Abstract
Chinese tongue sole (Cynoglossus semilaevis) is an economically important marine fish species with a ZZ/ZW sex determination mechanism, which can be influenced by temperature. Alternative splicing (AS) is an important mechanism regulating the expression of genes related to sex determination and gonadal differentiation, but has rarely been reported in fish. In this study, to explore the molecular regulatory mechanisms of sex determination and gonadal differentiation, we combined isoform and RNA sequencing (Iso-Seq and RNA-Seq) to perform transcriptome profiling of male and female gonads in C. semilaevis. In total, 81883 and 32341 full-length transcripts were obtained in males and females, respectively. A total of 8279 AS genes were identified, including 2 639 genes showing differential AS (DAS) between males and females. Many intersecting DAS genes and differentially expressed genes (DEGs) were enriched in the meiotic cell cycle pathway, and genes related to gonadal differentiation, such as esrrb and wt1a, were found to have sex-specific isoforms. Thus, this study revealed AS events in the gonadal transcriptomes of male and female C. semilaevis, described the characteristics of active transcription in the testes, and identified candidate genes for studying the regulatory mechanisms of AS during gonadal differentiation.
Keywords: Gonadal differentiation, Alternative splicing, Sex-specific splicing, Cynoglossus semilaevis, Iso-Seq, RNA-Seq
INTRODUCTION
Alternative splicing (AS) is a ubiquitous mechanism for regulating gene expression that allows the generation of multiple mRNAs from a single gene. During this process, RNA-binding proteins (RBPs) can influence the recognition efficiency of splicing complexes at splicing sites and thus modulate AS (Witten & Ule, 2011). Types of AS are categorized as skipped exons (SEs) (particularly exon removal), mutually exclusive exons (MXEs) (only one of two exons is retained after splicing), alternative 5’/3’ splice sites (A5SSs, A3SSs) (affecting boundaries between introns and exons), and retained introns (RIs) (Baralle & Giudice, 2017). AS can increase the diversity of mRNA and protein isoforms and alter the expression of gene isoforms. For example, 90%–95% of human genes undergo AS, and approximately 37% of genes generate multiple protein isoforms (Kim et al., 2014; Pan et al., 2008). AS can also change translation reading frames to regulate gene expression, known as nonsense-mediated decay (NMD) (Isken & Maquat, 2008). In addition, the A5SSs and A3SSs of mRNA untranslated regions (UTRs) can affect the stability, localization, and translation efficiency of mRNAs (Licatalosi & Darnell, 2010). Thus, due to its ability to enrich different types of mRNA, further functional analysis of AS is necessary.
AS plays an important role in sexual development and gonadal differentiation in teleosts. In Chinese tongue sole (Cynoglossus semilaevis), the ovarian germ-cell-specific vasa gene has three isoforms in the gonad, including vas-s, which shows sexually dimorphic expression during early gonadal differentiation (Wang et al., 2014). Another germ-cell-specific gene, factor in the germline alpha (figla), expresses two isoforms, figla_tv1 and figla_tv2, which are separately expressed in the oocytes of female and germ cells of pseudomale testes (Li et al., 2016). Knockdown of figla_tv2 in pseudomale testes significantly up-regulates the expression of two steroid hormone-coding genes, suggesting the involvement of figla_tv2 in spermatogenesis via regulation of steroid hormone synthesis (Li et al., 2016). In Nile tilapia, SRY-box containing gene 30 (sox30) is expressed exclusively in the gonads, and its four AS-generated isoforms are expressed at different stages of gonadal differentiation, thus showing clear sexual dimorphism (Han et al., 2010). In seabream, four progesterone receptor gene (pgr) variants are co-expressed in the ovary, among which two isoforms show differential expression under gonadotropin and estrogen stimulation, suggesting that ovarian progestin responsiveness may be regulated by AS of pgr mRNA during early oogenesis (Zapater et al., 2013). Thus, AS appears to be a universal regulatory mechanism of sexual development and gonadal differentiation in teleosts.
The Chinese tongue sole is an economically important and sexually dimorphic marine species, with females showing faster growth than males. As such, sex determination and gonadal differentiation in this species have become important areas of research. However, while genes related to sex determination and gonadal differentiation have been investigated, few studies have explored the mechanisms underlying differences in gene expression in female and male gonads, especially AS of key genes during gonadal differentiation. To perform a comparative transcriptomic analysis of expression and AS regulation between Chinese tongue sole females and males, we combined PacBio isoform sequencing (Iso-Seq) and short-read RNA sequencing (RNA-Seq) to generate a comprehensive transcript dataset of the gonads after differentiation. Abundant differentially expressed genes (DEGs) and genes with differential alternative splicing (DAS) were obtained. Based on enrichment analysis, DAS in DEGs was related to mRNA splicing and germ cell development. Our results provide a foundation and potential candidate genes for further research on the mechanisms underlying differential expression between the ovary and testis of C. semilaevis.
MATERIALS AND METHODS
Ethics statement
All animal procedures followed the principles of the Guide for the Care and Use of Laboratory Animals at the Chinese Academy of Fishery Sciences and were approved by the Institutional Animal Care and Use Committee (IACUC) of the Yellow Sea Fisheries Research Institute (CAFS) (Qingdao, China)(Approval No.: YSFRI-2022016).
Sample collection of C. semilaevis
In this study, a batch of healthy Chinese tongue sole (at 6 months post-fertilization (mpf)) was sampled from Laizhou Mingbo Co., Ltd. (Yantai, Shandong, China). The gonads were collected and immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction. The caudal fins were collected and stored in ethanol for genetic sex identification.
Histological observation
The collected gonadal tissues were fixed in 4% paraformaldehyde (PFA) at 4 °C overnight, then soaked in 10 mmol/L phosphate-buffered saline (PBS) (Solarbio Science, China) for 1 h. The fixed samples were dehydrated through a series of graded ethanol concentrations, embedded in paraffin blocks, and cut into 6 μm sections. The sections were fixed on slides and stained with hematoxylin-eosin (Solarbio Science, China). The slides were photographed with a Leica DM4000 B light microscope (Leica Microsystems, Germany).
Genetic sex identification
Genomic DNA was extracted from the fins using the phenol-chloroform method. Genetic sex was identified by polymerase chain reaction (PCR) amplification of sex-specific simple sequence repeat (SSR) markers (Liu et al., 2014).
Total RNA extraction
Total RNA was isolated from each gonad sample using TRIzol reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. RNA sample quality was measured with an Agilent 2100 Bioanalyzer (Agilent, USA).
