Abstract
Background
Within the class Enoplea, the earliest-branching lineages in the phylum Nematoda, the relatively highly conserved ancestral mitochondrial architecture of Trichinellida is in stark contrast to the rapidly evolving architecture of Dorylaimida and Mermithida. To better understand the evolution of mitogenomic architecture in this lineage, we sequenced the mitogenome of a fish parasite Pseudocapillaria tomentosa (Trichinellida: Capillariidae) and compared it to all available enoplean mitogenomes.
Results
P. tomentosa exhibited highly reduced noncoding regions (the largest was 98 bp), and a unique base composition among the Enoplea. We attributed the latter to the inverted GC skew (0.08) in comparison to the ancestral skew in Trichinellidae (-0.43 to -0.37). Capillariidae, Trichuridae and Longidoridae (Dorylaimida) generally exhibited low negative or low positive skews (-0.1 to 0.1), whereas Mermithidae exhibited fully inverted low skews (0 to 0.05). This is indicative of inversions in the strand replication order or otherwise disrupted replication mechanism in the lineages with reduced/inverted skews. Among the Trichinellida, Trichinellidae and Trichuridae have almost perfectly conserved architecture, whereas Capillariidae exhibit multiple rearrangements of tRNA genes. In contrast, Mermithidae (Mermithida) and Longidoridae (Dorylaimida) exhibit almost no similarity to the ancestral architecture.
Conclusions
Longidoridae exhibited more rearranged mitogenomic architecture than the hypervariable Mermithidae. Similar to the Chromadorea, the evolution of mitochondrial architecture in enoplean nematodes exhibits a strong discontinuity: lineages possessing a mostly conserved architecture over tens of millions of years are interspersed with lineages exhibiting architectural hypervariability. As Longidoridae also have some of the smallest metazoan mitochondrial genomes, they contradict the prediction that compact mitogenomes should be structurally stable. Lineages exhibiting inverted skews appear to represent the intermediate phase between the Trichinellidae (ancestral) and fully derived skews in Chromadorean mitogenomes (GC skews = 0.18 to 0.64). Multiple lines of evidence (CAT-GTR analysis in our study, a majority of previous mitogenomic results, and skew disruption scenarios) support the Dorylaimia split into two sister-clades: Dorylaimida + Mermithida and Trichinellida. However, skew inversions produce strong base composition biases, which can hamper phylogenetic and other evolutionary studies, so enoplean mitogenomes have to be used with utmost care in evolutionary studies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08607-4.
Keywords: Compositional heterogeneity, GC skew, Inversion of the replication order, Gene rearrangement, Mitogenome, Pseudocapillaria tomentosa, Capillariidae, Phylogeny
Background
The class Enoplea comprises the earliest-branching lineages in the phylum Nematoda. It is divided into two subclasses: Dorylaimia (or Clade I) and Enoplia (Clade II) [1]. Dorylaimia exhibit a diverse life history range: the vertebrate-parasitic order Trichinellida, the insect-parasitic Mermithida, the plant-parasitic Dorylaimida, and the free-living Mononchida [1]. Previously sequenced enoplean mitochondrial genomes (mitogenomes) suggest that the evolution of mitochondrial architecture in this clade exhibits a rather stark discontinuity: Trichinellidae and Trichuridae families of the order Trichinellida appear to possess a perfectly conserved architecture [2–5], Dorylaimida (Longidoridae) exhibit a fast-evolving mitogenomic architecture [5, 6], and Mermithidae (Mermithida) exhibit a rampant gene order rearrangement rate, unmatched within the Bilateria [7, 8]. This pattern fits our previous observation that mitogenome architecture evolution is discontinuous (in nematodes) and that long evolutionary periods of stasis are interspersed with lineages that exhibit exponentially accelerated mitochondrial evolution rates [9]. However, several key enoplean lineages remain unrepresented in terms of available mitogenomes, e.g. the entire Enoplia and Mononchida lineages, which limits our understanding of the evolution of mitochondrial architecture within this nematode lineage.
The Nematoda has been recognised as a problematic lineage in bilaterian/ecdysozoan mitochondrial phylogenomics [10, 11]. The phylogeny of Enoplea also remains unresolved, largely because genomic data (both mitogenomic and nuclear) remain unavailable for many key enoplean lineages, but also because previous mitogenomic studies of Dorylaimia produced inconsistent results [6, 12, 13]. Base composition biases may cause strong artefacts in phylogenetic analyses [14–18], and mitochondrial genomes often exhibit strong compositional biases as a consequence of directional mutational pressures predominantly associated with mitochondrial replication [19, 20]. During the replication, the parental H-strand is left in a mutagenic single-stranded state for almost two hours [21], which may cause spontaneous hydrolytic deamination of A and C (into G and T respectively). Over multiple generations, this results in the accumulation of T and G on the H-strand, and A and C on the L-strand [19]. In some cases, mitochondrial architecture rearrangements may result in the inversion of the origin of replication, which then also causes an inversion of the direction of mutational pressures, i.e., now T and G accumulate on the L-strand, and A and C on the H-strand [22]. Such skew inversions can affect the branch length, mutational saturation, codon usage, protein properties, and cause reverse mutations, so they can interfere with a broad range of evolutionary studies [16].
Pseudocapillaria tomentosa is a widespread intestinal nematode parasite found in the intestines of a broad range of fish hosts in the Northern Hemisphere [23, 24]. Until relatively recently, this parasite was very poorly studied [23], but this species often parasitizes zebrafish, so along with the relatively rapid increase in the popularity of zebrafish as a model animal for molecular biology, this nematode concomitantly received somewhat increased scientific attention in recent years [25–27]. Despite this, molecular data for P. tomentosa remain almost completely unavailable. The only currently (July 2021) available GenBank entry is a partial 18S rRNA gene sequence (KU987805) [23]. Furthermore, at the onset of this study, there were no mitogenomes available for the entire family Capillariidae (Trichinellida). In the meantime, two sequences were submitted to the GenBank: the mitogenome of a fish parasite Eucoleus annulatus, collected in the USA [12], and the mitogenome of a cat parasite Capillaria sp. (Australia) (MH665363; unpublished).
For this study, we sequenced the complete mitogenome of Pseudocapillaria tomentosa (Dujardin, 1843) Moravec, 1987 (Dorylaimia: Trichinellida: Capillariidae) to generate molecular data necessary for the identification, population studies, and phylogeny of this species and family, as well as to test two working hypotheses: 1. mitogenomic architecture evolution is discontinuous in the Enoplea, and 2. base composition biases are hampering mitochondrial phylogenomic reconstruction in the Enoplea. Along with P. tomentosa, we used all mitogenomic data available for the Enoplea to conduct detailed comparative mitogenomic analyses and test the two hypotheses.
Results and Discussion
General mitogenomic features of P. tomentosa and Capillariidae
The complete mitogenome of P. tomentosa contained all 37 standard mitochondrial genes, which were encoded on both strands (Fig. 1; for additional results and discussion see Additional file 1). This is common for the Enoplea, whereas in Chromadorea genes are encoded on a single strand [28]. The strand distribution of protein-coding genes (PCGs) was conserved among the Trichinellida, with nad2, nad5, nad4 and nad4L consistently encoded on the minus strand, but variable among the Dorylaimida and Mermithida (Fig. 1). In the mitogenome of P. tomentosa, a majority of genes kept the transcription sense: a stretch of 16 genes on the minus strand was punctuated only by trnK and trnT. trnW was the only gene encoded on the minus strand that was isolated from this main stretch. Start codons (ATG, ATT, ATA) and stop codons (TAA and TAG) were standard. Gene overlaps were few (4) and small (1 to 2 bp). The distribution of tRNA genes was conserved among most Trichinellida, apart from the Capillariidae. In this family, the architecture was conserved between P. tomentosa and E. annulatus [12], whereas Capillaria sp. appears to exhibit a slightly rearranged architecture, but these may be sequencing and annotation artefacts as the mitogenome is almost certainly incomplete (Additional file 1). In comparison to E. annulatus, P. tomentosa exhibited an identical gene order and strand distribution, but only partially conserved codon usage, low gene sequence similarity (average identity = 64.65%), partially conserved distribution of intergenic regions, and almost no similarity in the distribution of gene overlaps (Table 1).
