Abstract
Lung adenocarcinoma is one of the most common malignant tumors worldwide. The purpose of this study was to construct a stable immune gene signature for prediction of prognosis (IGSPP) and response to immune checkpoint inhibitors (ICIs) therapy in LUAD patients. Five genes were screened by weighted gene coexpression network analysis, Cox regression and LASSO regression analyses and were used to construct the IGSPP. The survival rate of the IGSPP low-risk group was higher than that of the IGSPP high-risk group. Multivariate Cox regression analysis showed that IGSPP could be used as an independent prognostic factor for the overall survival of LUAD patients. IGSPP genes were enriched in cell cycle pathways. IGSPP gene mutation rates were higher in the high-risk group. CD4 memory-activated T cells, M0 and M1 macrophages had higher infiltration abundance in the high-risk group, which was associated with poor overall survival. In contrast, the abundance of resting CD4 memory T cells, monocytes, resting dendritic cells and resting mast cells associated with a better prognosis was higher in the low-risk group. TIDE scores and the expressions of different immune checkpoints showed that patients in the high-risk IGSPP group benefited more from ICIs treatment. In short, an IGSPP of LUAD was constructed and characterized. It could be used to predict the prognosis and benefits of ICIs treatment in LUAD patients.
Subject terms: Immunology, Immune evasion, Immunotherapy, Tumour immunology, Cancer, Tumour biomarkers, Tumour immunology
Introduction
Lung cancer is the leading cause of cancer death worldwide. According to global tumor statistics in 2020, the fatality rate of lung cancer accounts for 18% of the total cancer deaths1. Lung cancer can be divided into small cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC). NSCLC accounts for approximately 85% of all lung cancers and can be divided into lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) based on its pathological characteristics2. LUAD is the most common type of non-small-cell lung cancer3. It is difficult to diagnose LUAD in the early stage, and thus it is usually diagnosed in the late stage (> 75%). Although the 5-year survival of patients with clinical stage IA is about 60%, the 5-year survival rate with clinical stage II-IV ranges from 40 to 5%4,5. Therefore, there is an urgent need for a new index to predict the survival rate and guide the clinical management of patients with lung adenocarcinoma.
In recent years, immunotherapy has become a hot spot in the treatment of lung adenocarcinoma6. Cancer immunotherapy overcomes tumor immune escape and reactivates the antitumor immune response by regulating the immune system7. Early methods of cancer immunotherapy targeted cytokines that affect the function of immune cells8, while the new generation of immune checkpoint inhibitors (ICIs) therapy targets immune checkpoint molecules, including cytotoxic T lymphocyte antigen-4 (CTLA-4), programmed cell death protein 1 (PD-1) and PD-1 ligand (PD-L1)9. ICIs had been used in NSCLC therapy10. For example, in an early-phase study involving patients with advanced NSCLC, the median duration of overall survival of patients was 17.1 months with combination of anti-CTLA-4 and anti-PD-1 antibodies and 13.9 months with chemotherapy11. However, compared with LUSC patients, LUAD patients have a lower response to ICIs therapy11–13. The effect of ICIs treatment can be modulated by many factors, including the tumor microenvironment (TME). Analysis of the TME is helpful to improve the effect of cancer immunotherapy and the prognosis14. Therefore, it is urgent to construct an immune gene marker to predict the efficacy and prognosis of LUAD immunotherapy.
Several prognostic biomarkers of LUAD have been established15–18. In this study, we developed a new immune gene marker that can predict the response to immunotherapy. By using weighted gene co-expression network analysis (WGCNA), immune hub genes related to LUAD patient prognosis and an immune gene signature for prediction of prognosis (IGSPP) were constructed. The relationships between IGSPP, the pathological features and the overall survival of LUAD patients were then studied. The molecular and immune characteristics of IGSPP were analyzed. The prognostic ability of IGSPP in patients with immunotherapy was detected and compared with other prediction models. We hope that this IGSPP can be used as a new prognostic biomarker for patients with LUAD.
Methods
Data collection
RNA-seq data and related clinical information of 535 LUAD samples and 59 para-cancer samples were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). Samples with an unknown total survival time were excluded. The RNA-seq data and clinical information of 1134 LUAD patient samples (dataset GSE68465, GSE30219, GSE72094) were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Immune-related gene (IRG) lists were downloaded from the ImmPort (https://www.immport.org/home) and InnateBD (https://www.innatedb.ca/) databases.
Screening of immune hub genes
Gene expression data of 535 TCGA LUAD samples were extracted by Strawberry Perl (version 5.32.1.1). Differentially expressed genes (DEGs, |log2FC| > 1, q-value < 0.05) between tumors and para-cancer tissues were screened by using the R package "limma". A list of differentially expressed Immune-related gene (DEIRGs) was then obtained by intersecting the DEG and IRG lists.
WGCNA can be used to find clusters of highly related genes. We used WGCNA to find immune gene sets related to the LUAD patient prognosis and then constructed a similarity matrix based on our DEIRG expression data. The scale independence and average connectivity of the networks were tested with different power values (1–10). The appropriate power value was determined when the independent scale was greater than 0.85 and the connectivity was high. Then, the similarity matrix was transformed into a topological matrix with the topological overlap measure (TOM) describing the correlation between genes. The genes were clustered by using 1-TOM as the distance. The dynamic pruning tree was built, and the merging threshold function was set to 0.25 for module identification. Finally, the module with the highest correlation with the clinical features was selected as the key module. This module was used to identify immune-related hub genes19.
Construction of IGSPP
We used the TCGA dataset as a training cohort and three GEO dataset (GSE68465, GSE30219, GSE72094) as validation cohorts (Table 1). The TCGA cohort was used to identify prognostic immune genes and to establish a prognostic-immune risk model. Three GEO cohorts were used to validate the predictive ability of the risk model. We used the R package "survival" to analyze hub genes in the TCGA cohort by using univariate Cox regression and then screened candidate genes significantly related to overall survival (OS). We used Kaplan–Meier (KM) plots to visualize the survival curves. Candidate genes were then integrated into least absolute shrinkage and selection operator (LASSO) regression. To prevent overfitting of the model, the R package "glmnet" was used to find the best gene model. Multivariate Cox regression analysis was then carried out, and the R package "survival" was used to identify genes significantly affecting OS. These genes were used to construct IGSPP. The risk scores of each sample in the TCGA and GEO cohorts were calculated according to the Cox regression coefficients and gene expression20.
Table 1.
Clinical characteristics of patients with LUAD in each data set.
| Characteristics | Training cohort | Testing cohort | ||
|---|---|---|---|---|
| TCGA | GSE68465 | GSE30219 | GSE72094 | |
| n | 522 | 443 | 293 | 398 |
| State | ||||
| Alive | 334 | 207 | 93 | 285 |
| Dead | 188 | 236 | 200 | 113 |
| Age | ||||
| ≤ 65 | 241 | 231 | 179 | 118 |
| > 65 | 262 | 212 | 113 | 280 |
| Unknown | 19 | 0 | 1 | 0 |
| Gender | ||||
| Female | 280 | 220 | 43 | 222 |
| Male | 242 | 223 | 250 | 176 |
| Stage | ||||
| Stage I | 279 | 0 | 0 | 254 |
| Stage II | 124 | 0 | 0 | 67 |
| Stage III | 85 | 0 | 0 | 57 |
| Stage IV | 26 | 0 | 0 | 15 |
| Unknown | 8 | 443 | 293 | 5 |
| T | ||||
| T1 | 172 | 150 | 166 | 0 |
| T2 | 281 | 251 | 69 | 0 |
| T3 | 47 | 28 | 31 | 0 |
| T4 | 19 | 12 | 21 | 0 |
| Unknown | 3 | 2 | 6 | 398 |
| N (%) | ||||
| N0 | 335 | 299 | 198 | 0 |
| N1 | 98 | 88 | 53 | 0 |
| N2 | 75 | 53 | 30 | 0 |
| N3 | 2 | 0 | 10 | 0 |
| Unknown | 12 | 3 | 2 | 398 |
| M (%) | ||||
| M0 | 353 | 0 | 282 | 0 |
| M1 | 25 | 0 | 8 | 0 |
| Unknown | 144 | 443 | 3 | 398 |
According to the median risk score, the samples were divided into high-risk groups (TCGA cohort, n = 252; GSE68465 cohort, n = 209; GSE30219 cohort, n = 200; GSE72094 cohort, n = 190) and low-risk groups (TCGA cohort, n = 252; GSE68465 cohort, n = 233; GSE30219 cohort, n = 93; GSE72094 cohort, n = 208). KM survival curves were used to compare the OS between subgroups. Time-dependent ROC curves were drawn by using the R package "timeROC" according to the risk scores. The area under the curve (AUC) was calculated to verify the accuracy and prediction ability of the model. Univariate and multivariate Cox regression analyses were performed to verify the independent prognostic value of IGSPP and explore the potential clinicopathological factors related to the prognosis. A nomogram was constructed by the R package "rms", and the predictive variables in multivariate analysis were used. Calibration curves were used to evaluate the consistency between the predictions and the actual survival results.
Enrichment analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were analyzed by using the R package "clusterProfiler" (filtering condition: q-value < 0.05) to study the possible molecular mechanism of DEIRGs. The GO terms included biological process (BP), molecular function (MF) and cellular component (CC)21. The enrichment results were visualized with the R package "ggplot2".