RNA-Seq and data processing
The libraries of three females (F1–F3) and three males (M1–M3) were sequenced on the BGISEQ-500 platform. Raw reads were filtered using SOAPnuke v1.4.0 (Chen et al., 2018) and Trimmomatic v0.36 (Bolger et al., 2014) with default parameters to remove adapters, low-quality reads with more than 5% unknown nucleotides, and reads with more than 20% low-quality bases. The filtered clean data were subsequently mapped to the C. semilaevis genome (NCBI Cse_v1.0) with HISAT2 (v2.1.0) (Kim et al., 2019) and aligned with the reference transcript sequence using Bowtie2 (v2.2.5) (Langmead & Salzberg, 2012) to remove ribosomal RNA (rRNA) sequences and annotate genes.
Full-length transcriptome sequencing and data processing
Four full-length transcriptome libraries were constructed by using RNA samples from the gonads of each two females and two males. Total RNA was enriched with oligo (dT) magnetic beads and then reverse transcribed to produce cDNA using a SMARTer™ PCR cDNA Synthesis Kit (Takara Bio, USA). The full-length cDNA was amplified by PCR, and the BluePippin™ Size Selection System (Sage Science, USA) was then used for size selection to generate four libraries (insert size of 1–10 kb). After selection, the full-length cDNA was amplified again and end repaired, and SMRT sequencing adaptors were ligated to the cDNA to produce the SMRT bell libraries. The libraries were quantified using a Qubit RNA BR Assay Kit (Thermo Fisher Scientific, USA). The library insert size was checked with an Agilent 2100 Bioanalyzer, and sequencing primers and polymerase were bound to the SMRT template in appropriate proportions according to the PacBio calculator sequencing results obtained from the PacBio Sequel platform (Pacific Biosciences, USA). We first performed raw read quality control and filtration, removing reads with low quality and short length, and then generated circular consensus sequences (CCSs) by filtering based on number of full passes greater than zero and accuracy greater than 0.75. The CCSs were classified and clustered using SMRT Link (v5.1.0) software supported by Pacific Biosciences. For the clustered CCSs, we performed alignment to the C. semilaevis genome (NCBI Cse_v1.0) using GMAP software (v2017.06.20), and the final transcriptome sequences were generated using TOFU (v1.0) to remove redundant sequences. We annotated the full-length transcriptome and identified novel transcripts by matching the alignment results to the genome annotation file with MatchAnnot (v1.0), and then used TransDecoder (v3.0.0) to predict the open reading frames (ORFs) of full-length transcripts.
Expression analysis
The sequences aligned by Bowtie2 were quantified by RSEM (v2.2.5), and gene expression levels were normalized using the fragments per kilobase million (FPKM) method to eliminate the influence of sequencing depth and gene length. Differential expression analysis of normalized gene expression was performed using DEseq2 (v1.30.1) (Love et al., 2014), with fold-change≥2 and adjusted P (q-value)<0.05 indicating significant DEGs.
DAS analysis
We utilized the lr2rmats pipeline (v0.1) to integrate SMRT long-read and RNA-Seq short-read data, thus generating a new gene annotation file. Clean RNA-Seq reads were then aligned to the genome using STAR (v2.5.3a) (Dobin et al., 2013), resulting in an alignment file for each sample. The newly generated annotation and alignment files for each sample were fed into rMATS (v4.1.0) (Shen et al., 2014) for AS analysis, whereby short reads were used to compare differences in reads per kilobase million (RPKM) in specific regions of the transcripts derived from each gene. For each AS event, rMATS calculated percentage of exon inclusion (IncLevel) for each sample across the biological triplicates and detected differential IncLevel (IncLevelDifference) between the two sexes. The DAS events were screened and categorized using summary.py in rMATS based on a IncLevelDifference absolute value greater than 0.1 and false discovery rate (FDR) of less than 0.05. The splicing event categories included SEs, A5SSs, A3SSs, MXEs, and RIs.
Functional analysis of differential AS genes
Gene Ontology (GO) enrichment analysis was performed using Metascape (https://metascape.org). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed based on annotation results from the alignment of C. semilaevis protein sequences in the KEGG database (https://www.genome.jp/kegg/).
Visualization of AS events
Sashimi plots of AS events were generated using rmats2sashimiplot (v2.0.3). RNA-Seq and full-length transcript structures were viewed and visualized in Integrative Genomics Viewer (IGV) (v2.8.13) (Robinson et al., 2011).
Gene expression validation by quantitative real-time PCR (qRT-PCR)
To verify splicing regulator expression based on RNA-Seq data, eight candidate genes potentially involved in gonadal differentiation were selected. Primers for the selected genes were designed based on the longest mRNA sequences generated from the C. semilaevis genome sequence (Supplementary Table S1). The RNA-Seq samples of each group were employed for cDNA synthesis using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Japan). qRT-PCR was performed using a QuantiNova SYBR Green PCR Kit (Qiagen, Germany). Total qRT-PCR volume (20 μL) contained 10 µL of 2×SYBR Green PCR Master Mix, 2 µL of QN ROX reference dye, forward and reverse primers (each at 0.7 µmol/L), and 1 µL of cDNA. PCR amplification was performed using a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, USA) under the following conditions: 95 °C for 2 min, followed by 40 cycles at 95 °C for 5 s and 60 °C for 10 s, with final collection of the fluorescence signal of the dissolution curve. The PCR assays were performed in triplicate. The β-actin gene was used as an internal reference gene. The relative expression levels of eight genes were measured using the 2−ΔΔCt equation. The t-test was used for assessment of significance, and P<0.05 was considered statistically significant.
RESULTS
Histological observations of gonadal differentiation
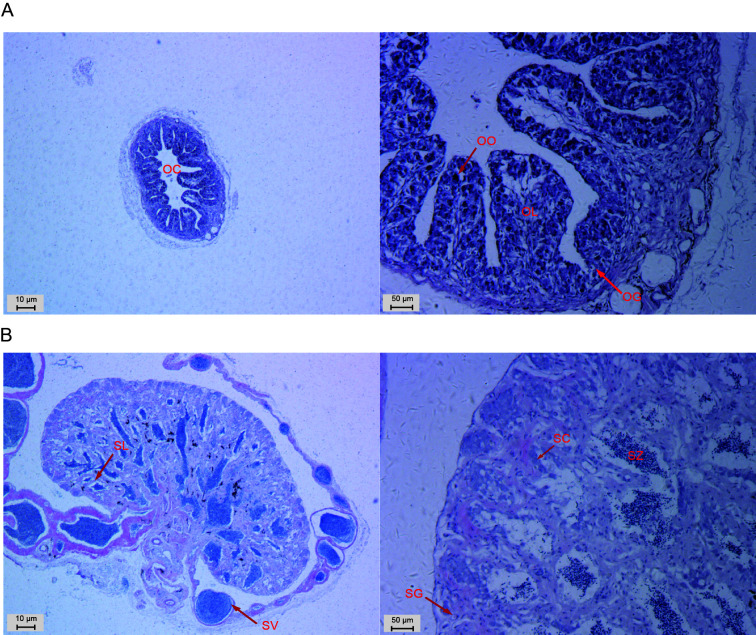
At 6 mpf, the ovarian cavity and seminal lobules were observed in the ovary and testis, respectively, indicating completion of gonadal differentiation. In the ovary, oocytes were orderly distributed along the ovarian lobules, with few oogonia (Figure 1A). In the testis, many seminal vesicles were wrapped around the testis periphery and filled with sperm. Germ cells, including spermatogonia, spermatocytes, and spermatozoa, were observed in the seminal lobules (Figure 1B).