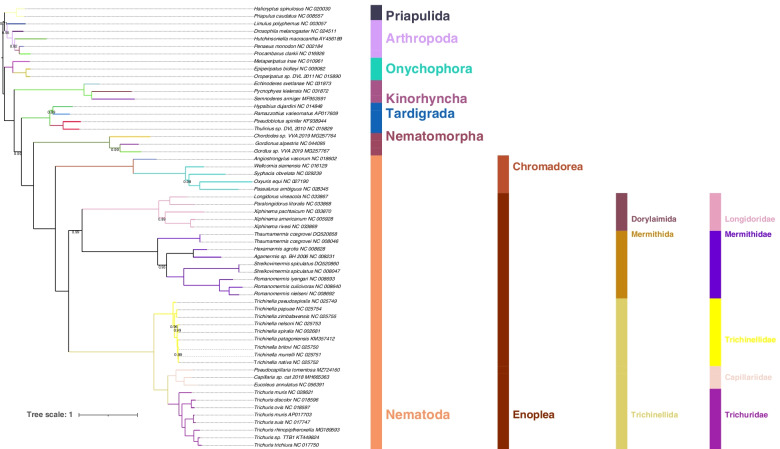
Fig. 1.
Mitochondrial architecture and skews of enoplean mitogenomes. GC skews on the plus strand are shown as yellow bars, where negative values are to the left from the axis and positive to the right. The legend for the mitochondrial architecture is shown in the figure, with only NCRs larger than 200 bp shown. Species names are given with GenBank accession numbers. The family and order-level taxonomic identity is shown to the right. Pseudocapillaria tomentosa is highlighted in blue
Table 1.
The comparison of mitochondrial architectures of Eucoleus annulatus (left) and Pseudocapillaria tomentosa (right)
| Gene | Position | Size | IGN | Codon | Strand | Identity | ||
|---|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | |||||
| cox1 | 1/1 | 1548/1548 | 1548/1548 | ATG/ATG | TAA/TAA | H/H | 76.42 | |
| cox2 | 1570/1554 | 2253/2234 | 684/681 | 21/5 | ATG/ATG | TAA/TAA | H/H | 68.13 |
| trnL2 | 2262/2252 | 2325/2319 | 64/68 | 8/17 | H/H | 70.59 | ||
| trnE | 2335/2319 | 2391/2374 | 57/56 | 9/-1 | H/H | 73.68 | ||
| nad1 | 2438/2400 | 3337/3293 | 900/894 | 46/25 | ATG/ATT | TAA/TAA | H/H | 68.56 |
| trnC | 3383/3392 | 3440/3441 | 58/50 | 45/98 | L/L | 58.62 | ||
| trnQ | 3423/3446 | 3498/3500 | 76/55 | -18/4 | L/L | 63.16 | ||
| trnG | 3546/3515 | 3620/3568 | 75/54 | 47/14 | L/L | 49.33 | ||
| nad2 | 3676/3619 | 4578/4518 | 903/900 | 55/50 | ATT/ATA | TAA/TAG | L/L | 65.15 |
| trnM | 4579/4519 | 4640/4580 | 62/62 | L/L | 85.48 | |||
| nad5 | 4655/4579 | 6214/6120 | 1560/1542 | 14/-2 | ATA/ATA | TAA/TAG | L/L | 62.18 |
| trnH | 6215/6121 | 6270/6177 | 56/57 | L/L | 64.41 | |||
| nad4 | 6275/6228 | 7228/7487 | 954/1260 | 4/50 | ATT/ATT | TAA/TAA | L/L | 48.57 |
| NCR | 7229/– | 7534/– | 306/– | NA | ||||
| trnL1 | 7535/7489 | 7599/7557 | 65/69 | –/1 | L/L | 71.43 | ||
| trnS2 | 7600/7576 | 7654/7629 | 55/54 | –/18 | L/L | 30.99 | ||
| trnI | 7654/7703 | 7722/7767 | 69/65 | -1/73 | L/L | 71.01 | ||
| trnY | 7728/7766 | 7786/7824 | 59/59 | 5/-2 | L/L | 62.71 | ||
| trnK | 7834/7849 | 7904/7911 | 71/63 | 47/24 | H/H | 68.06 | ||
| trnF | 7893/7927 | 7967/7982 | 75/56 | -12/15 | L/L | 61.33 | ||
| trnR | 7964/7988 | 8026/8054 | 63/67 | -4/5 | L/L | 64.71 | ||
| nad4L | 8032/8058 | 8268/8306 | 237/249 | 5/3 | ATA/ATT | TAA/TAA | L/L | 66.67 |
| trnT | 8282/8308 | 8338/8361 | 57/54 | 13/1 | H/H | 77.19 | ||
| trnP | 8328/8374 | 8399/8427 | 72/54 | -11/12 | L/L | 70.83 | ||
| nad6 | 8400/8429 | 8860/8884 | 461/456 | –/1 | TTG/ATT | TA/TAG | H/H | 66.67 |
| cytb | 8861/8898 | 9973/10010 | 1113/1113 | –/13 | ATG/ATG | TAA/TAA | H/H | 73.23 |
| trnS1 | 9974/10010 | 10,038/10063 | 65/54 | –/-1 | H/H | 55.07 | ||
| rrnS | 10,039/10064 | 10,852/10739 | 814/676 | H/H | 61.88 | |||
| trnV | 10,853/10740 | 10,909/10793 | 57/54 | H/H | 58.62 | |||
| rrnL | 10,910/10794 | 11,751/11734 | 842/941 | H/H | 60.78 | |||
| atp6 | 11,752/11735 | 12,540/12532 | 789/798 | ATA/ATT | TAA/TAA | H/H | 66.79 | |
| cox3 | 12,541/12538 | 13,317/13311 | 777/774 | –/5 | ATG/ATG | TAA/TAA | H/H | 66.41 |
| trnW | 13,336/13315 | 13,398/13376 | 63/62 | 18/3 | L/L | 84.13 | ||
| trnD | 13,402/13378 | 13,456/13432 | 55/55 | 3/1 | H/H | 72.73 | ||
| atp8 | 13,463/13433 | 13,603/13579 | 141/147 | 6/– | ATG/ATC | TAA/TAA | H/H | 51.35 |
| nad3 | 13,631/13588 | 13,960/13929 | 330/342 | 27/8 | ATA/ATA | TAA/TAA | H/H | 66.08 |
| trnN | 13,985/13934 | 14,044/13993 | 60/60 | 24/4 | H/H | 49.32 | ||
| trnA | 14,044/14009 | 14,099/14062 | 56/54 | -1/15 | H/H | 59.68 | ||
IGN column shows the sizes of intergenic regions (positive values) or gene overlaps (negative values) in base pairs
Gene order
We numerically assessed our observation that the evolution of mitochondrial architecture in enoplean nematodes appears to be discontinuous using the common intervals gene order similarity measure (where the value 0 indicates a complete absence of shared common intervals) [29]. Almost all Trichinellidae and Trichuridae exhibited an identical gene order (Table 2). The only exceptions were Trichinella zimbabwensis, which exhibited a translocation of the trnK gene, and Trichinella patagoniensis, which exhibited a loss of the trnK gene. Both of these might be sequencing or annotation artefacts. Trichinellidae exhibit a great similarity to the ancestral arthropod mitochondrial architecture (gene arrangement), which is also similar to the ancestral ecdysozoan mitogenome architecture [2, 30–32]. Trichinellidae are therefore considered to possess the most ancestral architecture among all nematodes, whereas all other lineages exhibit highly derived architectures, so we will treat this gene order as the ancestral for all Nematoda. In comparison to the ancestral gene order, Capillariidae exhibited a large number of rearrangements (Fig. 1, Additional file 1), but within the enoplean dataset, they exhibited an intermediate gene rearrangement rate (168—178). Strongly accelerated evolutionary rates were observed in Mermithidae, which exhibited very little similarity to the ancestral gene order (2—12), and Longidoridae, which exhibited almost no similarity to the ancestral gene order (0 – 2). This partially corroborates the results of visual assessment, but surprisingly indicates that Longidoridae, and not Mermithidae, possess the most highly rearranged gene orders. The explanation is that rearrangements in Mermithidae are concentrated in specific, hypervariable regions [7], whereas some segments remain conserved. This is not the case in Longidoridae, which do not exhibit any conserved segments. Intriguingly, Longidoridae (Xiphinema) species have some of the smallest metazoan mitochondrial genomes sequenced so far [5, 6], which contradicts the proposition that compact mitogenomes are structurally stable, i.e. that shorter mitogenomes exhibit fewer gene order rearrangements relative to the ancestral state [33]. Indeed, it is expected that rearrangements should produce an increase in the number of noncoding regions, regardless of whether the underlying mechanism is tandem duplication/random loss [34] of recombination [35]. In agreement with this, Mermithidae, where the mechanism producing the mitogenomic architecture variability is believed to be recombination, exhibit multiple large (> 100 bp) NCR and some of the largest mitogenomes among the Bilateria [7, 8]. Therefore, this is an intriguing discrepancy, which was not discussed by previous studies (to our knowledge) [5, 6]. There are two possible mechanisms to explain it: 1) Longidoridae is undergoing a strong purifying selection directed towards the reduction of mitogenome size, 2) The mechanism of rearrangement is unique in this lineage, and it does not produce multiple NCRs. It should be noted that Longidoridae also exhibit a high level of within-family rearrangement rates (Table 2), which implies that the first scenario would require very strong purifying selection levels. A possible explanation could be found in the ‘race for replication’ hypothesis, which proposes that high metabolic demands impose stronger purifying selection constraints for small genome size [36], but it is unclear why Longidoridae would have higher metabolic demands than other enoplean nematodes. Further studies, and more mitogenomes, are needed to elucidate the rearrangement mechanisms in this family and the evolutionary pressures producing the small mitogenome size.