Gene set enrichment analysis (GSEA) was used to analyze the enrichment differences of the functionally annotated gene sets according to the expression data. To explore the molecular mechanism of the difference in prognosis between the IGSPP subgroups, we used the R packages "limma" and "clusterProfiler" to analyze the GSEA enrichment in the hallmark gene sets (h.all.v7.4.symbols.gmt) and the KEGG subsets of the canonical pathway gene sets (c2.cp.kegg.v7.4.symbols.gmt).
Tumor mutation burden assay
Mutation data of LUAD patients were downloaded from the TCGA portal (https://portal.gdc.cancer.gov/). Tumor mutation burden (TMB) was calculated by Strawberry Perl. Mutations of all 30 DEIRGs related to OS were visualized by the R package "maftools". Gene mutation data of the IGSPP subgroups were also extracted by Perl. Ten genes with the highest mutation frequency in the IGSPP subgroups were then visualized as waterfall charts by the R package "maftools". The correlations between TMB and the IGSPP risk score were analyzed. KM survival curves were used to show the effects of TMB on the survival rate on the IGSPP subgroups.
Tumor immune microenvironment analysis
We used the CIBERSORT algorithm to estimate the immune cell infiltration22. According to the expression matrix of the TCGA cohort, the infiltration of 22 kinds of immune cells in 535 samples of LUAD patients was estimated. The relative ratios of the 22 types of immune cells between IGSPP subgroups were compared.
To further determine the immune and molecular characteristics between IGSPP subgroups, single sample GESA (ssGSEA) was performed on certain gene signatures, and the scores between two IGSPP subgroups were compared by using the R package "GSVA"23.
Analysis of the prognostic value of IGSPP after ICIs therapy
Tumor immune dysfunction and exclusion (TIDE) score files of LUAD patients were downloaded from the TIDE website (http://tide.dfci.harvard.edu). The R package "limma" was used to analyze the differences among the IGSPP subgroups. We calculated the tumor inflammation signature (TIS) score of the LUAD sample of the TCGA cohort. Time ROC curves were drawn according to the IGSPP risk score, TIDE and TIS score by using the R package "timeROC" to verify the accuracy of the prediction by our IGSPP model.
Cell lines and reagents
Human lung cancer cell lines (A549, H1299, PC9, H292, H1975) and human bronchial epithelial cell lines (Beas-2B) were purchased from the National Collection of Authenticated Cell Cultures (https://www.cellbank.org.cn, Shanghai, China). Cells were cultured in RPMI-1640 or F-12K medium (HyClone, Beijing, China) containing 10% fetal bovine serum (Ausbian, Australia) at 37 °C and 5% CO2, according to manufacturer’s instructions.
RNA extraction and real-time quantitative PCR (RT-qPCR)
Total RNA were extracted from cultured cells by TRIzol (Invitrogen, Shanghai, China). Reverse transcription reactions were then performed by using a first-strand cDNA synthesis kit (novoprotein, Shanghai, China). Real-time PCR system was configured according to an ABI SYBR Green Master Mix (Applied Biosystems, USA), and the mRNA expressions of IGSPP genes were detected by using a real-time fluorescent quantitative PCR instrument (QuantStudio 3, Thermo Fisher Scientific, USA). 2−ΔΔCt method was used to caculated the relative expression levels of the IGSPP candidate genes24. GAPDH was used as an internal reference. Primers used in RT-qPCR were listed in Table S1.
Statistical analysis
Statistical analyses were carried out with R 4.1.0 (https://www.R-project.org) and GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA). The differences in continuous variables between the two groups were tested by independent t-tests or nonparametric Wilcoxon tests. Categorical data were tested by chi-square tests. Univariate and multivariate Cox regression analyses were used to evaluate the effects of immune and clinicopathological factors on the prognosis of patients with LUAD. The Kaplan–Meier method and log-rank test were used for survival analysis. A two-sided p value < 0.05 was considered significant.
Results
Screening and characteristics of immune-related hub genes
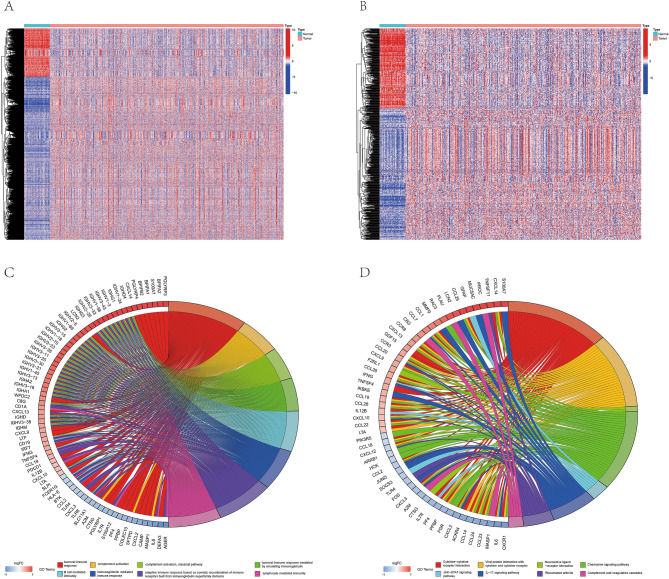
RNA sequencing data of 535 lung adenocarcinoma tissues and 59 normal tissues from the TCGA database were analyzed, and a total of 8191 differentially expressed genes (DEGs) (Table S2) were obtained. Among them, 6317 genes were upregulated and 1874 genes were downregulated (Fig. 1A). By overlapping DEGs and immune-related genes (IRGs) from ImmPort and InnateDB, 681 differentially expressed immune-related genes (DEIRGs) were obtained (Table S3). There were 258 downregulated and 423 upregulated genes among them (Fig. 1B).
Figure 1.
Differentially expressed immune-related genes in LUAD. (A) Differentially expressed genes (DEGs) (logFC filter = 1, FDR Filter = 0.05) between 535 LUAD samples (red) and 59 para-cancerous samples (blue). (B) Differentially expressed immune-related genes (DEIRGs) between 539 LUAD samples (red) and 59 para-cancerous samples (blue). (C) Gene Ontology (GO) enrichment analysis of DEIRGs (p < 0.05). (D) Kyoto Encyclopedia of Gene and Genome (KEGG) pathway analysis of DEIRGs (p < 0.05).
GO analysis showed that the DEIRGs were enriched in humoral immune response, complement activation, B cell-mediated immunity, immunoglobulin-mediated immune response, and adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains (Fig. 1C, Table S4). KEGG analysis25–27 showed that the DEIRGs were enriched in cytokine-cytokine receptor interaction, viral protein interaction with cytokines and cytokine receptors, neuroactive ligand-receptor interaction, chemokine signal pathway, JAK-STAT signal pathway, IL-17 signal pathway, rheumatoid arthritis and complement and coagulation cascade (Fig. 1D, Table S5).
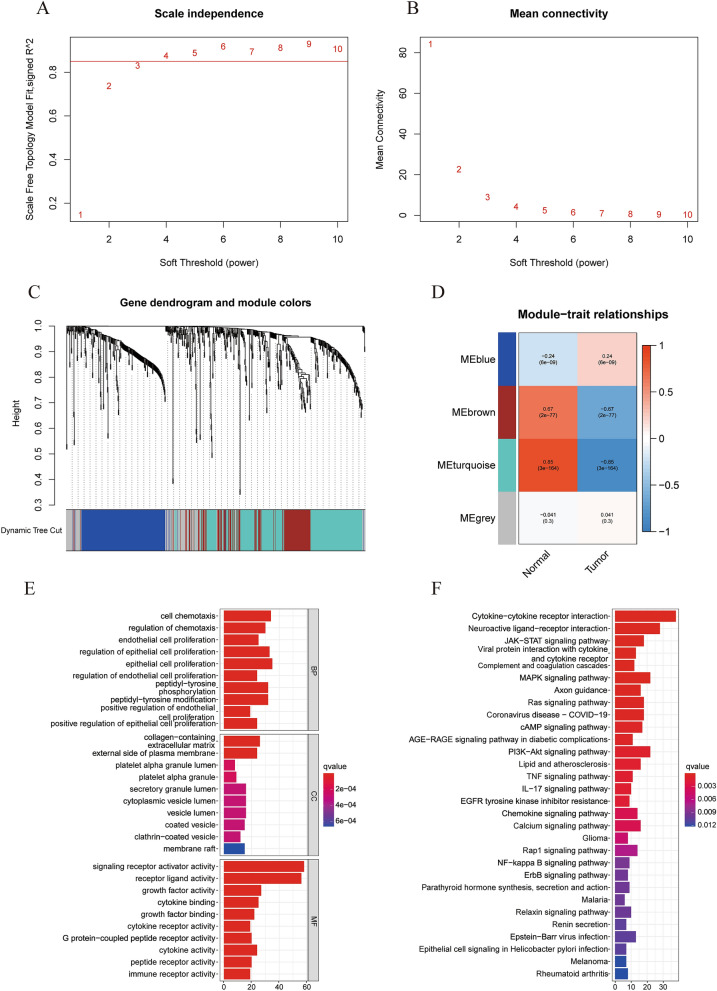
To identify the module most related to prognosis, we performed WGCNA on the obtained DEIRGs. The optimal soft-thresholding power was 4 based on the scale-free network (Fig. 2A,B). Four modules were determined according to the average linkage hierarchical clustering and the optimal soft-thresholding power. The DEIRGs obtained above were allocated to four modules. According to the Pearson correlation coefficient between the module and the sample characteristics of each module, the turquoise module was found to be closely related to the prognosis of LUAD tumors (Fig. 2C,D). Therefore, we identified the turquoise module as the key module. A total of 263 genes in the turquoise module were selected for GO and KEGG analysis. Based on the p value, the top 10 enriched GO terms of BP, MF and CC (Fig. 2E) and the top 30 KEGG pathways are shown (Fig. 2F). For more information, please see Tables S6 and Table S7.