Figure 1.
Light microscope images of C. semilaevis gonads at 6 mpf
A: Cross-section of ovary at different magnifications (left bar: 10 μm; right bar: 50 μm). B: Cross-section of testis at different magnifications (left bar: 10 μm; right bar: 50 μm). OC, ovarian cavity; OL, ovarian lobule; OG, oogonium; OO, oocyte; SL, seminal lobule; SV, seminal vesicle; SG, spermatogonium; SC, spermatocyte; SZ, spermatozoa.
AS events in gonads
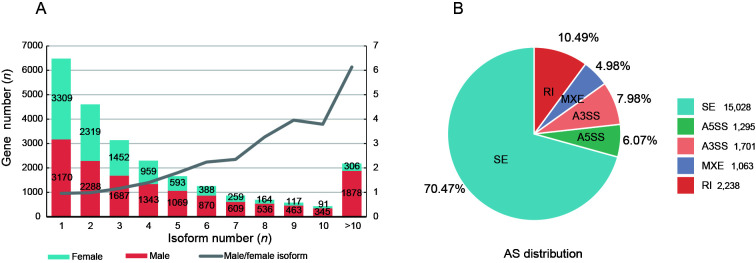
To fully characterize AS in the gonads, we performed Iso-Seq and RNA-Seq to obtain full-length transcripts and their relative expression levels. We identified 32 341 and 81 883 transcripts in the ovary and testis, with mean transcript lengths of 1 670 and 2 301 bp, respectively. In total, 9 957 and 14 258 genes were annotated, respectively. Among those transcripts, we identified 1 321 (ovary) and 4 038 (testis) novel transcripts, which were shorter in length than known transcripts in the gonads (Supplementary Figure S1A, B). Furthermore, 6 648 and 11 088 genes showed AS in the ovary and testis, respectively (isoform number≥2) (Figure 2A). The ratio of male to female genes increased with isoform number, indicating that genes expressed in the testis had more isoforms than those in the ovary, which was especially true for genes with more than 10 isoforms (1 878) (Figure 2A). AS events were predicted using short RNA-Seq reads, resulting in 21 325 AS events in the gonads (Figure 2B). The SE, A5SS, A3SS, MXE, and RI splicing types accounted for 70.47%, 6.07%, 7.98%, 4.98%, and 10.49% of all splicing events, respectively.
Figure 2.
Number and type distribution of AS events in gonads of C. semilaevis
A: Isoform numbers of expressed genes and ratio of gene numbers (male relative to female) in gonads of C. semilaevis. B: Number and proportion of splicing events in gonads of C. semilaevis.
Enrichment analysis of DAS genes between females and males
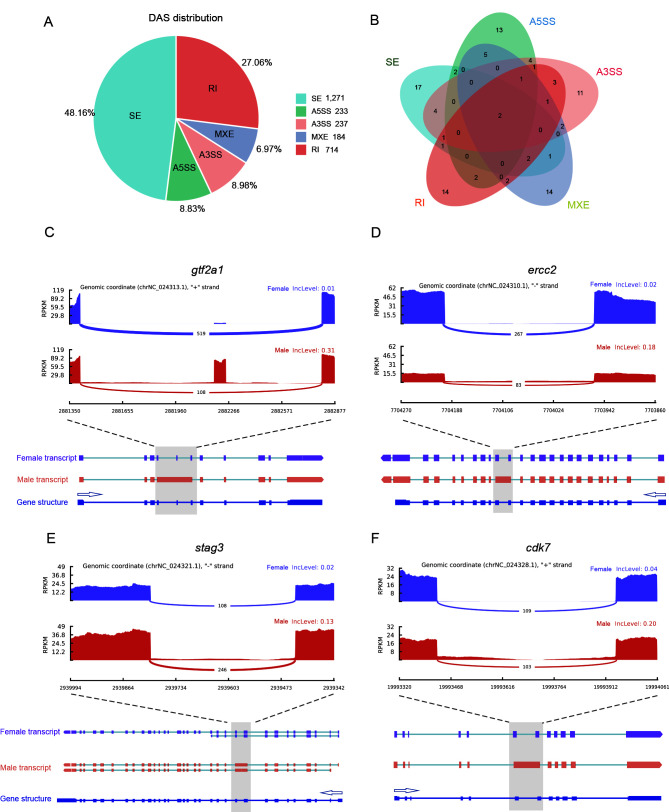
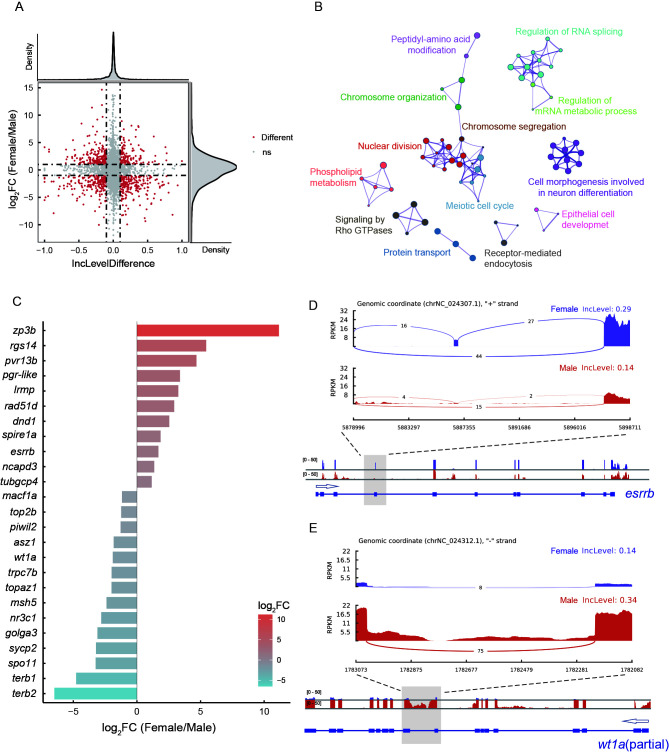
In total, 8 279 AS genes were identified in females and males. After filtering (|IncLevelDifference|>0.1 and FDR<0.05), 2 639 genes showing DAS were obtained (Supplementary Table S2). The SE, A5SS, A3SS, MXE, and RI slicing types accounted for 48.16%, 8.83%, 8.98%, 6.97%, and 27.06% of AS events, respectively (Figure 3A). KEGG enrichment analysis of these genes was performed (Supplementary Tables S3−S7), and a Venn diagram of the top 30 pathways of each type showed that the spliceosome and mRNA surveillance pathways were shared among the five types of DAS genes (Figure 3B). We also identified several other enriched pathways involved in gametogenesis, such as oocyte meiosis, basal transcription factors, and cell cycle. SEs were mainly found in the oocyte meiosis pathway. Structural maintenance of chromosomes protein 1B (LOC103379931, smc1b) and pgr-like (LOC103391230) contained sex-specific exons (Supplementary Figure S2A, B). In the basal transcription factor and cell cycle pathways, the RIs of these genes mainly occurred in males. In addition, TATA box-binding protein-like 2 (LOC103381809,tbpl2), transcription initiation factor TFIID subunit 9 (LOC103395620, taf9), transcription initiation factor IIA subunit 1 (gtf2a1), and TFIIH subunit XPD (ercc2) exhibited male-specific introns (Supplementary Figure S2C, D; Figure 3C, D). In the cell cycle pathway, male-specific RIs were also found in meiosis-specific stromal antigen 3 (stag3), cyclin-dependent kinase 7 (cdk7), and stromal antigen 2 (LOC103395545, stag2) (Figure 3E, F; Supplementary Figure S2E).