Table 2.
Gene order distances in the class Enoplea
| T + T | Tz | Tp | Csp | Pt | Xa | Xp | Lv | Pl | Tc1 | Tc2 | Ss1 | Ss2 | Asp | Rc | Rn | Ri | Ha | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trichinellidae and Trichuridae | ||||||||||||||||||
| Ancestral GO | 640 | 1254 | 178 | 168 | 2 | 0 | 2 | 0 | 8 | 8 | 6 | 6 | 2 | 8 | 4 | 12 | 8 | |
| Trichinella zimbabwensis | 640 | 1184 | 214 | 174 | 2 | 0 | 0 | 0 | 10 | 10 | 6 | 6 | 0 | 4 | 2 | 8 | 8 | |
| Trichinella patagoniensis | 1254 | 1184 | 206 | 168 | 2 | 0 | 0 | 0 | 8 | 8 | 6 | 6 | 0 | 8 | 4 | 12 | 8 | |
| Capillariidae | ||||||||||||||||||
| Capillaria sp | 178 | 214 | 206 | 872 | 4 | 2 | 0 | 2 | 10 | 10 | 10 | 10 | 2 | 2 | 2 | 6 | 4 | |
| Pseudocapillaria tomentosa | 168 | 174 | 168 | 872 | 6 | 2 | 0 | 2 | 10 | 10 | 12 | 12 | 0 | 2 | 2 | 8 | 0 | |
| Longidoridae | ||||||||||||||||||
| Xiphinema americanum | 2 | 2 | 2 | 4 | 6 | 50 | 36 | 28 | 6 | 10 | 4 | 2 | 0 | 12 | 8 | 8 | 6 | |
| Xiphinema pachtaicum | 0 | 0 | 0 | 2 | 2 | 50 | 30 | 20 | 2 | 0 | 10 | 6 | 0 | 24 | 10 | 6 | 8 | |
| Longidorus vineacola | 2 | 0 | 0 | 0 | 0 | 36 | 30 | 110 | 6 | 10 | 4 | 4 | 2 | 14 | 14 | 4 | 6 | |
| Paralongidorus litoralis | 0 | 0 | 0 | 2 | 2 | 28 | 20 | 110 | 8 | 12 | 6 | 6 | 4 | 6 | 14 | 6 | 4 | |
| Mermithidae | ||||||||||||||||||
| Thaumamermis cosgrovei 1 | 8 | 10 | 8 | 10 | 10 | 6 | 2 | 6 | 8 | 942 | 4 | 4 | 2 | 4 | 2 | 12 | 8 | |
| Thaumamermis cosgrovei 2 | 8 | 10 | 8 | 10 | 10 | 10 | 0 | 10 | 12 | 942 | 4 | 4 | 2 | 4 | 2 | 10 | 8 | |
| Strelkovimermis spiculatus 1 | 6 | 6 | 6 | 10 | 12 | 4 | 10 | 4 | 6 | 4 | 4 | 684 | 2 | 2 | 4 | 8 | 8 | |
| Strelkovimermis spiculatus 2 | 6 | 6 | 6 | 10 | 12 | 2 | 6 | 4 | 6 | 4 | 4 | 684 | 2 | 2 | 4 | 6 | 6 | |
| Agamermis sp | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 4 | 2 | 2 | 2 | 2 | 6 | 4 | 0 | 10 | |
| Romanomermis culicivorax | 8 | 4 | 8 | 2 | 2 | 12 | 24 | 14 | 6 | 4 | 4 | 2 | 2 | 6 | 130 | 28 | 6 | |
| Romanomermis nielseni | 4 | 2 | 4 | 2 | 2 | 8 | 10 | 14 | 14 | 2 | 2 | 4 | 4 | 4 | 130 | 58 | 4 | |
| Romanomermis iyengari | 12 | 8 | 12 | 6 | 8 | 8 | 6 | 4 | 6 | 12 | 10 | 8 | 6 | 0 | 28 | 58 | 4 | |
| Hexamermis agrotis | 8 | 8 | 8 | 4 | 0 | 6 | 8 | 6 | 4 | 8 | 8 | 8 | 6 | 10 | 6 | 4 | 4 | |
The distances were inferred using the Common Intervals measure in CREx, where a value of 0 indicates no similarity, whereas numbers over 1,000 indicate almost perfectly conserved GO. Column headers mirror row headers, but names are acronymic. Only unique architectures are shown, and species are clustered in the corresponding families. Ancestral GO row comprises all Trichinellidae and Trichuridae mitogenomes, apart from the two Trichinella species below. Pseudocapillaria tomentosa is equal to Eucoleus annulatus, and Xiphinema rivesi is equal to Xiphinema americanum
Until relatively recently the mitogenomic architectural variability was believed to be concentrated largely within the enoplean family Mermithidae, whereas Chromadorea, which contains the majority of Nematoda species, was believed to possess a relatively stable mitogenomic architecture [7, 13, 28, 37–39]. In our previous study, we have shown that some Chromadorean lineages also exhibit exponentially accelerated rates of mitogenomic architecture evolution, which prompted us to propose that the mitogenomic architecture evolution is discontinuous in nematodes [9]. Our analyses conducted for this study indicate that the evolution of mitogenomic architecture is nonlinear in the class Enoplea as well.
A strongly reduced size of noncoding regions
The newly sequenced mitogenome exhibited a remarkably small largest noncoding region of 98 base pairs (bp) (Fig. 1; Table 1). Normally, metazoan mitogenomes possess a large noncoding region, commonly spanning > 500 bp to a few thousand bp, which usually comprises the control region (CR). For example, in the closely related Trichinellidae, the putative CR spans 700 to 1600 bp, and it is duplicated in most species [2, 3] (Fig. 1; see Additional file 1 for discussion of putative sequencing artefacts). Mermithidae exhibited on average about eight large (> 100 bp) NCRs, but some Romanomermis species had more than ten of them [40]. This is attributed to the disrupted, rapidly evolving, mitogenomic architecture in this family, comprising multiple duplicated genes and some of the largest mitogenomes among the nematodes [7]. Surprisingly, the longest NCR (putative CR) in the mitogenome of T. cosgrovei is also reduced in size (401 bp) in comparison to standard metazoan mitogenomes [7]. Some Longidoridae (Xiphinema) species are known to have some of the smallest metazoan mitochondrial genomes [5, 6], as a result of shortened genes, short noncoding regions, and gene overlaps [5]. However, the mitogenome (14,118 bp) and protein-coding genes (PCGs) of P. tomentosa were not reduced in size in comparison to other Trichinellida species, and the overlaps were few and small. Therefore, only the noncoding regions are strongly reduced in size in P. tomentosa. In Capillariidae and Trichuridae, only E. annulatus and Trichuris rhinopiptheroxella had an NCR larger than 200 bp (306 and 208 bp, respectively; see Additional file 1 for additional discussion). However, despite perfectly matching gene arrangements between P. tomentosa and E. annulatus, the former mitogenome exhibited a negligible intergenic region of 1 bp in the location corresponding to the location of the putative CR of E. annulatus (306 bp NCR between nad4 and trnL1) (Fig. 1; Additional file 1). This indicates that the putative CR is rapidly evolving in this family, which makes its identification very difficult. Trichuris species largely exhibit two “major” NCRs, both of which are also smaller than 200 bp: the longer one tends to be in the range of 120–170 bp, whereas the short one tends to be 90—120 bp [4, 41]. In conclusion, NCRs are normal-sized in Trichinellidae, slightly reduced in size but multiplied due to rearranged architecture in Mermithidae, and strongly reduced in the Longidoridae, Capilariidae and Trichuridae.