Figure 2.
Identification of immune-related hub genes. (A) The horizontal threshold is 0.85. (B) The best soft threshold for WGCNA is 4. (C) DEIRG clustering tree based on dissimilarity measure (1-TOM). (D) The gene modules related to LUAD obtained by WGCNA. (E) Functional enrichment analysis of 263 genes in the turquoise module. (F) KEGG pathway enrichment analysis of 263 genes in the turquoise module.
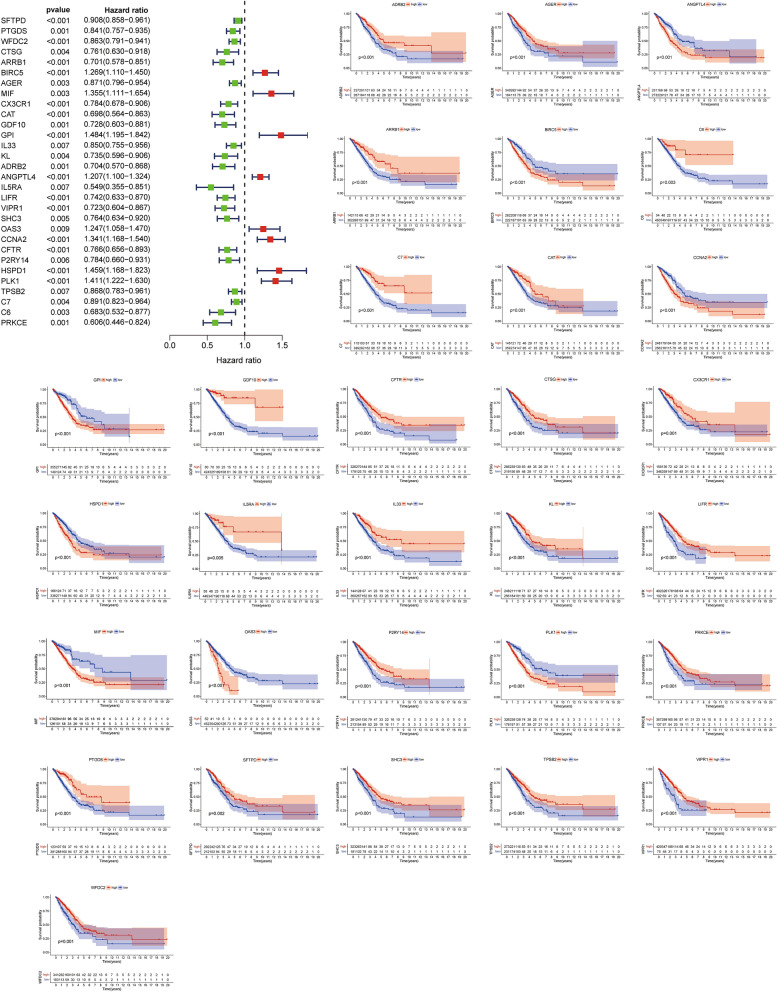
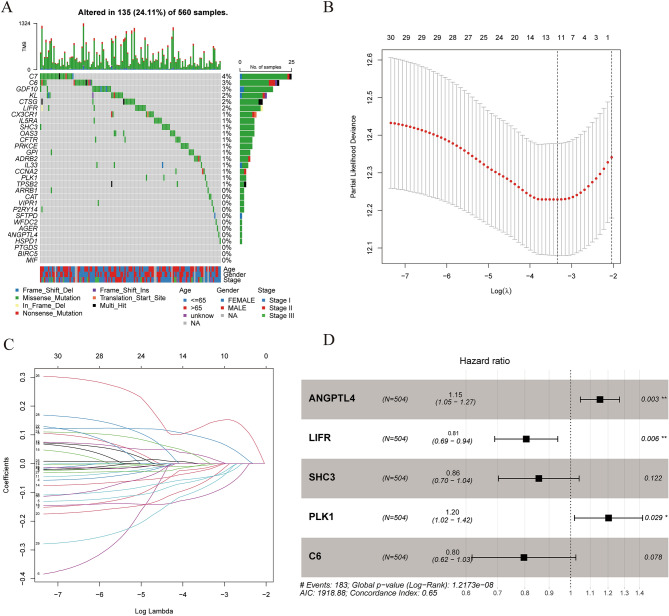
The expression of turquoise module genes was extracted from the TCGA, GSE68465, GSE30219, GSE72094 datasets. A total of 210 intersecting genes were obtained. With a p value < 0.01 as the filtering condition, 30 DEIRGs related to OS were screened by univariate Cox analysis in the TCGA cohort (Fig. 3). We also analyzed the mutations of these 30 DEIRGs. Most of these genes had missense mutations, but the mutation rate was no more than 5% (Fig. 4A). To determine the independent prognostic genes, LASSO regression and multivariate Cox regression analyses were performed with the 30 DEIRGs obtained above (Fig. 4B,C). As shown in Fig. 4D, five genes (ANGPTL4, LIFR, SHC3, PLK1 and C6) were obtained and used to construct a prognostic index. IGSPP was constructed by multiplying the expression level of these genes by the Cox regression coefficient. The formula was IGSPP = expression level of ANGPTL4 × 0.14 + expression level of LIFR × (− 0.22) + expression level of SHC3 × (− 0.16) + expression level of PLK1 × 0.19 + expression level of C6 × (− 0.23).
Figure 3.
Kaplan–Meier survival curves of 30 immune-related hub genes obtained by univariate Cox regression analysis.
Figure 4.
Construction of IGSPP for LUAD patients. (A) Mutation analysis of 30 immune-related hub genes. (B) The confidence interval of each λ. (C) Establishment of the LASSO regression model. (D) Multivariate Cox regression analysis was used to identify 5 immune-related hub genes used to construct the IGSPP.
mRNA expressions of five IGSPP genes in cultured lung carcinoma cells
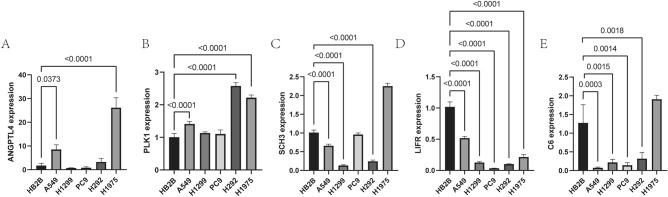
RT-qPCR was used to compare the expression levels of five genes used to construct IGSPP between five lung carcinoma cells and bronchial epithelial cell Beas-2B. Compared with the normal bronchial epithelial cell Beas-2B, the expression levels of ANGPTL4 is up-regulated in A549 and H1975 cells (Fig. 5A), PLK1 is up-regulated in A549, H292 and H1975 cells (Fig. 5B), SHC3 is down -regulated in A549 and H1299 cells (Fig. 5C), LIFR is down-regulated in all five cells (Fig. 5D), C6 is down-regulated in A549, H1299, PC9, and H292 cells (Fig. 5E). In general, the expression patterns of IGSPP genes in cultured lung carcinoma cells and LUAD tissues were consistent with TCGA data.
Figure 5.
The expression levels of five genes in the construction of IGSPP. A-E. The expression levels of five genes in cell lines.
