in the regulation of angiogenesis. J Clin Endocrinol Metab. 2010;95(5):2476-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiaotao L, Xiaoxia Z, Yue X, et al. Serum chemerin levels are associated with the presence and extent of coronary artery disease. Coron Artery Dis. 2012;23(6):412-416. [DOI] [PubMed] [Google Scholar]
- 54. Aksan G, İnci S, Nar G, et al. Association of serum chemerin levels with the severity of coronary artery disease in patients with metabolic syndrome. Int J Clin Exp Med. 2014;7(12):5461-5468. [PMC free article] [PubMed] [Google Scholar]
- 55. Er LK, Hsu LA, Juang JJ, et al. Circulating chemerin levels, but not the RARRES2 polymorphisms, predict the long-term outcome of angiographically confirmed coronary artery disease. Int J Mol Sci . 2019;20(5):1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gu P, Cheng M, Hui X, et al. Elevating circulation chemerin level is associated with endothelial dysfunction and early atherosclerotic changes in essential hypertensive patients. J Hypertens. 2015;33(8):1624-1632. [DOI] [PubMed] [Google Scholar]
- 57. Zhang O, Ji Q, Lin Y, et al. Circulating chemerin levels elevated in dilated cardiomyopathy patients with overt heart failure. Clin Chim Acta. 2015;448:27-32. [DOI] [PubMed] [Google Scholar]
- 58. van der Vorst EPC, Mandl M, Müller M, et al. Hematopoietic ChemR23 (Chemerin Receptor 23) fuels atherosclerosis by sustaining an M1 macrophage-phenotype and guidance of plasmacytoid dendritic cells to murine lesions-brief report. Arterioscler Thromb Vasc Biol. 2019;39(4):685-693. [DOI] [PubMed] [Google Scholar]
- 59. Chavarria N, Kato TS, Khan R, et al. Increased levels of retinol binding protein 4 in patients with advanced heart failure correct after hemodynamic improvement through ventricular assist device placement. Circ J. 2012;76(9):2148-2152. [DOI] [PubMed] [Google Scholar]
- 60. Li XZ, Zhang KZ, Yan JJ, et al. Serum retinol-binding protein 4 as a predictor of cardiovascular events in elderly patients with chronic heart failure. ESC Heart Fail. 2020;7(2):542-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu XM, Lin YH, Chen A, et al. Prognostic significance of adipocytokines in systolic heart failure patients. Eur J Clin Invest. 2012;42(10):1079-1086. [DOI] [PubMed] [Google Scholar]
- 62. Majerczyk M, Choręza P, Mizia-Stec K, et al. Plasma level of retinol-binding protein 4, N-terminal proBNP and renal function in older patients hospitalized for heart failure. CardioRenal Med. 2018;8(3):237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. von Jeinsen B, Short MI, Xanthakis V, et al. Association of circulating adipokines with echocardiographic measures of cardiac structure and function in a community-based cohort. J Am Heart Assoc. 2018;7(13):e008997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Farjo KM, Farjo RA, Halsey S, et al. Retinol-binding protein 4 induces inflammation in human endothelial cells by an NADPH oxidase- and nuclear factor kappa B-dependent and retinol-independent mechanism. Mol Cell Biol. 2012;32(24):5103-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gao W, Wang H, Zhang L, et al. Retinol-binding protein 4 induces cardiomyocyte hypertrophy by activating TLR4/MyD88 pathway. Endocrinology. 2016;157(6):2282-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang L, Li B, Zhao Y, et al. Prognostic value of adiponectin level in patients with coronary artery disease: a systematic review and meta-analysis. Lipids Health Dis. 2019;18(1):227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guo Y, Liu L, Wang J. Adiponectin and the risk of new-onset atrial fibrillation: a meta-analysis of prospective cohort studies. Biosci Rep. 2019;39(6):BSR20182284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bai W, Huang J, Zhu M, et al. Association between elevated adiponectin level and adverse outcomes in patients with heart failure: a systematic review and meta-analysis. Braz J Med Biol Res. 2019;52(7):e8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yuan HP, Sun L, Li XH, et al. Association of adiponectin polymorphism with metabolic syndrome risk and adiponectin level with stroke risk: a meta-analysis. Sci Rep. 2016;6:31945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Christen T, de Mutsert R, Lamb HJ, et al. Mendelian randomization study of the relation between adiponectin and heart function, unravelling the paradox. Peptides. 2021;146:170664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in the present study are based on publicly available summary level data from the GWAS catalog. Data generated during this study are available from the corresponding author on reasonable request.