Figure 3.
Number distribution and enriched pathway analysis of genes showing DAS in female and male gonads of C. semilaevis
A: Pie plot showing proportions of five genes exhibiting DAS events. B: Venn diagram showing intersection of KEGG signaling pathway categories associated with five types of DAS events. C–F: Sashimi plot depicting RNA-Seq and exon junction reads from females and males above gtf2a1, ercc2, stag3, and cdk7 full-length transcripts and gene structure. Arrows indicate direction of gene transcription.
Effect of AS on gene expression
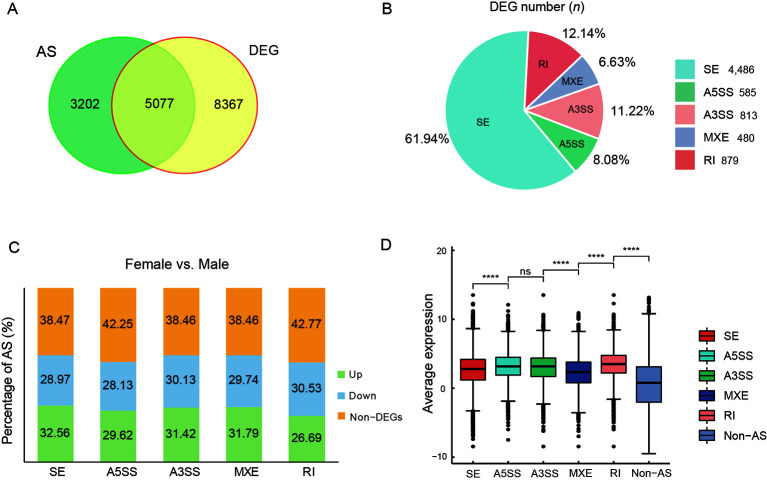
There were 5 077 intersecting genes between the 8 279 AS genes and 13 444 DEGs (Figure 4A). We counted the number of DEGs showing AS and found that SEs were the dominant AS type (Figure 4B). The proportions of differentially expressed AS genes containing SEs, A5SSs, A3SSs, MXEs, and RIs were 61.53%, 57.75%, 61.55%, 61.53%, and 57.22%, respectively (Figure 4C). More genes were up-regulated in females than in males in all AS categories, except RIs (Figure 4C). We also found that the expression levels of genes showing AS events were higher than the expression levels of genes not showing AS events (Figure 4D).
Figure 4.
Analysis of AS and DEG association in female and male gonads of C. semilaevis
A: Venn diagram showing overlap between DEGs and AS genes. B: Number of DEGs in which AS occurred and proportion of DEGs showing different AS types in gonads. C: Stacked histograms showing ratio of up- and down-regulated genes and non-DEGs among AS types in gonads. D: Box plots showing average expression levels (log2FPKM) of genes showing five splicing types or no splicing. ns: No significance. ****: P<0.0001.
AS regulates expression of genes related to gonadal differentiation
In total, 1 112 DEGs were screened from 2 639 DAS genes (Figure 5A). The network of enriched GO terms is shown in Figure 5B. Meiotic cell cycle-related GO terms, such as meiotic cell cycle process and homologous recombination, were enriched (Supplementary Table S8).
Figure 5.
Function and expression analysis of intersecting DAS genes and DEGs
A: Scatter plots showing DEGs that underwent DAS in gonads, with each dot representing a gene. Density plots above and to the right of scatter plots illustrate distribution of dots. B: Enriched GO terms of differentially expressed DAS genes. Network of enriched terms is colored according to cluster identity. Nodes sharing the same cluster identity are typically close to each other. C: Bar plot showing expression of genes involved in gonadal differentiation (log2FC>0 indicates up-regulated gene in females, log2FC<0 indicates up-regulated gene in males). D, E: RNA-Seq reads, exon junction reads, and RNA-Seq read coverage ofesrrb andwt1a. Lower box represents gene structure. Gray box indicates differentially spliced region. Arrows indicate direction of gene transcription.
A total of 25 genes related to gonadal differentiation were identified (Figure 5C). We focused on two genes in particular: i.e., steroid hormone receptor ERR2 (esrrb), which encodes a protein similar to the estrogen receptor that functions as a transcription factor (Festuccia et al., 2018), and WT1 transcription factor a (wt1a), which is involved in urogenital development (Perner et al., 2007). Our results showed that esrrb was significantly up-regulated in the ovary compared with the testis (q-value<0.01), and a female-specific 4th exon existed, which was missing in males (Figure 5D). In the testis, wt1a was markedly expressed (q-value<0.01) and an intron between the 9th and 10th exons was specifically retained (Figure 5E).