IGSPP can predict the survival of LUAD patients
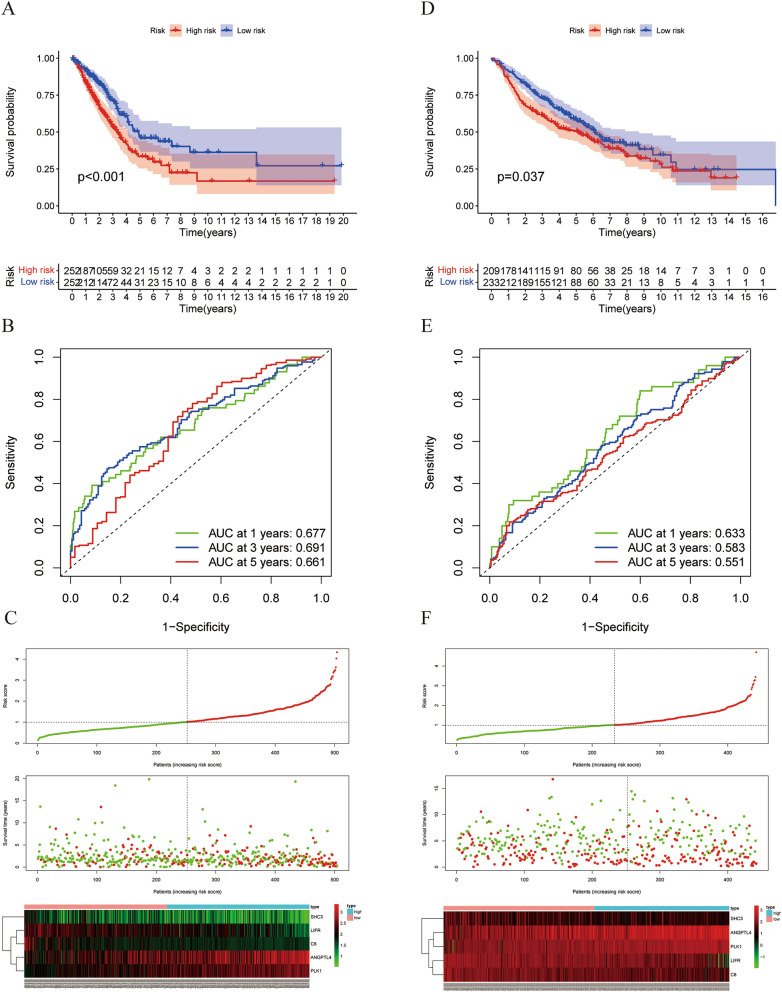
LUAD patients in the TCGA training cohort were divided into a low-risk group and a high-risk group according to the median risk score calculated by the IGSPP formula. As shown in Fig. 6A, the survival of the high-risk group of TCGA was significantly lower than that of the low-risk group (p < 0.001). IGSPP time-dependent ROC curves showed that the areas under the curves (AUCs) of IGSPP were 0.677, 0.691 and 0.661 for 1-, 3- and 5-year overall survival (OS), respectively (Fig. 6B). This result showed that the IGSPP we constructed had potential for monitoring survival. We ranked the risk score of each patient in the TCGA training cohort and used a dot map to show the survival status of the patient. As shown in Fig. 6C, the higher the risk score, the shorter the survival time. A heatmap describing the expression pattern of the DEIRGs in both subgroups was constructed. We further verified the predictive ability of IGSPP in the GSE68465 (Fig. 6D–F), GSE30219 (Fig. S1A–C) and GSE72094 (Fig. S1D–F) validation cohorts. The results of the validation cohort were highly consistent with those of the training cohorts. Therefore, the IGSPP had high accuracy in predicting OS in LUAD patients.
Figure 6.
The relationship between the IGSPP scores and the prognosis of patients. (A, D) KM survival curves of IGSPP high- and low-risk groups TCGA training cohort (A) and GEO validation cohort (D). (B, E) Time ROC curves of the TCGA cohort (B) and GEO cohort (E). (C, F) (From top to bottom) patient risk score distribution, scatter diagram of patient survival status, and expression patterns of risk genes.
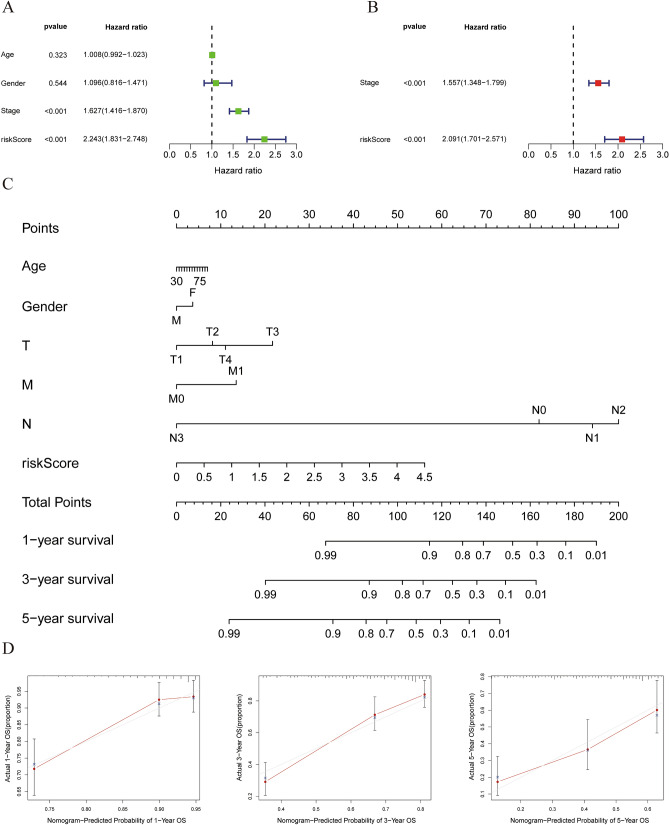
We evaluated the impact of clinicopathological factors and prognostic model risk scores on the OS of LUAD patients by performing univariate Cox regression analysis on the TCGA training cohort (Fig. 7A). Tumor stage and patient risk score were adverse factors of OS. We also performed multivariate Cox regression analysis (Fig. 7B). This confirmed that IGSPP could be used as an independent prognostic factor in LUAD patients after adjusting for other clinicopathological factors. To improve the prediction of survival of LUAD patients, we constructed a nomogram model of the TCGA cohort to quantitatively evaluate the OS of individual patients according to the variables related to OS (age, sex, tumor invasion, lymph node metastasis, distant metastasis and risk score) (Fig. 7C). Each patient received a total score from each prognostic parameter. The higher the total score, the lower the survival. The calibration curve of the prognostic nomogram showed good consistency between the prediction of the TCGA cohort and the actual 1-, 3- and 5-year survival rates (Fig. 7D). This result was further verified in the GSE68465 (Fig. S2A–D), GSE30219 (Fig. S3A–D) and GSE72094 (Fig. S4A–D) validation cohorts.
Figure 7.
Prognostic value of IGSPP in the TCGA cohort. (A) Univariate Cox regression analysis. (B) Multivariate Cox regression analysis. (C) OS nomograms of 1-, 3- and 5-year. (D) Consistency between predicted and observed 1-, 3- and 5-year survival rates.
We used the chi-square test to analyze the correlation between IGSPP risk scores and a variety of clinicopathological factors in the TCGA cohort. Correlations between tumor pathological stage, tumor invasion, lymph node metastasis, sex and age and the risk score of the patients were observed (Fig. S5A). We also analyzed the effect of IGSPP risk scores on the prognosis of patients with different clinical characteristics. We found that the OS of patients with high risk was lower than that of patients with low risk in different stages (Fig. S5B).
Molecular characteristics of the different groups of IGSPP
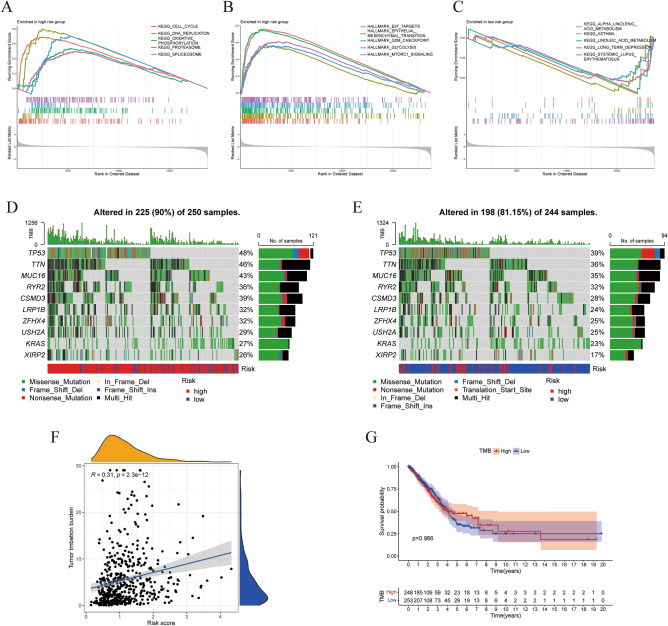
We carried out GSEA on different IGSPP subgroups to determine the enriched gene sets. The gene sets of the IGSPP high-risk subgroup were significantly enriched in cell cycle pathways, such as "Cell cycle", "DNA replication", "G2 M checkpoint" and "E2F targets" (Fig. 8A,B). This suggested that the cell cycle progression of LUAD patients in the high-risk IGSPP group was accelerated, which may lead to excessive proliferation of tumor cells. The gene sets in the low-risk group were significantly enriched in substance metabolism pathways (Fig. 8C). The detailed results of GSEA are listed in Tables S8 and Table S9.
Figure 8.
Molecular characteristics of IGSPP subgroups. (A, B) GSEA enrichment analysis in the IGSPP high-risk group (p < 0.05). (C) GSEA enrichment analysis in the IGSPP low-risk group (p < 0.05). (D) Gene mutations of patients within the IGSPP high-risk group. (E) Gene mutations of patients within the IGSPP low-risk group. (F) Correlation between the TMB and IGSPP score. (G) Correlation between the TMB and OS in patients with LUAD.
Gene mutation analysis showed that the mutation rate of the IGSPP high-risk group was significantly higher than that of the IGSPP low-risk group. Missense mutations were the most frequent mutation type, followed by multipoint mutations. We visualized the top ten genes with the highest mutation rate. The mutation rates of TP53, TTN, MUC16, RYR2, CSMD3, LRP1B, ZFHX4, USH2A, KRAS, and XIRP2 were all more than 15% in both IGSPP subgroups (Fig. 8D,E). We also explored the correlation between the tumor mutation burden (TMB) and IGSPP risk score. The IGSPP risk score was positively correlated with TMB (Fig. 8F). The effect of TMB on the prognosis of LUAD patients is shown as a KM survival curve. The results showed that there was no significant correlation between TMB and OS (p = 0.966) (Fig. 8G).