Base composition and skews
High AT content is common in the mitogenomes of nematodes [28], but P. tomentosa exhibited the highest AT bias among all available Trichinellida (79.3%). Among the Dorylaimia, only three Mermithidae species had marginally higher AT content (Agamermis and Romanomermis sp.; Table 3). As the content of A and G was average, the exceptional AT content is attributable to the very high T content (41.3%), with only three Mermithidae species exhibiting higher values, and to exceptionally low C content, which was the lowest in the entire dataset: 9.5%. This made us suspect that the unique base composition of this species may be associated with mutational pressures associated with the mitogenomic replication mechanism [16, 19, 22]. To assess this, we inspected their base composition skews (GC) [42]. All Trichinellidae mitogenomes exhibited high negative GC skews (skews were calculated for the entire plus strand, and we refer to skew magnitude in absolute terms, i.e. distance from zero, throughout the manuscript) from -0.431 to -0.368. The remaining Trichinellida lineages, comprising Trichuridae and Capillariidae, exhibited low negative skews (-0.073 to -0.007), with Trichuris muris (0.043) and P. tomentosa (0.083) even exhibiting fully inverted (positive) skews. Dorylaimida (Longidoridae) exhibited a very similar pattern: low negative skews (-0.075 to -0.018), with Xiphinema pachtaicum exhibiting a fully inverted skew (0.124). Finally, all available Mermithida (Mermithidae) species exhibited fully inverted (positive) but low skews in the range between 0.007 and 0.054. After X. pachtaicum, P. tomentosa exhibited the second-highest positive skew in the dataset. The patterns produced by cumulative skew plots were inconsistent, even among some closely related taxa, such as P. tomentosa and Capillaria sp. (Additional file 1). Apart from Trichinellidae, most species (including P. tomentosa and E. annulatus) exhibited rather noisy patterns, with skew plots switching between positive and negative values.
Table 3.
Comparative mitochondrial architecture in the Dorylaimia subclass
| Organism | ID | Length | A | T | C | G | A + T | G + C | GC | GC_NCR |
|---|---|---|---|---|---|---|---|---|---|---|
| Dorylaimida: Longidoridae | ||||||||||
| Xiphinema pachtaicum | NC_033870 | 12,489 | 29.4 | 39.1 | 13.7 | 17.6 | 68.5 | 31.3 | 0.124 | 0.115 |
| Paralongidorus litoralis | NC_033868 | 12,763 | 32.4 | 31.5 | 18.4 | 17.7 | 63.9 | 36.1 | -0.018 | 0.007 |
| Xiphinema rivesi | NC_033869 | 12,624 | 37.4 | 31.5 | 16.6 | 14.5 | 68.9 | 31.1 | -0.069 | 0 |
| Xiphinema americanum | NC_005928 | 12,626 | 36.6 | 29.9 | 18 | 15.5 | 66.5 | 33.5 | -0.075 | -0.127 |
| Longidorus vineacola | NC_033867 | 13,519 | 31.7 | 32 | 18.8 | 17.5 | 63.7 | 36.3 | -0.037 | -0.128 |
| Mermithida: Mermithidae | ||||||||||
| Strelkovimermis spiculatus | NC_008047 | 18,030 | 36 | 42.5 | 10.1 | 11.3 | 78.5 | 21.4 | 0.054 | 0.047 |
| Strelkovimermis spiculatus | DQ520860 | 17,118 | 35.9 | 42.7 | 10.2 | 11.2 | 78.6 | 21.4 | 0.049 | 0.047 |
| Romanomermis iyengari | NC_008693 | 18,919 | 39.3 | 40.2 | 10.1 | 10.4 | 79.5 | 20.5 | 0.016 | 0.03 |
| Thaumamermis cosgrovei | NC_008046 | 20,013 | 32.1 | 39.3 | 13.9 | 14.7 | 71.4 | 28.6 | 0.026 | 0.011 |
| Thaumamermis cosgrovei | DQ520858 | 21,506 | 31.9 | 39.4 | 14 | 14.7 | 71.3 | 28.7 | 0.025 | 0.008 |
| Hexamermis agrotis | NC_008828 | 24,606 | 41 | 37.4 | 10.7 | 10.9 | 78.4 | 21.6 | 0.009 | 0.002 |
| Romanomermis nielseni | NC_008692 | 15,546 | 38.8 | 40.3 | 10.3 | 10.5 | 79.1 | 20.8 | 0.009 | 0.001 |
| Romanomermis culicivorax | NC_008640 | 26,194 | 41.3 | 38 | 10.3 | 10.4 | 79.3 | 20.7 | 0.007 | -0.014 |
| Agamermis sp. | NC_008231 | 16,561 | 36.8 | 43.7 | 9.6 | 9.9 | 80.5 | 19.5 | 0.019 | -0.02 |
| Trichinellida: Capillariidae | ||||||||||
| Pseudocapillaria tomentosa | 14,062 | 38 | 41.3 | 9.5 | 11.2 | 79.3 | 20.7 | 0.083 | 0.032 | |
| Eucoleus annulatus | NC_056391 | 14,118 | 38.3 | 38.3 | 12.6 | 10.9 | 76.6 | 23.5 | -0.073 | -0.288 |
| Capillaria sp. cat-2018 | MH665363 | 13,624 | 37 | 39 | 12.1 | 11.9 | 76 | 24 | -0.007 | -0.391 |
| Trichinellida: Trichinellidae | ||||||||||
| Trichinella murrelli | NC_025751 | 16,592 | 40.7 | 26.8 | 22.9 | 9.7 | 67.5 | 32.6 | -0.404 | -0.37 |
| Trichinella spiralis | NC_002681 | 16,706 | 40.5 | 26.5 | 23 | 9.7 | 67 | 32.7 | -0.405 | -0.371 |
| Trichinella britovi | NC_025750 | 16,421 | 40.6 | 26.6 | 23.1 | 9.8 | 67.2 | 32.9 | -0.405 | -0.395 |
| Trichinella papuae | NC_025754 | 17,326 | 40.2 | 26.6 | 22.7 | 10.4 | 66.8 | 33.1 | -0.371 | -0.412 |
| Trichinella pseudospiralis | NC_025749 | 17,667 | 40.9 | 26.6 | 22.6 | 9.9 | 67.5 | 32.5 | -0.392 | -0.425 |
| Trichinella nelsoni | NC_025753 | 15,278 | 40.6 | 25.5 | 24.1 | 9.8 | 66.1 | 33.9 | -0.422 | -0.46 |
| Trichinella patagoniensis | KM357412 | 15,179 | 40.1 | 24.9 | 24.4 | 9.7 | 65 | 34.1 | -0.431 | -0.485 |
| Trichinella zimbabwensis | NC_025755 | 14,244 | 39.6 | 25.4 | 24 | 11.1 | 65 | 35.1 | -0.368 | -0.502 |
| Trichinella nativa | NC_025752 | 14,077 | 40.4 | 25.8 | 23.8 | 10 | 66.2 | 33.8 | -0.411 | -0.639 |
| Trichinellida: Trichuridae | ||||||||||
| Trichuris ovis | NC_018597 | 13,946 | 34.5 | 35.3 | 15.9 | 14.4 | 69.8 | 30.3 | -0.05 | 0.01 |
| Trichuris discolor | NC_018596 | 13,904 | 33.9 | 36 | 15.3 | 14.9 | 69.9 | 30.2 | -0.012 | -0.033 |
| Trichuris muris | NC_028621 | 14,105 | 35.6 | 37.8 | 12.7 | 13.8 | 73.4 | 26.5 | 0.043 | -0.089 |
| Trichuris muris | AP017703 | 14,297 | 35.5 | 36.4 | 14.7 | 13.5 | 71.9 | 28.2 | -0.043 | -0.131 |
| Trichuris suis | NC_017747 | 14,436 | 35.6 | 35.9 | 15.1 | 13.5 | 71.5 | 28.6 | -0.057 | -0.165 |
| Trichuris trichiura | NC_017750 | 14,046 | 33.6 | 34.5 | 16.9 | 15 | 68.1 | 31.9 | -0.059 | -0.263 |
| Trichuris rhinopiptheroxella | MG189593 | 14,186 | 33.4 | 36 | 15.4 | 15.2 | 69.4 | 30.6 | -0.007 | -0.315 |
| Trichuris sp. | KT449824 | 13,984 | 34.2 | 35 | 16.5 | 14.3 | 69.2 | 30.8 | -0.07 | -0.537 |
All base composition values are shown for the mitochondrial plus strand. The ‘length’ refers to the full length of the mitogenome in bases. Base composition is given in %. GC means GC skew, and GC_NCR is GC skew of all noncoding regions
This is indicative of inversions in the strand replication order, or otherwise disrupted replication mechanism (e.g. multiple origins of replication) in the lineages with reduced/inverted skews [16, 22]. As skews were not discussed in previous papers that reported clade I mitogenomes with inverted skews [6–8, 43–45], this is the first observation that some enoplean lineages underwent skew inversions. To get a better resolution we checked all other available Nematoda mitogenomes. This revealed that all chromadorean mitogenomes exhibit high positive GC skews on the plus strand: 0.18 to 0.64 (average = 0.40) (Additional file 2: panel D). As discussed above, Trichinellidae possess the most ancestral architecture among all nematodes, whereas all other lineages exhibit highly derived architectures. From this, we can infer that a high negative GC skew on the plus strand, similar to the one exhibited by most other Arthropoda [16, 46], is the ancestral skew for Nematodes. Therefore, chromadorean nematodes underwent a series of architectural rearrangements that resulted in all genes being encoded on a single strand and resulted in a full skew inversion on the plus strand (alternatively, but less likely, all genes from the plus strand may have migrated to the minus strand).