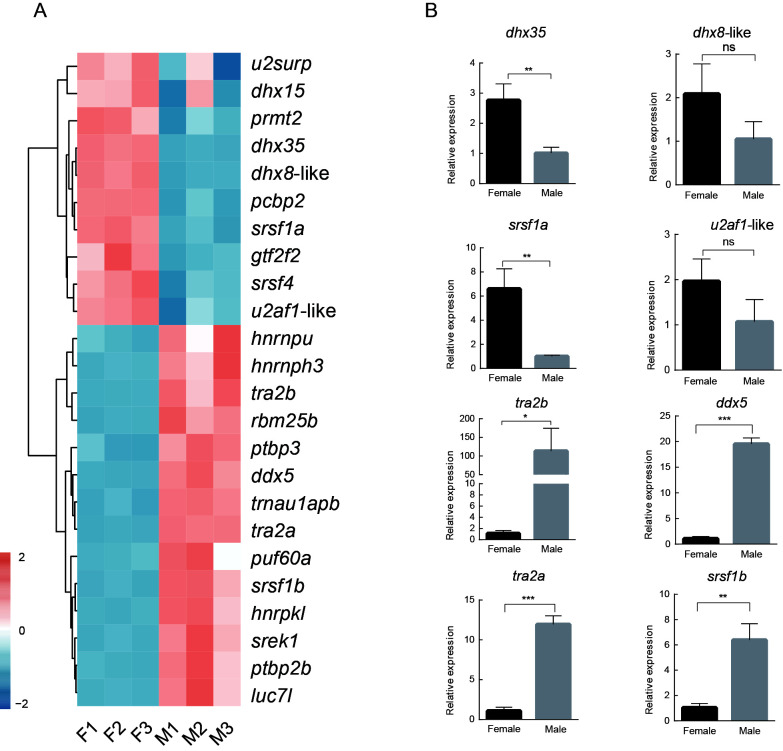
Role of splicing regulators in gonadal differentiation
Based on the enrichment results, we focused on the other significantly enriched GO term, i.e., regulation of RNA splicing (Figure 5B), which included 24 splicing regulator genes. These genes showed different expression patterns between females and males (Figure 6A). Certain genes were up-regulated in females, including DEAH-box helicase 35 (dhx35), DEAH-box helicase 8-like (LOC103381857, dhx8-like), serine/arginine-rich splicing factor 1a (LOC103378294, srsf1a), and splicing factor U2AF 35 kDa subunit-like (LOC103391595, u2af1-like), while certain other genes were up-regulated in males, including transformer 2 beta homolog (tra2b), DEAD-box helicase 5 (LOC103383782, ddx5), transformer 2 alpha homolog (tra2a), and serine/arginine-rich splicing factor 1b (LOC103395123, srsf1b).
Figure 6.
Gene expression and validation of splicing regulators in gonads of C. semilaevis
A: Expression of splicing regulators in gonads. Heatmap showing relative expression (Z-score) of splicing regulators between male and female gonads. B: qRT-PCR validation of gene expression of splicing regulators in gonads. ns: No significance;*: P<0.05;**: P<0.01;***: P<0.001.
To validate the RNA-Seq data, the expression levels of eight genes, i.e., dhx35, dhx8-like, srsf1a, u2af1-like, tra2b, ddx5, tra2a, and srsf1b, were measured by qRT-PCR (Figure 6B). Their expression trend patterns were consistent with the sequencing data.
DISCUSSION
While studies have implicated the involvement of AS in sex determination and gonadal differentiation in many species (Carreira-Rosario et al., 2016; Cesari et al., 2020; Gómez-Redondo et al., 2021; Kuravsky et al., 2011; Wang et al., 2014; Yu et al., 2021), knowledge on the regulatory mechanisms of AS in gonadal differentiation remains limited. Cynoglossus semilaevis is an economically important and sexually dimorphic fish species, with females considered more valuable than males in the aquaculture industry due to their faster growth. Research on sexual development and gonadal differentiation in C. semilaevis will provide theoretical support for the development of a monosex fish breeding industry. In this study, with the help of high-throughput sequencing technology, we explored the different regulatory mechanisms involved in gonadal differentiation in C. semilaevis mediated by AS.
AS events were more extensive in C. semilaevis testis
Based on Iso-Seq analysis, the number of full-length transcripts in the testis was 2.53 times higher than that in the ovary (81 883/32 341), and more genes were transcribed in the testis than in the ovary, indicating extensive and active gene transcription in the testis, as reported in mammals and birds (Soumillon et al., 2013). Here, many mRNAs were transcribed and novel transcripts identified in the C. semilaevis testes, consistent with previous study showing that the permissive environment of chromatin in the testis facilitates new genes, especially in spermatocytes and sperm cells (Kaessmann, 2010). In addition to the active transcription of genes in the testis, the average number of transcripts per gene in the testis (5.74) was higher than that in the ovary (3.25), thus reflecting more active AS in the testis. RNA-Seq showed more up-regulated genes in the testis than ovary, which may be related to the active transcription occurring in this organ. In mammals, RIs and SEs are the most common types of AS (Braunschweig et al., 2014; Kalsotra & Cooper, 2011). We identified 21 325 transcripts showing AS, with SEs and RIs also found to be the most common splicing types in C. semilaevis (accounting for 80.96%), thus suggesting conservation of AS type in mammals and teleosts.
Differential SEs and RIs were found in gametogenesis-related pathways
We compared DAS between the testis and ovary, with SEs and RIs again found to be the dominant DAS types (75.22%). Genes containing SEs, such as smc1b and pgr-like, were enriched in the oocyte meiosis pathway. The pgr gene is an important ovulation gene in preovulation follicular granulosa cells encoding the nuclear receptor transcription factor Pgr, which plays a key role in regulating the hypothalamic-pituitary-ovarian axis in reproduction (Natraj & Richards, 1993). Several genes with differential RIs, including taf9, gtf2a1, ercc2, and tbpl2, were enriched in basic transcription factor pathways. In the testis, these genes all contained RI regions, thus providing insight into the characteristics of RIs among transcription factors in this organ.Tbpl2 plays a key role in mouse oocyte development by regulating the transcription of oocyte-expressed genes (Gazdag et al., 2009; Yu et al., 2020). In fish, tbpl2 is indispensable for embryonic development, but its role in germ cell development has not yet been elucidated (Bártfai et al., 2004). AS in mouse meiotic spermatocytes favors RIs (Naro et al., 2017). In our study, most differential RIs observed in the gonads originated in the testis, and some genes, such as stag2, stag3, and cdk7, were enriched in the cell cycle pathway. Both stag2 and stag3 are components of the cohesion complex, which is necessary for sister chromatid cohesion (Nasmyth et al., 2000). STAG2 is required to repair DNA damage through homologous recombination in the S/G2 phase (Kong et al., 2014). Furthermore, stag3 is a meiosis-specific gene expressed in the gonads and plays an important role in gametogenesis and fertility (Garcia-Cruz et al., 2010). The protein encoded by cdk7, which is a member of the cyclin-dependent protein kinase (CDK) family and the catalytic subunit of CDK-activated kinase (CAK), is also involved in RNA polymerase II-mediated RNA transcription (Shiekhattar et al., 1995).