Immune characteristics of different groups of IGSPP
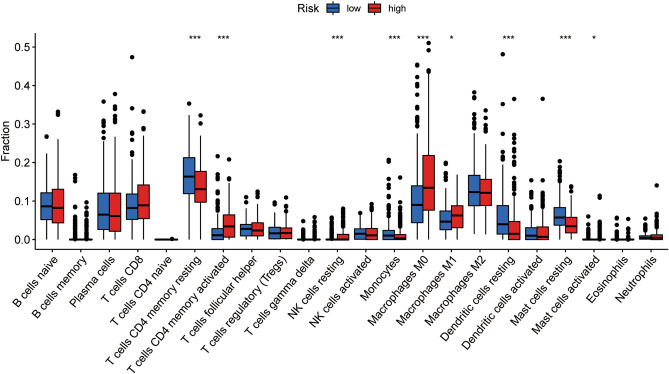
CIBERSORT is a tool for assessing immune cell infiltration. We used it to assess the relative proportion of 22 immune cells in all LUAD cases. CD4 memory activated T cells, activated NK cells, M0 macrophages, M1 macrophages and activated mast cells were more abundant in the high-risk subgroup of IGSPP, while CD4 memory resting T cells, monocytes, resting dendritic cells and resting mast cells were more abundant in the low-risk subgroup of IGSPP (Fig. 9). The content of immune cells in the high-risk group of IGSPP is shown in Fig. S6A.
Figure 9.
Immune cell infiltration in the IGSPP subgroups. Infiltration abundance of 22 immune cells in the IGSPP subgroups. Blue represents the low-risk group of IGSPP, and red represents the high-risk group. The horizontal line represents the median, and the bottom and top of the box are the 25th and 75th percentiles (quartile intervals), respectively. The Wilcoxon test was used to evaluate the differences between the two subgroups (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
We also discussed the prognostic value of 22 kinds of immune cells. Among them, the infiltration abundance of naive B cells, plasma cells, resting CD4 memory T cells, activated CD4 memory T cells, regulatory T cells (Tregs), monocytes, M0 macrophages, M1 macrophages, resting dendritic cells, activated dendritic cells, resting mast cells and neutrophils was significantly correlated with OS. The higher the infiltration abundance of naive B cells, resting dendritic cells, resting mast cells, monocytes, plasma cells, resting memory CD4 T cells and regulatory T cells (Tregs), the better the prognosis, while the higher the infiltration abundance of activated dendritic cells, M0 macrophages, M1 macrophages, neutrophils and memory CD4 T cells, the worse the prognosis (Fig. S6B). It seems that IGSPP is related to the infiltration levels of most immune cells and that IGSPP reflects the state of the tumor immune microenvironment.
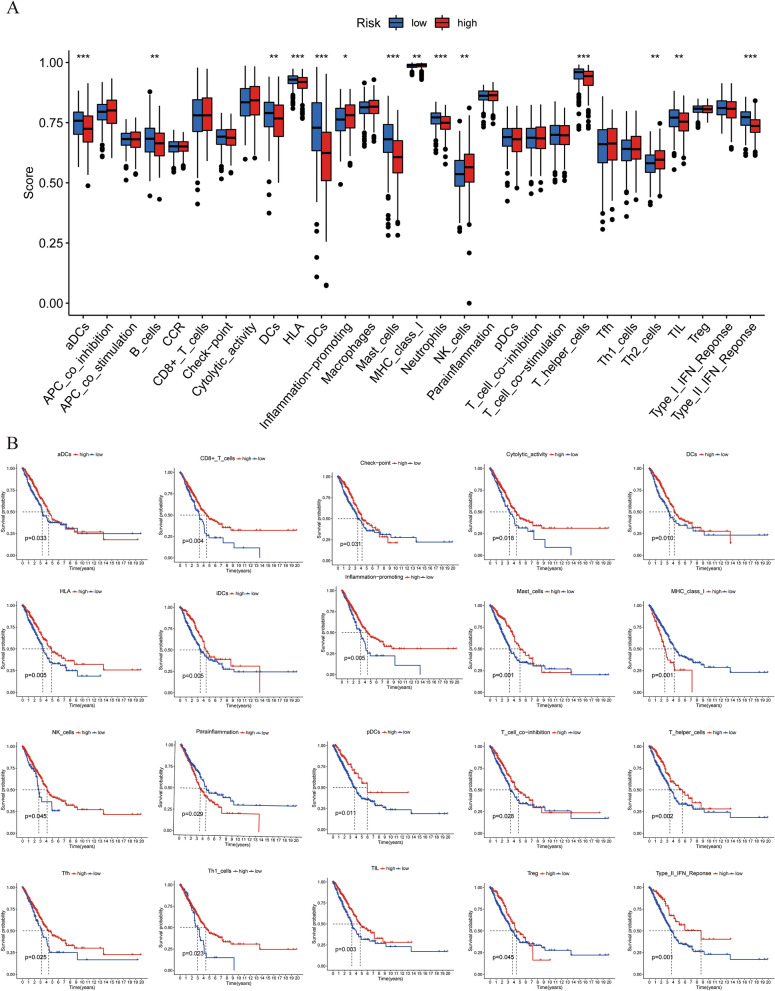
We used ssGESA to analyze the differences in immune cells and functions among the IGSPP subgroups. As shown in Fig. 10A, inflammation-promoting, MHC class I, NK cells and Th2 cells had higher scores in the IGSPP high-risk group, while aDCs, B cells, DCs, HLA, iDCs, mast cells, neutrophils, T helper cells, TILs and type II IFN response had higher scores in the IGSPP low-risk group. We also explored the effects of immune functions on prognosis. In a total of 20 immune cells and functions related to prognosis, 12 immune cells include aDCs, B cells, CD8+ T cells, DCs, iDCs, Mast_cells, NK_cells, pDCs, T_helper_cells, Tfh, Th1_cells, TIL and Treg, and 6 immune functions include Check-point, Cytolytic_activity, HLA, Inflammation-promoting, T cell co-inhibition and Type II IFN Response had a positive correlation with the prognosis of LUAD patients, while the other 2 immune functions of MHC_class_I and Parainflammation had a negative correlation with the prognosis (Fig. 10B). This result is consistent with the previous analysiswhich showed IGSPP was associated with the level of most immune cell infiltration and immune function, and means that our IGSPP can be used to reflect the state of the tumor microenvironment.
Figure 10.
Immune characteristics of the IGSPP subgroups. (A) Immune-related functions were enriched and analyzed by ssGSEA and then compared among different IGSPP subgroups. The scattered points represent the ssGSEA scores of the two subgroups, the horizontal line represents the median, and the bottom and top of the box represent the 25th and 75th percentiles (quartile intervals), respectively. The Wilcoxon test was used to evaluate the difference between two subgroups (*p < 0.05, **P < 0.01, ***P < 0.001). (B) Correlation between immune-related function score and overall survival time.
Six immune subtypes of cancer, including C1 (wound healing), C2 (IFN-γ dominant), C3 (inflammatory), C4 (lymphocyte depleted), C5 (immunologically quiet) and C6 (TGF-γ dominant), were identified to define immune response patterns impacting the prognosis28. We analyzed the distribution of various immune subtypes in different IGSPP subgroups by the chi-square test. The results showed that C1 and C2 subtypes accounted for more in the high-risk subgroup of IGSPP. The content of Th2 cells in the C1 subtype was higher28, and Th2 cells could promote the growth of tumor cells29, indicating that the high-risk subgroup of IGSPP had a higher tumor proliferation rate. The proportion of C3 subtype in the low-risk subgroup of IGSPP is significantly higher than that in the high-risk subgroup. The C3 subtype is defined by the elevated Th17 and Th1 genes28. Th1 has an inhibitory effect on tumor growth29. It indicates that the low-risk subgroup of IGSPP should have a lower tumor growth rate (Fig. S7). The above results further verify that the patients in the high-risk subgroup of IGSPP analyzed in our previous analysis have a worse prognosis.
Benefits of ICIs treatment in IGSPP subgroups
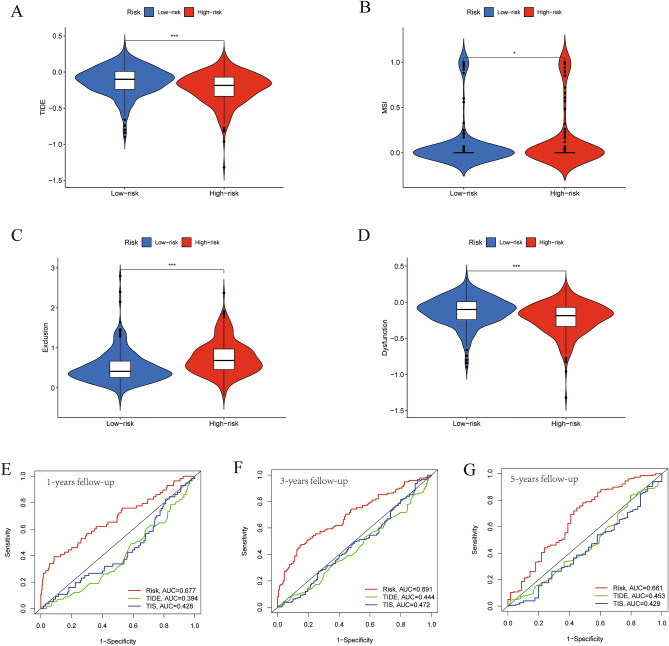
We used tumor immune dysfunction and exclusion (TIDE) to evaluate the potential clinical efficacy of ICIs therapy in IGSPP subgroups30. The results showed that the TIDE score of the IGSPP high-risk subgroup was lower than that of the low-risk subgroup. This means that patients within the IGSPP high-risk group would benefit more from ICIs treatment than those in the IGSPP low-risk group (Fig. 11A). In addition, we analyzed the relationship between the IGSPP subgroups and microsatellite instability (MSI), T cell exclusion and T cell dysfunction scores. We found that microsatellite instability (MSI) and T cell exclusion scores were higher in the high-risk IGSPP group (Fig. 11B,C), while T cell dysfunction scores were higher in the low-risk IGSPP group (Fig. 11D).