Theoretically, it may be possible to identify strand inversions of the origin of replication from conserved promoter sequence motifs [16]. For example, in Xiphinema americanum, a sequence motif 5’-GAGACCTGAGCCCAAGATA-3’ similar to the conserved promoter element sequence in the human mitogenome was found in the putative CR [5]. We assessed our dataset for the presence of this motif, but it matched to (highly derived) sequences in only four species: two Xiphinema and two Mermithidae species (Additional file 1: Figure S15). We also searched for other conserved motifs but, apart from several motifs that are common in Trichinellidae, other families did not exhibit any consistently conserved motifs (Additional file 3). This is in agreement with a previous study, which found no identifiable conserved motifs in Longidoridae [6]. Several common motifs were identified in the Mermithidae, but their apparent abundance was largely attributable to the multiplication of NCRs in this family. This further corroborates the fast and unique evolution of mitochondrial control regions in the Enoplea.
Phylogenetic analyses and implications of inverted skews for evolutionary analyses
The phylogeny of Enoplea remains unresolved, as different datasets produce incongruent results. Previous evidence that skew inversions affect phylogenetics and other evolutionary studies was limited to arthropods (mostly crustaceans), which often exhibit fully inverted skew biases (i.e. almost equal magnitude in opposite direction; e.g. -0.25 and + 0.25) [14, 16, 17]. However, skew inversion in enoplean nematodes is only partial, so effects on evolutionary analyses may be less strongly pronounced. We found that only two sequences (Capillaria sp. and T. muris) passed the compositional homogeneity test. Mutational saturation analyses indicated substantial or high levels of saturation (defined as transitions and transversions plateauing) for most individual genes and for the 3rd codon position dataset. However, concatenated 12 PCGs dataset and 1st and 2nd codon datasets did not exhibit strong levels of saturation (Additional file 1: Fig. S16). Also, ORI species did not appear to exhibit elevated levels of saturation. This indicates that concatenated mitogenomic datasets in combination with an algorithm that accounts for compositional heterogeneity may be a suitable tool for studying the evolutionary history of the Enoplea. However, previous (recent) studies that studied the enoplean mitochondrial phylogenomics relied on standard phylogenetic analysis algorithms: Maximum Likelihood (ML), Bayesian Inference (BI), and Maximum Parsimony (MP) [6, 12, 13]. Herein we tested the performance of the standard ML phylogenetic approach implemented in IQ-TREE, as well as the CAT-GTR algorithm designed to account for compositional heterogeneity [47]. The ML analysis produced paraphyletic Arthropoda, with Priapulida nested within, and a generally non-standard [48] topology of Ecdysozoa, but Nematoda and Nematomorpha were resolved as sister-phyla, which is accepted as the most likely evolutionary scenario [49]. Chromadorea and Enoplea were monophyletic sister-taxa, and Longidoridae were the basal Dorylaimia lineage (i.e. sister-clade to all remaining lineages) (Fig. 2). Mermithida and Trichinellida were sister-groups, and Capillariidae + Trichuridae exhibited a close sister-clade relationship in the latter clade (Fig. 2). In all crucial aspects, the tree produced using all available genes was identical (Additional file 4). The CAT-GTR model produced a similar topology, but with monophyletic Arthropoda (sister-group relationship with Priapulida), switched places between Kinorhyncha and Tardigrada, and with the Enoplean topology resolved into two major sister-clades: Dorylaimida + Mermithida and Trichinellida (Fig. 3). Support values for key nodes were higher in the CAT-GTR analysis than in the ML analysis, where Trichinellidae + (Mermithidae + Capillariidae) clade was weakly supported: 72% (100% in the CAT-GTR).
Fig. 2.
Mitochondrial phylogenomics of the Enoplea: Maximum Likelihood. The phylogram was inferred using the Maximum Likelihood methodology implemented in IQ-TREE and nucleotide sequences of 12 protein-coding genes. Bootstrap support values are shown on the braches (only the values < 100 are shown). Species names are given with GenBank accession numbers. The phylum, class, order, and family-level taxonomic identities are shown to the right
Fig. 3.
Mitochondrial phylogenomics of the Enoplea: CAT-GTR. The phylogram was inferred using the CAT-GTR algorithm designed for compositional heterogeneity implemented in PhyloBayes and amino acid sequences of 12 protein-coding genes. Posterior probability values are shown on the braches (only the values < 1.0 are shown). Species names are given with GenBank accession numbers. The phylum, class, order, and family-level taxonomic identities are shown to the right
With respect to the enoplean topology, 18S datasets appear to resolve Enoplia and Dorylaimia as sister clades, often with Trichinellidae and/or Dioctophymatida as the basal Dorylaimia clade, but results are inconsistent or topologies unresolved [1, 6, 50, 51]. The available nuclear genomic data resolve Mermithidae as the basal lineage [52], but many crucial lineages remain unrepresented. The available mitogenomic data mostly produce two different topologies across different studies: 1. Longidoridae as the basal clade, and 2. Enoplea comprised of two sister-clades: Dorylaimida + Mermithida and Trichinellida. Topology 1 was produced by the nucleotide dataset (NUC) in our ML analysis with high support (100%), and by two previous studies: ML AAs analysis (amino acids) [13], and ML and BI AAs [6]. Topology 2 was produced by our CAT-GTR analysis (100% support; AAs dataset), and by several previous studies: BI and ML AAs [12], BI AAs [13], BI and ML NUC [6], and ML, BI, and MP AAs [41]. The topology of the remaining enoplean lineages is remarkably consistent across all studies. In comparison, the available genomic data produce Mermithidae as the basal radiation, followed by the Dioctophymatida and finally Trichinellidae and Trichuridae as highly derived sister-families, but the absence of data for Longidoridae remains a major shortcoming [52].