RI genes were more up-regulated in testis compared to other AS types
A total of 61.32% (5 077/8 279) of the identified AS genes were differentially expressed in the gonads (Figure 4A), thus illustrating the close regulatory relationship between AS and gene expression. Our results showed that RI genes were more up-regulated (higher average expression) in the testis compared to other AS-type genes, which may be related to the process of meiosis in the testis. Recent studies on mouse testes have shown that meiotic intron-retaining transcripts (IRTs) are exclusively localized in the nucleus (possibly for later use) and show higher stability than other spliced transcripts (Naro et al., 2017). Thus, the up-regulation of RI genes in the testis of Chinese tongue sole may result from IRT storage. The effect of RIs on gene expression has also been illustrated by the down-regulation of non-physiologically relevant transcripts. For example, the steady-state expression levels of IRTs in mature neurons are significantly lower than that in murine embryonic stem cells during cell differentiation (Braunschweig et al., 2014).
AS may regulate differential expression of genes related to fish gonadal differentiation
Based on our results, 25 genes related to gonadal differentiation regulated by AS were screened, including esrrb and wt1a. The esrrb gene is known to encode estrogen-related receptor b, which belongs to the NR3B subgroup (Tremblay & Giguère, 2007). Furthermore, esrrb is a transcription factor related to the self-renewal of embryonic stem (ES) cells, and esrrb knockout embryos die by embryonic day 10.5 (E10.5) (Luo et al., 1997). As an important regulatory gene, esrrb also maintains the stemness of trophoblast stem cells (Latos et al., 2015). In addition, esrrb is involved in the proliferation of gonadal germ cells, with esrrb loss leading to a decrease in germ cell number (Mitsunaga et al., 2004). Our results showed that esrrb transcripts were highly expressed in the ovary of Chinese tongue sole and contained a female-specific exon (4th exon of esrrb). The role of esrrb in maintaining cell stemness suggests it may play a role in the development of ovarian germ cells. The Wilms’ tumor suppressor gene wt1 encodes a zinc-finger transcription factor and plays an essential role in the development of the urogenital system in mice and humans (Morrison et al., 2006; Smolen et al., 2004). Two wt1 paralogs, wt1a and wt1b, are found in fish. Both wt1a and wt1b are important in zebrafish kidney development (Bollig et al., 2006). Furthermore, wt1a plays crucial roles in kidney development and sex determination in Nile tilapia, with wt1a knockdown in the kidneys resulting in developmental failure and non-expression of sex-determining genes in the gonad (Jiang et al., 2017). The retention of the 9th intron of the wt1a transcript in the Chinese tongue sole testis may promote wt1a expression in this organ.
Regulation of gonadal differentiation by splicing regulators
Many splicing regulators in the gonads undergo sex-specific splicing. For example, in Caenorhabditis elegans, ~18% of splicing regulators are subject to AS during development (Barberan-Soler & Zahler, 2008). In Drosophila gonads, splicing regulators control cell type-specific splicing through sex- and stage-specific isoforms (Gan et al., 2010). Recent studies have shown that splicing regulators contribute to cell differentiation. Many splicing regulators are highly expressed in undifferentiated spermatogonia, indicating the active expression of splicing regulators and control of multiple processes by AS during cell differentiation (Liao et al., 2021). Splicing regulators, such as srsf1a, srsf1b, and u2af1, can modulate mRNA splicing. For example, the arginine/serine-rich domain of srsf1 can bind to U1 small nuclear ribonucleoprotein (snRNP) and help U1 snRNP to bind to the 5'-splice site containing pre-mRNA (Kohtz et al., 1994). In our study, two srsf1 paralogs were found in the gonads of Cynoglossus semilaevis, i.e., srsf1a and srsf1b, which showed different expression patterns. The DEAH/DEAD-box helicase family members dhx35, dhx8, and ddx5 function as adenosine triphosphate (ATP)-dependent RNA helicases (Bourgeois et al., 2016). In addition to its involvement in the splicing regulation of pre-mRNA, ddx5 plays a role in gonadal differentiation and is a novel androgen receptor-interacting protein (Clark et al., 2008). Furthermore, ddx5 and ddx17 are master regulators of the estrogen and androgen signaling pathways and affect steroid hormone synthesis by regulating the transcription and splicing of genes up- and downstream of estrogen and androgen (Samaan et al., 2014). Recent research has also shown that ddx5 is expressed in zebrafish gonads and most ddx5-deficient females develop into fertile males, with only a small proportion developing into infertile females, suggesting that ddx5 is essential for oocyte maturation (Sone et al., 2020). In addition, ddx5 also impacts male fertility, not only regulating the expression of cell cycle genes in spermatogonia but also regulating the proper splicing of genes required for spermatogenesis (Legrand et al., 2019). In the present study, ddx5 was highly expressed in the testes, indicating that ddx5 may also be involved in regulating testicular germ cell development and male fertility. We also found that tra2a and tra2b were expressed at significant levels in the testes and may play important roles in spermatogenesis. The Drosophila melanogaster homolog, tra2, is essential for female sexual differentiation and male spermatogenesis (Amrein et al., 1988). In medaka (Oryzias latipes), tra2a and tra2b have been detected in the germ cells of both sexes prior to sex differentiation, and may therefore be involved in this process (Shiraishi et al., 2004). The tra2 homolog (Mntra-2a) found in the oriental freshwater prawn (Macrobrachium nipponense) is highly expressed in the gonads of both sexes, mainly in oocytes and spermatocytes, and may play an important role in embryonic development and early gonadal development (Wang et al., 2019).
CONCLUSIONS
By sequencing the transcriptome of C. semilaevis, we revealed extensive transcription and novel gene generation in the testis. SEs and RIs were the most common types of AS in fish gonads, consistent with findings in mammalian and bird gonads, suggesting that splicing is conservatively regulated across species. The DAS genes identified between the testis and ovary of C. semilaevis were primarily related to RNA splicing activity, indicating differential effects of sex-specific regulation of AS on gonadal differentiation. Notably, we observed differential RIs in mitosis- and meiosis-related genes in the testis, suggesting that AS may also participate in the regulation of testicular germ cell development. Moreover, we identified several sex-specific isoforms related to gonad and germ cell development, which will facilitate research on the functions of spliced isoforms. In conclusion, AS in the gonads participates in sexual development by regulating splicing of sex-related genes and splicing regulators implicated in gonadal differentiation. Coordinated regulation of AS and splicing regulators contributes significantly to gonadal differentiation.