Figure 11.
The prognostic value of IGSPP for ICI treatment. (A–D) Scores of TIDE, MSI, T cell exclusion and T cell dysfunction in different IGSPP groups. The Wilcoxon test was used to evaluate the difference (*P < 0.05, **P < 0.01, ***P < 0.001). (E–G) The 1-, 3- and 5-year ROC curves based on the IGSPP, TIS and TIDE scores of the TCGA cohort.
The tumor inflammation signature (TIS) measures pre-existing but suppressed adaptive immune responses in tumors with an 18-gene signature, including genes associated with antigen presentation, chemokine expression, cytotoxic activity, and adaptive immune resistance31. We extracted TIS gene expression from TCGA LUAD patients and drew 1-, 3- and 5-year ROC curves based on TIS and TIDE scores. We compared these curves with our IGSPP time ROC curves. As shown in Fig. 11E–G, the predictive ability of IGSPP was significantly better than those of TIDE and TIS in the TCGA cohort.
Expressions of different immune checkpoints in IGSPP subgroups
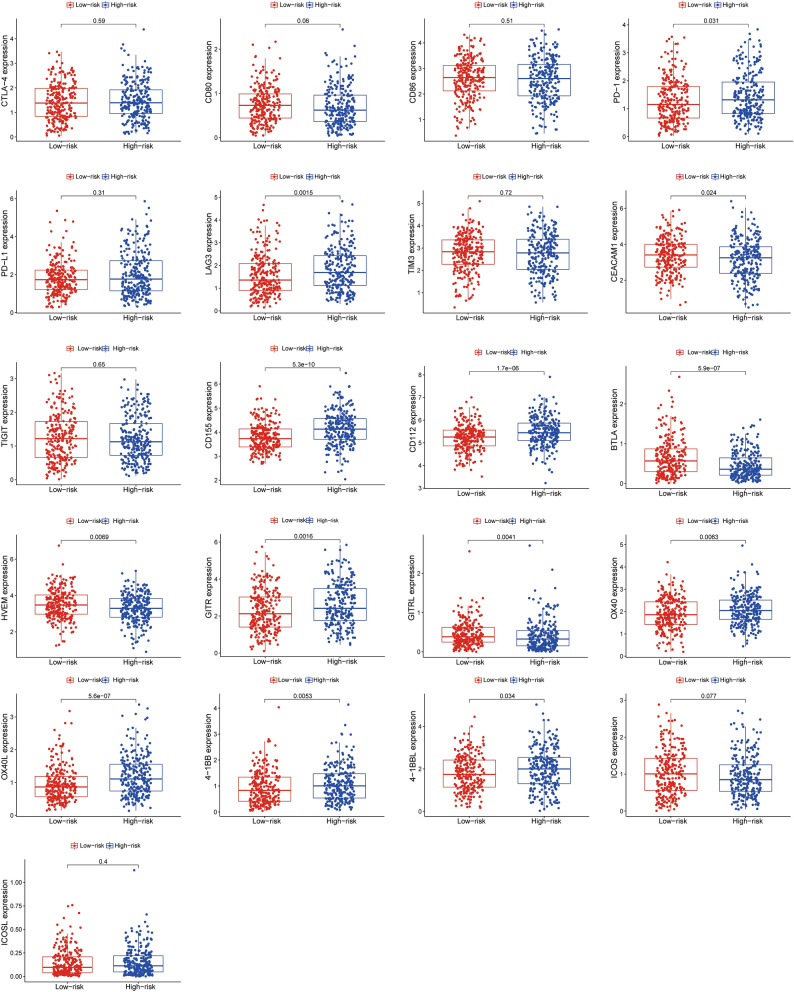
We compared the expressions of a group of co-stimulatory and/or co-inhibitory receptors of T cells as well as their ligands between IGSPP subgroups by using the TCGA database LUAD transcriptome expression data. As shown in Fig. 12, the mRNA expressions of co-inhibitory receptors/ligands, such as PD-1, LAG3, CD155, CD112, and BTLA, and co-stimulatory receptors/ligands, such as GITR, OX40, OX40L, 4-1BB, and 4-1BBL were higher in the high-risk subgroup. These results further suggested that patients in the high-risk subgroup should be more likely to benefit from ICIs therapy, not only in PD-1/PD-L1 treatment, but also in agents targeting emerging immune checkpoints, such as LAG3, GITR and OX40.
Figure 12.
The expression of some key immune checkpoint molecules.
Discussion
The oncogenesis and development of lung cancer depend not only on the changes in cancer genomes and the molecular characteristics of cancer cells but also on their interaction with the tumor environment, especially with the immune system6. Clinical applications of immune checkpoint inhibitors (ICIs), especially PD-1/PD-L1 antibodies, have improved the treatment of a variety of cancers, including NSCLCs32,33. The development of ICIs is also considered to be a milestone in immune oncology. However, the proportion of LUAD patients benefiting from ICIs is 25% lower than that of LUSC patients34. Novel biomarkers have been studied to predict the response of LUAD patients to ICIs therapy and further promote accurate immunotherapy35.
In this study, we integrated TCGA expression data and immune-related gene data and obtained 681 DEIRGs (Fig. 1B). Then, we used WGCNA and Cox regression analysis to screen DEIRGs related to the prognosis of LUAD patients. Finally, five genes were identified by LASSO and multivariate Cox regression analysis (Fig. 4B–D). These genes were used to construct the IGSPP. The prognostic value of the IGSPP was assessed in the TCGA training cohort and GEO validation cohort. The prognosis of patients within the IGSPP low-risk subgroup was better than that of patients within the IGSPP high-risk subgroup (Fig. 6A,D). IGSPP has been shown to be an immune-related biomarker. Its predictive value is reliable and accurate.
Five genes, including ANGPTL4, LIFR, SHC3, PLK1 and C6, were used to construct the IGSPP. Among them, ANGPTL4 and PLK1 are adverse prognostic genes in LUAD patients, while LIFR, SHC3 and C6 are favorable prognostic genes. ANGPTL4 has been identified as an oncogene in papillary thyroid carcinoma (PTC). Upregulation of ANGPTL4 promotes PTC cell proliferation and reduces PTC cell cycle arrest36. Overexpression of PLK1 promotes the proliferation of renal cell carcinoma (RCC) cells and inhibits apoptosis37. It also promotes the migration and invasion of NSCLC cells, thus reducing the survival rate of NSCLC patients38. PLK1 has also been confirmed as a key regulatory protein of the cell cycle39. In contrast, overexpression of LIFR in hepatocellular carcinoma (HCC) and breast cancer (BC) inhibited cancer cell migration and invasion in vivo and in vitro40,41. SHC3 has been used to construct other prognostic biomarkers in NSCLC42. NSCLC patients with higher expression of SHC3 have a better prognosis. This result is in agreement with our analysis. However, a higher expression level of SHC3 in HCC promotes the migration and invasion of HCC cells, leading to a worse prognosis of HCC patients43. It seems that SHC3 has different functions in various tumors. C6 is the sixth component of complement. Patients with C6 deficiency are susceptible to infection with Neisseria meningitidis44. C6 deficiency is common among African Americans in the southeastern United States45. However, there are few studies on the functions of complement C6 in lung cancer.
We verified the expression levels of five genes used to construct IGSPP in cultured lung cancer cell lines (Fig. 5A–E). The expression levels of these five genes in lung cancer cell lines are consistent with the results of our analysis. Since the expression levels of genes in each lung cancer cell line are different, the expression levels of genes in these five lung cancer cell lines are not all the same.
Abnormal cell proliferation is one of the most important phenomena of cancer46. Recent studies have shown that cell cycle-related genes are strongly associated with the prognosis of many malignant tumors. It is possible to evaluate tumor immunogenicity and response to ICIs therapy by analyzing cell cycle-related genes47. In this study, two genes (ANGPTL4 and PLK1) of the IGSPP that we constructed were involved in the regulation of the tumor cell cycle36,39. GSEA of the IGSPP subgroups showed that genes in the IGSPP high-risk group were significantly enriched in cell cycle-related pathways (Fig. 8A–B). These results suggest that the predictability and accuracy of the IGSPP in predicting the OS of LUAD patients are reasonable in the context of the molecular mechanism of cancer.