This uncertainty also makes it difficult to infer the evolutionary history of replication disruption events (producing changes in skew magnitude) in the enoplean nematodes. If Trichniellidae is the earliest-branching lineage, which is unlikely as it is partially supported only by the 18S data, and consistently rejected by mitogenomic and nuclear genomic data, a single disruption of the replication mechanism is sufficient to explain the observed pattern. The topology produced by the mitogenomic data requires a more complex evolutionary scenario. Topology 1 would require three independent disruptions: in the common ancestors of Longidoridae, Mermithidae, and Capillariidae + Trichuridae. Topology 2 would require two independent disruptions: in the common ancestors of Longidoridae + Mermithidae and Capillariidae + Trichuridae. Therefore, topology 2 is supported by the more reliable CAT-GTR analysis in our study, a majority of previous mitogenomic results, and by being the more parsimonious of the latter two skew disruption scenarios.
Conclusions
To improve our understanding of the discontinuity in the evolution of mitogenomes in the nematode class Enoplea, we sequenced and characterised the complete mitochondrial genome of P. tomentosa (Trichinellida: Capillariidae). As mitochondrial molecular data were previously unavailable for this species, the sequence will facilitate future molecular identification and evolutionary studies of this species. Similar to the Chromadorea, the evolution of mitochondrial architecture in the enoplean nematodes exhibits a strong discontinuity: lineages with relatively conserved architecture over tens of millions of years are interspersed with lineages exhibiting architectural hypervariability. Surprisingly, Longidoridae exhibited more highly rearranged mitogenomes than Mermithidae, which possess some of the fastest-evolving mitochondrial architecture among the Bilateria. This is in contradiction with the expectation that high rearrangement rates should produce multiple noncoding regions in mitogenomes. We provide the first observation of an inverted base composition skew in the enoplean lineages. Lineages exhibiting inverted skews appear to represent the intermediate phase between the Trichinellidae (ancestral) and fully derived (high positive) skews in Chromadorean mitogenomes. Our observations have important repercussions for future studies that aim to apply mitochondrial phylogenomics to nematodes. Despite the inverted skews and overall compositional heterogeneity, we found evidence that mitochondrial and nuclear phylogenomics might produce congruent topologies. In the absence of Enoplia and Mononchida representatives, multiple lines of evidence (CAT-GTR analysis in our study, a majority of previous mitogenomic results, and skew disruption scenarios) support the Dorlylaimia split into two sister-clades: Dorylaimida + Mermithida and Trichinellida. However, it is necessary to sequence data from the missing crucial lineages before this can be assessed with confidence, and future studies should pay close attention to putative artefacts caused by compositional heterogeneity and seek agreement between different types of data before any conclusions are made.
Methods
Sample collection and identification
Parasitic nematodes were obtained post mortem from bighead carp Hypophthalmichthys nobilis specimens caught by fishermen in the Bailianhe reservoir (Huanggang city, Hubei province, China) and bought from the local market on 2/Aug/2020. Live nematodes were removed from the fish intestines, and then taxonomically identified by their morphological characteristics [24] via dissecting microscopy. All nematodes were washed in 0.6% saline before being stored in the absolute ethanol in the Museum of Aquatic Organisms, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China. Further identification was conducted using the 18S gene sequence, for which primers (Table 4) were designed on the basis of the previously sequenced conspecific 18S sequence KU987805 [23].
Table 4.
Primers used for the amplification and sequencing of the mitochondrial genome of P. tomentosa. LR next to the fragment number means that long-range PCR was used to obtain the amplicon. See Additional file 1 for the full list of sequencing primers
| Fragment No | Gene or region | Primer name | Sequence (5’-3’) | Length (bp) |
|---|---|---|---|---|
| F1 | 16S | Eno 16SF | GTTTKTGACCTCGATGTTGN | 171 |
| Eno 16SR | CYTTTWGTTCCTTTCGTACT | |||
| F2 (LR) | 16S-cox1 | MX F1 | AACGTCTGTTCGACGTAAGA | 3395 |
| MX R1 | CTACATCCATACCTACGGTG | |||
| F3 | cox1 | Eno COX1F | GATTHTTNGGTCAYCCTGAAGT | 614 |
| Eno COX1R | ATACCGWCGNGGTATACCAT | |||
| F4 (LR) | cox1-tRNA-Thr | MX F2 | GATTGCCATGAATGATAGGA | 7150 |
| MX R2 | CAAAATCTATATTCTACTTAAAC | |||
| F5 (LR) | nadL-16S | MX F3 | CAAAACCAATAATTCTGTGTG | 3469 |
| MX R3 | TCTTACGTCGAACAGACGTT |
Genome sequencing and assembly
Genome sequencing and assembly were conducted as described before [9], so details are provided in Additional file 1. Briefly, DNA was isolated from a single specimen. Five primer pairs were designed on the basis of gene orthologues from Capillaria sp., and then used to amplify the entire mitogenome (Table 4). To avoid assembly artefacts, the amplified fragments were designed to overlap by approximately 100 bp. PCR products were sequenced using the Sanger method and an expanded set of primers, because fragments longer than 1kbp (amplified using long-range PCR) had to be sequenced in several steps (see Additional file 1 for full details). The mitogenome was assembled manually using DNAstar v7.1 [53], and roughly annotated using MITOS [54]. The annotation was further refined using several different methods: DNAstar, BLAST BLASTx (PCGs), orthologous sequences (PCGs and rRNAs), and ARWEN [55] (tRNAs).
Comparative mitogenomic and phylogenetic analyses
We downloaded all available Enoplea mitogenomes from GenBank. We removed all duplicates, two unannotated mitogenomes, and left only one mitogenome per species (unless we found indications that conspecific mitogenomes might exhibit a different architecture). PhyloSuite [56] was used to conduct these steps, as well as to parse and extract the annotation recorded in a Word (Microsoft Office) document, generate the GenBank format file, update the taxonomy from the NCBI database, standardise and extract data, generate comparative tables, and generate annotation files for visualisation in iTOL [57]. GC and AT base composition skews were also calculated by PhyloSuite, following the (G-C)/(G + C) and (A-T)/(A + T) formulas respectively [42]. PhyloSuite and its plug-in programs were also used to conduct all phylogenetic analysis steps, for which we used nucleotide sequences of concatenated 12 mitochondrial protein-coding genes (PCGs); atp8 was removed because it was absent from many species, and only one copy per gene was kept in the Mermithidae that exhibited duplicated PCGs (further details in Additional file 1). Sequence alignment in batches was conducted using the accurate E-ins-i strategy in MAFFT [58]. Alignments were concatenated by Phylosuite. Phylogenetic analyses were conducted using IQ-TREE (Maximum Likelihood – ML) [59] and the CAT-GTR site mixture model implemented in PhyloBayes-MPI 1.7a [47], which allows for site-specific rates of mutation, as there is evidence that in some cases it alleviates the base composition skew-driven long-branch attraction artefacts better than other standard phylogenetic algorithms, especially in combination with amino acid sequences [17]. PhyloBayes run parameters were burnin = 500, invariable sites automatically removed from the alignment, two MCMC chains, and the analysis was stopped when the conditions considered to indicate a good run were reached: maxdiff < 0.1 and minimum effective size > 300 (PhyloBayes manual). The best-fit partitioning strategy and models for partitions for IQ-TREE were inferred using the inbuilt functions of IQ-TREE and immediately followed by phylogenetic reconstruction with 50,000 ultrafast bootstrap replicates [60]. We also conducted an IQ-TREE analysis using the parameters described above to test whether the inclusion of all genes (PCGs + rRNAs + tRNAs) can stabilise the topology. IQ-TREE was also used to test for compositional heterogeneity. For mutational saturation analyses [61] and to generate cumulative skew plots for the entire plus strand, we used DAMBE 7.3.0 [62]. CREx was used to infer the GO distances [29]. All GenBank files were rearranged to start with the cox1 gene using the Reorder function in PhyloSuite, which was also used to generate the comparative mitogenomic architecture table for the P. tomentosa and E. annulatus mitogenomes. Conserved sequence motifs searches were conducted using MEME [63] and FIMO [64] tools.