DATA AVAILABILITY
The raw RNA-Seq reads are available in the NCBI SRA (BioProjectID PRJNA778900), GSA (accession No. CRA006156), and Science Data Bank databases (data doi: 10.11922/sciencedb.01543). The Iso-Seq reads are available in the SRA (BioProjectID PRJNA778651) and Science Data Bank databases (data doi: 10.11922/sciencedb.01543).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Q.W. and C.W.S. designed the research. Y.F.L. and K.Q.L. analyzed the data and wrote the manuscript. Q.L. and H.Y.W. performed histological sectioning and validation experiments. Y.F.L., Q.W., K.Q.L., H.Y.W., C.H.L., and C.W.S. revised the manuscript. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31722058, 31802275, and 31472269); National Key R&D Program of China (2018YFD0900301); AoShan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (2017ASTCP-ES06); Taishan Scholar Project Fund of Shandong of China to C.W.S.; National Ten-Thousands Talents Special Support Program to C.W.S.; Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2020TD19); Central Public-interest Scientific Institution Basal Research Fund, YSFRI, CAFS (20603022021018); China Agriculture Research System (CARS-47-G03); and Guangdong South China Sea Key Laboratory of Aquaculture for Aquatic Economic Animals, Guangdong Ocean University (KFKT2019ZD03)
Contributor Information
Qian Wang, Email: wangqian2014@ysfri.ac.cn.
Chang-Wei Shao, Email: shaocw@ysfri.ac.cn.
References
- 1.Amrein H, Gorman M, Nöthiger R The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein . Cell. 1988;55(6):1025–1035. doi: 10.1016/0092-8674(88)90247-4. [DOI] [PubMed] [Google Scholar]
- 2.Baralle FE, Giudice J Alternative splicing as a regulator of development and tissue identity. Nature Reviews Molecular Cell Biology. 2017;18(7):437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberan-Soler S, Zahler AM Alternative splicing regulation during C. elegans development: splicing factors as regulated targets . PLoS Genetics. 2008;4(2):e1000001. doi: 10.1371/journal.pgen.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bártfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, et al TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Current Biology. 2004;14(7):593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Bolger AM, Lohse M, Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollig F, Mehringer R, Perner B, Hartung C, Schäfer M, Schartl M, et al Identification and comparative expression analysis of a second wt1 gene in zebrafish . Developmental Dynamics. 2006;235(2):554–561. doi: 10.1002/dvdy.20645. [DOI] [PubMed] [Google Scholar]
- 7.Bourgeois CF, Mortreux F, Auboeuf D The multiple functions of RNA helicases as drivers and regulators of gene expression. Nature Reviews Molecular Cell Biology. 2016;17(7):426–438. doi: 10.1038/nrm.2016.50. [DOI] [PubMed] [Google Scholar]
- 8.Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, et al Widespread intron retention in mammals functionally tunes transcriptomes. Genome Research. 2014;24(11):1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carreira-Rosario A, Bhargava V, Hillebrand J, Kollipara RK, Ramaswami M, Buszczak M Repression of Pumilio protein expression by Rbfox1 promotes germ cell differentiation. Developmental Cell. 2016;36(5):562–571. doi: 10.1016/j.devcel.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesari E, Loiarro M, Naro C, Pieraccioli M, Farini D, Pellegrini L, et al Combinatorial control of Spo11 alternative splicing by modulation of RNA polymerase II dynamics and splicing factor recruitment during meiosis . Cell Death & Disease. 2020;11(4):240. doi: 10.1038/s41419-020-2443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YX, Chen YS, Shi CM, Huang ZB, Zhang Y, Li SK, et al SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7(1):gix120. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, et al The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Research. 2008;68(19):7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Festuccia N, Owens N, Navarro P Esrrb, an estrogen-related receptor involved in early development, pluripotency, and reprogramming. FEBS Letters. 2018;592(6):852–877. doi: 10.1002/1873-3468.12826. [DOI] [PubMed] [Google Scholar]
- 15.Gan Q, Chepelev I, Wei G, Tarayrah L, Cui KR, Zhao KJ, et al Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq . Cell Research. 2010;20(7):763–783. doi: 10.1038/cr.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Cruz R, Brieño MA, Roig I, Grossmann M, Velilla E, Pujol A, et al Dynamics of cohesin proteins REC8, STAG3, SMC1β and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes . Human Reproduction. 2010;25(9):2316–2327. doi: 10.1093/humrep/deq180. [DOI] [PubMed] [Google Scholar]
- 17.Gazdag E, Santenard A, Ziegler-Birling C, Altobelli G, Poch O, Tora L, et al TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes & Development. 2009;23(18):2210–2223. doi: 10.1101/gad.535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez-Redondo I, Planells B, Navarrete P, Gutiérrez-Adán A Role of alternative splicing in sex determination in vertebrates. Sexual Development. 2021;15(5-6):381–391. doi: 10.1159/000519218. [DOI] [PubMed] [Google Scholar]
- 19.Han F, Wang ZJ, Wu FR, Liu ZH, Huang BF, Wang DS Characterization, phylogeny, alternative splicing and expression of Sox30 gene . BMC Molecular Biology. 2010;11(1):98. doi: 10.1186/1471-2199-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isken O, Maquat LE The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nature Reviews Genetics. 2008;9(9):699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang DN, Chen JL, Fan Z, Tan DJ, Zhao JE, Shi HJ, et al CRISPR/Cas9-induced disruption of wt1a and wt1b reveals their different roles in kidney and gonad development in Nile tilapia . Developmental Biology. 2017;428(1):63–73. doi: 10.1016/j.ydbio.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Kaessmann H Origins, evolution, and phenotypic impact of new genes. Genome Research. 2010;20(10):1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalsotra A, Cooper TA Functional consequences of developmentally regulated alternative splicing. Nature Reviews Genetics. 2011;12(10):715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, et al A draft map of the human proteome. Nature. 2014;509(7502):575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, et al Protein-protein interactions and 5'-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368(6467):119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 27.Kong XD, Ball Jr AR, Pham HX, Zeng WH, Chen HY, Schmiesing JA, et al Distinct functions of human cohesin-SA1 and cohesin-SA2 in double-strand break repair. Molecular and Cellular Biology. 2014;34(4):685–698. doi: 10.1128/MCB.01503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuravsky ML, Aleshin VV, Frishman D, Muronetz VI Testis-specific glyceraldehyde-3-phosphate dehydrogenase: origin and evolution. BMC Evolutionary Biology. 2011;11(1):160. doi: 10.1186/1471-2148-11-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latos PA, Goncalves A, Oxley D, Mohammed H, Turro E, Hemberger M Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nature Communications. 