To further understand the immune characteristics of the different IGSPP subgroups, we analyzed gene mutations in the IGSPP subgroups (Fig. 8D–E). We found that missense mutations were the most common in LUAD patients’ genomes, followed by multiple point mutations. Several genomic alterations, such as alterations in EGFR, KRAS, STK11, ALK, JAK2, TP53, and ATM, have been shown to be associated with ICI efficacy. Among these mutations, co-occurring mutations in TP53 and other genes (EGFR, STK11, or KRAS) have been shown to have predictive value for immune checkpoint inhibitors10,48. Among the top 10 genes with the highest mutation rates we visualized, TP53 is the gene with the highest mutation frequency, and its mutation frequency is higher in the IGSPP high-risk subgroup than in the low-risk subgroup. It has been reported that mutant TP53 is an adverse prognostic factor of advanced NSCLC and head and neck squamous cell carcinoma (HNSCC)23,49. This result is consistent with our observation of a poor prognosis (OS) in the IGSPP high-risk subgroup. However, some studies have reported that mutant TP53 promotes antitumor immunity. For example, the antitumor immune signatures of TP53-mutated cancer are significantly higher than those of TP53 wild-type cancer in LUAD and breast invasive carcinoma (BRCA)50. This suggests that patients in the IGSPP high-risk subgroup should have better responses to immunotherapy. Other studies have shown that LUSC patients with high TP53 mutation rates had shorter overall survival times but were more responsive to ICIs treatment51. We also observed that the mutation rate of KRAS in the high-risk subgroup of IGSPP was also higher than that in the low-risk subgroup, with a mutation rate higher than 20%. KRAS has a high mutation rate in LUAD (20%-40%) and is associated with poor patient prognosis52,53. However, recent studies have shown that NSCLC patients with KRAS mutations have different degrees of sensitivity to ICIs treatment, and KRAS mutations are significant in enhancing PD-L1 expression, promoting T cell infiltration, and enhancing tumor immunogenicity52,54,55. Other common gene mutations associated with ICIs treatment, such as EGFR, STK11, ALK, JAK2, and ATM, had low mutation rates in this study. Considering that ICIs therapy had been advocated as standard second-line therapy for NSCLC patients without EGFR/ALK genetic alteration56,57, the cohorts in our study should not have contained many patients with EGFR/ALK mutations. The co-mutation of TP53 and KRAS suggests that the high-risk subgroup of IGSPP is more responsive to ICIs treatment. LUAD with KRAS and TP53 mutations shows a higher TMB58,59. We explored the correlation between IGSPP and known immunotherapy biomarkers. The IGSPP scores were positively correlated with TMB but not significantly correlated with the expression of PD-L1 and CTLA-4. NSCLCs usually have higher mutation burdens and can stimulate a stronger antitumor immune response (Fig. 8F). TMB can preferentially predict the clinical benefits of NSCLC immunotherapy regardless of the expression of PD-L160,61. This also suggests that the high-risk group of IGSPP may benefit more from ICIs therapy.
The role of immune cell infiltration in tumors has increasingly attracted attention in recent years. It has been shown that immune cell infiltration plays key roles in tumor progression and the response to immunotherapy in LUAD and colorectal cancer (CRC)62,63. In this study, we explored the potential relationship between IGSPP and immune cell infiltration (Fig. 9). We found that CD4+ T cells, NK cells, M0 macrophages, M1 macrophages and mast cells were enriched in the IGSPP high-risk subgroup23. This result is consistent with other immune gene prognostic index studies in HNSCC, which also showed that CD4+ T cells and M1 macrophages were abundant in the high-risk group. Cancer immunotherapy promotes the activity of cytotoxic T lymphocytes (CTLs) in tumors and assists in the activation of tumor-specific CTLs in lymphoid organs, establishing efficient and lasting antitumor immunity. CD4+ T cells enhance the antitumor activity of CTLs64. M1 proinflammatory macrophages can engulf tumor cells65. Macrophage-directed cancer immunotherapy, which modulates M2/repair-type macrophages into M1/kill-type macrophages, has been shown to slow or stop cancer growth66,67. These results suggest that patients within the IGSPP high-risk subgroup might have better responses to immunotherapy.
The tumor immune dysfunction and exclusion (TIDE) algorithm simulates two main immune escape mechanisms of tumors to predict the ICIs response23,30. The TIDE prediction score is positively correlated with the possibility of tumor immune escape. This indicates that patients with higher TIDE scores are less likely to benefit from ICIs therapy. In our study, the high-risk IGSPP subgroup had lower TIDE scores and higher microsatellite instability (MSI) and T cell exclusion scores, while the low-risk IGSPP subgroup had higher TIDE and T cell dysfunction scores (Fig. 11A–D). Therefore, patients with a low risk of IGSPP may have immune escape due to T cell dysfunction and have a poor response to ICIs treatment, while patients with a high risk of IGSPP could benefit more from ICIs treatment. Anti-PD-1/PD-L1 therapy for tumors with high MSI has shown lasting responses in prostate cancer and CRC68,69. Therefore, patients in the IGSPP high-risk group might have a better response to ICIs therapy because of MSI. Both TIDE and TIS could be used to predict the response of patients to immunotherapy30,31, but their foci were T cell functions and status and did not fully reflect the complexity of the TME involved in immunotherapy23. In addition, both markers focused on the patient's response to immunotherapy rather than the patient's survival time23. The advantages of our study are that IGSPP can distinguish the predicted survival time, molecular characteristics and the immune characteristics of patients.
The blockade of immune checkpoints can reduce the immune escape of tumor cells and activate immune reaction in tumor microenvironment70. At present, clinical trials of immune checkpoint blockade treatments mainly focus on the interaction between CTLA-4 and its ligands CD80/CD86, and the binding of PD-1 and PD-L171,72. However, compared with LUSC patients, LUAD patients have lower benefits from CTLA4 and anti-PD-1 or anti-PD-L1 therapy. Expanding ICI-based treatment approaches beyond CTLA4 and PD-1/PD- L1 pathways is therefore a clear clinical need. Antibodies targeting the co-inhibitory receptors of T cells, such as LAG3, TIM3, TIGIT and BTLA, as well as agonists of the co- stimulatory receptors, such as GITR, OX40, 4-1BB and ICOS, are undergoing clinical trials73–75. In this study, we analyzed the mRNA expression levels of these co-inhibitory receptors and their ligands by using the TCGA database LUAD transcriptome expression data (Fig. 12). It showed that the expressions of PD-1, LAG3, CD155, CD112, and BTLA, were increased in the high-risk subgroup of IGSPP. Co-stimulatory T cell receptors play critical roles in T cell activation, differentiation, effector function and survival75. Activating co-stimulatory T cell receptors is promising therapeutic strategy in clinical practice72,75. We analyzed the mRNA expression levels of targeted costimulatory receptors GITR, OX40, 4-1BB, ICOS and their ligands (Fig. 12). It showed that the expressions of GITR, OX40, OX40L, 4-1BB, 4-1BBL were higher in the high-risk IGSPP subgroup. It suggests that our IGSPP high-risk group should be also more suitable for co- stimulatory receptors targeting therapy.
In summary, we constructed an IGSPP that can be used to predict the survival rate, molecular and immune characteristics of LUAD patients and the benefits of ICIs treatment. The predictive ability of this prognostic index in LUAD needs to be further validated to determine whether it can improve patient outcomes and treatment responses by tailoring treatment to each individual patient.
Supplementary Information
Author contributions
H.C., R.L. and W.L. collected and analyzed the data, wrote the manuscript. Q.C., D.Y., J.L., J.F. and W.C. participated in the process of data analysis and table making. M.Z. and Y.Q. review and modify the manuscript. All authors listed have approved the manuscript that is enclosed.
Funding
The research was supported by the Grants from the National Natural Science Foundation of China (81773055), the Natural Science Foundation of Fujian Province (2018J01829, 2019J01293), the key talents training program of Fujian Provincial Health Commission (2017-ZQN-59 and 2017-ZQN-60) and the Joint Funds for the Innovation of Science and Technology of Fujian Province (2017Y9113).
Data availability
The datasets analyzed for this study can be found in the TCGA-LUAD project (https://portal.gdc.cancer.gov) and GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68465/GSE30219/GSE72094).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hongquan Chen, Renxi Lin and Weibin Lin.
Contributor Information
Mingfang Zhang, Email: mfzhang@fjmu.edu.cn.
Yuanlin Qi, Email: ylqi@fjmu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-12301-6.