Supplementary Information
Acknowledgements
We thank Rong Chen of the BT Lab for helping us with the molecular lab work, and mitogenome assembly and annotation.
Abbreviations
- AAs
Amino acids
- Bp
Base pair
- CR
Control region
- ML
Maximum likelihood
- MP
Maximum parsimony
- BI
Bayesian inference
- NCR
Noncoding region
- NUC
Nucleotides
- PCGs
Protein-coding genes
Authors’ contributions
HZ participated in the study conception and design, sample collection, molecular lab work, mitogenome assembly and annotation, data analysis, data visualisation, and manuscript preparation. FLC participated in the sample collection and molecular lab work. ML participated in the sample collection and molecular lab work. HPL participated in the data analysis and visualisation. DZ participated in the data analysis and provided software and codes. WXL participated in the study conception and design. IJ participated in the study conception, design, mitogenome annotation, data analysis, data visualisation, and drafted the manuscript. GTW participated in the study conception and design, and supervised the study. All authors have approved the submitted version and agree to be personally accountable for their own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Funding
This study was funded by the National Natural Science Foundation of China (31970408); the China Agriculture Research System of MOF and MARA (CARS-45); and the Start-up Funds of Introduced Talent in Lanzhou University (561120206). The funders had no role in the design of the study, collection, analysis and interpretation of data, and in writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files, as well as in the GenBank repository. The complete mitogenome of P. tomentosa is available under the accession number MZ708958 (https://www.ncbi.nlm.nih.gov/nuccore/MZ708958.1/) and the partial 18S sequence is available under MZ724160 (https://www.ncbi.nlm.nih.gov/nuccore/MZ724160).
Declarations
Ethics approval and consent to participate
All experimental procedures involving animals were reviewed, approved and supervised by the Animal Care Committee of the Institute of Hydrobiology, Chinese Academy of Sciences. As the study did not involve any live vertebrates, nor regulated invertebrates, no special permits were required to retrieve and process the samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ivan Jakovlić, Email: ivanjakovlic@yahoo.com.
Gui-Tang Wang, Email: gtwang@ihb.ac.cn.
References
- 1.Blaxter ML, Ley PD, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 2.Lavrov DV, Brown WM. Trichinella spiralis mtDNA: A nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics. 2001;157:621–637. doi: 10.1093/genetics/157.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohandas N, Pozio E, La Rosa G, Korhonen PK, Young ND, Koehler AV, et al. Mitochondrial genomes of Trichinella species and genotypes – a basis for diagnosis, and systematic and epidemiological explorations. Int J Parasitol. 2014;44:1073–1080. doi: 10.1016/j.ijpara.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Liu G-H, Gasser RB, Su A, Nejsum P, Peng L, Lin R-Q, et al. Clear Genetic Distinctiveness between Human- and Pig-Derived Trichuris Based on Analyses of Mitochondrial Datasets. PLoS Negl Trop Dis. 2012;6:e1539. doi: 10.1371/journal.pntd.0001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Y, Jones J, Armstrong M, Lamberti F, Moens M. The Mitochondrial Genome of Xiphinema americanum sensu stricto (Nematoda: Enoplea): Considerable Economization in the Length and Structural Features of Encoded Genes. J Mol Evol. 2005;61:819–833. doi: 10.1007/s00239-005-0102-7. [DOI] [PubMed] [Google Scholar]
- 6.Palomares-Rius JE, Cantalapiedra-Navarrete C, Archidona-Yuste A, Blok VC, Castillo P. Mitochondrial genome diversity in dagger and needle nematodes (Nematoda: Longidoridae) Scientific Reports. 2017;7:srep41813. doi: 10.1038/srep41813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyman BC, Lewis SC, Tang S, Wu Z. Rampant gene rearrangement and haplotype hypervariation among nematode mitochondrial genomes. Genetica. 2011;139:611–615. doi: 10.1007/s10709-010-9531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang S, Hyman BC. Mitochondrial genome haplotype hypervariation within the isopod parasitic nematode Thaumamermis cosgrovei. Genetics. 2007;176:1139–1150. doi: 10.1534/genetics.106.069518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou H, Jakovlić I, Chen R, Zhang D, Zhang J, Li W-X, et al. The complete mitochondrial genome of parasitic nematode Camallanus cotti: extreme discontinuity in the rate of mitogenomic architecture evolution within the Chromadorea class. BMC Genomics. 2017;18:840. doi: 10.1186/s12864-017-4237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernt M, Bleidorn C, Braband A, Dambach J, Donath A, Fritzsch G, et al. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 2013;69:352–364. doi: 10.1016/j.ympev.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Rota-Stabelli O, Kayal E, Gleeson D, Daub J, Boore JL, Telford MJ, et al. Ecdysozoan Mitogenomics: Evidence for a Common Origin of the Legged Invertebrates, the Panarthropoda. Genome Biol Evol. 2010;2:425–440. doi: 10.1093/gbe/evq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Y-P, Suleman. Zhang Y, Nie Y, Fu Y-T, Liu G-H. The complete mitochondrial genome of capillariid nematodes (Eucoleus annulatus): A novel gene arrangement and phylogenetic implications. Veterinary Parasitology. 2021;296:109476. doi: 10.1016/j.vetpar.2021.109476. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Lee S-HH, Gazi M, Kim T, Jung D, Chun J-YY, et al. Mitochondrial genomes advance phylogenetic hypotheses for Tylenchina (Nematoda: Chromadorea) Zoologica Scripta. 2015;44:446–62. doi: 10.1111/zsc.12112. [DOI] [Google Scholar]
- 14.Hassanin A. Phylogeny of Arthropoda inferred from mitochondrial sequences: Strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol. 2006;38:100–116. doi: 10.1016/j.ympev.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Helfenbein KG, Brown WM, Boore JL. The Complete Mitochondrial Genome of the Articulate Brachiopod Terebratalia transversa. Mol Biol Evol. 2001;18:1734–1744. doi: 10.1093/oxfordjournals.molbev.a003961. [DOI] [PubMed] [Google Scholar]
- 16.Jakovlić I, Zou H, Zhao X-M, Zhang J, Wang G-T, Zhang D. Evolutionary History of Inversions in Directional Mutational Pressures in Crustacean Mitochondrial Genomes: Implications for Evolutionary Studies. Mol Phylogenet Evol. 2021;164:107288. doi: 10.1016/j.ympev.2021.107288. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D, Zou H, Hua C-J, Li W-X, Mahboob S, Al-Ghanim KA, et al. Mitochondrial Architecture Rearrangements Produce Asymmetrical Nonadaptive Mutational Pressures That Subvert the Phylogenetic Reconstruction in Isopoda. Genome Biol Evol. 2019;11:1797–1812. doi: 10.1093/gbe/evz121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou H, Jakovlić I, Zhang D, Hua C-J, Chen R, Li W-X, et al. Architectural instability, inverted skews and mitochondrial phylogenomics of Isopoda: outgroup choice affects the long-branch attraction artefacts. Royal Society Open Science. 2020;7:191887. doi: 10.1098/rsos.191887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes A, Gissi C, Pesole G, Saccone C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 1998;15:957–966. doi: 10.1093/oxfordjournals.molbev.a026011. [DOI] [PubMed] [Google Scholar]
- 20.Xia X. DNA Replication and Strand Asymmetry in Prokaryotic and Mitochondrial Genomes. Curr Genomics. 2012;13:16–27. doi: 10.2174/138920212799034776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clayton DA. Transcription and replication of mitochondrial DNA. Human Reprod. 2000;15 suppl_2:11–7. doi: 10.1093/humrep/15.suppl_2.11. [DOI] [PubMed] [Google Scholar]
- 22.Hassanin A, Léger N, Deutsch J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst Biol. 2005;54:277–298. doi: 10.1080/10635150590947843. [DOI] [PubMed] [Google Scholar]