2015;6(1):7776. doi: 10.1038/ncomms8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legrand JMD, Chan AL, La HM, Rossello FJ, Änkö ML, Fuller-Pace FV, et al DDX5 plays essential transcriptional and post-transcriptional roles in the maintenance and function of spermatogonia. Nature Communications. 2019;10(1):2278. doi: 10.1038/s41467-019-09972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li HL, Xu WT, Zhang N, Shao CW, Zhu Y, Dong ZD, et al Two Figla homologues have disparate functions during sex differentiation in half-smooth tongue sole (Cynoglossus semilaevis) . Scientific Reports. 2016;6(1):28219. doi: 10.1038/srep28219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao JY, Suen HC, Luk ACS, Yang LL, Lee AWT, Qi HY, et al Transcriptomic and epigenomic profiling of young and aged spermatogonial stem cells reveals molecular targets regulating differentiation. PLoS Genetics. 2021;17(7):e1009369. doi: 10.1371/journal.pgen.1009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Licatalosi DD, Darnell RB RNA processing and its regulation: global insights into biological networks. Nature Reviews Genetics. 2010;11(1):75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Chen SL, Gao FT, Meng L, Hu QM, Song WT, et al SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cynoglossus semiliaevis) . Journal of Agricultural Biotechnology. 2014;22(6):787–792. [Google Scholar]
- 36.Love MI, Huber W, Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo JM, Sladek R, Bader JA, Matthyssen A, Rossant J, Giguère V Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-β. Nature. 1997;388(6644):778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- 38.Mitsunaga K, Araki K, Mizusaki H, Morohashi KI, Haruna K, Nakagata N, et al Loss of PGC-specific expression of the orphan nuclear receptor ERR-β results in reduction of germ cell number in mouse embryos. Mechanisms of Development. 2004;121(3):237–246. doi: 10.1016/j.mod.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Morrison AA, Venables JP, Dellaire G, Ladomery MR The Wilms tumour suppressor protein WT1 (+KTS isoform) binds alpha-actinin 1 mRNA via its zinc-finger domain. Biochemistry and Cell Biology. 2006;84(5):789–798. doi: 10.1139/o06-065. [DOI] [PubMed] [Google Scholar]
- 40.Naro C, Jolly A, Di Persio S, Bielli P, Setterblad N, Alberdi AJ, et al An orchestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Developmental Cell. 2017;41(1):82–93.e4. doi: 10.1016/j.devcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nasmyth K, Peters J-M, Uhlmann F Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288(5470):1379–1384. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 42.Natraj U, Richards JS Hormonal regulation, localization, and functional activity of the progesterone receptor in granulosa cells of rat preovulatory follicles. Endocrinology. 1993;133(2):761–769. doi: 10.1210/endo.133.2.8344215. [DOI] [PubMed] [Google Scholar]
- 43.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 44.Perner B, Englert C, Bollig F The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros . Developmental Biology. 2007;309(1):87–96. doi: 10.1016/j.ydbio.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 45.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al Integrative genomics viewer. Nature Biotechnology. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samaan S, Tranchevent LC, Dardenne E, Polay Espinoza M, Zonta E, Germann S, et al The Ddx5 and Ddx17 RNA helicases are cornerstones in the complex regulatory array of steroid hormone-signaling pathways. Nucleic Acids Research. 2014;42(4):2197–2207. doi: 10.1093/nar/gkt1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen SH, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(51):E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiekhattar R, Mermelstein F, Fisher RP, Drapkin R, Dynlacht B, Wessling HC, et al Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374(6519):283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 49.Shiraishi E, Imazato H, Yamamoto T, Yokoi H, Abe SI, Kitano T Identification of two teleost homologs of the Drosophila sex determination factor, transformer-2 in medaka (Oryzias latipes) . Mechanisms of Development. 2004;121(7-8):991–996. doi: 10.1016/j.mod.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 50.Smolen GA, Vassileva MT, Wells J, Matunis MJ, Haber DA SUMO-1 modification of the Wilms' tumor suppressor WT1. Cancer Research. 2004;64(21):7846–7851. doi: 10.1158/0008-5472.CAN-04-1502. [DOI] [PubMed] [Google Scholar]
- 51.Sone R, Taimatsu K, Ohga R, Nishimura T, Tanaka M, Kawahara A Critical roles of the ddx5 gene in zebrafish sex differentiation and oocyte maturation . Scientific Reports. 2020;10(1):14157. doi: 10.1038/s41598-020-71143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soumillon M, Necsulea A, Weier M, Brawand D, Zhang XL, Gu HC, et al Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Reports. 2013;3(6):2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 53.Tremblay AM, Giguère V The NR3B subgroup: an overrview. Nuclear Receptor Signaling. 2007;5(1):nrs.05009. doi: 10.1621/nrs.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang YB, Jin SB, Fu HT, Qiao H, Sun SM, Zhang WY, et al Molecular cloning, expression pattern analysis, and in situ hybridization of a Transformer-2 gene in the oriental freshwater prawn, Macrobrachium nipponense (de Haan, 1849) . 3 Biotech. 2019;9(6):205. doi: 10.1007/s13205-019-1737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang ZK, Gao JN, Song HY, Wu XM, Sun Y, Qi J, et al Sexually dimorphic expression of vasa isoforms in the tongue sole (Cynoglossus semilaevis) . PLoS One. 2014;9(3):e93380. doi: 10.1371/journal.pone.0093380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witten JT, Ule J Understanding splicing regulation through RNA splicing maps. Trends in Genetics. 2011;27(3):89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu CW, Cvetesic N, Hisler V, Gupta K, Ye T, Gazdag E, et al TBPL2/TFIIA complex establishes the maternal transcriptome through oocyte-specific promoter usage. Nature Communications. 2020;11(1):6439. doi: 10.1038/s41467-020-20239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu LP, Zhang HR, Guan XB, Qin DD, Zhou J, Wu X Loss of ESRP1 blocks mouse oocyte development and leads to female infertility. Development. 2021;148(2):dev196931. doi: 10.1242/dev.196931. [DOI] [PubMed] [Google Scholar]
- 59.Zapater C, Chauvigné F, Fernández-Gómez B, Finn RN, Cerdà J Alternative splicing of the nuclear progestin receptor in a perciform teleost generates novel mechanisms of dominant-negative transcriptional regulation. General and Comparative Endocrinology. 2013;182:24–40. doi: 10.1016/j.ygcen.2012.11.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The raw RNA-Seq reads are available in the NCBI SRA (BioProjectID PRJNA778900), GSA (accession No. CRA006156), and Science Data Bank databases (data doi: 10.11922/sciencedb.01543). The Iso-Seq reads are available in the SRA (BioProjectID PRJNA778651) and Science Data Bank databases (data doi: 10.11922/sciencedb.01543).