References
- 1.Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Blandin Knight S., et al. Progress and prospects of early detection in lung cancer. Open Biol.7 (2017). [DOI] [PMC free article] [PubMed]
- 3.Ma J, Ward EM, Smith R, Jemal A. Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer. 2013;119:1381–1385. doi: 10.1002/cncr.27813. [DOI] [PubMed] [Google Scholar]
- 4.Nooreldeen, R., Bach, H. Current and future development in lung cancer diagnosis. Int. J. Mol. Sci.22 (2021). [DOI] [PMC free article] [PubMed]
- 5.Reck M, et al. Atezolizumab in combination with bevacizumab, paclitaxel and carboplatin for the first-line treatment of patients with metastatic non-squamous non-small cell lung cancer, including patients with EGFR mutations. Expert Rev. Respir. Med. 2020;14:125–136. doi: 10.1080/17476348.2020.1701439. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch FR, et al. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y. Cancer immunotherapy: Harnessing the immune system to battle cancer. J. Clin. Invest. 2015;125:3335–3337. doi: 10.1172/JCI83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy LB, Salama AKS. A review of cancer immunotherapy toxicity. CA Cancer J. Clin. 2020;70:86–104. doi: 10.3322/caac.21596. [DOI] [PubMed] [Google Scholar]
- 9.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer. 2019;19:133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou F, Qiao M, Zhou C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell Mol. Immunol. 2021;18:279–293. doi: 10.1038/s41423-020-00577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellmann MD, et al. Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. N. Engl. J. Med. 2019;381:2020–2031. doi: 10.1056/NEJMoa1910231. [DOI] [PubMed] [Google Scholar]
- 12.Carbone DP, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y, Zhai X, Yan W, Zhu H, Yu J. Clinical outcomes of immune checkpoint blockades and the underlying immune escape mechanisms in squamous and adenocarcinoma NSCLC. Cancer Med. 2021;10:3–14. doi: 10.1002/cam4.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat. Med. 2019;25:389–402. doi: 10.1038/s41591-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng H, et al. Stemness related genes revealed by network analysis associated with tumor immune microenvironment and the clinical outcome in lung adenocarcinoma. Front. Genet. 2020;11:549213. doi: 10.3389/fgene.2020.549213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, et al. An immune-related signature predicts survival in patients with lung adenocarcinoma. Front. Oncol. 2019;9:1314. doi: 10.3389/fonc.2019.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Q, et al. Identification of an immune signature predicting prognosis risk of patients in lung adenocarcinoma. J. Transl. Med. 2019;17:70. doi: 10.1186/s12967-019-1824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi M, et al. Immune signature-based risk stratification and prediction of immune checkpoint inhibitor's efficacy for lung adenocarcinoma. Cancer Immunol. Immunother. 2021;70:1705–1719. doi: 10.1007/s00262-020-02817-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Z, et al. A 13-gene metabolic prognostic signature is associated with clinical and immune features in stomach adenocarcinoma. Front. Oncol. 2021;11:612952. doi: 10.3389/fonc.2021.612952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Li Z-Y, Zhou G-Q, Sun Y. An immune-related gene prognostic index for head and neck squamous cell carcinoma. Clin. Cancer Res. 2021;27:330–341. doi: 10.1158/1078-0432.CCR-20-2166. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruterbusch M, Pruner KB, Shehata L, Pepper M. In vivo CD4+ T cell differentiation and function: Revisiting the Th1/Th2 paradigm. Annu. Rev. Immunol. 2020;38:705–725. doi: 10.1146/annurev-immunol-103019-085803. [DOI] [PubMed] [Google Scholar]
- 30.Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danaher P, et al. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): results from The Cancer Genome Atlas (TCGA) J. Immunother. Cancer. 2018;6:63. doi: 10.1186/s40425-018-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forde PM, Kelly RJ, Brahmer JR. New strategies in lung cancer: translating immunotherapy into clinical practice. Clin. Cancer Res. 2014;20:1067–1073. doi: 10.1158/1078-0432.CCR-13-0731. [DOI] [PubMed] [Google Scholar]
- 33.Garon EB, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 34.Darvin P, Toor SM, Sasidharan NV, Elkord E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018;50:1–11. doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: Response evaluation and biomarker development. Nat. Rev. Clin. Oncol. 2017;14:655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, et al. ANGPTL4 promotes the proliferation of papillary thyroid cancer via AKT pathway. Onco Targets Ther. 2020;13:2299–2309. doi: 10.2147/OTT.S237751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Z, et al. PLK1 promotes proliferation and suppresses apoptosis of renal cell carcinoma cells by phosphorylating MCM3. Cancer Gene Ther. 2020;27:412–423. doi: 10.1038/s41417-019-0094-x. [DOI] [PubMed] [Google Scholar]
- 38.Shin S-B, Jang H-R, Xu R, Won J-Y, Yim H. Active PLK1-driven metastasis is amplified by TGF-β signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene. 2020;39:767–785. doi: 10.1038/s41388-019-1023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutteridge RE, Ndiaye MA, Liu X, Ahmad N. Plk1 inhibitors in cancer therapy: From laboratory to clinics. Mol. Cancer Ther. 2016;15:1427–1435. doi: 10.1158/1535-7163.MCT-15-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Q, et al. LIFR functions as a metastasis suppressor in hepatocellular carcinoma by negatively regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis. 2015;36:1201–1212. doi: 10.1093/carcin/bgv108. [DOI] [PubMed] [Google Scholar]
- 41.Wang M, et al. Long non-coding RNA-CTD-2108O9.1 represses breast cancer metastasis by influencing leukemia inhibitory factor receptor. Cancer Sci. 2018;109:1764–1774. doi: 10.1111/cas.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R, et al. Identification and validation of the prognostic value of immune-related genes in non-small cell lung cancer. Am. J. Transl. Res. 2020;12:5844–5865. [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, et al. Demethylation-induced overexpression of Shc3 drives c-Raf-independent activation of MEK/ERK in HCC. Cancer Res. 2018;78:2219–2232. doi: 10.1158/0008-5472.CAN-17-2432. [DOI] [PubMed] [Google Scholar]
- 44.Potter PC, et al. Prophylaxis against Neisseria meningitidis infections and antibody responses in patients with deficiency of the sixth component of complement. J. Infect. Dis. 1990;161:932–937. doi: 10.1093/infdis/161.5.932. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Z, et al. High prevalence of complement component C6 deficiency among African–Americans in the south-eastern USA. Clin. Exp. Immunol. 2000;119:305–310. doi: 10.1046/j.1365-2249.2000.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 47.Yin X, et al. Development of a novel gene signature to predict prognosis and response to PD-1 blockade in clear cell renal cell carcinoma. Oncoimmunology. 2021;10:1933332. doi: 10.1080/2162402X.2021.1933332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun H, et al. Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. EBioMedicine. 2020;60:102990. doi: 10.1016/j.ebiom.2020.102990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao X-D, Qin B-D, You P, Cai J, Zang Y-S. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer. 2018;123:70–75. doi: 10.1016/j.lungcan.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Li M, Wang X. Cancer type-dependent correlations between TP53 mutations and antitumor immunity. DNA Repair. (Amst). 2020;88:102785. doi: 10.1016/j.dnarep.2020.102785. [DOI] [PubMed] [Google Scholar]
- 51.Xu F, et al. A -associated gene signature for prediction of prognosis and therapeutic responses in lung squamous cell carcinoma. Oncoimmunology. 2020;9:1731943. doi: 10.1080/2162402X.2020.1731943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeanson A, et al. Efficacy of immune checkpoint inhibitors in KRAS-mutant non-small cell lung cancer (NSCLC) J. Thorac. Oncol. 2019;14:1095–1101. doi: 10.1016/j.jtho.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 53.Reck M, Carbone DP, Garassino M, Barlesi F. Targeting KRAS in non-small-cell lung cancer: Recent progress and new approaches. Ann. Oncol. 2021;32:1101–1110. doi: 10.1016/j.annonc.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Dong ZY, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 55.Uruga H, Mino-Kenudson M. Predictive biomarkers for response to immune checkpoint inhibitors in lung cancer: PD-L1 and beyond. Virchows Arch. 2021;478:31–44. doi: 10.1007/s00428-021-03030-8. [DOI] [PubMed] [Google Scholar]
- 56.Rittmeyer A, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herbst RS, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet. 2016;387:1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 58.Anichini, A., Perotti, V. E., Sgambelluri, F., Mortarini, R. Immune escape mechanisms in non small cell lung cancer. Cancers (Basel).12, (2020). [DOI] [PMC free article] [PubMed]
- 59.Skoulidis F, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan YH, et al. Predictive value of the TP53/PIK3CA/ATM mutation classifier for patients with bladder cancer responding to immune checkpoint inhibitor therapy. Front. Immunol. 2021;12:643282. doi: 10.3389/fimmu.2021.643282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai, X., et al. Development and validation of a genomic mutation signature to predict response to PD-1 inhibitors in non-squamous NSCLC: A multicohort study. J. Immunother. Cancer.8 (2020). [DOI] [PMC free article] [PubMed]
- 62.Zuo S, Wei M, Wang S, Dong J, Wei J. Pan-cancer analysis of immune cell infiltration identifies a prognostic immune-cell characteristic score (ICCS) in lung adenocarcinoma. Front. Immunol. 2020;11:1218. doi: 10.3389/fimmu.2020.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saito T, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 64.Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 65.Xia Y, et al. Engineering macrophages for cancer immunotherapy and drug delivery. Adv. Mater. 2020;32:e2002054. doi: 10.1002/adma.202002054. [DOI] [PubMed] [Google Scholar]
- 66.Mills CD, Lenz LL, Harris RA. A breakthrough: Macrophage-directed cancer immunotherapy. Cancer Res. 2016;76:513–516. doi: 10.1158/0008-5472.CAN-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klichinsky M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020;38:947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abida W, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 2019;5:471–478. doi: 10.1001/jamaoncol.2018.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kok M, Chalabi M, Haanen J. How I treat MSI cancers with advanced disease. ESMO Open. 2019;4:e000511. doi: 10.1136/esmoopen-2019-000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edner NM, Carlesso G, Rush JS, Walker LSK. Targeting co-stimulatory molecules in autoimmune disease. Nat. Rev. Drug Discov. 2020;19:860–883. doi: 10.1038/s41573-020-0081-9. [DOI] [PubMed] [Google Scholar]
- 73.Fang, J., et al. Prognostic value of immune checkpoint molecules in breast cancer. Biosci. Rep.40 (2020). [DOI] [PMC free article] [PubMed]
- 74.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013;13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kraehenbuehl L, Weng CH, Eghbali S, Wolchok JD, Merghoub T. Enhancing immunotherapy in cancer by targeting emerging immunomodulatory pathways. Nat. Rev. Clin. Oncol. 2022;19:37–50. doi: 10.1038/s41571-021-00552-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed for this study can be found in the TCGA-LUAD project (https://portal.gdc.cancer.gov) and GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68465/GSE30219/GSE72094).