- 23.Leis E, Easy R, Cone D. Report of the Potential Fish Pathogen Pseudocapillaria (Pseudocapillaria) tomentosa (Dujardin, 1843) (Nematoda) from Red Shiner (Cyprinella lutrensis) Shipped from Missouri to Wisconsin. copa. 2016;83:275–8. [Google Scholar]
- 24.Moravec F. Parasitic Nematodes of Freshwater Fishes of Europe. Netherlands: Springer; 1994. [Google Scholar]
- 25.Kent ML, Gaulke CA, Watral V, Sharpton TJ. Pseudocapillaria tomentosa in laboratory zebrafish Danio rerio: patterns of infection and dose response. Dis Aquat Org. 2018;131:121–131. doi: 10.3354/dao03286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norris L, Lawler N, Hunkapiller A, Mulrooney DM, Kent ML, Sanders JL. Detection of the parasitic nematode, Pseudocapillaria tomentosa, in zebrafish tissues and environmental DNA in research aquaria. J Fish Dis. 2020;43:1087–1095. doi: 10.1111/jfd.13220. [DOI] [PubMed] [Google Scholar]
- 27.Martins ML, Watral V, Rodrigues-Soares JP, Kent ML. A method for collecting eggs of Pseudocapillaria tomentosa (Nematoda: Capillariidae) from zebrafish Danio rerio and efficacy of heat and chlorine for killing the nematode’s eggs. J Fish Dis. 2017;40:169–182. doi: 10.1111/jfd.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- 29.Bernt M, Merkle D, Ramsch K, Fritzsch G, Perseke M, Bernhard D, et al. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics. 2007;23:2957–2958. doi: 10.1093/bioinformatics/btm468. [DOI] [PubMed] [Google Scholar]
- 30.Braband A, Cameron SL, Podsiadlowski L, Daniels SR, Mayer G. The mitochondrial genome of the onychophoran Opisthopatus cinctipes (Peripatopsidae) reflects the ancestral mitochondrial gene arrangement of Panarthropoda and Ecdysozoa. Mol Phylogenet Evol. 2010;57:285–292. doi: 10.1016/j.ympev.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Webster BL, Copley RR, Jenner RA, Mackenzie-Dodds JA, Bourlat SJ, Rota-Stabelli O, et al. Mitogenomics and phylogenomics reveal priapulid worms as extant models of the ancestral Ecdysozoan. Evol Dev. 2006;8:502–510. doi: 10.1111/j.1525-142X.2006.00123.x. [DOI] [PubMed] [Google Scholar]
- 32.Staton JL, Daehler LL, Brown WM. Mitochondrial gene arrangement of the horseshoe crab Limulus polyphemus L.: conservation of major features among arthropod classes. Molecular Biology and Evolution. 1997;14:867–74. doi: 10.1093/oxfordjournals.molbev.a025828. [DOI] [PubMed] [Google Scholar]
- 33.Lagisz M, Poulin R, Nakagawa S. You are where you live: parasitic nematode mitochondrial genome size is associated with the thermal environment generated by hosts. J Evol Biol. 2013;26:683–690. doi: 10.1111/jeb.12068. [DOI] [PubMed] [Google Scholar]
- 34.Boore JL. The Duplication/Random Loss Model for Gene Rearrangement Exemplified by Mitochondrial Genomes of Deuterostome Animals. In: Sankoff D, Nadeau JH, editors. Comparative Genomics: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment and the Evolution of Gene Families. Dordrecht: Springer, Netherlands; 2000. pp. 133–147. [Google Scholar]
- 35.Mao M, Austin AD, Johnson NF, Dowton M. Coexistence of Minicircular and a Highly Rearranged mtDNA Molecule Suggests That Recombination Shapes Mitochondrial Genome Organization. Mol Biol Evol. 2014;31:636–644. doi: 10.1093/molbev/mst255. [DOI] [PubMed] [Google Scholar]
- 36.Rand DM. Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol. 1993;37:281–295. doi: 10.1007/BF00175505. [DOI] [PubMed] [Google Scholar]
- 37.Hu M, Gasser RB. Mitochondrial genomes of parasitic nematodes–progress and perspectives. Trends Parasitol. 2006;22:78–84. doi: 10.1016/j.pt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Kang S, Sultana T, Eom KS, Park YC, Soonthornpong N, Nadler SA, et al. The mitochondrial genome sequence of Enterobius vermicularis (Nematoda: Oxyurida) - An idiosyncratic gene order and phylogenetic information for chromadorean nematodes. Gene. 2009;429:87–97. doi: 10.1016/j.gene.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Sultana T, Kim J, Lee S-H, Han H, Kim S, Min G-S, et al. Comparative analysis of complete mitochondrial genome sequences confirms independent origins of plant-parasitic nematodes. BMC Evol Biol. 2013;13:1–17. doi: 10.1186/1471-2148-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azevedo JLB, Hyman BC. Molecular characterization of lengthy mitochondrial DNA duplications from the parasitic nematode Romanomermis culicivorax. Genetics. 1993;133:933–942. doi: 10.1093/genetics/133.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu G-H, Wang Y, Xu M-J, Zhou D-H, Ye Y-G, Li J-Y, et al. Characterization of the complete mitochondrial genomes of two whipworms Trichuris ovis and Trichuris discolor (Nematoda: Trichuridae) Infect Genet Evol. 2012;12:1635–1641. doi: 10.1016/j.meegid.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Perna NT, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 43.Hunt VL, Tsai IJ, Coghlan A, Reid AJ, Holroyd N, Foth BJ, et al. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat Genet. 2016;48:299–307. doi: 10.1038/ng.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang S, Hyman BC. Rolling circle amplification of complete nematode mitochondrial genomes. J Nematol. 2005;37:236–241. [PMC free article] [PubMed] [Google Scholar]
- 45.Powers TO, Harris TS, Hyman BC. Mitochondrial DNA Sequence Divergence among Meloidogyne incognita, Romanomermis culicivorax, Ascaris suum, and Caenorhabditis elegans. J Nematol. 1993;25:564–572. [PMC free article] [PubMed] [Google Scholar]
- 46.Lavrov DV, Boore JL, Brown WM. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Biol Evol. 2000;17:813–824. doi: 10.1093/oxfordjournals.molbev.a026360. [DOI] [PubMed] [Google Scholar]
- 47.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laumer CE, Fernández R, Lemer S, Combosch D, Kocot KM, Riesgo A, et al. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proceedings of the Royal Society B: Biological Sciences. 2019;286:20190831. doi: 10.1098/rspb.2019.0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giribet G, Edgecombe GD. Current Understanding of Ecdysozoa and its Internal Phylogenetic Relationships. Integr Comp Biol. 2017;57:455–466. doi: 10.1093/icb/icx072. [DOI] [PubMed] [Google Scholar]
- 50.Bik HM, Lambshead PJD, Thomas WK, Lunt DH. Moving towards a complete molecular framework of the Nematoda: a focus on the Enoplida and early-branching clades. BMC Evol Biol. 2010;10:353. doi: 10.1186/1471-2148-10-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meldal BHM, Debenham NJ, De Ley P, De Ley IT, Vanfleteren JR, Vierstraete AR, et al. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol. 2007;42:622–636. doi: 10.1016/j.ympev.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 52.International Helminth Genomes Consortium Comparative genomics of the major parasitic worms. Nat Genet. 2019;51:163–174. doi: 10.1038/s41588-018-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burland TG. DNASTAR’s Lasergene sequence analysis software. In: Misener S, Krawetz SA, editors. Methods in Molecular Biology™. Totowa, NJ: Humana Press; 2000. pp. 71–91. [DOI] [PubMed] [Google Scholar]
- 54.Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 55.Laslett D, Canbäck B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 56.Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, et al. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 57.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 58.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol Phylogenet Evol. 2003;26:1–7. doi: 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 62.Xia X. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol Biol Evol. 2018;35:1550–1552. doi: 10.1093/molbev/msy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 64.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files, as well as in the GenBank repository. The complete mitogenome of P. tomentosa is available under the accession number MZ708958 (https://www.ncbi.nlm.nih.gov/nuccore/MZ708958.1/) and the partial 18S sequence is available under MZ724160 (https://www.ncbi.nlm.nih.gov/nuccore/MZ724160).